Summary
Bacillus subtilis GLB191 (hereafter GLB191) is an efficient biological control agent against the biotrophic oomycete Plasmopara viticola, the causal agent of grapevine downy mildew. In this study, we show that GLB191 supernatant is also highly active against downy mildew and that the activity results from both direct effect against the pathogen and stimulation of the plant defences (induction of defence gene expression and callose production). High‐performance thin‐layer chromatography analysis revealed the presence of the cyclic lipopeptides fengycin and surfactin in the supernatant. Mutants affected in the production of fengycin and/or surfactin were thus obtained and allowed us to show that both surfactin and fengycin contribute to the double activity of GLB191 supernatant against downy mildew. Altogether, this study suggests that GLB191 supernatant could be used as a new biocontrol product against grapevine downy mildew.
Keywords: Bacillus subtilis, defence, downy mildew, fengycin, lipopeptide, Plasmopara viticola, surfactin
Introduction
Grapevine (Vitis vinifera cvs), a major fruit crop worldwide, is susceptible to many cryptogamic diseases such as downy mildew (Plasmopara viticola), powdery mildew (Erysiphe necator) and grey mould (Botrytis cinerea). All of these can cause severe economic losses in both wine and table grape production. To ensure the quantity and quality of the harvest, the use of chemical fungicides remains the most common protection strategy. However, there is an increasing societal demand for human healthiness and environmentally friendly crop disease management. Alternative/complementary strategies to chemicals, such as biological control, have therefore been developed (Compant and Mathieu, 2016).
Biocontrol products protect plants against pathogens via diverse mechanisms such as direct antagonistic effect and/or activation of plant defences (Landy et al., 1948; Perazzolli et al., 2011; Shafi et al., 2017). They have various origins, including more or less purified plant extracts (Krzyzaniak et al., 2018) as well as living non‐pathogenic microorganisms (i.e. biological control agents, BCA) and their metabolites (Abdalla et al., 2011; Harwood et al., 2018; Ongena and Jacques, 2008). Studies have shown members of the Bacillus genus can be used as a versatile weapon against plant pathogens (Jacobsen et al., 2004; McSpadden Gardener and Fravel, 2002; Pérez‐García et al., 2011; Shafi et al., 2017). Bacillus is ubiquitous and widely distributed in both water and terrestrial ecosystems and even in environments under extreme conditions due to its ability to form endospores, which provide it with resistance and enhance its viability in different environmental conditions (Harwood et al., 2018; Nicholson, 2002; Villarreal‐Delgado et al., 2018). This is a technological advantage for the dryness step required for formulation into stable products and long shelf‐life (Keswani et al., 2016; Schisler et al., 2004). As a biocontrol agent, Bacillus uses various modes of action against different plant pathogens such as antagonism, competition for niche space and nutrients, and induction of host resistance (Santoyo et al., 2012; Villarreal‐Delgado et al., 2018). Bacillus‐based BCAs therefore represent a large range of microbe products used for crop protection (Fravel, 2005; Shafi et al., 2017).
B. subtilis is one of the most commercialized BCAs. It produces various bioactive compounds against a broad spectrum of pathogens. Some of the most prominent bioactive compounds for plant protection are cyclic lipopeptides (CLPs) (Stein, 2005). B. subtilis CLPs include the surfactin, iturin and fengycin (or plipastatin) families and they have different activities (Falardeau et al., 2013; Stein, 2005). Members of the surfactin family are biosurfactant molecules with antiviral and antibacterial activities but no marked fungitoxicity (Falardeau et al., 2013). For example, surfactin plays a major role in suppressing bacterial fruit blotch, but does not seem to have a direct toxic effect on Botrytis (Fan et al., 2017b; Farace et al., 2015). Besides the direct antagonistic activity, the surfactin family also induces resistance on a diversity of hosts against various diseases by stimulating plant immune responses. For example, purified surfactin triggers defence responses in grapevine and tobacco cell suspensions (Farace et al., 2015; Jourdan et al., 2009) and induces systemic resistance (ISR) against B. cinerea in tomato plants (Cawoy et al., 2014). The fengycin family mostly shows antifungal activity (Deleu et al., 2008). Unlike surfactin, fengycin‐induced defences are specific to certain plant species or host–pathogen systems. For example, fengycin produced by B. subtilis BBG111 plays an indispensable role in the induced defence state in rice (Oryza sativa L.) against Rhizoctonia solani (Chandler et al., 2015). However, plipastatin (fengycin family) does not induce defence gene expression in grapevine cell suspension (Farace et al., 2015). The iturin family possesses strong antifungal activity, but limited antiviral and antibacterial activity (Falardeau et al., 2013). No evidence exists for iturin‐induced ISR (Falardeau et al., 2013) except a recent report revealing that purified mycosubtillin (a member of the iturin family) activates defence responses in grapevine cell suspension and long‐lasting tolerance to B. cinerea in leaves (Farace et al., 2015).
According to the literature, different B. subtilis strains produce different types of CLPs. For example, B. subtilis BBG111 synthesizes surfactin and fengycin while B. subtilis RFB104 produces surfactin and mycosubtilin (Chandler et al., 2015). However, B. subtilis 916 produces surfactin, bacillomycin (iturin family), fengycin and a novel family of CLPs called locillomycin (Luo et al., 2015). Therefore, different strains have different activities even against the same pathogen. For example, B. subtilis BBG111 but not B. subtilis RFB104 protects rice against R. solani (Chandler et al., 2015). Moreover, the activity of the same strain can differ according to the plant/pathogen interaction considered. For example, B. subtilis BBG111 is capable of protecting rice against the necrotrophic R. solani, while it is ineffective against the hemibiotropic M. oryzae (Chandler et al., 2015). It is therefore necessary to unravel the modes of action in each pathosystem and identify the active compounds for each strain, especially for those which have the potential to be used commercially.
For grapevine, some B. subtilis strains are used for protection against grey mould and are currently available on the market (B. subtilis strain QST 713, Serenade®) (Rotolo et al., 2018). However, few have been developed for downy mildew. Recently, we found that the B. subtilis strain GLB191 (hereafter GLB191), isolated from grapevine leaves, is an efficient BCA against downy mildew in both controlled and field conditions (Zhang et al., 2017). However, the mechanisms of B. subtilis activity against downy mildew are not clear. The deciphering of these mechanisms and further identification of the active metabolites is crucial. In the present study, we investigated the mode(s) of action of this new BCA against downy mildew with a focus on the putative role played by CLPs. Supernatants of GLB191 and mutant strains affected in CLPs synthesis were therefore produced and used. We found that GLB191 has direct effect against the pathogen and stimulates the plant defences, which results from both fengycin and surfactin secreted in the supernatant. This is the first report showing that CLPs produced by B. subtilis have double effects in the biotrophic pathogen P. viticola/grapevine pathosystem.
Results
B. subtilis GLB191 supernatant protects grapevine plants against downy mildew
Foliar treatment with the supernatant of the B. subtilis strain GLB191 (hereafter called GLB191‐S, with S for supernatant) reduced the leaf sporulating area by 97.6%, compared to water treatment (Fig. 1). Non‐inoculated potato dextrose broth (PDB) culture medium also significantly reduced the pathogen sporulation (by 33.5% compared to the water control). However, the protective activity of GLB191‐S was markedly higher than that of the PDB medium (Fig. 1). The supernatant of GLB191 therefore plays a major role in the protection of grapevine against downy mildew.
Figure 1.
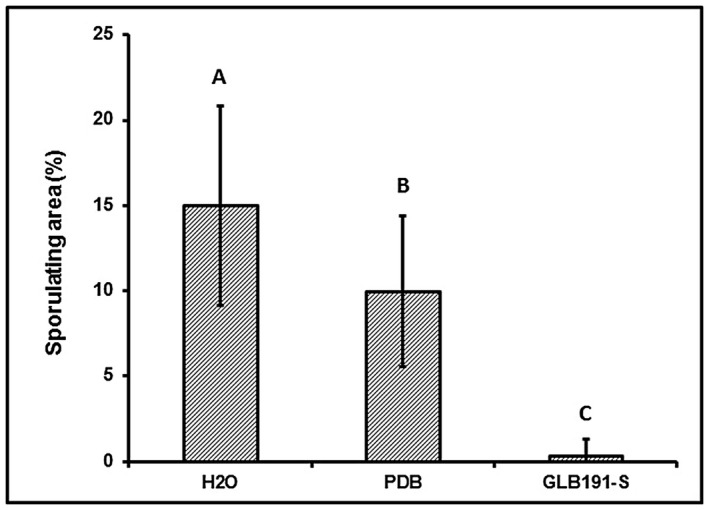
B. subtilis GLB191 supernatant‐induced protection of grapevine leaves against downy mildew. Sporulation of Plasmopara viticola was assessed with the downy mildew susceptible cultivar cv. Marselan treated with water (H2O), PDB medium (PDB) or the supernatant of B. subtilis GLB191 (GLB191‐S). Values correspond to the mean percentage obtained for 36 discs of six leaves from three plants (the second and third apical leaves from each plant). The data are representative of three independent experiments. Treatments were compared by means with the non‐parametric Kruskal–Wallis approach at the 5% significance level. Means with different letters are significantly different at P < 0.01 according to the Mann–Whitney pairwise post hoc test with application of the Bonferroni correction.
Direct effect of GLB191‐S on P. viticola zoospores
In order to further unravel the mode(s) of action of GLB191 to control downy mildew, the direct activity of GLB191‐S against P. viticola was first determined. The number of infection sites (i.e. stomata with encysted zoospores of P. viticola) was determined at 24 h post inoculation (hpi) by UV epifluorescence observations after aniline blue staining of the pathogen. The highest number of infection sites (9.1 ± 2.6 per observation field) was observed for the water control (Fig. 2). PDB treatment significantly reduced their number (2.1 ± 1.5), whereas almost none could be observed on leaves treated with GLB191‐S (Fig. 2). These results indicate that GLB191‐S has a significant direct effect on zoospores.
Figure 2.
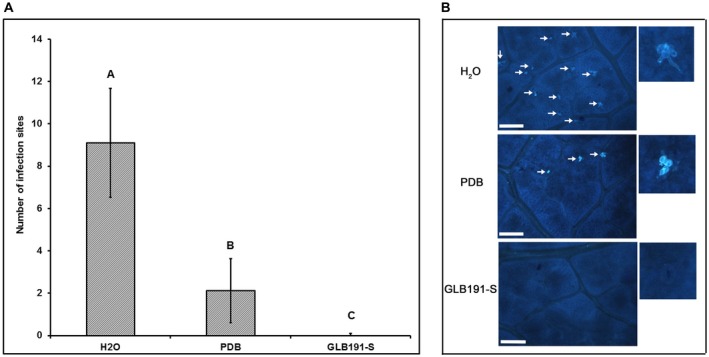
Direct effect of B. subtilis GLB191 supernatant on P. viticola zoospores. Grapevine (V. vinifera cv. Marselan) leaves were treated with water (H2O), PDB medium (PDB) or the supernatant of B. subtilis GLB191 (GLB191‐S) and inoculated with P. viticola sporangia 2 h later. At 24 h post inoculation, leaf discs were punched out from leaves and the number of infection sites (i.e. stomata with encysted zoospores of P. viticola) was determined by epifluorescence observations after aniline blue staining of the pathogen. Panel A: Values are the mean number of infection sites for ten discs of six leaves harvested from three plants (the second and third apical leaves from each plant). The data correspond to a representative of three independent experiments. Treatments were compared by means with the non‐parametric Kruskal–Wallis approach at the 5% significance level. Means with different letters are significantly different according to the Mann–Whitney pairwise post hoc test with application of the Bonferroni correction (P < 0.01). Panel B: Representative photographs of fluorescence microscopy observations. Arrows indicate the infection sites (encysted zoospores). Scale bars represent 100 μm.
GLB191‐S activates callose production and defence genes in grapevine leaves
Callose production was monitored by UV epifluorescence after aniline blue staining. The data revealed that the fluorescence of the spots was scarce for water‐treated leaves and slightly higher (2.7 ± 1.8) for PDB‐treated ones (Fig. 3A). In contrast, the number of spots was markedly increased after treatment by GLB191‐S (14.3 ± 7.4 per observation field) (Fig. 3A), indicating a strong callose production. Callose was localized at the level of stomata (Fig. 3B).
Figure 3.
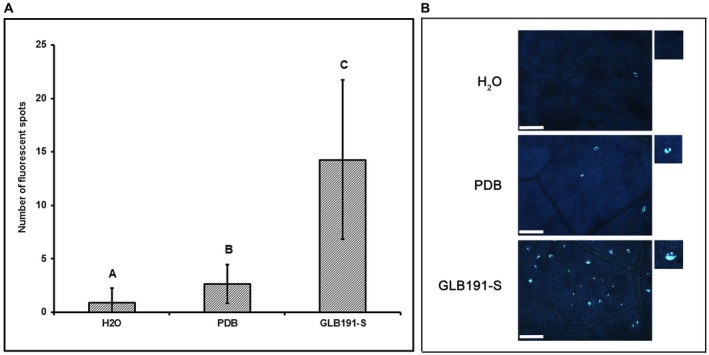
Callose production induced by B. subtilis GLB191 supernatant. Grapevine (V. vinifera cv. Marselan) leaves were treated by water (H2O), PDB medium (PDB) or the supernatant of B. subtilis GLB191 (GLB191‐S). Callose production (fluorescent spots) was assessed 3 days post treatment by epifluorescence observations after aniline blue staining. Panel A: Values are the mean number of fluorescent spots for six leaves from three plants (the second and third apical leaves from each plant). Treatments were compared by means with the non‐parametric Kruskal–Wallis approach at the 5% significance level. Means with different letters are significantly different according to the Mann–Whitney pairwise post hoc test with application of the Bonferroni correction (P < 0.01). The data is a representative of three independent experiments. Panel B: Representative photographs of fluorescence microscopy observations. Scale bars represent 100 μm.
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was next used to analyse the expression of a set of defence genes known to be involved in different defence pathways in grapevine: PAL (coding for phenylalanine ammonia lyase, a key enzyme of the phenylpropanoid pathway), STS (encoding stilbene synthase, downstream of PAL and responsible for the synthesis of resveratrol, the main phytoalexin produced by grapevine in response to biotic or abiotic stresses), PR1 (encoding the pathogenesis‐related (PR) protein 1 that is considered as a marker of the salicylic acid signalling pathway), PR2 (encoding the PR protein 2 β‐1,3‐glucanase), PR3 (encoding the PR protein 3 chitinase 4c), Lox9 (encoding the lipoxygenase 9, an enzyme involved in jasmonic acid signalling) and JAZ1 (encoding the jasmonate ZIM‐domain protein 1, another marker of the jasmonic acid signalling pathway). In response to GLB191‐S treatment, the expression of all genes tested was induced compared to the water control, except for Lox9, with difference of intensity (Fig. 4). STS was the most up‐regulated gene and JAZ1 the lowest one, with around 80‐ and 4‐fold relative expression to water, respectively. PDB medium also induced the expression of some defence genes, especially PR1, STS and PR3. However, the transcript accumulation of these defence genes was always lower in response to PDB treatment than that found in response to GLB191‐S. Moreover, results showed that PR2, STS and PR3 were the most up‐regulated genes by GLB191‐S with 15.4‐, 8.8‐ and 6.3‐fold relative to PDB, respectively.
Figure 4.
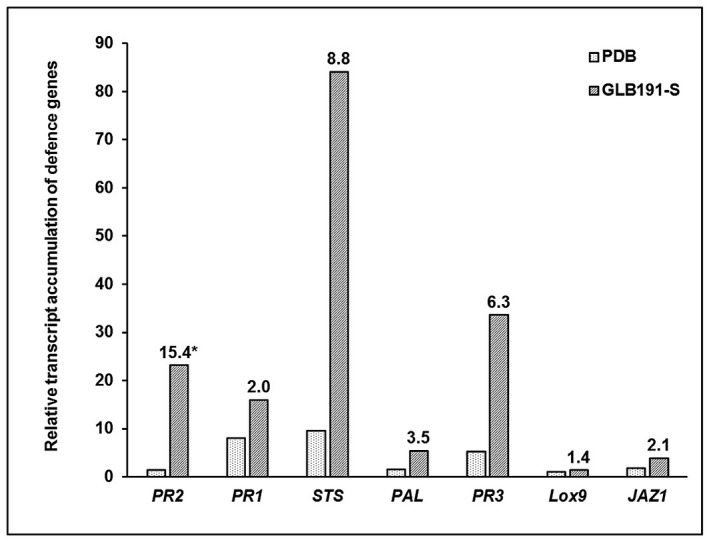
Defence‐related gene expression in grapevine leaves treated with B. subtilis GLB191 supernatant. The relative transcript accumulation of defence genes was determined by qRT‐PCR in grapevine (V. vinifera cv. Marselan) leaves 24 h post treatment with water (H2O), PDB medium (PDB) or the supernatant of B. subtilis GLB191 (GLB191‐S). Results represent relative fold expression calculated with the 2−ΔΔCt method, compared to the reference gene EF1γ and to the water control. The data are representative of three independent experiments. PR2, PR protein 2 β‐1,3‐glucanase; PR1, PR protein 1; STS, stilbene synthase; PAL, phenylalanine ammonia lyase; PR3, PR protein 3 chitinase 4c; Lox9, lipoxygenase 9; JAZ1, jasmonate ZIM‐domain protein. *The values above each column indicate the relative fold of transcript accumulation in leaves treated with GLB191‐S compared to that of PDB‐treated ones.
Altogether these results demonstrate that GLB191‐S stimulates various grapevine defence responses.
Both surfactin and fengycin contribute to GLB191 activity against downy mildew
In order to investigate whether CLPs contribute to the protection of GLB191‐S, deletion mutants affected in the synthesis of fengycin (∆ppsB), surfactin (∆srfAA) or both (∆ppsBsrfAA) were generated. First, the CLP concentration in the supernatant of GLB191 and its mutants affected in CLP production was determined by high‐performance thin‐layer chromatography (HPTLC) analysis. As expected, GLB191 produces fengycin and surfactin (56.2 and 52.1 mg/L, respectively) (Table 1). Deletion of srfAA resulted in no surfactin production and a level of fengycin similar to those of GLB191 (Table 1). Deletion of ppsB resulted in no fengycin production as expected but in a reduced surfactin production (22.5 mg/L) (Table 1). None of the analysed CLPs could be detected in the supernatant of the double mutant and no iturin A was detected in all strain supernatants tested (Table 1).
Table 1.
Production of cyclic lipopeptides (CLPs) by B. subtilis GLB191 and its derived mutants grown in PDB medium.*
| Strain | ||||
|---|---|---|---|---|
| CLPs | GLB191 | ΔppsB | ΔsrfAA | ΔppsBsrfAA |
| Surfactin | 52.1 ± 8.2† | 22.5 ± 12.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Fengycin | 56.2 ± 3.8 | 0.0 ± 0.0 | 54.6 ± 3.1 | 0.0 ± 0.0 |
| Iturin A | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
The values are means from three experiments.
Mean ± SD, mg/L.
We then investigated the protection efficacy of the supernatants of GLB191 mutants against downy mildew. Foliar treatment of Marselan with the supernatant of the single mutant ΔppsB (ΔppsB‐S) or ΔsrfAA (ΔsrfAA‐S) resulted in a significant increase of P. viticola sporulation compared to GLB191‐S (Fig. 5). Treatment with the supernatant of the double mutant ΔppsBsrfAA (ΔppsBsrfAA‐S) showed a significantly higher sporulation of P. viticola compared to the single mutants. It did not induce protection, as indicated by a level of sporulation similar to that of the water control (Fig. 5). A lower level of sporulation was observed in response to PDB treatment compared to the water control (as in Fig. 1) and with ΔppsBsrfAA‐S treatment.
Figure 5.
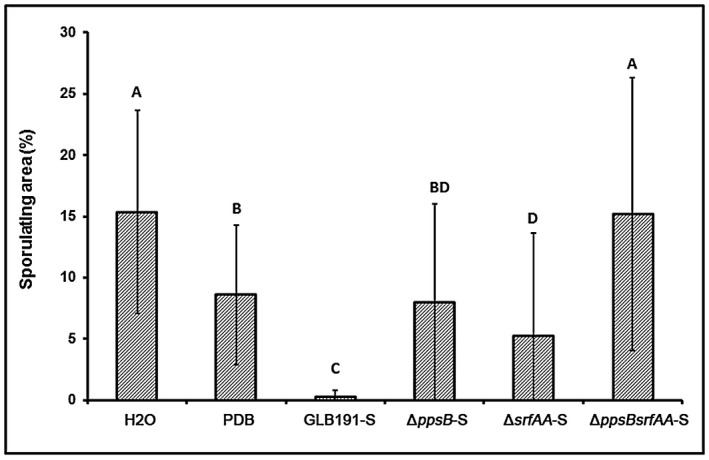
Protection of grapevine leaves against downy mildew induced by B. subtilis GLB191 and its derived mutants. Sporulation of Plasmopara viticola was assessed with the downy mildew susceptible cultivar cv. Marselan treated with water (H2O), PDB medium (PDB) or the supernatant of the wild‐type B. subtilis GLB191 (GLB191‐S) and its mutants ΔppsB (ΔppsB‐S), ΔsrfAA (ΔsrfAA‐S), ΔppsBsrfAA (ΔppsBsrfAA‐S). Values correspond to the mean percentage obtained for 36 discs of six leaves from three plants (the second and third apical leaves from each plant). The data are the mean of three independent experiments. Treatments were compared by means with the non‐parametric Kruskal–Wallis approach at the 5% significance level. Means with different letters are significantly different according to the Mann–Whitney pairwise post hoc test with application of the Bonferroni correction (P < 0.01).
Both surfactin and fengycin contribute to the direct effect of GLB191 on P. viticola zoospores
The supernatants of the different strains were next compared regarding their effects against P. viticola infection. Infection sites on leaves treated with GLB191‐S and ΔsrfAA‐S were scarce (Fig. 6). They seemed slightly more abundant in leaves treated with ΔppsB‐S but no statistically significant difference was found compared to GLB191‐S and ΔsrfAA‐S. Deletion of both ppsB and srfAA (ΔppsBsrfAA‐S) resulted in a significantly higher number of infection sites compared to the wild type and a similar level to the PDB medium (Fig. 6). These results indicate that both surfactin and fengycin contribute to the direct effect of GLB191‐S on P. viticola zoospores.
Figure 6.
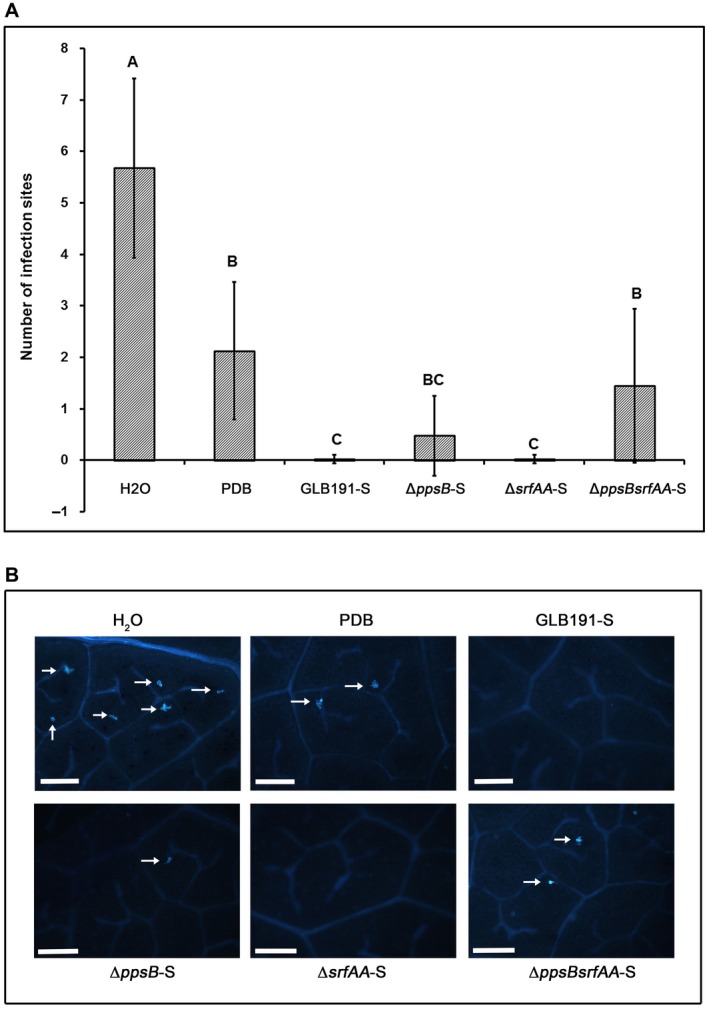
Direct effect of the supernatants of B. subtilis GLB191 and its derived mutants on P. viticola zoospores. Grapevine (V. vinifera cv. Marselan) leaves were treated with water (H2O), PDB medium (PDB) or the supernatant of B. subtilis GLB191 (GLB191‐S) and its mutants ΔppsB (ΔppsB‐S), ΔsrfAA (ΔsrfAA‐S), ΔppsBsrfAA (ΔppsBsrfAA‐S), and inoculated with P. viticola sporangia 2 h later. At 24 hpi, leaf discs were punched out from leaves and the number of infection sites (i.e. stomata with encysted zoospores of P. viticola) was determined by UV epifluorescence observations after aniline blue staining of the pathogen. Panel A: Values are the mean number of infection sites for ten discs of six leaves harvested from three cuttings (the second and third apical leaves from each plant). The data correspond to a representative of three independent experiments. Treatments were compared by means with the non‐parametric Kruskal–Wallis approach at the 5% significance level. Means with different letters are significantly different according to the Mann–Whitney pairwise post hoc test with application of the Bonferroni correction (P < 0.05). Panel B: Representative photographs of fluorescence microscopy observations. Arrows indicate the infection sites (encysted zoospores). Scale bars represent 100 μm.
Both surfactin and fengycin contribute to the stimulation of grapevine defences
We further investigated whether the supernatants of GLB191 mutants were still efficient to induce grapevine defences. The data showed that callose production in ΔppsB‐S‐ or ΔsrfAA‐S‐treated plants was significantly reduced compared to that in GLB191‐S‐treated plants but significantly higher than that of the PDB and water control (Fig. 7). Moreover, callose production in ΔppsBsrfAA‐S‐treated leaves was similar to that of ΔppsB‐S‐treated ones and significantly higher than that of the PDB control (Fig. 7). These results indicate that both surfactin and fengycin are main factors in the supernatant of GLB191 causing callose production in grapevine leaves.
Figure 7.
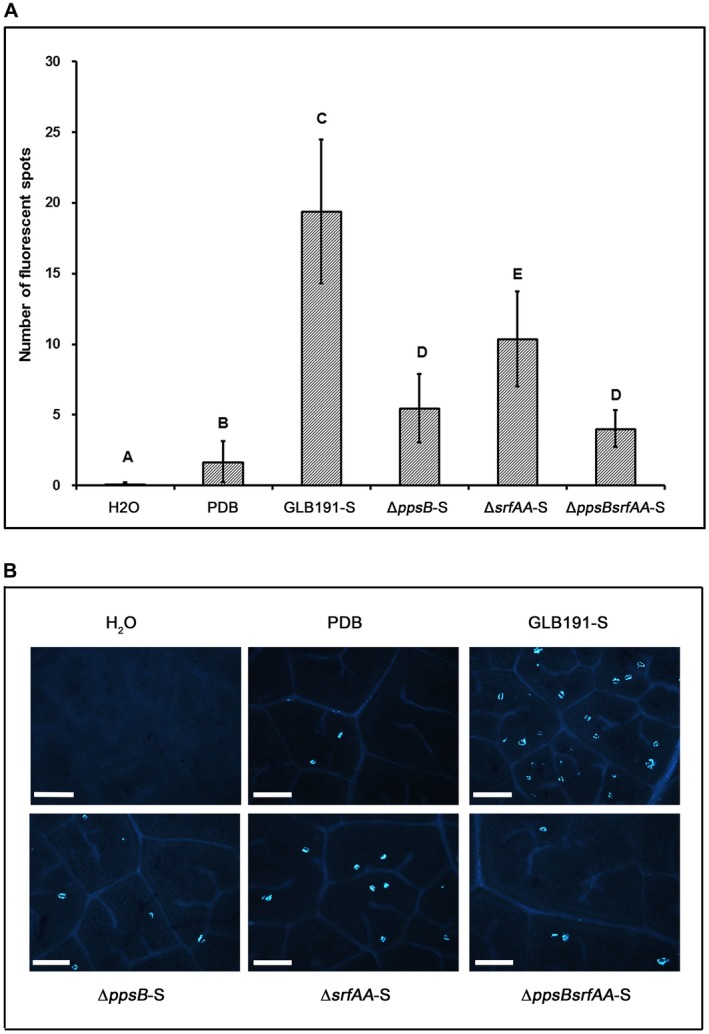
Callose production induced by the supernatant of B. subtilis GLB191 and its derived mutants. Callose deposition in grapevine (V. vinifera cv. Marselan) leaves treated by water (H2O), PDB medium (PDB) or the supernatant of the wild‐type B. subtilis GLB191 (GLB191‐S) and its mutants ΔppsB (ΔppsB‐S), ΔsrfAA (ΔsrfAA‐S), ΔppsBsrfAA (ΔppsBsrfAA‐S) was assessed 3 days post treatment by epifluorescence observations after aniline blue staining. Panel A: Values are the mean estimation for leaves from three plants (the second and third apical leaves from each plant) in the same treatment. Treatments were compared by means with the non‐parametric Kruskal–Wallis approach at the 5% significance level. Means with different letters are significantly different according to the Mann–Whitney pairwise post hoc test with application of the Bonferroni correction (P < 0.05). The data are representative of three independent experiments. Panel B: Representative photographs observed using a fluorescence microscope. Scale bars represent 100 μm.
For defence gene expression, we focused on PR2, PR3 and STS as they were highly induced by GLB191‐S. The qRT‐PCR data revealed that deletion of both ppsB and srfAA (ΔppsBsrfAA‐S) resulted in a stronger reduction of transcript accumulation of these defence genes than that observed for single mutants ppsB or srfAA (ΔppsB‐S or ΔsrfAA‐S) compared to the wild type (GLB191‐S) (Fig. 8). These results indicate again that both surfactin and fengycin contribute to the induction of defence gene expression after GLB191‐S treatment.
Figure 8.
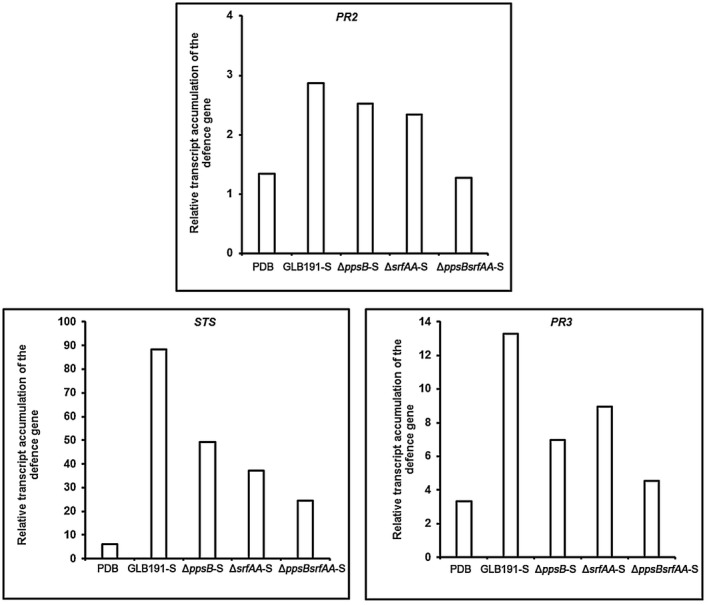
Defence‐related genes expression in grapevine leaves treated with supernatant of B. subtilis GLB191 and its derived mutants. The relative transcript accumulations of genes PR2 (encoding the pathogenesis‐related protein 2 β‐1,3‐glucanase), STS (encoding stilbene synthase) and PR3 (encoding the pathogenesis‐related protein 3 chitinase 4c) were determined by qRT‐PCR in leaves treated with water, PDB medium (PDB) or the supernatant of B. subtilis GLB191 (GLB191‐S) and its mutants ΔppsB (ΔppsB‐S), ΔsrfAA (ΔsrfAA‐S), ΔppsBsrfAA (ΔppsBsrfAA‐S). Results represent relative fold expression calculated with the 2−ΔΔCt method, compared to the reference gene EF1γ and to the water control. The data are representative of three independent experiments.
Discussion
We have previously observed that the endophytic bacterial strain B. subtilis GLB191 isolated from grapevine leaves has a strong preventive activity against P. viticola on the susceptible cultivars V. vinifera L. cv. Muscat Hamburg (a table grape cultivar) and cv. Cabernet Sauvignon (a wine grape cultivar) in both leaf disk assays and the field. Assays were performed with the culture broth containing both bacterial cells and their metabolites (Zhang et al., 2017). As B. subtilis is known to produce biologically active molecules, especially CLPs (Ongena and Jacques, 2008; Stein, 2005), experiments were performed in order to determine if these molecules were involved in GLB191 activity against P. viticola.
First, we demonstrated that the supernatant of GLB191, which is likely to contain CLPs, was also active against downy mildew. Preliminary experiments were performed with Luria‐Bertani (LB) and Landy media generally used for B. subtilis cultivation as they favour the production of CLPs (Fan et al., 2017a; Landy et al., 1948; Luo et al., 2015). However, their protective effect on P. viticola was stronger compared to PDB medium (data not shown). We therefore decided to use PDB as the culture medium for this study to reduce the effect of the medium itself.
The modes of action of GLB191‐S were next investigated. B. subtilis has been reported to protect plants against various phytopathogens by stimulating the plant host defences. Moreover, some CLPs are considered to be a new class of microbe‐associated molecular patterns (MAMPs) (Ongena and Jacques, 2008), i.e. as elicitors of plant immunity (Boutrot and Zipfel, 2017). GLB191‐S was therefore tested as putative defence inducer. In the present study, we showed that GLB191‐S induces callose production (Fig. 3A). In grapevine, this defence event was previously reported in response to several defence elicitors such as sulphated laminarin (PS3) or β‐aminobutyric acid (BABA), and was clearly associated with induced resistance against P. viticola (Hamiduzzaman et al., 2005; Trouvelot et al., 2008; Yu et al., 2016). Callose production is also involved in the natural resistance of grapevine species and hybrids to P. viticola. For example, Gindro et al., (2003) have reported callose production in leaf stomata of the downy mildew tolerant hybrid Solaris after challenge with P. viticola. Similarly, callose was localized at the level of stomata of GLB191‐S‐treated leaves (Fig. 3B), suggesting a role in the prevention of infection (Gindro et al., 2003; Trouvelot et al., 2008). GLB191‐S also induced the expression of several defence genes (Fig. 4), suggesting that different defence responses are activated. However, markers of SA‐ and JA‐signalling pathways were weakly induced (Fig. 4), suggesting a local induced resistance rather than an ISR. However, this hypothesis would need to be confirmed by further experiments.
GLB191‐S therefore induces grapevine resistance against downy mildew by both direct effect against the pathogen (we have shown a reduction in the number of infection sites) and plant defence stimulation. Since CLPs of B. subtilis are particularly important in plant protection (Stein, 2005), we next investigated whether they contribute to the activity of GLB191‐S. Deletion mutants affected in the synthesis of fengycin (∆ppsB), surfactin (∆srfAA) or both (∆ppsBsrfAA) were generated. Although iturin is known to display strong antifungal activity (Maget‐Dana and Peypoux, 1994), a search of iturin‐related genes in the GLB191 genome surprisingly yielded zero results. Thus, a mutant for iturin was not constructed. The CLP concentration in the supernatant of GLB191 and its mutants was determined. The absence of iturin A (Table 1) is consistent with the genome search that could not allowed us to identify an iturin‐related gene cluster. Deletion of ppsB resulted in no fengycin production and a reduced surfactin production (Table 1), which is contrary to previous reports (Coutte et al., 2010; Luo et al., 2015). In B. subtilis 916, deletion of fenA, which is involved in fengycin synthesis, resulted in increased surfactin production (Luo et al., 2015). Coutte et al. (2010) also observed an enhanced surfactin production after disruption of the plipastatin operon in B. subtilis 168 derivative BBG113, indicating interdependency of surfactin and plipastatin biosynthesis. This phenomenon could not be observed in the ppsB mutant of GLB191. The lower production of surfactin in ∆ppsB revealed in this study is surprising and would merit further investigation. It might be due to a specific crosstalk in the regulation of lipopeptide production in the GLB191 strain.
Using mutants, we observed that the protective activity against downy mildew was reduced in the absence of either surfactin or fengycin and it was lost in the absence of both of them (Fig. 5), indicating that both fengycin and surfactin are the main active compounds of GLB191‐S against P. viticola. Farace et al. (2015) observed that only surfactin and mycosubtilin (iturin family), but not plipastatin (i.e. fengycin), enhanced tolerance of grapevine leaves against the necrotrophic fungus B. cinerea. It therefore seems that the relative importance of these three CLPs families to induce resistance in grapevine could be pathogen dependent. The lower level of sporulation observed for the PDB medium treatment compared to the water control and ΔppsBsrfAA (Fig. 5) indicates that some components in the PDB medium have a protective activity against P. viticola and could probably be used by GLB191 bacteria for growth.
Surfactin is known to have a strong direct antibacterial activity and stimulate plant defences whereas fengycin has a strong direct antifungal effect and stimulates defence in certain plant species or specific host–pathogen systems (Ongena and Jacques, 2008). Therefore, reduction of P. viticola sporulation may be due to both direct antimicrobial effect and/or induced resistance resulting from surfactin and/or fengycin.
The direct antimicrobial effect of CLPs on phytopathogens is due to the interaction with the plasma membrane of the pathogen. CLPs can cause pores to form on the membrane, causing an osmotic imbalance that ultimately results in cell death (Pérez‐García et al., 2011). According to the literature, surfactin shows antiviral and antibacterial activity whereas fengycin has strong activity against fungi, especially filamentous ones (Vanittanakom et al., 1986). The differential activities of CLP families are linked to their respective mechanisms of action on phytopathogen biological members (Falardeau et al., 2013). Moreover, the antimicrobial activity of surfactin and iturin is dose‐dependent whereas it is all or none for fengycin (Falardeau et al., 2013). In our study, ΔppsB‐S, which produces surfactin but no fengycin (Table 1), and ΔsrfAA, which produces fengycin but no surfactin (Table 1), showed direct effect against P. viticola infection (Fig. 6), suggesting that both fengycin and surfactin have effect against this oomycete. There was no statistically significant difference observed in the number of infection sites on leaves treated with GLB191‐S and the single mutant ΔppsB‐S or ΔsrfAA‐S. The slightly higher and more variable number of infection sites in response to ΔppsB‐S might be due to the half concentration of surfactin present in this supermatant (Table 1). The double mutant ΔppsBsrfAA, which produces no fengycin and surfactin (Table 1), lost the inhibition of P. viticola zoospores (Fig. 6). These results suggest that the concentration of surfactin in ΔppsB‐S or fengycin in ΔsrfAA‐S is enough to inhibit zoospores of P. viticola.
The analysis of callose production and defence gene expression showed that surfactin and fengycin are the main factors in the supernatant of GLB191 responsible for stimulation of plant defences. However, it seems that the different defence genes are not regulated in a similar manner by the two CLPs. Previous studies have reported the role of surfactin in the activation of plant defences and suggested that it could result from its insertion into the plant plasma membrane (Henry et al., 2011). Using cell suspensions, Farace et al. (2015) have shown that purified surfactin, mycosubtillin and plipastatin differently activate defence responses in grapevine. In their conditions, surfactin and mycosubtilin stimulated grapevine innate immune responses whereas plipastatin perception only resulted in early signalling activation. Mycosubtilin activated the strongest gene expression, but it was associated with cell death.
Altogether, this study demonstrates that both surfactin and fengycin in the supernatant contribute to the protection of a natural strain GLB191 against downy mildew which resulted from both their direct anti‐oomycete activity against P. viticola and stimulation of the plant defences. However, different Bacillus strains are known to produce variable concentrations of each CLP in natural conditions when they interact with the plant at different times for ecological fitness (Debois et al., 2015). Further research is needed to determine which CLP is involved at a given time and the concentrations of each CLP when GLB191 is sprayed on the leaves of grapevine.
Experimental procedures
Plant material
The grapevine cultivar Vitis vinifera cv. Marselan (Cabernet sauvignon × Grenache), susceptible to P. viticola, was used in this study. Plants were grown in a glasshouse as described previously (Krzyzaniak et al., 2018). In brief, plants were produced from herbaceous cuttings planted in individual pots at 23 and 18 °C (day and night, respectively) with a photoperiod of 16 h of light until they developed six to eight leaves. Plants were watered daily and fertilized once a week (N/P/K 10‐10‐10, Plantin, Courthezon, France). The second and third youngest fully expanded leaves were used for experiments.
Bacterial strains and cultural conditions
The bacterial strains and plasmids used in this study are listed in Table 2. B. subtilis strains were stored at −80 °C in 15% glycerol. For experiments, they were grown in Potato Dextrose Broth (PDB) (Conda SA, Madrid, Spain) medium at 37 °C. E. coli strains were grown in LB at 37 °C. Antibiotics were added at the following concentrations when required: ampicillin (Ap, 100 μg/mL), erythromycin (Em, 5 μg/mL).
Table 2.
Strains and plasmids used in this study.
| Strains or plasmids | Characteristicsa | Sources or references |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| GLB191 | Wild‐type strain, isolated from grapevine leaves | Zhang et al. (2017) |
| ΔppsB | ppsB deletion mutant of GLB191, markerless | This work |
| ΔsrfAA | srfAA deletion mutant of GLB191, markerless | This work |
| ΔppsBsrfAA | ppsB and srfAA double mutant of GLB191, markerless | This work |
| E. coli | ||
| DH5α | F‐φ80 lac ZΔM15 Δ(lacZYA‐arg F) U169 endA1 recA1 hsdR17(rk‐,mk+) supE44λ‐thi ‐1 gyrA96 relA1 phoA | Life Technologies |
| EC135 | EC132 Δdam::FRT, genotype of R‐M systems: mcrA Δ(mrr‐hsdRMS‐mcrBC) Δdcm::FRT Δdam::FRT | Zhang et al. (2012) |
| Plasmids | ||
| pMAD | Shuttle vector for allele replacement; AmpR (E. coli), EmR (Bacillus); containing bgaB gene encoding a thermostable β‐galactosidase | Arnaud et al. (2004) |
| pMAD‐ppsB | A fusion of upstream and downstream of ppsB cloned into pMAD for allele replacement; AmpR (E. coli), EmR (Bacillus) | This work |
| pMAD‐srfAA | A fusion of upstream and downstream of srfAA cloned into pMAD for allele replacement; AmpR (E. coli), EmR (Bacillus) | This work |
Mutant construction
Markerless deletion mutants were constructed using the temperature‐sensitive suicide plasmid pMAD as described previously (Fan et al., 2017a). Upstream and downstream regions of the target gene (named X hereafter) were amplified from the genomic DNA of GLB191 using the primer pairs X‐up‐F/X‐up‐R and X‐dn‐F/X‐dn‐R, respectively. The two DNA fragments were joined together by PCR amplification using the primers X‐up‐F and X‐dn‐R. The resulting fragment was digested and cloned into pMAD, generating pMAD‐X in E. coli DH5α. The pMAD‐X plasmid was purified from E. coli DH5α and mobilized into E.coli EC135 by heat shock and then into GLB191 by electroporation. Erythromycin resistant (EmR) and blue transformants were obtained after incubation at 30 °C for 2 days on LB plates containing Em and X‐Gal (40 μg/mL), followed by incubation in LB broth containing Em at 42 °C with shaking at 180 r min−1 for 8–10 h for the first allelic exchange. EmR and blue transformants were obtained from LB plates supplemented with Em and X‐Gal and then incubated in LB broth at 25 °C with shaking at 180 r min−1 for 24 h for the second allelic exchange. Em‐sensitive and white clones were isolated and confirmed by PCR with primers X‐Up‐F and X‐Dn‐R and subsequently by sequencing. The primers used are listed in Table S1.
Preparation and application of the supernatant of B. subtilis GLB191 and its derivatives
Bacteria were grown in PDB medium at 37 °C with shaking at 180 r min−1 for 48 h. Cell‐free supernatants were obtained after centrifugation at 6000 × g for 20 min at 4 °C, and then filtration using a 0.22‐μm pore size filter. The supernatants were applied to leaves until run‐off using a manual sprayer. Plants sprayed with sterilized distilled H2O and the PDB medium, respectively, were used as controls.
P. viticola preparation
The P. viticola isolate used for this study was maintained on Marselan plants in the glasshouse as previously described (Trouvelot et al., 2008). To obtain sporangia, plants presenting oily spot symptoms were placed in the dark at > 95% relative humidity (RH) overnight to induce sporulation. Sporangia were then collected from the lower side of leaf using a brush and suspended in distilled water. The concentration was adjusted to 2.104 or 105 sporangia per milliliter (depending on experiments) using a haemocytometer.
Protection assays
Two days post‐treatment (dpt) with H2O, PDB or supernatant(s), the lower face of leaves was inoculated with a freshly prepared sporangia suspension (2.104 sporangia per milliliter) using a manual sprayer. Inoculated plants were then placed overnight in a humid chamber (RH > 95%) and then moved back to the glasshouse and grown as described above. Six days post‐inoculation, six discs were punched from each leaf and then placed on wet paper filter in a humid chamber overnight to provoke pathogen sporulation. Disease severity was then assessed by measuring the leaf area covered by sporulation using the image analysis Visilog 6.9 software (Noesis, Paris, France) (Kim Khiook et al., 2013). Three independent biological repeats were conducted.
Evaluation of the direct effect on P. viticola
The lower face of leaves which were treated with H2O, PDB or supernatant(s) was inoculated with a freshly prepared sporangia suspension (2.105 sporangia per milliliter) 2 h post treatment (hpt). This time is sufficient to highlight putative activity against P. viticola and too short to activate defence responses (Krzyzaniak et al., 2018). Leaves were harvested 24 h post inoculation (hpi) and discs (0.7 cm in diameter) were punched from each leaf (ten discs per leaf). They were subsequently bleached first with pure methanol at least 2 days and then chloral hydrate solution (1.0 g/L) for 12–24 h until they become completely transparent. Infection sites (corresponding to encysted zoospores) were detected after aniline blue staining (Gauthier et al., 2014). Four representative fields of each disc and ten discs per condition were observed by epifluorescence microscopy [magnification ×200, λexc = 340–380 nm, λem = 425 nm (long pass filter)]. Three independent biological repeats were conducted.
Quantification of callose deposition
Leaves were treated with H2O, PDB or supernatant(s) and harvested 3 dpt. Ten discs per leaf (0.7 cm in diameter) were punched. Callose deposition was revealed after tissue clearance and aniline blue staining as describe above. The number of deposits (fluorescent spots) was determined by epifluorescence microscopy observations. Three independent biological repeats were conducted.
RNA extraction and reverse transcription
The second‐youngest fully expanded leaves of plants (four plants/condition) were treated with H2O, PDB or supernatants and harvested at 24 hpt and pooled. They were ground in liquid nitrogen and total RNA was isolated using PureLink® Plant RNA Reagent (Invitrogen, Thermofisher, Carlsbad, USA) according to the manufacturer's instructions with minor modification. In brief, approximately 50 mg of leaf powder were incubated with 0.5 mL of Plant RNA Purification Reagent for 5 min at room temperature. After centrifugation at 12 000 × g for 2 min, the supernatant was transferred to a clean RNase‐free tube and equal volume of chloroform/IAA (24:1) was added to the sample. After thoroughly shaking, samples were centrifuged for 10 min at 12 000 × g at 4 °C and the upper aqueous phase was transferred to a clean RNase‐free tube. An equal volume of isopropanol and 40% volume of 5 M NaCl were then added to the sample. After incubation for 10 min at room temperature, samples were centrifuged for 10 min at 12 000 × g at 4 °C. The supernatant was removed and the pellet was washed with 1 mL of 75% ethanol. After centrifugation at 12 000 × g for 1 min, the supernatant was removed and the pellet was air dried and then suspended in 30 μL of distilled water. Then the RNA was treated with DNAse using DNA‐freeTM kit DNAse treatment and removal (Invitrogen, Thermofisher, Carlsbad, USA). The concentration of RNA extracts was determined by spectrophotometry. RNAs were reverse‐transcribed using the Superscript III Reverse Transcriptase kit (Invitrogen, Life Technologies, Saint Aubin, France), random hexamers, 1 mg of DNA‐free total RNA and anchored oligo‐dT 3:1 as primers according to the manufacturer's instructions.
Quantitative RT‐PCR
qRT‐PCR was used to measure transcript levels of target genes with the primers listed in Table S2. The qRT‐PCR experiments were performed with the ABsoluteTM SYBRGreen Low ROX qPCR Mix (Thermo Scientific, Waltham, MA, USA) in a LightCycler480 (Roche Applied Science, Penzberg, Germany) using a thermal cycling profile of 95 °C 15 min; 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 45 s. The melting curve of each reaction was produced to ensure a single amplicon. The absence of primer‐dimer formation was checked in no‐template controls. The mean cycle threshold (Ct) value of a sample's technical triplicates was used for further analysis. Relative gene expression was determined with the formula fold induction = 2−∆∆Ct (Livak and Schmittgen, 2001) where ∆∆Ct = ∆Ct (treated sample) – ∆Ct (control, i.e. water‐treated sample) and ∆Ct = Ct (target gene) – Ct (reference gene). EF1γ encoding the elongation factor 1 gamma was used as reference gene. Three independent biological repeats were conducted.
Quantification of cyclic lipopeptides
Supernatants of GLB191 and its derivatives were prepared as described above. Lipopeptides were quantified by HPTLC as previously described (Geissler et al., 2017). In brief, 2 mL cell‐free broth was extracted threefold with each 2 mL chloroform/methanol 2:1 (v/v). The solvent layers obtained after each extraction were pooled and evaporated to dryness in a rotary evaporator (RVC2‐25 Cdplus, Martin Christ Gefriertrocknungsanlagen GmbH, Osterode am Harz, Germany) at 10 mbar and 40 °C. For HPTLC analysis, samples were resuspended in 2 mL methanol and applied as 6 mm bands on HPTLC silica gel 60 plates from Merck (Darmstadt, Germany). A standard containing the cyclic lipopeptides surfactin, iturin A (both Sigma‐Aldrich Laborchemikalien GmbH, Seelze, Germany) and fengycin (Lipofabrik, Villeneuve d'Ascq, France) with a concentration of 0.1 mg/mL each was applied in a range from 30 ng/band to 600 ng/band. The first development was conducted using chloroform/methanol/water (65:25:4, v/v/v) and the second development using butanol/ethanol/0.1% acetic acid (1:4:1, v/v/v), both over a migration distance of 60 mm. After each development, the plate was scanned at 195 nm, and surfactin and iturin A were evaluated after the first and fengycin after the second development. Three independent biological repeats were conducted.
Statistical analysis
Treatments were compared by means of non‐parametric Kruskal–Wallis approaches at the 5% significance level. Data sets were grouped according to the Mann–Whitney pairwise post hoc test with application of Bonferroni correction. Statistical significance is acknowledged between two conditions if they have no letter (A, B, C etc.) in common.
Supporting information
Table S1 Primers used for mutant construction.
Table S2 Primers used for qRT PCR.
Acknowledgements
We would like to thank Dr Ching‐Hong Yang from the University of Wisconsin‐Milwaukee for the linguistic revision and critical review of the manuscript. Christelle Guillier and Annick Chiltz (Agroécologie, AgroSup Dijon, CNRS, INRA, Université Bourgogne Franche‐Comte) helped with growing plants and are gratefully acknowledged. This project was supported by grants from the National Key Research and Development Program of China (grant no. 2018YFD0201300, 2017YFD0201106), the China Agriculture Research system (29) (grant no. CARS‐29‐bc‐3), the Special Fund for Agro‐Scientific Research in the Public Interest of China (No. 201203035) and the State Scholarship Fund of the China Scholarship Council awarded to Yan Li.
Contributor Information
Qi Wang, Email: wangqi@cau.edu.cn.
Marielle Adrian, Email: marielle.adrian@u-bourgogne.fr.
References
- Abdalla, M.A. , Win, H.Y. , Islam, M.T. , Von Tiedemann, A. , Schüffler, A. and Laatsch, H. (2011) Khatmiamycin, a motility inhibitor and zoosporicide against the grapevine downy mildew pathogen Plasmopara viticola from Streptomyces sp. ANK313. J. Antibiot. 64, 655. [DOI] [PubMed] [Google Scholar]
- Arnaud, M. , Chastanet, A. and Debarbouille, M. (2004) New vector for efficient allelic replacement in naturally nontransformable, low‐GC‐content, gram‐positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot, F. and Zipfel, C. (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad‐spectrum disease resistance. Annu Rev Phytopathol. 55, 257–286. [DOI] [PubMed] [Google Scholar]
- Cawoy, H. , Mariutto, M. , Henry, G. , Fisher, C. , Vasilyeva, N. , Thonart, P. , Dommes, J. and Ongena, M. (2014) Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol. Plant‐Microbe Interact. 27, 87–100. [DOI] [PubMed] [Google Scholar]
- Chandler, S. , Van Hese, N. , Coutte, F. , Jacques, P. , Höfte, M. and De Vleesschauwer, D. (2015) Role of cyclic lipopeptides produced by Bacillus subtilis in mounting induced immunity in rice (Oryza sativa L.). Physiol. Mol. Plant P. 91, 20–30. [Google Scholar]
- Compant, S. and Mathieu, F. (2016) Biocontrol of Major Grapevine Diseases: Leading Research. Castanet‐Tolosan, France: University of Toulouse. [Google Scholar]
- Coutte, F. , Leclere, V. , Bechet, M. , Guez, J.S. , Lecouturier, D. , Chollet‐Imbert, M. , Dhulster, P. and Jacques, P. (2010) Effect of pps disruption and constitutive expression of srfA on surfactin productivity, spreading and antagonistic properties of Bacillus subtilis 168 derivatives. J. Appl. Microbiol. 109, 480–491. [DOI] [PubMed] [Google Scholar]
- Debois, D. , Fernandez, O. , Franzil, L. , Jourdan, E. , de Brogniez, A. , Willems, L. , Clément, C. , Dorey, S. , De Pauw, E. and Ongena, M. (2015) Plant polysaccharides initiate underground crosstalk with bacilli by inducing synthesis of the immunogenic lipopeptide surfactin. Env. Microbiol. Rep. 7, 570–582. [DOI] [PubMed] [Google Scholar]
- Deleu, M. , Paquot, M. and Nylander, T. (2008) Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophys. J. 94, 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falardeau, J. , Wise, C. , Novitsky, L. and Avis, T.J. (2013) Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. J. Chem. Ecol. 39, 869–878. [DOI] [PubMed] [Google Scholar]
- Fan, H.Y. , Ru, J.J. , Zhang, Y.Y. , Wang, Q. and Li, Y. (2017a) Fengycin produced by Bacillus subtilis 9407 plays a major role in the biocontrol of apple ring rot disease. Microbiol. Res. 199, 89–97. [DOI] [PubMed] [Google Scholar]
- Fan, H.Y. , Zhang, Z.W. , Li, Y. , Zhang, X. , Duan, Y.M. and Wang, Q. (2017b) Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin‐mediated antibacterial activity and colonization. Front. Microbiol. 8, 1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farace, G. , Fernandez, O. , Jacquens, L. , Coutte, F. , Krier, F. , Jacques, P. , Clément, C. , Barka, E.A. , Jacquard, C. and Dorey, S. (2015) Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Mol. Plant Pathol. 16, 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fravel, D. (2005) Commercialization and implementation of biocontrol. Annu. Rev. Phytopathol. 43, 337–359. [DOI] [PubMed] [Google Scholar]
- Gauthier, A. , Trouvelot, S. , Kelloniemi, J. , Frettinger, P. , Wendehenne, D. , Daire, X. , Joubert, J.M. , Ferrarini, A. , Delledonne, M. , Flors, V. and Poinssot, B. (2014) The sulfated laminarin triggers a stress transcriptome before priming the SA‐ and ROS‐dependent defenses during grapevine's induced resistance against Plasmopara viticola . PLoS One. 9, e88145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler, M. , Oellig, C. , Moss, K. , Schwack, W. , Henkel, M. and Hausmann, R. (2017) High‐performance thin‐layer chromatography (HPTLC) for the simultaneous quantification of the cyclic lipopeptides surfactin, iturin A and fengycin in culture samples of Bacillus species . J. Chromatogr. B. 1044, 214–224. [DOI] [PubMed] [Google Scholar]
- Gindro, K. , Pezet, R. and Viret, O. (2003) Histological study of the responses of two Vitis vinifera cultivars (resistant and susceptible) to Plasmopara viticola infections. Plant Physiol. Bioch. 41, 846–853. [Google Scholar]
- Hamiduzzaman, M.M. , Jakab, G. , Barnavon, L. , Neuhaus, J.M. and Mauch‐Mani, B. (2005) beta‐Aminobutyric acid‐induced resistance against downy mildew in grapevine acts through the potentiation of callose formation and jasmonic acid signaling. Mol. Plant‐Microbe Interact. 18, 819–829. [DOI] [PubMed] [Google Scholar]
- Harwood, C.R. , Mouillon, J.‐M. , Pohl, S. and Arnau, J. (2018) Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 42, 721–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, G. , Deleu, M. , Jourdan, E. , Thonart, P. and Ongena, M. (2011) The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune‐related defence responses. Cell Microbiol. 13, 1824–1837. [DOI] [PubMed] [Google Scholar]
- Jacobsen, B. , Zidack, N. and Larson, B. (2004) The role of Bacillus‐based biological control agents in integrated pest management systems: plant diseases. Phytopathology, 94, 1272–1275. [DOI] [PubMed] [Google Scholar]
- Jourdan, E. , Henry, G. , Duby, F. , Dommes, J. , Barthelemy, J.P. , Thonart, P. and Ongena, M.A.R.C. (2009) Insights into the defense‐related events occurring in plant cells following perception of surfactin‐type lipopeptide from Bacillus subtilis . Mol. Plant‐Microbe Interact. 22, 456–468. [DOI] [PubMed] [Google Scholar]
- Keswani, C. , Bisen, K. , Singh, V. , Sarma, B.K. and Singh, H.B. (2016) Formulation technology of biocontrol agents: present status and future prospects In: Bioformulations: For Sustainable Agriculture, (Arora N. K., Mehnaz S. and Balestrini R., eds), pp. 35–52. India: Springer. [Google Scholar]
- Kim Khiook, I.L. , Schneider, C. , Heloir, M.‐C. , Bois, B. , Daire, X. , Adrian, M. and Trouvelot, S . (2013) Image analysis methods for assessment of H2O2 production and Plasmopara viticola development in grapevine leaves: application to the evaluation of resistance to downy mildew. J. Microbiol. Meth. 95, 235–244. [DOI] [PubMed] [Google Scholar]
- Krzyzaniak, Y. , Trouvelot, S. , Negrel, J. , Cluzet, S. , Valls, J. , Richard, T. , Bougaud, A. , Jacquens, L. , Klinguer, A. , Chiltz, A. , Adrian, M. and Heloir, M.C. (2018) A plant extract acts both as a resistance inducer and an oomycide against grapevine downy mildew. Front. Plant Sci. 9, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy, M. , Warren, G.H. , Rosenman, S.B. and Colio, L.G. (1948) Bacillomycin – an antibiotic from Bacillus subtilis active against pathogenic fungi. P. Soc. Exp. Biol. Med. 67, 539–541. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luo, C.P. , Liu, X.H. , Zhou, H.F. , Wang, X.Y. and Chen, Z.Y. (2015) Nonribosomal peptide synthase gene clusters for lipopeptide biosynthesis in Bacillus subtilis 916 and their phenotypic functions. Appl. Environ. Microb. 81, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maget‐Dana, R. and Peypoux, F. (1994) Iturins, a special class of pore‐forming lipopeptides: biological and physicochemical properties. Toxicology, 87, 151–174. [DOI] [PubMed] [Google Scholar]
- McSpadden Gardener, B. and Fravel, D. (2002) Biological control of plant pathogens: research, commercialization, and application in the USA. Plant Health Progress, 10.1094/PHP-2002-0510-01-RV. [DOI] [Google Scholar]
- Nicholson, W. (2002) Roles of Bacillus endospores in the environment. Cell Mol Life Sci. 59, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena, M. and Jacques, P. (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. [DOI] [PubMed] [Google Scholar]
- Perazzolli, M. , Roatti, B. , Bozza, E. and Pertot, I. (2011) Trichoderma harzianum T39 induces resistance against downy mildew by priming for defense without costs for grapevine. Biol. Control. 58, 74–82. [Google Scholar]
- Pérez‐García, A. , Romero, D. and De Vicente, A. (2011) Plant protection and growth stimulation by microorganisms: biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotech. 22, 187–193. [DOI] [PubMed] [Google Scholar]
- Rotolo, C. , De Miccolis Angelini, R.M. , Dongiovanni, C. , Pollastro, S. , Fumarola, G. , Di Carolo, M. , Perrelli, D. , Natale, P. and Faretra, F. (2018) Use of biocontrol agents and botanicals in integrated management of Botrytis cinerea in table grape vineyards. Pest Manag. Sci. 74, 715–725. [DOI] [PubMed] [Google Scholar]
- Santoyo, G. , Orozco‐Mosqueda, M.D.C. and Govindappa, M. (2012) Mechanisms of biocontrol and plant growth‐promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci. Techn. 22, 855–872. [Google Scholar]
- Schisler, D. , Slininger, P. , Behle, R. and Jackson, M. (2004) Formulation of Bacillus spp. for biological control of plant diseases. Phytopathology, 94, 1267–1271. [DOI] [PubMed] [Google Scholar]
- Shafi, J. , Tian, H. and Ji, M. (2017) Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol. Biotec. Eq. 31, 446–459. [Google Scholar]
- Stein, T. (2005) Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56, 845–857. [DOI] [PubMed] [Google Scholar]
- Trouvelot, S. , Varnier, A.L. , Allegre, M. , Mercier, L. , Baillieul, F. , Arnould, C. , Gianinazzi‐Pearson, V. , Klarzynski, O., Joubert, J.M., Pugin, A. and Daire, X. (2008) A beta‐1,3 glucan sulfate induces resistance in grapevine against Plasmopara viticola through priming of defense responses, including HR‐like cell death. Mol. Plant‐Microbe Interact. 21, 232–243. [DOI] [PubMed] [Google Scholar]
- Vanittanakom, N. , Loeffler, W. , Koch, U. and Jung, G. (1986) Fengycin‐a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F‐29‐3. J. Antimicrobiot. 39, 888–901. [DOI] [PubMed] [Google Scholar]
- Villarreal‐Delgado, M.F. , Villa‐Rodríguez, E.D. , Cira‐Chávez, L.A. , Estrada‐Alvarado, M.I. , Parra‐Cota, F. and de los Santos‐Villalobos, S. (2018) The genus Bacillus as a biological control agent and its implications in the agricultural biosecurity. Mex. J. Phytopathol. 36, 95–130. [Google Scholar]
- Yu, Y. , Jiao, L. , Fu, S.F. , Yin, L. , Zhang, Y.L. and Lu, J. (2016) Callose synthase family genes involved in the grapevine defense response to downy mildew disease. Phytopathology, 106, 56–64. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , Wang, W. , Deng, A. , Sun, Z. , Zhang, Y. , Liang, Y. , Che, Y. and Wen, T . (2012) A mimicking of DNA methylation patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet. 8, e1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhou, Y.Y. , Li, Y. , Fu, X.C. and Wang, Q. (2017) Screening and characterization of endophytic Bacillus for biocontrol of grapevine downy mildew. Crop Prot. 96, 173–179. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers used for mutant construction.
Table S2 Primers used for qRT PCR.


