Currently, there is no licensed vaccine against CHIKV infection. An ideal CHIKV vaccine should generate an optimal balance between efficacy and safety. Live attenuated vaccines that can elicit strong immune responses often involve a trade-off of reduced safety. Here, a novel live attenuated vaccine candidate for CHIKV lacking the entire capsid gene, ΔC-CHIKV, was developed. It was demonstrated to be genetically stable, highly attenuated, immunogenic, and able to confer complete protection against lethal CHIKV challenge after a single dose of immunization. Such an infectious vaccine candidate devoid of capsid provides a novel strategy for the development of a live attenuated CHIKV vaccine.
KEYWORDS: chikungunya virus, alphavirus, capsid, vaccines
ABSTRACT
Chikungunya virus (CHIKV) is a mosquito-borne alphavirus that causes epidemics of debilitating disease worldwide. Currently, there are no licensed vaccines or antivirals available against CHIKV infection. In this study, we generated a novel live attenuated vaccine (LAV) candidate for CHIKV with a complete deficiency of capsid (ΔC-CHIKV). It could propagate in BHK-21 cells, and had antigenic properties similar to those of native CHIKV. Vaccination of either immunocompromised IFNAR−/− mice or immunocompetent C57BL/6 mice with a single dose of ΔC-CHIKV conferred complete protection upon challenge with wild-type (WT) CHIKV. Taken together, this vaccine candidate appeared to be safe and efficacious, representing a novel strategy for CHIKV vaccine design.
IMPORTANCE Currently, there is no licensed vaccine against CHIKV infection. An ideal CHIKV vaccine should generate an optimal balance between efficacy and safety. Live attenuated vaccines that can elicit strong immune responses often involve a trade-off of reduced safety. Here, a novel live attenuated vaccine candidate for CHIKV lacking the entire capsid gene, ΔC-CHIKV, was developed. It was demonstrated to be genetically stable, highly attenuated, immunogenic, and able to confer complete protection against lethal CHIKV challenge after a single dose of immunization. Such an infectious vaccine candidate devoid of capsid provides a novel strategy for the development of a live attenuated CHIKV vaccine.
INTRODUCTION
Chikungunya virus (CHIKV) is an important reemerging mosquito-transmitted pathogen within the Alphavirus genus of the Togaviridae family. Its genome is a single-stranded positive RNA of ∼12 kb with a 5′ cap and a 3′ poly(A) tail. Four nonstructural proteins (nsP1 to -4) encoded in the 5′ two-thirds of the CHIKV genome are responsible for viral replication and transcription. A 26S subgenomic mRNA is transcribed from the remaining 3′ one-third of the genome with a subgenomic promoter that is present on the full-length negative-stranded RNA replication intermediate. The subgenomic RNA (sgRNA) then encodes five structural proteins (capsid [C], E3, E2, 6K/TF, and E1) responsible for the production of infectious virions. CHIKV virions are spherical, enveloped particles of ∼70 nm in diameter. The E1 and E2 glycoproteins form heterodimers and assemble into spikes on the surface. At the center of the virion is the nucleocapsid (NC) core of ∼35 nm in diameter, which is composed of the C protein in complex with the viral genome (1).
CHIKV generally causes high fever, headache, rashes, myalgia, arthralgia, and occasionally crippling arthritis that may persist for months or even years (2). More severe symptoms, including encephalitis, hemorrhagic disease, and mortality, have also been reported during recent epidemics (3). Additionally, perinatal CHIKV infection with severe outcomes has been reported recently (4–7), such as neonatal encephalitis, microcephaly, long-term disabilities, and neonatal death. Before 2004, CHIKV was mainly restricted to sub-Saharan Africa and Southeast Asia. Since a large outbreak started in Kenya in 2004, it has been rapidly spreading throughout Asia, Africa, Europe, and the Americas (8–11), making it a global health threat. To date, neither licensed vaccines nor specific antiviral drugs are available to prevent CHIKV infection.
Different types of CHIKV vaccines have recently been developed, such as subunit vaccines (12), viruslike particles (VLPs) (13), formalin-inactivated vaccines (14), DNA vaccines (15), pseudoinfectious virus (16), a single-dose insect alphavirus-vectored vaccine (17), and different live attenuated vaccines (LAVs). Among these, as LAVs can mimic natural viral infection, they could efficiently stimulate a robust and sustained immune response in the body. Thus, typically, one dose is sufficient to confer long-term and even lifelong protection. But sometimes there are some safety issues associated with the use of LAVs. In an effort to achieve the optimal balance between immunogenetics and safety, researchers have recently taken different strategies for the development of live attenuated CHIKV vaccines. For instance, various live attenuated CHIKV vaccines (18–21) were constructed through the introduction of mutations or deletions within capsid (19), 6K (20), E2 (18, 22), and nsP3 (20) or the replacement of the subgenomic promoter with an internal ribosome entry site (IRES) in the CHIKV genome to drive viral structural protein expression (21). Another strategy for CHIKV live-vaccine development is the construction of chimeric viruses by utilizing other viral vaccine backbones or replication-defective virus. So far, insect-specific alphaviruses, including Eilat virus (17), Venezuelan equine encephalitis virus (23), vesicular stomatitis virus (24), modified vaccinia virus Ankara (MVA) (25–27), adenovirus (28), and the measles virus vaccine Schwarz strain (MV/Schw) (29), have been explored as potential viral vectors for chimeric CHIKV construction.
In this study, we produced and characterized a new type of infectious CHIKV particle with a complete viral capsid deletion (ΔC-CHIKV). It was found that the capsid protein, an exclusive requirement for NC formation of infectious wild-type (WT) CHIKV particles, was dispensable for the assembly of infectious ΔC-CHIKV particles. Although the underlying mechanism of ΔC-CHIKV formation was not clear, we found that ΔC-CHIKV was highly attenuated in immunocompromised IFNAR−/− mice compared with WT CHIKV. A single-dose immunization of ΔC-CHIKV protected immunocompetent C57BL/6 mice and immunocompromised IFNAR−/− mice from challenge with WT CHIKV; these mice developed no disease signs and no detectable viremia during viral infection. Our results show that ΔC-CHIKV is safe as a live attenuated vaccine and could be applied as a new strategy for CHIKV vaccine design.
RESULTS
ΔC-CHIKV can propagate in BHK-21 cells.
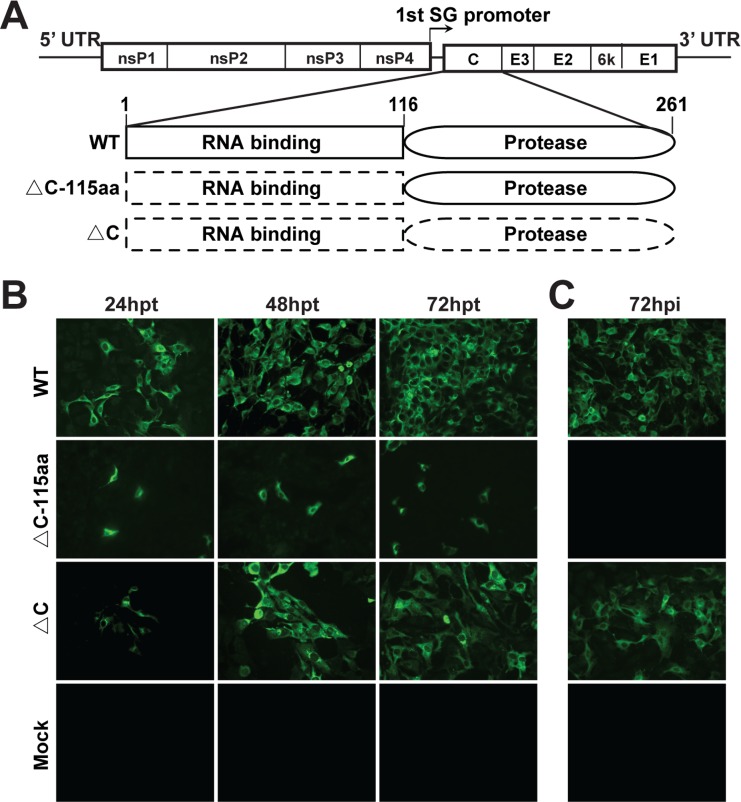
Several lines of evidence have shown that capsid is also a key determinant of alphavirus virulence (30–33). We wondered whether a CHIKV mutant with a partial or even a complete deletion of capsid could be a live attenuated vaccine candidate. Thus, a CHIKV mutant harboring a complete deletion of the capsid gene (ΔC-CHIKV) was constructed. In parallel, a CHIKV construct lacking the N-terminal RNA binding domain of capsid (ΔC-CHIKV-115aa) was also generated. Equal amounts of in vitro-transcribed WT and mutant genomic RNAs were transfected into BHK-21 cells. Viral replication and spread were evaluated by an immunofluorescence assay (IFA) using an anti-E2 polyclonal antibody. ΔC-CHIKV RNA produced increasing numbers of IFA-positive cells from 24 to 72 h posttransfection (hpt) although fewer than those produced by WT CHIKV RNA at each time point (Fig. 1B). The spread of IFA-positive cells indicated the production of infectious viruses from transfected cells. In contrast, only scattered IFA-positive cells were found in ΔC-CHIKV-115aa RNA-transfected cells at each time point (Fig. 1B), indicating that ΔC-CHIKV-115aa mutant RNA was replicative but failed to produce infectious viruses. To further analyze the infectivity of the supernatants from mutant RNA-transfected cells, the culture media obtained at 72 hpt were used to infect naive BHK-21 cells. As shown in Fig. 1C, only cells infected with WT CHIKV and ΔC-CHIKV supernatants produced IFA-positive cells. Our results indicate that ΔC-CHIKV RNA can produce infectious particles in BHK-21 cells.
FIG 1.
CHIKV lacking the full sequence of capsid (ΔC-CHIKV) is able to produce infectious viruses. (A) Schematic representation of different CHIKV capsid deletion mutants in the context of the CHIKV genome. The WT capsid contains an N-terminal RNA binding domain (represented by a rectangular box) and a C-terminal serine protease domain (represented by oval boxes). Dashed boxes indicate the deleted segments. Amino acid positions are indicated. SG, subgenomic. (B) IFA of RNA-transfected BHK-21 cells. Equal amounts of WT or mutant genomic RNAs were transfected into BHK-21 cells. At the indicated times posttransfection, the expression of viral E2 protein was detected by an IFA. (C) IFA of infected BHK-21 cells. Naive BHK-21 cells were infected with the supernatants harvested from WT or mutant genomic RNA-transfected BHK-21 cells at 72 hpt. At 72 hpi, the expression of viral E2 protein was analyzed by an IFA.
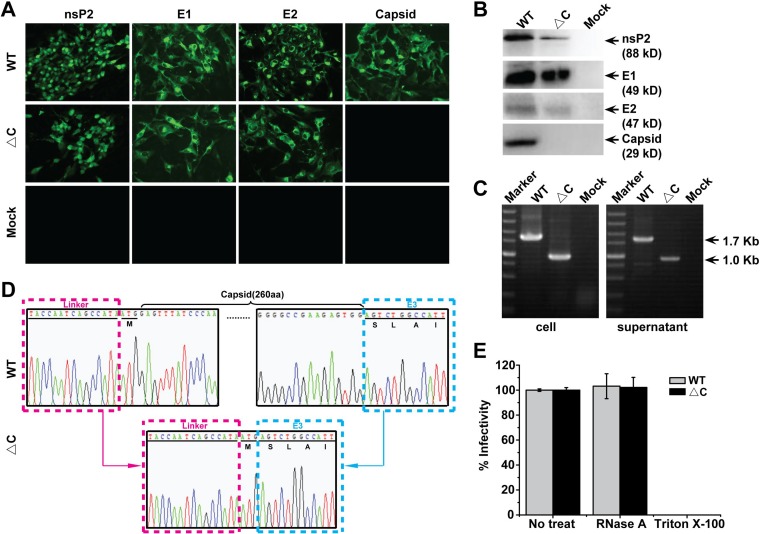
To exclude the possibility that the infectivity of ΔC-CHIKV might be WT CHIKV contamination, the absence of capsid in ΔC-CHIKV-infected cells was confirmed with different methods, as described below. IFAs and Western blot (WB) assays were first carried out using specific antibodies against different viral proteins. Capsid is the only viral protein that failed to be detected in ΔC-CHIKV-infected cells compared with WT CHIKV (Fig. 2A and B). Meanwhile, viral RNAs were extracted from culture media and infected cells for reverse transcriptase PCR (RT-PCR) spanning the region from the C terminus of nsP4 to the N terminus of E3 with specific primers that cover the complete coding sequence of the capsid gene. The expected 1.7- and 1.0-kb RT-PCR products were detected in WT and ΔC-CHIKV samples, respectively (Fig. 2C). Complete viral genome sequencing also provided evidence that complete capsid coding sequences are absent in the ΔC-CHIKV RNA genome (Fig. 2D). Additionally, we found that RNase A treatment had no effect on the infectivity of WT CHIKV and ΔC-CHIKV, and Triton X-100 treatment completely destroyed the infectivity of both viruses (Fig. 2E), which ruled out the possibility that RNA carryover from transfected cells generates virus transmission to new cultures. Overall, we confirm that ΔC-CHIKV is infectious.
FIG 2.
Confirmation of capsid deletion in infectious ΔC-CHIKV mutants. (A and B) Detection of viral protein expression (nsP2, E1, E2, and capsid) in infected cells by IFAs and Western blot assays. BHK-21 cells were infected with the supernatant that was collected from either WT CHIKV or ΔC-CHIKV RNA-transfected BHK-21 cells at 72 hpt. At 72 hpi, cells were subjected to IFAs (A) and Western blot assays (B) to analyze the expression of different viral proteins. (C) Detection of the expression of the capsid gene in infected cells and culture media by an RT-PCR assay. Viral RNAs were extracted from infected cells and culture media. RT-PCR was performed with the primer pair spanning the nsP4-E3 region. The resulting RT-PCR products were resolved by 1% agarose gel electrophoresis. The expected 1.7-kb and 1.0-kb bands were observed for WT CHIKV and ΔC-CHIKV, respectively. (D) Sequence chromatograms of the N terminus of capsid. (E) Effects of different agents on the infectivity of infectious WT CHIKV or ΔC-CHIKV. WT CHIKV-eGFP or ΔC-CHIKV-eGFP (MOI = 0.01) was incubated with 5 μg/ml RNase A during 20 min at 25°C or with 0.1% Triton X-100 during 1 h at 25°C, respectively. The percent infectivity in each case was calculated by dividing the infectivity of untreated samples (No treat). The data are the means ± standard deviations (SD) from at least three independent experiments.
Characterization of ΔC-CHIKV.
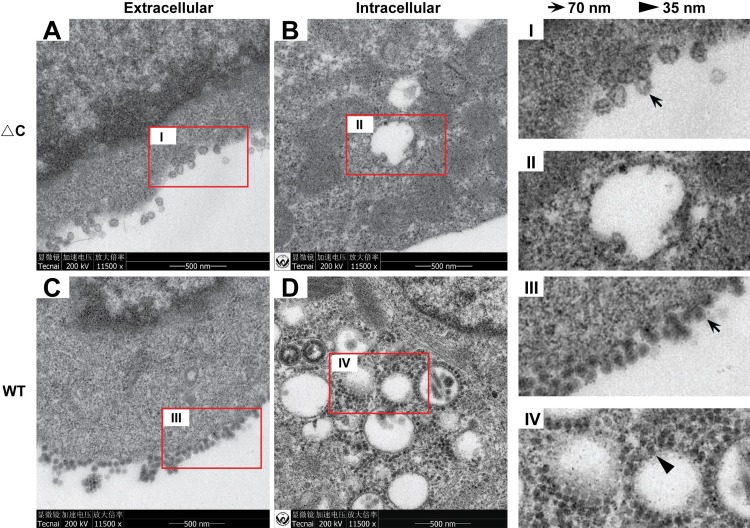
The morphologies of ΔC-CHIKV viral particles during viral infection were first examined by thin-section transmission electron microscopy (TEM) in BHK-21 cells (Fig. 3). Abundant viral particles were observed at the cell surface of ΔC-CHIKV (Fig. 3A)- and WT CHIKV (Fig. 3C)-infected cells. ΔC-CHIKV was irregular, although the particle size was similar to that of WT CHIKV. ΔC-CHIKV showed a core in the center that was much less electron dense, due to the lack of capsid protein, than the WT virus. Interestingly, typical NC structures (35 nm in diameter) were not detected within the cytoplasm of ΔC-CHIKV (Fig. 3B)-infected BHK-21 cells, in contrast to WT CHIKV (Fig. 3D), which was consistent with the capsid deletion in ΔC-CHIKV. Overall, our results demonstrate that the assembly of infectious ΔC-CHIKV particles can be completed without NC formation.
FIG 3.
TEM analysis of ΔC-CHIKV in infected BHK-21 cells. BHK-21 cells were infected with WT CHIKV or ΔC-CHIKV at an MOI of 0.01. WT CHIKV- and ΔC-CHIKV-infected cells were fixed for TEM analysis at 24 hpi and 84 hpi, respectively. Representative TEM images observed for WT (C and D)- or ΔC-CHIKV (A and B)-infected BHK-21 cells are shown. Budding viruses and NCs are indicated with arrows and arrowheads, respectively.
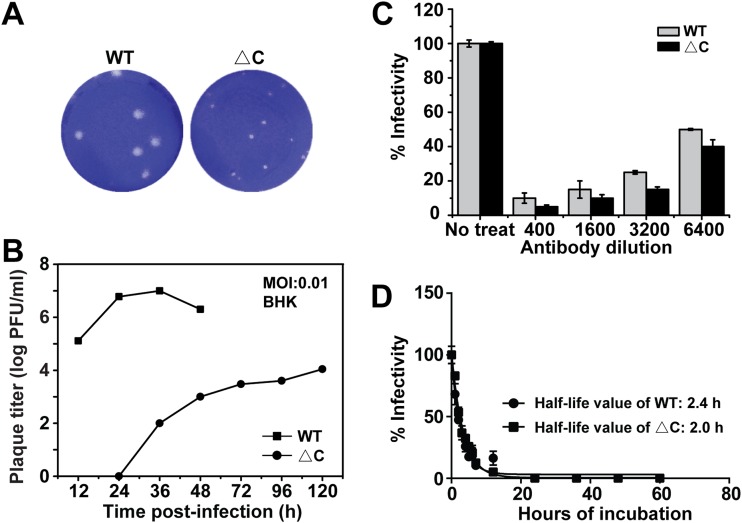
Viral growth and plaque morphology were then compared between the WT virus and ΔC-CHIKV. The culture media were collected from infected BHK-21 cells at different time points postinfection and subjected to a plaque assay. ΔC-CHIKV produced smaller plaques than the WT virus (Fig. 4A), with the highest viral titer of ∼104 PFU/ml at 120 h postinfection (hpi), about 1,000-fold lower than that of the WT (Fig. 4B). Neutralization assay showed that sera against CHIKV blocked viral infection by ΔC-CHIKV and WT CHIKV in a dose-dependent manner (Fig. 4C), indicating that ΔC-CHIKV had an antigenicity similar to that of WT CHIKV. Thermal stability assays illustrate that ΔC-CHIKV has a stability comparable to that of WT CHIKV at 37°C (Fig. 4D).
FIG 4.
Characterization of biophysical properties of ΔC-CHIKV virions. (A) Plaque morphology of WT CHIKV and ΔC-CHIKV on BHK-21 cells. (B) Growth kinetics of WT CHIKV and ΔC-CHIKV. BHK-21 cells were infected with WT CHIKV or ΔC-CHIKV mutants at an MOI of 0.01. Viral titers were measured by a plaque assay on BHK-21 cells. (C) Neutralization assay using mouse sera against WT CHIKV conducted with a PRNT. (D) Thermal stability of WT and ΔC-CHIKV. The corresponding eGFP reporter viruses, WT CHIKV-eGFP and ΔC-CHIKV-eGFP, were used as described in Materials and Methods. Shown are mean half-life values obtained from three independent experiments performed in triplicate. The bars indicate the standard deviations from three independent experiments.
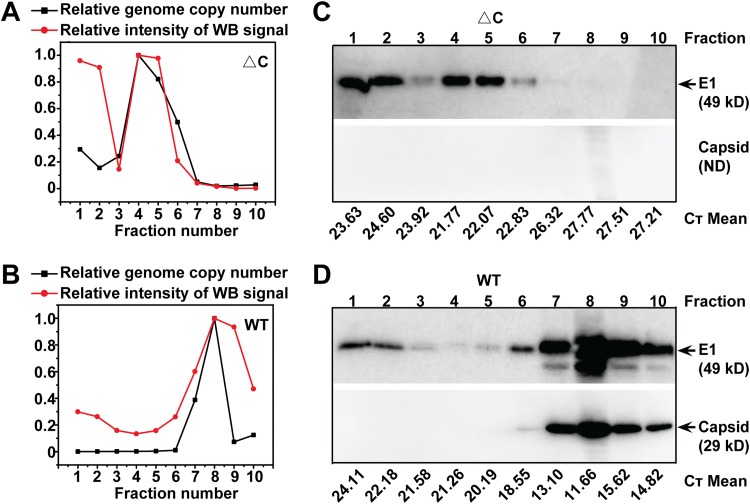
We also compared viral particle densities by sucrose density gradient ultracentrifugation analysis (Fig. 5). The supernatants from infected BHK-21 cells were collected at the indicated times postinfection for sedimentation in sucrose gradients. Each fraction from top to bottom was collected (fractions 1 to 10) and subjected to WB and quantitative RT-PCR (qRT-PCR) assays for quantification of viral proteins and RNA, respectively. As shown in Fig. 5A and B, ΔC-CHIKV had a lower density than WT CHIKV, residing in fraction 4, versus the WT residing in fraction 8, which also confirmed that viral RNAs were truly incorporated into viral particles of ΔC-CHIKV. On the other hand, it was found that viral envelope protein was present in fractions 1 and 2 in samples of both viruses (Fig. 5C and D). This suggested that there might always be some empty viruslike particle formation, concomitant with mature virion budding. Interestingly, the proportion of fraction 1/2 envelope proteins in WT CHIKV samples was lower than that for ΔC-CHIKV, indicating that the presence of capsid may guarantee efficient utilization of envelope proteins for the assembly of mature virions.
FIG 5.
Analysis of the density of purified ΔC-CHIKV virions. Shown are data for sucrose density gradient fractionation of WT and ΔC-CHIKV virions produced in BHK-21 cells. Cell-free virions were harvested, PEG 8000 concentrated, and separated on 20% to 60% linear sucrose density gradients. Ten fractions were harvested from the top (fraction 1) to the bottom (fraction 10) of the gradient. Each fraction from WT CHIKV (B and D) and ΔC-CHIKV (A and C) was subjected to qRT-PCR and E1-specific antibody-based Western blot assays to monitor the distribution of virions. ImageJ software was used to analyze the intensity of the viral E1 protein band in each fraction. The graphs (A and B) represent quantification of percent viral RNA/E1 protein in each fraction normalized to the largest amount of viral RNA/E1 protein. Three independent experiments were performed, with similar results, and one of the representative data sets is presented. CT, threshold cycle.
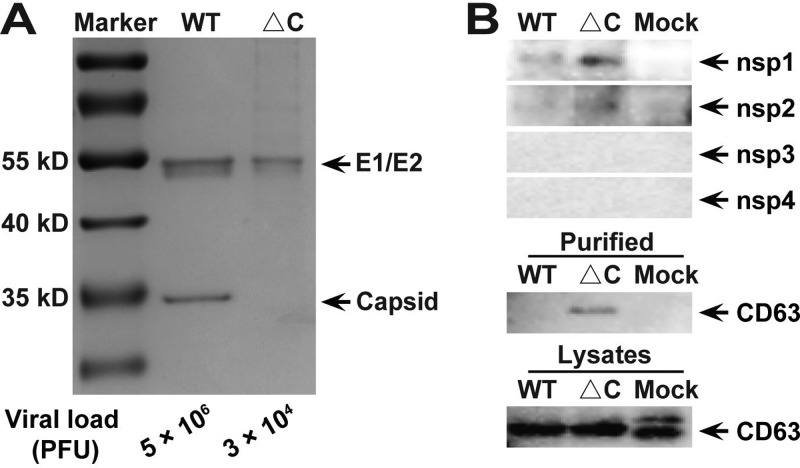
The compositions of purified viral particles were analyzed by SDS-PAGE using fraction 4 of ΔC-CHIKV and fraction 8 of WT CHIKV (Fig. 6A). As expected, capsid proteins were absent in ΔC-CHIKV, in contrast to WT CHIKV, and the envelope proteins E1 and E2 were detected in both WT CHIKV and ΔC-CHIKV. It is worth noting that different amounts of viruses (3 × 104 PFU ΔC-CHIKV versus 5 × 106 PFU WT CHIKV) were loaded onto the gel to produce similar signals in Fig. 6A. This suggested that the ratio of total particles to infectious particles for ΔC-CHIKV was higher than that for WT CHIKV, which indicated that many viral particles of ΔC-CHIKV are not infectious.
FIG 6.
Analysis of the composition of purified ΔC-CHIKV virions. (A) SDS-PAGE analysis followed by Coomassie staining for fraction 8 of WT CHIKV and fraction 4 of ΔC-CHIKV by sucrose density gradient centrifugation. Gels were loaded with purified ΔC-CHIKV at approximately 3 × 104 PFU or an equivalent volume of purified WT CHIKV at approximately 5 × 106 PFU. (B) Western blot assays to analyze viral nsPs with antiserum specifically targeting CHIKV nsP1 to nsP4 and exosomes with marker CD63 antibody of purified ΔC-CHIKV and WT CHIKV virions. Gels were loaded with purified ΔC-CHIKV or WT CHIKV as described above for panel A, and cell lysates for the detection of ΔC-CHIKV or WT CHIKV or mock-infected cells were adjusted for similar signals. Three independent experiments were performed, with similar results, and one of the representative data sets is presented.
We then analyzed all nsPs with purified ΔC-CHIKV and WT CHIKV. Only trace amounts of nsP1 and nsP2 were observed for both viruses (Fig. 6B), which excluded the possibility that the infectivity of ΔC-CHIKV was derived from viral replication complexes (RCs) containing viral nsPs and double-stranded RNA. Since the size of ΔC-CHIKV (70 nm) was within the size range of the exosome (50 to 100 nm) (34), we also analyzed ΔC-CHIKV with the exosome marker CD63. As shown in Fig. 6B, trace amounts of CD63 were detected in ΔC-CHIKV instead of WT CHIKV, which indicated that the exosome pathway may involve infectious particle formation by ΔC-CHIKV.
ΔC-CHIKV is considerably stable in cell culture.
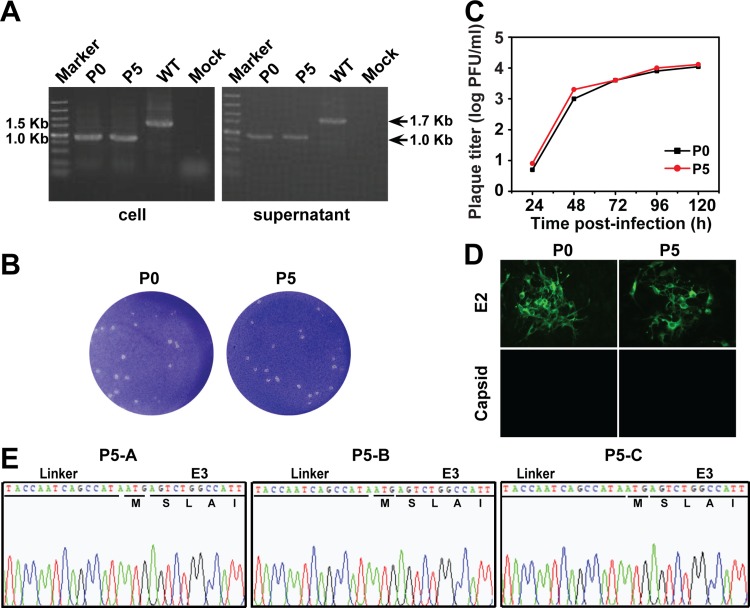
To analyze the genetic stability of ΔC-CHIKV, three independent passages (passages A, B, and C) were performed on BHK-21 cells (Fig. 7). After 5 rounds of passage, the growth curve (Fig. 7C), plaque morphology (Fig. 7B), and E2/capsid protein expression (Fig. 7D) of ΔC-CHIKV were not changed on BHK-21 cells. Using the primer pair spanning the nsP4 and E3 regions, an identical RT-PCR product was detected for both P0 (i.e., viruses in supernatant from ΔC-CHIKV RNA-transfected BHK-21 cells) and P5 virus samples (Fig. 7A). Moreover, the results of sequencing showed that the engineered capsid deletion still remained in all three passaged viruses (Fig. 7E). These results show that ΔC-CHIKV is relatively stable.
FIG 7.
Stability of ΔC-CHIKV in cell culture. (A) Detection of viral genome stability during passage by RT-PCR. Viral RNAs were extracted from infected cells or supernatants as described in Materials and Methods. RT-PCR was performed with a pair of primers spanning the nsP4-E3 region. (B) Plaque morphology comparison between P0 and P5 ΔC-CHIKV. (C) Comparison of growth kinetics of P0 and P5 ΔC-CHIKV mutants. Viral growth curves were conducted at an MOI of 0.01. Three independent experiments were performed in duplicate, and representative data are presented. (D) IFA of expression of different viral proteins in P0 and P5 ΔC-CHIKV using polyclonal antibodies against E2 and capsid. (E) Sequence chromatograms of all three passaged viruses (P5-A, P5-B, and P5-C). The initial amino acids of the E3 gene are indicated.
ΔC-CHIKV is highly attenuated in IFNAR−/− mice.
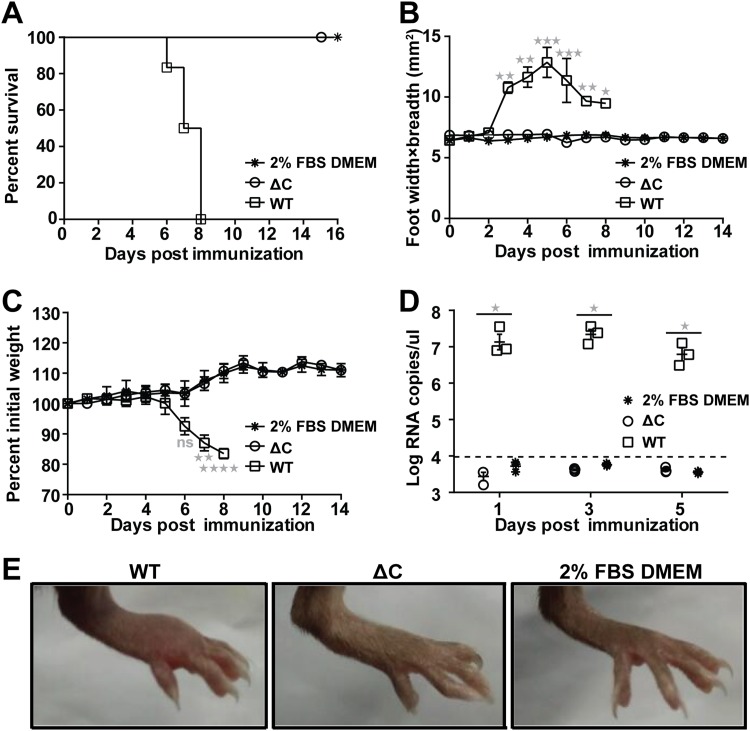
The IFNAR−/− mouse is a commonly used murine model for CHIKV pathogenesis studies (21). It has the advantage of demonstrating disease in adult mice and provides a lethal endpoint upon CHIKV challenge (17). Thus, 6-week-old IFNAR−/− mice were injected subcutaneously (s.c.) in the ventral/lateral side of the hind foot with 102 PFU of WT CHIKV and the highest dose of ΔC-CHIKV that we obtained, 105 PFU, respectively. Dulbecco’s modified Eagle’s medium (DMEM) with 2% fetal bovine serum (FBS) was inoculated as a negative control. All the WT-infected mice died within 8 days postinfection (Fig. 8A), with footpad swelling (Fig. 8B), weight loss (Fig. 8C), and significant viremia (Fig. 8D). In contrast, all the mice infected with ΔC-CHIKV survived (Fig. 8A), without any visible signs of disease during the 14-day observation period. There was no footpad swelling (Fig. 8B), weight loss (Fig. 8C), or viremia (Fig. 8D) observed, in contrast to those observed in WT CHIKV-infected mice. Overall, our results indicate that ΔC-CHIKV is highly attenuated in IFNAR−/− mice.
FIG 8.
Pathogenicity of ΔC-CHIKV determined by studies in the IFNAR−/− mouse model. IFNAR−/− mice (6 weeks old; n = 6 per group) were injected subcutaneously in the ventral/lateral side of the hind foot with 102 PFU of WT CHIKV, 105 PFU of ΔC-CHIKV, and 2% FBS–DMEM (negative control). (A to C) Animal survival (A), footpad swelling (n = 6 feet per group) (B), and weight loss (C) were monitored daily until 14 or 16 days postinoculation. (D) Viremia titer postinoculation. Viremia from day 1 to day 5 postinoculation was measured by qRT-PCR. (E) Images of feet on day 5 postinoculation. In panels B to D, data represent means ± SD for groups of mice, and the horizontal dotted line represents the limit of detection. Student’s t tests were used to determine differences between two groups, and a Kruskal-Wallis test followed by Dunn’s test was used for multiple comparisons. One, two, three, and four asterisks indicate statistical differences between indicated groups with P values of <0.05, <0.01, <0.001, and <0.0001, respectively. ns, not significant.
A single dose of ΔC-CHIKV can elicit protection against CHIKV infection in both IFNAR−/− and C57BL/6 mice.
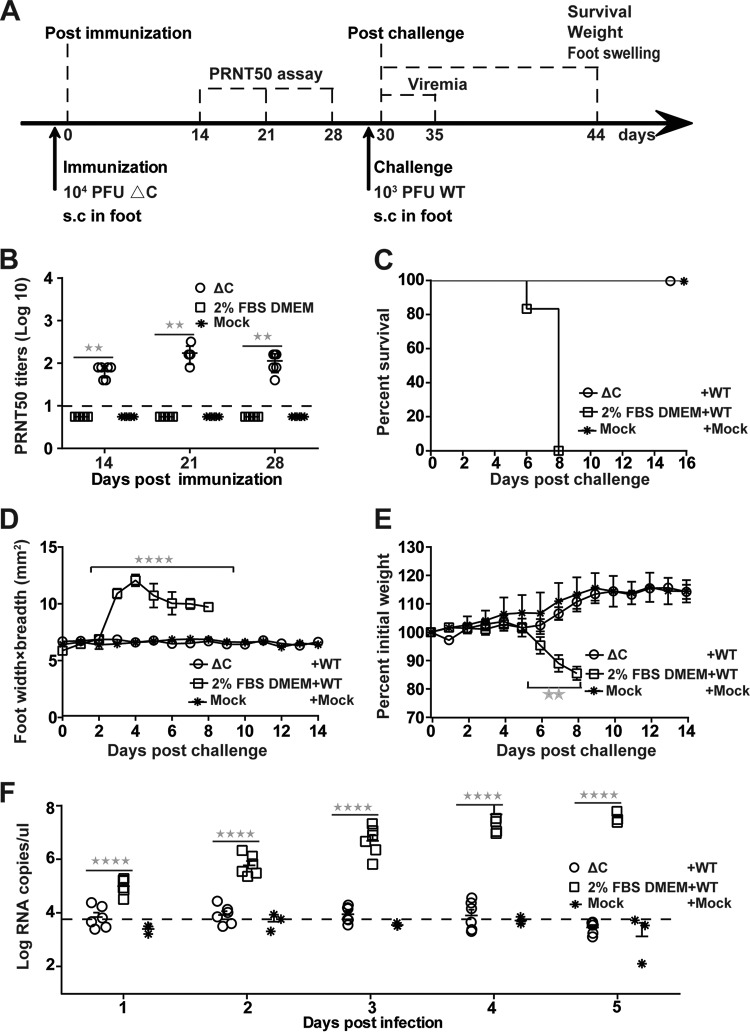
The dramatic attenuation of ΔC-CHIKV in IFNAR−/− mice indicates that ΔC-CHIKV may be a potential CHIKV vaccine candidate. Thus, IFNAR−/− mice were injected s.c. with 104 PFU of ΔC-CHIKV to test its prophylactic efficacy. The negative control was culture medium alone (2% FBS–DMEM immunization) or no treatment (mock immunization). Sera were collected from immunized mice at 7-day intervals over 28 days postimmunization. The serum neutralizing ability elicited by ΔC-CHIKV was assessed by a plaque reduction or neutralization test (PRNT). Apparent neutralizing antibodies were successfully induced at 14 days postimmunization and remained steady without a dramatic decline at 28 days postvaccination with 50% neutralization of CHIKV at a serum dilution of 1:160 (Fig. 9B), which was lower than other live-attenuated vaccines induced with an 80% plaque reduction/neutralization titer (PRNT80) of ≥1:160 in previous studies when mice were immunized with the same dose (21, 23).
FIG 9.
Single-dose immunization with ΔC-CHIKV protects IFNAR−/− mice from CHIKV infection. (A) Experimental design. Three groups of 6-week-old IFNAR−/− mice (n = 6 per group) were immunized subcutaneously in the ventral/lateral side of the hind foot once with 104 PFU of ΔC-CHIKV or 2% FBS–DMEM (culture medium control) or left untreated (mock). On day 30 postimmunization, mice were challenged with 103 PFU of WT CHIKV s.c. in feet. (B) Neutralizing antibody titers present in mouse sera on the indicated days postimmunization. (C) Animal survival postchallenge. (D) Footpad swelling (n = 6 feet per group) postchallenge. (E) Weight loss after challenge. (F) Viremia postchallenge. Viremia from day 1 to day 5 postchallenge was measured by qRT-PCR. In panels B to F, data represent means ± SD for groups of mice, and the horizontal dotted line represents the limit of detection. Student’s t tests were used to determine differences between two groups, and a Kruskal-Wallis test followed by Dunn’s test was used for multiple comparisons. Two and four asterisks indicate statistical differences between the indicated groups with P values of <0.01 and <0.0001, respectively.
Mice immunized with 2% FBS–DMEM and ΔC-CHIKV were subcutaneously challenged on the ventral/lateral side of the hind foot with 103 PFU of WT CHIKV at 30 days postimmunization and observed daily over a period of 14 days. The mice vaccinated with ΔC-CHIKV were fully protected against challenge with WT CHIKV (Fig. 9C) and remained healthy throughout the course of the experiment, without apparent footpad swelling (Fig. 9D) and weight loss (Fig. 9E). In contrast, 2% FBS–DMEM-vaccinated mice experienced substantial footpad swelling and weight loss beginning at day 3 and day 6 postchallenge, respectively, and died within 8 days postchallenge (Fig. 9C). At the same time, ΔC-CHIKV-immunized mice produced background levels of viremia compared with the 2% FBS–DMEM-immunized group at each time point postchallenge (Fig. 9F).
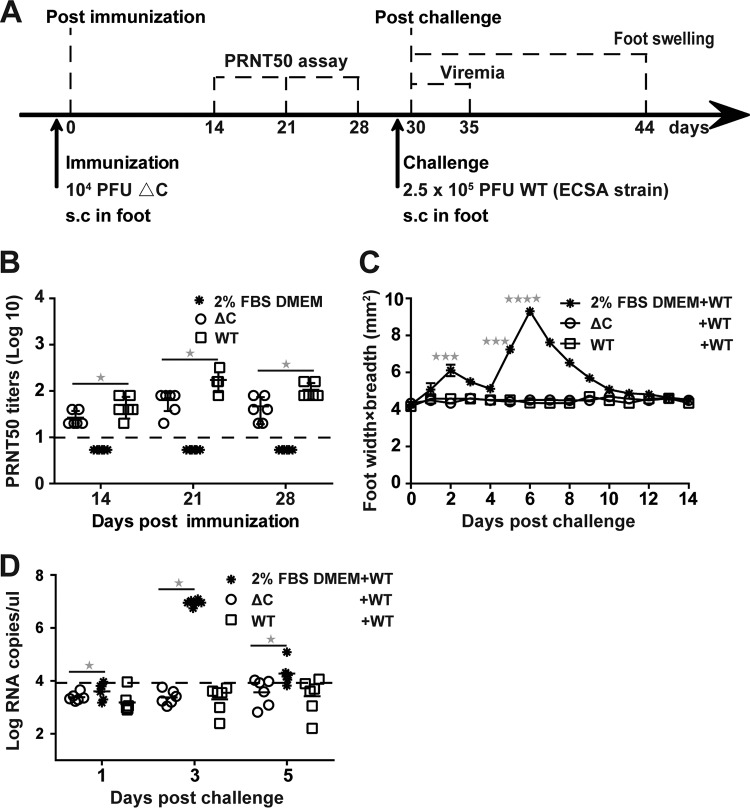
Next, an immunocompetent C57BL/6 mouse model was used to further evaluate the immunogenicity and efficacy of ΔC-CHIKV. Six-week-old C57BL/6 mice were inoculated by the s.c. route with 104 PFU of WT CHIKV or ΔC-CHIKV. Culture medium alone (2% FBS–DMEM) was used as a negative control. Sera were also collected at 7-day intervals over 28 days postimmunization for PRNTs (Fig. 10B). Apparent seroconversion was observed at 14 days postimmunization in all mice immunized with WT CHIKV or ΔC-CHIKV and appeared to increase thereafter until day 21. The neutralizing activities in sera from ΔC-CHIKV-immunized mice were slightly lower than those in sera from mice immunized with WT CHIKV; this may be due to less efficient in vivo spreading of ΔC-CHIKV to stimulate immunization. Nevertheless, ΔC-CHIKV could induce neutralizing antibody as efficiently as WT CHIKV in the C57BL/6 mouse model. At 30 days postimmunization, all the mice were challenged with 2.5 × 105 PFU of WT CHIKV (ECSA strain, a human isolate from Pakistan) (35). Signs of footpad swelling and viremia induced by CHIKV infection were also monitored. No footpad swelling was observed in both WT CHIKV- and ΔC-CHIKV-immunized mice, while two peaks of footpad swelling at approximately 2 and 6 days postchallenge were observed in mock-immunized mice (Fig. 10C). Swelling was fully recovered by day 14 postchallenge (Fig. 10C), which is consistent with previous observations in the CHIKV-infected C57BL/6 mouse model (19, 20, 25). Mock-immunized mice exhibited viremia, which reached a peak at day 3 postchallenge. All mice immunized with WT CHIKV and ΔC-CHIKV were completely protected from viremia (Fig. 10D). Collectively, these data demonstrate that single-dose immunization with ΔC-CHIKV could protect all mice against CHIKV infection.
FIG 10.
Single-dose immunization with ΔC-CHIKV protects C57BL/6 mice from CHIKV infection. (A) Experimental design. Three groups of 6-week-old C57BL/6 mice (n = 6 per group) were immunized subcutaneously in the ventral/lateral side of the hind foot once with either 104 PFU of WT CHIKV or ΔC-CHIKV, respectively. The culture medium (2% FBS–DMEM) was used as a negative control for mock immunization. On day 30 postimmunization, mice were challenged s.c. in feet with 2.5 × 105 PFU of WT CHIKV (ECSA strain). (B) Neutralizing antibody titers present in mouse sera on the indicated days postimmunization. (C) Footpad swelling postchallenge (n = 6 feet per group). (D) Viremia postchallenge. Viremia from day 1 to day 5 postchallenge was measured by qRT-PCR. All the data represent means ± SD for groups of mice, and the horizontal dotted line represents the limit of detection. Student’s t tests were used to determine differences between two groups, and a Kruskal-Wallis test followed by Dunn’s test was used for multiple comparisons. One, three, and four asterisks indicate statistical differences between the indicated groups with P values of <0.05, <0.001, and <0.0001, respectively.
DISCUSSION
In previous studies, viruslike replicon particle (VRP) vaccines were produced through a complementation/packaging system in which alphavirus replicons encoding envelope proteins are packaged into VRPs by capsid protein expressed in trans (36). Although the construct of ΔC-CHIKV is similar to that of replicon-expressing envelope proteins for VRP production (36), the ΔC-CHIKV described here differs from VRPs, because ΔC-CHIKV itself is infectious without capsid protein and the envelopes are expressed directly from ΔC-CHIKV and assembled on the surface of infectious particles. There is no requirement for structural proteins expressed in trans for packaging.
In this study, we found that ΔC-CHIKV genomes with a complete capsid deletion still produce infectious particles without NC structure assembly in BHK-21 cells, although their titers were lower than those of WT viruses (Fig. 4). The results of neutralization assays and Triton X-100 and RNase A treatment assays indicated that ΔC-CHIKV could pack viral genomes into infectious particles with the expression of envelope glycoproteins alone. Besides CHIKV, other alphaviruses, such as Semliki forest virus (SFV) and Sindbis virus (SINV), lacking a complete capsid also produced infectious particles called infectious microvesicles (iMVs) (37). Additionally, infectious viruslike vesicles (VLVs) were generated when cells were transfected with an SFV replicon without SFV structural proteins expressing only vesicular stomatitis virus glycoprotein (VSV-G) (38–40) or murine leukemia virus envelopes (41). The possible mechanism of VLV formation is vesiculation of the plasma membrane to generate particles that include VSV-G and SFV-G RNAs (38). During SFV replicon RNA replication, lightbulb-shaped replication complexes (spherules) assemble at the plasma membrane. VLVs containing SFV RNA and VSV-G bud from the cells and are likely derived from spherules containing VSV-G (42), and the fusion activity of VSV-G is essential for the generation of VLVs. A similar infectious particle without capsid protein has also been reported for murine coronavirus (43). Coronavirus-like particles were produced when envelope protein M was coexpressed with non-coronavirus RNA transcripts containing the short viral packaging signal in the absence of coronavirus capsid protein N. Overall, all these results, including those for ΔC-CHIKV, undoubtedly broaden our knowledge about viral infectious particle formation, showing that viral RNA could be packaged by envelope proteins in the absence of capsid.
CHIKV capsid contains two functional domains: the highly positively charged N-terminal RNA binding domain and the C-terminal serine protease domain (44). Following self-cleavage from the nascent structural polyprotein, the capsid protein specifically interacts with the replicated genomic RNA (49S RNA) to form NCs in the cytoplasm (45). The NCs are then transported to the plasma membrane, where they interact with envelope proteins to drive the budding of virus particles from the plasma membrane (46). Alternatively, previous studies also showed that NC preformation in the cytoplasm is not absolutely essential for virion assembly, and NC assembly might proceed at the plasma membrane concomitantly with virion budding (44, 47, 48). The underlying mechanism of ΔC-CHIKV formation is worth exploring in future studies.
Importantly, ΔC-CHIKV particles were nonpathogenic in a mouse model. The lack of pathogenesis can be interpreted in two ways. One is that infectious particle production by ΔC-CHIKV is not efficient without capsid protein compared with WT CHIKV, which in turn limits the spread of initial infection in cells. Alternatively, the attenuation of ΔC-CHIKV may be due to the absence of capsid. It has been demonstrated that capsid is one of the important virulence determinants of alphaviruses (30–33) and is necessary for the inhibition of host interferon (IFN) gene expression (30). Capsid has become a potential target of rationally designed LAVs for alphaviruses (16, 19, 30, 49), which are highly attenuated and immunogenic and protect hosts against viral infections. Consistent with data from previous studies, ΔC-CHIKV also protects immunized mice from disease after WT CHIKV challenge, demonstrating that ΔC-CHIKV might serve as an LAV candidate.
Stability is one of the greatest concerns for the development of LAVs (50). To assess the stability of ΔC-CHIKV, we passaged it in BHK-21 cells for five rounds. This showed that ΔC-CHIKV remained stable at least within five passages, without detectable genetic instability or phenotypic alterations of growth curves and plaque morphologies (Fig. 7). These results indicated that the risk of reversion to virulence of WT CHIKV from ΔC-CHIKV was low, thus guaranteeing the safety of its usage as a potential LAV.
One drawback of ΔC-CHIKV used as a vaccine in the present study is inefficient production. Only 104 PFU/ml ΔC-CHIKV was obtained in BHK-21 cells, and the production of ΔC-CHIKV in Vero cells was even less efficient (data not shown), which is not suitable for commercial vaccine production at the present stage. This limitation may be resolved by extensive passage in cell culture, which has been demonstrated for the efficient production of VLVs of SFV–VSV-G (40). After 50 rounds of serial passaging, the viral titer of VLV increased from 105 PFU/ml to 5 × 107 PFU/ml. Serial passaging of ΔC-CHIKV is under investigation in our laboratory. Overall, we provide a proof of concept for ΔC-CHIKV as a new type of vaccine.
MATERIALS AND METHODS
Animals.
Two kinds of mouse models were used in this study: 6-week-old immunocompetent C57BL/6 mice and immunocompromised IFNAR−/− mice were provided by the Animal Centre of Wuhan Institute of Virology. All the mice were cared for in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals (51). All animal experiments were conducted in an animal biosafety level 3 (ABSL-3) facility at Wuhan Institute of Virology under a protocol approved by the Laboratory Animal Ethics Committee of Wuhan Institute of Virology, Chinese Academy of Sciences (permit number WIVA26201701).
Cell lines and viruses.
BHK-21 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Germany) containing 10% fetal bovine serum (FBS), 100 U/ml of penicillin, and 100 μg/ml of streptomycin at 37°C with 5% CO2. The WT CHIKV stock was produced from the infectious cDNA clone (52) and stored in aliquots at −80°C.
Antibodies.
Murine polyclonal antiserum against CHIKV capsid proteins and E1 proteins as well as anti-CHIKV E2 rabbit polyclonal antibodies were generated in-house; rabbit polyclonal antisera specifically targeting CHIKV nsP1 to nsP4 were kindly provided by Andres Merits (Institute of Technology, University of Tartu, Estonia). Murine monoclonal antibodies specific for mouse CD63 were purchased from Abcam. Fluorescein isothiocyanate (FITC)/horseradish peroxidase (HRP)-conjugated goat anti-mouse/rabbit IgG was purchased from Proteintech (China).
Plasmid construction.
Standard recombinant DNA techniques were applied for the construction of all plasmids. The wild-type (WT) infectious cDNA clone of CHIKV (pACYC-CHIKV) and its enhanced green fluorescent protein (eGFP) reporter vector (eGFP-dual-sg-CHIKV) (52) were used as the backbone for engineering capsid deletion mutants. For the plasmid pACYC-ΔC-115aa-CHIKV, a fragment corresponding to the region from nsP4 to E2 lacking the RNA binding domain of capsid (amino acids [aa] 2 to 116) was amplified using overlap PCR and introduced into pACYC-CHIKV at the restriction enzyme sites RsrII and NdeI. Using the same strategy, the pACYC-ΔC-CHIKV plasmid with a whole deletion of the capsid gene (aa 2 to 261) was generated. The eGFP reporter virus vector lacking the full sequence of capsid (aa 2 to 261), ΔC-CHIKV-eGFP, was constructed as follows: a fragment containing a repeated subgenomic promoter and eGFP gene amplified from the eGFP-dual-sg-CHIKV plasmid was amplified via overlap PCR and introduced into the plasmid pACYC-ΔC-CHIKV between E2 and the 3′ untranslated region (UTR) at the restriction enzyme sites NdeI and BamHI. All the plasmids were confirmed by sequencing before the following assays.
RNA transcription and transfection.
The cDNA plasmids were subjected to sequential BamHI linearization and in vitro transcription using an mMessenger mMachine T7 kit (Ambion, USA) according to the manufacturer’s protocols. The recombinant viral RNAs were then transfected into BHK-21 cells with DMRIE-C (Invitrogen, USA) according to the manufacturer’s instructions. At different time points posttransfection, the supernatants containing viruses were collected, aliquoted, and stored at −80°C for the following experiments.
Plaque assay.
BHK-21 cells were seeded into 24-well plates at a density of 1 × 105 cells per well 1 day before plaque assays. A series of 1:10 dilutions was made by mixing 15 μl of the virus sample with 135 μl of DMEM. Next, 100 μl of each dilution was added to individual wells of 24-well plates containing confluent BHK-21 cells. The plates were incubated at 37°C with 5% CO2 for 1 h before a layer of 2% methylcellulose was added. After 3 days of incubation at 37°C with 5% CO2, the cells were fixed with 3.7% formaldehyde and then stained with 1% crystal violet. Plaque morphology and numbers were recorded after washing the plates with tap water.
Indirect immunofluorescence assay.
BHK-21 cells were seeded into six-well plates containing coverslips and transfected with CHIKV genomic RNA or infected with CHIKV at a multiplicity of infection (MOI) of 0.01. At the indicated time points, the coverslips containing transfected or infected cells were collected, washed with phosphate-buffered saline (PBS), and fixed with cold (−20°C) 5% acetic acid in acetone for 15 min at room temperature. After washing with PBS three times, the fixed cells were incubated with the primary antibodies diluted in PBS (CHIKV E2 and nsP2 at an ∼1:250 dilution; CHIKV E1 and capsid at an ∼1:200 dilution) for 1 h. The cells were washed three times with PBS and then incubated with goat anti-rabbit or anti-mouse IgG antibodies conjugated with FITC (1:125 dilution in PBS) at room temperature for another hour. The cells on the coverslips were mounted with 90% glycerol and examined under a fluorescence microscope. The fluorescent images were taken at a ×200 magnification with a Nikon (Tokyo, Japan) upright fluorescence microscope.
Quantitative reverse transcriptase PCR assay.
Viral RNA levels were quantified with a SYBR green real-time RT-PCR assay as described previously (52, 53). Total cellular RNAs were extracted using TRIzol reagent (TaKaRa, China) according to the manufacturer’s protocols. Viral genomic RNAs from 140-μl supernatants harvested from infected BHK-21 cells or each viral fraction by sucrose gradient centrifugation were extracted with a QIAamp viral RNA minikit (Qiagen, Germany) according to the instructions of the manufacturer.
Purification of ΔC-CHIKV virions.
BHK-21 cells were infected with WT CHIKV or ΔC-CHIKV at an MOI of 0.01. At 30 hpi (WT CHIKV) or 96 hpi (ΔC-CHIKV), the supernatants were collected through sequential centrifugation at 400 × g for 10 min and 5,000 × g for 20 min at 4°C to remove cells and cell debris. After filtration through a 0.22-μm filter (Millipore), the clarified supernatants were concentrated by polyethylene glycol 8000 (PEG 8000) precipitation at a final concentration of 8% at 4°C overnight. Following centrifugation at 14,000 × g for 1 h at 4°C, the pellet was gently resuspended in PBS using a pipette. Next, the suspension was overlaid on a 20 to 60% linear sucrose gradient in PBS and subjected to ultracentrifugation at 34,000 rpm for 3 h at 4°C using an MLS-50 rotor in an Optima MAX-XP ultracentrifuge (Beckman, USA). The virus fraction was recovered from the gradient. Ten 500-μl fractions were collected from the top to the bottom and analyzed by SDS-PAGE followed by either Coomassie staining or Western blotting and qRT-PCR analysis.
Western blotting.
WT CHIKV, ΔC-CHIKV, or mock-infected BHK cells were lysed with 200 μl lysis buffer containing 1% Triton X-100, 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (PMSF) on ice for 10 min. Purified WT CHIKV or ΔC-CHIKV particles were obtained from the supernatants of infected BHK-21 cells, which were clarified by low-speed centrifugation, filtered, and purified as described above. Following boiling at 95°C for 10 min, purified virus particles or cell lysates were separated by 10% SDS-PAGE and then electrotransferred onto a polyvinylidene difluoride (PVDF) membrane (0.2 μm; Millipore). The membranes were blocked with 5% skim milk in TBST (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20 [pH 7.4]) for 1 h at room temperature. The blocked membranes were then incubated with primary antibodies at room temperature for another 1 h. After washing three times with TBST, the membranes were incubated with HRP-conjugated secondary goat anti-mouse or goat anti-rabbit antibodies (Proteintech, China) at room temperature for 1 h, followed by washing three times with TBST. The protein bands were visualized with a chemiluminescent HRP-conjugated antibody detection reagent (Millipore, USA).
Plaque reduction or neutralization test.
The neutralizing activities of serum samples from WT CHIKV- or ΔC-CHIKV-infected mice were analyzed by a plaque reduction or neutralization test (PRNT) as described previously (29). Briefly, approximately 100 PFU of WT CHIKV was preincubated with 2-fold serial dilutions of heat-inactivated mouse sera (starting at a 1:10 dilution) for 1 h at 37°C, and the mixture was then added to BHK-21 cell monolayers in 12-well plates and removed after 1 h of incubation, followed by virus quantification by a plaque assay as described above. Neutralizing antibody titers (PRNT50) were determined to be the highest serial dilutions for which the virus plaque count was reduced by 50% compared with the control.
Thermal stability assay.
To facilitate the study of viral thermal stability, we used the corresponding eGFP reporter viruses (WT CHIKV-eGFP and ΔC-CHIKV-eGFP) to perform the assay. After equilibration at 37°C for 1 h, equal volumes of eGFP reporter viruses were collected periodically for the next 60 h. All the collected aliquots were stored at −80°C and thawed simultaneously to infect BHK-21 cells in triplicate. At 48 hpi, the fluorescence intensity was measured using both fluorescence microscopy and a microplate fluorimeter. Infection was normalized to the level acquired prior to incubation at 37°C and fitted with a one-phase exponential decay curve (GraphPad Prism software 6.0; GraphPad Software Inc.) to obtain the infectious half-life.
Thin-section electron microscopy.
BHK-21 cells monolayers were infected with WT CHIKV or ΔC-CHIKV at an MOI of 0.01. At 24 hpi (WT CHIKV) or 84 hpi (ΔC-CHIKV), the infected BHK-21 cells were prefixed with 2.5% glutaraldehyde at room temperature for 2 h before being scraped and pelleted by centrifugation at 400 × g for 10 min. Following rinsing with 0.1 M PBS, cells were postfixed with 1% OSO4 for at least 2 h at 4°C. All pellets were dehydrated stepwise in a graded series of ethanol and embedded in Epon 812. Ultrathin sections were cut on a Leica EM UC7 ultramicrotome and double stained with uranyl acetate and lead citrate. The grids containing ultrathin sections were examined with an FEI Tecnai G2 20 Twin microscope operated at 200 kV.
Mouse experiments.
For virulence experiments, cohorts of 6-week-old IFNAR−/− mice were infected subcutaneously (s.c.) in the ventral/lateral side of the hind foot with 102 PFU of WT CHIKV and 105 PFU of ΔC-CHIKV, respectively. The animals were monitored for survival, body weight changes, and viremia. Footpad swelling induced by WT CHIKV or ΔC-CHIKV was assessed by measuring the height and width of the perimetatarsal area of the hind foot using Kincrome digital vernier calipers.
For immunization and challenge experiments, cohorts of 6-week-old IFNAR−/− mice and C57BL/6 mice were immunized with 104 PFU of ΔC-CHIKV s.c. in the ventral/lateral side of the hind foot. On day 30 postimmunization, IFNAR−/− mice were challenged with 103 PFU of WT CHIKV, and C57BL/6 mice were challenged with 2.5 × 105 PFU of WT CHIKV (ECSA strain) s.c. in the ventral/lateral side of the hind foot. Animal survival, footpad swelling, and weight loss were monitored over 2 weeks. Viremia titers were detected on days 1 to 5 postchallenge.
Statistical analyses.
All data were analyzed using GraphPad Prism 6.0 software (GraphPad Software Inc.), and statistical significance was assigned when P values were <0.05. Kaplan-Meier tests were used for survival analysis, Student’s t tests were used to determine differences between two groups, and a Kruskal-Wallis test followed by Dunn’s test was used for multiple comparisons. For thermal stability assays, one-phase exponential decay curves were performed using GraphPad software.
ACKNOWLEDGMENTS
We are grateful to Core Facility and Technical Support staff (Pei Zhang, An-Na Du, and Ding Gao), Center for Animal Experiment staff (Xue-Fang An, Fan Zhang, He Zhao, and Li Li), and Biosafety Level 3 Laboratory staff (Hao Tang) at the Wuhan Institute of Virology and Wuhan Key Laboratory of Special Pathogens and Biosafety for their helpful support during the course of the work.
This work was supported by the National Key Research and Development Program of China (2018YFA0507201). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Metz SW, Gardner J, Geertsema C, Le TT, Goh L, Vlak JM, Suhrbier A, Pijlman GP. 2013. Effective chikungunya virus-like particle vaccine produced in insect cells. PLoS Negl Trop Dis 7:e2124. doi: 10.1371/journal.pntd.0002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol 8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 3.Das T, Jaffar-Bandjee MC, Hoarau JJ, Krejbich Trotot P, Denizot M, Lee-Pat-Yuen G, Sahoo R, Guiraud P, Ramful D, Robin S, Alessandri JL, Gauzere BA, Gasque P. 2010. Chikungunya fever: CNS infection and pathologies of a re-emerging arbovirus. Prog Neurobiol 91:121–129. doi: 10.1016/j.pneurobio.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Gerardin P, Samperiz S, Ramful D, Boumahni B, Bintner M, Alessandri JL, Carbonnier M, Tiran-Rajaoefera I, Beullier G, Boya I, Noormahomed T, Okoi J, Rollot O, Cotte L, Jaffar-Bandjee MC, Michault A, Favier F, Kaminski M, Fourmaintraux A, Fritel X. 2014. Neurocognitive outcome of children exposed to perinatal mother-to-child chikungunya virus infection: the CHIMERE cohort study on Reunion Island. PLoS Negl Trop Dis 8:e2996. doi: 10.1371/journal.pntd.0002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Enter BJD, Huibers MHW, van Rooij L, Steingrover R, van Hensbroek MB, Voigt RR, Hol J. 2018. Perinatal outcomes in vertically infected neonates during a chikungunya outbreak on the island of Curacao. Am J Trop Med Hyg 99:1415–1418. doi: 10.4269/ajtmh.17-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandeira AC, Campos GS, Sardi SI, Rocha VF, Rocha GC. 2016. Neonatal encephalitis due to chikungunya vertical transmission: first report in Brazil. IDCases 5:57–59. doi: 10.1016/j.idcr.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contopoulos-Ioannidis D, Newman-Lindsay S, Chow C, LaBeaud AD. 2018. Mother-to-child transmission of chikungunya virus: a systematic review and meta-analysis. PLoS Negl Trop Dis 12:e0006510. doi: 10.1371/journal.pntd.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiboutot MM, Kannan S, Kawalekar OU, Shedlock DJ, Khan AS, Sarangan G, Srikanth P, Weiner DB, Muthumani K. 2010. Chikungunya: a potentially emerging epidemic? PLoS Negl Trop Dis 4:e623. doi: 10.1371/journal.pntd.0000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. 2012. Chikungunya: a re-emerging virus. Lancet 379:662–671. doi: 10.1016/S0140-6736(11)60281-X. [DOI] [PubMed] [Google Scholar]
- 10.Dash AP, Bhatia R, Sunyoto T, Mourya DT. 2013. Emerging and re-emerging arboviral diseases in Southeast Asia. J Vector Borne Dis 50:77–84. [PubMed] [Google Scholar]
- 11.Morrison TE. 2014. Reemergence of chikungunya virus. J Virol 88:11644–11647. doi: 10.1128/JVI.01432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar M, Sudeep AB, Arankalle VA. 2012. Evaluation of recombinant E2 protein-based and whole-virus inactivated candidate vaccines against chikungunya virus. Vaccine 30:6142–6149. doi: 10.1016/j.vaccine.2012.07.072. [DOI] [PubMed] [Google Scholar]
- 13.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, Nabel GJ. 2010. A virus-like particle vaccine for epidemic chikungunya virus protects nonhuman primates against infection. Nat Med 16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edelman R, Ascher MS, Oster CN, Ramsburg HH, Cole FE, Eddy GA. 1979. Evaluation in humans of a new, inactivated vaccine for Venezuelan equine encephalitis virus (C-84). J Infect Dis 140:708–715. doi: 10.1093/infdis/140.5.708. [DOI] [PubMed] [Google Scholar]
- 15.Mallilankaraman K, Shedlock DJ, Bao H, Kawalekar OU, Fagone P, Ramanathan AA, Ferraro B, Stabenow J, Vijayachari P, Sundaram SG, Muruganandam N, Sarangan G, Srikanth P, Khan AS, Lewis MG, Kim JJ, Sardesai NY, Muthumani K, Weiner DB. 2011. A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Negl Trop Dis 5:e928. doi: 10.1371/journal.pntd.0000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atasheva S, Kim DY, Akhrymuk M, Morgan DG, Frolova EI, Frolov I. 2013. Pseudoinfectious Venezuelan equine encephalitis virus: a new means of alphavirus attenuation. J Virol 87:2023–2035. doi: 10.1128/JVI.02881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erasmus JH, Auguste AJ, Kaelber JT, Luo H, Rossi SL, Fenton K, Leal G, Kim DY, Chiu W, Wang T, Frolov I, Nasar F, Weaver SC. 2016. A chikungunya fever vaccine utilizing an insect-specific virus platform. Nat Med 23:192–199. doi: 10.1038/nm.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piper A, Ribeiro M, Smith KM, Briggs CM, Huitt E, Nanda K, Spears CJ, Quiles M, Cullen J, Thomas ME, Brown DT, Hernandez R. 2013. Chikungunya virus host range E2 transmembrane deletion mutants induce protective immunity against challenge in C57BL/6J mice. J Virol 87:6748–6757. doi: 10.1128/JVI.03357-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor A, Liu X, Zaid A, Goh LY, Hobson-Peters J, Hall RA, Merits A, Mahalingam S. 2017. Mutation of the N-terminal region of chikungunya virus capsid protein: implications for vaccine design. mBio 8:e01970-16. doi: 10.1128/mBio.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallengärd D, Kakoulidou M, Lulla A, Kümmerer BM, Johansson DX, Mutso M, Lulla V, Fazakerley JK, Roques P, Le Grand R, Merits A, Liljeström P. 2014. Novel attenuated chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol 88:2858–2866. doi: 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plante K, Wang E, Partidos CD, Weger J, Gorchakov R, Tsetsarkin K, Borland EM, Powers AM, Seymour R, Stinchcomb DT, Osorio JE, Frolov I, Weaver SC. 2011. Novel chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog 7:e1002142. doi: 10.1371/journal.ppat.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner CL, Hritz J, Sun C, Vanlandingham DL, Song TY, Ghedin E, Higgs S, Klimstra WB, Ryman KD. 2014. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis 8:e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang E, Kim DY, Weaver SC, Frolov I. 2011. Chimeric chikungunya viruses are nonpathogenic in highly sensitive mouse models but efficiently induce a protective immune response. J Virol 85:9249–9252. doi: 10.1128/JVI.00844-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay A, Wang E, Seymour R, Weaver SC, Rose JK. 2013. A chimeric vesiculo/alphavirus is an effective alphavirus vaccine. J Virol 87:395–402. doi: 10.1128/JVI.01860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Arriaza J, Cepeda V, Hallengärd D, Sorzano CÓS, Kümmerer BM, Liljeström P, Esteban M. 2014. A novel poxvirus-based vaccine, MVA-CHIKV, is highly immunogenic and protects mice against chikungunya infection. J Virol 88:3527–3547. doi: 10.1128/JVI.03418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Doel P, Volz A, Roose JM, Sewbalaksing VD, Pijlman GP, van Middelkoop I, Duiverman V, van de Wetering E, Sutter G, Osterhaus AD, Martina BE. 2014. Recombinant modified vaccinia virus Ankara expressing glycoprotein E2 of chikungunya virus protects AG129 mice against lethal challenge. PLoS Negl Trop Dis 8:e3101. doi: 10.1371/journal.pntd.0003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weger-Lucarelli J, Chu H, Aliota MT, Partidos CD, Osorio JE. 2014. A novel MVA vectored chikungunya virus vaccine elicits protective immunity in mice. PLoS Negl Trop Dis 8:e2970. doi: 10.1371/journal.pntd.0002970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dagley A, Ennis J, Turner JD, Rood KA, Van Wettere AJ, Gowen BB, Julander JG. 2014. Protection against chikungunya virus induced arthralgia following prophylactic treatment with adenovirus vectored interferon (mDEF201). Antiviral Res 108:1–9. doi: 10.1016/j.antiviral.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandler S, Ruffie C, Combredet C, Brault JB, Najburg V, Prevost MC, Habel A, Tauber E, Despres P, Tangy F. 2013. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine 31:3718–3725. doi: 10.1016/j.vaccine.2013.05.086. [DOI] [PubMed] [Google Scholar]
- 30.Aguilar PV, Leung LW, Wang E, Weaver SC, Basler CF. 2008. A five-amino-acid deletion of the Eastern equine encephalitis virus capsid protein attenuates replication in mammalian systems but not in mosquito cells. J Virol 82:6972–6983. doi: 10.1128/JVI.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilar PV, Weaver SC, Basler CF. 2007. Capsid protein of Eastern equine encephalitis virus inhibits host cell gene expression. J Virol 81:3866–3876. doi: 10.1128/JVI.02075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni P, Cheng Kao C. 2013. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology 446:123–132. doi: 10.1016/j.virol.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P, Kaslan M, Lee SH, Yao J, Gao Z. 2017. Progress in exosome isolation techniques. Theranostics 7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu SQ, Li X, Zhang YN, Gao AL, Deng CL, Li JH, Jehan S, Jamil N, Deng F, Wei H, Zhang B. 2017. Detection, isolation, and characterization of chikungunya viruses associated with the Pakistan outbreak of 2016-2017. Virol Sin 32:511–519. doi: 10.1007/s12250-017-4059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed DS, Glass PJ, Bakken RR, Barth JF, Lind CM, da Silva L, Hart MK, Rayner J, Alterson K, Custer M, Dudek J, Owens G, Kamrud KI, Parker MD, Smith J. 2014. Combined alphavirus replicon particle vaccine induces durable and cross-protective immune responses against equine encephalitis viruses. J Virol 88:12077–12086. doi: 10.1128/JVI.01406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Guillen M, Gabev E, Quetglas JI, Casales E, Ballesteros-Briones MC, Poutou J, Aranda A, Martisova E, Bezunartea J, Ondiviela M, Prieto J, Hernandez-Alcoceba R, Abrescia NG, Smerdou C. 2016. Capsid-deficient alphaviruses generate propagative infectious microvesicles at the plasma membrane. Cell Mol Life Sci 73:3897–3916. doi: 10.1007/s00018-016-2230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rolls MM, Webster P, Balba NH, Rose JK. 1994. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell 79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 39.Rose NF, Publicover J, Chattopadhyay A, Rose JK. 2008. Hybrid alphavirus-rhabdovirus propagating replicon particles are versatile and potent vaccine vectors. Proc Natl Acad Sci U S A 105:5839–5843. doi: 10.1073/pnas.0800280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose NF, Buonocore L, Schell JB, Chattopadhyay A, Bahl K, Liu X, Rose JK. 2014. In vitro evolution of high-titer, virus-like vesicles containing a single structural protein. Proc Natl Acad Sci U S A 111:16866–16871. doi: 10.1073/pnas.1414991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lebedeva I, Fujita K, Nihrane A, Silver J. 1997. Infectious particles derived from Semliki Forest virus vectors encoding murine leukemia virus envelopes. J Virol 71:7061–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds TD, Buonocore L, Rose NF, Rose JK, Robek MD. 2015. Virus-like vesicle-based therapeutic vaccine vectors for chronic hepatitis B virus infection. J Virol 89:10407–10415. doi: 10.1128/JVI.01184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narayanan K, Chen CJ, Maeda J, Makino S. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J Virol 77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsell K, Griffiths G, Garoff H. 1996. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J 15:6495–6505. doi: 10.1002/j.1460-2075.1996.tb01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown RS, Wan JJ, Kielian M. 2018. The alphavirus exit pathway: what we know and what we wish we knew. Viruses 10:E89. doi: 10.3390/v10020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lulla V, Kim DY, Frolova EI, Frolov I. 2013. The amino-terminal domain of alphavirus capsid protein is dispensable for viral particle assembly but regulates RNA encapsidation through cooperative functions of its subdomains. J Virol 87:12003–12019. doi: 10.1128/JVI.01960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H. 2000. Membrane proteins organize a symmetrical virus. EMBO J 19:5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garoff H, Sjoberg M, Cheng RH. 2004. Budding of alphaviruses. Virus Res 106:103–116. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Muthumani K, Lankaraman KM, Laddy DJ, Sundaram SG, Chung CW, Sako E, Wu L, Khan A, Sardesai N, Kim JJ, Vijayachari P, Weiner DB. 2008. Immunogenicity of novel consensus-based DNA vaccines against chikungunya virus. Vaccine 26:5128–5134. doi: 10.1016/j.vaccine.2008.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plante KS, Rossi SL, Bergren NA, Seymour RL, Weaver SC. 2015. Extended preclinical safety, efficacy and stability testing of a live-attenuated chikungunya vaccine candidate. PLoS Negl Trop Dis 9:e0004007. doi: 10.1371/journal.pntd.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 52.Deng CL, Liu SQ, Zhou DG, Xu LL, Li XD, Zhang PT, Li PH, Ye HQ, Wei HP, Yuan ZM, Qin CF, Zhang B. 2016. Development of neutralization assay using an eGFP chikungunya virus. Viruses 8:E181. doi: 10.3390/v8070181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu SQ, Li X, Deng CL, Yuan ZM, Zhang B. 2018. Development and evaluation of one-step multiplex real-time RT-PCR assay for simultaneous detection of Zika virus and chikungunya virus. J Med Virol 90:389–396. doi: 10.1002/jmv.24970. [DOI] [PubMed] [Google Scholar]