Mumps virus (MuV) infection is characterized by parotid gland swelling and can cause pancreatitis, orchitis, meningitis, and encephalitis. MuV-related hearing loss is also a serious complication because it is usually irreversible. MuV outbreaks have been reported in many countries, even in high-vaccine-coverage areas. MuV has tropism toward glandular tissues and the central nervous system. To understand the unique MuV tropism, revealing the mechanism of receptor recognition by MuV is very important. Here, using a large-scale glycan array and X-ray crystallography, we show that MuV recognizes sialyl LewisX and GM2 ganglioside as receptors, in addition to a previously reported MuV receptor, a trisaccharide containing an α2,3-linked sialic acid. The flexible recognition of these glycan receptors by MuV may explain the unique tropism and pathogenesis of MuV. Structures will also provide a template for the development of effective entry inhibitors targeting the receptor-binding site of MuV.
KEYWORDS: entry, glycans, hemagglutinin-neuraminidase, mumps, paramyxovirus
ABSTRACT
Mumps virus (MuV) is an important aerosol-transmitted human pathogen causing epidemic parotitis, meningitis, encephalitis, and deafness. MuV preferentially uses a trisaccharide containing α2,3-linked sialic acid as a receptor. However, given the MuV tropism toward glandular tissues and the central nervous system, an additional glycan motif(s) may also serve as a receptor. Here, we performed a large-scale glycan array screen with MuV hemagglutinin-neuraminidase (MuV-HN) attachment proteins by using 600 types of glycans from The Consortium for Functional Glycomics Protein-Glycan Interaction Core in an effort to find new glycan receptor motif(s). According to the results of the glycan array, we successfully determined the crystal structures of MuV-HN proteins bound to newly identified glycan motifs, sialyl LewisX (SLeX) and the oligosaccharide portion of the GM2 ganglioside (GM2-glycan). Interestingly, the complex structures showed that SLeX and GM2-glycan share the same configuration with the reported trisaccharide motif, 3′-sialyllactose (3′-SL), at the binding site of MuV-HN, while SLeX and GM2-glycan have several unique interactions compared with those of 3′-SL. Thus, MuV-HN protein can allow an additional spatial modification in GM2-glycan and SLeX at the second and third carbohydrates from the nonreducing terminus of the core trisaccharide structure, respectively. Importantly, MuV entry was efficiently inhibited in the presence of 3′-SL, SLeX, or GM2-glycan derivatives, which indicates that these motifs can serve as MuV receptors. The α2,3-sialylated oligosaccharides, such as SLeX and 3′-sialyllactosamine, are broadly expressed in various tissues, and GM2 exists mainly in neural tissues and the adrenal gland. The distribution of these glycan motifs in human tissues/organs may have bearing on MuV tropism.
IMPORTANCE Mumps virus (MuV) infection is characterized by parotid gland swelling and can cause pancreatitis, orchitis, meningitis, and encephalitis. MuV-related hearing loss is also a serious complication because it is usually irreversible. MuV outbreaks have been reported in many countries, even in high-vaccine-coverage areas. MuV has tropism toward glandular tissues and the central nervous system. To understand the unique MuV tropism, revealing the mechanism of receptor recognition by MuV is very important. Here, using a large-scale glycan array and X-ray crystallography, we show that MuV recognizes sialyl LewisX and GM2 ganglioside as receptors, in addition to a previously reported MuV receptor, a trisaccharide containing an α2,3-linked sialic acid. The flexible recognition of these glycan receptors by MuV may explain the unique tropism and pathogenesis of MuV. Structures will also provide a template for the development of effective entry inhibitors targeting the receptor-binding site of MuV.
INTRODUCTION
Mumps, characterized by parotid gland swelling, is caused by mumps virus (MuV), which is an important aerosol-transmitted pathogen for childhood illness. The reported cases decreased dramatically after a live attenuated vaccine was introduced in 1967, and its routine use as a part of the measles-mumps-rubella vaccine was recommended. However, sporadic outbreaks have still been reported in countries with high vaccination coverage (1–4).
MuV infection induces inflammation in the parotid and other salivary glands, pancreas, testis, ovary, and mammary glands. MuV also has tropism for the central nervous system and may cause encephalitis, meningitis, and sensorineural hearing loss (1). A recent study suggests that the frequency of MuV-caused deafness may be much higher, up to 1 in 1,000 cases (5), than the previous estimates of 1 in 15,000 to 20,000 cases (1, 6) although the estimates vary between studies, possibly because of the differences in the vaccination practice among nations (7). MuV typically affects glandular tissues and the central nervous system; however, the reason for the unique tropism is not fully understood.
Mumps virus is an enveloped, nonsegmented negative-strand RNA virus of the genus Rubulavirus of the family Paramyxoviridae. The genome contains 15,384 nucleotides with seven genes encoding the nucleocapsid, phospho-, matrix, fusion (F), small hydrophobic, hemagglutinin-neuraminidase (HN), and large proteins. The two envelope glycoproteins, HN and F, engage in receptor binding and mediate membrane fusion, respectively. Upon receptor binding, MuV-HN protein triggers a series of conformational changes of the adjacent MuV-F protein, resulting in membrane fusion between the viral envelope and the host cell membrane (1, 8).
Glycans are the common receptor for many viruses, including influenza virus (Flu), rotavirus, adeno-associated virus, JC virus (JCV), and MuV as well as other paramyxoviruses such as human parainfluenza virus types 1 to 4, parainfluenza virus type 5, Sendai virus, and Newcastle disease virus (8–15). Flu is well investigated in terms of its glycan-virus interaction in that its host ranges for birds and humans are highly dependent on the engagement between hemagglutinin (HA) protein and the α2,3- or α2,6-linked sialic acid (9). In addition, studies have indicated that additional carbohydrate moieties following the terminal sialic acid (Neu5Ac) linked to galactose (Gal) or N-acetylgalactosamine (GalNAc) are also involved in the binding with different Flu-HA proteins (10, 16–21). Other structural studies on polyomaviruses have also shown that JCV-VP1 and Simian virus 40 VP1, proteins responsible for the glycan receptor attachment, interact with the consecutive carbohydrates linked to the nonreducing terminal Neu5Ac-Gal of the pentasaccharide receptor fragment, lactoseries tetrasaccharide c, and the oligosaccharide portion of the GM1 ganglioside, respectively (13, 22). Such extensive interactions of carbohydrates beyond the nonreducing terminal Neu5Ac-Gal linkages with the viral proteins are substantially responsible for the observed binding specificity and affinity.
Recently, our study revealed that a trisaccharide containing an α2,3-linked sialic acid in the unbranched sugar chains constitutes a functional receptor for MuV (23). In the study, we found that the interaction between the tyrosine 369 residue of MuV-HN protein and the third carbohydrate from the nonreducing terminal of the trisaccharide contributes to the stability of the MuV-HN–glycan complex. The structural observations together with the binding analysis using representatives of the major sialylated glycans in the human respiratory tract demonstrated that the trisaccharide containing an α2,3-linked sialic acid, such as 3′-sialyllactose (3′-SL) or 3′-sialyllactosamine (3′-SLN), is the receptor motif for MuV. Glycans are a mixture of highly heterogeneous molecules composed of a variety of components, structures, and modifications. Since MuV shows unique tropism compared with the that of other sialic-acid using paramyxoviruses, an extensive/additional glycan receptor(s) for MuV may exist.
In this study, we performed a large-scale glycan array screen with MuV-HN protein using 600 types of glycans from The Consortium for Functional Glycomics (CFG) Protein-Glycan Interaction Core (Core H) in an effort to find the extensive/additional receptor motif(s) for MuV. Further, we determined the X-ray crystal structures of the MuV-HN protein in complex with the newly found glycan motifs including sialyl LewisX (SLeX) and the oligosaccharide portion of the GM2 ganglioside (GM2-glycan). In the structures thus obtained, MuV-HN protein is spatially allowed to bind with SLeX and GM2-glycan by structurally recognizing them as being similar to 3′-SL. We also show that the glycans with these motifs possess antiviral potencies with 50% inhibitory concentrations (IC50s) of as low as ∼80 μM in cultured cells. The distribution of glycan receptors in human tissues/organs may at least partly account for the unique MuV tropism.
RESULTS
A large-scale glycan array screen with MuV-HN proteins.
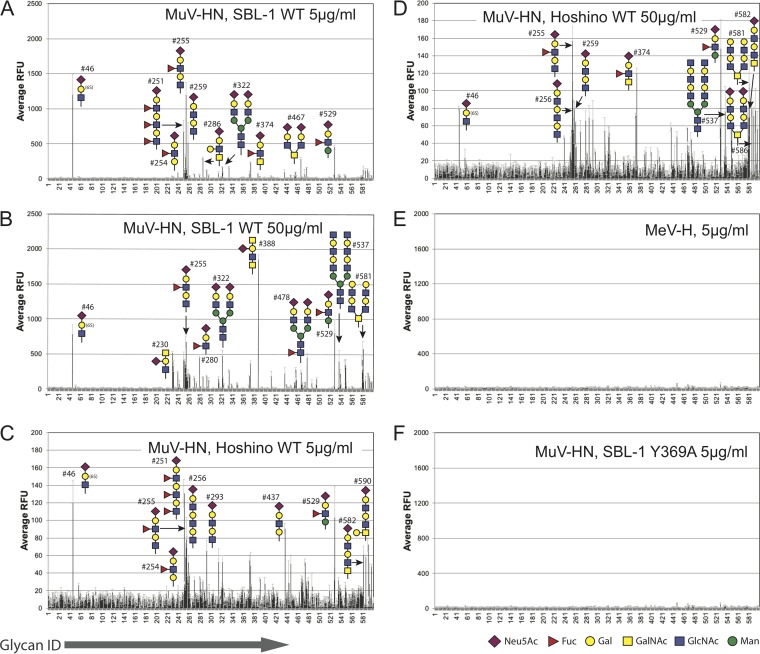
Purified MuV-HN proteins of the SBL-1 (genotype A) and Hoshino (genotype B) strains were reacted with the Mammalian Glycan Array (version 5.3; CFG) including 600 types of glycans. The most dominant motif observed in the top-ranked glycan molecules bound to the SBL-1 HN protein (5 μg/ml) was the trisaccharide structure of Neu5Acα2-3Galβ1-4GlcNAc (3′-SLN), in which the terminal N-acetylneuraminic acid (here referred to as Sia-1) is connected to the galactosyl unit (referred to as Gal-2) by the α2,3 linkage, and the Gal-2 is connected to the N-acetylglucose unit (referred to as GlcNAc-3) by the β1-4 linkage (Fig. 1 and Table 1). In contrast, the glycans containing an α2,6-linked sialic acid showed much lower binding with MuV-HN proteins than those of the glycans containing an α2,3-linked sialic acid. Polysialic acids also showed less reactivity against MuV-HN proteins (Fig. 1 and Table 1; see also Data Set S1 in the supplemental material; data are also available in the CFG database [http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh.shtml]. Additionally, the results of the glycan array demonstrated the preferred binding of unbranched oligosaccharides with MuV-HN proteins over branched ones although the MuV-HN proteins of the higher concentration (50 μg/ml) bound better to biantennary glycan structures than those of the lower concentration (5 μg/ml). The biantennary glycans containing the trisaccharide receptor motif ranked 8th and 10th (glycans 467 and 322), but the multiantennary glycan ranked 24th (glycan 457), which is below the threshold of the “binders” evaluated by GlycoPattern (24), a web-based bioinformatics tool used to analyze the glycan array data. The rest of the multiantennary glycans containing the terminal MuV receptor motif were below the background level (Fig. 1 and Table 1). The glycans with the Neu5Acα2-3Galβ1-4Glc (3′-SL) motif (glycans 261 and 262) also appeared in the binders list, albeit only in the high-concentration assays with both SBL-1 and Hoshino strains. These observations were consistent with our previous finding that a trisaccharide containing an α2,3-linked sialic acid, especially in unbranched sugar chains, preferentially functions as a receptor for MuV (23). The MuV-HN Y369A mutant protein showed poor binding to all glycans, which is similar to the results with the measles virus hemagglutinin (MeV-H), a receptor attachment glycoprotein (Fig. 1E and F). MeV-H was used as a negative control because MeV binds only to proteinaceous receptors. Therefore, the results of the glycan array support the idea that the receptor binding by MuV-HN protein is efficiently stabilized by the interaction between the Tyr369 residue of MuV-HN and the GlcNAc-3 of the trisaccharide motif (23). That the SBL-1 strain has a better signal-to-noise ratio than the Hoshino strain may suggest that the SBL-1 strain has a higher affinity to the glycans than the Hoshino strain. (Fig. 1 and Table 1). These observations confirmed that the CFG Core H glycan array experimental system is compatible with the systems (smaller-scale arrays, crystal structures, and fusion assays) used in our previous study (23).
FIG 1.
Large-scale glycan array analysis with MuV-HN proteins. (A to D) Glycan array 600 types of glycans from the Mammalian Glycan Array, version 5.3 (CFG resources), analysis with the purified MuV-HN proteins of the SBL-1 and Hoshino strains from a low-concentration assay (5 μg/ml) (A and C, respectively) and a high-concentration assay (50 μg/ml) (B and D, respectively). Glycan structures in the top 10th are drawn as schematics. (E) Glycan array analysis with the purified MeV-H as a negative control. (F) Binding of the MuV-HN Y369A mutant protein to the glycan array.
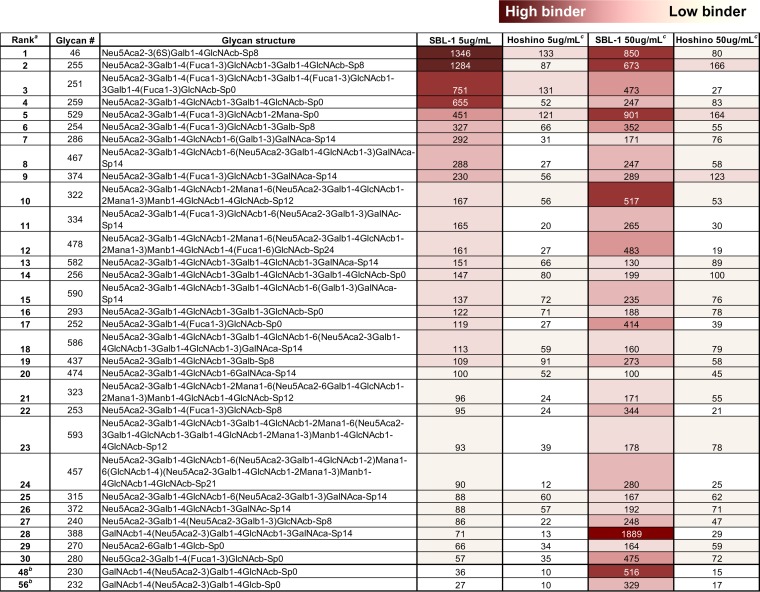
TABLE 1.
Glycan-binding specificity of MuV-HN proteins
The top 30 glycans from the results of glycan array analysis with the HN protein of SBL-1 strain (5 μg/ml) are listed in order of the highest binding. The degree of binding is indicated according to the color scale above the table.
Representative glycans of the GM2/Sda-glycan motif (glycans 230 and 232) are additionally listed at the bottom.
Binding (relative fluorescence units [RFU]) of the corresponding glycans for the MuV-HN protein of the Hoshino strain (5 μg/ml and 50 μg/ml) and SBL-1 strain (50 μg/ml) are also shown.
Identification of novel binding motifs for MuV-HN proteins.
In addition to the core trisaccharide structure described above, interestingly, the α1,3-fucosylated trisaccharide Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAc was newly detected in the top-ranked glycan molecules (Fig. 1A to D and Table 1). The fucose-modified motif accounted for 5 and 3 of the 10 highest-binding glycans in the assays with the low (5 μg/ml; glycans 255, 251, 529, 254, and 374) and high (50 μg/ml; glycans 529, 255, and 280) concentrations of the MuV-HN protein of the SBL-1 strain, respectively (Fig. 1A and B and Table 1). This motif also accounted for 4 and 3 of the 10 highest-binding glycans in the assays with the low (glycans 251, 529, 255, and 254) and high (glycans 255, 529, and 374) concentrations of the MuV-HN protein of the Hoshino strain, respectively (Fig. 1C and D and Table 1). This α1,3-fucosylated motif is generally known as an SLeX structure. Further, the GalNAc-modified trisaccharide at Gal-2 by the β1,4-linkage, Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAc, was also newly identified as the top-ranked glycan. This motif accounted for 2 of the 10 highest-binding glycans in the assay with a high concentration of the MuV-HN protein of the SBL-1 strain (glycans 388 and 230) (Fig. 1B and Table 1), while it first appeared to rank 28th in the assay with the low-MuV-HN protein concentration. This motif is known as Sda-glycan (Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAc-R), and the terminal branched glycan, in which GalNAc and Neu5Ac are both linked to the Gal, has the same structure as the oligosaccharide epitope of GM2 ganglioside (Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcCer). The oligosaccharide portion of GM2 ganglioside (glycan 232) ranked 20th in the assay with the high concentration of the MuV-HN protein of the SBL-1 strain. The glycans with nonsialylated motifs, GalNAcβ1,4-Gal and GlcNAcβ1,3-Gal (glycans 581, 537, and 579), were also newly listed in the top-ranked glycans, specifically in the assays with the high concentrations of the MuV-HN proteins of both strains. The data analysis using GlycoPattern suggested glycan structures equivalent to 3′-SLN and SLeX as binder motifs for the MuV-HN protein. Based on the frequency of appearance in the top ranks and the data analysis using GlycoPattern, we categorized the glycans that may be putative receptors for MuV into three new subgroups, the SLeX motif, the GM2/Sda-glycan motif, and the nonsialylated glycan motifs in addition to the 3′-SL (3′-SLN) motif that was previously reported as the core structure of a receptor for MuV (23).
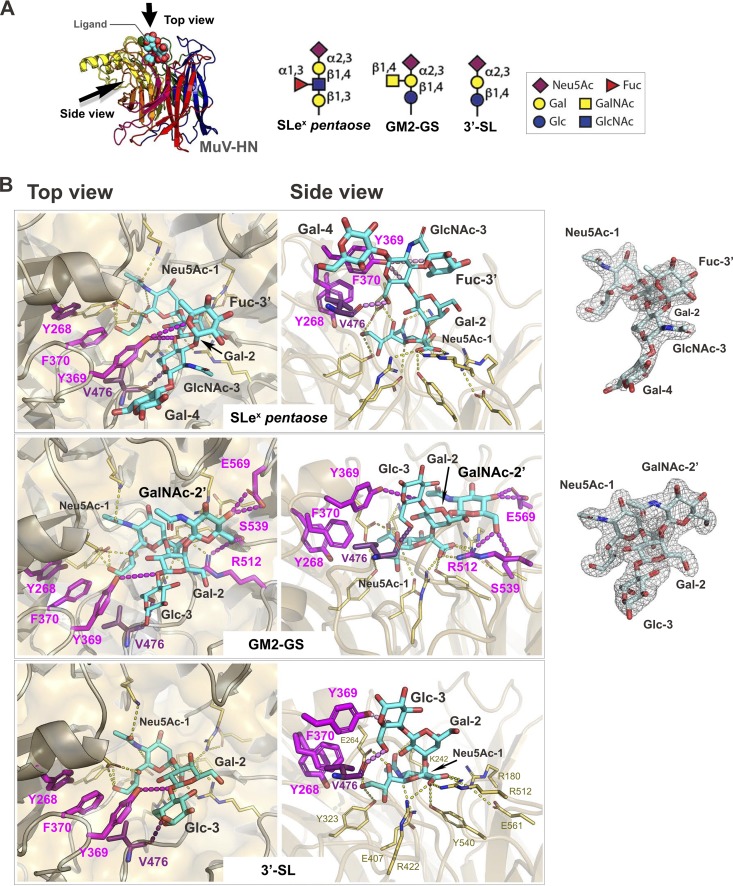
Interaction of MuV-HN with SLeX pentaose, a GM2-ganglioside sugar, or 3′-SL.
To gain insight into interactions of MuV-HN protein with the glycan motifs identified above, we attempted to determine the crystal structures of MuV-HN in complex with SLeX pentaose (Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Gal), GM2 ganglioside sugar (abbreviated as GM2-GS here; Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glc), or lacto-N-neotetraose (LnNT; Galβ1-4GlcNAcβ1-3Galβ1-4Glc). These are representatives of SLeX, the GM2-glycan, and the nonsialylated glycan motifs. The purified MuV-HN protein was cocrystalized with these glycans according to a previously described method (23). Diffractions to 2.5 Å and 2.05 Å were successfully obtained for the cocrystals of the MuV-HN with SLeX pentaose and GM2-GS, respectively (Table 2). Although MuV-HN was cocrystalized in the presence of a 5- to 50-fold excess molar concentration of LnNT, the electron density of LnNT was absent in the structure.
TABLE 2.
Data collection and refinement statistics (molecular replacement)
| Parametera | Value for the parameter in:b
|
|
|---|---|---|
| MuV-HN–GM2 complex | MuV-HN–SLeX complex | |
| Data collection | ||
| Space group | P61 | P61 |
| Cell dimensions | ||
| a, b, c (Å) | 137.26, 137.26, 178.01 | 137.42, 137.42, 177.81 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Resolution (Å) | 50–2.05 (2.09–2.05) | 50–2.5 (2.54–2.50) |
| Rsym | 0.111 (>1.00) | 0.20 (>1.00) |
| Rpim | 0.028 (0.376) | 0.05 (0.451) |
| CC1/2 | 0.999 (0.581) | 0.998 (0.609) |
| I/σI | 29.4 (1.56) | 19 (2.0) |
| Completeness (%) | 99.8 (97.1) | 100.0 (100.0) |
| Redundancy | 21.1 (7.6) | 20.8 (9.7) |
| Refinement | ||
| Resolution (Å) | 44.93–2.05 | 49.45–2.50 |
| No. of reflections | 118,595 | 65,599 |
| Rwork/Rfree | 18.3/20.1 | 17.4/20.5 |
| No. of atoms | ||
| Protein | 7,060 | 7,060 |
| Ligand/ion | 292 | 312 |
| Water | 467 | 470 |
| B factors (Å2) | ||
| Protein | 47.7 | 37.6 |
| Ligand/ion | 68.4 | 67.8 |
| Water | 50.1 | 40.2 |
| RMSD | ||
| Bond lengths (Å) | 0.003 | 0.003 |
| Bond angles (°) | 0.796 | 0.681 |
| Sugars in the minimal-energy conformation (%) | 90.0 | 90.9 |
CC1/2, Pearson correlation coefficient between two random half data sets; RMSD, root mean square deviation.
Single-crystal X-ray diffraction data were collected for each structure. Values in parentheses are for the highest-resolution shell.
In the previously reported crystal structure of MuV-HN bound to 3′-SL (23), the Sia-1 of 3′-SL interacts with the active-site residues, Arg180, Glu407, Arg422, Arg512, and Tyr540, along with Lys242, Glu264, and Tyr323 of the MuV-HN protein. Additionally, the Glc-3 of 3′-SL interacts with the residues Tyr369 and Val476 of MuV-HN to stabilize the protein-glycan interactions (Fig. 2A and B). The crystal structures demonstrated that these interactions are completely conserved in the binding with SLeX pentaose or GM2-GS (Fig. 2B) although the third sugar in SLeX pentaose is in the acetylated form, GlcNAc-3. Interestingly, the branched fucose (Fuc-3′) in SLeX pentaose interacts with Tyr369 of MuV-HN where the oxygen atom in the Fuc-3′ ring forms a hydrogen bond with the Oη oxygen atom of Tyr369 (Fig. 2B, top). GM2-GS also shows the unique interactions between the branched GalNAc (GalNAc-2′) and MuV-HN (Fig. 2B, middle). The O-4 and O-6 oxygen atoms of GalNAc-2′ form hydrogen bonds with the side chain of Glu569 and the side chains of Ser539 and Arg512 of MuV-HN, respectively.
FIG 2.
Interactions between the glycans and MuV-HN protein. (A) Overall structure of the MuV-HN−SLeX pentaose complex structure (left) and schematic of the glycan receptor derivatives for MuV (right). (B) Binding site of SLeX pentaose, GM2-GS, and 3′-SL in the MuV-HN−glycan complex structures. SLeX pentaose, GM2-GS, and 3′-SL are shown in cyan. Top (left) and side (middle) views are shown. The omit Fo – Fc (the observed and calculated structure factor amplitude, respectively) maps (3.0σ) for SLeX pentaose and GM2-GS are also shown (right). The MuV-HN residues directly involved in the interaction with Sia-1 are shown in yellow, whereas the ones interacting with the non-Sia-1 moieties of the glycans are indicated by magenta. Nitrogen atom, blue; oxygen atom, red.
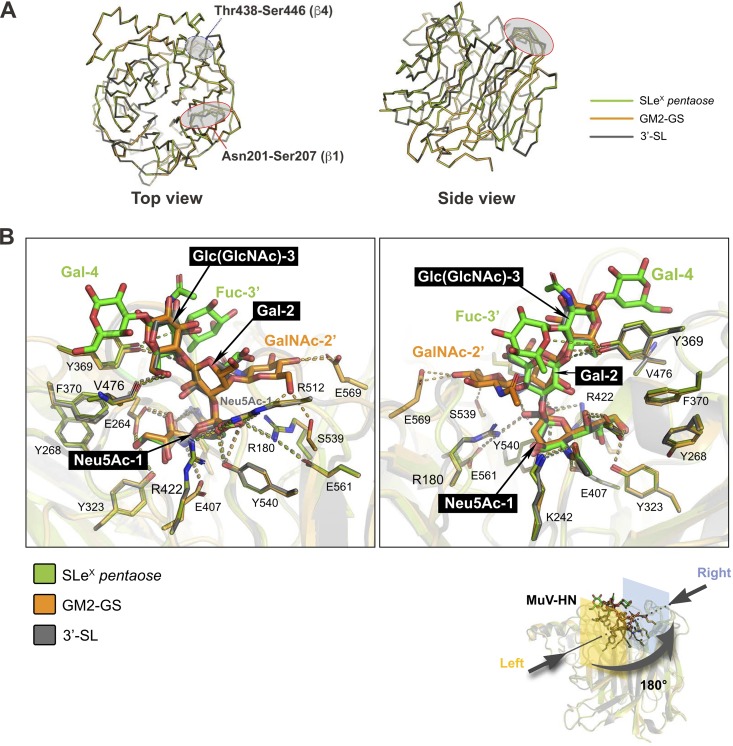
Structural basis for the flexible recognition of glycan receptors by MuV-HN.
Upon binding with SLeX pentaose, GM2-GS, or 3′-SL, no large structural differences were observed among the glycan-bound forms of MuV-HN (Fig. 3A). The residues ranging from Asn201 to Ser207 and from Thr438 to Ser446 in the interstrand loops of the β1 and β4 sheets of MuV-HN, respectively, showed relatively high flexibility. While the branched Fuc-3′ in SLeX pentaose and the GalNAc-2′ in GM2-GS individually formed unique interactions with the MuV-HN residues, as described in the above subsection, the core trisaccharide moiety in theses glycans had the same spatial configuration of 3′-SL (Fig. 3B). Thus, the additional modifications, such as Fuc-3′ or GalNAc-2′, support the binding of the receptors with MuV-HN without affecting the core trisaccharide configuration.
FIG 3.
Structures of MuV-HN in complex with SLeX pentaose or GM2-GS superimposed on the MuV-HN − 3′-SL complex. (A) Crystal structures of the SLeX pentaose-bound and the GM2-GS-bound MuV-HN proteins superimposed on the 3′-SL -bound MuV-HN. Only the main chains of the MuV-HN proteins are shown. (B) Crystal structures of the MuV-HN-bound SLeX pentaose and the MuV-HN-bound GM2-GS superimposed on the MuV-HN-bound 3′-SL. Right and left are the views of each rotated by 180°. Superimposition was performed for the core trisaccharide structure containing α2,3-linked sialic acid based on the root mean square deviation.
Inhibitory effect of glycans against MuV entry.
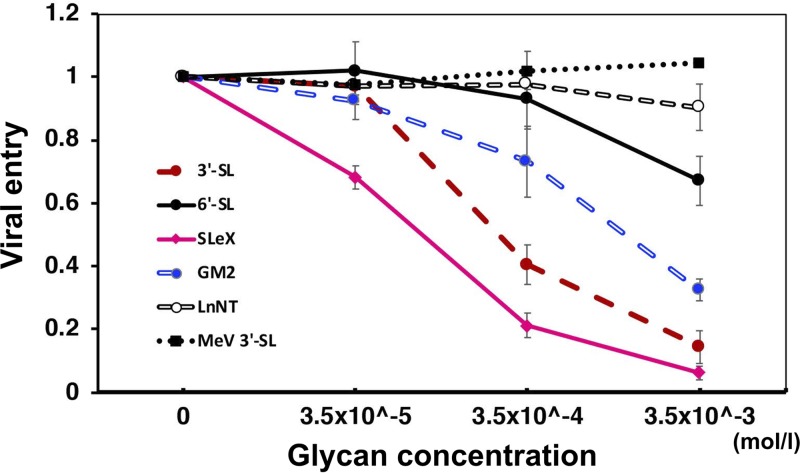
To assess the interaction of MuV virions with the glycans and the inhibitory potential of the glycans against MuV infection, enhanced green fluorescent protein (EGFP)-expressing recombinant MuV was preincubated with 3′-SL, SLeX pentaose, GM2-GS, LnNT, or 6′-SL at several concentrations created by serial dilutions and then used to infect Vero cells. EGFP-expressing recombinant MeV, bearing the hemagglutinin of the MeV Edmonston strain, was similarly preincubated with 3′-SL and then used to infect Vero cells as a negative control. The infected cells were overlaid with an agar-containing medium. The inhibitory effects of these glycans against MuV or MeV entry were evaluated by counting plaques at 2 to 3 days postinfection (dpi). Three binding motifs, SLeX pentaose, 3′-SL, and GM2-GS, significantly inhibited MuV entry, with IC50s of 78 μM, 270 μM, and 1.40 mM, respectively (Fig. 4). SLeX pentaose showed the best inhibitory effect, and the viral entry was greatly suppressed, especially at the higher glycan concentrations. In contrast, minimal inhibitory effects of LnNT and 6′-SL against MuV entry were observed. This result was compatible with the lack of electron densities for LnNT and 6′-SL in the complex structures with MuV-HN. LnNT might have nonspecifically bound to the MuV-HN protein in the glycan array assay with the high concentration of the MuV-HN protein.
FIG 4.
Inhibitory effect of the glycans against MuV entry. The GFP-recombinant MuV (1.2 × 102 PFU) was preincubated with the serially diluted 3′-SL, 6′-SL, LnNT, GM2-GS, or SLeX pentaose. Subsequently, Vero cells were infected with one of the viral combinations for 1 h at 37°C. After the aspiration of the viral solution, the infected cells were washed with phosphate-buffered saline and overlaid with 0.75% agar for the plaque assay. The EGFP-expressing recombinant MeV containing the Edmonston H protein was also preincubated with 3′-SL and then used in a similar fashion to infect Vero cells as a negative control. The viral entry was evaluated by counting GFP-positive plaques using fluorescence microscopy at 3 dpi for MuV and at 2 dpi for MeV (plaque sizes of MeV under the condition used were too large to count accurately at 3 dpi). The plaque numbers obtained from MuV or MeV preincubated with glycan-free medium are set to 1. Data are the means ± standard deviations of three samples. Data shown in this figure are representative of three independently performed experiments.
DISCUSSION
MuV causes systemic infection in humans and shows unique tropism for glandular tissues and the central nervous system, causing epidemic parotitis, meningitis, encephalitis, and deafness. However, the molecular mechanism of the unique MuV tropism is not well explained. To understand the unique MuV tropism, we investigated glycan receptor motifs for MuV using a glycan array screen from CFG Core H resources including 600 types of glycans. A large-scale glycan array analysis with viral attachment proteins is very useful to find new receptor motifs, but there are also limitations, for example, nonspecific binding, a nonlinear dose-response relationship because of binding saturation, and different optimal binding conditions for individual glycans. Indeed, MuV-HN proteins of Hoshino and SBL-1 strains showed partially different binding results in the array. To overcome the limitations attributed to single experimental methods, we employed the combined strategy of a large-scale glycan array, structural analysis, and a viral infection inhibition assay. These three different methods gave consistent results, allowing us to identify extensive/additional glycan receptor motifs for MuV. Based on the complex structure of MuV-HN protein and the glycan receptors, we further revealed the molecular mechanism that underlies the flexible recognition of glycan receptors by MuV.
Interestingly, fucose- or GalNAc-modified trisaccharides containing α2,3-linked sialic acid, SLeX, and GM2/Sda-glycan were identified as new MuV binding motifs. Both SLeX and GM2/Sda-glycan share a core structure with the previously reported 3′-SL (3′-SLN) motif. Thus, this core trisaccharide structure is likely critical for the receptor recognition by MuV. The MuV-HN Y369A mutant protein showed loss of binding ability to all glycans in the array (Fig. 1F). This result also supports the idea that the interaction between the Tyr369 residue of MuV-HN and the Glc/GlcNAc-3 moiety of the core glycan structure is crucial for the receptor recognition by MuV. Branched sugar chains showed reduced binding to the MuV-HN proteins, especially in more than triantennary glycans (Fig. 1A to D and Table 1). This may be caused by steric hindrance, as also discussed in our previous report (23).
In the crystal structures, SLeX and GM2-GS formed additional interactions with the residues of the MuV-HN protein compared with those of 3′-SL (Fig. 2). Nonetheless, all three glycan receptors exhibited the same binding configurations by maintaining all the linkages observed in 3′-SL (Fig. 3B). This observation indicates that the core glycan structure, Neu5Acα2-3Galβ1-4GlcNAc(Glc), is essential for the receptor recognition by the MuV-HN protein, while fucose or GalNAc modifications onto the core are spatially allowed to be positioned in the extra space of the receptor-binding pocket of the MuV-HN protein. The interstrand loops, ranging from Asn201 to Ser207 and Thr438 to Ser446 of the β1 and β4 sheets of MuV-HN, respectively, may partially contribute to the adjustment of the space of the receptor-binding pocket of the MuV-HN protein because these loops change their conformations depending on the ligands bound to the pocket (Fig. 3A).
SLeX constitutes the epitope of the carbohydrate antigen typically found in leukocytes and plays a role in cell attachment by being recognized by selectins expressed in vascular endothelia (25–27). SLeX is also one of the tumor-associated carbohydrate antigens found in various types of cancers, and its expression level is known to correlate with the development of metastatic lesions based on the cell adhesion mechanism mediated by selectins (25, 28). The SLeX structure is generally found on glycoproteins expressed in human tissues such as colon, heart, kidney, lung, and pancreas (http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh.shtml). It also exists as the terminal structure of some highly O-glycosylated mucins and sphingoglycolipids. 3′-SL (Neu5Acα2-3Galβ1-4Glc) constitutes the terminal structure of the sphingoglycolipid known as GM3, which is one of the main gangliosides in vertebrate extraneural tissues (29). 3′-SLN (Neu5Acα2-3Galβ1-4GlcNAc) exists as the terminal sugar of N-linked and O-linked glycoproteins (20, 23, 30). These broad distributions of SLeX, 3′-SL, and 3′-SLN may be the cause of the systemic infection by MuV. In contrast, GM2 exists as a component of glycolipids in the mammalian central nervous system, peripheral nerves, and adrenal gland (25, 31, 32). Thus, the distribution of GM2 may be related to the observed MuV tropism to the nervous tissues. MuV entry was efficiently inhibited by pretreating the virus with 3′-SL, SLeX pentaose, and GM2-GS (Fig. 4). This result strongly suggests that the glycans with the SLeX motif and the GM2-glycan motif as well as the core trisaccharide motif work as the functional receptors for MuV. The receptor function of the GM2-glycan motif may be dependent on the environment, such as the focal viral titer or the receptor density in the affected tissues or organs, because the GM2-GS concentration is required to be relatively high to achieve the equivalent inhibitory effect against MuV entry compared with that of 3′-SL or SLeX pentaose (Fig. 4). By comparison, the nonsialylated glycan motifs may not be efficient receptors for MuV because of the lack of electron density for LnNT in the complex structure with the MuV-HN protein and the absence of an inhibitory effect of LnNT against MuV entry (Fig. 4). Distributions of these glycan receptor motifs may account for the systemic infection as well as the unique MuV tropism for the nervous tissues.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Wako) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Sigma-Aldrich) and penicillin-streptomycin (Gibco) at 37°C and 5% CO2. The GFP-expressing recombinant MuV of the Hoshino strain (33) (MuV-GFP) and the EGFP-expressing recombinant measles virus of the IC323 strain (34), the hemagglutinin of which was replaced by that of the Edmonston strain (35) (MeV-EGFP [Edmonston H]), were recovered from their respective full-length cDNA plasmids using the support plasmids encoding the nucleocapsid and phospho- and large proteins of MuV and MeV, respectively. The Edmonston strain of MeV infects Vero cells via CD46 as a receptor, and MeV-EGFP (Edmonston H) was used as a control to infect Vero cells.
Protein expression and purification.
The expression plasmids encoding the MuV-HN proteins (amino acid positions 96 to 582) (23) of the wild-type (WT) Hoshino strain, WT SBL-1 strain, and Y369A mutant of the SBL-1 strain were transfected into HEK293S cells lacking N-acetylglucosaminyltransferase I [293S GnTI(−) cells] (36) using polyethyleneimine-MAX (Polysciences, Inc.). MuV-HN proteins were expressed as recombinant proteins containing the N-terminal secretion signal sequence and a C-terminal His6 tag sequence using the expression vector pHLsec (37, 38). At 6 days posttransfection, the supernatant containing the secreted MuV-HN protein was harvested and then centrifuged to eliminate cell components. MuV-HN proteins were purified using a Ni2+-nitrilotriacetic acid (NTA) affinity column (Cosmogel His-Accept; Nacalai Tesque) in purification buffer (50 mM NaH2PO4, 150 mM NaCl, 10 mM imidazole, pH 8.0) and then eluted with elution buffer (50 mM NaH2PO4, 150 mM NaCl, 500 mM imidazole, pH 8.0). The eluted MuV-HN proteins were further purified using a size exclusion column (Superdex 200 Increase GL 10/300; GE Healthcare) in buffer containing 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, and 2 mM MgCl2. MeV-H protein (amino acid positions 149 to 617) of the Edmonston strain (38) was also expressed and purified according to the same procedures described above for the MuV-HN proteins.
Glycan array analysis.
Purified MuV-HN proteins and MeV-H protein were diluted in binding buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween 20, and 1% bovine serum albumin [BSA]) and then sent to CFG’s Protein-Glycan Interaction Core (Core H [ http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh.shtml]). Purified viral glycoproteins were used with the Mammalian Glycan Array, version 5.3, at 5 μg/ml or 50 μg/ml. Binding of viral glycoproteins containing the C-terminal His6 tag sequence with glycans was evaluated by quantifying the fluorescence intensity using an anti-His6 tag Cy3-conjugated antibody. Six replicates of each glycan were analyzed, and the highest and the lowest signals were eliminated from the mean value (in relative fluorescence units [RFU]). The analysis of the SBL-1 WT HN protein at 5 μg/ml and 50 μg/ml was repeated to confirm the data. The obtained data were also analyzed using GlycoPattern (24) (https://glycopattern.emory.edu/index.html).
Crystallization, data collection, and structure determination.
To promote the crystallization of the MuV-HN protein, it was treated with 20 mM dimethylamine-borane complex (Sigma) and 40 mM formaldehyde at 4°C overnight to methylate lysine residues (39). Cocrystals with the 3′-SL sodium salt (Tokyo Chemical Industry), SLeX pentaose (Oligotech), GM2 ganglioside sugar (GM2-GS; Oligotech), or LnNT (Oligotech) were grown by hanging-drop vapor diffusion at 20°C in a drop containing 1 μl each of MuV-HN (10 mg/ml) and the crystallization buffer (1.95 M ammonium sulfate, 3% [vol/vol] glycerol, and 0.1 M sodium acetate, pH 5.0) containing 5- to 50-fold-higher molar concentrations of glycans than the concentration of the MuV-HN protein. For the data collection, the crystals were cryo-cooled (by a nitrogen gas stream; 100 K) in the original crystallization buffer containing 25% (vol/vol) glycerol. Crystals of MuV-HN bound to GM2-GS and SLeX pentaose were diffracted to 2.5 Å and 2.05 Å, respectively.
All of the diffraction data sets were collected on beamline BL26B2 (using an MX225 detector) at SPring-8 (Harima, Japan) and processed and scaled using HKL2000 (40). Structures of the MuV-HN head domain bound to GM2-GS and SLeX pentaose were solved by molecular replacement with Phaser (41) using the apo MuV-HN structure as a search model (PDB accession code 5B2C) (23). Further model refinement procedures were carried out using Phenix.refine (42). Interactive manual model building and correction were performed using Coot (43). Sugars were validated by the energy conformations using Privateer (44). Final structures of MuV-HN bound to GM2-GS or SLeX pentaose were refined to an Rwork of 18.2% and Rfree of 20.1% or to an Rwork of 17.5% and Rfree of 20.5%, respectively. Of the refined structure, 95.3% of residues were in favored regions, and 4.4% of the residues were in allowed regions in the Ramachandran plot for MuV-HN bound to GM2-GS. Of the refined structure, 95.1% of residues were in favored regions, and 4.6% of the regions were in allowed regions in the Ramachandran plot for MuV-HN bound to SLeX pentaose. Detailed data collection and crystallographic statistics are summarized in Table 2. Figures were produced using PyMOL (DeLano Scientific, LLC [http://www.pymol.org]).
Virus entry inhibition assay.
GFP-expressing recombinant MuV of the Hoshino strain was preincubated with the medium containing the serially diluted 3′-SL, SLeX pentaose, GM2-GS, 6′-SL, or LnNT at 37°C for 60 min. Vero cells cultured in a 96-well dish (Corning) were infected with the viruses (multiplicity of infection [MOI] of 0.01) for 60 min at 37°C. After the virus-containing medium was removed, cells were overlaid with 0.75% (w/vol) Sea Kem ME Agar (Invitrogen)-DMEM supplemented with 5% (vol/vol) FBS, 0.16% (w/vol) NaHCO3, and penicillin-streptomycin at 37°C and 5% CO2. MeV-EGFP (Edmonston H) was also preincubated with the serially diluted 3′-SL as a negative control and then was assayed using the same procedure as that employed for MuV. The plaque numbers obtained from MuV- or MeV-infected cells in the presence/absence of glycans were counted under a microscope (Keyence) at 2 to 3 dpi.
Data availability.
Coordinates for MuV-HN–SLeX pentaose and MuV-HN−GM2-GS have been deposited in the Protein Data Bank under accession codes 6JJN and 6JJM, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kaoru Takeuchi and Hiroaki Tokiwa for invaluable discussions.
We thank the Protein-Glycan Interaction Resource of the CFG (supporting grant R24 GM098791) and the National Center for Functional Glycomics (NCFG) at the Beth Israel Deaconess Medical Center and Harvard Medical School (supporting grant P41 GM103694) for glycan array resources and data analysis. The synchrotron radiation experiments were performed at BL26B2 in SPring-8 with the approval of RIKEN (proposal no. 20170098).
This study was supported by a grant for the Research Program on Emerging and Reemerging Infectious Diseases from AMED (JP18fk0108014h; to T.H.), by the Takeda Science Foundation (T.H.), by the Terumo Foundation for Life Sciences and Arts (T.H.), by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (24115005 and 17H05820; to Y.Y.), by AMED Platform Project Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS), and by a Japan Society for the Promotion of Science Postdoctoral Fellowship DC2 (M.K.).
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00344-19.
REFERENCES
- 1.Rubin SA, Sauder CJ, Carbone KM. 2013. Mumps virus, p 1024–1041. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6 ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.McLean HQ, Fiebelkorn AP, Temte JL, Wallace GS, Centers for Disease Control and Prevention. 2013. Prevention of measles, rubella, congenital rubella syndrome, and mumps, 2013: summary recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 62(RR-04):1–34. [PubMed] [Google Scholar]
- 3.Barskey AE, Glasser JW, LeBaron CW. 2009. Mumps resurgences in the United States: a historical perspective on unexpected elements. Vaccine 27:6186–6195. doi: 10.1016/j.vaccine.2009.06.109. [DOI] [PubMed] [Google Scholar]
- 4.Latner DR, Hickman CJ. 2015. Remembering mumps. PLoS Pathog 11:e1004791. doi: 10.1371/journal.ppat.1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto H, Fujioka M, Kinumaki H, Kinki Ambulatory Pediatrics Study Group. 2009. An office-based prospective study of deafness in mumps. Pediatr Infect Dis J 28:173–175. doi: 10.1097/INF.0b013e31818a8ca8. [DOI] [PubMed] [Google Scholar]
- 6.Hviid A, Rubin S, Muhlemann K. 2008. Mumps. Lancet 371:932–944. doi: 10.1016/S0140-6736(08)60419-5. [DOI] [PubMed] [Google Scholar]
- 7.Cohen BE, Durstenfeld A, Roehm PC. 2014. Viral causes of hearing loss: a review for hearing health professionals. Trends Hear 18:2331216514541361. doi: 10.1177/2331216514541361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb RA, Parks GD. 2013. Paramyxoviridae, p 957–995. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6 ed, vol I Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Wright PF, Neumann G, Kawaoka Y. 2013. Orthomyxoviruses, p 1186–1243. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6 ed, vol I Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Viswanathan K, Chandrasekaran A, Srinivasan A, Raman R, Sasisekharan V, Sasisekharan R. 2010. Glycans as receptors for influenza pathogenesis. Glycoconj J 27:561–570. doi: 10.1007/s10719-010-9303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulson BS. 2015. Expanding diversity of glycan receptor usage by rotaviruses. Curr Opin Virol 15:90–96. doi: 10.1016/j.coviro.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Mietzsch M, Broecker F, Reinhardt A, Seeberger PH, Heilbronn R. 2014. Differential adeno-associated virus serotype-specific interaction patterns with synthetic heparins and other glycans. J Virol 88:2991–3003. doi: 10.1128/JVI.03371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neu U, Maginnis MS, Palma AS, Stroh LJ, Nelson CD, Feizi T, Atwood WJ, Stehle T. 2010. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe 8:309–319. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tappert MM, Porterfield JZ, Mehta-D'Souza P, Gulati S, Air GM. 2013. Quantitative comparison of human parainfluenza virus hemagglutinin-neuraminidase receptor binding and receptor cleavage. J Virol 87:8962–8970. doi: 10.1128/JVI.00739-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stencel-Baerenwald JE, Reiss K, Reiter DM, Stehle T, Dermody TS. 2014. The sweet spot: defining virus-sialic acid interactions. Nat Rev Microbiol 12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen MB, Sabesan S, Skehel JJ, Wiley DC. 1997. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology 232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 17.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 18.Xiong X, McCauley JW, Steinhauer DA. 2014. Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr Top Microbiol Immunol 385:63–91. doi: 10.1007/82_2014_423. [DOI] [PubMed] [Google Scholar]
- 19.Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. 2008. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol 26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 20.Sriwilaijaroen N, Nakakita SI, Kondo S, Yagi H, Kato K, Murata T, Hiramatsu H, Kawahara T, Watanabe Y, Kanai Y, Ono T, Hirabayashi J, Matsumoto K, Suzuki Y. 2018. N-glycan structures of human alveoli provide insight into influenza A virus infection and pathogenesis. FEBS J 285:1611–1634. doi: 10.1111/febs.14431. [DOI] [PubMed] [Google Scholar]
- 21.Hiono T, Okamatsu M, Igarashi M, McBride R, de Vries RP, Peng W, Paulson JC, Sakoda Y, Kida H. 2016. Amino acid residues at positions 222 and 227 of the hemagglutinin together with the neuraminidase determine binding of H5 avian influenza viruses to sialyl Lewis X. Arch Virol 161:307–316. doi: 10.1007/s00705-015-2660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neu U, Woellner K, Gauglitz G, Stehle T. 2008. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc Natl Acad Sci U S A 105:5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubota M, Takeuchi K, Watanabe S, Ohno S, Matsuoka R, Kohda D, Nakakita SI, Hiramatsu H, Suzuki Y, Nakayama T, Terada T, Shimizu K, Shimizu N, Shiroishi M, Yanagi Y, Hashiguchi T. 2016. Trisaccharide containing α2,3-linked sialic acid is a receptor for mumps virus. Proc Natl Acad Sci U S A 113:11579–11584. doi: 10.1073/pnas.1608383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agravat SB, Saltz JH, Cummings RD, Smith DF. 2014. GlycoPattern: a web platform for glycan array mining. Bioinformatics 30:3417–3418. doi: 10.1093/bioinformatics/btu559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. 2009. Essentials of glycobiology, 2nd ed Cold Spring Harbor Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 26.Phillips ML, Nudelman E, Gaeta FC, Perez M, Singhal AK, Hakomori S, Paulson JC. 1990. ELAM-1 mediates cell adhesion by recognition of a carbohydrate ligand, sialyl-Lex. Science 250:1130–1132. doi: 10.1126/science.1701274. [DOI] [PubMed] [Google Scholar]
- 27.McEver RP. 2002. Selectins: lectins that initiate cell adhesion under flow. Curr Opin Cell Biol 14:581–586. doi: 10.1016/S0955-0674(02)00367-8. [DOI] [PubMed] [Google Scholar]
- 28.Kannagi R, Izawa M, Koike T, Miyazaki K, Kimura N. 2004. Carbohydrate-mediated cell adhesion in cancer metastasis and angiogenesis. Cancer Sci 95:377–384. doi: 10.1111/j.1349-7006.2004.tb03219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prokazova NV, Samovilova NN, Gracheva EV, Golovanova NK. 2009. Ganglioside GM3 and its biological functions. Biochemistry (Mosc) 74:235–249. doi: 10.1134/S0006297909030018. [DOI] [PubMed] [Google Scholar]
- 30.Walther T, Karamanska R, Chan RW, Chan MC, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, Haslam SM, Nicholls JM. 2013. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog 9:e1003223. doi: 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikami T, Kashiwagi M, Tsuchihashi K, Daino T, Akino T, Gasa S. 1998. Further characterization of equine brain gangliosides: the presence of GM3 having N-glycolyl neuraminic acid in the central nervous system. J Biochem 123:487–491. doi: 10.1093/oxfordjournals.jbchem.a021962. [DOI] [PubMed] [Google Scholar]
- 32.Sandhoff R, Sandhoff K. 2018. Emerging concepts of ganglioside metabolism. FEBS Lett 592:3835–3864. doi: 10.1002/1873-3468.13114. [DOI] [PubMed] [Google Scholar]
- 33.Ninomiya K, Kanayama T, Fujieda N, Nakayama T, Komase K, Nagata K, Takeuchi K. 2009. Amino acid substitution at position 464 in the haemagglutinin-neuraminidase protein of a mumps virus Urabe strain enhanced the virus growth in neuroblastoma SH-SY5Y cells. Vaccine 27:6160–6165. doi: 10.1016/j.vaccine.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Takeda M, Takeuchi K, Miyajima N, Kobune F, Ami Y, Nagata N, Suzaki Y, Nagai Y, Tashiro M. 2000. Recovery of pathogenic measles virus from cloned cDNA. J Virol 74:6643–6647. doi: 10.1128/jvi.74.14.6643-6647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enders JF, Peebles TC. 1954. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc Soc Exp Biol Med 86:277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- 36.Reeves PJ, Callewaert N, Contreras R, Khorana HG. 2002. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A 99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aricescu AR, Assenberg R, Bill RM, Busso D, Chang VT, Davis SJ, Dubrovsky A, Gustafsson L, Hedfalk K, Heinemann U, Jones IM, Ksiazek D, Lang C, Maskos K, Messerschmidt A, Macieira S, Peleg Y, Perrakis A, Poterszman A, Schneider G, Sixma TK, Sussman JL, Sutton G, Tarboureich N, Zeev-Ben-Mordehai T, Jones EY. 2006. Eukaryotic expression: developments for structural proteomics. Acta Crystallogr D Biol Crystallogr 62:1114–1124. doi: 10.1107/S0907444906029805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A 104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. 1993. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Agirre J, Iglesias-Fernandez J, Rovira C, Davies GJ, Wilson KS, Cowtan KD. 2015. Privateer: software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol 22:833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates for MuV-HN–SLeX pentaose and MuV-HN−GM2-GS have been deposited in the Protein Data Bank under accession codes 6JJN and 6JJM, respectively.