Skeletal muscle wasting causes both morbidity and mortality of cancer patients. Accumulating evidence suggests that the markers of endoplasmic reticulum (ER) stress and unfolded protein response (UPR) pathways are increased in skeletal muscle under multiple catabolic conditions, including cancer. However, the signaling mechanisms and the role of individual arms of the UPR in the regulation of skeletal muscle mass remain largely unknown.
KEYWORDS: atrophy, cancer cachexia, MyD88, skeletal muscle, XBP1, Toll-like receptors, unfolded protein response
ABSTRACT
Skeletal muscle wasting causes both morbidity and mortality of cancer patients. Accumulating evidence suggests that the markers of endoplasmic reticulum (ER) stress and unfolded protein response (UPR) pathways are increased in skeletal muscle under multiple catabolic conditions, including cancer. However, the signaling mechanisms and the role of individual arms of the UPR in the regulation of skeletal muscle mass remain largely unknown. In the present study, we demonstrated that gene expression of Toll-like receptors (TLRs) and myeloid differentiation primary response gene 88 (MyD88) was increased in skeletal muscle in a Lewis lung carcinoma (LLC) model of cancer cachexia. Targeted ablation of MyD88 inhibits the loss of skeletal muscle mass and strength in LLC tumor-bearing mice. Inhibition of MyD88 attenuates the LLC-induced activation of the UPR in skeletal muscle of mice. Moreover, muscle-specific deletion of X-box binding protein 1 (XBP1), a major downstream target of IRE1α arm of the UPR, ameliorates muscle wasting in LLC tumor-bearing mice. Our results also demonstrate that overexpression of an active form of XBP1 caused atrophy in cultured myotubes. In contrast, knockdown of XBP1 inhibits myotube atrophy in response to LLC or C26 adenocarcinoma cell conditioned medium. Collectively, our results demonstrate that TLR/MyD88-mediated activation of XBP1 causes skeletal muscle wasting in LLC tumor-bearing mice.
INTRODUCTION
Cancer cachexia is a devastating syndrome that is characterized by the progressive loss of skeletal muscle mass and strength. Cachexia afflicts a vast majority of cancer patients and is responsible for about 30% of all cancer-related deaths (1). In addition to the deterioration in the quality of life, patients with cachexia show intolerance to chemotherapy and other antineoplasm treatments (2). There is now evidence that increased protein degradation due to stimulation of the ubiquitin-proteasome system (UPS) and autophagy are prominent mechanisms for skeletal muscle wasting in many catabolic states, including cancer (3–5). Moreover, a systemic inflammatory response, triggered by factors generated by either tumor or nontumor cells, also plays an important role in skeletal muscle weakness in cancer-induced cachexia (6). While a number of drugs have been tested in clinical trials, none of them showed a significant impact on the quality of life or weight gain, emphasizing that a better understanding of the molecular mechanisms is paramount for the development of effective therapies to preserve skeletal muscle mass in cancer patients (1, 6, 7).
Proinflammatory cytokines are important mediators of skeletal muscle wasting in many chronic disease states, including cancer (1, 6). Furthermore, cancer cells generate danger-associated molecular patterns (DAMPs), as well as heat shock proteins (HSPs), that stimulate muscle proteolysis through binding of the Toll-like receptors (TLRs) (6, 8). Indeed, recent studies have suggested that the genetic ablation of TLR4 inhibits skeletal muscle wasting in response to microbial products, as well as in a model of cancer cachexia (9, 10). Myeloid differentiation primary response gene 88 protein (MyD88) is the key adaptor protein for the interleukin-1 (IL-1) receptor and for TLR-mediated activation of downstream signaling pathways (11–13). Accumulating evidence suggests that MyD88 plays an important role in the regulation of skeletal muscle mass under diverse conditions. For example, we previously reported that MyD88 promotes myoblast fusion in a cell-autonomous manner during postnatal muscle growth and overload-induced hypertrophy (14). In contrast, activation of the TLRs and MyD88 promotes inflammation, leading to a worsening of the muscle pathology in animal models of muscular dystrophy (15). However, the role of MyD88 and mechanisms by which it regulates skeletal muscle mass in cancer-induced cachexia remain to be investigated.
The endoplasmic reticulum (ER) is a major site for the synthesis and proper folding and maturation of cellular proteins in the mammalian cells. The ER also plays a critical role in the regulation of calcium levels in the cell. Accumulation of misfolded/unfolded proteins or disruption in calcium levels leads to stress in the ER. This stress is resolved through the activation of a signaling network commonly referred to as unfolded protein response (UPR). The UPR is initiated by three ER transmembrane sensors: protein kinase R-like endoplasmic reticulum kinase (PERK), inositol-requiring protein 1α (IRE1α), and activating transcription factor 6 (ATF6) (16–18). Under conditions of ER stress, PERK undergoes oligomerization and autophosphorylation, leading to its activation. PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α), which represses general translation while selectively augmenting the translation of ATF4 and gene expression of stress-responsive genes, such as the C/EBP homologous protein (CHOP) and GRP78/Binding immunoglobulin protein (BiP) genes (18–20). Similarly to PERK, IRE1α is activated through autophosphorylation. IRE1α possesses endonuclease activity that facilitates the splicing of a 26-base intron from X-box-binding protein 1 (XBP1) mRNA (21). Spliced XBP1 (sXBP1) acts as a potent transcription factor to induce the gene expression of several ER chaperones involved in protein folding or ER-associated protein degradation (22). Chronic ER stress also stimulates IRE1α kinase activity, leading to the activation of c-Jun N-terminal kinase (JNK) and nuclear factor kappa B (NF-κB) pathways (18). Finally, under stress conditions, ATF6 translocates from the ER to the Golgi apparatus, where it is cleaved by site 1 proteases (S1P), resulting in the formation of an active ATF6 transcription factor (23). The active ATF6 fragment is subsequently imported into the nucleus, where it increases the level of gene expression of various proteins that function to alleviate ER stress (18–21). While the main function of the UPR is to improve homeostasis, unmitigated ER stress and chronic activation of components of the UPR can also produce deleterious effects, such as inflammation and apoptosis (18). Interestingly, a few studies have suggested that some components of the UPR can be activated in the absence of ER stress (24–26). For example, agonists of TLRs activate IRE1α and its downstream target, the XBP1 transcription factor, to induce gene expression of various inflammatory molecules in macrophages (24).
Recent studies have suggested that the levels of markers of ER stress are increased in skeletal muscle under physiological and various pathological conditions (25, 26). We previously reported that the levels of several markers of ER stress and the activation of UPR pathways are significantly increased in the skeletal muscle of mice in response to starvation and during cancer-associated cachexia (27, 28). Intriguingly, our study also demonstrated that chronic administration of 4-phenylbutyrate (4-PBA), a chemical chaperone and inhibitor of ER stress, in wild-type mice leads to skeletal muscle weakness and atrophy. Moreover, treatment with 4-PBA aggravated muscle wasting in the Lewis lung carcinoma (LLC) model of cancer cachexia (27). These results highlighted that physiological levels of ER stress and the UPR may be important for the maintenance of skeletal muscle health both under naive conditions and in cancer-induced cachexia. It is also possible that individual arms of the UPR play distinct roles in the regulation of skeletal muscle mass in various catabolic states, including during tumor growth. However, the role of different arms of the UPR in the regulation of skeletal muscle mass has not yet been investigated. Moreover, the signaling mechanisms that regulate the activation of the UPR in skeletal muscle during cancer cachexia remain completely unknown.
In the present study, we demonstrated that the levels of several TLRs and MyD88 are increased in skeletal muscle of LLC tumor-bearing mice. The targeted ablation of MyD88 blocks LLC tumor-induced muscle wasting in mice. Our results demonstrate that TLR/MyD88 mediates the activation of the UPR in skeletal muscle during cancer cachexia. Similarly to MyD88, the skeletal muscle-specific deletion of XBP1 attenuates LLC tumor-induced loss of muscle mass both in vivo and in vitro. Finally, our results demonstrate that forced expression of sXBP1 is sufficient to cause atrophy and increases the gene expression of various proinflammatory cytokines, specific components of UPS, and autophagy in cultured myotubes.
RESULTS
Levels of TLRs and MyD88 are increased in skeletal muscle during cancer cachexia.
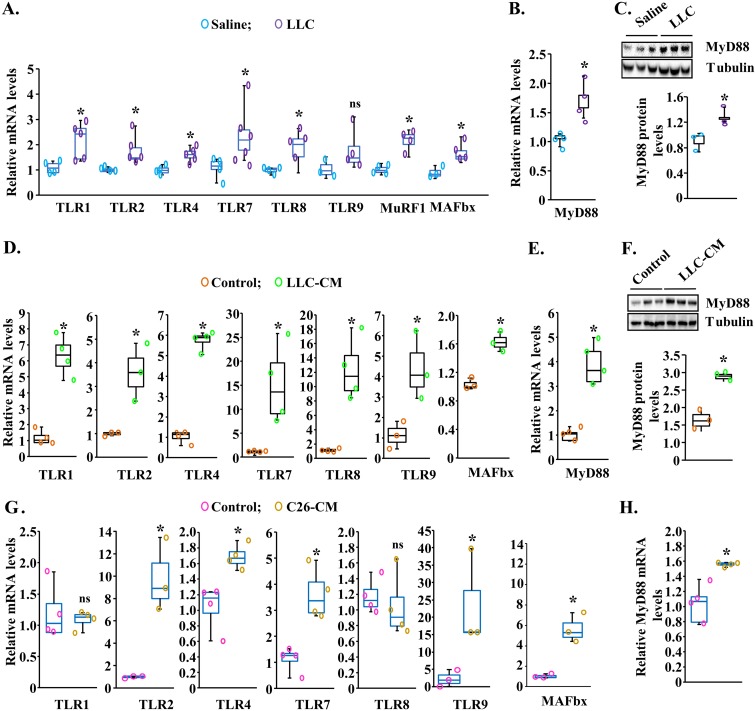
We first investigated how the levels of gene expression of various TLRs and MyD88 are regulated in skeletal muscle of mice in response to tumor growth. Adult C57BL/6J mice were inoculated with 2 × 106 LLC cells in the left flank. After 21 days, the mice were examined for body weight and grip strength. Consistent with our previous studies (27), there was a significant reduction in body weight and grip strength in LLC-bearing mice compared with control mice (see Fig. S1A and B in the supplemental material). The mice were then euthanized, and individual right hind limb muscles were isolated and analyzed by performing reverse transcription-quantitative PCR (QRT-PCR) analysis and Western blotting. We found that transcript levels of TLR1, TLR2, TLR4, TLR7, and TLR8 were significantly increased in the gastrocnemius (GA) muscle of LLC tumor-bearing mice compared with corresponding control mice. However, there was no significant difference in the mRNA levels of TLR9 in GA muscle of control and LLC tumor-bearing mice (Fig. 1A). Our analysis also showed that the mRNA levels of two muscle-specific E3 ubiquitin ligases (i.e., MuRF1 and MAFbx) were significantly higher in GA muscle of LLC-bearing mice (Fig. 1A). Moreover, both mRNA and protein levels of MyD88 were found to be significantly increased in the GA muscle of LLC tumor-bearing mice compared to control mice (Fig. 1B and C).
FIG 1.
Increased gene expression of TLRs and MyD88 during cancer cachexia. Three-month-old male C57BL/6J mice were injected with saline solution alone or with 2 × 106 Lewis lung carcinoma (LLC) cells in the left flank, and tumor growth was monitored. After 21 days, right-side hind limb muscles were isolated and analyzed by QRT-PCR and Western blotting. (A) Relative mRNA levels of TLR1, TLR2, TLR4, TLR7, TLR8, TLR9, MuRF1, and MAFbx in gastrocnemius (GA) muscle of control (saline solution alone injected) and LLC tumor-bearing mice. n = 4 to 6 in each group. (B and C) Relative mRNA (B) and protein (C) levels of MyD88 in GA muscle of control and LLC tumor-bearing mice. Data represent results of densitometry quantification of bands in immunoblots. n = 3 in each group. C2C12 myotubes were incubated in differentiation medium with or without LLC-conditioned medium (LLC-CM) or C26-CM for 24 h followed by performing QRT-PCR or Western blotting. (D and E) Transcript levels of TLR1, TLR2, TLR4, TLR7, TLR8, TLR9, and MAFbx (D) and MyD88 (E) in control and LLC-CM-treated myotubes. (F) Representative immunoblots and densitometry analysis of MyD88 protein in control and LLC-CM-treated myotubes. (G and H) Relative mRNA levels of TLR1, TLR2, TLR4, TLR7, TLR8, TLR9, and MAFbx (G) and MyD88 (H) in control and C26-CM-treated myotubes assayed by QRT-PCR. n = 3 or 4 in each group. Medians and 25th to 75th percentiles are shown. *, P < 0.05 (values significantly different from corresponding control values). ns, not significant.
To understand whether factors secreted by tumor cells induce the gene expression of TLRs and MyD88 in skeletal muscle, we first prepared LLC cell conditioned medium (LLC-CM) and validated that it causes atrophy in cultured C2C12 myotubes (Fig. S1C and D). Next, C2C12 myotubes were treated with LLC-CM in a 1:4 ratio for 24 h followed by performing QRT-PCR. Results showed that treatment with LLC-CM significantly increased the mRNA levels of TLR1, TLR2, TLR4, TLR7, TLR8, and TLR9 as well as MAFbx in C2C12 myotubes (Fig. 1D). Moreover, there was a significant increase in the mRNA and protein levels of MyD88 in C2C12 myotubes upon treatment with LLC-CM (Fig. 1E and F).
Mice implanted with C26 adenocarcinoma represent another extensively used model of cancer cachexia (29, 30). We investigated whether the factors produced by C26 adenocarcinoma also augment the expression of various TLRs and MyD88 in cultured myotubes. We first confirmed that C26 conditioned medium (C26-CM) causes atrophy in cultured C2C12 myotubes (Fig. S1C and D). We next examined the effects of C26-CM on the gene expression of various TLRs and MyD88 in C2C12 myotubes. As shown in Fig. 1G, mRNA levels of TLR2, TLR4, TLR7, and TLR9 were significantly increased in C2C12 myotubes upon treatment with C26-CM for 24 h. There was no significant difference between control and C26-CM-treated myotubes in mRNA levels of TLR1 (Fig. 1G). Moreover, treatment with C26-CM also increased the mRNA levels of MyD88 in C2C12 myotubes (Fig. 1H). Taken together, these results suggest that factors produced by tumor cells augment the gene expression of various TLRs and MyD88 in skeletal muscle during cancer cachexia.
Targeted deletion of MyD88 inhibits LLC tumor-induced skeletal muscle wasting in mice.
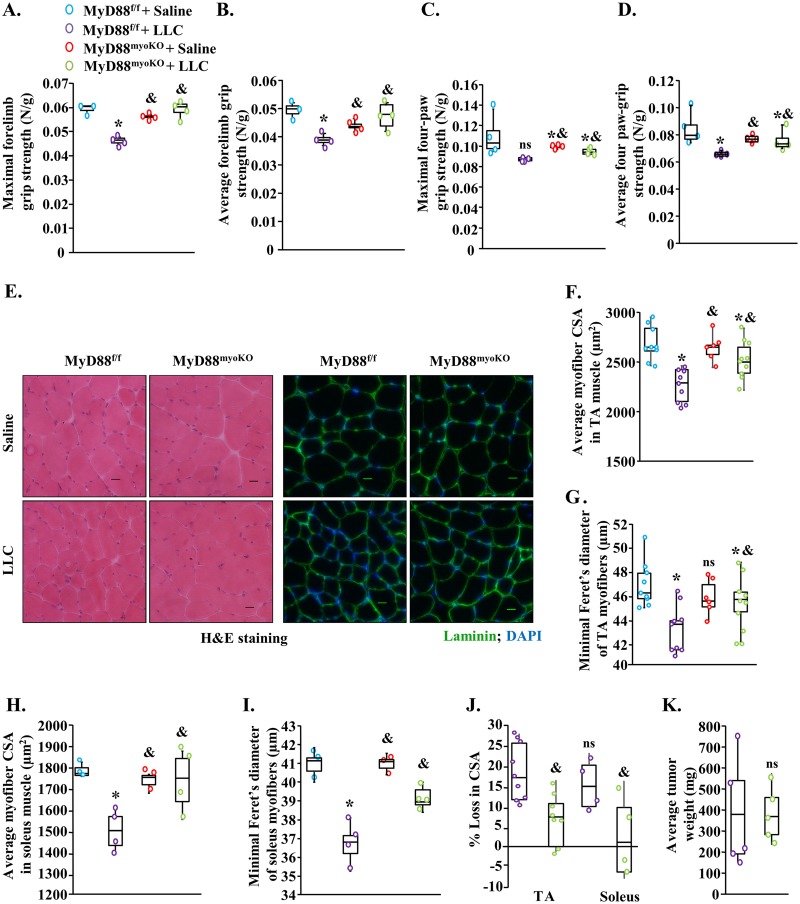
MyD88 is a critical adaptor protein that mediates the activation of downstream signaling from all TLRs, except TLR3 (11, 31). In fact, deletion of MyD88 has been a widely used approach to investigate the role of TLRs under both physiological and pathological conditions (31). To investigate the role of MyD88 in cancer cachexia, we used the Myod1-Cre line, which features deletions of the target gene in both myoblasts and differentiated skeletal muscle. Specifically, floxed MyD88 (MyD88f/f) mice were crossed with the Myod1-Cre line to generate littermate muscle-specific MyD88-knockout (MyD88myoKO) and control (MyD88f/f) mice as previously described (14). We have previously reported that the levels of MyD88 are drastically reduced in skeletal muscle of MyD88myoKO mice compared with littermate MyD88f/f mice (14). By performing immunoblotting, we confirmed that MyD88 levels were depleted in GA muscle of MyD88myoKO mice compared with littermate MyD88f/f mice (Fig. S2A). To understand the role of MyD88 in cancer cachexia, 3-month-old MyD88f/f and MyD88myoKO mice were injected with 2 × 106 LLC cells or saline solution alone (as a control) subcutaneously in the left flank and growth of LLC tumor was monitored every day. At day 20 post-LLC injection, the mice were analyzed for grip strength measurements. Results showed that maximal (peak) and average forelimb or total four-paw grip strength normalized with body weight was reduced in MyD88f/f mice in response to LLC tumor growth (Fig. 2A to D). Interestingly, MyD88myoKO mice were protected from LLC tumor-induced loss in grip strength. Specifically, maximal and average forelimb or total four-paw grip strength normalized with body weight was significantly higher in LLC tumor-bearing MyD88myoKO mice than in the corresponding MyD88f/f mice (Fig. 2A to D).
FIG 2.
Targeted ablation of MyD88 prevents skeletal muscle atrophy in LLC tumor-bearing mice. Three-month-old MyD88f/f and MyD88myoKO mice were inoculated with 2 × 106 LLC cells in the left flank and monitored for 21 days. (A and B) Quantification of the maximal (A) and average (B) forelimb strength of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice normalized by body weight. (C and D) Quantification of the maximal (C) and average (D) four-paw strength of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice normalized by body weight. (E) Representative photomicrographs of H&E-stained and antilaminin-stained sections of the TA muscle of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice. Bars, 50 μm. (F and G) Quantification of average cross-sectional area (CSA) (F) and minimal Feret’s diameter (G) of myofibers in TA muscle of control and tumor-bearing MyD88f/f and MyD88myoKO mice. (H and I) Quantification of average CSA (H) and minimal Feret’s diameter (I) of myofibers in soleus muscle of control and tumor-bearing MyD88f/f and MyD88myoKO mice. (J) Quantification of percentage of loss in myofiber CSA after implantation of LLC tumor in TA and soleus muscle of MyD88f/f and MyD88myoKO mice. (K) Average tumor wet weight in MyD88f/f and MyD88myoKO mice after 21 days of injection of LLC cells. n = 4 to 7/group. Medians and 25th to 75th percentiles are shown. *, P < 0.05 (values significantly different from those determined for MyD88f/f mice injected with saline solution alone); &, P < 0.05 values significantly different from those determined for LLC-bearing MyD88f/f mice; ns, not significant.
We next investigated whether genetic ablation of MyD88 has any effect on skeletal muscle wasting in response to LLC tumor growth. Our analysis showed that the wet weight of GA muscle, tibialis anterior (TA) muscle, and soleus muscle (normalized with body weight) was significantly higher in LLC tumor-bearing MyD88myoKO mice than in MyD88f/f mice (Fig. S2B to D). We next generated transverse sections of TA and soleus muscles and performed hematoxylin and eosin (H&E) staining or immunostaining for laminin protein. There was no overt difference in the skeletal muscle structure in control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice (Fig. 2E). Morphometric analysis of antilaminin-stained sections revealed a significant improvement in average cross-sectional area (CSA) as well as in the minimal Feret’s diameter in both TA and soleus muscle of LLC tumor-bearing MyD88myoKO mice compared with corresponding MyD88f/f mice (Fig. 2F to I). Our analysis also showed that there was a significant reduction in the LLC tumor-induced percentage of loss in myofiber CSA in MyD88myoKO mice compared with MyD88f/f mice (Fig. 2J). However, average tumor sizes were comparable between MyD88f/f and MyD88myoKO mice at day 21 after implantation of LLC cells (Fig. 2K), suggesting that the targeted deletion of MyD88 inhibited LLC tumor-induced muscle wasting without having any significant impact on tumor growth in mice.
MyD88 mediates the activation of catabolic pathways in skeletal muscle of LLC tumor-bearing mice.
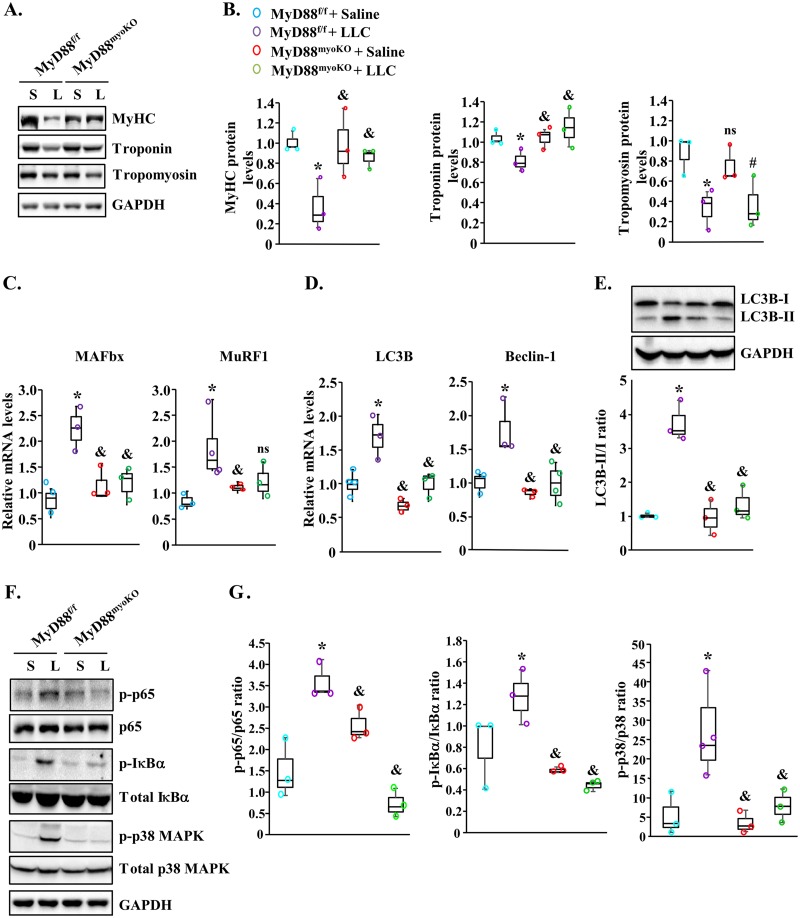
Skeletal muscle wasting under many catabolic conditions, including cancer growth, involves the degradation of specific muscle proteins (27, 30, 32). To investigate the biochemical mechanisms by which MyD88 mediates skeletal muscle wasting, we first measured levels of myosin heavy chain (MyHC), tropomyosin, and troponin protein in GA muscle of control and LLC tumor-bearing mice. There was a significant reduction in the levels of all three proteins in GA muscle of MyD88f/f mice compared with corresponding control mice without tumors. This analysis also showed that levels of MyHC and troponin but not tropomyosin were significantly higher in LLC tumor-bearing MyD88myoKO mice than in the corresponding MyD88f/f mice (Fig. 3A and B).
FIG 3.
Ablation of MyD88 inhibits catabolic pathways in skeletal muscle during cancer cachexia. (A) Representative immunoblots demonstrating levels of MyHC, troponin, tropomyosin, and unrelated protein GAPDH in GA muscle of control (S [saline solution]) and LLC tumor-bearing (L) MyD88f/f and MyD88myoKO mice. (B) Results of densitometry quantification of band intensities of MyHC, troponin, and tropomyosin from multiple immunoblots. (C) Relative mRNA levels of MAFbx and MuRF1 in GA muscle of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice. (D) Relative mRNA levels of autophagy genes LC3B and Beclin-1 in GA muscle of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice. (E) Representative immunoblots and quantification of ratio of LC3B-II and LC3B-I in GA muscle of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice. (F) Representative immunoblots demonstrating the levels of phosphorylated and total p65, IκBα, and p38 and unrelated protein GAPDH in GA muscle of control and LLC tumor-bearing mice. (G) Densitometry analysis of ratio of p-p65/p65, p-IκBα/IκBα, and p-p38/p38 from multiple immunoblots. n = 3 or 4 mice/group. Medians and 25th to 75th percentiles are shown. *, P < 0.05 (values significantly different from those determined for MyD88f/f injected with saline solution alone); &, P < 0.05 (values significantly different from those determined for LLC tumor-bearing MyD88f/f mice); #, P < 0.05 (values significantly different from those determined for MyD88myoKO mice injected with saline solution alone); ns, not significant.
Skeletal muscle wasting under multiple conditions is a result of accelerated protein degradation. The ubiquitin-proteasome system (UPS) and autophagy are two major proteolytic systems that cause the degradation of the bulk of muscle proteins in various catabolic states (3, 33, 34). Indeed, gene expression of various components of the UPS and autophagy is increased in skeletal muscle in response to a number of catabolic stimuli, including cancer. We next measured relative mRNA levels of select markers of UPS and autophagy in GA muscle of MyD88f/f and MyD88myoKO mice. Consistent with published reports (1, 27, 35), the mRNA levels of muscle-specific E3 ubiquitin ligases MAFbx and MuRF1 were found to be significantly increased in GA muscle of LLC tumor-bearing MyD88f/f mice compared with control mice (Fig. 3C). Importantly, mRNA levels of MAFbx were significantly reduced and a trend toward a decrease in MuRF1 mRNA levels was noticeable in GA muscle of MyD88myoKO mice compared with MyD88f/f mice (Fig. 3C). Similarly to the UPS, there was also a significant increase in transcript levels of autophagy markers LC3B and Beclin-1 in skeletal muscle of LLC-bearing MyD88f/f mice. However, transcript levels of both LC3B and Beclin-1 were found to be significantly reduced in GA muscle of MyD88myoKO mice compared with MyD88f/f mice in response to LLC tumor growth (Fig. 3D). Furthermore, the ratio of LC3B-II to LC3B-I (LC3B-II/I) was also found to be significantly reduced in the GA muscle of LLC-bearing MyD88myoKO mice compared with the corresponding MyD88f/f mice, further suggesting that the genetic ablation of MyD88 inhibits the activation of autophagy in skeletal muscle during cancer cachexia (Fig. 3E).
Activation of the UPS and autophagy in skeletal muscle involves the upstream activation of specific signaling pathways (34). Because MyD88 is a major adaptor protein involved in the activation of various signaling pathways, we next investigated whether MyD88 functions through the activation of specific signaling proteins in skeletal muscle of tumor-bearing mice. We first investigated whether MyD88 plays a role in the activation of nuclear factor kappa-B (NF-κB) in skeletal muscle of LLC tumor-bearing mice. By performing Western blotting, we first measured the phosphorylation of NF-κB subunit p65 (also called RelA). There was no significant difference in the levels of phosphorylated p65 in GA muscle of MyD88myoKO mice compared to MyD88f/f mice. Interestingly, LLC tumor-induced phosphorylation of p65 in GA muscle was significantly inhibited in MyD88myoKO mice compared with corresponding MyD88f/f mice (Fig. 3F and G). Inhibitor of κB (IκBα) protein masks the nuclear localization signals of NF-κB proteins and keeps them sequestered in an inactive state in the cytoplasm (36). In response to NF-κB-activating stimuli, IκBα is phosphorylated followed by its degradation through proteasome activity (36). Our results showed that the levels of phosphorylated IκBα protein were significantly reduced in the GA muscle of LLC tumor-bearing MyD88myoKO mice compared with MyD88f/f mice, further confirming that ablation of MyD88 inhibits LLC tumor-induced activation of NF-κB in skeletal muscle of mice (Fig. 3F and G). We also investigated whether MyD88 regulates the activation of p38 mitogen-activated protein kinase (MAPK) signaling in skeletal muscle of mice in response to tumor growth. Results showed that the levels of phosphorylated p38 MAPK were significantly reduced in GA muscle of LLC tumor-bearing MyD88myoKO mice compared with corresponding LLC tumor-bearing MyD88f/f mice (Fig. 3F and G). Taken together, these results suggest that deletion of MyD88 attenuates the activation of the proteolytic systems and catabolic signaling pathways in skeletal muscle of LLC tumor-bearing mice.
MyD88 mediates the activation of UPR pathways in skeletal muscle during cancer cachexia.
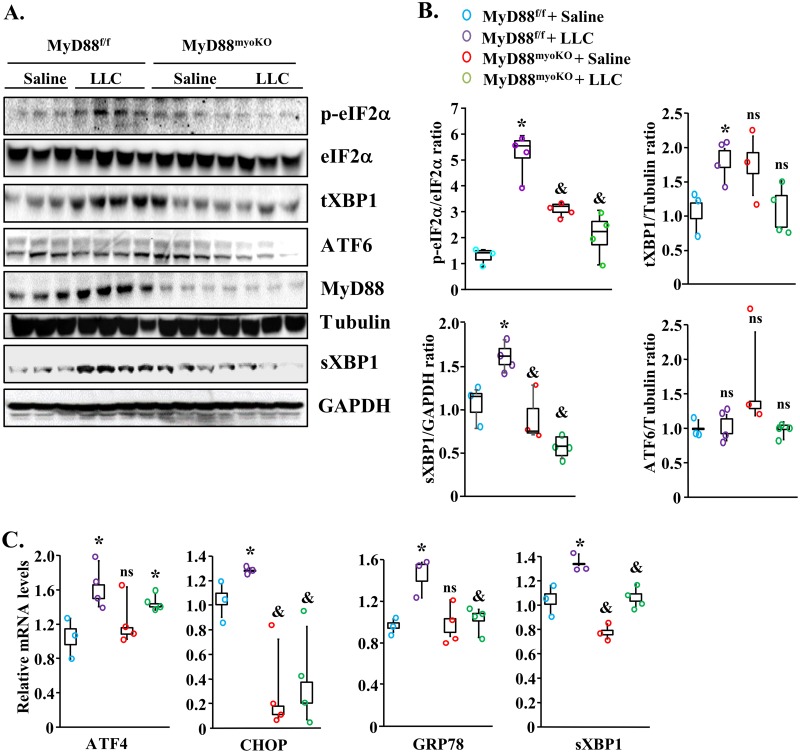
The UPR pathways are employed by cells as a corrective measure to avoid an increase in the unfolded-protein load. It is believed that the PERK and IRE1α arms of the UPR are involved in mediating deleterious effects, whereas the ATF6 arm mediates adaptive responses (25, 26). We investigated whether MyD88 has any role in the activation of the UPR pathways in skeletal muscle of LLC-tumor-bearing mice. Consistent with our previously published report (27), the markers of the PERK and IRE1α/XBP1 arms of the UPR pathway were found to be significantly increased in skeletal muscle of LLC tumor-bearing MyD88f/f mice. Interestingly, LLC-induced increases in the levels of p-eIF2α, total XBP1 (tXBP1), and sXBP1 were significantly inhibited in skeletal muscle of MyD88myoKO mice compared with MyD88f/f mice (Fig. 4A and B). We also compared mRNA levels of various markers of ER stress in skeletal muscle of MyD88f/f and MyD88myoKO mice. This analysis showed that LLC tumor-induced increases in ATF4, CHOP, GRP78, and sXBP1 mRNA levels were significantly diminished in skeletal muscle of MyD88myoKO mice compared with MyD88f/f mice (Fig. 4C).
FIG 4.
Genetic ablation of MyD88 in skeletal muscle inhibits the activation of UPR pathways during cancer cachexia. (A) Immunoblots demonstrating levels of p-eIF2α, total eIF2α, total XBP1 (tXBP1), spliced XBP1 (sXBP1), ATF6, MyD88, and unrelated protein tubulin in the GA muscle of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice. (B) Results of densitometry quantification of bands in immunoblots. (C) Relative mRNA levels of ATF4, CHOP, GRP78, and sXBP1 in GA muscle of control and LLC tumor-bearing MyD88f/f and MyD88myoKO mice. n = 3 or 4 in each group. Medians and 25th to 75th percentiles are shown. *, P < 0.05 (values significantly different from those determined for MyD88f/f injected with saline solution alone); &, P < 0.05 (values significantly different from those determined for LLC tumor-bearing MyD88f/f mice); ns, not significant.
A recent study provided evidence that tumor cells secrete heat shock protein 70 (Hsp70) and Hsp90, which cause muscle wasting through binding to TLRs in mouse models of cancer cachexia (8). We investigated whether recombinant Hsp70 protein can activate the PERK and IRE1α/XBP1 arms of the UPR in cultured myotubes. Results showed that treatment with Hsp70 significantly increased the levels of p-eIF2α and sXBP1 in cultured C2C12 myotubes (Fig. S3A and B). Furthermore, QRT-PCR analysis showed that mRNA levels of several ER stress markers, i.e., CHOP, GADD34, and sXBP1, were significantly increased in C2C12 myotubes upon treatment with Hsp70 protein (Fig. S3C). Collectively, these results suggest that tumor-derived factors activate the UPR through TLR/MyD88-dependent mechanisms in skeletal muscle.
Targeted ablation of XBP1 inhibits skeletal muscle wasting in LLC tumor-bearing mice.
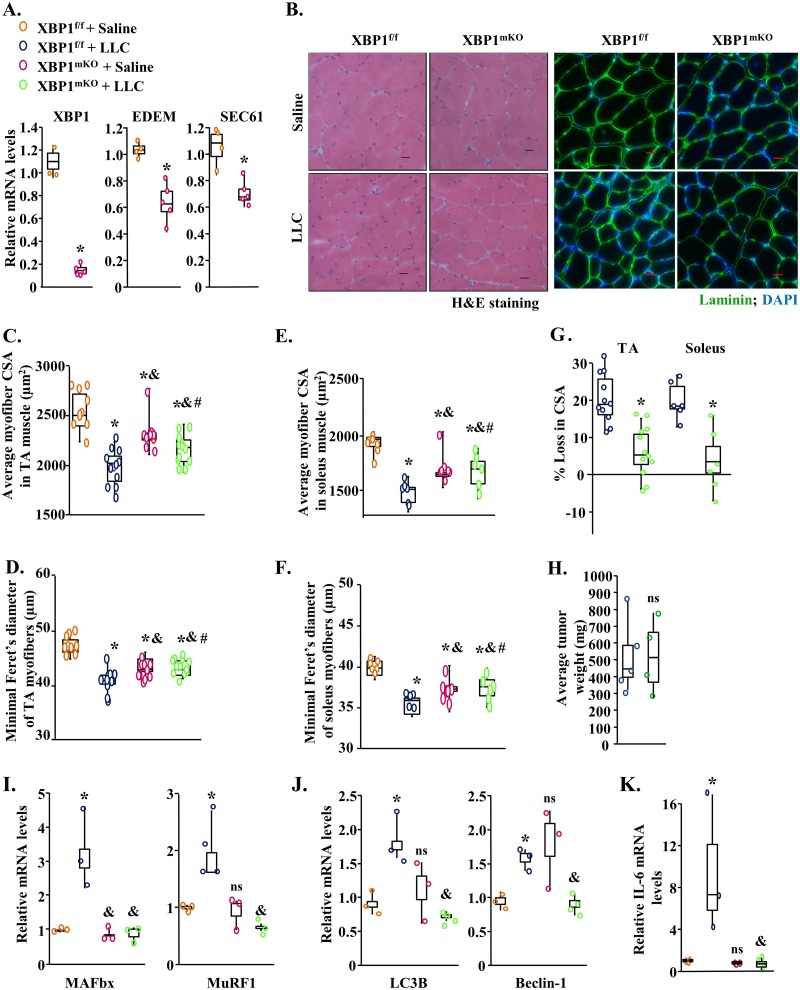
While both the PERK and IRE1α/XBP1 arms of the UPR are activated during cancer cachexia, we recently reported that targeted ablation of PERK does not attenuate skeletal muscle wasting in response to growth of LLC tumor in mice (37). We next investigated whether inhibition of XBP1, the major effector of the IRE1α arm of the UPR, can attenuate skeletal muscle wasting in an LLC model of cancer cachexia. Since XBP1 is also required for myogenic differentiation (38), we generated mice in which XBP1 is deleted only in differentiated skeletal muscle. Specifically, floxed XBP1 (XBP1f/f) mice were crossed with the muscle creatine kinase (MCK)-Cre line to generate muscle-specific XBP1-knockout mice (referred to here as XBP1mKO mice) and littermate XBP1f/f mice. Our QRT-PCR analysis confirmed that the mRNA levels of XBP1 were drastically reduced in GA muscle of XBP1mKO mice compared to littermate XBP1f/f mice. Furthermore, the mRNA levels of EDEM and SEC61, which are regulated by XBP1, were also found to be significantly reduced in GA muscle of XBP1mKO mice compared with XBP1f/f mice (Fig. 5A). It is notable that even though a drastic reduction in the mRNA levels of XBP1 in GA muscle of XBP1mKO mice was observed, the decrease in the levels of EDEM and SEC61 was not that dramatic. This finding could be attributed to the fact that in addition to XBP1, other transcription factors, such as ATF6, are required to induce gene expression of various ER stress-responsive molecules, including EDEM and SEC61, under stress conditions (18, 25, 39).
FIG 5.
Targeted deletion of XBP1 inhibits skeletal muscle wasting in LLC tumor-bearing mice. (A) Relative mRNA levels of XBP1, EDEM, and SEC61 in GA muscle of naive XBP1f/f and XBP1mKO mice. n = 4 in each group. Three-month-old XBP1f/f and XBP1mKO mice were inoculated with 2 × 106 LLC cells in the left flank and monitored for 21 days. (B) Representative photomicrographs of H&E-stained and antilaminin-stained sections of TA muscle of control and LLC tumor-bearing XBP1f/f and XBP1mKO mice. Bars, 50 μm. (C and D) Quantification of average myofiber CSA (C) and minimal Feret’s diameter (D) in TA muscle of control and LLC tumor-bearing XBP1f/f and XBP1mKO mice. (E and F) Quantification of average myofiber CSA (E) and minimal Feret’s diameter (F) in soleus muscle of control and LLC tumor-bearing XBP1f/f and XBP1mKO mice. (G) Quantification of percentage of loss in myofiber CSA in TA and soleus muscle of XBP1f/f and XBP1mKO mice after 21 days of inoculation with LLC cells. (H) Quantification of the average tumor wet weight in tumor-bearing XBP1f/f and XBP1mKO mice. n = 9 to 12 mice/group. (I to K) Relative mRNA levels of MAFbx and MuRF1 (I), LC3B and Beclin-1 (J), and IL-6 (K) in GA muscle of control and LLC-bearing XBP1f/f and XBP1mKO mice. n = 4 mice/group. Medians and 25th to 75th percentiles are shown. *, P < 0.05 (values significantly different from those determined for XBP1f/f mice injected with saline solution alone); &, P < 0.05 (values significantly different from those determined for LLC tumor-bearing XBP1f/f mice); #, P < 0.05 (values significantly different from those determined for XBP1mKO mice injected with saline solution alone); ns, not significant.
To understand the role of XBP1 in muscle wasting in cancer cachexia, 3-month-old XBP1f/f and XBP1mKO mice were implanted with 2 × 106 LLC cells. After 21 days, hind limb muscles of the mice were isolated and analyzed by morphometric and biochemical assays. Our analysis showed that the wet weight of GA, TA, and soleus muscle was significantly less in LLC tumor-bearing MyD88f/f mice than in control MyD88f/f mice. However, there was no significant reduction in the wet weight of GA, TA, or soleus muscle in XBP1mko mice in response to LLC tumor growth (Fig. S4A to C). To further understand the role of XBP1 in LLC tumor-induced muscle wasting, we generated transverse sections of TA and soleus and performed H&E staining or antilaminin staining (Fig. 5B). Intriguingly, our analysis showed that the average myofiber CSA and minimal Feret’s diameter in TA and soleus muscles were reduced in XBP1mKO mice compared with littermate XBP1f/f mice under naive conditions, suggesting that XBP1 may have a role in skeletal muscle growth (Fig. 5C to F). However, we found that XBP1mKO mice were protected from LLC-induced muscle wasting. The average CSA and minimal Feret’s diameter of myofibers were significantly higher in TA and soleus muscle of LLC tumor-bearing XBP1mKO mice than in the corresponding XBP1f/f mice (Fig. 5C to G). Our analysis also showed that the deletion of XBP1 in skeletal muscle had no effect on growth of LLC tumors in mice (Fig. 5H).
We next compared mRNA levels of components of UPS and autophagy in GA muscle of XBP1f/f and XBP1mKO mice. Results showed that LLC tumor-induced increases in the mRNA levels of MuRF1, MAFbx, LC3B, and Beclin-1 were significantly inhibited in GA muscle of XBP1mKO mice compared with XBP1f/f mice (Fig. 5I and J).
Inflammatory cytokines are some of the important mediators of skeletal muscle wasting in various catabolic states, including cancer (40, 41). Interestingly, sXBP1 has been reported to bind to the promoter region of the IL-6 gene to enhance the levels of IL-6 in macrophages (24). Thus, we next investigated whether inhibition of XBP1 would have any effect on the gene expression of IL-6 in LLC tumor-bearing mice. Our QRT-PCR analysis showed that the LLC tumor-induced increase in mRNA levels of IL-6 was significantly inhibited in GA muscle of XBP1mKO mice compared to the corresponding XBP1f/f mice (Fig. 5K). Collectively, these results suggest that the ablation of XBP1 inhibits LLC tumor-induced muscle wasting, potentially through the inhibition of various proteolytic pathways and repression of levels of IL-6 in skeletal muscle.
Activation of XBP1 induces atrophy in cultured myotubes.
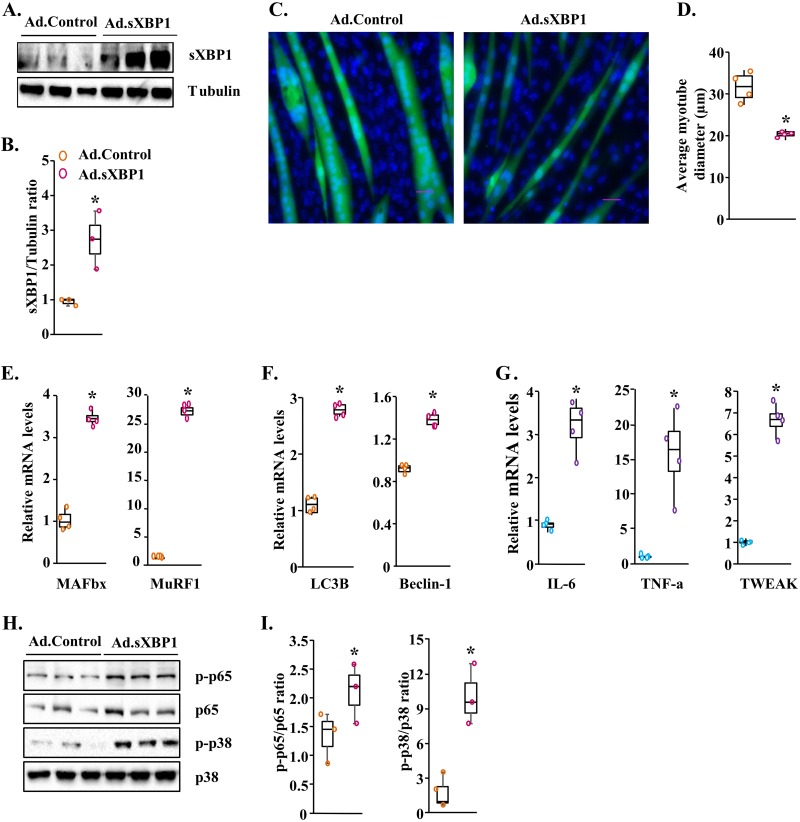
While our results demonstrated that the inhibition of XBP1 can prevent skeletal muscle wasting in response to LLC, whether forced activation of XBP1 is sufficient to cause skeletal muscle wasting remains unknown. To address this issue, we generated an adenoviral vector expressing spliced XBP1 (sXBP1) cDNA and green fluorescence protein (GFP). Western blot analysis showed that the levels of sXBP1 protein were significantly increased in C2C12 myotube cultures transduced with sXBP1-expressing adenoviral (Ad.sXBP1) vector compared with control adenoviral (Ad.Control) vector (Fig. 6A and B). In a parallel experiment, fully differentiated C2C12 myotubes were transduced with Ad.Control or Ad.XBP1 vector. After 48 h, the cultures of myotubes were fixed and stained with DAPI (4′,6-diamidino-2-phenylindole) (Fig. 6C). Interestingly, we found that the overexpression of sXBP1 led to a significant decrease in the diameter of myotubes (Fig. 6C and D).
FIG 6.
Overexpression of sXBP1 causes atrophy in cultured myotubes. C2C12 myotubes were transduced with control adenovirus (Ad.Control) or sXBP1-expressing (Ad.sXBP1) for 24 h. The cultures were incubated in differentiation medium for an additional 48 h. (A) Immunoblots presented here demonstrate the levels of sXBP1 and of an unrelated protein, tubulin, in C2C12 cultures transduced with Ad.Control and Ad.sXBP1. (B) Results of densitometry quantification of band intensity in immunoblots for sXBP1 and tubulin. (C) Representative images of control and sXBP1-overexpressing myotube cultures after staining with DAPI. Bar, 50 μm. (D) Quantification of average myotube diameter in control and sXBP1-overexpressing cultures. (E to G) Relative mRNA levels of MAFbx and MuRF1 (E), LC3B and Beclin-1 (F), and IL-6, TNF-α, and TWEAK (G) in control and sXBP1-overexpressing myotube cultures. (H) Immunoblots demonstrating levels of p-p65, p65, p-p38, and p38 in control and sXBP1-overexpressing cultures. (I) Results of densitometry quantification of band intensities in immunoblots. Ratios of p-p65/p65 and p-p38/p38 are presented here. n = 3 or 4 in each group. Medians and 25th to 75th percentiles are shown. *, P < 0.05 (values significantly different from those determined for cultures transduced with Ad.Control).
We next measured mRNA levels of markers of the UPS and autophagy in control and sXBP1-overexpressing myotubes. Our analysis showed that mRNA levels of MAFbx, MuRF1, LC3B, and Beclin-1 were significantly increased in cultures of myotubes transduced with sXBP1-expressing adenoviral vector (Fig. 6E and F). Moreover, mRNA levels of IL-6, TNF-α, and TWEAK were also significantly increased in sXBP1-overexpressing cultures compared to controls (Fig. 6G). We also measured whether overexpression of sXBP1 affects the activation of NF-κB and p38 MAPK pathways. Results showed that levels of phosphorylated p65 and phosphorylated p38 MAPK were significantly increased in sXBP1-overexpressing cultures compared with control cultures (Fig. 6H and I). Finally, Western blot analysis confirmed that protein levels of sXBP1 were significantly higher in cultures transduced with sXBP1-expressing adenoviral vector (Fig. 6H and I).
Knockdown of XBP1 attenuates myotube atrophy in response to tumor-derived factors.
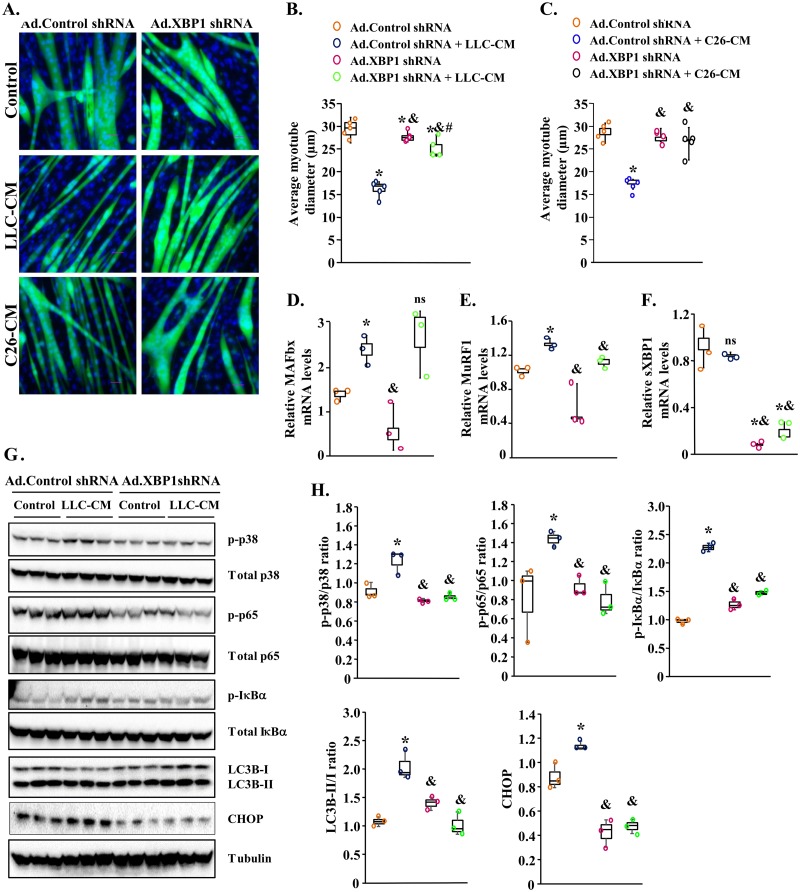
We next investigated whether the inhibition of XBP1 can attenuate myotube atrophy in response to treatment with LLC-CM or C26-CM. To address this issue, we first generated an adenoviral construct expressing either a scrambled (control) short hairpin RNA (shRNA) or XBP1 shRNA along with GFP. C2C12 myotubes were transduced with control shRNA- or XBP1 shRNA-expressing adenoviral vector. After 24 h, the myotubes were treated with LLC-CM or C26-CM for an additional 48 h. The cultures were then fixed and stained with DAPI (Fig. 7A). As expected, treatment with LLC-CM or C26-CM led to a significant reduction in the average myotube diameter (Fig. 7A to C). Interestingly, shRNA-mediated knockdown of XBP1 rescued the loss of myotube diameter after treatment with LLC-CM or C26-CM (Fig. 7A to C). Our QRT-PCR analysis also showed that mRNA levels of MuRF1 but not MAFbx were significantly reduced in Ad.XBP1 shRNA-expressing cultures compared with control cultures incubated with LLC-CM (Fig. 7D and E). Finally, QRT-PCR analysis also confirmed a drastic reduction in mRNA levels of XBP1 in C2C12 myotube transduced with Ad.shXBP1 (Fig. 7F).
FIG 7.
Knockdown of XBP1 inhibits atrophy of myotubes in response to tumor-derived factors. C2C12 myotubes were transduced with adenoviral vector expressing a scrambled shRNA (Ad.Control shRNA) or XBP1 shRNA (Ad.XBP1 shRNA) for 24 h. The cells were washed and incubated with or without LLC-CM or C26-CM at a 1:4 ratio for an additional 48 h. (A) Representative images of myotube cultures after staining with DAPI. (B and C) Quantification of average myotube diameters in control and XBP1 knocked-down myotube cultures incubated with normal medium or (B) LLC-CM or (C) C26-CM. (D to F) Relative mRNA levels of MAFbx (D), MuRF1 (E), and sXBP1 (F) in myotube cultures transduced with Ad.Control shRNA or Ad.XBP1 shRNA incubated with or without LLC-CM. n = 4 in each group. C2C12 myotubes were transduced with Ad.Control or Ad.XBP1 shRNA for 24 h followed by treatment with normal medium or LLC-CM for 3 h. (G) Immunoblots demonstrating the levels of phosphorylated and total p65, IκBα, and p38 protein and total LC3B-II/I and CHOP protein in control of XBP1 knocked-down cultures. (H) Results of densitometry quantification of band intensities. Data presented here show ratios of p-p65/p65, p-IκBα/IκBα, p-p38/p38, and LC3B-II/I and total levels of CHOP in control and XBP1 knocked-down myotubes incubated with normal medium or LLC-CM. Medians and 25th to 75th percentiles are shown. *, P < 0.05 (values significantly different from those determined for control cultures transduced with Ad.Control shRNA); &, P < 0.05 (values significantly different from those determined for LLC-CM-treated cultures transduced with Ad.Control shRNA); #, P < 0.05 (values significantly different from those determined for control cultures transduced with Ad.XBP1); ns, not significant.
We also investigated whether the knockdown of XBP1 has any effect on the activation of catabolic signaling pathways and markers of autophagy and ER stress in myotubes in response to LLC-CM. C2C12 myotubes were transduced with control or XBP1 shRNA-expressing adenoviral vectors for 24 h. The cultures were then treated with normal differentiation medium (control) or LLC-CM at a 1:4 ratio for 3 h followed by performing Western blot analysis. Results showed that treatment with LLC-CM increased the phosphorylation of p65, IκBα, and p38 protein in C2C12 myotubes transduced with Ad.Control shRNA. However, the LLC-CM-induced increase in phosphorylation of p65, IκBα, and p38 protein was significantly reduced in cultures transduced with Ad.XBP1 shRNA compared to those transduced with Ad.Control shRNA (Fig. 7G and H). Our Western blot analysis also showed that the ratio of LC3B-II/I and levels of CHOP protein were significantly reduced in XBP1 shRNA-expressing myotubes compared to controls in response to treatment with LLC-CM, suggesting that inhibition of knockdown of XBP1 inhibits autophagy and ER stress in response to tumor-derived factors (Fig. 7G and H). Taken together, these results indicate that while forced expression of XBP1 promotes atrophy, its knockdown preserves myotube size in response to treatment with LLC-CM and C26-CM.
DISCUSSION
Cancer cachexia is a complex syndrome that involves a severe loss of skeletal muscle mass (1). Tumor cells secrete a number of factors with significant impacts on muscle proteolysis and function (1, 6). The tumor itself or tumor-derived factors can also stimulate the production of certain molecules, such as proinflammatory cytokines, from nontumor cells, leading to systematic inflammation and metabolic dysfunction in various organs, including skeletal muscle (6, 29, 42). TLRs, which mediate downstream signaling through the recruitment of MyD88 protein, are important regulators of the inflammatory response under many pathological conditions. A number of molecules can bind to specific TLRs to initiate inflammatory response (11–13). Recently, it was reported that several types of cancer cells which cause cachexia constitutively secrete Hsp70 and Hsp90 proteins in circulation. These proteins are packed in extracellular vesicles and delivered to skeletal muscle, where they bind to TLR4 to activate catabolic signaling pathways and production of inflammatory cytokines (8). A recent study has also shown that skeletal muscle wasting is partially rescued in whole-body TLR4-KO mice in response to growth of LLC tumors (9). However, how gene expression of other TLRs as well as MyD88 is regulated and their potential role and mechanisms of action in cancer-associated skeletal muscle wasting remained largely unknown.
Our present study demonstrated that the levels of gene expression of several TLRs and MyD88 are considerably increased in skeletal muscle of mice after inoculation with LLC cells. Moreover, we found that conditioned media from LLC or C26 colon adenocarcinoma cells (i.e., LLC-CM or C26-CM, respectively) induced gene expression of several TLRs and MyD88 in cultured myotubes (Fig. 1), suggesting that factors of tumor origin induce the gene expression of TLRs and MyD88 in skeletal muscle during cancer cachexia. More importantly, our results showed that inhibition of TLR-mediated signaling through targeted ablation of MyD88 blunts the LLC tumor-induced loss of skeletal muscle strength and myofiber CSA in adult mice (Fig. 2). These results are consistent with a recently published report that also suggests that inhibition of MyD88 signaling attenuates pancreatic ductal adenocarcinoma-associated muscle wasting in mice (43). Our results further suggest that the inhibition of TLR/MyD88 signaling may prevent muscle wasting through the inhibition of the UPS and autophagy. It has been consistently observed that levels of gene expression of a number of components of the UPS and autophagy are increased through upstream activation of NF-κB and MAPK signaling pathways (36, 40). Consistent with reduced expression of markers of the UPS and autophagy, we found that the targeted ablation of MyD88 reduced the activation of NF-κB and p38 MAPK in skeletal muscle of LLC tumor-bearing mice (Fig. 3F and G).
It was previously reported that whole-body deletion of MyD88, but not skeletal-muscle-specific deletion, inhibits skeletal muscle atrophy after stimulation with lipopolysaccharide (LPS), an agonist of TLR4 (44). However, we found that muscle-specific inhibition of MyD88 effectively inhibited muscle wasting in LLC tumor-associated cachexia. While the exact reasons remain unknown, it is possible that LPS and tumor-derived factors induce muscle wasting through recruitment of distinct TLRs. Indeed, our results demonstrated that, in addition to TLR4, mRNA levels of several other TLRs were increased in the LLC model of cachexia (Fig. 1). TLR7 has been previously reported to mediate the loss of muscle mass during cancer cachexia (45, 46). Our results demonstrate that gene expression of TLR7 was also significantly increased in skeletal muscle in the LLC model of cancer cachexia and cultured myotubes treated with LLC-CM or C26-CM (Fig. 1), which may mediate muscle wasting through downstream recruitment of MyD88 protein.
Accumulating evidence suggests that the ER stress and UPR pathways are activated in skeletal muscle under diverse conditions (25, 26). We previously reported that the markers of ER stress were activated in skeletal muscle in two models of cancer cachexia: LLC mice and ApcMin/+ mice. We have also previously shown that the conditioned medium of LLC cells was able to induce the activation of the UPR pathways (27). Hsp70 protein is secreted by multiple tumor cells that are known to cause cachexia (8). Our results in the present investigation suggest that Hsp70 also activates markers of ER stress/UPR in cultured myotubes (see Fig. S3 in the supplemental material), suggesting that factors of tumor origin, such as Hsp70, are responsible for the activation of the UPR in skeletal muscle. Intriguingly, a previous study demonstrated that TLRs recruit IRE1α to stimulate the splicing and activation of XBP1 in macrophages to boost the innate immune response (24). Moreover, the TLR-mediated activation of IRE1α/XBP1 occurs in the absence of ER stress and does not lead to the induction of gene expression of ER chaperones. Instead, the activation of XBP1 leads to the induction of proinflammatory cytokines, including IL-6 and tumor necrosis alpha (TNF-α) in macrophages (24). Our results suggest that the TLR/MyD88 axis not only mediates the activation of IRE1α/XBP1 but also stimulates the activation of the PERK arm of the UPR in skeletal muscle in LLC tumor-bearing mice (Fig. 4A and B). However, the exact molecular interactions through which the TLR/MyD88 axis leads to the activation of the UPR in skeletal muscle remain to be investigated.
The role of ER stress and individual arms of the UPR in the regulation of skeletal muscle mass has just begun to be elucidated. We have recently demonstrated that pan-inhibition of ER stress produces deleterious effects on muscle mass and function during LLC-induced cancer cachexia both in vivo and in vitro (27). Recently, it was reported the inducible activation of PERK leads to the loss of skeletal muscle mass in adult mice (47, 48). While the PERK/eIF2α axis is activated in skeletal muscle in models of cancer cachexia, we found that targeted ablation of PERK does not have any significant effect on LLC tumor-induced muscle wasting in adult mice (37). These findings highlight the idea that different components of the UPR may have different roles in the regulation of skeletal muscle mass under catabolic conditions.
Activation of the IRE1α/XBP1 pathway causes insulin resistance through the activation of JNK in mice fed with a high-fat diet (25, 26). However, the role of the IRE1α/XBP1 pathway in skeletal muscle homeostasis remained unknown. Our results suggest that XBP1 may have some role in the regulation of skeletal muscle mass, evidenced by reduced myofiber size in XBP1mKO mice compared to littermate control mice, under naive conditions (Fig. 5B to F). However, the ablation of XBP1 does not induce an atrophy program in skeletal muscle. We did not find any significant differences in the levels of expression of the markers of UPS, autophagy, and IL-6 in skeletal muscle of control XBP1f/f and XBP1mKO mice (Fig. 5I to K). However, deficiency of XBP1 may affect muscle development programs, because similar differences in myofiber CSA were also notable in TA and soleus muscle of littermate 2-week-old XBP1f/f and XBP1mKO mice (data not shown). A previous study also demonstrated that XBP1 promotes fusion of cultured myoblasts (38), further suggesting that XBP1 may have a role in postnatal skeletal muscle growth. Interestingly, we found that myofiber-specific ablation of XBP1 considerably inhibited LLC tumor-induced skeletal muscle wasting in mice (Fig. 5B to G). Similarly, we found that shRNA-mediated knockdown of XBP1 inhibited atrophy in cultured myotubes in response to LLC-CM or C26-CM (Fig. 7A to C). In contrast, the forced expression of sXBP1 causes atrophy in cultured myotubes (Fig. 6A and B), suggesting that increased activation of XBP1 is sufficient to induce muscle wasting.
Our results also demonstrate that, similarly to innate immunity activity, XBP1 induced gene expression of proinflammatory cytokine IL-6, TNF-α, and TWEAK (Fig. 5K; see also Fig. 6E), which may be an important mechanism for skeletal muscle atrophy. Indeed, IL-6 is one of the important mediators of muscle wasting in cancer-associated cachexia (49–52). Increased expression of inflammatory cytokines can further stimulate muscle wasting through the activation of catabolic signaling pathways and proteolytic systems (6, 35, 36, 40). Consistently, we found that the forced activation of sXBP1 increased the activation of NF-κB and p38 MAPK (Fig. 6F and G) and that knockdown of XBP1 inhibited LLC-CM-induced activation of NF-κB and p38 MAPK (Fig. 7G and H) in cultured myotubes.
Our results also show that XBP1 regulated the gene expression of several components of UPS and autophagy in skeletal muscle. Given that it is a transcription factor, it is possible that sXBP1 directly binds to the promoter region of these genes to induce their expression. Alternatively, the increased expression of these molecules could have been a result of the activation of other catabolic pathways, including the NF-κB and p38 MAPK pathways. While our results demonstrate that ablation of XBP1 inhibited markers of autophagy in the skeletal muscle LLC model of cancer cachexia (Fig. 5J), a published report suggested that the genetic deletion of XBP1 ameliorates pathology of SOD1 mutant mice, a model of amyotrophic lateral sclerosis (ALS). Interestingly, it was reported that XBP1 deletion results in an increased activation of autophagy, which hastens the clearance of mutant SOD1 aggregates (53). Collectively, these findings further emphasize that, depending on the catabolic stimuli or pathological condition, inhibition of a specific arm of the UPR can have distinct effects on the activation of downstream pathways.
While targeted ablation of MyD88 or XBP1 significantly attenuated LLC tumor-induced muscle wasting in adult mice, it is noteworthy that the effect was more pronounced in MyD88myoKO mice. This could be attributable to the fact that, being an upstream adaptor protein, MyD88 directly mediates the activation of multiple signaling pathways upon stimulation of not only TLRs but also IL-1 receptor. Therefore, in addition to XBP1, the inhibition of MyD88 would result in the suppression of other catabolic signaling pathways, such as NF-κB and p38 MAPK in skeletal muscle. Indeed, our results demonstrate that LLC tumor-induced phosphorylation of p65, IκBα, and p38 protein was blunted in skeletal muscle of MyD88myoKO mice (Fig. 3F and G). In contrast, being a nuclear transcription factor, XBP1 might regulate the activation of catabolic pathways through indirect mechanisms such as augmentation of the gene expression of proinflammatory cytokines.
In summary, our present study identified the TLR/MyD88/XBP1 axis as an important mediator of skeletal muscle wasting in the LLC and, potentially, C26 models of cancer cachexia. However, it remains to be determined whether this pathway promotes muscle wasting under other conditions as well. Targeting components of this pathway may be an important approach to inhibit muscle wasting in various catabolic states, including cancer.
MATERIALS AND METHODS
Mice.
Floxed MyD88 [MyD88f/f; Jax strain: B6.129P2 (SJL)-MyD88tm1Defr/J] mice were purchased from Jackson Laboratory. Floxed XBP1 (XBP1f/f) mice prepared as previously described (18) were kindly provided by Laurie Glimcher. Myoblast-specific MyD88-knockout (MyD88myoKO) mice were generated by crossing MyD88f/f mice with MyoD-Cre (Jax strain: FVB.Cg-Myod1tm2.1(icre)Glh/J) mice. Skeletal-muscle-specific XBP1-knockout (XBP1mKO) were generated by crossing XBP1f/f mice with muscle creatine kinase (MCK)-Cre mice [Jax strain: B6.FVB(129S4)-Tg (Ckmm-cre)5Khn/J]. All of the mice were in the C57BL/6J background, and their genotype was determined by PCR from tail DNA. For cancer cachexia studies, LLC cells (2 × 106 cells in 100 μl saline) were injected subcutaneously into the left flank of 3-month-old mice using a method similar to one previously described (27, 32). Control mice were injected with saline solution only. The mice were weighed daily and euthanized on day 21 after injection of LLC cells or when the animal reached a predetermined endpoint. All experimental protocols with mice were approved in advance by the Institutional Animal Care and Use Committee (IACUC) and Institutional Biosafety Committee (IBC) at the University of Louisville.
Grip strength test.
We used a digital grip strength meter (Columbus Instruments, Columbus, OH) to measure forelimb or total four-paw grip strength of mice by following a previously described protocol (54). Mice were acclimatized for 5 min before the grip strength test began. The mouse was allowed to grab the metal pull bar with the forepaws and, in a separate experiment, with all 4 paws. The tail of the mouse was then gently pulled backward in the horizontal plane until it could no long grasp the bar. The force at the time of release was recorded as the peak tension. Each mouse was tested 10 times with a 20-s to 40-s break between tests. The average peak tension from the 10 repetitions was normalized against total body weight and was defined as the average grip strength. The maximum peak tension from the 10 repetitions was normalized against total body weight and was defined as the maximal grip strength.
Histology and morphometric analysis.
Individual TA and soleus muscles were isolated from mice, snap-frozen in liquid nitrogen, and sectioned with a microtome cryostat. For the assessment of muscle morphology, 10-μm-thick transverse sections of TA and soleus muscle were stained with hematoxylin and eosin (H&E) dye. Muscle sections were also processed for immunostaining for laminin protein to mark the boundaries of myofibers. The sections were examined under an Eclipse TE 2000-U microscope (Nikon, Tokyo, Japan). Average myofiber cross-sectional area (CSA) and minimal Feret’s diameter were analyzed in antilaminin-stained muscle sections using ImageJ software (NIH, Bethesda, MD). For each muscle, CSA was calculated by analyzing approximately 75 fibers per image (×20 magnification). Five images were randomly taken for each individual muscle, corresponding to 375 fibers measured in each muscle per mouse.
Immunohistochemistry.
For immunohistochemistry studies, frozen TA or soleus muscle sections were fixed in acetone, blocked in 2% bovine serum albumin–phosphate-buffered saline (PBS) for 1 h, and incubated with rabbit antilaminin in blocking solution at 4°C overnight under humidified conditions. The sections were washed briefly with PBS before incubation with goat anti-rabbit Alexa Fluor 468-conjugated secondary antibody for 1 h at room temperature and were then washed three times for 15 min each time with PBS. Finally, the fluorescence was captured with an Eclipse TE 2000-U microscope (Nikon). Averages of data representing CSA and minimal Feret’s diameter of myofibers in each muscle were determined using ImageJ software (NIH).
Cell culture.
C2C12, a myoblastic cell line, was purchased from the American Type Culture Collection (ATCC). These cells were grown in growth medium (GM; Dulbecco’s modified Eagle’s medium [DMEM] containing 10% FBS). To induce differentiation, the cells were incubated in differentiation medium (DM; DMEM supplemented with 2% horse serum) for 96 h. After appropriate treatments, cultures of myotubes were visualized at room temperature on an Eclipse TE 2000-U microscope equipped with a Digital Sight DS-Fi1 camera (Nikon). Images were captured, and the diameter of the myotubes was measured with ImageJ software (NIH). The myotube diameter was quantified as follows. Four fields were chosen randomly within each well, and 4 wells were measured. The average diameter per myotube was calculated as the mean of the 3 measurements taken along the length of the myotube. Myotubes were then collected, frozen, and stored at −80°C for further experiments.
Preparation of tumor cell conditioned medium.
LLC or C26 cells were seeded in 100-mm-diameter cell culture plates in growth medium (DMEM containing 10% fetal bovine serum) at a density of 5,000 cells/cm2. Additional growth medium was added to each plate after 48 h of plating. LLC cell cultures contain a heterogeneous mix of adherent and floating cells. After 4 days, floating cells were harvested by centrifugation at 800 rpm for 5 min. Pelleted cells and 10 ml differentiation medium were added back to the plate containing the adherent cells. After 24 h, conditioned medium (CM) was harvested and cleared of cells and debris by centrifugation (800 rpm, 5 min). The CM was passed through 0.45-μm-pore-size filters, and aliquots were frozen in liquid nitrogen for later use. The CM was diluted 1:4 with fresh differentiation medium (DM) for treatment of cultured myotubes and subsequent analysis of LLC-CM-induced or C26-CM-induced myotube atrophy.
Generation and use of adenoviral vectors.
Adenoviral vectors expressing XBP1 shRNA or XBP1 cDNA were generated following a protocol as previously described (55, 56). The target siRNA sequences for mouse XBP1 mRNA were identified using BLOCK-iT RNAi Designer online software (Life Technologies). The shRNA oligonucleotides were synthesized to contain the sense strand of target sequences for mouse XBP1 shRNA (GCCAAGCTGGAAGCCATTAAT), a short spacer (CTCGAG), and the reverse complement sequences followed by five thymidines as an RNA polymerase III transcriptional stop signal. Oligonucleotides were annealed and cloned into pLKO.1-Puro plasmid with AgeI/EcoRI sites. The insertion of shRNA sequence in the plasmid was confirmed by DNA sequencing. Adenovirus carrying XBP1 shRNA was generated (AdEasy adenoviral vector system; Agilent) following the manufacturer’s protocol. In brief, Xbp1 shRNA was PCR amplified from pLKO.1 plasmid and ligated into pAdTrack-CMV vector digested at KpnI and XbaI sites. For the generation of adenovirus expressing sXBP1, Flag-tagged sXBP1 cDNA was isolated from pCMV5-Flag-XBP1s plasmid (Addgene; plasmid catalog no. 63680) and ligated at KpnI and HindIII sites in pAdTrack-CMV plasmid. The resulting pAdTrack-CMV-XBP1 shRNA or pAdTrack-CMV-sXBP1 plasmid was linearized with PmeI and cotransformed into E. coli BJ5183 cells with the pAdEasy-1 plasmid. Clones undergoing Adtrack-Adeasy recombination were selected with kanamycin and confirmed by digestion with restriction endonuclease. The recombinant plasmid was linearized with PacI and transfected into the 293T cell line (ATCC) using Effectene transfection reagent (Qiagen) for packaging into active virus particles. Viruses were amplified by serial passage to concentrate viral titers. Titers were monitored under a microscope by visualizing the GFP marker coexpressed with XBP1 shRNA or sXBP1 cDNA in the Adtrack-Adeasy recombinants. Adenoviral vectors were transduced in cultured myotubes at multiplicity of infection (MOI) of 1:50.
Quantitative real-time PCR (QRT-PCR).
RNA isolation and QRT-PCR were performed following a previously described protocol (28, 32). In brief, total RNA was extracted from skeletal muscles of mice or cultured C2C12 myotubes using TRIzol reagent (Thermo Fisher Scientific Life Sciences) and an RNeasy minikit (Qiagen, Valencia, CA). First-strand cDNA was made with a commercially available kit (Thermo Fisher Scientific Life Sciences). Quantification of mRNA expression was performed using the SYBR green dye (Thermo Fisher Scientific Life Sciences) method on a sequence detection system (model 7300; Thermo Fisher Scientific Life Sciences). Primers were designed with Vector NTI software (Thermo Fisher Scientific Life Sciences). Primer sequences are available upon request.
Normalization of data was accomplished using the endogenous control β-actin, and the normalized values were subjected to a threshold cycle (2−ΔΔCT) formula to calculate the fold change between the control and experimental groups.
Western blotting.
Relative levels of various proteins were determined by performing Western blot analysis. Skeletal muscle of mice or cultured C2C12 myotubes were washed with sterile PBS and homogenized in lysis buffer consisting of the following: 50 mM Tris-Cl (pH 8.0), 200 mM NaCl, 50 mM NaF, 1 mM dithiothreitol (DTT), 1 mM sodium orthovanadate, 0.3% IGEPAL, and protease inhibitors. Approximately 100 μg protein was resolved in each lane on 10% to 12% SDS-polyacrylamide gels, electrotransferred onto nitrocellulose membranes, and probed with the following antibodies: anti-troponin T (Sigma-Aldrich, St. Louis, MO) (1:500), antitropomyosin (Sigma-Aldrich) (1:500), anti-sXBP-1 (Cell Signaling Technology) (1:1,000), anti-LC3B (Cell Signaling Technology) (1:500), anti-α-tubulin (Cell Signaling Technology) (1:1,000), and anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) (Cell Signaling Technology) (1:2,000). Bound antibodies were detected by secondary antibodies conjugated to horseradish peroxidase (Cell Signaling Technology). Signal detection was performed by the use of an enhanced chemiluminescence detection reagent (Bio-Rad). Approximate molecular masses were determined by comparison with the migration of prestained protein standards (Bio-Rad). Quantitative estimation of the intensity of each band was performed using ImageJ software (NIH).
Statistical analysis.
Data are presented in the form of box and whisker plots that include individual data points. Median values, 25th and 75th percentiles, and 10th and 90th percentiles (center line, box, and whiskers, respectively) are displayed. Graphs were plotted using Microsoft Office Excel 2010 software. One-way analysis of variance (ANOVA) was performed to analyze the data followed by Bonferroni’s multiple-comparison test (for comparisons of control groups versus all the treated groups). Statistical significance of differences in results of comparisons was set at a P value of <0.05 unless mentioned otherwise.
Data availability.
All relevant data related to the manuscript are available from us.
Supplementary Material
ACKNOWLEDGMENTS
We thank Laurie Glimcher of Cornell University for providing floxed XBP1 mice.
This work was supported by funding from National Institutes of Health grants AR059810, AR068313, and AG029623 to AK.
A.K. designed the work. K.R.B. and A.K. wrote the manuscript, and all of us edited the manuscript. K.R.B., P.G., A.R., A.K.S., G.X., and Y.S.G. performed all the experiments.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00184-19.
REFERENCES
- 1.Argiles JM, Busquets S, Stemmler B, Lopez-Soriano FJ. 2014. Cancer cachexia: understanding the molecular basis. Nat Rev Cancer 14:754–762. doi: 10.1038/nrc3829. [DOI] [PubMed] [Google Scholar]
- 2.Johns N, Stephens NA, Fearon KC. 2013. Muscle wasting in cancer. Int J Biochem Cell Biol 45:2215–2229. doi: 10.1016/j.biocel.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 3.Sandri M. 2016. Protein breakdown in cancer cachexia. Semin Cell Dev Biol 54:11–19. doi: 10.1016/j.semcdb.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Penna F, Costamagna D, Pin F, Camperi A, Fanzani A, Chiarpotto EM, Cavallini G, Bonelli G, Baccino FM, Costelli P. 2013. Autophagic degradation contributes to muscle wasting in cancer cachexia. Am J Pathol 182:1367–1378. doi: 10.1016/j.ajpath.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 5.Smith KL, Tisdale MJ. 1993. Mechanism of muscle protein degradation in cancer cachexia. Br J Cancer 68:314–318. doi: 10.1038/bjc.1993.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fearon KC, Glass DJ, Guttridge DC. 2012. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab 16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Baracos VE, Martin L, Korc M, Guttridge DC, Fearon K. 2018. Cancer-associated cachexia. Nat Rev Dis Primers 4:17105. doi: 10.1038/nrdp.2017.105. [DOI] [PubMed] [Google Scholar]
- 8.Zhang G, Liu Z, Ding H, Zhou Y, Doan HA, Sin KWT, Zhu ZJ, Flores R, Wen Y, Gong X, Liu Q, Li YP. 2017. Tumor induces muscle wasting in mice through releasing extracellular Hsp70 and Hsp90. Nat Commun 8:589. doi: 10.1038/s41467-017-00726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Liu Z, Ding H, Miao H, Garcia JM, Li YP. 2017. Toll-like receptor 4 mediates Lewis lung carcinoma-induced muscle wasting via coordinate activation of protein degradation pathways. Sci Rep 7:2273. doi: 10.1038/s41598-017-02347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle A, Zhang G, Abdel Fattah EA, Eissa NT, Li YP. 2011. Toll-like receptor 4 mediates lipopolysaccharide-induced muscle catabolism via coordinate activation of ubiquitin-proteasome and autophagy-lysosome pathways. FASEB J 25:99–110. doi: 10.1096/fj.10-164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Akira S, Takeda K. 2004. Toll-like receptor signalling. Nat Rev Immunol 4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 13.Janeway CA Jr, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 14.Hindi SM, Shin J, Gallot YS, Straughn AR, Simionescu-Bankston A, Hindi L, Xiong G, Friedland RP, Kumar A. 2017. MyD88 promotes myoblast fusion in a cell-autonomous manner. Nat Commun 8:1624. doi: 10.1038/s41467-017-01866-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hindi SM, Kumar A. 2016. Toll-like receptor signalling in regenerative myogenesis: friend and foe. J Pathol 239:125–128. doi: 10.1002/path.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Kaufman RJ. 2014. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer 14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Kaufman RJ. 2006. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ 13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 18.Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 19.Harding HP, Zhang Y, Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 20.Ma Y, Brewer JW, Diehl JA, Hendershot LM. 2002. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol 318:1351–1365. doi: 10.1016/S0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 21.Flamment M, Hajduch E, Ferre P, Foufelle F. 2012. New insights into ER stress-induced insulin resistance. Trends Endocrinol Metab 23:381–390. doi: 10.1016/j.tem.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Tirasophon W, Welihinda AA, Kaufman RJ. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinon F, Chen X, Lee AH, Glimcher LH. 2010. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol 11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afroze D, Kumar A. 2019. ER stress in skeletal muscle remodeling and myopathies. FEBS J 286:379–398. doi: 10.1111/febs.14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohnert KR, McMillan JD, Kumar A. 2018. Emerging roles of ER stress and unfolded protein response pathways in skeletal muscle health and disease. J Cell Physiol 233:67–78. doi: 10.1002/jcp.25852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohnert KR, Gallot YS, Sato S, Xiong G, Hindi SM, Kumar A. 2016. Inhibition of ER stress and unfolding protein response pathways causes skeletal muscle wasting during cancer cachexia. FASEB J 30:3053–3068. doi: 10.1096/fj.201600250RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul PK, Bhatnagar S, Mishra V, Srivastava S, Darnay BG, Choi Y, Kumar A. 2012. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol 32:1248–1259. doi: 10.1128/MCB.06351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seto DN, Kandarian SC, Jackman RW. 2015. A key role for leukemia inhibitory factor in C26 cancer cachexia. J Biol Chem 290:19976–19986. doi: 10.1074/jbc.M115.638411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acharyya S, Ladner KJ, Nelsen LL, Damrauer J, Reiser PJ, Swoap S, Guttridge DC. 2004. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J Clin Invest 114:370–378. doi: 10.1172/JCI20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 32.Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, Kumar A. 2010. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol 191:1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandri M. 2013. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol 45:2121–2129. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandri M. 2008. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 35.Egerman MA, Glass DJ. 2014. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol 49:59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Malhotra S, Kumar A. 2008. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J Mol Med (Berl) 86:1113–1126. doi: 10.1007/s00109-008-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallot YS, Bohnert KR, Straughn AR, Xiong G, Hindi SM, Kumar A. 2019. PERK regulates skeletal muscle mass and contractile function in adult mice. FASEB J 33:1946–1962. doi: 10.1096/fj.201800683RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Hetz C, Chevet E, Oakes SA. 2015. Proteostasis control by the unfolded protein response. Nat Cell Biol 17:829–838. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonaldo P, Sandri M. 2013. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech 6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spate U, Schulze PC. 2004. Proinflammatory cytokines and skeletal muscle. Curr Opin Clin Nutr Metab Care 7:265–269. doi: 10.1097/00075197-200405000-00005. [DOI] [PubMed] [Google Scholar]
- 42.Judge SM, Wu CL, Beharry AW, Roberts BM, Ferreira LF, Kandarian SC, Judge AR. 2014. Genome-wide identification of FoxO-dependent gene networks in skeletal muscle during C26 cancer cachexia. BMC Cancer 14:997. doi: 10.1186/1471-2407-14-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X, Burfeind KG, Michaelis KA, Braun TP, Olson B, Pelz KR, Morgan TK, Marks DL. 2019. MyD88 signalling is critical in the development of pancreatic cancer cachexia. J Cachexia Sarcopenia Muscle 10:378–390. doi: 10.1002/jcsm.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun TP, Grossberg AJ, Krasnow SM, Levasseur PR, Szumowski M, Zhu XX, Maxson JE, Knoll JG, Barnes AP, Marks DL. 2013. Cancer- and endotoxin-induced cachexia require intact glucocorticoid signaling in skeletal muscle. FASEB J 27:3572–3582. doi: 10.1096/fj.13-230375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He WA, Calore F, Londhe P, Canella A, Guttridge DC, Croce CM. 2014. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci U S A 111:4525–4529. doi: 10.1073/pnas.1402714111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calore F, Londhe P, Fadda P, Nigita G, Casadei L, Marceca GP, Fassan M, Lovat F, Gasparini P, Rizzotto L, Zanesi N, Jackson D, Mehta S, Nana-Sinkam P, Sampath D, Pollock RE, Guttridge DC, Croce CM. 2018. The TLR7/8/9 antagonist IMO-8503 inhibits cancer-induced cachexia. Cancer Res 78:6680–6690. doi: 10.1158/0008-5472.CAN-17-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyake M, Nomura A, Ogura A, Takehana K, Kitahara Y, Takahara K, Tsugawa K, Miyamoto C, Miura N, Sato R, Kurahashi K, Harding HP, Oyadomari M, Ron D, Oyadomari S. 2016. Skeletal muscle-specific eukaryotic translation initiation factor 2alpha phosphorylation controls amino acid metabolism and fibroblast growth factor 21-mediated non-cell-autonomous energy metabolism. FASEB J 30:798–812. doi: 10.1096/fj.15-275990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyake M, Kuroda M, Kiyonari H, Takehana K, Hisanaga S, Morimoto M, Zhang J, Oyadomari M, Sakaue H, Oyadomari S. 2017. Ligand-induced rapid skeletal muscle atrophy in HSA-Fv2E-PERK transgenic mice. PLoS One 12:e0179955. doi: 10.1371/journal.pone.0179955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonetto A, Aydogdu T, Jin X, Zhang Z, Zhan R, Puzis L, Koniaris LG, Zimmers TA. 2012. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am J Physiol Endocrinol Metab 303:E410–E421. doi: 10.1152/ajpendo.00039.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baltgalvis KA, Berger FG, Pena MM, Davis JM, Muga SJ, Carson JA. 2008. Interleukin-6 and cachexia in ApcMin/+ mice. Am J Physiol Regul Integr Comp Physiol 294:R393–R401. doi: 10.1152/ajpregu.00716.2007. [DOI] [PubMed] [Google Scholar]
- 51.Puppa MJ, White JP, Velazquez KT, Baltgalvis KA, Sato S, Baynes JW, Carson JA. 2012. The effect of exercise on IL-6-induced cachexia in the Apc (Min/+) mouse. J Cachexia Sarcopenia Muscle 3:117–137. doi: 10.1007/s13539-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White JP, Puppa MJ, Sato S, Gao S, Price RL, Baynes JW, Kostek MC, Matesic LE, Carson JA. 2012. IL-6 regulation on skeletal muscle mitochondrial remodeling during cancer cachexia in the ApcMin/+ mouse. Skelet Muscle 2:14. doi: 10.1186/2044-5040-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH. 2009. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev 23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato S, Ogura Y, Tajrishi MM, Kumar A. 2015. Elevated levels of TWEAK in skeletal muscle promote visceral obesity, insulin resistance, and metabolic dysfunction. FASEB J 29:988–1002. doi: 10.1096/fj.14-260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong G, Hindi SM, Mann AK, Gallot YS, Bohnert KR, Cavener DR, Whittemore SR, Kumar A. 23 March 2017, posting date The PERK arm of the unfolded protein response regulates satellite cell-mediated skeletal muscle regeneration. Elife doi: 10.7554/eLife.22871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhatnagar S, Kumar A, Makonchuk DY, Li H, Kumar A. 2010. Transforming growth factor-beta-activated kinase 1 is an essential regulator of myogenic differentiation. J Biol Chem 285:6401–6411. doi: 10.1074/jbc.M109.064063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data related to the manuscript are available from us.