Abstract
Thyroid hormone (TH) homeostasis is dependent upon coordination of multiple key events including iodide uptake, hormone synthesis, metabolism and elimination, to maintain proper TH signaling. Deiodinase enzymes catalyze iodide release from THs to interconvert THs between active and inactive forms, and are integral to hormone metabolism. The activity of deiodinases has been identified as an important endpoint to include in the context of screening chemicals for thyroid hormone disruption. To begin to address the potential for chemicals to inhibit these enzymes an adenovirus expression system was used to produce human deiodinase type 1 (DIO1) enzyme, established robust assay parameters for non-radioactive determination of iodide release by the Sandell-Kolthoff method, and employed a 96-well plate format for screening chemical libraries. An initial set of 18 chemicals was used to establish the assay, along with the known DIO1 inhibitor 6-propylthiouracil as a positive control. An additional 292 unique chemicals from the EPA’s ToxCast phase 1_v2 chemical library were screened. Chemicals were initially screened at a single high concentration of 200 μM to identify potential DIO1 inhibitors. There were 50 chemicals, or 17% of the TCp1_v2 chemicals tested, that produced >20% inhibition of DIO1 activity. Eighteen of these inhibited DIO1 activity >50% and were further tested in concentration-response mode to determine IC50s. This work presents an initial effort toward identifying chemicals with potential for affecting thyroid hormones via inhibition of deiodinases and sets the foundation for further testing of large chemical libraries against DIO1 and the other deiodinase enzymes involved in TH function.
Keywords: screening, throughput, thyroid, deiodinase, inhibition, in vitro
Introduction
The U.S. Environmental Protection Agency (U.S. EPA) is transitioning from a sole dependence upon whole animal in vivo studies for hazard assessment of chemicals, to an inclusion of in vitro based data. This pivot is underway in the U.S. EPA Endocrine Disruptor Screening Program (EDSP) which has been charged to evaluate chemicals for their potential to disrupt endocrine pathways (USEPA, 2014). Progress has been made advancing in vitro screening assays for prioritization of estrogen signaling disruptors (Schmieder et al., 2014; Browne et al., 2015); but inclusion of assays that cover relevant endpoints for potential disruption of the hypothalamic-pituitary-thyroid axis (HPT-axis) is not as mature.
It is well-established that a series of complex coordinated events within the HPT-axis and associated peripheral tissues are necessary to regulate thyroid hormone (TH) levels and ultimately TH signaling (Zoeller et al., 2007). Interference by environmental chemicals with TH action in humans and wildlife has been a topic of concern for some time (Brucker-Davis, 1998; DeVito et al., 1999). Efforts to define the important molecular initiating events around which to develop in vitro assays for chemical screening and prioritization has been of international regulatory interest and discussion (OECD, 2014; Murk et al., 2013). Yet, the in vitro screening assays for identifying potential TH disrupting chemicals are limited. Progress has recently been made on the development of higher throughput in vitro screening assays to assess chemicals for inhibition of thyroperoxidase (TPO) activity (Paul et al., 2014; Paul Freidman et al., 2016), inhibition of iodide uptake via the sodium iodide symporter (Hallinger et al., 2017; Lecat-Guillet et al., 2007), and inhibition of TH cellular uptake via monocarboxylate transporters such as MCT8 (Jayarama-Naidu et al., 2015; Dong and Wade, 2017).
Control of TH signaling in vertebrates is dependent upon maintaining sufficient circulating hormone levels in organs and tissues when and where their action is needed. Among the key events in regulating TH homeostasis, deiodinase enzyme activity has been identified as important to pursue in the context of screening for thyroid disrupting chemicals (Murk et al., 2013; OECD, 2014). The conversion of THs via deiodination, from the low affinity prohormone T4 to the more active T3, and also the conversion of T4 and T3 to inactivated forms of the hormone, are mediated by the three deiodinase enzymes: deiodinase type 1 (DIO1), type 2 (DIO2) and type 3 (DIO3). These three deiodinases have substrate specificities and differences in tissue specific expression that provides a fine level of control of TH concentrations at specific sites of action (Gereben et al., 2008). Despite the importance of deiodinases in TH metabolism, there is limited knowledge regarding the potential for chemicals to inhibit these enzymes (Schweizer and Steegborn, 2015).
The specific focus of this initial study was on screening chemicals for inhibitory activity toward DIO1. Functionally, DIO1 can remove iodine from either the inner tyrosyl or outer phenolic ring of the iodothyronines. A primary role for this enzyme in mammals is the deiodination of T4 to T3 in the liver with subsequent release of T3 to the blood to maintain its circulating levels (Larsen and Zavacki, 2012). Additionally, this enzyme may contribute to the conservation of iodide in the organism by releasing iodide from TH destined for elimination (Galton et al., 2009). The role of DIO1 in other vertebrates or its importance in regulating TH levels, especially the poikilothermic fish and amphibians, is less well understood (Darras and Van Herck, 2012).
Catalytic activity of the deiodinases can be measured by the release of free iodide from the substrate hormones. Several recent works have utilized the Sandell-Kolthoff (SK) reaction (Sandell and Kolthoff, 1937) for the non-radiometric detection of iodide in a 96-well plate format. These include assays for sodium iodide symporter (Waltz et al., 2010), DIO1 (Renko et al., 2012), DIO2 and DIO3 (Renko et al., 2015), iodotyrosine deiodinase (Renko et al., 2016), and TH transporters (Jayarama-Naidu et al., 2015). In the study reported here, robust assay parameters for non-radioactive determination of iodide release were developed and used in a 96-well plate format assay similar to that reported by Renko et al. (2015). An adenovirus recombinant expression system was used for production of the selenocysteine-containing human DIO1 enzyme against which the ToxCast Phase 1 version 2 (TCp1_v2) chemical library (Richard et al., 2016) was screened. The approach used an initial single high concentration to identify potential inhibitors and followed up with concentration-response assessment; similar to the approach used screening for TPO inhibition, which is also a cell-free loss-of-signal assay (Paul Friedman et al., 2016).
Materials and Methods.
Materials.
Chemical reagents, other than test chemicals described below, were purchased from Sigma Aldrich (St. Louis, MO, USA). Untreated polystyrene 96-well plates (360 μl/well) for DIO1 assays and SK reactions, and polypropylene 96-well plates for storage of diluted test chemical in dimethyl sulfoxide (DMSO) were obtained from Corning (Corning, NY, USA). Collection plates (1 ml) and pressure sensitive sealing tape were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Encapsulated pressure sensitive film to seal plates of test chemicals dissolved in DMSO was purchased from Phenix Research Products. (Candler, NC, USA). Polypropylene 96-well filtration plates (2 ml/well) with 20 μm polyethylene frit used to prepare Dowex columns and Pressure+96 positive pressure unit were purchased from Biotage USA (Charlotte, NC, USA). Liquidator™ 96 pipettors were obtained from Mettler-Toledo Rainin, LLC (Oakland, CA, USA).
Test chemicals.
The 292 chemicals that comprise the TCp1_v2 chemical library (Richard et al., 2016) were obtained through Dr. Ann Richard (US EPA, Research Triangle Park, NC, USA). The chemicals were supplied blinded in a 96-well plate format with one chemical per well at a target concentration of 20 mM in DMSO. Chemical plates were stored at −80 °C. The complete list of chemicals that were screened are provided in Supplemental Tables 1 and 2.
Six chemicals were included on multiple blinded test plates to evaluate assay repeatability across plates. There is not a standardized set of chemicals for DIO1 inhibition, thus these chemicals were selected from a set of replicated TCp1_v2 and TC Phase 2 chemicals that gave a range of responses in a TPO screening assay (Paul Friedman et al., 2016). Each test plate included at least three of these chemicals, with each chemical repeated two to five separate times across the plates.
Adenoviral expression of hDIO1.
The expression plasmid for human DIO1 (pD1CDM; Mandel et al., 1992), was a gift from P. Reed Larsen (Harvard Medical School, Boston MA.). Adenovirus plasmids pDeltaE1sp1A and pBHG10, and HEK293 cells were purchased from Microbix Biosystems Inc. (Toronto, Canada). The DIO1 expression cassette was liberated from parental plasmid with TaqII and Acc65I digests, end filled, gel purified and blunt end cloned into the EcoRV site of pDeltaE1sp1A. Subclones were isolated and identified by predicted restriction patterns. Adenoviruses expressing deiodinase were constructed by co-transfecting HEK293 cells with the subcloned gene and pBHG10 (Graham, 2000; Graham and Prevec, 1991; Hitt et al., 1994). FuGENE HD (Promega Corp.; Madison, WI, USA) liposome-mediated transfection was performed in accordance with manufacturer’s instructions and incubation was extended up to 6 weeks with weekly medium changes. Once viral cytopathology was noted, cultures were harvested and viral clones isolated by standard serial limiting dilution assays in HEK293 cells and concentrations determined by 50% tissue culture infective dose (TCID50). Deiodinase enzyme was produced by infecting HEK293 cells with a virion to cell ratio of five, followed by a 48 h incubation of medium containing 100 nM sodium selenate. After incubation, cells were scraped off the dishes, pelleted by centrifugation, and suspended in 100 mM phosphate buffer with 1 mM ethylenediaminetetraacetic acid (EDTA), 10mM dithiothreitol (DTT) and 0.25 M sucrose. The suspension was frozen at −80°C until use in the deiodinase inhibition assay. The specific activity was determined as described below (“Deiodinase Inhibition Assay” section) to be 9 pmole iodide/h/μg of protein. The protein concentration was determined using Bradford Reagent with bovine serum albumin as the standard (Sigma-Aldrich).
Test chemical plates.
Initial assay development test chemical set.
Prior to screening the TCp1_v2 chemicals described above, we performed preliminary experiments on a smaller set of chemicals to establish and verify the assay was performing in a manner capable of detecting enzyme inhibition in proportion to concentration of the model inhibitor. These chemicals included the model DIO1 inhibitor 6-propyl-thiouracil (PTU; CASRN 51–52-5) as the positive control. There is currently no set of established reference chemical inhibitors available for the deiodinases; therefore, a small set of chemicals available in-house was selected to initially test the assay. We selected methimazole and 2,2′,4,4′-polybrominated diphenyl ether (PBDE-47) as likely non-inhibitors. Methimazole has been shown to be inactive as a DIO1 inhibitor (Taurog et al., 1994) and the 5′-hydroxy metabolite of PBDE-47 has been shown to be inactive (Butt et al., 2011). We also selected a set of chemicals available in-house to match a subset of TCp1_v2 and TC Phase 2 chemicals that gave a range of responses in a TPO screening assay (Paul Friedman et al., 2016). Finally, a set of benzothiazoles for which we had in vitro TPO inhibition data and in vivo responses in an Xenopus laevis metamorphosis assay (Hornung et al., 2015) were also included, in part, to determine if they also might have activity as deiodinase inhibitors. These assay development test chemicals are included in Supplemental Table 1. Unlike the TCp1_v2 chemical library that were provided at 20 mM target concentrations in DMSO, this set of initial test chemicals was prepared in-house in DMSO at 100 mM, resulting in an initial single point test concentration of 1 mM. A subset of these was also run in triplicate in concentration-response mode at 1000, 200, 100, 20, 4, 0.8, and 0.16 μM.
Single-point screening for TCp1_v2 library.
For single-point screening of the TCp1_v2 chemical library, the source plates containing test chemicals at 20 mM in DMSO were thawed and mixed by pipetting action before use. The first column of wells in each chemical source plate was provided empty by the source lab (see Supplemental Figure 1 for an example plate layout). On the day of the first test run for that plate, a DMSO control and graded concentrations (100, 3.3, 1.0, 0.33, 0.10, 0.033, and 0.0033 mM) of PTU in DMSO were added to this column of wells. Additionally, DMSO and 20 mM PTU in DMSO were each plated into six randomly assigned empty wells per plate. Thus each plate contained six randomly assigned DMSO wells and one DMSO well in plate column 1 that were used to establish the uninhibited maximal deiodinase activity. The six randomly assigned wells with the high concentration PTU (20mM) were used to establish the maximal inhibition level (see example plate map Supplemental Fig 1). Transfer of 1.2 μl from each well of the chemical source plate to an assay plate resulted in final target concentration of test chemical of 200 μM, six randomly assigned wells with 200 μM PTU, and a PTU concentration-response curve of 1000, 33, 10, 3.3, 1, 0.33 and 0.033 μM final concentration. Each chemical source plate was tested on three different days to achieve n=3 data points for each chemical.
Concentration-response for TCp1_v2 with >50% inhibition in single point.
Chemicals that produced greater than 50% median inhibition in the initial single point screening were tested further in concentration-response mode. Those chemicals with greater than 50% inhibition were removed from the original source plate and added to a new polypropylene 96-well plate. Serial dilutions of chemicals in this plate were made with DMSO to prepare a new source plate of a series of seven graded concentrations such that targeted final tested concentrations in the assay were 200, 100, 20, 4.0, 0.8, 0.16, and 0.032 μM (see example plate map Supplemental Fig 2)
Deiodinase inhibition assay.
Inhibition assay and iodide extraction.
The method used for the deiodinase assay was largely based on Renko et al. (2012, 2015). These works describe the utilization of the SK reaction to measure deiodinase-liberated iodide in a 96-well-plate format. A summary diagram showing the steps involved in this assay are included in the Supplementary Materials online (Supplemental Figure 3). We modified their method by replacing the phosphate-EDTA buffer with 0.1 M 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), pH 7.0, and 1 mM EDTA (HEPES buffer) to avoid occasional precipitation problems. Other modifications are as described below. The HEK293 cell lysate with the expressed DIO1 was thawed and diluted in HEPES buffer so that the maximum desired uninhibited response was obtained when 8 μg protein (59.4 μl) were added per well of the 96-well assay plate. This response was equivalent to a net change of approximately 0.6 absorbance units at 420 nm between 1 and 10 minutes, and was similar to a response generated by 60 pmoles potassium iodide in the SK reaction. The presence of 60 pmoles of iodide represents the consumption of 19% of the rT3 substrate. To begin the assay, enzyme was added to a 96-well plate using an 8-channel pipet while keeping the suspension well-mixed. A Liquidator™ 96–20 pipettor was then used to add 1.2 μl of test chemical in DMSO from the chemical source plate to the enzyme in the 96-well assay plate. The reaction was initiated with 59.4 μl of 20.2 μM 3,3’,5’-triiodo-L-thyronine (reverse T3; rT3) and 80.8 mM DTT in HEPES buffer using a Liquidator™ 96–200 pipettor. The final assay conditions were 0.1 M HEPES, 1 mM EDTA, 10 μM rT3, and 40 mM DTT, 1% DMSO, and a final assay volume of 120 μl. The assay plate was sealed with an adhesive sheet, mixed in a plate shaker, and incubated at 37 °C for 3 hours. At the end of the incubation, 75 μl were removed with a multi-channel pipet and added to a previously prepared Dowex filtration plate. These plates were made using 2-ml filtration plates (Biotage). Each well was filled with Dowex 50 WX2, 100 to 200 mesh, to a packed bed height of 22 mm. The Dowex was equilibrated and regenerated before and after each use as described in the original method (Renko et al., 2015), except that the volumes of the acetic acid solutions were changed from 180 to 400 μl. Preparation and processing of the Dowex plates was performed in a Pressure+96 unit (Biotage). After the sample from the enzyme reaction was added, an additional 100 μl of 10 % acetic acid was applied and the free iodide was eluted into a 96-well collection plate.
SK reaction to detect iodide.
The SK reaction is based on the reduction of the yellow-colored cerium IV to the non-colored cerium III by arsenic, with the rate of this reaction increased in the presence of iodide in a concentration-dependent manner (Sandell and Kolthoff, 1937). The SK reaction was conducted by transferring 75 μl of eluent from the wells of the collection plate to another 96-well plate. Then 75 μl of arsenic reagent [25 mM NaAsO2, 0.8 M NaCl, 0.5 M H2SO4] was added and mixed well using the Liquidator™ 96–200 pipet. Finally, 75 μl of cerium reagent [20 mM (NH4)4Ce(SO4)4.2H20, 0.44 M H2SO4] was added with the liquidator pipet and immediately placed in a Synergy 4 plate reader (BioTek Instruments, Inc., Winooski, VT, USA). The plate reader was used to initially mix the plate for 3 seconds (fast mode) and then read absorbance at 420 nm every minute. The plate reader was used in sweep mode to measure the entire plate as fast as possible. Typically, the greatest range between uninhibited and inhibited reactions was obtained near the 10 min time point (see Supplemental Figure 4). Data was processed by determining the change in absorbance between the 1 and 10 min reading for each well. The initial zero time reading was not used due to higher variability. The net change in absorbance for each well was determined by subtracting the mean background change in absorbance defined by the completely inhibited reaction in the six wells in that plate containing 200 μM PTU. Finally, all data were normalized by converting to % control of the mean of seven uninhibited reactions representing the maximum DIO1 activity (net change in absorbance of the DMSO control wells). The median of the three replicates was calculated and test chemical results were reported as percent inhibition, which was calculated as 100% minus percent of control uninhibited reaction.
Data processing and analysis:
Data from the single-point screening were processed in an automated pipeline for normalization (percent of control, as described above), plate diagnostics, and assay-specific flags using the R data analysis software (version 3.3.1; R Core Team 2016). The response of the randomly assigned DMSO and PTU high concentration wells were used to calculate a Z’ factor for each plate. The Z’ factor is an indicator of the quality of the assay, based upon a measure of the dynamic range of the assay and the variability around the maximal and minimal response levels of the assay (Zhang et al., 1999). A Z’ factor value of >0.5 indicates good separation between the positive and negative controls, and was considered an acceptable plate run. A plate-wise DMSO median absolute deviation (DMSO-MAD) was calculated to define the variability around the uninhibited enzyme activity in the DMSO wells in each plate. A plate-wise positive median absolute deviation (PTU-MAD) was calculated to describe the variability around the fully inhibited assay response produced by the wells with 200 μM PTU. Data points or series of points that fell outside of acceptable parameters for the single concentration screen were automatically flagged for manual review. These included Z’ factor less than 0.5, wells that had no change in absorbance (which indicates that well missed receiving one or more reagent), high variability or outliers, potential assay interference indicated by absorbance outside of normal ranges, and noted observations or procedural errors. Individual assay runs that did not meet quality criteria (e.g. low Z’ factor, poor control data) were re-run. Wells with observed problems in the assay (e.g. observed low volume, or no change in absorbance) were excluded from analyses. Samples with high variability across replicates or other anomalous absorbance data were further evaluated in concentration-response mode. For chemicals replicated across multiple plates, median percent inhibition (from the single concentration assay) was compared across plates and then the median of the multiple samples was used in analyses. For those chemicals tested in concentration-response mode, data were analyzed with the ToxCast Analysis Pipeline (tcpl) package version 1.0 using R version 3.3.1 (Filer, 2015), which fits curves based on three models: constant, constrained Hill model, and constrained gain-loss model. The best model for each chemical was selected based on the model with the lowest Akaike Information Criterion (AIC) value. For chemicals that fit the robust Hill model, absolute IC20, absolute IC50, and Hill slope were calculated from the model fit parameters provided by the tcpl package. Concentration-response curves are displayed in the negative direction (inhibition from maximum response).
Results.
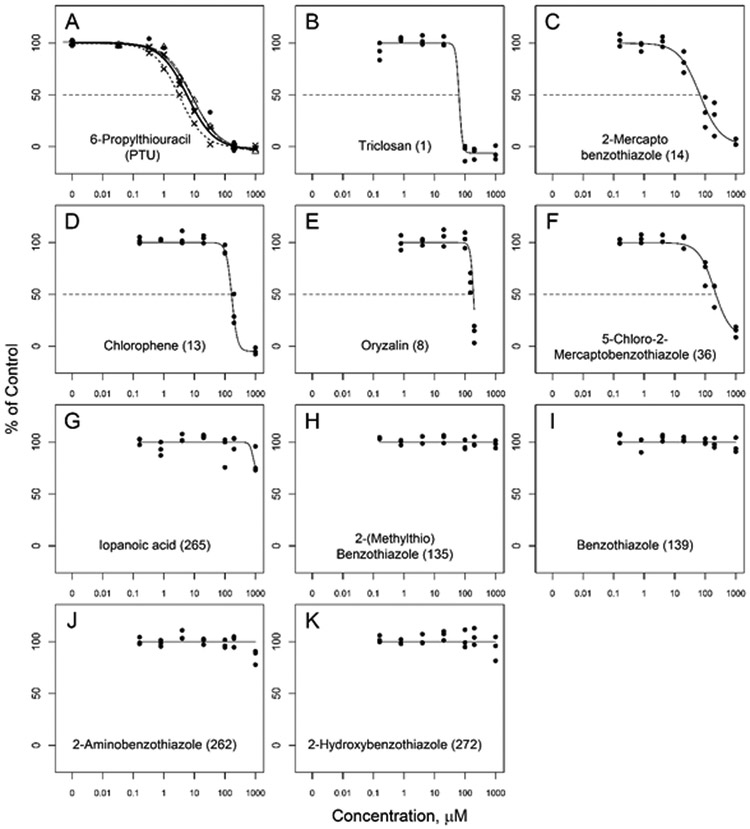
In the initial performance assessment of the DIO1 inhibition assay a set of 18 chemicals were tested in triplicate at a single high concentration of 1 mM. Four of the 18 fully inhibited DIO1 activity compared with PTU and one inhibited DIO1 by 85%. These chemicals are shown in Table 1 in descending rank-order from this initial screen. A subset of these chemicals were tested further in concentration-response mode. The absolute IC20 and IC50 from the concentration-response curves for the active chemicals of this group are included in Table 1. Iopanoic acid, a known DIO1 inhibitor, did not appear to be active and was further evaluated (see below in “Chemicals with potential false responses or interference” section).
Table 1.
Initial screen of assay development chemicals to establish assay performance for inhibition of DIO1 activity.
| Median % Inhibitiona |
Concentration-Responseb |
|||
|---|---|---|---|---|
| Chemical Name | 1 mM | IC20 (μM) |
IC50 (μM) |
Hill slope |
| 2-Mercaptobenzothiazole | 114.6 | 22.0 | 66.8 | −1.3 |
| Chlorophene | 112.6 | 124.3 | 165.3 | −4.7 |
| Triclosan | 112.2 | 53.2 | 63.0 | −8.0 |
| Oryzalin | 109.5c | 159.8 | 187.8 | −8.0 |
| 5-Chloro-2-mercaptobenzothiazole | 85.4 | 93.4 | 224.2 | −1.7 |
| 2-Aminobenzothiazole | 37.5 | NAd | NA | NA |
| tert-Butylhydroquinone | 34.5 | --e | -- | -- |
| Bisphenol A | 32.1 | -- | -- | -- |
| Benzotriazole | 28.0 | -- | -- | -- |
| 2-Hydroxybenzothiazole | 25.6 | NA | NA | NA |
| Dibutylphthalate | 23.9 | -- | -- | -- |
| Benzothiazole | 22.2 | NA | NA | NA |
| Methimazole | 16.8 | -- | -- | -- |
| 4-Pentylaniline | 14.8 | -- | -- | -- |
| 2-(Methylthio)benzothiazole | 4.9 | NA | NA | NA |
| 2,2',4,4'-TBDPE | −3.2 | -- | -- | -- |
| 4-Bromobiphenyl | −7.6 | -- | -- | -- |
| Iopanoic acid | −20.9 | 922.9 | NA | −7.8 |
Value shown is the median percent inhibition of n=3 replicates at the 1 mM.
Absolute IC20, absolute IC50, and Hill slope for chemicals with Hill model as best fit in ToxCast Pipeline (v1.0)
The 1 mM concentration was associated with observed and measured precipitate formation.
NA = not applicable. The chemical did not produce an inhibition curve or the curve did not reach 20% or 50% inhibition, and therefore an IC20 or IC50 could not be calculated.
--, chemicals not tested in concentration-response
The concentration-response curves for these chemicals, along with PTU that was run as a positive control, are shown in Figure 1. These chemicals produced a range of responses. The concentration-response inhibition curves for PTU produced consistent responses (Figure 1), with IC50 values from near 4 to 6.5 μM, and Hill slope values near −1 (Table 2 and Supplemental Figure 5). Two chemicals, 2-mercaptobenzothiazole and 5-chloro-2-mercaptobenzothiazole (Fig 1, C and F) produced complete or nearly complete inhibition curves with Hill slopes more similar to PTU. Three chemicals in this set that produced apparent full inhibition, triclosan, oryzalin, and chlorophene (Fig 1, B, D, E), also produced relatively steep inhibition curves with Hill slopes of −8.0, −8.0, and −4.7 respectively. The steepness of the curves suggests mechanism different from PTU, and could be related to undefined inhibition. Oryzalin was tested at 1 mM but precipitates were visually evident in these wells, so this data point was not included on the graph. The other five chemicals tested in concentration-response did not produce significant inhibition.
Figure 1.
Concentration-response curves for subset of the assay evaluation test chemicals. Symbols represent three independent observations at each concentration: in panel (A) each replicate plate is represented with a different symbol and curve fit for 6-propylthiouracil (PTU) in addition to the combined fit (solid line). Numbers in parentheses after the chemical name indicates their rank order efficacy within the full set of all chemicals tested at 200 μM (See Supplemental Table 1).
Table 2.
Summary of the 6-propylthioruracil (PTU) concentration response replicates and assay performance across the test chemical plates tested in single-point and concentration-response.
| PTU Curve |
Z’ factor |
DMSO-MADb | PTU-MADc | ||||
|---|---|---|---|---|---|---|---|
| Platea | IC50 (μM) | SE | Hill slope | Mean | SD | (median; n=3) | (median; n=3) |
| TP | 5.92 | 0.69 | −0.99 | 0.7862 | 0.0589 | 2.909 | 5.553 |
| SP1 | 4.70 | 0.36 | −1.13 | 0.7544 | 0.0682 | 5.148 | 3.775 |
| SP2 | 5.35 | 0.43 | −0.95 | 0.7735 | 0.0957 | 3.467 | 5.201 |
| SP3 | 4.63 | 0.49 | −0.94 | 0.6825 | 0.0883 | 12.81 | 2.601 |
| SP4 | 4.47 | 0.45 | −0.89 | 0.7324 | 0.0121 | 5.971 | 1.883 |
| CR1 | 3.98 | 0.39 | −0.98 | 0.7056 | 0.0547 | 3.299 | 1.787 |
| CR2 | 6.43 | 1.04 | −1.16 | 0.5985 | 0.1467 | 6.843 | 5.520 |
TP was the plate used to test initial chemicals in concentration-response when establishing the assay. Plates SP1 – SP4 were plates of TCp1_v2 chemicals tested in single-point screen. Plates CR1 and CR2 were plates with TCp1_v2 chemicals tested in concentration-response mode.
DMSO-MAD is the platewise median absolution deviation calculated from all the DMSO wells in each of the three replicated assay plates.
PTU-MAD is the platewise positive control median absolute deviation calculated from all the 200 μM PTU wells in each of the three replicated assay plates.
Assay Performance.
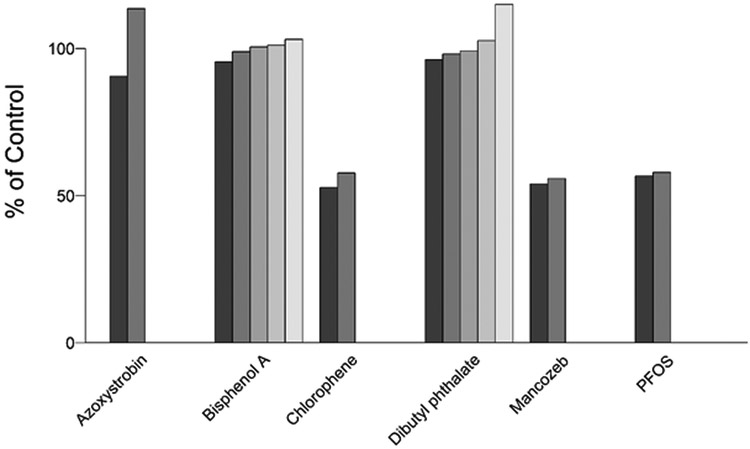
Performance of the assay across plates was monitored in each run. The assay performed reproducibly as observed by the consistent response of the PTU inhibition-response curve in each plate. The IC50 for PTU ranged from a low of 3.98 to a high of 6.43 μM, with Hill slopes consistently near −1 (Table 2). The Z’ factor was consistently above 0.5 and was generally near 0.7. The platewise median absolute deviations around the DMSO controls (DMSO-MAD) and the 200 μM PTU wells (PTU-MAD) were generally near 5 or lower with the exception of one of the single concentration plates for which the DMSO-MAD was 12.8 indicating greater variability in the DMSO control wells in this plate. The six chemicals used to evaluate intra-assay reproducibility had very consistent results across multiple plates, with most differences in median percent inhibition less than five percent (Figure 2).
Figure 2.
A subset of six test chemicals was randomly replicated across the source plates (n=2 or n=5 wells) to evaluate the plate-to-plate variability. These included azoxystrobin, bisphenol A, chlorophene, dibutylphthalate, mancozeb, and perfluorooctane sulfonic acid (PFOS).
TCp1_v2 Single Concentration Initial Screen.
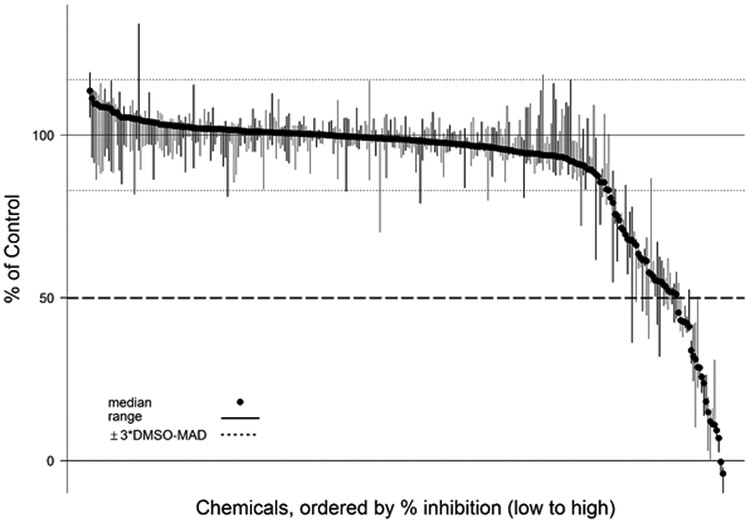
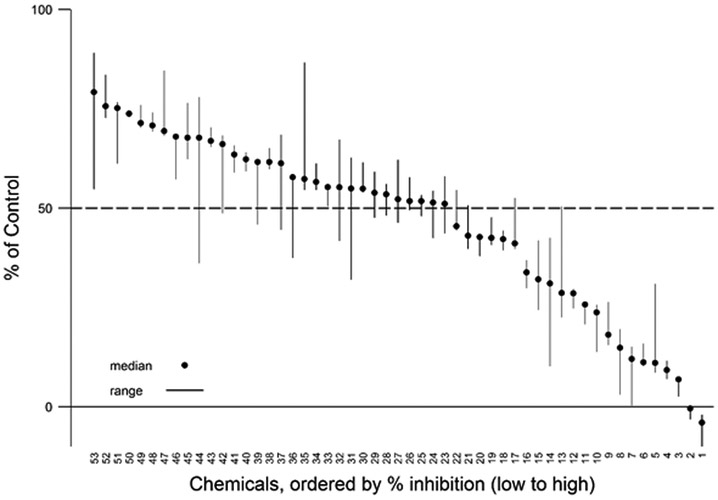
A total of 292 unique TCp1_v2 chemicals were tested in the screening assay. The majority of these chemicals produced less than 20% inhibition compared with activity of the DMSO controls (Figure 3). The median, maximum, and minimum response values for all chemicals tested in the single concentration screen are provided in Supplemental Table 1 available online. There were 50 TCp1_v2 chemicals that inhibited DIO1 activity by >20% of the background activity of the DMSO controls (Figure 4). This represents 17% of the chemical library tested. Twenty of these active chemicals produced greater than 50% inhibition of DIO1. Eighteen of these 20 are listed in Table 3 and were further tested in concentration response (Figure 5). The other two chemicals, 3-iodo-2-propynyl-N-butylcarbamate and methylene bis(thiocyanate), were determined to have potential false responses or interference in the assay, as described in more detail below (“Chemicals with potential false responses or interference” section).
Figure 3.
Inhibition activity of all chemicals tested in single concentration. Plot of the % inhibition of DIO1 (median and range; n=3) for chemicals from in-house and TCp1_v2 library tested at 200 μM. The dotted lines above and below the 100% control line show the bounds of 3 × DMSO MAD (median absolute deviation) around the normalized activity of the DMSO controls. The median, maximum and minimum % inhibition values for the three replicate runs with each chemical can be found in Supplemental Table 1.
Figure 4.
Expanded plot of chemicals with >20% inhibition. Plot of the % inhibition of DIO1 (median and range; n=3) for those 53 chemicals that produced >20% inhibition when initially screened at a single concentration (200 μM), includes 48 TCp1_v2 chemicals and 5 internally sourced chemicals. Chemicals are plotted by rank order with those producing the greatest inhibition to the right. The numbers on the x-axis correspond to the specific chemicals. Those with greater than 50% inhibition are shown in Table 3. Those with less than 50% inhibition can be found in Supplemental Table 1 along with all other chemicals.
Table 3.
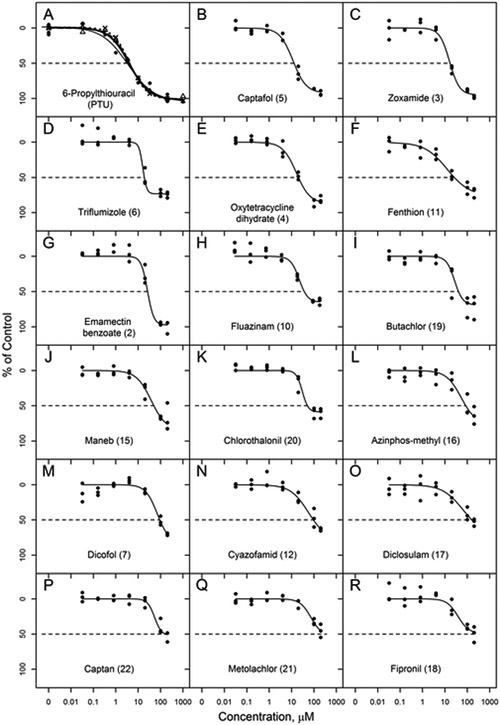
ToxCast p1_v2 chemicals that produced greater than 50% median inhibition (n=3) in the single point screen at 200 μM and were further tested in concentration-response.
| Single Point Screen |
Concentration Response |
||||
|---|---|---|---|---|---|
| Ranka | Chemical | Median % Inhibition | IC20 (μM) | IC50 (μM) | Hill slope |
| 2 | Emamectin benzoate | 100 | 16.68 | 26.41 | −3.06 |
| 3 | Zoxamide | 93 | 8.18 | 17.01 | −1.95 |
| 4 | Oxytetracycline | 91 | 7.00 | 20.24 | −1.42 |
| 5 | Captafol | 89 | 4.78 | 12.90 | −1.47 |
| 6 | Triflumizole | 89 | 13.28 | 18.81 | −5.01 |
| 7 | Dicofol | 88 | 35.75 | 91.95 | −1.67 |
| 9 | Oryzalinb | 82 | 117.64 | 145.11 | −7.97 |
| 10 | Fluazinam | 76 | 15.26 | 37.03 | −2.27 |
| 11 | Fenthion | 74 | 3.79 | 26.38 | −0.86 |
| 12 | Cyazofamid | 71 | 21.09 | 94.51 | −1.09 |
| 15 | Maneb | 68 | 15.52 | 50.10 | −1.31 |
| 16 | Azinphos-methyl | 66 | 24.09 | 85.76 | −1.31 |
| 17 | Diclosulam | 59 | 23.84 | 161.27 | −0.95 |
| 18 | Fipronil | 58 | 30.78 | NA | −1.78 |
| 19 | Butachlor | 58 | 18.63 | 40.57 | −2.39 |
| 20 | Chlorothalonil | 57 | 24.52 | 51.35 | −3.23 |
| 21 | Metolachlor | 57 | 55.73 | 370.84 | −1.79 |
| 22 | Captan | 55 | 45.26 | 166.10 | −2.77 |
Number corresponds to median inhibition efficacy rank in single point screen at 200 μM (with the exception of fluazinam, which was plated at 180 μM), shown in Figure 4 and Supplemental Table 1.
Concentration-response for TCp1_v2 oryzalin matched internally sourced oryzalin (IC50 = 187.8 μM and Hillslope = −7.97, see Figure 1E).
Figure 5.
Concentration-response curves for DIO1 inhibition by those TCp1_v2 chemicals that produced >50% median inhibition of DIO1 in the initial single point screen at 200 μM. 6-Propylthiouracil (PTU) in panel A is replicated PTU curves from plate CR1 (all PTU curves are shown in Supplemental Figure 5). (Note: Oryzalin is not shown as the results from the plated sample from the TCp1_v2 library closely match those from the internally sourced assay development chemical set shown in Figure 1. Also see footnote to Table 3.)
TCp1_v2 Concentration-Response Screen.
For all 18 of the chemicals tested in concentration-response mode, the robust Hill model was the best fit model. Hill slope, absolute IC20 and absolute IC50 for these chemicals are included in Table 3. The concentration-response curves are shown in Figure 5, in order of potency based on IC50s. None of the chemicals that inhibited DIO1 activity displayed greater potency than the positive control PTU. Captafol and zoxamide were closest in potency; however, PTU which generally had IC50 values ranging from 4 to 6 μM, was approximately 3-times more potent than these two chemicals. Other than oryzalin which produced a very steep slope for the inhibition curve as found when tested as part of the development chemical set (Table 1, Figure 1), the remaining 17 chemicals produced inhibition curves with Hill slopes from −0.86 to −5.0.
Chemicals with potential false responses or interference
Compounds that contain iodine can interfere with the assay. As reported by Renko et al. (2012), iopanoic acid is a known deiodinase inhibitor which was shown to act as a substrate for DIO1. We tested this compound in the absence of rT3 and confirmed that it was being deiodinated. Since the iodide released from iodinated test compounds is indistinguishable from that released from the rT3 substrate, it would be expected that other iodine containing compounds could give false negative responses if they are substrates for the enzyme. One iodinated compound, iodosulfuron-methyl-sodium, was tested further to confirm the results from the initial screen. The median DIO1 activity was 93% of control (min 89%; max 108%; n=3) in the 200 μM single concentration test so it appeared to not be a DIO1 inhibitor. To verify that this apparent lack of inhibition was not due to deiodination of this chemical by DIO1, as occurs with iopanoic acid, we further evaluated it in additional assays. This chemical did not act as a substrate for DIO1 when tested in the absence of rT3 since a similar SK response was seen whether or not enzyme was present. Another possible anomaly with iodine-containing compounds is that the presence of free iodide associated with the stock chemical could interfere with accurate interpretation of the SK reaction. This may explain the result observed with 3-iodo-2-propynyl-N-butylcarbamate. Testing this compound in single concentration mode produced an anomalous SK result whereby the change in A420 had gone to near completion by 1 min reading, though normally it is still changing well past 10 minutes. A further test was conducted on a dilution of this compound which showed the same elevated amount of iodide was detected in the presence or absence of enzyme when rT3 was also absent, indicating the compound was not being deiodinated enzymatically. The response in the SK reaction was likely due to the presence of iodide in the chemical stock solution that gave the false indication of deiodinase activity.
The non-iodinated compound, methylene bis(thiocyanate) also appeared to interfere with the assay. It appeared to produce greater than 100% inhibition in the initial single concentration screen. This is likely an interference with the SK reaction. Thiocyanate is known to interfere with the SK reaction (Sandell and Kolthoff, 1937) and it seems likely this thiocyanate-containing compound acts similarly. Methyl isothiocyanate is also in this chemical library, but it did not produce any indication of enzyme inhibition or assay interference when tested in the single point screen (median DIO1 activity was 100.9% of control). It may be that unlike the thiocyanates, the isothiocyanates do not interfere with the SK reaction. As stated earlier, there are several classes of compounds that are known to interfere with the SK reaction. These types of chemicals will need to be monitored for false responses.
Discussion.
The role of the deiodinases in regulation of TH signaling via their catalytic conversion of THs between inactive and active forms, makes it critical to understand the extent to which these enzymes may be potential targets for chemical inhibition. For the types of chemicals for which the US EPA is tasked with evaluating for endocrine disrupting activity, there is a lack of information in the published literature regarding chemicals with potential for inhibiting deiodinases. The relationship between deiodinase inhibition and alteration of TH levels and subsequent adverse effects is not well-established. Other than PTU which is the model inhibitor of DIO1 activity, and some structurally similar analogs to PTU including aromatic thiols (Harbottle and Richardson, 1984) and selenouracil compounds (Visser et al., 1992), there are limited reports of other chemical structure groups that have been tested for their potential to inhibit deiodinase activity. Inactivation by iodinated dyes has been demonstrated by the competitive inhibition of deiodinase by rose bengal (Mol et al., 1984) and inhibition of deiodinase activity in the rat by the tetra-iodinated color additive FD&C No. 3 (Capen and Martin, 1989). Other chemical groups shown to inhibit deiodinase activity include phenolics such as the plant-derived and synthetic flavonoids (Auf’mkolk et al., 1986; Ferreira et al., 2002; Spanka et al., 1990; Renko et al., 2015), halogenated phenolics such as the phenolphthalein dyes (Fekkes et al., 1982), and halogenated aromatic hydrocarbons and related chemicals such as the brominated and chlorinated diphenyl ethers and bisphenols (Butt et al., 2011).
The majority of chemicals tested in the present study include active pesticide ingredients that comprise much of the TCp1 chemical library, a few chemicals established as inactive against deiodinase, and a limited number of selected compounds suspected of thyroid disruption, covering a range of chemical structures. Within the TCp1 library, we identified 10 chemicals with median inhibition greater than 70% in single concentration screening and 10 chemicals with potency for inhibition of DIO1 activity within 10-fold of that of PTU (IC50 between 12.9 and 51.4 μM in concentration-response assay). Very little information on thyroid related endpoints for these chemicals could be found in the literature. Of those tested in our study and reported in literature to affect thyroid-axis endpoints, only butachlor and triclosan produced greater than 20% inhibition of DIO1 activity in this assay. The data for butachlor in the adult Chinese rare minnow (Gobiocyprus rarus) (Zhu et al., 2014) are inconclusive with regard to mechanism of interference with the thyroid axis. The TH serum levels were quite variable, and no determination of the effect of butachlor on the activity of DIO1 or DIO3 was measured. Triclosan has been shown to be a weak inhibitor of deiodinase activity using human liver microsomes, with IC50s in the 300–400 μM range (Butt et al., 2011). Another proposed mechanism for TH disruption by triclosan in the rat is via upregulation of liver phase 1 and phase 2 metabolism activity leading to increased elimination of TH (Paul et al., 2012). Additionally, methimazole which has been confirmed previously to not inhibit DIO1 (Tuarog et al., 1994) and PBDE47, the parent of the inactive 5’-OH-PBDE47 metabolite (Butt et al., 2011) were also verified as inactive in this assay. Iopanoic acid is a known competitive inhibitor of the deiodinase enzymes (Braga and Cooper, 2001), but in this assay as run in screening mode it did not identify this as an enzyme inhibitor. This is due to iopanoic acid being a substrate of the DIO1 enzyme with release of iodide from the iopanoic acid (Renko et al., 2012), essentially mimicking the iodide released from the endogenous assay substrate rT3.
A series of benzothiazoles were included in the test set that we previously tested in a TPO inhibition assay (Hornung et al., 2015). These benzothiazoles produced similar rank order potency as DIO1 inhibitors as we found for TPO inhibition, with 2-mercaptobenzothiazole the most potent, followed by the 5-chloro-2-mercaptobenzothiazole, and the remaining four which had low to no activity. Like PTU, the model chemical for DIO1 inhibition and which is also a potent TPO inhibitor, the DIO1 inhibition activity observed for two of these TPO-inhibiting benzothiazoles indicates there may be groups of structurally similar chemicals with the potential to interfere with TH signaling via multiple mechanisms.
The deiodinase assay in this study proved to be an efficient and reliable method to screen and prioritize chemicals for potential thyroid disruption. The adenoviral expression system provided a successful method for producing large quantities of active deiodinase enzyme. The assay modifications and optimization resulted in strong assay performance, with low variability in controls and reproducible results across replicates and plates. The consistent assay performance metrics (DMSO MAD, IC50 of PTU curve, % inhibition of replicated chemicals) and high Z’ factor (0.60–0.79) provide confirmation that this inhibition assay can be used for screening of large chemical libraries in a moderate-to-high throughput format. The results of the screening assay with only 17% of test chemicals displaying any inhibitory activity is an indication that there is specificity in this assay. Methimazole and 2,2’,4,4’-tetrabromodiphenylether that were selected as inactive chemicals to run in this assay based on literature information, did not produce any significant indication of inhibition of DIO1, further demonstrating assay specificity. Unlike for endpoints such as estrogen receptor activation for which there is abundant information regarding inactive, mildly active and strongly active chemicals from which to select reference chemicals, there is little information to use to select “reference” chemicals for positive and negative controls for deiodinases.
During assay development, some procedural details became apparent that are important for achieving robust results. The use of Liquidator pipets was found to be valuable in starting the SK reaction in all 96 wells simultaneously. Without this there was some column bias in the SK rate corresponding to the order of reagent addition. Likewise, it was determined that using the plate reader in sweep mode resulted in much faster measurements than in the standard mode. In standard mode a bias between wells was observed depending on the order that measurements were made. Achieving simultaneous assay start and rapid well absorbance reading was crucial for obtaining quality data because the initial A420 readings change very fast. Also, the PTU positive controls, and DMSO controls, were randomly distributed throughout the plate. Thus the initial A420 reading can vary significantly in replicates that are in different parts of the plate if the assay is not initiated simultaneously across the plate. If these methods are not employed there is larger variation in the control reactions, which decreases the useful range between uninhibited and inhibited reactions, and thereby the ability to detect inhibition by test chemicals.
Utilizing the SK reaction to screen large numbers of chemicals for inhibitory activity across the deiodinases has its advantages and disadvantages. An advantage of the SK reaction is that it is potentially available for use by more laboratories, where the use of radioisotopes is limited due to licensing limitations or expense. Disadvantages include possible interference with certain chemicals. Osmium, ruthenium, manganese (with bromides), nitrite, thiocyanate, iron, fluoride, silver, mercury, and cyanide have been shown to affect the reaction (Sandell and Kolthoff, 1937). Iodinated chemicals may produce apparent increased activity or mask enzyme inhibition. This may be due to contamination by free iodide, test chemical degradation that releases free iodide, deiodination of test chemical itself, or other confounding issues; they cannot be summarily ruled out as untestable and should be tested with the appropriate follow-up experiments when needed. Nonetheless, numerous methods utilizing the SK reaction have been developed to measure iodine in a variety of matrices, including blood, urine and food, as reviewed for example, by Shelor and Dasgupta (2011). Additionally, the method used here includes an ion-exchange purification step prior to the SK reaction, which is likely to remove many interfering compounds, and the test chemicals are present at much lower stoichiometric amounts than the reagents in the SK reaction. Furthermore, use of the SK reaction to measure deiodinase activity has previously been successfully demonstrated (Renko et al., 2012; 2015).
In summary, the adaptation of a 96-well plate assay for screening large chemical libraries for inhibition of DIO1 activity has been successfully demonstrated. The adenoviral expression system provided a successful method for producing large quantities of active selenocysteine-containing deiodinase enzyme. This provides a basis for further screening chemicals for DIO1 inhibitory activity and for adapting this assay to screen the other deiodinase enzymes, DIO2 and DIO3. Although the TCp1 chemical library is comprised primarily of pesticidal active ingredients, the full chemical set tested here expands the number and breadth of known chemicals screened for DIO1 inhibition. Importantly, the majority (83%) of the tested chemicals showed no indication of DIO1 enzyme inhibition when tested to 200 μM. These results set the groundwork for screening additional ToxCast chemical libraries and others to include a more diverse representation of the EDSP chemical universe. For those active chemicals, confirmation that chemical inhibition of DIO1 activity in vitro can indicate potential for adverse effects in vivo needs to be evaluated to provide greater confidence in the value of this assay as a screening tool and establish its predictive utility.
Supplementary Material
Acknowledgements:
The authors thank Dr. P. Reed Larson for sharing the DIO1 plasmids and gift of rat DIO1 enzyme for assay development, and Dr. Kostja Renko for valuable advice and gift of DIO1 enzyme while we optimized the methods in our lab. We also thank Dr. Eric Watt (Oak Ridge Institute for Science and Education, US EPA, NCCT) for R analysis guidance, and Drs. Ann Richard and Chris Grulke (US EPA, NCCT) for assistance in obtaining the ToxCast chemical library for screening. The authors thank Dr. Susan Laws and Richard Kolanczyk for providing comments on an early draft of the manuscript.
Funding Information: This work was supported by the US Environmental Protection Agency.
Footnotes
Disclaimer: The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Conflict of Interest: The authors claim no conflicts of interest.
Supplementary Data: Two supplementary files are available online. The Supplemental Tables file includes a list of all chemicals tested in the DIO1 inhibition assay, and their relative activity rank based upon inhibition activity at single target concentration of 200 μM. The Supplemental Figures file contains example plate layouts for single concentration and concentration-response plates, a simplified diagram of assay steps, example absorbance data and processing, and figures of the PTU concentration response curves run for each assay plate
References.
- Auf’mkolk M, Koehrle J, Hesch R-D, and Cody V (1986). Inhibition of rat liver iodothyronine deiodinase: Interaction of aurones with the iodothyronine ligand-binding site. J. Biol. Chem. 261, 11623–11630. [PubMed] [Google Scholar]
- Braga M and Cooper DS (2001). Clinical Review 129. Oral cholecystographic agents and the thyroid. J. Clin. Endocrinol. Metab. 86, 1853–1860. [DOI] [PubMed] [Google Scholar]
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, and Thomas RS (2015). Screening chemicals for estrogen receptor bioactivity using a computational model. Environ. Sci. Technol. 49, 8804–8814. [DOI] [PubMed] [Google Scholar]
- Brucker-Davis F (1998). Effects of environmental synthetic chemicals on thyroid function. Thyroid 8, 827–856. [DOI] [PubMed] [Google Scholar]
- Butt CM, Wang DL, and Stapleton HM (2011). Halogenated phenolic contaminants inhibit the in vitro activity of the thyroid-regulating deiodinases in human liver. Toxicol. Sci. 124, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capen CC and Martin SL (1989). The effects of xenobiotics on the structure and function of thyroid follicular and C-cells. Toxicol. Pathol. 17, 266–293. [DOI] [PubMed] [Google Scholar]
- Darras VM and Van Herck SLJ (2012). Iodothyronine deiodinase structure and function: from ascidians to humans. J. Endocrinol. 215, 189–206. [DOI] [PubMed] [Google Scholar]
- DeVito M, Biegel L, Brouwer A, Brown S, Brucker-Davis F, Cheek AO, Christensen R, Colborn T, Cooke P, Crissman J et al. (1999). Screening Methods for Thyroid Hormone Disruptors. Environ. Health Perspect. 107, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H and Wade MG (2017). Application of a nonradioactive assay for high throughput screening for inhibition of thyroid hormone uptake via the transmembrane transporter MCT8. Toxicol. In Vitro 40, 234–242. [DOI] [PubMed] [Google Scholar]
- Fekkes D, Hennemann G, and Visser TJ (1982). Inhibition of iodothyronine deiodinase by phenolphthalein dyes. FEBS Lett. 137, 40–44. [DOI] [PubMed] [Google Scholar]
- Ferreira ACF, Lisboa PC, Oliveira KJ, Lima LP, Barros IA, and Carvalho DP (2002). Inhibition of thyroid type 1 deiodinase activity by flavonoids. Food Chem. Toxicol. 40, 913–917. [DOI] [PubMed] [Google Scholar]
- Filer DL (2015). tcpl: ToxCast data analysis pipeline. R package version 1.0. URL https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data. Accessed 12/8/2017.
- Galton VA, Schneider MJ, Clark AS, and St. Germain DL (2009). Life without thyroxine to 3,5,3′-triiodothyronine conversion: Studies in mice devoid of the 5′-deiodinases. Endocrinology 150, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, Simonides WS, Zeold A, and Bianco AC (2008). Cellular and Molecular Basis of Deiodinase-Regulated Thyroid Hormone Signaling. Endocr. Rev. 29, 898–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL (2000). Adenovirus vectors for high-efficiency gene transfer into mammalian cells. Immunol. Today 21, 426–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL and Prevec L (1991). Manipulation of adenovirus vectors In Methods in Molecular Biology (Murray EJ, Ed.) pp. 109–128. The Humana Press, Inc: Clifton, NJ. [DOI] [PubMed] [Google Scholar]
- Hallinger DR, Murr AS, Buckalew AR, Simmons SO, Stoker TE, and Laws SE (2017). Development of a screening approach to detect thyroid disrupting chemicals that inhibit the human sodium iodide symporter (NIS). Toxicol. In Vitro 40, 66–78. [DOI] [PubMed] [Google Scholar]
- Harbottle R and Richardson SJ (1984). Structural requirements of thiol compounds in the inhibition of human liver iodothyronine 5′-deiodinase. Biochem. J. 217, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt MM, Ng P, and Graham FL (1994). Construction and propagation of Human Adenovirus vectors, in Cell Biology: A Laboratory Handbook, Celis JE, Editor. Academic Press: San Diego, CA, USA. [Google Scholar]
- Hornung MW, Kosian PK, Haselman JT, Korte JJ, Challis K, Macherla C, Nevalainen E, and Degitz SJ (2015). In vitro, ex vivo, and in vivo thyroid hormone modulating activity of benzothiazoles. Toxicol. Sci. 146, 254–264. [DOI] [PubMed] [Google Scholar]
- Jayarama-Naidu R, Johannes J, Meyer F, Wirth EK, Schomburg L, Köhrle J, and Renko K (2015). A nonradioactive uptake assay for rapid analysis of thyroid hormone transporter function. Endocrinology 156, 2739–2745. [DOI] [PubMed] [Google Scholar]
- Larsen PR and Zavacki AM (2012). Role of the iodothyronine deiodinases in the physiology and pathophysiology of thyroid hormone action. Euro Thyroid J, 1, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecat-Guillet N, Merer G, Lopez R, Pourcher T, Rousseau B and Ambroise Y (2007). A 96-well automated radioiodide uptake assay for sodium/iodide symporter inhibitors. Assay Drug Devel. Tech. 5, 535–540. [DOI] [PubMed] [Google Scholar]
- Mandel SJ, Berry MJ, Keiffer JD, Harney JW, Warne RL, and Larsen PR (1992). Cloning and in vitro expression of the human selenoprotein, type I iodothyronine deiodinase. J. Clin. Endocrinol. Metab. 75, 1133–1139. [DOI] [PubMed] [Google Scholar]
- Mol JA, Docter R, Hennemann G, and Visser TJ (1984). Modification of rat liver iodothyronine 5′-deiodinase activity with diethylpyrocarbonate and rose bengal; evidence of an active site histidine residue. Biochem. Biophys. Res. Comm. 120, 28–36. [DOI] [PubMed] [Google Scholar]
- Murk AJ, Rijntjes E, Blaauboer BJ, Clewell R, Crofton KM, Dingemans MM, Furlow JD, Kavlock R, Köhrle J, Opitz R, et al. (2013). Mechanism-based testing strategy using in vitro approaches for identification of thyroid hormone disrupting chemicals. Toxicol. In Vitro 27, 1320–1346. [DOI] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). (2014). New Scoping Document on In Vitro and Ex Vivo Assays for the Identification of Modulators of Thyroid Hormone Signalling In OECD Environment, Health and Safety Publications, Series on Testing and Assessment, No. 207. Paris, France. ENV/JM/MONO(2014)23 [Google Scholar]
- Paul KB, Hedge JM, Bansal R, Zoeller RT, Peter R, DeVito MJ, and Crofton KM (2012). Developmental triclosan exposure decreases maternal, fetal, and early neonatal thyroxine: A dynamic and kinetic evaluation of a putative mode-of-action. Toxicology 300, 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul KB, Hedge JM, Rotroff DM, Hornung MW, Crofton KM, and Simmons SO (2014). Development of a thyroperoxidase inhibition assay for high-throughput screening. Chem. Res. Toxicol. 27, 387–399. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K, Watt ED, Hornung MW, Hedge JM, Judson RS, Crofton KM, Houck KA, and Simmons SO (2016). Tiered high-throughput screening approach to identify thyroperoxidase inhibitors within the ToxCast phase I and II chemical libraries. Toxicol. Sci. 151, 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: URL https://www.R-project.org/. Accessed 12/8/2017. [Google Scholar]
- Renko K, Schäche S, Hoefig CS, Welsink T, Schwiebert C, Braun D, Becker NP, Köhrle J, and Schomburg L (2015). An improved nonradioactive screening method identifies genistein and xanthohumol as potent inhibitors of iodothyronine deiodinases. Thyroid 25, 1–7 [DOI] [PubMed] [Google Scholar]
- Renko K, Hoefig CS, Hiller F, Schomburg L, and Köhrle J (2012). Identification of iopanoic acid as substrate of type 1 deiodinase by a novel nonradioactive iodide-release assay. Endocrinology 153, 2506–2513. [DOI] [PubMed] [Google Scholar]
- Renko K, Hoefig CS, Dupuy C, Harder L, Schwiebert C, Köhrle J, and Schomburg L (2016). A nonradioactive DEHAL assay for testing substrates, inhibitors, and monitoring endogenous activity, Endocrinology 157, 4516–4525. [DOI] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin M, Wambaugh JF, et al. (2016). ToxCast chemical landscape: Paving the road to 21st century toxicology. Chem. Res. Toxicol. 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Sandell EB and Kolthoff IM (1937). Micro determination of iodine by a catalytic method. Michrochim. Acta, 1, 9–25. [Google Scholar]
- Schmieder PK, Kolanczyk RC, Hornung MW, Tapper MA, Denny JS, Sheedy BR, and Aladjov H (2014). A rule-based expert system for chemical prioritization. SAR QSAR Environ. Res. 25, 253–287. [DOI] [PubMed] [Google Scholar]
- Schweizer U and Steegborn C (2015). New insights into the structure and mechanism of iodothyronine deiodinases. J. Mol. Endocrin. 55, R37–R52. [DOI] [PubMed] [Google Scholar]
- Shelor CP and Dasgupta PK (2011). Review of analytical methods for the quantification of iodine in complex matrices. Anal. Chim. Acta, 702, 16–36. [DOI] [PubMed] [Google Scholar]
- Spanka M, Hesch RD, Irmscher K, and Köhrle J (1990). 5′-Deiodination in rat hepatocytes: Effects of specific flavonoid inhibitors. Endocrinology 126, 1660–1667. [DOI] [PubMed] [Google Scholar]
- Tuarog A, Dorris ML, Guziec LJ, and Guziec FS Jr. (1994). The selenium analog of methimazole. Measurement of its inhibitory effect on type 1 5′-deiodinase and of its antithyroid activity. Biochem. Pharmacol. 48, 1447–1453. [DOI] [PubMed] [Google Scholar]
- US Environmental Protection Agency (US EPA). (2014). U.S. Environmental Protection Agency Endocrine Disruptor Screening Program Comprehensive Management Plan. https://www.epa.gov/sites/production/files/2015-08/documents/edsp_comprehesive_management_plan_021414_f.pdf. Accessed 12/8/2017.
- Waltz F, Pillette L, and Ambroise Y (2016). A nonradioactive iodide uptake assay for sodium iodide symporter function. Anal. Biochem. 396, 91–95. [DOI] [PubMed] [Google Scholar]
- Visser TJ, Kaptein E, and Aboul-Enein Y (1992). Selenouracil derivatives are potent inhibitors of the selenoenzyme type I iodothyronine deiodinase. Biochem. Biophys. Res. Comm. 189, 1362–1367. [DOI] [PubMed] [Google Scholar]
- Zhang J-H, Chung TDY, and Oldenburg KR (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomolec. Screening. 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Zhu L, Li W, Zha J, Wang M, Yuan L and Wang Z (2014). Butachlor causes disruption of HPG and HPT axes in adult female rare minnow (Gobiocypris rarus). Chem. Biol. Interact. 221, 119–126. [DOI] [PubMed] [Google Scholar]
- Zoeller RT, Tan SW, and Tyl RW (2007). General background on the hypothalamic-pituitary-thyroid (HPT) axis. Crit. Rev. Toxicol. 37:11–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.