Abstract
A leukotoxin (LtxA) that is produced by Aggregatibacter actinomycetemcomitans (Aa) is an important virulence determinant in an aggressive form of periodontitis in adolescents. Understanding the function of this protein at the molecular level is critical to elucidating its role in the disease process. To accomplish genetic analysis of the protein structure and relating these observations to toxin function, we have developed an E. coli expression system for the generation and rapid purification of LtxA. Cloning the structural toxin gene, ltxA, from Aa strain JP2 under control of T7 promoter-1 of pCDFDuet-1 vector resulted in expression of a 114 KDa protein which could be easily purified by the presence of a carboxy-terminal engineered double hexahistidine (double–His6) tag and was immunologically reactive with an anti-LtxA monoclonal antibody, but was not cytotoxic. Cloning a second gene, ltxC, an acyltransferase gene, into the vector under control of T7 promoter-2, resulted in expression of the biologically active LtxA. The toxin was extracted from E. coli inclusion bodies, purified by immobilized metal affinity chromatography, and refolded by dialysis. When compared by circular dichroism (CD) spectroscopy analysis, acylated recombinant LtxA has a secondary structure consistent with wt LtxA, while variations in α-helical structure of nonacylated LtxA were observed. No modifications in α-helix were found upon the toxin’s binding with liposome-incorporated cholesterol. Our results suggest that pure, biologically active recombinant LtxA can be isolated by a one-step affinity chromatography from E. coli. The toxic and structural properties of the recombinant LtxA are similar to its wt counterpart.
Keywords: Aggregatibacter actinomycetemcomitans, RTX toxin, recombinant toxin, polyhistidine tag, localized aggressive periodontitis
INTRODUCTION
Aggregatibacter actinomycetemcomitans (Aa) is a common inhabitant of the upper aerodigestive tract of man (Henderson et al., 2003; Zambon, 1985, 1996) and is associated with various infections including endocarditis, meningitis, pneumonia, septicemia, urinary tract infections, vertebral osteomyelitis, and abscesses (Berbari et al., 1997; Das et al., 1997). The organism can also be cultured from adolescent children of African descent with a form of periodontal disease that is seen in known as localized aggressive periodontitis (LAP) (Haubek and Johansson, 2014). When Aa that is isolated from the gingival crevice of LAP patients is compared with organisms isolated from patients with adult periodontitis or distant infection sites, a 530 bp deletion mutation in the promoter region of leukotoxin (LtxA) operon is observed (Brogan et al., 1994). Organisms that contain this mutation produce 10- to 20-fold higher levels of LtxA. This deletion is not found in Aa that is cultured from adult periodontitis patients or infections at more distant sites, suggesting that it is playing a pivotal role of LtxA in the development of LAP. There is evidence for the correlation between leukotoxicity and the periodontal attachment loss (Hoglund Aberg et al., 2014).
LtxA is a member of the RTX (Repeats in ToXin) family of bacterial protein toxins. RTX are large (<100,000 kDa) proteins that are produced by a wide variety of genera that include Actinobacillus, Aggregatibacter, Bordetella, Escherichia, Kingella, Mannheimia, Moraxella, Morganella, Pasteurella, Proteus and Vibrio (Linhartova et al., 2010). RTX-containing organisms all share a common operon organization and the functional domains of its structural toxin gene product. LtxA is synthesized as a component of a five-gene operon (ltxC, ltxA, ltxB, ltxD) in transcriptional order and tdeA. The products of the ltxBD genes, and tdeA (Crosby and Kachlany, 2007), a tolC homolog (Balakrishnan et al., 2001) located 572 bp downstream of the Ltx operon, form a type I secretion system that facilitates the movement of the toxin from the bacterial cytoplasm directly into the extracellular environment without periplasmic intermediates.
RTX are transcribed as biologically inactive protoxins and undergo a subsequent modification to exert an effect on the host. While the most common post-translational modification in bacteria is either C-terminal or N-terminal proteolytic cleavage that produces a catalytic fragment or facilitates oligomerization and subsequent membrane damage, RTX toxins share all a unique mechanism of activation mechanism whereby acyl groups are covalently to linked to internal LtxA lysines to become biologically active (Balashova et al., 2009; Hackett et al., 1994; Hardie et al., 1991; Issartel et al., 1991).
RTX toxins also share both structural and organizational features structural toxin gene product. Most notable of these is the presence of varying numbers of a glycine-rich [(L/I/F)-X-G-G-X-G-(N/D)-D] repeat motif in the structural toxin gene. The sequence is known as a parallel β-roll motif (Baumann et al., 1993) is not unique to LtxA nor other RTX but rather it is associated proteins that utilize a type I secretion system. The β-roll is a calcium-induced switch that prevents protein folding of the protein until it is released into the external environment. LtxA kills leukocytes of humans and Old World Primates (Taichman et al., 1980; Taichman et al., 1987). LtxA requires LFA-1 (lymphocyte function-associated antigen 1) receptor on host cells (Lally et al., 1999) and cholesterol for the binding to host cell membrane (Brown et al., 2013).
Genetic systems for studying Aggregatibacter species or other RTX-containing bacteria are beginning to be developed. Initial attempts to investigate the mechanistic properties and the activities of LtxA isolated from the clinical isolates (Diaz et al., 2006) have been explored while a shuttle vector, pJAK16, that permitted autoregulated high-level expression in Gram-negative bacteria was used in Aa complementation experiments (Balashova et al., 2007; Balashova et al., 2009). Some other experiments were also performed in our lab using cell lysates of LtxA expressing E. coli constructs pSHH (Brown et al., 2013) and pMG1 (Lally et al., 1994). To facilitate rapid purification of LtxA for our structural and functional studies, we produced his-tagged toxin in E. coli. Here we report the development of an efficient system for the active and inactive recombinant LtxA purification in E. coli. In our approach the genes for the LtxA expression and post-translational modification were cloned under separate promoters, which allows a greater range for genetic manipulation in the future. The recombinant toxins are expressed as double–His6-tagged proteins and can be purified in one-step procedure. We characterized biological and physical properties of the different toxin forms and demonstrated that the recombinant LtxA closely resembles the properties of the native toxin suggesting usefulness of the recombinant protein for further structure/functional analysis of LtxA.
MATERIALS AND METHODS
Aa strains
Aa, strain JP2 (Tsai et al., 1984) and CS001, a non-acylated mutant (Balashova et al., 2009) were grown on solid AAGM medium (Fine et al., 1999) for 48 h at 37 °C in 10% CO2 atmosphere. Strain CS001 was also grown in the presence of kanamycin (35 μg/ml).
Wild type (wt) and CS001 toxin purification
JP2 and CS001 Aa strains were grown in 1.5 L of liquid AAGM medium for 18 h. The supernatant was collected and the toxins were purified by ammonium sulfate precipitation from the bacterial culture supernatants as described previously (Diaz et al., 2006).
Plasmids construction
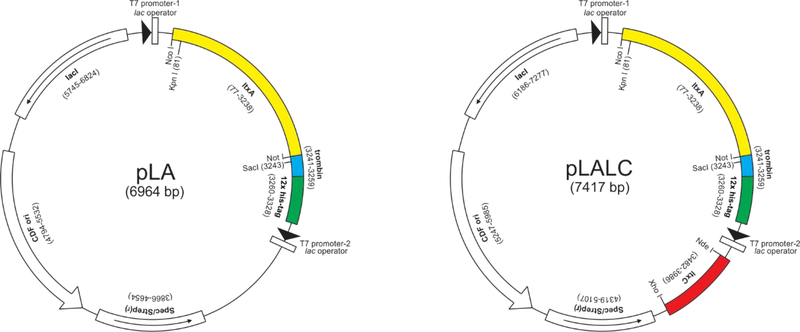
Genomic DNA was isolated from strain JP2 using DNeasy Blood & Tissue Kit™ (Qiagen, Thermo Fisher Scientific Inc., Waltham, MA). The ltxA and ltxC genes were amplified and cloned into pCDFDuet™−1 vector (Novagen, Madison, WI) using MCS sites (Nco I/Not I, ltxA; Nde I/Xho I, ltxC). Primers and oligonucleotides used for the cloning are listed in Table 1. The full sequences of plasmid expressing biologically inactive LtxA (pLA) and active LtxA (pLALC) are provided in Supplementary Materials.
Table 1. Primers used for the study.
The capital letters designate the gene/tag sequence.
| Gene/tag | Primer | Sequence (5’ to 3’) |
|---|---|---|
| ltxA | Forward | ttaactttaataaggagatataccatgGCAGGTACCACTACACTGCCAAATACAAAACAG |
| Reverse | tctgttcgacttaagcattatgcggccgcttagtggtgatgatggtgatggagctcAGCAGTAGTTGCTAACGAATTTGCGT | |
| ltxC | Forward | ttaagtataagaaggagatatacatATGGAGAAAAATAATAATTTTGAAGTGTTA |
| Reverse | cagcagcggtttctttaccagactcgagttaATTAATAGATATAACTTCAGATTTGT | |
| thrombin/double-His6 | Forward | ggccgcttaGTGATGATGGTGGTGGTGGCCCCCAAATTGGGGGTGACGCCAGGCGCGAGAGTGGTGATGGTGGTGATGCGAGCCACGTGGTACgagct |
| Reverse | cGTACCACGTGGCTCGCATCACCACCATCACCACTCTCGCGCCTGGCGTCACCCCCAATTTGGGGGCCACCACCACCATCATCACtaagc |
His-tagged LtxA expression and purification
Plasmids were transformed into E. coli BL21(λDE3) cells (Novagen, Madison, WI). Production of LtxA was achieved by inoculation of 200 ml of LB broth containing 50 μg/ml spectinomycin with overnight cultures at the ratio 1:100. Cells were cultivated at 37 °C in presence of autoinduction system ingredients (Novagen, Madison, WI) or 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) on the rotary shaker for 9 h-18 h at 200 rpm. The cell pellet was collected by centrifugation at 15,000 × g for 10 min, suspended in 8 ml 20 mM Tris, 250 mM NaCl and 200 μM CaCl2 pH 6.8 buffer containing protease inhibitors cocktail (Sigma-Aldrich, St. Louis, MO) and the cells were disrupted by sonication on ice for 6 cycles of 30 min on/30 min off using Virtis Virsonic 475 Sonicator (SP Scientific, Gardiner, NY). Insoluble components were collected by centrifugation at 18,000 × g for 20 min. The inclusion bodies were dissolved by agitating in 50 mM Tris, 5 mM imidazole, 8 M Urea, 0.5 M NaCl, pH 8.0 buffer for 1h. Then, the debris was precipitated by centrifugation at 18,000 x g for 10 min and the supernatant was used for the batch purification employing Ni-NTA His-Bind Resin (EMDMilipore, Danvers, MA). Two ml of Ni-NTA His-Bind Resin were loaded into 0.7 × 15 cm Econo-Column Chromatography column (Bio-Rad, Hercules, CA), washed with binding buffer (50 mM Tris, 5 mM imidazole, 8 M Urea, 0.5 M NaCl, pH 8.0) and the solution containing solubilized inclusion body proteins was passed through the column. The column was then washed with 6 ml binding buffer and followed with 6 ml wash buffer (50 mM Tris, 50 mM imidazole 8 M urea, 0.5 M NaCl, pH 8.0). The toxin was eluted in 1 ml fractions with elution buffer (50 mM Tris, 500 mM imidazole, 8 M Urea, 0.5 M NaCl, pH 8.0). The dialysis of 500 μl toxin samples was performed in dialysis cassette (10 kDa MWCO; Pierce, Rockford, IL) against 4 L of buffer containing 20 mM Tris, 250 mM NaCl and 200 μM CaCl2 pH 6.8 at 4°C, overnight. The eluted fractions were kept frozen.
Protein assay
Proteins were resolved by 4 to 20% SDS-PAGE and visualized by staining with GelCode blue stain reagent (Pierce, Rockford, IL). The protein concentration was determined by absorption at 280 nm on A1 NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Cytotoxicity assay
THP-1 cells obtained from ATCC were maintained in RPMI 1640 medium containing 10% FBS and 0.05 mM 2-mercaptoethanol at 37 °C under 5% CO2. For toxicity tests, 100 ng/ml purified LtxA was added to 1 × 106 THP-1 cells in RPMI with 10% FBS and incubated for 3 or 24 h. The cell membrane permeability was determined with Trypan blue assay using Vi-cell Cell Viability Analyzer (Beckman Coulter, Miami, FL). All reactions were run in duplicate; the assay was performed three independent times. Untreated cells were used as controls.
Western blot analysis
The protein samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were subjected to Western blot analysis with anti-LtxA monoclonal antibody, 107A3A3 (DiRienzo et al., 1985) in hybridoma supernatants (1:1,000 dilution) or anti His-tag monoclonal anti-body (Novagen, Madison, WI) (1:10,000 dilution) and then secondary horseradish peroxidase (HRP)-goat anti-mouse IgG (Fc) (1:10,000) (Pierce, Rockford, IL).
Analysis of LtxA acylation by liquid chromatography tandem mass spectrometry (LC-MS/MS)
LC-MS/MS analysis was performed using a Q Exactive™ HF Orbitrap Mass Spectrometer™ mass spectrometer (Thermo Fisher Scientific, Waltham, MA) coupled with a Nano-ACQUITY UPLC system (Waters Corporation, Milford, MA). Samples were digested ingel with trypsin and injected onto UPLC Symmetry trap column (180 μm i.d. × 2 cm packed with 5 μm C18 resin; Nano-ACQUITY UPLC system (Waters Corporation, Milford, MA). Tryptic peptides were separated by reversed phase HPLC on a BEH C18 nanocapillary analytical column (75 μm i.d. × 25 cm, 1.7 μm particle size; Waters Corporation, Milford, MA) using a gradient formed by solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). Peptides were eluted at 200 nl/min for 5–28% B over 42 min, 28–50% B over 25.5 min, 50–90% B over 5 min, 90% B for 22.5 min before returning to 5% B in 0.5 min. A 30-min blank gradient was run between sample injections to minimize carryover. Eluted peptides were analyzed by the mass spectrometer set to repetitively scan m/z from 300 to 2000 in positive ion mode. The full MS scan was collected at 70,000 resolution followed by data-dependent MS/MS scans at 17,500 resolution on the 20 most abundant ions exceeding a minimum threshold of 20,000. Peptide match was set as preferred, exclude isotopes option and charge-state screening were enabled to reject unassigned and singly charged ions.
Peptide sequences were identified using MaxQuant 1.5.2.8 (Cox and Mann, 2008). MS/MS spectra were searched against a UniProt E. coli database containing Aa LtxA protein sequence or NCBI Aa database using full tryptic specificity with up to two missed cleavages, static carboxamidomethylation of cysteine, and variable oxidation of methionine, and protein N-terminal acetylation. Additional variable modifications interrogated are saturated acyl group, hydroxylated acyl group, and monounsaturated hydroxylated acyl group with 8 to 18 carbons (Fong et al., 2011). Consensus identification lists were generated with false discovery rates of 1% at protein, peptide and site levels. MS/MS spectra of identified acylated peptides were manually verified.
Circular dichroism (CD) spectroscopy
To obtain the secondary and tertiary spectra of the proteins in solution at room temperature, a Jasco J-1500 CD spectrometer (Jasco Inc., Easton, MD) was used. Protein solutions at a concentration of 10 mM were subjected to far UV CD measurements using a quartz cell with a 0.05 cm path length.
Liposome preparation was described previously (Brown et al., 2015). To obtain the secondary structure of the proteins in a membrane environment, the proteins, at a concentration of 10 mM in the presence of liposomes at a concentration of 0.25 mg/mL were subjected to far UV CD measurements on a Jasco J-815 spectrometer using a quartz cell with a 0.01 cm path length. After subtracting the buffer spectrum, the CD data were converted to the mean residue ellipticity (expressed as Mol. Ellip.) using the path length of the cell, the protein concentration, and the mean residue weight of 108.
The secondary structure was determined with DichroWeb using CONTIN/LL and either the SP175 references set (for solutions containing only protein) or the SMP180 reference set (for solutions containing protein and liposomes). (Abdul-Gader et al., 2011; Lees et al., 2006; Provencher and Glockner, 1981; van Stokkum et al., 1990; Whitmore and Wallace, 2004, 2008) Only helical content was evaluated, as CD provides more reliable results for that structure.
Planar lipid bilayers
The methods used for the lipid bilayer measurements have been described previously in detail (Benz et al., 1978). A Teflon chamber divided into two 5 mL compartments. The compartments are connected by a small circular hole with a surface area of about 0.4 mm2 and were filled with 1 M KCl, 10 mM MES, pH 6.0. Black lipid bilayer membranes were obtained by painting onto the hole solutions of 1% (w/v) asolectin (phospholipids from soybean, Sigma-Aldrich) or diphytanoyl phosphatidylcholine (DiphPC; Avanti Polar Lipids, Alabaster, AL) in n-decane. All salts were analytical grade and the temperature was maintained at 20°C during all experiments. The current across the membrane was measured with a pair of Ag/AgCl electrodes with salt bridges switched in series with a voltage source and a highly sensitive current amplifier Keithley 427 (Keithley Instruments, INC. Cleveland, OH). The amplified signal was recorded by a strip chart recorder (Rikadenki Electronics GmbH, Freiburg, Germany).
Statistical Analysis
The statistical analyses were performed using either Student’s t test or one-way analysis of variance using SigmaPlot® (Systat Software, Inc. Chicago, IL). The following statistical criteria were applied: p < 0.001, p < 0.05, and p < 0.01.
RESULTS
Construction of plasmids for the recombinant toxins expression
To construct plasmid pLA for expression of the LtxA protoxin (proLtxA), the ltxA gene from JP2 strain was cloned into pCDFDuet-1 vector and under control of T7 promoter-1 using NcoI/NotI sites (Fig.1A). Unique KpnI and SacI sites were the introduced in the beginning of ltxA and before stop codon, respectively, for easy in frame exchange of wt toxin gene to mutated variants. Our preliminary experiments demonstrated that expression of ltxA with C-terminus hexahistidine tag did not allow efficient binding of the toxin to the Co2+, Ni2+, or Zn2+ containing matrix to perform purification by metal affinity chromatography (data not shown). Therefore, to improve the binding characteristics of the polyhistidine tag, the construct was modified to provide C-terminal in frame fusion of LtxA to the double–His6 tag (HHHHHHSRAWRHPQFGGHHHHHH) (Khan et al., 2006). To obtain LtxA without purification tag the thrombin cleavage site (LVPRGS) was genetically inserted prior polyhistidine tag between SalI/NotI sites. Thrombin can be used to selectively cut between the arginine and glycine residues of the cleavage site in order to remove the purification tag from the toxin (Waugh, 2011).
Figure 1. Maps of genetic constructs for LtxA expression.
(A) To construct plasmid (pLA) for expression of the proLtxA, the ltxA gene from JP2 strain was cloned into pCDFDuet-1 vector using NcoI/NotI sites. Unique KpnI and SacI sites were introduced in the beginning of ltxA and before stop codon, respectively. The thrombin cleavage site and double–His6 tag were inserted prior to stop codon. (B) To construct plasmid (pLALC) for expression of the biologically active recombinant LtxA, the ltxC gene amplified from chromosomal DNA of the JP2 strain and the PCR product was cloned under a separate promoter in pLA using NdeI/XhoI sites.
The acyltransferase converting proLtxA to mature LtxA was identified is a product of the ltxC gene that is located in the ltx locus (Balashova et al., 2009). To produce biologically active recombinant LtxA, the ltxC gene amplified from chromosomal DNA of the JP2 strain and the PCR product was cloned under a separate T7 promoter (T7 promoter-2) using NdeI/XhoI sites (Fig.1B). The resulting plasmid, pLALC, allowing co-expression of the ltxC and ltxA gene products and thereby production and purification of the acylated and therefore biologically active LtxA toxin (LtxAREC).
Purification of recombinant toxins
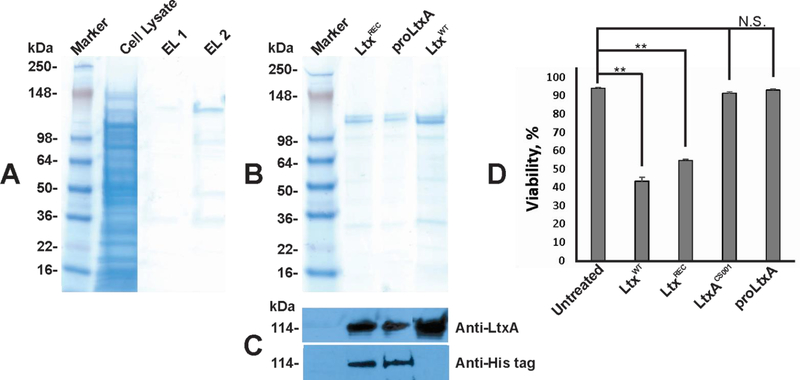
The LtxAREC and proLtxA proteins were produced as double–His6 tag proteins in E. coli BL21(λDE3) cells. When solubilized with 8 M urea, LtxA was observed in both the cytosol and inclusion bodies. Overnight Express™ Autoinduction System 1 (Novagen) was found to produce higher levels of the toxin expression than 1 mM IPTG and was therefore used in these studies. The toxins purified from the cell lysates on Ni-NTA agarose (Qiagen) typically yield ~20 mg/L per liter of culture. The toxin was bound to the column at 5 mM imidazole concentration and eluted with 500 mM imidazole. Figure 2A shows batch purification on LtxAREC on Ni-NTA column. Purified proLtxA and LtxAREC were recognized by both monoclonal anti-LtxA and anti His-tag antibody (Fig. 2C). The functionality of the recombinant toxins were tested in cytotoxicity assays on THP-1 cells. Neither exposure of the toxin to imidazole nor 8 M urea inhibited the activity. While purified LtxAREC was cytotoxic to a THP-1 cells, which correlates with previously observed toxicity of the native LtxA (LtxAWT) (Fig. 2D). No cytotoxicity was observed with proLtxA (Fig. 2D).
Figure 2. Toxins purification.
(A) SDS-PAGE of polyhistidine-tagged LtxA purification from pLALC. The inclusion bodies from E. coli BL21(λDE3) cells transfected with pLALC were solubilized and the toxins were purified as described in Materials and Methods. The cell lysates and the samples after elution after toxin samples were separated by SDS-PAGE. The major band of approximately 114 kDa was detected on SDS-PAGE in the purified active sample (EL1 and EL2). (B) SDS-PAGE and (C) Western blot analysis of dialyzed recombinant toxins samples and LtxAWT. The immunodetection of LtxA was performed with monoclonal antibody 107A3A3 (DiRienzo et al., 1985) and anti His-tag (Novagen). (D) In vitro assay of LtxA toxicity. For toxicity tests, 100 ng toxin samples were added to 0.5 × 106 THP-1 cells in RPMI with 10% FBS and incubated for 3 h at 37 °C. The cell viability was estimated by Trypan blue assay using Vi-cell counter. Untreated THP-1 cells served as control. The data in the figure are representative of the three experiments.
LtxAREC modification
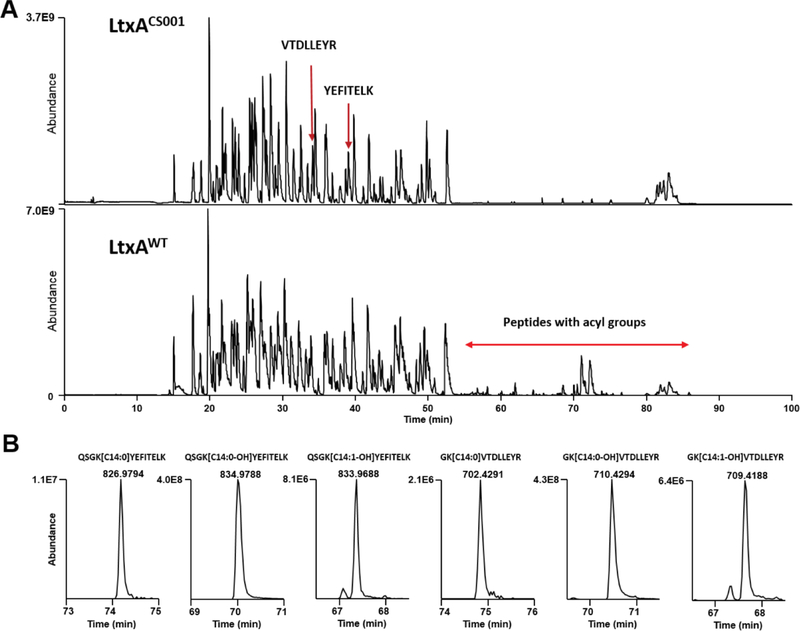
In previous studies we identified the sites for the fatty acid modification in LtxA (Balashova et al., 2009; Fong et al., 2011). Similar LC-MS/MS analyses were used to characterize the toxins acylation sites in this study. The reverse-phase LC gradient was adjusted to include a 23 min hold at 90% acetonitrile to ensure elution of very hydrophobic acylated peptides (Fig. 3A). LC-MS/MS of tryptic peptides from_both LtxAWT and LtxAREC confirmed that Lys562 and Lys687 are the sites of acyl group attachment and the type of the acylation detected is similar. Both LtxAWT and LtxAREC are modified with predominantly 14-carbon saturated acyl group, hydroxylated acyl group, and monounsaturated hydroxylated acyl group (Fig. 3B). No acylation was detected in LtxA purified from Aa CS001 (LtxACS001), containing transposon insertion in ltxC (Balashova et al., 2009) and proLtxA. Acyl groups greater than 16-carbon were not detected by LC-MS/MS even after replacing acetonitrile in LC solvent B with isopropanol which has a stronger elution strength.
Figure 3. LC-MS/MS analysis of leukotoxin proteins.
(A) Base peak chromatograms of LtxAWT, and LtxACS001 tryptic digests. The tryptic peptides VTDLLEYR and YEFITELK were observed in LtxACS001 because the unacylated preceding lysine residues were susceptible to trypsin cleavage. Peptides acylated at K562 and K687 were detected in LtxAWT at retention time 55–86 min. (B) Extracted ion chromatograms of doubly charged ions corresponding to the major types of acylated peptides from LtxAWT. These acylated peptides were not detected in LtxACS001.
Pore forming activity
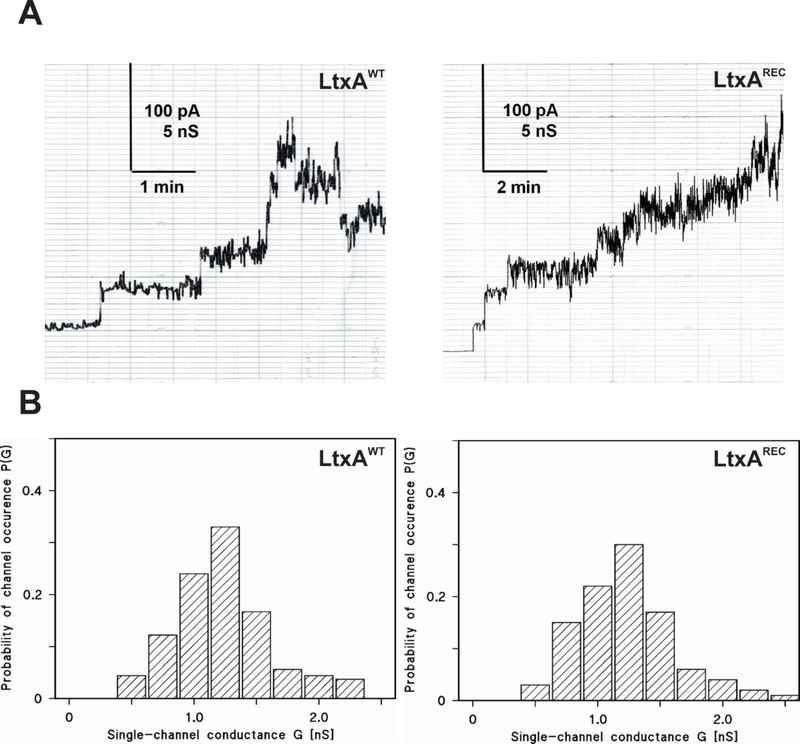
The pore forming capacity of LtxAWT and LtxAREC were compared using planar lipid bilayers. In initial experiments, membranes formed using DiphPC/n-decane had a relatively low pore-forming activity however, the results we obtained were similar to those reported for HlyA from E. coli and other RTX toxins such as Morganella morganii, Proteus vulgaris and Kingella kingae (Barcena-Uribarri et al., 2015; Benz, 2016; Benz et al., 1994a; Benz et al., 1994b; Benz et al., 1989; Schmidt et al., 1996). Lipid bilayer membranes formed with asolectin/n-decane, LtxAWT had a much higher pore-forming activity. Fig. 4A shows a typical current recording of an asolectin/n-decane membrane in the presence of wild type LtxAWT, added in small concentration (50 ng/ml) to the aqueous phase on one side, the cis-side of a black asolectin/n-decane membrane. Shortly after the addition of the toxin to the aqueous salt solution, the conductance started to increase in steps (Fig. 4A). An increase of the current noise indicated that the LtxAWT channels formed pores that showed a considerable current noise that increased with pore number making it difficult to ascertain the contribution of individual pores. Nevertheless, it was possible to calculate the average single-channel conductance by only taking membranes into account where up to approximately four to five LtxAWT pores were present in the membrane at the same time. Analysis of the LtxAWT pores, as shown in the histogram in Fig. 4B, exhibited a broad distribution of pores, possibly caused by the high current noise of the single conductance units. The conductance of the LtxAWT pores was about 1.2 nS in the buffer containing 1 M KCl, 10 mM MES, pH 6.0 (Fig. 4B).
Figure 4. Analysis of the LtxAWTand LtxAREC activity in lipid bilayer membranes.
(A) Single-channel recordings. Current recording of an asolectin/n-decane membrane performed in the presence of 50 ng/ml toxins added to the cis-side of the membrane. The aqueous phase contained 1 M KCl, 10 mM MES, pH 6. The applied membrane potential was 20 mV at the cis-side; T = 20°C. (B) Histograms of the probability P(G) for the occurrence of a given conductivity unit in membranes formed of 1% asolectin dissolved in n-decane in the presence of LtxAWT or LtxAREC. The histogram was calculated by dividing the number of fluctuations with a given conductance unit by the total number of conductance fluctuations. The average conductance of all conductance steps for LtxAWT was 1.20 nS for 95 conductance steps derived from 17 individual membranes. The average conductance of all conductance steps for LtxAREC was 1.30 nS for a total of about 74 single-channel events derived from 15 individual membranes.
“Noisy” pores were also encountered with LtxAREC at a similar concentration (50 ng/ml) to the cis-side of black asolectin/n-decane membranes (see Fig. 4A). Also for LtxAREC it was not possible because of the high current noise to clearly detect the pores when more than four to five pores were inserted into the membranes. In general, we were not able to detect any difference of the pores formed by the two versions of LtxA because also the histogram of LtxAREC showed a similar broad distribution as LtxAWT (see Fig. 4B). This was also valid for the single channel conductance of the LtxAREC pores, which had on average a conductance of about 1.3 nS under the same conditions as for LtxAWT. Both proLtxA and LtxACS001 did not express any pore-forming activity in asolectin membranes at the concentrations up to 500 ng/ml.
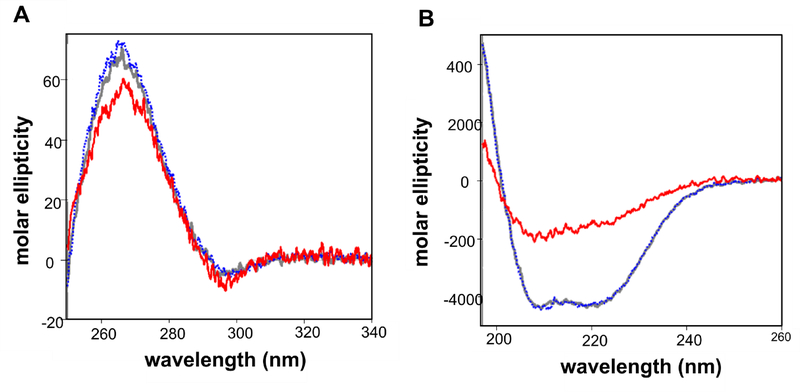
CD Spectroscopy
CD spectroscopy to investigate the structures of LtxAWT, LtxAREC and LtxACS001. Fig. 5 shows the near UV CD and the far spectra in mean residue ellipticity (expressed as Mol. Ellip.). The near UV CD spectra (Fig. 5A) of LtxAWT and LtxAREC are superimposable, indicting an identical tertiary structure. The spectra are highly similar to the spectrum of CS001, indicating its similar tertiary structure. The spectra are characterized by a negative peak at 296 nm arising from tryptophan residues and positive signals below 290 nm, indicating that aromatic amino acids and/or disulfide bonds are in folded environments. Thus, it is evident that these three proteins are folded.
Figure 5. CD spectra of leukotoxin proteins.
(A) Near UV spectra of LtxAWT (gray), LtxAREC (blue), and LtxACS001 (red). The spectra of all three proteins are nearly identical, indicating that these proteins are folded with the same tertiary structure. (B) Far UV spectra of LtxAWT (gray), LtxAREC (blue), and LtxACS001 (red). The spectra of LtxAWT and LtxAREC are superimposable, demonstrating that they have an identical secondary structure, while the spectra of LtxACS001 indicates a decrease in helical structure.
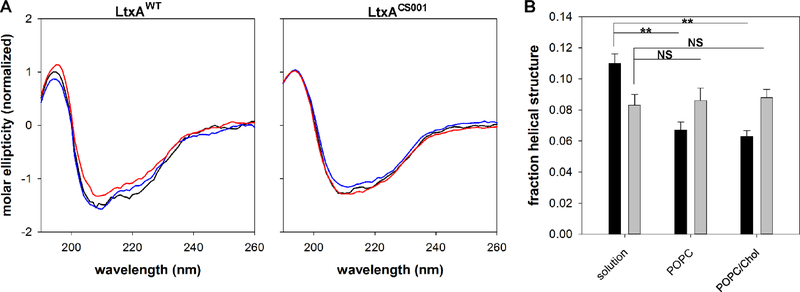
Fig. 5B shows the far UV CD spectra of the three proteins. The spectra of LtxAWT and LtxAREC are superimposable, suggesting that they have an identical secondary structure. Both spectra show signatures of helical structure, with double minima at 209 and 220 nm. Fig. 5 also shows the far UV CD spectrum of LtxACS001, which has a significantly different from the spectra of LtxAWT and LtxAREC. The negative signals are greatly reduced, consistent with a reduced helical content. Together with the near UV CD data, these results indicate that although LtxACS001 is folded, it has reduced helical content compared to LtxAWT and LtxAREC. We then performed CD spectroscopy analysis of LtxAWT and LtxACS001 upon incubation with membranes composed of 100% POPC, and 60% POPC/ 40% cholesterol (Fig. 6). As LtxAWT moves from solution to either a POPC or POPC/ Cholesterol membrane, the negative peak at 222 nm decreases (Fig. 6A), indicative of a loss in helicity. Deconvolution of the spectra demonstrates that the helical structure of LtxAWT decreases as the protein moves from solution to either type of membrane; the structure of LtxAWT in a POPC membrane is not significantly different from its structure in a POPC/cholesterol membrane (Fig. 6B). Conversely, LtxACS001 exhibits no change in structure as it moves from solution to either a POPC membrane or a POPC/chol membrane (Fig. 6).
Figure 6. CD spectra of leukotoxin proteins in membrane environments.
(A) The far UV spectra of LtxAWT (left) and LtxACS001 (right) in solution (black), POPC (blue), or POPC/Chol (red) membranes was recorded. A decrease in the peak at 222 nm was observed as LtxAWT moved from solution to either type of membrane, indicating an increase in helical structure. LtxACS001 exhibited no difference in spectra as the protein moved from solution to a membrane environment. (B) Deconvolution of the LtxAWT (black) and LtxACS001 (grey) spectra demonstrate that LtxAWT undergoes a significant decrease in helicity upon interacting with either type of membrane, relative to its solution structure, while LtxACS001 does not. ** p < 0.001, NS not significant.
DISCUSSION
Limitations in genetic manipulations in Aa did not allow structural characterization of LtxA that plays critical role in the organism virulence. Here we present a new E. coli system for the expression and recovery of recombinant LtxA and LtxA mutants. The development of such system enables further genetic analysis of the protein structure to identify the structural domains of the toxin. This work provides a basis for studies of LtxA activation or analysis of LtxA mutations.
LtxA is transcribed as proLtxA and is not nontoxic to target cells. To become biologically active posttranslational fatty acid acylation prior to secretion at Lys562 and Lys687 is necessary (Balashova et al., 2009; Fong et al., 2011). Acylation is not essential for LtxA secretion (Balashova et al., 2009). Compare to other available expression systems, pSHH (Brown et al., 2013) and pMG1 (Lally et al., 1994), ltxA and ltxC genes were cloned under different T7 promoters of pCDFDuet-1 vector, designed to coexpress two target proteins in E. coli. Our approach would allow to achieve optimal yield and would facilitate further manipulation with two genes to analyze of the amino acid substitutions, truncated, or hybrid versions of the toxin.
Multistep purification chromatography procedures have been employed for purification of untagged RTX toxins. CyaA of B. pertussis (Ostolaza et al., 1991) and FrpC of N. meningitidis (Sebo et al., 1991) have been expressed in E. coli. Among RTX proteins his-tagged toxins production in E. coli was reported for Apx proteins of Actinobacillus pleuropneumoniae. Purified unmodified Apx proteins of A. pleuropneumonia contained hexahistidine tags on C- and N- terminal ends and were applied for porcine vaccinations (Sadilkova et al., 2012). An expression of active ApxIA toxin was achieved by subclonning apxIC into the expression plasmid and the toxin was used purinergic signaling study (Masin et al., 2013).
Double–His6 tags were demonstrated order magnitude stronger binding to Ni-NTA-modified surfaces than conventional hexahistidine tags (Khan et al., 2006). In addition, these tags were detected with greater sensitivity by anti His-tag antibodies than the single–His6 (Khan et al., 2006). Double–His6 tag consists of two hexahistidine regions separated by an 11-amino acid spacer. The double–His6 tag has been used for purification of antibody fragments and other proteins on Ni-NTA resin after in vitro protein synthesis (He and Taussig, 2001). In our experimental system, purification of LtxA was achieved by addition of double–His6 tag to the C-terminal end of the protein. Utilization of a double–His6 tag has played a critical role for increasing LtxA binding to Ni-NTA resin since the purification of the toxin expressed with single His6-tag did not yield significant amounts of protein.
Using this system, we were able to generate and purify an unacylated toxin proLtxA, to begin to investigate the role of acylation in interaction the toxin membrane of LtxA with membranes. We have previously shown that although LtxA undergoes membrane composition-independent tertiary structural changes upon interacting with membranes, changes in secondary structure are small and membrane composition specific (Walters et al., 2013). The results obtained here demonstrate that the helical nature of LtxAWT decreases as the protein moves from solution to a membrane, but that change is independent of the presence of cholesterol. Unacylated toxin exhibited no change in secondary structure.
The planar lipid bilayer assay is a highly sensitive method, which allows characterization of membrane damaging activity in small amounts of RTX-toxin samples (Barcena-Uribarri et al., 2015). Previous studies demonstrated a strong correlation between the pore forming activity of RTX-toxins on erythrocytes and macrophages with that in planar lipid bilayers (Benz et al., 1994b; Masin et al., 2005; Menestrina et al., 1987). Using membranes formed with 1% asolectin/n-decane resulted in a much higher pore forming activity than DiphPC/n-decane. While DiphPC/n-decane is a pure lipid, asolectin is a heterogeneous mixture of lipids, mostly phosphatidylcholine (20%) as it is also the case in biological membranes but it contains also different phospholipids and some glycolipids, typical for plants (Scholfield, 1981). On the other hand, the single channel conductance observed here and elsewhere for LtxA was very similar to that measured previously for RTX-toxins from different Gram-negative bacteria in lipid bilayer membranes at similar conditions (Barcena-Uribarri et al., 2015; Benz et al., 1994a; Benz et al., 1989; Lear et al., 1995; Schmidt et al., 1996).
It is noteworthy, that the pores formed by the two forms of LtxA exhibited a much higher current noise than pores formed by other RTX-toxins, such as HlyA of E. coli or RtxA of K. kingae (Barcena-Uribarri et al., 2015; Benz et al., 1989). “Noisy” behavior of LtxA proteins in planar bilayers can be an indication of conformational changes in the protein secondary structure upon interaction with some lipids as we demonstrated in our previous work (Walters et al., 2013).
Supplementary Material
Highlights:
The expression system for production of RTX toxin (LtxA) was developed an E. coli
Active and inactive LtxAREC forms were produced with C-terminal double–His6 tag
LtxAREC has a secondary structure consistent with LtxAWT
Functional activity of LtxAREC and LtxAWT in planar lipid bilayers was similar
The expression system will allow analysis of the LtxA protein structure
ACKNOWLEDGEMENTS
This work was supported by the United States National Institute of Health grant R01DE009517 (ETL). The authors thank Oskar Laur (Integrated biology Inc.) and Juan Reyes-Reveles (JRRDesign Inc.) for technical assistance. The authors are grateful to Radim Osicka (Institute of Microbiology, Czech Republic) for helpful discussions and suggestions.
ABBREVIATIONS
- Aa
Aggregatibacter actinomycetemcomitans
- E. coli
Escherichia coli
- CD
circular dichroism
- double–His6
double hexahistidine tag
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- HRP
horseradish peroxidase
- LAP
localized aggressive periodontitis
- LC-MS/MS
liquid chromatography tandem mass spectrometry (LC-MS/MS)
- LFA-1
lymphocyte function-associated antigen 1
- LtxAWT
leukotoxin purified from Aggregatibacter actinomycetemcomitans
- LtxAREC
recombinant leukotoxin purified from E. coli
- LtxACS001
unmodified leukotoxin from Aggregatibacter actinomycetemcomitans
- MCS
multiple cloning site
- proLtxA
LtxA protoxin, unmodified recombinant leukotoxin purified from E. coli
- RTX
Repeats in ToXin
- wt
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abdul-Gader A, Miles AJ, Wallace BA, 2011. A reference dataset for the analyses of membrane protein secondary structures and transmembrane residues using circular dichroism spectroscopy. Bioinformatics 27, 1630–1636. [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, Hughes C, Koronakis V, 2001. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J Mol Biol 313, 501–510. [DOI] [PubMed] [Google Scholar]
- Balashova NV, Park DH, Patel JK, Figurski DH, Kachlany SC, 2007. Interaction between leukotoxin and Cu,Zn superoxide dismutase in Aggregatibacter actinomycetemcomitans. Infection and immunity 75, 4490–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashova NV, Shah C, Patel JK, Megalla S, Kachlany SC, 2009. Aggregatibacter actinomycetemcomitans LtxC is required for leukotoxin activity and initial interaction between toxin and host cells. Gene 443, 42–47. [DOI] [PubMed] [Google Scholar]
- Barcena-Uribarri I, Benz R, Winterhalter M, Zakharian E, Balashova N, 2015. Pore forming activity of the potent RTX-toxin produced by pediatric pathogen Kingella kingae: Characterization and comparison to other RTX-family members. Biochim Biophys Acta 1848, 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann U, Wu S, Flaherty KM, McKay DB, 1993. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J 12, 3357–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz R, 2016. Channel formation by RTX-toxins of pathogenic bacteria: Basis of their biological activity. Biochim Biophys Acta 1858, 526–537. [DOI] [PubMed] [Google Scholar]
- Benz R, Hardie KR, Hughes C, 1994a. Pore formation in artificial membranes by the secreted hemolysins of Proteus vulgaris and Morganella morganii. Eur. J. Biochem 220, 339–347. [DOI] [PubMed] [Google Scholar]
- Benz R, Janko K, Boos W, Lauger P, 1978. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim Biophys Acta 511, 305–319. [DOI] [PubMed] [Google Scholar]
- Benz R, Maier E, Ladant D, Ullmann A, Sebo P, 1994b. Adenylate cyclase toxin (CyaA) of Bordetella pertussis. Evidence for the formation of small ion-permeable channels and comparison with HlyA of Escherichia coli. J. Biol. Chem 269, 27231–27239. [PubMed] [Google Scholar]
- Benz R, Schmid A, Wagner W, Goebel W, 1989. Pore formation by the Escherichia coli hemolysin: evidence for an association-dissociation equilibrium of the pore-forming aggregates. Infect. Immun 57, 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari EF, Cockerill FR 3rd, Steckelberg JM, 1997. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clinic proceedings. Mayo Clinic 72, 532–542. [DOI] [PubMed] [Google Scholar]
- Brogan JM, Lally ET, Poulsen K, Kilian M, Demuth DR, 1994. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infection and immunity 62, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AC, Balashova NV, Epand RM, Epand RF, Bragin A, Kachlany SC, Walters MJ, Du Y, Boesze-Battaglia K, Lally ET, 2013. Aggregatibacter actinomycetemcomitans leukotoxin utilizes a cholesterol recognition/amino acid consensus site for membrane association. The Journal of biological chemistry 288, 23607–23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AC, Koufos E, Balashova N, Boesze-Battaglia K, Lally ET, 2015. Inhibition of LtxA Toxicity by Blocking Cholesterol Binding With Peptides. Molecular oral microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M, 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Crosby JA, Kachlany SC, 2007. TdeA, a TolC-like protein required for toxin and drug export in Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene 388, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Badley AD, Cockerill FR, Steckelberg JM, Wilson WR, 1997. Infective endocarditis caused by HACEK microorganisms. Annual review of medicine 48, 25–33. [DOI] [PubMed] [Google Scholar]
- Diaz R, Ghofaily LA, Patel J, Balashova NV, Freitas AC, Labib I, Kachlany SC, 2006. Characterization of leukotoxin from a clinical strain of Actinobacillus actinomycetemcomitans. Microbial pathogenesis 40, 48–55. [DOI] [PubMed] [Google Scholar]
- DiRienzo JM, Tsai CC, Shenker BJ, Taichman NS, Lally ET, 1985. Monoclonal antibodies to leukotoxin of Actinobacillus actinomycetemcomitans. Infection and immunity 47, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH, 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol 44, 1063–1076. [DOI] [PubMed] [Google Scholar]
- Fong KP, Tang HY, Brown AC, Kieba IR, Speicher DW, Boesze-Battaglia K, Lally ET, 2011. Aggregatibacter actinomycetemcomitans leukotoxin is post-translationally modified by addition of either saturated or hydroxylated fatty acyl chains. Molecular oral microbiology 26, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett M, Guo L, Shabanowitz J, Hunt DF, Hewlett EL, 1994. Internal lysine palmitoylation in adenylate cyclase toxin from Bordetella pertussis. Science 266, 433–435. [DOI] [PubMed] [Google Scholar]
- Hardie KR, Issartel JP, Koronakis E, Hughes C, Koronakis V, 1991. In vitro activation of Escherichia coli prohaemolysin to the mature membrane-targeted toxin requires HlyC and a low molecular-weight cytosolic polypeptide. Molecular microbiology 5, 1669–1679. [DOI] [PubMed] [Google Scholar]
- Haubek D, Johansson A, 2014. Pathogenicity of the highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans and its geographic dissemination and role in aggressive periodontitis. J Oral Microbiol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Taussig MJ, 2001. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method). Nucleic Acids Res 29, E73–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B, Nair SP, Ward JM, Wilson M, 2003. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annual review of microbiology 57, 29–55. [DOI] [PubMed] [Google Scholar]
- Hoglund Aberg C, Haubek D, Kwamin F, Johansson A, Claesson R, 2014. Leukotoxic activity of Aggregatibacter actinomycetemcomitans and periodontal attachment loss. PLoS One 9, e104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issartel JP, Koronakis V, Hughes C, 1991. Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature 351, 759–761. [DOI] [PubMed] [Google Scholar]
- Khan F, He M, Taussig MJ, 2006. Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on ni-nitrilotriacetic acid surfaces. Anal Chem 78, 3072–3079. [DOI] [PubMed] [Google Scholar]
- Lally ET, Golub EE, Kieba IR, 1994. Identification and immunological characterization of the domain of Actinobacillus actinomycetemcomitans leukotoxin that determines its specificity for human target cells. The Journal of biological chemistry 269, 31289–31295. [PubMed] [Google Scholar]
- Lally ET, Hill RB, Kieba IR, Korostoff J, 1999. The interaction between RTX toxins and target cells. Trends in microbiology 7, 356–361. [DOI] [PubMed] [Google Scholar]
- Lear JD, Furblur UG, Lally ET, Tanaka JC, 1995. Actinobacillus actinomycetemcomitans leukotoxin forms large conductance, voltage-gated ion channels when incorporated into planar lipid bilayers. Biochim. Biophys. Acta 1238, 34–41. [DOI] [PubMed] [Google Scholar]
- Lees JG, Miles AJ, Wien F, Wallace BA, 2006. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics 22, 1955–1962. [DOI] [PubMed] [Google Scholar]
- Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P, 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev 34, 1076–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masin J, Basler M, Knapp O, El-Azami-El-Idrissi M, Maier E, Konopasek I, Benz R, Leclerc C, Sebo P, 2005. Acylation of lysine860 allows tight binding and cytotoxicity of Bordetella adenylate cyclase on CD11b-expressing cells. Biochemistry 44, 12759–12766. [DOI] [PubMed] [Google Scholar]
- Masin J, Fiser R, Linhartova I, Osicka R, Bumba L, Hewlett EL, Benz R, Sebo P, 2013. Differences in purinergic amplification of osmotic cell lysis by the pore-forming RTX toxins Bordetella pertussis CyaA and Actinobacillus pleuropneumoniae ApxIA: the role of pore size. Infection and immunity 81, 4571–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menestrina G, Mackman N, Holland IB, Bhakdi S, 1987. Escherichia coli haemolysin forms voltage-dependent ion channels in lipid membranes. Biochimica et Biophysica Acta 905, 109–117. [DOI] [PubMed] [Google Scholar]
- Ostolaza H, Bartolome B, Serra JL, de la Cruz F, Goni FM, 1991. Alpha-haemolysin from E. coli. Purification and self-aggregation properties. FEBS letters 280, 195–198. [DOI] [PubMed] [Google Scholar]
- Provencher SW, Glockner J, 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37. [DOI] [PubMed] [Google Scholar]
- Sadilkova L, Nepereny J, Vrzal V, Sebo P, Osicka R, 2012. Type IV fimbrial subunit protein ApfA contributes to protection against porcine pleuropneumonia. Vet Res 43, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Maier E, Karch H, Benz R, 1996. Pore-forming properties of the plasmid-encoded hemolysin of enterohemorrhagic Escherichia coli O157:H7. Eur J Biochem 241, 594–601. [DOI] [PubMed] [Google Scholar]
- Scholfield C, 1981. Composition of soy bean lecithin. J Am Oil Chem Soc 58, 889–892. [Google Scholar]
- Sebo P, Glaser P, Sakamoto H, Ullmann A, 1991. High-level synthesis of active adenylate cyclase toxin of Bordetella pertussis in a reconstructed Escherichia coli system. Gene 104, 19–24. [DOI] [PubMed] [Google Scholar]
- Taichman NS, Dean RT, Sanderson CJ, 1980. Biochemical and morphological characterization of the killing of human monocytes by a leukotoxin derived from Actinobacillus actinomycetemcomitans. Infection and immunity 28, 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taichman NS, Simpson DL, Sakurada S, Cranfield M, DiRienzo J, Slots J, 1987. Comparative studies on the biology of Actinobacillus actinomycetemcomitans leukotoxin in primates. Oral microbiology and immunology 2, 97–104. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Shenker BJ, DiRienzo JM, Malamud D, Taichman NS, 1984. Extraction and isolation of a leukotoxin from Actinobacillus actinomycetemcomitans with polymyxin B. Infection and immunity 43, 700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC, 1990. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem 191, 110–118. [DOI] [PubMed] [Google Scholar]
- Walters MJ, Brown AC, Edrington TC, Baranwal S, Du Y, Lally ET, Boesze-Battaglia K, 2013. Membrane association and destabilization by Aggregatibacter actinomycetemcomitans leukotoxin requires changes in secondary structures. Molecular Oral Microbiology 28, 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh DS, 2011. An overview of enzymatic reagents for the removal of affinity tags. Protein Expr Purif 80, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA, 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA, 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400. [DOI] [PubMed] [Google Scholar]
- Zambon JJ, 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol 12, 1–20. [DOI] [PubMed] [Google Scholar]
- Zambon JJ, 1996. Periodontal diseases: microbial factors. Annals of periodontology / the American Academy of Periodontology 1, 879–925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.