Summary
Gasdermin D (GSDMD) is an effector molecule for pyroptosis downstream of canonical and noncanonical inflammasome signaling pathways. Cleavage of GSDMD by inflammatory caspases trigger the oligomerization and lipid-binding by its N-terminal domain that assembles membrane pores, whereas its C-terminal domain binds the N-terminal domain to inhibit pyroptosis. Despite recent progress in our understanding of the structure and function of the murine gasdermin A3 (mGSDMA3), the molecular mechanisms of GSDMD activation and regulation remain poorly characterized. Here we report the crystal structures of the full-length murine and human GSDMDs, which reveal architecture of the GSDMD N-terminal domains and demonstrate distinct and common features of autoinhibition among gasdermin family members utilizing their β1-β2 loops. Disruption of the intramolecular domain interface enhanced pyroptosis, whereas mutations at the predicted lipid-binding or oligomerization surface reduced cytolysis. Our study provides a framework for understanding the autoinhibition, lipid-binding and oligomerization of GSDMD using overlapping interfaces.
GRAPHICAL ABSTRACT
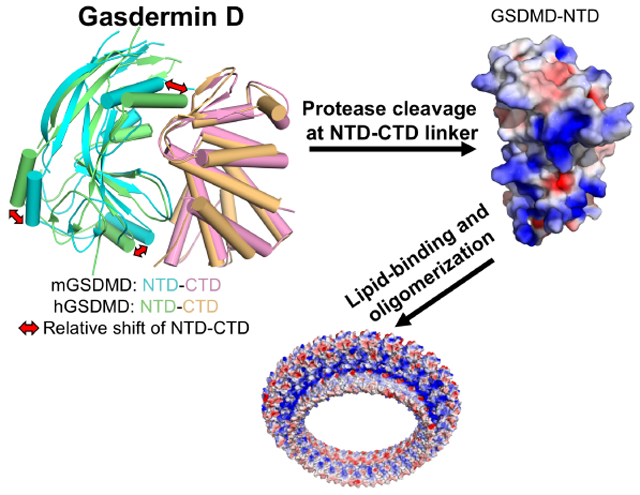
eTOC Blurb
The molecular mechanisms for activation and regulation of gasdermin D (GSDMD) are poorly characterized. Liu et al. report the crystal structures of the full-length murine and human GSDMDs, which reveal distinct and common features of autoinhibition among gasdermin family members, as well as overlapping surface for lipid-binding and oligomerization.
INTRODUCTION
Pyroptosis is an inflammatory form of programmed cell death that plays important roles in innate immune defense against microbial infections and in tissue damage from excessive inflammation (Cookson and Brennan, 2001; Latz et al., 2013; Miao et al., 2010). Gasdermin D (GSDMD) is an executor of pyroptosis downstream of canonical and noncanonical inflammasome pathways and is a substrate for inflammatory caspases-1,4, 5, and 11 (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015). It has also been reported to induce cell death upon cleavage by caspase-8 in a receptor9-interacting serine/threonine kinase 1 (RIPK1)-dependent manner (Chen et al., 2019; Orning et al., 2018; Sarhan et al., 2018), or by a neutrophil-specific serine protease (Kambara et al., 2018). The gasdermin family members contain N-terminal domains (NTDs) that are capable of forming membrane pores to induce cytolysis, whereas their C-terminal domains (CTDs) function as inhibitors of cell killing through intramolecular domain association (Aglietti et al., 2016; Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Rogers et al., 2017; Sborgi et al., 2016; Wang et al., 2017). It was proposed that upon protease cleavage of the linker region between gasdermin NTDs and CTDs, the disruption of the intramolecular domain interaction in the presence of lipids releases the NTDs to assemble oligomeric membrane pores that trigger cell death. This is consistent with the extensive intramolecular domain interactions between the NTD and CTD from the murine GSDMA3 (mGSDMA3) (Ding et al., 2016), and the recent structural studies of the murine and human GSDMD-CTDs (Liu et al., 2018, Kuang et al., 2017). Furthermore, cryo-electron microscopy (cryo-EM) study of the mGSDMA3 pore suggests large conformational changes in its NTD upon assembly of the oligomeric pore, with two membrane-spanning β-hairpins that are partially disordered in the autoinhibited structure (Ruan et al., 2018). Although the full-length mGSDMA3 and GSDMD possess minimal lipid-binding activities (Aglietti et al., 2016; Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Rogers et al., 2017; Sborgi et al., 2016), the full-length human GSDMB (hGSDMB) was reported to bind phospholipids (Chao et al., 2017), which suggests different regulatory mechanisms among gasdermin family members. To shed light on autoinhibition of GSDMD, its lipid-binding and oligomerization activities, we initiated structural studies of the full-length murine and human GSDMDs. Our work reveals architecture of the GSDMD N-terminal domains and demonstrate distinct and common features of autoinhibition among gasdermin family members utilizing their β 1-β2 loops. Disruption of the intramolecular domain interface enhanced pyroptosis, whereas mutations at the predicted lipid-binding or oligomerization surface reduced cytolysis. This study thus provides a molecular basis for understanding the mechanisms of GSDMD autoinhibition, lipid-binding, and oligomerization through overlapping interfaces.
RESULTS
The two domains from the GSDMDs adopt similar structures as other gasdermins
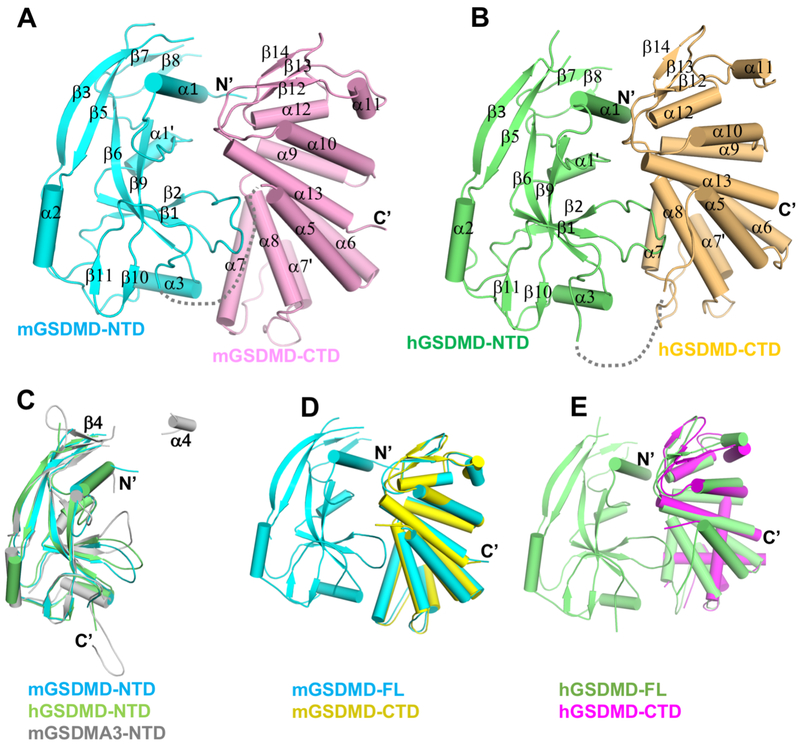
Crystal structures for murine GSDMD (mGSDMD) and human GSDMD (hGSDMD) were determined by molecular replacement method, using the NTD of mGSDMA3 and CTD of mGSDMD or hGSDMD (Liu et al., 2018) as search models (Figures 1A, 1B and S1A, Table S1). The two molecules of mGSDMD in the crystallographic asymmetric unit adopt same structure, so are the four hGSDMDs in the crystal lattice (Figure S1B). We will primarily focus on the mGSDMD structure since it is of higher resolution. Both mGSDMD and hGSDMD contain NTDs and CTDs connected by a linker region that is partially disordered, similar to the structure of the full-length mGSDMA3 (Ding et al., 2016). The NTDs from mGSDMD and hGSDMD superimpose onto that from mGSDMA3 (Ding et al., 2016) with a root mean square deviation (rmsd) of 1.4-1.8 Å (Figure 1C). The NTDs for both mGSDMD and hGSDMD are composed of three α helices and ten β strands, with the predicted α4 helix and β4 strand (in reference to the mGSDMA3 structure) not observed in the electron density (Figures 1C and S1C). The CTDs of GSDMD adopt globular folds consisting of nine helices capped by an anti-parallel three-stranded β-sheet in the order of β12-β14-β13. Superposition of the previous reported murine and human GSDMD CTD structures (Kuang et al., 2017; Liu et al., 2018) with the current structures demonstrated rmsd of 1.2-1.3 Å (Figures 1D and 1E). The above analysis suggests that there are no gross structural changes for the individual domains from the murine and human GSDMDs.
Figure 1. Overall structures of the murine and human GSDMDs.
(A-B) Overall structures of mGSDMD and hGSDMD with their NTDs and CTDs colored cyan or lime, and pink or orange, respectively. The gray dotted lines indicate the disordered 20 and 15 linker residues of mGSDMD and hGSDMD in the crystals.
(C) Superposition of the NTDs from mGSDMD (cyan), hGSDMD (lime), and mGSDMA3 (gray, 5B5R). The β4 strand and α4 helix from mGSDMA3 are not observed in the GSDMD structures.
(D) Superposition of the mGSDMD-CTD structure (yellow, 6AO3) onto the mGSDMD full-length structure (cyan).
(E) Superposition of the hGSDMD-CTD structure (magenta, 6AO4) onto the hGSDMD full-length structure (lime).
See also Figure S1.
A hydrophobic pocket in the CTD engages the β1-β2 loop to achieve autoinhibition
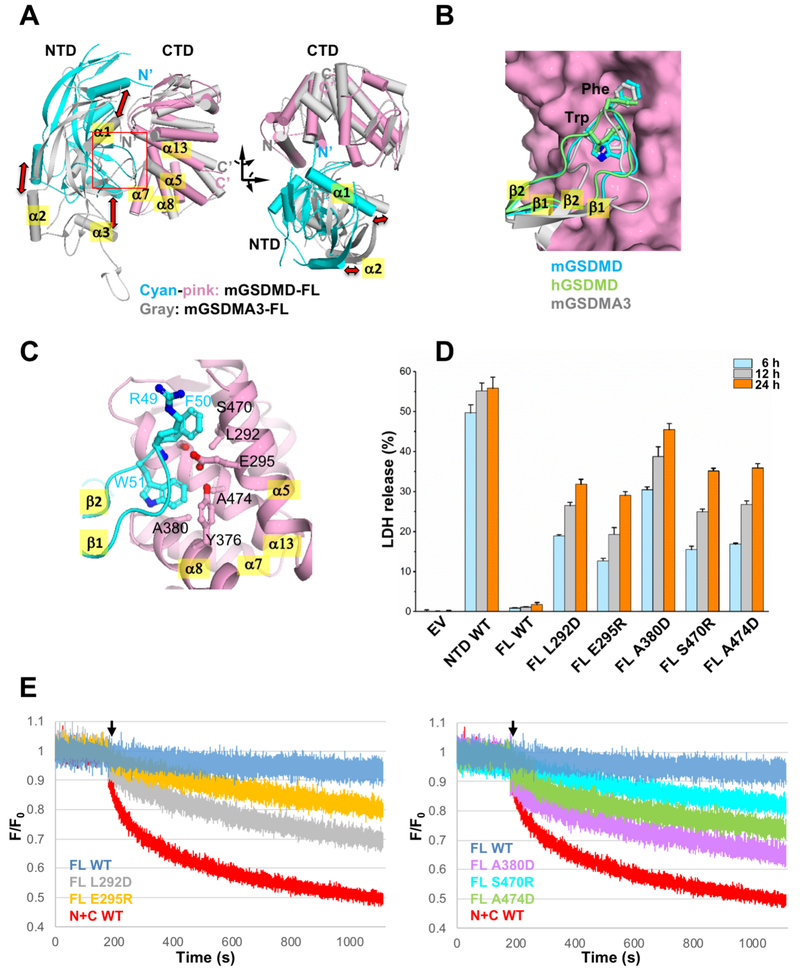
In contrast to the similar overall fold of individual NTDs and CTDs, significant shifts of the relative positions of the NTDs and CTDs are observed, in analogous to a hinged movement of the NTDs relative to the CTDs (Figures 2A and S2A). For example, the a2 helices from the GSDMD NTDs are shifted 15-20 Å compared with that from mGSDMA3 when the three CTDs are superimposed. Similar magnitude of shifts is observed for the α1 and α3 helices. Remarkably, the β1-β2 loops are docked onto a common pocket in the CTDs, even though traces of the β1-β2 loops are different among the gasdermin structures (Figure 2B). Analysis of the interface between the NTDs and CTDs demonstrates that 1700 Å2 and 2200 Å2 of solvent accessible surface area (Lee and Richards, 1971) is buried within the domain interface for mGSDMD and hGSDMD, respectively. Residues participating in the NTD-CTD interfaces are conserved among gasdermin family members (Figure S1C). In both structures, the β1-β2 loop contributes to the majority of the interface. In particular, two aromatic residues in the middle of the β1-β2 loop, F50 and W51 in mGSDMD, or F49 and W50 in hGSDMD, dock onto a hydrophobic pocket formed by residues L292, E295, Y376, A380, S470, and A474 from the α5, α7, α8, and α13 helices in mGSDMD-CTD (Figure 2C) or equivalent structural elements in hGSDMD-CTD (Figures S2B-S2C). Most of these residues are conserved among gasdermin family members (Figure S1C). To probe the role of such intramolecular domain interaction for GSDMD autoinhibition, wild type or mutant mGSDMD was expressed in HEK293T cells and an LDH assay was used as a read-out for pyroptosis. Expression of the full-length wild type mGSDMD led to minimum LDH release compared with the empty vector control (Figure 2D). By contrast, significant LDH release was observed upon expression of the mGSDMD-NTD, as well as full-length mGSDMD harboring mutations that disrupt the hydrophobic pocket at the NTD-CTD interface. This suggests that compromised autoinhibition in these mutants led to membrane permeabilization. In agreement, significant propidium iodide (PI) uptake was observed by cells expressing the same full-length mutant mGSDMDs, in contrast to the full-length wild type mGSDMD (Figure S2D). Furthermore, the full-length mutant mGSDMDs induced significant liposome leakage compared with the full-length wild type protein (Figures 2E and S2F). Collectively our data demonstrate distinct and common features of gasdermin intramolecular domain association through the binding of the NTD β1-β2 loops to a common hydrophobic pocket in the CTDs, and suggest that disruption of such domain interface compromised autoinhibition, promoted membrane permeabilization and cytolysis.
Figure 2. The NTD-CTD domain interface shares common and distinct features among gasdermins.
(A) Superposition of the full-length mGSDMD structure (NTD in cyan and CTD in pink) and the full-length mGSDMA3 structure (gray) through their respective CTDs. Shifts of their NTDs are indicated with red arrows.
(B) Close-up view of the NTD-CTD interface from (A). Two aromatic residues at the tip of the β1-β2 loops from mGSDMD (cyan), hGSDMD (lime), and mGSDMA3 (gray) are shown in ball-and-sticks. For clarity, only the mGSDMD-CTD is shown as pink surface.
(C) The domain interface near the β1-β2 loop of mGSDMD with the NTD in cyan and CTD in pink ribbons. Residues participating in NTD-CTD interface are shown in ball-and-sticks.
(D) LDH release induced by expression of wild type mGSDMD-NTD, wild type mGSDMD-FL, and mutant mGSDMD-FL, plotted as a percentage of total LDH content upon 1% Triton X-100 treatment of the cells. Data shown are mean ± SD of three independent experiments.
(E) The ability of the wild type and mutant mGSDMDs at their CTDs to induce liposome leakage was monitored through measuring the quench of the [Tb(DPA)3]3− fluorescent complex inside liposomes by external EDTA. The “N+C WT” denotes the wild type mGSDMD cleaved by caspase-1. The rest of the mGSDMD samples were full-length and not cleaved. “F” stands for fluorescence at each time point, whereas ”F0” stands for fluorescence in the first second when no mGSDMD was present. The black arrows mark the time point when mGSDMD samples were added to the liposomes. Data shown are representative of three independent experiments.
See also Figure S2.
The NTD α1 helix and β1-β2 loop are indispensable for lipid-binding and pyroptosis
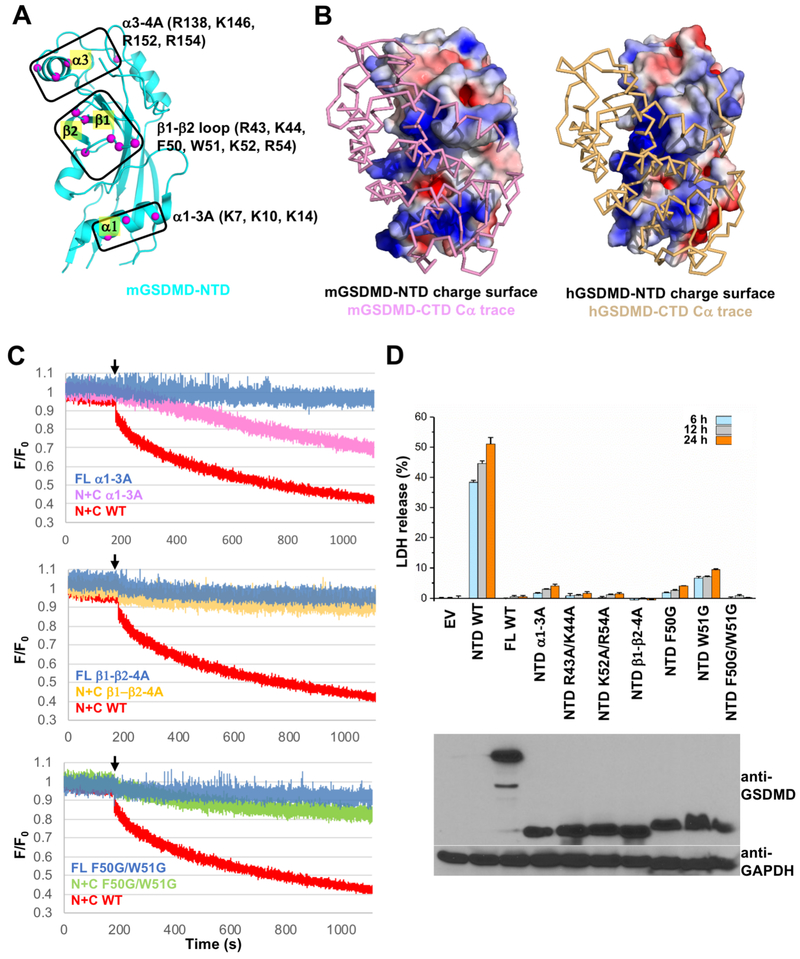
The NTDs from GSDMD and mGSDMA3 were shown to bind lipids and form membrane pores (Aglietti et al., 2016; Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Sborgi et al., 2016). Mutation of a cluster of four conserved positively-charged residues (R138, K146, R152 and R154) near the a3 helix in mGSDMD-NTD was shown to suppress lipid-binding and pyroptosis (Liu et al., 2016). A recent report of the cryo-EM structures of the mGSDMA3 membrane pore reveals large conformational changes in the NTD upon pore formation, perhaps partially induced by the binding of membrane lipids near the a1 helix (Ruan et al., 2018). Importantly, the a1 and a3 helices are located at or near the NTD-CTD domain interface in the autoinhibited structure (Ding et al., 2016). To investigate the association of GSDMD-NTD with lipids, we analyzed the surface feature of GSDMD-NTD and focused on the positively charged and hydrophobic residues located at the a1 helix and the adjacent β1-β2 loop (Figures 3A and 3B). These regions are masked by GSDMD-CTD in the full-length structure, which is consistent with the observation that GSDMD-NTD, not the full-length GSDMD or GSDMD-CTD, participates in lipid-binding and membrane-targeting. To test lipid-binding, mGSDMD mutants were generated at the α1 helix (K7A/K10A/K14A, named “α1-3A”), the β1-β2 loop basic residues (R43K/K44A/K52A/R54A, named “β1-β2-4A”) and hydrophobic residues (F50G/W51G). Upon cleavage of the full-length protein by caspase-1, the wild type “N+C” protein induced leakage of liposomes containing cardiolipin, a phospholipid reported to bind GSDMD (Ding et al., 2016; Liu et al., 2016). By contrast, the mutants harbor much reduced activities (Figures 3C and S3A). This is corroborated by the diminished ability of the NTD mutant “α1-3A” to induce lysis of bacterial protoplasts compared with the wild type protein (Figure S3B). In agreement, mutations of the above residues at either the α1 helix or β1-β2 loop significantly compromised the ability of the mGSDMD-NTD to induce pyroptotic cell death (Figure 3D). A model of the mGSDMD pore based on the mGSDMA3 pore structure reveals a positively charged surface patch on the outer rim of the pore formed by the α1 helix, β1-β2 loop and α3 helix (Figures S3C and S3D). Our data thus suggest that these structural elements may participate in lipid-binding and are indispensable for GSDMD-mediated pyroptosis.
Figure 3. Lipid-binding is critical for GSDMD-mediated pyroptosis.
(A) Three potential lipid-binding sites at mGSDMD-NTD are marked. The Cα atoms of potential lipid-binding residues are shown as magenta spheres.
(B) The GSDMD-FL structures are shown in electrostatic charge surface in the back for the NTDs and Cα trace in front for the CTDs of mGSDMD (left) and hGSDMD (right). The views of the NTDs are the same as that in (A).
(C) The ability of the wild type and mutant mGSDMD-NTDs α1-3A, β1-β2-4A and F50G/W51G to induce liposome leakage was monitored through measuring the quench of the [Tb(DPA)3]3− fluorescent complex inside liposomes by external EDTA. The “FL” mutants were not cleaved by caspase-1. The “N+C” samples were cleaved by caspase-1. “F” stands for fluorescence at each time point, whereas ”F0” stands for fluorescence in the first second when no mGSDMD was present. The black arrows mark the time point when mGSDMD samples were added to the liposomes. Data shown are representative of three independent experiments.
(D) Mutation of the predicted lipid-binding residues block the mGSDMD-NTD-mediated LDH release, Data shown are mean ± SD of three independent experiments. Expression of mGSDMD is shown on the bottom panel.
See also Figure S3.
Three adjacent interfaces are important for the oligomerization of the NTD
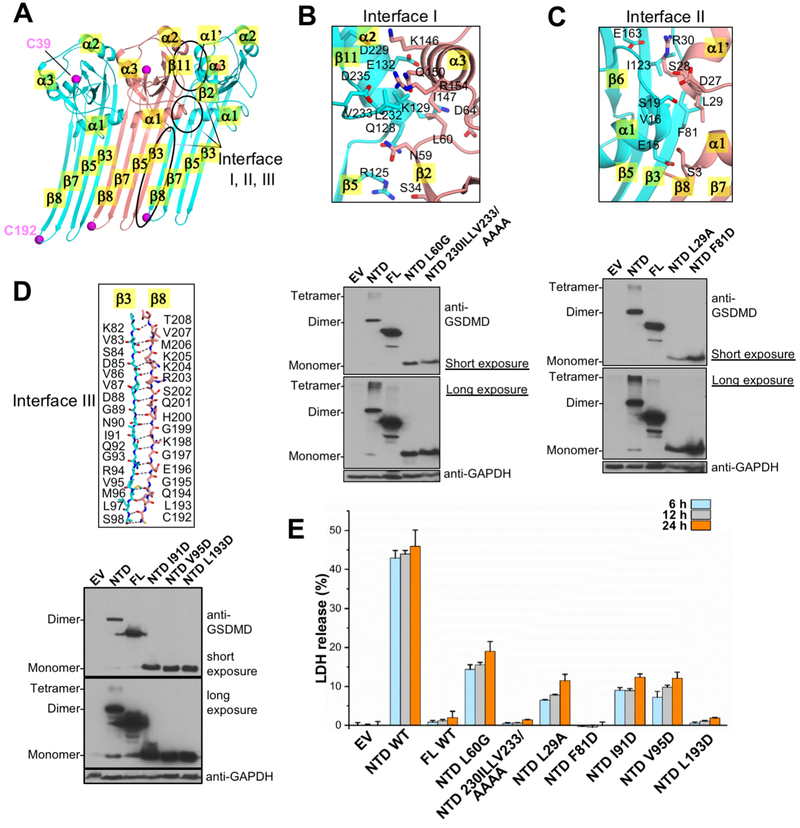
The NTDs of GSDMD share ~45% sequence similarity with the mGSDMA3-NTD, and their structures superimpose well using either the autoinhibited form (Figure 1C) or the pore form of mGSDMA3 (Figure S3C). We hypothesize that mGSDMD-NTD may undergo conformational changes in a similar manner as the mGSDMA3-NTD to oligomerize and form membrane pores. To investigate whether structural elements in the mGSDMD pore model indeed mediate oligomerization and assembly of the pore structure, we probed the oligomerization and pyroptosis-inducing activities of mGSDMD-NTD. Using the published structure of the GSDMA3 pore (Ruan et al., 2018) as a template, we generated a model for the mGSDMD-NTD in the pore form using UCSF Chimera (Pettersen et al., 2004). Three potential oligomerization interfaces in this model are identified (Figure 4A): interface I is located between the α2 helix and β11 strand from one subunit and α3 helix and β2 strand from its neighbor; the adjacent interface II is centered on the α1 helix and β3 strand from one subunit and the α1 and α1’ helices from the other; the anti-parallel β3 and β8 strands from neighboring subunits assemble the extensive interface III. As demonstrated previously, the wild type mGSDMD-NTD forms oligomers upon overexpression in HEK293T cells (Ding et al., 2016; Liu et al., 2016; Rathkey et al., 2018). By contrast, mutation of residues located at the center of interface I (L60G and 230-IILLV-233 to Ala), interface II (L29A and F81D), and interface III (I91D, V95D, and L193D) abolished mGSDMD-NTD oligomerization (Figures 4B-4D), and compromised mGSDMD-NTD-mediated pyroptotic cell death (Figure 4E). This is consistent with reports that mutations or truncations at interface II (E15K, α1 helix truncation) or interface III (L192D, I104N/I105N) of hGSDMD-NTD or mGSDMD-NTD decreased their liposome leakage activities and compromised pyroptosis (Aglietti et al., 2016; Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Rogers et al., 2017; Sborgi et al., 2016; Wang et al., 2017). In agreement, the crystallized mGSDMD, which harbors truncations at the interface III β8 and adjacent β7 strands, lost the ability to induce leakage of liposomes (Figure S4A) or lysis of bacterial protoplasts (Figure S3B). In short, our data suggest that protein-protein interaction between adjacent GSDMD-NTDs is indispensable for the oligomerization and pore formation activities in pyroptosis.
Figure 4. Oligomerization of mGSDMD-NTD is essential for its pyroptosis activity.
(A) A model of the mGSDMD-NTD in the pore form was generated based on the mGSDMA3 pore structure, and three contiguous mGSDMD-NTD are shown. The three NTDs are colored cyan, salmon, and cyan, respectively. The three inter-subunit interfaces I, II, and III are marked, as are important secondary structural elements participating in these interfaces. The magenta spheres mark the Cα atoms for the Cys39 and Cys192 residues.
(B, C, D) Detailed views of the three interfaces are shown in the top panels. The bottom panels illustrate the oligomerization of the wild type and mutant mGSDMD upon over-expression in HEK293T cells. The samples were run with reducing SDS-PAGE gels.
(E) Mutation at the predicted oligomerization interfaces compromised LDH release induced by expression of mGSDMD-NTD in HEK293T cells. Data shown are mean ± SD of three independent experiments.
See also Figure S4.
DISCUSSION
Activation and regulation of gasdermin D underlie the inflammatory responses downstream of both canonical and noncanonical inflammasome signaling pathways. Despite recent progress in our understanding of mGSDMA3 autoinhibition and pore formation, mechanistic understanding of GSDMD autoinhibition, lipid-binding and pore formation remain incomplete. Here we report the crystal structures of the full-length GSDMD from both mouse and human, which reveal structures of the GSDMD NTDs and the architecture of the GSDMD intramolecular domain association. Even though the overall structures of the individual NTDs and CTDs are similar, significant shifts of the relative positions of the two domains are observed among different gasdermins. Such shift is hinged upon two hydrophobic residues at the β1-β2 loop in the NTDs that dock onto a common hydrophobic pocket in the CTDs from gasdermins. Our structures thus demonstrate distinct and common features of intramolecular domain-domain association among different gasdermin family members.
The β1-β2 loop, along with the adjacent α1 helix and the α3 helix at or near the NTD-CTD domain interface, also play important roles in lipid-binding by GSDMD-NTD. The overlapping lipid-binding and NTD-CTD interface is consistent with the role of GSDMD-CTD as an autoinhibitory domain that blocks the lipid-binding surface of GSDMD-NTD, thus preventing spurious pore formation and cytolysis in the context of the full-length GSDMD without inflammasome activation. The presence of negatively charged phospholipids concomitant with caspase cleavage of the NTD-CTD linker may allow the lipid molecules to both release the autoinhibition and induce large conformational changes in NTD. This precedes the assembly of the NTD pore structures involved in pyroptosis.
The GSDMD pore is composed of oligomerized NTDs. Oligomerization of mGSDMD-NTD has been reported to require residues Cys39 and Cys192 (Liu et al., 2016). In agreement, mutation of the equivalent Cys191 in hGSDMD led to reduced pyroptosis by its NTD (Rathkey et al., 2018). However, mutation of Cys38 in hGSDMD did not reduce pyroptosis (Rathkey et al., 2018). In our crystal structures Cys39/Cys38 in mGSDMD/hGSDMD are exposed to the solvent, whereas Cys192/Cys191 in mGSDMD/hGSDMD are located in a disordered region, so presumably also exposed to the solvent (Figure S4B). Both Cys39 and Cys192 residues are exposed to the solvent in the model of the mGSDMD NTD pore structure as well (Figure 4A). It is not clear how these residues may mediate oligomerization since there is no adjacent Cys residues from neighboring NTD subunits. Clearly the role(s) of the cysteine residues in GSDMD oligomerization remain to be defined.
The mGSDMA3 pore is assembled through extensive interactions between adjacent subunits, utilizing many structural elements such as the α1, α1’ and α3 helices and the β1, β2, β3, β8 and β11 strands. Many of these structural elements also participate in lipid-binding, particularly within the α1- β2 fragment of the NTD. It is conceivable that lipid-binding by the NTDs from GSDMDs and their subsequent conformational changes facilitate their oligomerization and/or stabilizes the pore structures. This study thus reveals that overlapping interfaces are involved in GSDMD autoinhibition, lipid-binding, and oligomerization. Whether the same principles apply to other gasdermin family members remain to be determined through future studies of gasdermin-lipid complexes and pore structures.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tsan Sam Xiao, Ph.D. (tsx@case.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293T cells
HEK293T cells (ATCC CRL-3216) were originally derived from a female fetus. The cells were cultured at 37 °C, 5% CO2 in DMEM plus 10% fetal bovine serum (FBS) (Thomas Scientific, Swedesboro, NJ) supplemented with 100 U/ml penicillin and 100 U/ml streptomycin (Pen+Strep), and 2 mg/ml L-glutamine (Thermo Fisher Scientific, Waltham, MA).
Immortalized mouse macrophages
Immortalized mouse macrophages (iBMDM) were a gift from Dr. E. Latz (University of Bonn) and the sex was unknown. The cells were cultured in DMEM plus 10% SuperCalf serum (Gemini Bio-Products, West Sacramento, CA), 1X antibiotic-antimycotic (Thermo Fisher Scientific, Waltham, MA) and 3 mg/ml puromycin (InvivoGen, San Diego, CA).
Bacillus megaterium
Bacillus megaterium de Bary cells (ATCC 14581) were grown in Difco antibiotic medium 3 (Thermo Fisher Scientific, Waltham, MA) at 37 °C.
METHOD DETAILS
Protein Expression and Purification
The coding sequences for the murine (Q9D8T2) and human (P57764) GSDMD were cloned into a bacterial expression vector encoding a His6-SUMO tag (Mossessova and Lima, 2000). The expression constructs were transformed in BL21 (DE3) Codon Plus RIPL cells (Agilent Technologies, Santa Clara, CA), and the cells were grown at 37 °C until OD600 reached 0.6. Protein expression was induced at 18 °C overnight with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were harvested and lysed by sonication in a lysis buffer containing 25 mM Tris-HCl (pH 8.0), 300 mM NaCl. The recombinant SUMO-fusion protein in the cleared cell lysate was purified using Ni-NTA (Thermo Fisher Scientific, Waltham, MA) affinity chromatography. Elution fractions containing the purified SUMO-fusion proteins were pooled and incubated overnight with the Ulp1 protease (Mossessova and Lima, 2000) at 4 °C, followed by a second Ni-NTA ch romatography to remove the SUMO tag and uncleaved fusion protein. The GSDMD protein samples were further purified through S200 size-exclusion chromatography in a buffer containing the lysis buffer plus 5 mM DTT at 4 °C. The purified protein was concentrated to ~20 mg/ml before frozen in aliquots at −80 °C. The purified full-length GSDMD protein samples tended to aggregate which impeded crystallization. To mitigate this, and in light of previous crystal structures that contain multiple disordered regions among different gasdermin family members (Ding et al., 2016), several truncated versions of GSDMD with deletions in the common disordered regions were tested. Truncation of three loop regions of mGSDMD, including residues 182-187, 197-199 and 248-273 (Figure S1C), led to its successful purification and crystallization (see below). To obtain diffraction quality crystals for hGSDMD, residues 181-186 and residues 196-198 were truncated, and the NTD-CTD linker was substituted by that from the crystallized mGSDMD construct. In addition, the hGSDMD protein sample was treated with a reductive alkylation kit (HR2-434) from Hampton Research (Aliso Viejo, CA) following manufacturer’s instructions, and purified further through size-exclusion chromatography as described above. Human caspase-1 catalytic domain was expressed and purified as described (Yang et al., 2018), and was used to cleave mGSDMD in liposome leakage assay and protoplast lysis assay.
Crystallization, X-ray Diffraction and Structure Determination
Screening of crystallization conditions for both mGSDMD and hGSDMD was performed using commercial kits from Hampton Research (Aliso Viejo, CA) and Molecular Dimensions (Maumee, OH) with sitting drop vapor diffusion method at 20 °C or 4 °C. The mGSDMD sample was crystallized at 5 mg/ml using a reservoir solution containing 25% PEG3350, 0.1M Bis-Tris pH 6.5, and 10 mM DTT. The hGSDMD sample was crystallized at 10 mg/ml using a reservoir solution containing 25% PEG3350, 0.1M Bis-Tris pH 5.5, 200 mM NaCl, and 10 mM DTT. Crystals were frozen in the reservoir solutions plus 20-25% glycerol in liquid nitrogen. X-ray diffraction data were collected at beamlines SER-CAT (22-ID) and GM/CAT (23-ID) at the Advanced Photon Source, Argonne National Laboratory (Lemont, IL), and processed with the program XDS (Kabsch, 2010). The structures were determined using the GSDMD-CTD (6AO3 and 6AO4) and mGSDMA3-NTD (5B5R) structures as search models. Manual model building and refinement were performed with Coot (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010), respectively. The crystal structures were validated by the MolProbity server (Chen et al., 2010), which demonstrated MolProbity scores >2.0 at 100th percentile (best) among structures of similar resolutions. Figures were prepared using PyMol (The PyMOL Molecular Graphics System, Version 1.8.2.3 Schrodinger, LLC.). The electrostatic charge surface (Dolinsky et al., 2004) was calculated with PDB2PQR. Modeling of the mGSDMD-NTD pore structure was performed using UCSF Chimera (Pettersen et al., 2004).
Cell Culture and Cytotoxicity Assay
HEK293T cells grown in DMEM plus 10% serum were transfected with pCDNA4/TO expression vectors coding for mGSDMD using the T-REx expression system (Thermo Fisher Scientific, Waltham, MA). Co-transfection of the pCDNA6/TR vector coding for the Tet repressor and the pCDNA4/TO vectors containing mGSDMD coding sequences at 4:1 molar ratio was performed using 8 mg of plasmids mixed with calcium phosphate per 60 mm tissue culture plate. After overnight culture of the transfected cells, expression of mGSDMD was induced with 1 μg/ml tetracycline for 6, 12, and 24 hours followed by collection of culture supernatants that were centrifuged to pellet detached cells. To measure cytotoxicity upon inflammasome activation in macrophages, iBMDM cells grown in DMEM plus 10% SuperCalf Serum (Gemini Bio-Products, West Sacramento, CA) were stimulated with 200 ng/ml LPS (Sigma-Aldrich, St. Louis, MO). Four hours after LPS stimulation, the cells were treated with no stimulus (LPS only) or with 20 μM nigericin (Sigma-Aldrich, St. Louis, MO) for 30 minutes. Lactate dehydrogenase (LDH) activities in the cell culture supernatants were measured using a cytotoxicity detection kit from Roche (Indianapolis, IN) or Thermo Fisher Scientific (Waltham, MA) following the manufacturers’ instructions. The LDH release was expressed as a percentage of total LDH content upon 1% Triton X-100 treatment of the cells. For western blots, cells were lysed in RIPA buffer (Thermo Fisher, Waltham, MA) or 8 M urea plus 5% SDS, and cell extracts were used for SDS-PAGE. Monoclonal anti-mouse GSDMD antibody (A-7, sc-393656) and monoclonal anti-GAPDH antibody (sc-47724) were from Santa Cruz Biotechnology (Dallas, TX).
Microscopy Imaging
To examine cell morphology and propidium iodide uptake, bright field and fluorescent images of cells in 24-well plates were captured using a Leica DFC400 fluorescent microscope (Wetzlar, Germany).
Liposome Leakage Assay
The ability of mGSDMD to bind lipids and form pores in liposomes was determined by measuring the release of the encapsulated and highly fluorescent Tb3+-dipicolinic acid ([Tb(DPA)3]3−) complex into an external solvent containing 5 mM EDTA as described previously for a cytolysin (Ramachandran et al., 2002). Liposomes were produced by rehydrating a dried lipid mixture composed of 40:35:25 mol% of dioleoylphosphatidylcholine (DOPC), dioleoylphosphatidylethanolamine (DOPE), and bovine heart cardiolipin (CL), respectively, in a buffer containing 20 mM HEPES (pH 7.5), 150 mM KCl, 3 mM TbCl3 and 9 mM DPA. A trace amount (0.1 mol%) of rhodamine-PE was included to quantify lipid yield at the end of the procedure. The liposome suspension was subjected to three freeze-and-thaw cycles followed by extrusion through a 100-nm pore diameter polycarbonate membrane 21 times using the Avanti mini-extruder (Avanti Polar Lipids, Inc. Alabaster, AL). The extruded liposomes were then passed through a PD-10 desalting column (GE Healthcare Biosciences, Pittsburgh, PA) to remove unincorporated [Tb(DPA)3]3−. GSDMD variants (0.1 mM protein final) were added at a defined time point at room temperature to liposomes (50 μM total lipid) suspended in a buffer containing 20 mM HEPES (pH 7.5), 150 mM KCl, 5 mM EDTA and 1 mM DTT in a final volume of 2.1 ml. Fluorescence emission at 544 nm (2 nm bandpass) upon excitation at 278 nm (2 nm bandpass) was recorded continuously for 20 min at 0.1 second intervals using a Fluorolog 3-22 photon-counting spectrofluorometer (Horiba Ltd, Kyoto, Japan). Data were normalized using fluorescence values in the first second of recording prior to the addition of GSDMD samples.
Bacterial Protoplast Lysis Assay
The bacterial protoplast lysis assay was performed as previously described (Ding et al., 2016). Briefly, Bacillus megaterium cells (ATCC, Manassas, VA) were grown in Difco antibiotic medium 3 (Thermo Fisher Scientific, Waltham, MA) at 37°C overnight. The harvested cells were washed and resuspended in a buffer containing 20 mM sodium malate (pH 6.5), 20 mM MgCl2 and 500 mM sucrose. Mutanolysin (Sigma-Aldrich, St. Louis, MO) at 100 U/ml and lysozyme (Hampton Research, Aliso Viejo, CA) at 1 mg/ml were added to the cell suspension and incubated at 37°C until protoplasts were formed as judged under optical microscopy. Aliquots of the protoplasts (0.5 ml) in the above buffer at OD600 of ~1.0 were incubated with 5 μl of indicated GSDMD proteins at final concentrations of 0.1 μM, and OD600 was continuously monitored for 15 minutes at 1 min intervals before Triton X-100 was added at final concentrations of 0.1%.
Reconstitution of mGSDMD Expression in Macrophages
A murine immortalized bone-marrow derived macrophage (iBMDM) cell line was a gift from E. Latz (University of Bonn) (Stutz et al., 2013) and was cultured in DMEM plus 10% SuperCalf serum (Gemini Bio-Products, West Sacramento, CA), 1X antibiotic-antimycotic (Thermo Fisher Scientific, Waltham, MA) and 3 mg/ml puromycin (InvivoGen, San Diego, CA). Generation of the Gsdmd deficient iBMDMs was described previously (Russo et al., 2016). The wild type or truncated variant of Gsdmd was reconstituted in the Gsdmd deficient cells using a lentiviral construct published previously (Chirieleison et al., 2016). Reconstituted cells were selected in 1 mg/mL G418 (Thermo Fisher Scientific, Waltham, MA) before stimulation with LPS and nigericin and being assayed for cytotoxicity.
Quantification and Statistical Analysis
Data are represented as mean ± SD of three independent experiments, unless otherwise stated. Statistical analyses were performed with program Excel (Microsoft Corporation, Redmond, WA).
Data and Software Availability
Coordinates and structural factors have been deposited with the Protein Data Bank with accession codes 6N9N and 6N9O for murine and human GSDMD structures, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-mouse GSDMDC1 (A-7) | Santa Cruz Biotech | Cat#sc-393656; RRID: AB_2728694 |
| Rabbit monoclonal anti-mouse GSDMDC1 (EPR19828) | Abcam | Cat#ab209845; RRID: AB_2783550 |
| Rabbit polyclonal anti-human GSDMDC1 | Sigma-Aldrich | Cat#HPA044487; RRID: AB_2678957 |
| Mouse monoclonal anti-human GAPDH (0411) | Santa Cruz Biotech | Cat#sc-47724; RRID: AB_627678 |
| Bacterial and Virus Strains | ||
| BL21 (DE3) Codon Plus RIPL | Agilent | Cat#230280 |
| NEB® 5-alpha Competent E. coli | New England Biolabs | Cat#C2987I |
| Bacillus megaterium de Bary | ATCC | Cat#14581 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) | Avanti Polar Lipids | Cat#850725 |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) | Avanti Polar Lipids | Cat#850375 |
| Cardiolipin (bovine heart, CL) | Avanti Polar Lipids | Cat#840012 |
| 1,2-dioleoyl-sn-glycero-3- phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rhodamine-PE) | Avanti Polar Lipids | Cat#810150 |
| Dipicolinic acid (DPA, 2,6-Pyridinedicarboxylic acid) | Sigma-Aldrich | Cat#P63808 |
| Difco antibiotic medium 3 | Thermo Fisher Scientific | Cat#DF0243-17-8 |
| Mutanolysin | Sigma-Aldrich | Cat#M9901 |
| Lysozyme | Hampton Research | Cat#HR7-110 |
| Mouse full-length wild type GsdmD | This paper | N/A |
| Mouse GSDMD harboring truncations of residues 182-187, 197-199 and 248-273 | This paper | N/A |
| Human GSDMD harboring truncations of residues 181-186, 196-198, and its NTD-CTD linker replaced by that from mouse GsdmD | This paper | N/A |
| Mouse full-length GSDMD harboring L292D mutation | This paper | N/A |
| Mouse full-length GSDMD harboring E295R mutation | This paper | N/A |
| Mouse full-length GSDMD harboring A380D mutation | This paper | N/A |
| Mouse full-length GSDMD harboring S470R mutation | This paper | N/A |
| Mouse full-length GSDMD harboring A474D mutation | This paper | N/A |
| Mouse full-length GSDMD harboring α1-3A mutations | This paper | N/A |
| Mouse full-length GSDMD harboring β1-β2-4A mutations | This paper | N/A |
| Mouse full-length GSDMD harboring F50G/W51G mutations | This paper | N/A |
| Critical Commercial Assays | ||
| Cytotoxicity Detection Kit (LDH) | Roche | Cat#04744934001 |
| LDH cytotoxicity assay kit | Thermo Fisher Scientific | Cat#88953 |
| Quick Blunting Kit | New England Biolabs | Cat#E1201S |
| T4 DNA Ligase | New England Biolabs | Cat#M0202S |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs | Cat#M0530L |
| Gibson Assembly Cloning Kit | New England Biolabs | Cat#E5510S |
| Deposited Data | ||
| Crystal structure of the murine GSDMD | RCSB PDB | PDB ID:6N9N |
| Crystal structure of the human GSDMD | RCSB PDB | PDB ID: 6N9O |
| Experimental Models: Cell Lines | ||
| HEK293T | ||
| iBMDM expressing cerulean-ASC | Stutz et al., 2013 | N/A |
| iBMDM with Gsdmd knockout | Russo et al., 2016 | N/A |
| iBMDM Gsdmd knockout reconstituted with WT Gsdmd | Rathkey et al., 2017 | N/A |
| iBMDM Gsdmd knockout reconstituted with Gsdmd harboring truncations of residues 182-187, 197-199 and 248-273. | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides: Please see Supplemental Table S2 for oligo/primer sequences. | ||
| Recombinant DNA | ||
| pSMT3-full length hGSDMD | This paper | N/A |
| pSMT3-full length mGSDMD | This paper | N/A |
| pcDNA3.1(+)-full length mGSDMD | This paper | N/A |
| pCMV-3Tag-1-full length mGSDMD | This paper | N/A |
| pCDNA6/TR (T-REx expression system) | Thermo Fisher Scientific | Cat#V1025-20 |
| pCDNA4/TO (T-REx expression system) | Thermo Fisher Scientific | Cat#K1020-01 |
| pCDNA4/TO mGSDMD-NTD | This paper | N/A |
| pCDNA4/TO mGSDMD-FL WT | This paper | N/A |
| pCDNA4/TO mGSDMD-FL L292D | This paper | N/A |
| pCDNA4/TO mGSDMD-FL E295R | This paper | N/A |
| pCDNA4/TO mGSDMD-FL A380D | This paper | N/A |
| pCDNA4/TO mGSDMD-FL S470R | This paper | N/A |
| pCDNA4/TO mGSDMD-FL A474D | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD α1-3A | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD R43A, K44A | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD K52A, R54A | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD β1-β2-4A | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD F50G | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD W51G | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD F50G/W51G | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD L60G | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD 230ILLV233/AAAA | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD L29A | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD F81D | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD I91D | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD V95D | This paper | N/A |
| pCDNA4/TO mGSDMD-NTD L193D | This paper | N/A |
| Software and Algorithms | ||
| Microsoft Excel | Microsoft Corporation | Version 16.16.8 |
| UCSF Chimera | UCSF | Version 1.12 |
| Other | ||
Highlights.
Determined crystal structures of the murine and human GSDMDs
The N- and C-domain association shares features with the murine GSDMA3
Disruption of lipid-binding and oligomerization surface suppresses cell killing
Overlapping surfaces mediate autoinhibition, lipid-binding, and oligomerization
Acknowledgements
Z.L. is supported by a Careers in Immunology postdoctoral fellowship from the American Association of Immunologists. B.Z. is supported by NIH grant T32AI089474, R.R. is supported by NIH grant R01GM121583, D.W.A. is supported by NIH grants P01DK091222 and R01GM086550, and T.S.X. is supported by NIH grants R21AR069908 and R01GM127609. We are grateful for help from the Ramakrishnan lab on the use of the inducible expression system. We would like to thank the Xiao, Abbott, Dubyak, Ramakrishnan, and Adoro laboratories for insightful discussions. X-ray diffraction data were collected at the Advanced Photon Source (APS) SER-CAT beamline (Supporting institutions may be found at www.ser-cat.org/members.html) and GM/CA-CAT beamline, funded in whole or in part by funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the APS, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Molecular graphics and analyses performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH grant P41GM103311.
Footnotes
Declaration of interests: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, and Dueber EC (2016). GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. USA 113, 7858–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao KL, Kulakova L, and Herzberg O (2017). Gene polymorphism linked to increased asthma and IBD risk alters gasdermin-B structure, a sulfatide and phosphoinositide binding protein. Proc. Natl. Acad. Sci. USA 114, E1128–E1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KW, Demarco B, Heilig R, Shkarina K, Boettcher A, Farady CJ, Pelczar P, and Broz P (2019). Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. The EMBO Journal e101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, He W-T, Hu L, Li J, Fang Y, Wang X, Xu X, Wang Z, Huang K, and Han J (2016). Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Research 26, 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirieleison SM, Kertesy SB, and Abbott DW (2016). Synthetic Biology Reveals the Uniqueness of the RIP Kinase Domain. The Journal of Immunology 196, 4291–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson BT, and Brennan MA (2001). Pro-inflammatory programmed cell death. Trends in Microbiology 9, 113–114. [DOI] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang D-C, and Shao F (2016). Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- He W-T, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang Z-H, Zhong C-Q, and Han J (2015). Gasdermin D is an executor of pyroptosis and required for interleukin-1 β secretion. Cell Research 25, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Liu F, Zhang X, Liu P, Bajrami B, Teng Y, Zhao L, Zhou S, Yu H, Zhou W, et al. (2018). Gasdermin D Exerts Anti-inflammatory Effects by Promoting Neutrophil Death. Cell Reports 22, 2924–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, and O’Rourke K (2015). Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671. [DOI] [PubMed] [Google Scholar]
- Kuang S, Zheng J, Yang H, Li S, Duan S, Shen Y, Ji C, Gan J, Xu X-W, and Li J (2017). Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis. Proc. Natl. Acad. Sci. USA 114, 10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Xiao TS, and Stutz A (2013). Activation and regulation of the inflammasomes. Nature Reviews Immunology 13, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, and Richards FM (1971). The interpretation of protein structures: estimation of static accessibility. Journal of Molecular Biology 55, 379–400. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, and Lieberman J (2016). Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang C, Rathkey JK, Yang J, Dubyak GR, Abbott DW, and Xiao TS (2018). Structures of the Gasdermin D C-Terminal Domains Reveal Mechanisms of Autoinhibition. Structure 26, 778–784.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, and Aderem A (2010). Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nature Immunology 11, 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, and Lima CD (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Molecular Cell 5, 865–876. [DOI] [PubMed] [Google Scholar]
- Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, et al. (2018). Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Heuck AP, Tweten RK, and Johnson AE (2002). Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nature Structural Biology 9, 823–827. [DOI] [PubMed] [Google Scholar]
- Rathkey JK, Zhao J, Liu Z, Chen Y, Yang J, Kondolf HC, Benson BL, Chirieleison SM, Huang AY, Dubyak GR, et al. (2018). Chemical disruption of the pyroptotic pore-forming protein gasdermin D inhibits inflammatory cell death and sepsis. Science Immunology 3, eaat2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, and Alnemri ES (2017). Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nature Communications 8, 14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Xia S, Liu X, Lieberman J, and Wu H (2018). Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo HM, Rathkey J, Boyd-Tressler A, Katsnelson MA, Abbott DW, and Dubyak GR (2016). Active Caspase-1 Induces Plasma Membrane Pores That Precede Pyroptotic Lysis and Are Blocked by Lanthanides. The Journal of Immunology 197, 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhan J, Liu BC, Muendlein HI, Li P, Nilson R, Tang AY, Rongvaux A, Bunnell SC, Shao F, Green DR, et al. (2018). Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. USA 115, E10888–E10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborgi L, Ruhl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Muller DJ, Broz P, and Hiller S (2016). GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. The EMBO Journal 35, 1766–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665. [DOI] [PubMed] [Google Scholar]
- Stutz A, Horvath GL, Monks BG, and Latz E (2013). ASC speck formation as a readout for inflammasome activation. Methods in Molecular Biology (Clifton, N.J.) 1040, 91–101. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, and Shao F (2017). Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a Gasdermin. Nature 547, 99–103. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu Z, Wang C, Yang R, Rathkey JK, Pinkard OW, Shi W, Chen Y, Dubyak GR, Abbott DW, et al. (2018). Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor. Proc. Natl. Acad. Sci. USA 115, 6792–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structural factors have been deposited with the Protein Data Bank with accession codes 6N9N and 6N9O for murine and human GSDMD structures, respectively.