SUMMARY
CRISPR-Cas systems provide acquired immunity in prokaryotes. Upon infection, short sequences from the phage genome, known as spacers, are inserted between the CRISPR repeats. Spacers are transcribed into small RNA molecules that guide nucleases to their targets. The forces that shape the distribution of newly acquired spacers, which is observed to be uneven, are poorly understood. We studied the spacer patterns that arise after phage infection of Staphylococcus aureus harboring the Streptococcus pyogenes type II-A CRISPR-Cas system. We observed that spacer patterns are established early during the CRISPR-Cas immune response and correlate with spacer acquisition rates, but not with spacer targeting efficiency. The rate of spacer acquisition depended on sequence elements within the spacer, which in turn determined the abundance of different spacers within the adapted population. Our results reveal how the two main forces of the CRIPSR-Cas immune response, acquisition and targeting, affect the generation of immunological diversity.
INTRODUCTION
CRISPR (clustered regularly interspaced short palindromic repeats) loci and their CRISPR-associated (Cas) proteins protect prokaryotes against infection by viruses (Barrangou et al., 2007) and plasmids (Marraffini and Sontheimer, 2008). Upon phage infection, a low fraction of cells acquire and integrate short fragments of the invader’s DNA (known as spacers) between CRISPR repeat sequences (Barrangou et al., 2007). After integration, spacers are transcribed and processed into small CRISPR RNAs (crRNAs) (Brouns et al., 2008; Deltcheva et al., 2011) that are used by Cas nucleases to find their complementary sequences (protospacers) within the invading genetic element and cleave it. In the type II-A CRISPR-Cas system from Streptococcus pyogenes, cleavage is performed by the crRNA-guided nuclease Cas9 (Garneau et al., 2010; Jinek et al., 2012), whose catalytic activity depends on the recognition of a 5’-NGG-3’ protospacer adjacent motif (PAM) (Jiang et al., 2013; Jinek et al., 2012). Cas9 contains a PAM-interacting domain to recognize this motif (Anders et al., 2014; Jiang et al., 2016) that is not only required for target cleavage but also for the acquisition of spacers matching protospacers flanked by the appropriate PAM (Heler et al., 2015).
Besides the presence of a functional PAM, the rules that govern spacer acquisition in type II CRISPR-Cas systems are not completely understood. Multiple studies have shown an uneven pattern of spacer acquisition, where different spacer sequences have markedly different abundances within the population of cells that survive phage infection (Heler et al., 2015; Paez-Espino et al., 2013; Paez-Espino et al., 2015). This observation led to the hypothesis that some spacers become overrepresented because they are more effective at directing targeting and/or cleavage by Cas9 and therefore have a selective advantage (Paez-Espino et al., 2013). However, even when spacer acquisition was measured within 30 minutes of infection, i.e. before the viral lytic cycle is completed and the spacers cannot be selected for their abilities to guide DNA destruction, the pattern of spacer acquisition is constricted to the viral region that is first injected but with highly variable frequencies of acquisition for different spacers sequences within this genomic location (Modell et al., 2017). These data suggest that the abundance of a spacer in the bacterial population can be independent of its targeting properties and determined solely by its acquisition rate.
Here we used the type II-A CRISPR-Cas system from Streptococcus pyogenes expressed in Staphylococcus aureus RN4220 cells (Heler et al., 2015) to investigate the mechanisms behind the pattern of spacer acquisition when cells are infected with the staphylococcal phage ϕNM4γ4 (Goldberg et al., 2014; Heler et al., 2015). We found that this pattern is remarkably reproducible, and by measuring spacer abundance early and late during the CRISPR-Cas immune response, we showed that the frequency of individual spacers is mainly determined at the onset of infection and that there is little selection of spacer sequences thereafter. This led to the hypothesis that spacer abundance depends on the rate of acquisition rather than enhanced Cas9 cleavage activity. We tested this on selected spacer sequences at each end of the distribution spectrum by performing targeting assays and quantifying CRISPR acquisition of spacer-length oligonucleotides. These experiments demonstrated that high and low abundance spacers have similar targeting abilities but differ dramatically in their efficiency of acquisition. Our studies reveal that, for type II-A systems, spacer acquisition rates are fundamental to determine the distribution and diversity of the CRISPR-Cas immune response.
RESULTS
Acquired spacer sequences display a consistent distribution pattern.
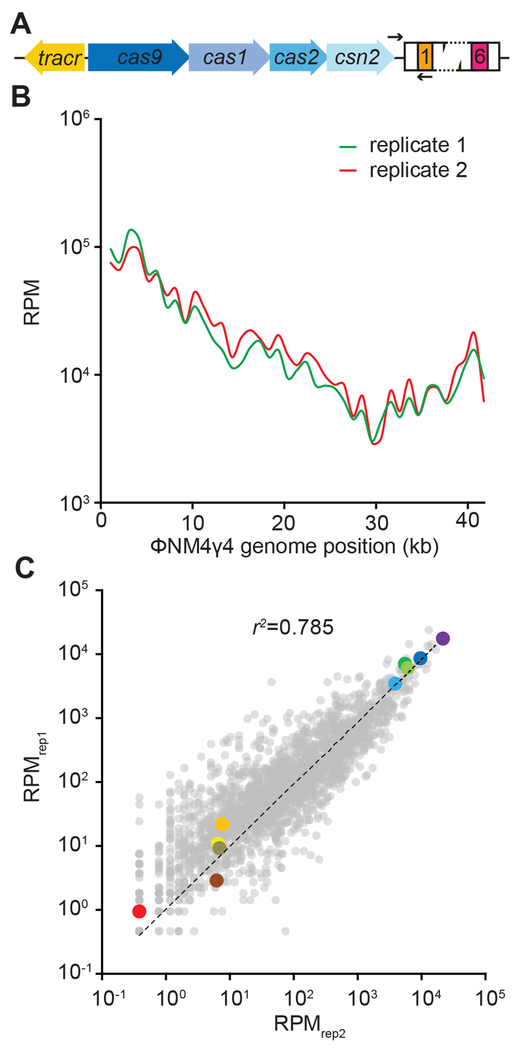
To analyze spacer distribution in the type II-A CRISPR-Cas system of S. pyogenes (Fig. 1A) we performed infection assays with lytic phage ϕNM4γ4, as described previously (Heler et al., 2017). DNA from surviving cells obtained 24 hours after infection was used to amplify the CRISPR array by PCR and perform next generation sequencing of newly acquired spacers. We performed the infection in duplicate and obtained two libraries of 2.52 and 2.28 million phage-mapping reads, respectively. Of all the possible 2,318 NGG-adjacent protospacers on the genome of ϕNM4γ4, 2,096 (>90%) were sampled in both libraries (Data S1). The frequency of each spacer was normalized as reads per million (RPM) and plotted across the phage genome (1 kb bins, Fig. 1B). We observed a similar pattern of spacer distribution for each duplicate experiment. This pattern was not a reflection of the PAM distribution across the phage genome (Data S1). To determine if the correlation is present not only in the groups of spacers within each 1 kb bin, but also at the level of the individual spacer sequences, we compared the RPM value for each of the 2,096 spacers (Fig. 1C). We found a remarkable correlation of the spacer frequencies in both replicas, particularly of the most abundant spacer sequences. We arbitrarily picked five spacer sequences with high and five with low RPM and marked them with different colors to follow their abundance over different experiments. This is an effort to illustrate the relative consistency in the distribution of individual spacer sequences, for example after mapping the spacers across the phage genome in our replicates (Fig. S1A–C). To test if this correlation extends to experiments using other phages and type II-A CRISPR-Cas systems, we performed duplicate infection experiments of S. aureus RN4220 containing the S. pyogenes type II-A system with the phage ϕ85 (Kwan et al., 2005) (Fig. S1D), or staphylococci harboring the type II-A (also known as CRISPR3 (Deveau et al., 2008)) from Streptococcus thermophilus with ϕNM4γ4 (Fig. S1E). Although we obtained only 50-100 different spacer sequences (Data S1) in both cases due to a low efficiency of spacer acquisition in these systems (Heler et al., 2015)), a very strong correlation for spacer abundance in the replicas was found. Altogether, these results indicate that the abundance of individual spacer sequences within the population of surviving cells is relatively constant after the type II-A CRISPR-Cas immune response.
Figure 1. Acquired spacer sequences display a consistent distribution pattern.
(A) Schematic diagram of the type II-A CRISPR-Cas system from Streptococcus pyogenes. Black arrows indicate the position of the PCR primers used to check for spacer integration. (B) Average abundance (in reads per million per 1-kb bins, RPM) of φNM4γ4 viral sequences incorporated as spacers into the CRISPR array, mapped against location on the phage genome, in duplicate (red and green traces). (C) Individual spacers common to the two data sets in (B) were plotted with RPM values for replicate 1 on the x axis and replicate 2 on the y axis. The dotted line represents the linear regression fit. Ten spacers were color-coded based on their abundance (warm colors for low abundance and cold colors for high abundance). See also Fig. S1.
Highly abundant spacer sequences have high rates of acquisition.
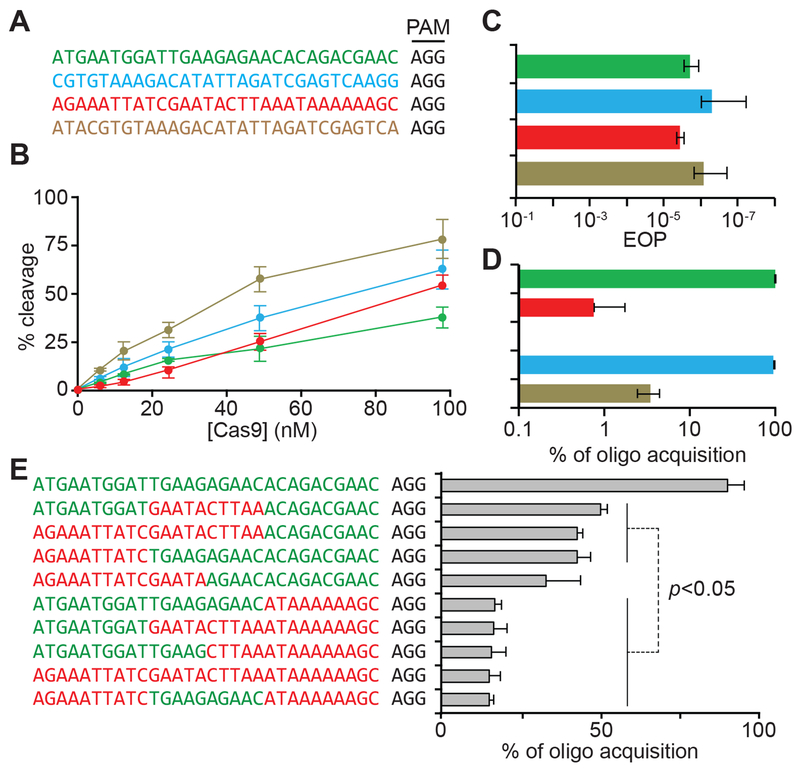
In principle, the different but reproducible abundance of spacers could be explained by two non-mutually exclusive forces that depend on their individual sequences: their efficiency of viral targeting and/or their inherent frequency of acquisition. We tested these possibilities using spacer sequences (Figs. 2A and S2A) that displayed markedly dissimilar abundances in both of our replicates (Fig. 1C): two highly abundant (“dark green” and “light blue”) and two consistently under-represented (the “red” and “tan”). First, we compared the efficiency of in vitro DNA cutting by Cas9 using each of these spacers as guides and we found similar cleavage properties (Figs. 2B and S2B–E). Second, we measured the targeting efficiency of each of these spacers in vivo, through the quantification of the reduction in phage propagation that they mediate, to determine if the frequency of phage escape correlated with spacer abundance (Fig. 2C). We did not detect substantial differences between the spacers, a result that demonstrates that not only in vitro, but also in vivo, these sequences provide similar levels of defense.
Figure 2. High and low abundance spacers have different rates of acquisition but similar targeting efficiencies.
(A) Sequences of the targets of select spacers from Figure 1 with either high abundance (dark green and light blue) or low abundance (red and tan); all containing an AGG PAM. (B) Quantification of in vitro cleavage (after 5 minutes) of a 2-kb phage target by various concentrations of Cas9 loaded with sgRNAs matching the protospacers shown in (A). (C) Phage propagation on strains harboring the spacers shown in (A), measured as the efficiency of plaquing (EOP) against propagation in non-CRISPR control staphylococci. (D). Relative acquisition rates (%) of spacers following electroporation of pairs of high/low abundance dsDNA oligonucleotides with the sequences shown in (A). (E) Relative acquisition rates (%) of spacers following electroporation of 10 dsDNA oligonucleotides with mixed sequences of the dark green and red targets shown in (A). See also Fig. S2.
Next, we tested the second variable that could impact the distribution pattern of spacers: their intrinsic rate of acquisition. We co-transformed cells with pairs of annealed, dsDNA oligonucleotides at equimolar concentrations, harboring the sequences over- and under-represented spacers. To increase the frequency of acquisition of the oligos, we used staphylococci carrying an engineered type II-A CRISPR-Cas locus in which expression of the cas1, cas2 and csn2 genes is controlled by an anhydrotetracycline-inducible promoter (Heler et al., 2015; Modell et al., 2017), allowing their over-expression to enhance spacer integration. Transformation was followed by next-generation sequencing of the amplified CRISPR array to quantify the relative frequency of acquisition for each transformed oligonucleotide. We compared the acquisition of the selected over- and under-represented sequences (Fig. 2A): “dark green” vs “red” and “light blue” vs. “tan”. We observed a striking difference in the number of reads, with ~ 96 % of the reads from oligo-derived spacers matching the highly abundant sequence (Data S1 and Fig. 2D). To corroborate this finding, we performed spacer-specific PCR after transformation using the “dark green” or “red” spacer sequences as reverse primers to amplify the CRISPR array. Consistent with our next generation sequencing data, we were able to detect a strong PCR product only when using the highly acquired spacer as reverse primer (Fig. S2F). Finally, we compared the frequency of acquisition of another high- and low-abundance spacer pair (the “light green” and “orange” spacers in Figure 1C, respectively), and observed the same differential integration into the CRISPR array (Data S1 and Fig. S2G). Altogether, these experiments demonstrate that for a given spacer sequence, its efficiency of acquisition but not its targeting capabilities, correlate with its abundance in the population of CRISPR-resistant cells.
PAM-proximal sequences determine the frequency of spacer acquisition.
The above results suggest that there must be elements within the sequence of high-abundance spacers that increase their rate of acquisition. This has been previously described for spacers acquired during the Escherichia coli type I CRISPR-Cas immune response (Shipman et al., 2017; Yosef et al., 2013). Applying bioinformatics analysis of spacer groups composed with the highest and lowest frequencies of acquisition, it was determined that the presence of the correct PAM at the 3’ end of the spacer (AAG) as well as an AA dinucleotide located 30 nt downstream of the AAG motif significantly enhance the rate of acquisition of the spacers they flank. To test for a possible role of the flanking sequences we added the 15 nt that precede and follow the “dark green” and “red” protospacers (Fig. S2H). We then transformed these extended oligonucleotides and looked for their acquisition both by spacer-specific PCR analysis (Fig. S2F) and next generation sequencing of the amplified CRISPR locus (Fig. S2H). Both assays showed a remarkable preference of acquisition of the highly abundant “green” spacer over the under-represented “red” one. To check for the importance of the flanking sequences the additional 15 nt were swapped (Fig. S2H) and the resulting dsDNA oligonucleotides were transformed into staphylococci carrying the S. pyogenes type II-A CRISPR-Cas system. Next generation sequencing of the expanded CRISPR arrays showed that the protospacer, but not its flanking sequences, is the main determinant of the efficiency of acquisition (Fig. S2H).
To look for the presence of protospacer sequence elements that affect its acquisition, we divided the 30-nt sequence of the “dark green” and “red” spacers into PAM-distal, middle and PAM-proximal 10-nt regions (Fig. S2I) and swapped these regions in the high and low abundance spacer sequences. Electroporation with different pairs of swapped oligos, followed by next generation sequencing of expanded CRISPR arrays revealed that the presence of the 10-nt PAM-proximal region of the high-abundance spacer was necessary and sufficient to ensure high levels of acquisition of a dsDNA oligo (Fig. S2I). Moreover, the addition of the 10-nt PAM-proximal region of the “dark green” highly acquired spacer, but not the middle or PAM-distal sequences, was also sufficient to increase the frequency of acquisition of the “orange” low-abundance spacer (Fig. S2J). To corroborate these findings, we co-transformed 10 different dsDNA oligonucleotides containing different combinations of 10-nt regions of the “dark green” and “red” spacer sequences (Fig. 2E). Again, we found that dsDNA oligos containing the 10-nt PAM-proximal sequence of the highly acquired spacer were integrated into the CRISPR array at significantly higher frequencies than those having the same region from the low-abundance spacer. Nevertheless, this sequence was not sufficient to make the acquisition of the “red” spacer as high as that of the “dark green” one, suggesting that there are additional stimulatory (in the “dark green”) or inhibitory (in the “red”) nucleotides that affect acquisition. Due to the impossibility of testing every acquired spacer via oligo transformation, we evaluated the importance of this sequence within the entire set of acquired spacers. To do this, we used kpLogo (Wu and Bartel, 2017) to look for a conserved motif in the PAM-proximal 10-nt sequence of the most abundant spacers (in the top 1 % of average spacer reads in Fig. 1C). We obtained two sets of sequences, corresponding to the enriched and depleted nucleotides at each position of the PAM-proximal region (Fig. S2K). Although the analysis did not yield any significant motif in this region, we picked the most conserved nucleotide in each position and appended the resulting sequences to the low abundance (“red”) spacer to check for their influence in spacer acquisition. We found that the addition of the enriched PAM-proximal nucleotides dramatically increased spacer acquisition (Fig. S2L). Finally, we investigated whether the rates of acquisition of different protospacers correlated with the kinetics of in vitro integration (Fig. S2M). While we did see variation in the rates of integration (Fig. S2N), the differences were unrelated to acquisition rate, with a poorly-acquired sequence supporting the fastest integration rate as a blunt protospacer. This, together with the primary importance of PAM-proximal sequences for selection in vivo, is consistent with a model where sequence preference is established at the protospacer selection stage, rather than during the integration reaction itself. The overall results of these experiments demonstrate that specific DNA sequences located immediately upstream of the PAM have important effects on the frequency of acquisition of the 30-nt spacer determined by that PAM.
The spacer distribution pattern is established early during infection.
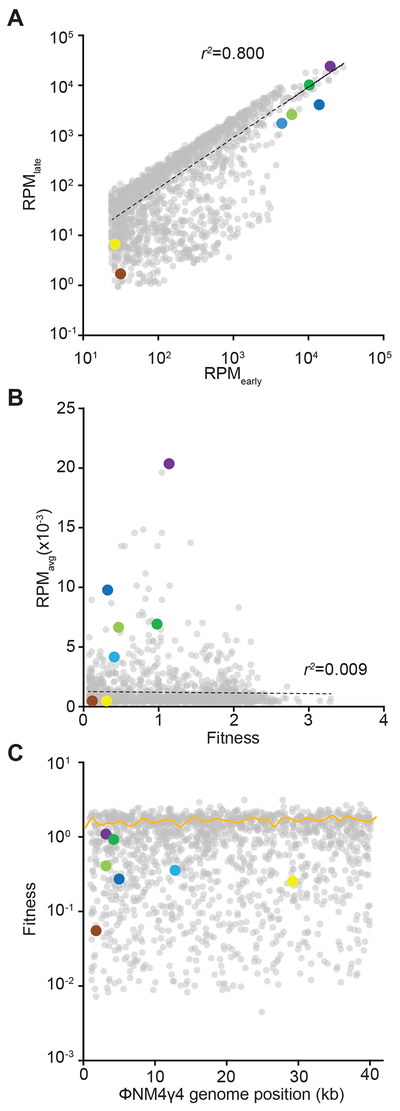
Our data that compared the targeting abilities and acquisition rates of a limited number spacer sequences showed that the latter, but not the former, correlates with the abundance of these spacers in the distribution pattern resulting after the type II-A CRISPR-Cas immune response. These results led us to formulate the hypothesis that the rate of acquisition of the different spacers early during infection, but not their subsequent selection for their phage cleavage properties, is the major force that shapes this pattern. To test this hypothesis, we compared the spacer distribution 30 minutes after infection, when the great majority of cells have not lysed yet [the ϕNM4γ4 viral cycle takes ~ 40 minutes (Modell et al., 2017)], with the distribution obtained after 16 hours of infection, a time during which the acquired spacers can be selected against or for their targeting properties (Fig. S3A). We analyzed over 0.72 million spacers for the early time point and 12.3 million spacers for the late time point, with 1,517 sequences shared between the two libraries (Data S1). We detected reads for 1614 spacers in the early time point and 2019 in the late one, with all of the non-detected spacers in the early time point having a very low number of reads (Data S1). This suggests that approximately 75% of the acquisition occurs in the first 30 minutes post-infection, and that the spacers acquired afterwards have a minimal contribution to the type II-A CRISPR-Cas immune response. When we compared the abundance of the spacers shared by both time points, we observed a strong correlation for the frequency of each individual spacer (Fig. 3A) and for their overall distribution across the phage genome (Fig. S3B–D). This result suggests that spacer abundance is determined early after infection, and selection throughout the re-growth of CRISPR-adapted cells has a minimal impact on shaping the spacer distribution. To explore this more directly, we calculated the fold-increase in abundance from the early time point to the late time point for each spacer. This value reflects the fitness of each sequence after its acquisition; i.e., the positive or negative selection suffered by a spacer due to its targeting abilities. We found that the fitness range of the entire spacer repertoire was narrow and did not correlate with the average spacer abundance obtained in the Figure 1C replicates (Fig. 3B). For example, our set of highly abundant spacers had average fitnesses close to 1, even though they were order of magnitudes more frequent than other spacers with similar fitnesses (Fig. 3B). Interestingly, we did not detect a strong positive selection for any spacer sequence (the maximum fitness value was 3.3, Data S1), but there were 14 that displayed more than a 100-fold negative selection (Data S1, Fig. 3C, Fig. S3E). On average, the acquired spacers have a fitness value close to 1 (Fig. 3C), with approximately half of them displaying fitness higher than 1 and half lower than 1 (Fig. S3E). These findings indicate that the relative abundance of spacer sequences is determined at their time of acquisition, early during the CRISPR-Cas immune response, and remains relatively constant during the targeting phase of CRISPR immunity.
Figure 3. The spacer distribution pattern is established early during infection.
(A) Individual spacers common to the early and late time point samples plotted as RPM values against each other. (B) Average spacer abundance obtained from the replicates of Figure 1C as a function of spacer fitness calculated as RPMlate/RPMearly). (C) Fitness mapped across the phage genome. The yellow curve represents average fitness in 1-kb bins. See also Fig. S3.
Spacer abundance is determined by the rate of acquisition.
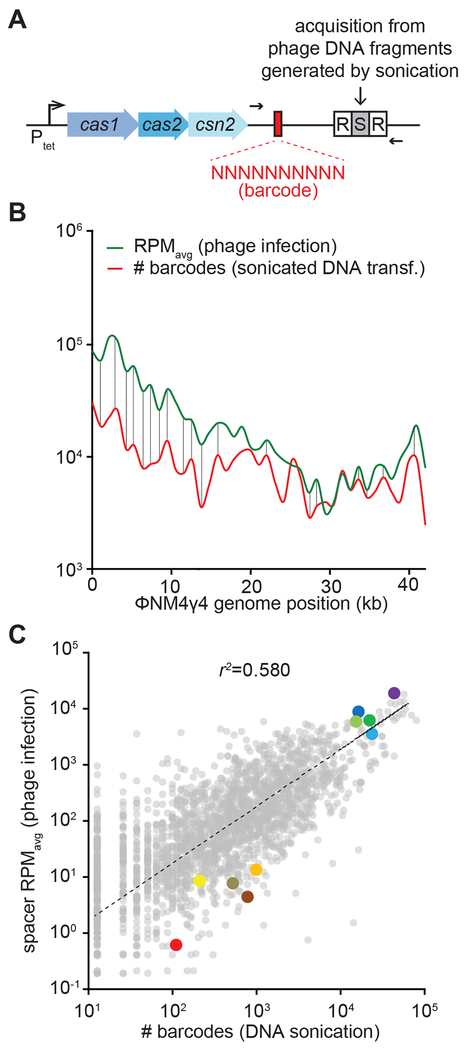
To test whether targeting efficiency affects the relative abundance of individual spacer sequences, we performed a barcoded, phage-free spacer acquisition experiment. For this we used a plasmid-based, modified type II-A locus (Fig. 4A) containing a random 10 nt sequence located 50 bp immediately upstream of a single repeat, a barcoding strategy we previously used to count independent acquisition events (Heler et al., 2017). In addition, expression of the cas1, cas2 and csn2 genes, essential for spacer acquisition, is controlled by an anhydrotetracycline-inducible promoter, allowing turning on and off spacer integration (Heler et al., 2015; Modell et al., 2017). Instead of using a replicating virus, cells harboring this engineered type II-A system were transformed via electroporation with ϕNM4γ4 phage DNA, sheared into ~150 bp fragments by sonication, in the presence of anhydrotetracycline. After two hours the inducer was washed off, DNA was extracted from cells and the new CRISPR loci along with barcoded leaders were amplified by PCR (Fig. 4A) and subjected to next-generation sequencing. We analyzed 2.00 million spacer reads each with its respective barcode that sampled almost all (2,274) of the existing protospacers on the ϕNM4γ4 genome (Data S1 and Fig. S4A–B). To test the barcoded system, we plotted the relative abundance versus the number of different barcodes for each individual sequence (Fig. S4C). Assuming that different barcode sequences in front of the same spacer are the result of independent events of integration, this value reflects how many times a given spacer was acquired during transformation. We detected a strong correlation between the abundance of a spacer and its number of barcodes, a result that validates the use of barcode count as an absolute measure of the acquisition of a given spacer sequence present in the ϕNM4γ4 genome.
Figure 4. Spacer abundance is determined by its rate of acquisition.
(A) Schematic diagram of the modified S. pyogenes CRISPR locus showing the location of the leader barcodes and primers (black arrows) used to quantify the number of independent spacer acquisition events from sheared phage DNA. (B) Overlap of spacer distribution during phage infection (Figure 1) and number of barcodes as a measure of acquisition frequency, both plotted in 1-kb bins. (C) Comparison between abundance of individual spacers during replicating phage infection and independent acquisition events from sheared phage DNA. See also Fig. S4.
We then compared the number of barcodes with the number of reads obtained for each spacer sequence in the experiment using replicating phage presented in Figure 1. In this way we can determine how much of the spacer distribution obtained after viral infection (measured as the average RPM of the replica experiments of Figure 1) can be explained by the intrinsic rate of acquisition of each viral spacer sequence (measured by the number of barcodes obtained in Figure S4). First we compared the distribution patterns across the ϕNM4γ4 genome (Fig. 4B). We found very similar distribution patterns, with a conservation of most peaks and valleys in both curves (note that the RPM and number of barcode values are intrinsically different and therefore the curves do not overlap). Next, we plotted both values against each other and found a good correlation, in which our ten selected spacers maintained their low or high abundance, and with an r2 value of 0.580 (Fig. 4C). This indicates that the distribution of more than half of the spacers acquired in response to viral infection can be explained by their intrinsic rate of acquisition; i.e. independent of the targeting abilities of the spacer sequence.
DISCUSSION
Early studies of the type II-A CRISPR-Cas response to phage infection have shown that the population of surviving bacteria has a diverse content of new spacer sequences, some much more abundant than others (Modell et al., 2017; Paez-Espino et al., 2013; Paez-Espino et al., 2015). In principle, the abundance of a spacer should be determined by two factors: its frequency of integration into the CRISPR array and its targeting capabilities (Bradde et al., 2017). Here we found that the abundance of most spacers is determined shortly after phage infection, when positive or negative selection for good or bad targeting, respectively, is still not a factor at play. In addition, there is a strong correlation between the abundance of most spacers acquired during infection with replicating phage and their abundance after transformation with sheared phage DNA, again, when targeting is not required for survival. Finally, we showed that the frequency of most spacers in the surviving population correlates directly with their frequency of acquisition.
The data presented here show that the spacer abundance that emerges after the type II CRISPR-Cas immune response is basically determined shortly after infection, depending mostly on the acquisition rate of each acquired sequence and not on its properties as a guide for Cas9 DNA cleavage. In support of our findings, modeling of the CRISPR-Cas immune response determined that high spacer acquisition probabilities will lead to greater diversity in the spacer distribution, while strong selection of spacers providing better phage clearance will tend to homogenize the population of spacers in favor of the most effective one (“winner takes all” situation) (Bradde et al., 2017). Since differences in the targeting efficiency between different spacers definitively exist, how is it possible that they do not play a significant role in the in the outcome of the CRISPR-Cas response? Previous studies in our lab showed that spacers at low concentrations within the host population have marked differences in targeting efficiency, provided equally strong immunity once they reached a certain threshold (McGinn and Marraffini, 2016; Modell et al., 2017). We found that spacers located downstream in the CRISPR array provide very weak protection when the cells that carry them are a minority within the population of infected cells, but they provide strong immunity when they constitute a bulk of the culture infected (McGinn and Marraffini, 2016). Likewise, spacers targeting regions of the phage genome that are injected last mediate a poor immune response when they are present in a small proportion of the population but enable robust protection when they are in the majority of the cells of the culture (Modell et al., 2017). We believe that a similar situation can occur during the infection of naïve cultures that acquired multiple (thousands) of new spacers. The high rate of acquisition of certain sequences would effectively create a high concentration of immune cells that will reach the concentration threshold necessary to provide most of the immunity to the population, and no further selection of these sequences due to their targeting efficiency will take place.
Our findings showed that spacer abundance is mostly determined at the acquisition stage of type II-A CRISPR-Cas immunity. The uneven distribution of different spacer sequences could be in principle explained by the existence of phage genomic regions that are better substrates for spacer acquisition. Indeed, this is the case for the regions proximal to the cos site in ϕ12γ3 that first enter the host cell (Modell et al., 2017) and is a possible explanation for the clustering of highly abundant spacers from the 5’ end of the ϕNM4γ4 genome (Fig. S1a–b). However, even within these regions there is a wide spectrum of spacer abundances. Here we showed that a key factor for these different abundances is the intrinsic frequency of acquisition of a given spacer sequence. Mutagenesis analysis revealed that the 10-nt sequence at the PAM end of a spacer is determinant for its frequency of acquisition. However we could not identify a critical motif within these 10 nucleotides. This differs from the findings for the acquisition of type I-A spacers in E. coli, where a strong motif with the PAM sequence (AAG), the “acquisition affecting motif” upstream of the spacer, as well as an AA dinucleotide 30 nucleotides downstream of this motif, were found to significantly enhance the rate of acquisition (Shipman et al., 2017; Yosef et al., 2013). Interestingly, the 10-nucleotides proximal to the PAM are also part of the target seed region and their complementarity to the crRNA is fundamental for Cas9 cleavage (Deveau et al., 2008; Jinek et al., 2012). Therefore the molecular mechanisms behind this preferential acquisition remain unknown. The current model of spacer acquisition by type II-A CRISPR-Cas systems involves three major steps. First, the injected DNA is degraded by the AddAB when the phage’s own mechanism that inhibit this host nuclease fail. This creates the spacer substrates (Levy et al., 2015; Modell et al., 2017), which are selected and processed by a Cas9-Cas1-Cas2-Csn2 complex (Heler et al., 2015). Finally the processed spacer sequence is integrated by the Cas1-Cas2 integrase into the CRISPR array (Wright and Doudna, 2016). Because the 10-nt PAM-proximal sequence did not affect the rate of insertion of the spacer by the Cas1-Cas2 integration complex, our data suggests that this sequence plays a role in determining the rate of acquisition in either of the first two steps. In summary, our study begins to uncover the rules that govern the generation of immunological diversity during the type II-A CRISPR-Cas response, revealing that spacer acquisition early during this process dominates over spacer-mediated targeting to determine the structure of the surviving population. This contrasts with mammalian adaptive immunity, in which the generation of diversity is the result of random V(D)J recombination to create millions of different antibodies that are then selected for their abilities to recognize and mediate the destruction of the foreign antigen.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Luciano Marraffini (marraffini@rockefeller.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The bacterial strain used in this study was Staphylococcus aureus RN4220. Cultivation of Staphylococcus aureus RN4220 was carried out in heart infusion broth (BHI) at 37°C. Whenever applicable, media were supplemented with chloramphenicol at 10 μg/mL, erythromycin at 10 μg/mL or spectinomycin at 250 μg/mL to ensure maintenance of pC194, pE194 and pLZ12 derived plasmids, respectively, or 5 mM CaCl2 for phage adsorption. The phages used were φNM4γ4, a φNM4 derivative containing a deletion within the lysogeny cassette (Goldberg et al., 2014) and staphylococcal phage ϕ85 (Kwan et al., 2005).
METHOD DETAILS
Bacterial Growth Curves
Overnight cultures were launched from single colonies and diluted 1:100 in BHI. After 1 hour of growth, optical density at 600nm (OD600) was measured for each culture, and samples were brought to equal cell densities. Immune cells carrying targeting spacers were diluted with cells lacking CRISPR-Cas to a 1:1000 ratio and loaded into 96-well plates along with φNM4γ4 at MOI = 1. Measurements were taken every 10 minutes for 24 hours.
Phage Interference Assay
Overnight cultures were launched from single colonies. Serial dilutions of a stock of phage φNM4γ4 (Goldberg et al., 2014) were spotted on fresh soft heart infusion agar (HIA) lawns of targeting cells containing chloramphenicol 10 μg/ml and 5 mM CaCl2. Plates were incubated at 37 °C overnight and interference efficiency was measured in plaque forming units (pfu).
Acquisition from live phage
Acquisition from live phage in cells harboring the CRISPR system of Streptococcus pyogenes (plasmid pWJ40) or CRISPR3 of Streptococcus thermophilus (pRH200) was performed as described previously (Heler et al., 2015). In Figure 2 and S2, plasmid pWJ40* containing randomized leader barcodes was used instead of pWJ40 (Heler et al., 2017). The unweighted probability Logo (Figure S2K) of the top 1% protospacers was generated using kpLogo (Wu and Bartel, 2017).
Acquisition from shredded phage DNA
Phage DNA was shredded by sonication to fragments of ~150bp as described in Modell (Modell et al., 2017). Following dialysis, 100μg of phage DNA was electroporated into competent S. aureus cells carrying plasmids pRH317 and pRH318*. Cells were recovered for 2h in BHI supplemented with anhydrotetracycline at 1μg/μl.
Acquisition from dsDNA oligonucleotides
dsDNA substrates were obtained by annealing ssDNA oligonucleotides in Duplex Buffer from IDT. Following dialysis, 100 nM of each competing dsDNA substrate were mixed and electroporated in competent S. aureus cells carrying plasmids pRH223 and pRH240 (Heler et al., 2015). Cells were recovered for 2h in BHI supplemented with anhydrotetracycline at 1μg/μl. In order of appearance, the annealed oligo pairs are H612-H613, H617-H618, H690-H691, H690-H691 (Figure 2D), H612-H613, H626-H627, H634-H635, H630-H631, H622-H623, H624-H625, H628-H629, H620-H621, H617-H618, H632-H633 (Figure 2E), H655-H656, H657-H658 (Figure S2G), H614-H615, H618-H619, H626-H627, H638-H639 (Figure S2H), H630-H631, H628-H629, H626-H627, H632-H633, H634-H635, H624-H625, H622-H623, H620-H621 (Figure S2I), H668-H669, H657-H658, H670-H671, H657-H658, H672-H673, H657-H658 (Figure S2J), H700-H701, H702-H703 (Figure S2L). The significance (p-value) of the results in Figure 2E was assessed using a two-tailed, unequal variance Student’s t-test. Oligonucleotide sequences are shown in Table S1.
High-throughput sequencing
Plasmid DNA was extracted from adapted cultures. 200 ng of plasmid DNA was used as template for Phusion PCR to amplify the CRISPR locus with primer pairs H370-H371 (Figure 1, 3), H180-B153 (Figure S1E), H372-H366 (Figure 4) and H186-H366 (Figure 2 and S2). Following gel extraction and purification of the adapted bands, samples were subject to Illumina MiSeq (Figures 1, 2, 4, S1, S2, S4) or NextSeq (Figures 3 and S3) sequencing. Data analysis was performed in Python: first, all newly acquired spacer sequences were extracted from raw MiSeq FASTA data files. Next, the frequency, number of different barcodes, the phage target location, and the flanking PAM were determined for each unique spacer sequence. Analysis was finished in Excel.
In vitro Cas9 target cleavage
Cleavage by Cas9 of various targets was assessed using the Guide-It Complete sgRNA Screening System from Clontech (Cat. No. 632636) with minor modifications. Cas9 and the sgRNAs were pre-incubated for 5 min at 37C in equimolar ratio and then diluted into the cleavage reaction to final concentrations of 100, 50, 25, 12.5 and 6.25nM. All reactions contained 10 nM of a phage-derived PCR template with the target site. All reactions were stopped after 5 minutes by heat inactivation at 80C for 5 minutes and stored at −80C until ready to be run on an agarose gel. Guides used in Figure 2B were transcribed from oligos H521 (blue, also S2D), H522 (brown, also S2E), H694 (green, also S2b), H695 (red, also S2C).
Plasmid Construction
Plasmid pRH317 was constructed by deleting the CRISPR leader and array from pRH223 (Heler et al., 2015) via a one-piece Gibson assembly reaction with primer pair JM126-JM127. Plasmid pRH318 was constructed by a two-piece Gibson assembly reaction from pRH240 (Heler et al., 2015) and pLZ12 with primer pairs H558-H559 and H555-H557, respectively. Plasmid pRH318* (containing randomized leader barcodes) was constructed by a two-piece Gibson assembly with primers pairs H378-H294 and H379-H293. Plasmid pRH248, pRH249, pRH328 and pRH329 were constructed via BsaI cloning as described in (Heler et al., 2015) with annealed oligonucleotide pairs H433-H434, H435-H436, H641-H642, and H643-H645, respectively. Oligonucleotide sequences are shown in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed in Microsoft Excel. Significance was calculated using Student’s paired t-Test, with a two-tailored distribution, assuming a two-sample unequal variance. Error bars represent mean ± standard deviation.
DATA AND SOFTWARE AVAILABILITY
Raw data for all figures: Data S1.
Supplementary Material
Data S1. Next generation sequencing data obtained in this study. Related to all figures.
Table S1. Oligonucleotides used in this study. Related to STAR methods.
ACKNOWLEDGEMENTS.
We would like to thank the Rockefeller University Genomics Resource Center for assistance with next generation sequencing experiments. L.A.M. is supported by a Burroughs Wellcome Fund PATH Award, an NIH Director’s Pioneer Award (DP1GM128184) and an HHMI-Simons Faculty Scholar Award. A.V.W. is a NSF Graduate Research Fellow. M.V. was supported in part by the NSF under award number NSF PHY-1748958. J.A.D. is supported by the NSF award number 1244557. L.A.M. and J.A.D. are investigators of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS. J.A.D. is a cofounder and Scientific Advisory Board member of Caribou Biosciences, Intellia Therapeutics, and Scribe Therapeutics; a cofounder of Editas Medicine and Mammoth Biosciences; and a Scientific Advisory Board member of eFFECTOR Therapeutics, Synthego, Metagenomi, and Inari, all of which develop CRISPR-based technologies. She is a Board of Directors member for Driver and Johnson & Johnson. She has Sponsored Research Projects with Pfizer, Inc, Roche Biopharma, and Biogen. L.A.M. is a cofounder and Scientific Advisory Board member of Intellia Therapeutics, and a co-founder of Eligo Biosciences.
REFERENCES
- Anders C, Niewoehner O, Duerst A, and Jinek M (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bradde S, Vucelja M, Tesileanu T, and Balasubramanian V (2017). Dynamics of adaptive immunity against phage in bacterial populations. PLoS Comput Biol 13, e1005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, and van der Oost J (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, and Charpentier E (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, and Moineau S (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, and Moineau S (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. [DOI] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, and Marraffini LA (2014). Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, and Marraffini LA (2015). Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Wright AV, Vucelja M, Bikard D, Doudna JA, and Marraffini LA (2017). Mutations in Cas9 Enhance the Rate of Acquisition of Viral Spacer Sequences during the CRISPR-Cas Immune Response. Mol Cell 65, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, and Doudna JA (2016). Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, and Marraffini LA (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, and Pelletier J (2005). The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A 102, 5174–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, and Sorek R (2015). CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, and Sontheimer EJ (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, and Marraffini LA (2016). CRISPR-Cas systems optimize their immune response by specifying the site of spacer integration. Mol Cell 64, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Jiang W, and Marraffini LA (2017). CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544, 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, and Banfield JF (2013). Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nature communications 4, 1430. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, and Banfield JF (2015). CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. MBio 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman SL, Nivala J, Macklis JD, and Church GM (2017). CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature 547, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, and Doudna JA (2016). Protecting genome integrity during CRISPR immune adaptation. Nat Struct Mol Biol 23, 876–883. [DOI] [PubMed] [Google Scholar]
- Wu X, and Bartel DP (2017). kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res 45, W534–W538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Shitrit D, Goren MG, Burstein D, Pupko T, and Qimron U (2013). DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc Natl Acad Sci USA 110, 14396–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Next generation sequencing data obtained in this study. Related to all figures.
Table S1. Oligonucleotides used in this study. Related to STAR methods.
Data Availability Statement
Raw data for all figures: Data S1.