Studies of SIV-infected macaques that deplete CD8+ T cells in vivo with monoclonal antibodies have provided compelling evidence for their direct antiviral role. These studies utilized CD8α-specific mAbs that target both the major (CD8αβ+) and minor (CD8αα+) populations of CD8+ T cells but additionally deplete non-CD8+ T cell populations that express CD8α, such as NK cells and γδ T cells. In the current study, we administered the CD8β-specific depleting mAb CD8β255R1 to cynomolgus macaques chronically infected with a LASIV to selectively deplete CD8αβ+ T cells without removing CD8αα+ lymphocytes. We evaluated the impact on control of virus replication and protection from pathogenic SIVmac239 challenge. These results underscore the utility of CD8β255R1 for studying the direct contribution of CD8αβ+ T cells in various disease states.
KEYWORDS: CD8αβ+ T cells, live-attenuated SIV, cynomolgus macaques, depletion
ABSTRACT
We evaluated the contribution of CD8αβ+ T cells to control of live-attenuated simian immunodeficiency virus (LASIV) replication during chronic infection and subsequent protection from pathogenic SIV challenge. Unlike previous reports with a CD8α-specific depleting monoclonal antibody (mAb), the CD8β-specific mAb CD8β255R1 selectively depleted CD8αβ+ T cells without also depleting non-CD8+ T cell populations that express CD8α, such as natural killer (NK) cells and γδ T cells. Following infusion with CD8β255R1, plasma viremia transiently increased coincident with declining peripheral CD8αβ+ T cells. Interestingly, plasma viremia returned to predepletion levels even when peripheral CD8αβ+ T cells did not. Although depletion of CD8αβ+ T cells in the lymph node (LN) was incomplete, frequencies of these cells were 3-fold lower (P = 0.006) in animals that received CD8β255R1 than in those that received control IgG. It is possible that these residual SIV-specific CD8αβ+ T cells may have contributed to suppression of viremia during chronic infection. We also determined whether infusion of CD8β255R1 in the LASIV-vaccinated animals increased their susceptibility to infection following intravenous challenge with pathogenic SIVmac239. We found that 7/8 animals infused with CD8β255R1, and 3/4 animals infused with the control IgG, were resistant to SIVmac239 infection. These results suggest that infusion with CD8β255R1 did not eliminate the protection afforded to LASIV vaccination. This provides a comprehensive description of the impact of CD8β255R1 infusion on the immunological composition in cynomolgus macaques, compared to an isotype-matched control IgG, while showing that the control of LASIV viremia and protection from challenge can occur even after CD8β255R1 administration.
IMPORTANCE Studies of SIV-infected macaques that deplete CD8+ T cells in vivo with monoclonal antibodies have provided compelling evidence for their direct antiviral role. These studies utilized CD8α-specific mAbs that target both the major (CD8αβ+) and minor (CD8αα+) populations of CD8+ T cells but additionally deplete non-CD8+ T cell populations that express CD8α, such as NK cells and γδ T cells. In the current study, we administered the CD8β-specific depleting mAb CD8β255R1 to cynomolgus macaques chronically infected with a LASIV to selectively deplete CD8αβ+ T cells without removing CD8αα+ lymphocytes. We evaluated the impact on control of virus replication and protection from pathogenic SIVmac239 challenge. These results underscore the utility of CD8β255R1 for studying the direct contribution of CD8αβ+ T cells in various disease states.
INTRODUCTION
Multiple lines of evidence suggest that CD8+ T cells contribute to control of virus replication and subsequently influence disease progression following human immunodeficiency virus (HIV) infection. For instance, the emergence of HIV-specific CD8+ cytotoxic T lymphocytes (CTLs) during acute infection is temporally associated with decreases in peak viral load and the decline of viral replication to set point viral load (1, 2). Indeed, CD8+ CTLs have been shown to lyse HIV-infected cells in vitro, and the selective pressure they exert in vivo often leads to the emergence of immune escape variants (3–7). The strongest argument comes from studies of macaques infected with simian immunodeficiency virus (SIV) that are infused with a monoclonal antibody (mAb) that is specific for the CD8α molecule of CD8+ lymphocytes. Following infusion with this antibody, depletion of CD8α+ cells persists for approximately 2 to 4 weeks and is accompanied by a transient increase in virus replication until control is regained coincident with the reemergence of CD8α+ lymphocytes (8–20). Of note, control of virus replication is lost following in vivo depletion of CD8+ lymphocytes even during antiretroviral therapy (ART), further suggesting that functional CD8+ T cells are needed to maintain effective viral control even while on ART (11, 12). Notably, however, CD8α-specific mAbs deplete not only CD8+ T cells but also a variety of cell populations that express the CD8α molecule.
The CD8 molecule is expressed as either a CD8αα homodimer or a CD8αβ heterodimer on the cell surface and is present on lymphocytes of both the innate and adaptive immune systems (21–24). The most common lymphocytes to express CD8 are conventional CD8+ T cells (TCRαβ+ CD3+), which can be divided into a major population that express CD8αβ and a minor population that express CD8αα (25). There also exist populations of TCRγδ+ CD3+ T cells and CD3− natural killer (NK) cells that express CD8αα (23, 26, 27). γδ T cells (TCRγδ+ CD3+ CD8αα+), which comprise ∼6% of CD3+ T cells (26), can block HIV-1 entry via the secretion of β-chemokines (28), enhance antibody-dependent cellular cytotoxicity (ADCC) (29), and directly lyse HIV-infected cells (30). NK cells (CD3− CD8αα+) comprise ∼16% of peripheral lymphocytes and have recently been reported to possess traits of adaptive immunity that may contribute to control of HIV-1 replication (31, 32). Accordingly, the contribution of conventional CD8+ T cells to viral control is complicated by the depletion of additional cell populations that express CD8α when using CD8α-depleting mAbs (10, 13, 19). One approach to better define the antiviral role of CD8+ T cells in vivo is to administer a CD8β-specific depleting mAb, as this should selectively deplete CD8αβ+ T cells without removing CD8αα+ lymphocytes or other non-T cell populations. Indeed, two recent studies using the CD8β-specific mAb CD8β255R1 in rhesus macaques provide evidence that CD8αβ+ T cells can be specifically depleted in vivo (33, 34).
Macaques vaccinated with SIVmac239Δnef, a live-attenuated SIV (LASIV) variant of pathogenic SIVmac239, are useful for evaluating the role of CD8αβ+ T cells in control of virus replication and protection from SIV challenge. Although rare hosts spontaneously control pathogenic HIV or SIV in a manner dependent on particular major histocompatibility complex (MHC) alleles, control of SIVmac239Δnef replication occurs in nearly every vaccinated animal, regardless of host MHC genetics (14, 35–38). These observations question whether the contribution of conventional CD8αβ+ T cells to control of SIVmac239Δnef is equivalent to their contribution to control of pathogenic SIV. Moreover, vaccination with SIVmac239Δnef is the most successful example of vaccine-induced protection from challenge with homologous SIV strains and, less frequently, from challenge with heterologous SIV strains (14, 39–41). After more than 25 years of effort, the precise immune mechanism(s) responsible for this protection is still under debate (42–46). Thus, defining whether SIVmac239Δnef-mediated vaccine protection requires CD8αβ+ T cells may also help inform either therapeutic or prophylactic HIV vaccine design.
In this study, we utilized the CD8β-specific mAb CD8β255R1 to specifically deplete CD8αβ+ T cells in LASIV-vaccinated Mauritian cynomolgus macaques (MCMs) (47) and then measure the impact on control of LASIV replication and protection from pathogenic SIV challenge. In contrast to two recent studies that evaluated the impact of CD8β255R1 in SIV-infected or SHIV-infected rhesus macaques (33, 34), we included animals treated with a control IgG to distinguish those immunological effects specific to CD8β depletion from those that are a result of infusion of a nonspecific IgG antibody. We also determined whether depletion of CD8αβ+ T cells affects the frequency and proliferative capacity of CD8αα+ T cells, NK cells, and γδ T cells. Thus, we expand the current knowledge of the immunological effects that follow infusion with CD8β255R1 and provide the first comprehensive comparison to an isotype-matched control IgG.
(This article was submitted to an online preprint archive [48].)
RESULTS
Infusion with CD8β255R1 selectively depletes CD8αβ+ T cells in naive cynomolgus macaques.
Previous studies of CD8β depletion following infusion with the anti-CD8β mAb CD8β255R1 were performed exclusively with rhesus macaques (33, 34). To determine whether CD8β255R1 can similarly deplete CD8αβ+ T cells in cynomolgus macaques, two naive animals (A12412 and A12372) were intravenously infused with a single 50-mg/kg dose of the anti-CD8β mAb CD8β255R1 and evaluated for 15 weeks. Flow cytometry was used to monitor CD8αβ+ T cells in peripheral blood mononuclear cells (PBMC) and peripheral lymph nodes (LNs) before and after infusion in both animals. Depletion of peripheral CD8αβ+ T cells was rapid and sustained (Fig. 1a, top), with the percentage of lymphocytes relative to baseline reaching its nadir at week 2 (A12412, 89%; A12372, 99%) and remaining substantially reduced at week 15 (A12412, 75%; A12372, 85%). The frequency of circulating CD4+ T cells as a fraction of total T cells increased transiently following infusion (Fig. 1a, middle), while circulating NK cells were relatively unchanged over the course of 15 weeks (Fig. 1a, bottom).
FIG 1.
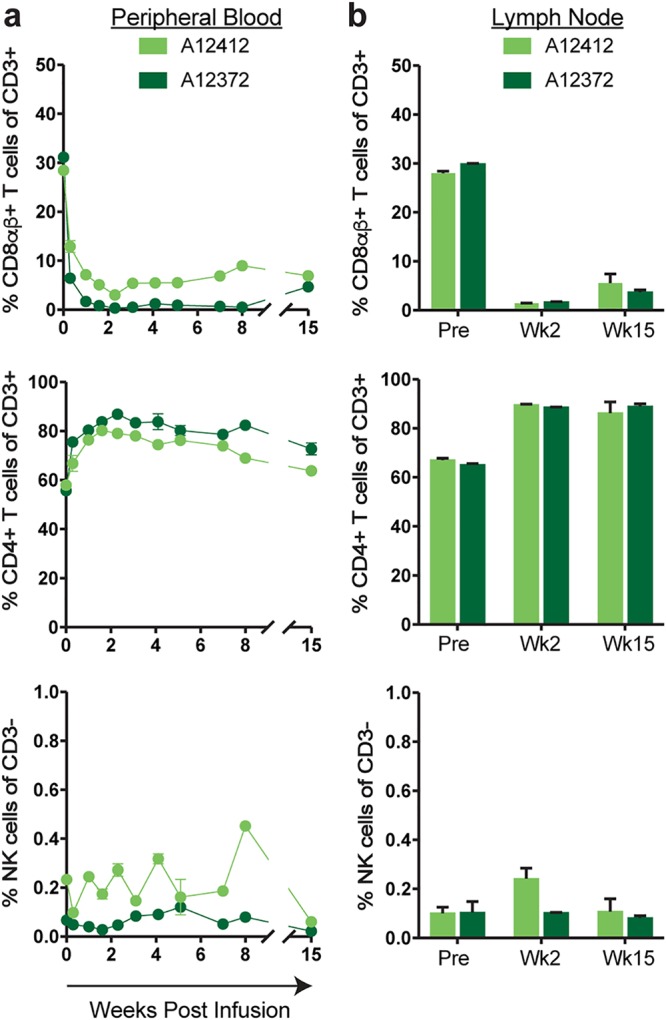
Infusion with CD8β255R1 selectively depletes CD8αβ+ T cells in naive cynomolgus macaques. Two naive cynomolgus macaques (A12412 and A12372) received a single 50-mg/kg intravenous infusion with CD8β255R1. The percentages of CD8β+ T cells (top), CD4+ T cells (middle), and CD16+ NK cells (bottom) in peripheral blood (a) and lymph nodes (b) were measured by multicolor flow cytometry. Data are shown as mean and standard deviation. Peripheral blood samples from each animal were analyzed in triplicate, while lymph node samples from each animal were analyzed in duplicate.
Depletion of LN CD8αβ+ T cells after administration of the CD8β255R1 mAb has been reported in rhesus macaques (http://www.nhpreagents.org/NHP/availablereagents.aspx?Cat=2). We also observed reductions in the frequency of LN CD8αβ+ T cells (Fig. 1b, top) in both cynomolgus macaques at week 2 (mean, 94.5%; range, 94% to 95%) that remained reduced by approximately 84% at week 15 (range, 80% to 88%) compared to the frequency predepletion. Similar to our observations in the blood, we detected an increased frequency of CD4+ T cells in LNs (Fig. 1b, middle) that persisted for 15 weeks following infusion, while the frequency of NK cells in LNs (Fig. 1b, bottom) remained largely unaffected for 15 weeks. These results are not surprising, as a decrease in the frequency of CD3+ T cells that are CD8+ will correspond to an increase in the frequency of CD3+ cells that are CD4+ in the same compartment. To our knowledge, this is the first assessment of the immunological effects following infusion with CD8β255R1 in cynomolgus macaques, and these results are similar to those observed in rhesus macaques (33, 34).
Impact of CD8β255R1 infusion on CD8αβ+ T cells during chronic LASIV infection.
Seven MCMs chronically infected with either SIVmac239Δnef (n = 2) or a SIVmac239Δnef variant (n = 5) (47) received a single 50-mg/kg intravenous infusion of the anti-CD8β mAb CD8β255R1. Four MCMs infected with the same SIVmac239Δnef variant received a single 50-mg/kg intravenous infusion of an isotype-matched rhesus recombinant IgG mAb. All animals had been infected with LASIV for different lengths of time, as they were used in previous studies of T cell-mediated control of acute SIV replication (47). Four weeks after the CD8β255R1 or IgG control antibody was administered, animals were challenged intravenously with 100 50% tissue culture infective doses (TCID50) (0.71 ng p27) SIVmac239. Animals were evaluated for an additional 14 weeks before euthanasia, with the exception of animal cy0690, which was sacrificed per veterinarian recommendation at 6 weeks after CD8β depletion due to non-SIV-related complications.
Flow cytometry was used to monitor CD8αβ+ T cells in PBMC and peripheral LNs of all 11 MCMs. Percent reductions in the absolute count of peripheral CD8αβ+ T cells in animals that received CD8β255R1 were most prominent at week 2 (median, 94%; range, 70% to 98%); CD8αβ+ T cells remained significantly depleted at the time of SIVmac239 challenge (median, 89%; range, 45% to 97%) through the time of necropsy (Fig. 2a, blue). In control IgG-treated animals, the absolute counts of peripheral CD8αβ+ T cells fluctuated around baseline levels for 16 weeks (Fig. 2a, black). Shortly after CD8β255R1 infusion, the reduction in peripheral CD8αβ+ T cell count was statistically significant compared to changes in the IgG control animals (data not shown). Remarkably, the absolute count of peripheral CD8αβ+ T cells at 16 weeks remained depleted by an average of 64% compared to baseline (median, 67%; range, 28% to 82%) in animals that received CD8β255R1. Persistent depletion of CD8αβ+ T cells after administration of CD8β255R1 is in stark contrast to the reemergence of CD8α+ lymphocytes within 2 to 4 weeks after administration of the anti-CD8α mAb cM-T807 that has been reported in other studies (8, 10, 13–20, 33). Our results in both SIV-naive and LASIV+ animals are, however, consistent with another report that observed sustained CD8αβ+ T cell depletion in two SIV+ rhesus macaques that were treated with CD8β255R1 and followed for approximately 18 weeks (33).
FIG 2.
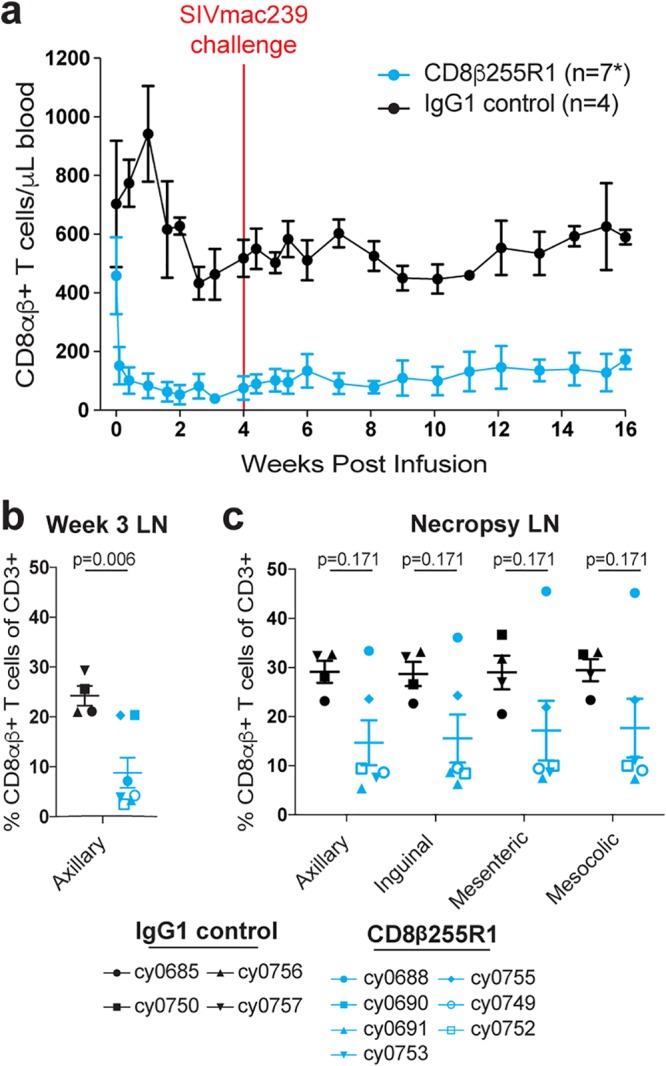
Depletion of CD8αβ+ T cells during chronic LASIV infection. (a) The absolute count of CD8αβ+ T cells in peripheral blood was assessed for 16 weeks following infusion with CD8β255R1 (blue) or control IgG (black). (b and c) Percent CD8αβ+ T cells from lymph node biopsy specimens at 3 weeks postinfusion (b) and at necropsy (week 17 to 18) (c). Data are represented as mean and standard error of the mean (SEM). A significant change (P < 0.05) from baseline in the absolute count of CD8αβ+ T cells (a) was observed at each time point following mAb infusion for animals that received CD8β255R1 but not animals that received control IgG. *, data shown include four IgG-treated animals and seven CD8β255R1-treated animals through week 6, at which point six CD8β255R1-treated animals are shown for the remainder of the study due to one animal (cy0690) requiring early euthanasia.
The extent to which CD8β255R1 depletes LN CD8αβ+ T cells in macaques during either chronic LASIV or pathogenic SIV infection has not yet been defined (33, 34). While technical limitations prevented the longitudinal analysis of cell populations in lymph nodes of individual LASIV+ animals, we compared the percentage of CD8αβ+ T cells in the lymph nodes of animals treated with CD8β255R1 to that in animals treated with the control IgG antibody (Fig. 2b). At week 3 postinfusion, there were 3-fold-fewer (P < 0.006) CD8αβ+ T cells as a proportion of CD3+ T cells in biopsy specimens of axillary lymph nodes isolated from animals infused with CD8β255R1 (median, 4%; range, 1.9% to 20%) than in those from animals receiving control IgG (median, 27%; range, 21% to 30%). When lymph nodes were assessed at 17 to 18 weeks after mAb infusion following necropsy, CD8αβ+ T cell frequencies remained substantially lower in the majority of CD8β255R1-treated animals than in control IgG-treated animals (Fig. 2c).
Plasma viral load increases transiently following CD8β255R1 infusion.
To determine the impact of CD8β depletion on control of chronic LASIV viremia, we measured levels of circulating virus after mAb infusion. The plasma viral load peaked at 3 to 11 days after infusion with CD8β255R1 and was significantly increased at day 7 (P < 0.001), day 10 (P < 0.001), and day 14 (P = 0.006) compared to baseline (Fig. 3a). Surprisingly, at weeks 3 and 4 after infusion with CD8β255R1, viral loads returned to levels that were not significantly different from baseline, despite diminished CD8αβ+ T cells in peripheral blood and LNs (Fig. 2). As expected, minimal changes to peripheral CD8αβ+ T cells during the first 4 weeks after administration of the control IgG antibody (Fig. 2) corresponded to viral loads that remained essentially unchanged (Fig. 3b).
FIG 3.
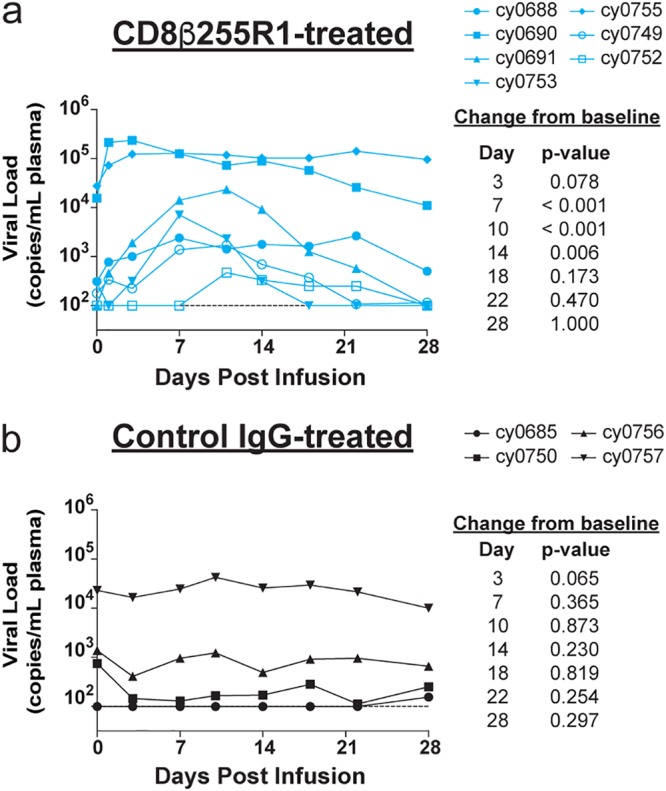
Regained control of viral recrudescence following depletion of peripheral CD8αβ+ T cells. The plasma viral load was measured for 4 weeks after mAb infusion. (a) The viral load initially increased following infusion with CD8β255R1 but returned to predepletion levels by day 18. (b) The viral load did not change significantly in animals that received control IgG. The limit of detection of the plasma viral load assay (100 vRNA copies/ml) is shown with a horizontal dashed line.
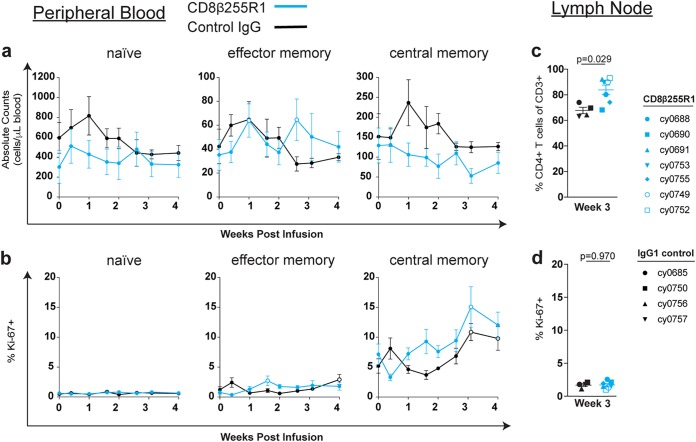
Minimal changes to CD4+ T cells following infusion with CD8β255R1.
One previously reported consequence of depletion of CD8α+ cells was a corresponding increase in the frequency or activation state of CD4+ T cell populations (12, 15–17). We monitored the frequency and proliferation of peripheral CD4+ naive (TN), CD4+ effector memory (TEM), and CD4+ central memory (TCM) T cells following CD8β255R1 infusion. Changes from baseline for both the absolute count (Fig. 4a) and percent Ki-67+ cells (Fig. 4b) were compared to baseline within and between each antibody-treated group. Although peripheral CD4+ TEM cells were significantly increased compared to baseline at two time points in the CD8β255R1-treated group (Fig. 4a, middle), these changes did not differ significantly compared to animals that received control IgG (data not shown). Similarly, CD4+ T cell subpopulations exhibited minimal changes in proliferation compared to baseline within and across groups (Fig. 4b and data not shown). We also compared changes in the frequency of total (Fig. 4c) and percent Ki-67+ (Fig. 4d) CD4+ T cells in the lymph nodes isolated at 3 weeks from animals in both groups. We found that the frequency of CD4+ T cells was higher (P = 0.029) (Fig. 4c) in the CD8β255R1-treated animals than in the control IgG-treated animals, but that may be attributed to a lower frequency of lymph node CD8αβ+ T cells detected at this time (Fig. 2b). The percent Ki-67+ of total CD4+ T cells was similar between groups at week 3 after mAb infusion (Fig. 4d). From these analyses, animals that received an infusion of CD8β255R1 exhibited minimal changes in the number and proliferative capacity of CD4+ T cells, even while plasma viremia was fluctuating.
FIG 4.
Minimal changes in the frequency and proliferation of CD4+ T cell populations following administration of CD8β255R1. The absolute cell count and percent Ki-67+ of CD4+ T cells from peripheral blood and lymph nodes were measured by multicolor flow cytometry for 4 weeks following mAb infusion. Within peripheral blood, individual time points represent mean ± SEM for animals that received CD8β255R1 (blue) or control IgG (black), and open circles represent a significant (P < 0.05) change from baseline. (a and b) Absolute cell count (a) and percent Ki-67+ (b) of naive (left panels), effector memory (middle panels), and central memory (right panels) CD4+ T cells in peripheral blood. Within the lymph nodes, individual animals are represented by a unique symbol. (c and d) Percent CD4+ T cells of CD3+ T cells (c) and percent Ki-67+ of CD4+ T cells (d) from axillary lymph nodes biopsied at week 3 after mAb infusion. In peripheral blood, paired difference comparisons of changes from baseline in animals that received CD8β255R1 or control IgG were statistically significant (P < 0.05) at day 10 for the percentage of effector memory CD4+ T cells expressing Ki-67.
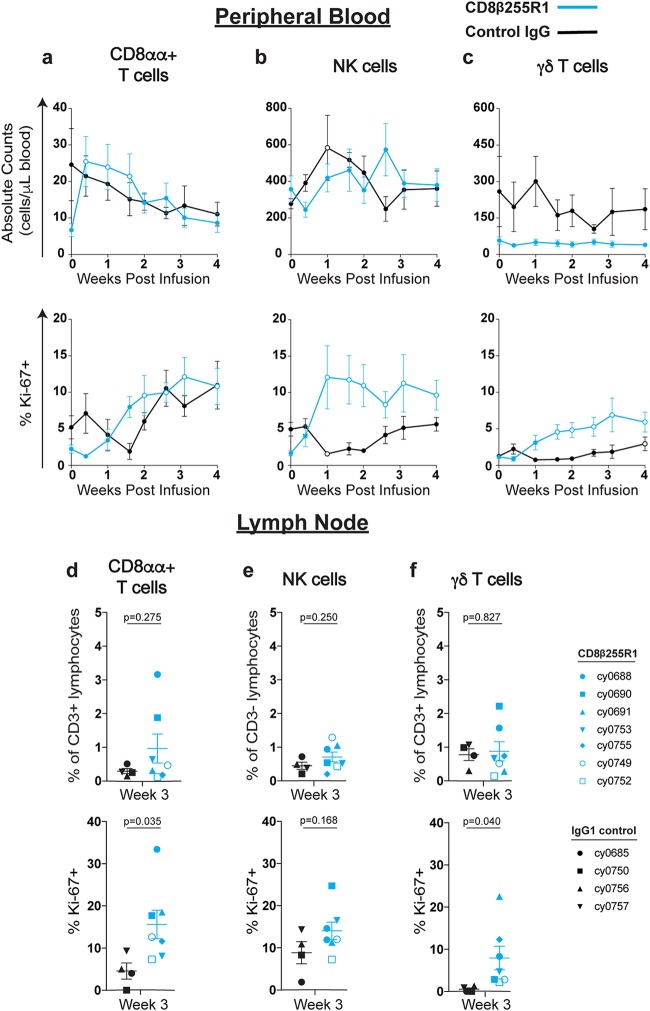
Proliferation of CD8αα+ T cells, NK cells, and γδ T cells after infusion with CD8β255R1.
Another consequence of using a CD8α-specific mAb is the simultaneous depletion of additional cell populations that express CD8α, such as CD8αα+ T cells, NK cells, and γδ T cells (10, 12–14, 17, 19, 33). Following infusion with CD8β255R1, we evaluated the absolute count and percent Ki-67+ of these three cell populations in the periphery. In animals that received CD8β255R1, there was a rapid increase in the absolute count of CD8αα+ T cells (median, 3.6-fold; range, 3.2-fold to 3.8-fold) that differed initially from that for control IgG-treated animals (Fig. 5a, top). This was followed by increases in the percentage of CD8αα+ T cells expressing Ki-67 (median, 4.5-fold; range, 4.2-fold to 5.3-fold) (Fig. 5a, bottom). Control IgG-treated animals did not exhibit significant differences from baseline in the absolute count of CD8αα+ T cells or percentage that express Ki-67. Few differences were detected in the absolute count of NK cells (Fig. 5b, top) or γδ T cells (Fig. 5c, top) in each group or between groups for 4 weeks after infusion. In contrast, we detected significant increases from baseline in the frequency of NK cells and γδ T cells expressing Ki-67 in the group treated with CD8β255R1 that peaked at approximately 7-fold (median, 4.3-fold; range, 1.8-fold to 17.7-fold) and 10-fold (median, 9.9-fold; range, 0.1-fold to 18.8-fold), respectively (Fig. 5b and c, bottom). The observed proliferation of NK cells in the CD8β255R1-treated animals was significantly greater than that in control IgG-treated animals at days 7, 10, and 14.
FIG 5.
Proliferation of CD8αα+, NK cells, and γδ T cells in peripheral blood following infusion with CD8β255R1. The absolute cell count and percent Ki-67+ of CD8α+ lymphocytes from peripheral blood and lymph nodes were measured by multicolor flow cytometry for 4 weeks after infusion with CD8β255R1 (blue) or control IgG (black). Within peripheral blood, individual time points represent mean ± SEM for animals, and open circles represent a significant change from baseline. Within the lymph nodes, individual animals are represented by a unique symbol. (a to c) Absolute count (top panels) and percent Ki-67+ (bottom panels) of CD8αα+ T cells (a), NK cells (b), and γδ T cells (c) in peripheral blood. (d) Percent CD8αα+ T cells of CD3+ lymphocytes (top) and percent Ki-67+ CD8αα+ T cells (bottom) in lymph nodes. (e) Percent NK cells of CD3− lymphocytes (top) and percent Ki-67+ NK cells (bottom) in lymph nodes. (f) Percent γδ T cells of CD3+ lymphocytes (top) and percent Ki-67+ γδ T cells (bottom) in lymph nodes. In peripheral blood, paired difference comparisons of changes from baseline in animals that received CD8β255R1 or control IgG were statistically significant (P < 0.05) at day 3, day 7, and day 18 for the absolute count of CD8αα+ T cells, at day 18 for the absolute count of γδ T cells, at day 7, day 10, and day 14 for the percentage of NK cells expressing Ki-67, and at day 22 for the percentage of γδ T cells expressing Ki-67.
We also evaluated changes to the frequency and proliferation of CD8αα+ T cells (Fig. 5d), NK cells (Fig. 5e), and γδ T cells (Fig. 5f) in lymph nodes of animals treated with CD8β255R1 or control IgG at week 3 postinfusion. Similar to cells in the periphery, the frequency of NK cells (Fig. 5e, top) and γδ T cells (Fig. 5f, top) in lymph nodes of animals treated with CD8β255R1 was comparable to that in control IgG-treated animals. At week 3 after CD8β255R1 infusion, the frequency of Ki-67+ CD8αα+ T cells (Fig. 5d, bottom) and γδ T cells (Fig. 5f, bottom) was significantly higher than that observed in control IgG-treated animals. Taken together, our data indicate that CD8αα+ T cells, NK cells, and γδ T cells exhibited rapid proliferation in either the PBMC or lymph nodes following CD8β255R1 infusion.
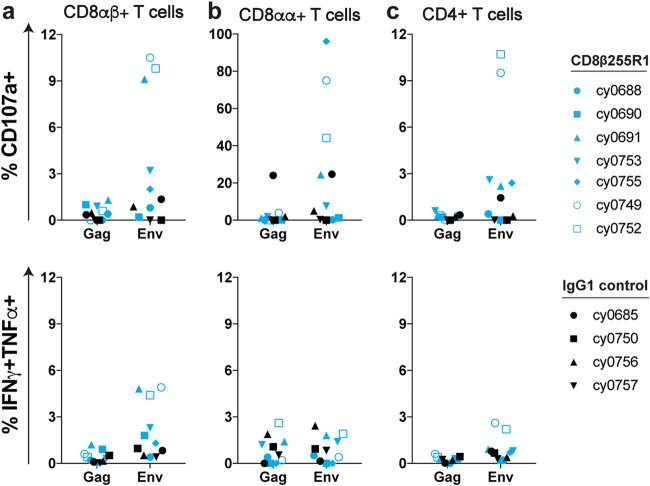
Limited SIV-specific T cell responses in LNs following CD8β255R1 infusion.
To determine the impact of CD8β255R1 infusion on SIV-specific T cells within LNs, we performed intracellular cytokine staining (ICS) with cells isolated from LN biopsy specimens taken 3 weeks after antibody infusion. Multiple pools containing overlapping peptides spanning the entire Gag protein and Env protein were used to stimulate cells, followed by staining with CD107a (Fig. 6, top) and IFN-γ/TNF-α (Fig. 6, bottom) to assess degranulation and proinflammatory cytokine secretion, respectively. None of the animals that received the control IgG antibody exhibited remarkable SIV-specific responses. This was not surprising, as these animals did not exhibit any marked changes in viremia. Animals that received CD8β255R1 exhibited a higher proportion of residual virus-specific CD8αβ+ T cells than animals that received the control IgG (Fig. 6). Of note, three animals (cy0691, cy0749, and cy0752) that received CD8β255R1 were each infected with a LASIV that contained known immunogenic epitopes. Thus, ongoing replication of a virus with intact M3 epitopes may have provided stronger antigenic stimulation to T cells and therefore driven proliferation in these three animals. The other four animals that received CD8β255R1 were homozygous for the M3 MHC haplotype and originally infected with a LASIV that contained point mutations in eight epitopes restricted by MHC molecules expressed by the M3 MHC haplotype (47). Even though it is likely that CD8β255R1 infusion alone cannot completely eliminate virus-specific CD8αβ+ T cells from lymph nodes, the combination of infecting animals with a minimally immunogenic LASIV and infusion of CD8β255R1 was able to limit the frequency of Gag- and Env-specific CD8β+ T cells at this important tissue site.
FIG 6.
Env-specific antiviral responses in lymph nodes following infusion with CD8β255R1. At 3 weeks postinfusion, we performed intracellular cytokine staining to assess Gag-specific and Env-specific responses from lymph node biopsy specimens for CD8αβ+ T cells (a), CD8αα+ T cells (b), and CD4+ T cells (c). We assessed degranulation (CD107a, top panels) and proinflammatory cytokine secretion (IFN-γ and TNF-α, bottom panels) for seven animals infused with CD8b255R1 (blue) and four animals infused with IgG1 control (black). Both single and double positive responses were included as percent IFN-γ+ TNF-α+. Symbols represent individual animals.
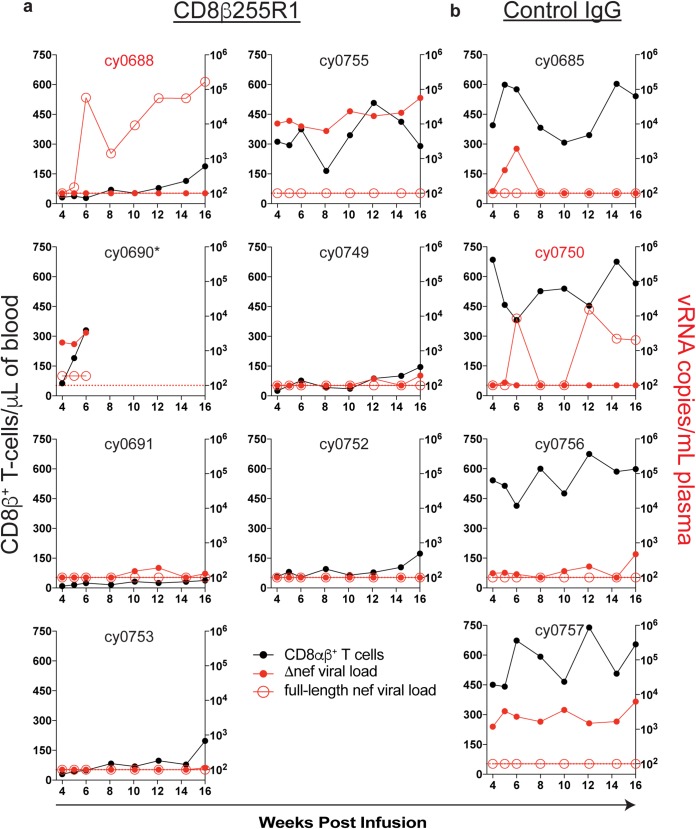
Protection from SIVmac239 challenge during depletion of CD8αβ+ T cells.
To determine whether CD8β255R1 infusion increased susceptibility to infection with pathogenic SIV, we performed a high-dose intravenous challenge in all 11 animals at 4 weeks after antibody infusion with SIVmac239 (49, 50). Previously, we found that 4/7 MCMs chronically infected with a minimally immunogenic LASIV were protected from intravenous challenge with the same dose of SIVmac239 (41). The 11 animals used in the current study were also chronically infected with a nef-deleted SIV, so discriminating reverse transcription-quantitative PCR (qRT-PCR) assays were used to differentiate previous Δnef circulating virus from full-length nef that was present in the challenge strain. Surprisingly, there was no replicating SIVmac239 in the plasma of six of the seven MCMs infused with CD8β255R1 (Fig. 7a), despite depletion of peripheral CD8β+ lymphocytes at the time of challenge (median, 87%; range, 45% to 95%) (Fig. 2a). Similarly, there was no replicating SIVmac239 detected in the plasma of three out of four MCMs that received the control IgG antibody (Fig. 7b), despite having CD8αβ+ T cell levels similar to those predepletion (Fig. 2a). It is possible that residual virus-specific CD8αβ+ T cells in the lymph nodes (Fig. 6) were sufficient to protect animals from challenge, as others have found that virus-specific CD8+ T cells in lymph nodes are an immune correlate of protection by LASIV (45). However, with so few animals becoming infected with SIVmac239, we could not determine if this was the main mechanism of protection.
FIG 7.
Protection from intravenous SIVmac239 challenge following infusion with CD8β255R1. At 4 weeks after infusion with CD8β255R1 or control IgG, animals were challenged intravenously with SIVmac239. Viral load assays were performed for full-length nef (red, open symbols) and Δnef (red, closed symbols) at the indicated time points for each animal and graphed in the context of absolute CD8αβ+ T cell count (black). The limit of detection for the assay (100 vRNA copies/ml) is represented by a horizontal red dotted line. Animals with identifications highlighted in red became infected following SIVmac239 challenge. *, cy0690 required necropsy at week 6 following CD8β255R1 infusion.
Neutralizing antibody titers to SIVmac239 do not predict challenge outcome.
We wanted to determine whether depletion of CD8β+ lymphocytes had a direct impact on neutralizing antibody titers, which may have contributed to protection from pathogenic SIVmac239 infection. To accomplish this, we performed in vitro antibody neutralization assays with plasma collected immediately before mAb infusion, as well as immediately before SIVmac239 challenge. We tested neutralization potency against SIV strains that are highly neutralization sensitive (tier 1, SIVmac251.30 and SIVsmE660.CP3C), moderately neutralization resistant (tier 2, SIVmac251.H9), and highly neutralization resistant (tier 3, SIVmac239) (51). In animals that received control IgG, neutralizing antibody titers prior to infusion (Fig. 8a, left) were similar to those detected prior to SIVmac239 challenge (Fig. 8a, right). Similarly, we did not detect substantial changes to neutralizing antibody titers prior to CD8β255R1 infusion (Fig. 8b, left) compared to the titers right before SIVmac239 challenge (Fig. 8b, right). Accordingly, we could not conclude that protection from challenge following infusion with CD8β255R1 was a result of increased in vitro SIVmac239 neutralization antibody titers.
FIG 8.
Plasma neutralization potency in vitro is not predictive of protection from challenge in vivo. Plasma samples collected immediately before mAb infusion and 4 weeks later immediately before intravenous SIVmac239 challenge were used to test antibody neutralization potency against select tier 1 (SIVmac239.H9 and SIVsmE660.CP3C), tier 2 (SIVmac251.30), and tier 3 (SIVmac239) viruses. (a) Neutralizing antibody titers were measured prior to infusion with control IgG (left panel) and prior to SIVmac239 challenge (right panel). (b) Neutralizing antibody titers were measured prior to infusion with CD8β255R1 (left panel and prior to SIVmac239 challenge (right panel). Titers are shown as 50% inhibitory dilution (ID50). Animals that became infected following SIVmac239 challenge are highlighted in red.
DISCUSSION
Studies of SIV-infected macaques using a CD8α-specific mAb to deplete CD8+ T cells in vivo have frequently been interpreted as evidence for a direct antiviral role of CD8+ T cells in HIV infection (8, 18). However, these results are complicated by the simultaneous depletion of CD8α-expressing NK cells and γδ T cells (10, 13, 21, 24). Here we showed that a single intravenous infusion of the CD8β-specific mAb CD8β255R1 selectively depleted CD8αβ+ T cells in peripheral blood, though to a lesser extent in lymph nodes, from SIV-naive and LASIV-infected cynomolgus macaques. Following depletion, we observed a transient rise in plasma viremia that was followed by reestablishment of viral control, even when CD8αβ+ T cells remained depleted in the peripheral blood. During the time of prolonged peripheral CD8αβ+ T cell depletion, we found that CD8αα+ T cells, NK cells, and γδ T cells exhibited markers of proliferation regardless of changes to their total frequencies in blood. These observations demonstrated that the CD8β255R1 antibody specifically depleted CD8αβ+ T cells. CD8β255R1 could not fully deplete CD8αβ+ T cells from tissue sites, as lymph nodes contained residual CD8αβ+ T cells. It is also possible that CD8αβ+ T cells from other tissue sites important for SIV replication that were not sampled in this study, such as the intestinal tract, were also incompletely depleted (52). Previously, administration of the anti-CD8α mAb cM-T807 did not deplete CD8+ cells in the jejunum of SIV-infected rhesus macaques, leaving open the possibility that CD8αβ+ T cells in gut tissues of the CD8β255R1-treated animals may have also played a critical role in the current study (53). While our results specifically demonstrated that administration of the CD8β255R1 depleting antibody led to a rise in LASIV plasma viremia, we could not conclusively determine whether the residual CD8αβ+ T cells in tissues or the persistence of CD8αα+ T cells, NK cells, and/or γδ T cells was responsible for reestablishment of LASIV control and subsequent protection from pathogenic SIVmac239 challenge. Because animals also had very low neutralizing antibody titers against SIVmac239 throughout the study period, it is unlikely that protection against challenge was mediated by neutralization of the challenge virus.
We found that animals treated with the CD8β255R1 antibody exhibited demonstrably fewer total CD8αβ+ T cells in the periphery and the lymph nodes than control IgG-treated animals. To determine if we depleted virus-specific CD8αβ+ T cells, we initially performed gamma interferon enzyme-linked immunospot (IFN-γ ELISPOT) assays with PBMC prior to and 3 weeks after infusion with CD8β255R1, but we were unable to confirm if the SIV-specific responses that were detected 3 weeks after infusion were attributable to CD8+ T cells or CD4+ T cells (data not shown). While numerous pieces of evidence point toward a critical role of CD8+ T cells in long-term viral control (8–10, 13, 14, 17, 19), one report suggests that virus-specific CD8+ T cells in peripheral blood may not be absolutely required for control of virus replication during chronic SIV infection, even if they are needed to initially suppress viremia (54). Tetrameric reagents are an alternative method to detect virus-specific T cells, but these reagents were not available for two reasons: (i) Many of the epitopes that would elicit CD8αβ+ T cells were rendered nonimmunogenic in the mutant LASIV used to infect several of these animals (47), and (ii) tetrameric reagents specific for the CD8 and CD4 T cells that developed in the animals infected with the mutant LASIV have not been produced. We did perform intracellular cytokine staining experiments with lymph nodes that were collected 3 weeks after antibody infusion. Interestingly, the three animals with the largest frequency of Env-specific CD8αβ+ T cells were the three animals that were originally infected with strains of LASIV containing immunogenic epitopes. Two animals were infected with wild-type SIVmac239Δnef, and the third animal was infected with the mutant LASIV but expressed some non-M3 MHC alleles. As a result, these three animals likely had the largest pool of virus-specific memory CD8αβ+ T cells that could emerge when viremia increased. Together, our data imply that the most effective way to eliminate virus-specific CD8αβ+ T cells with currently available technology is to infect animals with a virus whose T cell epitopes are rendered nonimmunogenic combined with a CD8β-specific depleting antibody. Even with this “double-knockout approach,” more sophisticated reagents are needed to improve CD8αβ+ T cell depletion in tissues to determine if they are required to reestablish viral control.
Even when we minimized the CD8αβ+ T cell populations with our interventions, we were surprised to find that six out of seven animals treated with CD8β255R1 were resistant to infection with pathogenic SIVmac239. This level of protection was on par with that for the animals treated with control IgG. Unfortunately, even when we examined non-CD8β+ immune cell populations and neutralizing antibody titers, there were no obvious immune correlates of protection. Protection afforded to LASIV has previously been associated with the presence of effector-differentiated T cell responses within the lymph node (44, 45), so it is entirely possible that the residual virus-specific T cells in the lymph nodes or tissues were sufficient to protect animals from SIVmac239 challenge. Continuing to improve the methods to deplete virus-specific immune cells in the LASIV model will be needed to directly demonstrate the immune correlates of protection mediated by LASIV.
One unique attribute of our study was the inclusion of animals treated with an isotype-matched IgG control antibody, which was absent in recent reports of CD8β255R1 (33, 34). By comparing paired differences of cell populations between animals treated with CD8β255R1 and control IgG, we accounted for potential artifacts that may result from infusion of a nonspecific IgG antibody. For example, increases over baseline in the percentage of central memory CD4+ T cells expressing Ki-67+ in both groups suggest that IgG infusion alone induces proliferation of these cells. Additionally, similar to a previous study using CD8β255R1 (33), we observed increased numbers of circulating NK cells in animals receiving CD8β255R1, though similar increases were also detected in the control IgG animals. These observations question whether changes to central memory CD4+ T cell populations were a direct consequence of administration of the specific CD8β255R1 monoclonal antibody or whether the administration of a control antibody was sufficient to induce this expansion. Thus, including an IgG control group in these types of antibody infusion studies is critical for identifying nonspecific depletion effects.
In this study, we provide a comprehensive comparison of the immunological impact that follows infusion with the anti-CD8β mAb CD8β255R1 compared to an isotype-matched control IgG. The persistent depletion of peripheral CD8αβ+ T cells following administration of CD8β255R1 is in stark contrast to the transient depletion of CD8αβ+ T cells following infusion with a CD8α-specific mAb. Unlike studies with the CD8α-depleting antibody that observed regained control of virus replication coincident with rebound of CD8+ T cells, we observed regained control of viral replication, even when peripheral CD8β+ T cells remained depleted. Moreover, protection from intravenous challenge with pathogenic SIVmac239 was achieved when the CD8αβ+ T cell magnitude was reduced by administration of CD8β255R1 and, in some cases, in combination with a mutant LASIV that failed to elicit many virus-specific T cells (47). Nonetheless, the data we provide demonstrate that the CD8β255R1 antibody can be used to specifically deplete CD8αβ+ T cells, while leaving other CD8α+ immune cell populations intact. This may be valuable in future studies evaluating the importance of CD8αβ+ T cells in diverse disease models.
MATERIALS AND METHODS
Animal care and use.
Two Vietnamese-origin cynomolgus macaques were purchased from Covance Inc. and included in the depletion study of naive cynomolgus macaques. Both animals were housed and cared for at the National Institutes of Health Animal Center (NIHAC) (Poolesville, MD) according to protocols approved by the Vaccine Research Center Animal Care and Use Committee. Both animals received a single 50-mg/kg intravenous infusion of the anti-CD8β monoclonal antibody (mAb) CD8β255R1.
Eleven Mauritian cynomolgus macaques (MCMs) were purchased from Bioculture Ltd. and included in the depletion/challenge study of cynomolgus macaques chronically infected with a LASIV. These MCMs were housed and cared for at the Wisconsin National Primate Research Center (WNPRC) according to protocols approved by the University of Wisconsin Graduate School Animal Care and Use Committee. All 11 animals were previously infected intravenously with 10 ng p27 of wild-type SIVmac239Δnef (animals cy0749 and cy0752) or a variant of SIVmac239Δnef containing 10 nonsynonymous mutations (cy0685, cy0688, cy0690, cy0691, cy0750, cy0753, cy0755, cy0756, and cy0757) for 34 to 73 weeks prior to the start of this study (47). All animals were homozygous for the M3 MHC haplotype, with the exception of cy0691, which was heterozygous for the M2 and M3 MHC haplotypes. Seven MCMs received a single 50-mg/kg intravenous infusion of the anti-CD8β mAb CD8β255R1, and four MCMs received a single 50-mg/kg intravenous infusion of the DSPR1 rhesus recombinant IgG control mAb, both of which were provided by the NIH Nonhuman Primate Reagent Resource (R24 OD010976 and U24 AI126683). All 11 MCMs were challenged intravenously with 100 TCID50 of SIVmac239 (0.71 ng p27) at 4 weeks following mAb infusion.
Antibodies.
CD3-AF700 (clone SP34-2; BD Biosciences), CD4-BV711 (clone OKT4; BioLegend), CD8α-phycoerythrin (PE) (clone DK25; Dako), CD8β-ECD (clone 2ST8-5H7; Beckman Coulter), TCRγδ-fluorescein isothiocyanate (FITC) (clone 5A6.E9; Invitrogen), CD95-PE/Cy5 (clone DX2; BD Biosciences), CD28-BV510 (clone CD28.2; BD Biosciences), CCR7-Pacific Blue (clone G043H7; BioLegend), NKG2a-PE/Cy7 (clone Z199; Beckman Coulter), CD16-BV650 (clone 3G8; BD Biosciences), CD16-BV786 (clone 3G8; BD Biosciences), and Ki-67-AF647 (clone B56; BD Biosciences) antibodies were used. Live/Dead fixable near-infrared dead cell stain (Invitrogen) was used to assess cell viability. All samples were run on an LSR II instrument (BD Biosciences) and analyzed using FlowJo, version 9.9.6 (BD Biosciences). The presence of the CD8β255R1 mAb does not cross-block binding with the anti-CD8β clone 2ST8-5H7 (33).
Peptides.
The NIH AIDS Research and Reference Reagent Program (Germantown, MD) provided 15-mer peptides overlapping by 11 amino acid positions that spanned the entire SIVmac239 proteome. Gag and Env peptides were combined into 2 and 3 pools, respectively, and used at a final pooled concentration of 5 μM during stimulation.
Lymphocyte isolation and phenotype staining.
In the depletion study of naive cynomolgus macaques, phenotype staining of peripheral blood mononuclear cells (PBMC) was performed in triplicate on whole blood and in duplicate on lymph node samples. Cells were incubated with a surface antibody mix for 25 min in the dark at room temperature. In the depletion/challenge study, PBMC were isolated from EDTA-anticoagulated blood by Ficoll-based density centrifugation as previously described (41). Cells were resuspended in RPMI 1640 (HyClone) supplemented with 10% fetal calf serum (FCS), 1% antibiotic-antimycotic (HyClone), and 1% l-glutamine (HyClone) (R10 medium). Approximately 0.5 to 1.0 × 106 PBMC were placed in cluster tubes (Fisher Scientific) and washed with 1× phosphate-buffered saline (PBS) prior to staining. Surface antibodies were added, and cells were incubated for 30 min in the dark at room temperature. Cells were then washed twice with 1× PBS supplemented with 10% FCS (10% FCS/PBS) and fixed with 2% paraformaldehyde (PFA) for a minimum of 30 min. Fixed cells were then washed once with 1× PBS and then permeabilized with medium B (Invitrogen, Carlsbad CA) and allowed to incubate with intracellular antibodies for 30 min in the dark at room temperature. Cells were then washed twice with 10% FCS/PBS and resuspended in 2% PFA prior to data collection. Following exclusion of doublets and dead cells, lymphocyte populations were defined as follows: CD8αβ+ T cells, CD3+ CD4− TCRγδ− CD8α+ CD8β+; CD8αα+ T cells, CD3+ CD4− TCRγδ− CD8α+ CD8β−; γδ T cells, CD3+ CD4− CD8β− CD8α+ TCRγδ+; NK cells, CD3− CD4− CD8α+ NKG2a+ CD16+; CD4 T cells (naive), CD3+ CD4+ CD8α− CD95− CD28+ CCR7+; CD4 T cells (effector memory), CD3+ CD4+ CD8α− CD95+ CD28− CCR7−; and CD4 T cells (central memory), CD3+ CD4+ CD8α− CD95+ CD28+ CCR7+.
ICS.
Flow cytometry was used to measure intracellular cytokine expression as previously described (55). Cryopreserved cells isolated from LNs were thawed, washed twice in warm R10 medium, and allowed to rest at 37°C overnight prior to stimulation. Ki-67 was used to measure the proliferative capacity of lymphocytes freshly isolated from blood, and IFN-γ, tumor necrosis factor alpha (TNF-α), and CD107a were used to measure SIV-specific responses in cryopreserved lymphocytes isolated from LNs at week 3 after mAb infusion. LN cells were incubated with CD107a and stimulated with Gag or Env peptide pools for a total of 6 h, with brefeldin A (Sigma-Aldrich) and monensin (BioLegend) added 1 h after stimulation. PBMC and LN cells were incubated with Live/Dead fixable near-infrared dead cell stain for 20 min, incubated with surface markers for 30 min, and fixed with 2% paraformaldehyde (PFA) for at least 20 min. Medium B (Invitrogen) was used to permeabilize PBMC, and 0.1% saponin was used to permeabilize LN cells. PBMC and LN cells were stained for 30 min with Ki-67 or with IFN-γ and TNF-α, respectively. Data were collected on an LSR II instrument (BD Biosciences) using 2% PFA-fixed cells and then analyzed using FlowJo version 9.9.6 (BD Biosciences).
Plasma viral load analysis.
Plasma was isolated alongside PBMC and cryopreserved at −80°C prior to analysis. SIV gag viral loads were determined as previously described (41). Briefly, viral RNA (vRNA) was isolated from plasma, reverse transcribed, and amplified with the Superscript III platinum one-step quantitative reverse-transcription-PCR (RT-PCR) system (Invitrogen). The detection limit of the assay was 100 vRNA copy equivalents per ml of plasma (copies/ml). When the viral load was at or below the limit of detection, the detection limit value of 100 was reported. Full-length nef and Δnef viral loads were determined as previously described (41). Briefly, highly specific real-time RT-PCR assays were used with primers that accurately differentiate viruses containing full-length nef from those that contain nef with a 182-bp deletion, using the methods described above. Serial dilutions of in vitro transcripts for both full-length nef and nef with a 182-bp deletion were used as internal standards for each run. The same machines and software used for the gag viral load assay were used to detect and quantify the nef and Δnef viral loads. The limit of detection was identical to that for the SIV gag viral load assay.
Virus neutralization assays.
SIV Env pseudoviruses were produced as previously described (51). Briefly, plasmid DNA encoding SIV gp160 was combined with a luciferase reporter plasmid containing the essential HIV structural genes to produce pseudoviruses expressing SIVmac251.H9, SIVmac251.30, SIVsmE660.CP3C, or SIVmac239 Env. Using TZM-bl target cells, virus neutralization was measured following incubation with SIV-Env pseudovirus and plasma collected from blood. The 50% inhibitory dilution (ID50) was defined as the plasma dilution that caused a 50% reduction in relative light units (RLU) compared to virus control wells following subtraction of background RLU. A nonlinear-regression 5-parameter Hill slope equation was used to calculate plasma ID50 values.
Statistical analyses.
Comparison of two groups in Fig. 2b and c, 4c and d, and 5d to f was conducted using Wilcoxon rank sum tests. Comparison of changes from baseline separately for each treatment group shown in Fig. 2a, 3, 4a and b, and 5a to c utilized repeated-measures analysis of variance (RM-ANOVA) to estimate the mean contrasts with animal as a random effect. Comparison between both groups in changes from pretreatment levels at multiple follow-up time points also utilized RM-ANOVA with treatment, time, and their interaction as fixed effects and animal as a random effect. The model assumptions of RM-ANOVA were examined and were not deemed to be grossly violated. Dunnett’s P value adjustment for multiple testing over multiple time points was used to keep a family-wise 0.05 type 1 error rate (56). All tests were conducted using a two-sided significance level of 0.05. All analyses were conducted using R for statistical computing version 3.3 (57).
ACKNOWLEDGMENTS
The research reported in this publication was supported in part by the Office of The Director, National Institutes of Health (NIH), under award number P51OD011106 to the Wisconsin National Primate Research Center (University of Wisconsin—Madison), as well as by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH. It was also supported in part by National Institutes of Health award number R01AI108415, as well as by the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM081061. Support was also received from the Clinical and Translational Science Award (CTSA) program through NIH National Center for Advancing Translational Sciences (NCATS) grant UL1TR002373 to Scott J. Hetzel. The research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01.
We thank staff at the Wisconsin National Primate Research Center (WNPRC) for veterinary care of the animals involved in the study. Special thanks go to Keith Reimann, Afam Okoye, Mauricio Martins, and Matthew Reynolds for helpful discussions.
The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol 68:4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol 68:6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker CM, Moody DJ, Stites DP, Levy JA. 1986. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science 234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 4.Gauduin MC, Glickman RL, Means R, Johnson RP. 1998. Inhibition of simian immunodeficiency virus (SIV) replication by CD8(+) T lymphocytes from macaques immunized with live attenuated SIV. J Virol 72:6315–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mutua G, Farah B, Langat R, Indangasi J, Ogola S, Onsembe B, Kopycinski JT, Hayes P, Borthwick NJ, Ashraf A, Dally L, Barin B, Tillander A, Gilmour J, De Bont J, Crook A, Hannaman D, Cox JH, Anzala O, Fast PE, Reilly M, Chinyenze K, Jaoko W, Hanke T, HIV-Core 004 Study Group. 2016. Broad HIV-1 inhibition in vitro by vaccine-elicited CD8(+) T cells in African adults. Mol Ther Methods Clin Dev 3:16061. doi: 10.1038/mtm.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, McNevin JP, Holte S, McElrath MJ, Mullins JI. 2011. Dynamics of viral evolution and CTL responses in HIV-1 infection. PLoS One 6:e15639. doi: 10.1371/journal.pone.0015639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMichael AJ, Rowland-Jones SL. 2001. Cellular immune responses to HIV. Nature 410:980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 9.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzner KJ, Jin X, Lee FV, Gettie A, Bauer DE, Di Mascio M, Perelson AS, Marx PA, Ho DD, Kostrikis LG, Connor RI. 2000. Effects of in vivo CD8(+) T cell depletion on virus replication in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Exp Med 191:1921–1931. doi: 10.1084/jem.191.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong JK, Strain MC, Porrata R, Reay E, Sankaran-Walters S, Ignacio CC, Russell T, Pillai SK, Looney DJ, Dandekar S. 2010. In vivo CD8+ T-cell suppression of SIV viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog 6:e1000748. doi: 10.1371/journal.ppat.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, Bosinger SE, Paiardini M, Chahroudi A, Vanderford TH, Estes JD, Lifson JD, Derdeyn CA, Silvestri G. 2016. CD8(+) lymphocytes are required for maintaining viral suppression in SIV-infected macaques treated with short-term antiretroviral therapy. Immunity 45:656–668. doi: 10.1016/j.immuni.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, León EJ, Soma T, Napoe G, Capuano SV, Wilson NA, Watkins DI. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J Virol 81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds MR, Weiler AM, Weisgrau KL, Piaskowski SM, Furlott JR, Weinfurter JT, Kaizu M, Soma T, León EJ, MacNair C, Leaman DP, Zwick MB, Gostick E, Musani SK, Price DA, Friedrich TC, Rakasz EG, Wilson NA, McDermott AB, Boyle R, Allison DB, Burton DR, Koff WC, Watkins DI. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J Exp Med 205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okoye A, Park H, Rohankhedkar M, Coyne-Johnson L, Lum R, Walker JM, Planer SL, Legasse AW, Sylwester AW, Piatak M, Lifson JD, Sodora DL, Villinger F, Axthelm MK, Schmitz JE, Picker LJ. 2009. Profound CD4+/CCR5+ T cell expansion is induced by CD8+ lymphocyte depletion but does not account for accelerated SIV pathogenesis. J Exp Med 206:1575–1588. doi: 10.1084/jem.20090356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller YM, Do DH, Boyer JD, Kader M, Mattapallil JJ, Lewis MG, Weiner DB, Katsikis PD. 2009. CD8+ cell depletion of SHIV89.6P-infected macaques induces CD4+ T cell proliferation that contributes to increased viral loads. J Immunol 183:5006–5012. doi: 10.4049/jimmunol.0900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, Apetrei C, Schmitz JE, Ribeiro RM, Perelson AS, Silvestri G. 2010. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog 6:e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin MA. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol 72:164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitz JE, Simon MA, Kuroda MJ, Lifton MA, Ollert MW, Vogel CW, Racz P, Tenner-Racz K, Scallon BJ, Dalesandro M, Ghrayeb J, Rieber EP, Sasseville VG, Reimann KA. 1999. A nonhuman primate model for the selective elimination of CD8+ lymphocytes using a mouse-human chimeric monoclonal antibody. Am J Pathol 154:1923–1932. doi: 10.1016/S0002-9440(10)65450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz JE, Johnson RP, McClure HM, Manson KH, Wyand MS, Kuroda MJ, Lifton MA, Khunkhun RS, McEvers KJ, Gillis J, Piatak M, Lifson JD, Grosschupff G, Racz P, Tenner-Racz K, Rieber EP, Kuus-Reichel K, Gelman RS, Letvin NL, Montefiori DC, Ruprecht RM, Desrosiers RC, Reimann KA. 2005. Effect of CD8+ lymphocyte depletion on virus containment after simian immunodeficiency virus SIVmac251 challenge of live attenuated SIVmac239delta3-vaccinated rhesus macaques. J Virol 79:8131–8141. doi: 10.1128/JVI.79.13.8131-8141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terry LA, DiSanto JP, Small TN, Flomenberg N. 1990. Differential expression and regulation of the human CD8 alpha and CD8 beta chains. Tissue Antigens 35:82–91. doi: 10.1111/j.1399-0039.1990.tb01761.x. [DOI] [PubMed] [Google Scholar]
- 22.Gan YH, Pauza CD, Malkovsky M. 2008. Gamma delta T cells in rhesus monkeys and their response to simian immunodeficiency virus (SIV) infection. Clin Exp Immunol 102:251–255. doi: 10.1111/j.1365-2249.1995.tb03773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster RL, Johnson RP. 2005. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology 115:206–214. doi: 10.1111/j.1365-2567.2005.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baume DM, Caligiuri MA, Manley TJ, Daley JF, Ritz J. 1990. Differential expression of CD8 alpha and CD8 beta associated with MHC-restricted and non-MHC-restricted cytolytic effector cells. Cell Immunol 131:352–365. doi: 10.1016/0008-8749(90)90260-X. [DOI] [PubMed] [Google Scholar]
- 25.Magalhaes I, Vudattu NK, Ahmed RK, Kühlmann-Berenzon S, Ngo Y, Sizemore DR, Wehlin L, Weichold F, Andersson J, Skeiky YA, Sadoff J, Gaines H, Thorstensson R, Spångberg M, Maeurer MJ. 2010. High content cellular immune profiling reveals differences between rhesus monkeys and men. Immunology 131:128–140. doi: 10.1111/j.1365-2567.2010.03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosub DA, Lehrman G, Milush JM, Zhou D, Chacko E, Leone A, Gordon S, Silvestri G, Else JG, Keiser P, Jain MK, Sodora DL. 2008. Gamma/delta T-cell functional responses differ after pathogenic human immunodeficiency virus and nonpathogenic simian immunodeficiency virus infections. J Virol 82:1155–1165. doi: 10.1128/JVI.01275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz JE, Forman MA, Lifton MA, Concepción O, Reimann KA, Crumpacker CS, Daley JF, Gelman RS, Letvin NL. 1998. Expression of the CD8alpha beta-heterodimer on CD8(+) T lymphocytes in peripheral blood lymphocytes of human immunodeficiency virus- and human immunodeficiency virus+ individuals. Blood 92:198–206. [PubMed] [Google Scholar]
- 28.Poccia F, Battistini L, Cipriani B, Mancino G, Martini F, Gougeon ML, Colizzi V. 1999. Phosphoantigen-reactive Vgamma9Vdelta2 T lymphocytes suppress in vitro human immunodeficiency virus type 1 replication by cell-released antiviral factors including CC chemokines. J Infect Dis 180:858–861. doi: 10.1086/314925. [DOI] [PubMed] [Google Scholar]
- 29.Poonia B, Pauza CD. 2012. Gamma delta T cells from HIV+ donors can be expanded in vitro by zoledronate/interleukin-2 to become cytotoxic effectors for antibody-dependent cellular cytotoxicity. Cytotherapy 14:173–181. doi: 10.3109/14653249.2011.623693. [DOI] [PubMed] [Google Scholar]
- 30.Gan YH, Malkovsky M. 1996. Mechanisms of simian gamma delta T cell cytotoxicity against tumor and immunodeficiency virus-infected cells. Immunol Lett 49:191–196. doi: 10.1016/0165-2478(96)02508-4. [DOI] [PubMed] [Google Scholar]
- 31.Marras F, Nicco E, Bozzano F, Di Biagio A, Dentone C, Pontali E, Boni S, Setti M, Orofino G, Mantia E, Bartolacci V, Bisio F, Riva A, Biassoni R, Moretta L, De Maria A. 2013. Natural killer cells in HIV controller patients express an activated effector phenotype and do not up-regulate NKp44 on IL-2 stimulation. Proc Natl Acad Sci U S A 110:11970–11975. doi: 10.1073/pnas.1302090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL, Varner V, Manickam C, Eslamizar L, Altfeld M, von Andrian UH, Barouch DH. 2015. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol 16:927–932. doi: 10.1038/ni.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martins MA, Tully DC, Shin YC, Gonzalez-Nieto L, Weisgrau KL, Bean DJ, Gadgil R, Gutman MJ, Domingues A, Maxwell HS, Magnani DM, Ricciardi M, Pedreño-Lopez N, Bailey V, Cruz MA, Lima NS, Bonaldo MC, Altman JD, Rakasz E, Capuano S, Reimann KA, Piatak M, Lifson JD, Desrosiers RC, Allen TM, Watkins DI. 2017. Rare control of SIVmac239 infection in a vaccinated rhesus macaque. AIDS Res Hum Retroviruses 33:843–858. doi: 10.1089/aid.2017.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura Y, Gautam R, Chun TW, Sadjadpour R, Foulds KE, Shingai M, Klein F, Gazumyan A, Golijanin J, Donaldson M, Donau OK, Plishka RJ, Buckler-White A, Seaman MS, Lifson JD, Koup RA, Fauci AS, Nussenzweig MC, Martin MA. 2017. Early antibody therapy can induce long-lasting immunity to SHIV. Nature 543:559–563. doi: 10.1038/nature21435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O’Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld R, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, Gulick RM, Shikuma CM, Haubrich R, Riddler S, Sax PE, Daar ES, Ribaudo HJ, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, Sette A. 2009. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol 182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol 80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris M, Burns CM, Becker EA, Braasch AT, Gostick E, Johnson RC, Broman KW, Price DA, Friedrich TC, O'Connor SL. 2013. Acute-phase CD8 T cell responses that select for escape variants are needed to control live attenuated simian immunodeficiency virus. J Virol 87:9353–9364. doi: 10.1128/JVI.00909-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 40.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, Desrosiers RC. 2006. HIV vaccine design: insights from live attenuated SIV vaccines. Nat Immunol 7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 41.Sutton MS, Burns CM, Weiler AM, Balgeman AJ, Braasch A, Lehrer-Brey G, Friedrich TC, O'Connor SL. 2016. Vaccination with live attenuated simian immunodeficiency virus (SIV) protects from mucosal, but not necessarily intravenous, challenge with a minimally heterologous SIV. J Virol 90:5541–5548. doi: 10.1128/JVI.00192-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alpert MD, Harvey JD, Lauer WA, Reeves RK, Piatak M, Carville A, Mansfield KG, Lifson JD, Li W, Desrosiers RC, Johnson RP, Evans DT. 2012. ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS Pathog 8:e1002890. doi: 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Zeng M, Duan L, Voss JE, Smith AJ, Pambuccian S, Shang L, Wietgrefe S, Southern PJ, Reilly CS, Skinner PJ, Zupancic ML, Carlis JV, Piatak M, Waterman D, Reeves RK, Masek-Hammerman K, Derdeyn CA, Alpert MD, Evans DT, Kohler H, Müller S, Robinson J, Lifson JD, Burton DR, Johnson RP, Haase AT. 2014. Live simian immunodeficiency virus vaccine correlate of protection: local antibody production and concentration on the path of virus entry. J Immunol 193:3113–3125. doi: 10.4049/jimmunol.1400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adnan S, Colantonio AD, Yu Y, Gillis J, Wong FE, Becker EA, Piatak M, Reeves RK, Lifson JD, O’Connor SL, Johnson RP. 2015. CD8 T cell response maturation defined by anentropic specificity and repertoire depth correlates with SIVΔnef-induced protection. PLoS Pathog 11:e1004633. doi: 10.1371/journal.ppat.1004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak M, Lifson JD, Sékaly RP, Picker LJ. 2012. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med 18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng M, Smith AJ, Shang L, Wietgrefe SW, Voss JE, Carlis JV, Li Q, Piatak M, Lifson JD, Johnson RP, Haase AT. 2016. Mucosal humoral immune response to SIVmac239Δnef vaccination and vaginal challenge. J Immunol 196:2809–2818. doi: 10.4049/jimmunol.1500156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton MS, Ellis-Connell A, Moriarty RV, Balgeman AJ, Gellerup D, Barry G, Weiler AM, Friedrich TC, O’Connor SL. 2018. Acute-phase CD4+ T cell responses targeting invariant viral regions are associated with control of live-attenuated simian immunodeficiency virus. J Virol 92:e00830-18. doi: 10.1128/JVI.00830-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutton MS, Ellis-Connell A, Balgeman AJ, Barry G, Weiler AM, Hetzel SJ, Zhou Y, Kilby A, Mason RD, Biris KK, Mascola JR, Sullivan NJ, Roederer M, Friedrich TC, O’Connor SL. 2019. Extensive CD8β depletion does not prevent control of viral replication or protection from challenge in macaques chronically infected with a live attenuated simian immunodeficiency virus. bioRxiv 10.1101/608554. [DOI] [PMC free article] [PubMed]
- 49.Mansfield K, Lang SM, Gauduin MC, Sanford HB, Lifson JD, Johnson RP, Desrosiers RC. 2008. Vaccine protection by live, attenuated simian immunodeficiency virus in the absence of high-titer antibody responses and high-frequency cellular immune responses measurable in the periphery. J Virol 82:4135–4148. doi: 10.1128/JVI.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans DT, Bricker JE, Sanford HB, Lang S, Carville A, Richardson BA, Piatak M, Lifson JD, Mansfield KG, Desrosiers RC. 2005. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J Virol 79:7707–7720. doi: 10.1128/JVI.79.12.7707-7720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mason RD, Welles HC, Adams C, Chakrabarti BK, Gorman J, Zhou T, Nguyen R, O’Dell S, Lusvarghi S, Bewley CA, Li H, Shaw GM, Sheng Z, Shapiro L, Wyatt R, Kwong PD, Mascola JR, Roederer M. 2016. Targeted isolation of antibodies directed against major sites of SIV Env vulnerability. PLoS Pathog 12:e1005537. doi: 10.1371/journal.ppat.1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 53.Veazey RS, Acierno PM, McEvers KJ, Baumeister SH, Foster GJ, Rett MD, Newberg MH, Kuroda MJ, Williams K, Kim EY, Wolinsky SM, Rieber EP, Piatak M, Lifson JD, Montefiori DC, Brown CR, Hirsch VM, Schmitz JE. 2008. Increased loss of CCR5+ CD45RA- CD4+ T cells in CD8+ lymphocyte-depleted Simian immunodeficiency virus-infected rhesus monkeys. J Virol 82:5618–5630. doi: 10.1128/JVI.02748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruel T, Hamimi C, Dereuddre-Bosquet N, Cosma A, Shin SY, Corneau A, Versmisse P, Karlsson I, Malleret B, Targat B, Barré-Sinoussi F, Le Grand R, Pancino G, Sáez-Cirión A, Vaslin B. 2015. Long-term control of simian immunodeficiency virus (SIV) in cynomolgus macaques not associated with efficient SIV-specific CD8+ T-cell responses. J Virol 89:3542–3556. doi: 10.1128/JVI.03723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donaldson MM, Kao S, Eslamizar L, Gee C, Koopman G, Lifton M, Schmitz JE, Sylwester AW, Wilson A, Hawkins N, Self SG, Roederer M, Foulds KE. 2012. Optimization and qualification of an 8-color intracellular cytokine staining assay for quantifying T cell responses in rhesus macaques for pre-clinical vaccine studies. J Immunol Methods 386:10–21. doi: 10.1016/j.jim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunnett CW. 1955. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121. doi: 10.2307/2281208. [DOI] [Google Scholar]
- 57.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]