Local transport of plant rhabdoviruses likely involves the passage of viral nucleocapsids through MP-gated plasmodesmata, but the molecular mechanisms are not fully understood. We have conducted complementation assays with MPs encoded by five distinct rhabdoviruses to assess their movement specificity. Each of the rhabdovirus MPs complemented the movement of MP-defective mutants of two positive-stranded RNA viruses that have different movement strategies. In marked contrast, cell-to-cell movement of two recombinant plant rhabdoviruses was highly specific in requiring their cognate MPs. We have shown that these rhabdovirus MPs are localized to the cell periphery and associate with cellular membranes, and that they interact only with their cognate nucleocapsid core proteins. These interactions are able to redirect viral nucleocapsid core proteins from their sites of replication to the cell periphery. Our study provides a model for the specific inter- and intracellular trafficking of plant rhabdoviruses that may be applicable to other negative-stranded RNA viruses.
KEYWORDS: cell-to-cell movement, movement protein, nucleocapsid, plant rhabdovirus, sonchus yellow net virus, tomato yellow mottle-associated virus, trans-complementation
ABSTRACT
Positive-stranded RNA virus movement proteins (MPs) generally lack sequence-specific nucleic acid-binding activities and display cross-family movement complementarity with related and unrelated viruses. Negative-stranded RNA plant rhabdoviruses encode MPs with limited structural and functional relatedness with other plant virus counterparts, but the precise mechanisms of intercellular transport are obscure. In this study, we first analyzed the abilities of MPs encoded by five distinct rhabdoviruses to support cell-to-cell movement of two positive-stranded RNA viruses by using trans-complementation assays. Each of the five rhabdovirus MPs complemented the movement of MP-defective mutants of tomato mosaic virus and potato X virus. In contrast, movement of recombinant MP deletion mutants of sonchus yellow net nucleorhabdovirus (SYNV) and tomato yellow mottle-associated cytorhabdovirus (TYMaV) was rescued only by their corresponding MPs, i.e., SYNV sc4 and TYMaV P3. Subcellular fractionation analyses revealed that SYNV sc4 and TYMaV P3 were peripherally associated with cell membranes. A split-ubiquitin membrane yeast two-hybrid assay demonstrated specific interactions of the membrane-associated rhabdovirus MPs only with their cognate nucleoproteins (N) and phosphoproteins (P). More importantly, SYNV sc4-N and sc4-P interactions directed a proportion of the N-P complexes from nuclear sites of replication to punctate loci at the cell periphery that partially colocalized with the plasmodesmata. Our data show that cell-to-cell movement of plant rhabdoviruses is highly specific and suggest that cognate MP-nucleocapsid core protein interactions are required for intra- and intercellular trafficking.
IMPORTANCE Local transport of plant rhabdoviruses likely involves the passage of viral nucleocapsids through MP-gated plasmodesmata, but the molecular mechanisms are not fully understood. We have conducted complementation assays with MPs encoded by five distinct rhabdoviruses to assess their movement specificity. Each of the rhabdovirus MPs complemented the movement of MP-defective mutants of two positive-stranded RNA viruses that have different movement strategies. In marked contrast, cell-to-cell movement of two recombinant plant rhabdoviruses was highly specific in requiring their cognate MPs. We have shown that these rhabdovirus MPs are localized to the cell periphery and associate with cellular membranes, and that they interact only with their cognate nucleocapsid core proteins. These interactions are able to redirect viral nucleocapsid core proteins from their sites of replication to the cell periphery. Our study provides a model for the specific inter- and intracellular trafficking of plant rhabdoviruses that may be applicable to other negative-stranded RNA viruses.
INTRODUCTION
Natural infections of plant viruses are initiated from one or a few primary infected cells, followed by cell-to-cell spread to adjacent cells and then to distal tissues via vascular networks. To accomplish systemic infections, plant viruses must circumvent formidable barriers imposed by rigid plant cell walls to move through plasmodesmata (PD) interconnections (1). PD normally permit the diffusion of small molecules, such as ions, hormones, and photosynthates, between adjacent cells but restrict the movement of larger nucleic acid and proteins (1). To facilitate PD movement of infectious entities, plant viruses have evolved to encode one or more movement proteins (MPs) that selectively increase the PD size exclusion limit (SEL). Additionally, viral MPs function to orchestrate the intracellular trafficking of movement entities from replication sites to PD at the cell periphery, followed by intercellular movement and long-distance transport through the vascular system to distal tissues and organs (2–4).
Many groups of plant RNA viruses encode a single MP, and the best studied example is the nonstructural 30-kDa protein (30K) encoded by tobacco mosaic virus (TMV) and related viruses in the genus Tobamovirus (reviewed in reference 5). The TMV MP is capable of targeting to and gating PD (6–9), associating with host membranes and cytoskeleton complexes (10–12), and binding nucleic acids nonspecifically (13, 14). Most of the TMV MP properties are shared by a group of viral MPs belonging to the so-called 30K superfamily, whose members exhibit little amino acid sequence similarity but share a secondary structural fold consisting of seven beta-strands (15–17). During virus infection, the TMV MP forms a ribonucleoprotein complex (RNP) with viral RNA, likely in association with host membranes and viral replication proteins to mediate passage of the RNP through gated PD (5). Apart from movement mechanisms involving the RNP forms, some 30K members modify PD and form hollow tubules through which virus particles are transported to neighboring cells (18).
Other groups of plant RNA viruses have cell-to-cell movement functions distributed among two or more MPs. The most extensively studied of these are the triple gene block (TGB) proteins TGB1, TGB2, and TGB3, encoded by potex-, hordei-, pomo-, and benyviruses (19, 20). TGB1, also known as p25 in potato virus X (PVX), is able to dilate the PD (21–23) and requires two integral transmembrane proteins, TGB2 and TGB3, for PD targeting (20, 23–25). TGB2 and TGB3 also recruit the PVX replicase complexes to endoplasmic reticulum-derived membranous caps at PD orifices, where nascent genomic RNAs are encapsidated by viral coat protein (CP) and the assembled virions are inserted into PD by the TGB1 protein (20, 26, 27). Although the movement of PVX and other potexviruses with flexuous virions requires viral CPs in addition to their TGB proteins, many hordei-, pomo-, and benyviruses with rod-shaped particles are able to dispense with the CP for cell-to-cell movement (19, 20).
Regardless of differences in MP sequences and movement strategies, widespread cross-complementarity has been documented between MPs of taxonomically distinct plant viruses (28), suggesting common functions among diverse viral MPs. For example, the MPs of tobamoviruses support cell-to-cell movement of potex-, alfamo-, tobra-, diantho-, cucumo-, hordei-, bromo-, and benyviruses (see reference 28 and references therein). These findings are in agreement with the fact that plant viral MPs generally bind to nucleic acids in a non-sequence-specific manner (2). trans-Complementation assays, in which a movement-defective virus is complemented by an ectopically expressed MP, are often used to assess movement compatibility between viruses. These assays also provide valuable tools to assign movement functions to particular protein(s) encoded by viruses that lack reverse genetics systems (29).
Unsegmented negative-stranded (NS) plant rhabdoviruses are currently classified into two genera, Nucleorhabdovirus and Cytorhabdovirus, according to their sites of replication and virion morphogenesis, i.e., the nucleus and cytoplasm, respectively (30, 31). As with animal-infecting counterparts in the family Rhabdoviridae, plant unsegmented rhabdoviruses encode five conserved structural proteins, the nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and the large RNA polymerase (L), in the order 3′-N-P-M-G-L-5′. In addition, unique accessory genes (here collectively designated P3 genes) located between the P and M genes of plant rhabdoviruses are thought to be required for movement (30–32). Several lines of evidence support a movement function for the plant rhabdovirus P3 homologs designated the sc4 in sonchus yellow net virus (SYNV) (33), Y in potato yellow dwarf virus (PYDV) (34) and eggplant mottle virus (35), 4b in lettuce necrotic yellows virus (LNYV) (36), and P3 in most other plant rhabdoviruses (31, 32). First, computational analyses predict that many rhabdovirus P3 homologs share secondary structural similarities to the 30K superfamily MPs (15, 16, 37, 38). Second, localization studies have shown that SYNV sc4, PYDV Y, LNYV 4b, and P3 from rice yellow stunt virus (RYSV), alfalfa dwarf virus (ADV), Datura yellow vein virus, and maize mosaic virus are targeted to the cell periphery or near the PD (34, 39–45). More direct evidence has come from trans-complementation studies of RYSV P3, LNYV 4b, and ADV P3, which have been shown to complement the cell-to-cell movement of the PVX or tobamovirus MP mutants (37, 38, 41). These studies revealed that MPs encoded by plant rhabdoviruses are compatible with cell-to-cell movement of positive-stranded RNA viruses, even though these groups of RNA viruses differ in fundamental replication strategies.
More relevant experimental evidence for plant rhabdovirus MP functions has been obtained by using the reverse genetics system recently developed for SYNV, which shows that the sc4 protein is indispensable for cell-to-cell movement (46). In this study, an SYNV sc4 deletion mutant was found to be unable to move from initially infected cells. However, an SYNV minigenome comprising the N, P, sc4, and L genes and terminal noncoding sequences was capable of limited autonomous localized movement, and the movement of an SYNV sc4 deletion mutant could be complemented in trans by transient expression of the sc4 protein. Collectively, these results suggest that SYNV nucleocapsids consisting of the viral genome encapsidated by the N, P, and L core proteins are the minimal infectious units that navigate intercellular connections (46).
In this study, we conducted trans-complementation assays with five distinct rhabdovirus MPs and showed that they all complement movement-defective mutants of tomato mosaic virus (ToMV), a closely related virus to TMV, and PVX. Unexpectedly, cell-to-cell movement of a recombinant SYNV (rSYNV) sc4 deletion mutant and a recombinant tomato yellow mottle-associated virus (rTYMaV) P3 deletion mutant were not rescued by heterologous MPs, whereas their cognate sc4 and P3 MPs specifically restored the movement functions of rSYNV and rTYMaV mutants, respectively. The SYNV sc4 and TYMaV P3 proteins were found to localize to the cell periphery and to associate with membranes. We have further demonstrated that the SYNV and TYMaV MPs interact only with their cognate nucleocapsid core proteins, and that these interactions are able to redirect viral nucleocapsid proteins from their sites of replication to the cell periphery.
RESULTS
Five plant rhabdovirus MPs are able to trans-complement movement-deficient ToMV and PVX.
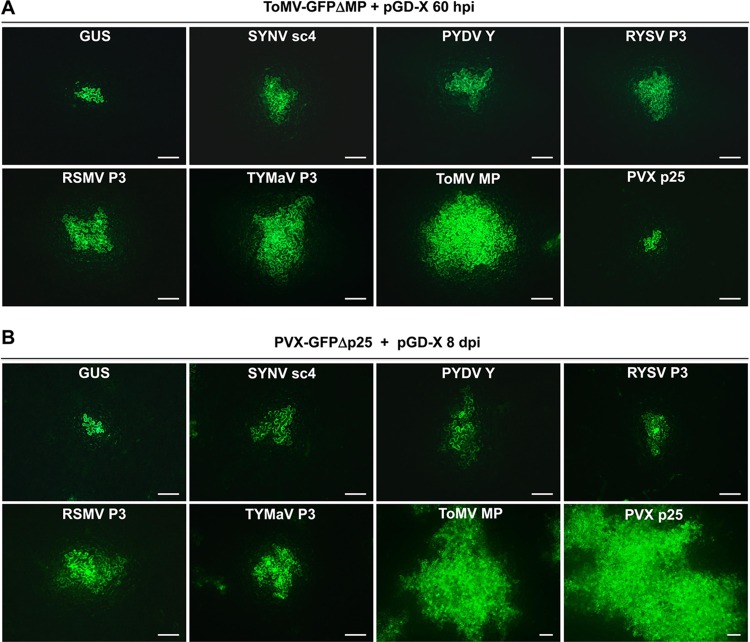
Since some of the rhabdovirus P3 homologs are structurally related to 30K superfamily proteins, we first set out to test whether there is a widespread complementation between P3 proteins and a ToMV MP mutant. The MP genes from three nucleorhabdoviruses, SYNV, PYDV, and RYSV, and two cytorhabdoviruses recently identified in China, rice stripe mosaic virus (RSMV) (47) and TYMaV (48), were cloned into the pGD binary vector (49) for transient expression. ToMV-GFPΔMP, an MP-defective ToMV clone carrying a green fluorescence protein (GFP) reporter gene substitution for the CP gene (50), was used to evaluate complementation by the rhabdovirus MPs. For these experiments, Agrobacterium tumefaciens cultures harboring the ToMV-GFPΔMP plasmid and individual pGD plasmids for the expression of rhabdoviral P3 proteins were coinfiltrated in Nicotiana benthamiana leaves. Notably, the ToMV-GFPΔMP bacterial cultures were diluted in this assay so that isolated infection foci could be visualized, and complementation of cell-to-cell movement was compared by analyzing the spread of GFP fluorescence to adjacent cells. At 50 h postinfiltration (hpi), predominantly single-cell GFP foci were observed in infiltrated leaf tissues, regardless of which viral MP was coexpressed (data not shown), indicating that cell-to-cell movement had not occurred at this time. However, by 60 hpi, efficient ToMV-GFPΔMP cell-to-cell movement was restored by coexpression of the ToMV MP, as large clusters of GFP-expressing cells were evident in coinfiltrated leaf tissues (Fig. 1A). In negative controls in which ToMV-GFPΔMP and beta-glucuronidase (GUS) were coexpressed in N. benthamiana leaves, GFP foci were confined to single cells. When SYNV sc4, PYDV Y, RYSV P3, RSMV P3, or TYMaV P3 was coexpressed individually, moderate cell-to-cell spread of ToMV-GFPΔMP was observed in all cases (Fig. 1A). In contrast, the ToMV-GFPΔMP movement defect was not rescued by the coexpressed PVX p25, as was expected because of the absence of TGB2 and TGB3, which are required for targeting TGB1 to the PD (19, 20, 24, 25). To compare movement complementation efficiencies, we counted the cells present in GFP clusters for each infection focus. In the GUS control leaves, the majority of the infection foci contained only one GFP-expressing cell (55 out of 60), with a few (5/60) foci possibly composed of two-cell clusters (Table 1). Therefore, infection foci with four or more cells expressing GFP at 60 hpi were considered to represent authentic movement under our experimental conditions. Using these criteria, we found that coexpression of each of the five rhabdovirus MPs resulted in localized ToMV-GFPΔMP movement in 43.3 to 60.0% of the infection foci (Table 1). In comparison, the cognate ToMV MP was substantially more efficiently in complementing ToMV-GFPΔMP movement, as 76.7% of the infection foci were classified as positive (Table 1), and the sizes of the foci were much larger than those expressing the plant rhabdovirus MPs (Fig. 1A). For example, out of 60 fluorescent foci, 22 containing 10 or more cells were recorded in the case of ToMV MP, compared to 0 to 11 for the rhabdovirus MPs (Table 1). Among the five rhabdovirus MPs, movement complementation by the two cytorhabdovirus MPs was 48.3% for RSMV P3 and 60.0% for TYMaV P3. These levels of trans-complementation appeared to be slightly higher than those by the three nucleorhabdovirus MPs, which ranged from 43.3 to 46.7% for SYNV sc4, PYDV Y, and RYSV P3 (Table 1). Whether the difference in complementation efficiencies was due to intrinsic MP functions or differential expression/accumulation levels of these viral MPs is not known. Our attempts to tag the SYNV sc4 and TYMaV P3 protein with epitopes or fluorescent proteins either as N- or C-terminal fusions resulted in a failure of movement complementation, precluding a direct comparison of their expression levels versus movement efficiencies (data not shown).
FIG 1.
trans-Complementation of movement-defective ToMV and PVX. (A and B) Leaves of N. benthamiana plants were agroinfiltrated with bacterial mixtures carrying transcription plasmid for ToMV-GFPΔMP (A) or PVX-GFPΔp25 (B), plus a binary plasmid (pGD-X) to express SYNV sc4, PYDV Y, RYSV P3, RSMV P3, TYMaV P3, ToMV MP, PVX p25, or GUS as a negative control, as indicated on the top of each panel. Images were taken with a fluorescence microscope at 60 hpi (A) or 8 dpi (B). Scale bars = 200 μm. Note that the bars in the ToMV MP and PVX p25 images in panel B are half the lengths of the bars in other panels due to more extensive movement with these treatments.
TABLE 1.
trans-Complementation of cell-to-cell movement of ToMV-GFPΔMP and PVX-GFPΔMP
| MP | No. of foci by no. of cells in clustera
|
Total no. of foci counted |
Movement efficiency (≥ 4 cells) (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
2–3 |
4–5 |
6–7 |
8–9 |
≥10 |
|||||||||||
| ToMV | PVX | ToMV | PVX | ToMV | PVX | ToMV | PVX | ToMV | PVX | ToMV | PVX | ToMV | PVX | ToMV | PVX | |
| GUS | 55 | 53 | 5 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 60 | 60 | 0 (0) | 0 (0) |
| SYNV sc4 | 13 | 32 | 21 | 14 | 13 | 11 | 8 | 3 | 5 | 0 | 0 | 0 | 60 | 60 | 26 (43.3) | 14 (23.3) |
| PYDV Y | 17 | 29 | 17 | 11 | 10 | 9 | 6 | 5 | 7 | 4 | 3 | 2 | 60 | 60 | 26 (43.3) | 20 (33.3) |
| RYSV P3 | 14 | 29 | 18 | 14 | 10 | 7 | 7 | 6 | 5 | 2 | 6 | 2 | 60 | 60 | 28 (46.7) | 17 (28.3) |
| RSMV P3 | 14 | 22 | 17 | 10 | 5 | 7 | 10 | 5 | 6 | 6 | 8 | 10 | 60 | 60 | 29 (48.3) | 28 (46.7) |
| TYMaV P3 | 12 | 25 | 12 | 13 | 9 | 8 | 10 | 3 | 6 | 5 | 11 | 6 | 60 | 60 | 36 (60.0) | 22 (36.7) |
| PVX P25 | 53 | 2 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 5 | 0 | 11 | 60 | 20 | 0 (0) | 17 (85.0) |
| ToMV MP | 4 | 3 | 10 | 2 | 7 | 1 | 6 | 0 | 11 | 4 | 22 | 10 | 60 | 20 | 46 (76.7) | 15 (75.0) |
ToMV, ToMV-GFPΔMP infection foci at 60 h postinfiltration. PVX, PVX-GFPΔMP infection foci at 8 days postinfiltration.
To test whether MPs of rhabdoviruses support the movement of additional positive-stranded RNA viruses outside the 30K superfamily, we conducted similar trans-complementation assays with PVX-GFPΔp25, a p25-defective PVX mutant containing a GFP reporter gene (51). Suspensions of agrobacteria harboring the PVX-GFPΔp25 plasmid were diluted similarly to those in the ToMV-GFPΔMP experiments to produce single-cell primary infection foci, and individual viral MPs were coexpressed in N. benthamiana leaves to assess movement complementation. As shown in Fig. 1B, coexpression of each of the five rhabdovirus MPs resulted in PVX-GFPΔp25 cell-to-cell movement by 8 days postinfiltration (dpi). However, the complementation efficiencies were generally low, as the GFP signals encompassed only a limited number of adjacent cells. In contrast, PVX p25 and, to a lesser extent, the ToMV MP, resulted in more extensive PVX-GFPΔp25 movement (Fig. 1B). Again, quantitative analyses of the cell numbers in the GFP-expressing clusters suggested that the cytorhabdovirus MPs were somewhat more efficient than were the nucleorhabdovirus MPs in complementing PVX-GFPΔp25 movement (Table 1). For example, 23.3 to 33.3% of the PVX infection foci exhibited authentic movement when coinfiltrated for expression of SYNV sc4, PYDV Y, or RYSV P3, compared to 36.7% and 46.7% for P3 of TYMaV and RSMV (Table 1). Taken together, our results show that the heterologous rhabdovirus MPs are able to support cell-to-cell movement of two distinct positive-stranded RNA viruses, albeit with relatively low efficiencies.
Cognate MPs are specifically required for cell-to-cell movement of a plant nucleorhabdovirus and cytorhabdovirus.
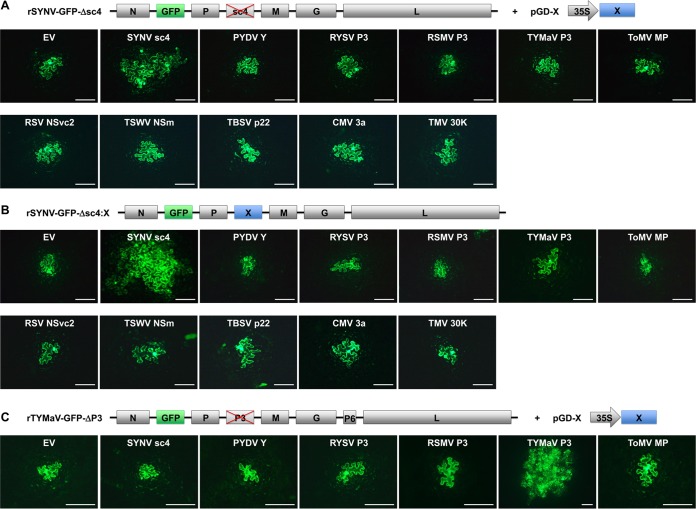
Having demonstrated that rhabdovirus MPs are able to complement the local movement of members of two well-studied positive-stranded RNA virus families, we next evaluated the abilities of these MPs to support cell-to-cell movement of NS plant rhabdoviruses. By taking advantage of our recently developed SYNV reverse genetics system (46), we used a GFP-tagged rSYNV with an sc4 deletion (rSYNV-GFP-Δsc4) for trans-complementation experiments with transiently expressed MPs (Fig. 2A). N. benthamiana leaves were infiltrated with mixtures of A. tumefaciens cultures carrying plasmids designed for the expression of rSYNV-GFP-Δsc4 RNA, the SYNV N, P, and L core proteins, and three viral RNA silencing suppressors (VSRs), i.e., p19 from tomato bushy stunt virus (TBSV), γb from barley stripe mosaic virus (BSMV), and P1/HC-Pro from tobacco etch virus (TEV), which are collectively required for the recovery of rSYNV (46). In addition, bacterial cultures containing the pGD plasmids encoding individual MPs were also included in the mixtures. As shown previously (46), GFP fluorescence from rSYNV-GFP-Δsc4 was restricted to single cells (Fig. 2A). Transient expression of the sc4 protein restored the cell-to-cell movement of the rSYNV-GFP-Δsc4 mutant, as GFP fluorescence was observed in adjacent cells at 12 dpi; however, none of the other four noncognate rhabdovirus MPs or the ToMV MP complemented rSYNV-GFP-Δsc4 movement (Fig. 2A). Analysis of the cell numbers in GFP clusters for each infection focus showed that 40% of the rSYNV-GFP-Δsc4 infection foci exhibited authentic movement when supported by the transiently expressed sc4 protein, whereas infection foci with 4 or more cells were not observed in leaf tissues expressing other MPs (Table 2). Notably, the same agrobacterial strains used for MP expression in the ToMV and PVX complementation experiments were used to assess SYNV movement, so complementation failures were unlikely to be due to the lack of expression of functional MPs.
FIG 2.
Inabilities of heterologous MPs to complement the movement of the rSYNV and rTYMaV MP deletion mutants. (A) trans-Complementation of the cell-to-cell movement of rSYNV-GFP-Δsc4 by transiently expressed MPs. Leaves of N. benthamiana plants were agroinfiltrated with bacterial mixtures carrying plasmids required for recovery of rSYNV-GFP-Δsc4 (see Materials and Methods for details) and a binary vector (pGD-X) designated to express individual viral MPs and empty vector (EV) control, as indicated on the panels. (B) Cell-to-cell movement of rSYNV-GFP-Δsc4 derivatives expressing heterologous MPs. The MP genes were each substituted for the sc4 gene in rSYNV-GFP genome, and the resulting rSYNV chimeras were recovered in N. benthamiana leaves after agroinfiltration and monitored for cell-to-cell movement. (C) trans-Complementation of rTYMaV-GFP-ΔP3 movement by transiently expressed MPs in experiments similar to those described in panel A. Genome structures of rSYNV-GFP and rTYMaV-GFP MP deletion mutants and expression strategies of viral MPs are diagramed on the top of each panel. (A to C) GFP fluorescence in infiltrated leaf epidermal cells was photographed with a fluorescence microscope at 12, 15, and 8 dpi, respectively. Scale bars = 200 μm.
TABLE 2.
Complementation of rSYNV-GFP-Δsc4 and rTYMaV-GFP-ΔP3 movement by cognate and noncognate MPs
| MP | No. of foci by no. of cells in clustera
|
Total no. of foci counted |
Movement efficiency (%) (no. of ≥4-cell foci/total no. of foci) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
2–3 |
≥4 |
|||||||||||||
| SYNV-Δsc4+X | SYNV-Δsc4::X | TYMaV-ΔP3+X | SYNV-Δsc4+X | SYNV-Δsc4::X | TYMaV-ΔP3+X | SYNV-Δsc4+X | SYNV-Δsc4::X | TYMaV-ΔP3+X | SYNV-Δsc4+X | SYNV-Δsc4::X | TYMaV-ΔP3+X | SYNV-Δsc4+X | SYNV-Δsc4::X | TYMaV-ΔP3+X | |
| None | 29 | 28 | 29 | 1 | 2 | 1 | 0 | 0 | 0 | 30 | 30 | 30 | 0 | 0 | 0 |
| SYNV sc4 | 10 | 6 | 29 | 8 | 5 | 1 | 12 | 19 | 0 | 30 | 30 | 30 | 40.0 | 63.3 | 0 |
| PYDV Y | 28 | 28 | 28 | 2 | 2 | 2 | 0 | 0 | 0 | 30 | 30 | 30 | 0 | 0 | 0 |
| RYSV P3 | 29 | 27 | 27 | 1 | 3 | 3 | 0 | 0 | 0 | 30 | 30 | 30 | 0 | 0 | 0 |
| RSMV P3 | 28 | 29 | 29 | 2 | 1 | 1 | 0 | 0 | 0 | 30 | 30 | 30 | 0 | 0 | 0 |
| TYMaV P3 | 29 | 29 | 5 | 1 | 1 | 4 | 0 | 0 | 21 | 30 | 30 | 30 | 0 | 0 | 70.0 |
| PVX P25 | 29 | 28 | 28 | 1 | 2 | 2 | 0 | 0 | 0 | 30 | 30 | 30 | 0 | 0 | 0 |
| ToMV MP | 27 | 29 | 28 | 3 | 1 | 2 | 0 | 0 | 0 | 30 | 30 | 30 | 0 | 0 | 0 |
SYNV-Δsc4+X, rSYNV-GFP-Δsc4 infection foci supported by individual MPs expressed in trans from a binary plasmid (pGD-X) at 12 dpi; SYNV-Δsc4::X, rSYNV-GFP-Δsc4 infection foci supported by individual MPs expressed in cis from the sc4 gene locus (Δsc4::X) at 15 dpi. TYMaV-ΔP3+X, rTYMaV-GFP-ΔP3 infection foci supported by individual MPs expressed in trans from a binary plasmid (pGD-X) at 8 dpi.
It is known that rSYNV cell-to-cell movement commences only after about 9 dpi (46), whereas agroinfiltration-mediated transient protein expression peaks at 2 to 3 dpi and then begins to decrease gradually. Therefore, it is possible that by 12 dpi, the transiently expressed MPs might be present at such low levels that they were unable to support rSYNV-GFP-Δsc4 movement in trans, although the positive results obtained with sc4 expression argue against this possibility. Nevertheless, to address this issue, we engineered chimeric SYNV genomes by inserting each of the MP genes into the sc4 locus, and the resulting rSYNV-GFP chimeras were infiltrated into N. benthamiana leaf tissues under conditions permitting recombinant virus recovery. At 8 dpi, isolated single cells exhibiting GFP fluorescence were observed in all infiltrated leaves (data not shown), indicating that these rSYNV derivatives were capable of replication. Moreover, by 15 dpi, in N. benthamiana leaves infected by rSYNV-GFP-Δsc4::sc4, in which the sc4 gene was reintroduced into rSYNV-GFP-Δsc4, extensive spread of GFP fluorescence was observed in adjacent cells (Fig. 2B), and symptoms of systemic infections were subsequently observed (data not shown). However, in marked contrast, all of the chimeric rSYNV-GFP mutants containing MP genes from each of the other four rhabdoviruses or from ToMV were mostly restricted to single cells at 15 dpi, with a few infection foci containing two GFP-expressing cells (Fig. 2B and Table 2).
We also tested additional MPs encoded by representative plant negative- and positive-stranded RNA viruses, namely, rice stripe virus NSvc4 (52), tomato spotted wilt virus NSm (53), tomato bushy stunt virus p22 (54), cucumber mosaic virus 3a (55), and TMV 30K (56). Again, none of these MPs was able to support rSYNV-GFP-Δsc4 movement when provided either in trans through transient expression (Fig. 2A) or in cis by rSYNV derivatives carrying relevant MP gene substitutions for the sc4 gene (Fig. 2B). These data collectively show that SYNV cell-to-cell movement is highly specific in requiring its cognate MP.
To determine whether or not additional rhabdoviruses exhibit similar movement specificity, we conducted trans-complementation assays to assess the movement of a recombinant cytorhabdovirus, rTYMaV, that we recently engineered (W. Lin, Q. Ling, Z. Li, unpublished data). TYMaV was identified recently by small RNA sequencing of tomato samples collected from Southwest China and has a genome structure similar to that of ADV (48). To evaluate movement compatibility, we constructed a GFP-tagged rTYMaV P3 deletion mutant (rTYMaV-GFP-ΔP3) for trans-complementation experiments with transiently expressed MPs similar to those described above for the rSYNV-GFP-Δsc4 experiments. The results of these experiments show that the rTYMaV-GFP-ΔP3 mutant was able to move from cell to cell at 8 dpi when supported in trans by TYMaV P3, the homologous MP (Fig. 2C). However, in marked contrast, none of the heterologous rhabdovirus MPs or the ToMV MP restored rTYMaV-GFP-ΔP3 movement (Fig. 2C and Table 2). In summary, these results collectively demonstrate that cognate MPs are specifically required for local movement of both a nucleorhabdovirus and a cytorhabdovirus.
MPs of SYNV and TYMaV are peripheral membrane proteins.
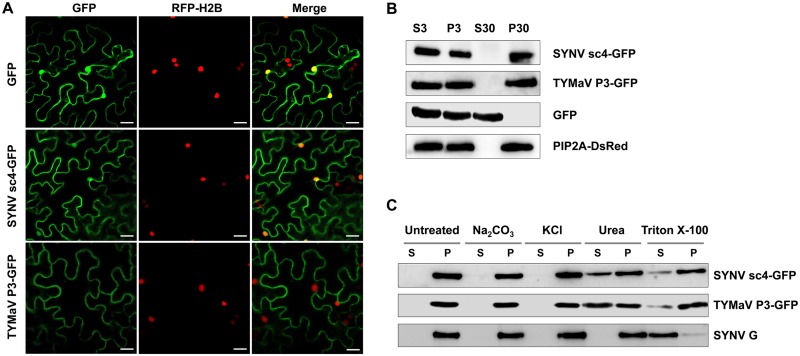
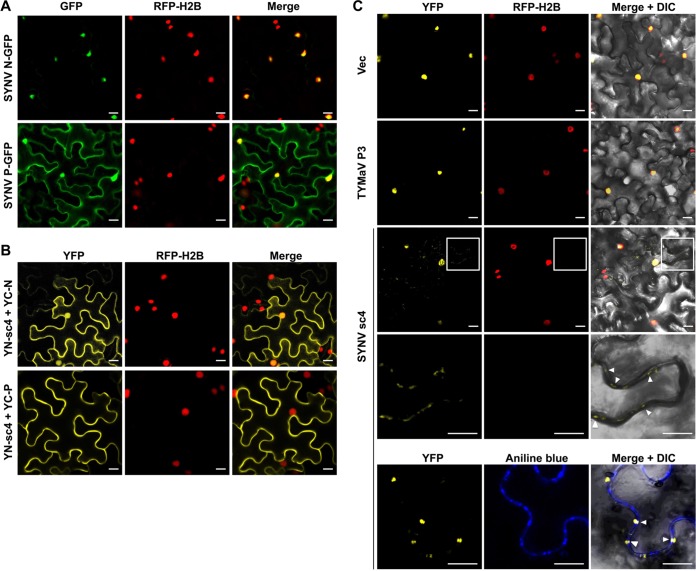
To further characterize the SYNV and TYMaV MPs, we next investigated their subcellular localizations. For this purpose, SYNV sc4 and TYMaV P3 were fused to the N terminus of GFP and transiently expressed in a transgenic N. benthamiana line expressing a red fluorescence protein fused to the histone 2B nuclear marker protein (RFP-H2B) (57). Both rhabdovirus MP fusions localized predominantly to the cell periphery (Fig. 3A), but, as shown previously (45, 49), a small proportion of the SYNV sc4-GFP was also present in the nucleus (Fig. 3A, top images). The localization patterns of these two MPs are generally consistent with previous studies conducted with other rhabdovirus P3 homologs (34, 39–45).
FIG 3.
Subcellular localization and microsomal association analyses of SYNV sc4-GFP and TYMaV P3-GFP. (A) Confocal micrographs of leaf epidermal cells of transgenic N. benthamiana plants expressing RFP fused to histone 2B (RFP-H2B) that had been infiltrated to express GFP, SYNV sc4-GFP, or TYMaV P3-GFP. Scale bars = 20 μm. (B) Protein extracts from N. benthamiana plants expressing either SYNV sc4-GFP, TYMaV P3-GFP, or the transmembrane SYNV G protein, together with GFP and PIP2A-DsRed marker proteins, were separated into supernatant (S3) and pellet (P3) fractions by centrifugation at 3,000 × g for 10 min, and the S3 fraction was further separated into the pellet (P30) and supernatant (S30) fractions by ultracentrifugation at 30,000 × g for 1 h. Protein gel blots revealed that SYNV sc4 and TYMaV P3 were present only in the P30 fraction. (C) The P30 fractions in (B) were divided into several aliquots and treated with either 0.1 M Na2CO3 (pH 11.5), 1 M KCl, 7 M urea, or 1% Triton X-100. Samples were again separated into S and P fractions by ultracentrifugation and subjected to protein gel blot analyses with anti-GFP antibodies or anti-G antibodies.
Mounting evidence suggests that MPs encoded by various plant viruses are associated with cellular membranes (58), but this has not been investigated extensively for plant rhabdovirus MPs. We therefore conducted subcellular fractionation assays with protein extracts isolated from N. benthamiana tissues transiently expressing GFP fusions of SYNV sc4 and TYMaV P3. After centrifugation at 3,000 × g for 10 min, tissue extracts were separated into pellet (P3) and supernatant (S3) fractions, and then the S3 fractions were centrifuged at 30,000 × g for 1 h to obtain pellet (P30) and supernatant (S30) fractions. Protein gel blots revealed that SYNV sc4-GFP and TYMaV P3-GFP were present only in the P30 fraction. Similarly, a membrane marker, the plasma membrane intrinsic protein 2A (PIP2A)-DsRed fusion, also partitioned into the P30 fraction. In contrast, soluble GFP was detected only in the S30 fraction (Fig. 3B). The fractionation results suggest that SYNV sc4 and TYMaV P3 are membrane-bound proteins.
To identify the types of membrane interactions, we treated the membrane-rich P30 fraction with either 0.1 M Na2CO3 (pH 11.5), 1 M KCl, 7 M urea, or 1% Triton X-100, followed by separation of these extracts into pellet (P) and supernatant (S) fractions by high-speed centrifugation. Note that the alkaline treatment releases soluble luminal protein from organelles by transforming microsomes into membranous sheets (59), and high KCl concentrations disrupt protein-lipid electrostatic interactions, whereas 7 M urea releases peripheral membrane proteins from the membrane but does not solubilize integral membrane proteins (60). As shown in Fig. 3C, both sc4 and P3 remained in the membrane-rich fraction after treatment with either sodium carbonate or KCl but were partially released into the soluble fractions after urea treatment. As a control experiment, the SYNV G protein, a type I transmembrane protein, can only be solubilized by treatment with 1% Triton X-100. Consistent with the chemical treatments, we failed to detect any transmembrane regions within the two MPs by using the membrane protein prediction algorithms TMHMM (61), TMMOD (62), ΔG Prediction (63), and DAS-TMfilter (64). These results suggest that SYNV sc4 and TYMaV P3 are peripherally associated with cellular membranes.
Rhabdovirus MPs specifically interact with their cognate N and P proteins.
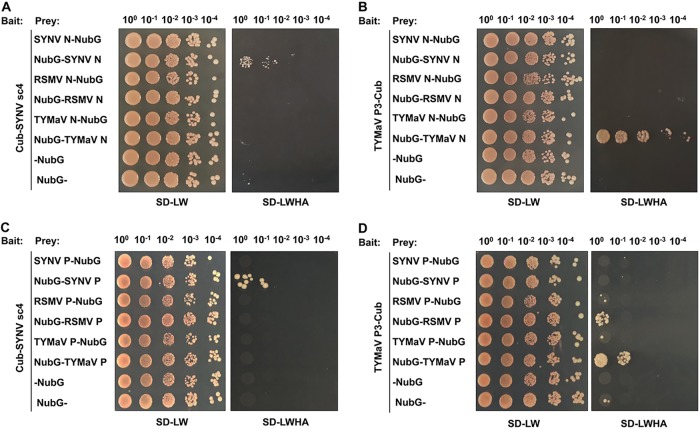
NS plant rhabdovirus genomic and antigenomic RNAs are encapsidated by N proteins that interact with the P and L proteins to form viral nucleocapsids, which are thought to represent the minimal infectious units that are transported through the PD (31, 46, 65). Therefore, the requirements of SYNV and TYMaV cognate MPs for cell-to-cell movement (Fig. 2) raised the possibility that specific interactions exist between rhabdovirus MPs and their nucleocapsid proteins. In light of the membrane-associated nature of the rhabdovirus MPs (Fig. 3), we used a split-ubiquitin-based membrane yeast two-hybrid (MYTH) assay to detect possible interactions between the MPs and the nucleocapsid core proteins. In this assay, interactions with a membrane protein are detected in situ at the cellular membrane, and the interactions captured by this assay are thought to be of more physiological relevance than interactions resulting from conventional yeast two-hybrid assays in which the interacting proteins require transport into the nuclei (66). The membrane-associated SYNV sc4 and TYMaV P3 proteins were each fused to the C-terminal half of ubiquitin (Cub) and the artificial transcription factor LexA-VP16, whereas the N proteins of SYNV, TYMaV, and RSMV were separately fused to the mutated N-terminal half of ubiquitin (NubG) either as N-terminal (X-NubG) or C-terminal (NubG-X) fusions. Using this assay, we found that sc4 interacted specifically with the SYNV N protein but not with the N proteins of TYMaV and RSMV (Fig. 4A). Likewise, TYMaV P3 interacted only with its cognate TYMaV N protein (Fig. 4B). In both cases, only the C-terminal fusion proteins (NubG-SYNV N and NubG-TYMaV N) were able to interact with their cognate MPs (Fig. 4A and B), suggesting that the N-terminal fusion derivatives (SYNV N-NubG and TYMaV N-NubG) affect their folding or function.
FIG 4.
Analyses of rhabdovirus MP-nucleocapsid core proteins interactions by split-ubiquitin membrane yeast two-hybrid (MYTH) assays. (A to D) The bait constructs contain the Cub fusions to either SYNV sc4 (A and C) or TYMaV P3 (B and D), and the prey constructs encode the N (A and B) and P (C and D) proteins of SYNV, RSMV, or TYMaV fused to the N terminus (N-NubG) or C terminus (NubG-N) of NubG. Yeast cells were cotransformed with the bait and prey plasmids. Transformants were spotted as 10-fold serial dilutions (top) on synthetic dropout medium lacking leucine and tryptophan (SD-LW) to confirm the presence of both plasmids. Medium lacking leucine, tryptophan, histidine and adenine (SD-LWHA) was used to screen for positive interactions.
Since the P protein is also a constituent of the viral nucleocapsid, we next tested whether or not the SYNV sc4 and TYMaV P3 proteins interact with their respective P proteins in MYTH assays. For these assays, the P proteins of SYNV, TYMaV, and RSMV were separately fused to NubG either as N-terminal (X-NubG) or C-terminal (NubG-X) fusions. The MYTH assays revealed that the SYNV sc4 interacted specifically with its cognate P proteins but not with noncognate P proteins (Fig. 4C). Similarly, cognate TYMaV P3-P interactions were also detected, although TYMaV P3 also appeared to have a weaker interaction with RSMV P protein (Fig. 4D). Again, only the NubG-P fusions but not the P-NubG fusions interacted with their respective cognate MPs. The specific rhabdovirus MP-N and MP-P interactions strongly reinforce the SYNV and TYMaV cell-to-cell movement experiments and suggest that cognate MP-nucleocapsid core protein interactions provide a mechanism to explain the rhabdovirus movement specificities.
Coexpressed sc4 facilitates redistribution of SYNV N-P complexes from the nuclei to the cell periphery.
To gain additional insights into the SYNV sc4-N and sc4-P interactions, we analyzed their subcellular distribution by using bimolecular fluorescence complementation (BiFC) assays. For this purpose, we coexpressed an SYNV sc4 protein fusion to the N-terminal fragment of the yellow fluorescence protein (YFP) (YN-sc4) and the N and P protein fusions to the C-terminal fragment of YFP (YC-N and YC-P, respectively) in leaves of N. benthamiana transgenic for RFP-H2B. The resulting BiFC signals revealed that sc4-N interacted at both the cell periphery and the nuclei (Fig. 5B, top images). These distributions resembled the sc4 subcellular localization patterns (Fig. 3A) but differed from the exclusive nuclear localization of SYNV N-GFP fusion protein (Fig. 5A) (49, 67). The sc4-P interaction signals were evenly distributed along the cell periphery (Fig. 5B, bottom images), despite localization of both proteins to the nuclei and the cytoplasm when expressed alone (Fig. 3A and 5A) (45, 49). No interactions were detected for any of negative-control combinations, i.e., YN-sc4 with YC, YC-N, or YC-P with YN (data not shown).
FIG 5.
Relocalization of SYNV N-P complexes from the nuclei to the cell periphery by specific interactions with sc4. (A). Confocal micrographs of SYNV N-GFP and P-GFP fusions expressed after agroinfiltration of leaf epidermal cells of H2B-RFP transgenic N. benthamiana. (B). Bimolecular fluorescence complementation (BiFC) assays to determine interactions between the sc4 and the N or P proteins. Leaf epidermal cells of N. benthamiana RFP-H2B transgenic plants were agroinfiltrated to express the YFP N-terminal-fusion to sc4 (YN-sc4) together with YFP C-terminal-fusions to N (YC-N) or P (YC-P). Confocal micrographs of YFP (BiFC), RFP-H2B, and the merged channel are shown. (C). Localization of SYNV N-P BiFC signals in the absence or presence of sc4. YC-N and YN-P fusions were coexpressed with either an empty vector control, TYMaV P3, or SYNV sc4 in transgenic RFP-H2B N. benthamiana epidermal cells. Note that the fourth row shows higher magnifications of the boxed areas to highlight the punctate BiFC foci along the cell wall (indicated by white arrowheads). In the bottom row, the infiltrated leaf tissues were stained with aniline blue fluorochrome to show colocalization of N-P BiFC signals with the PD, as indicated by the white arrowheads. A differential interference contrast (DIC) channel was included in the merged micrograph to illustrate the cell outline. (A to C) Scale bars = 20 μm.
Previous studies have shown that when SYNV N and P proteins are coexpressed, N interacts with and redirects P to the nuclei, where they form punctate subnuclear foci resembling viroplasms (67–69). Accordingly, our BiFC assays revealed that the N-P interactions were localized exclusively in subnuclear foci (Fig. 5C) (57, 70). Interestingly, when sc4 was coexpressed, although the majority of the N-P interaction signals continued to be evident in the subnuclear regions, faint, albeit obvious, BiFC signals were also detected in the cytoplasm. In some cases, the BiFC cytoplasmic signals localized to punctate loci at the cell periphery that were reminiscent of PD sites (Fig. 5C, third and fourth rows). The infiltrated leaf tissues were further stained with aniline blue fluorochrome, a callose-binding PD marker dye. Indeed, some of these BiFC foci colocalized with or were near the PD structures on the cell periphery (Fig. 5C, bottom row). Redistribution of the N-P complex from the nucleus to the cytoplasm is likely mediated by specific interactions with the SYNV sc4 protein, since coexpression of the noninteracting TYMaV P3 protein had no effect on the localization of the SYNV N-P BiFC signals (Fig. 5C, second row). Taken together, these data show that the SYNV sc4 protein interacts specifically with the N and P nucleocapsid core proteins and is able to redirect N-P complexes from nuclear replication sites to cell periphery near the PD.
DISCUSSION
Rhabdoviruses of plants and animals share conserved genome organization and five structural proteins, and our understanding of plant rhabdovirus replication and morphogenesis has benefited tremendously from extensively studied animal rhabdovirus models, such as that for vesicular stomatitis virus (31, 65, 71). However, plant rhabdoviruses also encode nonstructural proteins (P3 homologs) that are uniquely required for plant infections, but the functions of these putative MPs are poorly understood. Through secondary structure predictions, it has been suggested that the P3 homologs of plant rhabdoviruses belong to the 30K superfamily (15, 16, 37, 38). Direct experimental evidence to support a movement function was initially provided by trans-complementation experiments showing that RYSV P3, upon transient expression, restores cell-to-cell movement of a p25-defective PVX mutant (37). Subsequently, P3 of another strain of RYSV, designated rice transitory yellowing virus (41), has been shown to complement a movement-defective ToMV. Similarly, P3 proteins of two cytorhabdoviruses, namely, ADV P3 and LNYV 4b, also support the movement of a different tobamovirus, turnip vein-clearing virus (38).
In this study, we started with parallel analyses of five putative MPs of plant rhabdoviruses for their abilities to complement the movement of ToMV-GFPΔMP and PVX-GFPΔp25. These rhabdoviruses differ in infecting monocotyledonous (RSMV and RYSV) or dicotyledonous (SYNV, PYDV, and TYMaV) hosts, as well as in their nuclear (SYNV, PYDV, and RYSV) or cytoplasmic (RSMV and TYMaV) sites of replication (31, 47, 48). Despite these differences, all five rhabdovirus MPs complemented cell-to-cell movement of ToMV-GFPΔMP and PVX-GFPΔp25 when expressed in trans. ToMV and PVX differ in movement transport systems (single MP versus three TGB proteins) and requirement of the CP for movement (5, 20), but both MPs trigger PD gating and are able to move between cells upon injection or upon transient expression following biolistic bombardment (21–23, 72). Although having not been analyzed directly, MPs encoded by rhabdoviruses presumably have similar functional activities as judged from trans-complementation experiments with positive-strand RNA viruses. Nonspecific nucleic acid-binding activity is another hallmark feature of MPs encoded by various positive-stranded RNA viruses (4). Such activities have also been documented previously for RYSV P3 (37), and our unpublished data (Zhou et al.) reveal that SYNV sc4 also binds to RNA nonspecifically. However, naked genomic RNAs of rhabdoviruses and other NS RNA viruses are rarely present in infected cells; instead, they are encapsidated during replication to form nucleocapsids (73), which are the minimal units for movement and infection (46). Thus, the relevance of MP-RNA binding in rhabdovirus movement is not straightforward; however, this activity may account for their abilities to facilitate the movement of positive-stranded RNA viruses in trans-complementation experimental systems.
Although widespread compatibilities of rhabdovirus MPs with ToMV movement systems were observed, we should note that the efficiencies of ToMV-GFPΔMP movement are substantially lower when complemented by rhabdovirus MPs than by the cognate MP. The same conclusion holds true for PVX-GFPΔp25 complementation, but in this case, the movement differences were even greater. It was not until 8 dpi that limited PVX-GFPΔp25 movement became evident during complementation by rhabdovirus MPs, whereas the cognate PVX p25 protein supported efficient movement by about 3 dpi (data not shown). Thus, although the rhabdovirus MPs appear to be able to provide some minimal movement functions for the ToMV-GFPΔMP and PVX-GFPΔp25, they probably lack other functional activities associated with their native MPs. The MPs of both TMV and PVX associate with their own replicase complexes, and these interactions are believed to guide replication/movement complexes to the PD or near the PD orifices, thereby increasing the efficiency and specificity of movement (11, 27, 74). Such spatial links between replication and movement are likely absent or weak in the heterologous trans-complementation systems with positive-stranded RNA viruses, which could explain the low movement efficiencies of such studies.
Despite our findings that rhabdovirus MPs display cross-family movement complementarity with distinct positive-stranded RNA viruses, the movement defects of SYNV and TYMaV MP mutants were rescued only by their cognate MPs, but not by MPs of related rhabdoviruses and the distantly related tenuivirus and tospoviruses, as well as representative members of several positive-stranded RNA viruses. Our results suggest that the strict requirements of cognate MP for rhabdovirus movement can be explained by dependence on specific MP-nucleocapsid core protein interactions. Similar interactions have previously been shown for RYSV P3 and N proteins by using an in vitro glutathione S-transferase (GST) pulldown assay (37), but it is not known whether the proteins interact in vivo, and the species specificities of the interactions were not investigated. Using MYTH assays, we found that the SYNV sc4 and TYMaV P3 engage in species-specific interactions with their respective N and P core proteins in vivo. Therefore, movement specificity can be achieved through these interactions. In addition to unsegmented plant rhabdoviruses, MP-N interactions have been described for various tripartite NS tospoviruses (75–80), suggesting that conserved movement mechanisms may exist across NS RNA virus families. Notably, for positive-stranded RNA viruses requiring the CP for cell-to-cell transit, e.g., alfamo-, bromo-, como-, cucumo-, clostero-, potex-, and potyviruses, specific binding of MPs to cognate CPs or virions has also been reported (see reference 81 and references therein). Nevertheless, movement mutants of these viruses can still be trans-complemented by heterologous MPs (28). In these cases, the non-sequence-specific RNA binding activities of the complementing MPs may facilitate cell-to-cell transit of mutant virus genomic RNAs, thus bypassing CP interactions that are required in native infections. In contrast, the minimal infectious units of NS viruses are viral nucleocapsids that contain encapsidated genomic RNA and the core (N, P, and L polymerase) proteins, so passage of viral genomic RNA alone across the PD is not sufficient for productive infections.
Plant rhabdoviruses establish replication compartments through the formation of cytoplasmic (cytorhabdoviruses) or nuclear (nucleorhabdoviruses) viroplasms. Infectious nucleocapsids assembled in the viroplasms are thought to be transported to the PD, which facilitates passage of the movement complexes across cell boundaries. It has become apparent that for positive-stranded RNA viruses, membrane-associated MPs have important roles in orchestrating intracellular transit of infectious genomes toward the PD (reviewed in references 81 and 82). Membrane associations of a plant rhabdovirus MP were first suggested by Scholthof and coworkers, who found that SYNV sc4 is present in the P30 membrane fraction and cosediments in sucrose gradients with viral glycoprotein fractions (33). Here, we have extended the subcellular fractionation analyses to include SYNV sc4 and TYMaV P3, and our data reveal that both MPs are membrane associated. Moreover, our MYTH assays demonstrate that the membrane-associated SYNV sc4 and TYMaV P3 are able to interact with their cognate nucleocapsid proteins. The BiFC assays also showed that SYNV sc4-N and sc4-P interactions occurred predominantly at the cell periphery, and this pattern correlates with the sc4 subcellular localization. More significantly, transient expression of sc4 directed a portion of the SYNV N-P complexes from subnuclear foci to punctate structures spanning the cell wall that appear to be PD. Although these localization/BiFC experiments are performed with ectopically expressed viral proteins, our findings are consistent with several earlier observations made in the context of SYNV infections. First, upon ectopic expression, the SYNV N protein localizes exclusively with the nuclei (49, 67); however, in SYNV-infected cells, a small proportion of the N protein appears in the cell wall fractions (33). Accordingly, the SYNV sc4 protein localizes at the cell periphery in transient expression experiments (49) and forms punctate structures at the periphery of SYNV-infected cells (45). These observations suggest that the SYNV sc4 protein helps orchestrate intracellular trafficking of viral nucleocapsids. Previous studies have shown that SYNV sc4 (70), as well as LNYV 4b (38), interact with N. benthamiana vascular one-zinc-finger 1 (NbVOZ1), a microtubule-associated transcriptional activator. In the case of LYNV 4b, a mutation analysis showed that the movement activity correlated with the ability to interact with NbVOZ1 (38). It is therefore conceivable that both nucleorhabdovirus and cytorhabdovirus MPs may anchor to microtubule networks for intracellular trafficking of movement complexes. Taken together, these findings support a plant rhabdovirus movement model whereby nucleocapsid core proteins interact specifically with cognate MP proteins to form MP-nucleocapsid complexes (Fig. 6). Our model posits that the complexes may interact with host proteins to facilitate MP-nucleocapsid transit to peripheral membranes near the PD and, subsequently, to adjacent cells. To enable the onset of replication of the negative-sense genomic RNA in invaded cells, nucleocapsids must contain the N protein-encapsidated genomic RNA, as well as the P and L core proteins. Interactions of rhabdovirus MPs with both the N and P proteins may ensure intercellular passage of biologically active nucleocapsids, as the L protein is associated with the encapsidated genome through L-P and P-N protein interactions (31).
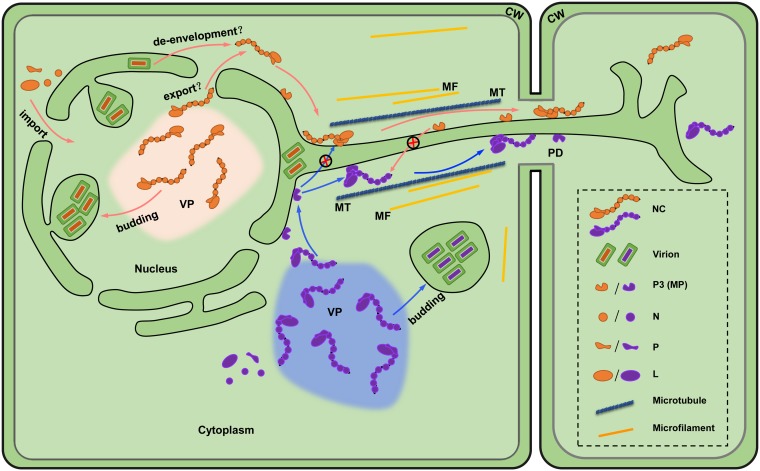
FIG 6.
Model for cell-to-cell movement of plant rhabdoviruses. Replication of nucleorhabdoviruses (tangerine) and cytorhabdoviruses (purple) occur in the nucleus or cytoplasm, respectively, and induce the formation of viroplasm (VP) sites of replication. Newly synthesized viral genomes in the VP are encapsidated by the nucleoprotein (N) and then associated with the phosphoprotein (P) and large polymerase protein (L) to form nucleocapsids (NCs), which are the minimal infectious units and movement entities. For nucleorhabdoviruses, the NCs must exit the nuclei prior to intercellular movement either by nuclear export activities or by de-envelopment of the intraluminal matured virions. The NC core N and P proteins then interact with membrane-associated P3 movement proteins (MPs) that guide intracellular NC trafficking to the cell periphery and intercellular transport through gated plasmodesmata (PD) across the cell wall (CW). Rhabdovirus movement is highly specific in requiring cognate MPs because of the species-specific MP-NC core protein interactions. Microtubule (MT)- and microfilament (MF)-associated proteins may also be involved in assisting NC trafficking.
Cytorhabdoviruses replicate in the cytoplasm, and the assembled nucleocapsids should be available for interactions with MPs that coordinate subsequent local movements. However, SYNV and other nucleorhabdoviruses undergo replication and morphogenesis in subnuclear viroplasms, so an unsolved issue is how the nucleocapsids egress from the nuclear viroplasms to gain access to the membrane-anchored sc4 protein. Whether this process is mediated by nuclear export activities of a viral nucleocapsid protein or via a budding mechanism awaits future studies. With the advent of plant rhabdovirus reverse genetics systems, it is now possible to tag viral proteins genetically with fluorescent proteins, as has been shown in several mammalian rhabdovirus studies (83–88). This approach might permit intracellular and intercellular tracking of the movement of infectious virus entities in live cells.
Although the movement of positive-stranded RNA viruses has been studied for nearly 3 decades, a functional plasmodesmatal localization sequence in the TMV 30K MP has only recently been identified (89) and shown to recognize the synaptotagmin A host protein that participates in tobamo-, poty-, and begomoviruses intracellular trafficking and promotes MP associations with the plasmodesmatal membrane (9). Because of the demonstrated abilities of various rhabdovirus MPs to support the movement of positive-stranded RNA viruses, we anticipate that rhabdovirus MPs likely contain related recognition sequences that have evolved to recognize proteins involved in PD targeting.
MATERIALS AND METHODS
Virus and plant.
Plasmids used to generate rSYNV-GFP and rSYNV-GFP-Δsc4 were described previously (46). The recovery of rTYMaV and rTYMaV-GFP-ΔP3 primarily followed procedures developed by Wang et al. (46) and will be described in detail elsewhere (Lin et al., unpublished data). Engineering of PVX-GFPΔp25 and GUS expression vectors were described previously (51). ToMV-GFPΔMP was generated essentially as described by Hiraguri et al. (50). Wild-type and RFP-H2B transgenic N. benthamiana plants were grown in enriched loam soil in an environment-controlled chamber at 25°C under a 16-h light/8-h dark cycle.
Plasmid construction.
To construct vectors for transient expression of rhabdovirus MPs, the coding sequences of the SYNV sc4 (GenBank accession no. L32604), PYDV Y (GenBank accession no. GU734660), RYSV P3 (GenBank accession no. NC_003746), RSMV P3 (GenBank accession no. KX525586), and TYMaV P3 (GenBank accession no. KY075646) MPs were amplified by PCR and inserted into the pGD binary vector (49) by using the In-Fusion HD PCR cloning kit (Clontech, Japan). ToMV 30K, PVX p25, TMV 30K, CMV 3a, TBSV p22, RSV NSvc4, and TSWV NSm genes were also engineered into pGD using similar strategies, and the primer sequences used for cloning will be made available upon request. The SYNV sc4 and TYMaV P3 genes were also cloned into the pGD-GFP to generate individual GFP fusion expression vectors.
To engineer plasmids for the recovery of chimeric rSYNV plasmids carrying foreign MP genes, we first replaced the sc4 gene in the pSYNV-GFP plasmid (46) with an SpeI restriction site to facilitate foreign gene insertion. For this purpose, an F1 fragment containing a portion of the P cistron was amplified from the pSYNV-GFP plasmid by using the P/NheI/F (5′-AAGAGAAGGGGCTAGCATGTC-3′) and SpeI/delsc4/R (5′-ACTAGTGATACCTGCATACAGAATATATATAAAATC-3′) primers, and an F2 fragment containing a portion of the M cistron was amplified by using the SpeI/delsc4/F (5′-TGCAGGTATCACTAGTTGGCTGGACCTCCGTATTAAG-3′) and M/PmlI/R (5′-CATCACGACTGACACGTGACCT-3′) primers. The two fragments were infused with NheI and PmlI doubly digested pSYNV-GFP to generate pSYNV-GFP-Δsc4::SpeI. Subsequently, the coding sequences of SYNV sc4, PYDV Y, RYSV P3, RSMV P3, TYMaV P3, ToMV MP, TMV 30K, CMV 3a, TBSV p22, RSV NSvc4, and TSWV NSm were amplified by PCR, and their respective PCR products were individually inserted to the SpeI-linearized plasmid pSYNV-GFP-Δsc4::SpeI by In-Fusion cloning to generate rSYNV derivatives containing heterologous MP gene substitutions for the sc4 gene.
For MYTH assay constructs, the SYNV sc4 gene was inserted into the bait plasmid pBT3-N (Dualsystems Biotech, Switzerland) as a C-terminal fusion, while the TYMaV P3 gene was cloned into the pBT3-C bait plasmid as an N-terminal fusion (Dualsystems Biotech). For the prey vectors, the N and P genes of SYNV, RSMV, and TYMaV were each cloned into both pPR3-C and pPR3-N prey plasmids (Dualsystems Biotech) to generate x-NubG and NubG-x fusions, respectively.
To generate BiFC plasmids, the coding sequences of the SYNV N, P, and sc4 proteins were fused to the N-terminal and C-terminal fragments of enhanced YFP gene in the p2YN and p2YC vectors (90) using In-Fusion cloning.
trans-Complementation of movement-defective ToMV and PVX.
Suspensions of A. tumefaciens EHA 105 strains carrying PVX-GFPΔp25 (optical density [OD] A600, 1.0) were diluted 1,000-fold and mixed in 1:1:1 ratios with Agrobacterium cultures containing the TBSV p19 VSR plasmid and each of the movement protein constructs to be tested for trans-complementation. The mixtures were agroinfiltrated into the leaves of 6-week-old N. benthamiana plants. GFP fluorescence in the leaves was photographed with an epifluorescence microscope first at 60 hpi and then at 8 dpi.
A. tumefaciens GV3101 strains harboring ToMV-GFPΔMP (OD A600, 1.0) were diluted 2,000-fold and mixed 1:1 with agrobacterial cultures carrying individual pGD plasmids designed for the expression of each of the movement proteins. Infiltrated leaves of N. benthamiana plants were photographed with an epifluorescence microscope first at 50 and 60 hpi.
SYNV and TYMaV movement complementation assays.
For SYNV and TYMaV trans-complementation assays, A. tumefaciens EHA 105 strains carrying each of the MP expression plasmids were mixed with agrobacterial cultures harboring either pSYNV-GFP-Δsc4 or pTYMaV-GFP-ΔP3, the pGD-NPL plasmid, and the TBSV p19, BSMV γb, and TEV P1/HC-Pro VSR plasmids (46) at a ratio of 1:9 (vol/vol) with a total OD A600 of 0.7. After agroinfiltration, leaves were observed at 8 and 12 dpi with a fluorescence microscope. For cis-complementation with the chimeric SYNV substitution mutants, A. tumefaciens EHA105 strains harboring the pSYNV-GFP derivatives, the pGD-NPL plasmid, and the three VSRs were coinfiltrated into N. benthamiana leaves.
BiFC, fluorescence, and confocal laser scanning microscopy.
Transient expression plasmids and BiFC plasmids were introduced into A. tumefaciens strain EHA105 by electroporation, followed by agroinfiltration of wild-type or transgenic RFP-H2B N. benthamiana plants. BiFC assays were carried out essentially as described previously (71). Epifluorescence microscopy was performed with a Zeiss Lumar V12 stereomicroscope. Images were captured with a Lumar 38 filter set for GFP detection (excitation, 470/40 nm; emission, 525/50 nm) and a Lumar 31 filter set for RFP detection (excitation, 565/30 nm; emission, 620/60 nm; Carl Zeiss, Germany). Confocal laser scanning micrographs were captured with a Zeiss 780 confocal microscope. GFP, RFP, and YFP were excited by using 488-, 561-, and 514-nm laser lines, respectively. All images were processed using LSM software Zen 2009 (Carl Zeiss, Germany) and Photoshop CS4 (Adobe, Mountain View, CA).
Isolation of microsomal fractions and protein analysis.
Plant materials were ground in lysis buffer (20 mM HEPES [pH 6.8], 150 mM potassium acetate, 250 mM mannitol, 1 mM MgCl2, 1 mM dithiothreitol, and a protease inhibitor cocktail [Sigma]), followed by centrifugation at 3,000 × g for 10 min at 4°C to separate the soluble (S3) and pellet (P3) fractions. The S3 fraction was then subjected to high-speed centrifugation (30,000 × g for 1 h) to obtain the soluble (S30) and pellet (P30) fractions. For chemical treatments, microsomal pellets (P30) were resuspended in either 10 volumes of lysis buffer, 100 mM Na2CO3 (pH 11.5), 1 M KCl, 7 M urea, or 1% Triton X-100. After incubation for 30 min on ice, the samples were centrifuged at 30,000 × g for 1 h. Protein samples in the pellet and supernatant fractions were separated by 12% SDS-PAGE gels and detected by Western blotting with antibodies against GFP, RFP, or SYNV G protein (Abcam, Cambridge, UK).
MYTH assays.
MYTH assays were performed essentially as described previously (91). Briefly, the yeast strain NMY 51 [MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 ura3::(lexAop)8-lacZ ade2::(lexAop)8-ADE2 GAL4] was cotransformed with the bait (pBT3-N or pBT3-C derivatives) and prey (pPR3-N or pPR3-C derivatives) plasmids. Transformants were plated onto a synthetic dropout medium lacking tryptophan and leucine (SD-LW) and a selective medium lacking tryptophan, leucine, histidine, and adenine (SD-LWHA).
ACKNOWLEDGMENTS
We thank Michael Goodin (University of Kentucky) for sharing RFP-H2B transgenic N. benthamiana seeds. We are grateful to Fei Yan (Ningbo University, China) for providing the PVX-GFPΔp25 construct and to Qiansheng Liao (Zhejiang Sci-Tech University, China) for providing the ToMV-GFPΔMP construct as a gift.
This work was supported by the National Natural Science Foundation of China (grants 31671996 and 31470255).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Lucas WJ, Wolf S. 1999. Connections between virus movement, macromolecular signaling and assimilate allocation. Curr Opin Plant Biol 2:192–197. doi: 10.1016/S1369-5266(99)80035-1. [DOI] [PubMed] [Google Scholar]
- 2.Lucas WJ. 2006. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344:169–184. doi: 10.1016/j.virol.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Scholthof HB. 2005. Plant virus transport: motions of functional equivalence. Trends Plant Sci 10:376–382. doi: 10.1016/j.tplants.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Taliansky M, Torrance L, Kalinina NO. 2008. Role of plant virus movement proteins. Methods Mol Biol 451:33–54. doi: 10.1007/978-1-59745-102-4_3. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Nelson RS. 2013. The cell biology of tobacco mosaic virus replication and movement. Front Plant Sci 4:12. doi: 10.3389/fpls.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waigmann E, Lucas WJ, Citovsky V, Zambryski P. 1994. Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci U S A 91:1433–1437. doi: 10.1073/pnas.91.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf S, Deom CM, Beachy R, Lucas WJ. 1991. Plasmodesmatal function is probed using transgenic tobacco plants that express a virus movement protein. Plant Cell 3:593–604. doi: 10.1105/tpc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf S, Deom CM, Beachy RN, Lucas WJ. 1989. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246:377–379. doi: 10.1126/science.246.4928.377. [DOI] [PubMed] [Google Scholar]
- 9.Yuan C, Lazarowitz SG, Citovsky V. 2018. The plasmodesmal localization signal of TMV MP is recognized by plant synaptotagmin SYTA. mBio 9:e01314-18. doi: 10.1128/mBio.01314-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN. 1998. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawakami S, Watanabe Y, Beachy RN. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc Natl Acad Sci U S A 101:6291–6296. doi: 10.1073/pnas.0401221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLean BG, Zupan J, Zambryski PC. 1995. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Citovsky V, Knorr D, Schuster G, Zambryski P. 1990. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60:637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- 14.Citovsky V, Wong ML, Shaw AL, Prasad BV, Zambryski P. 1992. Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell 4:397–411. doi: 10.1105/tpc.4.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melcher U. 2000. The '30K' superfamily of viral movement proteins. J Gen Virol 81:257–266. doi: 10.1099/0022-1317-81-1-257. [DOI] [PubMed] [Google Scholar]
- 16.Mushegian AR, Elena SF. 2015. Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology 476:304–315. doi: 10.1016/j.virol.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Yu C, Karlin DG, Lu Y, Wright K, Chen J, MacFarlane S. 2013. Experimental and bioinformatic evidence that raspberry leaf blotch emaravirus P4 is a movement protein of the 30K superfamily. J Gen Virol 94:2117–2128. doi: 10.1099/vir.0.053256-0. [DOI] [PubMed] [Google Scholar]
- 18.Ritzenthaler C, Hofmann C. 2007. Tubule-guided movement of plant viruses, p 63–83. In Waigmann E, Heinlein M (ed), Viral transport in plants. Springer Berlin Heidelberg, Berlin, Germany. [Google Scholar]
- 19.Jackson AO, Lim HS, Bragg J, Ganesan U, Lee MY. 2009. Hordeivirus replication, movement, and pathogenesis. Annu Rev Phytopathol 47:385–422. doi: 10.1146/annurev-phyto-080508-081733. [DOI] [PubMed] [Google Scholar]
- 20.Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. 2010. Varied movement strategies employed by triple gene block-encoding viruses. Mol Plant Microbe Interact 23:1231–1247. doi: 10.1094/MPMI-04-10-0086. [DOI] [PubMed] [Google Scholar]
- 21.Angell SM, Davies C, Baulcombe DC. 1996. Cell-to-cell movement of potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology 216:197–201. doi: 10.1006/viro.1996.0046. [DOI] [PubMed] [Google Scholar]
- 22.Howard AR, Heppler ML, Ju HJ, Krishnamurthy K, Payton ME, Verchot-Lubicz J. 2004. Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328:185–197. doi: 10.1016/j.virol.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Ding B, Baulcombe DC, Verchot J. 2000. Cell-to-cell movement of the 25K protein of potato virus X is regulated by three other viral proteins. Mol Plant Microbe Interact 13:599–605. doi: 10.1094/MPMI.2000.13.6.599. [DOI] [PubMed] [Google Scholar]
- 24.Haupt S, Cowan GH, Ziegler A, Roberts AG, Oparka KJ, Torrance L. 2005. Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17:164–181. doi: 10.1105/tpc.104.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju HJ, Samuels TD, Wang YS, Blancaflor E, Payton M, Mitra R, Krishnamurthy K, Nelson RS, Verchot-Lubicz J. 2005. The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol 138:1877–1895. doi: 10.1104/pp.105.066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lough TJ, Netzler NE, Emerson SJ, Sutherland P, Carr F, Beck DL, Lucas WJ, Forster RL. 2000. Cell-to-cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol Plant Microbe Interact 13:962–974. doi: 10.1094/MPMI.2000.13.9.962. [DOI] [PubMed] [Google Scholar]
- 27.Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN, Oparka KJ. 2013. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 201:981–995. doi: 10.1083/jcb.201304003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latham JR, Wilson AK. 2008. Transcomplementation and synergism in plants: implications for viral transgenes? Mol Plant Pathol 9:85–103. doi: 10.1111/j.1364-3703.2007.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiraguri A, Netsu O, Sasaki N, Nyunoya H, Sasaya T. 2014. Recent progress in research on cell-to-cell movement of rice viruses. Front Microbiol 5:210. doi: 10.3389/fmicb.2014.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietzgen RG, Kondo H, Goodin MM, Kurath G, Vasilakis N. 2017. The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res 227:158–170. doi: 10.1016/j.virusres.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. 2005. Biology of plant rhabdoviruses. Annu Rev Phytopathol 43:623–660. doi: 10.1146/annurev.phyto.43.011205.141136. [DOI] [PubMed] [Google Scholar]
- 32.Walker PJ, Dietzgen RG, Joubert DA, Blasdell KR. 2011. Rhabdovirus accessory genes. Virus Res 162:110–125. doi: 10.1016/j.virusres.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholthof KB, Hillman BI, Modrell B, Heaton LA, Jackson AO. 1994. Characterization and detection of sc4: a sixth gene encoded by sonchus yellow net virus. Virology 204:279–288. doi: 10.1006/viro.1994.1532. [DOI] [PubMed] [Google Scholar]
- 34.Bandyopadhyay A, Kopperud K, Anderson G, Martin K, Goodin M. 2010. An integrated protein localization and interaction map for potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology 402:61–71. doi: 10.1016/j.virol.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappi PG, Dovas CI, Efthimiou KE, Maliogka VI, Katis NI. 2013. A novel strategy for the determination of a rhabdovirus genome and its application to sequencing of eggplant mottled dwarf virus. Virus Genes 47:105–113. doi: 10.1007/s11262-013-0911-5. [DOI] [PubMed] [Google Scholar]
- 36.Dietzgen RG, Callaghan B, Wetzel T, Dale JL. 2006. Completion of the genome sequence of Lettuce necrotic yellows virus, type species of the genus Cytorhabdovirus. Virus Res 118:16–22. doi: 10.1016/j.virusres.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 37.Huang YW, Geng YF, Ying XB, Chen XY, Fang RX. 2005. Identification of a movement protein of rice yellow stunt rhabdovirus. J Virol 79:2108–2114. doi: 10.1128/JVI.79.4.2108-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann KS, Bejerman N, Johnson KN, Dietzgen RG. 2016. Cytorhabdovirus P3 genes encode 30K-like cell-to-cell movement proteins. Virology 489:20–33. doi: 10.1016/j.virol.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 39.Bejerman N, Giolitti F, de Breuil S, Trucco V, Nome C, Lenardon S, Dietzgen RG. 2015. Complete genome sequence and integrated protein localization and interaction map for alfalfa dwarf virus, which combines properties of both cytoplasmic and nuclear plant rhabdoviruses. Virology 483:275–283. doi: 10.1016/j.virol.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Dietzgen RG, Innes DJ, Bejerman N. 2015. Complete genome sequence and intracellular protein localization of Datura yellow vein nucleorhabdovirus. Virus Res 205:7–11. doi: 10.1016/j.virusres.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Hiraguri A, Hibino H, Hayashi T, Netsu O, Shimizu T, Uehara-Ichiki T, Omura T, Sasaki N, Nyunoya H, Sasaya T. 2012. The movement protein encoded by gene 3 of rice transitory yellowing virus is associated with virus particles. J Gen Virol 93:2290–2298. doi: 10.1099/vir.0.044420-0. [DOI] [PubMed] [Google Scholar]
- 42.Jang C, Wang R, Wells J, Leon F, Farman M, Hammond J, Goodin MM. 2017. Genome sequence variation in the constricta strain dramatically alters the protein interaction and localization map of Potato yellow dwarf virus. J Gen Virol 98:1526–1536. doi: 10.1099/jgv.0.000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin KM, Dietzgen RG, Wang R, Goodin MM. 2012. Lettuce necrotic yellows cytorhabdovirus protein localization and interaction map, and comparison with nucleorhabdoviruses. J Gen Virol 93:906–914. doi: 10.1099/vir.0.038034-0. [DOI] [PubMed] [Google Scholar]
- 44.Martin KM, Whitfield AE. 2018. Cellular localization and interactions of nucleorhabdovirus proteins are conserved between insect and plant cells. Virology 523:6–14. doi: 10.1016/j.virol.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Goodin MM, Chakrabarty R, Yelton S, Martin K, Clark A, Brooks R. 2007. Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant-adapted rhabdovirus. J Gen Virol 88:1810–1820. doi: 10.1099/vir.0.82698-0. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, Ma X, Qian S, Zhou X, Sun K, Chen X, Zhou X, Jackson AO, Li Z. 2015. Rescue of a plant negative-strand RNA virus from cloned cDNA: Insights into enveloped plant virus movement and morphogenesis. PLoS Pathog 11:e1005223. doi: 10.1371/journal.ppat.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Huang J, Liu C, Chen B, Zhang T, Zhou G. 2016. Rice stripe mosaic virus, a novel cytorhabdovirus infecting rice via leafhopper transmission. Front Microbiol 7:2140. doi: 10.3389/fmicb.2016.02140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu C, Sun X, Taylor A, Jiao C, Xu Y, Cai X, Wang X, Ge C, Pan G, Wang Q, Fei Z, Wang Q. 2017. Diversity, distribution, and evolution of tomato viruses in China uncovered by small RNA sequencing. J Virol 91:e00173-17. doi: 10.1128/JVI.00173-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. 2002. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31:375–383. doi: 10.1046/j.1365-313X.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- 50.Hiraguri A, Netsu O, Shimizu T, Uehara-Ichiki T, Omura T, Sasaki N, Nyunoya H, Sasaya T. 2011. The nonstructural protein pC6 of rice grassy stunt virus trans-complements the cell-to-cell spread of a movement-defective tomato mosaic virus. Arch Virol 156:911–916. doi: 10.1007/s00705-011-0939-6. [DOI] [PubMed] [Google Scholar]
- 51.Bayne EH, Rakitina DV, Morozov SY, Baulcombe DC. 2005. Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J 44:471–482. doi: 10.1111/j.1365-313X.2005.02539.x. [DOI] [PubMed] [Google Scholar]
- 52.Xiong R, Wu J, Zhou Y, Zhou X. 2008. Identification of a movement protein of the tenuivirus rice stripe virus. J Virol 82:12304–12311. doi: 10.1128/JVI.01696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kormelink R, Storms M, Van Lent J, Peters D, Goldbach R. 1994. Expression and subcellular location of the NSM protein of tomato spotted wilt virus (TSWV), a putative viral movement protein. Virology 200:56–65. doi: 10.1006/viro.1994.1162. [DOI] [PubMed] [Google Scholar]
- 54.Scholthof HB, Scholthof KB, Kikkert M, Jackson AO. 1995. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- 55.Vaquero C, Turner AP, Demangeat G, Sanz A, Serra MT, Roberts K, Garcia-Luque I. 1994. The 3a protein from cucumber mosaic virus increases the gating capacity of plasmodesmata in transgenic tobacco plants. J Gen Virol 75:3193–3197. doi: 10.1099/0022-1317-75-11-3193. [DOI] [PubMed] [Google Scholar]
- 56.Meshi T, Watanabe Y, Saito T, Sugimoto A, Maeda T, Okada Y. 1987. Function of the 30 kD protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J 6:2557–2563. doi: 10.1002/j.1460-2075.1987.tb02544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- 58.Pitzalis N, Heinlein M. 2017. The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. J Exp Bot 69:117–132. doi: 10.1093/jxb/erx334. [DOI] [PubMed] [Google Scholar]
- 59.Peremyslov VV, Pan YW, Dolja VV. 2004. Movement protein of a closterovirus is a type III integral transmembrane protein localized to the endoplasmic reticulum. J Virol 78:3704–3709. doi: 10.1128/jvi.78.7.3704-3709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaad MC, Jensen PE, Carrington JC. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J 16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 62.Kahsay RY, Gao G, Liao L. 2005. An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21:1853–1858. doi: 10.1093/bioinformatics/bti303. [DOI] [PubMed] [Google Scholar]
- 63.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 64.Cserzo M, Eisenhaber F, Eisenhaber B, Simon I. 2004. TM or not TM: transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics 20:136–137. doi: 10.1093/bioinformatics/btg394. [DOI] [PubMed] [Google Scholar]
- 65.Jackson AO, Li Z. 2016. Developments in plant negative-strand RNA virus reverse genetics. Annu Rev Phytopathol 54:469–498. doi: 10.1146/annurev-phyto-080615-095909. [DOI] [PubMed] [Google Scholar]
- 66.Stagljar I, Korostensky C, Johnsson N, Te Heesen S. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci U S A 95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodin MM, Austin J, Tobias R, Fujita M, Morales C, Jackson AO. 2001. Interactions and nuclear import of the N and P proteins of sonchus yellow net virus, a plant nucleorhabdovirus. J Virol 75:9393–9406. doi: 10.1128/JVI.75.19.9393-9406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martins CR, Johnson JA, Lawrence DM, Choi TJ, Pisi AM, Tobin SL, Lapidus D, Wagner JD, Ruzin S, McDonald K, Jackson AO. 1998. Sonchus yellow net rhabdovirus nuclear viroplasms contain polymerase-associated proteins. J Virol 72:5669–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deng M, Bragg JN, Ruzin S, Schichnes D, King D, Goodin MM, Jackson AO. 2007. Role of the sonchus yellow net virus N protein in formation of nuclear viroplasms. J Virol 81:5362–5374. doi: 10.1128/JVI.02349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Min BE, Martin K, Wang R, Tafelmeyer P, Bridges M, Goodin M. 2010. A host-factor interaction and localization map for a plant-adapted rhabdovirus implicates cytoplasm-tethered transcription activators in cell-to-cell movement. Mol Plant Microbe Interact 23:1420–1432. doi: 10.1094/MPMI-04-10-0097. [DOI] [PubMed] [Google Scholar]
- 71.Sun K, Zhou X, Lin W, Zhou X, Jackson AO, Li Z. 2018. Matrix-glycoprotein interactions required for budding of a plant nucleorhabdovirus and induction of inner nuclear membrane invagination. Mol Plant Pathol 19:2288–2301. doi: 10.1111/mpp.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamai A, Meshi T. 2001. Tobamoviral movement protein transiently expressed in a single epidermal cell functions beyond multiple plasmodesmata and spreads multicellularly in an infection-coupled manner. MPMI 14:126–134. doi: 10.1094/MPMI.2001.14.2.126. [DOI] [PubMed] [Google Scholar]
- 73.Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283:61–119. [DOI] [PubMed] [Google Scholar]
- 74.Szécsi J, Ding XS, Lim CO, Bendahmane M, Cho MJ, Nelson RS, Beachy RN. 1999. Development of tobacco nosaic virus infection sites in Nicotiana benthamiana. Mol Plant Microbe Interact 12:143–152. doi: 10.1094/MPMI.1999.12.2.143. [DOI] [Google Scholar]
- 75.Widana Gamage SMK, Dietzgen RG. 2017. Intracellular localization, interactions and functions of Capsicum chlorosis virus proteins. Front Microbiol 8:612. doi: 10.3389/fmicb.2017.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leastro MO, Pallas V, Resende RO, Sanchez-Navarro JA. 2015. The movement proteins (NSm) of distinct tospoviruses peripherally associate with cellular membranes and interact with homologous and heterologous NSm and nucleocapsid proteins. Virology 478:39–49. doi: 10.1016/j.virol.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 77.Leastro MO, Pallas V, Resende RO, Sanchez-Navarro JA. 2017. The functional analysis of distinct tospovirus movement proteins (NSM) reveals different capabilities in tubule formation, cell-to-cell and systemic virus movement among the tospovirus species. Virus Res 227:57–68. doi: 10.1016/j.virusres.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 78.Tripathi D, Raikhy G, Pappu HR. 2015. Movement and nucleocapsid proteins coded by two tospovirus species interact through multiple binding regions in mixed infections. Virology 478:137–147. doi: 10.1016/j.virol.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 79.Soellick T, Uhrig JF, Bucher GL, Kellmann JW, Schreier PH. 2000. The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc Natl Acad Sci U S A 97:2373–2378. doi: 10.1073/pnas.030548397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tripathi D, Raikhy G, Goodin MM, Dietzgen RG, Pappu HR. 2015. In vivo localization of iris yellow spot tospovirus (Bunyaviridae)-encoded proteins and identification of interacting regions of nucleocapsid and movement proteins. PLoS One 10:e0118973. doi: 10.1371/journal.pone.0118973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tilsner J, Oparka KJ. 2012. Missing links? The connection between replication and movement of plant RNA viruses. Curr Opin Virol 2:705–711. doi: 10.1016/j.coviro.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 82.Heinlein M. 2015. Plant virus replication and movement. Virology 479-480:657–671. doi: 10.1016/j.virol.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 83.Das SC, Nayak D, Zhou Y, Pattnaik AK. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J Virol 80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finke S, Brzozka K, Conzelmann KK. 2004. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J Virol 78:12333–12343. doi: 10.1128/JVI.78.22.12333-12343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klingen Y, Conzelmann KK, Finke S. 2008. Double-labeled rabies virus: live tracking of enveloped virus transport. J Virol 82:237–245. doi: 10.1128/JVI.01342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ruedas JB, Perrault J. 2009. Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro. J Virol 83:12241–12252. doi: 10.1128/JVI.01273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soh TK, Whelan SP. 2015. Tracking the fate of genetically distinct vesicular stomatitis virus matrix proteins highlights the role for late domains in assembly. J Virol 89:11750–11760. doi: 10.1128/JVI.01371-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Falzarano D, Groseth A, Hoenen T. 2014. Development and application of reporter-expressing mononegaviruses: current challenges and perspectives. Antiviral Res 103:78–87. doi: 10.1016/j.antiviral.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan C, Lazarowitz SG, Citovsky V. 2016. Identification of a functional plasmodesmal localization signal in a plant viral cell-to-cell-movement protein. mBio 7:e02052-15. doi: 10.1128/mBio.02052-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang X, Baliji S, Buchmann RC, Wang H, Lindbo JA, Sunter G, Bisaro DM. 2007. Functional modulation of the geminivirus al2 transcription factor and silencing suppressor by self-interaction. J Virol 81:11972–11981. doi: 10.1128/JVI.00617-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J Virol 82:6911–6926. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]