Abstract
Germinal centers (GCs) are important sites for antibody diversification and affinity maturation and are also a common origin of B-cell malignancies. Although made up of motile cells, GCs are tightly confined within B-cell follicles. The cues promoting GC B-cell confinement are incompletely understood. P2RY8 is a Gα13-coupled receptor that mediates migration inhibition and growth regulation of B cells in lymphoid tissues4,6. P2RY8 is frequently mutated in GC-derived diffuse large B-cell lymphoma (DLBCL) and Burkitt lymphoma (BL)1–5, and the ligand for this receptor has been undefined. In a search for P2RY8 ligands, we found P2RY8 bioactivity in bile and in culture supernatants of several cell lines. Using a seven-step biochemical fractionation procedure and a drop-out mass spectrometry approach, we identified a previously undescribed biomolecule, S-geranylgeranyl-L-glutathione (Ggg) as a potent P2RY8 ligand. Ggg was detectable in lymphoid tissues in the nanomolar range. Ggg inhibited chemokine-mediated migration of human GC B cells and follicular helper T cells and antagonized induction of pAkt in GC B cells. We found that gamma-glutamyltransferase-5 (Ggt5) metabolized Ggg to a form inactive on the receptor. Ggt5 was highly expressed by follicular dendritic cells (FDCs). Over-expression of this enzyme disrupted the ability of P2RY8 to promote B-cell confinement to GCs, indicating that it establishes a Ggg gradient in lymphoid tissues. This work defines Ggg as an intercellular signaling molecule involved in organizing and controlling GC responses. As well as DLBCL and BL the P2RY8 locus is modified in several other cancers and we speculate that Ggg has organizing and growth regulatory activities in multiple human tissues.
To establish a bioassay for P2RY8 we utilized the inferred ability of P2RY8 to support migration inhibition4. P2RY8 was expressed in a lymphoid cell line (WEHI-231) and the highest expressing cells were selected to maximize ligand sensitivity. Extracts were prepared from mouse tissues and tested for their ability to inhibit P2RY8+ cell migration to a chemokine, CXCL12 (Fig. 1a). We detected bioactivity in extracts from liver, but not from spleen, lymph nodes, thymus, brain, kidney or serum. Further analysis of hepatic tissues revealed that bile was a more potent source of activity (Fig. 1b).
Figure 1. Purification and identification of S-geranylgeranyl-L-glutathione as an endogenous compound active on P2RY8.
(a) Diagram of P2RY8 ligand bioassay, depicting migration inhibition of P2RY8+ WEHI-231 cells by extracts containing P2RY8 ligand. (b) Flow cytometry plots of cells from the bottom well of the bioassay described in (a), using mouse liver extract or diluted bile. (c) P2RY8 ligand bioassay of culture media from the indicated cell lines (n=5). (d) P2RY8 ligand bioassay of media from Hepa1-6 cells incubated with the indicated agents (10 μM statin, 100 μM mevalonate (MVA), 100 μM GG-PP or DMSO vehicle) (n=8, one-way ANOVA with Bonferroni’s multiple comparisons test). (e) Diagram of 7-step purification strategy to identify the bioactive compound in bile; asterisks indicate steps used for culture supernatants. Right panel shows scheme for MS detection of candidate ions. (f) Full MS scan (Q1) of purified fractions from the indicated conditions, in positive ion mode. (g) Chemical structure of S-geranylgeranyl-L-glutathione (Ggg). (h) Positive ion mode MS/MS spectra of the 580.3 ion from purified bile (left) and from synthesized Ggg (right). (i) LC-MS/MS quantification of Ggg in C18 solid phase extracts (SPE) of mouse spleen (n=8) and lymph node (n=5), human tonsil (n=6), or mouse bile (n=6). (j) P2RY8 ligand bioassay of C18 SPE concentrates from 500 mg of spleen or tonsil (n=5). Data are representative of or pooled from 3 (b,c,d,h,j,), 2 (i) or 1 (f) experiments. Graphs depict mean with s.d. and each point represents a biological replicate.
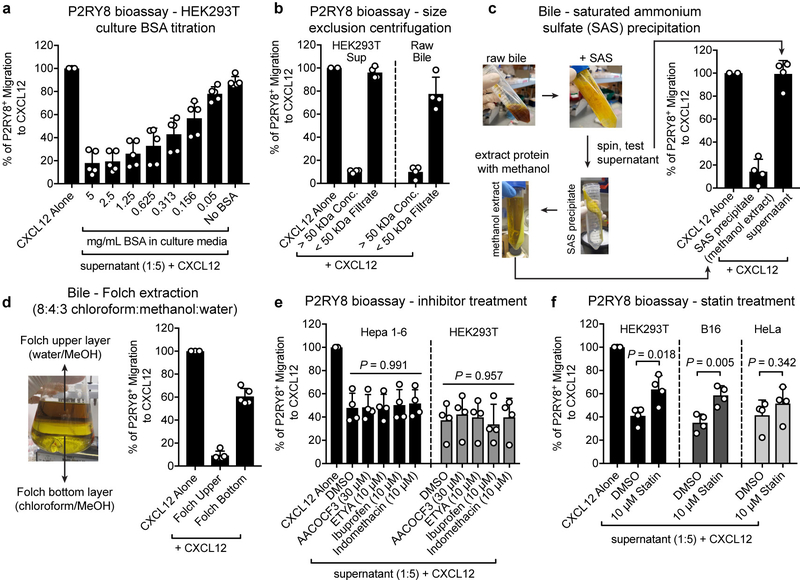
We then found that several adherent cell lines also produced bioactivity (Fig. 1c). The presence of bioactivity in the culture supernatants was enhanced by inclusion of albumin in the medium (Extended Data Fig. 1a). Separation of molecules greater than versus less than 50 kDa (bovine albumin, ~66.5 kDa) revealed that bioactivity was enriched in the >50 kDa fraction (Extended Data Fig. 1b). However, bioactivity could be extracted from the protein precipitate using methanol, suggesting that the bioactive compound was a metabolite that was associated with albumin (Extended Data Fig. 1c). Using a Folch extraction, the bioactivity partitioned with the methanol-water layer, suggesting it could be a polar lipid (Extended Data Fig. 1d).
Given this result, we asked whether inhibitors of lipid biosynthesis affected bioactivity production. Phospholipase, lipoxygenase and cyclooxygenase inhibitors were without effect (Extended Data Fig. 1e), but statins caused a marked reduction in bioactivity production (Fig. 1d and Extended Data Fig. 1f). Bioactivity production could be rescued by supplying statin-treated cells with mevalonate or geranylgeranyl-pyrophosphate (GG-PP), suggesting that the isoprenoid biosynthesis pathway contributed to ligand generation (Fig. 1d).
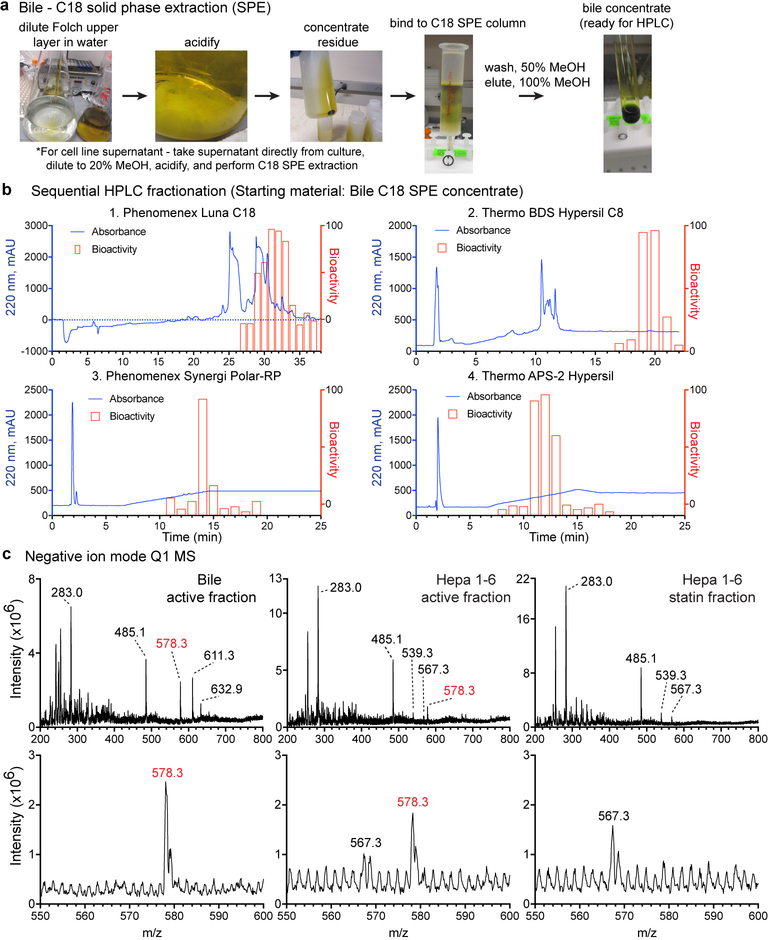
We developed a HPLC fractionation procedure to purify the bioactivity from bile and culture supernatants and used mass spectrometry (MS) to identify molecules common between the active fractions (Fig. 1e and Extended Data Fig. 2a,b). We also performed a drop-out MS analysis of side-by-side purified supernatants from control and statin-treated Hepa1–6 cells. The purified fractions were analyzed using positive ion mode Q1 MS scans, which identified a single ion with m/z 580.3 that was enriched in bioactive fractions, and was absent from the corresponding statin-treated fraction (Fig. 1f). Negative ion mode scans revealed drop-out of an ion with m/z 578.3 (Extended Data Fig. 2c). Given the 2-unit m/z difference, the positive ion candidate and negative ion candidate could be assigned, respectively, to [M+H]+ and [M-H]- ions of the same molecule. High-resolution LC-MS identified a positive ion with m/z 580.3435 (Extended Data Fig. 3a).
A positive ion with m/z 580.3435 did not match any known biological molecules in metabolite databases. Fragmentation of this ion produced an MS/MS spectra that shared similarity with glutathione7 (Extended Data Fig. 3b). Subtracting the monoisotopic mass of a glutathione conjugate from 580.3435 and accounting for the positive proton adduct yielded a monoisotopic mass of 274.2674, which possibly corresponded to a chemical formula of C20H34. This matched geranylgeranyl, an isoprenoid produced by cells in the form of GG-PP8. Comparison of the fragmentation patterns of GG-PP and the candidate ion revealed a shared product ion with m/z 273.1 that produced similar MS/MS/MS spectra, likely corresponding to a geranylgeranyl ion (Extended Data Fig. 3c).
Next, we chemically synthesized the glutathione-S-conjugate of geranylgeranyl (Ggg) (Fig. 1g). This compound had the same elution profile, mass, and fragmentation pattern as the m/z 580.3435 ion of purified bile (Fig. 1h and Extended Data Fig. 3d and e). Using synthesized Ggg as a reference standard, we developed an LC-MS/MS method to quantify Ggg in tissues. We detected low nanomolar amounts of Ggg in extracts from mouse spleen and lymph nodes as well as human tonsil, and low micromolar levels in mouse bile (Fig. 1i). Concentrated extracts from spleen and tonsil also showed P2RY8 bioactivity (Fig. 1j).
Ggg inhibited migration of P2RY8+ but not P2RY8– cells to CXCL12 and a second chemokine, CXCL13, with maximal activity between 10–100 nM (Fig. 2a and Extended Data Fig. 4a, b). Ggg had no migration inhibitory activity on several other receptors that can couple to Gα13 including S1PR2 and Gpr4 (Extended Data Fig. 4a, c). Ggg was potent in inhibiting the migration of tonsil GC B cells while having lower activity on naïve B cells (Fig. 2b) consistent with their lower P2RY8 expression (Extended Data Fig. 5a). Human Tfh cells express P2RY8 (Extended Data Fig. 5a) and Ggg was inhibitory for their migration, though with less potency than for GC B cells (Fig. 2c). Leukotriene C4 (LTC4), a ligand for Cysltr1 and Cysltr2, is also a glutathione-lipid conjugate though the lipid moiety (derived from arachidonic acid) is produced by a distinct pathway compared to geranylgeranyl11. LTC4 had measurable activity on P2RY8 but with 100-fold lower potency than Ggg (Fig. 2a). Ggg promoted internalization of P2RY8, but not of other receptors including S1PR2, Gpr55, Cysltr1 and Cysltr2 (Fig. 2d). These findings establish that Ggg is a potent and selective P2RY8 ligand.
Figure 2. Ggg inhibits the migration of P2RY8-expressing cells.
(a) P2RY8 ligand bioassay using the indicated concentrations of Ggg, glutathione (GSH), GG-PP, or LTC4 with 50 ng/mL CXCL12 (n=4 biological replicates). (b, c) Transwell migration assays of tonsil cells towards CXCL12 mixed with the indicated concentrations of Ggg. Left plots, representative flow cytometry of CD19+ cells showing the gate for CD38+ IgD- GC B cells (b) or of CD4+ cells showing the gate for PD-1+, CXCR5+ Tfh cells (c). Right graphs show summary data for indicated cell types. (n=3 tonsils, 2 technical replicates each). (d) Internalization assay using cells expressing OX56 epitope-tagged P2RY8, read by measuring surface OX56 levels (representative flow cytometry histogram, left). Right graphs show summarized data for the indicated receptors (n=6 biological replicates, one-way ANOVA with Bonferroni’s multiple comparisons test). Data are pooled from 3 experiments (a,b,c,d). Graphs depict mean with s.d.
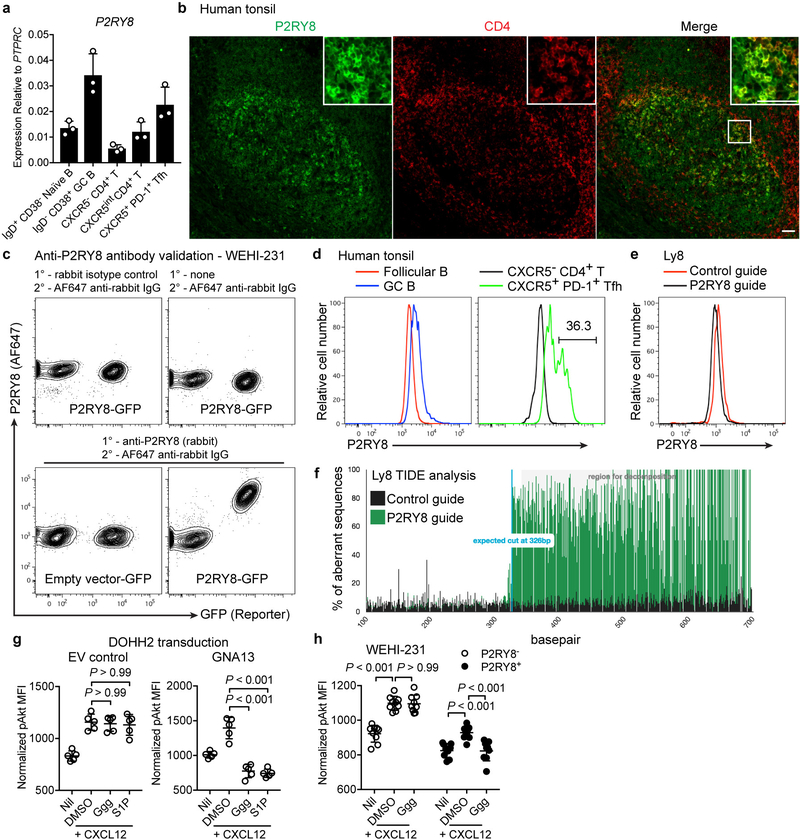
Tonsil tissue staining showed P2RY8 was present throughout the GC and revealed higher expression in a subset of GC-associated cells (Extended Data Fig. 5b). Many of the P2RY8-high cells co-stained for CD4 though not all GC-associated CD4+ cells were P2RY8-high (Extended Data Fig. 5b). Intracellular flow cytometry with this C-terminus specific antibody revealed expression in GC B cells and Tfh cells (Extended Data Fig. 5c and d). Consistent with the microscopy data, a subset of the Tfh cells had high P2RY8 expression (Extended Data Fig. 5d). P2RY8-high CD4+ T cells were not observed outside GCs, consistent with the notion that high receptor expression confined cells to the GC.
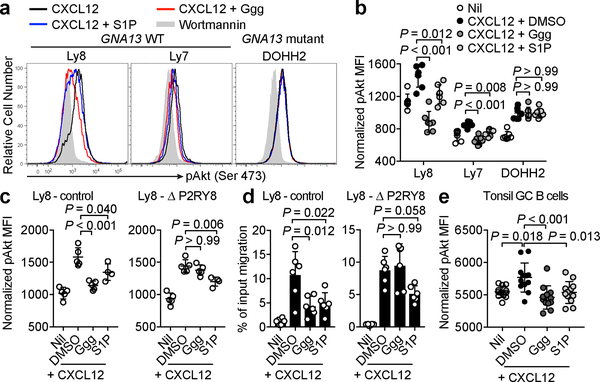
The frequent mutation of P2RY8 in DLBCL and BL is thought to reflect an ability of the receptor to function, analogously to S1PR2, as a repressor of Akt activation4,13. In accord with this model, Ggg antagonized chemokine-induced pAkt in GCB DLBCL lines with intact P2RY8 and GNA13 genes (Fig. 3a, b). To confirm that Ggg was acting via endogenously expressed P2RY8, CRISPR-Cas9 mediated gene editing was used to generate a line of Ly8 GCB DLBCL cells harboring ~80% mutant P2RY8 alleles (Extended Data Fig. 5e, f). Ggg no longer caused inhibition of CXCL12-mediated pAkt induction or migration in P2RY8 mutated cells (Fig. 3c, d). Moreover, restoration of Gα13 expression in the GNA13 mutant DOHH2 cell line rescued the ability of Ggg to repress pAkt (Extended Data Fig. 5g). Ggg also antagonized pAkt induction in P2RY8-transduced WEHI-231 cells without affecting control P2RY8– cells (Extended Data Fig. 5h). Importantly, Ggg antagonized pAkt induction in tonsil GC B cells to an extent similar to that caused by S1P (Fig. 3g).
Figure 3. Ggg suppresses chemokine-induced Akt phosphorylation in cell lines and tonsil GC B cells.
(a, b) Representative histograms (a) and summary data (b) showing pAkt levels in the indicated DLBCL lines treated with wortmannin (grey fill), CXCL12 (black), CXCL12 + S1P (blue), or CXCL12 + Ggg (red) (n=7). (c) pAkt levels in Ly8 cells edited using CRISPR-Cas9 with either a control guide or a guide targeting P2RY8, treated as in (a) (n=5, n=4 for S1P). (d) Transwell migration assay using edited Ly8 cells towards 5 ng/mL CXCL12 along with either 100 nM Ggg, 100 nM S1P, or vehicle (n=6). (e) pAkt levels in tonsil GC B cells, treated as indicated (n = 6 tonsils, 2 replicates each). pAkt MFI data were normalized based on the nil condition. Data are representative of or pooled from 4 (a,b,e) or 3 (c,d) experiments. Graphs depict mean with s.d. Points represent biological replicates. One-way ANOVA with Bonferroni’s multiple comparisons test (b, c, d, e).
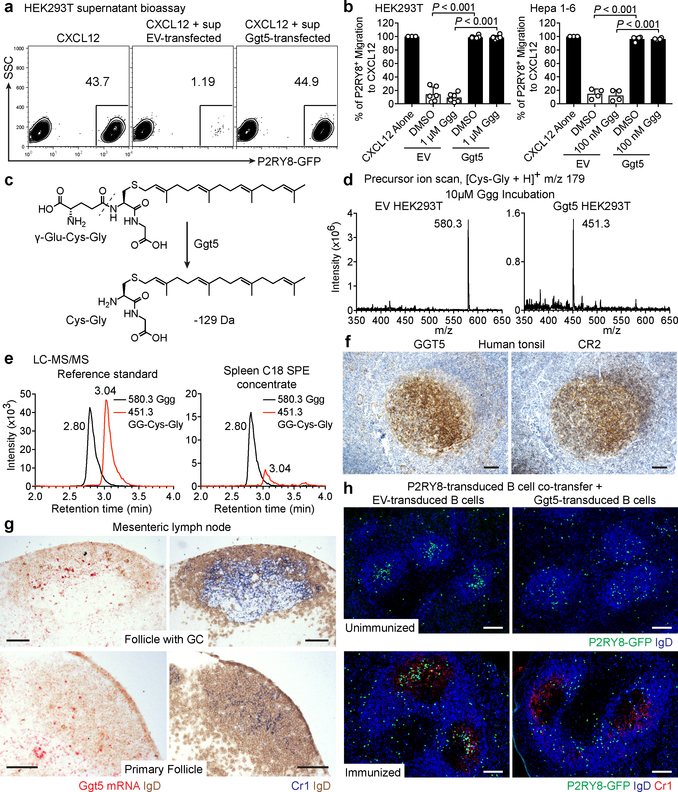
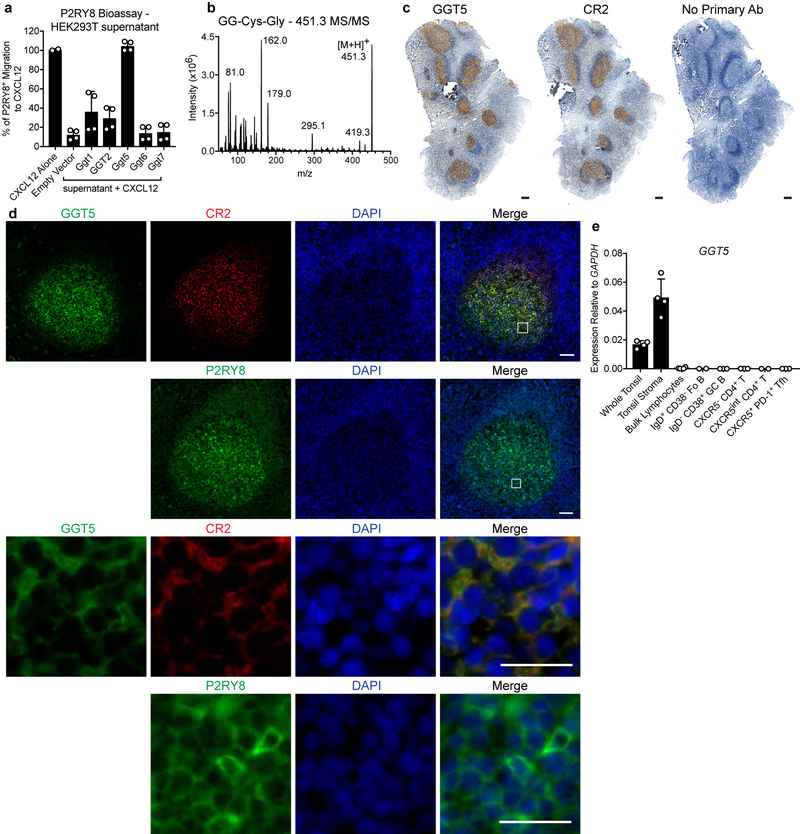
We speculated that Ggg, as a glutathione conjugate, might be metabolized by γ-glutamyltransferase-class enzymes14. We therefore tested the ability of this enzyme class to antagonize P2RY8 bioactivity production by cell lines. Overexpression of Ggt5, also known as γ-glutamyl leukotrienase due to its ability to metabolize LTC4 to LTD415,16, caused loss of bioactivity in culture supernatants (Fig. 4a, b). Moreover, Ggt5-expressing cells were capable of inactivating synthetic Ggg (Fig. 4b). The other mouse Ggt family members as well as GGT2, a human GGT, had either weak or no activity in metabolizing Ggg (Extended Data Fig. 6a). We speculated that Ggt5 was cleaving the γ-glutamyl moiety off of Ggg, resulting in a loss of 129 Da and forming S-geranylgeranyl-L-Cys-Gly (Fig. 4c). Indeed, Ggt5-transfected cells caused conversion of synthetic Ggg (m/z 580.3) to its predicted Cys-Gly metabolite (m/z 451.3) (Fig. 4d and Extended Data Fig. 6b). We detected LC-MS/MS signals corresponding to the Cys-Gly metabolite in extracts from mouse spleen (Fig. 4e), suggesting that this metabolic process occurs in vivo.
Figure 4. Ggt5 metabolizes Ggg and regulates P2RY8 function in vivo.
(a, b) Flow cytometry plots (a) and summary data (b) of P2RY8 ligand bioassay of supernatants from the indicated cells overexpressing Ggt5 or empty vector (EV), cultured for 18 hr with Ggg or vehicle (HEK293T n=6, Hepa1-6 n=4 biological replicates, one-way ANOVA with Bonferroni’s multiple comparisons test). Graphs depict mean with s.d. (c) Diagram of Ggg conversion into S-geranylgeranyl-L-Cys-Gly (GG-Cys-Gly). (d) Positive precursor ion scan for m/z 179 to identify ions producing [Cys-Gly]+ fragments, from purified supernatants of HEK293T cells overexpressing EV or Ggt5 and incubated with Ggg. (e) LC-MS/MS MRM for Ggg and GG-Cys-Gly in a mixture of 100 nM Ggg and GG-Cys-Gly, or in C18 SPE concentrates of mouse spleen. (f) Immunohistochemistry for GGT5 or CR2 (brown) in serial sections of tonsil counterstained with hematoxylin (blue). (g) RNAscope for Ggt5 mRNA (red) in mouse lymph node GCs and primary follicles, counterstained with IgD (brown). Serial sections were stained for Cr1 (blue) and IgD (brown). (h) Immunofluorescence for P2RY8-overexpressing B cells (GFP, green) co-transferred with EV or Ggt5-overexpressing B cells, in unimmunized mice without GCs (top), and in GCs (Cr1, red) of immunized mice (bottom), relative to endogenous B cells (IgD, blue). Data are representative of or pooled from 3 (a,b,e) or 2 (d,h) experiments, 3 (h) or 5 (f,g) biological replicates. Scale bars, 100 μm.
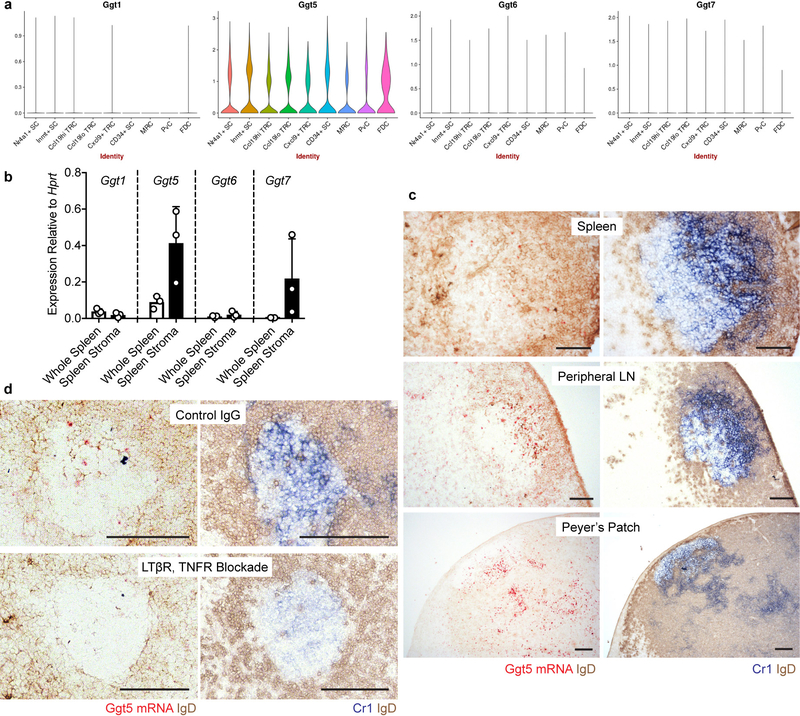
Gain of function studies in mice showed that P2RY8 promotes B-cell positioning in the center of follicles4,6. We speculated that Ggg would be strongly metabolized in the follicle center, resulting in higher Ggg levels in outer areas and confinement of P2RY8+ cells in the center. In accord with this model, Protein Atlas data provided evidence that GGT5 was expressed in GCs in human tonsil and lymph nodes12. Our staining of tonsil showed GGT5 was expressed in a pattern that matched with the FDC marker, CR2 (Fig. 4f and Extended Data Fig. 6c). Co-staining confirmed that GGT5 was expressed by CR2+ FDCs and not by GC B cells (Extended Data Fig. 6d), and qPCR showed high expression in tonsil stroma (Extended Data Fig. 6e). A scRNAseq analysis of mouse lymph node stromal cells18 showed Ggt5, but not Ggt1, 6 or 7, was enriched in FDCs (Extended Data Fig. 7a). By qPCR, Ggt5 was enriched in spleen stroma (Extended Data Fig. 7b) and RNAseq analysis has shown that the Ggt family is minimally expressed in GC B cells (Immgen.org). In situ hybridization revealed mouse Ggt5 expression in GCs and in primary follicles in a pattern similar to Cr1+ FDCs (Fig. 4g and Extended Data Fig. 7c). In human and mouse tissue, lower levels of expression were detectable in the T zone and expression was evident in some HEVs, in accord with microarray data19, but there was minimal expression in the outer regions of lymphoid follicles (Fig. 4f, g and Extended Data Fig. 6c and 7c). Treatment of mice with LTβR-Fc and TNFR-Fc to ablate FDCs20 caused loss of Ggt5 in GCs (Extended Data Fig. 7d).
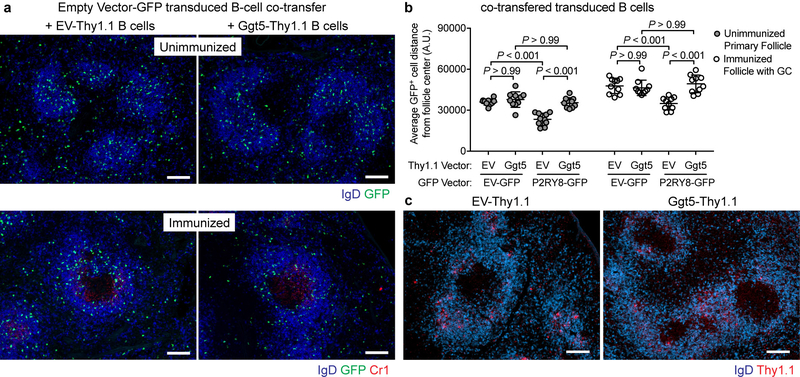
We hypothesized that if Ggt5 was involved in establishing Ggg gradients then increasing expression of Ggt5 throughout the follicle should disrupt the gradient and thus the ability of P2RY8 to confine cells to the follicle center. To test this idea, P2RY8-expressing B cells were transferred into mice along with large numbers of Ggt5-expressing B cells or, as a control, empty vector-expressing B cells. In the controls, P2RY8 caused B cells to localize within pre-existing GCs or in the central region of follicles that lacked GCs (Fig. 4h and Extended Data Fig. 8a, b) as expected4. In contrast, in recipients harboring Ggt5-expressing B cells, P2RY8 no longer caused B-cell confinement to the GC or follicle center (Fig. 4h and Extended Data Fig. 8a, b). The Ggt5-expressing B cells were scattered through the follicle and did not display altered positioning compared to control B cells (Extended Data Fig. 8c). These data provide in vivo evidence that Ggt5 controls P2RY8 ligand distribution and they are in accord with the finding that FDCs are required for P2RY8 function as a B-cell confinement receptor6.
P2RY8 is mutated in up to 20% of GCB DLBCL and BL cases and in some transforming follicular lymphomas1–5,21, providing strong evidence that the receptor and its ligand have an essential, non-redundant constraining function on human GC B cells. Our work here identified Ggg as an intercellular signaling molecule that engages P2RY8 to exert migration inhibitory and growth regulatory effects. The low abundance of Ggg in lymphoid tissue is consistent with the nM potency of Ggg as a P2RY8 ligand and with local metabolism of Ggg to establish organizing gradients. Our data indicate that albumin serves as a carrier for Ggg. We hypothesize that stromal cells in the outer regions of follicles are a source of extracellular Ggg. We do not have an explanation for the abundance of Ggg in bile but it may indicate a role in the hepatobiliary system. There are ~25 glutathione transferases that can conjugate glutathione to target molecules16 and many of these are expressed in lymphoid tissues (Immgen.org). Future studies will be needed to define the biosynthetic pathway for Ggg. P2RY8 is down-regulated on memory B cells and plasma cells23, likely helping these cells exit from GCs. P2RY8 is widely conserved in vertebrates, but puzzlingly it is not present in rodents4. We speculate that a non-orthologous Ggg receptor may exist in the mouse. Given that P2RY8 could respond to both Ggg and LTC4, P2RY8 or P2RY8-like receptors may have the ability to sense a range of glutathione-conjugated lipids. Our studies raise the possibility that Ggg derivatives might be useful as suppressors of GC B-cell growth, for example in the case of P2RY8+ DLBCL or BL. Given that Ggg is made by multiple tumor cell lines and the P2RY8 locus is modified in some other cancer types such as acute lymphoblastic leukemia24, prostate cancer and stomach cancer [www.cbioportal.org and www.intogen.org], we speculate that Ggg has organizing and growth regulatory activities in multiple human tissues.
Methods
Mice and treatments
C57BL/6J mice were bred in an internal colony and mice of both sexes were used between 7 and 12 weeks of age. CD19−/− mice on a B6 background were from Jax. Littermate controls were used for experiments, mice were allocated to control and experimental groups randomly, sample sizes were chosen based on previous experience to obtain reproducible results, and the investigators were not blinded. For SRBC immunization, mice were injected i.p. with SRBCs (Colorado Serum Company) once on day 0 and again on day 3. For FDC ablation, 100 μg of LTβR-Fc and TNFR-Fc or control IgG (provided by J. Browning) were injected i.v. and mice were analyzed on day 4 after injection. Animals were housed in a pathogen-free environment in the Laboratory Animal Resource Center at the University of California, San Francisco, and all experiments conformed to ethical principles and guidelines approved by the Institutional Animal Care and Use Committee.
Flow Cytometry and cell sorting
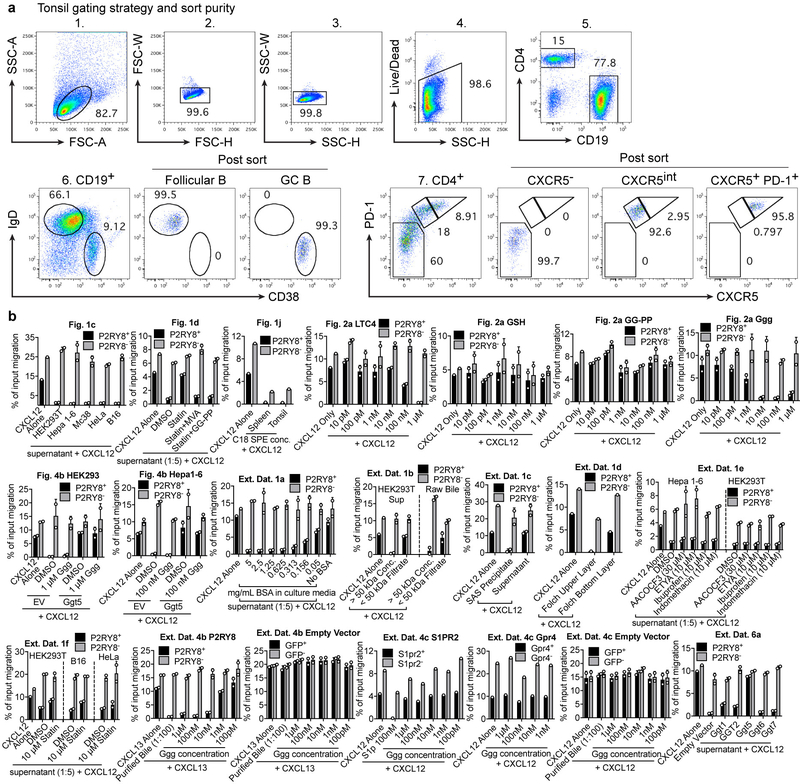
To identify human GC B cells and Tfh cells, the following antibodies were used: FITC-conjugated anti-human CD4 (Tonbo, RPA-T4, 35–0049-T100), PE-conjugated anti-human CXCR5 (ThermoFisher, MU5UBEE, 12–9185-41), PerCP-Cy5.5-conjugated anti-human CD38 (Biolegend, HIT2, 303518), PE-Cy7-conjugated anti-human CD19 (Biolegend, HIB19, 302216), APC-conjugated anti-human IgD (Biolegend, IA6–2, 348221), and pacific blue-conjugated anti-human PD-1 (Biolegend, EH12.2H7, 329916). Cells were placed in a 96 well round bottom plate and washed with staining buffer (PBS containing 2% FBS, 0.1% sodium azide and 1 mM EDTA). 25 μL of antibody cocktail was added to each sample for 20 min on ice. After incubation, cells were washed twice with staining buffer. For staining of OX56, a 1:200 dilution of a biotinylated OX56 antibody was placed on the cells for 25 min on ice, after which the cells were washed and a 1:200 dilution of streptavidin-AF647 (Invitrogen) was incubated with the cells for 20 min. To identify Thy1.1 reporter expression, PE-conjugated anti-mouse/rat CD90.1 (Biolegend, OX-7) was used. For staining of pAkt or P2RY8, intracellular flow cytometry was performed on fixed cells (see details in section below). Data were acquired on a BD LSR II flow cytometer or a BD FACS Calibur. A BD FACSAriaII was used to sort human tonsil subsets, and an example of the gating strategy and post-sort purity is provided in Extended Data Figure 9a. Flow cytometry data were analyzed using Flowjo (ver 9.7.6).
Generation of P2RY8-expressing WEHI-231 cells
P2RY8 was cloned into the MSCV-GFP retroviral vector (P2RY8-GFP). Retrovirus encoding P2RY8-GFP was produced using the PLAT-E packaging cell line. 5 × 105 WEHI-231 cells were placed in a 6-well plate along with retroviral supernatant and the cells were centrifuged at 2400 rpm for 2 hr at RT. The viral supernatant was aspirated, and the cells were resuspended in growth medium and returned to culture. This spinfection was repeated with fresh retrovirus for a second time 24 hrs later. 48 hrs after the second spinfection, the highest 5% of GFP-expressing cells were sorted using a BD FACSAriaII. These cells were combined with GFP-negative cells at a 1:1 ratio, and this mixture was maintained in culture for use in transwell bioassays.
Migration inhibition transwell bioassay
A confluent T25 flask containing a mixture of WT and P2RY8-GFP-transduced WEHI-231 cells was washed twice in pre-warmed migration media (RPMI containing 0.5% fatty acid-free BSA, 10 mM HEPES, and 50 IU penicillin/streptomycin). The cells were resuspended in migration media at 2×106 cells/mL and resensitized for 10 min in a 37°C water bath.
Recombinant human CXCL12 (Peprotech) was diluted to 50 ng/mL in migration media. Tissue extracts or purified compounds were diluted at varying concentrations in the CXCL12-containing migration media, and 600 μL of these mixtures were added to a 24-well tissue culture plate. Methanol-based extracts could be added to the media at a concentration of up to 1:100 without interfering with overall WEHI-231 migration. In some experiments, 500 ng/mL CXCL13 (Peprotech) was used instead of CXCL12. Transwell filters (6 mm insert, 5 μm pore size, Corning) were placed on top of each well, and 100 μL of P2RY8-GFP-expressing WEHI-231 cells (2 × 105 cells) were added to the transwell insert. The cells were allowed to migrate for 3 hr, after which the cells in the bottom well were enumerated by flow cytometry. To assess migration inhibition, the proportion of P2RY8-GFP+ cells that migrated for each well was divided by the proportion of P2RY8-GFP+ cells that migrated to CXCL12 alone. This normalized metric is plotted as “% of P2RY8+ migration to CXCL12” for each bioassay. Representative experiments for each bioassay are also plotted as % of input migration in Extended Data Figure 9b. The baseline migration between experiments differs based on the growth state of the WEHI-231 cells.
Cell lines and treatments
HEK293T, HeLa, Hepa 1–6, Mc38, and B16 cells were grown in 10 cm tissue culture dishes in DMEM containing 10% FBS, 10 mM HEPES, 2 mM glutamine, and 50 IU penicillin/streptomycin. WEHI-231, Ly7, Ly8, DOHH2, and M12 cells were grown in upright T25 flasks in RPMI containing 10% FBS, 10 mM HEPES, 2 mM glutamine, 55 μM 2-mercaptoethanol, and 50 IU penicillin/streptomycin. All cell lines were previously obtained from other laboratories and further authentication was not performed. The cell lines were not tested for mycoplasma contamination. For some experiments, DOHH2 cells were transduced with retrovirus encoding GNA13-IRES-GFP or empty vector-GFP.
For testing bioactivity production, cells were plated out in either 12-well or 6-well plates and allowed to reach confluence. The media was then replaced with serum free media (RPMI containing 0.5% fatty acid-free BSA, 10 mM HEPES, and 50 IU penicillin/streptomycin) at 750 uL per well for a 12-well plate or 1.5 mL per well of a 6-well plate, incubated for 16–18 hr, and tested in the bioassay. Culturing cells in this serum-free media resulted in greater bioactivity production compared to culturing cells in media containing FBS. To test whether bioactivity production was dependent on albumin, the amount of BSA in the serum-free media was titrated or removed entirely. The supernatant from these cultures was diluted 1:5 in migration assay media, mixed with CXCL12, and tested in the bioassay. For testing effects of small molecule inhibitors on bioactivity production, inhibitor-containing serum free media was used to replace the growth media. 16–18 hr later, the media was removed, centrifuged to remove cells and debris, and tested at varying dilutions in the P2RY8 bioassay.
For transfection of HEK293T cells, mouse Ggt1, Ggt5, Ggt6, Ggt7, and human GGT2 were cloned into an MSCV-Thy1.1 retroviral vector. HEK293T cells were seeded into 6-well tissue culture plates and grown until 75% confluent in antibiotic-free media. To prepare the transfection mixture, the plasmids were aliquoted in Opti-MEM and mixed with Lipofectamine 2000, at 6 μL per 3 μg plasmid, and allowed to sit for 25 min at room temperature. The mixtures were gently added dropwise to the HEK293T cells. 24 hr after transfection, the media was replaced with serum-free media containing 1 uM Ggg or DMSO (vehicle control) for 18 hr and the supernatants were tested in the bioassay. In some experiments, media containing 10 μM Ggg was placed on transfected HEK293T cells for 7 hr, after which the supernatant was purified for mass spectrometry analysis. Hepa 1–6 cells were retrovirally transduced with Ggt5 and incubated with serum-free media containing 100 nM Ggg or DMSO (vehicle control) for 18 hr, and the supernatants were tested in the bioassay.
Chemicals and reagents
Indomethacin, ibuprofen, mevalonic acid, mevastatin, S1P, and glutathione were purchased from Sigma. HPLC-grade solvents were purchased from Fisher. Leukotriene C4, D4, and geranylgeranyl-pyrophosphate was purchased from Cayman Chemical. LPI was purchased from Avanti Polar Lipids. ETYA and AACOCF3 were purchased from Biomol. Ggg was chemically synthesized using the protocol specified in the section below.
Transwell migration assay on human tonsil cells
Fresh de-identified human tonsil tissue was obtained through the UCSF Biospecimen Resources (BIOS) Program from donors undergoing tonsillectomies and analyzed within 4–6 hr after surgery (stored on ice in RPMI media). Informed consent was obtained by the BIOS program, which complied with all ethical and regulatory requirements and de-identified the samples upon collection. Tonsils were mashed through metal mesh to form a cell suspension and washed in migration media. The cells were RBC-lysed due to blood contamination during surgical removal, washed twice in warm migration media, and resensitized at 107 cells/mL in migration assay media in a 37°C water bath for 10 min. A titration of Ggg or 100 nM S1p was prepared with 100 ng/mL CXCL12, and the different mixtures were placed in the bottom wells of a 24-well plate. (106) cells were placed into 5 μm transwell filters (Corning Costar), allowed to migrate for 1.5 hr at 37°C, and migrated cells were stained with antibodies against GC B cell and Tfh markers and enumerated by flow cytometry.
Purification of P2RY8 ligand from pig bile
Frozen pig bile was purchased from Pel-Freez Biologicals. 60 mL of frozen pig bile was thawed in a 37°C water bath. Saturated ammonium sulfate (SAS), pH 7.4 was added to the pig bile to achieve a 70% SAS solution, resulting in a large amount of precipitate. This was centrifuged for 15 min at 8300 rcf. The liquid was decanted and the precipitate was mixed thoroughly with 20 mL of water. 120 mL of methanol was added and the mixture was vortexed vigorously, followed by centrifugation for 15 min at 8300 rcf. The supernatant was transferred into an Erlenmeyer flask. The pellet was washed with 40 mL of methanol to extract residual lipids, centrifuged for 15 min at 8300 rcf, and the supernatant was combined into the Erlenmeyer flask. To perform a Folch extraction (8:4:3 chloroform/methanol/water), 320 mL of chloroform was added to the flask, along with 100mL of water. The flask was vigorously shaken, and the resulting aqueous and organic layers were allowed to separate overnight. The upper (aqueous) layer was transferred to a separate Erlenmeyer flask. An additional 200 mL of 1:1 methanol:water was added to the bottom (chloroform) layer and vigorously shaken again to further extract polar compounds. The layers were allowed to separate for 1 hr, after which the upper aqueous layer was combined with the previous aqueous layer. To further remove non-polar compounds from this aqueous layer, 100 mL of chloroform was added to this aqueous layer, shaken, and allowed to separate. The aqueous layer was transferred to a 4 L Erlenmeyer flask. 3 L of water acidified with 27 mL of 1 M HCl was added to the bile extract, which caused a yellow-green precipitate to form. The solution was then divided into 500 mL Nalgene bottles and centrifuged at 7000 rpm in an ultracentrifuge. The supernatant was decanted and the green, wax-like pellet was dissolved in 400 mL of 50% methanol. This extract was bound to a 10 gram C18 solid phase extraction column (Waters) using a vacuum manifold (Agilent). The column was washed with 50 mL of 50% methanol, and then the compounds were eluted with 50 mL of 100% methanol. The methanol was evaporated under compressed air to produce 600 μL of a concentrated extract with potent bioactivity on P2RY8-transduced cells.
HPLC purification was performed using an Agilent 1220 Infinity HPLC coupled with an Agilent 1260 Infinity Fractionator. HPLC-grade solvents were purchased from Fisher. The max injection amount was 100 μL. For purifying larger amounts of sample, the sample was injected and run multiple times per column and the corresponding fractions per minute were pooled. For each column, solvent A: 100% Water+ 0.1% formic acid and solvent B: 100% methanol+0.1% formic acid. Fractions were collected every minute, concentrated via evaporation, and tested at a 1:100 dilution via bioassay. The bioactive fractions were pooled, concentrated, and run on the next column.
-
1st separation: Phenomenex Luna C18, 100A pore size, 250 × 10.00 mm, 10μm particle size, Part no. 00G-4094-N0.
Flow rate: 2mL/min. 0–2min, 50%B, 2–26.5 min, ramp to 95%B, 26.5–36.5min, 95%B, 36.5–37min ramp to 50%B, 37–38min, 50%B.
-
2nd separation: Thermo BDS Hypersil C8, 150 × 4.6mm, 5μm particle size, Part no. 28205–154630
Flow rate: 1mL/min. 0–2min, 50%B, 2–10 min, ramp to 90%B, 10–20min, 90%B, 20–20.5min ramp to 50%B, 20.5–22min, 50%B.
-
3rd separation: Phenomenex Synergi Polar-RP 80A pore size, 150 × 4.6 mm, 4μm particle size, Part no. 00F-4336-E0
Flow rate: 1mL/min. 0–4min, 50%B, 4–12 min, ramp to 95%B, 12–23min, 95%B, 23–23.5min ramp to 50%B, 23.5–25min, 50%B.
-
4th separation: Thermo APS-2 Hypersil, 150 × 4.6 mm, 5μm particle size, Part no. 30705–154630
Flow rate: 1mL/min. 0–4min, 50%B, 4–12 min, ramp to 95%B, 12–23min, 95%B, 23–23.5min ramp to 50%B, 23.5–25min, 50%B.
Purification of P2RY8 ligand from cell culture supernatants
Hepa 1–6 cells were grown in 16 T175 flasks using DMEM containing 10% FBS, 10 mM HEPES, 2 mM glutamine, and 50 IU penicillin/streptomycin. When cells were confluent, the media was replaced with RPMI containing 0.5% fatty acid-free BSA, 10 mM HEPES, 50 IU penicillin/streptomycin, and 50 μM nicardipine, a drug that we found increased bioactivity production specifically in Hepa 1–6 cells. To half of the flasks, 10 μM mevastatin was also added in order to inhibit bioactivity production. After 24 hr, the supernatant from each condition was collected from the cells and centrifuged at 2000 rpm to remove cell debris. Methanol was added to form a 20% methanol solution, and the solution was acidified using 1 M HCl to pH 3.5. For each condition, the solution was bound to a 10 gram C18 solid phase extraction column using a vacuum manifold. The columns were washed with 50 mL of 50% methanol, the compounds were eluted with 50 mL of 100% methanol, and evaporated to produce a 600 μL concentrated extract. The extracts from statin-treated Hepa 1–6 cells and control Hepa 1–6 cells were then simultaneously purified using the same HPLC and solvent system as for bile, using the Thermo BDS Hypersil C8, Phenomenex Synergi Polar-RP, and Thermo APS-2 Hypersil columns. Fractions were collected every minute, concentrated via evaporation, and tested at a 1:100 dilution via bioassay. The bioactive fractions for the control Hepa 1–6 cells were pooled, concentrated, and run on the next column. The corresponding fractions from the statin-treated Hepa 1–6 cells were also pooled, concentrated and run simultaneously.
Solid phase extraction
C18 solid phase extraction (SPE) columns were purchased from Waters (Sep-Pak, 10g-35cc and 500mg-6cc versions). The SPE columns were attached to a vacuum manifold (Vac Elut 20, Agilent) and pressure was maintained at 2–5psi. The column was washed with two column-volumes of methanol, then two volumes of water. Samples were typically diluted to 20% or less methanol content and acidified to pH 3.5 and loaded onto the column at a drop rate of 1–2 drops/sec. The column was washed with two volumes of water and one volume of 50% methanol. The compounds were then eluted with one volume of 100% methanol into glass test tubes. The solution was dried down using a Thermo Reacti-Vap apparatus under compressed air and the residue was dissolved in a small amount of 100% methanol.
Mass Spectrometry
An AB SCIEX QTRAP 6500 mass spectrometer was used to obtain full MS (Q1) spectra, MS/MS fragment ion spectra, and precursor ion spectra. HPLC fractions were diluted 1:10 in HPLC-grade methanol without any additives. The diluted samples were directly injected into the ion source via syringe at 10 μL/min and ionized using electrospray ionization (ESI). Mass spectra were acquired in both positive and negative ion mode. The ion source was maintained at 100°C, 20 CUR, 14 GS1, 8 GS2, and 5500 IS (positive mode) or −4500 IS (negative mode), 135 DP in positive mode or −60 DP in negative mode, EP 10, CXP 10, and a range of CE was used when performing fragmentation analysis. Positive and negative mode fragmentation spectra (MS/MS) were obtained for the candidate ion. The mass of each fragment ion in the MS/MS spectra was input into Google to search for publications reporting molecules with similar fragmentation patterns. This led to finding a publication containing the MS/MS spectra of glutathione7, which displayed many of the fragments observed in the MS/MS spectra of the candidate ion, suggesting the presence of similar chemical structures. Precursor ion scans in positive ion mode using m/z 179 were used to detect Cys-Gly conjugates. The data was analyzed using Analyst software.
To quantify Ggg in tissues, an LC-MS/MS method was developed using synthetic Ggg as a reference standard. Ggg was detected using multiple reaction monitoring (MRM) scans with ion pair 580.3/179.0, and GG-Cys-Gly was detected using ion pair 451.3/162.0. A reference standard for GG-Cys-Gly was produced by purifying supernatants from Ggt5-expressing HEK293T cells incubated with Ggg. 3 μL of each sample was injected into a Shimadzu Nexera X2 HPLC, with a Synergi Polar-RP column (75 × 4.6 mm) and a mobile phase gradient consisting of A: 100% H2O+0.1%FA and B: 100% ACN+0.1% FA. 0–1 min, 50%B; 1–4 min ramp to 80%B; 4–6 min, 80%B; 6–6.5 min, ramp to 50% B; 6.5–8 min, 40% B. The internal standard used was LTC4-d5, identified with ion pair 631.4/179.0. Peak area was integrated using Analyst software and referenced against a standard curve to calculate compound abundance.
High resolution LC-MS was performed using a Waters XEVO-G2 XS QTOF with an Acquity UPLC equipped with a BEH C18 column. The mobile phase was H2O with 0.05 % formic acid (A) and Acetonitrile with 0.05 % formic acid (B), 5–95 %B, 0.1–1.9 min, 95%B 1.9–2.2 min, ramp down to 5%B, 2.2–2.3 min, 5%B 2.3–2.6min. MS spectra were acquired using ESI in positive ion mode. Metabolite databases including HMDB, LipidMaps, LipidBank, and Chemspider were used to search for the identity of the m/z 580.3435 candidate ion, although each query led to 0 matches.
Chemical Synthesis
Unless otherwise noted, all materials used in chemical synthesis were obtained commercially from MilliporeSigma and were reagent grade. NMR spectra were obtained on a Bruker Avance III HD 400 MHz spectrometer.
Geranylgeranyl bromide - following the procedure outlined in Liu et al., 25 Triphenylphosphine (21.2 mg, 80.8 μmol, 1.3 eq.) was added to a solution of geranylgeraniol (20 mg, 68.9 μmol, 1 eq.) in 1 ml of dry DCM stirring at room temperature under an atmosphere of argon. Carbon tetrabromide (29.6 mg, 89.3 μmol, 1.3 eq.) was then added, and the reaction left stirring at room temperature for 4 hours. The reaction mixture was concentrated under reduced pressure and a small volume of n-hexane added. The resulting precipitate was removed by filtration and the filtrate concentrated again under reduced pressure. The unstable product was used in the next step without further purification.
S-geranylgeranyl-L-glutathione - Modifying the procedure used by Niegowski et al., 26 l-glutathione (23.0 mg, 74.9 μmol, 1.1 eq.) was dissolved in 0.5 ml of 2 M NaOH, and approximately 1 ml of EtOH added drop-wise until the solution started to become cloudy. Geranylgeranyl bromide (24.0 mg, 68.7 μmol, 1 eq.) was added drop-wise, and the reaction left stirring at room temperature overnight. The pH was then adjusted to 2 by addition of 1 M HCl, and the mixture cooled in an ice bath for 20 min. The resulting precipitate was collected by filtration, washed with ice-cold EtOH and water, and dried to afford geranylgeranyl glutathione as an off-white solid (6.0 mg, 10.4 μmol, 15 %).
1H NMR (δ ppm, DMSO-d6) 8.65 (1H, app. s, NH); 8.36 (1H, d, J = 8.17 Hz, NH); 5.20–5.14 (1H, m, CH); 5.12–5.04 (3H, m, CH); 4.48–4.39 (1H, m); 3.70 (3H, m); 3.32 (1H, m); 3.15 (3H, m); 2.89–2.80 (1H, m); 2.61–2.52 (1H, m); 2.42–2.22 (2H, m, CH); 2.10–1.90 (12H, m, 6×CH2); 1.64 (3H, s, CH3 ); 1.63 (3H, s, CH3); 1.56 (9H, s, 3×CH3).
ESI-HRMS (m/z) Calcd for chemical formula C30H49N3O6S [M + H]+, 580.3415; found 580.3435.
Crude tissue extract preparation
Crude tissue extracts were prepared by grinding mouse tissues into water (1:10 weight/volume) then diluting this lysate with 4 volumes of methanol. The mixture was centrifuged twice at 4000g for 5 min to remove precipitate. The supernatant was evaporated and the residue was dissolved in a small amount of 100% methanol. Raw mouse bile was collected directly from the gallbladder using a syringe.
To obtain C18 solid phase extracts of spleen, lymph node, and tonsil, tissue was homogenized in 66% methanol using a Precellys 24 bead homogenizer, 1:10 weight/volume. For mass spec analysis, 20–100 mg of tissue was homogenized along with 15 μL of a 150 nM solution of LTC4-d5 as an internal standard. The homogenate was transferred to a new tube. 500 μL of 66% methanol was used to wash the beads and was combined with the homogenate. The mixture was centrifuged for 10 min at 4000g in a microcentrifuge and the supernatant was diluted 10-fold in water containing 3 mM HCL. This was then bound to a 500 mg C18 SPE column, washed with 50% methanol, eluted with 100% methanol, and concentrated down to 100 μL by evaporation.
Size-exclusion centrifugal filtration of bile and cell culture supernatant
Amicon centrifugal filtration units with 50kD-cutoff membranes were purchased from Millipore. A 100-fold dilution of raw mouse bile in RPMI or undiluted HEK293T culture supernatant (serum starved, 0.5% BSA) was loaded into the top chamber of each type of centrifugal filtration unit. The unit was centrifuged at 7500rcf for 15 min in a fixed-angle rotor. The filtrate in the bottom chamber of the filtration unit and the concentrate in the upper chamber of the filtration unit was tested for P2RY8 bioactivity.
Internalization Assay
P2RY8, Gpr55, S1PR2, Cysltr1, and Cysltr2 were cloned into an MSCV-Thy1.1 retroviral vector with an OX56 (rat CD43-derived27) epitope tag in order to track surface expression levels of each receptor using the OX56 antibody. P2RY8-Ox56-Thy1.1 was retrovirally transduced into M12 cells, and the other GPCR constructs were transduced into WEHI-231 cells. Confluent cultures of each of the lines indicated above were washed twice in migration media, resuspended at 5×106 cells/mL and resensitized at 37°C for 10 min. For each line, 20 μL of cells were aliquoted into a 96 well plate. Ggg, LPI, S1P, LTC4, and LTD4 were prepared in migration media and 80μL aliquots were placed into a 96 well round bottom plate such that the concentration after adding the cells would be as indicated in the figures. Using a multichannel pipette, the compounds were placed on the cells, and the plate was placed in a 37°C cell culture incubator for 45 min. The plate was then placed on ice, washed with ice cold flow cytometry buffer, and stained for OX56 levels by flow cytometry. OX56 surface levels on transduced cells were assessed by drawing a gate on the top 40% of OX56-expressing cells in the control condition, then using the same gate on the transduced cells treated with various compounds to assess internalization.
pAkt stimulation, fixation, and intracellular staining
DLBCL lines, P2RY8-GFP WEHI231 cells, or human tonsil cells were washed twice in migration media and resensitized for 10 min at 37°C. In 5 mL polystyrene FACS tubes, 5 × 104 cells were diluted in 500 μL of migration media containing the indicated combinations of 100 ng/mL CXCL12, 100 nM Ggg, 100 nM S1P, and 200 nM Wortmannin, for either 10 min (human tonsil cells) or 5 min (DLBCL lines, WEHI-231 cells). Afterwards, 50 μL of 16% PFA was added to each tube. The cells were fixed at RT for 10 min, centrifuged, and 1 mL cold methanol was added to each tube while vortexing. The samples were placed at −20°C overnight, washed 3 times with FACS buffer, blocked 20 min at RT with 5% normal goat serum (Sigma) and 1:100 Fc-block, stained at RT for 1hr with a 1:100 dilution of rabbit anti-pAkt (Ser 473, clone D9E, Cell Signaling Technology), washed twice in FACS buffer, and stained for 1 hr at RT with 1:300 dilution of APC-conjugated goat anti-rabbit-IgG (Santa Cruz Biotechnologies), or AF647-conjugated goat anti-rabbit IgG in some experiments (Invitrogen). Unstimulated P2RY8+ WEHI-231 cells were noted to have a lower pAkt level that might reflect endogenous production of small amounts of Ggg. For human tonsil cells, antibodies towards GC B cell markers were added alongside the APC-conjugated goat anti-rabbit-IgG, and PE-conjugated anti-human IgD was used instead of APC-conjugated anti-human IgD. For staining of P2RY8 in DLBCL lines or human tonsil cells, the cells were fixed, permeabilized, blocked, and stained as above using a rabbit polyclonal anti-P2RY8 (Sigma - Atlas Antibodies, HPA003631) and AF647-conjugated goat anti-rabbit-IgG (Invitrogen).
CRISPR-Cas9 targeting of P2RY8 in Ly8 Cells
The LentiCRISPRv2 system (purchased from Addgene) was used to disrupt P2RY8 in Ly8 cells. A non-targeting control guide (sequence: GAGATGATAACTTAATTTGT) or a guide targeting P2RY8 (sequence: GATCATGAAGATGACCGACG) was cloned into the LentiCRISPRv2 plasmid. Lentivirus for each construct was produced in HEK293T cells and Ly8 cells were spinfected for 2 hr at RT. The infected Ly8 cells were allowed to recover for 2 days, after which the transduced cells were selected using puromycin (5 μg/mL, Invivogen) for 2 weeks. Targeting of P2RY8 was assessed by extracting genomic DNA from the culture and performing TIDE analysis 28 on a PCR product encompassing the expected cut-site, which indicated an editing efficiency of 83% (https://tide.deskgen.com/). P2RY8 protein levels were also assessed using flow cytometry.
Adoptive co-transfer of transduced B cells
B cells were enriched from spleens of mice using EasySep kits by removing T cells with biotin-conjugated anti-CD3e and streptavidin-conjugated beads. B cells were cultured in 6 well plates with a final concentration of 0.25 μg/mL (1:4000 dilution) of anti-CD180 (clone RP/14, BD) diluted in RPMI 1640 containing 10% FBS, 10mM HEPES, 55μM 2-mercaptoethanol, 2mM glutamine, and 50 IU penicillin/streptomycin. 24 hr after activation, the plate was centrifuged and the culture supernatant was saved. Retrovirus encoding MSCV-P2RY8-GFP, MSCV-EV-GFP, MSCV-EV-Thy1.1, or MSCV-Ggt5-Thy1.1 was produced using the PLAT-E packaging cell line and added to separate plates of activated B cells. The B cells were spinfected at 2400rpm for 2 hr at RT, the viral supernatant was aspirated, and the original culture supernatant was returned to the cells. This spinfection was repeated a second time 24 hr later. 24 hr after the second spinfection, cells were harvested from each plate and washed twice. qPCR analysis established that S1pr2 was not upregulated on the transduced cells. 2–3 × 107 P2RY8-GFP or EV-GFP B cells were mixed with either 4–5 × 107 EV-Thy1.1 B cells or Ggt5-Thy1.1 B cells, and adoptively transferred into unimmunized CD19−/− mice which lack germinal centers, mice day 6 after immunization with sheep red blood cells (SRBCs). Mice were analyzed 24 hr after transfer. Transduced cells comprised between 1–3% (GFP) or 3–5% (Thy1.1) of all B cells in the spleen by flow cytometry. Positioning of GFP-expressing and Thy1.1-expressing B cells was tracked by immunofluorescence.
RNAscope in situ hybridization
RNA in situ hybridization was performed using the RNAscope RED 2.5HD manual assay kit (Advanced Cell Diagnostics) The RNAscope probe used for mouse Ggt5 targeted region 996–2040 of NM_011820.5. Tissues were frozen in OCT. Within 1 hr, 10 μm cryosections were cut and slides were dried at −20°C for 20 min. Serial sections for each slide were stored at −20°C for IHC analysis of FDCs. Slides were fixed for 15 min with ice-cold 4% paraformaldehyde and washed in 50%, 70%, and 100% ethanol for 5 min each. After drying for 5 min, slides were treated with hydrogen peroxide (from kit) for 8 min and protease IV (from kit) for 12 min. Probes were allowed to hybridize in a HybEZ oven at 40°C for 3.5 hr. The following incubation times for the amplification steps were used: Amp 1, 35 min; Amp 2, 20 min; Amp 3, 35 min; Amp 4, 20 min; Amp 5, 40 min; Amp 6, 25 min. Slides were then developed with FastRed (from kit) for 15 min, washed in PBS, and counterstained for IgD using goat anti-mouse IgD (Cedarlane Laboratories) and HRP-conjugated donkey anti-goat IgG (Jackson Immunoresearch).
Immunohistochemistry and Immunofluorescence
Pieces of human tonsil tissue were fixed in 4% PFA for 2 hr at 4°C, washed with PBS, submerged in 30% sucrose overnight, and embedded in OCT. For staining human GGT5 and CD21, cryosections of 7 um were dried for 1 hr at RT and then subjected to heat-induced antigen retrieval (HIER) by placing the slide in a solution of 1X RNAscope target retrieval reagent (cat. no. 322000) at 95°C for 1 hr. Slides were allowed to cool for 20 min then placed in RT ddH2O for 10 min, and blocked in TBS containing 0.1% fatty acid-free BSA for 10 min. A 1:200 dilution of rabbit anti-human GGT5 (Thermo, PA5–52514) or biotin-conjugated anti-human CD21 (Biolegend, clone Bu32, 354913) along with 1% NMS and NDS (Sigma) was incubated with the slides for 2hr at RT. The slides were washed, and a 1:200 dilution of HRP-conjugated anti-rabbit-IgG or HRP-conjugated streptavidin (Jackson Immunoresearch) was incubated with the slides for 2 hr at RT. The slides were developed using Sigma DAB and counterstained with hematoxylin. For immunofluorescence, a 1:200 dilution of AF647-conjugated goat anti-rabbit-IgG (Invitrogen), streptavidin-conjugated AF555 (Invitrogen) or Cy3 (Jackson Immunoresearch), and DAPI was used to visualize co-localization of GGT5 and CD21 signals. For staining human P2RY8, cryosections were stained with a 1:200 dilution of rabbit anti-human P2RY8 (Sigma - Atlas Antibodies, HPA003631). In some experiments, non-HIER treated sections were co-stained with anti-human P2RY8 and biotin-conjugated anti-human CD4 (Biolegend, clone RPA-T4) since the CD4 epitope was degraded by heat treatment. The same secondary antibodies as above were used. The GGT5 stain required HIER, but the P2RY8 stain produced similar results with or without HIER.
To track positioning of GFP or Thy1.1 expressing B cells, mouse tissues were fixed in 4% PFA for 2 hr at 4°C, washed with PBS, submerged in 30% sucrose overnight, and embedded in OCT. Cryosections of 7 μm were dried for 1 hr at RT, and rehydrated in PBS containing 0.1% fatty acid-free BSA for 10 min. A 1:100 dilution of biotin-conjugated anti-Thy1.1 (eBioscience) or A488-conjugated rabbit anti-GFP (Invitrogen) was used. For immunized mice, biotin-conjugated anti-mouse CD35 was used to track FDC positioning. Endogenous naïve B cells were labeled using goat anti-mouse IgD (GAM/IGD(FC)/7S, Cedarlane Laboratories). Antibodies were diluted along with 1% NMS and NDS and incubated with the slides overnight at 4°C. The slides were then washed in PBS and stained with A647-conjugated streptavidin and AMCA-conjugated donkey anti-goat IgG for 2 hr at RT, and images were captured with a Zeiss AxioObserver Z1 inverted microscope.
For analysis of Cr1 (CD35) staining in serial sections from RNAscope tissues, the serial sections that were stored at −20°C were fixed in cold acetone for 10 min, and dried at RT for 1 hr. Slides were rehydrated for 10 min in TBS containing 0.1% fatty acid-free BSA. A 1:100 dilution of biotin-conjugated anti-mouse CD35 (8C12, BD Biosciences) and a 1:100 dilution of goat anti-mouse IgD along with 1% NMS and NDS was incubated with the slides overnight at 4°C. The slides were washed in TBS and stained with alkaline phosphatase (AP)-conjugated streptavidin and HRP-conjugated donkey anti-goat IgG (1:100 dilution, Jackson Immunoresearch) for 2 hr at RT. Slides were washed and sequentially developed using Sigma DAB and Fast Blue (Sigma).
Image quantification
Immunofluorescence images were imported into IMARIS software (ver. 7.4.2). Using the Spots function, single B-cell follicles were chosen as the region of interest (ROI) and GFP+ cells within these follicles were automatically labeled by the software. The center of the follicle was marked using the Measurement Points function. The distance of each labeled cell from the measurement point at the center of the follicle was calculated, and used to determine the average distance of GFP+ cells from the center of the follicle. 3–4 similarly-sized follicles from each biological replicate were chosen randomly and quantified for each condition tested.
Quantitative RT-PCR
Total RNA from tissues or sorted cells was extracted using an RNeasy kit (Qiagen) and reverse-transcribed. Tonsil and spleen stroma was prepared by gently mashing the tissue in a 70 micron cell strainer and using saline to wash away the lymphocytes. The tissue aggregates remaining in the strainer are enriched in stromal cells and were extracted for qPCR analysis. Quantitative PCR was performed using Power SYBR Green with an Applied Biosystems StepOnePlus instrument. Data were analyzed using the comparative CT (2-ΔΔCt) method using the housekeeping genes indicated in the figures.
Statistical Analysis
Prism software (GraphPad, ver. 7.0e) was used for all statistical analyses. The statistical tests used are specified in the figure legends. Two-tailed unpaired t-tests were performed when comparing only two groups, and ordinary one-way analysis of variance (ANOVA) using Bonferroni’s multiple comparisons test was performed when comparing one variable across multiple groups. P values less than 0.05 were considered significant. In summary graphs, points indicate individual samples, and horizontal lines indicate means. In bar graphs, bars indicate means, and error bars indicate standard deviation.
Data availability
The data that support the findings of this study are available from the authors upon reasonable request. Source data for experiments involving animal models or tonsil specimens is provided with the paper.
Extended Data
Extended Data Figure 1. Dependence of P2RY8 bioactivity production on albumin and the isoprenoid biosynthetic pathway.
(a) Serum-free media containing the indicated amounts of fatty acid-free BSA was placed on HEK293T cells for 16–18 hr. The supernatants from these cultures were combined with CXCL12 in migration media (1:5 dilution) and tested for P2RY8 bioactivity (n=5). (b) P2RY8 ligand bioassay on 50 kDa concentrate (molecules > 50 kDa) vs filtrate (molecules < 50 kDa) from serum-starved HEK293T supernatant (left) or raw mouse bile (right) (n=4). (c) Diagram of protein precipitation from pig bile using saturated ammonium sulfate (SAS) and methanol extraction of the SAS protein precipitate. Graph shows P2RY8 ligand bioassay of the SAS supernatant and methanol extracts from the protein precipitate, as indicated by arrows (n=4). (d) P2RY8 ligand bioassay of the two layers of a Folch extraction prepared by adding chloroform and water to the methanol extract of the SAS precipitate described in (c) (n=5). (e) P2RY8 ligand bioassay on supernatants from Hepa1-6 or HEK293T cells treated with the indicated inhibitors for 16hr (n=4, one-way ANOVA). (f) P2RY8 ligand bioassay on supernatants from HEK293T, HeLa, or B16 cells treated with 10 μM mevastatin or vehicle (DMSO) for 16hr (n=4, unpaired two-tailed t-test for the indicated comparisons). Data are pooled from 3 (a,b,c,d,e,f) independent experiments. Graphs depict mean with s.d. and each point represents a biological replicate.
Extended Data Figure 2. HPLC Fractionation of P2RY8 bioactivity from bile and Q1 MS candidate identification.
(a) Preparation of a concentrated bile extract from the Folch upper layer described in Extended Data Fig. 1d using acid precipitation, centrifugation, and C18 solid phase extraction. (b) HPLC chromatograms (blue) showing absorbance at 220 nm for each column used for fractionation. Columns were initially tested with a small amount of extract to determine the interval where bioactivity eluted. The bioactivity graphs (red) corresponding to 1 minute fractions are overlaid for the bioactive interval. (c) Full scan (Q1 MS) of purified fractions from the indicated conditions, in negative ion mode. Zoomed-in spectra between m/z 550–600 are shown directly below each Q1 scan. Data are representative of 2 (a,b) or 1 (c) independent experiments.
Extended Data Figure 3. High resolution MS and fragmentation analysis supports that the bioactive compound is a derivative of glutathione and geranylgeranyl.
(a) Positive ion mode LC-MS total ion chromatogram of purified bile bioactive fraction (red) overlaid with an adjacent non-bioactive fraction (black), on left. High-resolution MS spectra from time 1.79 of the active fraction, on right. (b) MS/MS fragmentation spectra of glutathione in positive ion mode (top left) and negative ion mode (top right), compared with MS/MS spectra of purified bile positive ion 580.3 (bottom left) and negative ion 578.3 (bottom right). CE, collision energy. (c) Positive ion mode MS/MS/MS fragmentation spectra of the 273.1 ion present in the MS/MS spectra of GG-PP (top) and purified bile ion 580.3 (bottom; zoom-in of spectra shown in (b)). (d) Positive ion mode LC-MS total ion chromatogram (left) and high-resolution MS spectra from time 1.79 of chemically synthesized Ggg (right). (e) Negative ion mode MS/MS spectra of the 578.3 ion from chemically synthesized Ggg. Compare to the MS/MS spectra for the 578.3 ion from purified bile in (b). Data are representative of 2 (b,c,e) or 1 (a,d) independent experiments.
Extended data Figure 4. Ggg specifically inhibits migration of P2RY8-expressing WEHI-231 cells.
(a) Representative flow cytometry plots of migration inhibition assays performed with 50 ng/mL CXCL12 and 100 nM Ggg on WEHI-231 cells transduced with empty vector-GFP, P2RY8-GFP, S1PR2-Thy1.1, or Gpr4-Thy1.1. (b) Transwell migration inhibition assay using 500 ng/mL CXCL13 and the indicated amounts of Ggg for P2RY8-GFP and empty vector-GFP transduced WEHI-231 cells (n=4). (c) Summarized data for S1PR2-Thy1.1, Gpr4-Thy1.1, and empty vector-GFP transduced WEHI-231 cells from assays of the type in (a) (S1PR2 and Gpr4 n = 3, empty vector n=4). Data are representative of 2 independent experiments in (a) and pooled from 2 independent experiments in (b, c). Graphs depict mean with s.d. and each point represents a biological replicate.
Extended Data Figure 5. P2RY8 expression and distribution in human tonsil.
(a) qPCR for expression of P2RY8 in the indicated subsets sorted from human tonsil, relative to PTPRC. (n=3) (b) Immunofluorescence for P2RY8 (green) and CD4 (red) in PFA-fixed human tonsil sections. Inset depicts P2RY8+ and P2RY8high expressing CD4+ cells within the GC and at the GC border. Scale bars, 50 μm. (c) Intracellular flow cytometry using the anti-P2RY8 antibody from (b), which binds the C-terminus of P2RY8, on empty vector-GFP or P2RY8-GFP transduced WEHI-231 (mouse) cells, compared to rabbit isotype control or no-primary antibody staining conditions. (d) Intracellular flow cytometry for P2RY8 in tonsil IgD+ CD38- follicular B cells, IgD- CD38+ GC B cells, CXCR5- CD4+ T cells, or CXCR5+ PD-1+ Tfh cells. (e) Intracellular flow cytometry for P2RY8 in Ly8 cells edited using CRISPR-Cas9 with a control non-targeting guide (red) or a guide targeting P2RY8 (black). (f) TIDE analysis of edited Ly8 cells showing editing efficiency around the expected cut site. (g) pAkt levels in DOHH2 cells transduced with either GNA13 or empty vector (EV), treated as in Figure 3a (n=5). (h) pAkt levels in P2RY8-expressing or control WEHI-231 cells, treated as indicated (n=9). Data are representative of or pooled from 3(a), 4(b) or 2 (d) tonsils, 4 (h), 2 (c,e,g) or 1 (f) experiments. Graphs depict mean with s.d. Points represent biological replicates. One-way ANOVA with Bonferroni’s multiple comparisons test (g,h).
Extended Data Figure 6. Expression of Ggt5 by human tonsil follicular dendritic cells and fragmentation pattern of S-geranylgeranyl-L-Cys-Gly.
(a) P2RY8 ligand bioassay on supernatants from HEK293T cells transfected with the indicated enzymes (n=4 biological replicates). (b) Positive ion mode MS/MS spectra of the m/z 451.3 metabolite from extracts of the type in Figure 4d, corresponding to S-geranylgeranyl-L-Cys-Gly. (c) Immunohistochemistry for GGT5 or CR2 (brown), in serial sections of human tonsil, counterstained with hematoxylin (blue). Scale bars, 200 μm. (d) Immunofluorescence for GGT5 (green), CR2 (red), and DAPI (blue) in tonsil sections. Serial sections were stained for P2RY8 (green) and DAPI (blue) to visualize the difference between FDC-extensions and GC B-cell membranes. The indicated regions in the upper panels (scale bars, 100 μm) are enlarged in the lower panels (scale bars, 25 μm). (e) qPCR for GGT5 in the indicated tissues and cells from human tonsil, relative to GAPDH. Points within each category represent individual tonsils (whole tonsil=4, tonsil stroma=4, bulk lymphocytes=4, FoB=2, GC B=3, CXCR5- CD4+ T=3, CXCR5int CD4+ T=2, Tfh=3). Data are representative of or pooled from 2 (a,b) independent experiments and data in (c, d) are representative of 4 tonsil specimens. Graphs depict mean with s.d.
Extended Data Figure 7. Ggt5 is expressed by mouse follicular dendritic cells.
(a) Violin plots from a single cell RNAseq dataset18 showing relative expression level of Ggt1, Ggt5, Ggt6, and Ggt7 in the indicated stromal cell subsets. (b) qPCR for expression of Ggt1, Ggt5, Ggt6, and Ggt7 in whole spleen tissue or spleen stroma, relative to Hprt. (n=3 biological replicates) (c, d) RNAscope detection of Ggt5 mRNA (red) counterstained with IgD (brown) in the indicated tissues in mice 8 days after immunization with SRBCs (c) or in lymph nodes from mice treated with LTβR-Fc and TNFR-Fc, or a control-IgG for 4 days (d). Serial sections are stained for Cr1 (blue) and IgD (brown). Scale bars, 100 μm. Each point in (b) corresponds to a biological replicate. Data are representative of 5 (c), 2 (d), or 1 (a) biological replicates per condition. Graphs depict mean with s.d. The violin plots in (a) were generated by a webtool (http://scorpio.ucsf.edu/shiny/LNSC/) which does not display the exact minima, maxima, centre, percentiles, or n for each group.
Extended Data Figure 8. Controls for transduced B-cell co-transfer experiments.
(a) Immunofluorescence images tracking positioning of adoptively transferred B cells overexpressing empty-vector-GFP (green), co-transferred with either empty-vector-Thy1.1 or Ggt5-Thy1.1 overexpressing B cells, in unimmunized (top) or SRBC-immunized (bottom) mice, relative to endogenous B cells (IgD, blue). (b) Quantitation of images of the type in (a) and in Figure 4h, measuring the average distance (A.U., arbitrary units) of GFP+ cells from the center of B-cell follicles using IMARIS software. Each point represents a B-cell follicle, and 3–4 similarly-sized follicles were chosen randomly from 3 mice per condition (n=10 follicles per condition). Graph depicts mean with s.d. One-way ANOVA with Bonferroni’s multiple comparisons test. (c) Immunofluorescence images tracking positioning of adoptively transferred B cells overexpressing Ggt5 or an empty vector control construct from immunized mice of the type in Figure 4h, by staining for Thy1.1 (red) relative to endogenous B cells (IgD, blue). Scale bars, 100 μm. Data are representative of 3 (a,c) biological replicates per condition.
Extended Data Figure 9. FACS gating strategy and purity.
(a) Flow cytometry plots showing gating scheme used to sort the indicated subsets from human tonsil, along with post-sort purity. (b) For each bioassay performed, representative experiments are graphed as % of input migration for both the transduced and un-transduced WEHI-231 subsets indicated. For Fig 1j, the C18 SPE concentrates exhibited inhibition of overall migration likely due to slight toxicity; however, P2RY8+ cells were more selectively inhibited compared to P2RY8- cells. The baseline migration across experiments differs based on the growth state of the WEHI-231 cells. Graphs depict mean with s.d.
Supplementary Material
Acknowledgements
We thank Elizabeth Bess and Peter Turnbaugh for providing access to their HPLC, Ziyang Zhang and Kevan Shokat for providing access to their chemistry equipment and QTof mass spec, Jillian Jespersen and Jody Baron for helping coordinate tonsil specimens, Louis Staudt for DLBCL cell lines, Neil Barclay for the OX56 hybridoma, Jinping An for assistance with mouse husbandry, and David Russell, Jeffrey McDonald, and Jiaxi Wu for helpful discussions. J.G.C is an investigator of the Howard Hughes Medical Institute. E.L. was supported by the UCSF Biomedical Sciences Graduate Program and NSF grant 1144247. This work was supported in part by NIH grant RO1 AI45073.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Lohr JG et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. U. S. A 109, 3879–3884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morin RD et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122, 1256–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forbes SA et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muppidi JR et al. Loss of signalling via Galpha13 in germinal centre B-cell-derived lymphoma. Nature 516, 254–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz R et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med 378, 1396–1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muppidi JR, Lu E & Cyster JG The G protein-coupled receptor P2RY8 and follicular dendritic cells promote germinal center confinement of B cells, whereas S1PR3 can contribute to their dissemination. J. Exp. Med 212, 2213–2222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieckhaus CM, Fernandez-Metzler CL, King R, Krolikowski PH & Baillie TA Negative ion tandem mass spectrometry for the detection of glutathione conjugates. Chem. Res. Toxicol 18, 630–638 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Wiemer AJ, Wiemer DF & Hohl RJ Geranylgeranyl diphosphate synthase: an emerging therapeutic target. Clin. Pharmacol. Ther 90, 804–812 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Xie C, Zhong D & Chen X A fragmentation-based method for the differentiation of glutathione conjugates by high-resolution mass spectrometry with electrospray ionization. Anal. Chim. Acta 788, 89–98 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Justus CR & Yang LV GPR4 decreases B16F10 melanoma cell spreading and regulates focal adhesion dynamics through the G13/Rho signaling pathway. Exp. Cell Res 334, 100–113 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Shimizu T Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol 49, 123–150 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Uhlen M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Green JA et al. The sphingosine 1-phosphate receptor S1P(2) maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat. Immunol 12, 672–680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heisterkamp N, Groffen J, Warburton D & Sneddon TP The human gamma-glutamyltransferase gene family. Hum. Genet 123, 321–332 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Carter BZ et al. Metabolism of leukotriene C4 in gamma-glutamyl transpeptidase-deficient mice. J. Biol. Chem 272, 12305–12310 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Hayes JD, Flanagan JU & Jowsey IR Glutathione transferases. Annu. Rev. Pharmacol. Toxicol 45, 51–88 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Green JA & Cyster JG S1PR2 links germinal center confinement and growth regulation. Immunol. Rev 247, 36–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodda LB et al. Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 48, 1014–1028 e1016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee M et al. Transcriptional programs of lymphoid tissue capillary and high endothelium reveal control mechanisms for lymphocyte homing. Nat. Immunol 15, 982–995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen CD & Cyster JG Follicular dendritic cell networks of primary follicles and germinal centers: phenotype and function. Semin. Immunol 20, 14–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kridel R et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 13, e1002197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proia RL & Hla T Emerging biology of sphingosine-1-phosphate: its role in pathogenesis and therapy. J. Clin. Invest 125, 1379–1387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brune V et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J. Exp. Med 205, 2251–2268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullighan CG et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat. Genet 41, 1243–1246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Zhang Y, Hou S & Zhao ZK Synthesis of isoprenoid chain-contained chemical probes for an investigation of molecular interactions by using quartz crystal microbalance. Tetrahedron Lett. 54, 6208–6210 (2013). [Google Scholar]
- 26.Niegowski D et al. Crystal structures of leukotriene C4 synthase in complex with product analogs: implications for the enzyme mechanism. J. Biol. Chem 289, 5199–5207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cyster JG, Shotton DM & Williams AF The dimensions of the T lymphocyte glycoprotein leukosialin and identification of linear protein epitopes that can be modified by glycosylation. EMBO J. 10, 893–902 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinkman EK, Chen T, Amendola M & van Steensel B Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the authors upon reasonable request. Source data for experiments involving animal models or tonsil specimens is provided with the paper.