Papillomaviruses (PVs) comprise a large family of nonenveloped DNA viruses that include HPV16, among other oncogenic types, the causative agents of cervical cancer. Delivery of the viral DNA into the host cell nucleus is necessary for establishment of infection. This was thought to occur via a subviral complex following uncoating of the larger viral capsid. In this study, we demonstrate that little disassembly of the PV capsid occurs prior to nuclear delivery. These surprising data reveal a previously unrecognized viral strategy to access the nuclear replication machinery. Understanding viral entry mechanisms not only increases our appreciation of basic cell biological pathways but also may lead to more effective antiviral interventions.
KEYWORDS: HPV, nuclear import/export, papillomavirus, uncoating, vesicle
ABSTRACT
Infectious human papillomavirus 16 (HPV16) L1/L2 pseudovirions were found to remain largely intact during vesicular transport to the nucleus. By electron microscopy, capsids with a diameter of 50 nm were clearly visible within small vesicles attached to mitotic chromosomes and to a lesser extent within interphase nuclei, implying nuclear disassembly. By confocal analysis, it was determined that nuclear entry of assembled L1 is dependent upon the presence of the minor capsid protein, L2, but independent of encapsidated DNA. We also demonstrate that L1 nuclear localization and mitotic chromosome association can occur in vivo in the murine cervicovaginal challenge model of HPV16 infection. These findings challenge the prevailing concepts of PV uncoating and disassembly. More generally, they document that a largely intact viral capsid can enter the nucleus within a transport vesicle, establishing a novel mechanism by which a virus accesses the nuclear cellular machinery.
IMPORTANCE Papillomaviruses (PVs) comprise a large family of nonenveloped DNA viruses that include HPV16, among other oncogenic types, the causative agents of cervical cancer. Delivery of the viral DNA into the host cell nucleus is necessary for establishment of infection. This was thought to occur via a subviral complex following uncoating of the larger viral capsid. In this study, we demonstrate that little disassembly of the PV capsid occurs prior to nuclear delivery. These surprising data reveal a previously unrecognized viral strategy to access the nuclear replication machinery. Understanding viral entry mechanisms not only increases our appreciation of basic cell biological pathways but also may lead to more effective antiviral interventions.
INTRODUCTION
Many viruses, including most DNA and some RNA viruses, depend upon nuclear cellular proteins for their replication. Transport of molecules across the nuclear envelope (NE) is tightly regulated, and these viruses have necessarily evolved a mechanism to deliver their genomes across this barrier. In cultured cells, the DNA genome of papillomaviruses (PVs), small (50 to 60 nm) naked icosahedron viruses, gains access to the nucleus during mitosis, when the NE is temporarily disassembled (1). This observation is consistent with the preferential in vivo infection of mitotically active basal keratinocytes within the stratified epithelium (reviewed in reference 2).
The PV capsid is composed of two proteins, the major capsid protein, L1, and the minor capsid protein, L2. L1 comprises the major structural component of the virion and is the basis for the current human papillomavirus (HPV) virus-like particle (VLP) vaccines, whereas L2 contributes little to the particle’s physical features but is essential for infection (3–5). In fact, studies focused on PV entry pathways have largely relegated L1’s role to the initial interaction of the capsid with the host cell and as an accompaniment through the early endosomes into multivesicular bodies (MVBs) or late endosomes, where it has long been considered that uncoating occurs, leading to a divergence in the trafficking pathways of L1 and L2 (reviewed in reference 6). The existing model proposes that during intracellular trafficking, L1 is partitioned away from a subviral particle that contains both L2 and the double-stranded circular DNA viral genome (vDNA) in a process that requires endosome acidification as well as host cyclophilin and γ-secretase activities. The majority of L1 is thought to remain in the late endosome/lysosome, whereas the L2/vDNA complex is transported to the trans-Golgi network (TGN) via the retromer and/or a Rab9- and Rab7b-dependent process, where it apparently resides until the initiation of mitosis (7, 8). However, recent studies challenging this paradigm have described the detection of L1 colocalizing with the L2/vDNA complex within the TGN, on the mitotic chromosomes, and transiently within the nucleus postmitosis (9, 10). It was suggested that residual L1 pentamers were anchored to the L2/vDNA complex via DNA-specific attachments. This subviral particle has been proposed to travel within vesicles that attach to the mitotic chromosomes and persist until entry into the nucleus (11). Sequestration within a vesicle might explain how PVs avoid triggering host innate immune nucleic acid sensors present in the cytoplasm (12). This model also addresses the lingering issue of how L2 could protect the viral DNA without adopting a higher-order structure. In the present study, we demonstrate that HPV16 pseudoviruses (PsVs) mediate their nuclear entry via chromatin-associated vesicles in an L2-dependent but vDNA-independent manner. Remarkably, we found that the virus was not substantially modified during trafficking, as 50-nm particles were evident within the vesicles. We propose that PVs utilize a novel nuclear entry pathway which entails vesicle-mediated nuclear delivery of a largely intact capsid followed by nuclear disassembly. This retention of assembled L1 during the later stages of PV trafficking, which stemmed from the analysis of newly generated as well as previously overlooked L1 monoclonal antibodies (mAbs) and transmission electron microscopy (TEM), challenges the prevailing model of PV infectious uncoating and disassembly.
RESULTS
A newly derived monoclonal antibody detects nuclear L1 during infection.
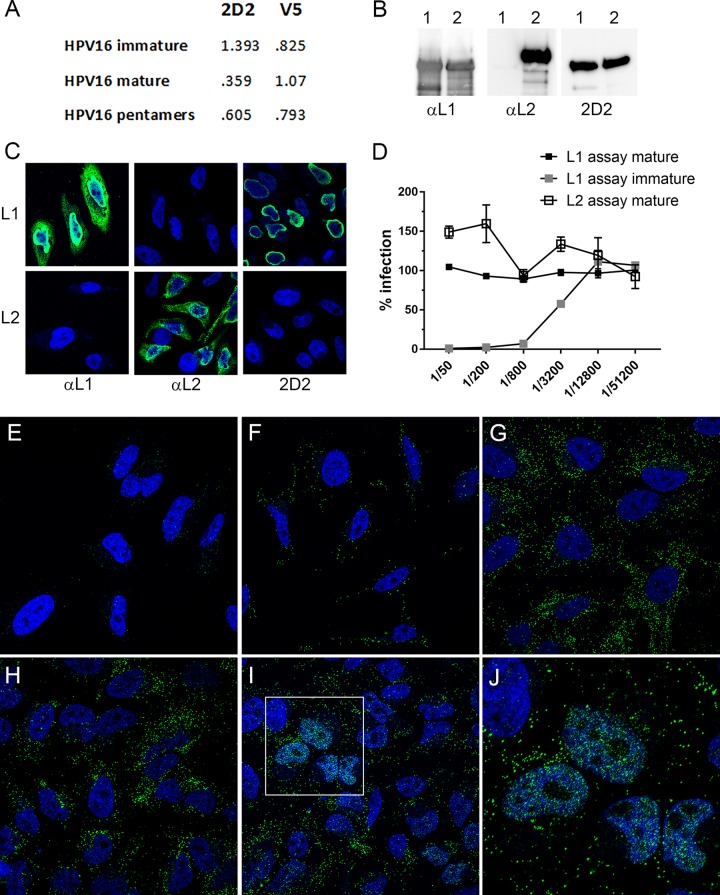
mAbs that preferentially recognize an open immature HPV16 capsid conformation were developed using a PsV immunogen comprised of a mutated L1 protein that lacked a cysteine residue necessary for maturation, assembled in association with L2. We utilized parallel enzyme-linked immunosorbent assay (ELISA) screening against both mature and immature HPV16 particles to find clones that displayed preferential recognition of the immature particles. From this screening, we derived a clone, 2D2, that merited further characterization. As shown in Fig. 1A, the reactivity of the 2D2 mAb was almost 4-fold higher for immature particles than for the mature counterpart. The mAb also showed strong recognition of pentamers. In contrast, H16.V5, a commonly utilized antibody raised against HPV16 VLPs, bound to mature capsids more strongly than to immature capsids or pentamers but did not demonstrate a strong preferential recognition among the preparations. As the immunogen contained both L1 and L2 proteins, we determined which component comprised the 2D2 epitope. 2D2 recognized L1 by both immunoblotting of either L1-only particles or L2-containing PsV (Fig. 1B) and immunostaining of L1-transfected cells (Fig. 1C). Neither method demonstrated 2D2 reactivity against the L2 protein.
FIG 1.
Characterization of 2D2 mAb. (A) Differential ELISA reactivities of 2D2 to HPV16 immature and mature L1/L2 capsids and pentamers. Capture ELISAs were performed by coating plates with a capture reagent, rabbit anti-HPV16 VLP. HPV16 preparations were applied at 150 ng/well. 2D2 or V5 antibody (anti-HPV16 L1 VLP) was added at a dilution of 1:10 (hybridoma supernatant) or 1:5,000 (ascites), respectively. Bound antibody was detected with horseradish peroxidase-conjugated secondary antibody. (B) Detection of L1 and L2 by Western blotting. In each instance, L1-only VLPs were run in lane 1, and L1 and L2 PsV was run in lane 2. Proteins were detected on the three blots with anti-HPV16 L1, anti-HPV16 L2, or 2D2. (C) HeLa cells that were transfected with either an L1 or an L2 expression plasmid were immunostained for the same antigens. (D) Neutralization of immature and mature PsVs with 2D2. Neutralization assays were performed in triplicate across a 4-fold dilution series of the 2D2 hybridoma supernatant starting at a 1:50 dilution. In the L1-based assay, antibody dilutions are applied to PsV prior to addition to 293TT cells, whereas in the L2-based assay, capsids are applied to an epithelial cell extracellular matrix and cleaved with furin prior to exposure of the capsids to the antibodies and target cells. (E to I) Time course of PsV detection. HeLa cells were either uninfected (E) or infected with HPV16 PsV for 1 h (F), 2 h (G), 7 h (H), or 24 h (I). (J) Magnification of the inset in panel I. L1 was stained with 2D2 and donkey anti-mouse IgG-Alexa Fluor 488 (green channel). Nuclei are stained with DAPI (4′,6-diamidino-2-phenylindole) (blue channel). This temporal exposure of the 2D2 epitope was observed in multiple time course experiments.
We found that the 2D2 antibody neutralized immature HPV16 PsV but not mature PsV in both L1-based and L2-based neutralization assays (Fig. 1D), indicating that the conformation of L1 recognized by 2D2 does not occur on the cell surface during normal infection, even following furin cleavage of L2, as mimicked by the L2-based assay (13). This initial lack of epitope accessibility was confirmed by immunofluorescence (IF) staining. PsV was added to HeLa cells for either 1 h (Fig. 1F), 2 h (Fig. 1G), 7 h (Fig. 1H), or 24 h (Fig. 1I) at 37°C. Uninfected cells (Fig. 1E) showed no specific staining. 2D2 staining of infected cells progressively increased throughout the time course. Surprisingly, at 24 h, we detected a strong nuclear signal in many cells. The nuclear L1 signal was generally diffuse, with nucleolar exclusion but with additional focal depositions dispersed throughout the nucleoplasm (see the magnified region in Fig. 1J). By using a different detection mAb, a recent report (9) suggested that a fraction of L1 may reach the nucleus with L2/vDNA, but that study proposed L1 instability in the nucleus following release from mitosis, seemingly inconsistent with our detection of a robust nuclear signal.
Nuclear L1 accumulation is an L2-dependent phenomenon.
The interaction of L2 with cellular factors during passage through the endocytic pathway and TGN prior to association with mitotic chromosomes is well established, and there is a strict requirement for L2 in infectious entry, culminating in the delivery of the viral genome to ND10 (reviewed in references 6 and 14). Consistent with this, we found that 2D2 detection of nuclear L1 was also an L2-dependent phenomenon (Fig. 2A and B), as no nuclear staining was evident following entry of L1-only VLPs.
FIG 2.
L1 nuclear delivery is L2 dependent and prevented with biochemical inhibition of infection. (A to F) Either HPV16 L1-only VLPs (A and B) or L1/L2 PsV (C to F) was applied to HeLa cells for 24 h and stained with 2D2 and donkey anti-mouse IgG-Alexa Fluor 488.(A and B) Duplicate images with the DAPI nuclear signal removed from the second panel to better visualize L1 nuclear localization. (C to F) The following inhibitors were added during PsV incubation: 10 μg cycloheximide (C), 10 μM RVKR-cmk (furin inhibitor) (D), 300 nM compound XXI (γ-secretase inhibitor) (E), and 10 μM cyclosporine (cyclophilin inhibitor) (F). (G to J) Nuclear L1 delivery does not require packaged DNA. Particles were confirmed to lack DNA by Western blot determination of histone H3 content. (G) Western blots of L1 (bottom bands) and L2 (top bands). Lane 1, full particles; lane 2, empty particles; lanes 3 and 4, Western blots for histone H3 in the same sample order. (H) DNA content extracted from these particles. DNA ladders are shown in the first two lanes, the third lane shows DNA extracted from full particles, and the fourth lane shows the extraction from the empty particles. (I and J) Cells that were infected in parallel with either full particles (I) or empty particles (J), stained with antibody H16.2F at 24 h postinfection. Nuclei are stained with DAPI.
The PsV preparation should not contain the L1/L2 expression plasmid used for PsV generation, as it exceeds the packaging size constraint (15). Nevertheless, we eliminated the possibility that the nuclear L1 signal was simply due to the delivery and expression of this plasmid, by prevention of de novo protein synthesis during PsV entry. As shown in Fig. 2C, cycloheximide treatment during PsV entry did not affect the intensity or character of nuclear L1 staining. We also examined the effect of biochemical inhibitors of HPV entry on L1 nuclear accumulation. Inhibition of furin processing, γ-secretase activity, and cyclophilin activity, which are well documented to result in vesicular retention of L2/vDNA, thereby preventing PsV infection (10, 16–18), all precluded nuclear detection of L1 (Fig. 2D to F). Taken together, these results support the conclusion that nuclear accumulation of L1 occurs by the pathway used for infectious viral entry.
Nuclear L1 delivery is not dependent upon vDNA.
A model delineating the vesicular delivery of a subviral complex to the mitotic chromosomes proposed that the association of residual capsomeric L1 with L2 was stabilized by interactions with the vDNA following release from the TGN (9). We addressed this hypothesis using DNA-free particles generated in a disassembly/reaction in the presence of an endonuclease (19) and confirmed the absence of encapsidated DNA in these particles by Western blot analysis of the DNA-associated histone H3 (Fig. 2G). The DNA-positive (full) and DNA-negative (empty) particle preparations had equal levels of L1 and L2 proteins (Fig. 2G, lanes 1 and 2), but only the DNA-containing particles had associated histone H3 (compare lanes 3 and 4). We also directly analyzed the amount of extracted DNA (Fig. 2H). Marker ladders are shown in lanes 1 and 2 of Fig. 2H, and the extracted content is shown in lanes 3 and 4 (full and empty particles, respectively). Microscopic evaluation of nuclear L1 showed equivalent nuclear staining following incubation with empty viral particles as with the DNA-containing particles (compare Fig. 2I and J), indicating that association with L2, but not the vDNA, is critical for the nuclear trafficking of L1.
Nuclear L1 localizes at ND10.
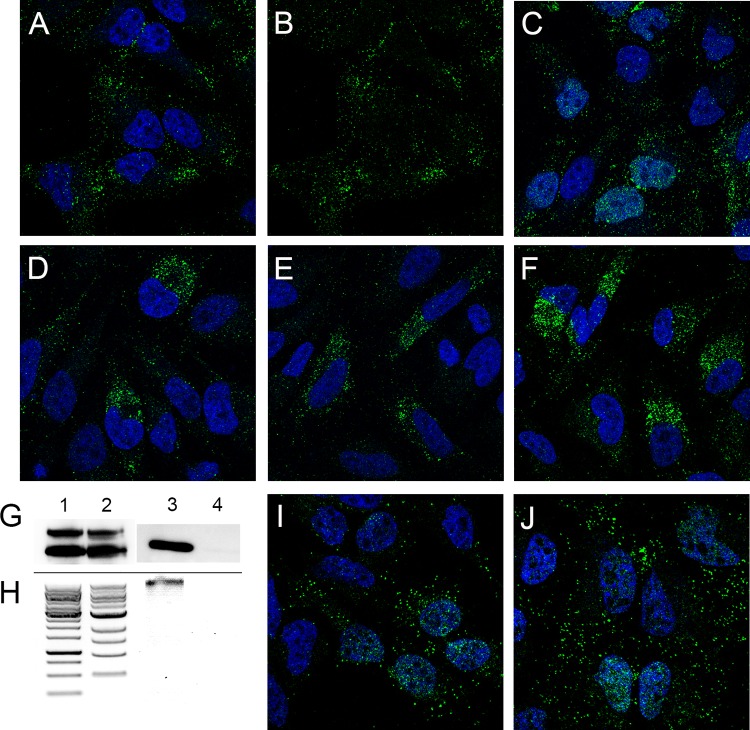
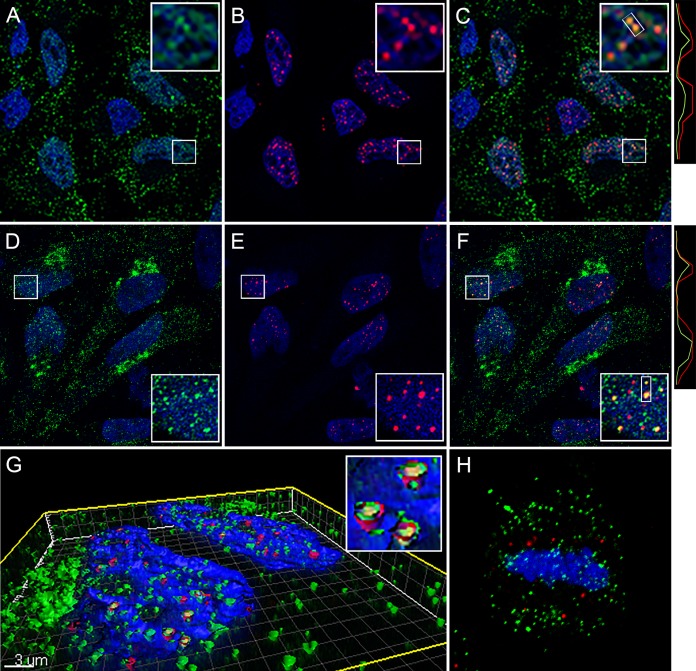
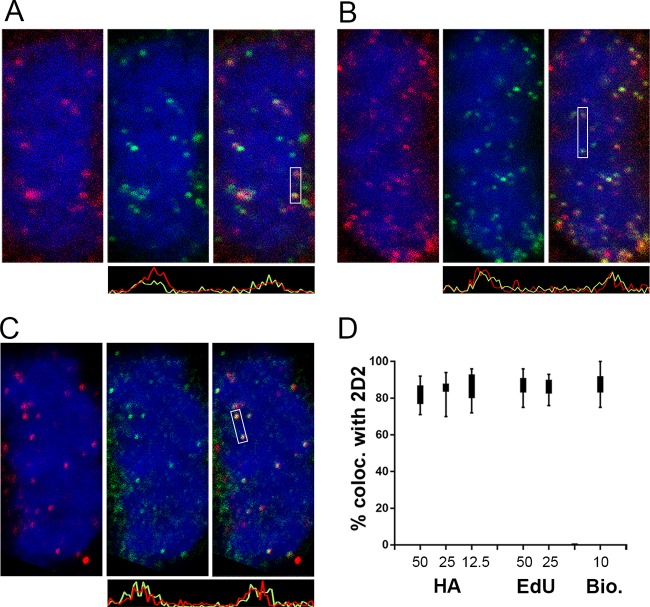
The partially punctate character of the nuclear L1 detected with 2D2 was reminiscent of L2 and vDNA localization at ND10 at this time point (20). Therefore, we evaluated the localization of L1 relative to that of PML, an ND10-resident protein. As evident in Fig. 3A to C, the nuclear L1 signal had a more diffuse character than PML staining, but some colocalization was apparent. The degree of colocalization was clearer when soluble proteins were extracted with NP-40 prior to fixation. As shown in Fig. 3D to F, the diffuse nuclear L1 signal was largely removed, revealing more pronounced PML colocalization. As previously described for L2 nuclear deposition, L1 appears to be preferentially associated with an ND10 subset (20). This close association was more evident following surface rendering of a Z-stack series, as shown in Fig. 3G, containing an inset of a magnified region. The extraction procedure also enabled clear detection of L1 association with mitotic chromosomes (Fig. 3H) and the TGN (colocalization data are not shown, but the pattern is evident in Fig. 3D). We also confirmed that this L1 signal colocalized with L2 and the vDNA on mitotic chromosomes. This analysis was performed with a titration of input virus. To obtain robust L2 staining, we utilized a multihemagglutinin (multi-HA) carboxyl-terminally tagged L2 protein. The colocalization of 2D2 staining and HA on the mitotic chromosome is shown in Fig. 4A. The localization relative to 5-ethynyl-2'-deoxyuridine (EdU)-labeled vDNA is shown in Fig. 4B. Additionally, we confirmed that the 2D2 antibody was specifically recognizing L1 on the mitotic chromosomes by colocalization of 2D2 with biotinylated PsV, detected with fluorescent streptavidin (Fig. 4C). The indicated regions of colocalization within the merged panels are illustrated in histograms of pixel intensity below the appropriate panels. To determine the extent of colocalization, 2D2 puncta through Z-stack series were counted (minimum of 600 puncta under each condition), and the presence of the second component was evaluated (Fig. 4D). Colocalization was strong for 2D2 and L2 down through the titration series. However, we did not achieve good vDNA detection with a smaller amount of virus (data not shown). Eighty to ninety percent of the 2D2 signal colocalized with the other viral component. This noncoincident staining could indicate non-L1-associated L2 and vDNA on the mitotic chromosome. However, a similar colocalization range was observed when 2D2 staining was correlated with biotinylated capsid detection, which represents detection of the same antigen. Therefore, the lack of 100% colocalization of viral components on the mitotic chromosome is more likely due to methodological limitations than detection of an L1-independent component. Arrival at the mitotic chromosomes is the penultimate step in PsV entry, which necessitates a relatively high number of virions to be used for microscopic detection. There is documentation of a substantial loss of virus during trafficking due to degradation and other physiological bottlenecks (21, 22).
FIG 3.
Nuclear localization of L1 at ND10. (A to C) Colocalization of 2D2 staining with PML at 24 h postinfection. (A) L1 was detected with 2D2 and donkey anti-mouse IgG-Alexa Fluor 488 (green). (B) PML was detected with a rabbit polyclonal antiserum and donkey anti-rabbit IgG-Alexa Fluor 594 (red). (C) Merged signals. Histograms illustrating the degree of colocalization through the indicated region are on the right. (D to F) The same staining in cells that were extracted with 1% NP-40 prior to fixation. (G) A Z-stack series from cells that underwent this treatment was surface rendered. (H) Detection of L1 on mitotic chromosomes in NP-40-extracted cells. Nuclei are stained with DAPI in all panels.
FIG 4.
Colocalization of 2D2-reactive L1 with L2, vDNA, and biotinylated pseudovirions on mitotic chromosomes. All panels show mitotic chromosomes from HeLa cell cultures at 24 h postinfection. (A) Colocalization of 2D2 staining with L2 in a single slice from a Z-stack series. 2D2 binding was detected with donkey anti-mouse IgG-Alexa Fluor 488 (green). HA-tagged L2 was detected with a goat polyclonal antiserum and donkey anti-goat IgG-Alexa Fluor 594 (red). (B) Colocalization of 2D2 staining with the vDNA in a single slice from a Z-stack series. HPV16 PsV containing EdU-labeled DNA is shown. L1 was stained with 2D2 and donkey anti-mouse-Alexa Fluor 594 (red). Cells were then processed for EdU detection with Click-iT Alexa Fluor 488 reagents (green). (C) Colocalization of 2D2 staining (red) with biotinylated HPV16 PsV particles, detected with Alexa 488-coupled streptavidin. Histograms illustrating the degree of colocalization through the indicated regions are shown below the corresponding images. (D) Percent colocalization between 2D2-stained puncta and HA, EdU, or biotin across a range of input concentrations, as indicated. The microscopic images shown correspond to 25 ng input PsV for L2 and EdU colocalization and 10 ng for colocalization with biotinylated capsids.
Additional anti-L1 reagents detect nuclear signals.
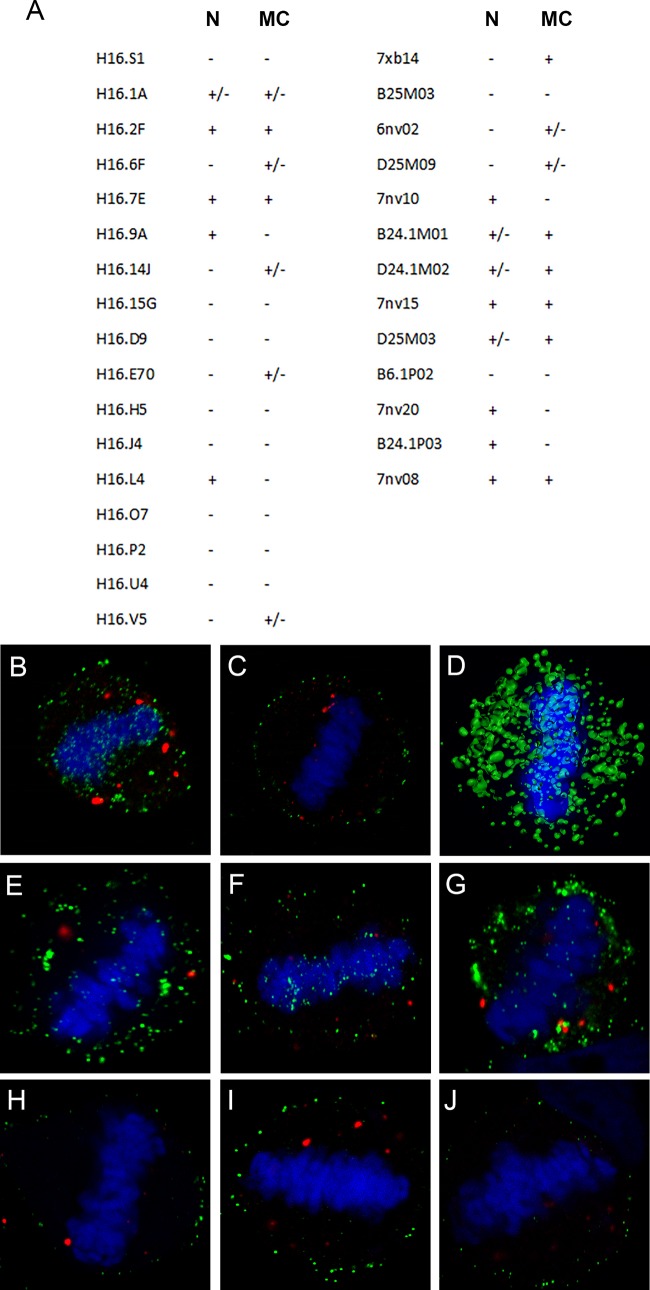
Many laboratories, including our own, have monitored PsV microscopically during the infectious process without detection of nuclear L1. This is probably due to the antibody reagents commonly used in these studies. In our hands, the often employed monoclonal antibody H16.V5 weakly detected L1 on mitotic chromosomes, but no L1 nuclear staining was observed. In a recent study, limited chromosomal staining and transient nuclear staining were described (9). As many other HPV16 L1-specific mAbs exist, we decided to examine a panel of these anti-VLP reagents for detection of L1 within the nucleus and on mitotic chromosomes by evaluation of 17 additional mouse mAbs (23, 24) and 13 human mAbs (25), (summarized in Fig. 5A). The immunogen for the mouse mAbs was HPV16 L1-only VLPs. The human antibodies were cloned from memory B cells isolated from quadrivalent HPV vaccinees. These data demonstrate that multiple anti-L1 reagents can detect nuclear L1 following endocytosis. However, the antibodies that do so are not among those commonly utilized in entry studies, which may explain why this phenomenon has not been previously described, although one previous report detected H16.V5-stained nuclear puncta (9). The four antibodies that recognized L1 in both locations (H16.2F, H16.7E, 7nv15, and 7nv08) were determined by an ELISA to recognize both immature and mature capsid conformations as well as pentamers (data not shown).
FIG 5.
Reactivity of a panel of anti-L1 reagents. (A) The evaluated monoclonal antibodies, with murine antibodies in the first column and human antibodies in the second column. Their recognition of either nuclear L1 (N) or L1 on mitotic chromosomes (MC) within HeLa cells at 24 h postinfection is indicated. (B and C) Differential permeabilization staining. Shown is the accessibility of H16.7E antibody to L1 on mitotic chromosomes following NP-40 permeabilization (B) or digitonin permeabilization (C) followed by the secondary antibody donkey anti-mouse-Alexa Fluor 488. PML was detected with a rabbit antiserum and donkey anti-rabbit-Alexa Fluor 594. (D) Surface-rendered image from a Z-stack series of an infected HeLa cell nucleus stained with 7nv15. (E to J) Differential permeabilization staining of HeLa cells that were infected with HPV31 PsV (E and H), HPV45 PsV (F and I), or authentic MusPV1 (G and J), with NP-40-permeabilized cells in the top panels and digitonin-permeabilized cells in the bottom panels. In all instances, PV L1 was detected with rabbit polyclonal serum raised against VLPs followed by the secondary antibody donkey anti-rabbit-Alexa Fluor 488. PML was detected with a mouse antibody and donkey anti-mouse-Alexa Fluor 594. Central sections from Z-stack series are shown.
The strong staining of L1 on mitotic chromosomes attained with antibody H16.7E allowed us to evaluate the accessibility of L1 following differential detergent permeabilization. This technique was previously used to assess the vesicular nature of the chromosomally associated HPV complex of L1/L2/vDNA (9, 11). Consistent with that study, we found that L1 on mitotic chromosomes was inaccessible to antibodies when membranes were not fully permeabilized. Compare the NP-40-permeabilized staining in Fig. 5B with that in cells that were permeabilized with a low-concentration digitonin solution (Fig. 5C), which would prevent access to luminal epitopes. As confirmation that L1 staining represented chromosomally associated protein, we constructed a surface-rendered view of a Z-stack image series of a metaphase chromosome from an infected cell. As shown in Fig. 5D, the L1 protein appears to be embedded within the chromatin structure.
Although it appears that PV types across genera utilize similar intracellular pathways (26, 27), different host proteins may be involved in this trafficking (28). We chose to examine PsV from two other oncogenic types for mitotic chromosome association. For HPV31 and HPV45, we found polyclonal sera that detected L1 associated with mitotic chromosomes following NP-40 but not digitonin permeabilization, indicative of vesicular localization (compare Fig. 5E and H for HPV31 and Fig. 5F and I for HPV45). We also took advantage of the availability of authentic Mus musculus PV 1 (MusPV1) purified from murine warts. Importantly, we identified L1-containing vesicles associated with condensed mitotic chromosomes, as indicated by the differential permeabilization (compare Fig. 5G and J). Thus, this process is also conserved between distantly related PV genera and is not an artifact of PsV technology.
L1 shows nuclear localization in HaCaT keratinocytes and in the murine genital tract model.
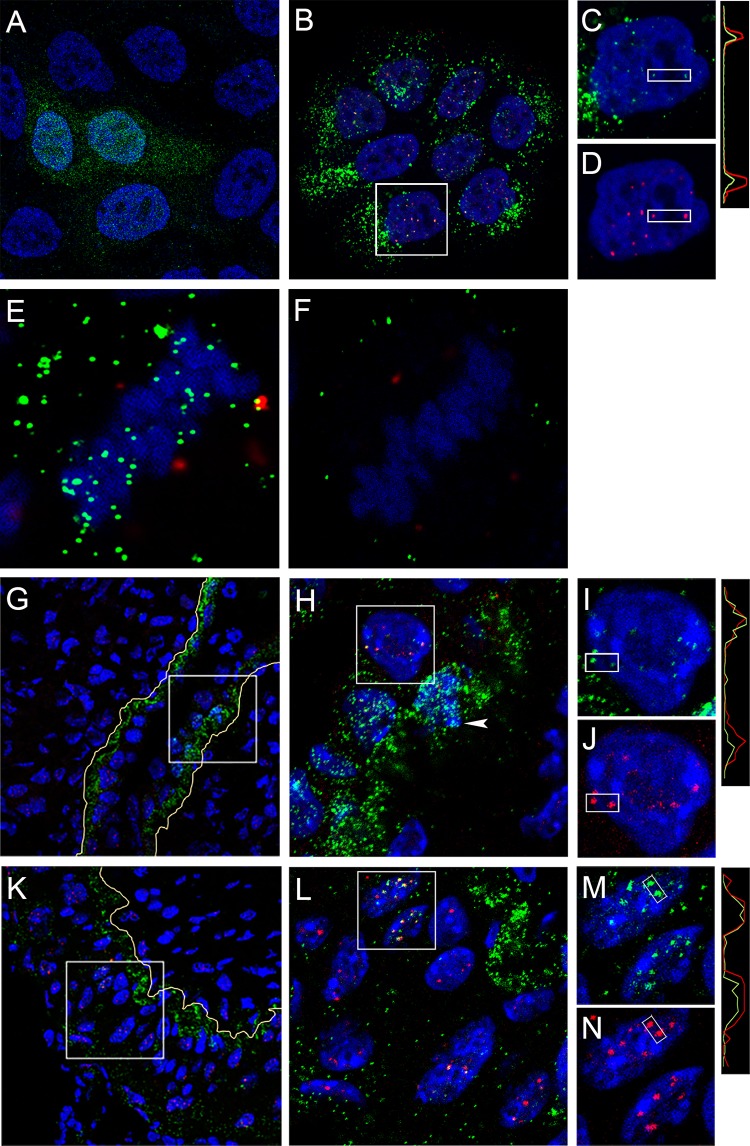
HeLa cells have been utilized for many HPV entry studies and have provided the foundation for many recent advances (8, 29, 30). Comparative analyses of HPV trafficking in HeLa and the normal human keratinocyte cell line HaCaT have revealed no differences (31–34). Nevertheless, we confirmed the observation of L1 nuclear deposition during PsV infection in HaCaT cells. Representative staining is shown in Fig. 6A (2D2) and Fig. 6B (7nv15). The latter reagent demonstrated predominantly punctate nuclear staining that partially colocalized with PML (Fig. 6C and D). We also observed chromatin-associated L1 within vesicles in HaCaT cells, as demonstrated by differential permeabilization staining with 7nv15, shown in Fig. 6E and F (NP-40 and digitonin permeabilization, respectively), demonstrating that this phenomenon is also not cell type specific.
FIG 6.
Nuclear L1 detected in keratinocytes. (A to D) HaCaT cells were infected with HPV16 PsV for 24 h and processed for IF with either 2D2 (A) or human monoclonal anti-L1 mAb 7nv15 (B and C) using either donkey anti-mouse IgG-Alexa Fluor 488 or donkey anti-human IgG-Alexa Fluor 488 secondary antibody conjugates, respectively. 7nv15 staining was performed in conjunction with PML protein detection (B and D) with rabbit anti-PML and donkey anti-rabbit IgG-Alexa Fluor 594. Colocalization through the indicated region is illustrated in the histogram. (E and F) Differential permeabilization staining. Shown is the accessibility of 7nv15 antibody to L1 on the mitotic chromosomes on HaCaT cells following NP-40 permeabilization (E) or digitonin permeabilization (F) followed by the secondary antibody donkey anti-mouse-Alexa Fluor 488. PML was detected with a rabbit antiserum and donkey anti-rabbit-Alexa Fluor 594. A central section obtained from a Z-stack series is shown for each. (G to N) In vivo staining. The presence of nuclear L1 following murine intravaginal infection was determined by staining tissues with 7nv15 and anti-PML. Tissues were prepared at 18 h (G to J) or 48 h (K to N) postinstillation. The boxed regions are magnified in the adjacent panels. The split panels show 7nv15 anti-L1 staining in green (I and M) or anti-PML staining in red (J and N). Nuclei are stained with DAPI (blue). Histograms illustrating the degree of colocalization through the indicated region are on the right. The basement membrane is traced in yellow in the lower-magnification images.
The murine cervicovaginal challenge (CVC) model has been useful in the in vivo analysis of early events in PV-host interactions (35, 36). To determine if nuclear L1 could be also be observed in this system, we processed tissue for IF staining following intravaginal delivery of HPV16 PsV. We utilized the human monoclonal antibody 7nv15 for this analysis to avoid the staining issues encountered when using mouse antibodies on murine tissue. Vaginal tracts were excised either 18 h or 48 h after PsV instillation to allow visualization of mitotic basal cells above the basement membrane (indicated as a yellow line) relatively soon after virus binding and of suprabasal cells later in the infectious process, respectively. The tissue shown in Fig. 6G, at the 18-h time point, contains a region that exemplifies the typical pattern of basement membrane association (37). Above the basement membrane lies an area in which we could visualize both positively stained mitotic chromosomes and ND10-localized L1, as determined by PML colocalization. These localizations are better appreciated in the higher-magnification image shown in Fig. 6H, in which a mitotic cell is indicated by an arrowhead. Figures 6I and J show the split-color images at an even higher magnification to allow visualization of the partial L1 and PML colocalization. Figures 6K and L show staining with the same antibodies at the 48-h time point, when strong ND10 localization of L1 in cells located above the basement membrane was evident. The split-color images of the magnified region are shown in Fig. 6M and N. The indicated regions of colocalization are demonstrated in the histograms of pixel intensity on the right side of the panels. In total, these data indicate that L1 traffics to the nucleus during HPV16 PsV infection in both normal human keratinocytes and cervicovaginal tissue in the in vivo model. ND10 localization was evident in both instances, tying the observations to the established biology of HPV nuclear establishment and early transcription.
Infected cells examined by TEM contain membrane-bound viral capsids.
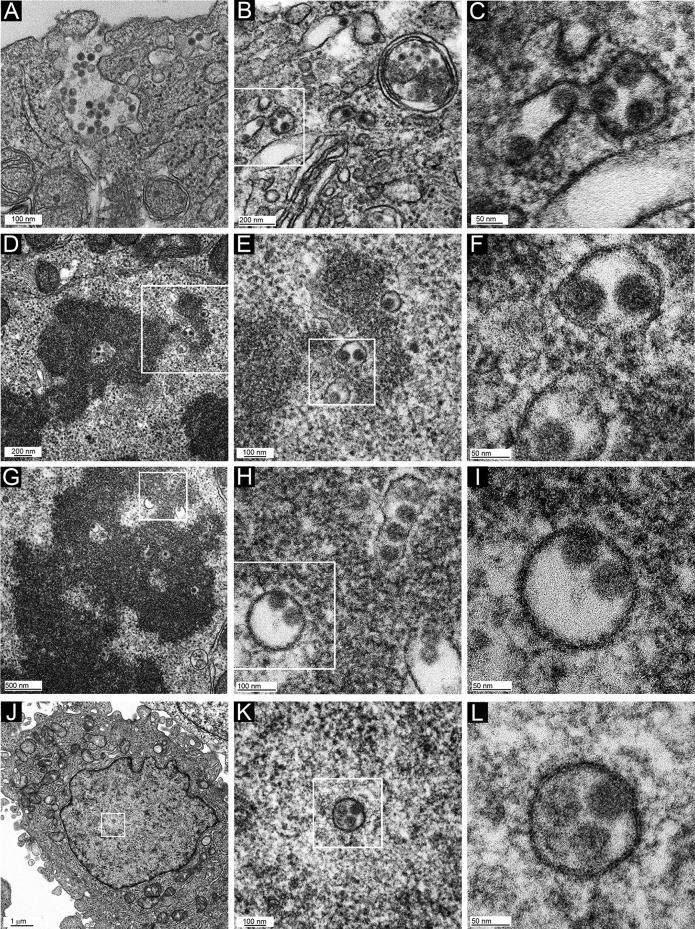
The differential exposure of L1 epitopes after NP-40 treatment suggests that the L1/L2/vDNA complex might be present on the mitotic chromosomes within a membrane-bound vesicle (9, 11). A structure with this character should be amenable to transmission electron microscopy (TEM) imaging. Therefore, we prepared samples from uninfected HeLa cells and those infected with HPV16 PsV or L1-only VLPs for 24 h and processed them for TEM (Fig. 7). As shown in Fig. 7A, even at this time postinoculation, some PsV capsids were evident proximal to the cell surface in some cells, reflecting the asynchronous nature of the infectious process. Unsurprisingly, capsids were also found in MVBs and small vesicles (Fig. 7B). A region of Fig. 7B is magnified in Fig. 7C to illustrate the presence of 50-nm particles within vesicles. It is likely that these particles represent relatively intact capsids, as the extracellular HPV capsid has an average diameter of 50 to 60 nm. The presence of unprocessed capsids within MVBs and lysosomes at 18 h postinfection was previously noted (38).
FIG 7.
TEM micrographs of HPV16-infected HeLa cells. Cells were infected for 24 h with HPV16 PsV and processed for TEM imaging. Panel inset boxes show the area magnified in the following panel. Two different mitotic cells were chosen for panels D to F and G to I. Cells shown in panels A to C and J to L are in interphase.
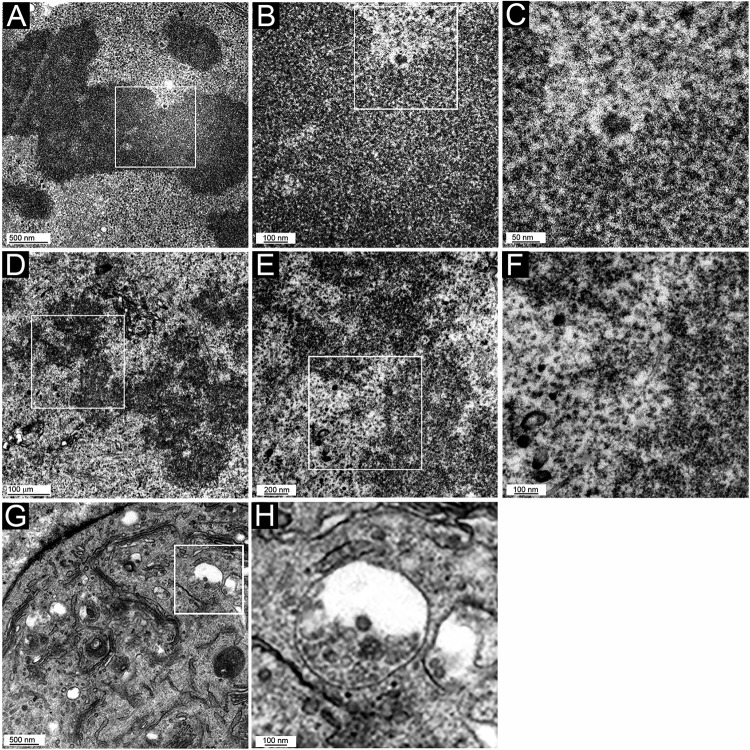
When we examined mitotic cells, distinguished by obvious chromatin condensation and a lack of a nuclear membrane, we could readily identify numerous small membranous vesicles associated with the mitotic chromosome in HPV16 PsV-infected cells (Fig. 7D to F and G to I show two examples from different cells), proving previous suggestions, based on indirect evidence of differential membrane permeabilization. Surprisingly, 50-nm particles were clearly visible within all vesicles. These particles were indistinguishable from those found in the larger cytoplasmic vesicles (Fig. 7A to C), indicating that extensive capsid processing had not occurred prior to mitosis. Intriguingly, many of the particles were situated asymmetrically within the vesicle, with the particle surface adjoined to the vesicular membrane. This abutment was also evident in small cytoplasmic vesicles in interphase cells and in some instances within MVBs (Fig. 7B and C). This could indicate visualization of the above-described transmembrane passage of L2 that enables interaction with cellular transport mediators. A chromosomal binding region of L2 that lies within the proposed cytosolically extruded region was recently defined (33). Our results are consistent with the data defining L2 as an essential mediator of infectious entry and chromosome tethering. No chromosome-associated vesicles (with or without internal particles) were seen in uninfected cell samples that had been processed in parallel (Fig. 8A to C) or in cells that had internalized L1-only particles (Fig. 8D to F). L1-only particles were found within, but not beyond, endosomal vesicles (Fig. 8G and H).
FIG 8.
TEM micrographs of control HeLa cells. Mitotic cells (A to F) and an interphase cell (G and H) are shown. (A to C) Uninfected cells; (D to H) cells incubated with L1-only VLPs for 24 h. Panel inset boxes show the area magnified in following panel. Note that there is some electron-dense material in the chromosomal region, but it is irregular in shape and not within vesicles. This is most apparent in panels B and C. Panel G and the magnification in panel H show the localization of L1-only particles within endosomal vesicles.
As numerous capsid-containing vesicles were associated with the condensed metaphase chromatin in PsV-infected cells, each daughter cell should receive multiple vesicles upon completion of mitosis. Scrutiny of interphase cells revealed the presence of intranuclear membrane-bound vesicles that contained 50-nm particles; however, there were fewer positive sections than would be expected based on the number of mitotic vesicles observed and the percentage of cells infected under these conditions. Infected interphase nuclei are shown in magnification series in Fig. 7J to L. We never detected more than one vesicle per nucleus, within the sectioned plane. Therefore, it is likely that the vesicle and particle are unstable after nuclear membrane reformation.
DISCUSSION
Policing of transport across the nuclear membrane likely provoked the evolution of multiple strategies by which viruses circumvent this host control. These strategies include transport through the nuclear pore complex (NPC), local disruption of the NE and nuclear lamina, or gaining access during mitosis (reviewed in references 39 and 40). The NPC can be commandeered either for the transport of viral components containing nuclear localization signals after virion disassembly in the cytoplasm, as seen for influenza A virus and HIV-1, or as a dock for capsids that then release the viral genome across the NPC, as described for herpesviruses and adenoviruses. Capsids with diameters smaller than the capacity of the NPC (39 nm for facilitated transport) can pass intact across the NE and undergo uncoating within the nucleus, e.g., hepatitis B virus and alphabaculoviruses. Parvoviruses have developed a strategy whereby the intact particle enters the nucleus through local NE disruption mediated by caspase-3. Gammaretroviruses and PVs are examples of viruses that gain access to the nucleus during mitosis, when the NE is temporarily disassembled. The preintegration complex (PIC) of Moloney murine leukemia virus (MMLV) contains the reverse-transcribed vDNA along with a number of viral proteins, including p12, which tethers the PIC to the mitotic chromatin (41). Our data lead us to conclude that PVs have developed a unique mechanism of depositing their genomes within the host cell nucleus. The reliance upon mitosis for PV infection has been well established in vitro (1) and meshes well with the differential in vivo infection of mitotically active basal keratinocytes within the stratified epithelium (reviewed in reference 42). Here we report the nuclear delivery of relatively intact HPV16 capsids within transport vesicles through an intermediary association with mitotic chromosomes.
Two proteolytic events occur early in the interaction of PV capsids with host cells. Kallikrein 8 cleavage of L1 and the subsequent furin cleavage of the L2 amino terminus are essential, in conjunction with exposure to low pH, for trafficking of the genome-containing complex beyond the endosomal compartment (16, 32). Additional proteolytic events occur during endocytosis but cannot be linked to infectious entry. Acid-dependent cathepsin proteases process the L1 capsid, as monitored by the generation of degradative products and by visualization of L1-7 monoclonal antibody reactivity (43). Exposure of this epitope occurs during the degradation of virions that are routed for disposal rather than for infection (32). The L1-7-visualized conformational change occurs with the same kinetics as for both the exposure of the N terminus of L2 and the accessibility of bromodeoxyuridine (BrdU)-labeled vDNA (20). The detection of buried L2 epitopes and vDNA was originally interpreted as evidence of endosomal uncoating, as neither was accessible to antibody detection while enclosed in the intact viral particle. However, our results suggest that this early vesicular BrdU positivity and L1-7 reactivity may, in fact, distinguish disassembled particles within a degradative pathway from the virions which are the true mediators of infection. Indeed, comparison of capsid levels found on the cell surface to those detected within the TGN suggests a substantial loss of L1 during trafficking (21, 22). The later detection of L2-HA and BrdU-labeled genomes at ND10 is consistent with exposure following intranuclear uncoating.
Through TEM, we readily identified numerous vesicles containing 50-nm particles associated with the mitotic chromosomes, indicating that extensive capsid processing had not occurred. The particles appear intact, but a selective loss of a specific subset of capsomers may not be detectable at the resolution reached in the present study. The detection of some slightly smaller and hazier particles was likely due to the location of the vesicle relative to the sectioned plane, but we cannot fully eliminate the possibility of the transport of a minority of lower-order structures. Particles were also detected in cytoplasmic transport vesicles and within the nucleus. Based on the number of particles visualized on mitotic chromosomes compared to within the nucleus, it is likely that the vesicles are dissolved and the particles are disassembled after nuclear delivery. These events would presumably be necessary in order to gain access to host transcription machinery. The robust immunofluorescent nuclear signal detected with numerous anti-L1 reagents could be largely attributed to the detection of pentamers or monomers following disassembly. The ELISA reactivity of these reagents with L1 in all of the assembly states tested prevented us from more definitively addressing this question.
Our results are consistent with the data defining L2 as an essential mediator of infectious entry. No intranuclear or mitotic chromosome-associated L1 was detected in the examination of L1-only VLPs. Many of the L1/L2 capsids observed by TEM were located asymmetrically within the vesicle, with one edge abutted alongside the membranous border. This orientation could indicate transmembrane passage of L2 to enable interactions with cellular transport mediators. We postulate a model in which the initial proteolytic events and γ-secretase interaction (34, 44) allow the correct positioning of L2, within an L1 shell, for partial translocation across the internal vesicle membrane, to allow interaction with cellular trafficking mediators. The capsid-containing vesicles traffic in a retrograde fashion to the TGN, where they reside until initiation of mitosis. Upon TGN fragmentation during mitosis, small capsid-containing vesicles form and become bound to the condensed mitotic chromosome via cytoplasmic L2 determinants. Upon completion of mitosis, the vesicles are delivered to the nuclear lumen via their association with chromosomes and are subsequently degraded by nuclear lipases and/or proteases, enabling the release of L2 and vDNA, which allows the transit of this complex to ND10, along with a subset of L1, now pentameric or monomeric, reflecting the IF staining distribution. It was previously surmised that intact capsids cannot enter the nucleus based on in vitro nuclear import assays (45). However, the mechanism by which we suggest that capsids enter the nucleus, via small vesicular intermediates after nuclear membrane breakdown during mitosis, would not be evaluated by this assay. It was previously demonstrated that a complex comprised of L2, vDNA, and L1 (proposed to be pentameric) associates with the mitotic chromosome (9). As we did not visualize any chromosome-associated vesicles that did not contain 50-nm particles, we conclude that this represents the predominant conformation of the PV infectious complex during mitosis. However, we cannot eliminate the possibility that some more fully uncoated lower-order complexes are also transferred within these vesicles and contribute to nuclear import and infection.
It has not been previously reported that a largely intact viral capsid can enter the nucleus within a transport vesicle. Therefore, our observations expand the repertoire of basic mechanisms used by viruses to establish infection. In addition, they potentially alter the concepts of PV uncoating and disassembly during this process, although biochemical confirmation would bolster these imaging studies. These findings are unexpected from a cell biological perspective, as neither intranuclear nor mitotic chromatin-associated transport vesicles have been previously described in any context. Vesicular transport of cargo into the nucleus may be a tightly regulated or induced event, rather than a constitutive process, and therefore may have been previously overlooked. Alternatively, these vesicles may transiently localize to the nucleus after mitosis under normal conditions but are stabilized by HPV capsid interactions with the membrane and so become more apparent. The origin and composition of these unique vesicles will be important to investigate, both for the added insight into PV trafficking and for a more complete understanding of the basic cell biological phenomenon.
MATERIALS AND METHODS
Pseudovirus and VLP production.
PsV preparations were produced according to the protocol and by utilizing the plasmids reported on the laboratory website (http://home.ccr.cancer.gov/lco/plasmids.asp). Briefly, 293TT cells were transfected with either a bicistronic plasmid, p16SheLl, encoding the HPV16 L1 and L2 proteins or two plasmids separately encoding the capsid proteins, together with a reporter gene plasmid. A plasmid encoding an L2 protein with five sequential carboxyl-terminal HA tags (L2-laugh) was constructed within the existing p16L2HA plasmid by standard cloning technologies. The packaged marker plasmid varied according to usage: pYSEAP for L1-based neutralization assays, ELISAs, the CVC mouse model, and microscopy; phGluc for L2-based neutralization assays; and pfwB for assessment of infection by flow cytometry. In accordance with convention, the encapsidated marker plasmids are generically termed vDNA. Plasmids encoding HPV16 L1 with a cysteine-to-serine conversion at residue 428 (pC428S) and HPV16 L2 (p16L2h) were used to produce particles for immunization. The immature capsids used for the ELISA were made using the bicistronic plasmid encoding HPV16 L1 with a cysteine-to-serine conversion at residue 175 and wild-type (WT) L2 (p16mL1L2). HPV16 PsV particles were conjugated to biotin according to an established protocol for dye conjugation (35), using the EZ-Link sulfo-NHS-LC biotinylation kit (Thermo Fisher). Briefly, approximately 750 μg of the lysate was mixed with 22 μg of biotin prior to purification. Particles lacking DNA were a gift from Aura Biosciences (19). Authentic MusPV1 virions were isolated from tail warts generated by previously described techniques (46).
Hybridoma production.
HPV16 L1 C428S and L2 particles were purified by agarose column filtration as described on the laboratory website. Seven-week-old BALB/c mice were immunized subcutaneously with 70 μg particles in Freund’s adjuvant. Mice were sacrificed at 2 weeks postinoculation, and hybridomas were created by fusion of spleen cells with P3 × 63Ag8.6S3 plasmacytoma cells (ATCC CRL1580) according to standard methodology (23). The fusions were plated into 96-well, flat-bottom microtiter plates in RPMI medium. Three days following fusion, medium was changed to RMPI supplemented with hypoxanthine, aminopterin, and thymidine. Wells were screened by an ELISA for antibody reactivity. Antibody-secreting clones were purified to a single population by limiting dilution. The isotype of 2D2 was determined to be IgG1 lambda by using an isotyping kit (Serotec).
Antibodies.
The H16 series of monoclonal antibodies raised against HPV16 VLPs was a gift from Neil Christensen (Pennsylvania State University, Hershey, PA) (23, 24). The human anti HPV16 antibodies, derived from cloned memory B cells isolated from Gardasil-4 vaccinees and selected for their ability to bind to HPV16 PsV, were gifts from Denise Galloway and Jody Carter (Fred Hutchinson Cancer Research Center, Seattle, WA) (25). Camvir-1 (Abcam) was used to detect L1 on Western blots. The rabbit polyclonal antisera recognizing PV capsids were previously described (13, 46). The anti-L2 monoclonal antibodies K1L2 (L2 amino acids 64 to 81), used for immunofluorescence detection, and K5L2 (amino acids 56 to 75), used for Western blot detection, were kind gifts from Martin Müller (DKFZ, Heidelberg, Germany). Rabbit anti-PML (PM001) was purchased from MBL International. Mouse anti-PML (PG-M3) was purchased from Santa Cruz Biotechnology. Goat anti-HA was purchased from GenScript (catalog number 00168). Alexa Fluor-conjugated secondary antibodies and streptavidin were purchased from Molecular Probes/Thermo Fisher Scientific.
ELISA and neutralization.
Monoclonal antibodies were screened for differential reactivity to immature (C175S L1) versus mature (WT) HPV16 particles by an ELISA using a capture technique. Immulon 2HB plates (Thermo Corporation) were coated with 500 ng/well of purified rabbit anti-HPV16 L1 antiserum (47) and incubated overnight at 4°C. Following three phosphate-buffered saline (PBS) washes, the plates were blocked in PBS containing 0.5% powdered milk and 0.1% fetal bovine serum (FBS) for 2 h at room temperature. Plates were rewashed, and 150 ng/well of VLPs was added in PBS–0.5% powdered milk for 2 h at room temperature. Unbound particles were removed by washing, and diluted mAbs were added for 2 h at room temperature. After washing, a horseradish peroxidase-conjugated goat anti-mouse IgG antiserum (Caltag) was added at a 1:2,000 dilution in PBS–0.5% milk, for 1 h at room temperature. The plates were washed again and developed with the peroxidase substrate 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) (Roche). L1- and L2-based neutralization assays were performed as previously described, except for the use of Gaussia luciferase instead of green fluorescent protein (GFP) as the infectious readout for the L2 assay (13, 48).
Immunofluorescence.
HeLa or HaCaT cells were seeded onto glass coverslips in 24-well plates at a density of 1 × 105 cells/well, cultured overnight, infected with 50 ng viral particles per well (or as indicated), and incubated for the times indicated. Cells were fixed with 2% paraformaldehyde for 20 min at room temperature, washed three times with 200 mM glycine in PBS, and processed for immunostaining. Briefly, cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature, blocked for 20 min in 5% normal donkey serum in PBS, and incubated with primary antibodies diluted in 2.5% normal donkey serum. Following this incubation and washing, appropriate secondary antibodies, as indicated, were applied. Following final washes, coverslips were inverted onto Prolong Gold mounting solution (Invitrogen) on a glass slide. For detergent extraction prior to fixation, cells were washed in Kern matrix (KM) buffer (10 mM N-morpholinoethanesulfonic acid, 10 mM NaCl, 1.5 mM MgCl2, 10% glycerol, protease inhibitor cocktail [Sigma]) and then extracted by incubation in KM buffer supplemented with 1% NP-40, 1 mM EGTA, and 5 mM dithiothreitol for 30 min on ice prior to fixation, as previously described (27). Differential permeabilization was performed as previously described (9). Briefly, cells were fixed with 4% paraformaldehyde. Following a glycine wash, cells were permeabilized with either 0.5% Triton X-100 or 0.625 μg/ml digitonin in PBS for 10 min. Prior to antibody incubation, cells were blocked for 20 min with 5% normal donkey serum. Detection of EdU-labeled packaged DNA was performed as previously described, using a Click-iT EdU Alexa Fluor 488 imaging kit (Thermo Fisher), except that 2D2 staining of L1 was completed prior to initiation of the EdU protocol (7). All images were acquired with a 63× objective on a Zeiss 780 confocal system interfaced with a Zeiss Axiovert 100M microscope. Images were collated with Adobe Photoshop software; adjustments were applied consistently across experimental groups. Surface-rendered images of Z-stack images were processed with Imaris software. Colocalization histograms were created by line profile analysis in Zen software (Zeiss). The pixel values were exported to Microsoft Excel.
Inhibitors.
The inhibitors decanoyl-RVKR-cmk (catalog number 344930), cyclosporine (catalog number 239835), and γ-secretase inhibitor compound XXI (catalog number 565790) were purchased from Calbiochem. Cycloheximide (catalog number C7698) was purchased from Sigma.
CVC mouse infection.
Six- to eight-week-old female BALB/cAnNCr mice were obtained from the National Institutes of Health and handled in accordance with their guidelines. Protocols were approved by the National Cancer Institute’s Animal Care and Use Committee. HPV16 PsV intravaginal instillation was performed as previously described (35). Genital tracts were excised at 18 or 48 h postinstillation, washed with PBS, and snap-frozen in tissue freezing medium (EMS). Six-micrometer tissue sections were cryosectioned, transferred to glass slides, and fixed for 10 min in 100% ethanol at −20°C. Tissues were processed for staining as described previously (37). Briefly, tissue sections were blocked with 10% donkey serum in PBS with 0.1% Brij 58 for 30 min at room temperature. Antibody 7nv15 was diluted to 1 μg/ml and incubated in conjunction with rabbit anti-PML antiserum. Bound antibody was detected with Alexa Fluor 488-conjugated donkey anti-human serum and Alexa Fluor 594-conjugated donkey anti-rabbit serum (Invitrogen). Antibodies were diluted in PBS with 0.1% Brij 58. Following staining, sections were mounted with Prolong Gold mounting solution.
Electron microscopy.
HeLa cells (2 × 106) were seeded in 100-mm plates, cultured overnight, and infected with 2,000 ng viral particles per well. At 24 h postinfection cells were fixed with 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) and processed as previously described (49). Figure 5A was obtained with a Hitachi H7600 electron microscope equipped with an AMT XL41M digital camera. All other EM micrographs were obtained with a Tecnai G2 Spirit transmission electron microscope (FEI/Thermo Fisher Scientific, Hillsboro, OR) fitted with a Gatan Orius charge-coupled-device (CCD) camera (Gatan, Inc., Warrendale, PA).
ACKNOWLEDGMENTS
We thank Neil Christensen from the University of Pennsylvania, Hershey, PA; Denise Galloway and Joseph Carter from the Fred Hutchinson Cancer Research Center; Martin Müller from DKFZ-Heidelberg; and Aura Biosciences for the generous sharing of reagents. We also thank the CCR-NCI-NIH confocal core facility for access to their facilities and Kunio Nagashima at the CCR-NCI-NIH Electron Microscopy Laboratory for processing the TEM samples. We appreciate Bernard Moss, NIAID, for the use of his TEM facilities for image acquisition.
Research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
REFERENCES
- 1.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. 2009. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog 5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 3.Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, Trus BL. 2008. Arrangement of L2 within the papillomavirus capsid. J Virol 82:5190–5197. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day PM, Roden RB, Lowy DR, Schiller JT. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol 72:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. 2005. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J Virol 79:3938–3948. doi: 10.1128/JVI.79.7.3938-3948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos SK. 2017. Subcellular trafficking of the papillomavirus genome during initial infection: the remarkable abilities of minor capsid protein L2. Viruses 9:E370. doi: 10.3390/v9120370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. 2013. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol 87:3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, DiMaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiGiuseppe S, Bienkowska-Haba M, Guion LGM, Keiffer TR, Sapp M. 2017. Human papillomavirus major capsid protein L1 remains associated with the incoming viral genome throughout the entry process. J Virol 9:e00537-17. doi: 10.1128/JVI.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Kazakov T, Popa A, DiMaio D. 2014. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires gamma-secretase activity. mBio 5:e01777-14. doi: 10.1128/mBio.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiGiuseppe S, Luszczek W, Keiffer TR, Bienkowska-Haba M, Guion LG, Sapp MJ. 2016. Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis. Proc Natl Acad Sci U S A 113:6289–6294. doi: 10.1073/pnas.1600638113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley MA. 2012. Epithelial cell responses to infection with human papillomavirus. Clin Microbiol Rev 25:215–222. doi: 10.1128/CMR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day PM, Pang YY, Kines RC, Thompson CD, Lowy DR, Schiller JT. 2012. A human papillomavirus (HPV) in vitro neutralization assay that recapitulates the in vitro process of infection provides a sensitive measure of HPV L2 infection-inhibiting antibodies. Clin Vaccine Immunol 19:1075–1082. doi: 10.1128/CVI.00139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JW, Roden RB. 2013. L2, the minor capsid protein of papillomavirus. Virology 445:175–186. doi: 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2004. Efficient intracellular assembly of papillomaviral vectors. J Virol 78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards RM, Lowy DR, Schiller JT, Day PM. 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A 103:1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bienkowska-Haba M, Patel HD, Sapp M. 2009. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog 5:e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karanam B, Peng S, Li T, Buck C, Day PM, Roden RB. 2010. Papillomavirus infection requires gamma secretase. J Virol 84:10661–10670. doi: 10.1128/JVI.01081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kines RC, Varsavsky I, Choudhary S, Bhattacharya D, Spring S, McLaughlin R, Kang SJ, Grossniklaus HE, Vavvas D, Monks S, MacDougall JR, de Los Pinos E, Schiller JT. 2018. An infrared dye-conjugated virus-like particle for the treatment of primary uveal melanoma. Mol Cancer Ther 17:565–574. doi: 10.1158/1535-7163.MCT-17-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day PM, Baker CC, Lowy DR, Schiller JT. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci U S A 101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bienkowska-Haba M, Williams C, Kim SM, Garcea RL, Sapp M. 2012. Cyclophilins facilitate dissociation of the human papillomavirus type 16 capsid protein L1 from the L2/DNA complex following virus entry. J Virol 86:9875–9887. doi: 10.1128/JVI.00980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiGiuseppe S, Bienkowska-Haba M, Hilbig L, Sapp M. 2014. The nuclear retention signal of HPV16 L2 protein is essential for incoming viral genome to transverse the trans-Golgi network. Virology 458–459:93–105. doi: 10.1016/j.virol.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 24.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 25.Scherer EM, Smith RA, Simonich CA, Niyonzima N, Carter JJ, Galloway DA. 2014. Characteristics of memory B cells elicited by a highly efficacious HPV vaccine in subjects with no pre-existing immunity. PLoS Pathog 10:e1004461. doi: 10.1371/journal.ppat.1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spoden G, Kuhling L, Cordes N, Frenzel B, Sapp M, Boller K, Florin L, Schelhaas M. 2013. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J Virol 87:7765–7773. doi: 10.1128/JVI.00370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Day PM, Thompson CD, Lowy DR, Schiller JT. 2015. The HPV16 and MusPV1 papillomaviruses initially interact with distinct host components on the basement membrane. Virology 481:79–94. doi: 10.1016/j.virol.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day PM, Thompson CD, Lowy DR, Schiller JT. 2017. Interferon gamma prevents infectious entry of human papillomavirus 16 via an L2-dependent mechanism. J Virol 91:e00168-17. doi: 10.1128/JVI.00168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. 2015. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog 11:e1004699. doi: 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddiqa A, Broniarczyk J, Banks L. 2018. Papillomaviruses and endocytic trafficking. Int J Mol Sci 19:E2619. doi: 10.3390/ijms19092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kuhbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A, Schelhaas M. 2014. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog 10:e1004162. doi: 10.1371/journal.ppat.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerqueira C, Samperio Ventayol P, Vogeley C, Schelhaas M. 2015. Kallikrein-8 proteolytically processes human papillomaviruses in the extracellular space to facilitate entry into host cells. J Virol 89:7038–7052. doi: 10.1128/JVI.00234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aydin I, Villalonga-Planells R, Greune L, Bronnimann MP, Calton CM, Becker M, Lai KY, Campos SK, Schmidt MA, Schelhaas M. 2017. A central region in the minor capsid protein of papillomaviruses facilitates viral genome tethering and membrane penetration for mitotic nuclear entry. PLoS Pathog 13:e1006308. doi: 10.1371/journal.ppat.1006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue T, Zhang P, Zhang W, Goodner-Bingham K, Dupzyk A, DiMaio D, Tsai B. 2018. Gamma-secretase promotes membrane insertion of the human papillomavirus L2 capsid protein during virus infection. J Cell Biol 217:3545–3559. doi: 10.1083/jcb.201804171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts JN, Buck CB, Thompson CD, Kines R, Bernardo M, Choyke PL, Lowy DR, Schiller JT. 2007. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat Med 13:857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- 36.Kines RC, Thompson CD, Lowy DR, Schiller JT, Day PM. 2009. The initial steps leading to papillomavirus infection occur on the basement membrane prior to cell surface binding. Proc Natl Acad Sci U S A 106:20458–20463. doi: 10.1073/pnas.0908502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson KM, Kines RC, Roberts JN, Lowy DR, Schiller JT, Day PM. 2009. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J Virol 83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schelhaas M, Shah B, Holzer M, Blattmann P, Kuhling L, Day PM, Schiller JT, Helenius A. 2012. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog 8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen S, Au S, Pante N. 2011. How viruses access the nucleus. Biochim Biophys Acta 1813:1634–1645. doi: 10.1016/j.bbamcr.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Fay N, Pante N. 2015. Nuclear entry of DNA viruses. Front Microbiol 6:467. doi: 10.3389/fmicb.2015.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elis E, Ehrlich M, Prizan-Ravid A, Laham-Karam N, Bacharach E. 2012. p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes. PLoS Pathog 8:e1003103. doi: 10.1371/journal.ppat.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day PM, Kines RC, Thompson CD, Jagu S, Roden RB, Lowy DR, Schiller JT. 2010. In vivo mechanisms of vaccine-induced protection against HPV infection. Cell Host Microbe 8:260–270. doi: 10.1016/j.chom.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calton CM, Schlegel AM, Chapman JA, Campos SK. 2013. Human papillomavirus type 16 does not require cathepsin L or B for infection. J Gen Virol 94:1865–1869. doi: 10.1099/vir.0.053694-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P, Monteiro da Silva G, Deatherage C, Burd C, DiMaio D. 2018. Cell-penetrating peptide mediates intracellular membrane passage of human papillomavirus L2 protein to trigger retrograde trafficking. Cell 174:1465.e13–1476.e13. doi: 10.1016/j.cell.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merle E, Rose R, LeRoux L, Moroianu J. 1999. Nuclear import of HPV11 L1 capsid protein is mediated by karyopherin alpha2beta1 heterodimers. J Cell Biochem 74:628–637. doi:. [DOI] [PubMed] [Google Scholar]
- 46.Handisurya A, Day PM, Thompson CD, Buck CB, Pang YY, Lowy DR, Schiller JT. 2013. Characterization of Mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J Virol 87:13214–13225. doi: 10.1128/JVI.02162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Day PM, Thompson CD, Buck CB, Pang YY, Lowy DR, Schiller JT. 2007. Neutralization of human papillomavirus with monoclonal antibodies reveals different mechanisms of inhibition. J Virol 81:8784–8792. doi: 10.1128/JVI.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pastrana DV, Buck CB, Pang YY, Thompson CD, Castle PE, FitzGerald PC, Kruger Kjaer S, Lowy DR, Schiller JT. 2004. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 321:205–216. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 49.Nagashima K, Zheng J, Parmiter D, Patri AK. 2011. Biological tissue and cell culture specimen preparation for TEM nanoparticle characterization. Methods Mol Biol 697:83–91. doi: 10.1007/978-1-60327-198-1_8. [DOI] [PubMed] [Google Scholar]