Abstract
Intensified treatment and control efforts since the early 2000s have dramatically reduced the burden of Plasmodium falciparum malaria. However, drug resistance threatens to derail this progress. In this review, we present four antimalarial resistance case studies that differ in timeline, technical approaches, mechanisms of action, and categories of resistance: chloroquine, sulfadoxine-pyrimethamine, artemisinin, and piperaquine. Lessons learned from prior losses of treatment efficacy, drug combinations, and control strategies will help advance mechanistic research into how P. falciparum parasites acquire resistance to current first-line artemisinin-based combination therapies. Understanding resistance in the clinic and laboratory is essential to prolong the effectiveness of current antimalarial drugs and to optimize the pipeline of future medicines.
In this review, Ross and Fidock examine Plasmodium resistance to antimalarials, notably chloroquine, sulfadoxine-pyrimethamine, artemisinin, and piperaquine. They discuss the lessons learned from prior losses of treatment efficacy, drug combinations, parasite resistance mechanisms, and implications for treatment and future research.
Introduction
Malaria, caused by infection with Plasmodium protozoan parasites, threatens over half the world’s population. The vast majority of deaths are from young children in Africa infected with P. falciparum. Malaria symptoms include cyclical chills and fever, anemia, and malaise. More severe cases can also have metabolic acidosis, respiratory distress, cerebral malaria, coma, and death (Phillips et al., 2017). The World Health Organization estimated ~216 million malaria cases in 2017, resulting in ~435,000 deaths. This number represents major progress from the million-plus deaths per year in the 1990s. Between 2000 and 2015, the widespread adoption of artemisinin-based combination therapies (ACTs) and the increased use of bed nets and insecticides against the Anopheles mosquito vector decreased the global number of malaria deaths by an estimated 37% (Gething et al., 2016). Recently, these fragile gains have lost ground, hampered by the rise of resistance in both the parasite and the mosquito vector to the current front-line drugs and insecticides (Blasco et al., 2017).
Plasmodium Biology and Propensity for Resistance
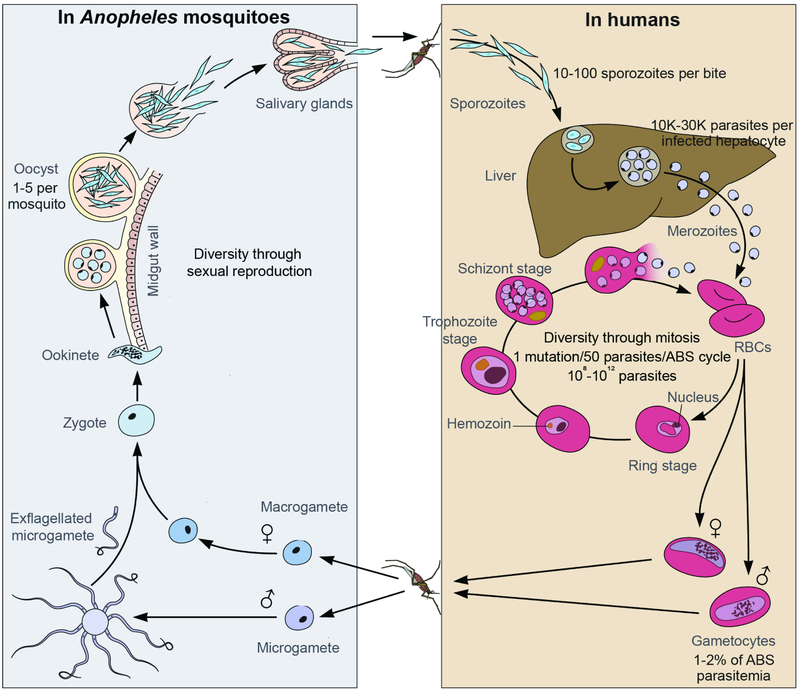
Plasmodium parasites have a sexual life cycle in Anopheles mosquitoes and an asexual cycle in vertebrates (Figure 1). In humans, sporozoites transmitted in a mosquito blood meal will travel through the skin into the bloodstream, where they quickly home in to the liver and invade hepatocytes. After one week of development, tens of thousands of P. falciparum merozoites will be released from each infected hepatocyte into the bloodstream, where they rapidly invade red blood cells (RBCs). Intraerythrocytic residence limits exposure to the immune system, and parasites also suppress the immune response through processes including antigenic variation, cytoadherence to the vascular endothelium to prevent splenic clearance, and induction of immunosuppressive cytokines (Gomes et al., 2016). P. falciparum parasites progress through an asexual blood stage (ABS) cycle lasting approximately 48 hours. An invading merozoite matures to the ring stage, then trophozoite, then schizont, and finally bursts out of the host RBC as 8-24 daughter merozoites that can reinitiate further ABS cycles. 1-2% of the ABS parasites will undergo sexual differentiation into male and female gametocytes. Gametocytes are taken up in a mosquito bloodmeal, where they undergo sexual recombination and develop into sporozoites, ready to infect another person (Phillips et al., 2017). The majority of current antimalarials focus on the ABS, and drug discovery efforts increasingly focus on compounds that also act on liver or mosquito transmission stages as the lower parasite numbers may decrease the potential for drug resistance (Burrows et al., 2017).
Figure 1. Plasmodium falciparum life cycle, shared between the human host and the Anopheles mosquito vector.
Adapted from (Lee et al., 2014). Sporozoites, gametocytes, and oocyst stages are genetic population bottlenecks. ABS, asexual blood stages; RBCs, red blood cells.
Plasmodium parasites are highly host-specific, and five species are known to infect humans: P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi (Phillips et al., 2017). The majority of infections are caused by P. falciparum and P. vivax, and P. falciparum causes the vast majority of deaths. P. vivax and P. ovale have a latent form called the hypnozoite, which resides in the liver and can reactivate months or years later to initiate a relapse infection (Wells et al., 2010). P. knowlesi infects macaques and is primarily a zoonosis. Years of repeated infections can give partial, nonsterilizing immunity that quickly wanes without exposure, and these individuals often become asymptomatic carriers (Gomes et al., 2016). These factors, coupled with the evolution of drug resistance, complicate treatment and eradication goals. Non-biological obstacles include affordability, distribution logistics, production volume, counterfeit or low-quality drugs, patient non-compliance, inappropriate use of monotherapies, and contraindications.
The complex P. falciparum life cycle leads to more pronounced random genetic drift and more efficient purifying selection than expected in classic population genetics, leading to more overall nonsynonymous mutations (Chang et al., 2013). Its ABS cycle is ~48 hours, and a typical expansion rate is 5-to-10 fold per cycle, so a parasite with a selective advantage can expand exponentially over the course of a typical 1-2 week infection (Figure 1). The mosquito stages are diploid during the oocyst stage and generate diversity through sexual reproduction. The human stages are all haploid and reproduce asexually. A typical infection will have an inoculum of 10-100 sporozoites per mosquito bite, then 105-106 liver-stage parasites (10,000-30,000 per infected hepatocyte), then 108-1012 ABS parasites. The Plasmodium nuclear genome is ~23 Mb, plus two small plastid genomes for the mitochondria and apicoplast, and the measured in vitro mutation rate is ~10−9 per base pair per mitotic division (Bopp et al., 2013). This translates to about 1 mutation for every 50 parasites per mitotic generation (Figure 1). Pseudo-polyploidy in segments of the nuclear or plastid genomes via copy number variation can help the parasite survive stochastic mutations in individual genes as well as increase gene dosage to counteract drug action (Guler et al., 2013; Nair et al., 2008).
The initial emergence of resistance in Southeast Asia, especially in the Greater Mekong Subregion (comprising Cambodia, Vietnam, Thailand, Laos, Myanmar, and southern China), has been observed for chloroquine (CQ), sulfadoxine-pyrimethamine (SP), artemisinin (ART), piperaquine (PPQ), and mefloquine (MFQ). Contributing factors could include the low transmission rates in Southeast Asia, which can allow relatively unfit mutations to spread due to the lack of within-host asexual parasite competition in a person and the lack of other parasite genomes to recombine with in the sexual mosquito stages. Low transmission is also associated with an older age range of infected patients, and adults often do not seek medical care at the same rate or as quickly as parents do for their young children (Carneiro et al., 2010). Additionally, Southeast Asian parasites have a strong population structure, characterized by marked differences in allele frequencies in subpopulations, leading to small effective population sizes from which resistance can quickly evolve on predisposing genetic backgrounds (Takala-Harrison and Laufer, 2015). Far lower rates of infection overall in Southeast Asia versus sub-Saharan Africa also translates into less robust acquired immunity, causing greater reliance on therapeutics that in turn exerts increased selection pressure on parasite populations.
The history of antimalarial chemotherapy shows repeated instances where the front-line therapy at the time has been compromised by resistance (Table 1). Drug resistance largely falls into four categories: removal or sequestration of drug, detoxification, mutation and/or amplification of the target(s), or stress response-based survival mechanisms. All phenotypes, including drug resistance, are limited by fitness constraints for survival, reproduction, and transmission. Below, we present four case studies, beginning with parasite resistance to chloroquine (CQ).
Table 1.
Antimalarial drugs and associated markers of resistance in Plasmodium falciparum asexual blood stage parasites.
| Antimalarial class | Antimalarial name (abbreviation) |
Major clinical use | Affected pathway(s), mechanism(s) |
Genetic change associated with clinical resistance |
Fitness cost of resistance determinant |
|---|---|---|---|---|---|
| Endoperoxides | Artemisinins (ARTs): artesunate (AS), artemether (ATM), dihydroartemisinin (DHA) | First-line treatment as part of ACTs; intravenous artesunate gold standard to treat severe malaria | Pleiotropic, triggers parasite stress response. Alkylates and oxidized heme, multiple proteins and lipids | Mutations in k13 | One single K13 mutation permitted at a time; nil to low C580Y in vitro fitness cost depending on genetic background |
| 4-aminoquinolines | Chloroquine (CQ) | Treatment of non-falciparum malaria | Heme detoxification in digestive vacuole | Mutations in pfcrt and pfmdr1 | Mutant pfcrt in Africa less fit, overtaken by wild-type allele upon removal of CQ pressure |
| Amodiaquine (AQ) | Partner drug for ACT (ASAQ) | Reduced fitness observed with mutant pfcrt and pfmdr1 | |||
| Piperaquine (PPQ) | Partner drug for ACT (DHA-PPQ) | plasmepsin II and III amplification, pfcrt mutations | In vitro fitness cost observed with novel pfcrt mutations | ||
| Pyronaridine (PND) | Partner drug for ACT (PA) | None observed | No published data | ||
| Aryl-amino alcohols | Quinine (QN) | Treatment of P. falciparum uncomplicated malaria in first trimester of pregnancy, or severe malaria | Might include inhibition of hemoglobin import and/or heme detoxification | pfcrt (QN), pfmdr1 amplification and sequence (LMF and MFQ) | pfmdr1 overexpression imparts fitness cost |
| Lumefantrine (LMF) | Partner drug for ACT (AL) | ||||
| Mefloquine (MFQ) | Partner drug for ACT (ASMQ) and prophylaxis (Lariam™ and generic) | ||||
| Antifolates | Pyrimethamine (PYR) + Sulfadoxine (SDX) | Combination (SP) used mostly for intermittent preventive treatment | Folate biosynthesis in parasite cytosol | Mutations in dhfr and dhps | Some DHFR mutations alter enzyme kinetics; pfgch1 amplification is a possible fitness-compensatory mechanism |
| Proguanil (PG) | see atovaquone-proguanil | ||||
| Naphthoquinones | Atovaquone (ATQ) | Used in combination with proguanil (Malarone™ and generic) | Mitochondrial electron transport chain required for pyrimidine biosynthesis | Mutation(s) in cytb | Y268S associated with decreased enzyme activity; cytb mutants failed to produce sporozoites in mosquitoes and therefore are non-transmissible |
| 8-aminoquinolines | Primaquine (PQ) | Radical cure and terminal prophylaxis of P. vivax and P. ovale; gametocytocidal drug for P. falciparum | Unknown | None observed | No published data |
| Tafenoquine | Radical cure, terminal prophylaxis, and gametocidal activity for P. vivax and P. falciparum | Unknown | None observed | No published data |
ACTs: artemisinin-based combination therapies; ASAQ: artesunate+amodiaquine; DHA-PPQ: dihydroartemisinin+piperaquine; PA: pyronaridine+artesunate; SP: sulfadoxine+pyrimethamine; AL: artemether+lumefantrine; ASMQ: artesunate+mefloquine; K13: Kelch-like gene; cytb cytochrome b; dhfr: dihydrofolate reductase; dhps: dihydropteroate synthase; gch1: GTP cyclohydrolase I; pfcrt: P. falciparum chloroquine resistance transporter; pfmdr1: P. falciparum multidrug resistance gene-1. Adapted in abbreviated form from Blasco et al., 2017.
The “Spent Magic Bullet” of Chloroquine
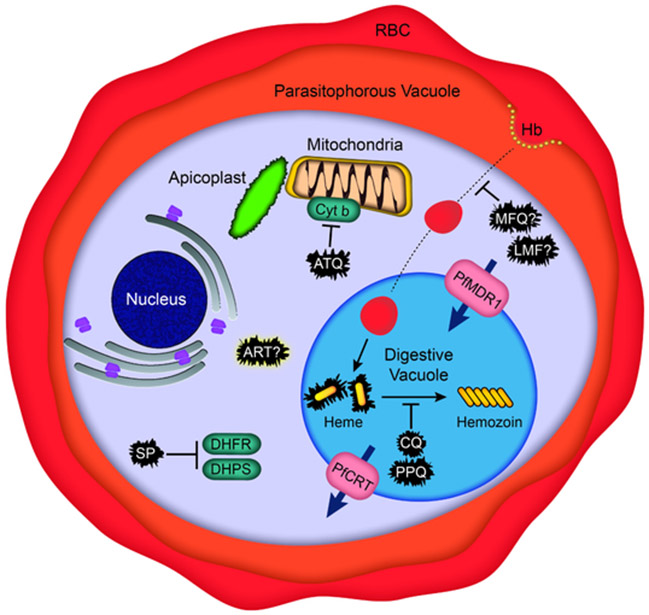
Chloroquine (CQ) was the first-line antimalarial drug for decades (1940s - late 1980s) by virtue of its efficacy, safety, and affordability. Resistance arose at least four independent times: once in Southeast Asia, once in Papua New Guinea, and twice in South America (Takala-Harrison and Laufer, 2015). Resistance is thought to have spread from Southeast Asia to Africa, resulting in a substantial increase in malaria deaths. The genetic basis of resistance was determined by a genetic cross between a CQ-resistant Asian parasite and a CQ-sensitive South American parasite using a splenectomized chimpanzee, which mapped the resistance determinant to a 50 kb parasite genomic region (Su et al., 1997). This region contains the P. falciparum chloroquine resistance transporter (PfCRT), whose causal role was subsequently demonstrated using allelic exchange (Sidhu et al., 2002). PfCRT is a 10-transmembrane protein located in the digestive vacuole (DV) membrane (Figure 2). Host cell hemoglobin is trafficked into this vacuole, wherein it is proteolytically degraded, releasing highly reactive, toxic heme. Parasites convert free heme into chemically inert hemozoin crystals. Multiple antimalarials interfere with this detoxification process, including CQ (Figure 2, Table 1, Table 2). Mechanistic studies provide evidence that mutations in PfCRT allow this transporter to efflux CQ across the DV membrane into the cytosol, away from its heme/hemozoin target (Roepe, 2011; Summers et al., 2012). While PfCRT is the major driver of CQ resistance, sequence variants of PfMDR1, an ABC transporter that also resides on the DV membrane, can modulate the degree of resistance (Veiga et al., 2016).
Figure 2. Schematic of a P. falciparum trophozoite indicating the intracellular targets of major classes of antimalarials and drug resistance determinants.
ART, artemisinin; ATQ, atovaquone; CQ, chloroquine; Cyt b, cytochrome b; DHFR, dihydrofolate reductase; DHPS, dihydropteroate synthase; Hb, hemoglobin; LMF, lumefantrine; MFQ, mefloquine; PfCRT, P. falciparum chloroquine resistance transporter; PfMDR1, P. falciparum multidrug resistance-1; PPQ, piperaquine; RBC, red blood cell; SP, sulfadoxine-pyrimethamine.
Table 2.
In vitro resistance to select antimalarials in the discovery or development pipeline in Plasmodium falciparum asexual blood stage parasites.
| Target location | Pathway/mechanism | Target molecule | Selection compound(s)a |
Resistance gene(s)b |
|---|---|---|---|---|
| Plasma membrane | Ion homeostasis | ATPase 4 | KAE609, (+)-SJ733 |
pfatp4 (PF3D7_1211900) |
| pH regulation | V-type H+-ATPase | AZ412 |
pfVATPase-D (PF3D7_1341900) |
|
| Cytoplasm | Protein synthesis | Translation elongation factor 2 | DDD107498 |
pfeEF2 (PF3D7_1451100) |
| Leucyl-tRNA synthetase | AN6426 |
pfLeuRS (PF3D7_0622800) |
||
| Cytoplasmic isoleucyl-tRNA synthetase | Thiaisoleucine |
pflleRS (PF3D7_1332900) |
||
| Prolyl-tRNA synthetase | Halofuginone |
pfProRS (PF3D7_1213800) |
||
| Lysyl-tRNA synthetase | Cladosporin |
pfKrs1 (PF3D7_1350100) |
||
| Phenylalanyl-tRNA synthetase | BRD1095 |
pfPheRS (PF3D7_0109800) |
||
| Mitochondrion | Electron transport | Cytochrome B | ELQ-300,decoquinate |
pfcytb (mal_mito_3) |
| Pyrimidine biosynthesis | Dihydroorotate dehydrogenase | DSM265 |
pfdhodh (PF3D7_0603300) |
|
| Endomembrane system | Membrane trafficking | Phosphatidylinositol-4-OH kinase | KAI407, MMV390048 |
pfpi4k (PF3D7_0509800) |
| Endoplasmic Reticulum and Golgi apparatus | Unknown | Unknown | KAF156 and analogues |
pfcarl (PF3D7_0321900) pfugt (PF3D7_1113300) pfact (PF3D7_1036800) |
| Apicoplast | Protein synthesis | Apicoplast rRNA | Azithromycin |
pfRpl4 (PF3D7_API00100) |
| Apicoplast isoleucyl-tRNA synthetase | Mupirocin |
pfapi-IRS (PF3D7_1225100) |
||
| Isoprenoid synthesis | 1-Deoxy-D-xylulose 5-phosphate reductoisomerase | Fosmidomycin |
pfdxr (PF3D7_1467300) |
|
| Proteasome | Ubiquitin-regulated protein degradation | Pf Proteasome | WLL-vs, WLW-vs, asparagine ethylenediamines |
20S b5 (PF3D7_1011400) 20S b6 (PF3D7_0518300) 20S b2 (PF3D7_1328100) 19S RPT4 (PF3D7_1306400) 19S RPT5 (PF3D7_1130400) 19S RPN6 (PF3D7_1402300) |
| Nucleus | mRNA processing | Cleavage and polyadenylation specificity factor subunit 3 | AN3661 |
pfcpsf3 (PF3D7_1438500) |
| Rhoptries and Exonemes | Egress and invasion of host cells | Plasmepsins IX and X | 49c, CWHM-117 |
pfPMIX (PF3D7_1430200) pfPMX (PF3D7_0808200) |
| DV membrane | Unknown | P. falciparum multidrug resistance-1c | ACT-451840 |
pfmdr1 (PF3D7_0523000) |
Many of these compounds are being developed with support from the Medicines for Malaria Venture (Geneva, Switzerland; www.mmv.org).
A large-scale analysis of antimalarial resistance determinants in P. falciparum is being undertaken by the Malaria Drug Accelerator Consortium (https://winzeler.ucsd.edu/malda), with supoprt from the Bill & Melinda Gates Foundation.
Whether this is the primary molecular target or the mechanism of resistance remains to be clarified.
Recent studies have leveraged gene-editing techniques to create panels of isogenic pfcrt-modified parasite lines. Their analysis has demonstrated how region-specific haplotypes modify parasite susceptibility to multiple antimalarial drugs whose modes of action intersect with hemoglobin import and heme detoxification. As an example, the “Dd2” CQ-resistant PfCRT haplotype common in Southeast Asia confers partial cross-resistance to amodiaquine (ADQ), but increases parasite sensitivity to MFQ and lumefantrine (LMF) (Blasco et al., 2017; Venkatesan et al., 2014). This dichotomous response supports the use of triple combination therapies that pair drugs with opposing selective pressures, such as the combination of ADQ + artemether (ATM) + LMF that is being evaluated in 17 sites (clinical trial number NCT02453308). Gene-editing studies also provided evidence that CQ-resistant PfCRT haplotypes, which require at least four point mutations and can have up to nine, likely evolved through rare mutational bursts that balance decreasing parasite susceptibility to CQ while minimizing fitness costs (Gabryszewski et al., 2016). The importance of fitness in dictating the prevalence of mutant pfcrt is best illustrated by data from Malawi, where resistant alleles dropped from 85% prevalence to 0% over the course of twenty years without CQ pressure, raising the possibility of recycling drug regimens (Frosch et al., 2014). Reintroducing CQ, however, would rapidly select for parasites that retained variant alleles conferring resistance, necessitating combination strategies that would also select against those parasites.
Sulfadoxine + Pyrimethamine (SP) − Preserving Usefulness Despite Drug Resistance
The loss of CQ efficacy led to a worldwide shift towards SP, which targets two enzymes in the folate synthesis pathway. Clinical resistance to SP was detected in the first year of rollout, and resulted from a series of point mutations in the targets dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS) (Figure 2; Table 1) (Gregson and Plowe, 2005). In Asian parasites, high-grade resistance is closely associated with the four point mutations N51I, C59R, S108M, and I164L in DHFR, of which I164L causes the greatest loss of susceptibility. This latter mutation also causes a significant fitness loss through impaired enzyme kinetics, and Asian parasites were found to have adapted by amplifying the GTP cyclohydrolase-1 gene, which increases the upstream substrate (Nair et al., 2008). The I164L mutation has been rarely observed in Africa, likely because its fitness cost is too significant in areas with frequent mixed infections and thus within-host competition amongst parasites. Genetic mapping of DHFR and DHPS across Africa has identified combinations of point mutations that cause varying levels of SP resistance and that serve as molecular markers to predict efficacy (Naidoo and Roper, 2013). Molecular genotyping is especially important given the widespread use of SP in intermittent preventive treatment (IPT) campaigns. An earlier study with a small patient cohort in Tanzania showed that the use of SP for standard intermittent preventive treatment in pregnancy (IPTp) selected for an increase in the within-host and population-level prevalence of SP-resistant parasites and more intense placental inflammation (Harrington et al., 2009). Those results highlight the risk of implementing IPTp with drugs that have already been compromised by widespread resistance. Meta-analysis of data from Africa nonetheless has shown an overall benefit of using more frequent doses of SP in IPTp, along with the use of insecticide-treated bed nets, and also suggests that dihydroartemisinin (DHA; the active ART metabolite) + PPQ on this continent could be an effective substitute (Desai et al., 2018). SP + ADQ has also proven to be effective in reducing the incidence of malaria when administered monthly as a 3-day course (Ba et al., 2018). Thus, while SP alone has lost considerable efficacy due to mutations in the target enzymes and pathway, it can still be beneficial as a well-tolerated and safe component of a combination therapy or with more frequent dosing.
Artemisinin (ART) − the Core Component of Current First-line Antimalarial Combinations
The diminished efficacy of CQ and SP forced a major reevaluation of treatment options, resulting in the selection of ART as the core chemical class in current first-line therapies. Its initial description dates back to a traditional Chinese medicine text published in AD 340, which reported the use of Artemisia annua plant extracts to treat cyclical fevers. The active natural product, ART, was identified in 1971 (Tu, 1999). This drug acts against rings as well as the later trophozoite stage of ABS development (Figure 1). Its mode of action appears to involve iron-catalyzed scission of its endoperoxide bridge, leading to widespread alkylation and oxidative damage inside the ABS parasite (Figure 2) (Ismail et al., 2016; Wang et al., 2015). The primary source of iron is thought to derive from parasite-mediated proteolysis of host hemoglobin (Xie et al., 2016). ART displays outstanding pharmacodynamic properties: its clinically-used derivatives DHA, ATM, and artesunate (AS) reduce the biomass of a drug-sensitive infection by up to 10,000-fold every ~48 hour ABS cycle. These derivatives, however, are highly labile, with plasma half-lives of only 1-2 hours (White, 1997, 2013). ART derivatives are therefore paired in ACTs with a longer-acting partner drug with a different mechanism of action. One drawback is that the partner drug is effectively a monotherapy after ART elimination, increasing the chance of partner drug resistance.
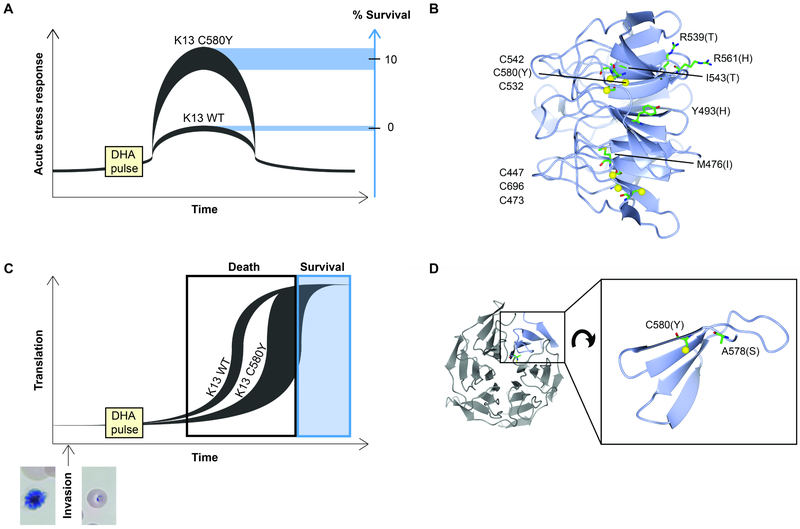
ACTs were adopted globally in the early 2000s. However, emerging resistance arose in Cambodia a decade ago, manifesting as delayed parasite clearance times in patients treated with AS monotherapy (Dondorp et al., 2009; Noedl et al., 2008). The main ART resistance determinant was identified by comparing sequence changes from an in vitro selection and clinical isolates representing a spectrum of parasite clearance times (Ariey et al., 2014). This work identified specific point mutations in the k13 gene (kelch13, PlasmoDB ID: Pf3D7_1343700) as an important determinant and biomarker of ART resistance. Several insights were key to this discovery. One, evidence that ART resistance manifests during the early ring stages and not throughout the entire ABS developmental cycle. Two, the subsequent development of the ring-stage survival assay (RSA0-3h), in which early rings (0-3 hours post-invasion) are exposed to a pharmacologically relevant 6 hour, 700 nM drug pulse of DHA, with survival measured one replication cycle later. Levels of parasite survival in this assay track well with clinical response (Witkowski et al., 2013). Resistance is generally benchmarked in vitro as a survival rate >1% and clinically as a parasite clearance half-life >5 hours (Ariey et al., 2014; Ashley et al., 2014). Three, insights into the Cambodian parasite population structure that was used to distinguish genetic background from potentially causal or compensatory fitness mutations (Miotto et al., 2013). Four, the use of grouped alterations in a gene or pathway. No single mutation in k13 rose to significance in the field study population, but a collection of k13 mutations in genetically-defined subpopulations did (Ariey et al., 2014). Several of these mutations (Y493H, R539T, I543T, and C580Y), all located in the six-bladed propeller region of the K13 protein (Figure 3B), have been confirmed to confer in vitro ART resistance through gene editing via CRISPR-Cas9 or zinc-finger nucleases (Ghorbal et al., 2014; Straimer et al., 2015). Studies with multiple strains and mutations revealed that a consistent subset of early ring parasites survives each drug pulse, with the genetic background influencing the degree of survival. For example, K13 C580Y or R539T mutants can display ~4-24% and 19-49% survival, respectively, compared with essentially no survival in K13 wild-type parasites, and with recent strains showing the most survival (Straimer et al., 2015). Population studies have shown the rapid fixation of the C580Y mutation throughout Southeast Asia (Amato et al., 2018; MalariaGEN, 2016; Menard et al., 2016). Not all K13 mutations lead to ART resistance, as exemplified by the A578S mutation that was observed at very low prevalence in several African countries and that upon gene editing conferred no survival (Menard et al., 2016) (Figure 3D).
Figure 3. Model for K13-mediated ART resistance.
(A) Schematic proposing differences in the acute stress response of P. falciparum K13 wild-type (WT) or K13 C580Y early ring-stage parasites exposed to DHA. Parasite genetic backgrounds contribute to variations in the levels of K13 isoform-mediated stress response and subsequent survival. (B) Side-view of the β-propeller of K13 (PDB: 4YY8, residues 444-726) showing cysteines and/or resistance-associated mutations. Cysteine sulfurs are indicated with yellow spheres. The putative oxidative sensor cysteines are all located in a vertical plane inside the β-propeller. (C) An alternate hypothesis of ART resistance is that eIF2α phosphorylation-mediated translational repression during the schizont-to-ring transition might be extended in K13 mutant, ART-resistant parasites. This delayed re-entry into active translation and growth could allow these parasites to survive ART pulses. Parasites could re-enter growth in a stochastic manner, with later recovery permitting survival. Schizont and early ring-stage parasites are shown below. (D) Close-up of one β-sheet (residues 577-615) showing spatial separation of C580 and A578, located on a structured β-strand and on an unstructured loop, respectively. C580Y is a mediator of ART resistance, whereas A578S is not (Menard et al., 2016). Images made with CCP4mg.
Investigations into K13-mediated ring-stage survival following ART treatment have been complicated by the high death rate even in resistant lines, and most research has therefore focused on constitutive differences between K13 mutant and wild-type parasites without exposure to drug. K13-mutant parasite appear to alter their ABS cell cycle to lengthen the ring stage and shorten the trophozoite stage (Hott et al., 2015; Mok et al., 2015). This may reduce exposure to drug-activating iron found in heme, whose levels are maximal in trophozoites (Cheng et al., 2012). Temporary dormancy would be of substantial benefit given the extremely short plasma half-life of ART derivatives (typically <1-2 hr). Transcriptomic analysis of over 1,000 patient isolates has linked ART resistance with increased expression of the unfolded protein response (UPR) pathway and decelerated development through the ring stage (Mok et al., 2015). Metabolomics showed a basal increase in antioxidant glutathione levels and a decrease in hemoglobin catabolism in ART-resistant versus sensitive lines (Siddiqui et al., 2017). The damaged proteome in ART-treated parasites leads to a high burden of ubiquitinated proteins, which is lower in ART-resistant parasites, perhaps because of enhanced stress responses that engage the ubiquitin/proteasome system (Dogovski et al., 2015). DHA-treated ART-sensitive 3D7 parasites were found to have reduced but not ablated glycolytic flux (Shivapurkar et al., 2018). These data point to ART resistance being an actively maintained, cytoprotective, metabolically dampened but not quiescent state that primarily protects early rings.
Several hypotheses have been put forth on the function of mutant and wild-type k13 in normal physiology and under ART pressure. One proposal is that this gene acts as a negative regulator of stress responses and that the mutations conferring ART resistance destabilize the K13 protein, leading to a greater induction of protective stress responses (Figure 3A). The K13 protein has significant structural and sequence similarity to human Keap1 (Ariey et al., 2014). Keap1 is a cysteine-based oxidative stress sensor that negatively regulates stress responses by controlling the turnover of many proteins, including the transcription factor Nrf2 that serves as the master regulator of oxidative stress responses. Studies with a mutant P. falciparum line harboring a piggyBac transposon inserted upstream of the k13 coding sequence identified reduced k13 transcription in early ring stages and upregulation in trophozoite stages, which was inversely associated with altered transcription levels in several DNA replication and repair genes, raising the possibility that K13 can also alter transcriptional responses in the parasite (Gibbons et al., 2018). Specific mutations in K13 may destabilize the protein, reduce its functional capabilities, or mimic damaged cysteine residues by preventing disulfide bond formation, thereby releasing the repression of protective stress responses. A different k13 study using diCre-driven “knock sideways” protein mislocalization and endogenous gene excision yielded parasites that arrested their growth at the ring stage. and later became condensed forms (Birnbaum et al., 2017). In that study, GFP-tagged K13 was observed in punctate foci in the parasite cytoplasm (Birnbaum et al., 2017). The development of an antibody specific to the native protein would allow epitope tag-free localization, examination of wild-type and mutant K13 turnover rates via pulse-chase experiments, and the identification of interacting partners and ubiquitination substrates. Mutating the putative sensor cysteines (including C580; Figures 3B, 3D) would also be a useful approach to testing their contribution to oxidative stress responses (Haldar et al., 2018). Faster induction, longer duration, and/or larger amplitude of stress responses may explain ART resistance, but excessive stress responses have repercussions in terms of energy usage and inflammation. Perhaps the K13 C580Y mutation dominates in the field, despite other mutations giving higher levels of survival, because the stress response is sufficiently enhanced to survive but is not so high that it results in major fitness costs. Another possibility is that, if the cysteines of K13 are oxidative damage sensors, mutating C580 to a tyrosine sensitizes or constitutively activates the sensor system.
Another hypothesis involves lipid signaling, with elevated levels of phosphatidylinositol-3-phosphate (PI3P) tracking closely with ART resistance in field isolates and conferring resistance in laboratory experiments (Mbengue et al., 2015). In this study, phosphatidylinositol-3-kinase (PfPI3K) was shown to be elevated in ART-resistant isolates with or without K13 mutations, and PfPI3K function was inhibited by ART. PfPI3K was shown to interact with and be polyubiquitinated by K13. Ubiquitination was decreased in a K13 C580Y mutant when compared to wild-type parasites, leading to increased PfPI3K protein, and thus increased PI3P. These resistant cells displayed endoplasmic reticulum (ER) hypervesiculation, with these vesicles being enriched in PI3P, K13, and a variety of protein homeostatic proteins, such as those involved in protein export and the UPR (Bhattacharjee et al., 2018). PI3P could provide a mechanistic link between the K13 protein’s putative sensor function and increased capacity of resistant cells to survive massive damage to the proteome via upregulation of protein homeostatic processes such as the UPR and proteasomal clearance of damaged proteins.
A separate mechanistic study showed that ART treatment in both P. falciparum and the rodent malaria P. berghei resulted in phosphorylation of the parasite eukaryotic initiation factor-2α (eIF2α), and that increased phosphorylation correlated with increased rates of ART recrudescence (Zhang et al., 2017). Phosphorylation of eIF2α represses global protein translation while enhancing the translation of a subset of mRNAs involved in stress responses. This translationally-repressed state protects against a variety of cell stresses and is a normal part of the Plasmodium life cycle in the transitions between RBCs and between mosquitoes and vertebrate hosts. In the former case, an ER-resident kinase called PK4 phosphorylates eIF2α during the schizont stage. PK4 inhibition blocked eIF2α-induced latency and ART survival in rodent malaria (Zhang et al., 2017). In the latter case, eIF-2a phosphorylation is regulated by the IK2 kinase, which enforces latency in Plasmodium salivary gland sporozoites and the subsequent de-repression of translation after liver stage formation in the vertebrate host (Zhang et al., 2010). ART resistance could be due to an elongation of schizont-stage eIF2α phosphorylation further into the subsequent ring stage, or a K13-mediated rapid reengagement of this protective state after stress (Figure 3C). As PK4 resides in the ER, perhaps ER stress and the subsequent UPR from the ART-damaged proteome triggers increased PK4 activity. PK4 may also act like its mammalian homolog, PERK, and be involved in maintaining physical links between the ER and mitochondrial membranes. These specialized contact sites are involved in reciprocal transfer of danger signals like calcium ions and also synthesize lipids, including PI3P, among other roles (Zhang et al., 2017). Although K13 mutations dominate in the field, ART resistance could result from multiple signaling pathways that activate proteostatic stress responses in the mitochondrial-ER tubulovesicular network. Not all of the data point to k13 as the primary or sole possible mediator of ART resistance. A genetic cross of P. falciparum lines in splenectomized Aotus monkeys revealed that although in vitro survival segregated with K13 C580Y mutant allele status, both K13 wild-type and mutant parasites recrudesced after AS treatment. These findings provided evidence that mutant K13 was insufficient in this model system to cause lengthened parasite clearance times following AS treatment (Sa et al., 2018). Recent evidence for an alternate path to resistance came from long-term DHA selection studies with two Senegalese isolates, which yielded resistant parasites carrying point mutations in the coronin protein, whose 7-bladed propeller domain is structurally similar to K13. CRISPR-Cas9 editing confirmed the casual role of these mutations (Demas et al., 2018), which have not been detected in field isolates. K13-independent low-grade ART resistance was also separately obtained in early rings following longterm gradual increases in DHA pressure, and was associated with enhanced adaptive responses against oxidative stress and protein damage (Rocamora et al., 2018).
With these occasionally overlapping and conflicting results, it may be useful to consider ART in a variety of systems. ART is being widely explored as a cancer treatment or adjuvant. Mechanistic studies in a variety of human cancer cells have converged on a role for the mitochondria, with reports of redox imbalances, increased reactive oxygen species (ROS), and iron-catalyzed outer mitochondrial membrane permeabilization leading to autophagy, mitophagy, and apoptosis (Efferth, 2017). In Plasmodium, ARTs have been found to trigger rapid ROS-dependent depolarization of mitochondrial and plasma membrane potentials (Antoine et al., 2014). Of note, Southeast Asian Cambodian parasites have been observed to have 3-4 fold more mitochondrial genomes than West African Ghanaian parasites, allowing for more genetic diversity through heteroplasmy and perhaps also cushioning ART-caused loss of mitochondrial function (Siegel et al., 2017). DHA-treated ART-sensitive W2 parasites were reported to have low levels of survival in a modified in vitro assay, attributed to dormant parasites whose subsequent recovery has been associated with the presence of transcriptionally active mitochondria (Peatey et al., 2015).
One can also draw insight from bacterial persister cells. Persisters are a subpopulation that transiently tolerates antibacterial stresses without being genetically resistant, leading to incomplete clearance and prolonged or relapsing infection. Persister formation is linked to various stress response pathways. Antibiotic tolerance is often accomplished via slowed or arrested growth and a corresponding decrease in metabolism and drug uptake (Cohen et al., 2013). Phenotypic heterogeneity in the population via stochastic gene expression, microenvironments, and/or asymmetric cell division increases the odds of successful adaptation via genetic resistance. Mutations that increase the probability of persister formation in a clonal population are called high-persistence mutations. k13 mutations might be analogous, with different mutations yielding different levels of acute stress response, correlating with different ranges of persister cell survival (Figure 3A). Exposure to ART tends to result in stalled cells, and some small but reproducible portion of ART-resistant parasites are able to stochastically re-enter active growth. Perhaps Plasmodium mitochondria have retained bacterial-like pathways for sensing oxidative damage and stimulating persister cell formation as a mechanism that underlies ART resistance. If so, this suggests treatment possibilities, as bacterial persister cells can be targeted by also inhibiting the integrated stress response (Cohen et al., 2013). Indeed, inhibiting the parasite proteasome can synergize with DHA and overcome ART resistance (Dogovski et al., 2015; Kirkman et al., 2018; Stokes et al., 2019). Inhibiting other stress response systems, such as protein folding chaperones, should also be tested for effective combination therapies.
ACT partner drug resistance: piperaquine
Parasites resistant to both DHA and PPQ emerged in the first year of roll-out in Cambodia, and within 5 years, over 50% of DHA+PPQ treatments were failing (Amato et al., 2018). Treatment failures were also observed in 26% of patients receiving DHA+PPQ in neighboring Vietnam (Thanh et al., 2017). Genome-wide association studies identified gene amplification of the hemoglobinases plasmepsins II and III as a molecular marker of DHA+PPQ treatment failures in patients and of PPQ resistance in vitro (Amato et al., 2017; Witkowski et al., 2017). These multi-drug resistant parasites may have been the result of genetic recombination events between lineages independently carrying mutant K13 and multicopy plasmepsins II and III. This “KEL1/PLA1” or “PfPailin” co-lineage has been observed throughout Southeast Asia and is predominant in western Cambodia (Amato et al., 2018; Imwong et al., 2017). Prior use of ART and PPQ as monotherapies might have separately selected for resistance with current parasites being a result of these two traits combining. Of note, the much longer half-life of PPQ (~30 days) versus DHA (~1 hour) gives a months-long tail of PPQ monotherapy that would have facilitated the selection of PPQ-resistant parasites (White, 1997, 2013). In vitro studies have been complicated by the unusual bi-phasic dose-response curves observed with resistant parasites, in which PPQ IC50 values are similar to sensitive parasites but IC90 values increase substantially. Analysis of dose-response data with Southeast Asian field isolates found that multicopy plasmepsins II and III were generally associated with reduced parasite killing at high PPQ concentrations, with some exceptions (Bopp et al., 2018). In a separate study, overexpression of these genes in a PPQ-sensitive background did not alter PPQ susceptibility (Loesbanluechai et al., 2018).
A separate analysis of Cambodian culture-adapted isolates observed that the association between elevated PPQ IC90 values and multicopy plasmepsins II and III was significantly increased when parasites also harbored a novel mutation, F145I, in PfCRT. Indeed, this novel PfCRT variant and multicopy plasmepsin II showed very similar hazard ratios for parasite recurrence in patients treated with DHA+PPQ (Agrawal et al., 2017). The ability of PfCRT variants including F145I, M343L, and G353V to confer varying levels of PPQ resistance was recently demonstrated via gene editing. Importantly, PPQ resistance was achieved in the Dd2 parasite line that carries a single copy of plasmepsins II and III (Ross et al., 2018). Studies also reveal a very recent and rapid increase in the proportion of novel PfCRT mutations on a Dd2 isoform background in Cambodia, where PPQ resistance first arose (Duru et al., 2015; Ross et al., 2018). We suspect that plasmepsin II and III gene amplification leads to low-grade PPQ resistance or in some way enables parasites to develop high-grade resistance via mutations in PfCRT. Variant isoforms of PfCRT have been shown to interfere with drug-mediated inhibition of heme detoxification by effluxing compound out of the digestive vacuole, with the drug specificity determined by the transporter’s sequence. Of note, the Cambodian PfCRT mutations that confer PPQ resistance sensitize the parasites to CQ, presumably by altering drug interactions with this pleotropic transporter (Ross et al., 2018). The emergence of a novel C350R mutation in a South American PfCRT isoform (7G8) was also recently implicated in decreased PPQ susceptibility in French Guiana, again rendering these parasites CQ-sensitive (Pelleau et al., 2015). Structural insights into this transporter and its interactions with various small molecules will be helpful in elucidating the underlying mechanisms by which PfCRT can mediate drug resistance and how alterations in selectivity could be exploited by partnering drugs with incompatible resistance mechanisms.
Implications for Treatment and Future Research Outlook
In 1955, The World Health Organization (WHO) spearheaded the Global Malaria Eradication Program. Culturally, the “golden age of antibiotics” led to an optimistic sense of invincibility against microbial pathogens. DDT (dichloro-diphenyl-trichloroethane) had been used with great success as an insecticide in World War II, and the antimalarial CQ was effective, easily available, and inexpensive. Malaria was eradicated from many locales with mild to moderate transmission with these tools, including the southern United States. However, the global program was abandoned after 14 years with the acknowledgement that the current tools were no longer effective or suitable, and that political will and financial support were dwindling. The price of dismantling elimination efforts was highlighted in Madagascar, which had eliminated malaria by 1960, but had a devastating rebound epidemic by the 1980s after withdrawing control efforts in a now-immunologically naive population (Randrianarivelojosia et al., 2009). An important objective of current malaria elimination and eradication efforts is to avoid similar scenarios.
Further research is necessary to determine how to best use existing treatments, spanning areas ranging from community engagement and training local health workers to identifying the most effective and tolerable drug combinations and dosing regimens. One promising new regimen is DHA + pyronaridine, with the partner drug having recently received regulatory approval and safety concerns broadly abetted. Separately, clinical trials show that triple ACTs (ATM+LMF plus ADQ or DHA+PPQ plus MFQ; clinical trial numbers NCT03355664 and NCT02453308, respectively) may be safe and more effective than the current two-component ACTs, even in areas with widespread ART resistance. Laboratory studies are useful here in assessing whether parasites can become resistant to all three agents, or whether certain combinations are better in exerting opposing selective pressures. For example, LMF and ADQ are known to exert opposing pressures on mutant versus wild-type isoforms of both PfCRT and PfMDR1 (Blasco et al., 2017; Venkatesan et al., 2014). PPQ and MFQ were also thought to potentially exert opposing pressures, based on the finding that pfmdr1 gene amplification, which contributes to MFQ resistance and treatment failures, was found to be substantially less prevalent in Cambodia several years after switching from MFQ to PPQ as first-line drugs (Amato et al., 2017; Witkowski et al., 2017). Studies with isogenic lines expressing one or two copies of pfmdr1 showed no difference in MFQ IC50 values (Dhingra et al., 2017), and some isolates have been found to carry multicopy pfmdr1 and plasmepsin II (Bopp et al., 2018), suggesting that resistance to both drugs in a single parasite is possible. Similarly, changing the dosing regimen to “sequential double ACT” may be sufficient to overpower transient dormancy-like ART resistance, and is also being tested in a clinical trial (Schallig et al., 2017). In contrast, “split dosing,” i.e., splitting a once-daily dose into a twice-daily dose to increase daily ART exposure, did not increase the efficacy of ART, likely because splenic parasite clearance was exceeded (White et al., 2017). A recent clinical trial in multiple villages in Myanmar, Vietnam, Cambodia, and Laos showed that despite the widespread presence of mutant K13 parasites, three monthly rounds of mass drug administration with DHA+PPQ reduced malaria prevalence and incidence over a one-year follow up period (von Seidlein et al., 2019). Further studies into the benefits versus risks of mass drug administration will be required in areas with pre-existing drug resistance, or where coverage would be incomplete.
Deepening the ties between clinical studies and experimental research would clearly be beneficial, especially in terms of linking treatment failures with molecular surveillance, spatial epidemiology, and complex covariables such as HIV or bacterial pathogen coinfections. Improved diagnostics would also be helpful in detecting and treating asymptomatic carriers as well as reducing overtreatment by identifying non-malaria causes of fever. The further integration of genomic data science and population biology will aid in determining which genomic regions show signs of selection in the field after drug treatment of patients, and in the lab after in vitro resistance selections. Performing large-scale selections of parasite lines resistant to a broad array of antimalarial agents and combining these with omics studies have been useful in delineating novel resistance pathways and candidate druggable targets and pathways (Allman et al., 2016; Cowell et al., 2018). Screening resistant parasites against panels of drugs with known mechanisms, or conversely small molecules against panels of parasites with known resistance determinants, can quickly narrow down areas of focus (Corey et al., 2016). Extensive screening efforts have yielded thousands of sub-micromolar inhibitors of P. falciparum ABS parasite growth, and subsequent investigations have identified a plethora of novel antimalarial targets with modes of action unrelated to current ACTs (Phillips et al., 2017); Table 2). Resistance continues to be problematic, however, with a number of antimalarials in development yielding resistance readily in culture (Blasco et al., 2017). A recent clinical trial showed that mutations affording moderate resistance to the plasmodial DHODH inhibitor DSM265 were identified in P. falciparum-infected patients who recrudesced following a single dose (Llanos-Cuentas et al., 2018). Resistance also arises readily to another mitochondrial inhibitor, atovaquone, which targets cytochrome b and inhibits electron transport. Cytochrome b mutations that afford atovaquone resistance were reported to have a substantial fitness cost during sexual stage development in Anopheles mosquitoes, suggesting that resistance is not transmissible in the field (Goodman et al., 2016). This result highlights the importance of considering fitness costs throughout the life cycle and the anticipated benefit of developing multistage-active antimalarials (Burrows et al., 2017).
Experimental research into antimalarial resistance also benefits from several technological advances in genetics and genomics, including CRISPR-Cas9 gene editing, tunable expression via aptamers, diCre recombinase-based conditional knockouts, “knock sideways” protein mislocalization approaches, transposon-generated saturation mutagenesis libraries, genetic crosses in non-human hosts, and the increasing accessibility and robustness of whole-genome sequencing (Birnbaum et al., 2017; Ghorbal et al., 2014; Goldfless et al., 2014; Vaughan et al., 2015; Zhang et al., 2018). These approaches would benefit from improved tools in other arenas, including the development of panels of antibodies specific to Plasmodium proteins, which would enable deeper cell biology-based investigations, and single-cell manipulation and analysis.
Malaria is a global public health threat, and drug resistance is a perennial issue. The former gold standard CQ eventually succumbed to resistance via parasite-mediated drug efflux. The next major therapeutic, SP, was compromised by mutation and copy number amplification in the targeted folate biosynthesis pathway. Pathways of resistance to the current first-line treatments, which combine ART derivatives with partner drugs, are less clear. ART resistance might be attributable to a stress response-based persister cell phenotype, and PPQ resistance appears to be due to changes in the hemoglobin degradation pathway and DV transport processes. New technologies are enabling previously infeasible studies, including in parasite genetics and genomics. Rethinking how drug combinations are chosen or dosed could help preserve drug efficacy under threat from parasite evolution. A better understanding of resistance to existing and in-development therapies presents a rational path to identify new means to regain the momentum in the fight to reduce the global impact of malaria.
Acknowledgements
The authors declare no competing interests. Funding for this work was provided by the NIH (R01 AI50234, R01AI124678 and R01 AI109023 to DAF). LSR gratefully acknowledges earlier funding from the NIH (NRSA T32 AI120578).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Agrawal S, Moser KA, Morton L, Cummings MP, Parihar A, Dwivedi A, Shetty AC, Drabek EF, Jacob CG, Henrich PH, et al. (2017). A novel mutation in the Plasmodium falciparum chloroquine resistance transporter is associated with decreased piperaquine sensitivity. J Infect Dis 216, 468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman EL, Painter HJ, Samra J, Carrasquilla M, and Llinas M (2016). Metabolomic profiling of the malaria box reveals antimalarial target pathways. Antimicrob Agents Chemother 60, 6635–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, et al. (2017). Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 17, 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato R, Pearson RD, Almagro-Garcia J, Amaratunga C, Lim P, Suon S, Sreng S, Drury E, Stalker J, Miotto O, et al. (2018). Origins of the current outbreak of multidrug-resistant malaria in southeast Asia: a retrospective genetic study. Lancet Infect Dis 18, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine T, Fisher N, Amewu R, O'Neill PM, Ward SA, and Biagini GA (2014). Rapid kill of malaria parasites by artemisinin and semi-synthetic endoperoxides involves ROS-dependent depolarization of the membrane potential. J Antimicrob Chemother 69, 1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, et al. (2014). A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, et al. (2014). Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba EH, Pitt C, Dial Y, Faye SL, Cairns M, Faye E, Ndiaye M, Gomis JF, Faye B, Ndiaye JL, et al. (2018). Implementation, coverage and equity of large-scale door-to-door delivery of Seasonal Malaria Chemoprevention (SMC) to children under 10 in Senegal. Sci Rep 8, 5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Coppens I, Mbengue A, Suresh N, Ghorbal M, Slouka Z, Safeukui I, Tang HY, Speicher DW, Stahelin RV, et al. (2018). Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood 131, 1234–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum J, Flemming S, Reichard N, Soares AB, Mesen-Ramirez P, Jonscher E, Bergmann B, and Spielmann T (2017). A genetic system to study Plasmodium falciparum protein function. Nat Methods 14, 450–456. [DOI] [PubMed] [Google Scholar]
- Blasco B, Leroy D, and Fidock DA (2017). Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 23, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp S, Magistrado P, Wong W, Schaffner SF, Mukherjee A, Lim P, Dhorda M, Amaratunga C, Woodrow CJ, Ashley EA, et al. (2018). Plasmepsin II-III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun 9, 1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, et al. (2013). Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet 9, e1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows JN, Duparc S, Gutteridge WE, Hooft van Huijsduijnen R, Kaszubska W, Macintyre F, Mazzuri S, Mohrle JJ, and Wells TNC (2017). New developments in anti-malarial target candidate and product profiles. Malar J 16, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro I, Roca-Feltrer A, Griffin JT, Smith L, Tanner M, Schellenberg JA, Greenwood B, and Schellenberg D (2010). Age-patterns of malaria vary with severity, transmission intensity and seasonality in sub-Saharan Africa: a systematic review and pooled analysis. PLoS One 5, e8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Moss EL, Park DJ, Ndiaye D, Mboup S, Volkman SK, Sabeti PC, Wirth DF, Neafsey DE, and Hartl DL (2013). Malaria life cycle intensifies both natural selection and random genetic drift. Proc Natl Acad Sci USA 110, 20129–20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Kyle DE, and Gatton ML (2012). Artemisinin resistance in Plasmodium falciparum: A process linked to dormancy? Int J Parasitol Drugs Drug Resist 2, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Lobritz MA, and Collins JJ (2013). Microbial persistence and the road to drug resistance. Cell Host Microbe 13, 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey VC, Lukens AK, Istvan ES, Lee MC, Franco V, Magistrado P, Coburn-Flynn O, Sakata-Kato T, Fuchs O, Gnadig NF, et al. (2016). A broad analysis of resistance development in the malaria parasite. Nat Commun 7, 11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell AN, Istvan ES, Lukens AK, Gomez-Lorenzo MG, Vanaerschot M, Sakata-Kato T, Flannery EL, Magistrado P, Owen E, Abraham M, et al. (2018). Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 359, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demas AR, Sharma AI, Wong W, Early AM, Redmond S, Bopp S, Neafsey DE, Volkman SK, Hartl DL, and Wirth DF (2018). Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc Natl Acad Sci USA 115, 12799–12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M, Hill J, Fernandes S, Walker P, Pell C, Gutman J, Kayentao K, Gonzalez R, Webster J, Greenwood B, et al. (2018). Prevention of malaria in pregnancy. Lancet Infect Dis 18, e119–e132. [DOI] [PubMed] [Google Scholar]
- Dhingra SK, Redhi D, Combrinck JM, Yeo T, Okombo J, Henrich PP, Cowell AN, Gupta P, Stegman ML, Hoke JM, et al. (2017). A variant PfCRT isoform can contribute to Plasmodium falciparum resistance to the first-line partner drug piperaquine. MBio 8, e00303–00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogovski C, Xie SC, Burgio G, Bridgford J, Mok S, McCaw JM, Chotivanich K, Kenny S, Gnadig N, Straimer J, et al. (2015). Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol 13, e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, et al. (2009). Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duru V, Khim N, Leang R, Kim S, Domergue A, Kloeung N, Ke S, Chy S, Eam R, Khean C, et al. (2015). Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med 13, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efferth T (2017). From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin Cancer Biol 46, 65–83. [DOI] [PubMed] [Google Scholar]
- Frosch AE, Laufer MK, Mathanga DP, Takala-Harrison S, Skarbinski J, Claassen CW, Dzinjalamala FK, and Plowe CV (2014). Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J Infect Dis 210, 1110–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabryszewski SJ, Modchang C, Musset L, Chookajorn T, and Fidock DA (2016). Combinatorial genetic modeling of pfcrt-mediated drug resistance evolution in Plasmodium falciparum. Mol Biol Evol 33, 1554–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething PW, Casey DC, Weiss DJ, Bisanzio D, Bhatt S, Cameron E, Battle KE, Dalrymple U, Rozier J, Rao PC, et al. (2016). Mapping Plasmodium falciparum mortality in Africa between 1990 and 2015. N Engl J Med 375, 2435–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, and Lopez-Rubio JJ (2014). Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32, 819–821. [DOI] [PubMed] [Google Scholar]
- Gibbons J, Button-Simons KA, Adapa SR, Li S, Pietsch M, Zhang M, Liao X, Adams JH, Ferdig MT, and Jiang RHY (2018). Altered expression of K13 disrupts DNA replication and repair in Plasmodium falciparum. BMC Genomics 19, 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfless SJ, Wagner JC, and Niles JC (2014). Versatile control of Plasmodium falciparum gene expression with an inducible protein-RNA interaction. Nat Commun 5, 5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes PS, Bhardwaj J, Rivera-Correa J, Freire-De-Lima CG, and Morrot A (2016). Immune escape strategies of malaria parasites. Front Microbiol 7, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, Siregar JE, Mollard V, Vega-Rodriguez J, Syafruddin D, Matsuoka H, Matsuzaki M, Toyama T, Sturm A, Cozijnsen A, et al. (2016). Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science 352, 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson A, and Plowe CV (2005). Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev 57, 117–145. [DOI] [PubMed] [Google Scholar]
- Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, Phillips MA, DeRisi J, and Rathod PK (2013). Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS Pathog 9, e1003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar K, Bhattacharjee S, and Safeukui I (2018). Drug resistance in Plasmodium. Nat Rev Microbiol 16, 156–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington WE, Mutabingwa TK, Muehlenbachs A, Sorensen B, Bolla MC, Fried M, and Duffy PE (2009). Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc Natl Acad Sci USA 106, 9027–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hott A, Casandra D, Sparks KN, Morton LC, Castanares GG, Rutter A, and Kyle DE (2015). Artemisinin-resistant Plasmodium falciparum parasites exhibit altered patterns of development in infected erythrocytes. Antimicrob Agents Chemother 59, 3156–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Hien TT, Thuy-Nhien NT, Dondorp AM, and White NJ (2017). Spread of a single multidrug resistant malaria parasite lineage (PfPailin) to Vietnam. Lancet Infect Dis 17, 1022–1023. [DOI] [PubMed] [Google Scholar]
- Ismail HM, Barton V, Phanchana M, Charoensutthivarakul S, Wong MH, Hemingway J, Biagini GA, O'Neill PM, and Ward SA (2016). Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc Natl Acad Sci U S A 113, 2080–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman LA, Zhan W, Visone J, Dziedziech A, Singh PK, Fan H, Tong X, Bruzual I, Hara R, Kawasaki M, et al. (2018). Antimalarial proteasome inhibitor reveals collateral sensitivity from intersubunit interactions and fitness cost of resistance. Proc Natl Acad Sci USA 115, E6863–E6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Symington LS, and Fidock DA (2014). DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol Mol Biol Rev 78, 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos-Cuentas A, Casapia M, Chuquiyauri R, Hinojosa JC, Kerr N, Rosario M, Toovey S, Arch RH, Phillips MA, Rozenberg FD, et al. (2018). Antimalarial activity of single-dose DSM265, a novel plasmodium dihydroorotate dehydrogenase inhibitor, in patients with uncomplicated Plasmodium falciparum or Plasmodium vivax malaria infection: a proof-of-concept, open-label, phase 2a study. Lancet Infect Dis 18, 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesbanluechai D, Kotanan N, de Cozar C, Kochakarn T, Ansbro MR, Chotivanich K, White NJ, Wilairat P, Lee MCS, Gamo FJ, et al. (2018). Overexpression of plasmepsin II and plasmepsin III does not directly cause reduction in Plasmodium falciparum sensitivity to artesunate, chloroquine and piperaquine. Int J Parasitol Drugs Drug Resist 9, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MalariaGEN (2016). Genomic epidemiology of artemisinin resistant malaria. Elife 5, e08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, et al. (2015). A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520, 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, et al. (2016). A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374, 2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, et al. (2013). Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, et al. (2015). Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 347, 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo I, and Roper C (2013). Mapping 'partially resistant', 'fully resistant', and 'super resistant' malaria. Trends Parasitol 29, 505–515. [DOI] [PubMed] [Google Scholar]
- Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, Newton P, Nosten F, Ferdig MT, and Anderson TJ (2008). Adaptive copy number evolution in malaria parasites. PLoS Genet 4, e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, and Artemisinin Resistance in Cambodia 1 Study, C. (2008). Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359, 2619–2620. [DOI] [PubMed] [Google Scholar]
- Peatey CL, Chavchich M, Chen N, Gresty KJ, Gray KA, Gatton ML, Waters NC, and Cheng Q (2015). Mitochondrial membrane potential in a small subset of artemisinin-induced dormant Plasmodium falciparum parasites in vitro. J Infect Dis 212, 426–434. [DOI] [PubMed] [Google Scholar]
- Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, Volkman SK, Wirth DF, Legrand E, Fidock DA, et al. (2015). Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA 112, 11672–11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MA, Burrows JN, Manyando C, van Huijsduijnen RH, Van Voorhis WC, and Wells TNC (2017). Malaria. Nat Rev Dis Primers 3, 17050. [DOI] [PubMed] [Google Scholar]
- Randrianarivelojosia M, Raveloson A, Randriamanantena A, Juliano JJ, Andrianjafy T, Raharimalala LA, and Robert V (2009). Lessons learnt from the six decades of chloroquine use (1945-2005) to control malaria in Madagascar. Trans R Soc Trop Med Hyg 103, 3–10. [DOI] [PubMed] [Google Scholar]
- Rocamora F, Zhu L, Liong KY, Dondorp A, Miotto O, Mok S, and Bozdech Z (2018). Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLoS Pathog 14, e1006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepe PD (2011). PfCRT-mediated drug transport in malarial parasites. Biochemistry 50, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kumpornsin K, Takala-Harrison S, Witkowski B, Fairhurst RM, Ariey F, et al. (2018). Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 9, 3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa JM, Kaslow SR, Krause MA, Melendez-Muniz VA, Salzman RE, Kite WA, Zhang M, Moraes Barros RR, Mu J, Han PK, et al. (2018). Artemisinin resistance phenotypes and K13 inheritance in a Plasmodium falciparum cross and Aotus model. Proc Natl Acad Sci USA 115, 12513–12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallig HD, Tinto H, Sawa P, Kaur H, Duparc S, Ishengoma DS, Magnussen P, Alifrangis M, and Sutherland CJ (2017). Randomised controlled trial of two sequential artemisinin-based combination therapy regimens to treat uncomplicated falciparum malaria in African children: a protocol to investigate safety, efficacy and adherence. BMJ Glob Health 2, e000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivapurkar R, Hingamire T, Kulkarni AS, Rajamohanan PR, Reddy DS, and Shanmugam D (2018). Evaluating antimalarial efficacy by tracking glycolysis in Plasmodium falciparum using NMR spectroscopy. Sci Rep 8, 18076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui G, Srivastava A, Russell AS, and Creek DJ (2017). Multi-omics based identification of specific biochemical changes associated with PfKelch13-mutant artemisinin-resistant Plasmodium falciparum. J Infect Dis 215, 1435–1444. [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Verdier-Pinard D, and Fidock DA (2002). Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298, 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel SV, Rivero AV, Adapa SR, Wang C, Manetsch R, Jiang RH, and Kyle DE (2017). Mitochondrial heteroplasmy is responsible for atovaquone drug resistance in Plasmodium falciparum. bioRxiv 10.1101/232033. [DOI] [Google Scholar]
- Stokes BH, Yoo E, Murithi JM, Luth MR, Afanasyev P, da Fonseca PC, Winzeler EA, Ng CL, Bogyo M, and Fidock DA (2019). Covalent Plasmodium falciparum-selective proteasome inhibitors exhibit a low propensity for generating resistance in vitro and synergize with multiple antimalarial agents. PLoS Pathog in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, et al. (2015). K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347, 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Kirkman LA, Fujioka H, and Wellems TE (1997). Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell 91, 593–603. [DOI] [PubMed] [Google Scholar]
- Summers RL, Nash MN, and Martin RE (2012). Know your enemy: understanding the role of PfCRT in drug resistance could lead to new antimalarial tactics. Cell Mol Life Sci 69, 1967–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S, and Laufer MK (2015). Antimalarial drug resistance in Africa: key lessons for the future. Ann N Y Acad Sci 1342, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh NV, Thuy-Nhien N, Tuyen NT, Tong NT, Nha-Ca NT, Dong LT, Quang HH, Farrar J, Thwaites G, White NJ, et al. (2017). Rapid decline in the susceptibility of Plasmodium falciparum to dihydroartemisinin-piperaquine in the south of Vietnam. Malar J 16, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y (1999). The development of new antimalarial drugs: qinghaosu and dihydro-qinghaosu. Chin Med J (Engl) 112, 976–977. [PubMed] [Google Scholar]
- Vaughan AM, Pinapati RS, Cheeseman IH, Camargo N, Fishbaugher M, Checkley LA, Nair S, Hutyra CA, Nosten FH, Anderson TJ, et al. (2015). Plasmodium falciparum genetic crosses in a humanized mouse model. Nat Methods 12, 631–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnadig N, Uhlemann AC, Martin RE, Lehane AM, and Fidock DA (2016). Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun 7, 11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M, Gadalla NB, Stepniewska K, Dahal P, Nsanzabana C, Moriera C, Price RN, Martensson A, Rosenthal PJ, Dorsey G, et al. (2014). Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemetherlumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 91, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Seidlein L, Peto TJ, Landier J, Nguyen TN, Tripura R, Phommasone K, Pongvongsa T, Lwin KM, Keereecharoen L, Kajeechiwa L, et al. (2019). The impact of targeted malaria elimination with mass drug administrations on falciparum malaria in Southeast Asia: A cluster randomised trial. PLoS Med 16, e1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang CJ, Chia WN, Loh CC, Li Z, Lee YM, He Y, Yuan LX, Lim TK, Liu M, et al. (2015). Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat Commun 6, 10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells TN, Burrows JN, and Baird JK (2010). Targeting the hypnozoite reservoir of Plasmodium vivax: the hidden obstacle to malaria elimination. Trends Parasitol 26, 145–151. [DOI] [PubMed] [Google Scholar]
- White NJ (1997). Assessment of the pharmacodynamic properties of antimalarial drugs in vivo. Antimicrob Agents Chemother 41, 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ (2013). Pharmacokinetic and pharmacodynamic considerations in antimalarial dose optimization. Antimicrob Agents Chemother 57, 5792–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ, Watson J, and Ashley EA (2017). Split dosing of artemisinins does not improve antimalarial therapeutic efficacy. Sci Rep 7, 12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, et al. (2017). A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 17, 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B, Khim N, Chim P, Kim S, Ke S, Kloeung N, Chy S, Duong S, Leang R, Ringwald P, et al. (2013). Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother 57, 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie SC, Dogovski C, Hanssen E, Chiu F, Yang T, Crespo MP, Stafford C, Batinovic S, Teguh S, Charman S, et al. (2016). Haemoglobin degradation underpins the sensitivity of early ring stage Plasmodium falciparum to artemisinins. J Cell Sci 129, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fennell C, Ranford-Cartwright L, Sakthivel R, Gueirard P, Meister S, Caspi A, Doerig C, Nussenzweig RS, Tuteja R, et al. (2010). The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J Exp Med 207, 1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Gallego-Delgado J, Fernandez-Arias C, Waters NC, Rodriguez A, Tsuji M, Wek RC, Nussenzweig V, and Sullivan WJ Jr. (2017). Inhibiting the Plasmodium eIF2alpha kinase PK4 Prevents artemisinin-induced latency. Cell Host Microbe 22, 766–776.e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang C, Otto TD, Oberstaller J, Liao X, Adapa SR, Udenze K, Bronner IF, Casandra D, Mayho M, et al. (2018). Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]