Summary
In the heart, primary microRNA-208b (pri-miR-208b) and Myheart (Mhrt) are long non-coding RNAs (lncRNAs) encoded by the cardiac myosin heavy chain genes. Although preclinical studies have shown that lncRNAs regulate gene expression and are protective for pathological hypertrophy, the mechanism underlying sex-based differences remains poorly understood. In this study, we examined DNA- and RNA-methylation-dependent regulation of pri-miR-208b and Mhrt. Expression of pri-miR-208b is elevated in the left ventricle of the female heart. Despite indistinguishable DNA methylation between sexes, the interaction of MeCP2 on chromatin is subject to RNase digestion, highlighting that affinity of the methyl-CG reader is broader than previously thought. A specialized procedure to isolate RNA from soluble cardiac chromatin emphasizes sex-based affinity of an MeCP2 co-repressor complex with Rest and Hdac2. Sex-specific Mhrt methylation chromatinizes MeCP2 at the pri-miR-208b promoter and extends the functional relevance of default transcriptional suppression in the heart.
Subject Areas: Molecular Physiology, Molecular Genetics, Molecular Mechanism of Gene Regulation
Graphical Abstract
Highlights
-
•
Mechanisms underlying sex-based gene expression are poorly understood
-
•
Expression of primary miR-208b is independent of DNA methylation in the heart
-
•
Sex-specific methylation of the long non-coding RNA Mhrt distinguishes MeCP2
-
•
Procedures assessing soluble chromatin emphasize RNA-dependent affinities
Molecular Physiology; Molecular Genetics; Molecular Mechanism of Gene Regulation
Introduction
Long non-coding RNAs (lncRNAs) have emerged as important regulators of heart development and disease (Mathiyalagan et al., 2014a, Schonrock et al., 2012). Several lncRNAs such as Braveheart (Klattenhoff et al., 2013), Fendrr (Grote et al., 2013), Upperhand (Anderson et al., 2016), and Carmen (Ounzain et al., 2015) are differentially activated at the fetal and adult stages of cardiac development. Although the precise roles of these lncRNAs still remain poorly understood, recent evidence suggests that lncRNAs not only control mRNA expression during the critical transition stages from fetal to adult heart but also function with mRNA stability, decay, and as regulatory sponges (Wang et al., 2014). Not entirely unexpected, in the fetal heart, lncRNAs associate more with genes implicated in processes that involve development and programming, whereas in the adult heart lncRNAs associate more with genes involved in disease. For example, in the adult heart the expression of lncRNAs Chast (Viereck et al., 2016), Chaer (Wang et al., 2016), and Wisper (Micheletti et al., 2017) are linked with cardiac hypertrophy and fibrosis. The sarcomeric myosin heavy chain (MHC) isogenes, originally designated α and β (recently renamed Myh6 and Myh7) are predominantly expressed in the heart (Mahdavi et al., 1982, Mahdavi et al., 1984). During fetal life Myh7 is principally expressed, whereas Myh6 and Myh7 are simultaneously expressed shortly after birth, with Myh6 dominantly expressed in adulthood (Chizzonite and Zak, 1984, Lompre et al., 1981, Lompre et al., 1984). During myocardial hypertrophy Myh7 is selectively expressed under hemodynamic and pressure overload (Izumo et al., 1987, Litten et al., 1982, Lompre et al., 1979).
Previously known as antisense β-MHC (Haddad et al., 2003), Myosin heavy-chain-associated RNA transcript (Myheart, or Mhrt) is a myocardium-specific lncRNA regulated by antisense transcription of a specific intergenic promoter that originates from the antisense strand of Myh7 that serves as a switch for Myh6/7 gene expression (Han et al., 2014). Recent studies have shown that Mhrt expression is cardioprotective and behaves as a decoy lncRNA involved in a negative feedback circuit with chromatin remodeling. Interestingly, cardiac hypertrophy induced by pressure overload in a mouse model progressively reduced Mhrt expression. Experiments have shown that restoring Mhrt to prestress levels can attenuate cardiac damage and rescue the heart from pathological hypertrophy (Han et al., 2014). It is hypothesized that under cardiac stress Mhrt restores the Myh6/7 gene shift by virtue of its interaction with the remodeling enzyme, Brg1, an ATP-dependent DNA helicase and a subunit of a much larger SWI/SNF complex. We have recently shown that pri-miR-208b, an lncRNA that originates from Myh7, regulates gene expression during cardiac hypertrophy (Mathiyalagan et al., 2014b). Upon transverse aortic constriction (TAC) in a mouse model of pathological hypertrophy, the left ventricular tissue shows elevated expression of pri-miR-208b that interacts with a Polycomb-group protein, Ezh2, and is part of a Hdac complex that regulates the Myh6/7 gene shift (Mathiyalagan et al., 2014b). The precise mechanisms regulating sex-based expression of lncRNAs such as pri-miR-208b and Mhrt remain poorly characterized.
Gender disparity in cardiovascular health and disease is well documented with females generally regarded less vulnerable to pathological cardiac remodeling (Maas and Appelman, 2010). Pathogenic processes such as cardiomyocyte apoptosis and necrosis characteristic of hypertrophy occur predominantly in males (Guerra et al., 1999). Pathological cardiac hypertrophy is characterized by reactivation of fetal gene programming that involves activation of fetal genes and the suppression of genes expressed in adults (Frey and Olson, 2003). This pattern of expression is common to both sexes, whereas molecular remodeling induced by pressure overload hypertrophy occurs predominantly in males (Weinberg et al., 1999, Zhong et al., 2003). Sex differences in the heart are largely determined by steroid hormones; however, several histone modifiers and methyl-binding proteins as well as promoter DNA methylation are closely linked to sex-based gene expression during development and in adult heart (Kurian et al., 2010, Ratnu et al., 2017). The role of lncRNAs in regulating sex-based gene expression by methylation in the heart remain poorly characterized. Because lncRNAs can interact with epigenetic modifiers, our aim was to determine the mechanism of lncRNA methylation in regulating sex-based gene expression in the heart.
MeCP2 is a reader of DNA methylation, a component of a co-repressor complex (Harikrishnan et al., 2005) and recently shown to regulate gene expression in chronic heart failure (Mayer et al., 2015). In addition to its high affinity for methylated cytosine sites on DNA, MeCP2 is recognized for its ability to bind RNA and regulate alternative splicing events by interacting with YB-1 (Young et al., 2005, Long et al., 2011, Jeffery and Nakielny, 2004). Although MeCP2 was initially characterized as a reader with high affinity for methylated cytosine in DNA (Bird and Wolffe, 1999), more recent studies have shown that MeCP2 can interact with other RNAs (Maxwell et al., 2013, Khan et al., 2017b, Khan et al., 2017c). For example, MeCP2 has been shown to interact with mRNAs (Long et al., 2011); non-coding RNAs (ncRNAs) such as let-7i, miR-375, and miR-126 (Khan et al., 2017c); as well as Rncr3 and Malat1 (Maxwell et al., 2013). Although these studies suggest that MeCP2 interacts with RNA to suppress gene expression, the mechanism mediated by lncRNA in the myocardium remains poorly understood.
One mechanism recently identified in the epigenetic control of lncRNAs is the deposition of 5-methylcytosine (5mC) (Squires et al., 2012, Amort et al., 2013). This epigenetic determinant occurs on coding RNA and ncRNA, and several lncRNAs such as Xist, Hotair, and Malat1 contain 5mC sites and have been shown to regulate ncRNA function (Amort et al., 2013, Amort et al., 2017). The purpose of this study was to explore the sex-based differences in lncRNA expression in the heart. Recent studies have shown that RNA-dependent MeCP2 binding on genes is independent of DNA methylation (Khan et al., 2017b). Therefore, we hypothesized that sex-based interaction of MeCP2 at the pri-miR-208b promoter is associated with Mhrt lncRNA methylation. Close examination of DNA methylation at the pri-miR-208b promoter using bisulfite sequencing identified nine sites of cytosine methylation at the CpG island (CGI). We observed no difference in DNA methylation between the sexes but show dramatic changes in the binding of the methylation reader, MeCP2, on the pri-miR-208b promoter using chromatin immunoprecipitation (ChIP). We identified that sex-specific expression of pri-miR-208b in the female heart is independent of DNA methylation. We provide proof of concept that MeCP2 affinity to chromatin is subject to RNA-methylation-dependent regulation. Sex-specific methylation of Mhrt distinguishes MeCP2 chromatinization of the pri-miR-208b promoter and gene regulation in the female heart.
Results
Sex Differences in Primary miR-208b Expression in the Heart
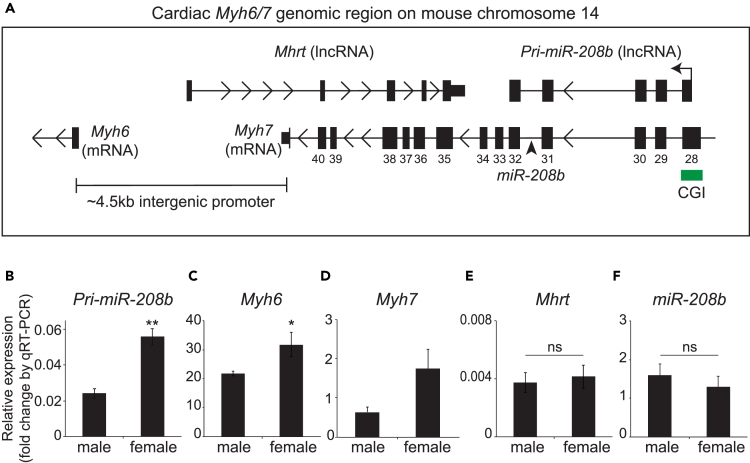
In the heart, cardiac myosin heavy chain (Myh) genes are transcriptionally regulated by distinct promoters that give rise to protein-coding genes and ncRNA production (Figure 1A). Located between Myh6 and Myh7 genes (Han et al., 2014, Mathiyalagan et al., 2014b) the promoter serves to regulate the transcription of the antisense lncRNA, Mhrt. To investigate gender differences in ncRNA expression we used qRT-PCR. Myh-encoded RNAs show sex-based differences in the expression of pri-miR-208b and Myh6 in female left ventricles when compared with males (Figures 1B and 1C). In contrast, we observe no significant differences in the expression of Myh7, Mhrt, and mature miR-208b transcripts (Figures 1D–1F).
Figure 1.
Elevated Primary miR-208b (pri-miR-208b) Transcript in Female Left Ventricle
(A) Genomic organization of myosin heavy chain coding and non-coding genes on chromosome 14 of mouse. qRT-PCR quantification of (B) pri-miR-208b; (C) Myh6; (D) Myh7, p = 0.065; (E) Mhrt (also known as antisense β-MHC); and (F) miR-208b. Normalized to Gapdh. CGI, CpG island; TSS, transcription start site; ns, not significant. *p < 0.05, **p < 0.01. n = 5 mice per group. Data are represented as mean ± SEM.
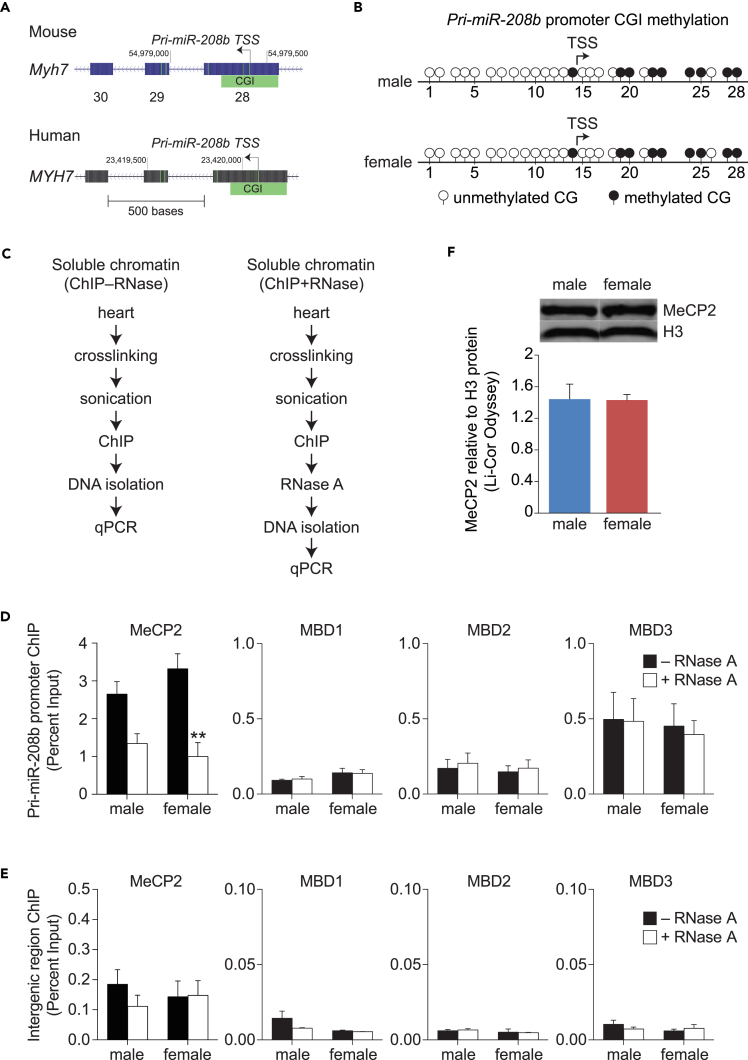
To understand the regulation of pri-miR-208b transcript, we examined its promoter located within exon 28 of mouse Myh7 gene (Figure 2A) (Monteys et al., 2010, van Rooij et al., 2009). The majority of RNA Pol II promoters are marked by CGI (Lujambio and Esteller, 2007, Carninci et al., 2006, Ozsolak et al., 2008) and served by specific bidirectional promoters (bdPs) (Trinklein et al., 2004). The promoter initiating pri-miR-208b transcription has a CGI that is conserved between mouse and human Myh orthologs (Figure 2A, CGI shown in green). Bisulfite sequencing identified nine CG sites (located at positions 14, 19, 20, 22, 23, 24, 25, 27, and 28) that are methylated in the promoter but were indistinguishable between male and female heart tissues (Figure 2B). Although these results suggest that pri-miR-208b promoter is methylated at specific CG dinucleotides, DNA methylation alone does not explain sex-based differences in pri-miR-208b expression.
Figure 2.
Interaction of the MeCP2 Reader on the pri-miR-208b Promoter in Female Left Ventricle Is Independent of DNA Methylation and Involves RNA Interactions
(A) Exon 28 of Myh7 gene showing pri-miR-208b promoter with CpG Island (CGI). CGI sequence within pri-miR-208b promoter is conserved between mice and humans.
(B) Bisulfite conversion and DNA methylation show site-specific CG methylation in male and female left ventricle (LV).
(C) Procedure of ChIP coupled with ribonuclease degradation (+RNase A) to assess cardiac RNA-dependent interactions.
(D and E) (D) Specific binding of MeCP2 at pri-miR-208b promoter in male and female LV in the presence (+) or absence (−) of RNase A. (E) Intergenic promoter is shown as a control for methyl CpG-binding proteins.
(F) Representative immunoblots of MeCP2 protein expression in male and female LV. Bar graph represents mean values of LI-COR Odyssey quantification of protein blots normalized to histone H3.
**p < 0.01. n ≥ 4 mice per group. Data are represented as mean ± SEM. See also Figure S1.
Sex-Specific Affinity for MeCP2 with Pri-miR-208b Chromatin Is RNA Dependent
Methyl-CpG-binding protein-mediated gene suppression involves the recognition of DNA methylation at CG dinucleotides at gene promoters, and recent experimental evidence implicates a regulatory role for MeCP2 in the mouse heart (Mayer et al., 2015). As pri-miR-208b promoter is methylated we assessed expression and chromatin interaction of methyl CpG-binding proteins in male and female hearts. qRT-PCR analysis shows no significant differences in the expression of Mecp2, Mbd1, Mbd2, and Mbd3 transcripts between the sexes (Figure S1). As the pri-miR-208b promoter is methylated albeit indistinguishably between sexes, we examined the binding specificities of protein readers of DNA methylation, members of the methyl-CpG-binding domain assessed by ChIP assays. Antibodies that specifically recognize MeCP2, MBD1, MBD2, and MBD3 were used to immunopurify soluble chromatin, followed by nucleic acid purification and quantification by qPCR. This technique specifically detects chromatin-bound MeCP2 fractions, whereas the protocol does not distinguish RNA-dependent interactions (Figure 2C, –RNase A). The specificity of antibody enrichment for pri-miR-208b chromatin was also assessed using IgG antibody controls (data not shown). As shown in Figure 2D, soluble chromatin was significantly enriched for MeCP2 on the pri-miR-208b promoter when compared with antibodies that recognize the other methyl-CpG-binding proteins. We also observed sex-based differences in chromatinized MeCP2 on the pri-miR-208b promoter (Figure 2D, –RNase A). We confirm that the affinity for MeCP2 was specific for the pri-miR-208b promoter when compared with the intergenic region (Figure 2E). Because these results were inconsistent with DNA methylation observed at the pri-miR-208b promoter (Figure 2B) we proposed that sex-based differences in MeCP2 affinity on soluble chromatin could be RNA dependent.
To test this hypothesis further, we devised a ChIP protocol that involves ribonuclease degradation (Figure 2C, +RNase A) to assess cardiac RNA-dependent MeCP2 affinity with the pri-miR-208b promoter (Khan et al., 2017b, Khan et al., 2017c, Maxwell et al., 2013, Mathiyalagan et al., 2014b). To test the feasibility of this approach, soluble MeCP2-associated chromatin fractions were subject to endoribonuclease A (+RNase A) to catalyze the cleavage of chromatin-associated RNA molecules. We observed significant reduction of MeCP2 affinity on the pri-miR-208b promoter in RNase-treated cardiac chromatin (Figure 2D, +RNase A). Furthermore, the qPCR signal for MeCP2 by ChIP was reduced for the pri-miR-208b promoter in the female heart. To confirm that the loss of MeCP2 binding following RNase exposure was specific for the pri-miR-208b promoter, we also assessed the intergenic region that is known to give rise to Myh6/Myh7 transcript diversity. Irrespective of sex we recovered the intergenic region equally well from chromatin isolates using MeCP2 antibody (Figure 2E). Furthermore, we assessed fractionated MeCP2 protein levels in the heart. Quantitative analysis of MeCP2 using LI-COR Odyssey imaging system show comparable protein expression from male and female heart tissue (Figure 2F), which is consistent with comparable mRNA expression (Figure S1). Taken together, these experimental results derived from soluble chromatin fractions using specific endoribonuclease assays further reinforce the view that pri-miR-208b promoter is subject to RNA-dependent MeCP2 binding.
Affinity of Rest and Hdac2 on Pri-miR-208b Chromatin Is RNA Dependent
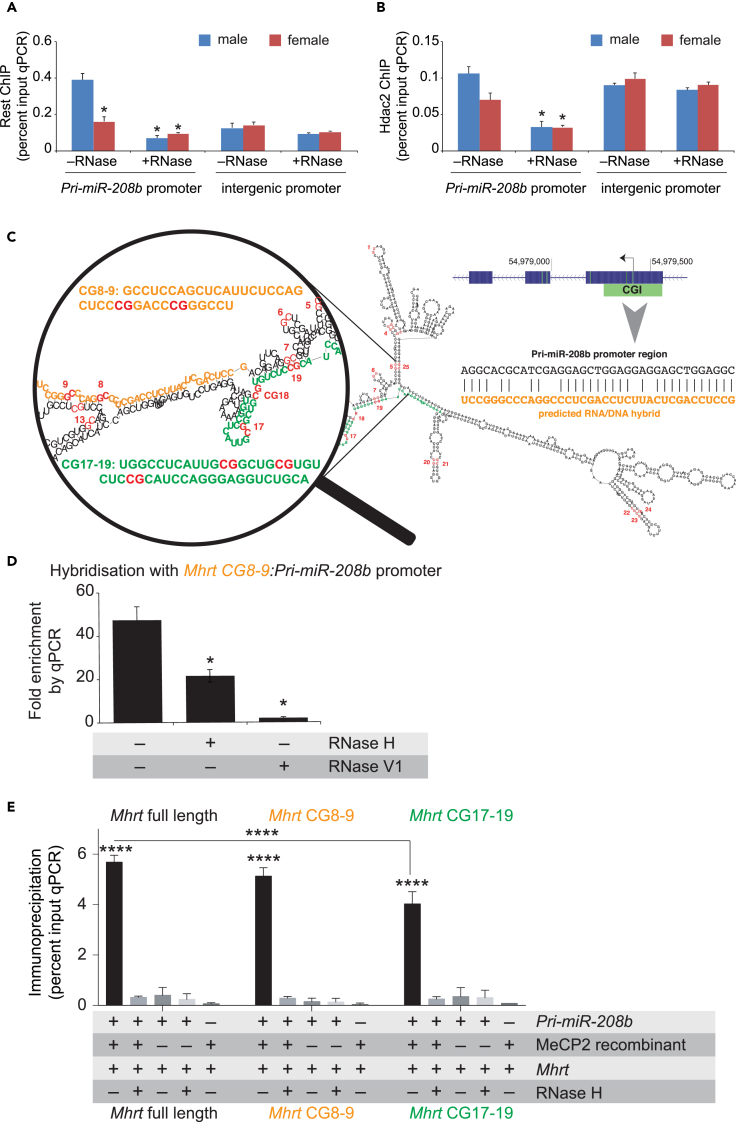
MeCP2 binds methylated DNA on assembled chromatin to recruit the RE1-silencing transcription factor (Rest) and histone deacetylase 2 (Hdac2) to repress gene transcription (Noh et al., 2012, Ooi and Wood, 2007). In silico analysis of pri-miR-208b promoter shows transcription factor-binding sites for Rest co-repressor protein (Figure S2). We assessed whether Rest and Hdac2 affinities for the pri-miR-208b promoter are subject to RNA-mediated interactions using specific ChIP assays. Consistent with previous results in the female heart, we observe reduced binding of Rest on the pri-miR-208b promoter by ChIP (–RNase A) and confirm that RNA is a substrate for binding (+RNase A), as shown in Figure 3A. Specificity for the pri-miR-208b promoter was shown by near-comparable Rest binding with the intergenic promoter in both sexes. Apart from the requirement for RNA to bind Rest to the pri-miR-208b promoter we observe no distinguishable difference at the intergenic promoter (Figure 3A). We also assessed Hdac2-associated chromatin fragments and confirm that affinity for the pri-miR-208b promoter is sensitive to nuclease degradation and in part mediated by RNA (Figure 3B, +RNase A). Taken together, these observations strengthen the view that sex-based affinity of the co-repressor complex at pri-miR-208b promoter is sensitive to endoribonuclease digestion and thus places specific emphasis on RNA-dependent interactions.
Figure 3.
MeCP2 Forms an RNA/dsDNA Hybrid with Mhrt and pri-miR-208b
(A and B) Chromatin immunoprecipitation of (A) Rest and (B) Hdac2 binding at pri-miR-208b promoter in male and female LV in the presence (+) or absence (−) of RNase A.
(C) Enlarged view of the RNA/DNA hybrid region in predicted secondary structure of full-length Mhrt (also shown in Figure S3). The predicted Mhrt lncRNA (shown in orange)/DNA hybrid sequence at the pri-miR-208b promoter (shown in black). Twenty-five CG dinucleotides of Mhrt lncRNA are shown in red.
(D) In vitro reconstitution assay showing qPCR amplification of pri-miR-208b promoter after hybridization with Mhrt oligomer in the presence of RNase H or RNase V1.
(E) Amplification of pri-miR-208b promoter by qPCR after MeCP2 immunopurification from the Mhrt RNA-dsDNA hybrid.
*p < 0.05. ****p < 0.0001. Data are represented as mean ± SEM. See also Figures S2–S4.
Mhrt Forms Stable RNA:dsDNA Hybrid with Pri-miR-208b Promoter
lncRNAs have emerged as a class of gene regulators that serve to alter chromatin structure and gene function (Mercer and Mattick, 2013, Mathiyalagan et al., 2014a). lncRNAs can form stable RNA-DNA hybrids and act as docking sites for transcription factors (Schmitz et al., 2010, Grote et al., 2013, Mathiyalagan et al., 2014b, Place et al., 2008). A recent study has shown that stress-induced interaction of Mhrt with the Brg1 complex regulates the Myh6/7 gene shift in the heart (Han et al., 2014). Therefore, we assessed whether Mhrt could bind the pri-miR-208b chromatin. In silico analysis identified a 37-nucleotide (nt) binding site for Mhrt RNA (CG8-9 correspond to positions 141–178: Figure S3) downstream of the pri-miR-208b transcription start site (Figure 3C). To assess whether Mhrt could form stable hybrid (RNA:dsDNA [double-stranded DNA]) with pri-miR-208b promoter, we performed binding assays using Mhrt RNA oligomer corresponding to the 37-nt sequence and dsDNA matching the pri-miR-208b promoter sequence (Grote et al., 2013, Mathiyalagan et al., 2014b). Because the RNA oligomer (RNAmer) was constructed with biotin label at the 3′ end of Mhrt this allowed for immunopurification using streptavidin beads. Incubation of RNA and dsDNA followed by streptavidin capture and qPCR amplification of pri-miR-208b promoter shows high affinity for the Mhrt oligomer (Figure 3D, –RNase). However, in the presence of RNase H, which specifically cleaves RNA in an RNA:dsDNA hybrid (Grote et al., 2013, Mathiyalagan et al., 2014b), the qPCR signal for pri-miR-208b was significantly reduced (Figure 3D, +RNase H). To further illustrate the specificity of RNA binding we used RNase V1, which cleaves base-paired RNA nucleotides (Grote et al., 2013, Lee et al., 2010), to confirm that Mhrt interacts directly with pri-miR-208b promoter (Figure 3D, +RNase V1).
Because MeCP2 binding is sensitive to RNA nuclease degradation we assessed binding to RNA:dsDNA hybrids. To do this, binding assays were performed using recombinant MeCP2 in the presence or absence of RNA (full-length Mhrt):dsDNA (Pri-miR-208b) hybrids. We made use of selective mixtures of RNA:dsDNA hybrids to test the sensitivity to RNase H nuclease digestion following immunopurification using an antibody that specifically recognizes MeCP2. Isolated DNA was assayed by qPCR detection of the pri-miR-208b promoter (Figure S4). Under optimal binding conditions, we observe no qPCR signal for the pri-miR-208b promoter using the MeCP2 antibody. Under identical reaction conditions, we found that MeCP2 was significantly enriched on the pri-miR-208b promoter in the presence of full-length Mhrt RNA (Figure 3E). RNA transcripts fold into secondary structures critical to form RNA-DNA interactions as predicted by the minimal free energy of Mhrt transcript (Figure S3). Interestingly, CG18 is the nearest dinucleotide site in the hybrid region (Figure 3C). To assess the hybrid region of Mhrt transcript, we performed MeCP2 binding assays using CG8-9 and CG17-19 Mhrt RNAmers as well as the full-length Mhrt transcript (Figure 3E). CG8-9 and CG17-19 displayed strong affinity for recombinant MeCP2 protein. To test the binding specificity under identical reaction conditions we also assessed the interaction of recombinant Set7 methyltransferase, which does not recognize methylated DNA. We confirm that Set7 does not interact with the RNA:dsDNA hybrid (personal observations). The interaction of MeCP2 with Mhrt and pri-miR-208b duplex led us to examine whether this affinity was sensitive to RNase H nuclease digestion. Following MeCP2 immunoprecipitation capture the signal for pri-miR-208b promoter was barely detectable. These results support the view that MeCP2 affinity for the pri-miR-208b promoter is mediated by CG8-9 and CG17-19 interaction for Mhrt lncRNA.
Mhrt Methylation Distinguishes Sex-Based Differences in MeCP2 Binding
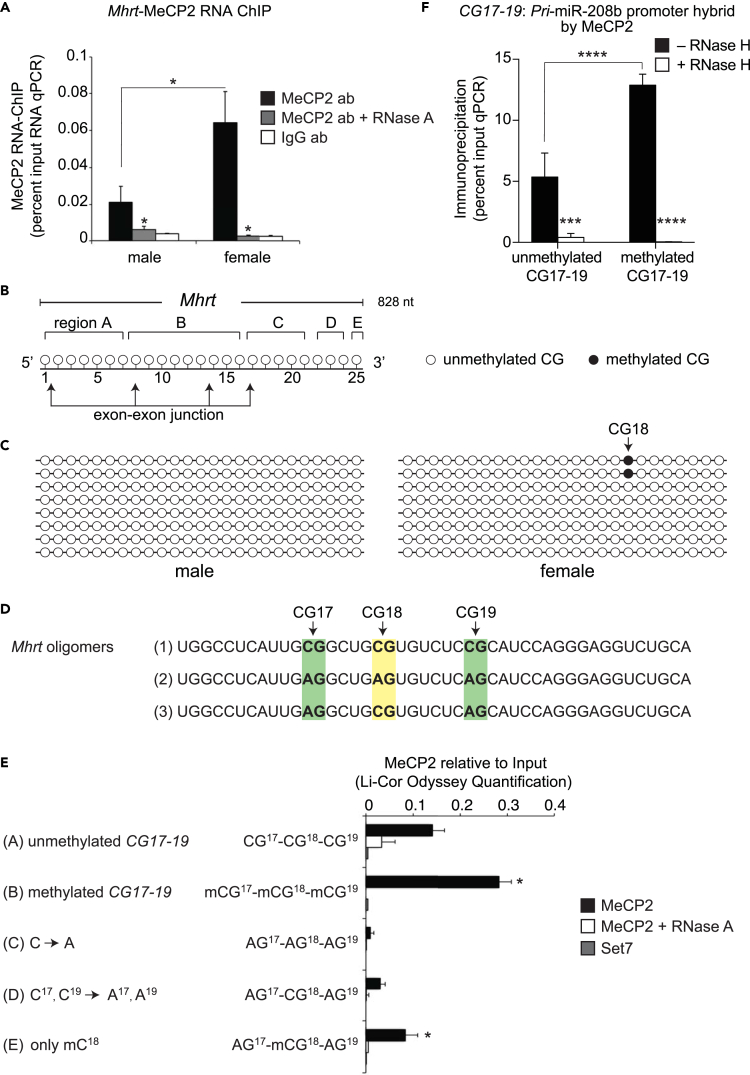
Because Mhrt has been shown to function as decoy lncRNA by interacting with Brg1 in cardiac hypertrophy (Han et al., 2014), we hypothesized that Mhrt could interact with MeCP2. To do this, we isolated RNA from soluble chromatin fractions immunopurified using anti-MeCP2 antibody, and tested binding affinities for Mhrt using qPCR detection. We observe sex-based differences for MeCP2 affinity on Mhrt using a specific RNA-ChIP procedure (Figures 4A and S5). The affinity of MeCP2 for Mhrt was significantly reduced in +RNase A-treated chromatin when compared with IgG antibody controls (Figure S5). Although these results imply MeCP2 interacts with Mhrt, the affinity for the lncRNA remains poorly characterized.
Figure 4.
Mhrt Is Methylated and Binds to MeCP2 in Female Left Ventricle
(A) RNA-ChIP using anti-MeCP2 antibody showing Mhrt interaction with MeCP2-bound chromatin in male and female left ventricle (LV). n = 6 mice per group.
(B) Illustration of 0.828 kb of Mhrt lncRNA with 25 CG dinucleotides and regions A-E targeted by bisulfite primers.
(C) Bisulfite-converted Mhrt clones showing methylation at CG18 of Mhrt lncRNA in female LV. n = 5 mice per group.
(D) Synthetic 45-nt-long Mhrt oligomer highlighting CG17-19 and variants with replaced cytosine to adenosine residues.
(E) Quantification of Mhrt-MeCP2 binding assay showing enhanced MeCP2 binding after methylation of synthetic Mhrt oligomer.
(F) RNA-dsDNA hybrid binding assay showing methylation-dependent affinity of MeCP2 for CG17-19 Mhrt RNAmer.
*p < 0.05, ***p < 0.001, ****p < 0.0001. Data are represented as mean ± SEM. See also Figures S5–S8.
Recent studies show that post-transcriptional RNA modifications are implicated in transcriptional control (Amort et al., 2013, Squires et al., 2012). We hypothesized that the affinity of the methylation reader MeCP2 is dependent on 5mC deposition on Mhrt lncRNA. To do this we examined the possible methylation sites at 25 CG dinucleotides (Figure S6) using bisulfite sequencing of five distinct regions labeled A-E that cover the Mhrt lncRNA (Figure 4B). RNA methylation analysis showed sex specific methylation at CG18 position detectable in 25% of total clones tested for this site in female heart tissues (Figure 4C). We observed no RNA methylation for other CG sites of the Mhrt transcript in male and female hearts. These results suggest that specific cytosine methylation at position CG18 of Mhrt could distinguish sex-specific affinity for MeCP2.
MeCP2 Binds Methylated Mhrt lncRNA
To investigate site-specific Mhrt methylation and MeCP2 interaction, we designed biotin-tagged synthetic RNAmers that include CG17, CG18, and CG19 sites of the Mhrt transcript (Figure 4D). The CG18 site of the Mhrt transcript is conserved in humans and mice, suggesting that it could be important for MeCP2 affinity (Figure S6). To specifically examine methylation at CG18 we designed RNAmers that replaced cytosine residues with adenosine (Figure 4D). Mhrt oligomer-1 contained three CGs intact (CG17-19), oligomer-2 contained “C” replaced with “A” at all three sites, and oligomer-3 contained “C” replaced with “A” only at CG17 and CG19, thus leaving CG18 intact. To assess the affinity for methylated CG18, RNAmers were reconstituted with recombinant MeCP2 followed by immunopurification using streptavidin beads. Quantitative protein blotting using LI-COR Odyssey was performed using an antibody that specifically recognizes MeCP2 (Figure S7). To assess MeCP2 affinity for the methylated and unmethylated RNAmers M.Sss1 methyltransferase (Flynn et al., 1996) was used to specifically methylate CG17-19 sites. We observed binding of recombinant MeCP2 when incubated with unmethylated CG17-19 Mhrt RNAmer (Figure 4E, panel A); however, the affinity for MeCP2 was significantly enriched in the presence of methylated CG17-19 Mhrt RNAmer (Figure 4E, panel B). Binding for MeCP2 was competed when incubated with oligomer-2, suggesting that the CG18 site is an important substrate for binding (Figure 4E, panel C). In support of this viewpoint, the affinity of MeCP2 was restored when site position CG18 was methylated using oligomer-3 (Figure 4E, panel E) when compared with the unmethylated version (Figure 4E, panel D). Furthermore, we confirm that MeCP2 affinity was RNA dependent using ribonuclease A (Figure 4E, MeCP2+RNase A). To illustrate the specificity of MeCP2 affinity for Mhrt we assessed binding of the lysine methyltransferase writer Set7, which neither distinguishes methylated DNA nor binds RNA. We confirm that Set7 does not interact with methylated RNAmers, confirming the affinity of MeCP2 for methylated Mhrt at CG18 site (Figure 4E). These results support the view that CG18 methylation could account for MeCP2 affinity to Mhrt lncRNA. Based on these results we examined the expression of known RNA methyltransferase transcripts (Nsun2, Nsun4, Nsun5, Rnmt1, Nop2, Trdmt1, Mrm1, Trmt1) in male and female tissues. There was no significant difference in the expression of RNA methyltransferases (Figure S8).
To assess whether Mhrt methylation influences the affinity of MeCP2-dependent formation with pri-miR-208b promoter, we performed RNA:dsDNA hybrid assays using methylated/unmethylated CG17-19 Mhrt RNAmers (oligomer-1 in Figure 4D). We made use of selective mixtures of RNA:dsDNA hybrids to test the sensitivity to RNase H digestion following immunopurification using an antibody that specifically recognizes MeCP2. Isolated DNA was assayed by qPCR detection of the pri-miR-208b promoter (Figure S4). Consistent with the binding affinity of MeCP2 with Mhrt in the female heart, MeCP2 was significantly enriched on the pri-miR-208b promoter when Mhrt is specifically methylated at CG17-19 following capture by RNA immunoprecipitation (Figure 4F –RNase H). The qPCR signal for the pri-miR-208b promoter was almost abolished using identical binding conditions and ribonuclease endonuclease H enzyme that catalyzes the cleavage of RNA in an RNA/DNA hybrid (Figure 4F + RNase H). Taken together, these results suggest that the affinity of MeCP2 for the RNA:dsDNA hybrid is likely to depend on Mhrt methylation to chromatinize the pri-miR-208b promoter to regulate gene transcription.
Discussion
Although sex-based differences in the incidence, prevalence, and severity of cardiovascular disease are well documented the functional mechanisms underlying sex-specific gene expression remains poorly understood. Whether sexual dimorphism is functionally regulated by methylation of DNA and RNA nucleobases remains uncharted in the heart.
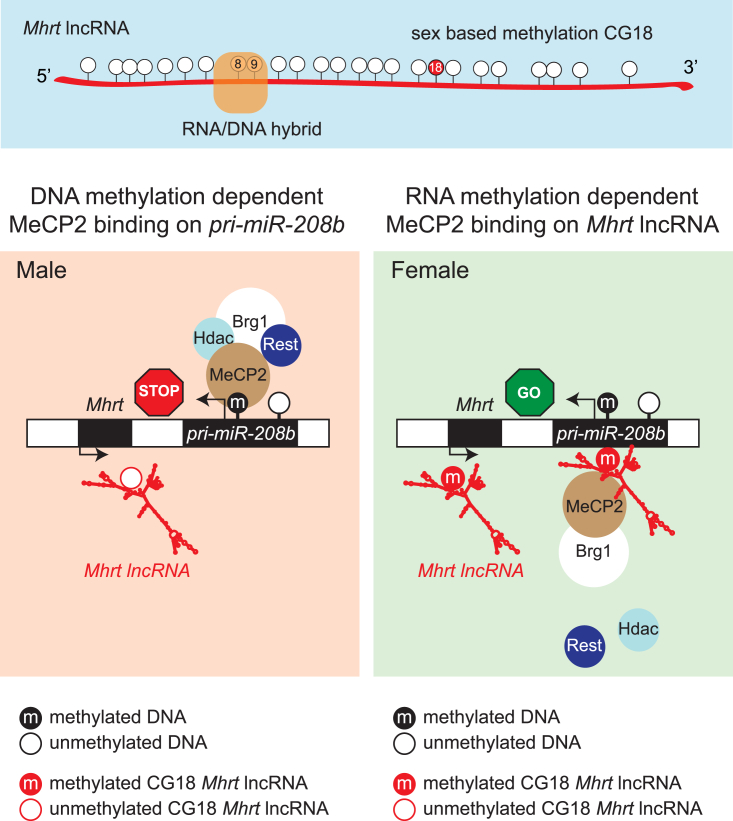
It is probable that methylation of Mhrt that we describe is responsible for sex-based differences in pri-miR-208b expression. The proposed model is mediated by DNA methylation, which we speculate is functioning in a context-dependent manner on the pri-miR-208b promoter and distinct from the exclusive RNA methylation observed in Mhrt in females. This is because, irrespective of sex, the pri-miR-208b promoter is characterized by CGI methylation at nine identical sites. Close inspection of Mhrt and pri-miR-208b lncRNAs shows several interesting features. First, Mhrt is methylated in female heart tissue and the presence of RNA methylation at the CG18 site may indicate regulatory function that is associated with MeCP2 affinity. Second, irrespective of sex the pri-miR-208b promoter is methylated at nine identical CG sites and represents default repression. We believe that interactions mediated by DNA and RNA methylation are context specific, meaning MeCP2 assembles on methyl-CG sites and cooperatively serves to suppress gene expression. It should be stressed, however, that we are not suggesting a hierarchy of affinity, rather a cooperative association. It has been commonly assumed that the 5mC determinant on DNA is the predominant target of methylation readers (Harikrishnan et al., 2005). We propose that the 5mC-dependent reader, MeCP2 co-repressor complex, targets the pri-miR-208b promoter to suppress expression in the male heart. Although methylation-mediated transcriptional silencing by MeCP2 is well documented (Nan et al., 1997), recent studies in animal and human cells have shown broader binding preferences that include mRNA (Long et al., 2011), double-stranded RNA (Jeffery and Nakielny, 2004), short-(Khan et al., 2017b) and long-ncRNAs (Maxwell et al., 2013), as well as RNA-dependent spliceosome and miRNA-processing determinants (Young et al., 2005, Cheng et al., 2014). Thus, although the conventional view of MeCP2-mediated suppression parallels our observations of DNA methylation in the heart, we hypothesize that RNA methylation could signal MeCP2. So how does Mhrt alleviate pri-miR-208b suppression in the female heart? As a resolution to this problem, rather than querying MeCP2 interaction solely with methylated DNA, we also assessed the affinity of MeCP2 with Mhrt lncRNA as a prime candidate for transcriptional control. This mode of regulation requires specialization, and Mhrt is a prime substrate for MeCP2, which is exclusively methylated in females (Figure 5). This context-dependent default repression (Tsang et al., 2007) ensures that pri-miR-208b is continually turned off when the promoter is methylated in the (male) heart but turned on when Mhrt lncRNA is methylated at CG18 (in the female heart). This is an argument supported by emerging experimental evidence showing RNA methylation as a functional regulatory signal in gene control (Amort et al., 2013, Amort et al., 2017, Squires et al., 2012) and in keeping with observations we describe for MeCP2-mediated binding of the lncRNA, Mhrt. At the molecular level, this mode of regulation could account for the transcriptional robustness in pri-miR-208b expression in the female heart as well as the capacity for sex-based expression, notwithstanding identical CGI methylation at the pri-miR-208b promoter.
Figure 5.
Working Model Based on Current Knowledge
Cooperative DNA and sex-based RNA methylation in the heart. In this study, we identify the pri-miR-208b promoter hybrid region and sex-based methylation site (CG18) of Mhrt lncRNA. We postulate that the methylation reader MeCP2 and the co-repressor protein complex interact with the pri-miR-208b promoter because the Mhrt lncRNA is unmethylated at CG18 in the male heart. Methylation at position CG18 of Mhrt lncRNA is observed with enriched MeCP2 interaction in the female heart allowing pri-miR-208b transcription. Mhrt lncRNA is also linked with the SWI/SNF-binding determinant Brg1 (Han et al., 2014).
RNA methylation of cardiac Mhrt extends the functional importance of MeCP2-mediated gene silencing. Nowhere is this more evident than in the signaling molecules that assemble on, and serve to, chromatinize the regulatory cross talk between DNA and RNA (Khan et al., 2017b, Khan et al., 2017c). There are several advantages to MeCP2 coordinating context-dependent regulation. First, 5mC is an effective target for MeCP2. Binding experiments have shown remarkable specificity for the nuclear protein, revealing that a single symmetrical methylated CG site is sufficient for MeCP2 binding (Nan et al., 1996, Meehan et al., 1992) and that hemi-methylated DNA is a poor substrate (Lewis et al., 1992). Second, on methylated CG sites MeCP2 forms a stable complex with HDAC activity to repress transcription. Of note, methylated DNA assembled into chromatin binds MeCP2 and resides in a complex that co-purifies with members of the DNA helicase/ATPase family such as ATRX (Nan et al., 2007), Brahma (Harikrishnan et al., 2005), Brg1 (Hwang et al., 2009), and Mi-2 (Wade et al., 1998). The ability of MeCP2 to bind to methylated nucleobases to form cell- and tissue-specific regulatory complexes serves to precisely control gene regulation in the heart. In the above examples, CG methylation is the predominant determinant. In other cases, MeCP2 can impart specificity by virtue of interactions with the DNA methylation writing enzyme, DNMT1, to target candidate sequence elements to repress transcription (Robertson et al., 2000). Once again, the effectiveness of MeCP2 to achieve gene control relies on its capacity to interpret and tightly bind methylated CG sites. Clearly, methylation-mediated regulation is complicated; however, the diversity in the mechanism of action by the methyl moiety interpreted by MeCP2 represents an attractive solution to connect what appear to be distinct and even seemingly unconnected signaling pathways (Simpson, 2015). This is strikingly reminiscent of the norepinephrine pathway leading to MeCP2 repression upon activation of α1- and β1-adrenoceptors in cardiomyocytes (Mayer et al., 2015). In a mouse model of pathological hypertrophy using TAC the specific CG methylation sites of target genes under direct regulation by MeCP2 remain unaltered. Cardiomyocytes derived from Sham animals show methylation-mediated suppression of Ppargc1a by MeCP2, whereas in TAC animals the induction of miR-212/132 removes MeCP2 from the same methyl-CG sites. Although that study (Mayer et al., 2015) did not examine sexual dimorphism in sympathetic cardioprotective pathways, it shows that dynamic signaling gradients can shape MeCP2 regulation and default repression mechanisms in hypertrophic hearts.
Important clinically, pathological signaling can integrate regulatory pathways to connect methylation of assembled chromatin with ncRNAs that silence gene expression by MeCP2. This is highlighted by the chromatinization of the norepinephrine transporter (NET) gene by MeCP2 in postural tachycardia syndrome (Khan et al., 2017b). Bisulfite sequencing studies in mouse and human cells show NET silencing by MeCP2 independent of CGI methylation changes (Harikrishnan et al., 2010, Bayles et al., 2012). Paradoxical to previous knowledge and against the paradigm at the time, these studies showed that altered chromatin states remodeled by helicase proteins together with MeCP2 serve to autonomously silence NET expression. To assess the impact of NET chromatinization, a protocol was developed to capture RNA-isolated chromatin followed by deep sequencing. NET-interacting RNAs identified let-7i as a prime target of MeCP2 (Khan et al., 2017b). At the molecular level, these studies support the view that DNA helicase or ATPase proteins connect ncRNAs and HDAC activity either to suppress gene transcription (Khan et al., 2017a) or to regulate chromatin architecture critical to the nuclear functions associated with gene control.
The regulatory capacity of MeCP2 to co-exist with distinct components on CG methylation sites may be exploited to connect otherwise dipartite regulatory pathways. Here, we report the myocardium-specific lncRNA, Mhrt, as a target for MeCP2 recognizing RNA methylation and regulating sex-specific expression of pri-miR-208b in the female heart. Binding studies show that the affinity of Mhrt RNAmer for MeCP2 is dependent on methylation at CG18, implicating probable role in gene regulation. Therefore, this study is potentially original. We have identified MeCP2 as a physiological target for a cardiac-specific lncRNA. Because methylation of the Mhrt lncRNA at CG18 is a substrate for MeCP2, we hypothesize that this affinity for methylated RNA safeguards sex-based cardiac gene suppression.
Recent transcriptome studies have shown widespread RNA methylation in mRNAs and lncRNAs; however, RNA methylation is less abundant than DNA methylation (Squires et al., 2012). Furthermore, RNA methylation at cytosine sites have been shown to influence post-transcriptional gene regulation (Amort et al., 2017, Yang et al., 2017, Aguilo et al., 2016, Shafik et al., 2016, Zhao et al., 2017). In our study, we observed RNA methylation of Mhrt from 25% bisulfite-sequenced clones in the female heart. This resembles the observed frequencies of RNA methylation for the lncRNAs, HOTAIR and XIST (Schaefer et al., 2009, Amort et al., 2013). Indeed, low-level methylation of XIST serves to functionally distinguish methylated from unmethylated versions that bind chromatin-modifying enzymes such as EZH2 and PRC2. In that study (Amort et al., 2013), the reported frequency of XIST methylation was approximately 20%. In another study, low-frequency RNA methylation was also observed at specific CG sites in tRNAs (Schaefer et al., 2009). Based on our experimental observations in the female heart we postulate that the affinity of MeCP2 on methylated Mhrt could serve to target specific CG sites for gene regulation.
Although the above example clearly indicates that XIST methylation influences the interaction of polycomb-group proteins such as EZH2, their effectiveness to bind chromatinized templates is also subject to histone modifications. Indeed, chromatin content as well as loading of specific co-regulatory complexes serve to integrate regional structures to regulate transcriptional events. For example, in cardiac hypertrophy the activation of α-MHC and coordinated suppression of β-MHC events are mediated by direct interaction of the pri-miR-208b lncRNA to the intergenic bdP (Mathiyalagan et al., 2014b). Specifically, α/β-MHC transcriptional responses are regulated by the EZH2/pri-miR-208b co-repressive complex to form a suppressive environment at the bdP together with the SWI/SNF chromatin-remodeling determinant Brg1. These distinct changes to the intergenic region mean enhanced H3K27me3 by EZH2 and reduced gene-permissive marks such as H3K4me3 and H3K9/14ac. Thus, we postulate that the importance of sex-based Mhrt methylation may serve as an effective regulator of pri-miR-208b gene expression for maintaining homeostatic transcriptional expression in the adult heart and serve to integrate these events in pathological remodeling of ventricular hypertrophy.
Limitations of the Study
Although the properties of MeCP2 affinity with methylated Mhrt are considered consistent with the idea of sexual dimorphism regulating pri-miR-208b expression, this idea has not yet been tested completely. First, sex-specific methylation of the lncRNA Mhrt at CG18 in the female heart remains poorly understood. The challenge now is to understand how the cardiovascular phenotype with respect to hormonal regulation could control gene expression and sex differences (Mosca et al., 2011). Second, we also acknowledge that although this study emphasizes RNA methylation-dependent MeCP2 binding in the heart, it now becomes apparent that methylation of other RNA sites could also be prime candidates for default repression by methylation that need to be tested. Although lncRNAs interacting with DNMTs have been documented to regulate DNA methylation (Di Ruscio et al., 2013), our results show proof of concept that Mhrt methylation is a target for MeCP2 to regulate pri-miR-208b expression independent of differential DNA methylation. Furthermore, although we appreciate that CG18 methylation of Mhrt was not critically tested using animal models of cardiac disease, future studies should allow us to test the generalizability of site-specific RNA methylation and protein interaction using in vitro and in vivo experimental models.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
Professor Assam El-Osta is a National Health and Medical Research Council (NHMRC) Senior Research Fellow (1154650), and this work is supported by NHMRC-NSFC International Joint Grant (1113188 and 15070000000090844) and NHMRC grants (1032687 and 526635).
Author Contributions
H.K.N., J.O., and P.M. performed the study design, validation, and data representation; H.K.N. and P.M. performed the ChIP, bisulfite sequencing, and in vitro binding assays; J.O. cloned and synthesized Mhrt full-length transcript; J.O., P.M., and A.W.K. prepared the figures; S.A.J., G.S., and X.-J.D. were involved in mouse tissue handling and resources; M.Z. analyzed bisulfite sequencing data; and A.E.-O. was involved in the design, supervision, funding, and writing of the manuscript, which has been reviewed and edited by the co-authors.
Declaration of Interests
The authors declare there is no competing interests.
Published: July 26, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.06.031.
Supplemental Information
References
- Aguilo F., Li S., Balasubramaniyan N., Sancho A., Benko S., Zhang F., Vashisht A., Rengasamy M., Andino B., Chen C.H. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1alpha. Cell Rep. 2016;14:479–492. doi: 10.1016/j.celrep.2015.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort T., Rieder D., Wille A., Khokhlova-Cubberley D., Riml C., Trixl L., Jia X.Y., Micura R., Lusser A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18:1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort T., Souliere M.F., Wille A., Jia X.Y., Fiegl H., Worle H., Micura R., Lusser A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013;10:1003–1008. doi: 10.4161/rna.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.M., Anderson D.M., Mcanally J.R., Shelton J.M., Bassel-Duby R., Olson E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles R., Harikrishnan K.N., Lambert E., Baker E.K., Agrotis A., Guo L., Jowett J.B., Esler M., Lambert G., El-Osta A. Epigenetic modification of the norepinephrine transporter gene in postural tachycardia syndrome. Arterioscler. Thromb. Vasc. Biol. 2012;32:1910–1916. doi: 10.1161/ATVBAHA.111.244343. [DOI] [PubMed] [Google Scholar]
- Bird A.P., Wolffe A.P. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cheng T.L., Wang Z., Liao Q., Zhu Y., Zhou W.H., Xu W., Qiu Z. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev. Cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- Chizzonite R.A., Zak R. Regulation of myosin isoenzyme composition in fetal and neonatal rat ventricle by endogenous thyroid hormones. J. Biol. Chem. 1984;259:12628–12632. [PubMed] [Google Scholar]
- Di Ruscio A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J., Glickman J.F., Reich N.O. Murine DNA cytosine-C5 methyltransferase: pre-steady- and steady-state kinetic analysis with regulatory DNA sequences. Biochemistry. 1996;35:7308–7315. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- Frey N., Olson E.N. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Blass G., Kellis M., Werber M., Herrmann B.G. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra S., Leri A., Wang X., Finato N., Di Loreto C., Beltrami C.A., Kajstura J., Anversa P. Myocyte death in the failing human heart is gender dependent. Circ. Res. 1999;85:856–866. doi: 10.1161/01.res.85.9.856. [DOI] [PubMed] [Google Scholar]
- Haddad F., Bodell P.W., Qin A.X., Giger J.M., Baldwin K.M. Role of antisense RNA in coordinating cardiac myosin heavy chain gene switching. J. Biol. Chem. 2003;278:37132–37138. doi: 10.1074/jbc.M305911200. [DOI] [PubMed] [Google Scholar]
- Han P., Li W., Lin C.H., Yang J., Shang C., Nurnberg S.T., Jin K.K., Xu W., Lin C.Y., Lin C.J. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan K.N., Bayles R., Ciccotosto G.D., Maxwell S., Cappai R., Pelka G.J., Tam P.P., Christodoulou J., El-Osta A. Alleviating transcriptional inhibition of the norepinephrine slc6a2 transporter gene in depolarized neurons. J. Neurosci. 2010;30:1494–1501. doi: 10.1523/JNEUROSCI.4675-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan K.N., Chow M.Z., Baker E.K., Pal S., Bassal S., Brasacchio D., Wang L., Craig J.M., Jones P.L., Sif S., El-Osta A. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- Hwang C.K., Song K.Y., Kim C.S., Choi H.S., Guo X.H., Law P.Y., Wei L.N., Loh H.H. Epigenetic programming of mu-opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodelling factor. J. Cell Mol. Med. 2009;13:3591–3615. doi: 10.1111/j.1582-4934.2008.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo S., Lompre A.M., Matsuoka R., Koren G., Schwartz K., Nadal-Ginard B., Mahdavi V. Myosin heavy chain messenger RNA and protein isoform transitions during cardiac hypertrophy. Interaction between hemodynamic and thyroid hormone-induced signals. J. Clin. Invest. 1987;79:970–977. doi: 10.1172/JCI112908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery L., Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J. Biol. Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- Khan A.W., Corcoran S.J., Esler M., El-Osta A. Epigenomic changes associated with impaired norepinephrine transporter function in postural tachycardia syndrome. Neurosci. Biobehav. Rev. 2017;74:342–355. doi: 10.1016/j.neubiorev.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Khan A.W., Ziemann M., Corcoran S.J., K N.H., Okabe J., Rafehi H., Maxwell S.S., Esler M.D., El-Osta A. NET silencing by let-7i in postural tachycardia syndrome. JCI Insight. 2017;2:e90183. doi: 10.1172/jci.insight.90183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.W., Ziemann M., Rafehi H., Maxwell S., Ciccotosto G.D., El-Osta A. MeCP2 interacts with chromosomal microRNAs in brain. Epigenetics. 2017;12:1028–1037. doi: 10.1080/15592294.2017.1391429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian J.R., Olesen K.M., Auger A.P. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.C., Aalto A.P., Yang Q., Chang S.S., Huang G., Fisher D., Cha J., Poranen M.M., Bamford D.H., Liu Y. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Litten R.Z., 3rd, Martin B.J., Low R.B., Alpert N.R. Altered myosin isozyme patterns from pressure-overloaded and thyrotoxic hypertrophied rabbit hearts. Circ. Res. 1982;50:856–864. doi: 10.1161/01.res.50.6.856. [DOI] [PubMed] [Google Scholar]
- Lompre A.M., Mercadier J.J., Wisnewsky C., Bouveret P., Pantaloni C., D'albis A., Schwartz K. Species- and age-dependent changes in the relative amounts of cardiac myosin isoenzymes in mammals. Dev. Biol. 1981;84:286–290. doi: 10.1016/0012-1606(81)90396-1. [DOI] [PubMed] [Google Scholar]
- Lompre A.M., Nadal-Ginard B., Mahdavi V. Expression of the cardiac ventricular alpha- and beta-myosin heavy chain genes is developmentally and hormonally regulated. J. Biol. Chem. 1984;259:6437–6446. [PubMed] [Google Scholar]
- Lompre A.M., Schwartz K., D'albis A., Lacombe G., Van Thiem N., Swynghedauw B. Myosin isoenzyme redistribution in chronic heart overload. Nature. 1979;282:105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- Long S.W., Ooi J.Y., Yau P.M., Jones P.L. A brain-derived MeCP2 complex supports a role for MeCP2 in RNA processing. Biosci. Rep. 2011;31:333–343. doi: 10.1042/BSR20100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A., Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- Maas A.H., Appelman Y.E. Gender differences in coronary heart disease. Neth. Heart J. 2010;18:598–602. doi: 10.1007/s12471-010-0841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi V., Chambers A.P., Nadal-Ginard B. Cardiac alpha- and beta-myosin heavy chain genes are organized in tandem. Proc. Natl. Acad. Sci. U S A. 1984;81:2626–2630. doi: 10.1073/pnas.81.9.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi V., Periasamy M., Nadal-Ginard B. Molecular characterization of two myosin heavy chain genes expressed in the adult heart. Nature. 1982;297:659–664. doi: 10.1038/297659a0. [DOI] [PubMed] [Google Scholar]
- Mathiyalagan P., Keating S.T., Du X.J., El-Osta A. Interplay of chromatin modifications and non-coding RNAs in the heart. Epigenetics. 2014;9:101–112. doi: 10.4161/epi.26405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiyalagan P., Okabe J., Chang L., Su Y., Du X.J., El-Osta A. The primary microRNA-208b interacts with Polycomb-group protein, Ezh2, to regulate gene expression in the heart. Nucleic Acids Res. 2014;42:790–803. doi: 10.1093/nar/gkt896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell S.S., Pelka G.J., Tam P.P., El-Osta A. Chromatin context and ncRNA highlight targets of MeCP2 in brain. RNA Biol. 2013;10:1741–1757. doi: 10.4161/rna.26921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S.C., Gilsbach R., Preissl S., Monroy Ordonez E.B., Schnick T., Beetz N., Lother A., Rommel C., Ihle H. Adrenergic repression of the epigenetic reader MeCP2 facilitates cardiac adaptation in chronic heart failure. Circ. Res. 2015;117:622–633. doi: 10.1161/CIRCRESAHA.115.306721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R.R., Lewis J.D., Bird A.P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T.R., Mattick J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Micheletti R., Plaisance I., Abraham B.J., Sarre A., Ting C.C., Alexanian M., Maric D., Maison D., Nemir M., Young R.A. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteys A.M., Spengler R.M., Wan J., Tecedor L., Lennox K.A., Xing Y., Davidson B.L. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca L., Barrett-Connor E., Wenger N.K. Sex/gender differences in cardiovascular disease prevention: what a difference a decade makes. Circulation. 2011;124:2145–2154. doi: 10.1161/CIRCULATIONAHA.110.968792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X., Campoy F.J., Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X., Hou J., Maclean A., Nasir J., Lafuente M.J., Shu X., Kriaucionis S., Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl. Acad. Sci. U S A. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X., Tate P., Li E., Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh K.M., Hwang J.Y., Follenzi A., Athanasiadou R., Miyawaki T., Greally J.M., Bennett M.V., Zukin R.S. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc. Natl. Acad. Sci. U S A. 2012;109:E962–E971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L., Wood I.C. Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- Ounzain S., Micheletti R., Arnan C., Plaisance I., Cecchi D., Schroen B., Reverter F., Alexanian M., Gonzales C., Ng S.Y. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell Cardiol. 2015;89:98–112. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Poling L.L., Wang Z.X., Liu H., Liu X.S., Roeder R.G., Zhang X.M., Song J.S., Fisher D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnu V.S., Emami M.R., Bredy T.W. Genetic and epigenetic factors underlying sex differences in the regulation of gene expression in the brain. J. Neurosci. Res. 2017;95:301–310. doi: 10.1002/jnr.23886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D., Ait-Si-Ali S., Yokochi T., Wade P.A., Jones P.L., Wolffe A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Schaefer M., Pollex T., Hanna K., Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K.M., Mayer C., Postepska A., Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock N., Harvey R.P., Mattick J.S. Long noncoding RNAs in cardiac development and pathophysiology. Circ. Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- Shafik A., Schumann U., Evers M., Sibbritt T., Preiss T. The emerging epitranscriptomics of long noncoding RNAs. Biochim. Biophys. Acta. 2016;1859:59–70. doi: 10.1016/j.bbagrm.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Simpson P.C. A New pathway for sympathetic cardioprotection in heart failure. Circ. Res. 2015;117:592–595. doi: 10.1161/CIRCRESAHA.115.307246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J., Zhu J., Van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Quiat D., Johnson B.A., Sutherland L.B., Qi X.X., Richardson J.A., Kelm R.J., Olson E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viereck J., Kumarswamy R., Foinquinos A., Xiao K., Avramopoulos P., Kunz M., Dittrich M., Maetzig T., Zimmer K., Remke J. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Jones P.L., Vermaak D., Veenstra G.J., Imhof A., Sera T., Tse C., Ge H., Shi Y.B., Hansen J.C., Wolffe A.P. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harb. Symp. Quant. Biol. 1998;63:435–445. doi: 10.1101/sqb.1998.63.435. [DOI] [PubMed] [Google Scholar]
- Wang K., Liu F., Zhou L.Y., Long B., Yuan S.M., Wang Y., Liu C.Y., Sun T., Zhang X.J., Li P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang X.J., Ji Y.X., Zhang P., Deng K.Q., Gong J., Ren S., Wang X., Chen I., Wang H. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E.O., Thienelt C.D., Katz S.E., Bartunek J., Tajima M., Rohrbach S., Douglas P.S., Lorell B.H. Gender differences in molecular remodeling in pressure overload hypertrophy. J. Am. Coll. Cardiol. 1999;34:264–273. doi: 10.1016/s0735-1097(99)00165-5. [DOI] [PubMed] [Google Scholar]
- Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y., Li A., Wang X., Bhattarai D.P., Xiao W. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.I., Hong E.P., Castle J.C., Crespo-Barreto J., Bowman A.B., Rose M.F., Kang D., Richman R., Johnson J.M., Berget S., Zoghbi H.Y. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Reiser P.J., Matlib M.A. Gender differences in myosin heavy chain-beta and phosphorylated phospholamban in diabetic rat hearts. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H2688–H2693. doi: 10.1152/ajpheart.00547.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.