Foot-and-mouth disease virus (FMDV) is a highly contagious virus of cloven-hoofed animals and is recognized as the most important constraint to international trade in animals and animal products. African buffaloes (Syncerus caffer) are efficient carriers of FMDV, and it has been proposed that new virus variants are produced in buffalo during the prolonged carriage after acute infection, which may spread to cause disease in livestock populations. Here, we show that despite an accumulation of low-frequency sequence variants over time, there is no evidence of significant antigenic variation leading to immune escape. Therefore, carrier buffalo are unlikely to be a major source of new virus variants.
KEYWORDS: African buffalo, FMDV, foot-and-mouth disease virus, evolution, immune escape, persistence
ABSTRACT
African buffaloes (Syncerus caffer) are the principal “carrier” hosts of foot-and-mouth disease virus (FMDV). Currently, the epithelia and lymphoid germinal centers of the oropharynx have been identified as sites for FMDV persistence. We carried out studies in FMDV SAT1 persistently infected buffaloes to characterize the diversity of viruses in oropharyngeal epithelia, germinal centers, probang samples (oropharyngeal scrapings), and tonsil swabs to determine if sufficient virus variation is generated during persistence for immune escape. Most sequencing reads of the VP1 coding region of the SAT1 virus inoculum clustered around 2 subpopulations differing by 22 single-nucleotide variants of intermediate frequency. Similarly, most sequences from oropharynx tissue clustered into two subpopulations, albeit with different proportions, depending on the day postinfection (dpi). There was a significant difference between the populations of viruses in the inoculum and in lymphoid tissue taken at 35 dpi. Thereafter, until 400 dpi, no significant variation was detected in the viral populations in samples from individual animals, germinal centers, and epithelial tissues. Deep sequencing of virus from probang or tonsil swab samples harvested prior to postmortem showed less within-sample variability of VP1 than that of tissue sample sequences analyzed at the same time. Importantly, there was no significant difference in the ability of sera collected between 14 and 400 dpi to neutralize the inoculum or viruses isolated at later time points in the study from the same animal. Therefore, based on this study, there is no evidence of escape from antibody neutralization contributing to FMDV persistent infection in African buffalo.
IMPORTANCE Foot-and-mouth disease virus (FMDV) is a highly contagious virus of cloven-hoofed animals and is recognized as the most important constraint to international trade in animals and animal products. African buffaloes (Syncerus caffer) are efficient carriers of FMDV, and it has been proposed that new virus variants are produced in buffalo during the prolonged carriage after acute infection, which may spread to cause disease in livestock populations. Here, we show that despite an accumulation of low-frequency sequence variants over time, there is no evidence of significant antigenic variation leading to immune escape. Therefore, carrier buffalo are unlikely to be a major source of new virus variants.
INTRODUCTION
Foot-and-mouth disease virus (FMDV) is a single-stranded, positive-sense RNA virus in the Aphthovirus genus of the family Picornaviridae. Although clinically indistinguishable, FMDV exists as seven serotypes, i.e., A, O, C, Asia-1, and Southern African territories (SAT) types SAT1, SAT2, and SAT3, with multiple genetic and antigenic subtypes (1). FMDV causes a highly contagious, acute, vesicular disease in more than 70 ungulate species, including domestic livestock and numerous wildlife species (2). FMDV is endemic in large parts of Asia, Africa, and South America (3). The debilitating effects of foot-and-mouth disease (FMD) cause severe productivity losses and is the most important constraint to international trade of animals and animal products throughout the world (4).
In African savannah ecosystems, African buffalo (Syncerus caffer) appears to be the primary maintenance host of FMDV (5), although clinical FMD in buffalo is mild, and in most cases, naturally infected buffalo do not develop obvious clinical signs of FMD (6). Buffalo are considered to be efficient carriers of FMDV; studies under field conditions have reported virus recovery from individuals for 5 years (7), indicating that FMDV can perpetuate long-term in buffalo without reintroduction from neighboring populations. However, the frequency and titer of virus recovered decrease over time and carrier buffalo can clear virus over a 15-month period (8). Field studies support these experimental observations, showing a significant number of animals fail to maintain persistent infection for prolonged periods and the proportion of persistently infected animals decreases after reaching a peak in the 1- to 3-year age group (9). In the Kruger National Park (KNP; South Africa), serological surveys indicate that over 98% of buffalo have been exposed to all three local serotypes (SAT1, -2, and -3) of FMDV by the time they are 2 years old (6).
Despite significant recent advances in our knowledge, the anatomical and cellular sites where FMDV persists in vivo and the origin of virus detected in oropharyngeal fluid remain to be elucidated. There is evidence that virus can be detected in epithelial cells in close proximity to lymphoid follicles in tissues of the oropharynx and induce specific cellular responses in pharyngeal epithelial cells in combination with a cytokine-mediated response (2, 10, 11). It has also been hypothesized that the maintenance of nonreplicating FMDV by follicular dendritic cells (FDCs) represents a source of persistent infectious virus and contributes to the generation of long-lasting antibody responses against FMDV (12). FDCs are immune cells that regulate the humoral response and are located in secondary lymphoid tissues where they trap, retain, and multimerize antigens (Ag) and present Ag periodically to B cells (13). Following primary infection in cattle and sheep, it has been demonstrated that intact, nonreplicating FMDV particles or immune complexes are trapped on the surface of FDCs and maintained in the light zones within the germinal centers (GCs) of mucosal-associated lymphoid tissue and lymph nodes (12, 14). The FMDV genome has also been localized to similar sites in African buffalo (9). Several other viruses have been shown to be trapped and retained by FDC in lymph follicles, including human Immunodeficiency virus type 1 (HIV-1) (15), bovine viral diarrhea virus (16), bovine herpesvirus 1 (17, 18), Epstein-Barr virus (19), porcine circovirus type 2 (20), and classical swine fever virus (21). After being captured upon the FDC surface, these pathogens may remain viable, infecting and replicating in the lymphoid cells that collect and transport immune complexes during their passage through the lymph tissue and along the extensive processes of FDCs. This process may support intermittent virus replication cycles, despite the presence of high titers of neutralizing antibodies (22).
Populations of RNA viruses with high mutation rates, such as FMDV, are often composed of a viral swarm, i.e., a cloud of viral genotypes differing from the consensus sequence by a few mutations (23). The existence of complex populations in FMDV infections is well known (24, 25). Furthermore, the population structure can be influenced by extrinsic factors, such as the presence of virus-neutralizing antibodies (26).
The main objective of the current work was to determine the detailed sequence variation and evolution of the viral populations present in oropharyngeal tissues at different times during persistent infection of buffalo with FMDV after experimental challenge. Currently, two sites of FMDV persistence have been identified, namely, the epithelia of the oropharynx and nasopharynx and the light zone of germinal centers in lymphoid tissue of the head and neck (9). It is not known whether similar or distinct virus populations are present within these different sites. The sequence data reveal a complex structure, with multiple subpopulations and recombinants coexisting both in the inoculum and in infected buffaloes. However, there was limited variation in the viral sequences in samples from different individual animals and in epithelial and lymphoid tissues within the same animal. Therefore, even though the genetic structure of the virus populations is highly dynamic during persistent infections, we observed no evidence of significant antigenic variation and escape from antibody responses.
RESULTS
Epithelium (Epi) and lymphoid tissue samples and viruses used in this study were obtained from a previously described animal challenge experiment carried out at the KNP (9) in which African buffaloes (Syncerus caffer) were coinoculated with representatives of the three FMDV SAT serotypes. Viral persistence in vivo was not homogenous and only SAT1 persisted for 400 days postinfection (dpi); for this reason, subsequent sample analysis focused on this FMDV serotype.
The inoculum presents a complex genetic structure comprising two predominant subpopulations.
The genetic composition of the SAT1 virus component of the inoculum used in the challenge studies was explored by deep sequencing of the P1 coding region, including the 3′ end of the l-protease coding region. The consensus nucleotide sequence was almost identical to the published sequence of SAT1/KNP/196/91 (GenBank accession number KR108948) (9). The P1 coding region sequences showed the same population structure as VP1 both in the inoculum and subsequent samples. Therefore, all sequence analyses were focused on the outer capsid region VP1 (1D), which represents the highest variation and the major antigenic determining regions (27).
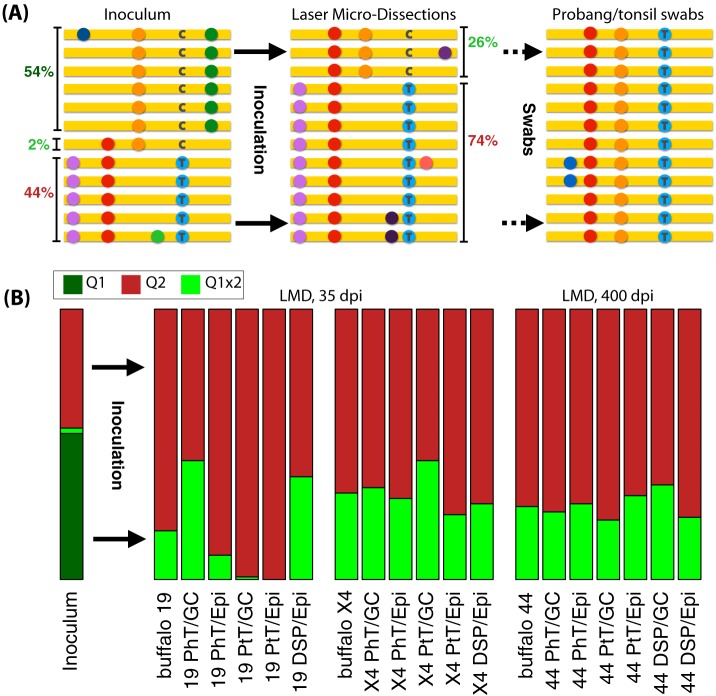
The inoculum contains at least two main subpopulations (denoted Q1 and Q2 in Fig. 1A) with approximate frequencies 54% and 44%, respectively. The VP1 (1D) coding sequences of the two subpopulations differ by 22 single nucleotide variants (SNVs), i.e., by 3% nucleotide divergence. The majority of the SNVs in the VP1 (1D) sequences (18 out of 22 SNVs; P = 4 × 10−3) were synonymous changes, most likely a signature of purifying selection during divergent evolution of subpopulations. We also detected reads of VP1 (1D) coding sequences from the inoculum that are possible recombinants of Q1 and Q2.
FIG 1.
(A) Graphical illustration of the structure and abundance of the genetic composition of the VP1 (1D) genomic region of the inoculum in terms of subpopulations (left) and their shifts in frequency in laser microdissected samples after inoculation (center) and in swab sample at day 280 postinoculation (right). The colored dots represent different mutations with respect to the reference sequence (in yellow). As an example, a C/T nucleotide variant is illustrated. (B) Frequencies of the different subpopulations in the inoculum and in LMDs from different individuals, tissues, and locations postinoculation. Samples are labeled by animal number (19, X4, or 44), tissue type (PhT, pharyngeal tonsils; PtT, palatine tonsils; DSP, dorsal soft palate), and germinal center (GC) or epithelium (Epi).
One of the putative recombinant sequences comprises about 2% of the inoculum and it is similar to the Q1 subpopulation but contains two nonsynonymous mutations that are found only in the population Q2. This third minor subpopulation could have been generated by recombination (four events between Q1 and Q2) or by convergent evolution (two mutation events on Q1). For this reason, we defined a third subpopulation which was named Q1x2.
This preexisting variability is a strong confounding factor for diversity analyses. Hence, in the next analysis of nucleotide diversity, we separate the effect of SNVs differing between Q1 and Q2 from other SNVs that were either present at very low frequencies or were a result of new mutations.
The population of viruses evolve under selection after inoculation.
To investigate sequence diversity and evolution of the virus in different tissues, we produced libraries of the VP1 (1D) coding region of SAT1 genomes and sequenced individual clones via Sanger sequencing. There was insufficient material from laser microdissected (LMD) samples to perform deep sequencing. Prior analysis of samples from this study by Maree et al. (9) showed that SAT1 RNA was detected in oropharyngeal tissue samples and tonsil swab samples until 400 dpi. However, at 400 dpi, SAT1 RNA was essentially limited to palatine tonsil (PtT), pharyngeal tonsil (PhT), and dorsal soft palate (DSP) samples. For this reason, PtT, PhT, and DSP tissues were targeted for LMD in two buffalo culled at 35 dpi (animals 19 and X4) and one at 400 dpi (animal 44).
Overall, 56 LMD tissue samples, which were positive for FMDV genome following reverse transcriptase PCR (RT-PCR) amplification of the VP1 (1D) coding region, were cloned and sequenced using Sanger sequencing. From these 56 samples, 674 nucleotides of the VP1 (1D) coding region of 569 individual clones out of 43 samples (12 for buffalo 19, 14 for buffalo X4, and 17 for buffalo 44) were subsequently analyzed. The sequences of the viral populations from these samples were compared with the nucleotide variability and subpopulation structure observed in the viral inoculum. In total, 57 variable nucleotide positions were found among sequences from the LMD tissue samples.
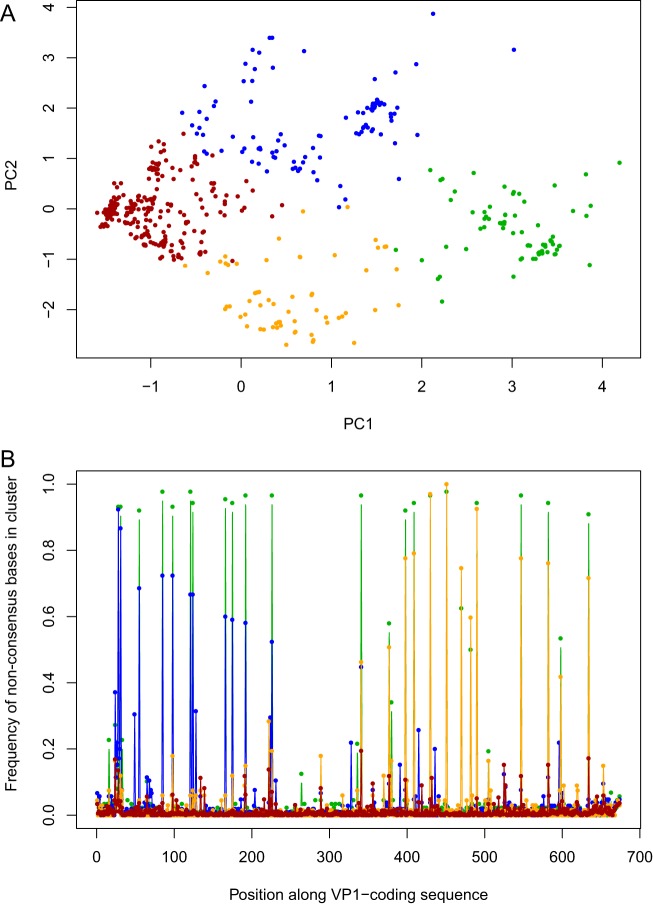
We observed the two main precharacterized SAT1 subpopulations Q1x2 and Q2 within the LMD tissue samples as well as recombinants between the subpopulations. The major virus subpopulation identified in buffaloes corresponds to the minor population Q2 of the inoculum, while the second most frequent population in buffaloes corresponds to the recombinant sequence Q1x2 in the inoculum (Fig. 1B). The frequencies of the two subpopulations Q2 and Q1x2 in buffalos are approximately 74% and 26%, respectively, across multiple tissues and all three animals, as shown in Fig. 1B. Therefore, there was a large and consistent shift in the frequency of these subpopulations in tissues compared with the inoculum, which was 44% and 2%, respectively. To confirm the presence of discrete subpopulations in the LMD samples, the sequences were analyzed using K-means clustering (Fig. 2). In addition to the two major subpopulations, two minor populations were also identified, both composed of recombinants of Q2 and Q1x2 (Fig. 2). Similar changes in virus subpopulation frequencies across multiple animals are a clear signal of strong consistent selective pressures, possibly due to the transition from passage in culture to infection in vivo. Furthermore, the consistency of the final frequencies across different tissues, time postinfection, and locations within tissues suggests that selection occurred in the early stages of the infection.
FIG 2.
(A) Subpopulation structure of VP1 sequences from LMDs. All sequences are projected on the first two principal components PC1 and PC2 of sequence space. The four clusters are illustrated by different colors. The two main subpopulations correspond to the clusters on the left and right of the plot, on opposite sides of PC1 (brown and green). (B) Genetic content of the clusters. The fraction of sequences with a nucleotide difference with respect to the global consensus is shown for each nucleotide position and cluster. The two main subpopulations differ by the 20 positions shown at high frequency in the plot. The other two clusters (blue and orange) are centered around two sets of recombinants corresponding to a major breakpoint in the middle of the VP1 sequence.
The simplest interpretation of the shifts in sequence frequencies is that different viruses have different intrahost fitness and, hence, different selective advantage with respect to each other. The subpopulation Q1 is probably less fit within buffalo hosts; hence, it is strongly selected against during the infection and disappears in favor of the subpopulations Q2 and Q1x2. Given the importance of VP1 for the immune response to the virus, we conjecture that the targets of this selective pressure could be one of the two nonsynonymous mutations in VP1 (1D) that differ between Q1 and Q1x2. In fact, these two mutations are the only variants in the VP1 (1D) coding sequence that become fixed among the virus population sequences from buffalo tissues. We can speculate the functional significance of one of these fixed mutations, G112R in VP1, which is associated with binding heparan sulfate and adaptation to cultured cells. However, there could be selected variants in other coding regions. The frequency of Q1x2 changed consistently from about 2% to 26% in all animals at day 35 dpi, showing that selective pressure drives the increase in frequency of Q1x2 (Fig. 1A).
Recombination occurred at a high rate during the acute phase of infection postinoculation and at a lower rate during the persistent phase, as discussed in detail in a companion publication by Ferretti and colleagues (28).
Viral population diversity is similar in individual animals and tissues.
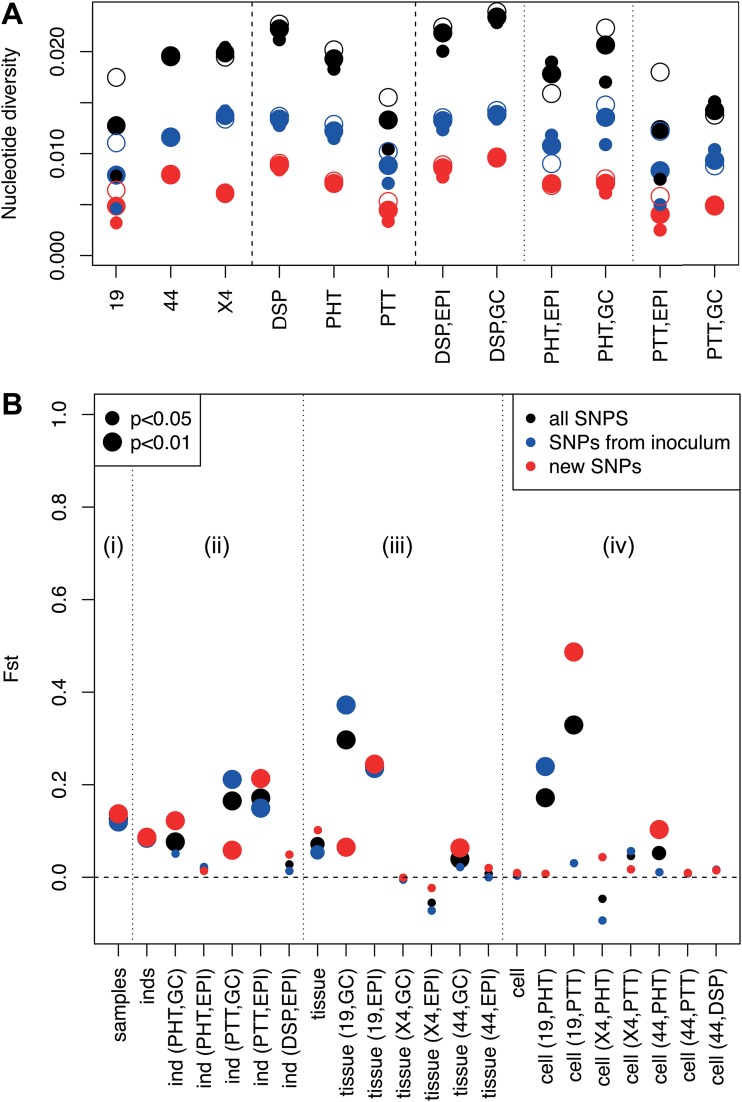
The nucleotide diversity of SAT1 VP1 (1D) coding sequences within different animals and tissues is illustrated in Fig. 3A. The average nucleotide diversity among sequences belonging to the same individual, tissue, and location is in the range 0.01 to 0.025 nucleotide differences per position. SNVs from the inoculum account for about two-thirds of this diversity, while the other one-third corresponds to variants that were absent or at very low frequency in the inoculum. There are minor but significant differences in the diversity of sequences within different animals or tissues. In particular, viruses in the dorsal soft palate tend to be approximately 35% more diverse than in tonsils, especially palatine tonsils (P < 0.001). When only new variants are considered, we found that viruses sampled at 400 dpi from buffalo 44 have an average diversity of 0.008, which is higher than the average diversity of 0.005 for viruses sampled at 35 dpi from buffalo 19 and X4 (P > 0.001), as expected since there was more time to accumulate new mutations. On the other hand, virus populations in epithelium (Epi) or GCs from the same animal and tissue show comparable levels of diversity generated by new variants, while diversity from preexisting variants is slightly higher in GCs. Overall, similar levels of nucleotide diversity between GCs and Epi were observed irrespective of the tissue or the buffalo analyzed.
FIG 3.
(A) Pairwise nucleotide diversity of VP1 viral sequences (defined as the average number of nucleotide differences between any pair of sequences) per base, within animals, tissues, and locations. Small dots represent diversity between sequences from the same sample, while empty dots represent diversity between sequences from different samples from the same animal/tissue/location. (B) Genetic differentiation between samples, animals, tissues, and locations within tissue as expressed by the Fst statistics (Fst of 0, no differentiation at all; Fst of 1, strong differentiation). The four subparts estimate differentiation (i) between samples within the same animal, tissue, and cell type; (ii) between animals within the same tissue and cell type; (iii) between tissues within the same animal and cell type; and (iv) between cell types within the same animal and tissue.
The patterns of genetic variation within the viral sequences between buffaloes, tissues, and Epi/GC were examined with several analysis of molecular variance (AMOVA)-like analyses. Results are shown in Fig. 3B in terms of Fst, a classical measure of genetic differentiation between populations (29). This measure can be interpreted as the fraction of diversity due to the molecular evolution within each population.
The diversity of viral VP1 (1D) coding sequences from different individuals is significant but not strong, with an Fst of ∼0.07 being slightly higher among sequences from PtT (Fst, ∼0.16). The evidence is significant but weaker for diversity between different tissues within the same animal, with a similar value of Fst of ∼0.06. No evidence for overall sequence differentiation between GCs and Epi within the same animal was found. These results hold both for preexisting variants and new variants. Only buffalo 19 shows a strong differentiation of virus population sequence between tissues and anatomical locations within tissues. Finally, sequences from different microdissections from the same animal, tissue, and cell type show a stronger genetic differentiation, namely, an Fst of ∼0.13, suggesting that local genetic heterogeneity within tissues can be more relevant than differences between tissues or animals.
The immediate interpretation of these findings is that the intrahost molecular evolution of the virus occurred mostly during the early acute infection phase. In this phase, high viral load and high replication rate favored the rapid evolution of the viral population, which contrast with the low viral replication in oropharyngeal tissues during persistent infection, which may be responsible for the limited diversity of sequences in different animals, tissues, and time points.
Viruses found in oropharyngeal samples suggest the presence of a reservoir and ongoing viral replication during persistence.
Six SAT1 viruses were isolated from the probangs and tonsil swabs from four animals (buffaloes 26, 44, 61, and X3) during persistent infection (213 to 400 dpi) (9). Surprisingly, SAT1 VP1 (1D) sequences derived from deep sequencing of the probang and swab samples did not share the same genetic subpopulation structure as viruses from PtT of the microdissected animals, despite being sampled from locations in close proximity. The buffalo sampled at 400 dpi (number 44) was the only one for which VP1 (1D) coding sequences are available from both microdissections and swabs; the viral sequences from the tonsil swab differed significantly from the viruses found inside the tonsillar tissue (P < 10−100). In all animals, viral sequences from the probangs and swabs showed a reduced within-sample variability. At the consensus level, these sequences are recombinants of the sequences identified in the inoculum, with a dominant contribution of subpopulation Q2 (Fig. 4). In addition, there are a large number of substitutions with respect to the inoculum. The average estimate for the substitution rate in these sequences is 1.2% per base per year, which is comparable with the substitution rate of FMDV from previous studies (30). There is a significant excess of synonymous substitutions (P < 10−9), suggesting that despite the high substitution rates, these sequences are evolving under purifying selection.
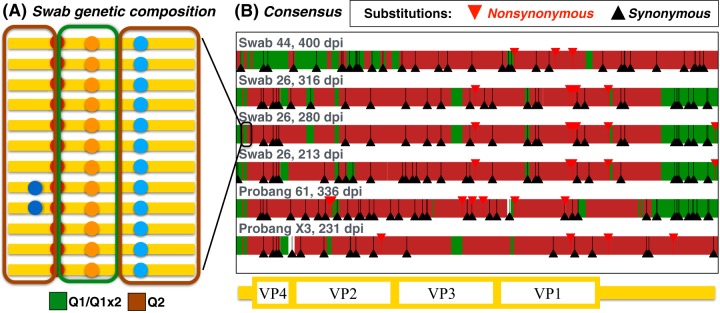
FIG 4.
Genetic composition of the viral sequences from tonsil swabs and probangs. (A, left) Illustration of the genetic composition of P1 and flanking regions of the viral populations in the tonsil swabs and probang samples. The origin of each sequence fragment is inferred from the presence/absence of variants characterizing the original inoculum composition (see also Fig. 1). (B, right) Consensus sequences in terms of the original inoculum composition. The colors of the sequences illustrate their composition in terms of fragments of the two main subpopulations found in the inoculum, while triangles represent the positions of substitutions with respect to the inoculum.
One of the animals (buffalo 26) was sampled via tonsil swabs at 213, 280, and 316 dpi during persistent infection. As shown in Fig. 4, the composition of the consensus sequence from this animal differed at different times postinfection, strengthening the evidence that the virus was still replicating in this phase.
There were signatures of parallel evolution of the viruses sampled from probang and swabs. At the consensus level, the population of sequences in probang/swabs from different animals showed signs of convergence in terms of recombination fragments and substitutions (Fig. 4). Recombination fragments from Q1 shared among three or more animals cover about 5% of the sequence, versus 1.7% expected if the fragments were randomly distributed. Furthermore, there were 54 substitutions shared between sequences from two or more animals, a strong excess with respect to the random expectation of less than 18 shared substitutions. While parallel selection in different animals is the most plausible explanation, other explanations include mutation/recombination “hotspots” and the presence of these variants at low frequency in the inoculum. More interestingly, no polymorphisms with frequencies higher than 0.1 in VP1 (1D) could be found in probang samples. Therefore, we extended the analysis to the Leader and P1 coding region which revealed 5 SNVs per sample above that frequency. The most frequent variant was a C/T mutation near the 3′ end of the l-protease coding region, which appears to be already present in the inoculum with a frequency of 0.16 and was present in all tonsil swab samples at frequencies of approximately 0.45. The consistency in the final frequency of this variant across two different animals and three different time points is likely to be a consequence of tissue-specific intrahost selective pressures acting on the virus.
Taken together, these results suggest that the virus replicates in discrete loci near lymphoid tissue of the oropharynx during the carrier phase. Genetic evolution in these loci is shaped by strong purifying selection pressure as well as intrahost adaptation.
Sera collected at early and late time points have an equal capacity to neutralize viruses at all time points.
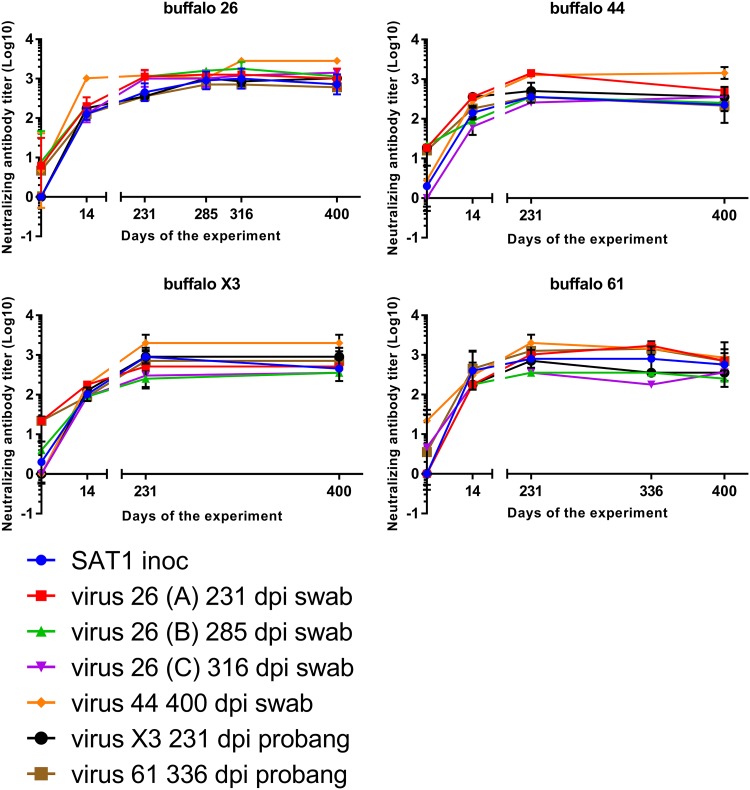
To determine differences in antigenicity between the SAT1 variants, six oropharyngeal SAT1 virus isolates obtained between 213 and 400 dpi and the SAT1 challenge virus were analyzed in cross-neutralization assays against serum samples collected from four buffalo at 0, 14, 231, 285 to 336, and 400 dpi (Fig. 5). Neutralizing antibody titers of >1.65 log10 on 14 dpi and >2.00 log10 50% tissue culture infective dose (TCID50) at later time points were present in all four buffalo against both the SAT1 challenge virus as well as the six SAT1 oropharyngeal variants.
FIG 5.
Evaluation of serological cross-neutralization between SAT1 viruses isolated from four buffaloes at different time points postinfection. Serum samples from 4 animals (buffalos 26, 44, X3, and 44), obtained at days 0, 14, 231, 285, 316, 336, and 400 dpi were evaluated on an antibody cross-neutralization assay using 6 different SAT1 isolates obtained from the oropharynx (by tonsil swab or probang sampling) from the same animals. Neutralizing antibody titers are expressed as the inverse dilution of serum yielding a 50% reduction of virus titer (Log10). The assay was repeated 3 times, results represent the mean of 3 repetitions of the assay, and bars represent the standard error of the mean (SEM). All animals are protected from day 14 to day 400 dpi against all virus isolates (Log 10 Ab titers, >1.65).
Statistical analysis (Kruskal-Wallis test) was also performed to evaluate the temporal differences in humoral response, but no significant differences (P = 0.12) among time points were observed, indicating the efficiency of the humoral response to neutralize virus did not change during the study. In agreement, no difference (Mann-Whitney U test, P = 0.11) was observed between the ability of each of the different sera harvested between 14 and 400 dpi to neutralize the inoculum or viruses isolated at later time points (>231 dpi) in the study.
In order to investigate if the neutralizing antibody responses to the SAT1 viruses isolated from different animals varied, the homologous-r1 (Ho-r1) and heterologous-r1 (He-r1) ratios were calculated and statistically analyzed using the Mann-Whitney U test. As expected, the Ho-r1 values were significantly (P = 0.0004) higher than the He-r1, confirming similar reactivity of sera collected at both early and late time points, to virus that was isolated from the same animal. However, even though there were significant differences between the groups, the titer of neutralizing antibodies of the heterologous sera was still greater than 1.65 log10 TCID50 which is consistent with protective immune responses (31).
DISCUSSION
The mechanism of FMDV persistence in vivo remains largely unknown. Which cell types harbor the virus particles, how virus is maintained, and precisely where low-level replication occurs are still open questions. Juleff et al. (12), showed that FDCs in the light zones of the GC located in lymph nodes and lymphoid tissue associated with the head and neck trap and retain intact FMDV particles. Also, viral genomes were detected in epithelial tissue in close proximity to these GCs. The authors concluded this may be a source of the virus detected during the persistent infection (9, 12).
In this study, we sequenced the VP1 (1D) region of viruses present at the two anatomical sites. All analyses presented in this paper are based on the frequencies of the SNVs in the challenge virus and in LMDs; only relatively common variants were analyzed, i.e., variants with a frequency of greater than 10%. In the analyses of intrahost evolution and selection, we compared variant frequencies obtained through different technologies; the results of these comparisons are reliable because there are no relevant biases in variant calling and frequency estimation of common variants. In fact, biases on frequency estimation for Sanger or for deep sequencing are of the order of the sequencing errors, i.e., about 0.5% (32), while the smallest change in frequency that we observe is more than 20%. For the samples sequenced both by Sanger and deep sequencing, the consensus sequences were consistent between the two approaches.
Sequencing the probang and swab samples required a single passage in cell culture, which in principle could introduce bias in the genetic composition of the viral population. We believe a single passage in cell culture is unlikely to result in significant changes. Indeed, when the virus SAT1/KNP/196/91 was originally isolated, the P1 region of the clinical sample and the virus after one passage in IB-RS-2 cells (33) was Sanger sequenced, and residue changes were detected at only three sites, namely, E133D and Q169H/Q [both VP2 (1B)] and E135K/D/N/E [VP3 (1C)]. The strong reduction in genetic variability in the tonsil swabs is also unlikely to be caused by sampling or sequencing issues; such issues would skew variant frequencies toward low values, while we observe SNVs in a wide range of frequencies up to about 50% in multiple samples.
There is a practical constraint when challenging numerous animals with a consistent inoculum of live virus, that the challenge material has to be expanded in cell culture. Even though the inoculum was passaged in IB-RS-2 cells, Q1 remains the major population after passage, as determined by consensus sequence analysis. Interestingly, after inoculation, the mutation G112R in VP1 (1D), which is usually associated with cell culture adaptation, becomes fixed in some virus subpopulations. Clearly, viruses containing this mutation have an advantage in vivo. In contrast, viruses containing other uncharacterized changes associated with cell culture are likely disadvantaged; hence, the recombinant subpopulation Q1x2 (which is still very similar to the original strain) has a fitness advantage over the subpopulation adapted to culture (Q2) and grows more rapidly than Q2 within hosts, therefore increasing in frequency. To further validate this conclusion, we removed VP1 residue 112 from our analysis, and there was no change in the subpopulation analysis in the LMD or swab or probang samples.
The analyses of genetic differentiation did not find any significant difference in the genetic composition of the sequences present in GCs and Epi and only small differences among tissues (PhT, PtT, and DSP), both at 35 and 400 dpi. For example, analysis of tissues from animal 44 at 400 dpi showed similar estimations of diversity both among the three tissues and between their respective GCs and epithelium. These results suggested that viruses with very similar sequences were circulating within and among all the tissues at the initial time points and subsequent low-level replication did not greatly influence the overall structure of those populations. Taken together, the results indicated the repertoire of FMDV trapped in the FDC for a period of more than a year were comprised of genetically similar variants because the majority of this diversity was generated during the acute, viremic phase of infection. Also, the similarity between the viral sequences in Epi and GCs suggest there is a link between the two sites. These results are in agreement with the idea that virus replicating in the epithelium is transferred to FDC in adjacent GCs, where there is no evidence of further viral replication (34). It is possible viruses from the GCs are transported to the Epi by immune cells, for example B cells (35), to seed new infections.
Deep sequencing revealed that the SAT1 inoculum was a complex mixture that could be grouped by the presence of SNV in VP1 (1D) into two major populations and their recombinants. Although such a genetic structure has only recently been described for FMDV (36), its occurrence has been previously discussed for other viruses (37–39).
Interestingly, after inoculation we observed a consistent change in the population structure in different animals, tissues, and time points. This finding shows that strong selective pressures shaped the intrahost evolution of the viral population during the acute phase of the infection. Our results suggest the composition of FMDV populations evolves in a highly dynamic fashion by mutation, selection, and recombination, but despite all these changes, the viruses do not escape neutralization by circulating anti-FMDV polyclonal sera. Specifically, there was no significant difference in the capacity of sera harvested between day 14 and day 400 postinfection to neutralize the inoculum or viruses isolated at later time points from the same animal.
Therefore, our analyses of virus evolution during persistent infection are consistent with the lack of immune escape reported and contrast with those described for HIV-1 in humans, in which FDCs were shown to act as viral reservoirs that accumulated different genetic variants during a period of 22 months (15). The population genetics data reported in this study do not support the existence of intense FMDV replication in the oropharyngeal tissue during persistent infection, at least not at a level that can significantly influence the population diversity (40, 41). Similarly, Juleff and colleagues (12) detected nonreplicating virus in GCs and only rare cells expressing viral nonstructural proteins, indicating virus replication in cattle at 38 dpi. Similar studies have demonstrated low levels of virus replication within pharyngeal epithelial cells (10, 34, 42, 43) during persistent infection. Gebauer et al. (44) proposed the selection of FMDV antigenic variants occurs during the first 2 months of infection in cattle and that the number of amino acid variants fixed after a few days of replication and after several years of evolution was similar. In the same way, other studies did not detect significant levels of virus replication in the oropharyngeal tissues during persistent FMDV infection in cattle (45).
These in vivo situations are not comparable with in vitro cell passage experiments, where cells are repeatedly infected with FMDV (46, 47) and replicate at high rates. Deep sequencing of tonsil swabs and probang samples in our study revealed a large number of substitutions with respect to the challenge virus, with rates comparable to those usually associated with FMDV evolution at epidemic scales; however, these substitutions are not driven by immune escape but rather by purifying selection. These findings support the existence of viral replication within a small reservoir during persistent infection. In agreement with these results, the analysis of probang samples in other in vivo studies has identified increasing levels of divergence during persistence in cattle (44), water buffalo (30, 48), and African buffalo (49).
In summary, despite the inherent tendency of the FMDV genome to evolve over time due to mutation and recombination, during experimental persistent SAT1 infections in African buffalo there is limited sequence variation detected and no evidence of immune escape.
MATERIALS AND METHODS
Animal samples and viruses.
All samples used in this study came from a previously described animal challenge experiment carried out at the Kruger National Park, South Africa (9). In brief, African buffalo (Syncerus caffer) were coinoculated with representatives of the three FMDV SAT serotypes, namely, SAT1/KNP/196/91 (GenBank accession number KR108948), SAT2/KNP/19/89 (GenBank accession number KR108949), and SAT3/KNP/1/08 (GenBank accession number KR108950). These viruses used as inoculum originated from African buffalo and were amplified in cell culture using porcine cells PK15 (one passage) and IB-RS-2 cells (5 passages). Animals were sequentially culled at 35, 95, 185, and 400 days postinfection (dpi), and lymphoid tissue samples from the head and neck were collected in optimal cutting temperature compound (Tissue-Tek). As described by Maree et al., (9) viral persistence in vivo was not homogenous and the SAT1 isolate was the only one that persisted until 400 dpi in palatine tonsil (PtT), pharyngeal tonsil (PhT), and dorsal soft palate (DSP) lymphoid tissue. Therefore, the present study focused on the analysis of SAT1 serotype in PtT, PhT, and DSP tissues obtained from two buffaloes at 35 dpi (animals 19 and X4) and one buffalo at 400 dpi (animal 44).
SAT1 viruses from 4 animals (buffalo 26, 61, X4, and 44) were isolated from the palatine tonsil swabs using nylon brushes (Cytotak Transwab), at late time points postinfection (200 to 400 dpi), and passaged once in cell culture using ZZR-127 cells. Moreover, serum samples from the same animals were obtained at early (0 and 14 dpi) and late (213 to 400 dpi) time points after SAT1 exposure.
Laser-capture microdissection.
The membrane-based laser-capture microdissection (LCM) method was adapted from a previous described protocol (12). Briefly, 7-μm-thick cryosections of PtT, PhT, and DSP tissues from two buffalo culled at 35 dpi (19 and X4), and one at 400 dpi (44) were affixed to framed polyester (PET)-membrane slides (Leica, UK) fixed in 100% cold ethanol and stained with 1% wt/vol toluidine blue (Sigma-Aldrich, UK) to observe the anatomy of the tissue. After dehydrating the tissue in ethanol, GC or epithelium (Epi) samples from each of the tissues were targeted for microdissection using the AS-LMD7000 microscope (Leica, Germany). RNA was extracted from microdissected tissue samples using RNeasy micro kits (Qiagen, UK), and cDNA was synthetized using TaqMan reverse transcription reagents and random hexamers (Applied Biosystems).
PCR, cloning, and Sanger sequencing.
The presence of SAT1 in the cDNA synthetized from microdissected GCs and Epi was screened by PCR targeting a 681-bp fragment in the VP1 (1D) coding region of SAT1, amplified with primers F (5′-AGTGCTGGACCCGACTTCGA-3′) and R (5′-TGTAGCGATCCTTGCCACCGT-3′) using Platinum Taq hi-fidelity polymerase (Invitrogen). The amount of virus in PCR-positive dissected samples was quantified by means of an SAT1-specific reverse transcription-quantitative PCR (qRT-PCR) on MX3005P QPCR systems (Stratagene, USA) using TaqMan universal PCR master mix (Applied Biosystems) as previously described (9). Copies of viral RNA per microliter were calculated using the standard curve method and MxPro software (Stratagene, USA), and samples with no detectable fluorescence above the threshold after 50 cycles were considered negative (50).
Using the qRT-PCR results, the virus concentration in the positive samples was standardized at 100 viral RNA copies per μl (CT, ∼35). Then, the PCR was repeated on the positive samples using equal amounts of template. Amplified PCR products were separated by gel electrophoresis and purified using a QIAquick gel extraction kit (Qiagen).
The amplified PCR products were cloned using a TOPO TA cloning kit for sequencing (Life Technologies) and transformed into One Shot TOP10 competent Escherichia coli (Invitrogen). Cells were grown overnight in selective plates, and for every sample 24 positive recombinants were selected and the plasmid isolated using a QIAprep spin miniprep kit (Qiagen).
Cycle sequencing was carried out using plasmid primers M13F and M13R and the BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems) on an ABI Prism 3730 analyzer (14). The SeqMan Pro program from the Lasergene 11 package (DNAStar) was used for assembly and proofreading of sequence trace files. Alignment and manipulation of sequences were performed using BioEdit software.
Deep sequencing.
Probangs and tonsil swabs from 4 buffaloes at late time points after SAT1 infection were passaged once in ZZR-127 cells (51), and whole genomes were deep sequenced. In addition, a sample of the SAT1 inoculum was directly sequenced. One nanogram of each double-stranded DNA (dsDNA) sample was used to prepare sequencing libraries using the Nextera XT DNA sample preparation kit (Illumina) according to manufacturer’s instructions. Libraries were sequenced on a MiSeq instrument using 300-cycle v2 reagent cartridges (Illumina) to produce paired-end reads of approximately 150 bp each. High-throughput reads were aligned with GEM (52). A sensitive in-house pipeline (Ribeca et al., in preparation) based on SPAdes (53) and additional bespoke software were used for a de novo assembly of the consensus. Variant calling was performed using SiNPle, an in-house program based on an approximation of a Bayesian method (54) optimized for high sequence coverages, selecting only variants with a P value of <0.05.
Diversity and differentiation estimations.
Sequencing errors were filtered using an exponential error model as described in reference 36. Both the nucleotide diversity and the differentiation of the resulting set of SNVs were estimated using a statistical approach that weights contributions from all animals, tissues, and locations equally.
Any linear combination of the number of pairwise differences between sequences was an unbiased estimator of nucleotide diversity. However, the uncertainty of the estimate was complicated by the unequal representation of individuals, tissues, and locations (GC or Epi) in terms of the number of samples and number of sequences per sample. To correct for this, we estimated nucleotide diversity by a weighted mean of point estimators of nucleotide diversity within a given individual, tissue, and location. Our weighted estimator assigned equal weight for all sequences from the same sample; equal weight for all samples from the same individual, tissue, and location; and equal weight for all combinations of individual, tissue, and location.
To confirm the subpopulation structure, we clustered all VP1 sequences from LMDs by K-means clustering. The optimal number of clusters K = 4 was assessed by the elbow method. K-means was then performed with 4 clusters and 100 starting points.
Principal component analysis in sequence space was also performed by recoding each nucleotide in each sequence and position into its ranking in abundance (from 1 to 4) for the position considered and then running singular value decomposition on the resulting integer-valued matrix (55).
To estimate genetic differentiation, we defined Fst as (29), where was the average nucleotide diversity between sequences from the same group, while was the average nucleotide diversity between sequences from different groups. Groups are either animal, tissue, or cell type. Nucleotide diversities were computed according to the estimator described above.
The significance of differentiation between animals, tissues, and locations was evaluated by an AMOVA-style permutation test (55) of the weighted Fst values (based on 1,000 permutations). The permutation test was performed as follows. First, a random permutation (conserving all groupings of individual/tissue/location except the tested one) was extracted. Then, the weights were permuted according to the same permutation and then reweighted once more to satisfy the condition III again. Finally, the averages of the nucleotide differences were evaluated according to the final weights, and the randomized value of Fst was computed from these averages according to the formula above.
Virus isolates and cross-neutralization assay.
Sera obtained at days 0, 14, 231, 285 to 336, and 400 dpi from buffaloes 26, 61, X3, and 44 (9) were titrated by a standard virus neutralization test (VNT) (56) and cross-neutralization assay on porcine kidney (IB-RS2) cells as an indicator, using viruses isolated from probangs and tonsil swabs from the same animals at 231, 285, 316, 336, or 400 dpi (9). Each test was performed at least 3 times, and SAT1 challenge virus was used for comparison purposes in the assays. Neutralizing and cross-neutralizing antibody titers, calculated by the Spearman-Karber method, were expressed as the last dilution of serum that neutralizes 50% of the virus (100 TCID50).
The neutralizing antibody (Ab) titer of each serum against each isolate was then classified as homologous (sera and virus isolated from the same animal), heterologous (sera and virus isolated from different animals), or SAT1 inoculum (sera against inoculum). To investigate the differences between homologous and heterologous humoral responses to the SAT1 isolates, the homologous-r1 (Ho-r1) and heterologous-r1 (He-r1) ratios were calculated dividing the corresponding homologous or heterologous Ab titer by the neutralizing Ab titer of SAT1 inoculum.
The neutrality of the whole data set and every subset was checked with Kolmogorov-Smirnoff tests. Then, Mann-Whitney and Kruskal-Wallis tests were used to compare the following: (i) the homologous and heterologous r1-values obtained in the cross-neutralization assays, (ii) the r-1 values obtained at different time points along the experiment, and (iii) the r1-values obtained when they are standardized with the SAT1 inoculum (He/SAT1) or the homologous titers (He/Ho).
ACKNOWLEDGMENTS
We thank the animal unit staff at Kruger National Park for assisting with the in vivo experiment. We are also grateful to the World Reference Laboratory for FMDV (WRL) personnel at The Pirbright Institute for their support with FMDV sequencing.
We are grateful to the Wellcome Trust and the Biotechnology and Biological Sciences Research Council (BBSRC) of the United Kingdom and the United States Department of Agriculture (USDA) for their funding. Nicholas D. Juleff and Martí Cortey were funded by Wellcome Trust intermediate-level research fellowship 091726/Z/10/Z. Luca Ferretti, Eva Pérez-Martín, Graham Freimanis, Paolo Ribeca, Julian Seago, and Bryan Charleston are funded by the BBSRC Institute Strategic Program on Enhanced Host Responses for Disease Control at The Pirbright Institute (BBS/E/I/00007030, BBS/E/I/00007032, and BBS/E/I/00001942). Fuquan Zhang was supported by collaborative BBSRC-USDA funding grant BB/L011085/1.
None of the authors have a financial or personal conflict of interest that could inappropriately influence the content of the paper.
Specific author contributions are as follows: conceptualization of the study, Nicholas D. Juleff, Bryan Charleston, and Francois F. Maree; methodology and data collection, all; data analysis, Martí Cortey, Luca Ferretti, Paolo Ribeca, and Eva Pérez-Martín; writing of the manuscript, Martí Cortey, Luca Ferretti, Eva Pérez-Martín, Francois F. Maree, and Bryan Charleston; review and editing, all; and supervision, Bryan Charleston.
REFERENCES
- 1.Carrillo C, Tulman ER, Delhon G, Lu Z, Carreno A, Vagnozzi A, Kutish GF, Rock DL. 2005. Comparative genomics of foot-and-mouth disease virus. J Virol 79:6487–6504. doi: 10.1128/JVI.79.10.6487-6504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandersen S, Zhang Z, Donaldson AI. 2002. Aspects of the persistence of foot-and-mouth disease virus in animals—the carrier problem. Microbes Infect 4:1099–1110. doi: 10.1016/S1286-4579(02)01634-9. [DOI] [PubMed] [Google Scholar]
- 3.Alexandersen S, Zhang Z, Donaldson AI, Garland AJ. 2003. The pathogenesis and diagnosis of foot-and-mouth disease. J Comp Pathol 129:1–36. doi: 10.1016/S0021-9975(03)00041-0. [DOI] [PubMed] [Google Scholar]
- 4.Casey-Bryars M, Reeve R, Bastola U, Knowles NJ, Auty H, Bachanek-Bankowska K, Fowler VL, Fyumagwa R, Kazwala R, Kibona T, King A, King DP, Lankester F, Ludi AB, Lugelo A, Maree FF, Mshanga D, Ndhlovu G, Parekh K, Paton DJ, Perry B, Wadsworth J, Parida S, Haydon DT, Marsh TL, Cleaveland S, Lembo T. 2018. Waves of endemic foot-and-mouth disease in eastern Africa suggest feasibility of proactive vaccination approaches. Nat Ecol Evol 2:1449–1457. doi: 10.1038/s41559-018-0636-x. [DOI] [PubMed] [Google Scholar]
- 5.Thomson GR. 1995. Overview of foot and mouth disease in southern Africa. Rev Sci Tech 14:503–520. doi: 10.20506/rst.14.3.855. [DOI] [PubMed] [Google Scholar]
- 6.Thomson GR, Vosloo W, Esterhuysen JJ, Bengis RG. 1992. Maintenance of foot and mouth disease viruses in buffalo (Syncerus caffer Sparrman, 1779) in southern Africa. Rev Sci Tech 11:1097–1107. doi: 10.20506/rst.11.4.646. [DOI] [PubMed] [Google Scholar]
- 7.Condy JB, Hedger RS, Hamblin C, Barnett IT. 1985. The duration of the foot-and-mouth disease virus carrier state in African buffalo (i) in the individual animal and (ii) in a free-living herd. Comp Immunol Microbiol Infect Dis 8:259–265. doi: 10.1016/0147-9571(85)90004-9. [DOI] [PubMed] [Google Scholar]
- 8.Bengis RG, Thomson GR, Hedger RS, De Vos V, Pini A. 1986. Foot-and-mouth disease and the African buffalo (Syncerus caffer). 1. Carriers as a source of infection for cattle. Onderstepoort J Vet Res 53:69–73. [PubMed] [Google Scholar]
- 9.Maree F, de Klerk-Lorist LM, Gubbins S, Zhang F, Seago J, Perez-Martin E, Reid L, Scott K, van Schalkwyk L, Bengis R, Charleston B, Juleff N. 2016. Differential persistence of foot-and-mouth disease virus in African buffalo is related to virus virulence. J Virol 90:5132–5140. doi: 10.1128/JVI.00166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang ZD, Kitching RP. 2001. The localization of persistent foot and mouth disease virus in the epithelial cells of the soft palate and pharynx. J Comp Pathol 124:89–94. doi: 10.1053/jcpa.2000.0431. [DOI] [PubMed] [Google Scholar]
- 11.Stenfeldt C, Eschbaumer M, Smoliga GR, Rodriguez LL, Zhu J, Arzt J. 2017. Clearance of a persistent picornavirus infection is associated with enhanced pro-apoptotic and cellular immune responses. Sci Rep 7:17800. doi: 10.1038/s41598-017-18112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juleff N, Windsor M, Reid E, Seago J, Zhang Z, Monaghan P, Morrison IW, Charleston B. 2008. Foot-and-mouth disease virus persists in the light zone of germinal centres. PLoS One 3:e3434. doi: 10.1371/journal.pone.0003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukumar S, El Shikh ME, Tew JG, Szakal AK. 2008. Ultrastructural study of highly enriched follicular dendritic cells reveals their morphology and the periodicity of immune complex binding. Cell Tissue Res 332:89–99. doi: 10.1007/s00441-007-0566-4. [DOI] [PubMed] [Google Scholar]
- 14.Juleff N, Valdazo-Gonzalez B, Wadsworth J, Wright CF, Charleston B, Paton DJ, King DP, Knowles NJ. 2013. Accumulation of nucleotide substitutions occurring during experimental transmission of foot-and-mouth disease virus. J Gen Virol 94:108–119. doi: 10.1099/vir.0.046029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keele BF, Tazi L, Gartner S, Liu Y, Burgon TB, Estes JD, Thacker TC, Crandall KA, McArthur JC, Burton GF. 2008. Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J Virol 82:5548–5561. doi: 10.1128/JVI.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fray MD, Supple EA, Morrison WI, Charleston B. 2000. Germinal centre localization of bovine viral diarrhoea virus in persistently infected animals. J Gen Virol 81:1669–1673. doi: 10.1099/0022-1317-81-7-1669. [DOI] [PubMed] [Google Scholar]
- 17.Winkler MT, Doster A, Jones C. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsils of latently infected calves. J Virol 74:5337–5346. doi: 10.1128/JVI.74.11.5337-5346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melot F, Defaweux V, Jolois O, Collard A, Robert B, Heinen E, Antoine N. 2004. FDC-B1: a new monoclonal antibody directed against bovine follicular dendritic cells. Vet Immunol Immunopathol 97:1–9. doi: 10.1016/S0165-2427(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 19.Lindhout E, Lakeman A, Mevissen ML, de Groot C. 1994. Functionally active Epstein-Barr virus-transformed follicular dendritic cell-like cell lines. J Exp Med 179:1173–1184. doi: 10.1084/jem.179.4.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen MS, Pors SE, Bille-Hansen V, Kjerulff SK, Nielsen OL. 2010. Occurrence and tissue distribution of porcine circovirus type 2 identified by immunohistochemistry in Danish finishing pigs at slaughter. J Comp Pathol 142:109–121. doi: 10.1016/j.jcpa.2009.07.059. [DOI] [PubMed] [Google Scholar]
- 21.Susa M, Konig M, Saalmuller A, Reddehase MJ, Thiel HJ. 1992. Pathogenesis of classical swine fever: B-lymphocyte deficiency caused by hog cholera virus. J Virol 66:1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Shikh ME, Pitzalis C. 2012. Follicular dendritic cells in health and disease. Front Immunol 3:292. doi: 10.3389/fimmu.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauring AS, Andino R. 2010. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog 6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright CF, Morelli MJ, Thebaud G, Knowles NJ, Herzyk P, Paton DJ, Haydon DT, King DP. 2011. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J Virol 85:2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domingo E, Escarmís C, Baranowski E, Ruiz-Jarabo CM, Carrillo E, Núñez JI, Sobrino F. 2003. Evolution of foot-and-mouth disease virus. Virus Res 91:47–63. doi: 10.1016/S0168-1702(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 26.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King DJ, Freimanis GL, Orton RJ, Waters RA, Haydon DT, King DP. 2016. Investigating intra-host and intra-herd sequence diversity of foot-and-mouth disease virus. Infect Genet Evol 44:286–292. doi: 10.1016/j.meegid.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferretti L, Perez-Martin E, Zhang F, de Klerk-Lorist LM, Van Schalkwyk L, Maree F, Charleston B, Ribeca P. 2018. Pervasive within-host recombination and epistasis as major determinants of the molecular evolution of the foot-and mouth disease virus capsid. bioRxiv doi: 10.1101/271239v2. [DOI] [PMC free article] [PubMed]
- 29.Hudson RR, Slatkin M, Maddison WP. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez-Carvajal L, Pauszek SJ, Ahmed Z, Farooq U, Naeem K, Shabman RS, Stockwell TB, Rodriguez LL. 2018. Genetic stability of foot-and-mouth disease virus during long-term infections in natural hosts. PLoS One 13:e0190977. doi: 10.1371/journal.pone.0190977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnett PV, Statham RJ, Vosloo W, Haydon DT. 2003. Foot-and-mouth disease vaccine potency testing: determination and statistical validation of a model using a serological approach. Vaccine 21:3240–3248. doi: 10.1016/S0264-410X(03)00219-6. [DOI] [PubMed] [Google Scholar]
- 32.Orton RJ, Wright CF, Morelli MJ, King DJ, Paton DJ, King DP, Haydon DT. 2015. Distinguishing low frequency mutations from RT-PCR and sequence errors in viral deep sequencing data. BMC Genomics 16:229. doi: 10.1186/s12864-015-1456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman WG, Ramshaw IA. 1971. Growth of the IB-RS-2 pig kidney cell line in suspension culture and its susceptibility to foot-and-mouth disease virus. Appl Microbiol 22:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenfeldt C, Eschbaumer M, Rekant SI, Pacheco JM, Smoliga GR, Hartwig EJ, Rodriguez LL, Arzt J. 2016. The foot-and-mouth disease carrier state divergence in cattle. J Virol 90:6344–6364. doi: 10.1128/JVI.00388-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki K, Grigorova I, Phan TG, Kelly LM, Cyster JG. 2009. Visualizing B cell capture of cognate antigen from follicular dendritic cells. J Exp Med 206:1485–1493. doi: 10.1084/jem.20090209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferretti L, Di Nardo A, Singer B, Lasecka-Dykes L, Logan G, Wright CF, Perez-Martin E, King DP, Tuthill TJ, Ribeca P. 2018. Within-host recombination in the foot-and-mouth disease virus genome. Viruses 10:E221. doi: 10.3390/v10050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baccam P, Thompson RJ, Li Y, Sparks WO, Belshan M, Dorman KS, Wannemuehler Y, Oaks JL, Cornette JL, Carpenter S. 2003. Subpopulations of equine infectious anemia virus Rev coexist in vivo and differ in phenotype. J Virol 77:12122–12131. doi: 10.1128/JVI.77.22.12122-12131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baccam P, Thompson RJ, Fedrigo O, Carpenter S, Cornette JL. 2001. PAQ: partition analysis of quasispecies. Bioinformatics 17:16–22. doi: 10.1093/bioinformatics/17.1.16. [DOI] [PubMed] [Google Scholar]
- 39.Zagordi O, Klein R, Daumer M, Beerenwinkel N. 2010. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res 38:7400–7409. doi: 10.1093/nar/gkq655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pacheco JM, Brito B, Hartwig E, Smoliga GR, Perez A, Arzt J, Rodriguez LL. 2017. Early detection of foot-and-mouth disease virus from infected cattle using a dry filter air sampling system. Transbound Emerg Dis 64:564–573. doi: 10.1111/tbed.12404. [DOI] [PubMed] [Google Scholar]
- 41.Arzt J, Juleff N, Zhang Z, Rodriguez LL. 2011. The pathogenesis of foot-and-mouth disease I: viral pathways in cattle. Transbound Emerg Dis 58:291–304. doi: 10.1111/j.1865-1682.2011.01204.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Alexandersen S. 2004. Quantitative analysis of foot-and-mouth disease virus RNA loads in bovine tissues: implications for the site of viral persistence. J Gen Virol 85:2567–2575. doi: 10.1099/vir.0.80011-0. [DOI] [PubMed] [Google Scholar]
- 43.Burrows R. 1966. Studies on the carrier state of cattle exposed to foot-and-mouth disease virus. J Hyg (Lond) 64:81–90. doi: 10.1017/S0022172400040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gebauer F, de la Torre JC, Gomes I, Mateu MG, Barahona H, Tiraboschi B, Bergmann I, de Mello PA, Domingo E. 1988. Rapid selection of genetic and antigenic variants of foot-and-mouth disease virus during persistence in cattle. J Virol 62:2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stenfeldt C, Belsham GJ. 2012. Detection of foot-and-mouth disease virus RNA in pharyngeal epithelium biopsy samples obtained from infected cattle: investigation of possible sites of virus replication and persistence. Vet Microbiol 154:230–239. doi: 10.1016/j.vetmic.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Carrillo C, Lu Z, Borca MV, Vagnozzi A, Kutish GF, Rock DL. 2007. Genetic and phenotypic variation of foot-and-mouth disease virus during serial passages in a natural host. J Virol 81:11341–11351. doi: 10.1128/JVI.00930-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Donnell V, Pacheco JM, Larocco M, Gladue DP, Pauszek SJ, Smoliga G, Krug PW, Baxt B, Borca MV, Rodriguez L. 2014. Virus-host interactions in persistently FMDV-infected cells derived from bovine pharynx. Virology 468-470:185–196. doi: 10.1016/j.virol.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Barros JJ, Malirat V, Rebello MA, Costa EV, Bergmann IE. 2007. Genetic variation of foot-and-mouth disease virus isolates recovered from persistently infected water buffalo (Bubalus bubalis). Vet Microbiol 120:50–62. doi: 10.1016/j.vetmic.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 49.Vosloo W, Bastos AD, Kirkbride E, Esterhuysen JJ, van Rensburg DJ, Bengis RG, Keet DW, Thomson GR. 1996. Persistent infection of African buffalo (Syncerus caffer) with SAT-type foot-and-mouth disease viruses: rate of fixation of mutations, antigenic change and interspecies transmission. J Gen Virol 77:1457–1467. doi: 10.1099/0022-1317-77-7-1457. [DOI] [PubMed] [Google Scholar]
- 50.Quan M, Murphy CM, Zhang Z, Alexandersen S. 2004. Determinants of early foot-and-mouth disease virus dynamics in pigs. J Comp Pathol 131:294–307. doi: 10.1016/j.jcpa.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Brehm KE, Ferris NP, Lenk M, Riebe R, Haas B. 2009. Highly sensitive fetal goat tongue cell line for detection and isolation of foot-and-mouth disease virus. J Clin Microbiol 47:3156–3160. doi: 10.1128/JCM.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marco-Sola S, Sammeth M, Guigó R, Ribeca P. 2012. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods 9:1185–1188. doi: 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- 53.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raineri E, Ferretti L, Esteve-Codina A, Nevado B, Heath S, Pérez-Enciso M. 2012. SNP calling by sequencing pooled samples. BMC Bioinformatics 13:239. doi: 10.1186/1471-2105-13-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trevor H, Robert T, Jh F. 2009. The elements of statistical learning: data mining, inference, and prediction. Springer, New York, NY. [Google Scholar]
- 56.OIE. 2012. Foot and mouth disease, chapter 2.1.5 In OIE terrestrial manual, 7th ed, vol 1 OIE, Paris, France. [Google Scholar]