This study suggests that CD4+ T cells found throughout tissues in the body can contain replication-competent SIV and contribute to rebound of the virus after treatment interruption. In addition, this study demonstrates that macrophages in tissues are another cellular reservoir for SIV and may contribute to viral rebound after treatment interruption. This new insight into the size and location of the SIV reservoir could have great implications for HIV-infected individuals and should be taken into consideration for the development of future HIV cure strategies.
KEYWORDS: CD4+ T cells, latency, macrophages, quantitative viral outgrowth assay, reservoir, simian immunodeficiency virus
ABSTRACT
Understanding the cellular and anatomical sites of latent virus that contribute to human immunodeficiency virus (HIV) rebound is essential for eradication. In HIV-positive patients, CD4+ T lymphocytes comprise a well-defined functional latent reservoir, defined as cells containing transcriptionally silent genomes able to produce infectious virus once reactivated. However, the persistence of infectious latent virus in CD4+ T cells in compartments other than blood and lymph nodes is unclear. Macrophages (Mϕ) are infected by HIV/simian immunodeficiency virus (SIV) and are likely to carry latent viral genomes during antiretroviral therapy (ART), contributing to the reservoir. Currently, the gold standard assay used to measure reservoirs containing replication-competent virus is the quantitative viral outgrowth assay (QVOA). Using an SIV-macaque model, the CD4+ T cell and Mϕ functional latent reservoirs were measured in various tissues using cell-specific QVOAs. Our results showed that blood, spleen, and lung in the majority of suppressed animals contain latently infected Mϕs. Surprisingly, the numbers of CD4+ T cells, monocytes, and Mϕs carrying infectious genomes in blood and spleen were at comparable frequencies (∼1 infected cell per million). We also demonstrate that ex vivo viruses produced in the Mϕ QVOA are capable of infecting activated CD4+ T cells. These results strongly suggest that latently infected tissue Mϕs can reestablish productive infection upon treatment interruption. This study provides the first comparison of CD4+ T cell and Mϕ functional reservoirs in a macaque model. It is the first confirmation of the persistence of latent genomes in monocytes in blood and Mϕs in the spleen and lung of SIV-infected ART-suppressed macaques. Our results demonstrate that transcriptionally silent genomes in Mϕs can contribute to viral rebound after ART interruption and should be considered in future HIV cure strategies.
IMPORTANCE This study suggests that CD4+ T cells found throughout tissues in the body can contain replication-competent SIV and contribute to rebound of the virus after treatment interruption. In addition, this study demonstrates that macrophages in tissues are another cellular reservoir for SIV and may contribute to viral rebound after treatment interruption. This new insight into the size and location of the SIV reservoir could have great implications for HIV-infected individuals and should be taken into consideration for the development of future HIV cure strategies.
INTRODUCTION
The development of drugs that target multiple stages of the human immunodeficiency virus (HIV) life cycle and reduce viremia to undetectable levels have transformed HIV infection from a deadly disease to a manageable chronic illness (1–3). However, life-long antiretroviral therapy (ART) is necessary, since the regimen suppresses viral replication but does not eliminate the latent HIV reservoir (4–6), defined as cells that harbor integrated but transcriptionally silent HIV provirus that can be induced to produce infectious virus (7). Studies have demonstrated that multiple tissues harbor HIV provirus in ART-suppressed patients, suggesting that the latent HIV reservoir can be found throughout the body (8–14). Latently infected cells that harbor HIV serve as the source of viral rebound after ART interruption, and the half-life of latently infected cells precludes the elimination of the virus by ART alone (15). Therefore, understanding both the location and the types of cells that harbor latent HIV throughout the body is critical for HIV eradication.
HIV and the simian immunodeficiency virus (SIV) cause immunodeficiency disease as well as tissue-specific diseases in humans and macaques, respectively (16). CD4+ T cells isolated from peripheral blood and lymph nodes have been the most extensively characterized reservoirs due to the limitations associated with obtaining tissue samples from HIV-infected patients (9, 17–19). However, less than 2% of the total lymphocyte population resides in the peripheral blood, and CD4+ T cell populations are present in multiple tissues, potentially contributing to viral persistence (15). Additionally, not all CD4+ T cell populations contribute equally to the HIV reservoir; the central memory and transitional memory populations are thought to contain the majority of integrated HIV DNA compared to their effector memory counterpart (19, 20). SIV-infected macaques treated with ART have been used to examine the contribution of CD4+ T cells to the latent reservoir in other tissues (21–31). Additionally, this model provides the opportunity to study other cells that may also play a role in HIV persistence, such as monocytes, macrophages (Mϕ), astrocytes, and follicular cells (13, 15, 32). Tissue-resident Mϕs, such as splenic Mϕs, alveolar Mϕs, and microglia, are long-lived and relatively resistant to the cytopathic effects of HIV/SIV infection, potentially serving as stable viral reservoirs (33, 34).
Previous studies have assessed the frequency of HIV/SIV-infected cells in different tissues by quantifying HIV cell-associated RNA or DNA. These studies suggest that the presence of HIV DNA with little or no viral RNA is equivalent to latency (35–38). However, this approach overestimates the number of latently infected cells due to the presence of a large proportion of defective proviral DNA as well as intact genomes that do not express virus in vivo (39). To measure the functional latent reservoir, an assay that quantifies the number of latently infected resting CD4+ T cells, the quantitative viral outgrowth assay (QVOA), was developed and has been widely used to measure CD4+ T cell reservoirs in ART-suppressed HIV-infected patients (40, 41). We have adapted the HIV CD4+ T cell QVOA to be used in an SIV-infected macaque model (33) and also developed a QVOA for monocytes and tissue Mϕs (Mϕ QVOA) using the same SIV model (34). Using the Mϕ QVOA we have shown that monocytes from the blood and Mϕs from bronchoalveolar lavage fluid (BAL), lung, spleen, and brain of untreated SIV-infected macaques harbor replication-competent virus (33). Further, using this assay, we demonstrated that brain Mϕs constitute a functional latent reservoir in a model of ART-suppressed SIV-infected macaques (34).
To accelerate progress toward a cure, it is important to fully characterize the CD4+ T cell and Mϕ functional latent reservoirs throughout the body. In this study, we analyzed the reservoirs in spleen, lung, and blood of ART-suppressed SIV-infected macaques. We used the CD4+ T cell QVOA and Mϕ QVOA to determine the number of functionally latent cells in each compartment. Notably, latently infected CD4+ T cells, monocytes, and Mϕs were identified in all ART-suppressed macaques studied. This study and our previous findings demonstrate that CD4+ T cells and Mϕs represent functional latent reservoirs in many tissues.
RESULTS
Treatment regimen and characteristics of the SIV-pigtailed model.
Seven pigtailed macaques were inoculated with SIV/DeltaB670 and SIV/17E-Fr, an SIV model that uses macrophage-tropic viral strains to accurately reproduce the neuropathic and immunologic events identified in HIV-infected patients (42–45). Macaques were treated with ART at 12 days postinfection (dpi), when the reservoirs found in both the peripheral tissues and central nervous system (CNS) have been shown to be seeded in our model and, in the case of lymph nodes and peripheral blood mononuclear cells (PBMCs), in other SIV models as well (46, 47). All macaques were treated daily treatment with tenofovir (TFV), integrase inhibitor (INI), ritonavir (RTV), and darunavir (DRV) (Table 1). This ART regimen was chosen based on the CNS penetrance score (CPE) to fully suppress virus replication in both the CNS and the peripheral blood and tissues (48). The viral load in both plasma and cerebrospinal fluid (CSF) was measured longitudinally to demonstrate viral suppression in both the peripheral blood and the CNS (Fig. 1). Three of the seven suppressed animals (animals Pm12, Pm22, and Pm23) were also treated with latency-reversing agents (LRA) during ART suppression, although these treatments did not measurably alter the results reported here. All animals were suppressed (less than 50 copies per ml, as measured by digital droplet PCR [ddPCR]) for a minimum of 6 months and as long as 18 months in both CSF and plasma, as measured by an SIV gag RNA ddPCR. Two SIV-infected macaques, Pm26 and Pm27, the results for which are not depicted in the graphs, were used as untreated controls. Both of the animals had detectable plasma and CSF viral loads throughout the infection and at the terminal time point (Table 1).
TABLE 1.
Characterization of SIV-infected ART-suppressed macaques at the terminal time point
| Animal codea | ART duration (mo) | Cells count (no. of cells/μl blood) |
Viral load (no. of SIV RNA copies/ml) |
SIV infection status | ||
|---|---|---|---|---|---|---|
| CD4+ cells | Monocytes | Plasma | CSF | |||
| Pm11 | 20.5 | 474 | 1,092 | <LODb | <LOD | Suppressed |
| Pm12 | 20.5 | 1,074 | 485 | <LOD | <LOD | Suppressed |
| Pm21 | 13.0 | 280 | 340 | <LOD | <LOD | Suppressed |
| Pm22 | 13.0 | 324 | 2,310 | <LOD | <LOD | Suppressed |
| Pm23 | 11.5 | 331 | 410 | <LOD | <LOD | Suppressed |
| Pm24 | 6.0 | 291 | 350 | <LOD | <LOD | Suppressed |
| Pm25 | 6.0 | 790 | 310 | <LOD | <LOD | Suppressed |
| Pm26 | 0 | 481 | 710 | 11,993,458 | 15,364,703 | Viremic |
| Pm27 | 0 | 436 | 580 | 13,610,950 | 2,935 | Viremic |
Two viremic animals, Pm26 and Pm27, were used as positive controls in some experiments.
<LOD, less than the limit of detection.
FIG 1.
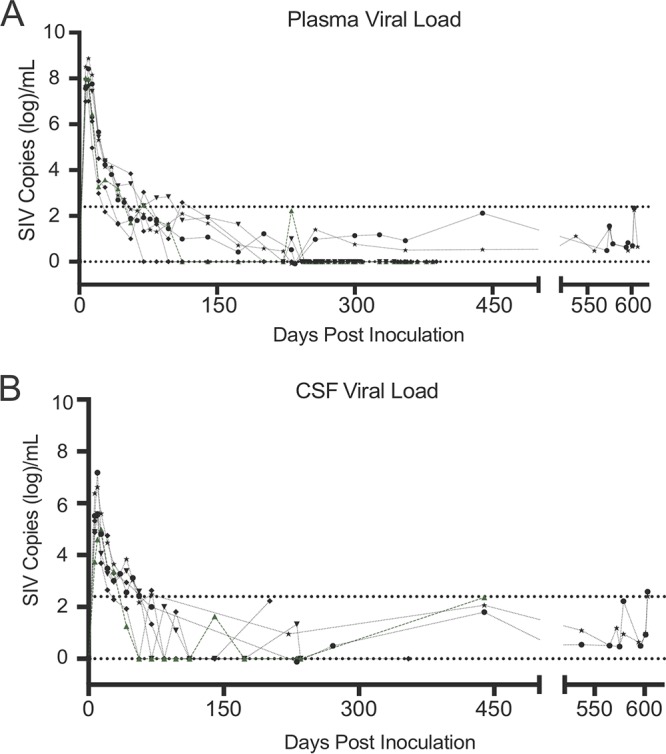
Viral load in plasma and CSF of seven suppressed SIV-infected ART-treated macaques (34). Seven SIV-infected pigtailed macaques were treated with similar ART regimens (tenofovir, darunavir, integrase inhibitor, and ritonavir). Symbols represent the plasma (A) and CSF (B) viral load values for each animal. Analyses of samples with values below the limit of quantitation (LOQ) for the SIV RT-qPCR assay (430 SIV RNA copies/ml; top dotted line) were repeated using RT-ddPCR (LOQ, 5 SIV RNA copies/ml; bottom dotted line).
SIV DNA but not RNA is detectable in whole tissue.
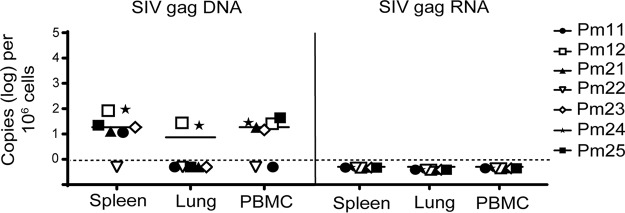
To evaluate whether the viral suppression observed in the plasma was reflected in PBMCs and tissues, such as spleen and lung, the levels of SIV gag DNA and RNA were measured in these compartments using reverse transcriptase (RT) quantitative PCR (qPCR) and RT-ddPCR. Of the seven ART-treated macaques studied, SIV gag DNA was detected in the spleens of six macaques, in the PBMCs of five macaques, and in the lungs of two macaques. Pm12 and Pm24 had detectable SIV DNA in all three compartments, with the highest level of DNA being in the spleen (Fig. 2, left). Pm21, Pm23, and Pm25 had similar levels of detectable SIV DNA in both the spleen and PBMCs, whereas Pm11 had detectable SIV DNA only in the spleen. Pm22 was undetectable in all three compartments. No correlation was observed between the amount of DNA detected in the tissues and the length of treatment. All seven ART-treated macaques had undetectable levels of SIV gag RNA in the PBMCs, spleens, and lungs, as measured by RT-ddPCR at the terminal time point (Fig. 2, right).
FIG 2.
Quantification of SIV DNA and SIV RNA from tissue samples. Cellular DNA and RNA were extracted from spleen, lung, and PBMCs from seven SIV-infected ART-suppressed macaques. SIV gag DNA (left) and SIV gag RNA (right) were measured by ddPCR, with the limit of quantitation (LOQ, dashed lines) being 1 copy per reaction mixture. Open symbols indicate animals that were treated with LRAs in vivo; closed symbols indicate animals that were not treated with LRAs.
SIV DNA is detectable in monocytes/Mϕs and CD4+ T cells isolated from blood and tissue.
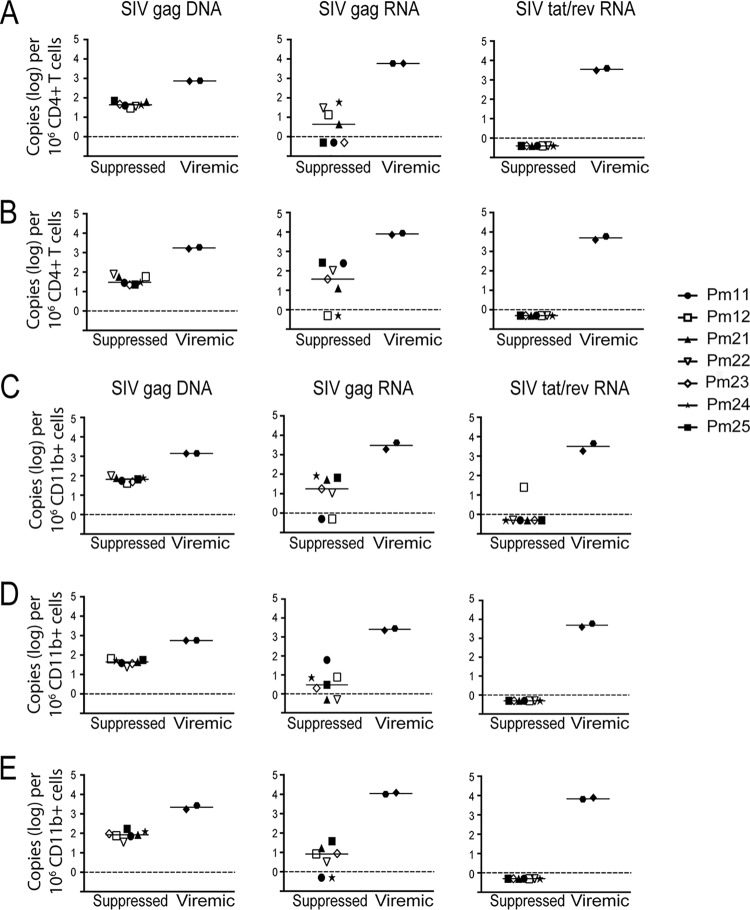
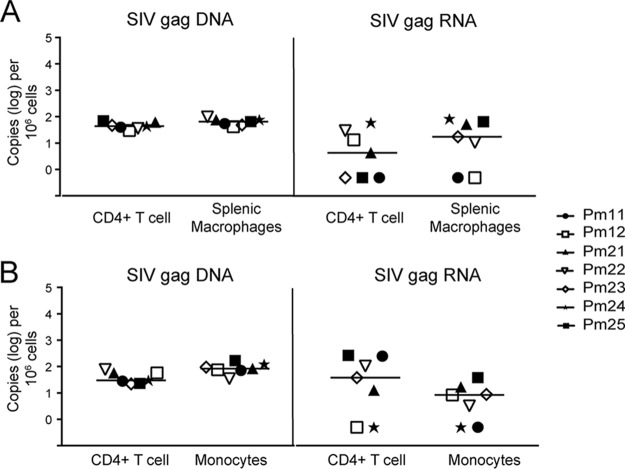
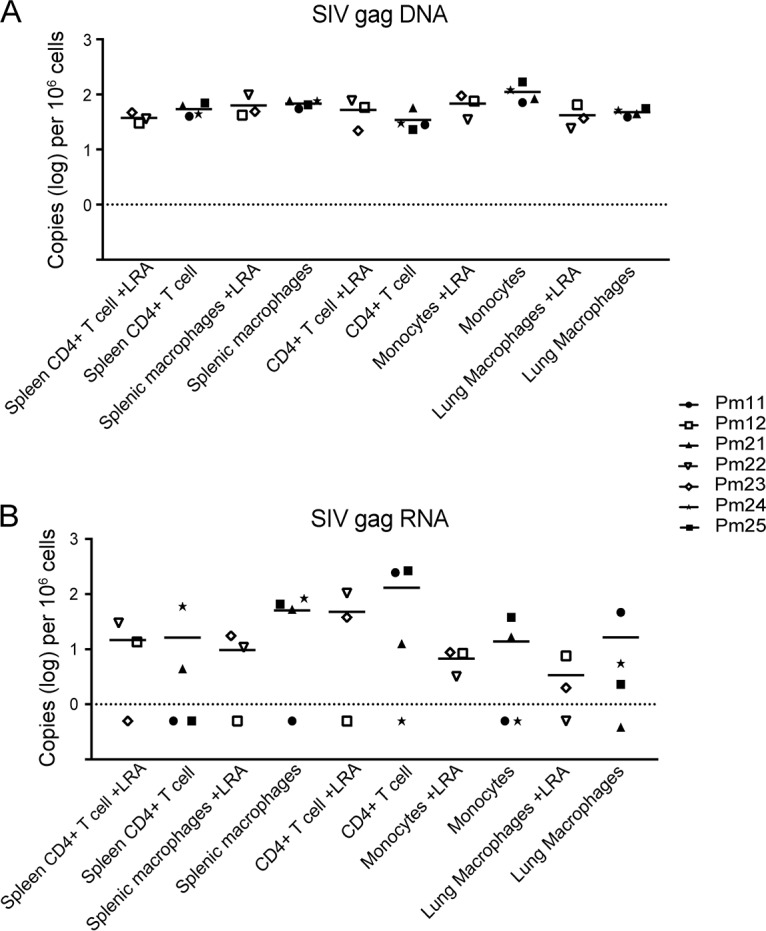
We hypothesized that SIV DNA is not measurable in the spleen, lung, and PBMCs of some animals due to the limit of quantitation for the RT-qPCR assay used to evaluate homogenized tissue (10 copies/μg total DNA or RNA). Therefore, to increase the sensitivity, we evaluated SIV DNA and RNA by ddPCR in enriched CD11b+ (monocytes/Mϕs) and CD4+ T cells isolated from these tissues and blood. CD11b+ cells were isolated from all three compartments, whereas CD4+ T cells, whose numbers are very low in the lung, were isolated from the spleen and blood. In contrast to whole tissue, all monocytes/Mϕs and CD4+ T cell isolations from the spleen, lung, and blood of the suppressed animals were positive for SIV DNA, with levels ranging from 23 to 168 copies per 106 cells (Fig. 3A to E). The viremic animals had detectable SIV DNA in all samples, with the levels ranging from 1,392 to 1,424 copies per 106 cells. Notably, the amounts of SIV DNA were similar between CD4+ T cells and monocytes/Mϕs isolated from spleen and blood (Fig. 4A and B). The length of ART treatment and LRA treatment and the duration of viral suppression did not appear to influence the levels of SIV DNA in either cell type (Fig. 5A).
FIG 3.
Quantification of SIV DNA and RNA from CD4+ T cells and CD11b+ cells isolated from tissues. CD4+ T cells (A and B) and CD11b+ cells (C, D, and E) were isolated from spleen, lung, and PBMCs from seven SIV-infected ART-suppressed macaques and two SIV-infected untreated macaques. Cellular DNA and RNA were then extracted and analyzed for SIV gag DNA (left), SIV gag RNA (middle), and SIV tat/rev RNA (right) by PCR. The limit of quantitation (LOQ) for ddPCR is 1 copy per reaction, and that for qPCR is 10 copies per reaction. The dashed lines represent the LOQ for ddPCR. Open symbols indicate animals that were treated with LRAs in vivo; closed symbols indicate animals that were not treated with LRAs.
FIG 4.
SIV DNA and RNA levels were similar between CD4+ T cells and macrophages or monocytes isolated from tissues. Comparison of SIV DNA and SIV RNA levels in CD4+ T cells and CD11b+ cells isolated from spleen (A) and blood (B). The dotted line represents the limit of quantitation for ddPCR. Each symbol represents an animal. Open symbols indicate animals that were treated with LRAs in vivo; closed symbols indicate animals that were not treated with LRAs.
FIG 5.
In vivo LRA treatment does not have a measurable effect on the level of SIV gag DNA or RNA in isolated cells. SIV gag DNA (A) and RNA (B) levels measured in cells isolated from SIV-infected macaques treated with LRA were compared to those measured in cells isolated from SIV-infected macaques without LRA treatment. CD4+ T cells were isolated from blood and spleen. CD11b+ cells (monocytes/macrophages) were isolated from blood, spleen, and lung.
Presence of residual SIV gag but not tat-rev multiply spliced RNA in monocytes/Mϕs and CD4+ T cells isolated from tissues and blood.
To evaluate transcriptional activity in monocytes/Mϕs and CD4+ T cells isolated from tissues, SIV cell-associated RNA (caRNA) was quantitated by RT-ddPCR and RT-qPCR. caRNA was isolated from the same samples from which DNA was isolated, as described above, and used for quantification of unspliced SIV gag RNA and multiply spliced SIV tat/rev RNA. SIV gag RNA was detectable in the spleen CD4+ T cells of four suppressed macaques and in the circulating CD4+ T cells of five suppressed macaques (Fig. 3A and B). In addition, SIV gag RNA was detectable in CD11b+ cells isolated from the spleen, lung, and blood of five suppressed macaques (Fig. 3C to E). Notably, the levels of SIV gag RNA were similar in both the CD11b+ and CD4+ T cells isolated from each comparable compartment (Fig. 4A and B). As a control, SIV gag RNA was also assessed in viremic macaques in CD11b+ and CD4+ T cells from the same compartments. The SIV gag RNA level was 100-fold higher in all cells isolated from viremic animals than in all cells isolated from suppressed animals (Fig. 3). Similar to SIV DNA levels, the length of ART treatment and LRA treatment and the duration of viral suppression did not appear to influence the levels of SIV RNA in either cell type (Fig. 5B).
Detection of SIV gag RNA does not necessarily indicate active transcription, nor does the presence of SIV tat/rev RNA (49, 50). Therefore, we also measured the presence of SIV tat/rev multiply spliced RNA in all isolated cells. All CD4+ T cell samples isolated from the spleen and blood of suppressed macaques were negative for SIV tat/rev RNA (Fig. 3A and B). In addition, all but one CD11b+ samples isolated from the spleen, lung, and blood were negative for SIV tat/rev RNA (Fig. 3C to E). The CD11b+ spleen sample (Pm12) with detectable SIV tat/rev RNA had 25 copies per 106 cells, which is close to the limit of quantitation for the assay (Fig. 3C) and substantially lower than the levels from any of the viremic animals (Fig. 3A to E). Although it is known that multiply spliced viral RNAs are usually detected at lower levels than the unspliced forms (51), these results suggest that the residual SIV gag RNA detected in the isolated CD11b+ cells (monocytes/Mϕs) and CD4+ T cells in suppressed macaques is not indicative of active replication but indicates expected stochastic oscillations (52). Therefore, detection of virus in the QVOA would suggest reactivation of latent viral genomes.
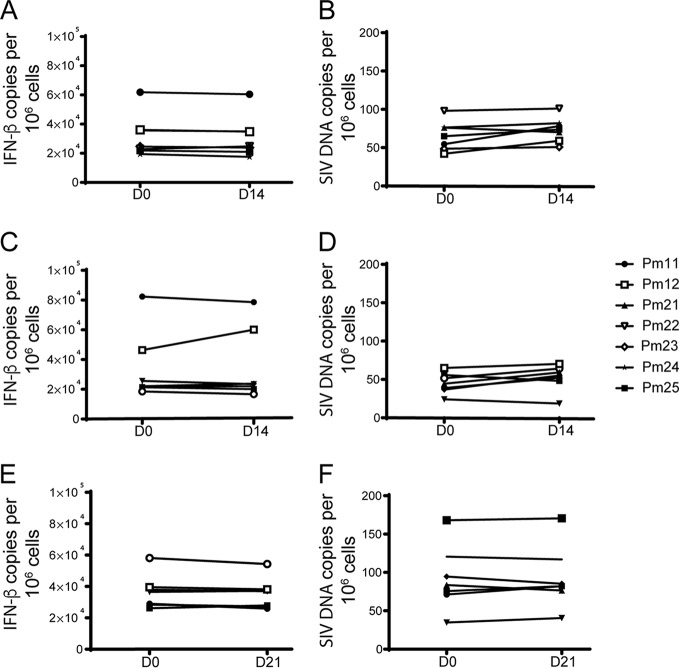
Macrophage numbers and SIV DNA levels are stable over time.
Mϕs, unlike CD4+ T cells, do not proliferate in vitro. Instead, the number of Mϕs remains stable and viable in culture for up to 3 weeks (33). To demonstrate that there was no expansion of the Mϕs derived from spleen, lung, and blood, beta interferon (IFN-β) gene expression was measured in Mϕs at days 0 and 14 or 21 in culture, and the number of cells in each well remained stable (Fig. 6A, C, and E). The IFN-β gene is a single-copy gene and is used to quantify the number of cells present in our assays. In parallel, SIV DNA was measured at days 0 and 14 or 21, and there was no significant change in the level of SIV DNA over time (Fig. 6B, D and F). These data corroborate that, unlike CD4+ T cells in culture, Mϕs do not proliferate and are resistant to cell death, confirming that these long-lived cells could constitute a viable reservoir in vivo.
FIG 6.
The macrophage population in culture is stable over time. IFN-β gene (A, C, and E) and SIV gag (B, D, and F) DNA copies were quantitated by qPCR in CD11b+ cells isolated from spleen, lung, and PBMCs before being plated for Mϕ QVOA and also from cells in the QVOA wells at 14 or 21 days postplating. Values are presented as the number of copies per 106 cells. Open symbols indicate animals that were treated with LRAs in vivo; closed symbols indicate animals that were not treated with LRAs. D0, D14, and D21, days 0, 14, and 21, respectively.
Functional latent virus is present in CD4+ T cells and Mϕs in the blood and spleen of suppressed SIV-infected macaques.
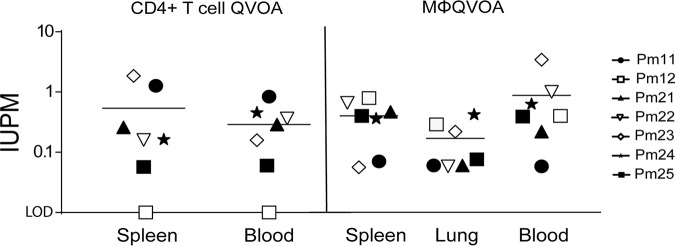
CD4+ T cells have been extensively characterized as the primary latent reservoir in peripheral blood and lymph nodes in ART-suppressed HIV-positive (HIV+) patients and ART-suppressed SIV-infected animals in our previous studies (17, 53–56). Previously, our studies showed that Mϕs also contribute to the reservoir in the brains of suppressed SIV-infected animals (34). We hypothesized that the spleen, an additional site that harbors large numbers of Mϕs and CD4+ T cells, also contributes to the SIV reservoir and may act as a sanctuary for latently infected cells. To estimate the frequency of latently infected CD4+ T cells and Mϕs in the spleen, we isolated both CD4+ lymphocytes and Mϕs from the spleen and used the quantitative viral outgrowth viral assays (QVOA) specific for each of these cells (33). Six of the seven suppressed macaques had quantifiable levels of latently infected CD4+ T cells in both the spleen and blood (Fig. 7 and Table 2). One suppressed animal, Pm12, had undetectable numbers of infectious units per million (IUPM) in the CD4 QVOA, which can be explained by the limited number of cells used for the assay compared to the number in the other samples (Fig. 7 and Table 2). All Mϕ QVOAs from the spleen and blood of the seven ART-suppressed macaques had measurable IUPM values (Fig. 7 and Table 2). The median Mϕ IUPM values from blood and spleen were similar, as were the median CD4 IUPM values from the blood and spleen, suggesting that the level in the blood may be a surrogate representation for the level of persistent virus in the spleen. Additionally, the median CD4 IUPM values and median Mϕ IUPM values from the same tissues were similar, regardless of the duration of ART treatment or cell-associated SIV gag DNA and RNA levels (Table 1 and Fig. 3), suggesting that both cell types have the potential to contribute equally to the SIV reservoir. Taken together, these data suggest that viral latency in the spleen and blood is maintained despite undetectable levels of viremia in the plasma and CSF for an extended time in the SIV-macaque model. Additionally, as seen with SIV DNA and RNA levels, the length of ART and LRA treatment and the duration of viral suppression did not appear to influence IUPM levels in either cell type (Fig. 8).
FIG 7.
Functional latent reservoirs are detected in both CD4+ T cells and monocytes/macrophages isolated from SIV-infected ART-suppressed animal tissues. The frequency of latently infected CD4+ T cells (left) and monocytes/macrophages (right) isolated from the spleen, lung, and PBMCs of suppressed ART-treated macaques were quantitated by cell-specific QVOAs. The horizontal black lines represent the median number of infectious units per million cells (IUPM), and each symbol represents an animal. Open symbols indicate animals that were treated with LRAs in vivo; closed symbols indicate animals that were not treated with LRAs. LOD, limit of detection.
TABLE 2.
Quantitation of IUPM in CD4+ T cell and monocyte/macrophage QVOA
| Animal code | IUPMa
|
||||
|---|---|---|---|---|---|
| CD4+ T cell QVOA |
Monocyte/macrophage QVOA |
||||
| Spleen | Blood | Spleen | Lung | Blood | |
| Pm11 | 1.27 | 0.84 | 0.07 | 0.06 | 0.06 |
| Pm12 | <LOD | <LOD | 0.79 | 0.29 | 0.40 |
| Pm21 | 0.26 | 0.29 | 0.47 | 0.06 | 0.22 |
| Pm22 | 0.16 | 0.37 | 0.66 | 0.06 | 1.00 |
| Pm23 | 1.86 | 0.16 | 0.06 | 0.22 | 3.44 |
| Pm24 | 0.16 | 0.46 | 0.36 | 0.42 | 0.63 |
| Pm25 | 0.06 | 0.06 | 0.40 | 0.08 | 0.39 |
IUPM was quantitated as described by Avalos et al. (33). IUPM values represent the presence or absence of positive wells with more than 50 SIV gag RNA copies/ml of supernatant as determined by RT-qPCR, as described in the legend to Fig. 5. Values reflect maximum likelihood estimates of the infection frequency (in numbers of IUPM). Given the resolution of the assay, the 95% confidence interval was typically 0.2 to 4 times the reported value. <LOD, less than the limit of detection.
FIG 8.
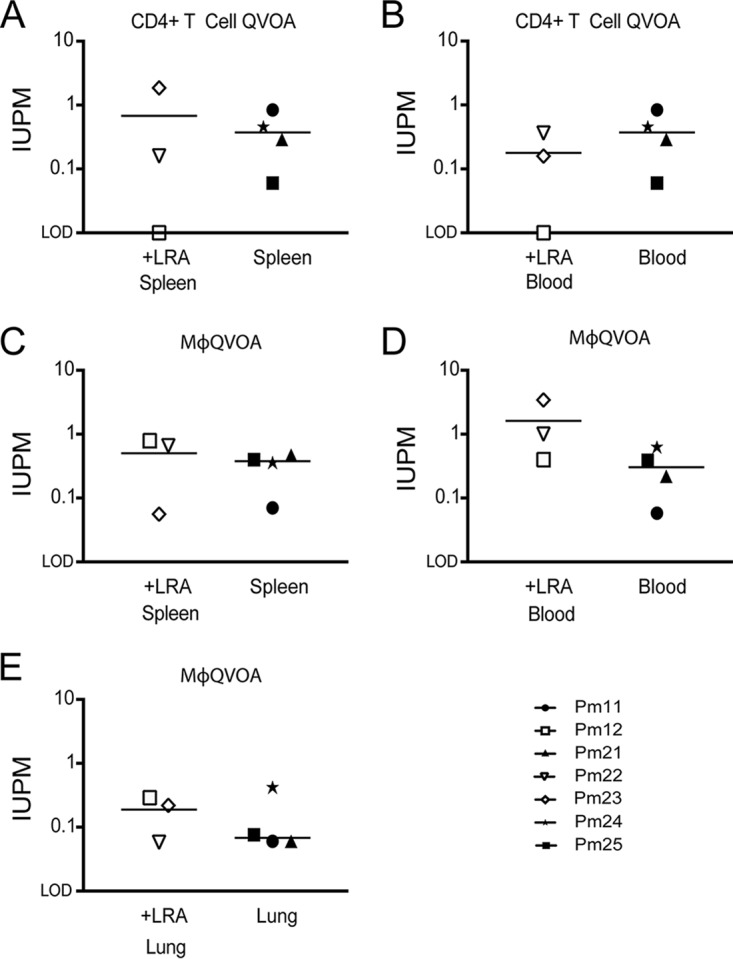
In vivo LRA treatment has no measurable effect on IUPM. A comparison of the IUPM values obtained from cells isolated from SIV-infected macaques treated with LRA compared to those obtained from cells isolated from SIV-infected macaques without LRA treatment is shown. (A) Number of IUPM from CD4+ T cells isolated from spleen; (B) number of IUPM from CD4+ T cells isolated from blood; (C) number of IUPM from macrophages isolated from spleen; (D) number of IUPM from monocyte-derived macrophages isolated from blood; (E) number of IUPM from alveolar macrophages isolated from lung.
Viral persistence in alveolar Mϕs in suppressed SIV-infected macaques.
Mϕs are present in every tissue in the body, suggesting that latent HIV/SIV could be detected in other organs. We examined alveolar Mϕs to determine whether they also harbored SIV DNA and whether these lung Mϕs could contribute to the SIV reservoir (Fig. 3D). All CD11b+ cells isolated from the lungs of suppressed macaques produced detectable and quantifiable levels of SIV in the QVOA (Fig. 7 and Table 2). Similar to the spleen and blood QVOAs, treatment duration, LRA treatment, and length of suppression did not correlate with IUPM values. These studies demonstrate that Mϕs in the lung and spleen are latently infected in ART-suppressed macaques and have the potential to contribute to the latent reservoir.
Reactivated latent viruses released from Mϕs are replication competent.
To confirm that the SIV RNA measured in the QVOA from spleen, lung, and blood Mϕs reflected infectious virus capable of de novo infection in cells, supernatants from Mϕ QVOA wells were used to infect activated healthy PBMCs from macaques. Virus derived from the Mϕ QVOA expanded exponentially in PBMCs by day 5 and continued to expand through day 14 (Fig. 9). These results demonstrate that virus derived from latently infected Mϕs in both blood and tissues is infectious and can contribute to the viral rebound once ART is interrupted.
FIG 9.
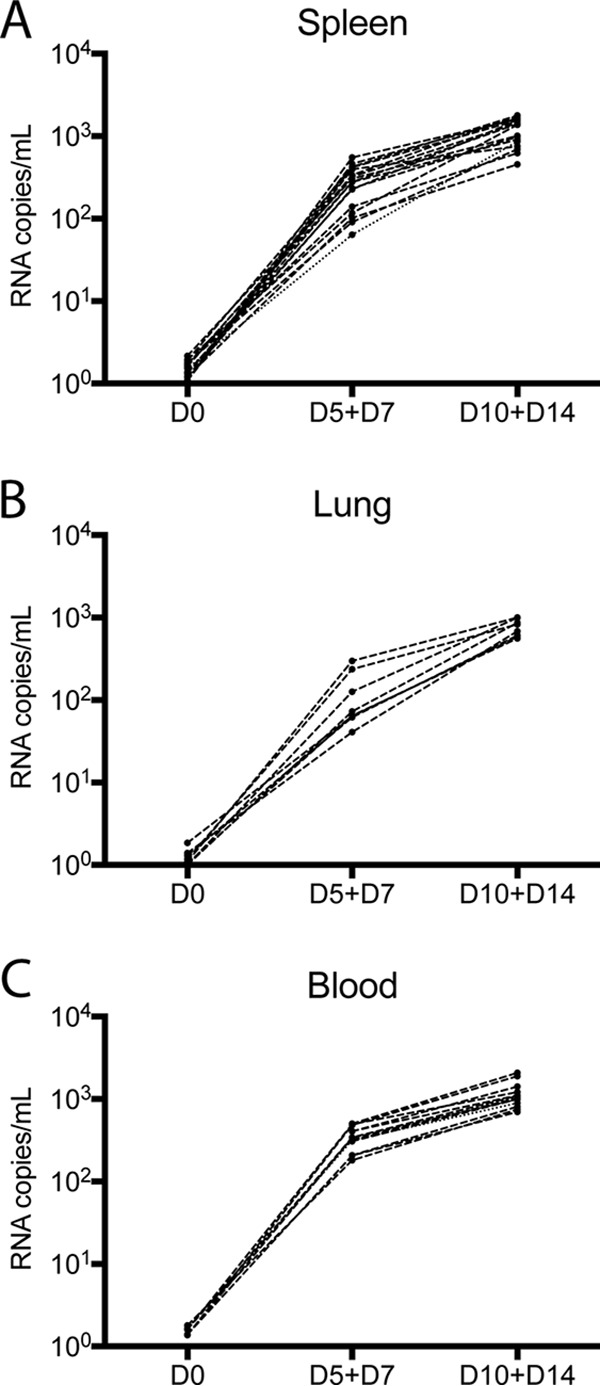
Supernatants from Mϕ QVOA cultures contain viruses that establish de novo infection in activated macaque PBMCs. Activated PBMCs from a healthy pigtailed macaque were spinoculated with culture supernatant from positive Mϕ QVOA wells with spleen tissue (A), lung tissue (B), and blood (C) from the seven suppressed macaques. The graphs show the viral kinetics over time postinfection, as measured by an SIV RNA RT-qPCR. Each line represents viral replication from one Mϕ QVOA well.
DISCUSSION
Despite continued improvements in antiretroviral therapy, HIV persists in millions of individuals worldwide. The greatest barrier to an HIV cure is the functional latent reservoir. Therefore, it is essential to understand the cell types that contribute to the latent reservoir, the anatomical locations, and the potential of these cells to contribute to viral resurgence after ART interruption. HIV cure strategies are aimed at either eliminating the reservoir, maintaining latency by immunologic control, or other approaches to suppress viral transcription (57, 58). The majority of previous studies have focused on latent HIV/SIV in CD4+ T cells in the tissues of ART-suppressed patients and animal models but have limited their studies to lymph nodes and blood, with little focus on other tissues, such as the spleen and lung (27, 49, 59). Monocytes and tissue-specific Mϕs have not been considered to contribute to the functional reservoir. The primary goal of this study was to examine the role of Mϕs as functional latent reservoirs in SIV-infected ART-suppressed macaques.
Current ART regimens used to treat HIV-infected patients and used in SIV-macaque studies are highly effective in reducing viremia to undetectable levels (21, 29). In this study, the viral loads in the plasma and CSF were measured longitudinally in seven SIV-infected pigtailed macaques, demonstrating that virus replication was controlled in both the periphery and the CNS. Plasma viral loads are easily accessible in HIV+ patients; however, whether the plasma viral load reflects the level of HIV in tissues is unknown. To address this question, we assessed SIV DNA and RNA levels in the PBMCs, spleens, and lungs of SIV-infected ART-suppressed macaques. All CD4+ T cells and myeloid cells isolated from tissues had detectable levels of SIV gag DNA (Fig. 2). Interestingly, no correlation was observed between the amount of DNA detected in the tissues and the length of treatment (data not shown). This may have been due to the short length of time that the macaques were suppressed compared to that for HIV-infected individuals (60). However, Whitney and colleagues showed that only SIVmac251-infected macaques started early on ART treatment (day 3 postinfection) and not macaques started on treatment at later time points (days 7 to 14) decreased the viral RNA and proviral DNA levels in peripheral blood, lymph nodes, and gastrointestinal mucosa (47). Additionally, the in vivo LRA treatments in three animals did not affect SIV DNA levels in the cells isolated from tissues (Fig. 5A). In contrast, all CD4+ T cells and most myeloid cells isolated from PBMCs, spleen, and lung had undetectable levels of SIV tat/rev RNA, and some had detectable gag RNA (Fig. 5B). Previous studies using cells from HIV-suppressed patients have shown that the predominant cellular HIV type 1 (HIV-1) RNA species detected during ART suppression is HIV gag, whereas HIV-1 tat/rev was found to be 10 to 100 times less abundant (61–63). The demonstration that SIV tat/rev RNA was undetectable, despite detectable levels of SIV gag DNA and RNA, supports the conclusion that SIV expression was transcriptionally silent in the tissues of these animals.
The presence of HIV DNA is not an accurate representation of the level of intact and activatable virus within tissues, since defective proviruses tend to outnumber intact proviruses 20 to 1 (64). In this study, we have shown that CD4+ T cells and myeloid cells isolated ex vivo can be reactivated to produce infectious virus in culture. These findings are novel and demonstrate that the Mϕ reservoir is not limited to the brain but that monocytes in blood and Mϕs in spleen and lung also harbor latent SIV genomes that, upon reactivation, can produce infectious virus ex vivo. Monocytes play an important role in immune surveillance and can enter tissues and mature into macrophages. The short half-life in monocytes suggests that these cells cannot represent a viral reservoir. However, entry into tissues of latently infected monocytes that mature into long-lived macrophages would constitute a reservoir. To determine the stability of the Mϕ reservoir, we assessed cell longevity and DNA stability in long-term ex vivo cultures. The CD4+ T cell reservoir is likely maintained by lymphocyte proliferation and the division of latently infected cells (55, 65–67). Similarly, macrophages directly originated from the yolk sac, such as red pulp and alveolar macrophages, may maintain their populations through cell division and self-renewal (68). In this study, we have demonstrated that ex vivo Mϕs are long-lived and maintain stable levels of proviral DNA in culture for at least 21 days (Fig. 6). Other groups have demonstrated that monocyte-derived macrophages with a shorter life span, such as interstitial macrophages in the lung, can also be infected (69), although the role that they play in the maintenance of the reservoir is still unexplored. Additionally, the viruses released in the Mϕ QVOAs are replication competent and infect PBMCs from healthy macaques (Fig. 9). These data suggest that although Mϕs do not release virus as robustly as CD4+ T cells, the virus that is released is infectious and capable of reestablishing infection after ART interruption.
The mechanism that establishes latency in macrophages is unclear but would be expected to be different from that in CD4+ T cells. The mechanisms by which CD4+ T cell establish latency are well-known and consist of two different processes. First, naive T cells carry the HIV-1 unintegrated DNA preintegration complex; therefore, naive T cells in preintegration latency are short-lived and may not contribute to long-term HIV-1 persistence. Second, memory T cells carry HIV-1 proviral DNA into the host’s genome postintegration and play an important role in long-term latency (70, 71). However, Kelly and colleagues have shown in an in vitro examination of HIV infection of macrophages that unintegrated viral DNA not only has an unusual stability but also maintains biological activity and may provide a mechanism of latency in macrophages (72).
The presence of persistent infectious virus in myeloid cells during ART remains controversial in HIV infection. Many HIV-1 strains replicate efficiently in macrophages independently of the presence of Vpx (73, 74), an auxiliary protein present in HIV-2 and SIV that specifically enhances viral replication in macrophages (75, 76). Comparably, the Vpr present in HIV-1 recruits UNG2 into virions and modulates viral mutation rates in macrophages (77–79), indicating that these long-lived cells may carry intact genomes in vivo. A recent study reported that resident urethral Mϕs isolated from penile tissues contained latent HIV and could be reactivated ex vivo (80). However, previous work suggested that though liver Mϕs contained HIV DNA, the cells could not be reactivated ex vivo and infect activating CD4 T cells, suggesting that the integrated DNA was not functional (81). These studies point to the possibility that functional Mϕ reservoirs may not be present in all tissues but also highlight the difficulty of working with macrophages isolated from tissue. A 2014 study of a Mississippi baby suggested that myeloid cells may contribute to viral persistence, since there was no detectable viral DNA in resting CD4+ T cells but HIV DNA was present in monocyte-derived adherent cells (82). At 2 years after treatment interruption, the child rebounded and was immediately placed on ART. In addition, two HIV+ individuals with lymphoma who received allogeneic hematopoietic stem cell transplants (HSCT) had undetectable HIV RNA by standard clinical assays. However, at 3 and 8 months after ART interruption, both patients experienced a spontaneous and rapid rebound of HIV RNA in the plasma and CSF, accompanied by neurological symptoms (83). These data suggest that HIV may rebound from alternative reservoirs found in monocytes and Mϕs, which are not typically assessed prior to or during treatment interruption. Despite the limited numbers of animals analyzed and the broad probability range associated with QVOAs, our study provides new insight into the size and location of the SIV reservoir within tissues as well as provides evidence that SIV may rebound from CD4+ T cells and Mϕs after ART interruption and that both should be considered in the development of future HIV cure strategies.
MATERIALS AND METHODS
Ethics statement.
Animal studies were conducted in accordance with the guidelines of the Weatherall report (84), the Guide for the Care and Use of Laboratory Animals (85), and the USDA Animal Welfare Act. All studies were approved by the Johns Hopkins University Institutional Animal Care and Use Committee under protocol PR12M310. Animals were monitored twice daily by trained technicians or veterinarians for clinical signs of disease, such as weight loss, intractable diarrhea, and opportunistic infection, so that an early endpoint could be performed if necessary. Macaques were housed in Johns Hopkins University facilities, which are fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC), and fed a balanced Purina Mills macaque chow (Gray Summit, MO, USA). All animals were housed in groups prior to infection and in cages providing 6 square feet of space with environmental enrichment (manipulanda and novel foodstuffs), as well as visual and auditory contact with conspecifics. As mandated by the USDA, animals were housed in pairs following infection, which helped ensure that the animals received the appropriate drug dosage.
Animal studies.
Seven juvenile male pigtailed macaques (Macaca nemestrina) were inoculated intravenously with the SIV/DeltaB670 swarm and the molecular clone SIV/17E-Fr as previously described (42–44). Macaques were treated at 12 days postinoculation (dpi) with 30 mg/kg of body weight of tenofovir (Gilead, Foster City, CA) for the first 2 weeks and then with 10 mg/kg of tenofovir once daily intramuscularly as previously described (34), as well as with 480 mg/kg darunavir (Janssen, Titusville, NJ), 10 mg/kg of integrase inhibitor L000870812 (Merck, Kenilworth, NJ), and 24 mg/kg ritonavir (AbbVie, North Chicago, IL); the last three drugs were administered orally twice daily. In addition to antiretroviral drugs, Pm12, Pm22, and Pm23 were treated with latency-reversing agents (LRA) after 550, 242, and 249 dpi, respectively (86). The LRA regimen consisted of the protein kinase C activator ingenol-B (Amazônia Fitomedicamentos Ltda., Brazil) and the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]; Merck). Blood and CSF were drawn from each animal monthly prior to necropsy. Viral loads in plasma and CSF as well as absolute CD4+ T cell and monocyte counts are reported in Table 1. These studies were performed in accordance with federal guidelines and institutional policies. All manipulations were done while the animals were anesthetized with ketamine-HCl (Parke-Davis). Macaques were euthanatized at 194 to 628 days dpi using sodium pentobarbital while the animals were under ketamine sedation (10-mg/kg intramuscular injection) prior to perfusion with phosphate-buffered saline (PBS) to remove blood from tissues as previously described (43).
Isolation of myeloid cells and lymphocytes from blood and tissue.
Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation on a 1.077-g/ml Percoll/Hanks gradient (GE Healthcare, Pittsburgh, PA) according to the manufacturer’s protocol. Spleen and lung cells were mechanically removed from tissues using an 18-gauge needle and passed through a 100-μm-mesh-size cell strainer. Blood, lung, and spleen Mϕs were cultured in Dulbecco modified Eagle medium (Life Technologies) supplemented with 20% heat-inactivated human type AB serum (Gemini Bio Products, West Sacramento, CA), 100 U/ml penicillin-streptomycin (Life Technologies), 20 μg/ml gentamicin (Life Technologies), 2 mM l-glutamine (Life Technologies), 2 mM sodium pyruvate (Sigma), 10 mM HEPES buffer (Life Technologies), and 50 ng/ml recombinant human Mϕ colony-stimulating factor (MCSF; R&D, Minneapolis, MN). CD4+ T cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated bovine serum (Atlanta Biologicals), 100 U/ml penicillin-streptomycin (Life Technologies), 1% T cell growth factor, and 100 U/ml interleukin-2 (IL-2; Novartis, New York, NY).
Spleen, lung, and PBMC monocyte/Mϕ QVOAs.
Mϕ quantitative viral outgrowth assay (QVOAs) were completed on myeloid cells isolated from PBMCs, spleen, and lung as previously described (33). In brief, myeloid cells were purified using nonhuman primate CD11b+ MicroBeads (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s recommendation. Plates were coated with poly-l-lysine solution (Sigma) for at least 30 min and washed twice with PBS prior to cell plating. Purified Mϕs were cultured in duplicate in a 10-fold limiting dilution in the presence of 10 μM zidovudine (Sigma), 5 nM raltegravir (Merck), and 25 nM darunavir (Janssen, Titusville, NJ). Mϕs isolated from the spleen and lung were incubated for 2 to 4 days to allow for adherence. Monocytes isolated from PBMCs were incubated for 7 days to allow for Mϕ differentiation. Cells were then rinsed once with PBS to remove nonadherent cells and replenished with activation medium containing 10 ng/ml tumor necrosis factor (TNF; ProSpec), 1 μg/ml Pam3CSK4 (Sigma), and 1 μg/ml prostaglandin E2 (Sigma). Between 106 and 104 CEMx174 cells were added to all wells except those with T cell receptor β (TCRβ) controls, as previously described (33). Supernatants were collected and replenished with TNF, Pam3CSK4, and prostaglandin E2 every 2 days and assessed for SIV RNA by RT-qPCR. Supernatants from early time points (days 2, 4, 6) and later time points (days 8, 10, 12) were each pooled and assessed for viral RNA as described below. Cells were collected at day 14 and lysed in Buffer RLT Plus (Qiagen) with β-mercaptoethanol for RNA and DNA isolation. The frequency of cells harboring replication-competent virus was determined using the IUPMStats (v1.0) infection frequency calculator and expressed as the number of infectious units per million (IUPM) (40, 41). All Mϕ QVOAs were assessed for CD3+ T cell contamination using RT-qPCR for TCRβ as previously described (Table 3) (33). The total number of monocytes/macrophages assessed in the Mϕ QVOAs is described in Table 4.
TABLE 3.
Frequency of infected CD4+ T cells among macrophages isolated from spleen, lung, and blood Mϕ QVOAs
| Compartment and animal code | No. of CD4+ T cell IUPMa | No. of CD3+ T cells in Mϕ QVOA well | % CD3+ CD4+ CD8− T cells in blood at necropsy | No. of CD4+ T cells in Mϕ QVOA well | Probability of an infected CD4+ T cell present in Mϕ QVOA well | % chance an infected CD4+ T cell is present in an Mϕ QVOA well | IUPM Mϕ QVOA |
|---|---|---|---|---|---|---|---|
| Spleen | |||||||
| Pm11 | 1.27 | 0.88 | 50.9 | 0.45 | 5.69E−07 | 0.00 | 0.07 |
| Pm12 | 0.00 | 0.53 | 60.2 | 0.32 | 0.00 | 0.00 | 0.79 |
| Pm21 | 0.26 | 0.56 | 55.2 | 0.31 | 0.00 | 0.00 | 0.47 |
| Pm22 | 0.16 | 0.46 | 56.8 | 0.26 | 0.00 | 0.00 | 0.66 |
| Pm23 | 1.86 | 0.12 | 56.4 | 0.07 | 0.00 | 0.00 | 0.06 |
| Pm24 | 0.16 | 0.52 | 45.8 | 0.24 | 0.00 | 0.00 | 0.36 |
| Pm25 | 0.06 | 0.54 | 48.9 | 0.26 | 0.00 | 0.00 | 0.40 |
| Lunga | |||||||
| Pm11 | 0.84 | 0.37 | 50.9 | 0.19 | 0.00 | 0.00 | 0.06 |
| Pm12 | 0.00 | 0.40 | 60.2 | 0.24 | 0.00 | 0.29 | |
| Pm21 | 0.29 | 2.40 | 55.2 | 1.32 | 0.00 | 0.00 | 0.06 |
| Pm22 | 0.37 | 0.36 | 56.8 | 0.20 | 0.00 | 0.00 | 0.06 |
| Pm23 | 0.16 | 0.46 | 56.4 | 0.26 | 0.00 | 0.00 | 0.22 |
| Pm24 | 0.46 | 0.58 | 45.8 | 0.27 | 0.00 | 0.00 | 0.42 |
| Pm25 | 0.06 | 0.28 | 48.9 | 0.14 | 0.00 | 0.00 | 0.08 |
| PBMC | |||||||
| Pm11 | 0.84 | 0.49 | 50.9 | 0.25 | 0.00 | 0.00 | 0.06 |
| Pm12 | 0.00 | 0.62 | 60.2 | 0.37 | 0.00 | 0.40 | |
| Pm21 | 0.29 | 0.64 | 55.2 | 0.35 | 0.00 | 0.00 | 0.22 |
| Pm22 | 0.37 | 0.19 | 56.8 | 0.11 | 0.00 | 0.00 | 1.00 |
| Pm23 | 0.16 | 0.15 | 56.4 | 0.09 | 0.00 | 0.00 | 3.44 |
| Pm24 | 0.46 | 0.36 | 45.8 | 0.16 | 0.00 | 0.00 | 0.63 |
| Pm25 | 0.06 | 0.15 | 48.9 | 0.07 | 0.00 | 0.00 | 0.39 |
CD4+ T cell IUPM values were based on blood QVOA results.
TABLE 4.
Total number of cells assessed in each QVOA
| Animal code | Total no. of cells |
|||||
|---|---|---|---|---|---|---|
| Spleen QVOA |
Lung QVOA |
PBMC QVOA |
||||
| Mϕ | CD4+ T cells | Mϕ | CD4+ T cells | Mϕ | CD4+ T cells | |
| Pm11 | 6.44E + 07 | 4.37E + 06 | 1.78E + 07 | NDa | 1.27E + 07 | 2.50E + 07 |
| Pm12 | 2.22E + 06 | 5.00E + 05 | 2.22E + 07 | ND | 2.22E + 07 | 5.00E + 06 |
| Pm21 | 1.38E + 07 | 4.50E + 07 | 1.11E + 07 | ND | 1.93E + 07 | 5.50E + 07 |
| Pm22 | 7.78E + 07 | 4.37E + 06 | 3.80E + 06 | ND | 5.11E + 06 | 2.50E + 07 |
| Pm23 | 7.78E + 07 | 4.37E + 06 | 3.80E + 06 | ND | 5.11E + 06 | 2.50E + 07 |
| Pm24 | 2.89E + 07 | 2.50E + 07 | 2.89E + 06 | ND | 6.67E + 06 | 2.50E + 07 |
| Pm25 | 4.07E + 07 | 2.50E + 07 | 1.30E + 07 | ND | 3.82E + 06 | 2.50E + 07 |
ND, not done.
CD4+ T cell QVOA.
CD4+ T cells were isolated from the remaining cells after CD11b+ selection, and the levels of latently infected CD4+ T cells in spleen and blood were assessed using the CD4+ T cell QVOA as previously described (33, 87). The total numbers of CD4+ T cells assessed in each QVOA are reported in Table 4.
Quantitation of SIV gag RNA.
Viral RNA was measured in plasma, CSF, cell culture supernatants, and tissues by reverse transcriptase (RT) qPCR (RT-qPCR) or RT-ddPCR as previously described (33, 34). In brief, viral RNA was isolated in duplicate from 140 μl of plasma or supernatant using a QIAamp viral RNA minikit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. For tissues, total RNA was isolated from 50 mg of tissue in singlet or triplicate using an RNeasy kit (Qiagen) according to the manufacturer's protocol. As suggested by the manufacturer’s protocol, an on-column DNase digestion was performed for all samples using an RNase-free DNase set (Qiagen) and 3 units of RQ1 DNase (Promega, Madison WI). Quantification of SIV RNA was performed by RT-qPCR using a QuantiTect virus kit (Qiagen) or RT-ddPCR with a ddPCR Supermix for Probes kit (Bio-Rad) and a primer/probe set for SIV gag: primer SIV21F (5′-GTCTGCGTCATCTGGTGCATTC-3′), primer SIV22R (5′-CACTAGGTGTCTCTGCACTATCTGTTTTG-3′), and probe SIV23 (FAM-5′-CTTCCTCAGTGTGTTTCACTTTCTCTTCTG-3′-BHQ1, where FAM is 6-carboxyfluorescein and BHQ1 is black hole quencher 1) (Integrated DNA Technologies, Coralville, IA, USA). A Rotor-Gene Q thermocycler (Qiagen) was used for RT-qPCRs, and a QX-100 droplet digital PCR system (Bio-Rad) was used for ddPCRs, as previously described (86). Samples were assessed by RT-ddPCR when there were less than 10 copies/PCR mixture detected by RT-qPCR.
Quantitation of SIV gag DNA.
DNA was isolated from tissues using an AllPrep DNA/RNA kit (Qiagen) according to the manufacturer’s recommendations. Viral DNA was measured in tissues by multiplex qPCR with a multiplex PCR kit (Qiagen) or ddPCR with a ddPCR Supermix for Probes kit (Bio-Rad) using primers in the SIV gag region and the macaque IFN-β gene for sample normalization and cellular quantitation. A Rotor-Gene Q thermocycler (Qiagen) was used for RT-qPCRs, and a QX-100 droplet digital PCR system (Bio-Rad) was used for ddPCRs, as previously described (53, 88). Samples were assessed by ddPCR when there were less than 10 copies detected by qPCR.
Quantification of cellular SIV tat/rev RNA.
Quantification of SIV tat/rev RNA was performed by RT-qPCR using a QuantiTect virus kit (Qiagen) and a primer/probe set for SIV tat/rev: forward primer 3′-CGMARGAGAAGAAGAACTCCGAARAAG-5′, reverse primer 3′-CTATCTGYCAAGGCCARGA-5′, and probe FAM-5′-AACCAGAGAAGGMRAAGAAGGAGACGGTGM-3′-BHQ1 (Integrated DNA Technologies, Coralville, IA, USA). Three reactions were performed for each sample. To control for DNA contamination, one reaction mixture without reverse transcriptase was analyzed. Reaction mixtures were analyzed using a CFX96 real-time PCR detection system (Bio-Rad) with the following cycles: 50°C for 30 min, 95°C for 5 min, and 40 cycles of 95°C for 15 s, 54°C for 30 s, and 60°C for 1 min.
Flow cytometry.
Isolated CD4+ T cell and monocyte purities were analyzed as previously described (33). In brief, the purities of cells from the spleen, lung, and PBMCs were assessed by comparing preselection suspensions, postselection CD11b+ cells, and postselection flowthrough. Samples were stained in 100 μl PBS–2% fetal bovine serum with mouse anti-human CD3-phycoerythrin (PE) (clone SP34-2; BD Biosciences, San Jose, CA), CD11b-fluorescein isothiocyanate (Beckman Coulter, USA), or CD4-Brilliant Violet 650 (clone OKT4; BioLegend, San Diego, CA) and LIVE/DEAD Aqua stain (Invitrogen). Whole blood was stained with CD3-PE (clone SP34-2; BD Biosciences, San Jose, CA), CD4-Brilliant Violet 650 (clone OKT4; BioLegend, San Diego, CA), and CD8-Brilliant Violet 570 (clone RPA-T8 BioLegend, San Diego, CA). Samples were incubated for 20 min at room temperature and fixed for 10 min with BD fluorescence-activated cell sorting lysing solution (BD Biosciences, San Jose, CA). Samples were acquired using a BD LSRFortessa flow cytometer (BD Biosciences). All data were analyzed using FlowJo software (FlowJo, Ashland, OR). The percentage of total CD3+ CD4+ CD8− cells in whole blood was used to calculate CD3+ T cell contamination in monocyte/Mϕ cultures of the Mϕ QVOA (Table 3).
In vitro infection of PBMCs.
Peripheral blood mononuclear cells (PBMCs) from uninfected pigtailed macaques were isolated by Percoll density gradient. Isolated PBMCs were cultured in 50 ml of RPMI medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin (Gibco), 1% l-glutamine (Gibco), and 1% HEPES 1 M (Gibco) and stimulated for 48 h with 2 μg/ml recombinant human IL-2 (Life Technologies, Inc.) and 2 μg/ml phytohemagglutinin, M form (Life Technologies, Inc.). The activated PBMCs were then washed with Dulbecco’s phosphate-buffered saline (DPBS) and counted. Activated cells (1 × 106) were spinoculated (2 h at 1,200 × g, room temperature) with 500 μl of supernatant from positive Mϕ QVOA wells from spleen, lung, and blood assays. After spinoculation, the cells were washed once with DPBS and resuspended in 1 ml of complete RPMI, plated in a 24-well plate, and incubated at 37°C for 14 days. Supernatants were collected at 5, 7, 10, and 14 days postinfection, and fresh medium was replaced at each time point. RNA was isolated from 1 ml of sample using a QIAamp MinElute virus vacuum kit (Qiagen), and SIV gag RNA was quantitated by RT-qPCR as described above.
Statistics.
Infected cell frequencies in limiting dilution assays were calculated using the IUPMStats (v1.0) infection frequency calculator (http://silicianolab.johnshopkins.edu) (89).
ACKNOWLEDGMENTS
We thank the Johns Hopkins School of Medicine Retrovirus Laboratory for technical assistance and guidance. We also thank Gilead, Merck, Bristol-Myers Squibb, AbbVie, Janssen, ViiV, and Amazônia Fitomedicamentos Ltda. for providing us with the antiretrovirals and LRAs.
This study was supported by NCRR and the Office of Research Infrastructure Programs (ORIP) of NIH (grant P40 OD013117) and NIH grants PPG MH070306, NS077869, NS076357, U19-0AI076113, R56 AI118753, and 1R01AI127142. This research has been facilitated by the infrastructure and resources provided by the Johns Hopkins University Center for AIDS Research, an NIH-funded program (1P30AI094189).
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Tyagi M, Bukrinsky MC. 2012. Human immunodeficiency virus (HIV) latency: the major hurdle in HIV eradication. Mol Med 18:1096–1108. doi: 10.2119/molmed.2012.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deeks SG, Lewin SR, Havlir D. 2013. The end of AIDS: HIV infection as a chronic disease. Lancet 382:1525–1533. doi: 10.1016/S0140-6736(13)61809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Picado J, Deeks SG. 2016. Persistent HIV-1 replication during antiretroviral therapy. Curr Opin HIV AIDS 11:417–423. doi: 10.1097/COH.0000000000000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun T-W, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Wong JK, Hezareh M, Günthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 7.Siliciano JD, Siliciano RF. 2005. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 8.Lamers SL, Rose R, Maidji E, Agsalda-Garcia M, Nolan DJ, Fogel GB, Salemi M, Garcia DL, Bracci P, Yong W, Commins D, Said J, Khanlou N, Hinkin CH, Valdes Sueiras M, Mathisen G, Donovan S, Shirimizu B, Stoddart CA, McGrath MS, Singer EJ. 2016. HIV DNA is frequently present within pathologic tissues evaluated at autopsy from combined antiretroviral therapy-treated patients with undetectable viral loads. J Virol 90:8968–8983. doi: 10.1128/JVI.00674-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrich TJ, Deeks SG, Pillai SK. 2017. Measuring the size of the latent human immunodeficiency virus reservoir: the present and future of evaluating eradication strategies. J Infect Dis 215:S134–S141. doi: 10.1093/infdis/jiw648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yukl SA, Shergill AK, Ho T, Killian M, Girling V, Epling L, Li P, Wong LK, Crouch P, Deeks SG, Havlir DV, McQuaid K, Sinclair E, Wong JK. 2013. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence. J Infect Dis 208:1212–1220. doi: 10.1093/infdis/jit308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belmonte L, Olmos M, Fanin A, Parodi C, Baré P, Concetti H, Pérez H, de Bracco MM, Cahn P. 2007. The intestinal mucosa as a reservoir of HIV-1 infection after successful HAART. AIDS 21:2106–2108. doi: 10.1097/QAD.0b013e3282efb74b. [DOI] [PubMed] [Google Scholar]
- 12.Licht A, Alter G. 2016. A drug-free zone—lymph nodes as a safe haven for HIV. Cell Host Microbe 19:275–276. doi: 10.1016/j.chom.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 13.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, Chung YS, Penugonda S, Chipman JG, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. 2016. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530:51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Marle G, Church DL, van der Meer F, Gill MJ. 2018. Combating the HIV reservoirs. Biotechnol Genet Eng Rev 8725:76–89. doi: 10.1080/02648725.2018.1478662. [DOI] [PubMed] [Google Scholar]
- 15.Svicher V, Ceccherini-Silberstein F, Antinori A, Aquaro S, Perno CF. 2014. Understanding HIV compartments and reservoirs. Curr HIV/AIDS Rep 11:186–194. doi: 10.1007/s11904-014-0207-y. [DOI] [PubMed] [Google Scholar]
- 16.Williams KC, Burdo TH. 2009. HIV and SIV infection: the role of cellular restriction and immune responses in viral replication and pathogenesis. APMIS 117:400–412. doi: 10.1111/j.1600-0463.2009.02450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 18.Chun TW, Fauci AS. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc Natl Acad Sci U S A 96:10958–10961. doi: 10.1073/pnas.96.20.10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee GQ, Lichterfeld M. 2016. Diversity of HIV-1 reservoirs in CD4+ T-cell subpopulations. Curr Opin HIV AIDS 11:383–387. doi: 10.1097/COH.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sékaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatziioannou T, Evans DT. 2012. Animal models for HIV/AIDS research. Nat Rev Microbiol 10:852–867. doi: 10.1038/nrmicro2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatziioannou T, Ambrose Z, Chung NPY, Piatak M, Yuan F, Trubey CM, Coalter V, Kiser R, Schneider D, Smedley J, Pung R, Gathuka M, Estes JD, Veazey RS, KewalRamani VN, Lifson JD, Bieniasz PD, Bieniasz PD. 2009. A macaque model of HIV-1 infection. Proc Natl Acad Sci U S A 106:4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddiqui S, Perez S, Gao Y, Doyle-Meyers L, Foley BT, Li Q, Ling B. 2019. Persistent viral reservoirs in lymphoid tissues in SIV-infected rhesus macaques of Chinese-origin on suppressive antiretroviral therapy. Viruses 11:E105. doi: 10.3390/v11020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck SE, Queen SE, Metcalf Pate KA, Mangus LM, Abreu CM, Gama L, Witwer KW, Adams RJ, Zink MC, Clements JE, Mankowski JL. 2017. An SIV/macaque model targeted to study HIV-associated neurocognitive disorders. J Neurovirol 24:204–212. doi: 10.1007/s13365-017-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clements JE, Gama L, Graham DR, Mankowski JL, Zink MC. 2011. A simian immunodeficiency virus macaque model of highly active antiretroviral treatment: viral latency in the periphery and the central nervous system. Curr Opin HIV AIDS 6:37–42. doi: 10.1097/COH.0b013e3283412413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.North TW, Higgins J, Deere JD, Hayes TL, Villalobos A, Adamson L, Shacklett BL, Schinazi RF, Luciw PA. 2010. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol 84:2913–2922. doi: 10.1128/JVI.02356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourry O, Mannioui A, Sellier P, Roucairol C, Durand-Gasselin L, Dereuddre-Bosquet N, Benech H, Roques P, Le Grand R. 2010. Effect of a short-term HAART on SIV load in macaque tissues is dependent on time of initiation and antiviral diffusion. Retrovirology 7:78. doi: 10.1186/1742-4690-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araínga M, Edagwa B, Mosley RL, Poluektova LY, Gorantla S, Gendelman HE. 2017. A mature macrophage is a principal HIV-1 cellular reservoir in humanized mice after treatment with long acting antiretroviral therapy. Retrovirology 14:17. doi: 10.1186/s12977-017-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar N, Chahroudi A, Silvestri G. 2016. Animal models to achieve an HIV cure. Curr Opin HIV AIDS 11:432–441. doi: 10.1097/COH.0000000000000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palesch D, Bosinger SE, Mavigner M, Billingsley JM, Mattingly C, Carnathan DG, Paiardini M, Chahroudi A, Vanderford TH, Silvestri G. 2018. Short-term pegylated interferon α2a treatment does not significantly reduce the viral reservoir of simian immunodeficiency virus-infected, antiretroviral therapy-treated rhesus macaques. J Virol 92:e00279-18. doi: 10.1128/JVI.00279-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mavigner M, Habib J, Deleage C, Rosen E, Mattingly C, Bricker K, Kashuba A, Amblard F, Schinazi RF, Jean S, Cohen J, McGary C, Paiardini M, Wood MP, Sodora DL, Silvestri G, Estes J, Chahroudi A. 2018. Simian immunodeficiency virus persistence in cellular and anatomic reservoirs in antiretroviral therapy-suppressed infant rhesus macaques. J Virol 92:e00562-18. doi: 10.1128/JVI.00562-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A, Abbas W, Herbein G. 2014. HIV-1 latency in monocytes/macrophages. Viruses 6:1837–1860. doi: 10.3390/v6041837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avalos CR, Price SL, Forsyth ER, Pin JN, Shirk EN, Bullock BT, Queen SE, Li M, Gellerup D, O'Connor SL, Zink MC, Mankowski JL, Gama L, Clements JE. 2016. Quantitation of productively infected monocytes and macrophages of simian immunodeficiency virus-infected macaques. J Virol 90:5643–5656. doi: 10.1128/JVI.00290-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avalos CR, Abreu CM, Queen SE, Li M, Price S, Shirk EN, Engle EL, Forsyth E, Bullock BT, Gabhann F, Wietgrefe SW, Haase AT, Zink MC, Mankowski JL, Clements JE, Gama L. 2017. Brain macrophages in simian immunodeficiency virus-infected, antiretroviral-suppressed macaques: a functional latent reservoir. mBio 8:e01186-17. doi: 10.1128/mBio.01186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIlroy D, Autran B, Cheynier R, Clauvel JP, Oksenhendler E, Debré P, Hosmalin A. 1996. Low infection frequency of macrophages in the spleens of HIV+ patients. Res Virol 147:115–121. doi: 10.1016/0923-2516(96)80225-1. [DOI] [PubMed] [Google Scholar]
- 36.Kinter A, Moorthy A, Jackson R, Fauci AS. 2003. Productive HIV infection of resting CD4+ T cells: role of lymphoid tissue microenvironment and effect of immunomodulating agents. AIDS Res Hum Retroviruses 19:847–856. doi: 10.1089/088922203322493012. [DOI] [PubMed] [Google Scholar]
- 37.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O’Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cribbs SK, Lennox J, Caliendo AM, Brown LA, Guidot DM. 2015. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses 31:64–70. doi: 10.1089/AID.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo Y-H, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 41.Laird GM, Rosenbloom DIS, Lai J, Siliciano RF, Siliciano JD. 2016. Measuring the frequency of latent HIV-1 in resting CD4+ T cells using a limiting dilution coculture assay. Methods Mol Biol 1354:239–253. doi: 10.1007/978-1-4939-3046-3_16. [DOI] [PubMed] [Google Scholar]
- 42.Mankowski JL, Flaherty MT, Spelman JP, Hauer DA, Didier PJ, Amedee AM, Murphey-Corb M, Kirstein LM, Muñoz A, Clements JE, Zink MC. 1997. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol 71:6055–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zink MC, Suryanarayana K, Mankowski JL, Shen A, Piatak M Jr, Spelman JP, Carter DL, Adams RJ, Lifson JD, Clements JE. 1999. High viral load in the cerebrospinal fluid and brain correlates with severity of simian immunodeficiency virus encephalitis. J Virol 73:10480–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babas T, Dewitt JB, Mankowski JL, Tarwater PM, Clements JE, Zink MC. 2006. Progressive selection for neurovirulent genotypes in the brain of SIV-infected macaques. AIDS 20:197–205. doi: 10.1097/01.aids.0000198078.24584.21. [DOI] [PubMed] [Google Scholar]
- 45.Amedee AM, Lacour N, Gierman JL, Martin LN, Clements JE, Bohm R, Harrison RM, Murphey-Corb M. 1995. Genotypic selection of simian immunodeficiency virus in macaque infants infected transplacentally. J Virol 69:7982–7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witwer KW, Gama L, Li M, Bartizal CM, Queen SE, Varrone JJ, Brice AK, Graham DR, Tarwater PM, Mankowski JL, Christine Zink M, Clements JE. 2009. Coordinated regulation of SIV replication and immune responses in the CNS. PLoS One 4:e8129. doi: 10.1371/journal.pone.0008129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DIS, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. 2014. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price RW, Spudich S. 2008. antiretroviral therapy and central nervous system HIV type 1 infection. J Infect Dis 197:S294–S306. doi: 10.1086/533419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deere JD, Kauffman RC, Cannavo E, Higgins J, Villalobos A, Adamson L, Schinazi RF, Luciw PA, North TW. 2014. Analysis of multiply spliced transcripts in lymphoid tissue reservoirs of rhesus macaques infected with RT-SHIV during HAART. PLoS One 9:e87914. doi: 10.1371/journal.pone.0087914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kauffman RC, Villalobos A, Bowen JH, Adamson L, Schinazi RF. 2014. Residual viremia in an RT-SHIV rhesus macaque HAART model marked by the presence of a predominant plasma clone and a lack of viral evolution. PLoS One 9:e88258. doi: 10.1371/journal.pone.0088258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, Kim P, Chen T, Milush J, Hunt PW, Deeks SG, Wong JK, Yukl SA. 2018. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog 14:e1007357. doi: 10.1371/journal.ppat.1007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, Li M, Clements JE, Siliciano RF. 2003. Resting CD4 T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol 77:4938–4949. doi: 10.1128/jvi.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chun T-W, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. 1998. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK. 2018. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 10:eaap9927. doi: 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chavez L, Calvanese V, Verdin E. 2015. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog 11:e1004955. doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kessing CF, Nixon CC, Li C, Tsai P, Takata H, Mousseau G, Ho PT, Honeycutt JB, Fallahi M, Trautmann L, Garcia JV, Valente ST. 2017. In vivo suppression of HIV rebound by didehydro-cortistatin A, a “block-and-lock” strategy for HIV-1 treatment. Cell Rep 21:600–611. doi: 10.1016/j.celrep.2017.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douek DC. 2018. HIV infection: advances toward a cure. Top Antivir Med 25:121–125. [PMC free article] [PubMed] [Google Scholar]
- 59.Almodovar S. 2014. The complexity of HIV persistence and pathogenesis in the lung under antiretroviral therapy: challenges beyond AIDS. Viral Immunol 27:186–199. doi: 10.1089/vim.2013.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, Kuritzkes DR, Lederman MM, Para M, Gandhi RT. 2016. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 30:343–353. doi: 10.1097/QAD.0000000000000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fischer M, Joos B, Hirschel B, Bleiber G, Weber R, Günthard HF. 2004. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J Infect Dis 190:1979–1988. doi: 10.1086/425983. [DOI] [PubMed] [Google Scholar]
- 62.Fischer M, Joos B, Wong JK, Ott P, Opravil M, Hirschel B, Weber R, Günthard HF. 2004. Attenuated and nonproductive viral transcription in the lymphatic tissue of HIV-1-infected patients receiving potent antiretroviral therapy. J Infect Dis 189:273–285. doi: 10.1086/380797. [DOI] [PubMed] [Google Scholar]
- 63.Pasternak AO, Lukashov VV, Berkhout B. 2013. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology 10:41. doi: 10.1186/1742-4690-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Persaud D, Zhou Y, Siliciano JM, Siliciano RF. 2003. Latency in human immunodeficiency virus type 1 infection: no easy answers. J Virol 77:1659–1665. doi: 10.1128/JVI.77.3.1659-1665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F. 2016. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DIS, Keele BF, Ho Y-C, Siliciano JD, Siliciano RF. 2017. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J Exp Med 214:959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Röszer T. 2018. Understanding the biology of self-renewing macrophages. Cells 7:E103. doi: 10.3390/cells7080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai Y, Sugimoto C, Arainga M, Midkiff CC, Liu DX, Alvarez X, Lackner AA, Kim W, Didier ES, Kuroda MJ. 2015. Preferential destruction of interstitial macrophages over alveolar macrophages as a cause of pulmonary disease in simian immunodeficiency virus-infected rhesus macaques. J Immunol 195:4884–4891. doi: 10.4049/jimmunol.1501194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol 76:8518–8531. doi: 10.1128/jvi.76.17.8518-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Machado Andrade V, Stevenson M. 2019. Host and viral factors influencing interplay between the macrophage and HIV-1. J Neuroimmune Pharmacol 14:33–43. doi: 10.1007/s11481-018-9795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelly J, Beddall MH, Yu D, Iyer SR, Marsh JW, Wu Y. 2008. Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology 372:300–312. doi: 10.1016/j.virol.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorry PR, Churchill M, Crowe SM, Cunningham AL, Gabuzda D. 2005. Pathogenesis of macrophage tropic HIV-1. Curr HIV Res 3:53–60. doi: 10.2174/1570162052772951. [DOI] [PubMed] [Google Scholar]
- 74.DiNapoli SR, Hirsch VM, Brenchley JM. 2016. Macrophages in progressive human immunodeficiency virus/simian immunodeficiency virus infections. J Virol 90:7596–7606. doi: 10.1128/JVI.00672-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharova N, Wu Y, Zhu X, Stranska R, Kaushik R, Sharkey M, Stevenson M. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog 4:e1000057. doi: 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Westmoreland SV, Converse AP, Hrecka K, Hurley M, Knight H, Piatak M, Lifson J, Mansfield KG, Skowronski J, Desrosiers RC. 2014. SIV Vpx is essential for macrophage infection but not for development of AIDS. PLoS One 9:e1000057. doi: 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. 2004. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J Biol Chem 279:28419–28425. doi: 10.1074/jbc.M403875200. [DOI] [PubMed] [Google Scholar]
- 78.Planelles V, Barker E. 2010. Roles of Vpr and Vpx in modulating the virus-host cell relationship. Mol Aspects Med 31:398–406. doi: 10.1016/j.mam.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller CM, Akiyama H, Agosto LM, Emery A, Ettinger CR, Swanstrom RI, Henderson AJ, Gummuluru S. 2017. Virion-associated Vpr alleviates a postintegration block to HIV-1 infection of dendritic cells. J Virol 91:e00051-17. doi: 10.1128/JVI.00051-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ganor Y, Real F, Sennepin A, Dutertre CA, Prevedel L, Xu L, Tudor D, Charmeteau B, Couedel-Courteille A, Marion S, Zenak AR, Jourdain JP, Zhou Z, Schmitt A, Capron C, Eugenin EA, Cheynier R, Revol M, Cristofari S, Hosmalin A, Bomsel M. 2019. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol 4:633–644. doi: 10.1038/s41564-018-0335-z. [DOI] [PubMed] [Google Scholar]
- 81.Kandathil AJ, Sugawara S, Goyal A, Durand CM, Quinn J, Sachithanandham J, Cameron AM, Bailey JR, Perelson AS, Balagopal A. 2018. No recovery of replication-competent HIV-1 from human liver macrophages. J Clin Invest 128:4501–4509. doi: 10.1172/JCI121678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M, Chun T, Strain M, Richman D, Luzuriaga K. 2013. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med 369:1828–1835. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. 2014. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weatherall DJ. 2006. The use of non-human primates in research. https://mrc.ukri.org/documents/pdf/the-use-of-non-human-primates-in-research/.
- 85.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 86.Gama L, Abreu CM, Shirk EN, Price SL, Li M, Laird GM, Pate KAM, Wietgrefe SW, O’Connor SL, Pianowski L, Haase AT, Van Lint C, Siliciano RF, Clements JE. 2017. Reactivation of simian immunodeficiency virus reservoirs in the brain of virally suppressed macaques. AIDS 31:5–14. doi: 10.1097/QAD.0000000000001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. 2013. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meulendyke KA, Pletnikov MV, Engle EL, Tarwater PM, Graham DR, Zink MC. 2012. Early minocycline treatment prevents a decrease in striatal dopamine in an SIV model of HIV-associated neurological disease. J Neuroimmune Pharmacol 7:454–464. doi: 10.1007/s11481-011-9332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rosenbloom DIS, Elliott O, Hill AL, Henrich TJ, Siliciano JM, Siliciano RF. 2015. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis 2:ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]