Abstract
Objective
To understand the relationships of Staphylococcus aureus (SA) bacteremic pneumonia (SABP) outcome with patient‐specific and SA‐specific variables.
Methods
We analysed SA bloodstream isolates and matching sera in SABP patients by sequencing SA isolates (n = 50) and measuring in vitro AT production, haemolytic activity and expression of ClfA and ClfB. Controls were sera from gram‐negative bacteremia patients with or without pneumonia and uninfected subjects. Levels of IgGs, IgMs and neutralizing antibodies (NAbs) against SA antigens were quantified and analysed by one‐way ANOVA. Associations of patient outcomes with patient variables, antibody levels and isolate characteristics were evaluated by univariate and multivariate logistic regression analyses.
Results
SABP patients had higher levels of IgGs against eight virulence factors and anti‐alpha toxin (AT) NAbs than uninfected controls. Levels of IgG against AT and IgMs against ClfA, FnbpA and SdrC were higher in clinically cured SABP patients than in clinical failures. Anti‐LukAB NAb levels were elevated in all cohorts. Increased odds of cure correlated with higher haemolytic activity of SA strains, longer time between surgery and bacteremia (> 30 days), longer duration of antibiotic therapy, lower acute physiology and total APACHE II scores, lack of persistent fever for > 72 h and higher levels of antibodies against AT (IgG), ClfA (IgM), FnbpA (IgM) and SdrC (IgM).
Discussion
Limitations included the cross‐sectional observational nature of the study, small sample size and inability to measure antibody levels against all SA virulence factors.
Conclusion
Our results suggest that SABP patients may benefit from immunotherapy targeting multiple SA antigens.
Keywords: antibody response, bacteremia, patient outcome, pneumonia, Staphylococcus aureus, virulence
Introduction
Staphylococcus aureus is a common bacterial pathogen that causes a multitude of life‐threatening infections.1 In the United States alone, S. aureus infections result in more than 11 000 deaths each year, along with an estimated annual cost of $14 billion, and S. aureus pneumonia accounts for an estimated 50 000 infections per year.2, 3, 4 S. aureus bacteremic pneumonia is associated with a high mortality rate (30‐day mortality, 46.9%).5 The increase in antibiotic resistance among S. aureus populations is an alarming concern.6, 7, 8
Staphylococcus aureus possesses an arsenal of virulence factors, many of which are employed to evade and counteract the host immune system.9 Recent studies have correlated high preexisting serum antibody levels against several S. aureus virulence factors, including alpha toxin (AT), with a decreased risk of S. aureus infections and improved patient outcomes.10, 11, 12
The aim of this study was to understand the relationships among patient variables, bacterial strain characteristics, antibody response and clinical outcomes by analysing S. aureus bloodstream isolates along with matching sera and clinical data in a cohort of patients with S. aureus bacteremic pneumonia. For comparators, we included cohorts of patients with gram‐negative bacteremic pneumonia, patients with gram‐negative bacteremia without pneumonia and uninfected control subjects who were matched to the cohort with S. aureus bacteremic pneumonia by the number of samples and patient demographic characteristics such as age, gender and race.
Results
Comparison of serum IgG and NAb levels among study cohorts
Our recent studies demonstrated elevated serum levels of anti‐AT immunoglobulin G (IgG) and neutralising antibodies (NAbs) in haemodialysis and surgery patients with S. aureus bacteremia in comparison with healthy control subjects.13, 14 To perform a similar assessment in this study and to expand the analysis beyond AT, we measured the serum levels of IgG and IgM against additional S. aureus secreted toxins and surface proteins, such as delta toxin, clumping factor A (ClfA), ClfB, fibronectin‐binding protein (FnbpA), leukocidin AB (LukAB), lipoprotein component C of ABC manganese transporter (MntC), serine‐aspartate repeat‐containing protein C (SdrC) and toxic shock syndrome toxin‐1 (TSST‐1). These antigens were previously validated as important virulence factors involved in adhesion, evasion of immune responses, cell and tissue damage, biofilm formation and inflammation.15, 16, 17, 18, 19, 20, 21, 22 As shown in Figure 1, patients with S. aureus bacteremic pneumonia had elevated levels of IgGs against eight of nine measured antigens as compared with uninfected control subjects: AT (3.2‐fold, P = 0.0002), delta toxin (2.95‐fold, P = 0.0003), ClfA (2.51‐fold, P = 0.0006), ClfB (1.9‐fold, P = 0.01), FnbpA (2.12‐fold, P = 0.0042), SdrC (1.74‐fold, P = 0.0225), LukAB (3.9‐fold, P < 0.0001) and MntC (7.28‐fold, P < 0.0001). In contrast, serum S. aureus‐specific IgG levels in patients with gram‐negative bacteremia (with and without pneumonia) were similar to those in uninfected subjects. Levels of IgG against AT, as well as IgM against ClfA, FnbpA and SdrC, were higher in patients with cured S. aureus bacteremic pneumonia than in those with clinical failure.
Figure 1.
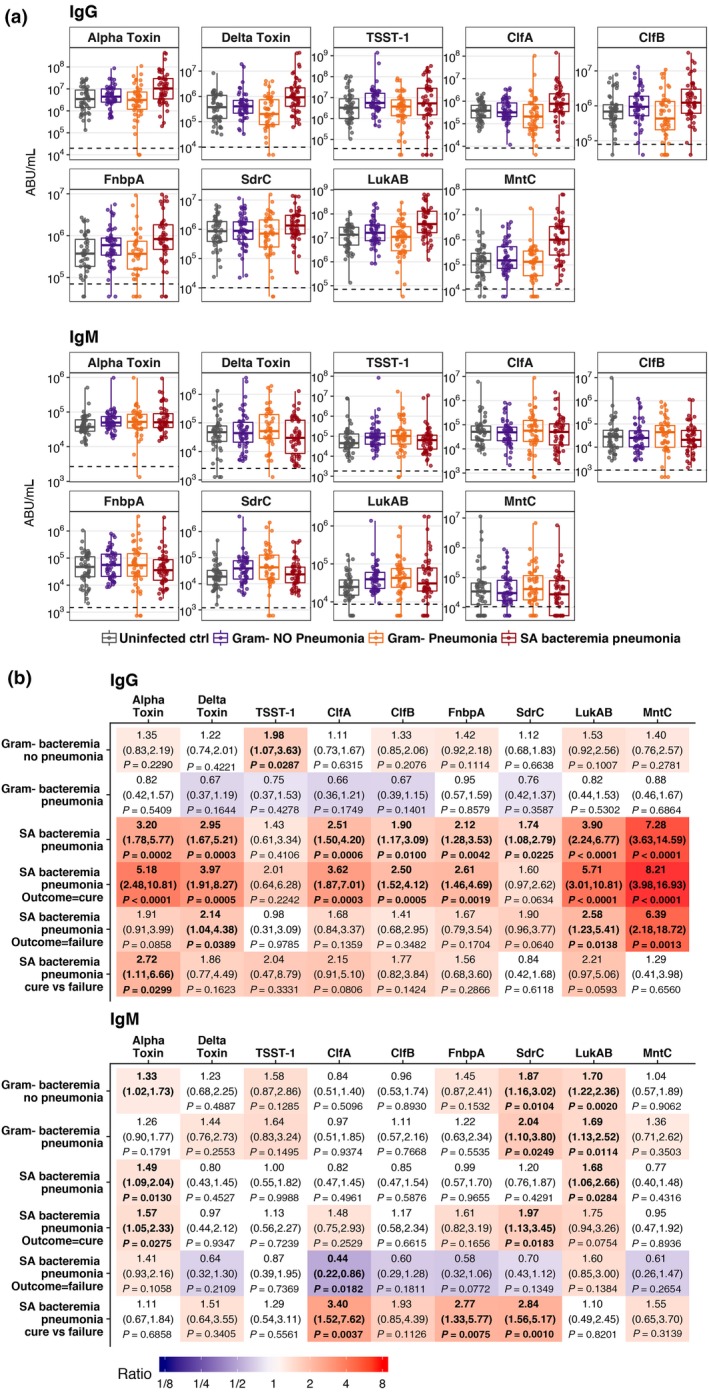
Serum anti‐AT IgG and IgM levels. (a) A box plot is presented for each cohort and is overlaid with values from individual subjects. The dashed line represents the assay's lower limit of quantitation (LLOQ). Values below the LLOQ were imputed with LLOQ/2. Each sample was tested in duplicate. (b) Ratios of geometric mean level for each disease cohort relative to uninfected control group and clinical outcome for the cohort with Staphylococcus aureus bacteremic pneumonia relative to uninfected control group and cure versus failure, with 95% CIs and P‐values. Fill colours ranging from purple to red represent lower to higher compared with the uninfected control group.
Next, we determined serum levels of anti‐AT and anti‐LukAB NAbs (Figure 2). Anti‐AT NAb levels were higher in patients with S. aureus bacteremic pneumonia than in uninfected subjects (2.12‐fold, P = 0.0236 overall, and 3.11‐fold, P = 0.0101 in cured patients), whereas no such differences were observed in patients with gram‐negative bacteremia. Interestingly, compared with uninfected subjects, the levels of anti‐LukAB NAbs were higher in patients with S. aureus bacteremic pneumonia (2.66‐fold, P = 0.0041) and were elevated to an even greater extent in those with gram‐negative bacteremia without pneumonia and those with gram‐negative bacteremic pneumonia (4.26‐fold, P < 0.0001, and 5.32‐fold, P = 0.001, respectively). We did not observe significant differences in either anti‐AT or anti‐LukAB NAb levels between cured patients and those with clinical failure in any of the study cohorts. We observed a high correlation between levels of IgG and NAbs against both AT (r = 0.8698; 0.7952 and 0.7911) and LukAB (r = 0.9044; 0.8259 and 0.7649) in patients with S. aureus bacteremic pneumonia, gram‐negative bacteremic pneumonia and gram‐negative bacteremia without pneumonia, respectively. There was no correlation between levels of IgG and NAbs against either AT (r = 0.0671) or LukAB (r = 0.2409) in uninfected control subjects.
Figure 2.
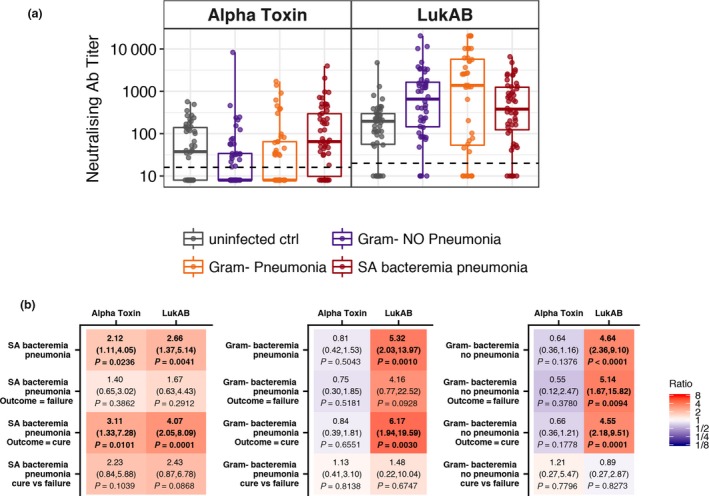
Serum anti‐AT and anti‐LukAB NAb levels. (a) A box plot is presented for each cohort and is overlaid with values from individual subjects. The dashed line represents the assay's LLOQ. Values below the LLOQ were imputed with LLOQ/2. Each sample was tested in duplicate. (b) Ratios of geometric mean level of anti‐AT and anti‐LukAB NAbs for each cohort relative to the uninfected control group and clinical outcome relative to uninfected control group and cure versus failure, with 95% CIs and P‐values. Fill colours ranging from purple to red represent lower to higher compared with the uninfected control group.
Characterisation of S. aureus isolates from patients with S. aureus bacteremic pneumonia
To assess the relationship between S. aureus‐specific factors and clinical outcomes, we characterised S. aureus bloodstream isolates from patients with S. aureus bacteremic pneumonia collected during the acute phase of infection. AT production was heterogeneous (96% positive by enzyme‐linked immunosorbent assay [ELISA]; range, 0–70 μg mL−1), and the majority of strains (n = 47) co‐expressed ClfA and ClfB (data not shown). The phylogenetic relatedness, AT production, haemolytic activity and genotypic characteristics of S. aureus isolates in relation to S. aureus bacteremic pneumonia cure or failure are summarised in Figure 3. All isolates carried hld, clfA, clfB, sdrC, lukAB and mntC genes. We identified three major groups of isolates based on their position on the phylogenetic tree, multilocus sequence types (MLSTs) and presence or absence of genes of interest. The first group (Figure 3, top; n = 23) is predominantly composed of ST8 mecA‐positive strains that carry the fnbpA gene. A majority of ST8 strains in this study (15 of 18) had elevated haemolytic activity above 100 haemolytic units (HU) per millilitre, which did not always correspond to high AT production and suggests that additional toxins other than AT contribute to haemolytic activity in these strains. The second group (Figure 3, middle; n = 9) includes a cluster of seven strains that have a Q113B stop codon in the hla gene, belong to sequence types 30 and 36 and carry the tst gene, which is consistent with previously reported findings.13, 23, 24, 25 The third group (Figure 3, bottom; n = 18) combines tst‐negative and predominantly fnbpA‐negative strains of various sequence types. One of the strains from this group (and the only strain in the collection) is null for the hla gene, and its MLST type (ST 225) is related to previously described hla gene‐null isolates.26 We did not observe any definitive association of patient outcomes with either phylogenetic grouping or genotypic characteristics of S. aureus isolates from the cohort of patients with S. aureus bacteremic pneumonia.
Figure 3.
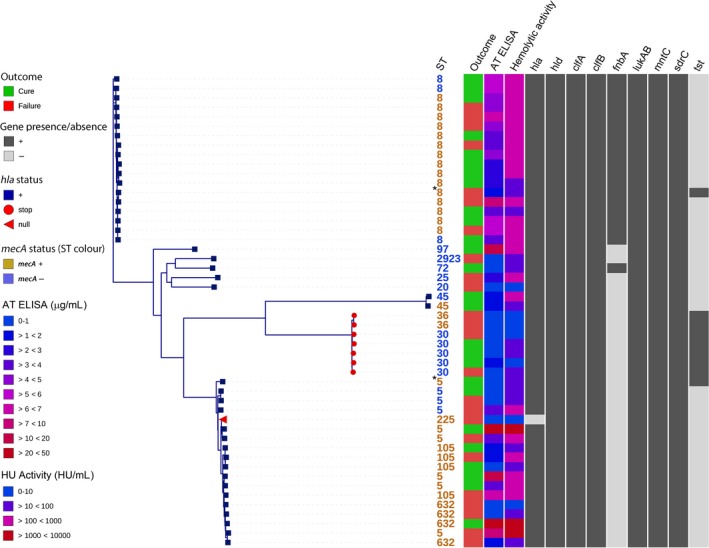
Phylogenetic relatedness, AT production, haemolytic activity and gene content of isolates from patients with Staphylococcus aureus bacteremic pneumonia. Whole‐genome‐based tree shows phylogenetic relatedness of S. aureus isolates. Isolates are labelled by MLST‐STs in orange or blue to indicate the presence or absence of the mecA gene, respectively. Shown are patient outcomes of cure (green) or failure (red), AT production (AT ELISA, in micrograms per millilitre) and haemolytic activity (HU activity, in HU per millilitre) of S. aureus isolates, and a panel displaying gene content. The hla gene‐null isolate is labelled with a red triangle. Isolates that contain hla Q113 stop codon are labelled with red circles. De novo assembly metrics of samples marked with an asterisk (*) suggest that these samples are probably mixed and their ST assignments and tree positions are unreliable.
We explored the relationships between certain phenotypic characteristics of these isolates, such as methicillin susceptibility, haemolytic activity and AT production, with patient outcomes. A correlation was observed between AT production and haemolytic activity (Figure 4), which was consistent with methicillin‐resistant S. aureus and methicillin‐susceptible S. aureus. Paradoxically, higher haemolytic activity and in vitro AT production were both associated with patient cure (Figure 5), which agrees with our previous observations in haemodialysis and postsurgical patients with S. aureus bacteremia.13 Both AT production and haemolytic activity were higher in methicillin‐resistant than in methicillin‐susceptible S. aureus isolates.
Figure 4.
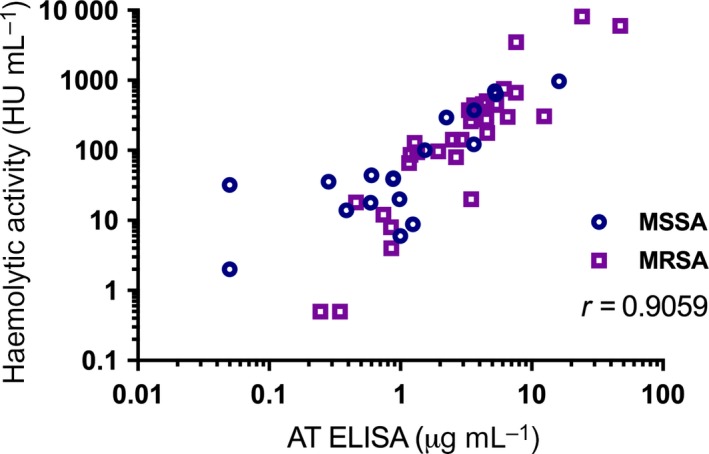
Correlation between haemolytic activity and AT production level. Spearman correlation coefficient is provided. MRSA = methicillin‐resistant Staphylococcus aureus; MSSA = methicillin‐susceptible S. aureus.
Figure 5.
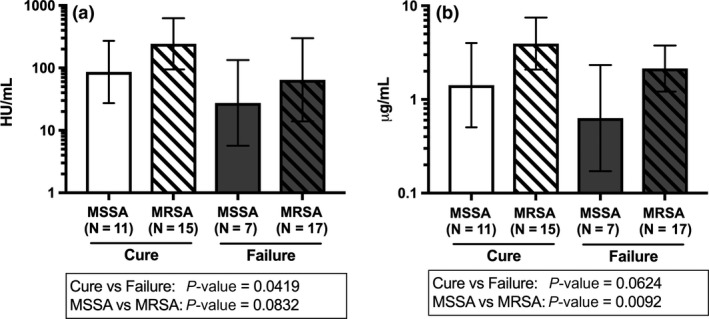
Haemolytic activity and AT production in Staphylococcus aureus bacteremic pneumonia isolates. Haemolytic activity, in HU per millilitre (a), and AT production, in micrograms per millilitre (b), were summarised in geometric mean and associated 95% CI in relation to methicillin susceptibility status of S. aureus isolates and patient outcomes. Each sample was tested in duplicate. MRSA = methicillin‐resistant S. aureus; MSSA = methicillin‐susceptible S. aureus.
Patient outcome analyses
To understand associations of S. aureus‐ and patient‐specific factors with clinical outcomes, we performed univariate analyses of patient demographics, clinical variables and S. aureus factors in relation to cure or failure (Tables 1 and 2; Figure 6a–c). Increased odds of cure were associated with longer time between surgical procedure and bacteremia (> 30 days) (odds ratio [OR], 3.93; 95% confidence interval [CI], 1.03–14.99; P = 0.0452); longer duration of antibiotic therapy (OR, 1.27; 95% CI, 1.03–1.56; P = 0.0259); lower acute physiology score (OR, 0.32; 95% CI, 0.12–0.84; P = 0.0215); total Acute Physiologic Assessment and Chronic Health Evaluation II (APACHE II) score (OR, 0.24; 95% CI, 0.09–0.67; P = 0.0064); lack of persistent fever for more than 72 h (OR, 0.26; 95% CI, 0.07–0.96; P = 0.0424); and higher levels of anti‐AT IgG (OR, 2.64; 95% CI, 1.05–6.63; P = 0.0392) and IgM against ClfA (OR, 4.32; 95% CI, 1.45–12.85; P = 0.0086), FnbpA (OR, 4.65; 95% CI, 1.35–16.07; P = 0.0151) and SdrC (OR, 10.10; 95% CI, 2.04–49.92; P = 0.0046). In contrast, no significant associations were observed between odds of cure and characteristics of S. aureus isolates.
Table 1.
Clinical measurements and outcomes in patients with S. aureus bacteremic pneumonia
| Variable | Cure (n = 26) | Failure (n = 24) | P |
|---|---|---|---|
| Age, years (mean ± SD) | 57.0 ± 14.5 | 63.5 ± 9.6 | 0.0760 |
| Gender, n (%) | |||
| Female | 12 (57.1) | 9 (42.9) | 0.5362 |
| Male | 14 (48.3) | 15 (51.7) | |
| Race, n (%) | |||
| Black/African American | 3 (30.0) | 7 (70.0) | 0.3117 |
| White | 22 (57.9) | 16 (42.1) | |
| Other | 1 (50.0) | 1 (50.0) | |
| Length of stay, days (mean ± SD) | 30.1 ± 39.0 | 37.4 ± 42.8 | 0.5265 |
| Days with symptoms of infection, mean ± SD | 5.0 ± 9.0 | 2.4 ± 4.0 | 0.2573 |
| Route of infection, n (%) | |||
| Hospital acquired | 9 (40.9) | 13 (59.1) | 0.2538 |
| Community acquired, healthcare associated | 12 (66.7) | 6 (33.3) | |
| Community acquired, non‐healthcare associated | 4 (44.4) | 5 (55.6) | |
| Surgery performed in 30 days before bacteremia, n (%) | |||
| No | 22 (61.1) | 14 (38.9) | 0.0452 |
| Yes | 4 (28.6) | 10 (71.4) | |
| Haemodialysis dependent, n (%) | |||
| No | 25 (53.2) | 22 (46.8) | 0.5144 |
| Yes | 1 (33.3) | 2 (66.7) | |
| Diabetic, n (%) | |||
| No | 15 (48.4) | 16 (51.6) | 0.5144 |
| Yes | 11 (57.9) | 8 (42.1) | |
| APACHE II score, mean ± SD | |||
| Age score | 3.0 ± 2.3 | 3.8 ± 1.6 | 0.1521 |
| Chronic health | 4.0 ± 1.6 | 4.6 ± 1.0 | 0.1143 |
| Acute physiology | 9.6 ± 6.2 | 14.2 ± 6.5 | 0.0215 |
| Total | 16.6 ± 6.7 | 22.6 ± 6.4 | 0.0064 |
| Corticosteroid use, n (%) | |||
| No | 15 (46.9) | 17 (53.1) | 0.3356 |
| Yes | 11 (61.1) | 7 (38.9) | |
| Transplant recipient, n (%) | |||
| No | 22 (51.2) | 21 (48.8) | 0.7693 |
| Yes | 4 (57.1) | 3 (42.9) | |
| Patient being treated for neoplasm/cancer, n (%) | |||
| No | 21 (53.8) | 18 (46.2) | 0.6238 |
| Yes | 5 (45.5) | 6 (54.5) | |
| Fever resolved within 72 h, n (%) | |||
| No | 7 (38.9) | 11 (61.1) | 0.0424 |
| Yes | 17 (70.8) | 7 (29.2) | |
| NA | 2 (25.0) | 6 (75.0) | |
| Main antibiotic regimen, n (%) | |||
| β‐lactamase‐resistant penicillin | 4 (80.0) | 1 (20.0) | 0.6581 |
| Cefazolin/first‐generation cephalosporin | 3 (100.0) | 0 (0.0) | |
| Ceftriaxone | 1 (100.0) | 0 (0.0) | |
| Daptomycin | 1 (100.0) | 0 (0.0) | |
| Linezolid | 3 (75.0) | 1 (25.0) | |
| Vancomycin | 13 (40.6) | 19 (59.4) | |
| Other | 1 (25.0) | 3 (75.0) | |
| Duration of antibiotic therapy, days (mean ± SD) | 36.4 ± 23.0 | 19.2 ± 24.9 | 0.0259 |
SD, standard deviation.
Clinical measurements were summarised as number and percentage for categorical variables; in median, minimum and maximum for ordinal variables; and as mean ± standard deviation for continuous variables. P‐values for the association between clinical measurements and patient outcome were calculated from logistic regression.
Figure 6.
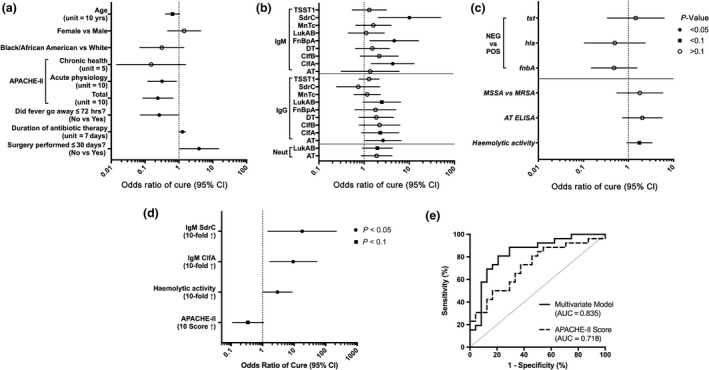
Associations of Staphylococcus aureus‐specific and patient‐specific factors with clinical outcome in patients with S. aureus bacteremic pneumonia. The OR of cure and associated 95% CI from univariate logistic regression analysis are shown for (a) clinical characteristics, (b) serum antibody levels and (c) S. aureus isolate characteristics. The ORs for antibody levels, AT production and haemolytic activity correspond to a 10‐fold increase. As shown in Figure 3, hld, clfA, clfB, sdrC, lukAB and mntC genes were present in all isolates and therefore were not included in the univariate analysis. (d) OR and associated 95% CI from the multivariate logistic model with independent predictors. (e) Receiver operating characteristics curve of the multivariate logistic model, using leave‐one‐out cross‐validation. The associated area under the curve (AUC) is shown.
In addition to univariate analyses, we performed multivariate modelling to identify a combination of independent variables associated with clinical outcomes. Increased odds of cure were associated with higher levels of IgM against SdrC (OR, 13.95; 95% CI, 1.18–164.49; P = 0.0363) or ClfA (OR, 5.72; 95% CI, 1.16–28.24; P = 0.0322), elevated haemolytic activity of S. aureus isolates (OR, 2.94; 95% CI, 1.05–8.27; P = 0.0409) and lower total APACHE II score (OR, 0.34; 95% CI, 0.11–1.05; P = 0.0605) (Table 2; Figure 6d, e). Total APACHE II score alone was predictive of clinical outcome, and predictive performance was further improved by additional variables in this multivariate model.
Table 2.
Results of statistical analyses
| Effect/variable | OR | 95% CI | P |
|---|---|---|---|
| Univariate model | |||
| Clinical | |||
| Age (units = 10 years) | 0.64 | 0.40–1.05 | 0.0760 |
| Gender (female vs male) | 1.43 | 0.46–4.42 | 0.5362 |
| Race (black/African American vs white) | 0.31 | 0.07–1.39 | 0.1272 |
| Chronic Health Points score (units = 5) | 0.15 | 0.01–1.58 | 0.1143 |
| Acute Physiology score (units = 10) | 0.32 | 0.12–0.84 | 0.0215 |
| Total APACHE II score (units = 10) | 0.24 | 0.09–0.67 | 0.0064 |
| Did fever go away ≤ 72 h? (No vs Yes) | 0.26 | 0.07–0.96 | 0.0424 |
| Was surgery performed ≤ 30 days? (No vs Yes) | 3.93 | 1.03–14.99 | 0.0452 |
| Duration of antibiotic treatment (units = 7 days) | 1.27 | 1.03–1.56 | 0.0259 |
| Serologya | |||
| NAb_AT | 1.90 | 0.87–4.15 | 0.1085 |
| NAb_LukAB | 1.97 | 0.91–4.26 | 0.0857 |
| IgG_AT | 2.64 | 1.05–6.63 | 0.0392 |
| IgG_ClfA | 2.28 | 0.88–5.93 | 0.0899 |
| IgG_ClfB | 2.20 | 0.78–6.23 | 0.1386 |
| IgG_DT | 1.86 | 0.77–4.50 | 0.1658 |
| IgG_FnbpA | 1.79 | 0.63–5.08 | 0.2751 |
| IgG_LukAB | 2.47 | 0.95–6.42 | 0.0637 |
| IgG_MntC | 1.17 | 0.60–2.31 | 0.6420 |
| IgG_SdrC | 0.74 | 0.25–2.24 | 0.5977 |
| IgG_TSST1 | 1.29 | 0.77–2.16 | 0.3284 |
| IgM_AT | 1.37 | 0.31–5.99 | 0.6785 |
| IgM_ClfA | 4.32 | 1.45–12.85 | 0.0086 |
| IgM_ClfB | 2.17 | 0.82–5.73 | 0.1175 |
| IgM_DT | 1.53 | 0.64–3.65 | 0.3378 |
| IgM_FnbpA | 4.65 | 1.35–16.07 | 0.0151 |
| IgM_LukAB | 1.12 | 0.44–2.81 | 0.8159 |
| IgM_MnTc | 1.60 | 0.65–3.96 | 0.3044 |
| IgM_SdrC | 10.10 | 2.04–49.92 | 0.0046 |
| IgM_TSST1 | 1.31 | 0.55–3.13 | 0.5467 |
| Staphylococcus aureus isolate | |||
| Haemolytic activity (10‐fold increase) | 1.75 | 0.92–3.33 | 0.0903 |
| AT ELISA (10‐fold increase) | 2.02 | 0.73–5.57 | 0.1731 |
| MSSA vs MRSA | 1.78 | 0.55–5.77 | 0.3356 |
| fnbA (negative vs positive) | 0.48 | 0.15–1.55 | 0.2195 |
| hla (negative vs positive) | 0.50 | 0.10–2.35 | 0.3763 |
| tst (negative vs positive) | 1.45 | 0.34–6.18 | 0.6175 |
| Multivariate model | |||
| Total APACHE II (score increase by 10) | 0.34 | 0.11–1.05 | 0.0605 |
| Haemolytic activity (10‐fold increase) | 2.94 | 1.05–8.27 | 0.0409 |
| IgM SdrC (10‐fold increase) | 13.95 | 1.18–164.49 | 0.0363 |
| IgM ClfA (10‐fold increase) | 5.72 | 1.16–28.24 | 0.0322 |
MRSA, methicillin‐resistant S. aureus; MSSA, methicillin‐susceptible S. aureus.
Serology OR corresponds to a 10‐fold increase in antibody levels.
Discussion
Our results demonstrated associations of clinical cure in patients with S. aureus bacteremic pneumonia with increased haemolytic activity of the infecting strains and various patient characteristics (longer time between surgery and onset of bacteremia, lower APACHE II scores, shorter persistent fever, longer duration of antibiotic therapy). Higher levels of IgG against AT and IgM against ClfA, FnbpA and SdrC were observed in cured patients than in patients with clinical failure. We hypothesise that elevated levels of antibodies specific to these antigens may correlate with protection against S. aureus bacteremic pneumonia.
In this study, irrespective of any specific S. aureus clonal lineage or clade, higher haemolytic activity and in vitro AT production were both associated with patient cure, in agreement with our prior findings in haemodialysis and postsurgical patients with S. aureus bacteremia.13 We previously hypothesised that the paradoxical correlation between AT production and patient survival in S. aureus bacteremia resulted from a stronger overall immune response against S. aureus because of AT proinflammatory properties.13 In agreement with this hypothesis, our results showed higher levels of IgG against a variety of S. aureus secreted toxins and surface proteins in patients with S. aureus bacteremic pneumonia than in uninfected control subjects and patients with gram‐negative bacteremia. Reduced virulence of AT‐hyperproducing S. aureus strains has been observed in a rabbit model of experimental endocarditis, and such a paradoxical association between high AT production and decreased virulence may result from higher AT‐mediated release of cationic platelet microbicidal proteins.27
Staphylococcal surface proteins such as ClfA, FnbpA and SdrC have been shown to facilitate attachment of S. aureus to host tissue cells and initiation of infection.16, 28, 29, 30 We observed direct associations between elevated serum levels of IgM against ClfA, FnbpA and SdrC and increased odds of cure, which may suggest the importance of targeting these adhesins early at the onset of infection for favorable clinical outcomes in patients with S. aureus bacteremia pneumonia.
LukAB (LukGH) is a bicomponent leukotoxin that is essential for S. aureus escape from within polymorphonuclear leucocytes (PMNLs) and S. aureus‐mediated killing of PMNLs.31, 32 Both S. aureus components and lipopolysaccharide of gram‐negative bacteria induce high sensitivity of human PMNLs to LukAB.33 We hypothesise that the elevated serum levels of anti‐LukAB NAbs that were observed in this study in patients with S. aureus bacteremic pneumonia and in those with gram‐negative bacteremia may be important in protecting PMNLs from LukAB‐mediated killing.
We observed a high correlation between levels of IgG and NAbs against both AT and LukAB in patients with S. aureus bacteremic pneumonia, confirming our previous findings in other patient populations with S. aureus bacteremia.13, 14 We observed a similar correlation in patients with gram‐negative bacteremic pneumonia and gram‐negative bacteremia without pneumonia, indicating that this phenomenon is not specific to S. aureus infection and includes gram‐negative infection groups. Interestingly, no such correlation was observed in uninfected subjects, suggesting that high correlation between levels of IgG and NAbs against both AT and LukAB may be an attribute of an acute infection.
This study had several limitations. All samples were collected at a single hospital, and the sample size was relatively small. The samples were collected over a 13‐year period, and study results might have been affected by changes in antibiotic treatment or other aspects of clinical care during this time. The customised multiplex assay used in this study could accommodate a maximum of only nine antigens, and therefore, we were not able to measure antibody levels against all of the numerous S. aureus virulence factors. Because of the cross‐sectional observational nature of the study, we could measure antibody levels only in the acute stage of disease and were unable to correlate antibody levels before S. aureus exposure or infection with clinical outcome. Nevertheless, the antibody levels in the uninfected control subjects were also measured as part of this study, providing a range of the preexisting antibody levels. The antibody levels in S. aureus‐infected patients were generally higher than those in uninfected subjects (i.e. preexisting antibody levels), suggesting that elevated antibody levels resulted from the acute infection.
Despite its limitations, the current study offered several advantages, including well‐matched study cohorts; availability of matched serum samples, S. aureus isolates and clinical data; and minimal interhospital variability, as all samples came from the same location. The use of the S. aureus multiplex serology assay allowed us to measure IgM and IgG against nine S. aureus antigens. Our results suggest that, in addition to AT, other S. aureus virulence factors, such as ClfA, FnbpA and SdrC, may serve as promising targets for immunotherapy or vaccination, and that IgM or IgG against multiple S. aureus antigens (both secreted toxins and surface proteins) may benefit patients with S. aureus bacteremic pneumonia.
Methods
Subject selection and ethics statement
The Duke University Institutional Review Board approved this investigation. Eligible patients met the following criteria: (1) adults hospitalised at Duke University Medical Center with monomicrobial S. aureus bacteremia; (2) no neutropenia (absolute neutrophil count of > 100 neutrophils μL−1); (3) availability of clinical data, bloodstream S. aureus isolate and acute‐phase sera in the S. aureus Bacteremia Group biorepository; and (4) availability of S. aureus bacteremic pneumonia samples collected from 2002 to 2015. After informed consent was obtained, blood was drawn from the hospitalised patients within 1–3 days after the first S. aureus‐positive blood culture. Blood was allowed to clot by incubating for 30–60 min at room temperature, and serum was then separated by centrifugation at 1000–2000×g for 10 min and then divided into aliquots, which were stored at –80°C. A single serum aliquot per patient was used for serological assays. A total of 50 samples (both isolates and sera) from patients with S. aureus bacteremic pneumonia were included in the study, along with sera from patients with gram‐negative bacteremic pneumonia (n = 50), patients with gram‐negative bacteremia without pneumonia (n = 50) and uninfected control subjects (n = 50) who were matched with S. aureus bacteremic pneumonia patients by the number of samples and demographic characteristics such as age, gender and race.
Clinical outcomes and definitions
Clinical outcomes were defined as either ‘Cure’ or ‘Failure’ by site investigators as described previously.13 ‘Cure’ was defined as occurring when a patient was alive with no evidence of recurrent S. aureus infection at 12 weeks after the initial positive blood culture. ‘Failure’ was defined as occurring when a patient died for any reason or experienced culture‐confirmed recurrent S. aureus infection within 12 weeks of the initial positive blood culture. ‘S. aureus bacteremic pneumonia’ was defined as bacteremia with an initial source of S. aureus pneumonia, as diagnosed by the physician providing care to the patient. ‘Gram‐negative bacteremic pneumonia’ was defined as bacteremia with an initial source of pneumonia caused by gram‐negative bacteria, as diagnosed by the physician providing care to the patient. In patients with gram‐negative bacteremia without pneumonia, the initial sources of bacteremia were endovascular, skin, soft tissue, joint or bone, gastrointestinal, genitourinary or unknown excluding pneumonia. The uninfected control subjects were hospitalised patients who had no infection at the time of screening and enrolment and no history of S. aureus or gram‐negative infections.
Staphylococcus aureus isolates
Staphylococcus aureus bloodstream isolates were identified from patients who provided written informed consent and were linked to the clinical details via a unique study number. For patients with multiple positive blood cultures, the initial isolate was used for all characterisations.
Genotypic characterisation of S. aureus isolates
Whole‐genome sequencing of S. aureus isolates was performed via MiSeq 2 × 250 runs (Illumina, San Diego, CA) with a targeted depth of 150‐fold. S. aureus genomic DNA was purified from bacterial cultures via bead beating, followed by extraction with a PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, Waltham, MA). Sequencing libraries were prepared by Covaris mechanical shearing, followed by a NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA). MLSTs were assigned to each genome according to the S. aureus MLST database (available from PubMLST at http://www.pubmlst.org) by mapping to known alleles, using SRST2.34 The sequence type was calculated by using the best‐scoring alleles. Read sets were screened for virulence genes with a direct‐read mapping approach implemented in SRST2, using a 90% coverage cut‐off.34 Sequences were assembled de novo with SPAdes 3.11.1.35 Phylogenetic trees were generated with kSNP336 and were visualised and annotated with ITOL v3.37
Serological assays
Serum levels of IgG against nine S. aureus antigens were measured with a customised multiplex assay (Meso Scale Discovery; Meso Scale Diagnostics, Rockville, MD) according to the manufacturer's instructions. Sulfo tag‐labelled HyTest 2A11 anti‐human IgG (2 μg mL−1) or anti‐human/nonhuman primate IgM (1 μg mL−1) was used for detection. Anti‐AT and anti‐LukAB NAb titres were measured with a red blood cell haemolytic assay and an assay with human monocytic cell line HL‐60 (American Type Culture Collection, Manassas, VA), respectively, as described previously.26, 38
Preparation of S. aureus supernatants
For each S. aureus isolate, a single colony from a tryptic soy agar plate was inoculated into 3 mL of tryptic soy broth and grown overnight with 250‐rpm shaking at 37°C. The overnight culture was diluted 1:100 in 10 mL of tryptic soy broth, incubated with shaking for an additional 16 h and centrifuged at 3700 × g, yielding a culture supernatant that was used to quantify AT production and haemolytic activity.
AT ELISA
AT production was measured by ELISA as described previously.26, 38 Ninety‐six‐well Maxisorp plates (Thermo Fisher Scientific) were coated overnight with anti‐AT monoclonal antibody LC1039 (0.1 μg mL−1) in 0.2 M carbonate/bicarbonate buffer. Plates were then washed with phosphate‐buffered saline (PBS)‐Tween (PBS‐T), blocked for 1 h at room temperature with PBS plus 5% bovine serum albumin (Sigma‐Aldrich, St. Louis, MO) and then incubated after three washes in PBS‐T for 1 h at room temperature, using serial dilutions of supernatants in PBS or purified AT as a reference standard. After washes in PBS‐T, rabbit polyclonal anti‐AT IgG was added at 2 μg mL−1 in PBS for 1 h at room temperature. After washes in PBS‐T, rabbit IgG was detected by addition of goat anti‐rabbit IgG horseradish peroxidase conjugate (Jackson Laboratories, Bar Harbor, ME) diluted at 1:10 000 in PBS. After 1 h of incubation at room temperature, plates were washed in PBS‐T and IgG binding was detected by adding 100 μL of 3,3′,5,5′‐tetramethylbenzidine substrate (KPL, Gaithersburg, MD) per well, followed by 100 μL of 0.2 M H2SO4. Optical density at 450 nm was measured with a spectrophotometer (Molecular Devices, San Jose, CA).
Haemolytic activity assay
Haemolytic activity was measured by haemolytic assay with rabbit red blood cells as described previously.26, 38 A total of 50 μL of washed red blood cells (Pel‐Freez Biologicals, Rogers, AR) was incubated with 50 μL of serially diluted supernatant for 2 h at 37°C in U‐bottomed, 96‐well plates (VWR, Radnor, PA). Plates were centrifuged for 3 min at 930 × g, and 50 μL of supernatant was transferred to a 96‐well, flat‐bottomed plate. One hundred per cent haemolysis was obtained by incubating red blood cells with 0.1% sodium dodecyl sulphate. Optical density at 450 nm was measured with a spectrophotometer (Molecular Devices), and haemolytic activity, expressed as HU per millilitre, was calculated as inverse of the dilution resulting in 50% haemolysis.
Cytofluorimetry
Expression of ClfA and ClfB in S. aureus isolates was measured by cytofluorimetry as described previously.26, 38 S. aureus isolates were grown to mid‐log phase, washed with fluorescence‐activated cell sorting (FACS) buffer (0.1% Tween 20, 0.1% bovine serum albumin in 1 × PBS) and incubated with irrelevant human IgG1 R34738 for 60 min at 4°C to block nonspecific binding of primary antibodies by protein A. Cells were then washed with FACS buffer and incubated with purified anti‐ClfA or anti‐ClfB polyclonal rabbit IgG for 30 min at 4°C, followed by washing with FACS buffer. Finally, the cells were incubated for 30 min at 4°C with goat anti‐rabbit Alexa Fluor 647 conjugate (Abcam, Cambridge, MA). Plates were then washed three times with FACS buffer, and cells were resuspended in 200 μL of FACS buffer. Data were acquired with a FACSCanto II analyzer (Becton Dickinson, Franklin Lakes, NJ). A total of 30,000 events were collected for each sample. The acquired data were analysed with FlowJo software (FlowJo, Ashland, OR).
Statistical analyses
One‐way analysis of variance with heterogeneous variance was applied for comparison of antibody levels in disease cohorts with the uninfected control cohort. Logistic regression was used to evaluate the univariate association of patient outcomes with clinical variables, antibody levels and characteristics of S. aureus isolates. For variables that significantly correlated with patient outcomes in the univariate assessment, a multivariate logistic regression model was applied to identify independent variables to predict patient outcomes. The predictive performance of the multivariate logistic regression model was assessed through a receiver operating characteristic curve and associated area under the curve. The receiver operating characteristic curve was constructed from the probability value of each patient predicted from the multivariate logistic regression model estimated with the remaining N–1 patients, that is, leave‐one‐out cross‐validation, to assess the predictive performance of the model. Spearman correlation coefficient was used to evaluate correlations.
Disclosures
This study was funded by AstraZeneca.
CT, JB, LY, AT, DET, MTE, BRS and AR are or were employees of AstraZeneca/MedImmune at the time of this study with stock ownership, interests and/or options in AstraZeneca. FR has nothing to disclose. VGF served as Chair of the V710 Scientific Advisory Committee (Merck); has received grant support from Basilea, Cerexa/Actavis, Pfizer, Advanced Liquid Logics, the National Institutes of Health (NIH), MedImmune, Cubist/Merck, Karius, Contrafect, Regeneron and Genentech; has NIH Small Business Technology Transfer/Small Business Innovation Research grants pending with Affinergy, Locus, and Medical Surface, Inc; has been a paid consultant for Achaogen, Astellas, Arsanis, Affinergy, Basilea, Bayer, Cerexa, Contrafect, Cubist, Debiopharm, Durata, Grifols, Genentech, MedImmune, Merck, Medicines Co., Pfizer, Novartis, Novadigm, Theravance, xBiotech, Regeneron, and Destiny; has received honoraria from Theravance and Green Cross; and has a patent pending in sepsis diagnostics. BSH and LP have received grant and/or salary support from MedImmune.
Acknowledgments
This article is dedicated to the memory of our deceased colleague Vicky Gunther, who contributed to this study. We acknowledge Anastasia Aksyuk, Jenny Ly and Pankaj Oberoi from Meso Scale Discovery for their guidance and efforts in developing the customised S. aureus multiplex serology assay.
Editorial support was provided by Deborah Shuman of AstraZeneca.
References
- 1. David MZ, Daum RS. Community‐associated methicillin‐resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 2010; 23: 616–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin‐resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis 2007; 13: 1840–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klevens RM, Morrison MA, Nadle J et al Invasive methicillin‐resistant Staphylococcus aureus infections in the United States. JAMA 2007; 298: 1763–1771. [DOI] [PubMed] [Google Scholar]
- 4. Kuehnert MJ, Hill HA, Kupronis BA et al Methicillin‐resistant Staphylococcus aureus hospitalizations, United States. Emerg Infect Dis 2005; 11: 868–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De la Calle C, Morata L, Cobos‐Trigueros N et al Staphylococcus aureus bacteremic pneumonia. Eur J Clin Microbiol Infect Dis 2016; 35: 497–502. [DOI] [PubMed] [Google Scholar]
- 6. Boucher HW, Talbot GH, Bradley JS et al Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48: 1–12. [DOI] [PubMed] [Google Scholar]
- 7. Fowler VG Jr, Boucher HW, Corey GR et al Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus . N Engl J Med 2006; 355: 653–665. [DOI] [PubMed] [Google Scholar]
- 8. Sanchez Garcia M, de la Torre MA, Morales G et al Clinical outbreak of linezolid‐resistant Staphylococcus aureus in an intensive care unit. JAMA 2010; 303: 2260–2264. [DOI] [PubMed] [Google Scholar]
- 9. Thammavongsa V, Kim HK, Missiakas D et al Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 2015; 13: 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jacobsson G, Colque‐Navarro P, Gustafsson E et al Antibody responses in patients with invasive Staphylococcus aureus infections. Eur J Clin Microbiol Infect Dis 2010; 29: 715–725. [DOI] [PubMed] [Google Scholar]
- 11. Adhikari RP, Ajao AO, Aman MJ et al Lower antibody levels to Staphylococcus aureus exotoxins are associated with sepsis in hospitalized adults with invasive S. aureus infections. J Infect Dis 2012; 206: 915–923. [DOI] [PubMed] [Google Scholar]
- 12. Fritz SA, Tiemann KM, Hogan PG et al A serologic correlate of protective immunity against community‐onset Staphylococcus aureus infection. Clin Infect Dis 2013; 56: 1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma‐Kuinkel BK, Wu Y, Tabor DE et al Characterization of alpha‐toxin hla gene variants, alpha‐toxin expression levels, and levels of antibody to alpha‐toxin in hemodialysis and postsurgical patients with Staphylococcus aureus bacteremia. J Clin Microbiol 2015; 53: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y, Liu X, Akhgar A et al Prevalence of IgG and neutralizing antibodies against Staphylococcus aureus alpha‐toxin in healthy human subjects and diverse patient populations. Infect Immun 2018; 86: e00671–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bubeck Wardenburg J, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun 2007; 75: 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foster TJ, Geoghegan JA, Ganesh VK et al Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus . Nat Rev Microbiol 2014; 12: 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alonzo FI, Torres VJ. The bicomponent pore‐forming leucocidins of Staphylococcus aureus . Microbiol Mol Biol Rev 2014; 78: 199–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diep BA, Phung Q, Date S et al Identifying potential therapeutic targets of methicillin‐resistant Staphylococcus aureus through in vivo proteomic analysis. J Infect Dis 2014; 209: 1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomsen IP, Dumont AL, James DB et al Children with invasive Staphylococcus aureus disease exhibit a potently neutralizing antibody response to the cytotoxin LukAB. Infect Immun 2014; 82: 1234–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Handke LD, Hawkins JC, Miller AA et al Regulation of Staphylococcus aureus MntC expression and its role in response to oxidative stress. PLoS One 2013; 8: e77874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jenkins A, Diep BA, Mai TT et al Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. MBio 2015; 6: e02272–02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus . Clin Microbiol Rev 2000; 13: 16–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Reilly M, Kreiswirth B, Foster TJ. Cryptic alpha‐toxin gene in toxic shock syndrome and septicaemia strains of Staphylococcus aureus . Mol Microbiol 1990; 4: 1947–1955. [DOI] [PubMed] [Google Scholar]
- 24. DeLeo FR, Kennedy AD, Chen L et al Molecular differentiation of historic phage‐type 80/81 and contemporary epidemic Staphylococcus aureus . Proc Natl Acad Sci USA 2011; 108: 18091–18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nienaber JJ, Sharma Kuinkel BK, Clarke‐Pearson M et al Methicillin‐susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J Infect Dis 2011; 204: 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tabor DE, Yu L, Mok H et al Staphylococcus aureus alpha‐toxin is conserved among diverse hospital respiratory isolates collected from a global surveillance study and is neutralized by monoclonal antibody MEDI4893. Antimicrob Agents Chemother 2016; 60: 5312–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bayer AS, Ramos MD, Menzies BE et al Hyperproduction of alpha‐toxin by Staphylococcus aureus results in paradoxically reduced virulence in experimental endocarditis: a host defense role for platelet microbicidal proteins. Infect Immun 1997; 65: 4652–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patti JM, Allen BL, McGavin MJ et al MSCRAMM‐mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 1994; 48: 585–617. [DOI] [PubMed] [Google Scholar]
- 29. Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus . Trends Microbiol 1998; 6: 484–488. [DOI] [PubMed] [Google Scholar]
- 30. Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus . Science 1995; 268: 103–106. [DOI] [PubMed] [Google Scholar]
- 31. Dumont AL, Nygaard TK, Watkins RL et al Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol 2011; 79: 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DuMont AL, Yoong P, Surewaard BG et al Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect Immun 2013; 81: 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Janesch P, Rouha H, Weber S et al Selective sensitization of human neutrophils to LukGH‐mediated cytotoxicity by Staphylococcus aureus and IL‐8. J Infect 2017; 74: 473–483. [DOI] [PubMed] [Google Scholar]
- 34. Inouye M, Dashnow H, Raven LA et al SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 2014; 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bankevich A, Nurk S, Antipov D et al SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J Comput Biol 2012; 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015; 31: 2877–2878. [DOI] [PubMed] [Google Scholar]
- 37. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 2016; 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tkaczyk C, Hua L, Varkey R et al Identification of anti‐alpha toxin monoclonal antibodies that reduce the severity of Staphylococcus aureus dermonecrosis and exhibit a correlation between affinity and potency. Clin Vaccine Immunol 2012; 19: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hua L, Hilliard JJ, Shi Y et al Assessment of an anti‐alpha‐toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus‐induced pneumonia. Antimicrob Agents Chemother 2014; 58: 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]


