Abstract
Background
The objective of this study was to determine the main risk factors of Pseudomonas aeruginosa mutation as well as the mechanisms of acquired resistance.
Methods
We conducted a 2-year prospective study in patients who were carriers of a Pseudomonas aeruginosa strain and who had been admitted to a medical/surgical ICU.
Results
Of the 153 patients who were included, 34 had a mutation in their strain. In a multivariate analysis, a duration of ventilation > 24 days was a risk factor for mutation (risk ratio 4.29; CI 95% 1.94–9.49) while initial resistance was a protective factor (RR 0.36; CI 95% 0.18–0.71). In a univariate analysis, exposure of P. aeruginosa to ceftazidime was associated with an over-production of AmpC cephalosporinase and exposure to meropenem was associated with impermeability. A segmentation method based on the duration of ventilation (> 24 days), initial resistance, and exposure of strains to ceftazidime made it possible to predict at 83% the occurrence of mutation.
Conclusion
The duration of ventilation and the presence of resistance as soon as P. aeruginosa is identified are predictive factors of mutation in ICU patients.
Keywords: Pseudomonas aeruginosa, Mutation, Antibiotic, Resistance mechanism, ICU (intensive care unit), Segmentation tree
Background
Pseudomonas aeruginosa is the primary microorganism responsible for nosocomial infections in the ICU [1, 2]. The prognosis for these infections is very poor with a mean mortality of 30% [3–5] and can be as high as 50 to 60% [6]. The prevalence of P. aeruginosa infections and the rates of antipyocyanic antibiotic resistance are on a rise internationally [7, 8].
P. aeruginosa is characterized by a high level of natural resistance to antibiotics and by its capacity to acquire new resistance mechanisms by chromosomal mutations or horizontal transmission of genetic materials [9], with the resulting risk of an unadapted antibiotic therapy [5, 10]. Mutation can also occur during treatment and generate therapeutic failure. According to studies, 6 to 53% of the P. aeruginosa infections that are treated will become resistant to one or more antipyocyanic antibiotics [11, 12]. This acquisition of resistance is associated with an increase in mortality, the duration of stay, and cost [1, 13, 14].
Exposure to antibiotics is one of the primary risk factors for the acquisition of resistance that is studied, notably to the fluoroquinolones and carbapenems. Policies that restrict the use of certain antibiotics including the fluoroquinolones have shown a decrease in the rate of resistance to these antibiotics as well as to the other classes [15]. Therefore, it would appear that it is important to address the risk factors for mutation and the resistance mechanisms induced by the various antipyocyanic antibiotics, in order to limit the use of those that contribute the most to resistance, in keeping with the experts’ recommendations [16].
Therefore, we conducted a study for which the main objective was to identify risk factors for the mutation of Pseudomonas aeruginosa strains in patients hospitalized on our unit.
The secondary objectives were to identify the resistance mechanisms induced by these antibiotics and to identify any correlation between the main antipyocyanic antibiotics prescribed and the different resistance mechanisms acquired by Pseudomonas aeruginosa.
Methods
We conducted a 2-year open prospective observational study in the Medical/Surgical ICU of Rangueil tertiary hospital, Toulouse.
This project was approved by the Toulouse Hospital Ethics and Research Committee (n° 85- 1114). The data were collected anonymously from computerized records. The patients’ informed consent was not required.
Study population
The patients who were included had at least one positive Pseudomonas aeruginosa bacterial culture during their hospitalization in the department either for colonization or infection. Colonization was defined by the presence of a positive culture of the specimen and the absence of general signs of infection. An infection was defined by a positive culture, quantitatively significant, and local or general signs of infection meeting the criteria in effect [17].
The exclusion criteria were being under 18 years of age and hospitalization in the department for less than 5 days. The patients who were included were monitored until their discharge from the department.
Clinical data collection
For each patient, the following data were collected: age, gender, SAPS II, the reason for hospitalization, mortality in the department, prior hospitalization within the 3 previous months, Pseudomonas aeruginosa colonization or infection, and the antibiotics received within the previous month.
Bacteriological eradication, defined as the absence of the microorganism from bacterial cultures, and clinical symptoms corresponding to the resolution of signs of sepsis related to P. aeruginosa infection were also noted.
Antibiotics examined
The following antibiotics were examined:
Amoxicillin, amoxicillin/clavulanic acid, piperacillin/tazobactam, ceftriaxone, ceftazidime, cefepime, imipenem, meropenem
Tobramycin, amikacin
Ciprofloxacin
For each prescribed antibiotic, the duration of treatment, the time of initiation in relation to the presence of P. aeruginosa, and whether or not there was an association were noted. It was also specified whether the treatment was probabilistic or documented and whether it was adapted.
Bacterial cultures
Samples were taken by tracheal aspiration (TA) on admission to the department for patients on invasive mechanical ventilation and then repeated twice per week. Other samples were from bronchoalveolar lavages (BAL), hemocultures, or even peroperative samples, according to the context. Pseudomonas aeruginosa was identified by mass spectrometry (BRUCKER).
Resistance studies
The minimum inhibitory concentrations (MICs) of the antibiotics that were examined were defined for each P. aeruginosa strain that was isolated, after antimicrobial susceptibility testing in a liquid medium by automated method on a Biomerieux Vitek 2 appliance. MICs were interpreted according to the CA-SFM/EUCAST recommendations that were applicable at the time of the study. The β-lactamine-resistance mechanisms and their changes in the same patient were inferred based on antimicrobial susceptibility testing when this was possible.
For P. aeruginosa strains resistant to ceftazidime, several complementary tests were carried out to differentiate resistance due to AmpC cephalosporinase hyper-production from that due to acquisitions by carbapenemases, extended spectrum oxacillinases (ES-OXA), or extended spectrum beta-lactamases (ESBL). These tests were:
✓ A comparison of the inhibition zone diameter on Mueller-Hinton agar and Mueller-Hinton agar with 2000 mg/l of added cloxacillin.
- ✓ Synergy tests on agar with cloxacillin in case of AmpC hyper-production:
- Between ticarcillin/clavulanic acid and ceftazidime or cefepime and between imipenem and cefepime and/or ceftazidime: if positive in favor of ESBL or ES-OXA
✓ Identification of a metallo-beta-lactamase (MBL): by imipenem alone/imipenem + EDTA E-test.
Suspicious strains of ESBL, oxacillinase, or any other carbepenemase were sent to the National Research Center (CNR) to identify these mechanisms. Only the first mutation was taken into account in our work.
The different resistance mechanisms
These are the acquired resistance mechanisms that were studied:
Penicillinase acquisition (difficult to differentiate from certain resistance mechanisms by over-expression of efflux with current laboratory techniques)
Hyper-production of inducible cephalosporinases, AmpC
Impermeability through the loss of the porin OprD
Over-expression of the efflux system
The mechanisms of resistance to fluoroquinolones and aminoglycosides were not examined because they were not detectable on antimicrobial susceptibility testing alone.
Statistical study
After the first stage of descriptive statistics and verification of the distribution of values (Kolmogorov-Smirnov test), the study population was separated into 2 groups according to the occurrence or non-occurrence of mutation.
The characteristics of the patients in the different groups were compared using non-parametric tests (Mann-Whitney test) for the continuous variables due to the lack of homogeneity of the total numbers in the groups. The results are expressed in median and confidence interval at 95% (CI 95). The categorical variables for the 2 groups were compared using Fischer’s exact test. The discriminant value of the covariates of interest, according to the occurrence of mutation, was evaluated by examination of the ROC (receiver operating characteristic) curves and their associated areas under curve (AUC).
In a second step, a multivariate analysis was used to evaluate the association between the different covariates (p < 0.2) and the variable explained (mutation of P. aeruginosa strains) by the risk ratio measurement. On a statistical level, this information is censored. Therefore, we used a survival model, the COX model, described before [5]. After excluding the covariates with co-linearity, we used a stepwise regression (backward elimination) procedure by including all the selected variables then progressively eliminating those that were non-significant. Several models were tested by selecting the one for which the AUC was highest and excluding the models with an AUC < 0.8.
In the final step, a division of the population was illustrated with the use of a segmentation tree. The aim of this technique was to describe the methods of population distribution in homogeneous groups according to the existence of mutation and covariates previously selected for multidimensional analysis. We thereby tested several growth methods including the so-called CHAID (CHAID: CHi-squared Automatic Interaction Detection) method and the CRT (classification and regression tree) method, using the one for which the predicted percentages were the highest.
The analyses were done on the MedCalc® statistical software, version 15 (Mariakerke, Belgium). The segmentation tree method was carried out on the software IBM® SPSS Statistics Version 23 (Chicago, IL). p < 0.05 was considered to be statistically significant.
Results
Description of the study population
One hundred fifty-three patients were included, for whom the characteristics are indicated in Table 1. They had a median age of 64 years (95% CI 62–66). Median SAPS II was 58 (95% CI 55–63), and ICU mortality was 34%. Median length of stay was 21 days (95% CI 18–25), and the duration of mechanical ventilation was 16 days (95% CI 14–20.6).
Table 1.
Clinical characteristics of the population and comparison of the patients with P. aeruginosa (P. a) without mutation vs with mutation
| Overall population | No mutation n= 119 |
Mutation n=34 |
p | ||||
|---|---|---|---|---|---|---|---|
| Median | 95% CI | Median | 95% CI | Median | 95% CI | ||
| Age (years) | 64 | 62 - 66 | 65 | 63 - 67 | 61 | 56 - 65 | 0.189 |
| SAPS II | 58 | 55 - 63 | 58 | 56 - 63 | 51 | 41 - 67 | 0.356 |
| Length of stay (days) | 21 | 18 - 25 | 18 | 16 - 23 | 34 | 26 - 40 | 0.0002* |
| Duration of ventilation (days) | 16 | 14 - 20.6 | 14.5 | 13 - 17 | 31.5 | 25 - 35 | 0.0001* |
| Prior hospitalization period | 6 | 4 - 8 | 5 | 3 - 8 | 9 | 5 - 18 | 0.094 |
| Colonization period | 9 | 7 - 11 | 9 | 7 - 11 | 9.5 | 3 - 15 | 0.981 |
| Infection period | 10.5 | 7 - 13 | 10.5 | 6 - 13 | 10.5 | 2 - 15.8 | 0.625 |
| Duration 1st treatment P.a 1(n: 68/30) | 4 | 4 - 5 | 4.0 | 3 – 5.6 | 4.5 | 3 – 6 | 0.656 |
| Duration 2nd treatment P.a 2(n: 41/26) | 6 | 5 - 9 | 5.0 | 3 – 7 | 8.5 | 5.6 - 12 | 0.041* |
| Duration 3rd treatment P.a (n: 13/15) | 8 | 2 – 10.6 | 6.0 | 1.5 – 11 | 9.0 | 2.5 - 13 | 0.579 |
| Number of ATB before P. aeruginosa in ICU | 2 | 1 - 2 | 2 | 1 - 2 | 2 | 1 - 2 | 0.598 |
| Gender M/F | 94 (79%)/25 (21%) | 22 (64.7%)/12 (35.3%) | 0.112 | ||||
| Clinical recovery no/yes | 50(32.7%)/103(67.3%) | 34 (28.6%) / 85 (71.4%) | 16 (47.1%)/18 (52.9%) | 0.061 | |||
| Bacteriological recovery no/yes | 121(79.1%)/32(20.9%) | 89 (74.8%) / 30 (25.2%) | 32 (94.1%) / 2 (5.9%) | 0.016* | |||
| Death no/yes | 101 (66%)/52 (34%) | 79 (66.4%) / 40 (33.6%) | 22 (64.7%) / 12 (35.3%) | 0.840 | |||
| Reason for hospitalization | |||||||
| Multiple trauma | 7 (4.6%) | 7 (5.9%) | 0 (0 %) | 0.351 | |||
| Medical | 75 (49%) | 58 (48.7%) | 17 (50 %) | ||||
| Surgical | 71 (46.4%) | 54 (45.4%) | 17 (50%) | ||||
| Sampling type | |||||||
| Tracheal aspiration | 77 (64.7%) | 21 (61.8%) | 0,698 | ||||
| Bronchoalveolar lavage | 18 (15.1%) | 6 (17.6%) | |||||
| Blood cultures | 17 (14.3%) | 5 (14.7%) | |||||
| Intraperitoneal sample | 3 (2.5%) | 2 (5.9%) | |||||
| Prior hospitalization no/yes | 41 (26.8%)/112(73.2%) | 33 (27.7%) / 86 (72.3%) | 8 (23.5%) / 26 (76.5%) | 0.826 | |||
* signify that it is considered to be statistically significant with p < 0.05
73.2% of the patients had been hospitalized before admission to the ICU, and 18.3% were P. aeruginosa carriers on admission. The “mutation” group included 34 patients and the “no mutation” group 119 patients.
Predictive factors of mutation among the clinical characteristics of the patients
The duration of stay was significantly longer, in the univariate analysis, in the “mutation” group than in the “no mutation” group (median of 34 days versus 18 days, p = 0.0002), as well as the duration of mechanical ventilation (31.5 days versus 14.5 days, p = 0.0001). On the three lines of antibiotic treatment, the duration of the second antipyocyanic treatment was significantly longer in the “mutation” group (8.5 days versus 5 days, p = 0.041).
Predictive factors of mutation among bacteriological data
Bacteriological eradication was significantly less frequent in the “mutation” group (5.9% versus 25.2%, p = 0.016). There was no difference in terms of clinical recovery or mortality between the 2 groups (Table 1).
In the univariate analysis, P. aeruginosa was significantly more often responsible for an infection than simple colonization in the “mutation” group (88.2% versus 61.3% of infection, p = 0.003). Initial resistance was more frequently identified in the “no mutation” group (80.7% versus 55.9%, p = 0.006). The type of initial resistance to the β-lactamines was distributed differently in the two groups (Table 2). Initial resistance to the aminoglycosides was only identified in the “no mutation” group (p = 0.001).
Table 2.
Comparison of patients with P. aeruginosa without mutation vs with mutation Bacteriological data
| No mutation n= 119 |
Mutation n=34 |
p | |
|---|---|---|---|
| Prior ICU colonization no/yes | 98 (82.4%) / 21 (17.6%) | 27 (79.4%) / 7 (20.6%) | 0.801 |
| Infection with P. aeruginosa no/yes | 46 (38.7%) / 73 (61.3%) | 4 (11.8%) / 30 (88.2%) | 0.003* |
| Other infections no/yes | 36 (30.3%) / 83 (69.7%) | 5 (14.7%) / 29 (85.3%) | 0.081 |
| Initial resistance no/yes | 23 (19.3%) / 96 (80.7%) | 15 (44.1%) / 19 (55.9%) | 0.006* |
| Type of initial resistance to β-lactamines | |||
| 1= impermeability | 1 (0.8%) | 2 (5.9%) | 0.005* |
| 3= Penicillinase and/or efflux | 45 (37.8%) | 11 (32.4%) | |
| 4= AmpC over-production | 10 (8.4%) | 0 (0%) | |
| 5= impermeability +/- Over-production of efflux | 9 (7.6%) | 4 (11.8%) | |
| 6= 4+5 | 25 (21%) | 1 (2.9%) | |
| Fluoroquinolone resistance no/yes | 74 (62.2%) / 45 (37.8%) | 25 (73.5%) / 9 (26.5%) | 0.309 |
| Aminoglycoside resistance no/yes | 94 (79%) / 25 (21%) | 34 (100%) / 0 (0%) | 0.001* |
| Antibiotic therapy after isolation of P. aeruginosa | |||
| P. aeruginosa ATB 1 adapted (n=97) no/yes | 10 (14.9%) / 57 (85.1%) | 2 (6.7%) / 28 (93.3%) | 0.332 |
| P. aeruginosa ATB 2 adapted (n=66) no/yes | 2 (5%) / 38 (95%) | 0 (0%) / 26 (100%) | 0.515 |
| P. aeruginosa ATB 3 adapted (n=28) no/yes | 0 (0%) / 13 (100%) | 2 (13.3%) / 13 (86.7%) | 0.484 |
| Tazocillin after P. aeruginosa no/yes | 94 (79%) / 25 (21%) | 23 (67.6%) / 11 (32.4%) | 0.176 |
| Ceftazidime after P. aeruginosa no/yes | 95 (79.8%) / 24 (20.2%) | 19 (55.9%) / 15 (44.1%) | 0.007* |
| Cefepime after P. aeruginosa no/yes | 101 (84.9%) / 18 (15.1%) | 30 (88.2%) /4 (11.8%) | 0.785 |
| Meropenem after P. aeruginosa no/yes | 79 (66.4%) / 40 (33.6%) | 15 (44.1%) / 19 (55.9%) | 0.027* |
| Fluoroquinolone after P. aeruginosa no/yes | 115 (96.6%) / 4 (3.4%) | 34 (100%) / 0 (0%) | 0.576 |
| Aminoglycosides after P. aeruginosa no/yes | 57 (47.9%) / 62 (52.1%) | 8 (23.5%) / 26 (76.5%) | 0.017* |
* signify that it is considered to be statistically significant with p < 0.05
Predictive factors of mutation among antibiotic therapies
Of the antibiotic therapies prescribed after isolation of P. aeruginosa, ceftazidime, meropenem, and the aminoglycosides were prescribed significantly more often in the “mutation” group with respectively 44.1% versus 20.2% (p = 0.007), 55.9% versus 33.6% (p = 0.027), and 76.5% versus 52.1% (p = 0.017). Receiving antibiotic therapy adapted to the P. aeruginosa strain was not significantly associated with the absence of mutation.
Predictive value of mutation among the variables of interest
The highest sensitivity and specificity as well as the negative predictive value (NPV) and positive predictive value (PPV) among the variables were those concerning the duration of ventilation. By including the influence of the duration of ventilation and of stay on the occurrence of mutation in the multivariate analysis (logistic regression), a duration of ventilation > 24 days was significant (p = 0.0084) while a duration of stay > 27 days was eliminated (p = 0.513). The initial multivariate analysis using the Cox model taking into account the thresholds of all the covariates (while eliminating those that are related) is presented in Table 3.
Table 3.
Multivariate analysis using the Cox model
| Significant covariates | Risk ratio | CI 95% | p |
|---|---|---|---|
| Duration of ventilation > 24 days | 4.29 | 1.94- 9.49 | 0.0003* |
| Initial resistance | 0.36 | 0.18- 0.71 | 0.0031* |
| Non-model variables | |||
| Ceftazidime after P. aeruginosa | 1.66 | 0.82- 3.37 | 0.160 |
| Meropenem after P. aeruginosa | 1.17 | 0.57- 2.40 | 0.674 |
* signify that it is considered to be statistically significant with p < 0.05
The AUC of the model is equal to 0.83 (CI 95% 0.758–0.884), sensitivity at 71%, and specificity at 82% (PPV at 53% and NPV at 91%). Therefore, this multidimensional analysis shows that the covariates associated with the risk of mutation are:
A duration of ventilation > 24 h with a risk ratio of 4.29
The existence of initial resistance with a risk ratio of 0.36.
However, for the prescription of ceftazidime after the identification of P. aeruginosa, the antibiotic therapy emerges from the model with a value of p < 0.2.
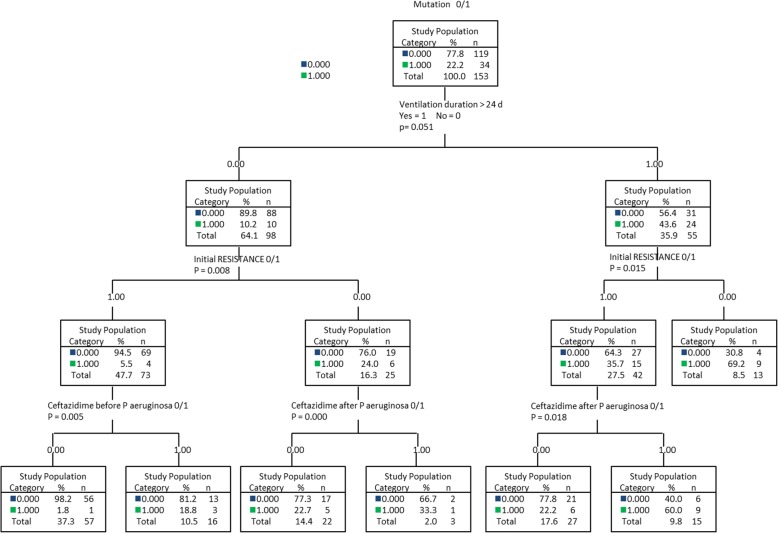
The description of the methods of population distribution in homogeneous groups according to the existence of mutation and previously selected covariates (p < 0.2) is illustrated in Fig. 1.
Fig. 1.
Distribution of the population according to the existence of mutation and the duration of ventilation (> or < 24 days), initial resistance, and the prescription of ceftazidime after identification of P. aeruginosa
The segmentation method (CRT) illustrates the dividing of individuals based on these 3 factors with a predictive value of 83%.
Examination of the various resistance mechanisms according to the antibiotic therapies
We analyzed the relationship between the 2 antipyocyanic β-lactamines prescribed which appeared to be related to the occurrence of mutation and the different mechanisms of acquired resistance (Table 4).
Table 4.
The relation between the 2 anti P. aeruginosa β-lactamines capable of generating a mutation (ceftazidime and meropenem) and the different acquired resistance mechanisms
| Acquired resistance mechanisms | Ceftazidime after P. aeruginosa | Meropenem after P. aeruginosa | ||||
|---|---|---|---|---|---|---|
| no | yes | p | no | yes | p | |
|
Impermeability no/yes |
108 (94.7%) / 6 (5.3%) | 38 (97.4%) / 1 (2.6%) | 0.679 | 93 (98.9%) / 1 (1.1%) | 53 (89.8%) / 6 (10.2%) | 0.0135* |
|
Over-expression of efflux no/yes |
108 (94.7%) / 6 (5.3%) | 37 (94.9%) / 2 (5.1%) | 0.999 | 92 (97.9%) / 2 (2.1%) | 53 (89.8%) / 6 (10.2%) | 0.055 |
|
Penicillinase and/or efflux no/yes |
111 (97.4%) / 3 (2.6%) | 38 (97.4%) / 1 (2.6%) | 0.999 | 92 (97.9%) / 2 (2.1%) | 57 (96.6%) / 2 (3.4%) | 0.640 |
|
AmpC over-expression no/yes |
105 (92.1%) / 9 (7.9%) | 30 (76.9%) / 9 (23.1%) | 0.019* | 84 (89.4%) / 10 (10.6%) | 51 (86.4%) / 8 (13.6%) | 0.613 |
|
Impermeability +/- Over-production of efflux no/yes |
113 (99.1%) / 1 (0.9%) | 37 (94.9%) / 2 (5.1%) | 0.160 | 93 (98.9%) / 1 (1.1%) | 57 (96.6%) / 2 (3.4%) | 0.559 |
* signify that it is considered to be statistically significant with p < 0.05
The use of ceftazidime was significantly associated with the occurrence of an over-expression of inducible (AmpC) cephalosporinase; 23.1% of the patients who received ceftazidime developed an over-expression of cephalosporinase, compared to 7.9% of the patients who did not receive ceftazidime (p = 0.019). The use of meropenem was significantly associated with the occurrence of impermeability (p = 0.0135).
Discussion
Our prospective study concretely demonstrates a mutation in the initial strain in 34 patients out of 153 Pseudomonas aeruginosa carriers during hospitalization in intensive care. The main risk factor for mutation identified is the duration of mechanical ventilation. The existence of initial resistance might protect against the risk of mutation.
Severity, mortality, the duration of ventilation and of stay, and the rate of mutation of strains in our department were similar to the previous studies on the subject [1, 18]. The rate of prior colonization and initial resistance to antibiotics are higher due to the method of recruiting patients who were often transferred to other units. In most cases, the literature is focused on identifying risk factors for the acquisition of resistant strains to a given antibiotic, especially according to prior exposure to antibiotics. We differentiated between the use of antibiotics before and during the presence of P. aeruginosa in order to determine the impact of exposure to the antibiotic on the microorganism and the mechanism of resistance induced.
Severity of the patients and so the durations of hospitalization and mechanical ventilation are risk factors often described as being associated with the acquisition of P. aeruginosa as well as resistant strains [8, 19–21]. All patients studied were severe with no significant difference between the two groups. In our study, only the duration of mechanical ventilation is significant in the multivariate analysis and seems to be the most predictive factor of the acquisition of new resistance mechanisms. The duration of any prior hospitalization and prior colonization were not identified as risk factors for mutation in our work. In most of the studies, there was a greater association between these factors and the risk of Pseudomonas aeruginosa infection or resistance of the strain as soon as it appears, than with the emergence of resistance [18, 22, 23].
The fact that as of its identification P. aeruginosa is already a carrier of resistance was noted as a protective factor against mutation. To our knowledge, this has never been described before. We also noted that the resistance mechanism that was most often acquired was an overproduction of AmpC cephalosporinase, followed by impermeability and over-expression of efflux, which corresponds with the data in the literature [3, 24]. Therefore, it would appear that a strain that is already resistant has a lower risk of developing new resistance mechanisms than a wild strain. The occurrence of a mutation was most often identified in P. aeruginosa-infected rather than P. aeruginosa-colonized patients, with less frequent bacteriological eradication. Naturally, these patients more frequently received an antipyocyanic antibiotic therapy and were therefore subject to a higher selection pressure which could explain the more frequent occurrence of mutation in these patients [25].
Most of the studies found a correlation between mortality and an infection by a resistant or even multi-resistant strain, in comparison with a sensitive strain [10, 14, 26]. However, they did not take into account the capacity of resistant strains to evolve, which our study did. Moreover, Park et al. [27] found that mortality was mainly related to the adequacy of initial treatment rather than the level of resistance of the strains. The increase in the mortality rate of patients who were carriers of resistant strains would in fact appear to be related to an adapted treatment given later than if they were carriers of a sensitive strain [5, 13]. In our study, the levels of adapted initial antibiotic therapy were relatively high. We identified a maximum of 15% of inappropriate antipyocyanic treatments in the “no mutation” group (for which the initial resistance levels were higher), compared to 25 to 35% in the literature [28].
The extent of prior antibiotic therapies is widely described in the literature [22, 23, 29]. A univariate analysis showed that of the antibiotics administrated to treat P. aeruginosa infections, ceftazidime, meropenem, and the aminoglycosides were significantly associated with the occurrence of mutation. This association was not identified in the multivariate analysis, possibly due to a lack of power of our study considering the low number of mutations. A recent study showed that exposure to meropenem, ceftazidime, or ciprofloxacin during the presence of P. aeruginosa was responsible for the emergence of resistance [11]. In compliance with the recommendations, in our study, only 4 patients received ciprofloxacin [16, 30]. In a previous study, imipenem, piperacillin-tazobactam, and cefotaxime were identified as risk factors in the emergence of resistance, but not ceftazidime [3]. However, other studies showed that ceftazidime was a risk factor for the appearance of resistance [31]. In general, the carbapenems, including imipenem and more recently meropenem, are among the antibiotics that are most often associated with the emergence of resistant strains, along with the fluoroquinolones [3, 20, 25, 30].
A factor, which might be astounding, is the frequency at which aminoglycosides are prescribed for infection. Although the multivariate analysis showed no significance in our work, some studies associated them with a risk factor for their own resistance [19, 32] or to other antibiotics [23, 33]. Piperacillin-tazobactam was not identified as a risk factor for mutation in our cohort. Its responsibility for mutation varies widely from one study to another [32, 34].
The risk of AmpC cephalosporinase induction by amoxicillin/clavulanic acid or ceftriaxone was not identified in this study [3, 35].
For acquired resistance mechanisms, our study identified a relationship between the use of ceftazidime and an over-production of inducible AmpC cephalosporinase, and between the use of meropenem and the acquisition of impermeability by loss of the porin OprD. It is responsible for the resistance attributed to imipenem, and according to the literature, it is found in 86 to 100% of the carbapenem-resistant strains [36, 37]. This acquisition, associated with exposure to the carbapenems, concerns imipenem and meropenem [19]. Several studies also describe an association between the use of ceftazidime and the over-production of inducible AmpC cephalosporinase [38]. This mechanism might explain the phenomena of co-resistance found between ceftazidime and cefepime [39], and ceftazidime and piperacillin-tazobactam [40] or of cross-resistance with other cephalosporins [41]. Exposure of P. aeruginosa to meropenem also tends to be associated with the acquisition of an over-expression of the efflux system.
By sending strains to the CNR (French national reference center) that were suspected of having emerging resistance mechanisms, we objectively identified a single carbapenemase carrier strain and no ESBL strain. These data are consistent with recent studies which estimate the prevalence in France of ESBL strains at 0.55% and carbapenemase carrier strains at 0.86% [24, 42].
The original nature of this work lies in the creation of a segmentation tree which enables the distribution of the study population according to the existence of mutation and covariates of interest, including a ventilation period of more than 24 days, initial resistance of the strain, and the prescription of ceftazidime after the appearance of P. aeruginosa with a predictive value of 83%. This tool seems useful to predict the risk of the occurrence of an event, in this case mutation, according to the variables of interest.
However, our study has several limits. When there is resistance to antipyocyanic penicillins alone, we could not differentiate the presence of penicillinase from an efflux system. It was also difficult at times to differentiate the presence or absence of over-expression of an efflux system associated with impermeability, when there was a resistance to carbapenems, if the strain also over-produced inducible AmpC cephalosporinase. The combination of an over-expression of AmpC cephalosporinase and impermeability might be enough to acknowledge that there is an associated resistance to meropenem [36–38, 43]. If such was the case, they were classified in a separate category called “impermeability±over-expression of efflux”. Therefore, the importance of an over-expression of the efflux system might be underestimated in this study. Secondly, the clonality of the different strains found in the same patient over time could not be examined, which did not make it possible to eliminate the acquisition of a new strain, if any, that might explain the change in the resistance profile. However, the literature is rather in favor of a change in the resistance of P. aeruginosa by mutation of the initial strain than by the acquisition of a different strain [3, 24]. When considering the segmentation tree, it does not predict a priori, from the first day of mechanical ventilation, the occurrence of a mutation. Indeed, the length of mechanical ventilation is a recognized risk factor for multidrug-resistant organisms or Pseudomonas aeruginosa [44].
Conclusion
Our study confirms that the duration of mechanical ventilation of Pseudomonas aeruginosa carrier patients is a risk factor for strain mutation. However, the notion of resistance as soon as the strain appears is a protective factor. In a univariate analysis, of all the antibiotics examined, ceftazidime, meropenem, and the aminoglycosides seem to be risk factors for mutation. However, these results were not confirmed in a multivariate analysis. The study of resistance mechanisms acquired when ceftazidime or meropenem are prescribed has concretely demonstrated that exposure of a strain of this bacillus to ceftazidime is significantly associated with the over-expression of an inducible AmpC cephalosporinase. Exposure to meropenem is significantly associated with the acquisition of impermeability by loss of the porin OprD. For now, the emergence of carbapenemase-secreting strains or ESBL remains marginal in this population. The original nature of our work lies in the segmentation according to the predictive factors of the occurrence of resistance, i.e., the duration of artificial ventilation, the prior existence or absence of resistance, and the prescription of ceftazidime.
Abbreviations
- ATB
Antibiotic
- AUIC
Area under the inhibitory curve
- BAL
Bronchoalveolar lavage
- CA-SFM/EUCAST
Comité de l’Antibiogramme de la Société Française de Microbiologie/European Committee on Antimicrobial Susceptibility Testing
- CHU
University hospital center
- CI
Confidence interval
- CNR
National Research Center
- ESBL
Extended spectrum β-lactamase
- MIC
Minimum inhibitory concentration
- NPV
Negative predictive value
- P. a
Pseudomonas aeruginosa
- P. aeruginosa
Pseudomonas aeruginosa
- PPV
Positive predictive value
- ROC
Receiver operating characteristic
- TA
Tracheal aspiration
- VAP
Ventilator-acquired pneumopathies
- VS
Versus
Authors’ contributions
BG, SD, JMC, VM, and MG contributed to the conception and design. BG, FBV, JMC, FL, MG, SR, and TS contributed to the analysis and interpretation. BG, OF, and SR edited the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding
None
Availability of data and materials
All the samples were sent to Laboratory of Bacteriology, Federative Institute of Biology of our institution. The MICs of the antibiotics were examined in this unit. Data can be found in our intensive care unit or in the Laboratory of Bacteriology.
Ethics approval and consent to participate
Approved by the Toulouse Hospital Ethics and Research Committee (n° 85- 1114)
Consent for publication
All authors consent for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stéphanie Druge, Email: steph.druge@gmail.com.
Stéphanie Ruiz, Email: ruiz.stephanie@chu-toulouse.fr.
Fanny Vardon-Bounes, Email: bounes.f@chu-toulouse.fr.
Marion Grare, Email: grare.m@chu-toulouse.fr.
François Labaste, Email: labaste.f@chu-toulouse.fr.
Thierry Seguin, Email: seguin.t@chu-toulouse.fr.
Olivier Fourcade, Email: fourcade.o@chu-toulouse.fr.
Vincent Minville, Email: minville.v@chu-toulouse.fr.
Jean-Marie Conil, Email: conil.jm@chu-toulouse.fr.
Bernard Georges, Phone: +33 5 61 32 23 41, Email: georges.b@chu-toulouse.fr.
References
- 1.Yoon YK, Kim HA, Ryu SY, Lee EJ, Lee MS, Kim J, et al. Tree-structured survival analysis of patients with Pseudomonas aeruginosa bacteremia: a multicenter observational cohort study. Diagn Microbiol Infect Dis. 2017;87(2):180–187. doi: 10.1016/j.diagmicrobio.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Kollef MH, Chastre J, Fagon J-Y, François B, Niederman MS, Rello J, et al. Global prospective epidemiologic and surveillance study of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit Care Med. 2014;42(10):2178–2187. doi: 10.1097/CCM.0000000000000510. [DOI] [PubMed] [Google Scholar]
- 3.Georges B, Conil J-M, Dubouix A, Archambaud M, Bonnet E, Saivin S, et al. Risk of emergence of Pseudomonas aeruginosa resistance to beta-lactam antibiotics in intensive care units. Crit Care Med. 2006;34(6):1636–1641. doi: 10.1097/01.CCM.0000215517.51187.CA. [DOI] [PubMed] [Google Scholar]
- 4.Pena C, Suarez C, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, et al. Effect of adequate single-drug vs combination antimicrobial therapy on mortality in Pseudomonas aeruginosa bloodstream infections: a post hoc analysis of a prospective cohort. Clin Infect Dis. 2013;57(2):208–216. doi: 10.1093/cid/cit223. [DOI] [PubMed] [Google Scholar]
- 5.Morata L, Cobos-Trigueros N, Martínez JA, Soriano A, Almela M, Marco F, et al. Influence of multidrug resistance and appropriate empirical therapy on the 30-day mortality rate of Pseudomonas aeruginosa bacteremia. Antimicrob Agents Chemother. 2012;56(9):4833–4837. doi: 10.1128/AAC.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumbarello M, De Pascale G, Trecarichi EM, Spanu T, Antonicelli F, Maviglia R, et al. Clinical outcomes of Pseudomonas aeruginosa pneumonia in intensive care unit patients. Intensive Care Med. 2013;39(4):682–692. doi: 10.1007/s00134-013-2828-9. [DOI] [PubMed] [Google Scholar]
- 7.Hong DJ, Bae IK, Jang I-H, Jeong SH, Kang H-K, Lee K. Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47(2):81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira ML, Dantas RC, Ribas RM, Gontijo-Filho PP. Pseudomonas aeruginosa bacteraemia: independent risk factors for mortality and impact of resistance on outcome. J Med Microbiol. 2014;63(12):1679–1687. doi: 10.1099/jmm.0.073262-0. [DOI] [PubMed] [Google Scholar]
- 9.Breidenstein EBM, de la Fuente-Núñez C, REW H. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Tam VH, Rogers CA, Chang K-T, Weston JS, Caeiro J-P, Garey KW. Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrob Agents Chemother. 2010;54(9):3717–3722. doi: 10.1128/AAC.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ong DSY, Jongerden IP, Buiting AG, Leverstein-van Hall MA, Speelberg B, Kesecioglu J, et al. Antibiotic exposure and resistance development in Pseudomonas aeruginosa and Enterobacter species in intensive care units. Crit Care Med. 2011;39(11):2458–2463. doi: 10.1097/CCM.0b013e318225756d. [DOI] [PubMed] [Google Scholar]
- 12.Zanetti G, Bally F, Greub G, Garbino J, Kinge T, Lew D, et al. Cefepime versus imipenem-cilastatin for treatment of nosocomial pneumonia in intensive care unit patients: a multicenter, evaluator-blind, prospective, randomized study. Antimicrob Agents Chemother. 2003;47(11):3442–3447. doi: 10.1128/AAC.47.11.3442-3447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodise TP, Patel N, Kwa A, Graves J, Furuno JP, Graffunder E, et al. Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob Agents Chemother. 2007;51(10):3510–3515. doi: 10.1128/AAC.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3(1):32. doi: 10.1186/2047-2994-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasink LB, Fishman NO, Weiner MG, Nachamkin I, Bilker WB, Lautenbach E. Fluoroquinolone-resistant Pseudomonas aeruginosa: assessment of risk factors and clinical impact. Am J Med. 2006;119(6):526.e19–526.e25. doi: 10.1016/j.amjmed.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 16.Bretonnière C, Leone M, Milési C, Allaouchiche B, Armand-Lefevre L, Baldesi O, et al. Strategies to reduce curative antibiotic therapy in intensive care units (adult and paediatric) Intensive Care Med. 2015;41(7):1181–1196. doi: 10.1007/s00134-015-3853-7. [DOI] [PubMed] [Google Scholar]
- 17.Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41(2):580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 18.Venier A-G, Leroyer C, Slekovec C, Talon D, Bertrand X, Parer S, et al. Risk factors for Pseudomonas aeruginosa acquisition in intensive care units: a prospective multicentre study. J Hosp Infect. 2014;88(2):103–108. doi: 10.1016/j.jhin.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 19.Mladenovic-Antic S, Kocic B, Velickovic-Radovanovic R, Dinic M, Petrovic J, Randjelovic G, et al. Correlation between antimicrobial consumption and antimicrobial resistance of Pseudomonas aeruginosa in a hospital setting: a 10-year study. J Clin Pharm Ther. 2016;41(5):532–537. doi: 10.1111/jcpt.12432. [DOI] [PubMed] [Google Scholar]
- 20.Holt VI ’t, AF SJA, Lesaffre EMEH, Vos MC. A systematic review and meta-analyses show that carbapenem use and medical devices are the leading risk factors for carbapenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(5):2626–2637. doi: 10.1128/AAC.01758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez JA, Delgado E, Martí S, Marco F, Vila J, Mensa J, et al. Influence of antipseudomonal agents on Pseudomonas aeruginosa colonization and acquisition of resistance in critically ill medical patients. Intensive Care Med. 2009;35(3):439–447. doi: 10.1007/s00134-008-1326-y. [DOI] [PubMed] [Google Scholar]
- 22.Fernández-Barat L, Ferrer M, De Rosa F, Gabarrús A, Esperatti M, Terraneo S, et al. Intensive care unit-acquired pneumonia due to Pseudomonas aeruginosa with and without multidrug resistance. J Infect. 2017;74(2):142–152. doi: 10.1016/j.jinf.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Cobos-Trigueros N, Solé M, Castro P, Torres JL, Hernández C, Rinaudo M, et al. Acquisition of Pseudomonas aeruginosa and its resistance phenotypes in critically ill medical patients: role of colonization pressure and antibiotic exposure. Crit Care Lond Engl. 2015;19:218. doi: 10.1186/s13054-015-0916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vettoretti L, Floret N, Hocquet D, Dehecq B, Plésiat P, Talon D, et al. Emergence of extensive-drug-resistant Pseudomonas aeruginosa in a French university hospital. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2009;28(10):1217–1222. doi: 10.1007/s10096-009-0767-8. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Zorrilla S, Camoez M, Tubau F, Periche E, Canizares R, Dominguez MA, et al. Antibiotic pressure is a major risk factor for rectal colonization by multidrug-resistant Pseudomonas aeruginosa in critically ill patients. Antimicrob Agents Chemother. 2014;58(10):5863–5870. doi: 10.1128/AAC.03419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furtado GHC, Bergamasco MD, Menezes FG, Marques D, Silva A, Perdiz LB, et al. Imipenem-resistant Pseudomonas aeruginosa infection at a medical-surgical intensive care unit: risk factors and mortality. J Crit Care. 2009;24(4):625.e9–625.14. doi: 10.1016/j.jcrc.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Park S-Y, Park HJ, Moon SM, Park K-H, Chong YP, Kim M-N, et al. Impact of adequate empirical combination therapy on mortality from bacteremic Pseudomonas aeruginosa pneumonia. BMC Infect Dis. 2012;12:308. doi: 10.1186/1471-2334-12-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhat S, Fujitani S, Potoski BA, Capitano B, Linden PK, Shutt K, et al. Pseudomonas aeruginosa infections in the intensive care unit: can the adequacy of empirical beta-lactam antibiotic therapy be improved? Int J Antimicrob Agents. 2007;30(5):458–462. doi: 10.1016/j.ijantimicag.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Cillóniz C, Gabarrús A, Ferrer M, Puig de la Bellacasa J, Rinaudo M, Mensa J, et al. Community-acquired pneumonia due to multidrug- and non-multidrug-resistant Pseudomonas aeruginosa. Chest. 2016;150(2):415–425. doi: 10.1016/j.chest.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 30.Falagas ME, Kopterides P. Risk factors for the isolation of multi-drug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: a systematic review of the literature. J Hosp Infect. 2006;64(1):7–15. doi: 10.1016/j.jhin.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Solé M, Fàbrega A, Cobos-Trigueros N, Zamorano L, Ferrer-Navarro M, Ballesté-Delpierre C, et al. In vivo evolution of resistance of Pseudomonas aeruginosa strains isolated from patients admitted to an intensive care unit: mechanisms of resistance and antimicrobial exposure. J Antimicrob Chemother. 2015;70(11):3004–3013. doi: 10.1093/jac/dkv228. [DOI] [PubMed] [Google Scholar]
- 32.Lee H-S, Loh Y-X, Lee J-J, Liu C-S, Chu C. Antimicrobial consumption and resistance in five Gram-negative bacterial species in a hospital from 2003 to 2011. J Microbiol Immunol Infect Wei Mian Yu Gan Ran Za Zhi. 2015;48(6):647–654. doi: 10.1016/j.jmii.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50(1):43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel N, McNutt L-A, Lodise TP. Relationship between various definitions of prior antibiotic exposure and piperacillin-tazobactam resistance among patients with respiratory tract infections caused by Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52(8):2933–2936. doi: 10.1128/AAC.00456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grohs P, Kernéis S, Sabatier B, Lavollay M, Carbonnelle E, Rostane H, et al. Fighting the spread of AmpC-hyperproducing Enterobacteriaceae: beneficial effect of replacing ceftriaxone with cefotaxime. J Antimicrob Chemother. 2014;69(3):786–789. doi: 10.1093/jac/dkt403. [DOI] [PubMed] [Google Scholar]
- 36.Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother. 2014;69(7):1804–1814. doi: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 37.Fournier D, Richardot C, Müller E, Robert-Nicoud M, Llanes C, Plésiat P, et al. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J Antimicrob Chemother. 2013;68(8):1772–1780. doi: 10.1093/jac/dkt098. [DOI] [PubMed] [Google Scholar]
- 38.Castanheira M, Mills JC, Farrell DJ, Jones RN. Mutation-driven β-lactam resistance mechanisms among contemporary ceftazidime-nonsusceptible Pseudomonas aeruginosa isolates from U.S. hospitals. Antimicrob Agents Chemother. 2014;58(11):6844–6850. doi: 10.1128/AAC.03681-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riou M, Carbonnelle S, Avrain L, Mesaros N, Pirnay J-P, Bilocq F, et al. In vivo development of antimicrobial resistance in Pseudomonas aeruginosa strains isolated from the lower respiratory tract of Intensive Care Unit patients with nosocomial pneumonia and receiving antipseudomonal therapy. Int J Antimicrob Agents. 2010;36(6):513–522. doi: 10.1016/j.ijantimicag.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Wong PHP, von Krosigk M, Roscoe DL, Lau TTY, Yousefi M, Bowie WR. Antimicrobial co-resistance patterns of gram-negative bacilli isolated from bloodstream infections: a longitudinal epidemiological study from 2002-2011. BMC Infect Dis. 2014;14:393. doi: 10.1186/1471-2334-14-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carsenti-Etesse H, Cavallo JD, Roger PM, Ziha-Zarifi I, Plesiat P, Garrabé E, et al. Effect of beta-lactam antibiotics on the in vitro development of resistance in Pseudomonas aeruginosa. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2001;7(3):144–151. doi: 10.1046/j.1469-0691.2001.00225.x. [DOI] [PubMed] [Google Scholar]
- 42.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45(6):568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 43.McCarthy K. Pseudomonas aeruginosa: evolution of antimicrobial resistance and implications for therapy. Semin Respir Crit Care Med. 2015;36(1):44–55. doi: 10.1055/s-0034-1396907. [DOI] [PubMed] [Google Scholar]
- 44.Parker Chris M., Kutsogiannis Jim, Muscedere John, Cook Deborah, Dodek Peter, Day Andrew G., Heyland Daren K. Ventilator-associated pneumonia caused by multidrug-resistant organisms or Pseudomonas aeruginosa: Prevalence, incidence, risk factors, and outcomes. Journal of Critical Care. 2008;23(1):18–26. doi: 10.1016/j.jcrc.2008.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the samples were sent to Laboratory of Bacteriology, Federative Institute of Biology of our institution. The MICs of the antibiotics were examined in this unit. Data can be found in our intensive care unit or in the Laboratory of Bacteriology.