Abstract
Existing single cell mass spectrometry (SCMS) sampling platforms are largely designed to work only with immobilized cells and not the suspended cells isolated from patient samples. Here, we present a novel method that integrates a commercially available cell manipulation system commonly used for in vitro fertilization with the Single-probe SCMS sampling technology. The combined Single-probe SCMS/cell manipulating platform is capable of rapidly analyzing intracellular species in real time from a suspension leukemia cell line. A broad range of molecular species was detected, and species of interest were verified using tandem MS (MS/MS). Experimental results were analyzed utilizing statistical analyses such as principle component analysis (PCA) and t-tests. The developed SCMS/cell manipulation system is a versatile tool to provide rapid single cell analysis of broad types of patient cell samples.
Graphical abstract
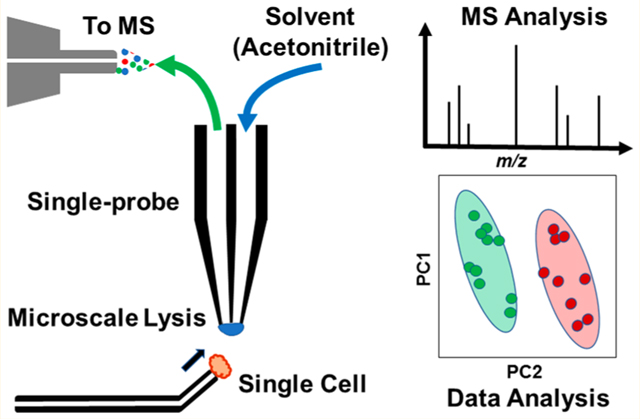
Liquid biopsy samples from patients, such as blood plasma, saliva, urine, or fine needle aspiration, are a complex milieu of different types of individual cells, and therefore any bioanalysis of the multicellular patient sample would likely be extremely heterogeneous.1 Moreover, patient-derived cells are typically converted into lysate and then analyzed using a bulk sampling technique, such as liquid chromatography mass spectrometry (LCMS). However, bulk analysis masks cellular heterogeneity, and the cellular characteristics, metabolites, and responses to drug administration in an abnormal subpopulation would be mixed with signals from healthy cells.2,3 A better approach would be to focus the bioanalysis on the individual patient cell levels, but the limited volume of an individual cell (i.e., ~1–3 pL),4 along with spectral interferences from the sampling medium make analysis on the single-cell level extremely challenging. Additionally, many cellular bioanalytical methods, including some single cell methods, require complex sample preparation prior to measurement.5 Extensive sample preparation, including cell attachment and detachment, centrifugation, and changes to the cellular environment can alter cellular metabolites and morphologies.6 Therefore, rapid, real-time analysis is necessary to study cells in a near-native state.2,3 Advancements in instrumentation over the past decade have allowed single cell analysis (SCA) to become increasingly popular as a means to characterize heterogeneity that is unable to be accounted for using bulk sampling techniques. Current methods of SCA include flow cytometry,7–10 microfluidics,11,12 RNA sequencing,13,14 and mass spectrometry (MS) methods of analysis.10,15–17
Single cell mass spectrometry (SCMS) is becoming popular due to its label-free approach and ability to detect broad ranges of analytes, including lipids, peptides, and metabolites.17,18 SCMS can be characterized into two groups, ambient and nonambient analysis, according to their sampling and ionization environment. Nonambient techniques are vacuum-based and require sample pretreatment, while ambient techniques are often performed in a near-native environment with minimal sample preparation. A pitfall of nonambient SCMS techniques is the inability to study live cells in real time in native cellular environments. Changes to the cellular environment could alter cellular metabolites. Thus, a number of ambient SCMS techniques have been developed for the measurement of live mammalian cells. These techniques analyze cellular contents obtained from individual cells using microprobe extraction11,19–34 or laser desorption/ionization.18,35–48 Among these ambient SCMS techniques, the Single-probe SCMS technique allows for real-time, in situ analysis of live cells, including mammalian and plant cells.49–55 Microscale separation methods, such as capillary electrophoresis (CE), and postionization methods, such as ion mobility (IM),17 have been coupled to MS to minimize spectral interferences during single cell analysis.56,57 A prerequisite of many nonambient SCMS techniques is the need for an immobilized substrate to secure cells, such as preparing a frozen, hydrated sample or utilizing micropatterned polystencil film.17 Because cellular metabolites are sensitive to the microenvironment, any perturbation, including cell attachment and changing culture medium, may potentially affect the molecular composition inside cells. A bioanalytical method capable of rapidly measuring drug pharmacology on single cells isolated from patients under ambient conditions could be used to develop personalized drug therapeutic regimens tailored to the individual’s response to a drug.58
SCMS techniques are often limited by the amount of content available from an individual cell. Single cells have a much smaller sample volume than bulk samples used in traditional analytical techniques but still maintain extremely complex compositions without extensive sample pretreatment. For example, typical cultured mammalian cells contain only a few picoliters of solution (individual cell size ~10–20 μm in diameter), but it is estimated that more than 109 proteins and 1010 lipids are present in one single cell.59 Limited amount and complex composition of cellular contents from individual cells lead to challenges generally present in SCMS studies, particularly for cancer cells with small sizes (e.g., ~10–13 μm).29,60–66
Despite the small sampling size of cultured cells, the Singleprobe SCMS technique has demonstrated its success qualitatively with metabolic profiling in individual mammalian cancer cell lines. In addition, this technique has been used for other applications, including MS tissue imaging28,30 and analysis of extracellular molecules inside live multicellular spheroids.56,57 There are a number of advantageous features unique to the Single-probe SCMS technique, including versatile selection and composition of the sampling solvent, capabilities for real-time and in situ analysis, and potential compatibility with broad types of mass spectrometers. However, this technique was previously used only for analysis of cells on an immobilized substrate (e.g., on glass coverslips).
In this work, we report the development of an integrated cell manipulation platform (ICMP), which combines the Single-probe SCMS method with a single cell manipulation platform, to analyze individual, suspended cancer cells in real time in a near-native environment with minimal sample pretreatment. This new instrumental setup allows for the direct selection and analysis of individual cells present in their original complex matrixes. Because matrix molecules in the sample solution are generally eliminated upon analysis, the matrix effect, which results in reduced ionization efficiency and detection sensitivity, is minimized.
To conduct SCMS of suspended cells, this ICMP setup is coupled to a high-resolution Thermo LTQ Orbitrap XL mass spectrometer, and a Single-probe is used as the sampling and ionization device. This multifunctional cell manipulation system (Figure 1) consists of two Eppendorf TransferMan cell micromanipulation devices (to control spatial movements of both the glass cell-selection device and Single-probe), a Nikon Eclipse TE300 inverted microscope (for cell monitoring), and a Tokai Hit ThermoPlate system (to mimic the temperature of the cells’ natural environment). A cell-selection probe, which is a glass tubing with a small tip size, is connected to a microinjector and coupled with one of the Eppendorf manipulation systems to capture target cells. The other Eppendorf manipulation system is modified to allow a Single-probe interface with the mass spectrometer via an extended ion transfer tube, which is used to replace the standard one. Cells are randomly selected with the cell- selection device using a digital stereomicroscope and then transferred to the Single-probe tip for real-time microscale lysis followed by MS analysis (Figure S1). Detailed experimental information can be found in the Supporting Information.
Figure 1.
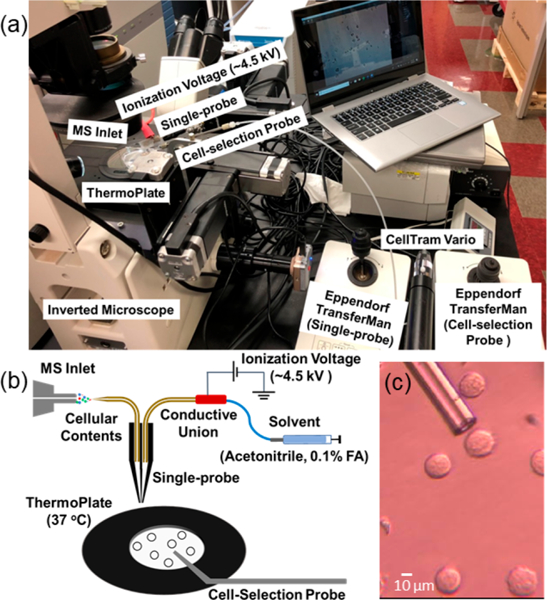
Experimental setup for suspension single cell MS experiments: (A) integrated cell manipulation platform (ICMP) coupled with a mass spectrometer, (B) schematic for analysis of suspended cells, and (C) experimental view of K562 cells to be selected using the cell-selection probe.
K562 cells in both control and drug-treated groups were subjected to the SCMS measurement. Three abundant cellular lipid(phosphatidylcholine (PC)) peaks (PC(34:4), PC(36:4), and PC(38:5) at m/z 754.536, 782.567, and 808.583, respectively (Figure 2)) were monitored throughout the experiment to ensure cells were transferred and analyzed.49,67,68 These common species have been observed in our previous studies using adherent cell lines.49,50,69
Figure 2.
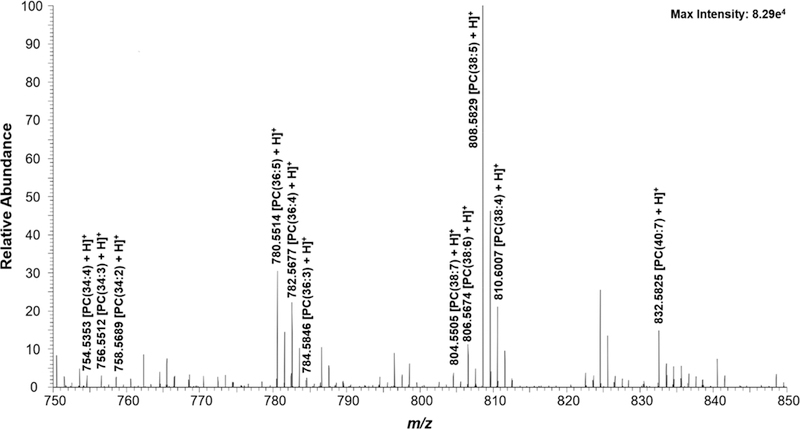
Zoomed-in mass spectrum from a single cell showing the representative species (m/z 750–850). Structure confirmation of the labeled ions were performed through MS/MS analysis (Figure S3).
K562 cells were subjected to treatment with various anticancer drug compounds (gemcitabine, Taxol, and OSW-1) to demonstrate the versatility of analyte that could be extracted from individual suspended cells. Gemcitabine (1 μM) and Taxol (1 μM) were incubated with cells for 1 h. OSW-1 (100 nM and 1 μM) was incubated with cells for 4 and 2 h, respectively. In addition to a large number of cellular metabolites, all three drug compounds were detected from individual cells (Figure S2, Table S1). The drug compounds were unable to be detected in the PBS in which the K562 cells were suspended, indicating that they were released from cells during cell lysis and not from the sampling solution.
The ICMP coupled to the Single-probe SCMS was also utilized for the analysis of global metabolic changes in K562 cells following a 24-h treatment with Taxol (100 nM). The SCMS data were subjected to pretreatment, including background removal, noise reduction, peak alignment, and intensity normalization using Geena2 prior to analysis using MetaboAnalyst.70,71 Under our experimental conditions, the physiological profiles of cells were not significantly changed by drug treatment; however, their molecular compositions were drastically changed (Figure S4). Principle component analysis (PCA) was performed to illustrate the differences of molecular profiles of cells between control and treatment groups (Figure 3). In addition, the within-group spread of SCMS data points in the PCA plot reflects the heterogeneity of cells in the same group.72 In total, the abundances of 78 metabolites were significantly changed (p < 0.05 from t-test) upon drug treatment. For further identification of compounds of interest determined from SCMS experiments, cell lysates were prepared using drug treated cells (100 nM Taxol for 24 h) and subjected to the complementary MS/MS analysis. Monoglycerides (MG) and diglycerides (DG) comprised many of the significantly changed metabolites. For example, upon treatment with Taxol, DG(44:6) is significantly down-regulated, while MG(16:0) and MG(18:0) are significantly up-regulated (Figure S5). These results illustrate a stark difference between these two groups, including multiple significantly up/down-regulated metabolites, which demonstrates the capability of this technology to identify global metabolic changes within individual, live suspension cells.
Figure 3.
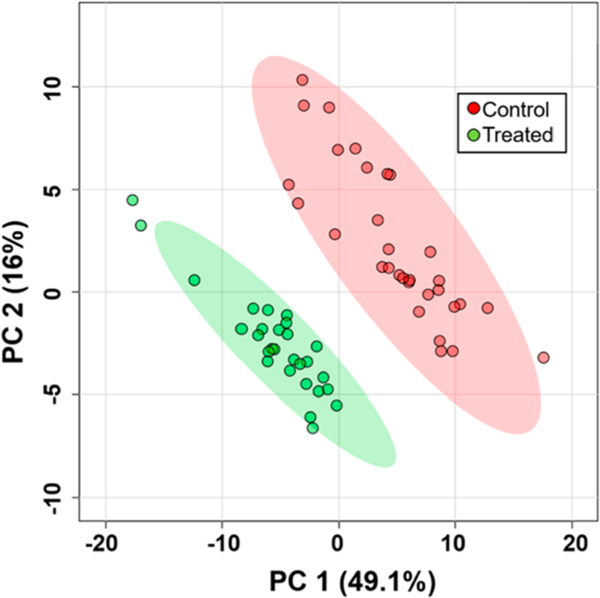
PCA showing the overall difference of metabolomic compositions of single K562 cells in the control and drug treatment (100 nM Taxol for 24 h) groups.
In summary, the Single-probe SCMS technique has successfully been coupled with an integrated cell manipulation platform for the analysis of suspended cells (K562) in solution. This integrated system allows us to observe and select individual cells suspended in solution. The selected cell is transported to the Single-probe tip, where a rapid single cell lysis occurs in an acetonitrile droplet. Released cellular contents are then immediately analyzed by a high-resolution mass spectrometer. More than 30 total cells were analyzed using this integrated SCMS system, and different cellular metabolites were found between treated and control cell groups. Complementary MS analysis of cell lysate can be performed for identification of species of interest obtained from the SCMS experiments.
This integrated system is suitable for the analysis of cells present in solutions, such as bodily fluids, with minimum sample preparation. The complex matrix is largely excluded from MS analysis by selecting a cell of interest for analysis with a probe not utilized for analysis, so minimal or no solution is extracted. With the ability to analyze small sampling volumes, it is feasible that the integrated cell manipulation platform can be used for rapid analysis of individual cells derived from patients in situ, which could allow for unprecedented personalization of drug administration.
Safety Considerations.
Both glass and silica capillaries pose a needle-stick hazard, so these items must be handled with caution. Standard safety protocols were enforced for the handling of chemicals and culturing and treating of cell lines.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the National Institutes of Health (Grants R01GM116116 and R21CA204706).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.8b05774.
Fabrication protocols; sample preparation; experimental parameters; and additional characterization data (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Neumann MHD; Bender S; Krahn T; Schlange T Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Jelonek K; Ros M; Pietrowska M; Widlak P Clin. Lipidol. 2013, 8 (1), 137–150. [Google Scholar]
- (3).Bendall SC; Simonds EF; Qiu P; Amir ED; Krutzik PO; Finck R; Bruggner RV; Melamed R; Trejo A; Ornatsky OI; et al. Science 2011, 332 (6030), 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Moran U; Phillips R; Milo R Cell 2010, 141 (7), 1262–1262.e1. [DOI] [PubMed] [Google Scholar]
- (5).Kataoka H TrAC, Trends Anal. Chem. 2003, 22 (4), 232–244. [Google Scholar]
- (6).Mao S; Zhang W; Huang Q; Khan M; Li H; Uchiyama K; Lin J-M Angew. Chem., Int. Ed. 2018, 57 (1), 236–240. [DOI] [PubMed] [Google Scholar]
- (7).Wang D; Bodovitz S Trends Biotechnol. 2010, 28 (6), 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Davey HM; Kell DB Microbiol. Rev. 1996, 60 (4), 641–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Andersson H; van den Berg A Curr. Opin. Biotechnol. 2004, 15 (1), 44–49. [DOI] [PubMed] [Google Scholar]
- (10).Schmid A; Kortmann H; Dittrich PS; Blank LM Curr. Opin. Biotechnol. 2010, 21 (1), 12–20. [DOI] [PubMed] [Google Scholar]
- (11).Sims CE; Allbritton NL Lab Chip 2007, 7 (4), 423–440. [DOI] [PubMed] [Google Scholar]
- (12).Lecault V; White AK; Singhal A; Hansen CL Curr. Opin. Chem. Biol. 2012, 16 (3–4), 381–390. [DOI] [PubMed] [Google Scholar]
- (13).Yan L; Yang M; Guo H; Yang L; Wu J; Li R; Liu P; Lian Y; Zheng X; Yan J; et al. Nat. Struct. Mol. Biol. 2013, 20 (9), 1131–1139. [DOI] [PubMed] [Google Scholar]
- (14).Kalisky T; Quake SR Nat. Methods 2011, 8 (4), 311–314. [DOI] [PubMed] [Google Scholar]
- (15).Boggio KJ; Obasuyi E; Sugino K; Nelson SB; Agar NY; Agar JN Expert Rev. Proteomics 2011, 8 (5), 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rubakhin SS; Romanova EV; Nemes P; Sweedler JV Nat. Methods 2011, 8 (4), S20–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhang L; Vertes A Angew. Chem., Int. Ed. 2018, 57 (17), 4466–4477. [DOI] [PubMed] [Google Scholar]
- (18).Yang Y; Huang Y; Wu J; Liu N; Deng J; Luan T TrAC, Trends Anal. Chem. 2017, 90, 14–26. [Google Scholar]
- (19).Tsuyama N; Mizuno H; Tokunaga E; Masujima T Anal. Sci. 2008, 24 (5), 559–561. [DOI] [PubMed] [Google Scholar]
- (20).Phelps M; Hamilton J; Verbeck GF Rev. Sci. Instrum. 2014, 85 (12), 124101. [DOI] [PubMed] [Google Scholar]
- (21).Phelps MS; Verbeck GF Anal. Methods 2015, 7 (9), 3668–3670. [Google Scholar]
- (22).Pan N; Rao W; Kothapalli NR; Liu R; Burgett AWG; Yang Z Anal. Chem. 2014, 86 (19), 9376–9380. [DOI] [PubMed] [Google Scholar]
- (23).Pan N; Rao W; Standke SJ; Yang Z Anal. Chem. 2016, 88 (13), 6812–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Rao W; Pan N; Yang ZJ Visualized Exp. 2016, No. 112, 53911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gong X; Zhao Y; Cai S; Fu S; Yang C; Zhang S; Zhang X Anal. Chem. 2014, 86 (8), 3809–3816. [DOI] [PubMed] [Google Scholar]
- (26).Mizuno H; Tsuyama N; Harada T; Masujima TJ Mass Spectrom. 2008, 43, 1692. [DOI] [PubMed] [Google Scholar]
- (27).Lorenzo Tejedor M; Mizuno H; Tsuyama N; Harada T; Masujima T Anal. Chem. 2012, 84 (12), 5221–5228. [DOI] [PubMed] [Google Scholar]
- (28).Fujii S; Matsuda S; Tejedor ML; Esaki T; Sakane I; Mizuno H; Tsuyama N; Masujima T Nat. Protoc. 2015, 10 (9), 1445. [DOI] [PubMed] [Google Scholar]
- (29).Shimizu T; Miyakawa S; Esaki T; Mizuno H; Masujima T; Koshiba T; Seo M Plant Cell Physiol. 2015, 56 (7), 1287–1296. [DOI] [PubMed] [Google Scholar]
- (30).Yamamoto K; Takahashi K; Mizuno H; Anegawa A; Ishizaki K; Fukaki H; Ohnishi M; Yamazaki M; Masujima T; Mimura T Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (14), 3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Date S; Mizuno H; Tsuyama N; Harada T; Masujima T Anal. Sci. 2012, 28 (3), 201. [DOI] [PubMed] [Google Scholar]
- (32).Hiyama E; Ali A; Amer S; Harada T; Shimamoto K; Furushima R; Abouleila Y; Emara S; Masujima T Anal. Sci. 2015, 31 (12), 1215–1217. [DOI] [PubMed] [Google Scholar]
- (33).Ali A; Abouleila Y; Amer S; Furushima R; Emara S; Equis S; Cotte Y; Masujima T Anal. Sci. 2016, 32 (2), 125–127. [DOI] [PubMed] [Google Scholar]
- (34).Fujita H; Esaki T; Masujima T; Hotta A; Kim SH; Noji H; Watanabe TM RSC Adv. 2015, 5 (22), 16968–16971. [Google Scholar]
- (35).Ferreira CR; Eberlin LS; Hallett JE; Cooks RG J. Mass Spectrom. 2012, 47 (1), 29–33. [DOI] [PubMed] [Google Scholar]
- (36).Lee JK; Jansson ET; Nam HG; Zare RN Anal. Chem. 2016, 88 (10), 5453–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chen F; Lin L; Zhang J; He Z; Uchiyama K; Lin J-M Anal. Chem. 2016, 88 (8), 4354–4360. [DOI] [PubMed] [Google Scholar]
- (38).Bergman H-M; Lanekoff I Analyst 2017, 142 (19), 3639–3647. [DOI] [PubMed] [Google Scholar]
- (39).Duenas ME; Essner JJ; Lee YJ Sci. Rep. 2017, 7 (1), 14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yin R; Prabhakaran V; Laskin J Anal. Chem. 2018, 90 (13), 7937–7945. [DOI] [PubMed] [Google Scholar]
- (41).Gonzalez-Serrano AF; Pirro V; Ferreira CR; Oliveri P; Eberlin LS; Heinzmann J; Lucas-Hahn A; Niemann H; Cooks RG PLoS One 2013, 8 (9), No. e74981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Liu Y; Zhang J; Nie H; Dong C; Li Z; Zheng Z; Bai Y; Liu H; Zhao J Anal. Chem. 2014, 86 (14), 7096–7102. [DOI] [PubMed] [Google Scholar]
- (43).Shrestha B; Vertes A Anal. Chem. 2009, 81 (20), 8265–8271. [DOI] [PubMed] [Google Scholar]
- (44).Shrestha B; Nemes P; Vertes A Appl. Phys. A: Mater. Sci. Process. 2010, 101 (1), 121–126. [Google Scholar]
- (45).Stolee JA; Shrestha B; Mengistu G; Vertes A Angew. Chem., Int. Ed. 2012, 51 (41), 10386–10389. [DOI] [PubMed] [Google Scholar]
- (46).Shrestha B; Sripadi P; Reschke BR; Henderson HD; Powell MJ; Moody SA; Vertes A PLoS One 2014, 9 (12), No. e115173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Shrestha B; Patt JM; Vertes A Anal. Chem. 2011, 83 (8), 2947–2955. [DOI] [PubMed] [Google Scholar]
- (48).Stolee JA; Vertes A Anal. Chem. 2013, 85 (7), 3592–3598. [DOI] [PubMed] [Google Scholar]
- (49).Pan N; Rao W; Kothapalli NR; Liu R; Burgett AWG; Yang Z Anal. Chem. 2014, 86 (19), 9376–9380. [DOI] [PubMed] [Google Scholar]
- (50).Sun M; Tian X; Yang Z Anal. Chem. 2017, 89 (17), 9069–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Rao W; Pan N; Tian X; Yang ZJ Am. Soc. Mass Spectrom. 2016, 27 (1), 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Rao W; Pan N; Yang ZJ Visualized Exp. 2016, No. 112, 53911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Pan N; Rao W; Standke SJ; Yang Z Anal. Chem. 2016, 88 (13), 6812–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Rao W; Pan N; Tian X; Yang ZJ Am. Soc. Mass Spectrom. 2016, 27 (1), 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Rao W; Pan N; Yang ZJ Am. Soc. Mass Spectrom. 2015, 26 (6), 986–993. [DOI] [PubMed] [Google Scholar]
- (56).Onjiko RM; Moody SA; Nemes P Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (21), 6545–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhang L; Foreman DP; Grant PA; Shrestha B; Moody SA; Villiers F; Kwak JM; Vertes A Analyst 2014, 139 (20), 5079–5085. [DOI] [PubMed] [Google Scholar]
- (58).Hu P; Zhang W; Xin H; Deng G Front. Cell Dev. Biol. 2016, 4, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chen X; Love JC; Navin NE; Pachter L; Stubbington MJT; Svensson V; Sweedler JV; Teichmann SA Nat. Biotechnol. 2016, 34, 1111. [DOI] [PubMed] [Google Scholar]
- (60).Zhao L; Kroenke CD; Song J; Piwnica-Worms D; Ackerman JJH; Neil JJ NMR Biomed. 2008, 21 (2), 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Tahara M; Inoue T; Miyakura Y; Horie H; Yasuda Y; Fujii H; Kotake K; Sugano K Biochem. Biophys. Res. Commun. 2013, 434 (4), 753–759. [DOI] [PubMed] [Google Scholar]
- (62).Gregg JL; McGuire KM; Focht DC; Model MA Pfluegers Arch. 2010, 460 (6), 1097–1104. [DOI] [PubMed] [Google Scholar]
- (63).Boss D; Kuhn J; Jourdain P; Depeursinge C; Magistretti P; Marquet PJ Biomed. Opt. 2013, 18, 036007. [DOI] [PubMed] [Google Scholar]
- (64).Masuda K; Abouleila Y; Ali A; Yanagida T; Masujima T; Anton io C Methods Mol. Biol 2018, 1778, 269–282. [DOI] [PubMed] [Google Scholar]
- (65).Mizuno H; Tsuyama N; Harada T; Masujima TJ Mass Spectrom. 2008, 43 (12), 1692–1700. [DOI] [PubMed] [Google Scholar]
- (66).Shashni B; Ariyasu S; Takeda R; Suzuki T; Shiina S; Akimoto K; Maeda T; Aikawa N; Abe R; Osaki T; et al. Biol. Pharm. Bull. 2018, 41 (4), 487–503. [DOI] [PubMed] [Google Scholar]
- (67).Schober Y; Guenther S; Spengler B; Rompp A Anal. Chem. 2012, 84 (15), 6293–6297. [DOI] [PubMed] [Google Scholar]
- (68).Pulfer M; Murphy RC Mass Spectrom. Rev. 2003, 22 (5), 332–364. [DOI] [PubMed] [Google Scholar]
- (69).Liu R; Pan N; Zhu Y; Yang Z Anal. Chem. 2018, 90 (18), 11078–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Chong J; Soufan O; Li C; Caraus I; Li S; Bourque G; Wishart DS; Xia J Nucleic Acids Res. 2018, 46, W486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Romano P; Profumo A; Rocco M; Mangerini R; Ferri F; Facchiano A BMC Bioinf. 2016, 17 (4), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Huang Q; Mao S; Khan M; Zhou L; Lin J-M Chem. Commun. 2018, 54 (21), 2595–2598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


