Abstract
The use and advantages of high-resolution mass spectrometry as a discovery tool for environmental chemical monitoring has been demonstrated for environmental samples but not for biological samples. We developed a method using liquid chromatography-quadrupole time-of-flight mass spectrometry (LC-QTOF/MS) for discovery of previously unmeasured environmental chemicals in human serum. Using non-targeted data acquisition (full scan MS analysis) we were able to screen for environmental organic acids (EOAs) in 20 serum samples from second trimester pregnant women. We define EOAs as environmental organic compounds with at least one dissociable proton which are utilized in commerce. EOAs include environmental phenols, phthalate metabolites, perfluorinated compounds (PFCs), phenolic metabolites of polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs), and acidic pesticides and/or predicted acidic pesticide metabolites. Our validated method used solid phase extraction, reversed phase chromatography in a C18 column with gradient elution, electrospray ionization in negative polarity, and automated MS/MS data acquisition to maximize true positive rates. We identified “suspect EOAs” using Agilent MassHunter Qualitative Analysis software to match chemical formulas it generated from each sample run with molecular formulas in our unique database of 693 EOAs assembled from multiple environmental literature sources. We found potential matches for 282 (41%) of the EOAs in our database. Sixty-five of these suspect EOAs were detected in at least 75% of the samples, only 19 of these compounds are currently biomonitored in NHANES. We confirmed two of three suspect EOAs by LC-QTOF/MS using a targeted method developed through liquid chromatography-tandem mass spectrometry, reporting the first confirmation of benzophenone-1 and bisphenol S in pregnant women’s sera. Our suspect screening workflow provides an approach to comprehensively scan environmental chemical exposures in humans. This can provide a better source of exposure information to help improve exposure and risk evaluation of industrial chemicals.
INTRODUCTION
There are an estimated 82,000 chemicals currently registered for use in the United States; approximately 8,000 of these are manufactured in excess of 25,000 pounds per year and thus subject to the Environmental Protection Agency Chemical Data Reporting (CDR) rule (1, 2). Extensive manufacturing, release and commercial use of thousands of industrial chemicals have led to their widespread presence in air, food, water and consumer products, with subsequent ubiquitous human exposure and potential for adverse health effects (3–9). Yet, only about 250 chemicals (<0.3% of registered chemicals and 3% of chemicals produced in excess of 25,000 pounds per year) are currently measured in large-scale human biomonitoring studies, largely due to limitations in the number of available chemical reference standards and the development of targeted analytic methods (2, 3). Due to the lack of access to data on production volume, manufacturing and usage for many chemicals, it is unknown whether the approximately 250 chemicals that have been selected for targeted method development represent the most important exposures with respect to human health (10). Thus, there is a need to supplement traditional biomonitoring methods with high resolution mass spectrometry and other non-targeted analysis techniques that do not require chemical reference standards and can rapidly screen for potentially relevant chemicals for targeted biomonitoring (11–13).
Full scan mass spectral analysis, facilitated by time-of-flight (TOF) mass spectrometers, is increasingly being recognized and applied as a discovery-driven screening tool that identifies potential exposure to a wide array of chemicals in biological specimens, and thereby can inform and guide the selection of chemicals for which targeted analysis methods are developed (11, 14–15). During the last decade, quadrupole time-of-flight mass spectrometry (QTOF/MS) and other high resolution mass spectrometry (HRMS) techniques have been used in the screening, confirmation, quantification and structure elucidation of chemicals such as organic contaminants, pharmaceuticals and their metabolites, and pesticides in environmental samples such as dust, water, sewage effluent and food (16–22). One study has applied HRMS approach in fish tissue samples (23), while another, proof of concept study, examined a limited version of this approach in one infant brain tissue sample (14); in both studies, a gas chromatography-quadrupole time-of-flight mass spectrometry (GC-QTOF/MS) platform was used. Platforms such as GC-QTOF/MS are compatible with highly non-polar compounds abundant in fatty tissues like the brain that are not amenable to commonly used ion sources used in liquid chromatography-quadrupole time-of-flight mass spectrometry (LC-QTOF/MS). Polar and slightly polar compounds, on the other hand, more abundantly found in common biological matrices like blood and urine may require derivatization for GC-QTOF/MS, an additional step that may cause partial loss of analytes and require additional time for analysis. Recent improvements in LC-QTOF/MS optics, flight tube design and detector capabilities have allowed this platform to attain a mass resolution of 20,000 to 40,000, with sub-2 ppm mass accuracy even with product ions (16), making it well-suited for comprehensive and unbiased environmental chemical screening of polar and slight polar organic contaminants. LC-QTOF/MS is starting to be applied to untargeted analysis of chemicals in human biological samples such as serum and urine.
The goal of our study was to apply LC-QTOF/MS technology to screen for environmental organic acids (EOAs) in human serum, and to evaluate the accuracy of this method through several validation studies, including the development of targeted analytic methods to confirm 3 novel suspect EOAs. EOAs are environmental organic compounds which are utilized in commerce and their known or predicted metabolites with at least one dissociable proton such as environmental phenols, phthalate metabolites, phenolic metabolites of polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs), perfluorinated compounds (PFCs), and phenolic and acidic pesticides and their predicted acidic and phenolic metabolites. Because EOAs comprise a smaller yet very commonly used group of environmental chemicals, we considered them ideal chemicals for a pilot study on this novel general suspect screen.
METHODS
Chemicals and Reagents
We obtained the following standards for LC-QTOF/MS method validation studies and targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses: 4-amino-2-nitrophenol, benzophenone-1 (Bzp-1), bisphenol A-d16 (BPA-d16), bisphenol S (BPS), methyl paraben and monopentyl phthalate (MPP) (Sigma, St. Louis, MO); and monobutyl phthalate and perfluorooctanoic acid (Toronto Research Chemicals, Toronto, ON). We purchased BPA-free water from Aqua Solutions (Deer Park, TX) and analytical grade methanol and acetonitrile were obtained from Honeywell Burdick and Jackson (Muskegon, MI). We prepared stock solutions of standards and internal standards at 1 mg/mL, aliquoted to 1mL portions in amber vials and stored at −80° C. All calibration standards, ranging in concentration from 0.01 to 80 ng/mL, were prepared from the stock solution by serial dilution with synthetic human serum.
Sample Preparation
We analyzed 20 banked (−80° C) serum samples from an existing study of chemical exposures in pregnant women, which was approved by the University of California, San Francisco Committee on Human Research (24). We thawed each 250 uL serum sample, spiked it with 2.5 uL of 1 ug/mL internal standard (2.5 ng BPA-d16) and centrifuged it at 3000 rpm for 10 minutes before preparing it for LC-QTOF/MS and LC-MS/MS analyses by solid phase extraction (SPE) using Waters Oasis HLB cartridge (10 mg, 1 cc). We washed each SPE cartridge with 5 column volumes of methanol to eliminate possible environmental chemical contamination, and then activated the cartridge with water before loading 250 uL of serum. We then washed the column with 5% methanol before eluting each analyte by methanol. We evaporated the methanol eluates under a stream of nitrogen gas, then reconstituted them in 250 uL of 10% methanol for column injection.
We also prepared split serum samples for LC-QTOF/MS method validation studies using protein precipitation as follows: we added 750 uL of 95:5 (v/v) acetonitrile:methanol to 250 uL of serum sample, centrifuged the resulting mixture at 3000 rpm for 5 minutes, separated the supernate, dried it under a gentle stream of nitrogen and reconstituted the residue in 10% methanol for column injection.
LC-QTOF/MS Instrumental Analysis
Separation of analytes in each sample was achieved by liquid chromatography using an Agilent LC 1260 (Sta. Cruz, CA). A 50 uL aliquot of the extract was used for each of the duplicate injections of the sample into an Agilent Poroshell 120 C18 column (2.1 × 100 mm, 2.7 um) maintained at 55° C. Chromatographic separation of the analytes was achieved by gradient elution using water with 0.05% ammonium acetetate (pH=7.8) as mobile phase A and methanol with 0.05% ammonium acetate (pH=7.8) as mobile phase B. The use of higher pH aids in further ionizing acidic compounds, thus enhancing the sensitivity of the assay. The elution gradient employed was: 0 – 0.5 min, 5% B; 1.5 min, 30% B; 4.5 min, 70% B; 7.5 – 10 min, 100% B; 10.01 – 14 min, 5%B.
The LC system was connected to an Agilent QTOF 6550 (Sta. Cruz, CA), which collects both accurate mass precursor ion and product ion scans using an Agilent Jetstream electrospray ionization (ESI) source operated in the negative polarity, a mode that facilitates better ionization of acidic compounds such as EOAs. A TOF-MS scan across the range of 80 – 600 m/z was collected at high resolution for eluates coming out of the LC from 1 – 12 min. Using the Auto MS/MS mode (information dependent acquisition), a product ion scan (MS/MS) of the three most abundant peaks at high resolution was triggered each time a precursor ion with an intensity of ≥ 500 counts per second was generated in the TOF-MS scan.
The LC-QTOF/MS run produces a total ion chromatogram (TIC) for each sample, which includes: the accurate mass of each unique compound (expressed as m/z of their corresponding anion); peak area; retention time; and spectral data on the parent ion and fragment ions, including isotopic pattern.
General Suspect Screen Data Analysis
EOA Database
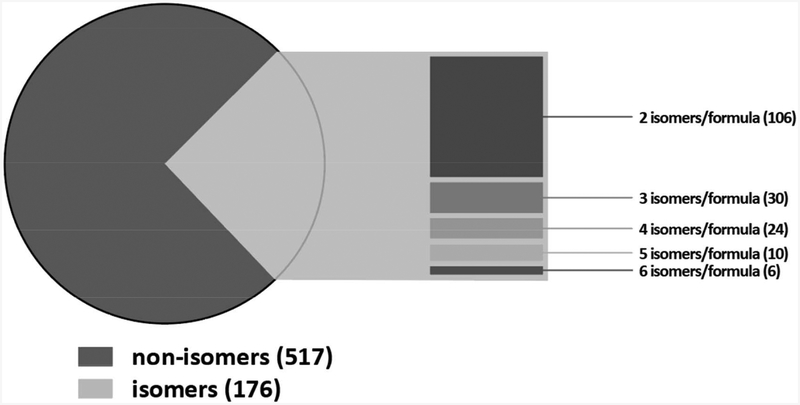
We assembled a database containing the molecular formula, name and chemical class of 693 EOAs using the following data sources: the Environmental Health Protection Agency’s Toxic Substances Control Act Inventory (25), ToxCast Chemicals (26), and High Production Volume (2), Inventory Update Reporting (27) and Chemical Data Reporting (28) chemical lists; the National Health and Nutrition Examination Survey (NHANES) 2009 biomonitoring chemicals list (29); the TEDx emerging environmental chemical list (30); the California Environmental Protection Agency’s Proposition 65 List of Chemicals (31) and Department of Pesticides Regulation reports; the Agilent Pesticides Database (32); and PubMed literature searches of environmental chemical biomonitoring studies. We also predicted phenolic and acidic metabolites of pesticides and included them in the database. The majority of EOAs in the database are phenolic and acidic pesticides or their predicted phenolic and acidic metabolites (60%) and environmental phenols (24%), and only a small portion (11% on average) is currently biomonitored by NHANES (Table 1). One hundred four of the EOAs in the database are isomers, with between 2 and 6 isomeric forms each; thus the number of unique molecular formulas in the EOA database is 589 (Figure 2). We utilized the Agilent Personal Compound Database software to automatically calculate the exact formula masses of the compounds in the EOA database.
Table 1.
Number of suspect Environmental Organic Acids (EOA) detected in 20 mid-gestation maternal serum samples from the EOA database, compared to the number biomonitored in the National Health and Nutrition Survey (NHANES), by chemical class
| N (%) biomonitored in NHANES |
N (%) detected in ≥75% of samples and biomonitored in NHANES |
||||
|---|---|---|---|---|---|
| Phenolic and acidic pesticides and their predicted phenolic and acidic metabolites | 416 | 20 (5) | 116 (28) | 11 (3) | 2 (0.5) |
| Phenols | 168 | 29 (17) | 113 (67) | 35 (21) | 9 (5) |
| Phthalate metabolites |
36 | 13 (36) | 31 (86) | 12 (33) | 4 (11) |
| PFCs | 49 | 13 (27) | 22 (45) | 7 (14) | 4 (8) |
| Phenolic metabolites of PBDEs and PCBs | 24 | 0 | 0 | 0 | 0 |
| Total | 693 | 75 (11%) | 282 (41%) | 65 (9%) | 19 (3%) |
Figure 2.
Number of non-isomer and isomer EOAs and number of isomers per molecular formula for the 176 isomers in the EOA Database (n= 693 EOAs).
Identification of suspect EOAs in maternal serum
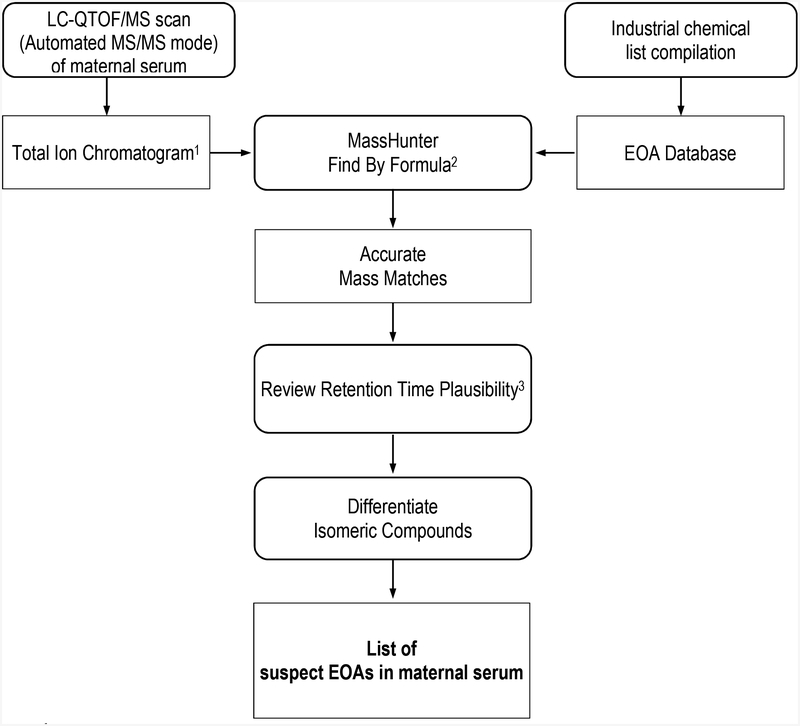
We identified suspect EOAs in maternal serum through a 3-step process diagramed in Figure 1. We used the Agilent MassHunter Qualitative Analysis software “Find By Formula” algorithm (FBF) to generate a list of accurate mass matches – compounds whose accurate masses (acquired in the LC-QTOF/MS analysis) matched the exact masses of chemicals in the EOA database. The FBF first calculates the monoisotopic mass and isotope pattern of a chemical formula in the EOA database, and then selects an extracted ion chromatogram (EIC) from the LC-QTOF/MS TIC data file based on the most abundant isotopes for an (−H) ion, extracts and examines the averaged spectra from the top 50% of the integrated peaks, and assigns each species a probability match score (target score) according to it’s concordance with accurate mass, isotopic abundance and spacing (isotope fitting) of chemicals in the EOA database. We selected the following criteria for accurate mass matches:
Figure 1.
Overview of General Suspect Screening Data Inputs and Workflow to identify suspect Environmental Organic Acids in pregnant women serum.
1 Total Ion Chromatogram contains accurate mass, retention time, and peak areas of all precursor and product ions detected in each serum sample
2 Criteria for accurate mass match: 1) Formula mass match ±10 ppm error; 2) Peak area >500 arbitrary units; 3) Signal-to-noise ratio ≥ 3; 4) Target score (accurate mass and isotopic pattern) ≥ 70.
3 Criteria for Retention Time Plausibility: 1) ± 0.5 min, accounting for RT drift; 2) consistent with chemical structure and predicted polarity.
Target score ≥ 70. In a previous analysis of a training set of known reference standards spiked into synthetic human serum, we empirically determined that selecting compounds with a target score of ≥ 70 would minimize false positives and false negatives.
Formula mass match within 10 ppm error. Using a formula match within 10 ppm is the current consensus for mass error criterion in LC-QTOF/MS analysis, as it weeds out non-specific formula matches without losing too many matches for compounds that have larger mass errors due to peak distortions that result from their large peak areas or detector saturation. We validated our selection of 10 ppm mass error by running matrix blanks spiked with a range of concentrations of known EOA reference standards.
Peak area >500 arbitrary units (AU). We selected a minimum peak area of 500 AU based on the background signals we observed in the serum sample TICs, most of which were below 500 AU.
Signal-to-noise ratio ≥ 3. We imposed a minimum signal-to-noise ratio of 3:1, as that is the accepted criterion for defining the limit of detection (LOD).
Review Retention Time Plausibility:
We reviewed and confirmed that the LC-QTOF/MS retention times for each suspect EOA were consistent across the 20 maternal serum samples, allowing for ± 0.5 min variation to account for the shift in retention times that occurs as samples are run through the LC-QTOF/MS. We also reviewed and confirmed the feasibility of each suspect EOA’s retention time based on the compound’s expected polarity.
Differentiate Isomeric Compounds:
We treated suspect EOAs with distinct plausible retention times as structural isomers, accounting for retention time drift. When retention times of two isomers were very similar, we looked at the fragment ions generated from the product ion scan to distinguish between isomers. We were unable to assign a chemical identity to the suspect isomeric EOA due to lack of reference standard-based retention time data and mass spectral data. Therefore, we report all isomeric compound names for a given molecular formula in our final list of suspect EOAs in maternal serum.
Using the final list of suspect EOAs in maternal serum, we generated suspect EOA exposure profiles for each of the 20 subjects. We also calculated the detection frequency of each of the 693 suspect EOAs across the 20 serum samples and ranked these suspect EOAs according to the frequency of their detection in the study sample.
LC-QTOF/MS Method Validation
To optimize the number of compound hits detected by our LC-QTOF/MS method, we validated the following method parameters: sample extraction method and LC-QTOF/MS sensitivity, precision and recovery.
Sample Extraction:
Sample extraction eliminates highly abundant proteins and easily ionizable inorganic ions that often interfere in mass spectral analysis. For sample extraction method, we compared the two most commonly used sample extraction methods in LC-MS analysis – protein precipitation (PP) and solid phase extraction (SPE) – to determine the effect of extraction method on the array of suspect chemicals detected. Specifically, we split five randomly selected serum samples, processed one aliquot by PP and the other by SPE (see Sample Preparation, above), then compared the suspect EOAs detected in each of the split samples.
Method Sensitivity:
A significant number of EOAs are present in pg/mL to ng/mL concentrations in biological matrices. Therefore, we compared the sensitivity of the LC-QTOF/MS method to that of LC-MS/MS, the current gold standard for quantitative analysis of most environmental chemicals. Specifically, we spiked known concentration ranges (0.001 – 100 ng/mL) of bisphenol A-d16, bisphenol A, methyl paraben, monobutyl phthalate, perfluorooctanoic acid, and 4-amino-2-nitrophenol into double charcoal stripped, drug-free serum and injected these samples into the LC-QTOF/MS. We assessed signals obtained for each of the six compounds and we established the LOD of each compound as the lowest concentration of the compound that has a signal-to-noise ratio of ≥ 3. We then compared these LODs to those obtained using the AB Sciex Triple Quadrupole 5500, one of the most sensitive LC-MS/MS platforms at the time we conducted this analysis.
Method Precision:
The inherent stochastic nature of mass detection in LC-MS methods can affect the coverage of compounds detected. Therefore, we assessed the method’s precision by injecting five randomly selected serum samples five times into the LC-QTOF/MS in a random order. We then calculated the reproducibility of the suspect EOAs for all five runs of the same serum sample.
Method Recovery:
We spiked low (1 ng/mL) and high (40 ng/mL) concentrations of bisphenol A-d16, bisphenol A, monobutyl phthalate and perfluorooctanoic acid into double charcoal-stripped, drug-free serum samples and injected them into the LC-QTOF/MS. We calculated the recovery of each compound in each run by comparing the signals obtained from an injection of extracted double charcoal-stripped serum spiked with the same amounts of the representative compounds immediately before injection into the LC-QTOF/MS.
LC-MS/MS Instrumental Analysis
Using Agilent LC 1260 and AB Sciex 5500 platforms, we developed an LC-MS/MS assay to confirm the presence and quantify the levels of three of the suspect EOAs detected in the 20 maternal serum samples. Our selection criteria for the three suspect EOAs were: 1) identified in at least 20% of the samples; 2) popular substitute chemical; and 3) not yet measured in large biomonitoring studies.
We prepared each sample by solid phase extraction as described above. We injected a 25 uL aliquot of each extract into the LC-MS/MS for analysis. We separated the three analytes by gradient elution chromatography through a Phenomenex Kinetex C18 column (Torrance, CA) using 0.5% ammonium acetate in water and 0.5% ammonium acetate in methanol as mobile phases at a flow rate of 0.5 mL/min. We applied the gradient elution as follows: 0–0.5 min, 30% B; 4.5 min, 70% B; 7–10 min, 100% B; 10.01–12.0 min, 30% B. Eluates coming out of the chromatographic column were ionized using an ESI source in the negative polarity. We monitored the three analytes by multiple reaction monitoring using the following transitions: benzophenone-1: 213.1–90.0 and 213.1–134.9; bisphenol-S; 249.1–108.0 and 249.1–91.9; monopentyl phthalate: 235.1–77.0 and 235.1–85.0; and bisphenol A–d16: 241.1–142.1 and 241.1–222.1. Labeled standards were not available for the three analytes; therefore we used BPA-d16 as an internal standard.
LC-MS/MS Method Validation
We assessed the precision, recovery, linearity and sensitivity of our developed LC-MS/MS method as follows. We spiked the matrix blank with low (0.05 ng/mL) and high (10 ng/mL) concentrations of analytes and ran 5 samples of each concentration in one batch to calculate within-run precision, and repeated this run on three separate days to calculate between-run precision. We also used the results of these three runs to calculate recovery rate. We evaluated linearity by running the calibration standards (0.01–100 ng/mL) five separate times on separate days and then assessing the linear regression coefficient for the calibration plot for each analyte in each run. We determined the sensitivity of the assay by establishing its LOD and Limit of Quantification (LOQ). We assessed the LOD for each analyte by running a series of calibration standards (0.001–100 ng/mL), established as the lowest concentration of the analyte that gives a signal/noise ratio of ≥3, and we defined the LOQ as the lowest concentration with signal/noise ratio of ≥10 that also keeps the linear regression coefficient of the standard curve ≥0.95.
Analysis of General Suspect Screen Performance
We developed our LC-QTOF/MS method to serve as a general suspect screening tool that can inform the selection of chemicals for which to develop targeted methods. Therefore, we evaluated the degree to which the LC-QTOF/MS accurately identified the presence or absence of the three suspect EOAs we confirmed via LC/MS-MS analysis by calculating the true and false positive rates, false negative rate, and the accuracy of the LC-QTOF/MS, treating the results from the LC-MS/MS as the gold standard. We define accuracy as the ability of the LC-QTOF/MS to correctly predict the presence and absence of a suspect compound in sample using the LC-MS/MS targeted results as the reference.
RESULTS
LC-QTOF/MS Method Validation
Sample Extraction:
We detected an average of 110 and 135 suspect EOAs when five randomly selected samples were prepared using protein precipitation and solid phase extraction, respectively; the majority (58%) of these suspect EOAs were detected when both sample preparation methods were used (Supporting Information, Figure S1).
LC-QTOF/MS Sensitivity, Precision and Recovery:
The LC-QTOF/MS was 4 – 40 times less sensitive than the LC-MS/MS for the six reference standards we tested in the LOD validation study (Supporting Information, Table S1). In the 5 consecutive test samples we ran for precision validation, we found that 74, 90, 85, 82 and 87% of the EOAs were the same (Supporting Information, Table S2). Lastly, we achieved recoveries typical of LC-MS methods for both low (80–85%) and high (87–90%) concentrations of the four compounds we tested (Supporting Information, Table S3).
LC-QTOF/MS Analysis of EOAs
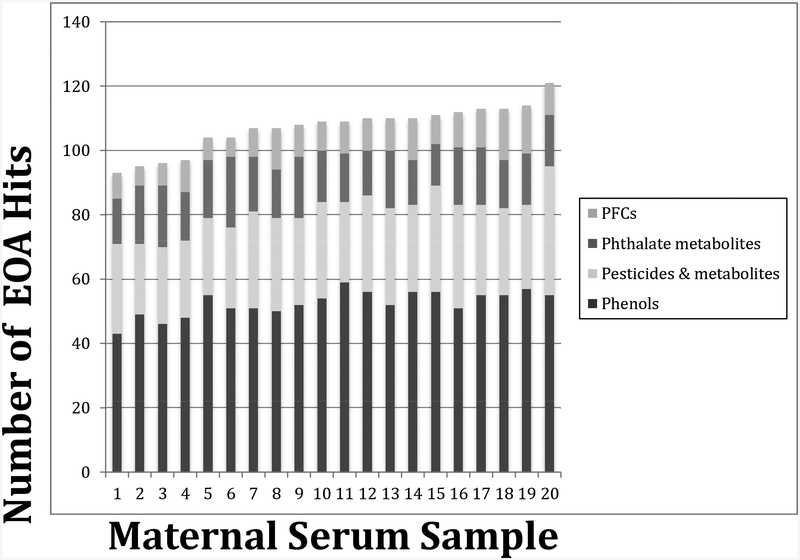
We detected 282 distinct suspect EOAs (41%) of the 693 EOAs in our database in our cohort, with the number of suspect EOAs detected in each serum sample ranging between 93 and 121 (mean= 107) (Figure 3). The distribution of suspect EOA chemical classes detected was similar among each serum sample, with phenolic and acidic pesticides and/or their predicted acidic and phenolic metabolites (n=116) and environmental phenols (n=113) predominating (Table 1). The three other classes of EOAs were detected less frequently (n= 0 – 31, see Supporting Information Table S4 for frequency of all 282 suspect EOAs). We detected 65 of the suspect EOAs in at least 75% of the 20 serum samples; only 19 of these are biomonitored in NHANES.
Figure 3.
Number of suspect EOAs, by compound class, in 20 maternal serum samples
LC-MS/MS Confirmation of suspect EOAs
From the group of compounds that met our criteria for confirmation via LC-MS/MS analysis, we selected monopentyl phthalate (MPP), benzophenone-1 (Bzp-1) and bisphenol S (BPS) because they are structurally related to three recognized endocrine disrupting chemicals that were not biomonitored at the time of analysis: monobutyl phthalate, benzophenone-3 and bisphenol A, respectively. We selected MPP for the additional reason that MPP and endogenous compounds share the same common elemental composition (carbon, hydrogen and oxygen); thus, evaluating the false positive rate for MPP would yield important information about the limitations of the LC-QTOF/MS screen.
We detected MPP, Bzp-1 and BPS in 15%, 100% and 85% of serum samples, with geometric means of 0.016, 0.097, 0.087 ng/mL, respectively (Table 2). We found much greater detects with Bzp-1 and BPS than in the suspect screen; we found similar detection frequency for MPP in the suspect screen and confirmatory analysis..
Table 2.
Detection frequency (DF) and mean, median and geometric mean (GM) serum levels (ng/mL) of bisphenol S, benzophenone-1 and monpentyl phthalate in twenty maternal sera analyzed by LC-MS/MS.
| LC-QTOF/MS | LC-MS/MS | ||||
|---|---|---|---|---|---|
| Analyte | DF (%) | DF (%) | Mean | Median | GM |
| Bisphenol S* | 50 | 85 | 0.153 | 0.153 | 0.087 |
| Benzophenone-1* | 30 | 100 | 0.110 | 0.095 | 0.097 |
| Monopentyl phthalate** | 20 | 15 | 0.017 | 0.014 | 0.016 |
LOD: 0.005 ng/mL
LOD: 0.01 ng/mL
LC-MS/MS Method Validation
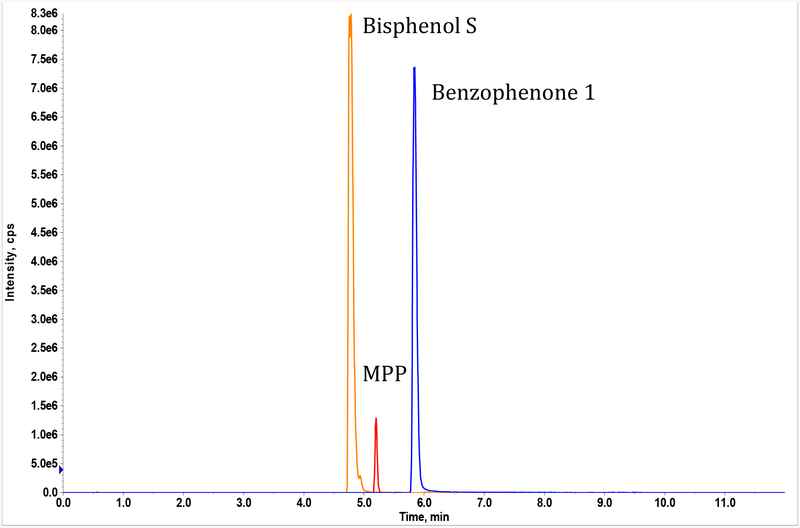
We observed coefficients of variation (CVs) of 1 – 8% for within-run precision (Table 3) and 3 – 10% for between-run precision for Bzp-1, BPS and MPP (Table 3). These results are within the acceptable precision prescribed for validated methods (< 20% CV) (33). We also obtained a narrow range of recoveries (85 – 94.5%) for the three analytes. BPS, Bzp-1 and MPP all ionized very well (Figure 4), which allowed us to establish very low LODs and LOQs. For BPS and Bzp-1, we established LODs and LOQs of 0.005 and 0.01 ng/mL, respectively. We obtained a slightly higher LOD and LOQ (0.01 and 0.05 ng/mL, respectively) for MPP. In establishing the linearity of the assay, we were able to consistently obtain linear regression coefficients of > 0.97 for all analytes, from their respective LOQs up to 100 ng/ml, in five separate trials.
Table 3.
Within-run and Between-run Coefficients of Variation and Recovery Rates for LC-MS/MS analysis of benzophenone-1, bisphenol S and monopentyl phthalate in spiked serum samples.
| Within Run (n=5) | Between Run (n=15) | |||
|---|---|---|---|---|
| 0.1 ng/mL | 10 ng/mL | 0.1 ng/mL | 10 ng/mL | |
| Monopentyl phthalate | 8.0 | 5.4 | 10.0 | 7.5 |
| Monopentyl phthalate | 88.2 | 90.5 | 85.0 | 89.4 |
Figure 4.
Typical chromatogram obtained for the targeted analysis of bisphenol S, benzophenone-1 and monopentyl phthalate using liquid chromatography-tandem mass spectrometry (LC-MS/MS)
LC-QTOF/MS General Suspects Screen performance
Our LC-QTOF/MS-based general suspect screen correctly detected the presence or absence of Bzp-1, BPS and MPP in 30%, 55% and 75% of samples, respectively (Table 4). The suspect screen differed in terms of true positive and false positive rates for these compounds (Table 4). Our method had false positive rates of 33% and 18% for BPS and MPP, respectively, but had no false positives for Bzp-1. Alternatively, the true positive rate for MPP, Bzp-1, and BPS were 33%, 30% and 53%, respectively.
Table 4.
True Positive Rate, False Positive Rate and Accuracy of LC-QTOF/MS assay for detecting three Environmental Organic Acids in 20 maternal serum samples.*
| Analyte | True Positive Rate |
False Positive Rate |
False Negative Rate |
Accuracy |
|---|---|---|---|---|
| Bisphenol S (BPS) | 53% | 33% | 47% | 55% |
| Benzophenone-1 (Bzp-1) | 30% | N/A | 70% | 30% |
| Monopentyl phthalate (MPP) | 33% | 18% | 67% | 75% |
Results of the validated LC-MS/MS targeted analysis was used as basis for TRUE positives and negatives for evaluating these parameters.
DISCUSSION
We report the first application and evaluation of the LC-QTOF/MS platform as a tool for the general suspect screening of environmental chemicals in human serum. We detected 234 suspect EOA molecular formulas, representing 282 suspect EOAs in the 20 serum samples in our pilot study, with an average of 107 suspect EOAs per sample (range 93–121) and a similar distribution pattern for the five classes of EOAs. Because each of the 282 suspect EOA molecular formulas we detected in our study sample may have either EOA or non-EOA isomers and chemical reference standard-based retention data are not available, targeted analysis is required to confirm chemical identities. We confirmed three of the suspect EOAs we detected through LC-MS/MS analysis; BPS and Bzp-1 were detected more frequently in the targeted analysis compared to the LC-QTOF/MS while the opposite is true for MPP.
This pilot study demonstrates the utility of the LC-QTOF/MS platform in identifying novel chemical targets for targeted biomonitoring studies. LC-QTOF/MS requires only 250 uL serum and yet provides a substantially more complete scan of potential chemical exposures, the results of which can be used to select and prioritize chemicals for targeted analysis and thus improve the efficiency of targeted biomonitoring studies. Currently, the number of chemical suspects that this approach can detect is limited by the number of chemicals for which compound specific information (molecular formula and chemical name) are available. However, as chemical identity databases expand, qualitative analysis of the TIC obtained from the LC-QTOF/MS run can be repeated to identify additional potential chemical exposures in the analyzed samples. Biomonitoring and epidemiologic studies can also utilize previously generated LC-QTOF/MS data sets in order to maximize sample sizes and expand the scope of their research to include new chemicals of interest.
Validation of the LC-QTOF/MS assay allowed us to optimize parameters that maximize the number of suspect EOAs for each sample and to assess the sensitivity and precision of our assay. We obtained a higher number of suspect EOAs using SPE as sample preparation method compared to protein precipitation. Because SPE is an established platform for cleaning up sample matrices like serum, it allows better elimination of background signals from the matrix and thus improves the signal-to-noise ratio of compounds that are present at lower concentrations in the sample. Seventy four to ninety percent of suspect EOAs detected in five samples run five times were the same; this high reproducibility indicates that the compounds we detected in the assay are not random masses picked up by the QTOF/MS detector and are most likely a true representation of the actual chemical profile of a sample.
There are several limitations and challenges to using the LC-QTOF/MS platform. First is the inability to use a universal MS/MS library that can be used across various QTOF/MS platforms such as is available for GC/MS. Unlike GC-MS, fragmentation achieved in QTOF/MS is platform and method specific; thus, preventing the use of MS/MS library collected in one QTOF/MS platform in other QTOF/MS platforms. Additionally, we ran the LC-QTOF/MS in the negative mode, which improves our ability to detect polar compounds with acidic functional groups, but limits our ability to detect basic compounds and less polar compounds (an example would be PBDEs or PCBs, we did not detect their polar metabolites, which have been shown to be present via targeted studies) (34). The latter was likely the reason we found a higher percentage of phenols, phthalate metabolites and pesticides detected in our samples compared to the hydroxy metabolites of PBDEs and PCBs. The relative ease of ionization of each of the classes of compounds may have contributed to this observed trend. Because hydoxylated PBDEs/PCBs and higher congeners of PFCs are more hydrophobic, they are expected to ionize less readily in an ESI source than most phenols, phthalate metabolites, and pesticides. While a next step of this method is to run the LC-QTOF/MS in the positive mode, which would improve our ability to detect additional compounds, there are still chemicals that will not be detected including metals, highly non-polar compounds like polyaromatic hydrocarbons, and very small highly polar compounds (e.g. dimethylphosphate, aminomethylphosphonic acid). However, each MS platform including GC-MS is limited in the breadth of chemical classes they can analyze.
Another criticism of the suspect screening approach and LC-QTOF/MS platform is that the LOD is higher than for targeted analysis. We did observe that this may influence the absolute rate at which we detected compounds. Our LC-QTOF/MS platform was 55% and 30% accurate in screening for BPS and Bzp-1, respectively, with similar or poorer performance for false negatives identifying the lack, versus the presence, of a chemical in serum. Our failure to detect the presence of BPS and Bzp-1 in 47 and 70%, respectively, of samples in which these compounds were detected via LC-MS/MS is likely explained by the LC-QTOF/MS having a higher LOD than the LC-MS/MS. Unlike LC-MS/MS, which is highly selective of the ions it allows through the quadrupole, the LC-QTOF/MS collects full scan MS data the majority of the time and is only selective for high intensity masses for a significantly shorter period of time. This results in a significantly lower sampling of a given mass in a sample and thus poorer sensitivity. In our validation studies, the LC-QTOF/MS was 4 to 40 times less sensitive than the LC-MS/MS for the six EOAs we analyzed; we anticipate that the comparative sensitivity of the LC-QTOF/MS and LC-MS/MS for BPS, Bzp-1 and MPP is similar to these results. However, we were able to detect the presence of these compounds, so the higher LOD did not hinder our ability to find the presence of these chemicals in biological samples. Additionally, caution should be applied to findings of no hits as these may be due to analytic reasons (e.g. PBDE metabolites) or other aspects of the method, and thus findings of no hits should not be interpreted as no chemicals present in the tissue. Finally, we have shown that the QTOF can perform well to detect abundant chemicals present in tissue.
An additional limitation is the extent to which we can identify industrial chemicals in biological samples given the ~700 chemicals in our database. The false positives rates we observed (33% for BPS and 18% for MPP) are much lower than false negative rates. The detection of false positives are likely explained by the presence, in maternal serum, of isomers that were not represented in the EOA database and thus not captured in the data analysis phase. These isomers could be other environmental chemicals or endogenous compounds, or both. Compounds comprised of C, O and H (such as MPP and other phthalate metabolites) may have a large number of isomers that are either endogenous human metabolites or other environmental chemicals. Thus the likelihood of false positives is high. It is also possible that the mass assigned to MPP corresponds to an isobaric compound that has a very close exact mass to it or a wrong formula assignment was made by the software which can still happen despite the criteria we impose to minimize this on compound matching.
Thirty-nine of the suspect EOA molecular formulas we detected are isomers that have between two and five isomeric forms each and thus represent 87 suspect EOAs. Due to the lack of reference standard-based retention time data, the FBF algorithm is not able to determine the chemical identify of the isomers; however, in all cases the retention time plausibility filter allowed distinguishing isomeric forms of each molecular formula.
Despite these limitations, we still found results consistent with the hypothesis that humans are potentially exposed to a broader range of industrial chemicals than previously documented in biomonitoring studies. For example, we detected 245 suspect EOAs that are not evaluated in NHANES; 43 of these were detected in ≥ 50% of samples. It is important to note that for compounds with lower detection frequency in the suspect screen, the false negative rates may be significant due to the lower sensitivity of QTOF/MS compared to LC-MS/MS, the current gold standard for targeted analysis.
Our analysis shows that the LC-QTOF/MS platform can be successfully applied as a general suspect screen for potential environmental chemical exposures that have not been previously measured in human populations. This allows us to increase our ability to identify and monitor chemical exposures, and ultimately prevent the most important exposures that may be adversely impacting health.
Supplementary Material
ACKNOWLEDGMENT
We thank the staff and faculty at San Francisco General Hospital Women’s Options Center for their assistance in blood sample collection. We also thank Katie Stevenson, Dylan Atchley, Mei-Lani Bixby, and Cynthia Megloza for their assistance in recruitment and data collection. This project was supported by NIH grants P20 ES018135, R21 ES017763, R01 ES013527, R01HD31544, P01 ES022841 (funded jointly by NIEHS and the U.S. Environmental Protection Agency), California EPA P0050869, and the Passport Foundation.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Muir DC, Howard PH. Are there other persistent organic pollutants? A challenge for environmental chemists. Environ Sci Technol. 2006;40(23):7157–66. [DOI] [PubMed] [Google Scholar]
- 2.U.S. Environmental Protection Agency, Chemical data reporting fact sheet: chemicals snapshot, U.S.E.P. Agency, Editor. 2012. [Google Scholar]
- 3.Centers for Disease Control and Prevention. National Health and Nutrition Examination Survey Hyattsville, MD2013 [cited 2013 July 30th], Available from: http://www.cdc.gov/nchs/nhanes.htm.
- 4.Kumar E, Holt WV. Impacts of endocrine disrupting chemicals on reproduction in wildlife. Advances in Experimental Medicine and Biology. 2014;753:55–70. [DOI] [PubMed] [Google Scholar]
- 5.Rogers JM, Ellis-Hutchings RG, Grey BE, Zucker RM, Norwood J Jr., Grace CE, et al. Elevated blood pressure in offspring of rats exposed to diverse chemicals during pregnancy. Toxicol Sci. 2014;137(2):436–46. [DOI] [PubMed] [Google Scholar]
- 6.Marmugi A, Lasserre F, Beuzelin D, Ducheix S, Hue L, Polizzi A, et al. Adverse effects of long-term exposure to bisphenol A during adulthood leading to hyperglycaemia and hypercholesterolemia in mice. Toxicology. 2014;325:133–43. [DOI] [PubMed] [Google Scholar]
- 7.Knower KC, To SQ, Leung YK, Ho SM, Clyne CD. Endocrine disruption of the epigenome: a breast cancer link. Endocr Relat Cancer. 2014;21(2):T33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kay VR, Bloom MS, Foster WG. Reproductive and developmental effects of phthalate diesters in males. Crit Rev Toxicol. 2014;44(6):467–98. [DOI] [PubMed] [Google Scholar]
- 9.Sobolewski M, Conrad K, Allen JL, Weston H, Martin K, Lawrence BP, et al. Sex-specific enhanced behavioral toxicity induced by maternal exposure to a mixture of low dose endocrine-disrupting chemicals. Neurotoxicology. 2014;45:121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, et al. The toxicity data landscape for environmental chemicals. Environ Health Perspect. 2009;117(5):685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andra SS, Austin C, Patel D, Dolios G, Awawda M, Arora M. Trends in the application of high-resolution mass spectrometry for human biomonitoring: An analytical primer to studying the environmental chemical space of the human exposome. Environ Int. 2017. pii: S0160–4120(16)30902–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aceña J, Stampachiacchiere S, Pérez S, Barceló D. Advances in liquid chromatography-high-resolution mass spectrometry for quantitative and qualitative environmental analysis. Anal Bioanal Chem. 2015. 407(21):6289–99. [DOI] [PubMed] [Google Scholar]
- 13.Schymanski EL, Singer HP, Slobodnik J, Ipolyi IM, Oswald P, Krauss M, Schulze T, Haglund P, Letzel T, Grosse S, Thomaidis NS, Bletsou A, Zwiener C, Ibáñez M, Portolés T, de Boer R, Reid MJ, Onghena M, Kunkel U, Schulz W, Guillon A, Noyon N, Leroy G, Bados P, Bogialli S, Stipaničev D, Rostkowski P, Hollender J. Non-target screening with high-resolution mass spectrometry: critical review using a collaborative trial on water analysis. Anal Bioanal Chem. 2015. 407(21): 6237–55. [DOI] [PubMed] [Google Scholar]
- 14.Cappiello A, Famiglini G, Palma P, Termopoli V, Lavezzi AM, Matturri L. Determination of selected endocrine disrupting compounds in human fetal and newborn tissues by GC-MS. Anal Bioanal Chem. 2014;406(12):2779–88. [DOI] [PubMed] [Google Scholar]
- 15.Hsu JY, Hsu JF, Chen YR, Shih CL, Hsu YS, Chen YJ, Tsai SH, Liao PC. Urinary exposure marker discovery for toxicants using ultra-high pressure liquid chromatography coupled with Orbitrap high resolution mass spectrometry and three untargeted metabolomics approaches. Anal Chim Acta. 2016. 939:73–83. [DOI] [PubMed] [Google Scholar]
- 16.Lacorte S, Fernandez-Albaz AR. Time of flight mass spectrometry applied to the liquid chromatographic analysis of pesticides in water and food. Mass Spectrometry Reviews. 2006;25(6):866–80. [DOI] [PubMed] [Google Scholar]
- 17.Chiaia-Hemandez AC, Krauss M, Hollender J. Screening of lake sediments for emerging contaminants by liquid chromatography atmospheric pressure photoionization and electrospray ionization coupled to high resolution mass spectrometry. Environ Sci Technol. 2013;47(2):976–86. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez F, Portoles T, Ibanez M, Bustos-Lopez MC, Diaz R, Botero-Coy AM, et al. Use of time-of-flight mass spectrometry for large screening of organic pollutants in surface waters and soils from a rice production area in Colombia. Sci Total Environ. 2012;439:249–59. [DOI] [PubMed] [Google Scholar]
- 19.Pitarch E, Portoles T, Marin JM, Ibanez M, Albarran F, Hernandez F. Analytical strategy based on the use of liquid chromatography and gas chromatography with triple-quadrupole and time-of-flight MS analyzers for investigating organic contaminants in wastewater. Anal Bioanal Chem. 2010;397(7):2763–76. [DOI] [PubMed] [Google Scholar]
- 20.Rager JE, Strynar MJ, Liang S, McMahen RL, Richard AM, Grulke CM, Wambaugh JF, Isaacs KK, Judson R, Williams AJ, Sobus JR. Linking high resolution mass spectrometry data with exposure and toxicity forecasts to advance high-throughput environmental monitoring. Environ Int. 2016. 88:269–80. [DOI] [PubMed] [Google Scholar]
- 21.Hogenboom AC, van Leerdam JA, and de Voogt P, Accurate mass screening and identification of emerging contaminants in environmental samples by liquid chromatography-hybrid linear ion trap Orbitrap mass spectrometry. J Chromatogr A, 2009. 1216(3): p. 510–9. [DOI] [PubMed] [Google Scholar]
- 22.Pugajeva I, Rusko J, Perkons I, Lundanes E, Bartkevics V. Determination of pharmaceutical residues in wastewater using high performance liquid chromatography coupled to quadrupole-Orbitrap mass spectrometry. J Pharm Biomed Anal. 2017. January 30;133:64–74. [DOI] [PubMed] [Google Scholar]
- 23.Nacher-Mestre J, Serrano R, Portoles T, Berntssen MH, Perez-Sanchez J, Hernandez F. Screening of pesticides and polycyclic aromatic hydrocarbons in feeds and fish tissues by gas chromatography coupled to high-resolution mass spectrometry using atmospheric pressure chemical ionization. J Agric Food Chem. 2014;62(10):2165–74. [DOI] [PubMed] [Google Scholar]
- 24.Gerona RR, Woodruff TJ, Dickenson CA, Pan J, Schwartz JM, Sen S, et al. Bisphenol-A (BPA), BPA glucuronide, and BPA sulfate in midgestation umbilical cord serum in a Northern and Central California population. Environ Sci Technol. 2013;47(21): 12477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.U.S. Environmental Protection Agency. ToxCast 2010. [cited 2010 August 1]. Available from: http://epa.gov/ncct/toxcast/. [PubMed] [Google Scholar]
- 26.U.S. Environmental Protection Agency. TSCA Chemical Substance Inventory Washington, DC2011 [cited 2011 April 29]. Available from: http://www.epa.gov/oppt/existingchemicals/pubs/tscainventory/basic.html. [Google Scholar]
- 27.U.S. Environmental Protection Agency. Non-confidential 2006 IUR Company/Chemical Records Washington, DC: U.S. Environmental Protection Agency,; 2010. [cited 2014 5/20]. Available from: http://cfpub.epa.gov/iursearch/. [Google Scholar]
- 28.U.S. Environmental Protection Agency. Chemical Data Reporting (CDR): U.S. Environmental Protection Agency; 2012. [cited 2014 12/17/2014], Available from: http://www.epa.gov/cdr/. [Google Scholar]
- 29.CDC (Centers for Disease Control and Prevention). Fourth national report on human exposure to environmental chemicals. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2009. [Google Scholar]
- 30.TEDx. TEDX List of Potential Endocrine Disruptors. 2011 [8/30/2013]. Available from: http://www.endocrinedisruption.com/endocrine.TEDXList.overview.php.
- 31.Office of Environmental Health and Hazard Assessment. Safe Drinking Water and Toxic Enforcement Act of 1986. Chemicals Known to the State to Cause Cancer or Reproductive Toxicity (Proposition 65 List) [cited 2010 April 14]. Available from: http://www.oehha.org/prop65/prop65_list/files/P65single040210.pdf.
- 32.Agilent Pesticides Personal Compound Database Library v 4.1, 2012. [Google Scholar]
- 33.U.S. Department of Health and Human Services FaDA, Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CVM),. Guidance for Industry: Bioanalytical Method Validation Silver Spring, MD: U.S. Department of Health and Human Services, Food and Drug Administration; 2001. [Google Scholar]
- 34.Zota AR, Mitro SD, Robinson JF, Hamilton EG, Park J-S, Parry E, Zoeller T and Woodruff TJ. 2017. Polybrominated diphenyl ether (PBDE) and OH-PBDE concentrations in maternal and fetal tissues, and associations with fetal cytochrome P450 gene expression. Envionmental Health Perspectives, manuscript accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.