Abstract
Previously, by targeting penicillin-binding protein 3, Pseudomonas-derived cephalosporinase (PDC), and MurA with ceftazidime-avibactam-fosfomycin, antimicrobial susceptibility was restored among multidrug-resistant (MDR) Pseudomonas aeruginosa. Herein, ceftazidime-avibactam-fosfomycin combination therapy against MDR P. aeruginosa clinical isolate CL232 was further evaluated. Checkerboard susceptibility analysis revealed synergy between ceftazidime-avibactam and fosfomycin. Accordingly, the resistance elements present and expressed in P. aeruginosa were analyzed using whole-genome sequencing and transcriptome profiling. Mutations in genes that are known to contribute to β-lactam resistance were identified. Moreover, expression of blaPDC, the mexAB-oprM efflux pump, and murA were upregulated. When fosfomycin was administered alone, the frequency of mutations conferring resistance was high; however, coadministration of fosfomycin with ceftazidime-avibactam yielded a lower frequency of resistance mutations. In a murine infection model using a high bacterial burden, ceftazidime-avibactam-fosfomycin significantly reduced the P. aeruginosa colony-forming units (CFUs), by approximately 2 and 5 logs, compared with stasis and in the vehicle-treated control, respectively. Administration of ceftazidime-avibactam and fosfomycin separately significantly increased CFUs, by approximately 3 logs and 1 log, respectively, compared with the number at stasis, and only reduced CFUs by approximately 1 log and 2 logs, respectively, compared with the number in the vehicle-treated control. Thus, the combination of ceftazidime-avibactam-fosfomycin was superior to either drug alone. By employing a "mechanism-based approach" to combination chemotherapy, we show that ceftazidime-avibactam-fosfomycin has the potential to offer infected patients with high bacterial burdens a therapeutic hope against infection with MDR P. aeruginosa that lack metallo-β-lactamases.
Keywords: Pseudomonas aeruginosa, β-lactams, fosfomycin, combination therapy
Multidrug-resistant (MDR) Pseudomonas aeruginosa is a serious public health threat. The ceftazidime-avibactam-fosfomycin combination possessed synergistic activity against MDR P. aeruginosa. A murine infection model validated the efficacy of this combination for MDR P. aeruginosa infections with high bacterial burdens.
Pseudomonas aeruginosa causes nearly 10% of hospital-acquired infections and is the most prevalent gram-negative pathogen isolated from patients with ventilator-associated pneumonia [1, 2]. Additionally, P. aeruginosa is difficult to treat with currently available antibiotics. Cationic antimicrobial peptides (eg, colistin) are often the only agents that remain effective against multidrug-resistant (MDR) P. aeruginosa [3]. Therefore, new therapeutic strategies against MDR P. aeruginosa are essential.
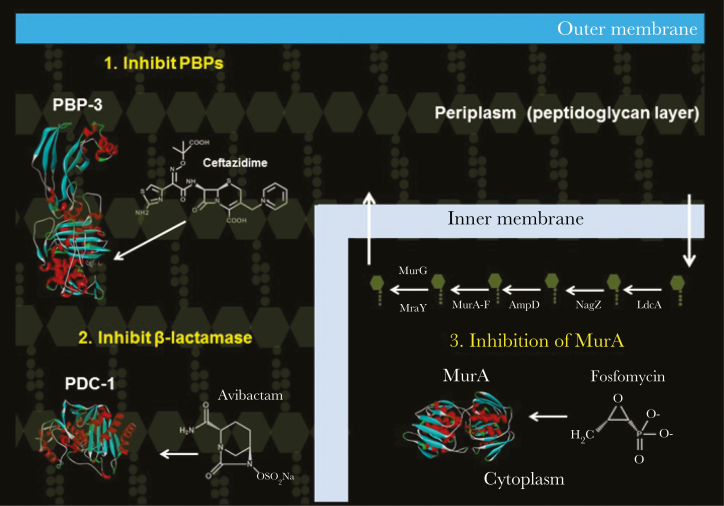
Ceftazidime-avibactam was approved for clinical use for the treatment of urinary tract infections caused by Enterobacteriaceae. Avibactam is a novel β-lactamase inhibitor shown to be effective against the class A and C β-lactamases expressed in P. aeruginosa [4]. However, the activity of avibactam toward class D β-lactamases (ie, OXA-2, OXA-5/10, and OXA-50 [poxB] families) from P. aeruginosa is more limited [5–7]. Winkler and Papp-Wallace et al demonstrated that ceftazidime-avibactam possesses potent activity against MDR P. aeruginosa, raising optimism regarding its potential use in the treatment of infections [8]. However, 18% of the MDR P. aeruginosa clinical isolates collected from a decade ago were resistant to ceftazidime-avibactam (Table 1) [8]. This observation was consistent with data obtained from the International Network for Optimal Resistance Monitoring Program, in the United States [9]. Importantly, by combining ceftazidime-avibactam with fosfomycin and targeting penicillin-binding protein 3 (PBP-3), the β-lactamases (eg, Pseudomonas-derived cephalosporinase [PDC] also known as Pseudomonas AmpC), and MurA (a UDP-N-acetylglucosamine-1-carboxyvinyltransferase), susceptibility was restored in vitro to most ceftazidime-avibactam–resistant P. aeruginosa isolates in the collection (Figure 1 and Table 1) [8].
Table 1.
Minimum Inhibitory Concentrations of Various Pseudomonas aeruginosa Clinical Isolates
| Strain | MEM | TZP | CIP | COL | CAZ | CAZ-AVIa | FOS | CAZ- AVI-FOSa,b | CAZ- AVI-COLa,c |
|---|---|---|---|---|---|---|---|---|---|
| PA01 | 0.5 | 1 | 1 | 0.5 | 2 | 1 | 64 | 1 | 0.5 |
| CL232 | 64 | >128 | 16 | 2 | >32 | 32 | 32 | 4 | 32 |
| 715 | 32 | >128 | 32 | 2 | >32 | 32 | 8 | 8 | 32 |
| 716 | 64 | 16 | 64 | 2 | >32 | >32 | 128 | 64 | 4–8 |
| 776 | 64 | 128 | 64 | 2 | >32 | >32 | 128 | 4 | 2 |
| 795 | 32 | >128 | 128 | 2 | >32 | >32 | 128 | 4 | 128 |
| 835 | 16 | 16 | 2 | 2 | >32 | 32 | 64 | 4 | 8 |
| 839 | 16 | 128 | >128 | 2 | >32 | >32 | 64 | 4 | 2–4 |
| 834 | 16 | 16 | >128 | 2 | >32 | 16 | >128 | 4 | 8–16 |
| 851 | 32 | 128 | 64 | 0.25 | >32 | 16 | >128 | 8 | <0.06 |
Data are mg/L and were modified from [8].
Abbreviations: AVI, avibactam; CIP, ciprofloxacin; COL, colistin; CAZ, ceftazidime; FOS, fosfomycin MEM, meropenem; TZP, piperacillin-tazobactam.
aAVI was maintained at 4 mg/L in combinations.
bFOS and CAZ were used at the same concentration.
cCOL was added to all plates at 0.5 mg/L.
Figure 1.
Structures of ceftazidime, avibactam, and fosfomycin and their targets, penicillin-binding protein 3 (PBP-3; Protein Data Bank identifier 3OCN), Pseudomonas-derived cephalosporinase 1 (PDC-1; 4HEF), and MurA (5BQ2), respectively.
The oral formulation of fosfomycin is approved in the United States for the treatment of urinary tract infections caused by gram-positive and gram-negative bacteria, including P. aeruginosa. Fosfomycin possesses bactericidal action against gram-negative bacteria, and an intravenous formulation is currently in development for US application and has completed phase I and II/III clinical trials (NCT02753946, NCT03709927, and NCT02178254 [10]). In some countries, intravenously administered fosfomycin is already used to treat a wide range of infections, including pneumonia [11, 12]. Interest in the possible use of fosfomycin in combination with β-lactams against infections due to MDR P. aeruginosa is increasing. Indeed, several recent studies revealed synergism between fosfomycin and β-lactams (ie, carbapenems and ceftolozane-tazobactam) [13–19].
The decision to combine ceftazidime-avibactam with fosfomycin was based on the rationale that even though fosfomycin can downregulate expression of PBP-3 and induce PDC expression, avibactam is so potent that it can significantly hinder the hydrolytic activity of PDC, as well as other class A and C β-lactamases present in P. aeruginosa [5, 8, 20]. Notably, many MDR P. aeruginosa already possess a derepressed blaPDC [8]. With avibactam targeting the class A and C β-lactamases, ceftazidime and fosfomycin are free to disrupt the cell wall recycling pathway, using 2 different strategies to kill the bacteria. One limitation of ceftazidime-avibactam-fosfomycin is the lack of coverage against P. aeruginosa producing metallo-β-lactamases; fortunately, the prevalence of metallo-β-lactamase–producing P. aeruginosa in United States remains low [21, 22].
Here, we determined the activity of ceftazidime-avibactam combined with fosfomycin against P. aeruginosa strain CL232, which was chosen because of its MDR phenotype and limited susceptibility profile to currently available agents (ie, colistin). In addition, the resistance determinants present and expressed in P. aeruginosa CL232 were identified. The mutation frequency of the single agents and the combinations were tested using this isolate. Finally, the in vivo activity of ceftazidime-avibactam combined with fosfomycin was assessed in a neutropenic mouse thigh infection model, using a high bacterial burden of P. aeruginosa CL232.
METHODS
Critical Reagents
P. aeruginosa stains PA01 and CL232 were used in this study [8]. For in vitro assays, ceftazidime and fosfomycin were purchased from Sigma-Aldrich, pharmaceutical-grade ceftazidime-avibactam was also used, and avibactam was provided by Allergan. For in vivo experiments, pharmaceutical-grade ceftazidime was used, while fosfomycin and avibactam were obtained from Zavante and Allergan, respectively.
Growth Curve Analysis
P. aeruginosa strains were grown in Mueller-Hinton (MH) broth with or without ceftazidime-avibactam (16 μg/mL–4 μg/mL), fosfomycin (16 μg/mL), or ceftazidime-avibactam-fosfomycin (16 μg/mL–4 μg/mL–16 μg/mL) at 37°C with shaking for 11 hours. At selected time points, the OD600 was measured.
Checkerboard Analysis
The minimum inhibitory concentrations (MICs) were determined for the combinations, and a fractional inhibitory concentration (FIC) index was calculated using checkerboard analysis in MH broth as previously described for P. aeruginosa CL232 [23].
Whole-Genome Sequencing (WGS)
Genomic DNA was purified from P. aeruginosa PA01, P. aeruginosa CL232, and its fosfomycin-resistant mutant (CL232FR) recovered from treated mice, using the MasterPure gram-positive DNA purification kit as recommended by the manufacturer. The genome was sequenced by Illumina NextSeq 500 Mid Output Kit or HiSeq X with 300 cycles (2 × 150 bp). Paired-end libraries were constructed using Illumina NexteraXT kits. Sequence reads were generated with a target average read depth of approximately 100-fold coverage. Sequence reads for each isolate were assembled individually, using Velvet v. 1.2.07 [24], and annotated using the National Center for Biotechnology Information’s Prokaryotic Genome Annotation Pipeline [25]. Single-nucleotide polymorphisms and insertion and deletion calling was performed using BWA, Samtools, and Vcftools, followed by annotation using SnpEff [26–29]. Raw DNA sequence reads were submitted to the National Center for Biotechnology Information Sequence Read Archive, and annotated genomes were deposited in the GenBank WGS repository (BioSample identifiers SAMN05774260 and SAMN10485642 and GenBank accession number MPVE00000000).
RNA Sequencing (RNAseq)
P. aeruginosa PA01 and CL232 were grown in lysogeny broth at 37°C until an OD600nm of 0.6 was reached. One milliliter of cells in log-phase growth was collected, mixed with 500 μL of RNAprotect Bacteria Reagent, and incubated at room temperature for 15 minutes. The RNeasy Mini Kit protocol was followed to purify the RNA from the samples. An on-column DNAse digestion using the RNAse-Free DNAse Set was conducted. In addition, a second step to remove residual DNA was conducted using the Turbo DNA-free kit protocol. RNAseq and data analysis were conducted by the Genomics Core facility and the Institute for Computational Biology Core, respectively, at Case Western Reserve University, including quality control of the submitted RNA specimens; library preparation, using the Illumina ScriptSeq Complete Gold (Bacteria) kit; quality control of the library preparation; use of the Illumina HiSeq 2500 Rapid Run flow cell system (2 × 75 bp); generation of the reads per kilobase million data set, including assessment of the statistical significance of gene expression changes between P. aeruginosa PA01 and CL232; and cluster of orthologous groups analysis. The RNAseq data set and cluster of orthologous groups analysis are presented in Supplementary Table 1 and Supplementary Figure 1, respectively.
Determination of Mutation Frequency
To determine the mutation frequency in P. aeruginosa CL232 after exposure to ceftazidime-avibactam, fosfomycin, and ceftazidime-avibactam-fosfomycin, P. aeruginosa CL232 was grown in MH broth and concentrated to a high inoculum (1010 colony-forming units [CFU]/mL). Cells were plated onto MH agar plates containing drug concentrations 4 times greater than the MIC and on MH agar only, to enumerate the total number of viable cells. The mutation frequency was calculated by dividing the number of cells on the drug-containing plates by the number of cells on the control plates.
Mice
Outbred 6–8-week-old female CD-1 mice were purchased from Charles River Laboratories. Mice were cared for according to the Institutional Animal Care and Use Committee guidelines of Rutgers University. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animal experiments and was adherent to principles stated in the National Research Council’s 1996 Guide for the Care and Use of Laboratory Animals. The facilities where this research was conducted are fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Fosfomycin Tolerance
As the humanized doses of fosfomycin were very high, a tolerance study was conducted in both immunocompetent and neutropenic mice. The dosing regimen for fosfomycin consisted of 6 doses administered subcutaneously at 0, 3, 8, 11, 16, and 19 hours at alternating doses of 70.4 mg and 46.9 mg. Outbred 6–7-week-old female CD-1 mice weighing 23–26 g were used with the dosing regimen stated above (adjusted for a body weight of 25 g). For neutropenic mice, a subset of CD-1 mice were administered 150 mg/kg and 100 mg/kg of cyclophosphamide via intraperitoneal injection 4 days and 1 day, respectively, before the administration of fosfomycin.
Neutropenic Thigh Infection Model and Bacterial Burden
In all studies, recumbent animals were considered moribund and humanely euthanized. Female outbred 6–8-week-old CD-1 mice (18 per treatment condition) were rendered neutropenic as described above and challenged with 2.0 × 107 CFU/thigh of P. aeruginosa CL232. The mice were treated with either vehicle (water for injection) or the following dosing regimen. A single 60/40 split dose was administered. One hour after infection, 9.54 mg of ceftazidime and/or 3.11 mg of avibactam and/or 70.4 mg of fosfomycin was given, while 4 hours after infection, 6.36 mg of ceftazidime and/or 2.08 mg of avibactam and/or 46.9 mg of fosfomycin was given. For measurement of bacterial burden, mice were euthanized by carbon dioxide narcosis at the end point of 24 hours after initiating infection. The thighs were harvested, and tissue homogenates were serially diluted in sterile phosphate-buffered saline, plated on MH agar, and incubated at 37°C for up to 48 hours. CFUs were counted. To screen for development of drug resistance, selected colonies from the groups that received ceftazidime-avibactam or fosfomycin only for 24 hours were replated on MH agar containing 192 and 12 µg/mL ceftazidime-avibactam or 192 µg/mL fosfomycin, respectively, and incubated at 37°C for up to 48 hours. The concentrations used are approximately 3 times the MIC for the original P. aeruginosa CL232 strain used.
Statistical Analysis
For neutropenic thigh infection models, mean log CFUs/thigh were compared across treatment groups and to stasis, using analysis of variance and Tukey post hoc tests, with adjusted P values calculated to assess differences among treatment pairs. All analyses were performed in R, version 3.5.0 [29, 30].
RESULTS AND DISCUSSION
In Vitro Efficacy of Ceftazidime-Avibactam-Fosfomycin
Previous analysis found that 9 out of 50 strains in a panel of archived clinical isolates of P. aeruginosa tested resistant to ceftazidime-avibactam (Table 1) [8]. The addition of fosfomycin to the ceftazidime-avibactam combination restored susceptibility to approximately 90% of the isolates (MIC range, 4–8 μg/mL) with the exception of strain 716 (MIC, 64 μg/mL). However, isolate 716 was susceptible to piperacillin-tazobactam (MICs, 16 and 4 μg/mL, respectively).
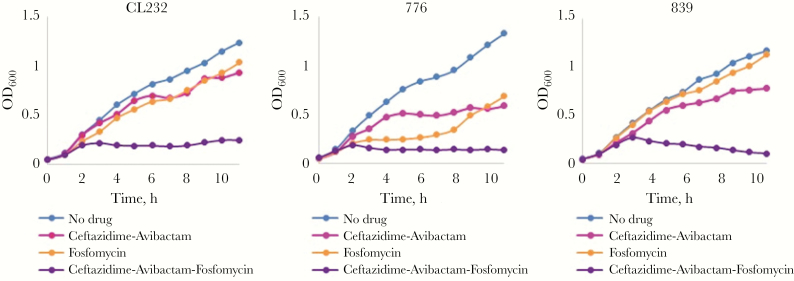
In this study, growth curve analysis was completed with ceftazidime-avibactam–resistant P. aeruginosa strains CL232, 776, and 839, using ceftazidime-avibactam, fosfomycin, ceftazidime-avibactam-fosfomycin and a no drug control (Figure 2). The presence of ceftazidime-avibactam-fosfomycin in the medium prevented the growth of all MDR P. aeruginosa strains for up to 11 hours, compared with treatment with ceftazidime-avibactam and fosfomycin separately. Based on the consistent growth pattern obtained for the strain, P. aeruginosa CL232 was selected for subsequent in vitro and in vivo studies. P. aeruginosa CL232 is highly resistant to all β-lactams and β-lactam–β-lactamase inhibitor combinations, as well as to fluoroquinolones, fosfomycin, and tigecycline (Table 1) [8].
Figure 2.
Growth of Pseudomonas aeruginosa strains CL232, 776, and 839 in Mueller-Hinton broth with or without ceftazidime-avibactam (16 and 4 μg/mL, respectively), fosfomycin (16 μg/mL), or ceftazidime-avibactam-fosfomycin (16, 4, and 16 μg/mL, respectively) over 11 hours.
Synergy of the Ceftazidime-Avibactam-Fosfomycin Combination
To assess whether ceftazidime-avibactam and fosfomycin were synergistic, a checkerboard analysis was conducted using P. aeruginosa CL232. The MICs were determined for the combinations, and a fractional inhibitory concentration (FIC) index was calculated. An FIC index of ≤0.5 was obtained for one of the combinations tested (Table 2). The combination of 25 μg/mL and 6.25 μg/mL of ceftazidime and avibactam, respectively, combined with 12.5 μg/mL of fosfomycin possessed an FIC index of 0.5. The addition of a β-lactamase inhibitor, avibactam, reversed the antagonism that was previously reported with fosfomycin and β-lactams [20]. Our data showed that β-lactamase inhibition is a vital component of this combination.
Table 2.
Results of Checkerboard Analyses of Ceftazidime-Avibactam-Fosfomycin (CAZ-AVI-FOS) in Mueller-Hinton Broth, Using Pseudomonas aeruginosa CL232
| Variable | CAZ-AVI MIC, mg/L | FOS MIC, mg/L | FIC CAZ-AVI | FOS FIC | FIC Index | Result |
|---|---|---|---|---|---|---|
| Alone | 50–12.5 | 400 | ||||
| Combined | 12.5–3.12 | 800 | 0.25 | 2 | 2.3 | No interaction |
| Combined | 25–6.25 | 400 | 0.5 | 1 | 1.5 | No interaction |
| Combined | 25–6.25 | 200 | 0.5 | 0.5 | 1 | No interaction |
| Combined | 25–6.25 | 100 | 0.5 | 0.25 | 0.8 | No interaction |
| Combined | 25–6.25 | 50 | 0.5 | 0.13 | 0.6 | No interaction |
| Combined | 25–6.25 | 25 | 0.5 | 0.06 | 0.6 | No interaction |
| Combined | 25–6.25 | 12.5 | 0.5 | 0.03 | 0.5 | Synergy |
| Combined | 50–12.5 | 6.25 | 1 | 0.02 | 1.0 | No interaction |
Synergy is defined by an FIC index ≤ 0.5
Abbreviations: FIC, fractional inhibitory concentration; MIC, minimum inhibitory concentration.
Resistome of P. aeruginosa CL232
To identify which determinants are contributing to the ceftazidime-avibactam and fosfomycin resistance phenotypes, the genome of P. aeruginosa CL232 was analyzed and compared to the P. aeruginosa PA01 genome. The list of genes involved in β-lactam and fosfomycin resistance in P. aeruginosa is displayed in Table 3 [31, 32].
Table 3.
Genetic and Transcriptome Analysis of β-Lactam and Fosfomycin Resistance in Pseudomonas aeruginosa CL232
| Gene, Function, Mutation(s)/Substitution(s) | Log2 Fold Change |
|---|---|
| mexABoprM | |
| β-lactam RND transporter | |
| MexA substitutions: none | +1.7 |
| MexB substitutions: none | +3.1 |
| OprM substitutions: none | +2.6 |
| mexXYoprM | |
| β-lactam RND transporter | |
| MexX substitutions: A30T | No change |
| MexY substitutions: I536V, T543A, G589A, Q840E, N1036T, Q1039R | No change |
| OprM substitutions: none | +2.6 |
| mexCDoprJ | |
| β-lactam RND transporter | |
| MexC substitutions: P47S, R76Q, H310R, S330A, A378T, P383S, A384Y | No change |
| MexD substitutions: T87S, A155T, E257Q, V660I, N669D, S685G, I701V, S845A, S915A, I982V, K1031R, S1040T | No change |
| OprJ substitutions: D68G, M69V | No change |
| oprD | |
| Porin | |
| Mutation: frameshift | −2.2 |
| oprF | |
| Porin | |
| Substitutions: none | No change |
| mrcB (ponB, pbpF) | |
| PBP1b: transglycolyase-transpeptidase | |
| Substitutions: V185A, Y568H, T911A | No change |
| pbpA | |
| PBP2: transglycolyase-transpeptidase | |
| Substitutions: none | No change |
| ftsI | |
| PBP3: transpeptidase | |
| Substitutions: none | No change |
| pbpC | |
| PBP3A | |
| Substitutions: A104P | No change |
| dacB | |
| PBP4: D-alanyl-D-alanine carboxypeptidase | |
| Substitutions: Q156H | No change |
| dacC | |
| PBP5: D-alanyl-D-alanine carboxypeptidase | |
| Substitutions: none | No change |
| pbpG | |
| PBP7 D-alanyl-D-alanine endopeptidase | |
| Substitutions: none | No change |
| bla OXA-488 (poxB) | |
| Class D β-lactamase | |
| Substitutions: T16LA, Q29R | No change |
| bla PDC-34 | |
| Class C β-lactamase | |
| G1D, A29T, T79A, Q129R, V179L, G363A | +6.6 |
| ampR | |
| Transcriptional regulator of blaPDC | |
| Substitutions: E114A, D135A, G283E, M288R | No change |
| fosA | |
| Glutathione transferase: enzymatically inactivates fosfomycin | |
| Substitutions: none | Not present in RNA sequencing data set |
| glpT | |
| Glycerol-3-phosphate transporter: used for entry of fosfomycin | |
| Substitutions: none | No change |
| oprO | |
| Porin: used for entry of fosfomycin | |
| Substitutions: none | No change |
| oprP | |
| Porin: used for entry of fosfomycin | |
| Substitutions: A98V | No change |
| murA | |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase | |
| Substitutions: none | +2.3 |
The PBPs in P. aeruginosa CL232 were examined, and single amino acid substitutions were found in PBP1B, PBP3A, and PBP4; the contribution of these changes to β-lactam resistance remains to be determined. Nevertheless, knock out of the gene encoding PBP4 (dacB) was shown to increase resistance to β-lactams [33]. Importantly, ftsI, which encodes PBP-3, the target of ceftazidime, was unaltered in P. aeruginosa CL232. Moreover, P. aeruginosa CL232 possesses 2 chromosomal β-lactamase genes, blaPDC-34 and blaOXA-488, of which the latter is a blaOXA-50 family member (Table 3). The expression of blaPDC-34 and blaOXA-488 is linked to cell wall recycling and is induced by exposure to certain β-lactams [33–35]. PDC-34, the dominant β-lactamase in P. aeruginosa CL232, possessed several amino acid substitutions, including G1D, A29T, T79A, Q128R, V178L, and G364A, which are not located within the active site motifs of AmpCs [36]. Thus, these changes likely do not result in an expanded-spectrum phenotype. β-lactamases in the OXA-50 family play a minor role in β-lactam resistance and, thus, are unlikely to significantly influence the resistance phenotype of this strain [35]. AmpR is the transcriptional regulator that positively regulates blaPDC-34 expression, but it negatively regulates blaOXA-488 expression [37]. ampR in P. aeruginosa CL232 carries a mutation that corresponds to an amino acid substitution at position D135, which was previously shown to lead to overexpression of blaPDC (Table 3) [38]. Additionally, a frameshift mutation was also identified in oprD of P. aeruginosa CL232; OprD is the major porin for the entry of carbapenems into P. aeruginosa (Table 3). The mutation in pbps and derepressed blaPDC, as well as the mutation in oprD, likely impact resistance to β-lactams in P. aeruginosa CL232 [39].
Alterations in 4 different resistance determinants (murA, glpT, oprO, and oprP) in P. aeruginosa can influence susceptibly to fosfomycin. murA, which encodes MurA, the target of fosfomycin, was unaltered in P. aeruginosa CL232 (Table 3). The entry of fosfomycin into the periplasmic space of P. aeruginosa is not well understood, and to date, 3 proteins, GlpT, a glycerol-3-phosphate transporter, and the OprO and OprP porins have been shown to contribute to drug entry [40, 41]. In P. aeruginosa CL232, only oprP possessed a mutation that resulted in an A98V amino acid change; however, the contribution of this substitution on fosfomycin entry remains unknown (Table 3).
To evaluate which resistance determinants are expressed by P. aeruginosa CL232, RNAseq experiments were conducted on P. aeruginosa CL232, and findings were compared to those for P. aeruginosa PA01 (Table 3). Results of transcriptome analysis was consistent with the predictions gleaned from WGS data, because mexABoprM and blaPDC-34 were upregulated (an immunoblot conducted previously confirmed upregulation of blaPDC-34 [8]) and oprD was downregulated (Table 3). These 3 modifications likely contribute to β-lactam resistance in P. aeruginosa CL232. Analysis of the upstream promoter of mexABoprM revealed that mutations were not present between mexA and mexR, the gene encoding the transcriptional regulator of the mexABoprM operon. Expression of murA, which encodes the target for fosfomycin, was also upregulated, while fosA, encoding a glutathione-S-transferase known to inactivate fosfomycin, was not expressed by either strain (Table 3) [42]. One could hypothesize that increased expression of murA could lead to fosfomycin resistance, because more MurA would be present in the cytoplasm and, thus, higher concentrations of fosfomycin would be required to inhibit the enzyme.
Ceftazidime-Avibactam Decreases the Mutation Frequency of Fosfomycin Alone In Vitro
To assess the contribution of the evolution of mutations in P. aeruginosa CL232 to the development of resistance to ceftazidime-avibactam, fosfomycin, and ceftazidime-avibactam-fosfomycin, mutation frequencies were determined. When P. aeruginosa CL232 was plated on agar containing fosfomycin at 4 times the MIC, a mutation frequency of 1.9 × 10-5 was obtained (Table 4). When P. aeruginosa CL232 was grown on 4 times the MIC of ceftazidime-avibactam, minor growth was observed, corresponding to a mutation frequency of 9.1 × 10-9 (Table 4). Finally, the cells plated on the ceftazidime-avibactam-fosfomycin combination demonstrated the same mutation frequency as those on the ceftazidime-avibactam combination (Table 4). Our data suggest that ceftazidime-avibactam diminished the selection of fosfomycin-resistant mutants of P. aeruginosa CL232 by >4-log orders.
Table 4.
Mutation Frequency for Pseudomonas aeruginosa CL232 After Exposure to Fosfomycin (FOS), Ceftazidime-Avibactam (CAZ-AVI), and CAZ-AVI-FOS
| Condition | Mutation Frequency | MIC(s),a mg/L |
|---|---|---|
| FOS | 1.9 × 10−5 | 64 |
| CAZ-AVI | 9.1 × 10−9 | 64 and 4, respectively |
| CAZ-AVI-FOS | 9.1 × 10−9 | 16, 4, and 16, respectively |
Abbreviation: MIC, minimum inhibitory concentration.
aAgar dilution.
High Doses of Fosfomycin Causes Injection Site Side Effects in Neutropenic Mice
Because the humanized doses of fosfomycin were very high, a tolerance study was conducted in immunocompetent and neutropenic mice. Adverse reactions were not observed in immunocompetent animals during the drug administrations, and mice were alert and responsive until the study end date (5 days after administration of the study drugs). However, in cyclophosphamide-induced neutropenic mice that were administered humanized doses of fosfomycin alone or in combination exhibited signs of drug toxicity after the fourth dose. Three mice (30%) became severely moribund and were humanely euthanized. The euthanized mice were necropsied, and gross evaluation showed mild inflammation of the internal organs; however, the injection sites had severe edema-like structures that were filled with fluid. The disodium salt composition of the fosfomycin, in combination with the antiinflammatory effect of cyclophosphamide, were likely important in contributing to these fluid-filled masses in the 3 euthanized mice. The remaining mice had ruffled fur and were slightly lethargic. Irritation was observed in 4 of 7 mice at the site of injection (the scruff). The mice were fully recovered at 24 hours and remained bright and alert for the next 4 days.
Despite the toxicity observed with fosfomycin in the neutropenic mouse model, fosfomycin is often used in combination with a variety of drugs, including β-lactams, for treatment of infections. In a meta-analysis of the clinical literature, the safety of fosfomycin was assessed alone and in combination [43]. Moreover, the results of the ZEUS clinical trial (clinical trials registration NCT02753946) were aligned with what has been observed outside the United States, where there is a long history of fosfomycin at dosages up to 24 g/day [44]. However, since intravenous fosfomycin contains a high salt load inherent to the disodium formulation, each gram contains 330 mg of sodium; thus, hypokalemia may be a concern in some patients.
Ceftazidime-Avibactam-Fosfomycin Decreases the Bacterial Burden in a Neutropenic Murine Thigh Infection Model
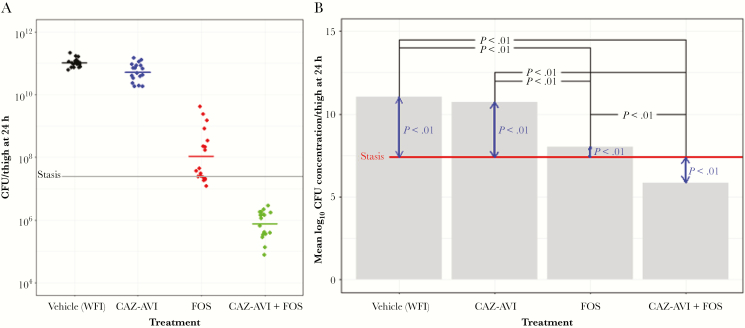
CD-1 mice were rendered neutropenic and challenged with 2.0 × 107 colony-forming units (CFU)/thigh of P. aeruginosa CL232. The mice were treated with vehicle, ceftazidime-avibactam, fosfomycin, or ceftazidime-avibactam-fosfomycin, using a single 60/40 split dose, 1 hour after infection. Human pharmacokinetics values were used to perform a 5000-iteration Monte Carlo simulation. The schedules, dosing amounts, and split were chosen to humanize the exposures. The murine peaks were calculated to not exceed the upper 95% confidence bound from the human-data Monte Carlo simulation. The results in Figure 3A show the end point at 24 hours as compared to stasis. This analysis revealed that ceftazidime-avibactam-fosfomycin was the most effective combination, with CFUs/thigh in the 105–6 range, compared with 1011 for the vehicle-treated mice. Specifically, the ceftazidime-avibactam-fosfomycin combination was found to significantly reduce CFUs by approximately 2 logs below those at stasis (Figure 3B). Ceftazidime-avibactam and fosfomycin dosed separately increased CFUs from those at stasis by approximately 3 logs and 1 log, respectively (Figure 3B). Among paired comparisons of treatments to each other and to stasis, all combinations except vehicle compared to the ceftazidime-avibactam treatment arm differed significantly (adjusted P <.01).
Figure 3.
CD-1 mice (18 mice per treatment condition) were rendered neutropenic and challenged with 2.0 × 107Pseudomonas aeruginosa CL232 colony-forming units (CFUs)/thigh. The mice were treated with either vehicle (water for injection [WFI]) or the following dosing regimen. A single 60/40 split dose was administered subcutaneously. One hour after infection, 9.54 mg of ceftazidime (CAZ) and/or 3.11 mg of avibactam (AVI) and/or 70.4 mg of fosfomycin (FOS) were administered, whereas 4 hours after infection, 6.36 mg of CAZ and/or 2.08 mg of AVI and/or 46.9 mg of FOS was given. A, CFUs/thigh for each mouse at an end point of 24 hours is presented for all treatment conditions; stasis is represented by the gray line. B, Mean log10 CFU concentration/thigh at an end point of 24 hours after initiating infection is represented. The red line represents stasis. In black are the statistical differences (expressed as P values) between treatment conditions, and in blue are the statistical differences (expressed as P values) between stasis and the treatment conditions.
Given the minor impact of ceftazidime-avibactam alone and fosfomycin alone in the mouse model, the bacteria isolated from the mouse thighs were selected on plates containing each drug, to assess for potential resistance acquired in vivo. In mice treated with ceftazidime-avibactam, observed regrowth of the P. aeruginosa CL232 strain was likely due to insufficient drug exposure, because the bacteria were not resistant to ceftazidime-avibactam. Conversely, the isolates obtained from the mice treated with fosfomycin alone were determined to be resistant; the fosfomycin MIC increased from 64 mg/L to 2048 mg/L. The fosfomycin-resistant isolate was subjected to WGS, and a single nucleotide change (C228A) was identified that resulted in the introduction of a stop codon in glpT, which encodes a glycerol-3-phosphate transporter that is used by fosfomycin for entry into P. aeruginosa [41]. Thus, in vivo, P. aeruginosa CL232 acquired resistance to fosfomycin. Elimination of fosfomycin may be involved in the acquisition of fosfomycin resistance.
Why Not Other Combinations of β-Lactam–β-Lactamase Inhibitor– Fosfomycin?
Ceftolozane-tazobactam-fosfomycin and aztreonam-avibactam-fosfomycin were also considered for testing. Indeed, ceftolozane-tazobactam-fosfomycin was previously found to be synergistic against MDR P. aeruginosa via time-kill analysis [17]. However, a major caveat of the ceftolozane-tazobactam-fosfomycin combination is that tazobactam is not as potent of a β-lactamase inhibitor as avibactam and cannot target PDC or class A carbapenemases [4]. In addition, several analyses have described PDC variants that are able to robustly hydrolyze ceftolozane, further diminishing interest in this pairing [45–49]. The aztreonam-avibactam-fosfomycin combination possesses other liabilities. Even though avibactam can target the class A and C β-lactamases and aztreonam would extend coverage to class B metallo-β-lactamases, the aztreonam-avibactam combination is not very effective in vitro (90% MIC, 32 μg/mL) against MDR P. aeruginosa as compared to carbapenem-resistant Enterobacteriaceae (90% MIC, 1 μg/mL) [50].
In conclusion, despite the introduction of highly effective therapies, MDR P. aeruginosa is still a significant threat to vulnerable and immunocompromised patients. Here, using a “mechanism-based” approach targeting important cell wall enzymes and resistance determinants, we showed that MDR P. aeruginosa can be readily overcome. In a high-bacterial-burden infection model, we showed that CFUs were reduced by approximately 2 logs, relative to CFUs at stasis. These findings have significant implications for further studies directed at clinical applications. Such an approach may be beneficial even when treating more-susceptible P. aeruginosa. By lowering CFUs and mutation frequency, we hope to eradicate the infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Allergan, for supplying avibactam powder as part of an investigator-initiated trial (CAZ-IT-24); Zavante, for supplying the fosfomycin powder used in this study; and Dr Ricky Chan (Case Western Reserve University’s Institute for Computational Biology Core), for assistance with the RNAseq data analysis.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Department of Veterans Affairs, or the US government.
Financial support. This work was supported “in whole or part” by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (awards U19-AI109713-SRP, to K. M. P.-W. and D. S. P.; R21AI114508, R01AI100560 and R01AI063517, and R01AI072219, to R. A. B.; R01AI090155 to B.N.K. and R01AI121430 to G.L.D. and U19AI110819, to J. C. V. I.); Louis Stokes Cleveland VA Medical Center; the Veterans Affairs Merit Review Program, Department of Veterans Affairs Biomedical Laboratory Research and Development Service (BX002872 to K. M. P.-W. and BX001974 to R. A. B.); and the Geriatric Research Education and Clinical Center (VISN 10 to R. A. B.).
Potential conflicts of interest. E. J. E.-G. was previously employed by Zavante. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: ASM Microbe 2017, New Orleans, Louisiana, 1–5 June 2017; IDWeek 2018, San Francisco, California, 3–7 October 2018.
References
- 1. Flamm RK, Nichols WW, Sader HS, Farrell DJ, Jones RN. In vitro activity of ceftazidime/avibactam against Gram-negative pathogens isolated from pneumonia in hospitalised patients, including ventilated patients. Int J Antimicrob Agents 2016; 47:235–42. [DOI] [PubMed] [Google Scholar]
- 2. Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalised with pneumonia in US and European hospitals: results from the SENTRY Antimicrobial Surveillance Program, 2009-2012. Int J Antimicrob Agents 2014; 43:328–34. [DOI] [PubMed] [Google Scholar]
- 3. El Solh AA, Alhajhusain A. Update on the treatment of Pseudomonas aeruginosa pneumonia. J Antimicrob Chemother 2009; 64:229–38. [DOI] [PubMed] [Google Scholar]
- 4. Papp-Wallace KM, Bonomo RA. New β-Lactamase Inhibitors in the Clinic. Infect Dis Clin North Am 2016; 30:441–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ehmann DE, Jahic H, Ross PL, et al. Kinetics of avibactam inhibition against Class A, C, and D β-lactamases. J Biol Chem 2013; 288:27960–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keepers TR, Gomez M, Celeri C, Nichols WW, Krause KM. Bactericidal activity, absence of serum effect, and time-kill kinetics of ceftazidime-avibactam against β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014; 58:5297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fraile-Ribot PA, Mulet X, Cabot G, et al. In vivo emergence of resistance to novel cephalosporin-β-lactamase inhibitor combinations through the duplication of amino acid D149 from OXA-2 β-lactamase (OXA-539) in sequence type 235 Pseudomonas aeruginosa. Antimicrob Agents Chemother 2017; 61:pii: e01117-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Winkler ML, Papp-Wallace KM, Hujer AM, et al. Unexpected challenges in treating multidrug-resistant Gram-negative bacteria: resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2015; 59:1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sader HS, Huband MD, Castanheira M, Flamm RK. Pseudomonas aeruginosa antimicrobial susceptibility results from four years (2012 to 2015) of the international network for optimal resistance monitoring program in the United States. Antimicrob Agents Chemother 2017; 61:pii: e02252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wenzler E, Ellis-Grosse EJ, Rodvold KA. Pharmacokinetics, safety, and tolerability of single-dose intravenous (ZTI-01) and oral fosfomycin in healthy volunteers. Antimicrob Agents Chemother 2017; 61:pii: e00775-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents 2009; 34:111–20. [DOI] [PubMed] [Google Scholar]
- 12. Falagas ME, Giannopoulou KP, Kokolakis GN, Rafailidis PI. Fosfomycin: use beyond urinary tract and gastrointestinal infections. Clin Infect Dis 2008; 46:1069–77. [DOI] [PubMed] [Google Scholar]
- 13. Perdigao Neto LV, Oliveira MS, Martins RCR, et al. Fosfomycin in severe infections due to genetically distinct pan-drug-resistant Gram-negative microorganisms: synergy with meropenem. J Antimicrob Chemother 2018. doi: 10.1093/jac/dky406 [DOI] [PubMed] [Google Scholar]
- 14. Frattari A, Savini V, Polilli E, et al. Ceftolozane-tazobactam and Fosfomycin for rescue treatment of otogenous meningitis caused by XDR Pseudomonas aeruginosa: case report and review of the literature. IDCases 2018; 14:e00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drusano GL, Neely MN, Yamada WM, et al. The combination of fosfomycin plus meropenem is synergistic for Pseudomonas aeruginosa PA01 in a Hollow Fiber Infection Model (HFIM). Antimicrob Agents Chemother 2018. doi: 10.1128/AAC.01682-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khawcharoenporn T, Chuncharunee A, Maluangnon C, Taweesakulvashra T, Tiamsak P. Active monotherapy and combination therapy for extensively drug-resistant Pseudomonas aeruginosa pneumonia. Int J Antimicrob Agents 2018; 52:828–34. [DOI] [PubMed] [Google Scholar]
- 17. Monogue ML, Nicolau DP. Antibacterial activity of ceftolozane/tazobactam alone and in combination with other antimicrobial agents against MDR Pseudomonas aeruginosa. J Antimicrob Chemother 2018; 73:942–52. [DOI] [PubMed] [Google Scholar]
- 18. Hamou-Segarra M, Zamorano L, Vadlamani G, et al. Synergistic activity of fosfomycin, β-lactams and peptidoglycan recycling inhibition against Pseudomonas aeruginosa. J Antimicrob Chemother 2017; 72:448–54. [DOI] [PubMed] [Google Scholar]
- 19. Asuphon O, Montakantikul P, Houngsaitong J, Kiratisin P, Sonthisombat P. Optimizing intravenous fosfomycin dosing in combination with carbapenems for treatment of Pseudomonas aeruginosa infections in critically ill patients based on pharmacokinetic/pharmacodynamic (PK/PD) simulation. Int J Infect Dis 2016; 50:23–9. [DOI] [PubMed] [Google Scholar]
- 20. Reguera JA, Baquero F, Berenguer J, Martinez-Ferrer M, Martinez JL. Beta-lactam-fosfomycin antagonism involving modification of penicillin-binding protein 3 in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1990; 34:2093–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 2011; 11:381–93. [DOI] [PubMed] [Google Scholar]
- 22. Kazmierczak KM, Rabine S, Hackel M, et al. Multiyear, multinational survey of the incidence and global distribution of Metallo-β-Lactamase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 2016; 60:1067–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsieh MH, Yu CM, Yu VL, Chow JW. Synergy assessed by checkerboard. A critical analysis. Diagn Microbiol Infect Dis 1993; 16:343–9. [DOI] [PubMed] [Google Scholar]
- 24. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tatusova T, DiCuccio M, Badretdin A, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 2016; 44:6614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Danecek P, Auton A, Abecasis G, et al. ; 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics 2011; 27:2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2018. [Google Scholar]
- 31. Mesaros N, Nordmann P, Plésiat P, et al. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 2007; 13:560–78. [DOI] [PubMed] [Google Scholar]
- 32. Wolter DJ, Lister PD. Mechanisms of β-lactam resistance among Pseudomonas aeruginosa. Curr Pharm Des 2013; 19:209–22. [PubMed] [Google Scholar]
- 33. Zamorano L, Reeve TM, Deng L, et al. NagZ inactivation prevents and reverts beta-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2010; 54:3557–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dik DA, Fisher JF, Mobashery S. Cell-wall recycling of the gram-negative bacteria and the nexus to antibiotic resistance. Chem Rev 2018; 118:5952–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zincke D, Balasubramanian D, Silver LL, Mathee K. Characterization of a carbapenem-hydrolyzing enzyme, PoxB, in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 2016; 60:936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev 2009; 22:161–82, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Balasubramanian D, Kumari H, Mathee K. Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator. Pathog Dis 2015; 73:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caille O, Zincke D, Merighi M, et al. Structural and functional characterization of Pseudomonas aeruginosa global regulator AmpR. J Bacteriol 2014; 196:3890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vu H, Nikaido H. Role of beta-lactam hydrolysis in the mechanism of resistance of a beta-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum beta-lactams. Antimicrob Agents Chemother 1985; 27:393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Citak F, Ghai I, Rosenkötter F, Benier L, Winterhalter M, Wagner R. Probing transport of fosfomycin through substrate specific OprO and OprP from Pseudomonas aeruginosa. Biochem Biophys Res Commun 2018; 495:1454–60. [DOI] [PubMed] [Google Scholar]
- 41. Castañeda-García A, Rodríguez-Rojas A, Guelfo JR, Blázquez J. The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J Bacteriol 2009; 191:6968–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beharry Z, Palzkill T. Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J Biol Chem 2005; 280:17786–91. [DOI] [PubMed] [Google Scholar]
- 43. Grabein B, Graninger W, Rodríguez Baño J, Dinh A, Liesenfeld DB. Intravenous fosfomycin-back to the future. Systematic review and meta-analysis of the clinical literature. Clin Microbiol Infect 2017; 23:363–72. [DOI] [PubMed] [Google Scholar]
- 44. Kaye KS, Rice LB, Dane A, et al. Fosfomycin for injection (ZTI-01) vs Piperacillin-Tazobactam (PIP-TAZ) for the treatment of Complicated Urinary Tract Infection (cUTI) Including Acute Pyelonephritis (AP): ZEUS, a phase 2/3 randomized trial. Clin Infect Dis 2019. doi: 10.1093/cid/ciz181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fraile-Ribot PA, Cabot G, Mulet X, et al. 2017. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother doi: 10.1093/jac/dkx424. [DOI] [PubMed] [Google Scholar]
- 46. MacVane SH, Pandey R, Steed LL, Kreiswirth BN, Chen L. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother 2017; 61:pii: e01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berrazeg M, Jeannot K, Ntsogo Enguéné VY, et al. Mutations in β-Lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 2015; 59:6248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cabot G, Bruchmann S, Mulet X, et al. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 2014; 58:3091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barnes MD, Taracila MA, Rutter JD, et al. Deciphering the evolution of cephalosporin resistance to ceftolozane-tazobactam in Pseudomonas aeruginosa. MBio 2018; 9:e02085–02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karlowsky JA, Kazmierczak KM, de Jonge BLM, Hackel MA, Sahm DF, Bradford PA. In vitro activity of aztreonam-avibactam against Enterobacteriaceae and Pseudomonas aeruginosa isolated by clinical laboratories in 40 countries from 2012 to 2015. Antimicrob Agents Chemother 2017; 61:pii: e00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.