Abstract
Genetic factors play a substantial role in the etiology of skeletal diseases, which involve 1) defects in skeletal development, including intramembranous ossification and endochondral ossification; 2) defects in skeletal metabolism, including late bone growth and bone remodeling; 3) defects in early developmental processes related to skeletal diseases, such as neural crest cell (NCC) and cilia functions; 4) disturbance of the cellular signaling pathways which potentially affect bone growth. Efficient and high-throughput genetic methods have enabled the exploration and verification of disease-causing genes and variants. Animal models including mouse and zebrafish have been extensively used in functional mechanism studies of causal genes and variants. The conventional approaches of generating mutant animal models include spontaneous mutagenesis, random integration, and targeted integration via mouse embryonic stem cells. These approaches are costly and time-consuming. Recent development and application of gene-editing tools, especially the CRISPR/Cas9 system, has significantly accelerated the process of gene-editing in diverse organisms. Here we review both mice and zebrafish models of human skeletal diseases generated by CRISPR/Cas9 system, and their contributions to deciphering the underpins of disease mechanisms.
Graphical Abstract
1. Introduction
The development of human skeletal system originates from the early embryonic period, followed by highly regulated growth and remodeling along the human life [1]. The disturbance of formation or maintenance of the skeletal system leads to a number of skeletal diseases, such as osteogenesis imperfecta, congenital scoliosis and osteoporosis.
Over the past few decades, it has been recognized that genetic factors play a substantial role in the etiology of human skeletal diseases [2]. Genetic defects can lead to skeletal diseases by interfering two processes: 1) prenatal development of the skeletal system or 2) postnatal bone metabolism. Representative diseases in the first category are osteogenesis imperfecta (OI) caused by COL1A1/COL1A2 etc. and achondroplasia caused by FGFR3 mutations [3,4], and congenital scoliosis resulted from compound inheritance of a rare null mutation and a hypomorphic allele of TBX6 in a considerable proportion of disease populations (about 10%) [[5], [6], [7]]. Typical skeletal diseases in the second category include osteopetrosis and osteoporosis leading to an abnormal bone density [8,9]. Osteopetrosis is mainly caused by defects of osteoclast, accompanied by a decreased bone resorption activity. Genetic variants affecting osteoclast function are often correlated with osteopetrosis, such as variants in TCIRG1, CLCN7, OSTM1, TNFSF11, and TNFRSF11A (RANK) [[10], [11], [12], [13]]. On the contrary, osteoporosis often occurs with decreased bone mass and low bone mineral density (BMD) [9].
Sequencing of human genome was accomplished in 2005, which has significantly driven the development of human genetics research [14]. Exome sequencing, as an efficient and cost-effective method, has promoted identification of novel genes and variants associated with genetic diseases [[15], [16], [17]]. Recently, whole-genome sequencing (WGS) emerged as a more powerful sequencing technique due to its whole coverage of human genome [18]. Although exome sequencing and WGS have generated a large number of potential candidate variants, many of them actually do not contribute to the disease. Even rare homozygous loss-of-function variants identified by WGS can sometimes appear to be clinically irrelevant to the corresponding diseases [19]. Therefore, function studies are indispensable to validate uncertain results from human genetic analysis and identify the true disease-causing variants/genes [20].
For the validation of a specific genetic variant, in vitro gene-modification techniques, including transgenesis, knock-out, and knock-in could be used [21,22]. Besides in vitro studies, animal models including mouse and zebrafish are also extensively used in studies of genetic diseases. Gene-modified animal models provide approaches for researchers to functionally verify the pathogenicity of candidate variants/genes and explore the disease mechanism [23,24].
The following part will review the advantages and disadvantages of several frequently-used animal models in the studies of skeletal diseases, advances in gene manipulation methods, and specific gene-edited animal models generated by CRISPR/Cas9 system which recapitulated various human skeletal diseases.
2. Animal Models in the Studies of Skeletal Diseases
In the studies of genetic skeletal diseases, mouse (Mus musculus) and zebrafish (Danio rerio) represent two important model animals.
Mouse is the most frequently used model animal for biomedical research [25]. Mouse is easy and inexpensive to breed. It takes 6–8 weeks to achieve sex maturity and 19–20 days to complete gestation. A female mouse can produce several litters per year, with approximately 5 cubs per litter. The skeletal phenotypes of mice and humans are comparable both physiologically and anatomically. Micro-CT, digital X-ray microradiography and other techniques enable the characterization of the skeletal structure of mice [26,27]. An important advantage of the mouse model is the high homology between its genome and the human genome, which enables functional studies of human genes in this model [25].
Zebrafish is a small tropical freshwater fish, the skeleton of which is also similar to that of human. Key genes regulating skeletal development are conservative across zebrafish and human. Therefore, most human genes can be studied and validated in zebrafish models and the same bio-markers can be used. For example, sp7 and cathepsin K can be used as the bio-marker of osteoblasts and osteoclasts respectively in both organisms [28,29]. Besides, easy maintenance and short life cycle of zebrafish can lower the cost and increase the efficiency of genetic research. The large brood size of zebrafish can offer a greater sample size. If properly bred, a female zebrafish can spawn every week or even at a shorter interval and generate hundreds of eggs in each clutch [30]. Further more, the zebrafish larvae are transparent and develop in vitro, which allows convenient observation and direct imaging of its skeletal development, making it an efficient tool to study skeletal diseases during the developmental process. Various techniques, such as live imaging, calcein staining, Alcian blue staining, micro-CT, have been developed to identify the skeletal phenotypes of both zebrafish larvae and adult fishes [31].
In comparison with mouse, zebrafish is more inexpensive and easier to breed. Its large brood size offers a large sample size for screening. In mouse genetic manipulation, labor-intensive embryo transplantation is usually required after microinjection. In contrast, the in vitro development of zebrafish can spare the procedure. These advantages make zebrafish especially suitable for high-throughput manipulation and screening.
Nevertheless, zebrafish genome usually has more than one orthologue for one human gene. In zebrafish knock-out models, the phenotypes of null mutants could be normal due to this genetic redundancy [32]. In contrast, mouse genome mostly has only one orthologous gene for each human gene. In addition, the skeletal structure of mouse is more similar to that of human compared to zebrafish [33].
3. CRISPR/Cas9 System in Animal Model Generation
Before gene manipulation became possible, the forward genetic approach was widely used to generate disease models. This approach requires random mutagenesis, which is whether spontaneous or induced by N-ethyl-N-nitrosourea (ENU), followed by high throughput forward screening. Since the mutagenesis is random and infrequent, it would take a long time to generate and validate an animal model with desired gene mutation [25,34]. In the early 1980s, scientists found that foreign DNA injected into zygotes can be integrated into the host genome via non-random microhomology-driven integration and nonhomologous end joining (NHEJ). In the following decades, microinjection of purified DNA into zygotes became the most common way to generate transgenic mouse or zebrafish models. Since the integration is unpredictable and inefficient, intensive screening is needed to select the subline with desired mutation, which is still costly and inefficient [35]. In the late 1980s, researchers made targeted integration possible via homologous recombination in mouse embryonic stem (ES) cells [36]. This approach requires microinjection of purified DNA into mouse ES cells and subsequent transplantation of ES cells into wild-type embryos. This approach increases the specificity of editing, but could only yield offspring with mosaic mutations. Several rounds of backcrosses are needed to yield a subline with germline transmission of the desired mutation, which makes it even more time-consuming than the random integration approach. Moreover, only a few ES cell lines are available, which limits the application of genome targeting via mouse ES cells [25].
In the last decade, the development of targeted gene editing tools, like zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and the CRISPR/Cas9 system, has revolutionized the process of model generation. Among the new tools, the CRISPR/Cas9 system has emerged as the most promising one. This system originates from the adaptive immune system of prokaryotes. Under the condition of infection, a fragment of the phage genome is taken up and integrated into the spacers of the CRISPR motif by the prokaryotes. The motif is then transcribed and processed into crRNA and tracrRNA. These transcription products can guide the Cas9 protein to the matching sequence of invading phage and cut it, generating double-strand breaks (DSBs). Researchers fused the crRNA and tracrRNA into a single guide RNA (sgRNA), which functions in the same way as the separated components do and has been widely used for its simplicity [37]. To direct the Cas9 nuclease to the specific sequence, the 20 nt at the 5′ end of the sgRNA should be homologous to the target sequence and immediately followed by an NGG protospacer adjacent motif (PAM) sequence [38]. In 2013, researchers demonstrated that the CRISPR-Cas9 system can be used to edit multiple genes simultaneously in eukaryotic cells [39]. In the same year, researchers successfully generated mouse and zebrafish models with multiple mutations [40,41]. Since then, various disease models have been generated using the CRISPR/Cas9 system. Some adaptations, including using organism-specific codons in Cas9 RNA, were made to the CRISPR/Cas9 system to increase its activity in different organisms [[41], [42], [43], [44], [45]]. The RNA-guided targeting activity of the CRISPR/Cas9 system also potentiates it for other sequence-specific activities besides generating DSBs, such as transcription regulation, epigenomic modification, and base editing [[46], [47], [48], [49]].
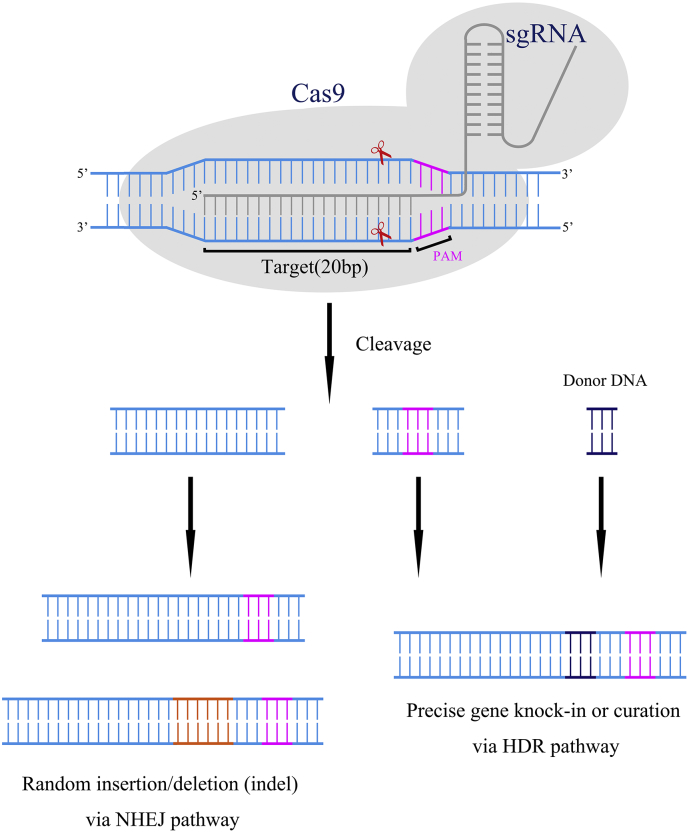
Like CRISPR/Cas9, ZFN and TALEN also have a targeting domain that directs them to a specific sequence and a nuclease domain capable of generating DSBs. The difference is their recognition of the specific sequence is based on protein-DNA interaction. But all these genome-editing tools achieve genome-editing by generating DSBs. Through microinjection into zygotes, these targeted nucleases can induce DSBs in a sequence-specific manner. As shown in Fig. 1, the DSBs can trigger endogenous DNA repair systems, including the NHEJ pathway and homology-directed repair (HDR) pathway [50]. The NHEJ pathway can repair the DSBs by directly joining the ends. This process is error-prone and may induce frame-shifting insertion or deletion (indel) at the excised site, thus knocking out the targeted gene. In contrast, the HDR pathway requires DNA templates to help repair the DSBs. When the targeted nucleases are co-injected with single-stranded oligodeoxynucleotides (ssODNs) or double-strand DNA (dsDNA) as donors for HDR, precise gene knock-in or curation can be achieved via the HDR pathway.
Fig. 1.
Mechanism of the CRISPR-Cas9 targeting system.
1. A sgRNA matches and binds to a 20-nt DNA sequence immediately upstream of an NGG DNA motif (Protospacer-Associated Motif, PAM).
2. The Cas9 protein is guided to the loci by the sgRNA and cuts both strands 3 bp upstream of the NGG.
3. The double-stranded DNA breaks activate the cellular DNA repair machinery, resulting in nonhomologous end joining (NHEJ) or homology-directed repair (HDR).
Precise gene knock-in and curation via the HDR pathway are usually preferable to the random indels generated via the NHEJ pathway, especially in clinical application. However, the efficiency of HDR is relatively low because most DSBs are repaired via the NHEJ pathway. Therefore, much effort has been made to increase the efficiency of the HDR pathway. Recent work has achieved that through optimization of templates, modification of the Cas9 protein, stimulation of the HDR pathway, and inhibition of the NHEJ pathway [[51], [52], [53], [54]].
The simplicity, relatively low cost, and high efficiency are the most important advantages of CRISPR/Cas9 system. It only takes one day to assemble the CRISPR/Cas9 system. In contrast, both ZFN and TALEN rely on DNA-protein interaction to recognize specific sequences. The design of a ZFN and TALEN requires the assembly of various protein motifs and subsequent validation, which is very complicated and requires a high level of experiment skills. In the current protocol, it takes seven days to design and assemble a TALEN system [[55], [56], [57], [58]]. Although the time required might be reduced to only one day, the complexity of the process is still a major drawback [59]. The simplicity and high efficiency of the CRISPR/Cas9 system also enable simultaneous editing of multiple loci and the introduction of different types of mutations. Besides indels, other types of mutations can also be achieved by targeting two loci simultaneously, which would be very complicated and time-consuming via the ZFN or TALEN approach. Small insertions can be achieved by co-injection of a small dsDNA or ssODNs. Large deletions can be achieved via NHEJ by co-injection of two sgRNAs targeting the two ends of the deletion. Co-injection of two sgRNAs and a circular DNA vector can trigger homologous recombination and introduce large insertions [25].
Nevertheless, the CRISPR/Cas9 system also has some limitations. The major limitation that restricts its clinical application is the off-target effect [60]. A few mismatches distal to the PAM region does not affect the activity of the CRISPR/Cas9 system. Therefore, sgRNAs may guide the Cas9 nuclease to the loci with sequences similar to the targeted loci and generate undesired mutations. As possible solutions to this drawback, optimization of sgRNA sequence and modification of Cas9 protein that degrades faster can both reduce the off-target effect [61]. Recent studies have also reported variant strains of Cas9 proteins with higher specificity [62,63]. Screening and engineering of Cas protein orthologues can also generate novel Cas protein orthologues with increased specificity and targeting range [47,[64], [65], [66], [67]]. Recent studies have also reported many techniques to precisely detect off-target sites in vivo, which makes the potential consequences of the off-target activities more manageable [68]. Another major limitation of the CRISPR/Cas9 system is the mosaicism in founders (F0). This could be caused by the delayed transcription of Cas9 RNA, the persistent activity of the CRISPR/Cas9 system after the one-cell stage, or the variety of DNA-repair mechanisms. Mosaicism can help avoid the lethality caused by homozygous null mutations, and can also lead to the generation of various mutant strains with different nucleotide changes and different mutation copies. However, in order to generate offspring with stable germline transmission of mutations, subsequent crosses and screening are needed, which is costly and time-consuming. Multiple improvements, such as using purified Cas9 protein instead of Cas9 RNA, engineering of Cas9 protein to accelerate its degradation, and embryo splitting, have been found to reduce mosaicism [69].
4. Skeletal Diseases and Corresponding Animals Model Generated by the CRISPR/Cas9 System
In recent years, the CRISPR/Cas9 system has been extensively used for eukaryotic genome editing and has led to the generation of numerous animal models of skeletal diseases. Existing disease animal models cover a range of skeletal diseases, revealing the pathogenic processes and factors involved in the embryonic skeletal development and bone metabolism.
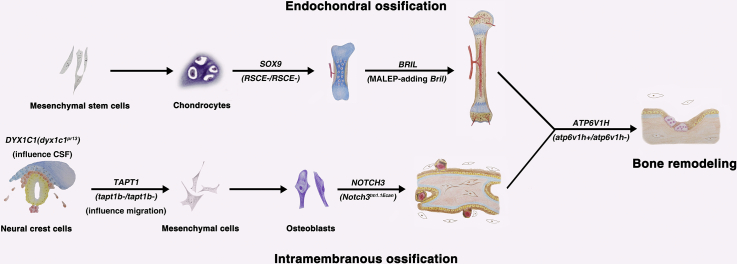
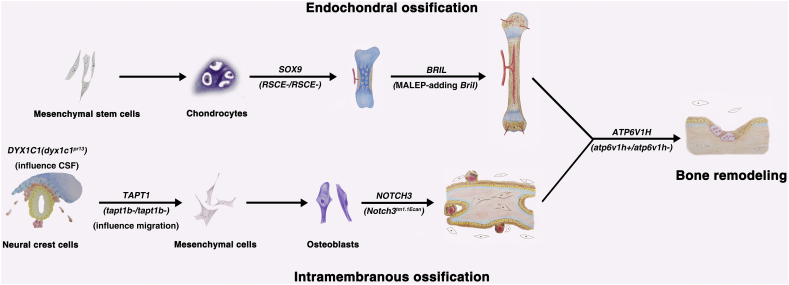
Here we review both mice and zebrafish models of human skeletal diseases generated by CRISPR/Cas9 system (Table 1), the etiologies of which could be divided into several categories: 1) defects in skeletal development, including intramembranous ossification defects and endochondral ossification defects; 2) defects in skeletal metabolism, including late bone growth defects and bone remodeling defects; 3) defects in early developmental processes related to skeletal diseases, like neural crest cell (NCC) and cilia defects; 4) disturbance of the cellular signaling pathways which potentially affect bone growth. Besides, human genes and the genotypes of corresponding animal models reviewed in this article are shown in Fig. 2 which illustrates the main processes of skeleton development and remodeling.
Table 1.
The contribution of CRISPR and zebrafish/mice/rat model to skeletal diseases.
| Animal model | Mutated animal gene | CRISPR/Cas9 interventions | Animal phenotype | Modeling disease | Mutated human gene |
|---|---|---|---|---|---|
| Zebrafish | mapk7 | Knock-out | Spinal curvature | AIS | MAPK7 |
| Rat | Bglap | Knock-out | Increased trabecular bone, increased bone strength | NA | NA |
| Mice | Notch3 | Knock-out | Marked osteopenia (decreased trabecular volume) | Lateral meningocele syndrome | NOTCH3 |
| Zebrafish | esf1 | Knock-out | Missing pharyngeal cartilages | Birth defects | NA |
| Zebrafish | atp6v1h | Knock-out | Bone loss and decreased bone formation | Osteoporosis | ATP6V1H |
| Mice | Atp6v1h | Knock-out | Bone loss and decreased bone formation | Osteoporosis | ATP6V1H |
| Zebrafish | dyx1c1 | Knock-out | Spinal curvature | NA | NA |
| Zebrafish | tapt1b | Knock-out | Malformation of the cranial cartilage, disorganization of chondrocytes in the ceratohyal and ceratobranchial cartilage | Osteochondrodysplasic | TAPT1 |
| Mice | Bril | Knock-in | Hypomineralization of the skull bones, bend limbs, wavy and thin dorsal ribs and wavy and thin dorsal ribs | Osteogenesis imperfecta | BRIL (IFITM5) |
| Mice | Sox9 | Enhancer elimination | Narrower and shorter rib cage | Campomelic dysplasia | SOX9 |
Fig. 2.
Manipulation of genes participating in various physiological processes
Human genes and genotypes/mutated-alleles of corresponding skeletal diseases related animal models are shown in the main physiological processes of bone development and remodeling. CRISPR/Cas9 system contributes to the establishment of those animal models.
Abbreviations: CSF, cerebrospinal fluid.
4.1. Skeletal Diseases Caused by Defects in Osteogenesis
In the early stages of skeletal development, mesenchymal cells aggregate into condensations, then differentiate into multiple cell types, including osteogenic and chondrogenic lineages which play critical roles in osteogenesis [70]. Osteogenesis occurs by one of the two processes: intramembranous ossification and endochondral ossification, both of which are accompanied by simultaneous bone resorption and remodeling [71].
4.1.1. Intramembranous Ossification Defects
Cranial bones, as well as some other flat bones, are formed through intramembranous ossification. As the basis of intramembranous ossification, mesenchymal condensation and ossification center formation first take place in embryonic mesenchymal tissue. Then, osteoprogenitor cells differentiated from the condensed mesenchymal cells proliferate and form osteoblast layers, which secret osteoid components [71]. Osteoblasts act as the builder in intramembranous ossification. Abnormalities in osteoblast can directly lead to several human skeletal diseases. For instance, the lateral meningocele syndrome (LMS) is a rare skeletal disease associated with osteoporosis [72]. Common clinical manifestations of LMS include the thickening of the cranial vault and craniofacial defects [73,74], which are etiologically related to abnormal intramembranous ossification. In 2015, Gripp et al. identified five novel de novo NOTCH3 mutations in six unrelated patients. All the mutations were located in exon33 of NOTCH3, leading to truncated proteins without the sequence required for protein degradation. Therefore, they attributed the etiology of LMS to truncating variants in exon33 of NOTCH3 [72]. However, there was limited knowledge about the mechanisms or the actions of NOTCH3 in the skeleton. Hence, in vivo functional study of those truncating variants was required to further explore the disease mechanism that how such variants affect human skeleton system.
Canalis et al. utilized CRISPR/Cas9 system to generate heterozygous Notch3tm1.1Ecan mice strain [75]. Notch3 sgRNA was designed to cleave between nucleotide 6691 and 6692 of exon 33 of Notch3 gene in mice genome. A nonsense mutation was introduced into the Notch3 gene of progenies. This mutation resulted in a truncated protein which deleted the sequence required for NOTCH3 degradation, leading to an upregulated NOTCH3 level. In Notch3tm1.1Ecan mice, the researchers observed a 35%–60% decrease in cancellous bone volume along with a reduction in trabecular number, which mimics the skeletal manifestation of human LMS [75]. Despite the skeletal manifestations reported in LMS, the mechanisms under LMS and the actions of NOTCH3 were also studied and discussed with those CRISPR-generated mice. Activated Notch signaling, increased cell proliferation, and enhanced Tnfsf11 expression were observed in osteoblasts extracted from LMS mice [75]. Those results indicated an enhanced osteoblast number and function in LMS mice. However, enhanced bone formation was not sufficient to maintain skeletal homeostasis because of the simultaneous increase in osteoclast surface/bone surface, and the bone loss still occurred. In that study, the phenotype of the Notch3tm1.1Ecan mutant mouse was congruent with the bone loss observed in humans with LMS. Such similarity strongly supported the pathogenicity of corresponding human variants. Additionally, CRISPR-mediated variant knock-in provided opportunities to investigate the molecular mechanism of LMS in a mice model, promoting the research of disease pathogenesis.
4.1.2. Endochondral Ossification Defects
Endochondral ossification is responsible for the initiation of osteogenesis in most components of human skeletal system, especially the long bones [76,77]. The most distinguished step of the endochondral ossification is the formation of hyaline cartilage models. After the hyaline cartilage models are formed, osteoblasts form a bone collar. The cartilage in the center starts to disintegrate, leading to a porous central region with a calcified remnant. Then, blood vessels invade and bring osteoprogenitor into the porous central region. After that, the primary and second ossification centers appear successively and promote the osteogenesis process. In the endochondral ossification process, the formation of hyaline cartilage is mostly studied [71]. Abnormalities of chondrocyte development could impede the endochondral ossification and cause various human skeletal diseases [78].
Campomelic dysplasia (CD) is a severe congenital disease characterized by impaired cartilage development. Common symptoms of CD include bowing and shortening of long bones, a bell-shaped thorax, narrow iliac wings, respiratory distress and other sexual problems [[79], [80], [81]]. As a subtype of CD, acampomelic campomelic dysplasia (ACD) shares all the other clinical phenotypes of CD except abnormal curvature of long bones [82]. Haploinsufficiency of SOX9 has been identified in many ACD patients, implicating its crucial role in this disease [83]. SOX9 is an important regulator of cartilage development [84,85], with a long enhancer region shared by different tissues [[86], [87], [88]]. Although the cartilage-specific SOX9 enhancer has been expected to be 1 Mb upstream of SOX9 based on genomic analyses of ACD patients [82,89,90]. More precise identification and characterization of the far upstream enhancer were still limited by the lack of successful and available approaches.
In recent years, the development of highly sequence-specific CRISPR/Cas9 system has offered researchers a better choice to localize and characterize the cartilage-specific SOX9 enhancer. Mochizuk et al. utilized SIN3A-dCas9-mediated epigenetic silencing to further screen for the candidate enhancer range. A conserved region in 1.7 Mb Sox9 upstream region was identified as cartilage-specific SOX9 enhancer, named as rib cage-specific enhancer (RCSE). CRISPR/Cas9 system was then used to eliminate RCSE region and construct an ACD/CD disease model in mice. Two different sgRNAs respectively matched with the sequence upstream and downstream RCSE sequence were designed. Co-injection of those two sgRNAs and hCas9 mRNA successfully deleted the RCSE sequence, generating RCSE+/− and RCSE−/− mice strains. Distinctly defective rib cages were observed in RCSE+/− mice and RCSE−/− mice, which appear to be significantly shorter and narrower compared to those of wild-type mice [91] phenotypes observed in RCSE null mice were consistent with the clinical phenotypes of human ACD/CD patients, providing opportunities to further study the mechanism and explore potential targeted therapies for ACD/CD.
In the former study, combinatorial analysis using the CRISPR/Cas9 system offered another approach for detailed investigation of the transcriptional mechanisms of inherited diseases. The identification of this new RCSE enhancer and its corresponding molecular functions will aid in the establishment of definitive diagnoses and identification of potential targets for the treatment of the ACD/CD. Moreover, CRISPR-generated RSCE null mouse could act as an animal model with impaired cartilage development, promoting the research focused on the skeletal diseases related to cartilage defect.
After the cartilage models are formed, during the process of endochondral ossification, bone collar and the primary ossification center play crucial roles. The formation of bone collar is accompanied by the impediment of nutrient supply and can lead to the hypertrophy and apoptosis of the chondrocytes, enlarging the lacunae and offering a porous structure for osteoblast covering. Simultaneously, calcification of cartilage matrix occurs, and the osteoblasts adhere to the cartilage remnant, forming primary ossification center. Disturbance of these normal processes will lead to serious skeletal defects [71].
Osteogenesis imperfecta (OI), clinically characterized by brittle bone [92], is mostly caused by autosomal dominant variants in COL1A1 and COL1A2 [3]. Those two genes code type I procollagen, which provides strength and resiliency to tissues such as bone. The pathogenic variants in COL1A1 and COL1A2 could decrease the expression of type I collagen or disrupt its structure, further affecting normal skeleton development [93]. The cartilage mineralization impediment is a classical phenotype of some subtypes of OI [94], which indicates abnormal endochondral ossification in such disease. As a specific type of OI, OI type V not only has common OI features like low bone mass and scoliosis, but also shows some particular features associated with ectopic mineralization such as the formation of “hyperplastic callus” and interosseous calcification of the forearm [[95], [96], [97]]. OI type V is caused by a single recurrent heterozygous mutation in the 5′-untranslated region (5’UTR) of BRIL gene. A novel translation starting site is created, which adds 5 amino acid residues (Met-Ala-Leu-Glu-Pro, denoted MALEP) on N-terminus of natural BRIL protein [98]. BRIL, almost exclusively expresses in osteoblasts, is a small 132-amino-acid transmembrane protein [99]. Although the MALEP-BRIL was involved in defective mineralization in vitro [99,100], its physiological functions in vivo still remain to be explored with the aid of animal models. Transgenic mice expressing OI type V MALEP-BRIL were generated by conventional techniques before [100,101]. However, CRISPR/Cas9 system offered more efficient access to create knock-in mice model which perfectly mimicked the BRIL mutation of OI V patients.
Rauch et al. generated a knock-in mice strain through the CRISPR/Cas9 system. They synthesized gRNA targeted to 5’UTR of mice Bril gene and injected it into mouse embryos together with spCas9 mRNA. Two mosaic knock-in founders were screened through PCR genotyping, which contained the dominant MALEP-adding variant [102]. The CRISPR generated MALEP-BRIL heterogeneous mice offered a reliable animal model for OI type V. Striking skeletal anomalies such as hypomineralized skulls, short and bent long bones, and frail and wavy ribs were all observed in the MALEP-BRIL heterogeneous mice. The hypertrophic chondrocytes filling the midshaft of long bones were also observed, which indicated the abnormality of chondrocyte hypertrophy and the following cell apoptosis. MALEP-BRIL heterogeneous mice also exhibited less mineralization and excessive cartilaginification, which were derived from the defective osteoblast differentiation and bone collar formation. Gene expression monitored in those knock-in mice embryos showed decreased osteoblast markers and overexpressed pro-inflammatory reaction related genes [102]. Such phenomena were consistent with the phenotypes observed in OI V patients, and indicated possible pathways (inflammation) which were abnormal in MALEP-BRIL heterogeneous mice.
4.2. Skeletal Disease Caused by Defects in Bone Growth and Remodeling
After osteogenesis and early bone growth, the newly formed bone stays in a dynamic state, in which the resorption and formation of bone tissue keep proceeding. The bone shape is generally maintained or slightly changed, while the bone mass keeps increasing [71]. During the bone remodeling and growth, osteoblasts and osteoclasts undertake the primary responsibility [103]. Defects in osteoblasts and/or osteoclasts could break the balance between osteoblastic formation and osteoclastic resorption, causing diseases with abnormality of bone growth and remodeling [71], such as osteoporosis and osteosclerosis.
Osteoporosis is characterized by lower-than-normal maximum bone mass [104], elevating the risk of fracture. The disturbance of osteoblast and osteoclast activities could result in a decrease of bone density, giving rise to a strong propensity of osteoporosis [105]. Similarly, CRISPR/Cas9 generated animal models also provided novel information about osteoporosis. In the research mentioned above, CRISPR generated Notch3tm1.1Ecan mice also showed BMD-related phenotype mimicking human osteopenia [75]. Furthermore, ATP6V1H, a subunit of V-ATPase which plays important roles in the biological and physiological functions of osteoclasts [106], is also a candidate gene associated with osteoporosis [107]. In the early years, research works on this gene were based on large-scale retroviral insertional mutagenesis screen. A variant in ATP6V1H was screened from hundreds of zebrafish mutants and identified as pathogenic mutation in the visual system [[108], [109], [110], [111]].
However, this forward genetics approach was unsuitable in the study of variants gained from clinical patients with specific diseases. The appearance of the CRISPR/Cas9 system supplied a powerful reverse genetic method, and offered better approaches for ATP6V1H function study in skeletal development and metabolism. Based on deleterious variants in ATP6V1H from clinical patients with osteoporosis (or low spine BMD), two groups generated Atp6v1h(atpv61h)-deficient mice and zebrafish respectively using CRISPR/Cas9 in the past five years. Frameshift mutations were introduced into both model animals and produced truncated proteins. The atp6v1h+/− zebrafish showed a distinct reduction of bone mineral density, bone volume and bone surface, which were basically consistent with the human osteoporosis phenotypes. Atp6v1h+/− mice also had decreased bone remodeling and a net bone matrix loss. Furtherly, both studies focused on the pathogenic mechanism of Atp6v1h(atpv61h) variants. In vitro analysis targeted to the osteoclast and osteoblast from atp6v1h+/− zebrafish revealed a high level of expression mmp9 and mmp13, which played a necessary role in extracellular matrix remodeling and possibly affected bone density. In Atp6v1h+/− mouse model, osteoclasts showed increased bone resorption. The TGF-β1 activation was downregulated on account of increased intracellular pH, thereby reducing the induction of osteoblast formation. Hence, the bone formation was reduced more than bone resorption in Atp6v1h deficient mice, possibly leading to osteoporosis [112,113].
In these studies, high throughput gene knock-out using CRISPR/Cas9 followed by extensive phenotyping could help elucidate the genetic basis of common skeletal diseases. CRISPR/Cas9 generated animal models play critical roles in the exploration of osteoporosis pathogenesis. Osteoblasts and osteoclasts extracted from CRISPR generated model animals with specific genotypes assisted the research of osteoporosis in cellular and molecular level.
4.3. NCC and Cilia-Related Skeletal Diseases
Besides osteogenesis, bone growth and remodeling, defects in earlier embryo components such as neural crest cells (NCC) could also lead to skeletal malformations. The development of NCC is the foundation of the whole craniofacial skeleton/cartilage development [114]. As a necessary component of NCC, Cranial neural crest cells have the potential to make most of the cranial bones [115]. Hence the defect of neural crest cells can lead to significant craniofacial malformations [116], which account for about 75% of all human birth defects [115]. Relevant studies have been focused on the development and migration of neural crest cells to reveal the mechanism of craniofacial defects [117,118].
In the field of craniofacial defect research, zebrafish model is extensively used as it has relative transparent larvae and efficient cartilage/skeleton stain methods [119,120]. The disturbance of NCC development and migration had been induced separately in CRISPR-generated mutated zebrafish, offering insight into the mechanism of NCC related human skeletal disease. For instance, development of NCC is impeded in CRISPR-generated esf1 mutated zebrafish strain. As an essential nucleolar protein, Esf1 is involved in 18sRNA biogenesis [121] and partially expressed in pharyngeal primordia and pharyngeal arch of zebrafish. Chen et al. designed gRNA to knockout two conserved sequences in C terminus of esf1, in order to interrupt the function of Esf1 protein. The esf1 mutated zebrafish model exhibited severe NCC-derived pharyngeal cartilage loss and defects in the eyes, brain and heart. Such an animal model supported a cellular requirement for Esf1 during neural crest survival and development [122]. In esf1-/esf1- zebrafish, NCC development was dramatically impaired, which made this zebrafish strain a suitable animal model for the research of cranial cartilage-related human diseases.
In recent years, the role of cilium in the process of NCC migration has been widely studied. Cilia are highly specialized microtubule-based organelles distributed on the cell membrane. They are responsible for signaling transduction, left-right (L-R) symmetry formation and central nervous system homeostasis [[123], [124], [125], [126], [127]]. Cilia also act as motile organelles, driving fluid movement and cell migration [122,124]. Symoens et al. analyzed two consanguineous families diagnosed as lethal osteogenesis imperfecta [126,128]. TAPT1 homozygous variants were screened out from patients' genome. TAPT1 encodes the evolutionary highly conserved transmembrane anterior posterior transformation 1 protein in centrosome and ciliary basal body. In the mechanism study, the TAPT1 variants were found to disturb the Golgi morphology and trafficking and normal primary cilium formation. Therefore, the homologous gene, tapt1b, was knocked out in the zebrafish through CRISPR/Cas9 system. CRISPR generated tapt1b−/− zebrafish model presented severe craniofacial cartilage malformations and delayed ossification. Such craniofacial cartilage defects were closely related to NCC migration dysfunction [126,128].
Apart from the establishment of NCC defect animal models, CRISPR/Cas9 was also used to explore the roles of cilia defect in human skeletal diseases. As shown in the former study, cilia were involved in the craniofacial cartilage development and potentially contributed to lethal osteogenesis imperfect [128]. Aside from that, ependymal cell cilia lining brain ventricles can generate cerebrospinal fluid (CSF) by the polarized beating [129]. Multiple evidence supports a conserved role of CSF in human spine development [[130], [131], [132], [133]]. In former studies, irregularities in CSF flow caused by cilia dysfunction have been demonstrated to lead to spine curvature and idiopathic scoliosis (IS) [130,[134], [135], [136]]. IS affects about 3% of children worldwide, causing spine deformities (scoliosis), reduced pulmonary function, and chronic pain [[137], [138], [139]]. Genetic factors play critical roles in the pathogenesis of IS, especially adolescent idiopathic scoliosis (AIS) [138,140]. As the etiology of IS is still unclear, the ‘cilia-CSF’ theory stands as an interesting hypothesis of IS pathogenesis [131,141]. Grimes et al. verified the critical role of cilia-driven CSF flow in spine development [130]. Besides, they also knocked out several cilia-related genes in zebrafish models through CRISPR/Cas9 system, which further proved the association between IS and cilia defect.
During the processes of mutant fish strains establishment, aberrant cilia motility caused a characteristic series of embryonic phenotypes that usually lead to death in 1 to 2 weeks of development, precluding further studies [142]. The phenomenon of lethality indicated the limitation of CRISPR, and could be restored with co-injection of WT mRNA. Transgenic cilia-gene mutants, similar phenotypes like severe three-dimensional spinal curvatures were also observed in the CRISPR-generated mutants [130]. Higher efficiency and greater convenience made CRISPR more accessible in the establishment of cilia defect zebrafish model, which benefited the research of cilia-related human skeletal diseases.
4.4. Skeletal Disease and Cellular Signaling Pathway Disturbance
Genes participating in some typical signaling pathways, like the Notch pathway, BMP pathway, and FGFR pathway are often associated with skeletal system development impediment [72,[143], [144], [145], [146], [147]]. For instance, as mentioned above, variants in SOX9 and NOTCH3 which are located on typical signaling pathways both contribute to human skeletal diseases [75,91]. CRISPR/Cas9 system–mediated gene knock-out could disturb skeletal system related signaling pathways and generate mutated animal models with abnormal phenotypes of skeletal diseases. Analyses of transcriptome and proteome were accessible in animal cells, which help researchers explore the changes of signaling pathways in molecular level and dramatically assist the research about molecular mechanism of skeletal diseases. In recent years, some novel cellular signaling pathways have also been related to skeletal development with the help of CRISPR. In Gao and his colleagues' work, a MAPK7 variant was screened from a three-generation Han Chinese family affected with AD-AIS. RAS/MAPK signally pathway has been known to regulate cell proliferation and differentiation [148]. CRISPR technique was utilized for in vivo validation of MAPK7 gene. Gao et al. designed a sgRNA targeted to mapk7 and generated chimeric mapk7-disrupting indel zebrafish mutants. Zebrafish mutants presented severe spinal curvature and recapitulated human disease phenotypes, which indicated the important role of RAS/MAPK signaling pathway in the etiology of adolescent idiopathic scoliosis [24].
5. Summary and Outlook
The extensive utilization of next-generation sequencing and genomic association analysis has generated plentiful candidate genes and variants in congenital and adult skeletal diseases, which warrant function test and validation. Due to its high efficiency and specificity, CRISPR/Cas9 system has stimulated the generation of animal models for human skeletal diseases. CRISPR-generated animal models have enabled mechanism explorations of various skeletal diseases, which are mainly related to skeletal development and metabolism processes. Based on the elucidation of pathogenic mechanisms, CRISPR-generated animal models could be coupled with drug screening and other therapeutic studies in the future [149].
However, there are still limitations to CRISPR/Cas9 system. Off-target gene knock-out effect would disturb the establishment of animal models and lead to potential false-positive results. Although the sequences and functions between human candidate genes and common model animals' orthologues are highly conserved, there are still natural genomic differences, impeding the confidence level of the proper skeletal disease model. Moreover, the cellular and molecular level changes caused by similar genomic operation can be dramatically different even opposite between human and model animals [75].
In conclusion, CRISPR/Cas9-editing is a powerful tool for genetic studies of human skeletal diseases. Extensive utilization of such efficient gene editing method will contribute to the future for mechanism exploration and therapeutic development of human skeletal diseases.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
This research was funded in part by the National Natural Science Foundation of China (81822030, 81772299, 81772301 & 81672123), Beijing Natural Science Foundation (7172175), the National Key Research and Development Program of China (No. 2018YFC0910506), the Central Level Public Interest Program for Scientific Research Institute (2018RC31003), CAMS Initiative Fund for Medical Sciences (2016-I2M-3-003, 2016-I2M-2-006 & 2017-I2M-2-001).
Contributor Information
Zhihong Wu, Email: orthoscience@126.com.
Jianguo Zhang, Email: zhangjianguo@pumch.cn.
References
- 1.Ljunggren O., Ljunghall S., Lerner U. Continuous remodeling of the skeleton. Growth factors and cytokines direct the activity. Lakartidningen. 1995;92(2094–2096):2099–2100. [PubMed] [Google Scholar]
- 2.McCarthy E.F. Genetic diseases of bones and joints. Semin Diagn Pathol. 2011;28:26–36. doi: 10.1053/j.semdp.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Forlino A., Marini J.C. Osteogenesis imperfecta. Lancet. 2016;387:1657–1671. doi: 10.1016/S0140-6736(15)00728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horton W.A., Hall J.G., Hecht J.T. Achondroplasia. Lancet. 2007;370:162–172. doi: 10.1016/S0140-6736(07)61090-3. [DOI] [PubMed] [Google Scholar]
- 5.Wu N., Ming X., Xiao J., Wu Z., Chen X. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang N., Wu N., Zhang L., Zhao Y., Liu J. TBX6 compound inheritance leads to congenital vertebral malformations in humans and mice. Hum Mol Genet. 2019;28:539–547. doi: 10.1093/hmg/ddy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Wu N., Yang N., Takeda K., Chen W. TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: further evidence supporting the compound inheritance and TBX6 gene dosage model. Genet Med. 2019;21:1548–1558. doi: 10.1038/s41436-018-0377-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobacchi C., Schulz A., Coxon F.P., Villa A., Helfrich M.H. Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat Rev Endocrinol. 2013;9:522–536. doi: 10.1038/nrendo.2013.137. [DOI] [PubMed] [Google Scholar]
- 9.Al-Barghouthi B.M., Farber C.R. Dissecting the genetics of osteoporosis using systems approaches. Trends Genet. 2019;35:55–67. doi: 10.1016/j.tig.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frattini A., Pangrazio A., Susani L., Sobacchi C., Mirolo M. Chloride channel ClCN7 mutations are responsible for severe recessive, dominant, and intermediate osteopetrosis. J Bone Miner Res. 2003;18:1740–1747. doi: 10.1359/jbmr.2003.18.10.1740. [DOI] [PubMed] [Google Scholar]
- 11.Pangrazio A., Poliani P.L., Megarbane A., Lefranc G., Lanino E. Mutations in OSTM1 (grey lethal) define a particularly severe form of autosomal recessive osteopetrosis with neural involvement. J Bone Miner Res. 2006;21:1098–1105. doi: 10.1359/jbmr.060403. [DOI] [PubMed] [Google Scholar]
- 12.Schinke T., Schilling A.F., Baranowsky A., Seitz S., Marshall R.P. Impaired gastric acidification negatively affects calcium homeostasis and bone mass. Nat Med. 2009;15:674–681. doi: 10.1038/nm.1963. [DOI] [PubMed] [Google Scholar]
- 13.Guerrini M.M., Sobacchi C., Cassani B., Abinun M., Kilic S.S. Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. Am J Hum Genet. 2008;83:64–76. doi: 10.1016/j.ajhg.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finishing the euchromatic sequence of the human genomeNature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 15.Do R., Kathiresan S., Abecasis G.R. Exome sequencing and complex disease: practical aspects of rare variant association studies. Hum Mol Genet. 2012;21:R1–R9. doi: 10.1093/hmg/dds387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welter D., MacArthur J., Morales J., Burdett T., Hall P. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. New Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoppers B.M., Zawati M.H., Senecal K. Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet. 2015;16:553–559. doi: 10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 19.Narasimhan V.M., Hunt K.A., Mason D., Baker C.L., Karczewski K.J. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du J., Feng W., Sun J., Kang C., Amizuka N. Ovariectomy upregulated the expression of Peroxiredoxin 1 &5 in osteoblasts of mice. Sci Rep. 2016;6:35995. doi: 10.1038/srep35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farr J.N., Weivoda M.M., Nicks K.M., Fraser D.G., Negley B.A. Osteoprotection through the deletion of the transcription factor Rorbeta in mice. J Bone Miner Res. 2018;33:720–731. doi: 10.1002/jbmr.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egunsola A.T., Bae Y., Jiang M.M., Liu D.S., Chen-Evenson Y. Loss of DDRGK1 modulates SOX9 ubiquitination in spondyloepimetaphyseal dysplasia. J Clin Invest. 2017;127:1475–1484. doi: 10.1172/JCI90193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao W., Chen C., Zhou T., Yang S., Gao B. Rare coding variants in MAPK7 predispose to adolescent idiopathic scoliosis. Hum Mutat. 2017;38:1500–1510. doi: 10.1002/humu.23296. [DOI] [PubMed] [Google Scholar]
- 25.Gurumurthy C.B., Lloyd K.C.K. Generating mouse models for biomedical research: technological advances. Dis Model Mech. 2019;12 doi: 10.1242/dmm.029462. (dmm029462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freudenthal B., Logan J., Croucher P.I., Williams G.R., Bassett J.H. Rapid phenotyping of knockout mice to identify genetic determinants of bone strength. J Endocrinol. 2016;231:R31–R46. doi: 10.1530/JOE-16-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackert-Bicknell C.L., Hibbs M.A. The need for mouse models in osteoporosis genetics research. Bonekey Rep. 2012 doi: 10.1038/bonekey.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay E.W., Apschner A., Schulte-Merker S. A bone to pick with zebrafish. Bonekey Rep. 2013;2:445. doi: 10.1038/bonekey.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Zhou Y., Qi X., Chen J., Chen W. CRISPR/Cas9 in zebrafish: an efficient combination for human genetic diseases modeling. Hum Genet. 2017;136:1–12. doi: 10.1007/s00439-016-1739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergen D.J.M., Kague E., Hammond C.L. Zebrafish as an emerging model for osteoporosis: a primary testing platform for screening new Osteo-active compounds. Front Endocrinol. 2019;10 doi: 10.3389/fendo.2019.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kok F.O., Shin M., Ni C.W., Gupta A., Grosse A.S. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon R.Y., Watson C.J., Karasik D. Using zebrafish to study skeletal genomics. Bone. 2019 doi: 10.1016/j.bone.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varshney G.K., Lu J., Gildea D.E., Huang H., Pei W. A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res. 2013;23:727–735. doi: 10.1101/gr.151464.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan B.W., Zhao Y.F., Cao W.G., Li N., Gou K.M. Mechanism of random integration of foreign DNA in transgenic mice. Transgenic Res. 2013;22:983–992. doi: 10.1007/s11248-013-9701-z. [DOI] [PubMed] [Google Scholar]
- 36.Doetschman T., Gregg R.G., Maeda N., Hooper M.L., Melton D.W. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 37.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, NY) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishino Y., Krupovic M., Forterre P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol. 2018:200. doi: 10.1128/JB.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cong L., Ran F.A., Cox D., Lin S., Barretto R. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jao L.E., Wente S.R., Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vejnar C.E. 2017. Optimized CRISPR–Cas9 system for genome editing in zebrafish. [DOI] [PubMed] [Google Scholar]
- 44.Modzelewski A.J., Chen S., Willis B.J., Lloyd K.C.K., Wood J.A. Efficient mouse genome engineering by CRISPR-EZ technology. Nat Protoc. 2018;13:1253–1274. doi: 10.1038/nprot.2018.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J., Zhou Y., Liu J., Chen J., Chen W. Progress and application of CRISPR/Cas Technology in Biological and Biomedical Investigation. J Cell Biochem. 2017;118:3061–3071. doi: 10.1002/jcb.26198. [DOI] [PubMed] [Google Scholar]
- 46.Tak Y.E., Kleinstiver B.P., Nunez J.K., Hsu J.Y., Horng J.E. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods. 2017;14:1163–1166. doi: 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuo E., Sun Y., Wei W., Yuan T., Ying W. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu W., Yang Y., Lei H. Progress in the application of CRISPR: from gene to base editing. Med Res Rev. 2019;39:665–683. doi: 10.1002/med.21537. [DOI] [PubMed] [Google Scholar]
- 50.Porteus M.H. A new class of medicines through DNA editing. New Engl J Med. 2019;380:947–959. doi: 10.1056/NEJMra1800729. [DOI] [PubMed] [Google Scholar]
- 51.Liu M., Rehman S., Tang X., Gu K., Fan Q. Methodologies for improving HDR efficiency. Front Genet. 2018;9:691. doi: 10.3389/fgene.2018.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boel A., De Saffel H., Steyaert W., Callewaert B., De Paepe A. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis Model Mech. 2018;11 doi: 10.1242/dmm.035352. (dmm035352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prykhozhij S.V., Fuller C., Steele S.L., Veinotte C.J., Razaghi B. Optimized knock-in of point mutations in zebrafish using CRISPR/Cas9. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., Zhang Z., Ge W. An efficient platform for generating somatic point mutations with germline transmission in the zebrafish by CRISPR/Cas9-mediated gene editing. J Biol Chem. 2018;293:6611–6622. doi: 10.1074/jbc.RA117.001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nerys-Junior A., Braga-Dias L.P., Pezzuto P., Cotta-de-Almeida V., Tanuri A. Comparison of the editing patterns and editing efficiencies of TALEN and CRISPR-Cas9 when targeting the human CCR5 gene. Genet Mol Biol. 2018;41:167–179. doi: 10.1590/1678-4685-GMB-2017-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ata H., Clark K.J., Ekker S.C. The zebrafish genome editing toolkit. Methods Cell Biol. 2016;135:149–170. doi: 10.1016/bs.mcb.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 58.Yasue A., Mitsui S.N., Watanabe T., Sakuma T., Oyadomari S. Highly efficient targeted mutagenesis in one-cell mouse embryos mediated by the TALEN and CRISPR/Cas systems. Sci Rep. 2014;4:5705. doi: 10.1038/srep05705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S., Chen H., Wang J. Generate TALE/TALEN as easily and rapidly as generating CRISPR. Mol Ther Methods Clin Dev. 2019;13:310–320. doi: 10.1016/j.omtm.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koo T., Lee J., Kim J.-S. Measuring and reducing off-target activities of programmable nucleases including CRISPR-Cas9. Mol Cells. 2015;38:475–481. doi: 10.14348/molcells.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kleinstiver B., Pattanayak V., Prew M., Tsai S., Nguyen N. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J.-J., Orlova N., Oakes B.L., Ma E., Spinner H.B. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566:218–223. doi: 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strecker J., Jones S., Koopal B., Schmid-Burgk J., Zetsche B. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10:212. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teng F., Li J., Cui T., Xu K., Guo L. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biol. 2019;20:15. doi: 10.1186/s13059-019-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen B., Zhang W., Zhang J., Zhou J., Wang J. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 68.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mehravar M., Shirazi A., Nazari M., Banan M. Mosaicism in CRISPR/Cas9-mediated genome editing. Dev Biol. 2019;445:156–162. doi: 10.1016/j.ydbio.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 70.Ju C., Liu R., Zhang Y.W., Zhang Y., Zhou R. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed Pharmacother. 2019;115:108912. doi: 10.1016/j.biopha.2019.108912. [DOI] [PubMed] [Google Scholar]
- 71.Mescher A.L. McGraw-Hill; 2013. Junqueira's basic histology: text and atlas. [Google Scholar]
- 72.Gripp K.W., Robbins K.M., Sobreira N.L., Witmer P.D., Bird L.M. Truncating mutations in the last exon of NOTCH3 cause lateral meningocele syndrome. Am J Med Genet A. 2015;167a:271–281. doi: 10.1002/ajmg.a.36863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Avela K., Valanne L., Helenius I., Makitie O. Hajdu-Cheney syndrome with severe dural ectasia. Am J Med Genet A. 2011;155a:595–598. doi: 10.1002/ajmg.a.33510. [DOI] [PubMed] [Google Scholar]
- 74.Gripp K.W., Scott C.I., Jr., Hughes H.E., Wallerstein R., Nicholson L. Lateral meningocele syndrome: three new patients and review of the literature. Am J Med Genet. 1997;70:229–239. [PubMed] [Google Scholar]
- 75.Canalis E., Yu J., Schilling L., Yee S.P., Zanotti S. The lateral meningocele syndrome mutation causes marked osteopenia in mice. J Biol Chem. 2018;293:14165–14177. doi: 10.1074/jbc.RA118.004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brighton C.T., Sugioka Y., Hunt R.M. Cytoplasmic structures of epiphyseal plate chondrocytes. Quantitative evaluation using electron micrographs of rat costochondral junctions with special reference to the fate of hypertrophic cells. J Bone Joint Surg Am. 1973;55:771–784. [PubMed] [Google Scholar]
- 77.Long F., Ornitz D.M. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013;5:a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aghajanian P., Mohan S. The art of building bone: emerging role of chondrocyte-to-osteoblast transdifferentiation in endochondral ossification. Bone Res. 2018;6:19. doi: 10.1038/s41413-018-0021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foster J.W., Dominguez-Steglich M.A., Guioli S., Kwok C., Weller P.A. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 80.Meyer J., Sudbeck P., Held M., Wagner T., Schmitz M.L. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: lack of genotype/phenotype correlations. Hum Mol Genet. 1997;6:91–98. doi: 10.1093/hmg/6.1.91. [DOI] [PubMed] [Google Scholar]
- 81.Wagner T., Wirth J., Meyer J., Zabel B., Held M. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 82.Fonseca A.C., Bonaldi A., Bertola D.R., Kim C.A., Otto P.A. The clinical impact of chromosomal rearrangements with breakpoints upstream of the SOX9 gene: two novel de novo balanced translocations associated with acampomelic campomelic dysplasia. BMC Med Genet. 2013;14:50. doi: 10.1186/1471-2350-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bi W., Huang W., Whitworth D.J., Deng J.M., Zhang Z. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 85.Wright E., Hargrave M.R., Christiansen J., Cooper L., Kun J. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 86.Bagheri-Fam S., Barrionuevo F., Dohrmann U., Gunther T., Schule R. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol. 2006;291:382–397. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 87.Sekido R., Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 88.Yao B., Wang Q., Liu C.F., Bhattaram P., Li W. The SOX9 upstream region prone to chromosomal aberrations causing campomelic dysplasia contains multiple cartilage enhancers. Nucleic Acids Res. 2015;43:5394–5408. doi: 10.1093/nar/gkv426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benko S., Fantes J.A., Amiel J., Kleinjan D.J., Thomas S. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 90.Leipoldt M., Erdel M., Bien-Willner G.A., Smyk M., Theurl M. Two novel translocation breakpoints upstream of SOX9 define borders of the proximal and distal breakpoint cluster region in campomelic dysplasia. Clin Genet. 2007;71:67–75. doi: 10.1111/j.1399-0004.2007.00736.x. [DOI] [PubMed] [Google Scholar]
- 91.Mochizuki Y., Chiba T., Kataoka K., Yamashita S., Sato T. Combinatorial CRISPR/Cas9 approach to elucidate a far-upstream enhancer complex for tissue-specific Sox9 expression. Dev Cell. 2018;46 doi: 10.1016/j.devcel.2018.07.024. (794-806.e796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bardai G., Moffatt P., Glorieux F.H., Rauch F. DNA sequence analysis in 598 individuals with a clinical diagnosis of osteogenesis imperfecta: diagnostic yield and mutation spectrum. Osteoporos Int. 2016;27:3607–3613. doi: 10.1007/s00198-016-3709-1. [DOI] [PubMed] [Google Scholar]
- 93.Gajko-Galicka A. Mutations in type I collagen genes resulting in osteogenesis imperfecta in humans. Acta Biochim Pol. 2002;49:433–441. [PubMed] [Google Scholar]
- 94.Glorieux F.H., Ward L.M., Rauch F., Lalic L., Roughley P.J. Osteogenesis imperfecta type VI: a form of brittle bone disease with a mineralization defect. J Bone Miner Res. 2002;17:30–38. doi: 10.1359/jbmr.2002.17.1.30. [DOI] [PubMed] [Google Scholar]
- 95.Blouin S., Fratzl-Zelman N., Glorieux F.H., Roschger P., Klaushofer K. Hypermineralization and high osteocyte lacunar density in osteogenesis imperfecta type V bone indicate exuberant primary bone formation. J Bone Miner Res. 2017;32:1884–1892. doi: 10.1002/jbmr.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Glorieux F.H., Rauch F., Plotkin H., Ward L., Travers R. Type V osteogenesis imperfecta: a new form of brittle bone disease. J Bone Miner Res. 2000;15:1650–1658. doi: 10.1359/jbmr.2000.15.9.1650. [DOI] [PubMed] [Google Scholar]
- 97.Liu G., Chen J., Zhou Y., Zuo Y., Liu S. The genetic implication of scoliosis in osteogenesis imperfecta: a review. J Spine Surg. 2017;3:666–678. doi: 10.21037/jss.2017.10.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cho T.J., Lee K.E., Lee S.K., Song S.J., Kim K.J. A single recurrent mutation in the 5'-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012;91:343–348. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moffatt P., Gaumond M.H., Salois P., Sellin K., Bessette M.C. Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res. 2008;23:1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 100.Hanagata N., Takemura T., Monkawa A., Ikoma T., Tanaka J. Phenotype and gene expression pattern of osteoblast-like cells cultured on polystyrene and hydroxyapatite with pre-adsorbed type-I collagen. J Biomed Mater Res A. 2007;83:362–371. doi: 10.1002/jbm.a.31240. [DOI] [PubMed] [Google Scholar]
- 101.Lietman C.D., Marom R., Munivez E., Bertin T.K., Jiang M.M. A transgenic mouse model of OI type V supports a neomorphic mechanism of the IFITM5 mutation. J Bone Miner Res. 2015;30:489–498. doi: 10.1002/jbmr.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rauch F., Geng Y., Lamplugh L., Hekmatnejad B., Gaumond M.H. Crispr-Cas9 engineered osteogenesis imperfecta type V leads to severe skeletal deformities and perinatal lethality in mice. Bone. 2018;107:131–142. doi: 10.1016/j.bone.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 103.Sims N.A., Martin T.J. Coupling the activities of bone formation and resorption: a multitude of signals within the basic multicellular unit. Bonekey Rep. 2014;3:481. doi: 10.1038/bonekey.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rachner T.D., Khosla S., Hofbauer L.C. Osteoporosis: now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brown C. Osteoporosis: staying strong. Nature. 2017;550:S15–s17. doi: 10.1038/550S15a. [DOI] [PubMed] [Google Scholar]
- 106.Yao G., Feng H., Cai Y., Qi W., Kong K. Characterization of vacuolar-ATPase and selective inhibition of vacuolar-H(+)-ATPase in osteoclasts. Biochem Biophys Res Commun. 2007;357:821–827. doi: 10.1016/j.bbrc.2007.04.082. [DOI] [PubMed] [Google Scholar]
- 107.Xu J., Cheng T., Feng H.T., Pavlos N.J., Zheng M.H. Structure and function of V-ATPases in osteoclasts: potential therapeutic targets for the treatment of osteolysis. Histol Histopathol. 2007;22:443–454. doi: 10.14670/HH-22.443. [DOI] [PubMed] [Google Scholar]
- 108.Gross J.M., Perkins B.D., Amsterdam A., Egana A., Darland T. Identification of zebrafish insertional mutants with defects in visual system development and function. Genetics. 2005;170:245–261. doi: 10.1534/genetics.104.039727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Amsterdam A., Nissen R.M., Sun Z., Swindell E.C., Farrington S. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nuckels R.J., Ng A., Darland T., Gross J.M. The vacuolar-ATPase complex regulates retinoblast proliferation and survival, photoreceptor morphogenesis, and pigmentation in the zebrafish eye. Invest Ophthalmol Vis Sci. 2009;50:893–905. doi: 10.1167/iovs.08-2743. [DOI] [PubMed] [Google Scholar]
- 111.Amsterdam A., Burgess S., Golling G., Chen W., Sun Z. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Duan X., Liu J., Zheng X., Wang Z., Zhang Y. Deficiency of ATP6V1H causes bone loss by inhibiting bone resorption and bone formation through the TGF-beta1 pathway. Theranostics. 2016;6:2183–2195. doi: 10.7150/thno.17140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y., Huang H., Zhao G., Yokoyama T., Vega H. ATP6V1H deficiency impairs bone development through activation of MMP9 and MMP13. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Minoux M., Rijli F.M. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- 115.Cordero D.R., Brugmann S., Chu Y., Bajpai R., Jame M. Cranial neural crest cells on the move: their roles in craniofacial development. Am J Med Genet A. 2011;155a:270–279. doi: 10.1002/ajmg.a.33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chai Y., Maxson R.E., Jr. Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- 117.Figueiredo A.L., Maczkowiak F., Borday C., Pla P., Sittewelle M. PFKFB4 control of AKT signaling is essential for premigratory and migratory neural crest formation. Development. 2017;144:4183–4194. doi: 10.1242/dev.157644. [DOI] [PubMed] [Google Scholar]
- 118.Smith T.G., Laval S., Chen F., Rock M.J., Strachan T. Neural crest cell-specific inactivation of Nipbl or Mau2 during mouse development results in a late onset of craniofacial defects. Genesis. 2014;52:687–694. doi: 10.1002/dvg.22780. [DOI] [PubMed] [Google Scholar]
- 119.Walker M.B., Kimmel C.B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- 120.Du S.J., Frenkel V., Kindschi G., Zohar Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev Biol. 2001;238:239–246. doi: 10.1006/dbio.2001.0390. [DOI] [PubMed] [Google Scholar]
- 121.Peng W.T., Krogan N.J., Richards D.P., Greenblatt J.F., Hughes T.R. ESF1 is required for 18S rRNA synthesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:1993–1999. doi: 10.1093/nar/gkh518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen J.Y., Tan X., Wang Z.H., Liu Y.Z., Zhou J.F. The ribosome biogenesis protein Esf1 is essential for pharyngeal cartilage formation in zebrafish. FEBS J. 2018;285:3464–3484. doi: 10.1111/febs.14622. [DOI] [PubMed] [Google Scholar]
- 123.Essner J.J., Vogan K.J., Wagner M.K., Tabin C.J., Yost H.J. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- 124.Simon M.J., Iliff J.J. Regulation of cerebrospinal fluid (CSF) flow in neurodegenerative, neurovascular and neuroinflammatory disease. Biochim Biophys Acta. 2016;1862:442–451. doi: 10.1016/j.bbadis.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yuan X., Serra R.A., Yang S. Function and regulation of primary cilia and intraflagellar transport proteins in the skeleton. Ann N Y Acad Sci. 2015;1335:78–99. doi: 10.1111/nyas.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singla V., Reiter J.F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 127.Goetz S.C., Anderson K.V. The primary cilium: a signalling Centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Symoens S., Barnes A.M., Gistelinck C., Malfait F., Guillemyn B. Genetic defects in TAPT1 disrupt Ciliogenesis and cause a complex lethal Osteochondrodysplasia. Am J Hum Genet. 2015;97:521–534. doi: 10.1016/j.ajhg.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee L. Riding the wave of ependymal cilia: genetic susceptibility to hydrocephalus in primary ciliary dyskinesia. J Neurosci Res. 2013;91:1117–1132. doi: 10.1002/jnr.23238. [DOI] [PubMed] [Google Scholar]
- 130.Grimes D.T., Boswell C.W., Morante N.F., Henkelman R.M., Burdine R.D. Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science. 2016;352:1341–1344. doi: 10.1126/science.aaf6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Engesaeth V.G., Warner J.O., Bush A. New associations of primary ciliary dyskinesia syndrome. Pediatr Pulmonol. 1993;16:9–12. doi: 10.1002/ppul.1950160103. [DOI] [PubMed] [Google Scholar]
- 132.Ohata S., Alvarez-Buylla A. Planar Organization of Multiciliated Ependymal (E1) cells in the brain ventricular epithelium. Trends Neurosci. 2016;39:543–551. doi: 10.1016/j.tins.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Knowles M.R., Daniels L.A., Davis S.D., Zariwala M.A., Leigh M.W. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Milhorat T.H., Chou M.W., Trinidad E.M., Kula R.W., Mandell M. Chiari I malformation redefined: clinical and radiographic findings for 364 symptomatic patients. Neurosurgery. 1999;44:1005–1017. doi: 10.1097/00006123-199905000-00042. [DOI] [PubMed] [Google Scholar]
- 135.Ozerdemoglu R.A., Denis F., Transfeldt E.E. Scoliosis associated with syringomyelia: clinical and radiologic correlation. Spine (Phila PA 1976) 2003;28:1410–1417. doi: 10.1097/01.BRS.0000067117.07325.86. [DOI] [PubMed] [Google Scholar]
- 136.Verhoef M., Barf H.A., Post M.W., van Asbeck F.W., Gooskens R.H. Secondary impairments in young adults with spina bifida. Dev Med Child Neurol. 2004;46:420–427. doi: 10.1017/s0012162204000684. [DOI] [PubMed] [Google Scholar]
- 137.Liu G., Liu S., Lin M., Li X., Chen W. Genetic polymorphisms of GPR126 are functionally associated with PUMC classifications of adolescent idiopathic scoliosis in a northern Han population. J Cell Mol Med. 2018;22:1964–1971. doi: 10.1111/jcmm.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu S., Wu N., Zuo Y., Zhou Y., Liu J. Genetic polymorphism of LBX1 is associated with adolescent idiopathic scoliosis in northern Chinese Han population. Spine (Phila PA 1976) 2017;42:1125–1129. doi: 10.1097/BRS.0000000000002111. [DOI] [PubMed] [Google Scholar]
- 139.Cheng J.C., Castelein R.M., Chu W.C., Danielsson A.J., Dobbs M.B. Adolescent idiopathic scoliosis. Nat Rev Dis Primers. 2015;1:15030. doi: 10.1038/nrdp.2015.30. [DOI] [PubMed] [Google Scholar]
- 140.Liu G., Liu S., Li X., Chen J., Chen W. Genetic polymorphisms of PAX1 are functionally associated with different PUMC types of adolescent idiopathic scoliosis in a northern Chinese Han population. Gene. 2019;688:215–220. doi: 10.1016/j.gene.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 141.Wang W.J., Yeung H.Y., Chu W.C., Tang N.L., Lee K.M. Top theories for the etiopathogenesis of adolescent idiopathic scoliosis. J Pediatr Orthop. 2011;31:S14–S27. doi: 10.1097/BPO.0b013e3181f73c12. [DOI] [PubMed] [Google Scholar]
- 142.Hjeij R., Onoufriadis A., Watson C.M., Slagle C.E., Klena N.T. CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am J Hum Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Coffin J.D., Homer-Bouthiette C., Hurley M.M. Fibroblast growth factor 2 and its receptors in bone biology and disease. J Endocr Soc. 2018;2:657–671. doi: 10.1210/js.2018-00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Canalis E. Notch in skeletal physiology and disease. Osteoporos Int. 2018;29:2611–2621. doi: 10.1007/s00198-018-4694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao B. TNF and bone remodeling. Curr Osteoporos Rep. 2017;15:126–134. doi: 10.1007/s11914-017-0358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tan Z., Ding N., Lu H., Kessler J.A., Kan L. Wnt signaling in physiological and pathological bone formation. Histol Histopathol. 2018;18062 doi: 10.14670/HH-18-062. [DOI] [PubMed] [Google Scholar]
- 147.Chen J., Liu J., Zhou Y., Liu S., Liu G. Molecular therapeutic strategies for FGFR3 gene-related skeletal dysplasia. J Mol Med (Berl) 2017;95:1303–1313. doi: 10.1007/s00109-017-1602-9. [DOI] [PubMed] [Google Scholar]
- 148.Stevenson D.A., Yang F.C. The musculoskeletal phenotype of the RASopathies. Am J Med Genet C Semin Med Genet. 2011;157c:90–103. doi: 10.1002/ajmg.c.30296. [DOI] [PubMed] [Google Scholar]
- 149.Brommage R. Genetic approaches to identifying novel osteoporosis drug targets. J Cell Biochem. 2015;116:2139–2145. doi: 10.1002/jcb.25179. [DOI] [PubMed] [Google Scholar]