Fanconi anemia (FA) is an inherited disease characterized by bone marrow failure and increased cancer risk. FA is caused by mutation of any 1 of 22 genes, and the FA proteins function cooperatively to repair DNA interstrand cross-links (ICLs). A central step in the activation of the FA pathway is the monoubiquitination of the FANCD2 and FANCI proteins, which occurs within chromatin. How FANCD2 and FANCI are anchored to chromatin remains unknown.
KEYWORDS: chromatin, DNA repair, Fanconi anemia, genome instability, ubiquitin
ABSTRACT
Fanconi anemia (FA) is an inherited disease characterized by bone marrow failure and increased cancer risk. FA is caused by mutation of any 1 of 22 genes, and the FA proteins function cooperatively to repair DNA interstrand cross-links (ICLs). A central step in the activation of the FA pathway is the monoubiquitination of the FANCD2 and FANCI proteins, which occurs within chromatin. How FANCD2 and FANCI are anchored to chromatin remains unknown. In this study, we identify and characterize a FANCD2 histone-binding domain (HBD) and embedded methyl-lysine-binding domain (MBD) and demonstrate binding specificity for H4K20me2. Disruption of the HBD/MBD compromises FANCD2 chromatin binding and nuclear focus formation and its ability to promote error-free DNA interstrand cross-link repair, leading to increased error-prone repair and genome instability. Our study functionally describes the first FA protein chromatin reader domain and establishes an important link between this human genetic disease and chromatin plasticity.
INTRODUCTION
Nucleosomes, the fundamental unit of chromatin, play a dynamic and instructive role in many cellular processes, including transcription, replication, and DNA repair. One mechanism by which nucleosomes instruct DNA repair is via the posttranslational modification (PTM) of histone tails. These modifications modulate the strength of noncovalent interactions between histones and DNA and serve as binding sites for chromatin-interacting proteins, also known as chromatin readers. Many important DNA repair proteins have been shown to harbor reader domains that are critical for their repair function, examples of which include the tandem Tudor domains of 53BP1 and the chromodomain of KAT5/TIP60 (1–3).
Fanconi anemia (FA) is a genetic disease characterized by congenital abnormalities, progressive pediatric bone marrow failure, and a heightened cancer risk in early adulthood (4). FA is caused by mutation of any 1 of 22 genes (5, 6). The FA proteins orchestrate the repair of DNA interstrand cross-links (ICLs), lesions that block the replication and transcription machineries and that can lead to structural and numerical chromosome aberrations if repaired erroneously (7, 8). A central step in the activation of the FA pathway is the site-specific monoubiquitination of the FANCD2 and FANCI proteins, which occurs within chromatin (9–13). FANCD2 has been shown to bind directly to DNA via two binding domains, and this activity stimulates FANCD2 monoubiquitination (14–17). However, the mechanism(s) by which FANCD2 is tethered to chromatin and whether FANCD2 displays specificity for particular histone PTMs are unknown. Indeed, no reader domains have been identified for any of the FA proteins to date. Intriguingly, recent studies have shown that FANCD2 possesses histone chaperone activity and that FANCD2 function can be modulated by changes in chromatin state (18–21).
In this study, we describe the identification and functional characterization of a FANCD2 histone-binding domain (HBD) and an embedded methyl-lysine (Kme)-binding domain (MBD). We establish that the FANCD2 MBD can bind to mono-, di-, and trimethylated H4K20 in vitro and exhibits cellular specificity for H4K20me2. Knockdown of KMT5A, the histone methyltransferase responsible for H4K20 monomethylation, which primes H4K20 for subsequent di- and trimethylation, results in decreased FANCD2 focus formation and increased sensitivity to the DNA interstrand cross-link (ICL)-inducing agent mitomycin C (MMC). Guided by sequence conservation and in silico modeling, we generated several HBD/MBD missense variants and stably transduced FA-D2 (FANCD2−/−) patient-derived cells. Our functional analyses reveal that disruption of the HBD/MBD decreases the affinity of FANCD2 for chromatin and impairs its ability to assemble into nuclear foci and to effectively promote ICL repair. Consequently, upon exposure to ICL-inducing agents, error-prone DNA repair pathways, including nonhomologous DNA end joining (NHEJ), are employed, resulting in increased cytotoxicity and chromosome structural aberrations. Our studies uncover a novel mechanism by which FANCD2 is anchored to chromatin and functionally link this important human genetic disease to chromatin plasticity.
RESULTS
FANCD2 has a histone-binding domain and embedded methyl-lysine-binding domain.
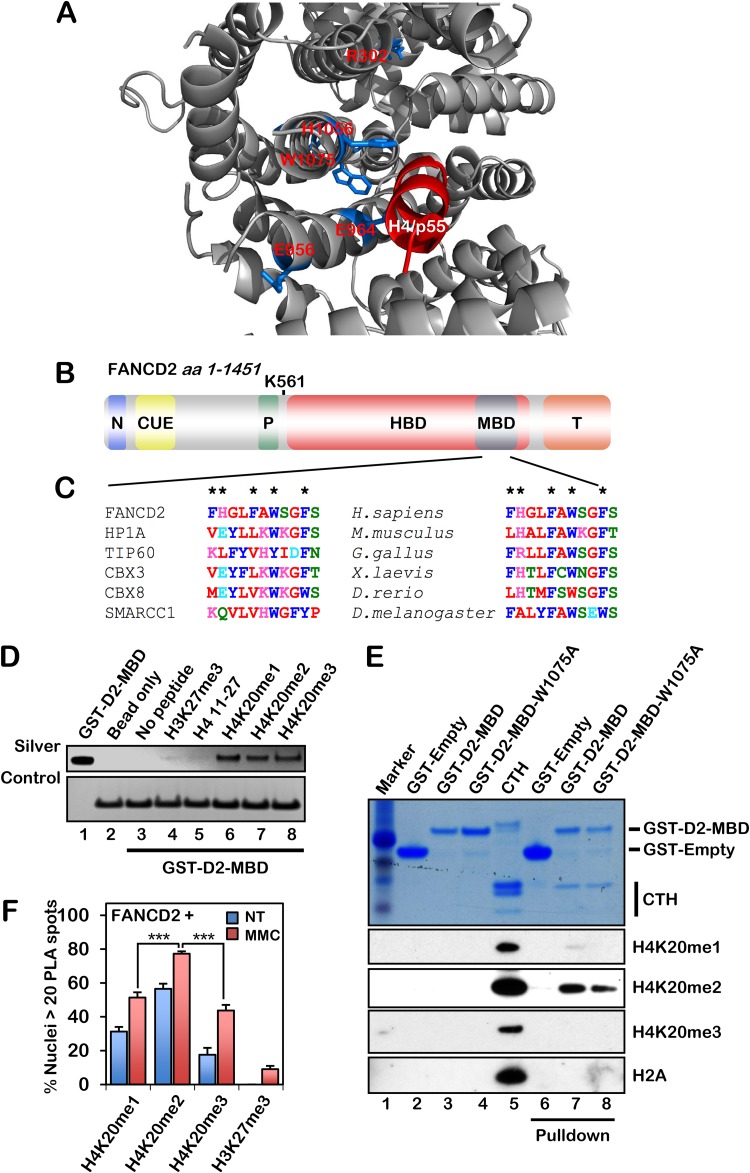
A BLASTp search using short fragments of human FANCD2 uncovered amino acid sequence homology between FANCD2 (amino acids [aa] 953 to 968) and the Drosophila melanogaster p55 protein (aa 358 to 373), a histone H4 binding protein and component of the NuRD, NuRF, and CAF1 nucleosome remodeling complexes (22–25). This region of FANCD2 is highly evolutionarily conserved among vertebrates. Using in silico molecular modeling, the histone H4 tail from the p55-H4 structure (PDB ID 3C9C) was docked into murine Fancd2 (PDB ID 3S4W) using the AutoDock Vina tool (26), illustrating favorable predicted binding energies between the H4 tail and the histone-binding domain (HBD) (Fig. 1A). Further examination of the FANCD2 HBD uncovered a highly conserved putative methyl-lysine (Kme)-binding domain (MBD) with sequence homology to the methyl-binding chromodomains of HP1α, TIP60, and CBX8 (Fig. 1B and C). A glutathione S-transferase (GST)-tagged FANCD2-HBD/MBD fragment (amino acids 604 to 1194) was purified and, in a histone peptide array screen, bound to an H4 17-mer that was either unmodified or harboring K20me1, K20me2, or K20me3. In an in vitro histone peptide pulldown assay, a GST-tagged FANCD2-MBD fragment (amino acids 1069 to 1142) bound to H4K20me1, H4K20me2, and H4K20me3 but not unmodified H4 or H3K27me3 (Fig. 1D). MBD-Kme binding involves the docking of Kme into an aromatic cage and the formation of cation-π interactions with delocalized electrons of aromatic residues (27). Highly conserved aromatic amino acids within the FANCD2 Kme binding cage include F1073, W1075, and F1078 (Fig. 1C). Using calf thymus histones, we observed preferential binding of the MBD to H4K20me2 and a modest reduction in the binding of a mutated MBD-W1075A fragment to H4K20me2 compared to the wild-type MBD (Fig. 1E). Using a proximity ligation assay (PLA), we observed the preferential binding of FANCD2 to H4K20me2 in cells and stimulation of H4K20me binding upon MMC exposure (Fig. 1F). Taken together, our results demonstrate that FANCD2 interacts directly with methylated H4 via its HBD/MBD and suggest that the chromatin recruitment of FANCD2 is mediated via an interaction between the HBD/MBD and H4K20me2, similar to that recently described for other important DNA repair proteins, e.g., 53BP1 and TIP60 (1–3, 28). Underscoring the clinical significance of the HBD/MBD, ∼69% of reported FA-D2 patient mutations and ∼46% of somatic FANCD2 mutations reported in the Memorial Sloan-Kettering Cancer Center cBioPortal for the analysis of TCGA integrated data sets map to this region (see Tables S1 and S2 in the supplemental material) (29–31).
FIG 1.
FANCD2 contains an HBD/MBD that exhibits specificity for H4K20me2. (A) AutoDock Vina molecular modeling was used to dock the H4 tail (taken from the p55-H4 crystal structure [PDB ID 3C9C]) into the histone-binding domain (HBD) of murine Fancd2 (PDB ID 3S4W), indicating favorable H bonding and electrostatic interactions between the H4 tail and the FANCD2 HBD. Residues R302, E956, E964, H1056, and W1075 are highlighted in marine blue. (B) Schematic of the known FANCD2 domains: N, nuclear localization signal (aa 1 to 58) (67); CUE, coupling of ubiquitin conjugation to endoplasmic reticulum degradation (aa 191 to 238) (58); P, PCNA-interaction motif (PIP box) (aa 527 to 534) (10); HBD, histone-binding domain (aa 604 to 1194); MBD, methyl-lysine-binding domain (aa 1069 to 1142); T, Tower domain (aa 1146 to 1451) (11). (C) Clustal Omega multiple-sequence alignment (MSA) of the FANCD2 MBD (aa 1069 to 1079) with known methyl-lysine-binding chromodomains (left) and with other FANCD2 orthologs (right). Predicted key aromatic residues are indicated by an asterisk. H. sapiens; Homo sapiens, M. musculus, Mus musculus; G. gallus, Gallus gallus; X. laevis, Xenopus laevis; D. rerio, Danio rerio; D. melanogaster, Drosophila melanogaster. (D) Streptavidin-Sepharose pulldown of biotinylated histone peptides incubated with purified GST-FANCD2-MBD. A streptavidin silver stain signal serves as a loading control. (E) Glutathione-agarose pulldown of GST-tagged wild-type FANCD2-MBD (GST-D2-MBD) and MBD-W1075A (GST-D2-MBD-W1075A) fragments incubated with purified histones from calf thymus (CTH). (F) Quantification of proximity ligation assay (PLA) results with FANCD2 and H4K20me1, H4K20me2, H4K20me3, and H3K27me3 in U2OS cells. Binding of 53BP1 to H4K20me was used as a positive control for our PLA. Nuclei with >20 PLA spots were considered positive. Experiments were performed three times with similar results. Error bars represent the standard errors of the means from three independent experiments. At least 300 nuclei were scored per biological replicate. ***, P < 0.001. NT, not treated.
The histone methyltransferase KMT5A is necessary for efficient activation of the FA pathway and ICL repair.
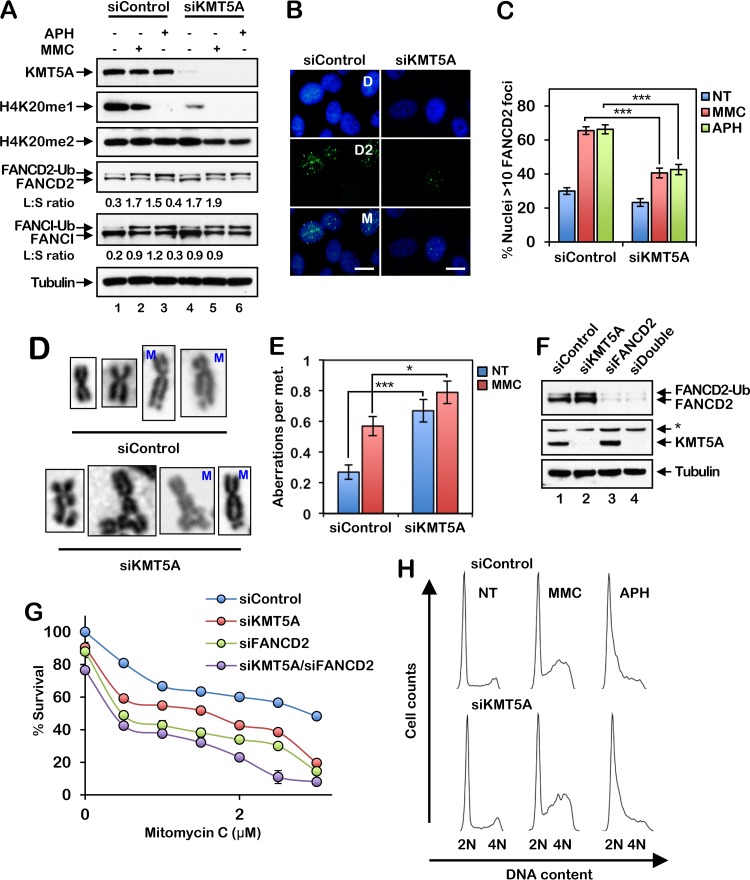
The KMT5A histone methyltransferase catalyzes the monomethylation of H4K20, a prerequisite for di- and trimethylation (32, 33). To determine if KMT5A plays a role in the regulation of the activation of the FA pathway, we depleted KMT5A in HeLa cells using small interfering RNA (siRNA) and examined FANCD2 monoubiquitination and nuclear focus formation. Depletion of KMT5A resulted in a reduction in the levels of H4K20me1 and a modest concomitant reduction in the levels of H4K20me2 (Fig. 2A). Intriguingly, treatment with DNA-damaging agents, in particular, the DNA polymerase inhibitor aphidicolin (APH), also led to a reduction in H4K20me1 levels, in the absence of a corresponding decrease in H4K20me2 (Fig. 2A). While depletion of KMT5A did not measurably impact spontaneous or DNA damage-inducible FANCD2 or FANCI monoubiquitination (Fig. 2A), KMT5A knockdown resulted in a significant decrease in FANCD2 nuclear focus formation following exposure to MMC and APH (Fig. 2B and C). In addition, similar to FA patient-derived cells, cells depleted of KMT5A exhibited increased levels of chromosome aberrations, including gaps, breaks, and radial formations, and decreased cell survival following ICL exposure (Fig. 2D to G). Combined depletion of FANCD2 and KMT5A resulted in a modest further increase in ICL sensitivity over that for cells with each single knockdown (Fig. 2G). However, unlike FA patient cells, under the experimental conditions described, we did not observe any appreciable differences in cell cycle profiles in the absence or presence of KMT5A following exposure to MMC or APH (Fig. 2H). Increased S-phase checkpoint activation has previously been observed in KMT5A-depleted cells (34). Very similar findings were observed for the nontransformed mammary epithelial line MCF10A (data not shown). Our results demonstrate an important role for the KMT5A histone methyltransferase in the activation of the FA pathway and indicate overlapping and unique functions for FANCD2 and KMT5A in the maintenance of genome stability.
FIG 2.
The histone methyltransferase KMT5A is necessary for efficient activation of the FA pathway and ICL repair. (A) HeLa cells were incubated with control nontargeting siRNA (siControl) or siRNA targeting KMT5A (siKMT5A) for 64 h. The cells were then treated with 200 nM mitomycin C (MMC) or 1 μM aphidicolin (APH) for 16 h, and whole-cell lysates were analyzed by immunoblotting. Ub, ubiquitin; L:S ratio, ratio of monoubiquitinated to nonubiquitinated FANCD2 or FANCI. (B) Representative images of FANCD2 foci in MMC-treated HeLa cells. Cells were treated as described in the legend to panel A. D, DAPI; D2; FANCD2; M, merge. Bars, 10 μm. (C) Quantification of FANCD2 focus formation for the assay whose results are presented in panel B. Nuclei with >10 FANCD2 foci were considered positive. At least 300 nuclei were scored per biological replicate. (D and E) HeLa cells were incubated with siControl or siKMT5A for 64 h. Cells were then treated with 10 nM MMC for 16 h, and metaphase (met.) chromosomes were analyzed for the presence of structural aberrations. Representative chromosome images are shown in panel D. M, mitomycin C. Fifty metaphases were scored per treatment. These experiments were performed twice with similar results. Error bars represent the standard errors of the means. *, P < 0.05; ***, P < 0.001. (F) HeLa cells were incubated with siControl, siKMT5A, siFANCD2, or siKMT5A plus siFANCD2 (siDouble) for 64 h, and whole-cell lysates were analyzed by immunoblotting. *, nonspecific band. (G) Cells were treated with increasing concentrations of MMC for 48 h, and percent survival was determined using an 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium survival assay. Error bars represent the standard errors of the means from four replicates. Experiments were performed twice with similar results. (H) HeLa cells were incubated with siControl or siKMT5A for 64 h and then treated with 200 nM MMC or 1 μM APH for 16 h. Cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. Cell cycle analysis was performed using FlowJo (v10.2) software.
The FANCD2 HBD/MBD is required for efficient chromatin binding and nuclear focus formation.
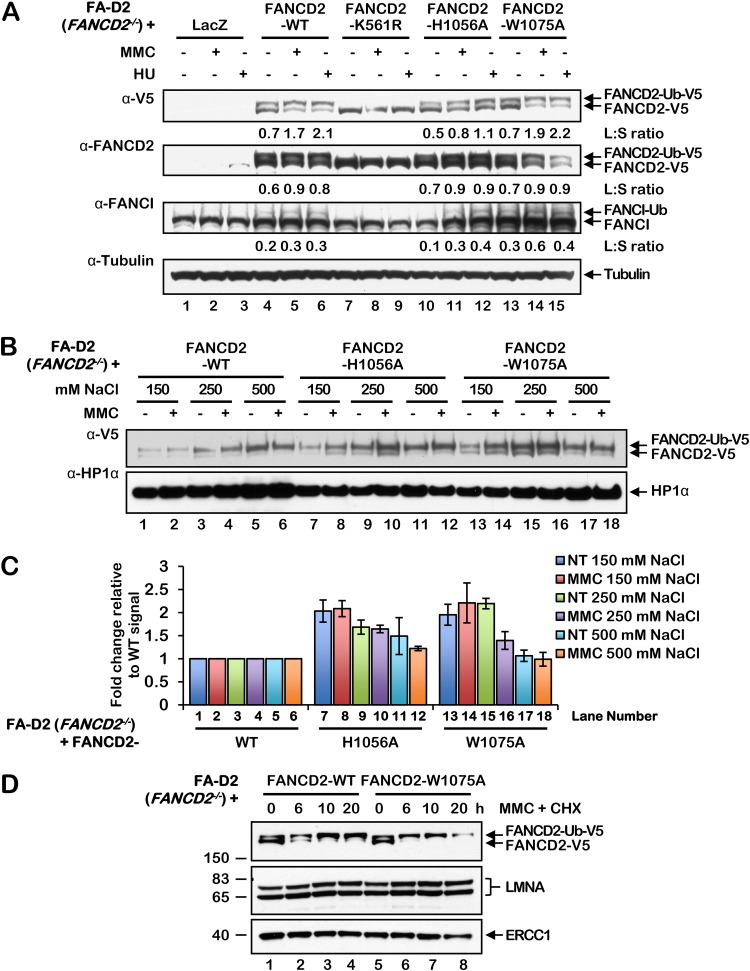
To determine the importance of the HBD/MBD for efficient FANCD2 monoubiquitination, nuclear focus formation, and its retention in chromatin, we generated a series of FANCD2 variants harboring missense mutations in the HBD/MBD and stably expressed these mutants in FA-D2 (FANCD2−/−) patient-derived cells (35). FANCD2 H1056 is predicted to play a role in histone tail binding, based on our molecular modeling analysis, while W1075 is a highly conserved aromatic amino acid within the predicted Kme binding cage (Fig. 1A). In contrast to the monoubiquitination-defective mutant FANCD2-K561R (36), the HBD/MBD missense mutants remained competent for DNA damage-inducible monoubiquitination (Fig. 3A). Importantly, these findings demonstrate that our HBD/MBD mutations do not perturb overall protein structure or stability or the interaction with the multisubunit FA core complex ubiquitin ligase. FANCI monoubiquitination was also restored, albeit weakly, in FA-D2 cells expressing the FANCD2 wild type (WT) and the HBD/MBD mutants but not in cells expressing FANCD2-K561R (Fig. 3A). To analyze the chromatin binding affinity of the HBD/MBD mutants, we used a salt chromatin extraction assay adapted from Xu et al. (2010) (37). The HBD/MBD mutants were released from chromatin at lower salt concentrations than wild-type FANCD2, indicating a reduced affinity of the mutants for chromatin (Fig. 3B and C). In an alternative approach, compared to wild-type FANCD2, FANCD2-W1075A exhibited reduced retention in chromatin when cells were cotreated with MMC and the protein synthesis inhibitor cycloheximide (Fig. 3D).
FIG 3.
FANCD2 HBD/MBD mutants are proficient for monoubiquitination but exhibit a decreased binding affinity for chromatin. (A) FA-D2 cells stably expressing LacZ, wild-type (WT) FANCD2, FANCD2-K561R, FANCD2-H1056A, and FANCD2-W1075A were incubated in the absence or presence of 200 nM mitomycin C (MMC) or 200 μM hydroxyurea (HU) for 24 h, and whole-cell lysates were analyzed by immunoblotting. L:S ratio, ratio of monoubiquitinated to nonubiquitinated FANCD2 or FANCI. (B) FA-D2 cells stably expressing wild-type FANCD2, FANCD2-H1056A, and FANCD2-W1075A were incubated in the absence or presence of 200 nM MMC for 24 h, and nuclear fractions were extracted in buffers containing the indicated NaCl concentrations. (C) Quantification of the protein band intensities from the assay whose results are presented in panel B calculated from three independent experiments. The y axis depicts the ratios of band intensities compared to the band from FA-D2 cells expressing wild-type FANCD2 at the equivalent NaCl concentration. (D) FA-D2 cells stably expressing wild-type FANCD2 or FANCD2-W1075A were incubated in the absence or presence of 200 nM MMC and 50 μg/ml (216 μM) cycloheximide (CHX) for the indicated times, and chromatin-associated proteins were analyzed by immunoblotting. The numbers to the left of the gel are molecular masses (in kilodaltons).
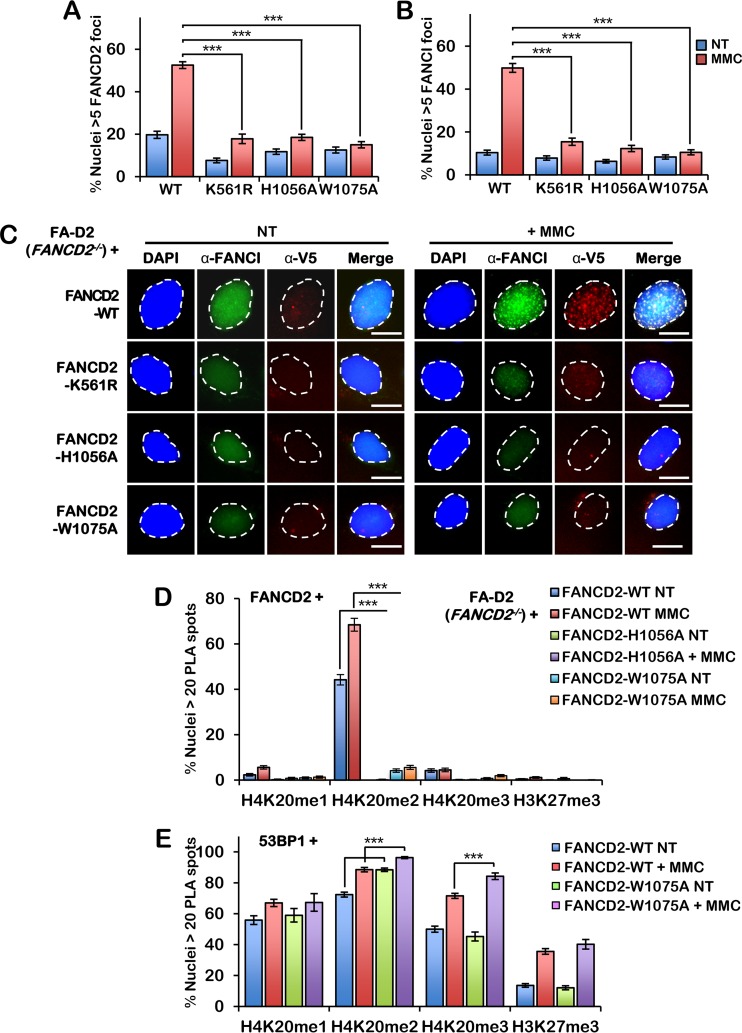
Moreover, similar to FANCD2-K561R and unlike wild-type FANCD2, the FANCD2 HBD/MBD mutants failed to efficiently assemble into discrete nuclear foci following ICL exposure (Fig. 4A and C). Consistent with previous studies showing a dependency between FANCD2 and FANCI nuclear focus formation (12, 13), FANCI nuclear focus formation was also markedly impaired in cells expressing the HBD/MBD mutants (Fig. 4B and C). We next used PLA to examine the interaction between wild-type FANCD2 and the HBD/MBD mutants and H4K20me1, -me2, and -me3 in FA-D2 cells, using methylation-specific antibodies. While wild-type FANCD2 interacted strongly with H4K20me2—and much less so with H4K20me1, H4K20me3, or H3K27me3—this interaction was markedly impaired for both the H1056A and W1075A mutants (Fig. 4D). Similar results were obtained with a FANCD2-EE956,964AA mutant; E956 and E964 are highly conserved residues of the p55 homology region (Fig. 1A and results not shown). We also performed PLA with 53BP1 and H4K20me1, H4K20me2, H4K20me3, and H3K27me3 in FA-D2 cells expressing wild-type FANCD2 and the W1075A mutant and observed a modest, yet statistically significant, increase in 53BP1 binding to H4K20me2 and -me3 in FA-D2 cells expressing FANCD2-W1075A compared to cells expressing wild-type FANCD2 (Fig. 4E). Taken together, our results indicate that FANCD2 preferentially binds to H4K20me2 in cells, with binding being mediated by the HBD/MBD domain. In addition, our results suggest that decreased binding of H4K20me2 by FANCD2 may lead to increased H4K20me2 binding by 53BP1.
FIG 4.
FANCD2 HBD/MBD mutants fail to assemble into nuclear foci or interact with H4K20me2. (A and B) FA-D2 cells stably expressing wild-type FANCD2, FANCD2-K561R, FANCD2-H1056A, and FANCD2-W1075A were incubated in the absence or presence of 200 nM mitomycin C (MMC) for 24 h. Cells were fixed and stained with anti-FANCI (green) and anti-V5 (red) antibodies and counterstained with DAPI (blue). (A) Quantification of FANCD2 nuclear foci. (B) Quantification of FANCI nuclear foci. Nuclei with >5 V5 (FANCD2) or FANCI foci were considered positive. (C) Representative images from the assay whose results are presented in panels A and B. Bars, 10 μm. (D) Quantification of proximity ligation assay (PLA) results with FANCD2 and H4K20me1, H4K20me2, H4K20me3, and H3K27me3 in FA-D2 cells stably expressing wild-type FANCD2, FANCD2-H1056A, and FANCD2-W1075A. (E) Quantification of PLA results with 53BP1 and H4K20me1, H4K20me2, H4K20me3, and H3K27me3 in FA-D2 cells stably expressing wild-type FANCD2 or FANCD2-W1075A. Nuclei with >20 PLA spots were considered positive. Experiments were performed three times with similar results. At least 300 nuclei were scored per biological replicate. Error bars represent the standard errors of the means from three independent experiments. P < 0.001.
The FANCD2 HBD/MBD is required for efficient conservative ICL repair.
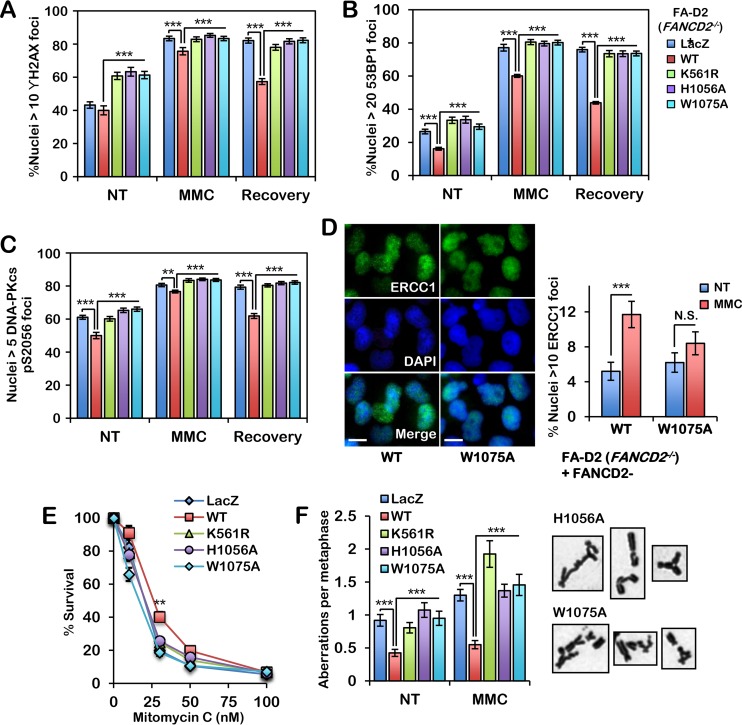
Next, we examined the impact of disruption of the HBD/MBD on ICL repair. We cultured our FA-D2 patient cell series in the absence and presence of MMC and examined γH2AX nuclear focus formation, a marker of DNA double-strand break (DSB) formation (38), immediately after exposure and following a recovery period. Unlike FA-D2 cells expressing wild-type FANCD2, cells expressing FANCD2-H1056A and -W1075A exhibited persistent elevated γH2AX nuclear focus formation following ICL exposure (Fig. 5A). We also examined 53BP1 and DNA-PKcs pS2056 nuclear focus formation in our FA-D2 cell series. 53BP1 is also a DSB marker and blocks 5′-3′ DNA strand resection, a critical initiating step of homologous recombination (HR) (39, 40), while DNA-PKcs phosphorylated at S2056 is a marker of error-prone NHEJ DSB repair (41, 42). Similar to the findings observed for γH2AX, persistent elevated levels of 53BP1 and DNA-PKcs pS2056 nuclear foci were observed in cells expressing FANCD2-H1056A and -W1075A, in contrast to cells expressing wild-type FANCD2 (Fig. 5B and C). Previous studies using Xenopus egg extracts have shown that FANCD2 promotes the recruitment of the XPF/ERCC1 endonuclease to sites of DNA damage (43). Consistent with these findings, we observed a defect in ERCC1 nuclear focus formation in cells expressing FANCD2-W1075A compared to cells expressing wild-type FANCD2 following MMC exposure (Fig. 5D). We also measured cell survival and metaphase chromosome aberrations in HBD/MBD mutant-expressing cells. Unlike wild-type FANCD2 and similar to FANCD2-K561R, the FANCD2-H1056A and -W1075A mutants failed to fully rescue the ICL sensitivity of FA-D2 patient cells (Fig. 5E). Chromosomes from cells expressing FANCD2-H1056A and -W1075A also exhibited greater numbers of aberrations, including gaps, breaks, and radial formations, than cells expressing wild-type FANCD2 (Fig. 5F). Stable expression of FANCD2-H1056A and -W1075A had no observable impact on the cellular growth rate. Taken together, our findings demonstrate that H4K20me2 binding by the FANCD2 HBD/MBD is essential for the promotion of error-free conservative ICL repair and link chromatin plasticity to activation of an important tumor suppressor pathway.
FIG 5.
The FANCD2 HBD/MBD is required for efficient conservative ICL repair. (A to C) FA-D2 cells stably expressing LacZ, wild-type FANCD2, FANCD2-K561R, FANCD2-H1056A, and FANCD2-W1075A were incubated in the absence or presence of 200 nM mitomycin C (MMC) for 24 h and allowed to recover for an additional 24 h. Cells were fixed and stained with anti-γH2AX (A), anti-53BP1 (B), or anti-DNA-PKcs pS2056 (C) antibodies, and the numbers of nuclei with >10 γH2AX (A), >20 53BP1 (B), or >5 DNA-PKcs pS2056 (C) foci were scored. At least 300 nuclei were scored per biological replicate. (D) FA-D2 cells stably expressing wild-type FANCD2 or FANCD2-W1075A were incubated in the absence (not treated [NT]) or presence of 200 nM MMC for 24 h. Cells were fixed and stained with anti-ERCC1, and the numbers of nuclei with >10 large foci were scored. At least 300 nuclei were scored per sample. Experiments were performed twice with similar results. Error bars represent the standard errors of the means from two independent experiments. ***, P < 0.001; N.S., not significant. (E) The same cells used in the assay whose results are presented in panels A to C were incubated in the presence of various concentrations of MMC for 7 to 10 days, the surviving cells were stained with crystal violet, and percent survival relative to the number of surviving untreated cells was scored. (F) The same cells were incubated in the absence (nontreated) or presence of 16 nM MMC for 24 h, and metaphase chromosomes were analyzed for structural aberrations, including gaps, breaks, and radial formations. Representative chromosome aberrations from MMC-treated FA-D2 cells expressing FANCD2-H1056A and FANCD2-W1075A are shown. All experiments were performed three times with similar results, except for those whose results are presented in panel F, which were performed twice. Error bars represent the standard errors of the means. For the assay whose results are presented in panel F, 80 metaphases were scored per treatment. **, P < 0.01; ***, P < 0.001.
DISCUSSION
In this study, we describe the identification and functional characterization of a methyl-lysine-binding domain in the FANCD2 protein that exhibits specificity for H4K20me2. Disruption of this domain results in a decreased affinity for chromatin and an inability to assemble into discrete nuclear foci, presumed sites of active ICL repair (36). Consequently, cells expressing FANCD2 HBD/MBD mutants demonstrate evidence of persistent DSBs and an increased dependence on error-prone ICL repair pathways, including NHEJ. This, in turn, leads to increased sensitivity to ICL-inducible chromosome structural aberrations and cytotoxicity. A role for the FA proteins in suppressing erroneous NHEJ repair has previously been described (41). Consistent with an important role for the H4K20me2 chromatin mark in facilitating efficient activation of the FA pathway and ICL repair, depletion of the KMT5A H4K20 monomethyltransferase reduced ICL-inducible FANCD2 nuclear focus formation and resulted in ICL sensitization. Mutation of the HBD/MBD or depletion of KMT5A did not, however, impact FANCD2 or FANCI monoubiquitination. This is consistent with several reports demonstrating the uncoupling of monoubiquitination from nuclear focus formation and chromatin binding (44–47). For example, in the absence of the histone variant H2AX, FANCD2 fails to assemble into DNA damage-inducible nuclear foci yet remains competent for monoubiquitination (44). Similarly, while Usp1−/− murine embryonic fibroblasts exhibit constitutively elevated Fancd2 monoubiquitination and chromatin localization, they fail to form Fancd2 nuclear foci and exhibit ICL hypersensitivity (46). Collectively, our results support a model whereby FANCD2 monoubiquitination and chromatin localization are necessary, but not sufficient, for effective ICL repair. The ability of FANCD2 to assemble into nuclear foci, however, is essential for effective ICL repair. Our study describes one critical determinant of FANCD2 nuclear focus formation: the ability of FANCD2 to interact directly with H4K20me2 via the HBD/MBD.
The loss of KMT5A (PR-SET7/SET8) or the KMT5B (SUV4-20H1) and KMT5C (SUV4-20H2) methyltransferases, which mediate the di- and trimethylation of H4K20, results in widespread genome instability across the evolutionary spectrum, even in the absence of exogenous DNA-damaging agents (34, 48–50). A DNA repair-independent role for FANCD2 in the protection of stalled replication forks has also been established (51). The importance of the H4K20me2 chromatin mark for the maintenance of genome stability has become increasingly well recognized (52, 53). H4K20me2 has been shown to be an important factor for the chromatin recruitment of 53BP1. 53BP1 promotes NHEJ and suppresses homologous recombination (HR) by negatively regulating 5′-3′ DNA strand resection, a critical initiating step of HR (39, 40). 53BP1 binds to H4K20me2 via its tandem Tudor domains (1). Similar to our findings for the FANCD2 MBD, disruption of the Tudor folds compromises the recruitment of 53BP1 to sites of DNA damage (54). However, while delayed, Suv4-20h-double-null mouse embryo fibroblasts support 53bp1 nuclear focus formation following exposure to ionizing radiation (IR) (50), highlighting the multifactorial nature of its chromatin recruitment. For example, binding to H2AK15ub via its ubiquitination-dependent recruitment motif (UDR) also promotes 53BP1 chromatin binding (55). Conversely, TIP60-mediated H2AK15 and H4K16 acetylation inhibits 53BP1 chromatin binding (56, 57). While much less is known about the mechanistic aspects of FANCD2 chromatin recruitment, we predict a similar scenario with the existence of multiple determinants of efficient chromatin targeting.
Our studies indicate that FANCD2 binding to H4K20me2 is necessary for stable association and site-specific accumulation within chromatin. We also previously established that FANCD2 has an ubiquitin-binding domain (UBD): an amino-terminal CUE (for coupling of ubiquitin conjugation to endoplasmic reticulum degradation) domain (58). While the FANCD2 UBD is required for efficient chromatin targeting, the ubiquitinated protein with which this domain interacts has yet to be identified. The RNF8 and BRCA1-BARD1 E3 ubiquitin ligases have also been shown to be required for the efficient chromatin recruitment of FANCD2 (36, 45, 59, 60), leading us to predict that FANCD2, like 53BP1, may also function as a bivalent chromatin reader. Therefore, despite the widespread abundance of H4K20me2 (50), only a subset of H4K20me2-marked nucleosomes is likely to support efficient FANCD2 recruitment. Interestingly, we observed a modest increase in 53BP1 binding to H4K20me2 upon mutation of the FANCD2 MBD, suggesting that FANCD2 and 53BP1 may compete for binding to the H4K20me2 mark. FANCD2 has been shown to promote the recruitment of TIP60 and the acetylation of H4K16 (61), which would be expected to decrease the affinity of 53BP1 for chromatin binding. The relationship between 53BP1 and FANCD2 remains to be clearly elucidated: while FANCD2 is generally thought to promote HR and restrict NHEJ (41, 62–64), combined deletion of murine 53bp1 and Fancd2 results in increased ICL-inducible genomic instability compared to deletion of Fancd2 alone (45). Studies of ICL repair using Xenopus egg extracts have established that FANCD2 is required for nucleolytic incisions proximal to the ICL, most likely by promoting the recruitment of SLX4 and XPF/ERCC1, consistent with our findings (43, 65). Thus, the absence of FANCD2 may preclude the generation of an optimal initiating structure for both HR and NHEJ. Future studies on the interplay between FANCD2 and 53BP1, the chromatin modifications that dictate their functions, and the chromatin remodeling complexes with which they interact will be essential for improving our understanding of ICL repair, the molecular basis of FA, and the development of effective therapeutic options for FA.
MATERIALS AND METHODS
Cell culture.
PD20 FA-D2 (FANCD2−/−) (35) and HeLa cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 15% (vol/vol) fetal bovine serum, 1% (vol/vol) l-glutamine, and 1% (vol/vol) penicillin-streptomycin. PD20 FA-D2 cells harbor a maternally inherited A-to-G change at nucleotide 376 that leads to the production of a severely truncated protein and a paternally inherited missense hypomorphic mutation leading to an R1236H change (35). PD20 FA-D2 cells were stably infected with pLenti6.2/V5-DEST (Invitrogen) harboring wild-type or mutant FANCD2 cDNAs. Stably infected cells were grown in DMEM complete medium supplemented with 2 μg/ml blasticidin. MCF10A cells were grown in DMEM–Ham’s F-12 medium supplemented with 5% (vol/vol) horse serum, 20 ng/ml epidermal growth factor, 0.5 mg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, 1% (vol/vol) l-glutamine, and 1% (vol/vol) penicillin-streptomycin.
siRNA, immunoblotting, and antibodies.
ON-TARGETplus SMARTpool siRNA against KMT5A (L-031917-00-0005; Dharmacon) was used for siRNA studies. siRNA targeting FANCD2 has previously been described (66). Cells were plated in six-well dishes at a density of 2 × 105 cells per well. On the following day, cells were transfected with siRNA specific for FANCD2 and/or KMT5A and control nontargeting siRNA using the Lipofectamine 2000 reagent. At 64 h following transfection, cells were incubated in the absence or presence of 200 nM MMC or 1 μM APH for 16 h and harvested for analysis. For immunoblotting analysis, cell suspensions were washed in ice-cold phosphate-buffered saline (PBS) and lysed in SDS lysis buffer (2% [vol/vol] SDS, 50 mM Tris-HCl, pH 7.4, 10 mM EDTA) with sonication. Proteins were resolved on NuPAGE 3 to 8% (wt/vol) Tris-acetate or 4 to 12% (wt/vol) bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes. For flow cytometry analysis, cells were washed in PBS and incubated in 50 μg/ml propidium iodide (PI) (Sigma) and 30 U/ml RNase A for 10 min at 37°C, followed by analysis using a BD FACSVerse flow cytometer. The percentages of cells in G1, S, and G2/M phase were determined by analyzing PI histograms with FlowJo (v10.2) software. The following mouse monoclonal antibodies were used: anti-ERCC1 (D-10; sc-17809; Santa Cruz Biotechnology), anti-GST (136700; Invitrogen), anti-γH2AX (05-636; Millipore), anti-H3K27me3 (ab6002; Abcam), anti-H4K20me1 (39727; Active Motif), anti-H4K20me2 (GTX630545; GeneTex), anti-H4K20me3 (39671; Active Motif), anti-HP1α (05-689; Millipore), anti-α-tubulin (MS-581-P1; NeoMarkers), and anti-V5 (R960-25; Invitrogen). The rabbit polyclonal antibodies used were anti-53BP1 (sc-22760; Santa Cruz Biotechnology), anti-DNA-PKcs pS2056 (ab18192; Abcam), anti-FANCD2 (NB100-182; Novus Biologicals), anti-FANCI (A300-212A and A300-254A; Bethyl Laboratories), anti-V5 (D3H8Q; Cell Signaling Technology), anti-H2A (07-146; Upstate), and anti-H3 (ab1791; Abcam).
Plasmids.
Mutant cDNAs were generated using a QuikChange II site-directed mutagenesis kit (Stratagene). The forward and reverse oligonucleotide sequences used were as follows: H1056A FP 5′-CCAGGAGTGAAAGTTCAGGAGTACGCCATAATGTCTTCCTGC-3′, H1056A RP 5′-GCAGGAAGACATTATGGCGTACTCCTGAACTTTCACTCCTGG-3′, W1075A FP 5′-CATGGGCTTTTTGCTGCGAGTGGATTTTCTCAACCTG-3′, and W1075A RP 5′-CAGGTTGAGAAAATCCACTCGCAGCAAAAAGCCCATG-3′. The histone-binding domain (HBD) and methyl-lysine-binding domain (MBD) fragments were cloned into the pGEX-6P-1 plasmid using restriction enzyme cloning. The forward and reverse oligonucleotide sequences used were as follows: HBD FP 5′-ATAGAATTCATGGATGAGCAGTGCACACAG-3′, HBD RP 5′-TATCTCGAGTCACTCTGTGTGCTCCAGGTA-3′, MBD FP 5′-ATAGAATTCATGTTTCATGGGCTTTTTG-3′, and MBD RP 5′-TATCTCGAGTCACAAAATAACCATCAAAAG-3′.
Protein purification.
GST fusion proteins were expressed in BL21 Rosetta2(DE3)pLysS cells following induction with isopropyl-β-d-1-thiogalactopyranoside (IPTG) at 16°C. Cells were pelleted, lysed in buffer A (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol, and 0.5% [vol/vol] NP-40 plus protease inhibitor cocktail tablets [Roche]), and flash frozen. On the following day, the cells were thawed on ice and lysed using a French press. Lysates were spun at 12,000 × g for 30 min at 4°C, and the supernatant was filtered using a 0.45-μm-pore-size filter. Glutathione-agarose (Invitrogen) was applied to a column and washed with deionized H2O and buffer A. Filtered supernatant was applied to the column. The column was washed in buffer B (50 mM Tris-HCl, pH 8.0, 750 mM NaCl, 0.5% [vol/vol] NP-40) and then buffer A. Proteins were eluted at room temperature in buffer C (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 0.5% [vol/vol] NP-40, 20 mM reduced glutathione). Elution samples were resolved on NuPAGE 4 to 12% (wt/vol) bis-Tris gels and stained with SimplyBlue SafeStain (Invitrogen). Protein-containing fractions were pooled and dialyzed against buffer D (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% [vol/vol] NP-40).
Immunofluorescence microscopy.
Cells were plated at a density of 3 × 105 cells per well of culture slides (BD Falcon). On the following day, cells were treated with 200 nM MMC for 24 h. Cells were prepermeabilized in permeabilization buffer (0.3% [vol/vol] Triton X-100 in PBS, pH 7.4) and fixed on ice in fixing buffer (4% [wt/vol] paraformaldehyde and 2% [wt/vol] sucrose in PBS, pH 7.4) for 15 min. Cells were permeabilized for 10 min and blocked in antibody dilution buffer (ADB; 5% [vol/vol] goat serum and 0.1% [vol/vol] NP-40 in PBS, pH 7.4) for 30 min. Cells were stained for 1 h in primary antibody diluted in ADB, washed with PBS and permeabilization buffer, and stained with fluorescent secondary antibodies for 30 min in ADB. Cells were washed in PBS and permeabilization buffer and counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). Foci were analyzed using a Zeiss AxioImager.A1 upright epifluorescence microscope with AxioVision LE (v4.6) image acquisition software. Proximity ligation assays (PLA) were performed similarly to the immunofluorescence microscopy protocol up to the secondary antibody step. The remainder of the PLA protocol was carried out according to the manufacturer’s instructions (DUO92101; Sigma-Aldrich).
MMC cell survival assay.
Cells were plated at a density of 1.5 × 104 cells per well in 6-well dishes. On the following day, the cells were treated with various concentrations of mitomycin C (MMC) for 7 to 10 days. The cells were washed in PBS and fixed in fixing buffer (10% [vol/vol] methanol, 10% [vol/vol] acetic acid). The cells were then stained with crystal violet (1% [wt/vol] crystal violet in methanol). On the following day, the crystal violet was dissolved in dissolving solution (0.02% [vol/vol] SDS in methanol) for 2 h. The solutions were transferred to 96-well dishes, and the optical density at 570 nm was read using a 96-well Bio-Rad 680 microplate reader.
Chromosome breakage assay.
For chromosome breakage assays, cells were grown in the absence or presence of 10 or 16 nM MMC for 24 h. Prior to harvesting, cells were treated with 0.1 μg/ml colcemid (Gibco/Invitrogen) for 2 h. Cell pellets were incubated in 0.075 M KCl at 37°C for 18 min, followed by fixation in Carnoy’s fixative (3:1 methanol-glacial acetic acid) with multiple changes. Cells were dropped onto chilled slides and air dried prior to staining with 3% (vol/vol) Giemsa solution (Sigma). Metaphases were analyzed using a Zeiss AxioImager.A1 upright epifluorescence microscope with AxioVision LE (v4.6) image acquisition software.
In vitro bulk histone-binding assays.
Glutathione-agarose (Invitrogen) was blocked in NETN150 (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.25% [vol/vol] NP-40, 1 mM EDTA) plus 1% (wt/vol) bovine serum albumin, washed in NETN150, and added to GST fusion proteins overnight at 4°C. Histones purified from calf thymus (Sigma) were precleared with glutathione-agarose at 4°C overnight. Precleared calf thymus histones were boiled for 10 min and added to bead-bound GST fusion proteins, and the mixture was incubated at 4°C for 1 h. The beads were washed 4 times in NETN200 (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.25% [vol/vol] NP-40, 1 mM EDTA), and bound proteins were eluted in 2× lithium dodecyl sulfate (LDS)–10% (vol/vol) β-mercaptoethanol with boiling. Proteins were resolved on 4 to 12% (wt/vol) bis-Tris gels (Invitrogen) and stained with SimplyBlue SafeStain (Invitrogen) or H4K20me1, H4K20me2, H4K20me3, and H2A antibodies.
NaCl extraction assay.
Cells were plated at a density of 3 × 106 cells in 15-cm2 dishes. Cells were treated with 200 nM MMC for 24 h. Cells were harvested in ice-cold PBS, and a portion was set aside for the whole-cell lysate. The remaining pellet was lysed in CSK buffer {10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.8), 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% (vol/vol) Triton X-100} for 10 min at 4°C, and the supernatant containing soluble proteins was collected. The remaining pellet was split into three and lysed in salt extraction buffer {20 mM HEPES (pH 7.9), 0.5 mM TCEP [Tris(2-carboxyethyl)phosphine hydrochloride], 1 mM PMSF, 1.5 mM MgCl2, 0.1% Triton X-100} containing 150, 250, or 500 mM NaCl.
In vitro histone peptide binding assays.
GST fusion proteins were incubated with biotinylated histone peptides (Epicypher) overnight at 4°C in binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% [vol/vol] NP-40). Samples were incubated for 1 h at 4°C with streptavidin-Sepharose (GE) (previously washed in binding buffer). After incubation, samples were washed 4 times with wash buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 0.1% [vol/vol] NP-40) and eluted in 2× LDS–10% (vol/vol) β-mercaptoethanol with boiling. Proteins were resolved on 4 to 12% (wt/vol) bis-Tris gels (Invitrogen) and either silver stained using a silver stain for mass spectrometry kit (Thermo) or immunoblotted with antibodies against GST (Invitrogen).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the N. G. Howlett, J. L. Camberg, and A. Dutta laboratories for critical discussion.
This work was supported by an American Society of Hematology Bridge Grant to N.G.H., Rhode Island IDeA Network of Biomedical Research Excellence (RI-INBRE) grant P20GM103430 from the National Institute of General Medical Sciences, and Rhode Island Experimental Program to Stimulate Competitive Research (RI-EPSCoR) grant number 1004057 from the National Science Foundation.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00194-19.
REFERENCES
- 1.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. 2009. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farf I. 2014. Fanconi anemia: guidelines for diagnosis and management, 4th ed Fanconi Anemia Research Fund, Inc, Eugene, OR. [Google Scholar]
- 5.Mamrak NE, Shimamura A, Howlett NG. 2017. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev 31:93–99. doi: 10.1016/j.blre.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knies K, Inano S, Ramirez MJ, Ishiai M, Surralles J, Takata M, Schindler D. 2017. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J Clin Invest 127:3013–3027. doi: 10.1172/JCI92069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kottemann MC, Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walden H, Deans AJ. 2014. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys 43:257–278. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- 9.Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. 2007. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol 27:8421–8430. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howlett NG, Harney JA, Rego MA, Kolling FW IV, Glover TW. 2009. Functional interaction between the Fanconi anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J Biol Chem 284:28935–28942. doi: 10.1074/jbc.M109.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang C-C, Li Z, Lopez-Martinez D, Nicholson WV, Vénien-Bryan C, Cohn MA. 2016. The FANCD2-FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat Commun 7:12124. doi: 10.1038/ncomms12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims AE, Spiteri E, Sims RJ III, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT. 2007. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol 14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 13.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, Elledge SJ, Pavletich NP. 2011. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science 333:312–316. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niraj J, Caron MC, Drapeau K, Berube S, Guitton-Sert L, Coulombe Y, Couturier AM, Masson JY. 2017. The identification of FANCD2 DNA binding domains reveals nuclear localization sequences. Nucleic Acids Res 45:8341–8357. doi: 10.1093/nar/gkx543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park WH, Margossian S, Horwitz AA, Simons AM, D'Andrea AD, Parvin JD. 2005. Direct DNA binding activity of the Fanconi anemia D2 protein. J Biol Chem 280:23593–23598. doi: 10.1074/jbc.M503730200. [DOI] [PubMed] [Google Scholar]
- 17.Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. 2012. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res 40:4553–4561. doi: 10.1093/nar/gks053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgs MR, Sato K, Reynolds JJ, Begum S, Bayley R, Goula A, Vernet A, Paquin KL, Skalnik DG, Kobayashi W, Takata M, Howlett NG, Kurumizaka H, Kimura H, Stewart GS. 2018. Histone methylation by SETD1A protects nascent DNA through the nucleosome chaperone activity of FANCD2. Mol Cell 71:P25–41.e6. doi: 10.1016/j.molcel.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato K, Ishiai M, Toda K, Furukoshi S, Osakabe A, Tachiwana H, Takizawa Y, Kagawa W, Kitao H, Dohmae N, Obuse C, Kimura H, Takata M, Kurumizaka H. 2012. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J 31:3524–3536. doi: 10.1038/emboj.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vierra DA, Garzon JL, Rego MA, Adroved MM, Mauro M, Howlett NG. 2017. Modulation of the Fanconi anemia pathway via chemically induced changes in chromatin structure. Oncotarget 8:76443–76457. doi: 10.18632/oncotarget.19470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Chang JF, Sun J, Chen L, Yang XM, Tang HY, Jing YY, Kang X, He ZM, Wu JY, Wei HM, Wang DL, Xu RG, Zhu RB, Shen Y, Zeng SY, Wang C, Liu KN, Zhang Y, Mao ZY, Jiang CZ, Sun FL. 2018. Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J Cell Sci 131:jcs215525. doi: 10.1242/jcs.215525. [DOI] [PubMed] [Google Scholar]
- 22.Marhold J, Brehm A, Kramer K. 2004. The Drosophila methyl-DNA binding protein MBD2/3 interacts with the NuRD complex via p55 and MI-2. BMC Mol Biol 5:20. doi: 10.1186/1471-2199-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Balbas MA, Tsukiyama T, Gdula D, Wu C. 1998. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci U S A 95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowak AJ, Alfieri C, Stirnimann CU, Rybin V, Baudin F, Ly-Hartig N, Lindner D, Müller CW. 2011. Chromatin-modifying complex component Nurf55/p55 associates with histones H3 and H4 and polycomb repressive complex 2 subunit Su(z)12 through partially overlapping binding sites. J Biol Chem 286:23388–23396. doi: 10.1074/jbc.M110.207407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song JJ, Garlick JD, Kingston RE. 2008. Structural basis of histone H4 recognition by p55. Genes Dev 22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blus BJ, Wiggins K, Khorasanizadeh S. 2011. Epigenetic virtues of chromodomains. Crit Rev Biochem Mol Biol 46:507–526. doi: 10.3109/10409238.2011.619164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du LL, Nakamura TM, Russell P. 2006. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalb R, Neveling K, Hoehn H, Schneider H, Linka Y, Batish SD, Hunt C, Berwick M, Callen E, Surralles J, Casado JA, Bueren J, Dasi A, Soulier J, Gluckman E, Zwaan CM, van Spaendonk R, Pals G, de Winter JP, Joenje H, Grompe M, Auerbach AD, Hanenberg H, Schindler D. 2007. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet 80:895–910. doi: 10.1086/517616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karachentsev D, Sarma K, Reinberg D, Steward R. 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9:1201–1213. doi: 10.1016/S1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 34.Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS. 2007. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D'Andrea A, Moses R, Grompe M. 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell 7:241–248. doi: 10.1016/S1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7:249–262. doi: 10.1016/S1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. 2010. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol 191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 39.FitzGerald JE, Grenon M, Lowndes NF. 2009. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans 37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann M, de Lange T. 2014. 53BP1: pro choice in DNA repair. Trends Cell Biol 24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 42.Yajima H, Lee KJ, Chen BP. 2006. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol 26:7520–7528. doi: 10.1128/MCB.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. 2014. XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogliolo M, Lyakhovich A, Callen E, Castella M, Cappelli E, Ramirez MJ, Creus A, Marcos R, Kalb R, Neveling K, Schindler D, Surralles J. 2007. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J 26:1340–1351. doi: 10.1038/sj.emboj.7601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, Chen HT, Fernandez-Capetillo O, D'Andrea A, Nussenzweig A. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD. 2009. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vuono EA, Mukherjee A, Vierra DA, Adroved MM, Hodson C, Deans AJ, Howlett NG. 2016. The PTEN phosphatase functions cooperatively with the Fanconi anemia proteins in DNA crosslink repair. Sci Rep 6:36439. doi: 10.1038/srep36439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. 2009. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakaguchi A, Steward R. 2007. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J Cell Biol 176:155–162. doi: 10.1083/jcb.200607178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, Espejo A, Bedford MT, Nussenzweig A, Busslinger M, Jenuwein T. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen S, Schotta G, Sorensen CS. 2013. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paquin KL, Howlett NG. 2018. Understanding the histone DNA repair code: H4K20me2 makes its mark. Mol Cancer Res 16:1335–1345. doi: 10.1158/1541-7786.mcr-17-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 55.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert J-P, Roques C, Pandita RK, Paquet E, Herst P, Gingras A-C, Pandita TK, Legube G, Doyon Y, Durocher D, Côté J. 2016. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol Cell 62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rego MA, Kolling FW, Vuono EA, Mauro M, Howlett NG. 2012. Regulation of the Fanconi anemia pathway by a CUE ubiquitin-binding domain in the FANCD2 protein. Blood 120:2109–2117. doi: 10.1182/blood-2012-02-410472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bick G, Zhang F, Meetei AR, Andreassen PR. 2017. Coordination of the recruitment of the FANCD2 and PALB2 Fanconi anemia proteins by an ubiquitin signaling network. Chromosoma 126:417–430. doi: 10.1007/s00412-016-0602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long DT, Joukov V, Budzowska M, Walter JC. 2014. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell 56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Renaud E, Barascu A, Rosselli F. 2016. Impaired TIP60-mediated H4K16 acetylation accounts for the aberrant chromatin accumulation of 53BP1 and RAP80 in Fanconi anemia pathway-deficient cells. Nucleic Acids Res 44:648–656. doi: 10.1093/nar/gkv1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M. 2011. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol 18:500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A 102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. 2010. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 65.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. 2005. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet 14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 67.Boisvert RA, Rego MA, Azzinaro PA, Mauro M, Howlett NG. 2013. Coordinate nuclear targeting of the FANCD2 and FANCI proteins via a FANCD2 nuclear localization signal. PLoS One 8:e81387. doi: 10.1371/journal.pone.0081387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.