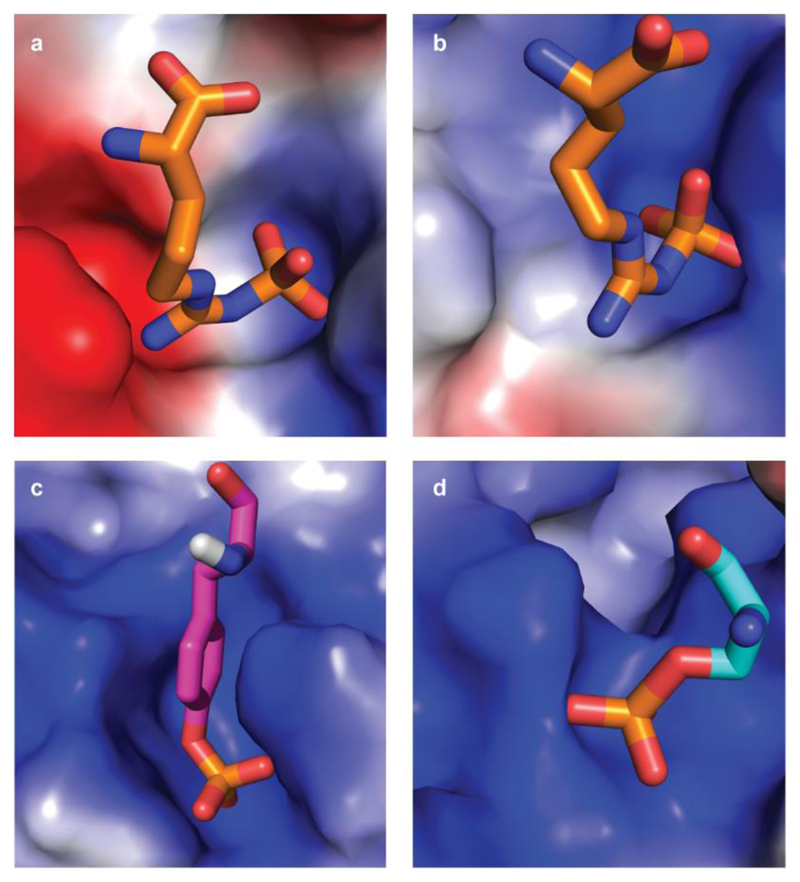
Extended Data Figure 5. Binding pockets of pArg, pTyr and pSer/Thr.
The binding pockets are shown as surface representation and coloured according to their electrostatics as calculated with PyMol (blue: positive, red: negative). Bound phosphoamino acids are presented as sticks with nitrogens and oxygens coloured blue and red, respectively. a, b, pArg-binding sites 1 and 2, respectively, of the ClpCNTD domain. The sites are characterized by a ‘bipolar’ architecture with both a positive and a negative area, jointly required to recognize a pArg side chain. c, d, pTyr-binding site of the Src SH2 domain (PDB code 1SPS; ref. 59) and pSer/Thr-binding site of the 14-3-3 domain (PDB code 1QJB; ref. 60). Both pTyr and pSer were part of a peptide but are shown in isolation for clarity. In contrast to the pArg-binding site, pTyr- and pSer/Thr-specific pockets are uniformly positively charged.