Abstract
Heme is an essential cofactor in metazoans that is also toxic in its free state. Heme is synthesized by most metazoans and must be delivered to all cellular compartments for incorporation into a variety of hemoproteins. The heme biosynthesis enzymes have been proposed to exist in a metabolon, a protein complex consisting of interacting enzymes in a metabolic pathway. Metabolons enhance the function of enzymatic pathways by creating favorable microenvironments for pathway enzymes and intermediates, facilitating substrate transport, and providing a scaffold for interactions with other pathways, signaling molecules, or organelles. Herein we detail growing evidence for a mitochondrial heme metabolon and discuss its implications for the study of heme biosynthesis and cellular heme homeostasis.
Keywords: Heme, Metabolon, Heme Biosynthesis, Porphyria, Anemia
Introduction
Heme is an indispensable cofactor for metazoan life. It consists of a protoporphyrin IX macrocycle with a centrally coordinated iron ion. Heme is utilized in a diverse array of cellular processes including oxygen binding, redox reactions, drug detoxification, and management of reactive oxygen species, as well as regulation of transcription and translation for a variety of genes. The necessity of heme represents an interesting challenge for organisms since not only is heme an essential prosthetic group required in all cellular compartments, but heme, as well as its biosynthetic intermediates, are highly reactive molecules that can be toxic to the cell[1, 2]. Thus, both the production and distribution of heme must be tightly regulated.
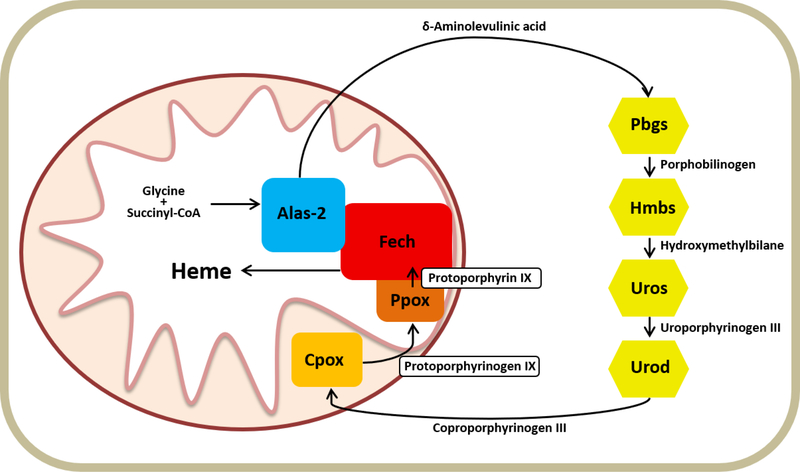
In metazoans, heme is synthesized by a highly conserved pathway. The canonical metazoan heme synthesis pathway consists of eight enzymes and begins in the mitochondrial matrix with the synthesis of 5-aminolevulinic acid (ALA) from succinyl-CoA and glycine catalyzed by the enzyme 5-aminolevulinate synthase (Alas). ALA is then transported to the cytosol where two ALA molecules are combined to form the monopyrrole porphobilinogen (PBG) by porphobilinogen synthase (Pbgs, previously called aminolevulinate dehydratase: Alad). Four PBGs are then cyclized and modified by the next three enzymes in the pathway, hydroxymethylbilane synthase (Hmbs, previously called porphobilinogen deaminase: Pbgd), uroporphyrinogen III synthase (Uros), and uroporphyrinogen III decarboxylase (Urod), to form the tetrapyrrole coproporphyrinogen III. Coproporphyrinogen III is then transported back into the mitochondria for further modification by coproporphyrinogen oxidase (Cpox) and protoporphyrinogen oxidase (Ppox), and finally iron is inserted by ferrochelatase (Fech)(Figure 1). Following its synthesis, heme must be distributed to all cellular compartments, yet details regarding this trafficking as well as incorporation into hemoproteins are greatly lacking. Additionally, mechanisms governing the regulation of heme synthesis remain relatively poorly understood. Disorders impacting heme biosynthesis can have a wide array of downstream consequences due to the requirement for heme in numerous cellular processes. This, coupled with the fact that heme synthesis lies at the convergence of diverse metabolic pathways including overall carbon metabolism, iron acquisition and transport, as well as the aforementioned variety of destinations for heme post production, argues for the necessity of well controlled regulation and facilitation of heme synthesis.
Figure 1. The Mammalian Heme Synthesis Pathway.
This diagram depicts the eight enzymes of the heme biosynthetic pathway and their intermediates as localized in the mitochondria and cytosol.
In metazoans with blood, developing erythrocytes represent the highest demand for heme anywhere in the body due to hemoglobin production[2]. The large amount of heme required, as well as the necessity for rapid production, results in a uniquely high level of heme synthesis in these cells. This presents a distinct set of regulatory challenges relative to other cell types which require and synthesize heme at much lower levels. One means by which this level of regulation occurs is via the first enzyme in the pathway. In mammalian developing erythroid cells, the first step in the pathway is catalyzed by Alas-2, which is encoded on the X-chromosome. Other cells utilize the housekeeping form of Alas, Alas-1. Second to developing erythroid cells, the liver requires high levels of heme synthesis for cytochrome P450 function. It is from these tissues with high levels of heme production that symptoms associated with the misregulation of heme synthesis arise, which include anemias and porphyrias[2] (Table 1).
Table 1.
Heme Synthesis Enzymes and Associated Disorders
| Enzyme | Mutation (Gain/Loss of Function) | Disorder |
|---|---|---|
| Alas-2 | Gain | X-Linked Protoporphyria |
| Loss | X-Linked sideroblastic anemia | |
| Pbgs | Loss | Aminolevulinate dehydratase porphyria |
| Hmbs | Loss | Acute intermittent porphyria |
| Uros | Loss | Congenital erythropoietic porphyria |
| Urod | Loss | Porphyria cutanea tarda |
| Cpox | Loss | Hereditary coproporphyria |
| Ppox | Loss | Variegate porphyria |
| Fech | Loss | Erythropoietic protoporphyria |
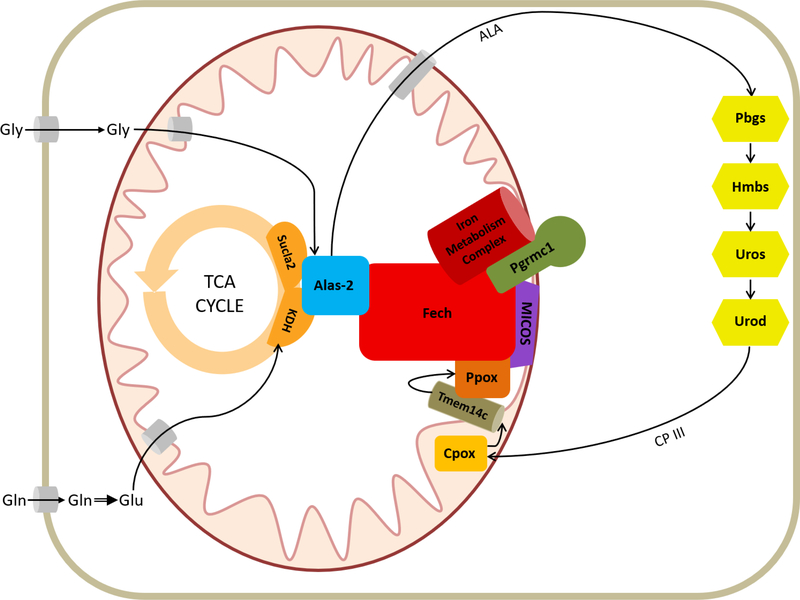
Recently, we presented evidence for a mitochondrial heme synthesis protein complex[3], or metabolon. In this metabolon, several heme synthesis enzymes, mitochondrial transporters, adapter and structural proteins, and intermediary metabolic enzymes involved in upstream substrate synthesis, interact with one another in a complex spanning both mitochondrial membranes (Figure 2). It is clear that regulation of both the synthesis of heme and its trafficking throughout the cell are controlled via proteins within this complex. Herein we discuss the role of the metabolon with regard to heme synthesis and transport and consider the direction of future studies in the field.
Figure 2. The Heme Synthesis Metabolon.
Representation of heme synthesis metabolon as localized in the mitochondria. Iron metabolism proteins found to interact with the metabolon include Abcb7, Abcb10, Mfrn1, and Tfrc. Interacting members of the MICOS include Mic60, Mic27, and Opa1. The cytoplasmic enzymes of the heme synthesis pathway are also shown.
Metabolons and the role of protein interactions in Heme synthesis
A metabolon is defined as a complex of interacting enzymes within a metabolic pathway[4]. Metabolons have been described for a wide range of processes including the tricarboxylic acid (TCA) cycle[5], glycolysis[6], the urea cycle[7], fatty acid synthesis[8], amino acid metabolism[9], and transmembrane transport[10]. Additionally, there is evidence to support the existence of metabolons in various cofactor biosynthesis pathways including those for vitamin B12[11] and coenzyme F430[12], demonstrating a precedent for such complex formation in tetrapyrrole metabolism. The close interactions of pathway proteins can provide a number of advantages for the synthesis of a given product, often through the creation of favorable micro-environments where conditions such as pH, hydrophobicity, reactant and product concentrations, or even the availability of various binding partners and cofactors can be modulated independently from the overall cellular environment. Metabolons can aid in acquisition, movement, and/or delivery of pathway metabolites, thereby increasing flux through the pathway, as well as shielding reactive or unstable intermediates from contact with the cellular environment at large. Metabolons allow for modulation of pathway activity via interactions with regulatory factors/proteins or by providing a means to sense overall cell status. These protein complexes can aid in positioning of pathway components with respect to both proximity to other proteins as well as positioning either within or between organelles.
For heme synthesis, a metabolon offers several advantages. First, many of the pathway intermediates as well as the ultimate product, heme, are chemically reactive when free in the cell[1, 2]. Thus, sequestration of these compounds from the cellular milieu is essential to reduce cytotoxicity of the molecules in question, as well as to protect intermediates from loss to extraneous reactions. Substrate channels or close associations between proteins where intermediates could be channeled directly circumvent this problem. Second, as the pathway progresses, intermediates must cross both mitochondrial membranes and the end product, heme, must be exported from the mitochondrial matrix for delivery to diverse cellular compartments. This movement argues for the presence of proteins and/or channels to facilitate transport. Third, the demand for heme synthesis is dynamic between cell types and dependent on cell status. For example, in liver cells, heme production must be upregulated in response to cytochrome P450 synthesis for xenobiotic detoxification[13]. Likewise, erythroid heme synthesis presents several unique challenges, foremost of which is the dramatically increased demand for heme production[2]. The potential for increased speed and efficiency of heme production conferred by a metabolon further add to the attractiveness of the hypothesis.
The idea of a metabolon for the mitochondrially localized enzymes was first proposed by Grandchamp et al. in 1978[14] and later expanded by others[15, 16]. This idea received support from in silico modeling based upon the crystal structures of Ppox[17] and Fech[18]. Published kinetic data are consistent with some level of substrate channeling, although these studies did not demonstrate the existence of a stable complex of the mitochondrial enzymes[15, 19]. However, available data clearly are consistent with the existence of a dynamic complex involving the terminal enzymes of the pathway. Similar proposals have been made for the formation of a complex involving at least some of the cytosolic enzymes, particularly Hmbs and Uros to prevent the non-productive cyclization of tetrapyrrole isomers which occurs in the absence of Uros [1]. Although complexes of heme synthesis enzymes seem likely in both the cytosol and mitochondria, no direct evidence in metazoa had demonstrated physical interactions between any of the enzymes.
The (Mitochondrial) Heme Synthesis Metabolon
As part of a comprehensive study, Medlock et al. conducted a series of experiments designed to elucidate proposed protein-protein interactions among heme biosynthesis enzymes and other peripheral enzymes such as mitochondrial transporters and potential regulatory/adapter proteins[3]. In these experiments, FLAG-tagged human heme synthesis enzymes were expressed in a murine erythroid model cell line. Cells were fractionated and affinity purification was carried out with the cytosolic and mitochondrial fractions. Proteins interacting with the tagged enzyme were identified by Multidimensional Protein Identification Technology mass spectrometry (MudPIT-MS)[20]. There were no interactions detected between the cytosolic enzymes; Pbgs, Hmbs, Uros, and Urod; nor was a common protein with which all of these enzymes might interact identified. Previously, potential proteinprotein interactions of Uros via nuclear magnetic resonance (NMR) experiments on purified protein were carried out and did not show any interaction between Uros and either Hmbs or Urod[21]. Together these results do strongly argue against direct protein-protein interactions between the cytosolic heme synthesis enzymes, though the lack of demonstrated interactions in these experiments does not necessarily preclude the existence of a cytosolic complex indirectly linked by cytoskeletal structures. Additional experiments such as proximity labeling may help to shed light on these possible indirect and/or transient interactions.
In contrast to the cytosolic enzymes, interactions were detected between several of the mitochondrial heme synthesis enzymes. Affinity purification of FLAG tagged Fech and reciprocal experiments with FLAG tagged Ppox were consistent with each protein interacting with the other. In addition, both untagged endogenous Fech and Ppox were found to be part of larger molecular weight complexes present in mitochondrial preparations of wild type cells[3]. A surprising finding was that the erythroid-specific form of the first enzyme in the pathway, Alas-2, interacts with the both Fech and Ppox[3]. It is interesting to speculate about the function this interaction may have in regulating production of ALA and subsequent intermediates. In the context of the most recently identified porphyria (Table 1), X-linked protoporphyria (XLP), which results from increased activity of Alas-2[22], determining whether XLP variants of the enzyme retain this interaction will be important in understanding the pathophysiology of this disease.
As well as enzymes with a known role in heme biosynthesis, many additional proteins were found to interact with the mitochondrial heme metabolon, including proteins involved in succinyl-CoA synthesis, iron metabolism, and mitochondrial metabolite transport (Table 2). The presences of proteins involved in iron metabolism in affinity purification data[3] indicates that the heme synthesis metabolon may have a role in regulating cellular iron homeostasis or vice versa. Metabolon interaction partners involved in iron transport include Abcb7, a mitochondrial transporter which facilitates cytosolic Fe-S cluster synthesis[23]; Abcb10, an ATP-binding cassette transporter shown to stabilize the iron importer mitoferrin(Mfrn1)[24, 25]; and transferrin receptor, a protein important for cellular iron import and potentially direct delivery of iron to the mitochondria via a kiss and run mechanism[26–28]. Also found in the metabolon is the protoporphyrinogen transporter Tmem14c. This protein is responsible for the transport of protoporphyrinogen IX into the mitochondrial matrix following conversion of coproporphyrinogen III to protoporphyrinogen IX by Cpox in the mitochondrial inner membrane space. Disruption of Tmem14c in mice results in the accumulation of porphyrins in the fetal liver, arrest of erythroid maturation and profound anemia[29]. Proper facilitation and coordination of iron delivery and porphyrin transport within the heme synthesis pathway is necessary for not only heme synthesis itself, but also for preventing detrimental accumulation of un/mismetallated porphyrins.
Table 2.
Components of the Mitochondrial Heme Synthesis Metabolon
| Protein | Function |
|---|---|
| Fech | Conversion of Protoporphyrin IX to Heme (Iron insertion) |
| Ppox | Conversion of Protoporphyrinogen IX to Protoporphyrin IX |
| Cpox | Conversion of Coproporphyrinogen III to Protoporphyrinogen IX |
| Alas-2 | 5-aminolevulinate synthesis |
| Sucla2 | Subunit of succinyl-CoA synthetase |
| Ogdh | Subunit of α-ketoglutarate dehydrogenase complex |
| Pgrmc1 | Heme binding |
| Pgrmc2 | Heme binding |
| Mic60 | MICOS complex subunit |
| Mic27 | MICOS complex subunit |
| Opa1 | Mitochondrial fusion and MICOS complex |
| Ymel1 | Opa1 cleavage |
| Ant1 | Membrane transport |
| Ant2 | Membrane transport |
| Abcb7 | Fe-S cluster synthesis |
| Abcb10 | Membrane transport and stabilizes mitoferrin |
| Glrx5 | Fe-S cluster synthesis |
| Tfrc | Iron import |
| Tmem14c | Mitochondrial Protoporphyrinogen IX import |
| Aralar1 | Mitochondrial membrane transport |
| Mfrn1 | Mitochondrial iron import |
The interaction between Alas-2 and Ppox/Fech suggests a wider level of communication/coordination among all mitochondrially-located heme synthesis enzymes. Additionally relevant to the integrated regulation of both ends of the pathway is the interaction found between the metabolon enzymes and enzymes responsible for the synthesis of the ALA precursor succinyl-CoA. The interaction between Sucla2, the ATP-utilizing subunit of succinyl CoA synthetase (SCS), with Alas-2[30], Fech, and Ppox[3] provides an intriguing link between the TCA cycle and the heme synthesis metabolon. It had long been presumed that succinyl-CoA used in heme synthesis is provided by the TCA cycle via the ATP-driven reverse Sucla2-SCS reaction. Recent work by Burch et. al[31] demonstrated that the main carbon source for ALA in developing erythroid cells is instead glutamine. Glutamine destined for ALA synthesis must first be converted to α-ketoglutarate, then to succinyl-CoA by the α-ketoglutarate dehydrogenase complex (KDH). They proposed that the observed interaction between the Sucla2 subunit of SCS and heme synthesis proteins may function to stabilize the apoproteins of Alas-2 and Fech prior to cofactor assembly or serve to sequester Sucla2 to prevent the ATP-utilizing reverse SCS reaction. Interestingly, subunits of KDH were also found to interact with the metabolon in developing erythroid cells[31].
It is becoming evident that one function of the heme synthesis metabolon is to mediate carbon flux in concert with the TCA cycle for the purpose of initial substrate synthesis. It should be noted that the interactions described for Sucla2 and KDH are with Alas-2, the erythroid specific form of ALA synthase. Additionally, the interaction with Sucla2 has been previously shown to only occur with Alas-2 and not Alas-1[30, 32]; however, it is unclear whether any interaction for KDH occurs in non-erythroid cells with Alas-1. It is tempting to speculate that mechanisms of regulation and possibly carbon source may be distinct between erythroid and nonerythroid heme synthesis. It stands to reason that, given the enormous demand for heme production during erythroid differentiation, it would be inefficient and energetically expensive to produce succinyl-CoA through the ATP dependent SCS path. Furthermore, withdrawing the large amount of succinate required for heme synthesis in developing erythroid cells from the TCA cycle could have detrimental effects on cell metabolism as a whole. The use of glutamine as a substrate avoids these problems as it appears that KDH may serve a moonlighting role in its interactions with Alas-2 during late erythropoiesis to facilitate succinyl-CoA synthesis for highly upregulated heme synthesis independent of TCA cycle intermediate pools. This circumvention may not be necessary for the drastically lower levels of heme synthesis present in non-erythroid cells. Continued characterization of the heme synthesis metabolon and its interplay with the TCA cycle in non-erythroid cells will be necessary to answer these intriguing questions.
Novel Heme Synthesis Metabolon Proteins
As noted, one role of the mitochondrial heme metabolon is the downstream distribution of heme post synthesis. Heme is both essential in all compartments and can be highly cytotoxic if not properly contained[1, 2]; thus, well regulated, timely, and efficient trafficking is a necessity. Intriguingly, Opa1, a GTPase required for mitochondrial fusion[33], was present in the mitochondrial co-immunoprecipitation studies performed by Piel et al.[34]. In mammalian cells, Opa1 is not only required for fusion but has also been identified as part of the mitochondrial contact site and cristae organizing system (MICOS)[35, 36], a protein complex responsible for anchoring junction points between the inner and outer mitochondrial membranes and stabilizing cristae structure[37]. There have been several proposed mechanisms for heme trafficking that involve mitochondrial fusion/fission, direct inter-organelle contacts, or mitochondrially derived vesicle transport[38]. Such mechanisms would allow for heme to be distributed in bulk throughout the cell while remaining sequestered from the larger cellular environment. Both Mic60 (mitofilin, Immt) and Mic27 (Apool), also members of the MICOS complex[39–41], were likewise found to interact with the heme synthesis metabolon[34]. These data suggest a direct interaction of the heme synthesis metabolon and the MICOS complex to localize the metabolon for efficient import of porphyrinogens and iron and export of heme. Further studies to understand the role of mitochondrial structure and dynamics in porphyrin and heme trafficking will shed light on to the role of these in heme synthesis.
In addition to enzymes with well-characterized functions, several proteins with no previously described role in heme synthesis were found to be present in the metabolon. One such protein was progesterone receptor membrane component 1 (Pgrmc1)[34], a heme binding protein that has been reported in a variety of cellular compartments and cell types. Pgrmc1 has many reported functions including binding and activation of cytochrome P450s[42–45], promotion of autophagy[46] and endocytosis[47], and regulation of iron metabolism in concert with hepcidin[48]. Pgrmc1 was also found to be enriched at ER-mitochondria contact sites[49]. In the context of heme synthesis, recent studies have demonstrated that Pgrmc1 can inhibit Fech activity in vitro and that treatment of a mammalian erythroid cell model with AG-205, a small molecule which perturbs heme binding in Pgrmc1, results in decreased hemoglobin production during differentiation[34]. At present, the mechanism by which Pgrmc1 influences heme synthesis is unclear. One possibility is that Pgrmc1 functions as a heme chaperone and influences flux through the synthesis pathway by facilitating release of heme from its interaction partner, Fech. Adding to the credibility of this model is the fact that, when tested against several conformation-specific variants of Fech, Pgrmc1 interacts most strongly with the variant in the release/heme bound conformation[34]. An alternative interpretation of this finding is that Pgrmc1 interacts differently with different conformations of Fech for the purpose of regulating enzyme activity through stabilization of particular conformations. Recently, Pgrmc1 has been shown to exhibit heme dependent dimerization, with dimerization altering binding affinity for other proteins[50]. It is conceivable that the effect of Pgrmc1 on Fech activity may be further mediated by cellular heme status, as detected by Pgrmc1’s ability to bind heme with moderate affinity[34, 51]. Notably, Pgrmc1 was also shown to interact with Mic60, Mic27, and Opa1[34], which are involved in mitochondrial structure and dynamics as discussed previously. The presence of these interactions hint at a function for Pgrmc1 beyond its interaction with Fech alone. A homologue of Pgrmc1, Pgrmc2 was also found in the heme synthesis metabolon[34], though its role has been less well described. While the exact functions and mechanisms of these proteins remain elusive at present, they serve as intriguing evidence that examining heme synthesis and regulation from a metabolon-centric point of view has the potential to reveal previously undescribed mechanisms of regulation for heme synthesis and its coordination with other aspects of cell metabolism.
Dynamic and Transient Nature of the Complex
The mitochondrial heme synthesis metabolon is almost certainly a highly dynamic entity, with protein-protein interactions forming and dissociating depending on a number of factors including cell type, stage in development, induction of heme synthesis, response to various cell states and signaling pathways, as well as conformational changes in the enzymes themselves as they progress through their catalytic cycles. The most immediately apparent evidence in support of a dynamic metabolon is the sheer number of interaction partners documented for each protein examined[3]. For some protein types such as transporters and adapters, this finding could be rationalized by claiming that a protein species may fill several distinct niches simultaneously, each with its own set of interactions, resulting in diverse binding partners in coimmunoprecipitation assays. However, this argument is less compelling for enzymes with a more singular function such as Fech or Ppox. A more generally plausible explanation is that a given protein will interact with many different partners depending on the current demands of the cell or progress through the enzyme’s own catalytic cycle. Many of the relevant assays, including coimmunoprecipitation experiments, are, by necessity, conducted on heterogeneous populations of cells and mitochondria. Though they are of the same cell type, individual cells will exist at varying stages of differentiation. Additionally, individual mitochondria will exist in at least slightly disparate conditions, and the individual enzymes will certainly not be synchronized in their activity. This results in data representing interactions from various cellular and enzymatic states and stages. The relative amounts of a particular protein-protein interaction found will thus represent not only the strength of the interaction, but also relative abundance or temporal persistence of any given state. This view of a dynamic metabolon is further supported by the fact that the interaction between Fech and Pgrmc1 has been shown to be conformation dependent[34]. Since Fech assumes several distinct conformations over the course of its catalytic cycle[52–54], this suggests that the Fech-Pgrmc1 interaction is broken and reformed repeatedly during heme synthesis. This is likely the case for other interactions as well. Given the variety of downstream destinations for heme and the variety of cellular signals heme synthesis must respond to, a dynamic heme synthesis metabolon exhibiting many transient interactions seems likely.
As noted with respect to the cytosolic heme synthesis enzymes, the lack of interaction partners in co-immunoprecipitation assays is not sufficient to rule out all types of interactions or complexes. It is likely that a portion of interactions in a complex such as this will be either very transient, or too low affinity to persist through immunoprecipitation conditions. It is also likely that some interactions will be indirect; mediated by either adapter proteins, membrane lipids, or cytoskeletal components. To further investigate weak, transient, or indirect interactions, conditions within the immunoprecipitation assays can be further modulated, i.e. affinity tag type, detergent concentrations, crosslinking, etc. or alternative methods can be attempted. Attractive alternate methods include those in which interactions are characterized by proximity of the components rather than direct interaction, including proximity labeling, proximity ligation assays or bi-molecular fluorescence assays, though the later require a putative interaction to be identified prior to testing.
Conclusion
While originally proposed four decades ago[14], it is only recently that substantial data have been presented that are consistent with the existence of a transient complex centered around the mitochondrial heme biosynthesis enzymes. This metabolon appears to play a role in the facilitation and regulation of heme synthesis via modulation of enzyme activity, metabolite transport, and pathway response to cell status and other signals. Further studies to elucidate the nature and individual functions of these interactions will help to broaden our understanding of the porphyrias, anemias, and other heme related disease states. The identification and characterization of the mitochondrial heme synthesis metabolon provides a new platform from which to examine the essential and multifaceted processes of heme synthesis and distribution.
Acknowledgements
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants DK111653 (A.E.M.), DK096501 (H.A.D.) and support from the U54 DK110858.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hamza I and Dailey HA, One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta, 2012. 1823(9): p. 1617–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dailey HA and Meissner PN, Erythroid heme biosynthesis and its disorders. Cold Spring Harb Perspect Med, 2013. 3(4): p. a011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medlock AE, et al. , Identification of the Mitochondrial Heme Metabolism Complex. PLoS One, 2015. 10(8): p. e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srere PA, The Metabolon. Trends in Biochemical Sciences, 1985. 10(3): p. 109–110. [Google Scholar]
- 5.Robinson JB Jr. and Srere PA, Organization of Krebs tricarboxylic acid cycle enzymes in mitochondria. J Biol Chem, 1985. 260(19): p. 10800–5. [PubMed] [Google Scholar]
- 6.Campanella ME, Chu H, and Low PS, Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc Natl Acad Sci U S A, 2005. 102(7): p. 2402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung CW, Cohen NS, and Raijman L, Channeling of urea cycle intermediates in situ in permeabilized hepatocytes. J Biol Chem, 1989. 264(7): p. 4038–44. [PubMed] [Google Scholar]
- 8.Srere PA and Sumegi B, Processivity and fatty acid oxidation. Biochem Soc Trans, 1994. 22(2): p. 446–50. [DOI] [PubMed] [Google Scholar]
- 9.Hyde CC, et al. , Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem, 1988. 263(33): p. 17857–71. [PubMed] [Google Scholar]
- 10.Moraes TF and Reithmeier RA, Membrane transport metabolons. Biochim Biophys Acta, 2012. 1818(11): p. 2687–706. [DOI] [PubMed] [Google Scholar]
- 11.Deery E, et al. , An enzyme-trap approach allows isolation of intermediates in cobalamin biosynthesis. Nat Chem Biol, 2012. 8(11): p. 933–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng K, et al. , The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science, 2016. 354(6310): p. 339–342. [DOI] [PubMed] [Google Scholar]
- 13.Correia MA, Sinclair PR, and De Matteis F, Cytochrome P450 regulation: the interplay between its heme and apoprotein moieties in synthesis, assembly, repair, and disposal. Drug Metab Rev, 2011. 43(1): p. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grandchamp B, Phung N, and Nordmann Y, The mitochondrial localization of coproporphyrinogen III oxidase. Biochem J, 1978. 176(1): p. 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira GC, et al. , Organization of the terminal two enzymes of the heme biosynthetic pathway. Orientation of protoporphyrinogen oxidase and evidence for a membrane complex. J Biol Chem, 1988. 263(8): p. 3835–9. [PubMed] [Google Scholar]
- 16.Dailey HA, Dailey TA, in The Porphyrin Handbook, S.K.M., Kadish KM, Guilard R, Editor. 2003, Academic: New York: p. 93–121. [Google Scholar]
- 17.Koch M, et al. , Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. EMBO J, 2004. 23(8): p. 1720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu CK, et al. , The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol, 2001. 8(2): p. 156–60. [DOI] [PubMed] [Google Scholar]
- 19.Proulx KL, Woodard SI, and Dailey HA, In situ conversion of coproporphyrinogen to heme by murine mitochondria: terminal steps of the heme biosynthetic pathway. Protein Sci, 1993. 2(7): p. 1092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schirmer EC, Yates JR 3rd, and Gerace L, MudPIT: A powerful proteomics tool for discovery. Discov Med, 2003. 3(18): p. 38–9. [PubMed] [Google Scholar]
- 21.Cunha L, et al. , Human uroporphyrinogen III synthase: NMR-based mapping of the active site. Proteins, 2008. 71(2): p. 855–73. [DOI] [PubMed] [Google Scholar]
- 22.Whatley SD, et al. , C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload. Am J Hum Genet, 2008. 83(3): p. 408–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bekri S, et al. , Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic ironsulfur protein maturation. Blood, 2000. 96(9): p. 3256–64. [PubMed] [Google Scholar]
- 24.Chen W, et al. , Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc Natl Acad Sci U S A, 2009. 106(38): p. 16263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W, Dailey HA, and Paw BH, Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood, 2010. 116(4): p. 628–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheftel AD, et al. , Direct interorganellar transfer of iron from endosome to mitochondrion. Blood, 2007. 110(1): p. 125–32. [DOI] [PubMed] [Google Scholar]
- 27.Richardson DR, Ponka P, and Vyoral D, Distribution of iron in reticulocytes after inhibition of heme synthesis with succinylacetone: examination of the intermediates involved in iron metabolism. Blood, 1996. 87(8): p. 3477–88. [PubMed] [Google Scholar]
- 28.Zhang AS, Sheftel AD, and Ponka P, Intracellular kinetics of iron in reticulocytes: evidence for endosome involvement in iron targeting to mitochondria. Blood, 2005. 105(1): p. 368–75. [DOI] [PubMed] [Google Scholar]
- 29.Yien YY, et al. , TMEM14C is required for erythroid mitochondrial heme metabolism. J Clin Invest, 2014. 124(10): p. 4294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuyama K and Sassa S, Interaction between succinyl CoA synthetase and the heme-biosynthetic enzyme ALAS-E is disrupted in sideroblastic anemia. J Clin Invest, 2000. 105(6): p. 757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burch JS, Marcero JR, Maschek JA, Cox JE, Jackson LK, Medlock AE, Phillips JD, Dailey HA Jr., Glutamine via α-ketoglutarate dehydrogenase provides succinyl-CoA for heme synthesis during erythropoiesis. Blood, 2018. 132(10): p. 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labbe RF, Kurumada T, and Onisawa J, The role of succinyl-CoA synthetase in the control of heme biosynthesis. Biochim Biophys Acta, 1965. 111(2): p. 403–15. [DOI] [PubMed] [Google Scholar]
- 33.Cipolat S, et al. , OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A, 2004. 101(45): p. 15927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piel RB 3rd, et al. , A Novel Role for Progesterone Receptor Membrane Component 1 (PGRMC1): A Partner and Regulator of Ferrochelatase. Biochemistry, 2016. 55(37): p. 5204–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olichon A, et al. , Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem, 2003. 278(10): p. 7743–6. [DOI] [PubMed] [Google Scholar]
- 36.Frezza C, et al. , OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell, 2006. 126(1): p. 177–89. [DOI] [PubMed] [Google Scholar]
- 37.Kozjak-Pavlovic V, The MICOS complex of human mitochondria. Cell Tissue Res, 2017. 367(1): p. 83–93. [DOI] [PubMed] [Google Scholar]
- 38.Reddi AR and Hamza I, Heme Mobilization in Animals: A Metallolipid’s Journey. Acc Chem Res, 2016. 49(6): p. 1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.John GB, et al. , The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell, 2005. 16(3): p. 1543–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie J, et al. , The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett, 2007. 581(18): p. 3545–9. [DOI] [PubMed] [Google Scholar]
- 41.Weber TA, et al. , APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS One, 2013. 8(5): p. e63683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oda S, et al. , Progesterone receptor membrane component 1 modulates human cytochrome p450 activities in an isoform-dependent manner. Drug Metab Dispos, 2011. 39(11): p. 2057–65. [DOI] [PubMed] [Google Scholar]
- 43.Szczesna-Skorupa E and Kemper B, Progesterone receptor membrane component 1 inhibits the activity of drug-metabolizing cytochromes P450 and binds to cytochrome P450 reductase. Mol Pharmacol, 2011. 79(3): p. 340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes AL, et al. , Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab, 2007. 5(2): p. 143–9. [DOI] [PubMed] [Google Scholar]
- 45.Mallory JC, et al. , Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol Cell Biol, 2005. 25(5): p. 1669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mir SU, et al. , Progesterone receptor membrane component 1/Sigma-2 receptor associates with MAP1LC3B and promotes autophagy. Autophagy, 2013. 9(10): p. 1566–78. [DOI] [PubMed] [Google Scholar]
- 47.Hand RA, et al. , Saccharomyces cerevisiae Dap1p, a novel DNA damage response protein related to the mammalian membrane-associated progesterone receptor. Eukaryot Cell, 2003. 2(2): p. 306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, et al. , Progesterone receptor membrane component-1 regulates hepcidin biosynthesis. J Clin Invest, 2016. 126(1): p. 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho IT, et al. , Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum-mitochondrial contacts. J Biol Chem, 2017. 292(39): p. 16382–16392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabe Y, et al. , Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat Commun, 2016. 7: p. 11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh K, et al. , Spectroscopic and biochemical characterization of heme binding to yeast Dap1p and mouse PGRMC1p. Biochemistry, 2005. 44(50): p. 16729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medlock A, et al. , Substrate interactions with human ferrochelatase. Proc Natl Acad Sci U S A, 2007. 104(6): p. 1789–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Medlock AE, et al. , A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase. J Mol Biol, 2007. 373(4): p. 1006–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medlock AE, et al. , Product release rather than chelation determines metal specificity for ferrochelatase. J Mol Biol, 2009. 393(2): p. 308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]