HSV entry is a multistep process that requires the actions of four glycoproteins, gD, gH/gL, and gB. Our current model predicts that gD must interact with both its receptor and gH/gL to promote viral entry. Although we know a great deal about how gD binds its receptors, until now we have been unable to demonstrate a direct interaction between gD and gH/gL. Here, we used a highly sensitive surface plasmon resonance technique to clearly demonstrate that gD and gH/gL interact. Furthermore, using multiple MAbs with defined epitopes, we have delineated a domain on gD that is independent of that used for receptor binding and which likely represents the gH/gL interaction domain. Targeting this interaction to prevent fusion may enhance both therapeutic and vaccine strategies.
KEYWORDS: glycoproteins, herpes simplex virus, surface plasmon resonance, virus entry
ABSTRACT
Herpes simplex virus (HSV) requires fusion between the viral envelope and host membrane. Four glycoproteins, gD, gH/gL, and gB, are essential for this process. To initiate fusion, gD binds its receptor and undergoes a conformational change that hypothetically leads to activation of gH/gL, which in turn triggers the fusion protein gB to undergo rearrangements leading to membrane fusion. Our model predicts that gD must interact with both its receptor and gH/gL to promote fusion. In support of this, we have shown that gD is structurally divided into two “faces”: one for the binding receptor and the other for its presumed interaction with gH/gL. However, until now, we have been unable to demonstrate a direct interaction between gD and gH/gL. Here, we used surface plasmon resonance to show that the ectodomain of gH/gL binds directly to the ectodomain of gD when (i) gD is captured by certain anti-gD monoclonal antibodies (MAbs) that are bound to a biosensor chip, (ii) gD is bound to either one of its receptors on a chip, and (iii) gD is covalently bound to the chip surface. To localize the gH/gL binding site on gD, we used multiple anti-gD MAbs from six antigenic communities and determined which ones interfered with this interaction. MAbs from three separate communities block gD-gH/gL binding, and their epitopes encircle a geographical area on gD that we propose comprises the gH/gL binding domain. Together, our results show that gH/gL interacts directly with gD, supporting a role for this step in HSV entry.
IMPORTANCE HSV entry is a multistep process that requires the actions of four glycoproteins, gD, gH/gL, and gB. Our current model predicts that gD must interact with both its receptor and gH/gL to promote viral entry. Although we know a great deal about how gD binds its receptors, until now we have been unable to demonstrate a direct interaction between gD and gH/gL. Here, we used a highly sensitive surface plasmon resonance technique to clearly demonstrate that gD and gH/gL interact. Furthermore, using multiple MAbs with defined epitopes, we have delineated a domain on gD that is independent of that used for receptor binding and which likely represents the gH/gL interaction domain. Targeting this interaction to prevent fusion may enhance both therapeutic and vaccine strategies.
INTRODUCTION
Herpes simplex virus (HSV) entry into a cell requires four glycoproteins, gD, gH/gL, and gB, to promote fusion between the viral envelope and host membrane (1–4). In our ongoing studies to determine the mechanistic role for these proteins in viral entry, we have developed a model that posits that fusion is driven by a cascade of protein-protein interactions, initiated by the binding of gD to one of its receptors, HVEM or nectin-1 (5–7). Such binding induces a conformational change in gD (8) that somehow allows it to communicate with gH/gL (9–11), thereby activating gH/gL into a form that in turn triggers the fusogenic activity of gB (12–14).
Crystal structures of gD with and without the cellular receptors HVEM and nectin-1 (8, 15–18) show that the C terminus of the gD ectodomain “tail” must vacate the receptor binding site before the interaction with the receptor can occur, demonstrating that there is a necessary structural change in gD required to initiate fusion. We postulated that this conformational change in gD structure and/or the subsequent exposure of the underlying residues renders gD capable of interacting with and activating the gH/gL heterodimer (9–11). In support, we have shown that gD is structurally divided into two “faces”: one for receptor binding and the other for its presumed interaction with gH/gL. However, despite numerous studies, both from our laboratory and others (19–25), that indirectly support the ability of gD to interact with gH/gL, before now we have been unable to show that gD and gH/gL bind to each other directly.
The critical requirement for an interaction between gD and gH/gL in viral fusion is supported by several studies. First, in Epstein-Barr virus (EBV), cryoelectron microscopy and crystallography have revealed the structure of a gp42-gH/gL complex (26, 27). Like gD, gp42 is a receptor binding protein for EBV and is considered a functional (albeit not structural) homologue of gD. Second, complexes of gH/gL with other viral proteins have been visualized for human cytomegalovirus (28, 29). Third, when gB, gD, and gH/gL were swapped between HSV-1 and the closely related saimiriine herpesvirus 1 (SaHV-1), cell-cell fusion was observed only when gD and gH/gL were both from the same virus, suggesting that the two proteins interact with each other in a species-specific manner (19, 20). Fourth, gH/gL and gD were coimmunoprecipitated from transfected and HSV-infected cells as well as from virions (21, 25), suggesting, but not directly demonstrating, that such interactions took place during fusion.
Our laboratory has tried several ways to examine this issue. First, we found that gD and gH are sufficiently close together in HSV virions to be cross-linked to each other (24). Second, we used a bimolecular complementation assay in which complementary halves of the yellow fluorescent protein (YFP) were engineered onto the C termini of the genes of gD and gH (30). We found that when cells were transfected with plasmids expressing half-YFP-tagged gD and half-YFP-tagged gH/gL, YFP fluorescence was restored (22, 30), indicating an interaction between gD and gH. However, when coexpressed with gB and a gD receptor, this YFP-tagged complex was not functional for fusion (30). We proposed that the two half-YFP tags stabilized a normally transient interaction between gD and gH/gL but that cell-cell fusion was blocked because gH/gL was not released from gD for the next step in the pathway (presumably its interaction with and activation of gB).
In this study, we readdressed this issue using surface plasmon resonance (SPR) as the most sensitive way to determine if there is even a transient interaction between gD and gH/gL. We bound anti-gD MAbs to a biosensor chip to capture soluble gD and then added soluble gH/gL. We used forms of each protein that would maximize even a modest interaction, i.e., gD2(285t) and gH2t/gL2 (12, 31). We looked for three things: (i) direct evidence of a gD-gH/gL interaction, (ii) evidence that this interaction has functional consequences, and (iii) data that would reveal portions of gD that constitute the site of interaction. To address points ii and iii, we used our panel of virus-neutralizing and fusion-blocking anti-gD monoclonal antibodies (MAbs) with known epitopes to observe which ones might prevent the interaction. Using this strategy, we found that gH/gL binds directly to gD. This binding is specific, as we found no interactions between gD and gB or between gB and gH/gL. Furthermore, this interaction is blocked by certain anti-gD MAbs that mapped to defined epitopes. These epitopes outlined a potential gH/gL binding region on gD that spanned a face adjacent to, but distinct from, the receptor binding site. Thus, our results show that gH/gL does indeed interact physically with gD, defining a critical role for this step in HSV entry.
RESULTS
gH/gL binds to gD when gD is immobilized via certain anti-gD MAbs.
Since we could only detect a direct interaction between gD and gH/gL by either cross-linking the proteins on the virion surface (24) or by stabilizing the endodomains of gD and gH/gL through bimolecular complementation (30), we postulated that the gD-gH/gL interaction was transient, as suggested by Heldwein (32). Therefore, we decided to use SPR, which monitors binding of proteins in real time and is sensitive enough to detect weak or transient interactions. To maximize our ability to detect such interactions, we chose the HSV-2 forms of gD and gH/gL, as we previously showed that HSV-2 forms are more than twice as efficient at activating gB than HSV-1 forms (12), potentially due to a tighter interaction between these proteins. In addition, we used the truncated form of the gD ectodomain [gD2(285t)], as the C-terminal ectodomain tail (amino acids 286 to 306) of the longer form [gD2(306t)] can obstruct receptor binding (8, 10, 31) and may also interfere with gD’s interaction with gH/gL (9). For gH/gL, we used a construct consisting of the gH2 ectodomain truncated just before the transmembrane region, coexpressed with full-length gL2 (gH2t/gL2). We examined each soluble glycoprotein as both a ligand (i.e., first protein added to the chip containing a coupled MAb) and as an analyte (i.e., second protein added).
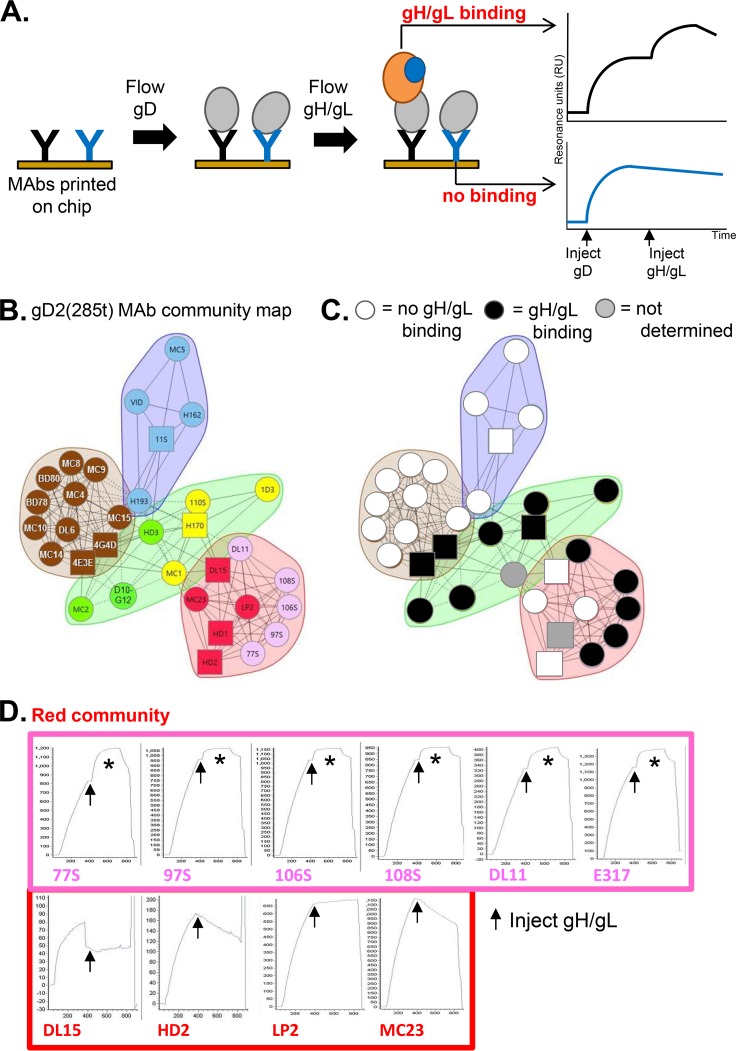
In our initial experiments, we used the Carterra continuous-flow microspotter (CFM)/IBIS SPR system (33–35) to print a large panel of previously characterized gD MAbs (33) onto an SPR chip (Fig. 1A). gD2(285t) then was flowed across this array, followed by gH2t/gL2. Our working hypothesis was that if a particular MAb captured gD and allowed gH/gL binding, it meant that the site for gD-gH/gL binding was not blocked by the MAb. Conversely, if there was no gH/gL binding, it might mean that the gH/gL binding site on gD was blocked. This system allowed us to evaluate 31 MAbs in one experiment.
FIG 1.
gD2(285t) binds gH2t/gL2 when gD is captured via certain anti-gD MAbs. (A) Graphical representation of the Carterra CFM/SPR protocol. A panel of anti-gD MAbs was printed to a CDM200M sensor chip. Soluble gD (light gray) next was flowed across the chip surface and captured by the MAbs. This can be seen on the binding curve as an increase in resonance units upon gD injection. Soluble gH/gL (orange and blue) then was flowed across the surface; some MAbs (e.g., black) allowed for the binding of gH/gL, while others (e.g., blue) did not. If gH/gL bound to gD, it generated a further increase in RU (black curve). If gH/gL did not bind, only a decrease in RU would be observed, representing the off-rate of the first glycoprotein-MAb complex (blue curve). Theoretical binding curves are shown on the right. (B) The gD2(285t) MAb community plot, as first illustrated in Cairns et al. (33). gD2(285t) was separated into four MAb communities: blue, brown, green, and red. The red community is subdivided into red and pink. The pink spheres within the red community highlight the group Ib MAbs. Likewise, the green community is subdivided into green and yellow. The yellow spheres/squares within the green community highlight group VII MAbs (N-terminal, linear epitopes). Spheres indicate that competition was measured as both a ligand and an analyte; squares indicate that competition was measured as either a ligand or an analyte only. Solid connecting lines specify that competition between the two MAbs was measured in both directions (each as a ligand and analyte). However, dashed connecting lines identify that the competition between MAbs was measured in one direction only. (Republished from PLOS Pathogens [33].) (C) Lensed view of the community map. Black, MAbs that do not permit gD-gH/gL binding. White, MAbs that permit gD-gH/gL binding. Gray, MAbs that exhibit partial gH/gL binding (see curves in panels D to G). Two MAbs, MC1 (yellow) and HD1 (red), did not yield binding data (MC1 exhibited poor binding to gD, while HD1 was sensitive to the acidic regeneration solution [33]). (D to G) gD and gH/gL binding curves. MAbs from each community/subcommunity are indicated by color. MAb E317 has been assigned to the pink subcommunity based on MAb competition, epitope mapping, and functional data (Table 1). The start of the gH/gL injection (arrow) is indicated on each curve. An asterisk indicates gH/gL binding (increase in RU upon gH/gL injection). This experiment was performed across the anti-gD MAb panel three times, with a single representative experiment shown for each MAb. x axis, time (seconds). y axis, RU.
The gD2(285t) community map (Fig. 1B) (33) illustrates the competitive relationships between the MAbs; those that reside in the same color community have epitopes that are located near one another. Real-time binding of gD2(285t) to a fixed MAb is illustrated in Fig. 1A by the initial increase in response units (RU). When we flow gH2t/gL2 across the gD-MAb complex, binding of gH/gL would be seen as a second increase in RU (Fig. 1A). The actual binding curves for gD-gH/gL as captured via each MAb are arranged by community color in Fig. 1D to G.
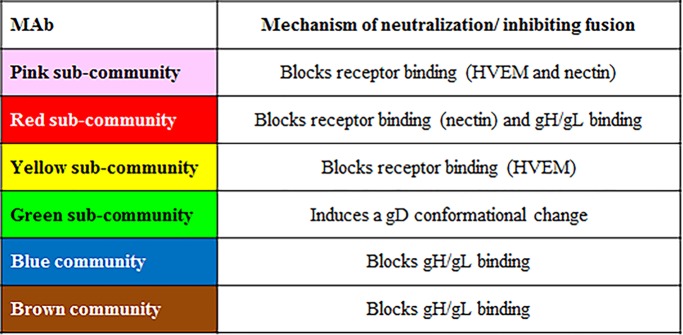
From previous work, we knew that all members of the red community strongly compete with each other for binding to gD, and each MAb has potent neutralizing activity (11, 33, 36, 37). This community is subdivided into red and pink subcommunities based on properties such as receptor blocking (nectin-1 only for red, both nectin-1 and HVEM for pink) (33). Here, we observed that when gD was presented via any one of the six pink subcommunity MAbs, there was a significant increase in RU upon addition of gH/gL, providing the first evidence that gD can directly bind gH/gL (Fig. 1D). In contrast, when gD was presented by any one of the four red subcommunity MAbs, there was no increase in RU upon injection of gH/gL (Fig. 1D), suggesting that these MAbs blocked gH/gL binding. Red-subcommunity MAbs block both gH/gL binding and receptor binding (11), suggesting that these MAbs bind to residues at the interface of these two interaction sites. Alternatively, red-subcommunity MAbs could induce a conformational change, preventing gH/gL binding.
The green community was also divided into subcommunities (yellow and green) based on epitope mapping and competition data (11, 33, 38). We detected a gD-gH/gL interaction when gD was captured by any of the members of the green/yellow communities (Fig. 1E). In contrast, when gD was captured via the four MAbs within the blue community (Fig. 1F) or nine of the eleven MAbs within the brown community (Fig. 1G), gH/gL failed to bind. The two exceptions within the brown community were MAbs 4E3E and 4G4D, both of which showed an increase in RUs upon gH/gL injection (Fig. 1G). These two MAbs are located on the periphery of the brown community that borders the green community (Fig. 1B), likely explaining why they behave more like green MAbs than brown MAbs in terms of gH/gL binding. However, their gH/gL binding curves exhibited a different pattern of gH/gL binding than what was seen with other MAbs (Fig. 1G). Instead of a rapid increase in RUs that quickly levels off (e.g., pink-community MAbs [Fig. 1D]), there is a slow, steady rise in RUs that continues throughout the injection (Fig. 1G). We propose that 4E3E and 4G4D partially block the gH/gL binding site on gD (Table 1).
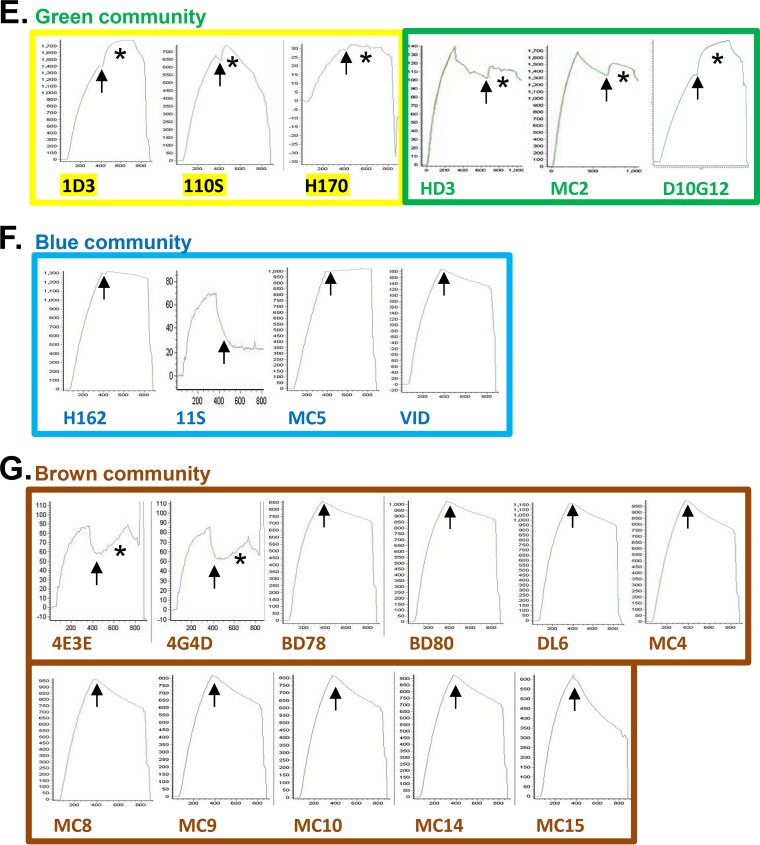
TABLE 1.
Properties of anti-gD MAbs
As the yellow-subcommunity MAbs bind to linear epitopes spanning residues 1 to 23 at the N terminus of gD (33, 39, 40), their ability to bind gH/gL binding suggests that the N terminus of gD is not involved in this interaction. At least one MAb within the green subcommunity (MC2) binds a discontinuous epitope located on the face of the gD structure adjacent to that of yellow MAb epitopes (11, 41), identifying another region that is not involved with gH/gL binding. By overlaying all the data onto the gD2(285t) MAb community map, we observed a striking picture of the localization of positive (black) and negative (white) gH/gL binding among the MAb communities (Fig. 1C). As gH/gL binding is allowed by all members of the green and pink communities, we conclude that these regions are not part of the interface between gD and gH/gL. Since these two groups of MAbs include ones that block HVEM or nectin-1 binding (or both), the data suggest that gH/gL binds a different face of gD than that used to bind receptors.
Complex formation is specific for gD and gH/gL, but only when gD is the ligand.
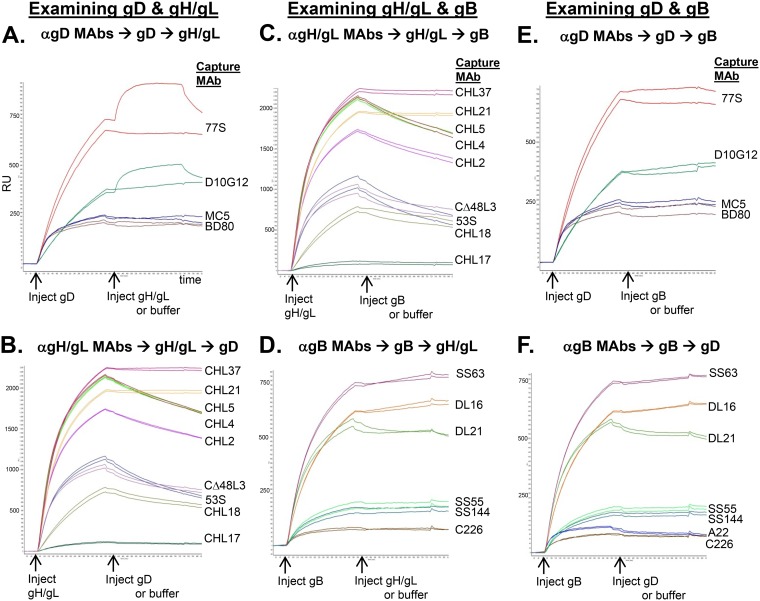
To test for the specificity of the gD-gH/gL interaction, we once again used the Carterra SPR system to analyze possible binding between different combinations of purified, soluble forms of gD, gH/gL, and the HSV fusion protein, gB (Fig. 2). Each glycoprotein was tested as both a ligand and an analyte. For each MAb used for capture, we show two curves: the control (glycoprotein 1 only) and the experimental (glycoproteins 1 and 2). We then stacked the curves for each MAb onto one graph, with the name of the MAbs used to capture glycoprotein 1 listed to the right. In the case of the control curve, we flowed buffer in lieu of a second glycoprotein in order to observe any decrease in binding of glycoprotein 1 to the capture MAb.
FIG 2.
Screening soluble HSV-2 glycoproteins for binding using CFM/SPR (IBIS MX96). Experiments were performed using the Carterra, Inc., CFM/SPRi system. (A and E) gD as the ligand. A panel of anti-gD MAbs was printed to a CDM200M sensor chip. Soluble gD next was flowed across the chip surface and captured by the MAbs. Soluble gH/gL (A), soluble gB (E), or buffer (negative-control curve for each printed MAb) then was flowed across the chip surface. The printed MAbs are listed to the right of each set of binding curves. Times of gD, gH/gL, gB, and buffer injections are indicated on the x axis by arrows. (B and C) Same protocol as that for panels A and E but with anti-gH/gL MAbs printed and gH/gL as the ligand (first injection). (D and F) Same protocol as that for panels A and E but with anti-gB MAbs printed and gB as the ligand. Each set of MAbs was tested a minimum of two times. x axis, time (seconds). y axis, RU.
As shown in Fig. 2A, gH/gL bound to gD when gD was captured by certain MAbs (e.g., 77S and D10G12) but not by others (e.g., BD80 and MC5). We next reversed the orientation by capturing gH/gL via an anti-gH/gL MAb (making it the ligand) and then flowing gD across the chip surface (making gD the analyte). For this experiment, we used multiple MAbs against gH/gL that bound to distinct, nonoverlapping epitopes across the protein (11, 33, 42–47). Interestingly, gH/gL was unable to bind to gD regardless of which anti-gH/gL MAb was used for capture (Fig. 2B). Therefore, we conclude that the binding between gD and gH/gL as detected via SPR is unidirectional, i.e., it occurs when gD is the ligand and gH/gL is the analyte (Fig. 2A) but not the reverse (Fig. 2B). Such results imply that gH/gL requires a conformational change to accommodate gD binding, and this cannot happen when gH/gL is immobilized by the capturing MAbs. Another possible explanation is that all of the anti-gH/gL MAbs tested could block gD binding. However, we find this explanation unlikely, since the MAbs bind to several different regions of gH/gL.
We next examined gH/gL and gB for binding. For gB capture, we used multiple MAbs that bind to distinct epitopes (42, 44). We were unable to detect any interaction between gH/gL and gB regardless of which protein was presented as the analyte or the ligand or which MAb was used for capture (Fig. 2C and D). Likewise, we detected no binding between gD and gB (Fig. 2E and F). One caveat is that the gB used here [gB2(727t)] is in its postfusion form (48, 49), and this might account for its inability to bind to the other HSV core glycoproteins.
Our overall conclusion is that gD and gH/gL can interact when gD is captured and presented by a subset of anti-gD MAbs; conversely, gD and gH/gL do not bind when gD is captured by another subset of MAbs, giving us some sense of where the interface might be. Furthermore, gH/gL may require a conformational change to accommodate gD binding, as capturing MAbs prevented binding when gH/gL was presented as a ligand. Finally, we failed to detect any direct interaction between gH/gL and gB or between gD and gB.
gH/gL can bind directly to gD.
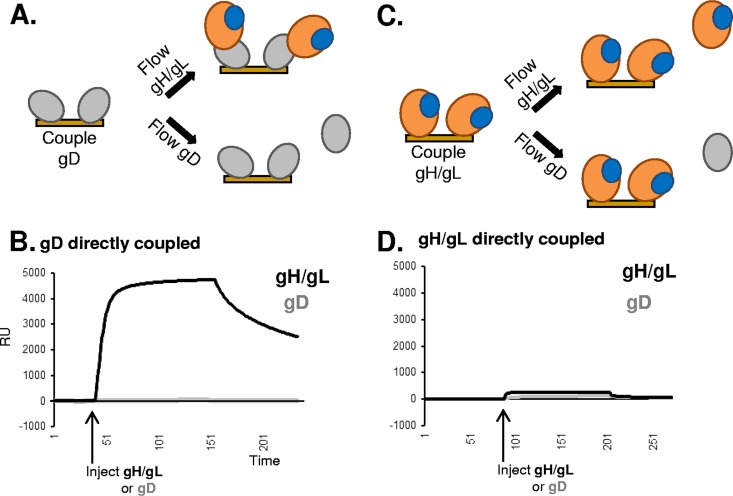
While we have posited that anti-gD MAbs that allow for gH/gL binding do so by presenting (and not blocking) an available binding site on gD, it remained possible that capture by these anti-gD MAbs induced a conformational change in gD which enabled it to bind gH/gL. To distinguish between these possibilities, we next asked whether gD could bind gH/gL when gD was directly coupled to the biosensor chip (Fig. 3A). In this case, we used the Biacore 3000 biosensor system, as we were studying a limited set of interactions. gD2(285t) was amine coupled to the CM5 SPR chip, and then gH2t/gL2 was flowed across the chip surface as an analyte (Fig. 3A). As with the Carterra system, gH/gL binding would be indicated by an increase in RU. We found that gD2(285t) directly coupled to the chip surface was capable of binding to gH2t/gL2 (Fig. 3B, black curve). This interaction was specific, as gD did not bind to itself (Fig. 3B, gray line) or other non-HSV control proteins (data not shown). These data indicate that MAbs are not required to induce conformational changes to allow gD-gH/gL binding and that the interaction site on gD2(285t) is open.
FIG 3.
gD2(285t) directly coupled to a biosensor chip can bind gH2t/gL2. (A) Schematic of the Biacore 3000 protocol for analyzing gD-gH/gL binding. gD2(285t) was attached directly to the biosensor chip via amine coupling. Either soluble gH/gL (top) or gD itself (bottom) then was injected across the chip surface. The chip was monitored for protein-protein binding, which would result in an increase in RU. (B) Binding curves. Black curve, response after gH/gL injection; gray curve, response after gD injection. x axis, time (seconds). y axis, RU. (C) Schematic of the Biacore 3000 protocol for analyzing gD-gH/gL binding, reverse orientation. Soluble gH/gL was attached directly to the biosensor chip via amine coupling. Either soluble gH/gL itself (top) or gD (bottom) then was injected across the chip surface. (D) Binding curves. Binding was tested a minimum of three times. Black curve, response after gH/gL injection; gray curve, response after gD injection.
In contrast, when gH/gL was coupled to the chip (Fig. 3C) with gD as the analyte, no interaction was observed between the two glycoproteins (Fig. 3D). Thus, in two different formats (Fig. 2 and 3) gD-gH/gL binding was unidirectional, occurring only when gD was presented as the ligand and when gH/gL was presented as the analyte. Such results suggest that gH/gL requires a conformational change to accommodate gD binding, and this cannot happen when gH/gL is immobilized on a biosensor chip, either directly (Fig. 3D) or by capture with MAbs (Fig. 2B).
In summary, we have shown that gH/gL does bind gD. This interaction can be observed in at least two ways: when gD is directly coupled to the biosensor chip or when gD is first captured by certain anti-gD MAbs. Our next goal was to carry out a more detailed study of the nature of the interaction.
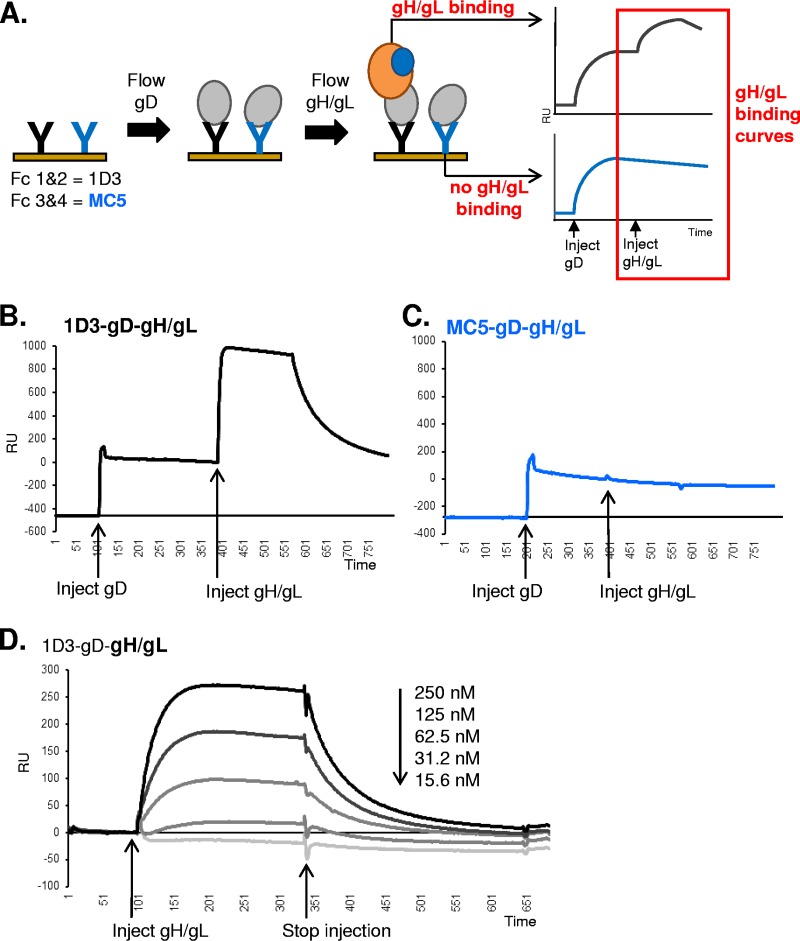
Analysis of the gD-gH/gL interaction.
When gD was directly coupled to the Biacore CM5 sensor chip, regeneration proved to be difficult, possibly due to inactivation of gD (see Materials and Methods for details). Therefore, the next experiments were performed using the MAb capture method (Fig. 4A). To ensure that there were no significant differences between the MAb capture results using the Carterra and Biacore systems of SPR, we chose one representative MAb that allowed gH/gL binding via Carterra (1D3; yellow subcommunity) and one that did not (MC5; blue community) and tested gD-gH/gL binding using the Biacore system for SPR. First, the two MAbs were coupled to separate flow cells of a CM5 chip. gD2(285t) next was injected across the chip surface and captured via MAb 1D3 (Fig. 4B) or MAb MC5 (Fig. 4C). We then injected gH/gL and monitored for binding. As in the Carterra SPR system, we found that gH/gL bound to gD when it was captured by 1D3 (Fig. 4B) but not when it was captured by MC5 (Fig. 4C). This result gave us confidence that the two SPR systems were interchangeable.
FIG 4.
gD-gH/gL binding can be recapitulated on the Biacore 3000 biosensor. (A) Diagram outlining the Biacore 3000 protocol. Anti-gD MAbs 1D3, which is permissive for gH/gL binding, and MC5, which is nonpermissive, were coupled to a CM5 biosensor chip on separate flow cells (Fc1-2 for 1D3, Fc3-4 for MC5). Sequential injections of soluble gD and soluble gH/gL were performed. An increase in RU after the gH/gL injection indicated gD-gH/gL binding (red box). Theoretical binding curves are shown on the right. (B and C) Experimental data for gD captured via 1D3 (B) or MC5 (C). The RU from a control Fc where only gH/gL was injected was subtracted from each. The gD-gH/gL binding resulting from gD capture via MAb 1D3 was observed many times and is used as a control in follow-up experiments (see Fig. 5). Inhibition of the gD-gH/gL interaction by MAb MC5 was performed three times. (D) The binding of gH/gL to gD is dose dependent. Serial dilutions of soluble gH/gL were injected across a CM5 chip where gD was captured via 1D3. Only the gH/gL binding curve is shown. The curves were stacked from highest concentration of gH/gL (250 nM; black curve) to lowest (15.6 nM; light gray curve). This experiment was repeated twice, and a representative experiment is shown. x axis, time (seconds). y axis, RU.
To determine if the binding of gH/gL to gD was dose dependent and to begin to look at the kinetics of the interaction, we flowed serial dilutions of soluble gH/gL over a fixed concentration of gD2(285t), which had been captured by MAb 1D3, and binding was recorded (Fig. 4D). We observed that binding was dose responsive, i.e., the higher the concentration of gH/gL that was flowed across gD the greater its binding. However, we were unable to calculate an affinity constant for gH/gL binding to gD because the data did not fit any of the models in the BIAevaluation software package. This may be due to either the nature of the binding interaction or the inclusion of the 1D3-gD binding step prior to gD-gH/gL binding. However, the shape of the binding curves suggests that gH/gL not only binds to gD rapidly (fast on-rate) but also rapidly dissociates from it (fast off-rate). We estimate that binding of gH/gL to gD is in the nanomolar range.
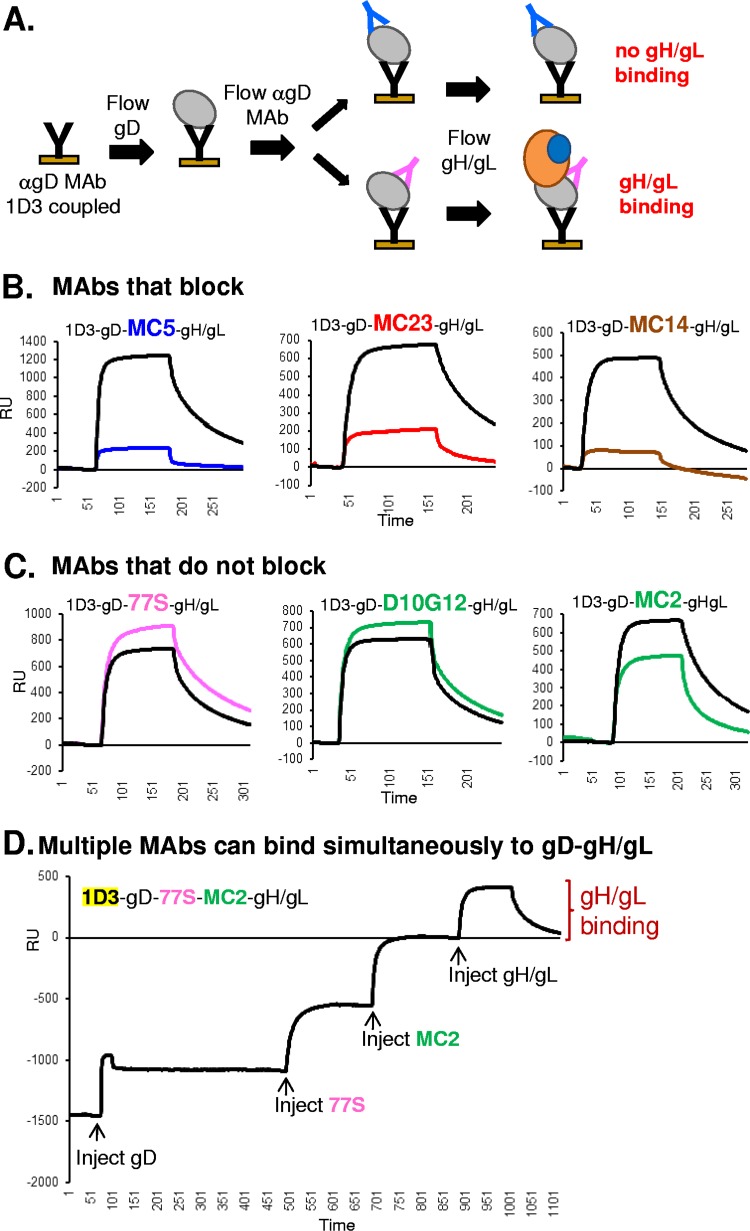
We next tested MAbs for their ability to block gD-gH/gL binding (Fig. 5A). Thus far, we showed that when gD is presented by MAbs from the blue, brown, or red subcommunities, gH/gL is unable to bind. However, we wondered if these MAbs would block gH/gL binding to preexisting MAb/gD complexes such as 1D3/gD, which can bind gH/gL. To answer this question, we captured gD with 1D3 (yellow subcommunity) and then flowed a second noncompeting MAb, thereby sandwiching the gD molecule between the two MAbs. We then flowed gH/gL across this “sandwich” (Fig. 5A). We defined blocking as at least a 50% reduction in gH/gL binding. For clarity, only gH/gL binding curves are shown in Fig. 5B and C. We chose MAbs MC5 (blue), MC23 (red), and MC14 (brown) to represent the three communities that blocked gH/gL binding (Fig. 1). Likewise, we chose MAbs 77S (pink) and D10G12 and MC2 (green) to represent the communities that failed to block gH/gL binding.
FIG 5.
Certain anti-gD MAbs block gD-gH/gL binding. (A) Diagram outlining the Biacore 3000 protocol. Anti-gD MAb 1D3, which is permissive for gH/gL binding, was coupled to a CM5 biosensor chip. Sequential injections of soluble gD, anti-gD MAb, and soluble gH/gL were performed. An increase in RU after the gH/gL injection indicated gD-gH/gL binding. (B and C) gH/gL binding curves. (B) If the anti-gD MAb blocked the gD-gH/gL interaction, a decrease in RUs upon gH/gL injection, compared to when no MAb was injected (black curve, control), was observed. (C) If the gH/gL binding curve mirrored that of the control, then no blocking was observed. Each MAb was tested a minimum of two times and is colored according to its MAb community, as shown in Fig. 1. (D) Multiple anti-gD MAbs can bind to gD simultaneously with gH/gL. The protocol was performed as outlined in panel A, with multiple injections of noncompeting, gH/gL-permissive anti-gD MAbs. Injection points for glycoproteins and MAbs are highlighted with arrows. x axis, time (seconds). y axis, RU.
We found that MAbs MC5 (blue curve), MC23 (red curve), and MC14 (brown curve) all inhibited the binding of gH/gL to the 1D3/gD complex compared to that of 1D3/gD alone (black curves) (Fig. 5B). We postulate that the blocking MAb prevents the interaction between gD and gH/gL. In concordance with the results shown in Fig. 1D and E, MAbs D10G12 (green) and 77S (pink) did not interfere with gH/gL binding (Fig. 5C). Green-community MAb MC2 caused a modest reduction in gH/gL binding (28% of that of the no MAb control) that was still above our 50% cutoff for blocking (Fig. 5C). Thus, gH/gL binding can be blocked in two ways: (i) when gD is captured to the sensor chip via an inhibitory MAb (Fig. 1) and (ii) when an inhibitory MAb is added to a preexisting, binding-competent MAb/gD complex (Fig. 5B).
Since the anti-gD MAbs that allow gH/gL binding map to three distinct antigenic communities (green, yellow, and pink), we wondered whether we could sequentially bind MAbs from these three communities and still be able to detect gH/gL binding. We used 1D3 (yellow) to capture gD and then flowed MAb 77S (pink), followed by MAb MC2 (green). Each of these MAbs bound to gD and did not interfere with each other (Fig. 5D). Finally, we flowed gH/gL across this multi-MAb/gD complex, and remarkably, it bound (Fig. 5D). We conclude that the site on gD for gH/gL binding excludes the residues in the epitopes of each of these MAbs.
gH/gL can bind to a gD/receptor complex.
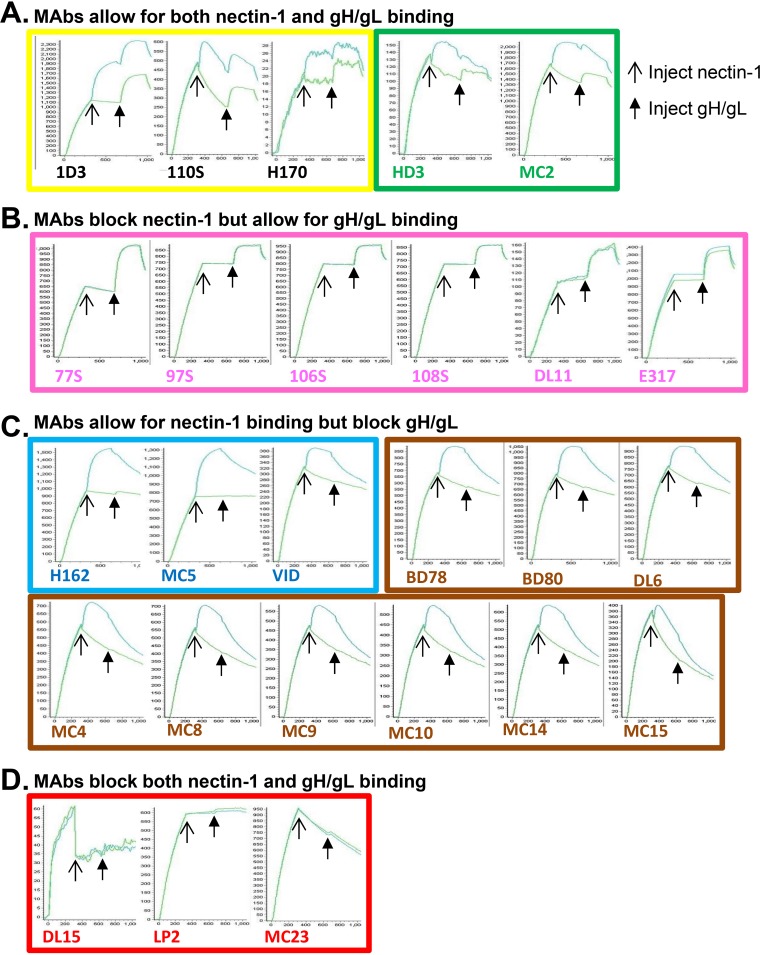
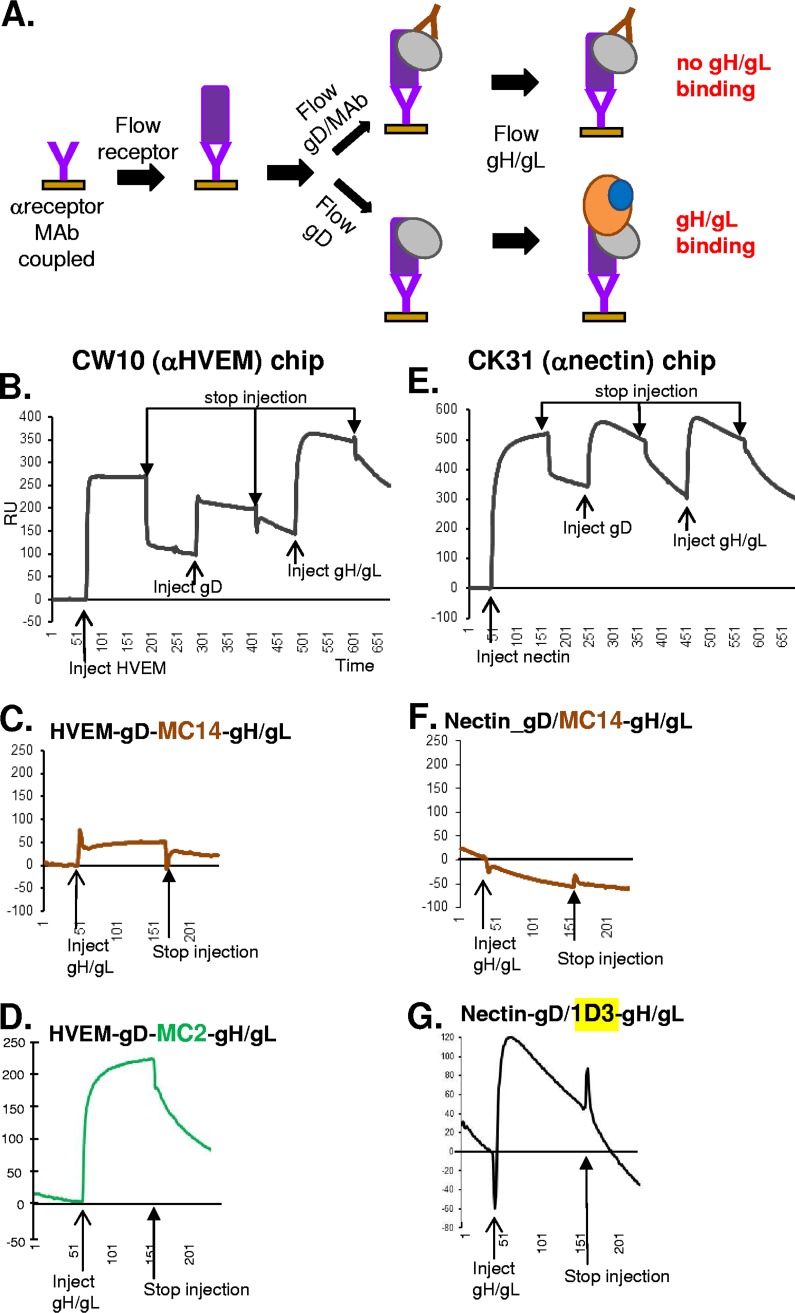
Our hypothesis that the gH/gL binding site occupies a face of gD that differs from the face that is used for binding nectin-1 and HVEM (10, 11) is supported by our finding that MAbs from the pink subcommunity block nectin-1 and HVEM binding yet do not interfere with gH/gL binding (Fig. 1). A direct prediction from these findings is that gD should be able to bind both receptor and gH/gL simultaneously. To formally test this concept, we used the Carterra SPR system (Fig. 1A) to screen a panel of MAb/gD complexes for their ability to bind both nectin and gH/gL (Fig. 6). We first printed our panel of anti-gD MAbs and then captured gD. We then flowed either nectin-1 (top curve) or buffer (bottom curve, control) across the chip surface, followed by gH/gL. We observed four outcomes that depended on which MAb was used for gD capture: (i) gD presented by MAbs in the yellow and green subcommunities bound both nectin and gH/gL (Fig. 6A), demonstrating that nectin and gH/gL occupy distinct binding sites; (ii) gD presented by pink-subcommunity MAbs did not bind nectin but did bind gH/gL (Fig. 6B), demonstrating that these MAbs interfere only with receptor binding; (iii) gD presented by blue- and brown-community MAbs bound nectin but not gH/gL (Fig. 6C), demonstrating that these MAbs occupied the site(s) needed for gH/gL binding; and (iv) gD presented by red-subcommunity MAbs were unable to bind either nectin or gH/gL (Fig. 6D). In this last case, the data indicate that there is a subset of residues involved in both receptor and gH/gL binding. These amino acids may define an “edge” between the two functional faces of gD. Alternatively, MAbs that simultaneously interfere with receptor binding and gH/gL binding may preclude access to different residues required for nectin-1 and gH/gL binding, respectively. To probe this question further, we tested gH/gL binding conditions where gD is first bound to and presented by a receptor (Fig. 7A). This format of three proteins (receptor-gD-gH/gL) should more closely reflect the complex that forms prior to fusion. In Fig. 7C, D, F, and G, only the gH/gL binding curves are shown for clarity. The full set of binding curves, showing all three injections (receptor, gD, and gH/gL), are shown in Fig. 7B and E. We used this format to examine gH/gL binding to gD when the latter was bound to truncated, purified forms of both HVEM and nectin-1.
FIG 6.
Screening gD for both nectin-1 and gH/gL binding using the Carterra CFM/SPR protocol. A panel of anti-gD MAbs was printed to a CDM200M sensor chip as explained in the legend to Fig. 2. Two curves are shown for each printed MAb. In both cases, soluble gD2(285t) is first injected across the chip surface and bound by the printed MAb (as seen by the initial increase in RU). The top curve shows protein binding after an injection of soluble nectin-1 (thin arrow) followed by an injection of soluble gH2t/gL2 (thick arrow). The bottom curve (control) shows protein binding after an injection of buffer (no increase in RU) followed by an injection of soluble gH2t/gL2 (thick arrow). In cases where no nectin-1 binding was observed, the two curves overlapped. MAbs are designated by the color of their antibody community as shown in Fig. 1. (A) MAbs that allow both nectin-1 and gH/gL binding. (B) MAbs that allow gH/gL binding but not nectin-1. (C) MAbs that allow nectin-1 binding but not gH/gL. (D) MAbs that do not permit either nectin-1 or gH/gL binding. x axis, time (seconds). y axis, RU.
FIG 7.
Receptor-bound gD is capable of binding gH/gL. (A) Diagram outlining the Biacore 3000 protocol. An anti-receptor MAb (CW10 for HVEM or CK31 for nectin-1) was coupled to a CM5 biosensor chip. Sequential injections of soluble receptor, gD or gD preincubated with an anti-gD MAb, and soluble gH/gL were performed a minimum of two times. An increase in RU after the gH/gL injection indicated gD-gH/gL binding. (B to D) gH/gL binding curves for when gD was captured via its receptor HVEM. (B and E) Sequential injections of soluble receptor, soluble gD, and soluble gH/gL were performed. Each injection start is demarked by a thin arrow under the binding curve, while each injection stop is demarked by a thick arrow above the curve. (C) Anti-gD MAb MC14, which blocks gH/gL binding, was prebound to gD. Only the gH/gL binding curve is shown. (D) Anti-gD MAb MC2, which does not block gH/gL binding, was prebound to gD. Only the gH/gL binding curve is shown. Neither MC14 nor MC2 block the binding of gD to HVEM. (E to G) gH/gL binding curves for when gD was captured via its receptor, nectin-1. (F) Anti-gD MAb MC14 was prebound to gD. (G) Anti-gD MAb 1D3, which does not block gH/gL binding, was prebound to gD. 1D3 does not block the binding of gD to HVEM. x axis, time (seconds). y axis, RU.
We first coupled the anti-HVEM MAb CW10, which does not block receptor binding (50), to the biosensor chip and used it to capture HVEM (Fig. 7A). We next flowed gD, which was captured by HVEM, and then flowed gH/gL across the CW10/HVEM/gD complex and monitored binding. The complex bound gH/gL in a stable fashion (Fig. 7B). Moreover, in agreement with what was seen in Fig. 5B, gH/gL binding to gD was blocked by MAb MC14 (brown community) (Fig. 7C). In contrast, MAb MC2 (green subcommunity) did not block gD-gH/gL binding (Fig. 7D). Because neither MC14 nor MC2 interfere with the binding of gD to HVEM (11), we conclude that MC14 but not MC2 blocks the site of the gD-gH/gL interaction. Since MC14 recognizes a linear epitope (Table 1), we presume that some or all of these gD residues are involved in binding to gH/gL.
To examine binding of gH/gL to a complex of gD and nectin-1, we used the anti-nectin MAb CK31 (51) to capture the receptor. gD next was flowed across the chip surface, followed by gH/gL. As expected from previous results (Fig. 7), the CK31/nectin/gD complex bound gH/gL (Fig. 7E). The steep slope of the gH/gL curve is reflective of the rapid dissociation of gD/nectin from MAb CK31 (Fig. 7E). As we found with gD/HVEM (Fig. 7C), MAb MC14 blocked gH/gL binding (Fig. 7F) but MAb 1D3 did not (Fig. 7G). Neither MC14 nor 1D3 interferes with the binding of nectin to gD (7, 11) (Fig. 7A and C).
We conclude that the gH/gL binding site occupies a portion of gD that differs from either of the receptor binding domains (i.e., nectin-1 and HVEM) (17, 18, 52, 53). The residues that constitute the binding interfaces between both receptors and gD are known from crystallographic studies (17, 18). Hence, these amino acids can be excluded from those that interact with gH/gL. Likewise, MAbs that block gD-gH/gL binding (Table 1) have enabled us to determine a possible gH/gL binding region on gD (Fig. 8) (see Discussion).
FIG 8.
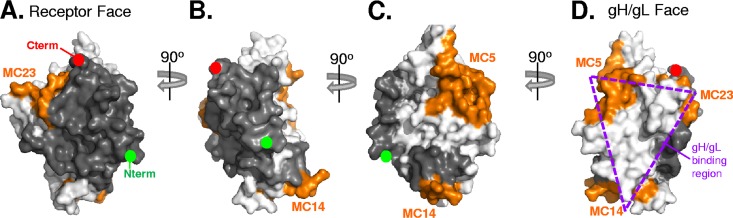
Epitope mapping of gD reveals a potential gH/gL binding face. Surface representation of the gD crystal structure is shown in white (PDB entry 2C36). Antibody-resistant mutations (Table 1) were used to position MAb epitopes onto the structure. We chose three sentinel MAbs, MC5, MC14, and MC23, to represent each of the three MAb communities that contain antibodies that block the gD-gH/gL interaction; these MAb footprints are colored orange. Dark gray residues denote areas that, when bound by receptor or MAbs, do not block gH/gL binding. White residues denote areas for which we have no binding or blocking information. The gD structure is rotated 90° from panels A to B, B to C, and C to D. We define the gH/gL binding face as the point when the epitopes for all three blocking MAbs (MC5, MC23, and MC14) are visible on the same face and have marked this area with a dashed purple triangle in panel D.
DISCUSSION
gH/gL binds directly to gD.
Based on multiple studies (9–11, 13, 23, 30), we proposed that HSV entry and cell-cell fusion occur in a cascade of events involving the four essential glycoproteins: gD (the receptor binding protein), gH/gL (the key regulator of fusion), and gB (the fusogen). One of the two gD receptors, HVEM or nectin-1, is also required. As stressed by Heldwein (32), “Determining how the triggering signal reaches the fusogen gB represents the next frontier in structural biology of herpesvirus entry.”
In thinking about the fusion cascade and the role of gD in it, we proposed that gD would have distinct sides (11), one of which would bind HVEM/nectin-1 and the other would bind gH/gL. We have extensively studied side one (receptor binding) using epitope mapping and mutational analysis (7, 11, 37, 38, 45, 50, 52–54). Crystallographic studies illuminated the sites of receptor binding (16–18) as well as the amino acids recognized by the HSV neutralizing MAb E317 (15), which is a member of the pink subcommunity and blocks gD-receptor binding (Fig. 1B).
However, until now, we had no direct evidence that gD physically interacts with gH/gL in a functionally relevant fashion. The major accomplishment of the work reported in this paper is the demonstration of a physical interaction between HSV gD and gH/gL, in real time, that can be blocked by neutralizing and fusion-blocking MAbs. We showed that the interaction between gD and gH/gL is specific and that certain anti-gD MAbs can block it while others cannot. Furthermore, we found that gD can be presented to gH/gL for binding in several different ways: when captured by an anti-gD MAb, when coupled directly to a biosensor chip, or when first captured by one of its receptors. We also found that the interaction between gD and gH/gL is unidirectional, i.e., gH/gL binds to gD when gD is presented as a ligand, but there is no interaction when gH/gL is the ligand and gD is the analyte. Such results suggest gH/gL requires a conformational change to accommodate gD binding, and this cannot happen when gH/gL is immobilized to a biosensor chip.
Defining the site on gD to which gH/gL binds.
By mapping the epitopes of anti-gD MAbs that either allowed or blocked gD-gH/gL binding, we identified the potential interaction site on gD (Fig. 8). We found that gH/gL can bind gD simultaneously with MAbs in the pink, yellow, and green subcommunities (Fig. 1). It is noteworthy that the epitopes for pink- and yellow-subcommunity MAbs overlap the receptor binding sites, as we also found that receptor and gH/gL can bind to gD simultaneously (Fig. 5). Since the gD-nectin-1 and gD-HVEM interfaces are known from crystallographic studies (16–18), none of these residues are part of the gH/gL binding site. Together, we excluded multiple sites from being involved in gH/gL binding (Fig. 8, gray).
Conversely, MAbs in three other communities (red, blue, and brown) all blocked gD-gH/gL binding (Fig. 1). The epitopes for these three communities are distinct and noncompeting and encircle a large area of gD (Fig. 8, orange), which is separate from the receptor-binding face. Since binding by any one of these MAbs blocks gH/gL binding, we suggest that the gH/gL interface on gD spans a large region encompassing these epitopes and the area between them (Fig. 8D, triangle). Our inability to fit the dose-response data to a 1:1 binding model remains an issue. Perhaps it means that there is more than one site for gH/gL to bind to gD, although the MAb blocking data argue that this is likely not the case. It could also be due, at least in part, to the configuration of the experiment, i.e., that gD is held by a MAb prior to testing gH/gL binding. The best way to resolve this issue relies on structural data obtained by cryoelectron microscopy or crystallography, and that is a consideration for future studies.
The properties of MC2 (green) and MC5 (blue) are distinct.
Both MC2 (green community) and MC5 (blue community) are potent virus-neutralizing MAbs (11). They do not block receptor binding but do block membrane fusion (41, 55) (Table 1). Our initial hypothesis was that MC2 and MC5 neutralize virus by occluding gD residues needed to trigger later events, presumably the interaction of gD with gH/gL (9, 11). In support, we show that the binding of gD to MC5 prevents the binding of gH/gL to gD (Fig. 1F and 4C and Table 2). To our surprise, binding of MC2 to gD does not prevent gH/gL from binding (Fig. 1E and 7D). Therefore, MAb MC2 must neutralize virus and block membrane fusion in another, undefined way. We know that the binding of MC2 induces the C terminus of gD2(306) to assume an open conformation similar to that caused by the receptor (11). One possibility is that MC2 neutralizes virus by triggering this conformation too soon, starting the fusion cascade prematurely (Table 2).
TABLE 2.
Mechanism of action of inhibitory anti-gD MAbs
Brown-community MAbs that block gH/gL binding but do not neutralize virus.
We know from earlier studies (11, 56) that the linear epitopes of brown-community MAbs overlap a region of gD within the C-terminal tail previously termed the “profusion domain” (57, 58). This domain was defined as a region of gD discrete from the receptor binding sites that is required for HSV entry (57). The profusion domain is located near the C terminus of the gD ectodomain (residues 260 to 285). It was suggested that the profusion domain includes a binding site for gB, gH/gL, or both (25, 57). Here, we show that these MAbs block gD-gH/gL binding (Fig. 1G). However, MAbs mapped to epitopes outside the profusion domain (e.g., MC5 and MC23) also block gH/gL binding (Fig. 1D and F), suggesting either a large gD-gH/gL interface on gD or multiple gH/gL binding sites. Interestingly, MAbs whose epitopes overlap the profusion domain, such as MC4, do not neutralize virus but do inhibit glycoprotein-mediated cell-cell fusion (11, 41) (Table 1), which is perplexing since gH/gL binding is required for both processes. One explanation for this unique phenotype may be the differential accessibility of these gD epitopes in virions versus that when gD is presented on the cell membrane.
Ultimately, the question of where gD physically binds to gH/gL will be determined by structural studies. Based on data presented in this paper, we now identify three regions on gD (Fig. 8): (i) the region colored dark gray, which is not part of the gD-gH/gL contact site and includes the nectin-1 and HVEM binding sites and the epitopes for MAbs from the green, yellow, and pink communities; (ii) the region colored orange, which may be part of the gD-gH/gL contact site and is defined by the epitopes for MAbs from the brown, blue, and red communities that block gD-gH/gL binding; and (iii) the regions colored white, containing the rest of the gD molecule for which no data are available. Our challenge now is to develop methods to stabilize the gD-gH/gL interaction for crystallography or cryoelectron microscopy studies so that this interaction site can be directly visualized. In addition, targeting the precise site of the gD-gH/gL interaction to prevent virus entry may enhance both therapeutic and vaccine strategies.
MATERIALS AND METHODS
Cells and soluble proteins.
All soluble proteins used in this study were purified from baculovirus-infected insect (Sf9) cells. HSV-2 gD(285t) was purified using a DL6 immunosorbent column as described previously (11, 31, 59). HSV-2 gB(727t) was purified by use of a DL16 immunosorbent column (60). The hexahistidine-tagged protein gH2t/gL2 (containing the full-length gH2 ectodomain) was purified via nickel affinity chromatography (13, 61). Soluble nectin-1(346t) and HVEM(200t) were purified as described earlier (7, 62).
Antibodies.
The following anti-gD MAbs were used in this study: 1D3 (11, 63, 64); DL6, DL11, and DL15 (11, 37, 46, 56, 65); MC1, MC2, MC4, MC5, MC8, MC9, MC10, MC14, MC15, and MC23 (11, 54); LP2 (kindly provided by A. Minson and H. Browne) (36); HD1, HD2, HD3, H162, H170, and H193 (66, 67); 11S, 12S, 45S, 77S, 97S, 106S, 108S, and 110S (63, 68); BD78 and BD80 (kindly supplied by Becton, Dickinson Co.) (11, 56); 4E3E, 4G4D, and 11B3AG (gifts of R. N. Lausch) (33); E317 (15); and D10G12 (from Chiron Corporation, now Novartis) (33). The following MAbs were used for gB: C226 (provided by Becton, Dickinson Co.) (44) and DL16, DL21, SS55, SS63, and SS144 (69). The following MAbs were used for gH/gL: 53S (61, 68); CHL2, CHL4, CHL5, CHL17, CHL18, CHL21, and CHL37 (43); and CΔ48L3 (13). MAbs CK31 (51) and CW10 (50) were generated against nectin-1 and HVEM, respectively.
Screening for protein-protein binding using CFM/SPRi.
Glycoprotein binding experiments were performed using the Carterra, Inc., CFM/SPR imaging (SPRi) system. We used a method described previously (33–35), with the following modifications. A CFM 2 was used to create a 48-spot microarray of amine-coupled anti-HSV glycoprotein MAbs on a CDM200M sensor chip (Xantec GmbH). Upon docking the printer chip into the SPR imager (IBIS MX96), the chip was blocked with ethanolamine and the system primed with a running buffer of phosphate-buffered saline–0.01% Tween 20. Protein binding was performed in a classical sandwich assay format using 100 nM soluble gD, gB, or gH/gL as the antigen, followed by 100 nM soluble gD or gB or 400 nM soluble gH/gL as the analyte and 1 M glycine, pH 2.0, for regeneration. Sensorgrams showing the gD, gB, or gH/gL capture followed by a buffer analyte instead of a glycoprotein analyte were included as negative controls. All experiments were done at room temperature.
Glycoprotein binding using the Biacore 3000.
Glycoprotein binding experiments were carried out on a Biacore 3000 optical biosensor at 25°C by following previous guidelines (11, 70, 71). All injections were performed at a flow rate of 5 μl/min using HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20). After each experiment, the chip surface was treated with brief pulses of 0.2 M Na2CO3 (pH 10) until the RU signal returned to baseline (regeneration).
First, soluble, purified gD2(285t) was amine coupled directly to a CM5 sensor chip (GE Healthcare Bio-Sciences, Pittsburgh, PA). Purified gH2t/gL2 (0.4 mg/ml) was then injected for 240 s (analyte) and the binding was recorded. The experiment was also performed in the reverse orientation, with soluble gH2t/gL2 amine coupled to the chip (ligand) and gD2(285t) injected across the surface (analyte). To regenerate the chip surface, 0.2 M Na2CO3 (pH 10) was injected until the RU signal returned to baseline. We found that the gD-coupled surface could only be regenerated a limited number of times before a new chip became necessary. Therefore, additional experiments were performed by first capturing soluble gD to the chip surface via the amine-coupled anti-gD MAb 1D3.
For kinetic analysis of gD-gH/gL binding, gD2(285t) was first captured via immobilized 1D3. Serial 2-fold dilutions of gH2t/gL2 in HBS-EP next were flowed across the chip surface for 240 s, with an additional 240 s allowed for disassociation. Sensorgrams were corrected for nonspecific binding by subtracting the signal achieved on Fc2 (gH/gL injection only) from that on Fc1 (gD injection followed by gH/gL). The signal obtained when buffer alone was injected was then subtracted from the total signal. Sensorgrams were analyzed using BIAevaluation software, version 4.1.
To test for blocking of gD-gH/gL binding via anti-gD MAbs, a gH/gL binding-permissive (1D3) or nonpermissive (MC5) anti-gD IgG was amine coupled to a CM5 sensor chip, and then 0.05 mg/ml purified gD2(285t) was injected across the chip until approximately 300 response units (RU) were captured by the coupled anti-gD MAb. Purified anti-gD IgG (0.1 mg/ml) was then injected for 120 s, followed by soluble gH2t/gL2 (0.2 mg/ml) for 120 s.
To test for receptor-gD-gH/gL binding, anti-receptor MAb CW10 (HVEM) or CK31 (nectin-1) was amine coupled to a CM5 sensor chip, and then 0.1 mg/ml soluble receptor was injected for 120 s, followed by 0.05 mg/ml purified gD2(285t) for 120 s and then 0.2 mg/ml gH2t/gL2 for 120 s.
ACKNOWLEDGMENTS
This research was supported by NIH grant R01-AI-018289 (to G.H.C.).
We thank Leslie King for critical readings of the manuscript and Claude Krummenacher for helpful discussions.
B.D.B. and N.T.D. are employed by Carterra, Inc.
REFERENCES
- 1.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner A, Bruun B, Minson T, Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a cos cell transfection system. J Virol 72:873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muggeridge MI. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J Gen Virol 81:2017–2027. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- 4.Browne H, Bruun B, Minson T. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J Gen Virol 82:1419–1422. doi: 10.1099/0022-1317-82-6-1419. [DOI] [PubMed] [Google Scholar]
- 5.Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the tnf/ngf receptor family. Cell 87:427–436. doi: 10.1016/S0092-8674(00)81363-X. [DOI] [PubMed] [Google Scholar]
- 6.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 7.Krummenacher C, Nicola AV, Whitbeck JC, Lou H, Hou W, Lambris JD, Geraghty RJ, Spear PG, Cohen GH, Eisenberg RJ. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol 72:7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J 24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher JR, Saw WT, Atanasiu D, Lou H, Eisenberg RJ, Cohen GH. 2013. Displacement of the C-terminus of herpes simplex virus gD is sufficient to expose the fusion activating interfaces on gD. J Virol 87:12656–12666. doi: 10.1128/JVI.01727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazear E, Carfi A, Whitbeck JC, Cairns TM, Krummenacher C, Cohen GH, Eisenberg RJ. 2008. Engineered disulfide bonds in herpes simplex virus type 1 gD separate receptor binding from fusion initiation and viral entry. J Virol 82:700–709. doi: 10.1128/JVI.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazear E, Whitbeck JC, Ponce-de-Leon M, Cairns TM, Willis SH, Zuo Y, Krummenacher C, Cohen GH, Eisenberg RJ. 2012. Antibody-induced conformational changes in herpes simplex virus glycoprotein gD reveal new targets for virus neutralization. J Virol 86:1563–1576. doi: 10.1128/JVI.06480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atanasiu D, Saw WT, Eisenberg RJ, Cohen GH. 2016. Regulation of herpes simplex virus glycoprotein-induced cascade of events governing cell-cell fusion. J Virol 91:10535–10544. doi: 10.1128/JVI.01501-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atanasiu D, Cairns TM, Whitbeck JC, Saw WT, Rao S, Eisenberg RJ, Cohen GH. 2013. Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors. mBio 4:00046-13. doi: 10.1128/mBio.00046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 15.Lee CC, Lin LL, Chan WE, Ko TP, Lai JS, Wang AH. 2013. Structural basis for the antibody neutralization of herpes simplex virus. Acta Crystallogr D Biol Crystallogr 69:1935–1945. doi: 10.1107/S0907444913016776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu G, Zhang N, Qi J, Li Y, Chen Z, Zheng C, Gao GF, Yan J. 2014. Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition. J Virol 88:13678–13688. doi: 10.1128/JVI.01906-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. 2001. Herpes simplex virus glycoprotein D bound to the human receptor hvea. Mol Cell 8:169–179. doi: 10.1016/S1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 18.Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C, Carfi A. 2011. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog 7:e1002277. doi: 10.1371/journal.ppat.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Q, Longnecker R, Connolly SA. 2014. Substitution of herpes simplex virus 1 entry glycoproteins with those of saimiriine herpesvirus 1 reveals a gD-gH/gL functional interaction and a region within the gD profusion domain that is critical for fusion. J Virol 88:6470–6482. doi: 10.1128/JVI.00465-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Q, Longnecker R, Connolly SA. 2015. A functional interaction between herpes simplex virus 1 glycoprotein gH/gL domains I and II and gD is defined by using alphaherpesvirus gH and gL chimeras. J Virol 89:7159–7169. doi: 10.1128/JVI.00740-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Romero P, Perez A, Capul A, Montgomery R, Fuller AO. 2005. Herpes simplex virus entry mediator associates in infected cells in a complex with viral proteins gD and at least gH. J Virol 79:4540–4544. doi: 10.1128/JVI.79.7.4540-4544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avitabile E, Forghieri C, Campadelli-Fiume G. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol 81:11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol 84:12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handler CG, Eisenberg RJ, Cohen GH. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol 70:6067–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gianni T, Amasio M, Campadelli-Fiume G. 2009. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL through the C-terminal profusion. J Biol Chem 284:17370–17382. doi: 10.1074/jbc.M109.005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sathiyamoorthy K, Jiang J, Hu YX, Rowe CL, Mohl BS, Chen J, Jiang W, Mellins ED, Longnecker R, Zhou ZH, Jardetzky TS. 2014. Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog 10:e1004309. doi: 10.1371/journal.ppat.1004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sathiyamoorthy K, Hu YX, Mohl BS, Chen J, Longnecker R, Jardetzky TS. 2016. Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat Commun 7:13557. doi: 10.1038/ncomms13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciferri C, Chandramouli S, Donnarumma D, Nikitin PA, Cianfrocco MA, Gerrein R, Feire AL, Barnett SW, Lilja AE, Rappuoli R, Norais N, Settembre EC, Carfi A. 2015. Structural and biochemical studies of hCMV gH/gL/gO and pentamer reveal mutually exclusive cell entry complexes. Proc Natl Acad Sci U S A 112:1767–1772. doi: 10.1073/pnas.1424818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandramouli S, Malito E, Nguyen T, Luisi K, Donnarumma D, Xing Y, Norais N, Yu D, Carfi A. 2017. Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Sci Immunol 2:eaan1457. doi: 10.1126/sciimmunol.aan1457. [DOI] [PubMed] [Google Scholar]
- 30.Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, Eisenberg RJ. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A 104:18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rux AH, Willis SH, Nicola AV, Hou W, Peng C, Lou H, Cohen GH, Eisenberg RJ. 1998. Functional region iv of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpesvirus entry mediator. J Virol 72:7091–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heldwein EE. 2016. gH/gL supercomplexes at early stages of herpesvirus entry. Curr Opin Virol 18:1–8. doi: 10.1016/j.coviro.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cairns TM, Ditto NT, Lou H, Brooks BD, Atanasiu D, Eisenberg RJ, Cohen GH. 2017. Global sensing of the antigenic structure of herpes simplex virus gD using high-throughput array-based spr imaging. PLoS Pathog 13:e1006430. doi: 10.1371/journal.ppat.1006430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdiche YN, Harriman R, Deng X, Yeung YA, Miles A, Morishige W, Boustany L, Zhu L, Izquierdo SM, Harriman W. 2016. Assessing kinetic and epitopic diversity across orthogonal monoclonal antibody generation platforms. MAbs 8:264–277. doi: 10.1080/19420862.2015.1118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J, Rajpal A. 2014. High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS One 9:e92451. doi: 10.1371/journal.pone.0092451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minson AC, Hodgman TC, Digard P, Hancock DC, Bell SE, Buckmaster EA. 1986. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J Gen Virol 67:1001–1013. doi: 10.1099/0022-1317-67-6-1001. [DOI] [PubMed] [Google Scholar]
- 37.Muggeridge MI, Isola VJ, Byrn RA, Tucker TJ, Minson AC, Glorioso JC, Cohen GH, Eisenberg RJ. 1988. Antigenic analysis of a major neutralization site of herpes simplex virus glycoprotein D using deletion mutants and monoclonal antibody-resistant mutants. J Virol 62:3274–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicola AV, Ponce de Leon M, Xu R, Hou W, Whitbeck JC, Krummenacher C, Montgomery RI, Spear PG, Eisenberg RJ, Cohen GH. 1998. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol 72:3595–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietzschold B, Eisenberg RJ, Ponce de Leon M, Golub E, Hudecz F, Varrichio A, Cohen GH. 1984. Fine structure analysis of type-specific and type-common antigenic sites of herpes simplex virus glycoprotein D. J Virol 52:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen GH, Dietzschold B, Ponce de Leon M, Long D, Golub E, Varrichio A, Pereira L, Eisenberg RJ. 1984. Localization and synthesis of an antigenic determinant of herpes simplex virus glycoprotein D that stimulates the production of neutralizing antibody. J Virol 49:102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atanasiu D, Saw WT, Lazear E, Whitbeck JC, Cairns TM, Lou H, Eisenberg RJ, Cohen GH. 2018. Using antibodies and mutants to localize the presumptive gH/gL binding site on HSV gD. J Virol 92:e01694-18. doi: 10.1128/JVI.01694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cairns TM, Fontana J, Huang Z-Y, Whitbeck JC, Atanasiu D, Rao S, Shelly SS, Lou H, Ponce de Leon M, Steven AC, Eisenberg RJ, Cohen GH, Hutt-Fletcher L. 2014. Mechanism of neutralization of herpes simplex virus by antibodies directed at the fusion domain of glycoprotein B. J Virol 88:2677–2689. doi: 10.1128/JVI.03200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cairns TM, Shaner MS, Zuo Y, Ponce-de-Leon M, Baribaud I, Eisenberg RJ, Cohen GH, Whitbeck JC. 2006. Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function. J Virol 80:2596–2608. doi: 10.1128/JVI.80.6.2596-2608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol 81:3827–3841. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitbeck JC, Muggeridge MI, Rux AH, Hou W, Krummenacher C, Lou H, van Geelen A, Eisenberg RJ, Cohen GH. 1999. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J Virol 73:9879–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisenberg RJ, Long D, Ponce de Leon M, Matthews JT, Spear PG, Gibson MG, Lasky LA, Berman P, Golub E, Cohen GH. 1985. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol 53:634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muggeridge MI, Roberts SR, Isola VJ, Cohen GH, Eisenberg RJ. 1990. Herpes simplex virus, p 459–481. In van Regenmortel MHV, Neurath AR (ed), Immunochemistry of viruses, II. The basis for serodiagnosis and vaccines. Elsevier Science Publishers, Amsterdam, the Netherlands. [Google Scholar]
- 48.Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 49.Vitu E, Sharma S, Stampfer SD, Heldwein EE. 2013. Extensive mutagenesis of the HSV-1 gB ectodomain reveals remarkable stability of its postfusion form. J Mol Biol 425:2056–2071. doi: 10.1016/j.jmb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitbeck JC, Connolly SA, Willis SH, Hou W, Krummenacher C, Ponce de Leon M, Lou H, Baribaud I, Eisenberg RJ, Cohen GH. 2001. Localization of the gD-binding region of the human herpes simplex virus receptor, HveA. J Virol 75:171–180. doi: 10.1128/JVI.75.1.171-180.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krummenacher C, Baribaud I, Ponce de Leon M, Whitbeck JC, Lou H, Cohen GH, Eisenberg RJ. 2000. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J Virol 74:10863–10872. doi: 10.1128/JVI.74.23.10863-10872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Cohen GH, Eisenberg RJ. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J Virol 77:8127–8140. doi: 10.1128/JVI.77.14.8127-8140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Connolly SA, Landsburg DJ, Carfi A, Whitbeck JC, Zuo Y, Wiley DC, Cohen GH, Eisenberg RJ. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J Virol 79:1282–1295. doi: 10.1128/JVI.79.2.1282-1295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lazear E, Whitbeck JC, Zuo Y, Carfi A, Cohen GH, Eisenberg RJ, Krummenacher C. 2014. Induction of conformational changes at the N-terminus of herpes simplex virus glycoprotein D upon binding to HVEM and nectin-1. Virology 448:185–195. doi: 10.1016/j.virol.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saw WT, Matsuda Z, Eisenberg RJ, Cohen GH, Atanasiu D. 2015. Using a split luciferase assay (SLA) to measure the kinetics of cell-cell fusion mediated by herpes simplex virus glycoproteins. Methods 90:68–75. doi: 10.1016/j.ymeth.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isola VJ, Eisenberg RJ, Siebert GR, Heilman CJ, Wilcox WC, Cohen GH. 1989. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol 63:2325–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, Cohen GH, Campadelli-Fiume G. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc Natl Acad Sci U S A 101:7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zago A, Jogger CR, Spear PG. 2004. Use of herpes simplex virus and pseudorabies virus chimeric glycoprotein D molecules to identify regions critical for membrane fusion. Proc Natl Acad Sci U S A 101:17498–17503. doi: 10.1073/pnas.0408186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicola AV, Willis SH, Naidoo NN, Eisenberg RJ, Cohen GH. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol 70:3815–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bender FC, Whitbeck JC, Ponce de Leon M, Lou H, Eisenberg RJ, Cohen GH. 2003. Specific association of glycoprotein B with lipid rafts during herpes simplex virus entry. J Virol 77:9542–9552. doi: 10.1128/JVI.77.17.9542-9552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol 17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitbeck JC, Peng C, Lou H, Xu R, Willis SH, Ponce de Leon M, Peng T, Nicola AV, Montgomery RI, Warner MS, Soulika AM, Spruce LA, Moore WT, Lambris JD, Spear PG, Cohen GH, Eisenberg RJ. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol 71:6083–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eisenberg RJ, Long D, Pereira L, Hampar B, Zweig M, Cohen GH. 1982. Effect of monoclonal antibodies on limited proteolysis of native glycoprotein gD of herpes simplex virus type 1. J Virol 41:478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedman HM, Cohen GH, Eisenberg RJ, Seidel CA, Cines DB. 1984. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the c3b complement component on infected cells. Nature 309:633–635. doi: 10.1038/309633a0. [DOI] [PubMed] [Google Scholar]
- 65.Cohen GH, Isola VJ, Kuhns J, Berman PW, Eisenberg RJ. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with western blotting. J Virol 60:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pereira L, Klassen T, Baringer JR. 1980. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect Immun 29:724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pereira L, Dondero DV, Gallo D, Devlin V, Woodie JD. 1982. Serological analysis of herpes simplex virus types 1 and 2 with monoclonal antibodies. Infect Immun 35:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Showalter SD, Zweig M, Hampar B. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein icp 4. Infect Immun 34:684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bender FC, Whitbeck JC, Lou H, Cohen GH, Eisenberg RJ. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J Virol 79:11588–11597. doi: 10.1128/JVI.79.18.11588-11597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aldaz-Carroll L, Whitbeck JC, Ponce de Leon M, Lou H, Hirao L, Isaacs SN, Moss B, Eisenberg RJ, Cohen GH. 2005. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein b5r. J Virol 79:6260–6271. doi: 10.1128/JVI.79.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Connolly SA, Whitbeck JJ, Rux AH, Krummenacher C, van Drunen Littel-van den Hurk S, Cohen GH, Eisenberg RJ. 2001. Glycoprotein d homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC (nectin-1) with different affinities. Virology 280:7–18. doi: 10.1006/viro.2000.0747. [DOI] [PubMed] [Google Scholar]