Abstract
The Jun dimerization protein 2 (JDP2) is part of the family of stress-responsible transcription factors such as the activation protein-1, and binds the 12-O-tetradecanoylphorbol-13-acetateresponse element and the cAMP response element. It also plays a role as a histone chaperone and participates in diverse processes, such as cell-cycle arrest, cell differentiation, apoptosis, senescence, and metastatic spread, and functions as an oncogene and anti-oncogene, and as a cellular reprogramming factor. However, the molecular mechanisms underlying these multiple functions of JDP2 have not been clarified. This review summarizes the structure and function of JDP2, highlighting the specific role of JDP2 in cellular-stress regulation and prevention.
Keywords: ARE, AP-1 repressor, Function, Histone chaperone, JDP2, ROS, Structure
1. Introduction
The Jun dimerization protein 2 (JDP2) is a member of the activating protein-1 (AP-1) family of transcription factors, detected using the Sos recruitment system, and dimerizes with c-Jun to repress AP-1-mediated activation (Aronheim et al., 1997). Based on a yeast two-hybrid system with the activation transcription factor 2 (ATF2) as bait, it was later shown to repress ATF-mediated transcriptional activation (Jin et al., 2001). The gene that encodes JDP2 is located on human chromosome 14 (Blazek et al., 2003) and is composed of four exons; its transcriptional start site is located in exon 1 and the translational start site is located in exon 2. Although JDP2 is transcribed ubiquitously (Jin et al., 2001), it is predominantly enriched in the mouse lung and brain. The alternative splicing of JDP2 generates at least seven transcripts, including two unprocessed transcripts and five coding transcripts that generate two isoforms. The canonical sequence of JDP2 is 163 amino acids long (molecular weight, 18,704 Da), and isoform 2 contains 174 amino acids, with an extra 11 amino acids located at the amino terminus (molecular weight, 19,783 Da) (Fig. 1). This family is a group of basic leucine zipper (bZIP) proteins (Aronheim et al., 1997; Jin et al., 2001; http://www.ncbi.nlm.nih.gov/gene/122953), that bind the 12-O-tetradecanoylphorbol-13-acetate (TPA) response element (TRE) and the cAMP response element (CRE) via heterodimerization with c-Jun or ATF-2 (Aronheim et al., 1997; Jin et al., 2001). JDP2 binds not only to DNA cis elements, but also to histones and the nucleosome, indicating that JDP2 possesses histone chaperone activity and inhibition of histone acetyl transferase activity (INHAT) (Jin et al., 2006). JDP2 is involved in multiple and diverse processes. Knockout mice for Jdp2 exhibit a shorter tail and small size, increased cell proliferation and differentiation (Pan et al., 2010), and a lower number of neutrophils and osteoclasts (for bone homeostasis) (Maruyama et al., 2012). Transgenic mice with JDP2 specifically expressed in the heart acquire massive atrial dilatation and lethal phenotype (Kehat et al., 2006). In this review, we focus on the structure and functions of JDP2, such as cell differentiation, apoptosis, cell-cycle arrest, senescence, and antioxidation. Depending on the context, JDP2 displays both oncogenic and tumor-suppressive properties. Recently, JDP2 was shown to play a role in the cellular reprogramming of somatic (Liu et al., 2015) and cancer (Chiou et al., 2013) cells. These functions will be addressed and targeted for future use as therapies in regenerative medicine and aging.
Fig. 1.
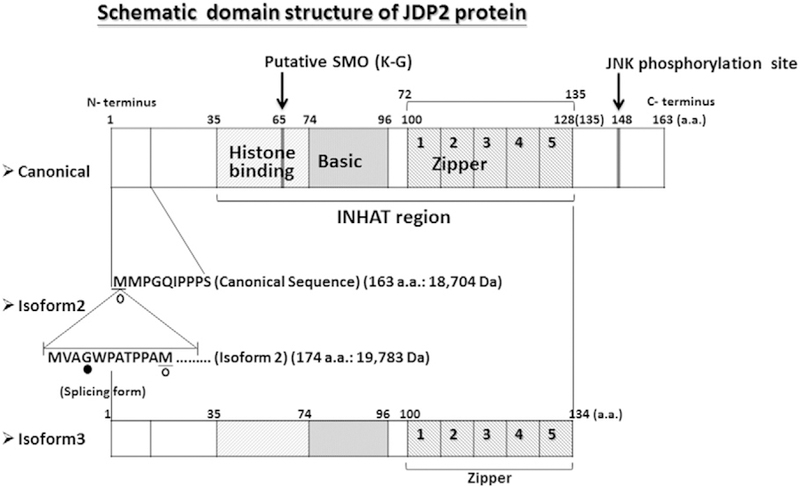
Schematic domain structure of Jun dimerization protein 2. The models of three Jun dimerization protein 2 (JDP2) isoforms such as canonical protein (163 amino acids) and isoform 2 (174 amino acids) and isoform 3 (134 amino acids) were presented [http://www.ncbi.nlm.nih.gov/gene/122953 LocusLink Report (LocusID: 122953), Aronheim et al., 1997; Jin et al., 2001; Kawaida et al., 2003]. The histone binding, basic region and leucine zipper region were listed (Jin et al., 2006). The first to fourth leucine zipper regions are L–L type and the 5th zipper region is L–H type.
2. JDP2 gene and expression
The human and mouse JDP2 genes are located on chromosome 14q3 and chromosome 12D2, respectively. JDP2 consists of four exons and spans about 46.4 kb and 39 kb, respectively (chr 14: 75,474,111 to 75,427,716; chr 12: 85,599,105 to 85,639,878). Its coding-region products produce a canonical protein of 163 amino acids (Aronheim et al., 1997), one variant of 174 amino acids, and one truncated protein of 134 amino acids that originates from the truncated transcript of the 3′-terminus. The single-nucleotide polymorphism (SNP) of JDP2 gene that correlates with intracranial aneurysms was detected among Japanese, Korean and Dutch cohorts (Krischek et al., 2010). In the mouse, SL-3–3 MLV-induced T-cell lymphomas show the insertional mutagenesis into the 250 kb locus of Fos/Jdp2/Batf locus (Rasmussen et al., 2005).
Seven transcripts of JDP2 have been identified, two of which are expressed pseudo-transcripts, which do not encode the JDP2 protein. The three transcripts of 5.4, 3.78, and 0.972 kb encoded one canonical JDP2 protein with 163 amino acids, the 1.7 kb transcript encodes a JDP2 spliced protein with 174 amino acids, and the 0.631 kb transcript leads to a truncated JDP2 protein with 134 amino acids and the remaining two transcripts (0.518 kb and 0.454 kb) are not encoding. The expression of JDP2 transcripts seems to be ubiquitous.
3. JDP2 protein structure
3.1. Domain structure
JDP2 belongs to the family of AP-1, which is a member of basic Zipper (bZIP) protein family. It contains a basic region from amino acid residues 74 to 96, and four (or five) zipper regions are located at the amino acid residues from 100 to 128 (or 135). The histone-binding region is located at amino acid residues from 35 to 74, just before the DNA-binding domain, and inhibition of histone acetyltransferase activity has been mapped within residues from 35 to 135 (Fig. 1).
3.2. Posttranscriptional modification
JDP2 is phosphorylated at threonine 148 by JNK in response to different stress conditions, such as UV irradiation, oxidative stress, and anisomycin treatment (Katz et al., 2001; Katz and Aronheim, 2002; Weidenfeld-Baranboim et al., 2011). Phosphorylated ATF2 inhibits the formation of heterodimers with JDP2 in vitro (Murata et al., 2008) while phosphorylated JDP2 undergoes the proteosomal degradation (Weidenfeld-Baranboim et al., 2011). JDP2 is also phosphorylated by other kinase such as doublecortin-like protein kinases (Nagamine et al., 2014). Interferon regulatory factor-2-binding protein-1 (IRF2-BP1) is a JDP2 E3 ligase and inhibits the ATF2-dependent transcription (Kimura, 2008). A putative SUMO site is also present at amino acid 65 (Fig. 1).
3.3. Dimer formation
JDP2 forms homodimers and heterodimerizes with c-Jun, JunD, JunB, Fra2, ATF2, the CCAAT/enhancer-binding protein C/EBPγ, and the C/EBP homologous protein (CHOP) (Aronheim et al., 1997; Broder et al., 1998; Jin et al., 2001; Cherasse et al., 2008; Weidenfeld-Baranboim et al., 2008). The JDP2 CHOP enhances TRE, but not CRE-dependent transcription (Weidenfeld-Baranboim et al., 2008). JDP2 also associates directly with the progesterone receptor (PR) and potentiates ligand-dependent PR-mediated transactivation (Edwards et al., 2002; Wardell et al., 2002, 2005; Hill et al., 2009). Finally, JDP2 interacts with IRF2-BP1 (Kimura, 2008) and HDAC3 (Jin et al., 2002), HDAC1, 2, and 6 (Darlyuk-Saadon et al., 2012; Heideman et al., 2013), and ATF-3 (Weidenfeld-Baranboim et al., 2009; Darlyuk-Saadon et al., 2012).
4. Transcriptional regulation
4.1. Histone chaperone
It is reported that histone acetylation by p300 is inhibited by exogenous JDP2 in a dose-dependent manner (Jin et al., 2006). An inhibitory effect of JDP2 was detected on histone acetylation induced by p300, the CREB-binding protein (CBP), the p300/CBP-associated factor (PCAF), and general control nonrepressive 5 (GCN5). The overexpression of JDP2 apparently represses the retinoic acid (RA)-induced acetylation of lysines 8 and 16 of histone H4 and some amino-terminal lysine residues of histone H3. The template activating factor-1β (TAF-Iβ), a component of the inhibition of histone acetyltransferase (INHAT) complex identified by Seo et al. (2001, 2002), is a histone chaperone that binds directly to core histones and facilitates the assembly of nucleosomes in vitro. Similarly, JDP2 interacts directly with all the core histones tested and inhibits the p300-mediated acetylation of those histones. Moreover, JDP2 also introduced supercoils into circular DNA in the presence of core histones, to levels that were similar to those observed for the yeast CCG1-interacting factor 1 protein (yCia1p) and CCG1-interacting factor (CIA1). Therefore, JDP2 appears to have significant histone chaperone activity in vitro (Jin et al., 2006). The HAT-inhibitory activity of JDP2 is involved, to some extent, in the repression of transcription by JDP2, whereas the maximal capacity of JDP2 to suppress the RA-mediated activation of the c-Jun promoter (Jin et al., 2002) and to suppress adipocyte differentiation (Nakade et al., 2007) requires the indirect recruitment of histone deacetylases (HDAC).
4.2. AP-1 repressor
JDP2 represses the transcription of the TRE-dependent gene, c-JUN (Aronheim et al., 1997) and the CRE-dependent gene, ATF-2 (Jin et al., 2001). Thus, JDP2 is a well-known AP-1 repressor that interacts with other transcription factors, such as JUNB, JUND, C/EBPγ (Broder et al., 1998), and CHOP (Cherasse et al., 2008; Weidenfeld-Baranboim et al., 2008). It has been shown that JDP2 was recruited to the promoter of the ccna2 gene at the AP-1 site to suppress cell cycle progression by downregulating cyclin-A2 (Pan et al., 2010). Previous work in fibroblast also showed that the expression of ATF3, a stress induced transcription factor sharing high homology with JDP2, was suppressed by JDP2 binding to its promoter region (Weidenfeld-Baranboim et al., 2009). Subsequently, it was demonstrated that the repression of ATF3 by JDP2 is via recruitment of multiple HDACs and the inhibition of histone acetylation (Darlyuk-Saadon et al., 2012; Maruyama et al., 2012). JDP2 inhibits p300/ATF2-mediated transactivation of c-Jun upon retinoic acid-induced commitment of murine F9 cells by recruiting histone deacetylase 3 (HDAC3) to the promoter of c-Jun (Jin et al., 2002), and inhibits the histone acetyltransferase activity (Jin et al., 2006). This inhibition of histone acetyltransferase (INHAT) activity is associated with the N-terminal domain, which is encoded by exon 2. Moreover, JDP2 inhibits the Epstein–Barr virus (EBV) immediate early gene BZLF1 promoter for the regulation of the latent-lytic switch in EBV infection (Murata et al., 2011).
4.3. Enhancer
In contrast, JDP2 is a coactivator of the progesterone receptor (Edwards et al., 2002; Wardell et al., 2002, 2005; Hill et al., 2009) and facilitates the soluble receptor activator of the nuclear-factor kappa-B ligand (sRANKL)-mediated activation of tartrate-resistant acid phosphatase (TRAP) and cathepsin K gene promoters in RAW264.7 cells (Kawaida et al., 2003). JDP2 also stimulates antioxidant response element (ARE)-dependent genes (Chiou et al., 2013; Tanigawa et al., 2013). Its function as an enhancer or repressor might depend on the gene context.
5. Cell differentiation, apoptosis, and senescence
JDP2 is involved in the diverse processes of cell differentiation. It plays a role in retinoic acid-induced F9 cell differentiation (Jin et al., 2002), in the terminal differentiation of C2 myoblasts and rhabdomyosarcoma cells via MyoD1 (Ostrovsky et al., 2002; Blum and Dynlacht, 2013). Jdp2 knockout mice have shown that JDP2 acts as a repressor of adipocyte differentiation (Nakade et al., 2007). The underlying mechanism was shown to involve the inhibition of histone H3 acetylation in the promoter of the adipogenesis-related gene C/EBPdelta (Nakade et al., 2007). JDP2 also participates in osteoclast differentiation through RANKL (Kawaida et al., 2003) and neutrophil differentiation (Maruyama et al., 2012) for bone homeostasis, as well as in bacterial immunity (Maruyama et al., 2012) and metastatic spread (Barbarov et al., 2015). Methylome mapping data suggest that JDP2 plays a role in progenitor cell differentiation in megakaryocytes, but not in lymphoid cells (Ji et al., 2010). Thus, JDP2 is one of the critical factors in the control of the differentiation of cells such as adipocytes, cardiomyocytes, myoblasts, osteoclast, neutrophils, megakaryocytes, and embryonic stem cells.
Specific depletion of JDP2 results in a p53-independent cell death that resembles apoptosis (Lerdrup et al., 2005). Chronic and acute overexpression of JDP2 represses cardiomyocytes against hypertrophic growth and apoptosis induction (Hill et al., 2013). In addition to these protective effects of JDP2 for cardiac remodeling, JDP2 is also required for maintaining a proper contractile function in cardiomyocytes (Hill et al., 2013).
JDP2-deficient mouse embryonic fibroblasts (MEFs) are resistant to replicative senescence through the polycomb repressive complex (PRC)-Ink4a (Nakade et al., 2009; Huang et al., 2010; Wang et al., 2011). The Jdp2KO MEFs continued to divide, even after 6 weeks, whereas the wild-type (WT) MEFs almost stopped proliferation and entered senescence under normoxia condition. Conversely, both WT MEFs and Jdp2KO MEFs did not yield to replicative senescence under hypoxia conditions (Nakade et al., 2009). These results demonstrate that MEFs lacking Jdp2 can escape from the irreversible growth arrest caused by environmental oxygen. The expressions of p16Ink4a and Arf were repressed in aged Jdp2KO MEF compared with the levels observed in WT MEFs. In hypoxia condition (3% oxygen), at the equivalent time at 40 days, WT MEFs expressed lower levels of p16Ink4a and Arf compared with those grown in normoxia, whereas Jdp2KO MEFs maintained low-level expression of p16Ink4a and p19Arf. Thus, these data indicate that the expression of p16Ink4a and p19Arf is dependent on oxygen stress, and that JDP2 controls the expression of both p16Ink4a and p19Arf. Studies based on a chromatin immunoprecipitation assay (ChIP assay) demonstrated that the methylation of H3K27 at the p16Ink4a/p19Arf locus is greater in Jdp2KO MEFs than it is in WT MEFs. The binding of PRC1 and PRC2 to the p16Ink4a and p19Arf promoters is more efficient in Jdp2KO MEFs than it is in WT MEFs. These observations suggest that H3K27 is methylated by PRC2 in the absence of JDP2, and the p16Ink4a/p19Arf locus is silenced by PRC1, whereas the increased expression of JDP2 helps to release PRC1 and PRC2 from the p16Ink4a/p19Arf locus, thereby reducing H3K27 methylation. Therefore, JDP2 may acts as a critical factor in the control of cellular senescence. The loss of JDP2 allows MEFs to escape senescence and, conversely, the overexpression of JDP2 induces cell-cycle arrest. The absence of JDP2 reduces the expression of both p16Ink4a and Arf, which inhibit cell-cycle progression.
6. Oncogene or tumor-suppressor gene
Depending on the context, JDP2 might display oncogenic or tumor suppressive functions. Thus, the regulatory effect of JDP2 during oncogenesis remains elusive. JDP2 inhibits cell transformation and tumor suppression in prostate cancer cell lines (Heinrich et al., 2004). It also induces partial oncogenic transformation in chicken embryonic fibroblasts (Blazek et al., 2003). In contrast, the insertional mutagenesis of JDP2 showed that it is an oncogene (Hwang et al., 2002; Rasmussen et al., 2005; Stewart et al., 2007; Sauvageau et al., 2008). Jdp2 transgenic mice display that expression of JDP2 during the promotion stage is found to be important for potentiation of liver cancer by acting as a tumor promoter (Bitton-Worms et al., 2010). JDP2 exhibits gene amplification in head and neck squamous cell carcinoma (Jarvinen et al., 2008). Downregulation of JDP2 is associated with tumor metastasis and poor prognosis in patients with pancreatic carcinoma (Yuanhong et al., 2010). Overexpression of JDP2 reverses the epithelial–mesenchymal transition (EMT) that is induced by co-treatment with TGF-β1 and EGF in human pancreatic cancer cell lines (Liu et al., 2012). Microarray and qPCR studies of patients with ST-segment elevation myocardial infarction (STEMI) suggest that JDP2 is a possible prognostic biomarker of the progression of heart-failure (Maciejak et al., 2015).
Moreover, JDP2 induces cell-cycle arrest via the inhibition of the expressions of cyclin D (Ostrovsky et al., 2002) and cyclin A2 (Pan et al., 2010) and the enhancement of the expression of p53 (Pan et al., 2010). JDP2 increases the expression of JUNB, JUND, and Fra2, and decreases the expression of c-JUN (Heinrich et al., 2004). In contrast, Piu et al. reported that JDP2 down-regulates expression of p53 gene and thereby protects cells from UV-mediated programmed cell death (Piu et al., 2001). Subsequently, it was demonstrated that JDP2 downregulates Trp53 transcription to promote leukemogenesis (van der Weyden et al., 2013). The mouse p53 protein negatively regulates the Jdp2 promoter in F9 cells (Xu et al., 2014), indicating the existence of a JDP2–p53 autoregulatory circuit. However, an opposite finding that Jdp2KO mice exhibited downregulation of the p53 and p21Cip1 proteins was reported (Pan et al., 2010). Thus, the possibility of a dependence on cell context cannot be ruled out. This differential regulation should be clarified in the future.
7. Cellular reprogramming
WNT/GSK3β signaling to the estrogen-related receptor beta (Esrrb) is critical for the stemness characteristics of iPSCs (Li et al., 2009; Stadtfeld and Hochedlinger, 2010). Previously, we showed that the expression of JDP2 is reduced significantly in hypoxia compared with normoxia (Nakade et al., 2009), and that JDP2 knockout upregulates the expression of Wnt/β-catenin pathway (Pan et al., 2010). The increase in the levels of reactive oxygen species (ROS) by metabolic changes in iPSCs may hinder the survival of reprogrammed cells, as suggested by observations of iPSC generation under hypoxia conditions (Yoshida et al., 2009; Saito et al., 2015). In addition, mitochondrial contents are also repressed in induced pluripotent stem cells (iPSCs) or human embryonic stem cells (hESCs) (Mah et al., 2011), suggesting that ROS generation by reprogramming factors is unfavorable to iPSCs. Thus, the introduction of JDP2 may be useful in triggering iPSC generation, because JDP2 inhibits ROS production (Tanigawa et al., 2013). Accordingly, JDP2 might play a critical role in nuclear reprogramming. Indeed, a medulloblastoma cell line (Chiou et al., 2013) and mouse embryonic fibroblasts (Liu et al., 2015) were reported to be induced into the iPSCs or iPSCs-like cells by JDP2. Moreover, Pei and colleagues showed that JDP2 anchors five non-Yamanaka factors (Id1, Jhdm1b, Lrh1, Sall4, and Glis1) to reprogram mouse embryonic fibroblasts into iPSCs (Liu et al., 2015). Consequently, JDP2 might be the guardian of somatic cell fate to a stem cell lineage. However, many questions remain unanswered.
8. ROS homeostasis and antioxidation control by JDP2
JDP2 controls the antioxidative response via an association with the nuclear-factor erythroid 2-related factor 2 (Nrf2) and the small musculoaponeurotic fibrosarcoma oncogene homolog (sMaf); moreover, it also reduces the amounts of ROS (Tanigawa et al., 2013). Thus, JDP2 plays a role in oxygen control to maintain the ROS homeostasis: (1) enhancement of the antioxidation, and (2) prevention of the ROS production (Fig. 2).
Fig. 2.
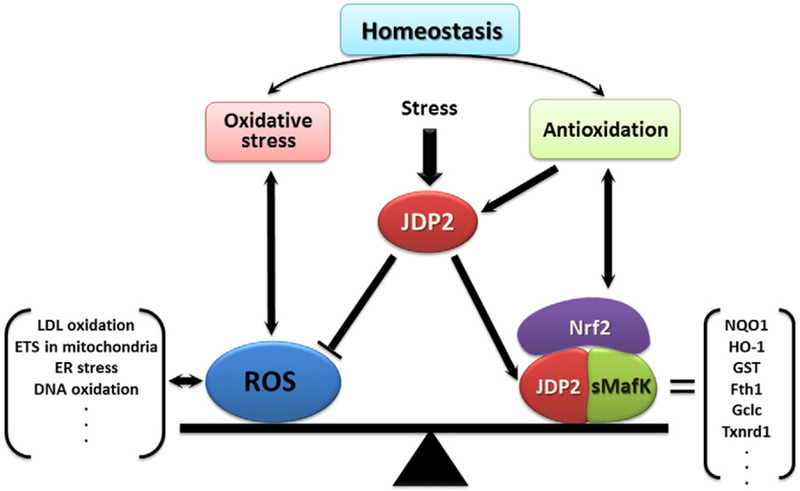
Schematic representation of the role of JDP2 on oxidative stress and antioxidation. Various stresses regulate the expression of JDP2. JDP2 associates with sMaf family and Nrf2 to induce the ARE response of variety of antioxidation response related genes, and at the same time JDP2 inhibits the ROS production to maintain the ROS homeostasis against oxidative stress (Tanigawa et al., 2013; Tanigawa and Yokoyama, 2013). ROS are generated by the oxidation of lipid and DNA via ER stress. Therefore, JDP2 controls the balance of ROS production through the transcriptional control with respective partner molecules for cytoprotection.
JDP2 is known as an AP-1 repressor and is a member of the stress protein family. Here, the working hypothesis of cross-talk between the stress response and protection response via a coupling factor such as JDP2 is presented (Fig. 2). Cytoprotection is mediated by cellular defense systems such as antioxidant enzymes and detoxification enzymes, which are regulated by anti-oxidant responsive element (ARE) in their gene-promoter regions. ARE was initially identified as an electrophile response element located in the promoter region of the mouse glutathione S-transferase alpha 1 (GSTA1) gene, and can be activated by β-naphthoflavone (β-NF), tert-butyl-1,4-hydroquinone (tBHQ), and di-phenols that can mimic the redox cycle (Friling et al., 1990). Consensus sequences of ARE (5′-TMAnnRTGAYnnnGCR-3′, where M = A/C, R = A/G, and Y = C/T, and the 9 bp core ARE is indicated in bold) is found in most of antioxidation related genes. By contrast, environmental stressors, such as UV radiation and 12-O-tetra-decanoylphorbol-13-acetate (TPA), activate ARE-containing genes that encode via the AP1 site (‘NNN’) (Angel et al., 1987). Thus, ARE-regulatory transcription factors have been identified, including c-JUN (Venugopal and Jaiswal, 1996, 1998), JUNB (Venugopal and Jaiswal, 1998; Yeligar et al., 2010), JUND (Venugopal and Jaiswal, 1998; Tsuji, 2005; Yeligar et al., 2010), c-FOS (Venugopal and Jaiswal, 1998), FRA1 (Venugopal and Jaiswal, 1996), Nrf1 (Venugopal and Jaiswal, 1996; Zhang et al., 2006), Nrf2 (Itoh et al., 1997; Dhakshinamoorthy et al., 2005), ATF3 (Brown et al., 2008; Kim et al., 2010), and ATF4 (He et al., 2001), which bind to ARE-containing genes that encode various cytoprotective enzymes. JDP2 is also included in this category. Moreover, JDP2 not only enhances the ARE response with Nrf2, the cap ‘n’ collar (CNC) family, and sMaf family, but also inhibits the ROS production (Tanigawa et al., 2013). Although the exact target of JDP2 for inhibition of ROS production has not been clarified yet, it might be possible that some transcriptional regulatory factors controlling ROS homeostasis are the downstream targets of JDP2.
The level of ROS is tightly controlled by an inducible antioxidant program that responds to cellular stressors and is regulated predominantly by Nrf2 and its repressor protein, Keap 1 (Newman and Keating, 2003; Kobayashi et al., 2004; Motohashi and Yamamoto, 2004; Zhang et al., 2005; Hayes and McMahon, 2009; Suzuki and Yamamoto, 2015). In contrast to the acute response of Nrf2, in the steady state, some somatic mutations cause destabilization of Nrf2 and decrease the constitutive transcription of its target genes, indicating that enhanced ROS detoxification and additional Nrf2 functions may be critical for the induction of the antioxidant response (Tebay et al., 2015). Because JDP2 is a member of the stress-induced AP-1 protein family (Aronheim et al., 1997), examining the role of JDP2 in cell proliferation, ROS production, and antioxidant response have identified JDP2 transcription factor as a cofactor that enhances ARE activity. JDP2 binds to ARE and regulates the ARE-mediated transcription in association with the Nrf2/MafK factors. Nrf2 is known as a central regulator of the induction of many antioxidant-responsive genes and genes that encode phase II detoxification enzymes. However, Nrf2 is not a DNA-binding protein and the addition of Nrf2 and MafK led to the repression of ARE reporter genes. Thus, the real target molecules that enhance the ARE response after oxidative stress remain to be identified. We proposed that JDP2 is one of the molecules that enhances the transcriptional activity of ARE reporter genes and inhibits ROS production to form a positive complex with Nrf2/MafK via leucine zipper domains. Thus, JDP2 not only acts as an AP-1 repressor protein to suppress cell proliferation during cancer progression, but also participates in the maintenance of ROS homeostasis to prevent cell damage by ROS.
9. Conclusion and perspectives
Histone-modification enzymes may not access to the nucleosome in vitro because JDP2 binds to not only the specific AP-1/ATF site, but also the core histone or nucleosome partially in a DNA-sequence-specific manner or histone-subset-specific manner. JDP2 inhibits at least the acetylation of histones H4K8 and H4K16, although other precise residues of histone H3 acetylation are not determined yet (Jin et al., 2006; Huang et al., 2010). Moreover, JDP2 associates with methylated lysine residues of histone H3K27 and blocks the methylation of histones in vitro (Nakade et al., 2009; Huang et al., 2010). Accordingly, it raises the possibility that the interaction between JDP2 and DNA and nucleosomes is in a sequence-specific manner ora specific interaction of acetylated or methylated lysine residues of certain restricted sets of histones. Addressing these precise functions in the context of epigenesis will help us understand the regulation of senescence, differentiation, and viral infection in a broader context. JDP2 is not a simple DNA-binding transcription factor; rather, it is a chromatin modifier. In the presence of antioxidants, JDP2 is a cofactor that reinforces the formation of the possible small Maf/Nrf2 complex and then activates the ARE (Tanigawa et al., 2013). It is also possible that JDP2 helps to stimulate the ARE activity by converting the repressor function of MafK/Nrf2 to the active stimulatory function. However, the molecular mechanism of this conversion of JDP2 from AP-1 repressor or chromatin compaction to positive expression of ARE/AP-1 or chromatin opening remains to be solved (Fig. 3). Moreover, the exact ligands required for JDP2 activation are not known. Stress inducers, morphogens, hormones, disruptors, and oxygen might be candidates for JDP2-inducing reagents. Moreover, the identification of the direct targets of JDP2 in the cell-cycle regulation and in specific tissues, such as the cerebellum and lung, is required for understanding the real role of JDP2. The manner in which antioxidants induce the expression of the JDP2 is also interesting. We propose a model for the induction of cellular senescence by JDP2: aging or oxygen stress upregulates JDP2 expression in primary untransformed cells.
Fig. 3.
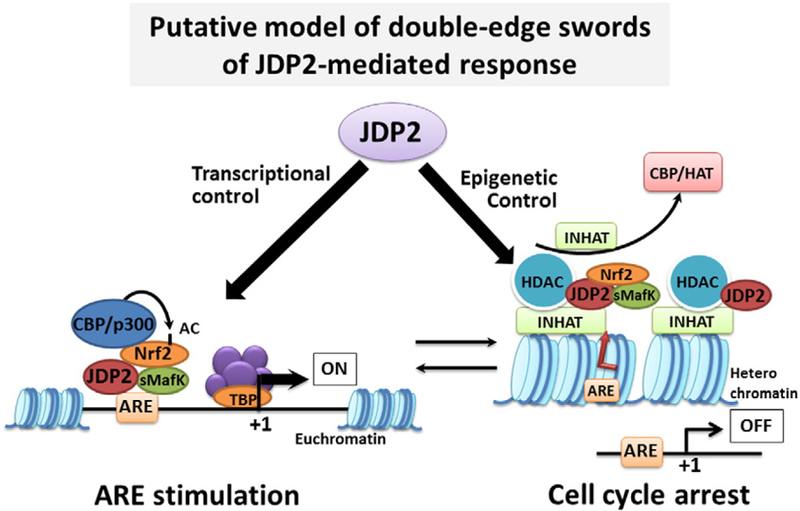
Putative models of JDP2 to play the double-edge swords to control the transcription and epigenetic regulation. Possible positive and negative transcriptional regulations of JDP2 are presented in this model. JDP2 might form the complex with sMafK and/or Nrf2 and bind to the ARE element to increase the transcription of ARE related genes (Tanigawa et al., 2013) via recruitment of CBP/p300 coactivator-dependent Nrf2 acetylation (Kawai et al., 2011) or possibly histone acetylation. On the other hand, JDP2 might recruit the HDAC family (Jin et al., 2002; Darlyuk-Saadon et al., 2012; Maruyama et al., 2012) or INHAT activity (Jin et al., 2006) to repress the transcription of cell cycle related genes (Pan et al., 2010).
One hint of the molecular conversion might be explained by the context of the epigenetic regulation of the p16Ink4a/Arf locus. There is a balance between ARE and ROS. The oxidative stress induces the expression of JDP2, and then forms a triple complex to stimulate the ARE; conversely, JDP2 inhibits ROS production to reduce the response to oxidative stress (Fig. 2). Similarly, Jenkins et al. reported that the loss of p16Ink4a increased the intracellular level of ROS, which can be lost upon the re-expression of p16Ink4a (Jenkins et al., 2011). This novel alternative Rb-independent regulation of p16Ink4a might be functional in the case of JDP2. JDP2 also stimulates the expression of the INK4a/ARF locus to exclude the polycomb complexes PRC1 and PRC2 from the INK4a/ARF loci. This coordination of the activations of ARE and the INK4a/ARF locus induced by JDP2 results in cellular senescence and protection of ROS production to maintain ROS/ARE homeostasis. These findings suggest that JDP2 plays a potential role in protecting cells against the malicious attack of carcinogens and endogenous reactive oxygen/nitrogen species, by inducing several detoxifying/antioxidant enzymes. Thus, JDP2 plays a role of not only the basic regulation of cell cycle, growth and differentiation but also the control of ROS homeostasis involving in tumorigenesis, stemness and pluripotency, and probably aging.
Acknowledgements
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series—a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The authors would like to thank Ms. Chia-Chen Ku for editing and drawing figures and table. This work was supported by the grants from the Ministry of Science and Technology (MOST-104–2320-B-037–033-My2; MOST-104–2314-B-037–002; MOST104–2811-B-037–015); the National Health Research Institutes (NHRI-Ex104–10416SI); and a Kaohsiung Medical University grant (KMU-TP103A04, KMU-TP103G03, KMU-TP104E23, KMU-DT104001, KMU-DT104001). The corresponding Gene Wiki entry for this review can be found here: https://en.wikipedia.org/wiki/Jun_dimerization_protein.
Abbreviations
- ATF-2
activation transcription factor-2
- cAMP
cyclic adenosine monophosphate
- ARE
antioxidant-responsive element
- AP-1
activation protein-1
- bZIP
basic leucine zipper protein
- CBP
CREB-binding protein
- C/EBP
CAAT enhancer-binding protein
- CHOP
C/EBP homologous protein
- CRE
cAMP response element
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- iPSCs
induced pluripotent stem cells
- INHAT
inhibition of histone acetyltransferase
- JDP2
Jun dimerization protein 2
- Keap1
Kelch-like ECH-associated protein 1
- MafK
musculoaponeurotic fibrosarcoma oncogene homolog K
- MEFs
mouse embryonic fibroblasts
- Nrf2
nuclear-factor erythroid 2-related factor 2
- PR
progesterone receptor
- PRC
polycomb repressive protein complex
- RANKL
receptor activator of nuclear-factor kappa-B ligand
- ROS
reactive oxygen species
- SNP
single-nucleotide polymorphism
- TPA
12-O-tetradecanoylphorbol-13-acetate
- TRE
TPA response element.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- http://www.ncbi.nlm.nih.gov/gene/122953, (LocusLink Report (LocusID: 122953)).
- Angel P, Imagawa M, Chiu R, Stein B, Imbra RJ, Rahmsdorf HJ, Jonat C, Herrlich P,Karin M, 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49, 729–739. [DOI] [PubMed] [Google Scholar]
- Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M, 1997. Isolation of an AP-1 repressor by a novel method for detecting protein–protein interactions. Mol. Cell. Biol 17, 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarov Y, Timaner M, Alishekevitz D, Hai T, Yokoyama KK, Shaked Y, Aronheim A, 2015. Host JDP2 expression in the bone marrow contributes to metastatic spread. Oncotarget 6, 37737–37749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton-Worms K, Pikarsky E, Aronheim A, 2010. The AP-1 repressor protein, JDP2, potentiates hepatocellular carcinoma in mice. Mol. Cancer 9, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek E, Wasmer S, Kruse U, Aronheim A, Aoki M, Vogt PK, 2003. Partial oncogenic transformation of chicken embryo fibroblasts by Jun dimerization protein 2, a negative regulator of TRE- and CRE-dependent transcription. Oncogene 22, 2151–2159. [DOI] [PubMed] [Google Scholar]
- Blum R, Dynlacht BD, 2013. The role of MyoD1 and histone modifications in the activation of muscle enhancers. Epigenetics 8, 778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder YC, Katz S, Aronheim A, 1998. The ras recruitment system, a novel approach to the study of protein–protein interactions. Curr. Biol 8, 1121–1124. [DOI] [PubMed] [Google Scholar]
- Brown SL, Sekhar KR, Rachakonda G, Sasi S, Freeman ML, 2008. Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2-related factor 2 (Nrf2)-regulated stress pathway. Cancer Res 68, 364–368. [DOI] [PubMed] [Google Scholar]
- Cherasse Y, Chaveroux C, Jousse C, Maurin AC, Carraro V, Parry L, Fafournoux P, Bruhat A, 2008. Role of the repressor JDP2 in the amino acid-regulated transcription of CHOP. FEBS Lett 582, 1537–1541. [DOI] [PubMed] [Google Scholar]
- Chiou SS, Wang SS, Wu DC, Lin YC, Kao LP, Kuo KK, Wu CC, Chai CY, Lin CL, Lee CY, Liao YM, Wuputra K, Yang YH, Wang SW, Ku CC, Nakamura Y, Saito S, Hasegawa H, Yamaguchi N, Miyoshi H, Lin CS, Eckner R, Yokoyama KK, 2013. Control of oxidative stress and generation of induced pluripotent stem cell-like cells by Jun dimerization protein 2. Cancers (Basel) 5, 959–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlyuk-Saadon I, Weidenfeld-Baranboim K, Yokoyama KK, Hai T, Aronheim A, 2012. The bZIP repressor proteins, c-Jun dimerization protein 2 and activating transcription factor 3, recruit multiple HDAC members to the ATF3 promoter. Biochim. Biophys. Acta 1819, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK, 2005. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem 280, 16891–16900. [DOI] [PubMed] [Google Scholar]
- Edwards DP, Wardell SE, Boonyaratanakornkit V, 2002. Progesterone receptor interacting coregulatory proteins and cross talk with cell signaling pathways. J. Steroid Biochem. Mol. Biol 83, 173–186. [DOI] [PubMed] [Google Scholar]
- Friling RS, Bensimon A, Tichauer Y, Daniel V, 1990. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. U. S. A 87, 6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, McMahon M, 2009. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci 34, 176–188. [DOI] [PubMed] [Google Scholar]
- He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, Alam J, 2001. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem 276, 20858–20865. [DOI] [PubMed] [Google Scholar]
- Heideman MR, Wilting RH, Yanover E, Velds A, de Jong J, Kerkhoven RM, Jacobs H, Wessels LF, Dannenberg JH, 2013. Dosage-dependent tumor suppression by histone deacetylases 1 and 2 through regulation of c-Myc collaborating genes and p53 function. Blood 121, 2038–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich R, Livne E, Ben-Izhak O, Aronheim A, 2004. The c-Jun dimerization protein 2 inhibits cell transformation and acts as a tumor suppressor gene. J. Biol. Chem 279, 5708–5715. [DOI] [PubMed] [Google Scholar]
- Hill KK, Roemer SC, Jones DN, Churchill ME, Edwards DP, 2009. A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain. J. Biol. Chem 284, 24415–24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Wurfel A, Heger J, Meyering B, Schluter KD, Weber M, Ferdinandy P, Aronheim A, Schulz R, Euler G, 2013. Inhibition of AP-1 signaling by JDP2 overexpression protects cardiomyocytes against hypertrophy and apoptosis induction. Cardiovasc. Res 99, 121–128. [DOI] [PubMed] [Google Scholar]
- Huang YC, Saito S, Yokoyama KK, 2010. Histone chaperone Jun dimerization protein 2 (JDP2): role in cellular senescence and aging. Kaohsiung J. Med. Sci 26, 515–531. [DOI] [PubMed] [Google Scholar]
- Hwang HC, Martins CP, Bronkhorst Y, Randel E, Berns A, Fero M, Clurman BE, 2002. Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc. Natl. Acad. Sci. U. S. A 99, 11293–11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y, 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun 236, 313–322. [DOI] [PubMed] [Google Scholar]
- Jarvinen AK, Autio R, Kilpinen S, Saarela M, Leivo I, Grenman R, Makitie AA, Monni O, 2008. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosom. Cancer 47, 500–509. [DOI] [PubMed] [Google Scholar]
- Jenkins NC, Liu T, Cassidy P, Leachman SA, Boucher KM, Goodson AG, Samadashwily G, Grossman D, 2011. The p16(INK4A) tumor suppressor regulates cellular oxidative stress. Oncogene 30, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP, 2010. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 467, 338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Kato K, Chimura T, Yamasaki T, Nakade K, Murata T, Li H, Pan J, Zhao M, Sun K, Chiu R, Ito T, Nagata K, Horikoshi M, Yokoyama KK, 2006. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nat. Struct. Mol. Biol 13, 331–338. [DOI] [PubMed] [Google Scholar]
- Jin C, Li H, Murata T, Sun K, Horikoshi M, Chiu R, Yokoyama KK, 2002. JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Mol. Cell. Biol 22, 4815–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Ugai H, Song J, Murata T, Nili F, Sun K, Horikoshi M, Yokoyama KK, 2001. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett 489, 34–41. [DOI] [PubMed] [Google Scholar]
- Katz S, Aronheim A, 2002. Differential targeting of the stress mitogen-activated protein kinases to the c-Jun dimerization protein 2. Biochem. J 368, 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S, Heinrich R, Aronheim A, 2001. The AP-1 repressor, JDP2, is a bona fide substrate for the c-Jun N-terminal kinase. FEBS Lett 506, 196–200. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Garduno L, Theodore M, Yang J, Arinze IJ, 2011. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J. Biol. Chem 286, 7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaida R, Ohtsuka T, Okutsu J, Takahashi T, Kadono Y, Oda H, Hikita A, Nakamura K, Tanaka S, Furukawa H, 2003. Jun dimerization protein 2 (JDP2), a member of the AP-1 family of transcription factor, mediates osteoclast differentiation induced by RANKL. J. Exp. Med 197, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I, Heinrich R, Ben-Izhak O, Miyazaki H, Gutkind JS, Aronheim A, 2006. Inhibition of basic leucine zipper transcription is a major mediator of atrial dilatation. Cardiovasc. Res 70, 543–554. [DOI] [PubMed] [Google Scholar]
- Kim KH, Jeong JY, Surh YJ, Kim KW, 2010. Expression of stress-response ATF3 is mediated by Nrf2 in astrocytes. Nucleic Acids Res 38, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, 2008. IRF2-binding protein-1 is a JDP2 ubiquitin ligase and an inhibitor of ATF2-dependent transcription. FEBS Lett 582, 2833–2837. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M, 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol 24, 7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krischek B, Tajima A, Akagawa H, Narita A, Ruigrok Y, Rinkel G, Wijmenga C, Feigl GC, Kim CJ, Hori T, Tatagiba M, Kasuya H, Inoue I, 2010. Association of the Jun dimerization protein 2 gene with intracranial aneurysms in Japanese and Korean cohorts as compared to a Dutch cohort. Neuroscience 169, 339–343. [DOI] [PubMed] [Google Scholar]
- Lerdrup M, Holmberg C, Dietrich N, Shaulian E, Herdegen T, Jaattela M, Kallunki T, 2005. Depletion of the AP-1 repressor JDP2 induces cell death similar to apoptosis. Biochim. Biophys. Acta 1745, 29–37. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M, 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Du R, Long J, Dong A, Fan J, Guo K, Xu Y, 2012. JDP2 inhibits the epithelial-to-mesenchymal transition in pancreatic cancer BxPC3 cells. Tumour Biol 33, 1527–1534. [DOI] [PubMed] [Google Scholar]
- Liu J, Han Q, Peng T, Peng M, Wei B, Li D, Wang X, Yu S, Yang J, Cao S, Huang K, Hutchins AP, Liu H, Kuang J, Zhou Z, Chen J, Wu H, Guo L, Chen Y, Chen Y, Li X, Wu H, Liao B, He W, Song H, Yao H, Pan G, Chen J, Pei D, 2015. The oncogene c-Jun impedes somatic cell reprogramming. Nat. Cell Biol 17, 856–867. [DOI] [PubMed] [Google Scholar]
- Maciejak A, Kiliszek M, Michalak M, Tulacz D, Opolski G, Matlak K, Dobrzycki S, Segiet A, Gora M, Burzynska B, 2015. Gene expression profiling reveals potential prognostic biomarkers associated with the progression of heart failure. Genome Med 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah N, Wang Y, Liao MC, Prigione A, Jozefczuk J, Lichtner B, Wolfrum K, Haltmeier M, Flottmann M, Schaefer M, Hahn A, Mrowka R, Klipp E, Andrade-Navarro MA, Adjaye J, 2011. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS One 6, e24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Fukasaka M, Vandenbon A, Saitoh T, Kawasaki T, Kondo T, Yokoyama KK, Kidoya H, Takakura N, Standley D, Takeuchi O, Akira S, 2012. The transcription factor Jdp2 controls bone homeostasis and antibacterial immunity by regulating osteoclast and neutrophil differentiation. Immunity 37, 1024–1036. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M, 2004. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med 10, 549–557. [DOI] [PubMed] [Google Scholar]
- Murata T, Noda C, Saito S, Kawashima D, Sugimoto A, Isomura H, Kanda T, Yokoyama KK, Tsurumi T, 2011. Involvement of Jun dimerization protein 2 (JDP2) in the maintenance of Epstein–Barr virus latency. J. Biol. Chem 286, 22007–22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Shinozuka Y, Obata Y, Yokoyama KK, 2008. Phosphorylation of two eukaryotic transcription factors, Jun dimerization protein 2 and activation transcription factor 2, in Escherichia coli by Jun N-terminal kinase 1. Anal. Biochem 376, 115–121. [DOI] [PubMed] [Google Scholar]
- Nagamine T, Nomada S, Onouchi T, Kameshita I, Sueyoshi N, 2014. Nuclear translocation of doublecortin-like protein kinase and phosphorylation of a transcription factor JDP2. Biochem. Biophys. Res. Commun 446, 73–78. [DOI] [PubMed] [Google Scholar]
- Nakade K, Pan J, Yamasaki T, Murata T, Wasylyk B, Yokoyama KK, 2009. JDP2 (Jun Dimerization Protein 2)-deficient mouse embryonic fibroblasts are resistant to replicative senescence. J. Biol. Chem 284, 10808–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade K, Pan J, Yoshiki A, Ugai H, Kimura M, Liu B, Li H, Obata Y, Iwama M, Itohara S, Murata T, Yokoyama KK, 2007. JDP2 suppresses adipocyte differentiation by regulating histone acetylation. Cell Death Differ 14, 1398–1405. [DOI] [PubMed] [Google Scholar]
- Newman JR, Keating AE, 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300, 2097–2101. [DOI] [PubMed] [Google Scholar]
- Ostrovsky O, Bengal E, Aronheim A, 2002. Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. J. Biol. Chem 277, 40043–40054. [DOI] [PubMed] [Google Scholar]
- Pan J, Nakade K, Huang YC, Zhu ZW, Masuzaki S, Hasegawa H, Murata T, Yoshiki A, Yamaguchi N, Lee CH, Yang WC, Tsai EM, Obata Y, Yokoyama KK, 2010. Suppression of cell-cycle progression by Jun dimerization protein-2 (JDP2) involves downregulation of cyclin-A2. Oncogene 29, 6245–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piu F, Aronheim A, Katz S, Karin M, 2001. AP-1 repressor protein JDP-2: inhibition of UV-mediated apoptosis through p53 down-regulation. Mol. Cell. Biol 21, 3012–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MH, Sorensen AB, Morris DW, Dutra JC, Engelhard EK, Wang CL, Schmidt J, Pedersen FS, 2005. Tumor model-specific proviral insertional mutagenesis of the Fos/Jdp2/Batf locus. Virology 337, 353–364. [DOI] [PubMed] [Google Scholar]
- Saito S, Lin YC, Tsai MH, Lin CS, Murayama Y, Sato R, Yokoyama KK, 2015. Emerging roles of hypoxia-inducible factors and reactive oxygen species in cancer and pluripotent stem cells. Kaohsiung J. Med. Sci 31, 279–286. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Miller M, Lemieux S, Lessard J, Hebert J, Sauvageau G, 2008. Quantitative expression profiling guided by common retroviral insertion sites reveals novel and cell type specific cancer genes in leukemia. Blood 111, 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SB, Macfarlan T, McNamara P, Hong R, Mukai Y, Heo S, Chakravarti D, 2002. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J. Biol. Chem 277, 14005–14010. [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D, 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104, 119–130. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K, 2010. Induced pluripotency: history, mechanisms, and applications. Genes Dev 24, 2239–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Mackay N, Hanlon L, Blyth K, Scobie L, Cameron E, Neil JC, 2007. Insertional mutagenesis reveals progression genes and checkpoints in MYC/Runx2 lymphomas. Cancer Res 67, 5126–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Yamamoto M, 2015. Molecular basis of the Keap1–Nrf2 system. Free Radic. Biol. Med 88, 93–100. [DOI] [PubMed] [Google Scholar]
- Tanigawa S, Yokoyama KK, 2013. In: Wan Jun (Ed.), Regulation of Nrf2-ARE Signaling and Cellular Senesence by Jun Dimerization Protein 2. Introduction to Genetics (DNA Methyaltion, Histone Modification and Gene Regulation) iConcept press Ltd, USA, pp. 201–220. [Google Scholar]
- Tanigawa S, Lee CH, Lin CS, Ku CC, Hasegawa H, Qin S, Kawahara A, Korenori Y, Miyamori K, Noguchi M, Lee LH, Lin YC, Steve Lin CL, Nakamura Y, Jin C, Yamaguchi N, Eckner R, Hou DX, Yokoyama KK, 2013. Jun dimerization protein 2 is a critical component of the Nrf2/MafK complex regulating the response to ROS homeostasis. Cell Death Dis 4, e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, Hayes JD, 2015. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med 88, 108–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, 2005. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene 24, 7567–7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK, 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H: quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. U. S. A 93, 14960–14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK, 1998. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene 17, 3145–3156. [DOI] [PubMed] [Google Scholar]
- Wang SW, Lee JK, Ku CC, Chiou SS, Steve Lin CL, Ho MF, Wu DC, Yokoyama KK, 2011. Jun dimerization protein 2 in oxygen restriction; control of senescence. Curr. Pharm. Des 17, 2278–2289. [DOI] [PubMed] [Google Scholar]
- Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP, 2002. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Mol. Cell. Biol 22, 5451–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardell SE, Kwok SC, Sherman L, Hodges RS, Edwards DP, 2005. Regulation of the amino-terminal trenacription activation domain of pregesterone receptor by a cofactor-induced protein folding mechanism. Mol. Cell. Biol 25, 8792–8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenfeld-Baranboim K, Bitton-Worms K, Aronheim A, 2008. TRE-dependent transcription activation by JDP2–CHOP10 association. Nucleic Acids Res 36, 3608–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenfeld-Baranboim K, Hasin T, Darlyuk I, Heinrich R, Elhanani O, Pan J, Yokoyama KK, Aronheim A, 2009. The ubiquitously expressed bZIP inhibitor, JDP2, suppresses the transcription of its homologue immediate early gene counterpart, ATF3. Nucleic Acids Res 37, 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenfeld-Baranboim K, Koren L, Aronheim A, 2011. Phosphorylation of JDP2 on threonine-148 by the c-Jun N-terminal kinase targets it for proteosomal degradation. Biochem. J 436, 661–669. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Rust AG, McIntyre RE, Robles-Espinoza CD, del Castillo Velasco-Herrera M, Strogantsev R, Ferguson-Smith AC, McCarthy S, Keane TM, Arends MJ, Adams DJ, 2013. Jdp2 downregulates Trp53 transcription to promote leukaemogenesis in the context of Trp53 heterozygosity. Oncogene 32, 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jin C, Liu Z, Pan J, Li H, Zhang Z, Bi S, Yokoyama KK, 2014. Cloning and characterization of the mouse JDP2 gene promoter reveal negative regulation by p53. Biochem. Biophys. Res. Commun 450, 1531–1536. [DOI] [PubMed] [Google Scholar]
- Yeligar SM, Machida K, Kalra VK, 2010. Ethanol-induced HO-1 and NQO1 are differentially regulated by HIF-1alpha and Nrf2 to attenuate inflammatory cytokine expression. J. Biol. Chem 285, 35359–35373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S, 2009. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 5, 237–241. [DOI] [PubMed] [Google Scholar]
- Yuanhong X, Feng X, Qingchang L, Jianpeng F, Zhe L, Kejian G, 2010. Downregulation of AP-1 repressor JDP2 is associated with tumor metastasis and poor prognosis in patients with pancreatic carcinoma. Int. J. Biol. Markers 25, 136–140. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Crouch DH, Yamamoto M, Hayes JD, 2006. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J 399, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M, 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem 280, 30091–30099. [DOI] [PubMed] [Google Scholar]


