Abstract
In biological cells, chemical processes often occur inside specialized subcellular compartments or organelles. For instance, in eukaryotes mRNA is transcribed and processed inside the nucleus, exported to the endoplasmic reticulum, and translated into the encoded protein. Inspired by this high degree of intracellular organization, we here develop gel-based artificial organelles that enable sequence-specific and programmable localization of cell-free transcription and translation reactions inside an artificial cellular system. To this end, we utilize agarose microgels covalently modified with DNA templates coding for various functions and encapsulate them into emulsion droplets. We show that RNA signals transcribed from transcription organelles can be specifically targeted to capture organelles via hybridization to the corresponding DNA addresses. We also demonstrate that mRNA molecules, produced from transcription organelles and controlled by toehold switch riboregulators, are only translated in translation organelles containing their cognate DNA triggers. Spatial confinement of transcription and translation in separate organelles is thus superficially similar to gene expression in eukaryotic cells. Combining communicating gel spheres with specialized functions opens up new possibilities for programming artificial cellular systems at the organelle level.
Keywords: Cell-free gene expression, artificial cells, compartments, microgel, organelle
In a variety of efforts to mimic biological self-organization processes occurring in cellular systems, researchers have created artificial cell-scale compartments by encapsulating biochemical machinery into membranous or membrane-free structures.
Important examples of membranous compartments are lipid bilayer vesicles[1], water-in-oil emulsion droplets[2] and proteinosomes[3]. In order to achieve higher order self-organization among these artificial cell-mimicking structures, it is both desirable to separate the processes occurring in them (to create a functional identity), as well as to let them interact and communicate. Hence, communication channels based on small molecule signals have been established to mediate interactions between different types of artificial cells[4] and between artificial and bacterial cells[5]. However, membranous encapsulation typically prohibits the exchange of macromolecules as signal carriers with larger (information) channel capacity.
In this context, membrane-free compartments such as chip-based gene expression systems[6], bead-based systems[7], coacervates[8] or hydrogels[9] have been more successfully utilized to employ systems dynamics involving diffusible nucleic acid signals. Among other applications, such systems were used to engineer biochemical reaction networks that exhibit a variety of spatial patterns and wave-like dynamics [7, 10], or to mimic predatory behavior[11].
In the present work, we develop a protocol for covalent immobilization of gene-length DNA molecules inside of agarose microgel spheres and demonstrate their compatibility with in vitro transcription (IVT) and cell-free protein expression (CFPE) reactions. Encapsulating several of the spheres into water-in-oil emulsion droplets results in artificial cellular systems composed of microgel organelles with dedicated functions and tunable stoichiometry. We show that RNA transcribed from one microgel organelle can be selectively captured by another organelle via hybridization to a sequence-encoded address tag. We also demonstrate spatial separation of transcription and translation processes. To this end, we utilize mRNAs controlled by toehold switch riboregulators[12], whose translation is activated in the presence of specific nucleic acid trigger molecules. The mRNA molecules are sent out from transcription organelles, and then captured by translation organelles equipped with the corresponding DNA trigger sequence. We note that RNA address tagging and spatial organization of gene expression in our in vitro system is superficially similar to mRNA localization in eukaryotic cell organelles.[13].
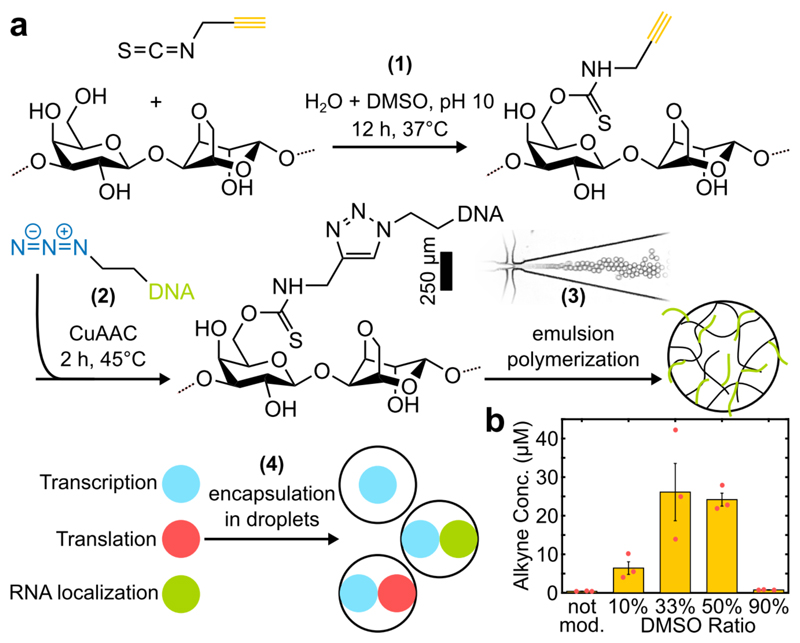
The experimental workflow (Figure 1a, SI Section 1) was adapted from Thiele et al.[9] and consists of chemical functionalization of the hydrogel (step 1) for subsequent covalent modification with PCR amplified DNA (step 2). The resulting DNA hydrogel is first encapsulated in emulsion droplets of diameter d ≈ 30 µm employing microfluidic flow focusing and afterwards gelled (step 3). In step 4, the recovered microgel spheres are added to the reaction mix and loaded into larger emulsion droplets (d ≈ 50-60 µm) for observation via time-lapse Epifluorescence microscopy.
Figure 1.
a) Workflow for production of DNA-functionalized microgel spheres. Agarose is first modified with PITC at mild conditions (1) and subsequently coupled to azide-modified DNA (2). The product is used to generate monodisperse microspheres with different functions via emulsion polymerization using a microfluidic flow-focusing device (3). Different types of microgels are added to the reaction mixture and encapsulated into emulsion droplets (4). b) Agarose functionalization was tested in solvent with varying DMSO content. Error bars represent standard deviations of 3 technical replicates.
We chose agarose as the hydrogel matrix, as it is charge neutral, biocompatible, has a high polymer fiber stiffness and correspondingly large pore sizes of >100 nm[14], which permits the diffusion of large biomolecules such as ribosomes (25-30 nm[15]) for CFPE reactions.
For covalent coupling of DNA to the agarose we chose to rely on copper catalyzed azide-alkyne cycloaddition ‘click chemistry’ due to its high specificity and efficiency[16]. We developed a single-step protocol for functionalization of agarose with alkyne groups at mild aqueous conditions using propargyl-isothiocyanate (PITC). To improve the solubility of PITC, we added varying fractions of DMSO to the solvent (Figure 1b). The degree of substitution, i.e. the amount of terminal alkynes per agarose subunit, was estimated to be ≈ 0.1%, which equals 25 µM of alkynes when dissolved at 1%wt (Figure S1). PITC-agarose was then dissolved in ddH2O, clicked to azide-modified DNA oligos or PCR amplified templates, mixed with dye labelled agarose for barcoding, emulsified and gelled overnight at 4°C. Following this procedure we manufactured different types of microgel spheres with DNA encoded functions such as i) sequence-orthogonal capturing of nucleic acids, ii) in vitro transcription of functional RNA, and iii) cell-free protein expression. To probe these functions, the organelles are added to either a T7 RNA polymerase based IVT mix or the PURExpress® system[17] and encapsulated in emulsion droplets.
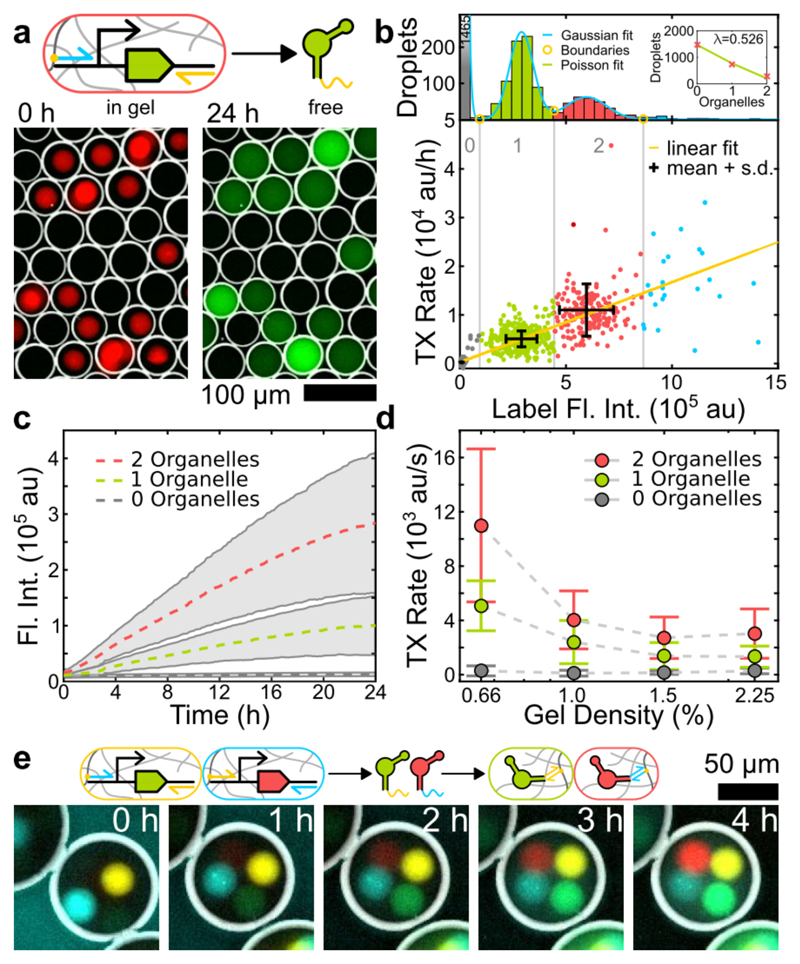
We first studied transcription from gel organelles equipped with DNA templates encoding for the fluorescent dBroccoli (dB) aptamer[18]. Accumulation of the reporter RNA can be observed only in droplets containing organelles (Figure 2a). Image analysis (SI Section 1.8) of droplets with different organelle stoichiometry reveals that the transcription rate depends linearly on organelle content (Figure 2b). The reaction is found to proceed over 24 hours (Figure 2c), which is considerably longer than in typical bulk experiments. As partitioning of T7 RNA polymerase into the gel is expected to depend on the gel mesh size, we produced organelles with varying gel densities (Figure 2d, Figure S2). Indeed, transcription was faster for lower gel densities, indicating that transcription occurs inside the organelles.
Figure 2.
a) Fluorescence false color, inverted bright field (BF) overlay images of droplets containing 1-3 Cy5 labelled transcription organelles (red) producing dB RNA (green) that distributes in solution. b) Droplets were clustered by fitting a sum of Gaussians to the distribution of the label intensity (top). The number of organelles per droplet roughly follows a Poisson distribution (inset). The rate is proportional to the number of sender spheres. c) Average time traces showing the total dB fluorescence corresponding to the populations in b). d) Transcription rate increases with decreasing gel density. Data shown in b) and c) corresponds to 0.66% gel density. All error bars and shaded areas represent standard deviations. e) Fluorescence overlay time series showing the transcription of RNA from dB (yellow) and MG (cyan) transcription organelles and orthogonal RNA localization in dB (green) and MG (red) capture organelles.
For the realization of spatially distributed reaction networks, it is desirable to target the RNA products to different locations in a programmable manner. To this end, we extended the aptamers with a 20 bp long linker sequence that is complementary to a single stranded DNA capture sequence immobilized in specialized receiver organelles. In Figure 2e we tested the simultaneous transcription of the fluorescent dB and the Malachite Green (MG) aptamer[19] from separate organelles and their subsequent sorting into dedicated ‘capture organelles’. As designed, the localization of reporter RNA species systematically depends on the stoichiometry of the organelles (Figure S3).
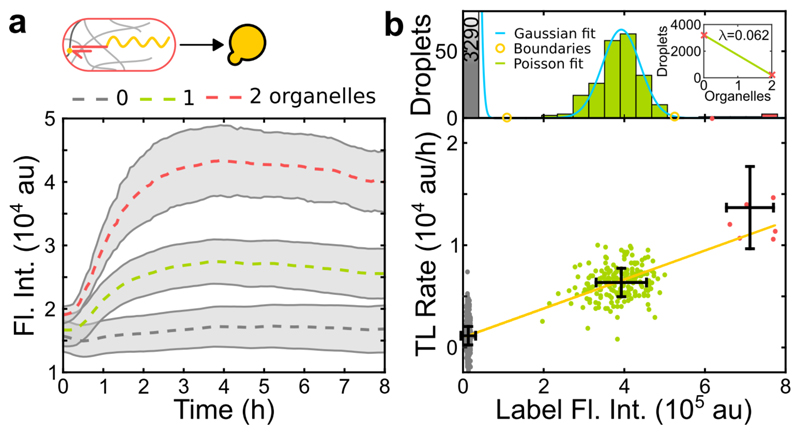
Next, we investigated cell-free protein expression in gel organelles by capturing mRNA molecules encoding mVenus as a reporter protein. To ensure that translation can only occur when the mRNA is localized in the gel, we added a toehold switch riboregulator to the 5’ untranslated region of the mRNA, which suppresses translation in the absence of appropriate RNA or DNA trigger molecules[12]. We immobilized the corresponding trigger DNA in the gel, which served both to localize the mRNA and to activate its translation. In contrast to the transcription reaction, translation activity ceased after 3-4 hours (Figure 3a). This is similar to what is found in bulk expression experiments, which are limited by the finite lifetime of components of the cell-free gene expression system. Clustering of droplets with the same organelle content shows a linear correlation of organelle number and expression level as for the transcription experiments (Figure 3b).
Figure 3.
a) Average time traces showing the total fluorescence in droplets containing a varying number of translation organelles. b) Clustering based on label intensity as in Figure 2. All error bars and shaded areas represent standard deviations.
We also tested the case when the transcription reaction is confined in the gel, but translation occurs in the surrounding solution (Figure S4). Here, the apparent expression rate was ≈ 2-fold higher than in translation organelles, which is consistent with the lower reaction volume of the organelle compared to the surrounding droplet.
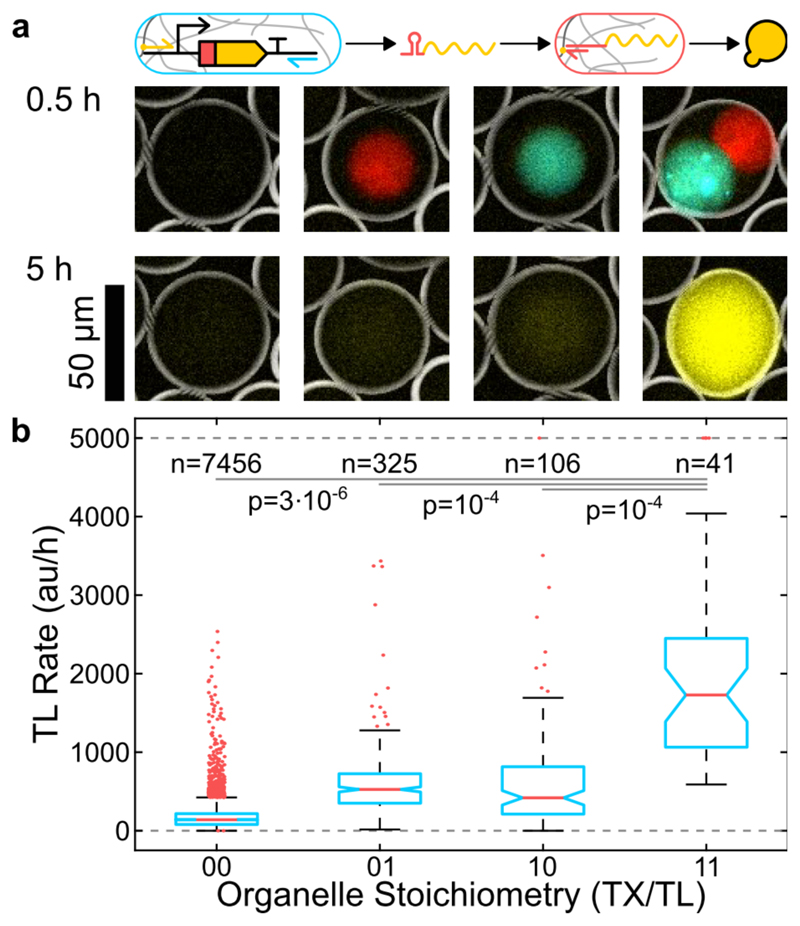
We finally combined transcription and translation organelles in one compartment. As illustrated in Figure 4a, mRNA is first transcribed in the transcription organelle, from which it diffuses into the surrounding solution. Blocked by a toehold switch, it is translationally inactive. Once the mRNA diffuses into a translation organelle it is captured and activated by the trigger DNA, resulting in protein expression. Consistent with this process, we observe a significantly increased reporter signal only in droplets containing both gel organelles (Figure 4b, Figure S5).
Figure 4.
Combination of localized transcription and localized translation. a) Inverted BF, fluorescence overlay images showing transcription (cyan) and translation organelles (red) after 0.5 and the reporter protein (yellow) after 5 hours. b) Boxplots illustrating the reporter expression rate for different organelle content as shown in a). Boxes are 25% and 75% percentiles, notches are 95% confidence intervals, outliers were clipped at 5000 au/h.
The small background signals observed in droplets with only transcription or translation organelles are likely caused by the leakiness of the toehold switch and mRNA transcribed prior to compartmentalization into droplets, respectively. We also note that the variance in the expression rate in compartments with separate transcription and translation organelles is increased compared to the isolated cases. For a discussion of experimental uncertainties see SI Section 2.
Quantitatively, the expression rate is ˜8-fold reduced compared to the case, where mRNA is transcribed in the gel and translated in solution (Figure S4) and ˜4-fold reduced compared to translation in the gel (Figure 3). It has been previously reported that separating the transcription and translation process reduces the efficiency of CFPE reactions.[20] However, already the use of toehold switches decouples the two processes – ribosomes cannot directly bind to the nascent mRNA -, and therefore the reduction in expression rate in our case is probably simply due to the reduced reaction volume.
We have developed hydrogel-based reaction compartments as organelles for artificial cells, which facilitate localization and spatial organization of transcription and translation reactions and also sequence-addressable exchange and sorting of RNA signals or cargoes. Using different compositions of functionally distinct gel-based organelles allows programming of biochemical reactions networks at a higher hierarchical level, i.e., at the level of functional modules[21]. An important aspect of gel-based organelles is their nature as open reaction compartments, which allows exchange of reactants and therefore the sustained maintenance of chemical non-equilibrium conditions. This should be of great interest for the generation of more sophisticated reaction dynamics and spatially extended dynamical systems, possibly supported by 3D bioprinting techniques.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge financial support European Research Council (grant agreement no. 694410 - AEDNA). We thank Elisabeth Falgenhauer for providing the toehold switch construct and Dr. Sandra Sagredo for T7 RNA polymerase.
References
- [1].Noireaux V, Libchaber A. Proceedings of the national academy of sciences of the United States of America. 2004;101:17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tawfik DS, Griffiths AD. Nature biotechnology. 1998;16:652. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- [3].Huang X, Li M, Green DC, Williams DS, Patil AJ, Mann S. Nature Communications. 2013;4 doi: 10.1038/ncomms3239. [DOI] [PubMed] [Google Scholar]
- [4].a Torre P, Keating CD, Mansy SS. Langmuir. 2014;30:5695–5699. doi: 10.1021/la404146g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Adamala KP, Martin-Alarcon DA, Guthrie-Honea KR, Boyden ES. Nature Chemistry. 2016 doi: 10.1038/nchem.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Tang TYD, Cecchi D, Fracasso G, Accardi D, Coutable-Pennarun A, Mansy SS, Perriman AW, Anderson JLR, Mann S. ACS Synthetic Biology. 2017 doi: 10.1021/acssynbio.7b00306. [DOI] [PubMed] [Google Scholar]
- [5].a Schwarz-Schilling M, Aufinger L, Mückl A, Simmel FC. Integr Biol. 2016;8:564–570. doi: 10.1039/c5ib00301f. [DOI] [PubMed] [Google Scholar]; b Lentini R, Martín NY, Forlin M, Belmonte L, Fontana J, Cornella M, Martini L, Tamburini S, Bentley WE, Jousson O, Mansy SS. ACS Central Science. 2017 doi: 10.1021/acscentsci.6b00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a Karzbrun E, Tayar AM, Noireaux V, Bar-Ziv RH. Science. 2014;345:829–832. doi: 10.1126/science.1255550. [DOI] [PubMed] [Google Scholar]; b Pardatscher G, Schwarz-Schilling M, Daube SS, Bar-Ziv RH, Simmel FC. Angewandte Chemie International Edition. 2018;57:4783–4786. doi: 10.1002/anie.201800281. [DOI] [PubMed] [Google Scholar]
- [7].Gines G, Zadorin AS, Galas JC, Fujii T, Estevez-Torres A, Rondelez Y. Nature Nanotechnology. 2017 doi: 10.1038/nnano.2016.299. [DOI] [PubMed] [Google Scholar]
- [8].a Tang TYD, van Swaay D, deMello A, Ross Anderson JL, Mann S. Chem Commun. 2015;51:11429–11432. doi: 10.1039/c5cc04220h. [DOI] [PubMed] [Google Scholar]; b Sokolova E, Spruijt E, Hansen MMK, Dubuc E, Groen J, Chokkalingam V, Piruska A, Heus HA, Huck WTS. Proceedings of the National Academy of Sciences. 2013;110:11692–11697. doi: 10.1073/pnas.1222321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thiele J, Ma Y, Foschepoth D, Hansen MMK, Steffen C, Heus HA, Huck WTS. Lab on a chip. 2014;14:2651–2656. doi: 10.1039/c3lc51427g. [DOI] [PubMed] [Google Scholar]
- [10].a Tayar AM, Karzbrun E, Noireaux V, Bar-Ziv RH. Proceedings of the National Academy of Sciences. 2017;114:11609–11614. doi: 10.1073/pnas.1710620114. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tayar AM, Karzbrun E, Noireaux V, Bar-Ziv RH. Nature Physics. 2015;11:1037–1041. [Google Scholar]; c Zadorin AS, Rondelez Y, Gines G, Dilhas V, Urtel G, Zambrano A, Galas J-C, Estevez-Torres A. Nature Chemistry. 2017 doi: 10.1038/nchem.2770. [DOI] [PubMed] [Google Scholar]
- [11].Qiao Y, Li M, Booth R, Mann S. Nature Chemistry. 2016 doi: 10.1038/nchem.2617. [DOI] [PubMed] [Google Scholar]
- [12].Green Alexander A, Silver Pamela A, Collins James J, Yin P. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a Holt CE, Bullock SL. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Martin KC, Ephrussi A. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pluen A, Netti PA, Jain RK, Berk DA. Biophysical Journal. 1999;77:542–552. doi: 10.1016/S0006-3495(99)76911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gabler R, Westhead EW, Ford NC. Biophysical journal. 1974;14:528. doi: 10.1016/S0006-3495(74)85933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angewandte Chemie. 2002;114:2708–2711. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; b Tornøe CW, Christensen C, Meldal M. The Journal of organic chemistry. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- [17].Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Nature biotechnology. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- [18].Filonov GS, Moon JD, Svensen N, Jaffrey SR. Journal of the American Chemical Society. 2014;136:16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Babendure JR, Adams SR, Tsien RY. Journal of the American Chemical Society. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- [20].Hansen MMK, Ventosa Rosquelles M, Yelleswarapu M, Maas RJM, van Vugt-Jonker AJ, Heus HA, Huck WTS. ACS Synthetic Biology. 2016;5:1433–1440. doi: 10.1021/acssynbio.6b00010. [DOI] [PubMed] [Google Scholar]
- [21].Rollié S, Mangold M, Sundmacher K. Chemical Engineering Science. 2012;69:1–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.