Summary
The hypersensitive defence response is found in all higher plants and is characterized by a rapid cell death at the point of pathogen ingress. It is usually associated with pathogen resistance, though, in specific situations, it may have other consequences such as pathogen susceptibility, growth retardation and, over evolutionary timescales, speciation. Due to the potentially severe costs of inappropriate activation, plants employ multiple mechanisms to suppress inappropriate activation of HR and to constrain it after activation. The ubiquity of this response among higher plants despite its costs suggests that it is an extremely effective component of the plant immune system.
Keywords: hypersensitive response
Introduction
Simply put, the plant hypersensitive response (HR) is a rapid localized cell death that occurs at the point of pathogen penetration and is associated with disease resistance. While others had noted similar phenomena previously (Mur et al., 2008), it is believed that E.C. Stakman (Stakman, 1915) was the first to use the term ‘hypersensitive’. Discussing the extreme resistance displayed by certain grass hosts to Puccinia graminis, he noted that the ‘host plant in such cases is hypersensitive to the fungus’. Gauman (1950) defined HR similarly: ‘the hypersensitive reaction implies to us, a priori, rapid plant cell death following elicitation by a pathogen or metabolite thereof, reflecting electrolyte leakage due to membrane damage, the cessation of cyclosis and cellular collapse … subsequent to this … is … necrosis of the collapsed cells and localization of the eliciting pathogen’. Goodman and Novacky (1994) describe HR by the following criteria: ‘A rapid cell death localized at the area of pathogen infection that results in some suppression of disease progress (sometimes although not always total)’.
HR is a widespread phenomenon, found in most if not all higher plants and induced by a number of classes of pathogen. HRs induced by fungi, oomycetes, bacteria and viruses are the most commonly observed, but HR can be induced by other organisms, such as insects (Rossi et al., 1998) and nematodes (Dropkin, 1969), that form sustained intimate interactions with the host plant. Cell death has also been observed in resistant interactions between parasitic plants and their hosts (Mohamed et al., 2003), though whether this represents an HR is unclear (Swarbrick et al., 2008). HR is generally associated with race‐specific resistance to biotrophic pathogens (pathogens which derive nutrition from living tissues). It is generally less effective against, and may actually be beneficial to, necrotrophs which require dead host tissue to complete their life cycle (see below). The situation is more complex in the case of hemibiotrophs such as Phytophthora infestans or Colletotrichum graminicola (respectively, the causal agents of potato late blight and anthracnose in many cereals) in which early interactions with the host are biotrophic but subsequently switch to a necrotrophic lifestyle. In these cases HR may be beneficial to the host early but not late in the interaction (Jupe et al., 2013; Münch et al., 2008).
HR and associated responses have been reviewed before (Goodman and Novacky, 1994; Greenberg, 1997; Heath, 2000b; Kamoun et al., 1999; Künstler et al., 2016; Lam et al., 2001; Mur et al., 2008). For the most part these reviews have focused on the literature on the ultrastructure, biochemistry, genetics and physiology of HR and how it relates to other kinds of programmed cell death. This review does not dwell on these previously addressed subjects. Rather it emphasizes aspects of HR that have been the subject of less past attention: the concept, the control and the consequences of HR.
Concepts
The genetics and molecular genetics of HR
Our understanding of HR is linked to the gene‐for‐gene concept discovered by H. H. Flor in the 1930s to the 1950s (Flor, 1971; Loegering and Ellingboe, 1987). Working on the flax/flax rust system (Linum margina/ Melampspora lini), Flor discovered a ‘gene‐for‐gene’ relationship between the plant and pathogen that determined whether the interaction resulted in disease or resistance/HR. Each dominant resistance gene (R‐gene) in the host corresponded with a dominant avirulence gene (Avr gene) in the pathogen. Resistance was only conferred if both the R‐gene and the corresponding Avr gene were present in the same interaction. The gene‐for‐gene relationship was subsequently determined to be broadly applicable across most species of higher plants that have been studied. Many R‐gene and Avr genes have been cloned (Kourelis and van der Hoorn, 2018). In the large majority of cases, R‐gene‐mediated resistance is associated with HR.
Most (not all) R‐genes encode cytoplasmically located proteins possessing both a nucleotide‐binding site (NBS) and leucine‐rich (LRR) repeat domains known as NBS‐LRR proteins or NLRs. Furthermore, most NLRs carry either coiled coil (CC) or toll‐interleukin receptor (TIR) domains at their N‐terminal. While the similarities in NLR structure and function suggest a high level of conservation of mechanism, this is not necessarily the case. In particular, their mechanisms of activation seem to vary quite considerably (Wang and Balint‐Kurti, 2015). Nevertheless, in general the N‐terminal domains appear to be responsible for activating cell death pathways while the NBS and LRR domains are generally associated with regulating the activity of the R‐protein (Kourelis and van der Hoorn, 2018; Wang and Balint‐Kurti, 2015; Wang et al., 2015).
A related class of proteins known as pattern recognition receptors (PRRs) encode membrane‐bound proteins that recognize broadly conserved microbial molecules known as microbe‐ or pathogen‐associated molecular patterns (MAMPs or PAMPs). Upon recognition they activate a defence response known as MAMP/PAMP‐triggered immunity (MTI/PTI) that is qualitatively similar to that induced by NLR, though quantitatively reduced and usually not including programmed cell death (Feechan et al., 2015).
NLRs originated in the earliest ancestors of photosynthetic plants and have been identified in basal‐branching streptophytes (charophytes, liverworts and mosses) and all the land plants (Gao et al., 2018; Shao et al., 2019). While the functions of most plant NLRs have not been defined, the only function that has been definitively associated with them is as R‐proteins. It is therefore not strictly correct to use the terms NLR protein and R‐protein interchangeably as is often seen in the literature (e.g. Gao et al., 2018) since it is likely that many NLRs are not R‐proteins. Furthermore some R‐proteins are not NLRs; there are a number of classes of R‐proteins that do not have the NLR structure (Kourelis and van der Hoorn, 2018).
Avr proteins are now considered to be a class of pathogen effector proteins; proteins that are produced by the pathogen and secreted into the host cells or apoplast specifically to enable the pathogenesis process (Lo Presti et al., 2015). Avr proteins are simply effectors whose presence can be detected by specific R‐proteins leading to the activation of the defence response. Resistance triggered by effectors in this way has been termed effector‐triggered immunity (ETI) (Jones and Dangl, 2006). The various mechanisms of Avr recognition by R‐proteins and subsequent activation of the defence response have been reviewed extensively (Bonardi et al., 2011, 2012; Kourelis and van der Hoorn, 2018). In some cases the R‐ and Avr proteins directly interact while in others the action of the Avr protein on another host protein (known variously as the guardee or decoy) is responsible for activating the R‐protein (Kourelis and van der Hoorn, 2018).
How should HR be defined?
While the HR is the most obvious manifestation of the defence response associated with NLRs, there are many other aspects which have been reviewed previously (Hammond‐Kosack and Jones, 1996; Heath, 2000b; Mur et al., 2008). Bursts of production of reactive oxygen species (ROS) and nitric oxide (NO) usually occur very early in the defence response and are often important for the initiation of HR (Delledonne et al., 2001). Other features commonly described include lipid peroxidation, transcriptional reprogramming, ion fluxes and cell wall fortification.
Host–pathogen interactions inevitably drive evolutionary arms races such that genes involved in these interactions tend to evolve at a relatively rapid rate (Tiffin and Moeller, 2006). It is therefore likely that mechanisms of NLR–effector protein interaction and resulting resistance mechanisms including the HR may have diverged substantially between species and between interactions over evolutionary time. It has been noted previously that aspects of HR differ substantially in different interactions (Mur et al., 2008). Nevertheless, the mechanisms underlying HR and its physiological and ultrastructural consequences have often explicitly or implicitly been assumed to be highly conserved across higher plants between hosts and interactions. It is perhaps ironic that a group of interactions each of which are defined by exquisite specificity between individual R‐genes and particular races of particular pathogens are nevertheless often assumed to function in identical ways after the initial recognition event. This review will likewise summarize findings made over a broad array of systems that induce HR in an attempt to make overarching conclusions; however, the reader is reminded of the likely variation in underlying mechanisms and effects of different HRs in different species and in different interactions involving the same host species.
The fact that cell death is a common phenomenon in plants with many different causes has also complicated our understanding of HR. The term HR has been invoked to describe a large range of phenomena whose only common feature may be the superficial macroscopic appearance of cell death. The term ‘hypersensitive‐like cell death’ or similar is often used simply when cell death is observed without any explicit link to R‐genes, the defence response or disease resistance (e.g. Chen et al., 2016; Király et al., 2008; Kumar and Kirti, 2015; Na et al., 2015; Wei et al., 2016).
This review will take a rather narrow view of HR, confined to the localized rapid cell death and excluding the other associated responses induced by the activation of NLR proteins. In most cases this NLR activation is due to the presence of the cognate pathogen‐encoded effector proteins and is part of a defence response. However, this definition also includes categories in which HR is ‘illegitimately’ activated through NLRs, due to either mutation of the NLRs or manipulation by plant pathogens. While the cell death in these cases is associated with detrimental phenotypes (susceptibility or reduced growth) rather than resistance, here they will be considered to be legitimate examples of HR as they are triggered by activation of NLRs.
Excluded by this definition is the HR caused by a small class of R‐genes called ‘executor R‐genes’. These genes are activated at the transcriptional level by a class of effectors known as transcription activator‐like effectors (TALEs). None of the six executor genes cloned so far are related to NLRs; two encode a putative flavin monooxygenase and four others encode proteins with predicted transmembrane domains (Kourelis and van der Hoorn, 2018; Zhang et al., 2015). The mechanism(s) by which executor R‐proteins cause cell death have not been characterized but, due to their lack of structural or sequence homology to NLRs, it seems unlikely that they act through similar mechanisms to NLRs. This may be an example of convergent evolution; a rapid localized cell death in response to infection may be such an efficient method of containing the spread of biotrophic pathogens that several mechanisms to achieve this have evolved independently.
Can we learn about the control of HR from the study of lesion mutants?
A large class of mutations known variously as lesioned, lesion mimic, accelerated cell death, spotted leaf or lesions simulating disease mutants display lesions, or spontaneously forming patches of dead cells on their leaves. This class of mutants (here referred to as LES) have been characterized in several species, notably Arabidopsis, maize and rice (Fig. 1A). They were named based on their superficial similarity to disease‐related phenotypes, specifically HR, though in most cases at the time of discovery there was no direct evidence for a mechanistic link. In fact the large diversity in lesion phenotypes (Fig. 1A and Johal, 2007) seemed to suggest a diversity of underlying mechanisms.
Figure 1.
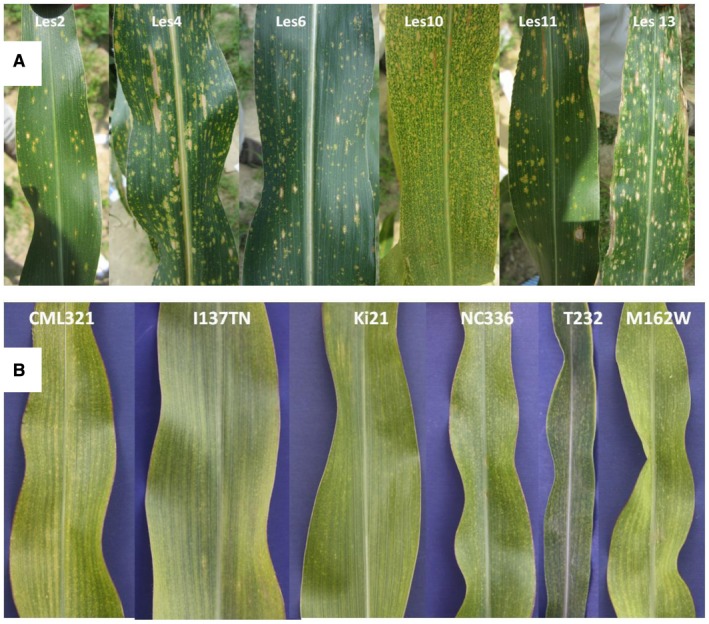
(A) The phenotypes caused by various LES genes in the background of the maize inbred line Mo20W. (B) Examples of inbred lines showing a mild ‘flecking’ phenotype. Source: P. Balint‐Kurti/B. Olukolu. Adapted from Fig. 1 in Vontimitta et al. (2015).
As the genes underlying LES phenotypes have been cloned it has become clear that a significant proportion of them are indeed associated with HR and the defence response. Bruggemen et al. (2015) lists about 80 genes conferring or suppressing LES phenotypes. A few are auto‐active NLRs which usually occur when mutations in the NLR gene itself disrupts delicate auto‐inhibitory intramolecular interactions causing inappropriately and apparently spontaneous activation in the absence of the cognate effector. Another group of LES genes appear to mediate their phenotype via interaction with the plant defence machinery, in some cases through direct interaction with NLRs (e.g. Li et al., 2009). Several others affect the processes regulating ROS in the plant. Therefore, in some cases at least, the phenotypes of LES mutants can inform us about aspects of HR. Some of these are discussed below.
Leaf flecking, a mild lesion phenotype observed on the leaves of some maize lines, is a trait familiar to most corn breeders (Fig. 1B), appearing in several widely used lines as well as in several other species such as barley (Makepeace et al., 2007), wheat (Nair and Tomar, 2001) and oats (Ferdinandsen and Winge, 1930). Recent work has indicated that in some cases flecking seems to be a mild form of misregulated HR (Olukolu et al., 2016; Vontimitta et al., 2015).
HR as a form of programmed cell death
One common definition of programmed cell death (PCD) is the death of a cell in any form, mediated by an intracellular programme. By this definition HR is clearly a form of PCD. A number of reviews have examined the symptomology of HR compared with much better understood forms of PCD (Birch et al., 2018; Goodman and Novacky, 1994; Mur et al., 2008). Many features of HR have been noted in apoptotic and/or autophagic cell death, types of PCD that have been best characterized in animal systems but which are believed to exist in some form in plants (Birch et al., 2018; Liu et al., 2005). These include cytoplasmic/protoplast collapse (Heath, 2000a), cessation of cytoplasmic streaming (Bushnell, 1981; Kitazawa et al., 1973; Naton et al., 1996) and disruption of the cytoskeleton (Chen and Heath, 1991; Gross et al., 1993; Kobayashi et al., 1994; Schadt et al., 2003), DNA laddering (Ryerson and Heath, 1996), formation of large vesicles (Liu et al., 2005) and involvement of the mitochondria (Xie and Chen, 2000). A nice summary of these and other features is given in Table 1 of Mur et al. (2008). The general consensus is that HR shares features with apoptosis and other forms of PCD but that it should be regarded as a specialized form of cell death (Coll et al., 2011; Mur et al., 2008; Van Doorn et al., 2011). Again, the reader should bear in mind that all HR is not identical (Mur et al., 2008).
Control
HR control mediated through NLR expression and protein accumulation
HR needs to be tightly regulated. It must be completely suppressed under non‐disease conditions since improper activation will lead to a spontaneous cell death phenotype which can be very detrimental to plant growth (e.g. Chintamanani et al., 2010). Conversely, it needs to be rapidly activated when required. These constraints have led to the evolution of multiple layers of control.
At the transcript level, NLR expression is generally low, often tissue‐specific and can be induced during the defence response in many cases (Mohr et al., 2010; Tan et al., 2007). The various mechanisms by which NLR gene transcript abundance is controlled have been recently reviewed (Lai and Eulgem, 2018). They include regulation at the level of chromatin structure, by transcription factors, alternative splicing, post‐transcriptional regulation by small RNAs and possibly by alternative polyadenylation, and nonsense‐mediated decay. The presence of transposons in or near specific NLR genes is also likely to regulate transcription in certain cases.
Control of HR is also exercised at the level of protein accumulation and stability. In particular, the molecular chaperone HSP90 and two interacting co‐chaperones RAR1 and SGT1 form a complex that interacts with many NLRs, stabilizing them and allowing their proper maturation and function (Zhang et al., 2010). Within this complex, SGT1 plays a central role: it recruits the NLR and also interacts with the E3 ubiquitin ligase subunit Skp1 (Catlett and Kaplan, 2006). The important roles that E3 ubiquitin ligases and the ubiquitin‐mediated protein degradation pathway play in plant immunity are becoming increasingly clear (Zhou and Zeng, 2017). While this is still an active area of research, there is some evidence that specific E3 ligases are recruited to direct the degradation of specific NLRs (Huang et al., 2016). The regulation of NLR turnover by the ubiquitin‐mediated protein degradation system has been reviewed in detail elsewhere (Dong et al., 2018; Feechan et al., 2015).
Silencing or mutation of any one of RAR1, SGT1 or HSP90 in many interactions is sufficient to abolish HR and to cause the reduction of NLR protein levels (e.g. Azevedo et al., 2006; Bieri et al., 2004; Hubert et al., 2003; Kim et al., 2018; Scofield et al., 2005). The role of SGT1 is not completely understood. In Arabidopsis, inactivation of one of the two functional SGT1 genes led to increased accumulation of the RPS5 NBS‐LRR R‐protein, suggesting that SGT1 antagonized the effect of RAR1 (Holt et al., 2005), acting to reduce levels of NLR proteins. In another case in Nicotiana benthamiana, silencing of SGT1 caused a reduction in steady‐state levels of the R‐protein, Rx (Azevedo et al., 2006). These contradictions have not been fully resolved, but the role of SGT1 may vary depending on the species and the particular NLR or its activation state (Azevedo et al., 2006). Translational control has recently been demonstrated to play an important part in regulating the plant defence system (Xu et al., 2017). While translational control of NLRs has not been demonstrated, this possibility does not seem to have been fully explored. Some studies have suggested that protein synthesis is required for HR (Keen et al., 1981; Nozue et al., 1977), while others have suggested the opposite (Doke and Tomiyama, 1975). The consensus is that protein synthesis is required (Greenberg, 1997) but it is of course quite possible that different cases of HR in different species may have somewhat different requirements.
Control of NLR activation mediated by intra‐ and intermolecular interactions
Arguably the most basic and conserved mode of HR control is the intramolecular interactions that occur between domains of the NLR. These have been well documented in a number of NLRs and while the specifics vary, in general the LRR domain usually plays an inhibitory role by suppressing the activity of the CC or TIR N‐terminal domains which, when expressed alone, induce a constitutive HR (e.g. Wang et al., 2015).
As well as the interactions with SGT1, HSP90 and RAR1 discussed above, NLRs interact with a number of other proteins in the cell that modulate their activity. NLR activation is often associated with the formation of homo‐ and hetero‐oligomers (Mestre and Baulcombe, 2006; Schreiber et al., 2016). A class of NLRs termed helper NLRs has been defined that is required for downstream signalling, including HR, initiated by effector recognition by so‐called ‘sensor’ NLRs (Baggs et al., 2017). An extensive network of helper and sensor NLRs was characterized in tomato in which multiple complex and redundant interactions mediated immunity to oomycetes, bacteria, viruses, nematodes and insects (Wu et al., 2017). In some cases the genes encoding interacting helper and sensor NLRs are located together in the genome and are termed ‘paired NLRs’ (Baggs et al., 2017). In at least one case these paired NLRs physically interact (Williams et al., 2014). In a groundbreaking study Wang et al. (2019a, b) demonstrated that, upon activation, the Arabidopsis ZAR1 NLR forms a homo‐pentamer which is assumed to be able to initiate the defence response.
In many cases interactions between NLRs and their specific guardee or decoy proteins are required for inhibition of HR. For example, the guardee protein RIN4 interacts with a number of NLRs across different species and disruptions in RIN4 can induce inappropriate activation of HR (Toruño et al., 2019). The activity of the maize autoactive NLR Rp1‐D21 is modulated by interactions with at least two enzymes in the lignin biosynthesis pathway as well by interactions with genes encoded by other Rp1 alleles (Olukolu et al., 2014; Wang and Balint‐Kurti, 2016; Wang et al., 2015, 2016).
Control at the population level
In a randomly mating population, the negative effects of inappropriately activated HR are also mediated at the level of allele frequency. For example, the Arabidopsis NLR RPM1 has been shown to confer yield penalties in the absence of particular strains of Pseudomonas syringae to which it confers resistance (Tian et al., 2003), presumably because it triggers an inappropriate low‐level defence response. It appears to have been maintained in the wild population through balancing selection due to its contrasting beneficial and detrimental effects, depending on the external conditions (Stahl et al., 1999). This may be the case for a number of R‐genes that confer benefits in the presence of the appropriate pathogen but confer growth/yield penalties in their absence (Karasov et al., 2014).
Thermoregulation
Plant–pathogen interactions that induce HR are commonly temperature sensitive such that HR is not induced at temperatures above 20–30 °C (Bromfield, 1961; Goodman and Novacky, 1994). Examples include wheat/P. graminis (Harder et al., 1979), oat/Puccinia coronata (Zimmer and Schafer, 1961), the tobacco NLR R‐gene N/tobacco mosaic virus (TMV; Jockusch, 1966), soybean/Pseudomonas spp. (Keen et al., 1981) and the tomato NLR R‐gene Mi/nematode (Abdul‐Baki et al., 1996; Branch et al., 2004; Dropkin, 1969).
This phenomenon is also commonly observed for auto‐active NLRs (Heidrich et al., 2013; Negeri et al., 2013; discussed more below) and in hybrid necrosis controlled by NLRs (Bomblies et al., 2007; also discussed below; Muralidharan et al., 2014). In some cases chilling sensitive phenotypes, where leaf yellowing and cell death are observed at temperatures under 16 °C, were shown to be caused by mutated versions of NLRs (Bao et al., 2014; Huang et al., 2010).
In most reported cases, elevated temperature suppressed disease resistance as well as HR. For example, temperatures above 30 °C abolish HR and resistance to TMV mediated by the N gene (Whitham et al., 1996). In another example Wang et al. (2009) observed that, compared to 22 °C conditions, Arabidopsis grown at 28 °C exhibited slower HR and compromised resistance mediated by the NLR resistance genes RPS2, RPM1 and RPS4 in response to infection with strains of P. syringae pv. tomato (Pto) DC3000. However, in some cases this was not true; elevated temperatures of 30 °C suppressed HR mediated by the NLRs ZAR1 and RPS2 in Arabidopsis infected with PtoDC3000 and in most, but not all, accessions growth of the pathogen was still suppressed (Menna et al., 2015). More examples of disconnects between HR and disease‐resistance phenotypes are discussed below.
The mechanisms underlying NLR temperature sensitivity are not completely understood. In some cases, protein levels may be important. The levels of the MLA NLR proteins conferring resistance to Blumeria graminis in barley were highly reduced when the temperature was shifted from 18 °C to 37 °C (Bieri et al., 2004), with a significant reduction observable within 30 min of the temperature change. Interestingly B. graminis is unable to colonize barley at these elevated temperatures so MLA proteins are likely not required under these conditions in any case (Aust, 1974). In most examples of temperature‐sensitive HR, NLR protein levels cannot be measured since the necessary antibodies are not available. However, the levels of the NLR rpp4 that conferred chilling sensitivity in Arabidopsis and of the autoactive NLR SNC1 were not altered by temperature (Bao et al., 2014; Zhu et al., 2010), indicating that mechanisms other than those affecting protein accumulation may be important for NLR temperature sensitivity.
The autoactive NLR snc1‐1 confers a dwarf phenotype and constitutive HR that is relieved at 28 °C and above (Zhang et al., 2003). In a seminal paper Zhu et al. (2010) showed that a mutation in the LRR motif of snc1‐1 abolished the temperature‐sensitive phenotype. As noted above, protein levels of snc1 did not appear to be altered by temperature, but the localization of the protein to the nucleus was absolutely correlated with the HR/dwarf phenotype (Zhu et al., 2010). Similar mutations in the N gene also abolished temperature sensitivity and that activity was again correlated with nuclear localization. In related work, also with the N gene, Padgett et al. (1997) showed that the temperature at which HR was abrogated depended on the precise sequence of the cognate Avr gene, suggesting that, in addition to the NLR structure itself and its localization, the precise nature of the NLR–effector interaction can influence the temperature sensitivity of HR.
As discussed above, NLRs appear to often function as oligomeric complexes. It has been suggested that higher temperatures may disfavour these interactions between proteins (Jones et al., 2016). While still untested, this may provide some explanation for the pervasive nature of the temperature‐sensitivity phenomenon. The self‐association of NLRs involves multiple domains, including the LRR (Schreiber et al., 2016; Wang et al., 2015). The mutation in snc1‐1 that abolished the temperature‐sensitive phenotype may have enhanced the ability of the protein to self‐associate, rendering the oligomerized state more resilient to the effects of high temperature.
Cheng et al. (2013) showed that while the Arabidopsis ETI response, including HR, is suppressed at temperatures above around 20 °C, the strength of the PTI response increased from 20 °C to 30 °C. Additionally, they examined plants mutant in the arp6 and hta9/hta11 genes, the wild‐type versions of which are associated with nucleosome assembly. These lines have been reported to phenocopy warm‐grown plants when grown at cooler temperatures (Kumar and Wigge, 2010). These mutants showed higher PTI and lower ETI (including HR) than wild‐type plants when grown at low temperatures. These data suggest that, as well as being an intrinsic property of NLRs themselves, the wider plant thermosensing mechanism may also be involved in controlling the reduction in HR at higher temperatures.
Since it is so widespread, the temperature sensitivity of HR is very likely to be an adaptive trait (Alcázar and Parker, 2011; Cheng et al., 2013). In many cases pathogens do not grow well at elevated temperatures (temperatures above the normal ambient levels of their immediate environment). For instance, optimal growth rates of bacteria and fungi in soils from southern Sweden were reported to drop off above 25–30 °C (Pietikäinen et al., 2005). As discussed above, NLRs can have negative growth consequences in the absence of the pathogens to which each confer resistance, probably due to low‐level activation of the defence response. It seems likely that the cost–benefit balance for NLR function might become more unfavourable at temperatures above which the pathogens cannot grow well, therefore the activity of NLRs might be down‐regulated at these higher temperatures in order to mitigate potential negative growth effects. The observation that B. graminis is unable to colonize barley at the elevated temperatures at which MLA proteins are non‐functional (Aust, 1974) supports this hypothesis. Another example is the maize NLR Rp1 which does not confer HR above 30 °C; Puccinia sorghi, the pathogen to which it confers resistance, does not infect well above 25 °C (Pryor, 1994). At substantially elevated temperatures the plant may be under considerable stress and down‐regulating defence responses, in part by using temperature‐sensitive NLRs, may be beneficial in order to maximize resources for survival growth and reproduction (Ramegowda and Senthil‐Kumar, 2015).
Of course, the most predictable temperature fluctuations occur over the 24‐h diurnal cycle. The Arabidopsis basal defence response is regulated in a circadian manner (Wang et al., 2011). The expression levels of several genes important for resistance peak around dawn, the time of peak spore dissemination. In this way, it was hypothesized, the plant is able to synchronize its maximal defence responsiveness to the period of maximal need. Dawn is also the coldest period of the day and so perhaps the temperature sensitivity of ETI is another way of fine‐tuning this system.
An interesting approach to test the hypothesis that the temperature sensitivity of HR is adaptive would be to compare the temperature sensitivity profiles of NLRs derived from plants that evolved in different environments. One might expect that NLRs derived from plants that evolved in cooler environments might show temperature sensitivity at lower temperatures. To our knowledge this type of approach has not been pursued. The temperature sensitivity of NLR‐based resistance has implications for agriculture in the face of climate change. Crops like wheat which are beset by a number of biotrophic pathogens and employ a large number of temperature‐sensitive R‐genes to combat them (Gousseau et al., 1985) may be particularly affected, though the exact ramifications are hard to predict (Garrett et al., 2016).
Other environmental requirements for HR
Light dependence of HR has often been observed (e.g. Chandra‐Shekara et al., 2006; Guo et al., 1993; Negeri et al., 2013; Zeier et al., 2004). This is assumed to be linked to the light‐dependent generation of ROS by chloroplasts (Allen et al., 1999; Ishikawa et al., 2014; Liu et al., 2007). The mitochondria are the other major source of ROS in the cell and the function of mitochondria, in particular the mitochondrial electron transport chain, has been linked to HR in a number of studies (Cvetkovska and Vanlerberghe, 2013; He et al., 2019; Xie and Chen, 2000). High humidity has also been shown to suppress or delay HR in some cases (Klement and Goodman, 1967; Wang et al., 2005), though this might simply be due to delayed dehydration of the cells.
Signal transduction, propagation and containment
Signal transduction
It is remarkable for such a well‐studied phenomenon that we do not understand the direct cause of cell death during HR. In mammalian apoptosis, a class of cysteine proteases called caspases orchestrate apoptotic cell death. These proteins have not been identified in plants (Dickman et al., 2017). It is generally assumed that R‐gene activation initiates a signalling cascade that leads to, among other things, HR. The components of this cascade have proved to be elusive, however. Repeated screens for mutants compromised in HR have generally identified the same few genes: RAR1, SGT1, HSP90 and the R‐gene itself (e.g. Hubert et al., 2009). Since RAR1, SGT1 and HSP90 are believed to work together to stabilize R‐proteins, the implication from these studies is that the pathway from the NLR to cell death is extremely short and/or there is significant genetic or physiological redundancy among the components of the HR signalling pathway such that mutagenesis screens are ineffective in identifying them. The recent finding that upon activation the NLR ZAR1 forms a homo‐pentamer that may form membrane pores suggests that the signalling pathway may indeed be very short or non‐existent (Wang et al., 2019a, b).
Cell‐to‐cell propagation and containment of the signal
In many cases HR is manifested as a single‐cell phenotype (e.g. Thordal‐Christensen et al., 1997; Hückelhoven et al., 1999), with the invaded cell being the only cell to show visible cell death (Fig. 2A). In other cases of HR initiated by incompatible interactions, the region of cell death appears to encompass tens or hundreds of cells, giving rise to spots on the leaf that are visible to the naked eye (Fig. 2B). In the case of multicellular HR, the question is whether HR has initiated in a single cell from which the cell death has then propagated, or whether each cell in the cluster of dead cells has independently initiated HR due to direct interaction with the effector protein. In the latter case this could be caused by a heavy initial inoculation such that many neighbouring cells may interact independently with a pathogen cell, or it may be caused by a phenomenon known as trailing necrosis in which HR does not completely stop pathogen growth, allowing the pathogen to grow or move from cell to cell, initiating HR in each cell in turn (Fig. 2C). This phenomenon has been documented in mutants or with treatments that partially suppress the defence response (Chivasa and Carr, 1998; Morel and Dangl, 1999).
Figure 2.

(A) Incompatible interaction between Colletotrichum lindemuthianum, causal agent of bean anthracnose and bean (Phaseolus vulgaris). HR is observed only in the cell which is directly invaded by the infection vesicle. Picture source: G. Johal (Purdue University). (B) Macroscopic HR symptoms during the interaction of Puccinia sorghi with a maize line carrying the Rp1D resistance gene. Source: Saet‐Byul Kim (NC State University). (C) Hyphae of the oomycete downy mildew Hyaloperonospora parasitica growing on resistant Arabidopsis thaliana with the presence of trailing necrosis (staining is trypan blue). Picture source: Emmanuel Boutetvia, WikiMedia Commons.
It seems likely that the causes of multicellular HR and the precise appearance of the lesions vary depending on the interaction being studied and the precise circumstances, and are shaped by interaction‐specific evolutionary forces that balance the damage to the plant due to the HR with the need to effectively control the pathogen. However, it does seem clear that in many cases there is a signal that is transmitted from cells undergoing the HR to their neighbours and, under the right circumstances, that this signal can cause HR cell death to spread to neighbouring cells. Perhaps the most compelling evidence for this comes from a study which investigated the interaction between HR and autophagy, the process by which cells degrade and recycle organelles and other components. The authors found that silencing ATG6, an important component of the autophagy mechanism, in Arabidopsis and tobacco conferred a runaway HR phenotype in which, once HR was initiated, cell death spread over the whole leaf and to neighbouring leaves instead of being restricted to the region in which it was initiated as it was in wild‐type plants (Liu et al., 2005; Patel and Dinesh‐Kumar, 2008) (Fig. 3). The tobacco HR in this study was caused by the recognition of a TMV protein p50‐helicase by the N protein. Interestingly, in other work it was reported that TMV was detected in living cells on the edges of HR lesions induced by N (Wright et al., 2000). This suggests that the mechanism for suppressing the spread of HR may be strong enough in certain circumstances to inhibit HR even if the appropriate NLR and its cognate effector occur together in the cell.
Figure 3.
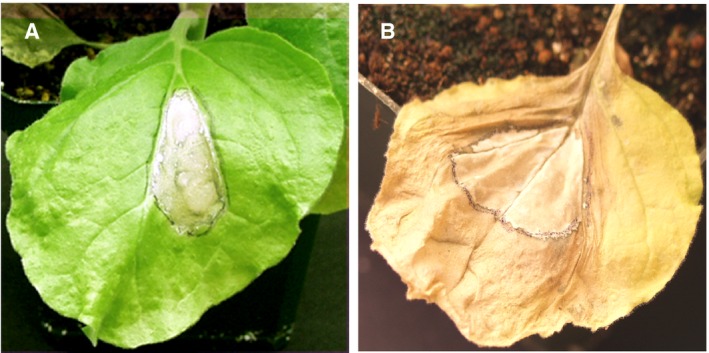
Transient expression of 50 kDa helicase domain (TMV‐p50) in Nicotiana benthamiana carrying the N gene which confers a HR in the presence of TMV‐p50. In panel B the NbBECLIN gene, which is required for autophagy, has been silenced by viral‐induced gene silencing (VIGS), causing HR to spread from the initial site of HR activation. Source: Dr. S.P. Dinesh‐Kumar (UC Davis).
Other evidence for the propagation of a cell death signal deriving from cells in which HR is initiated comes from the study of LES mutants caused by autoactive NLR mutants. In many cases spontaneous activation of HR in these mutants causes the production of macroscopic lesions (Bruggeman et al., 2015; Chintamanani et al., 2010). Genetic background has a profound effect on the size of the lesions induced by the autoactive maize NLR Rp1‐D21 (Fig. 4A). The major loci responsible for these background genetic effects have been mapped (Chaikam et al., 2011; Chintamanani et al., 2010; He et al., 2019; Olukolu et al., 2013, 2014) and several of the causal genes have been identified. They include components of the lignin biosynthesis pathway (Wang and Balint‐Kurti, 2016; Wang et al., 2016), the mitochondrial electron transport chain (He et al., 2019) and a polygalacturonase gene (Y. He et al., submitted). These genes are not absolutely required for HR, so they do not appear to be part of the primary signal transduction pathway but some may influence the strength of the response and its propagation from cell to cell.
Figure 4.

(A) Macroscopic lesions caused by the autoactive NLR Rp1‐D21 in maize. Upper image shows the phenotype conferred in a repressive background and the lower shows the phenotype in a permissive background. Source: P. Balint‐Kurti. (B) Leaf of a chimeric maize plant. The left sector carries the Rp1‐D21 autoactive NLR. The Rp1‐D21 gene has been mutated in the right sector so it is no longer functional. HR neither initiates nor propagates to the right sector. Source: P. Balint‐Kurti/S. Karre.
Chimeras in which Rp1‐D21 has been mutated in a section of the leaf show that HR neither initiates in or spreads to the sector that lacks Rp1‐D21 (author's unpublished data, Fig. 4B). This suggests that Rp1‐D21 is required for both the spontaneous initiation of HR and for sensitizing the cell so it is ‘competent’ to respond to a death signal propagated from the neighbouring cell. Interestingly, defence‐related gene expression was nevertheless elevated in the sectors carrying non‐functional Rp1‐D21, showing that Rp1‐D21 was not required for all aspects of the response to the signal. This is presumably the case with other autoactive NLR mutants (Bruggeman et al., 2015). In a related study, Bennetzen et al. (1988) used X‐ray mutagenesis and visible leaf colour mutations to generate marked leaf sectors lacking the maize NLR Rp1, which confers resistance to common rust and is the wild‐type allele of Rp1‐D21. They showed that HR induced by the incompatible rust pathogen isolate was cell autonomous. Sectors lacking Rp1 were susceptible to the fungus and HR induced in wild‐type sectors did not propagate to mutant sectors.
Taken together these data suggest that, once activated in a cell, the HR can transmit a signal inducing cell death in neighbouring cells. However, there appear to be other mechanisms responsible for the containment of this spreading cell death. The specific balance of these two processes may vary somewhat from situation to situation but in general HR lesions are restricted to one or just a few cells. When this balance is disrupted by genetical or experimental manipulation, larger necrotic lesions may develop.
These conclusions in turn suggest two further questions: What is the nature of the signal and what are the processes that act to suppress the cell death component of the response to the signal? As with many aspects of HR discussed here, the answer is unclear. It seems likely that the signal propagated from the cell undergoing HR usually includes ion fluxes and changes in the levels of ROS. Several LES and LES‐suppressor genes encode ion channel proteins, particularly for calcium (Bruggeman et al., 2015). For example, the BONZAI1 family genes BON1, 2 and 3 function in calcium signalling and their mutants exhibit LES phenotypes (Yang et al., 2006). Their phenotype is dependent on the action of EDS1 and PAD4 (Yang et al., 2006), which are involved in the function of many NLRs (Wiermer et al., 2005), suggesting a strong link to HR. BON1 and BON3 are negative regulators of several NLRs (Li et al., 2009) and the autoimmune phenotype of the bon1 mutant is dependent on the NLR SNC1 (Gou and Hua, 2012; Hua et al., 2001).
The role of ROS is suggested by several lines of evidence. ROS production from both the chloroplast and the mitochondria has been implicated in the initiation and spread of HR, and several LES genes are associated with chloroplast or mitochondrial function (Bruggeman et al., 2015; He et al., 2019). However, the best evidence for a role for ROS in propagation of a cell death signal may come from studies on the lesion simulating disease resistance 1 (LSD1) gene. Homozygous lsd1 mutants exhibit a spreading cell death phenotype which eventually encompasses the entire leaf (though not adjoining leaves) after initiation of cell death by treatment with incompatible pathogens or bioactive chemicals. Chemicals that generate ROS can initiate the spreading lesion phenotype in lsd1 mutants, and conditions that favour ROS accumulation can enhance it (Mateo et al., 2004), while addition of superoxide dismutase, an antioxidant enzyme that catalyses the dismutation of O2 −, suppresses lesion formation. LSD1 encodes a protein with three zinc finger domains which was shown to interact with and stimulates the activity of several catalases (Li et al., 2013). Its activity is dependent on the presence of the NLR ADR1 (Bonardi et al., 2011).
Consequences
Resistance
Perhaps surprisingly for what is the archetypal defence response, controversy remains on the precise role of HR in disease resistance. The argument for HR as a consequence or by‐product of the resistance response, rather than a cause of resistance, has been made a number of times over the last half‐century. Király et al. (1972) infected compatible hosts with the pathogenic oomycyte Phytophthora infestans and the fungi P. graminis and Uromyces phaseoli. They killed the pathogens but not the host tissue with chemical or heat treatments and observed host cell death in the infected host but not in uninfected controls. They surmised that cell death observed during HR was a consequence of the cessation of pathogen growth. In the intervening years of course, the characterization of multiple NLRs has shown that cell death can occur during HR independent of microbial presence.
However, other studies have from time to time indicated that resistance and HR can be uncoupled, for example:
Menna et al. (2015) reported that elevated temperature altered resistance and HR in quantitatively different and genotype‐dependent ways.
The TIR‐NBS‐LRR protein RPS6 confers resistance to P. syringae without visible cell death (Gassmann, 2005; Kim et al., 2009).
The RIN13 gene enhanced resistance conferred by RPM1 but abolished visible HR (Al‐Daoude et al., 2005).
The Arabidopsis NLR RPS4 conferred resistance to P. syringae in accessions Col‐0 and Ler, but while an HR is observed in Ler, no HR is observed in Col‐0 (Gassmann et al., 1999).
In cases where R‐proteins confer particularly rapid, high levels of resistance, HR may not occur. The tomato Cf‐9 gene for resistance to the fungus Cladosporium fulvum confers resistance in tomato to an avirulent C. fulvum isolate expressing the cognate Avr9 gene without HR (Hammond‐Kosack and Jones, 1994). However, in artificial circumstances when the Cf‐9 and Avr9 gene products are expressed together, high levels of cell death are observed (Hammond‐Kosack et al., 1994). This phenomenon has been called ‘extreme resistance’. Another example of this is the potato Rx R‐protein which confers resistance to potato virus X without HR (Tameling and Baulcombe, 2007). Much of this work and several further examples are summarized in a recent review (Künstler et al., 2016).
Ultimately, the question of whether HR cell death is a cause or consequence of resistance is perhaps not as interesting as it seems. While resistance and HR are unlinked in some situations, these remain the exception. The mechanisms of the resistance responses associated with activation of NLRs are multifaceted. In different cases it is likely that different aspects of the response are differentially effective in restricting pathogen growth and conferring resistance. In some of these cases cell wall fortification, ROS or calcium signalling responses may be the aspects that are most important and in those cases HR may not be necessary. In many other cases it is likely that HR is of primary importance. While the mechanisms of NLR activation vary between each NLR–effector interaction, they generally feed into common pathways in each particular host. It seems evolutionarily unfeasible that a host plant could evolve a series of subtly different NLR‐triggered defence responses perfectly honed to each pathogen encountered. Rather it seems likely that a common set of responses is invoked with different aspects of the response, including cell death, playing roles of varying importance in resistance, depending on the pathogen and environment.
Susceptibility
In several necrotrophic systems, most notably in the wheat/Parastagonospora nodorum interaction causing Septoria nodorum blotch (SNB) (Oliver et al., 2012), host‐specific toxins have been identified that cause cell death only on plants carrying dominant susceptibility (S) genes. In several cases the S‐genes have been shown to encode NLRs or proteins similar to PRRs (Faris et al., 2010; Lorang et al., 2007; Nagy and Bennetzen, 2008; Shi et al., 2016). The gene‐for‐gene paradigm is inverted in this case since, rather than being hindered by the HR, the necrotrophic pathogen is able to derive nutrition from the surrounding dead cells (Friesen et al., 2007; Lorang, 2018). It seems that in these cases HR is ‘deliberately’ invoked by the pathogen to induce the host to trigger PCD/HR and provide dead cells on which the necrotrophic pathogen can grow. It is not clear how many necrotrophic pathogens use this strategy beyond the four or five systems that have been characterized already (Lorang, 2018), but new cases are being discovered regularly.
Speciation
The activity of NLRs is often delicately regulated by a combination of intra‐ and interspecific protein interactions. Since the consequences of inappropriate activation are so profound, it seems likely that NLRs and the proteins that regulate their activity co‐evolve and that divergence of these proteins may occur within subpopulations of a species with low population gene flow. Within the genus Arabidopsis, the progeny of some crosses between species, or between distant accessions within a species, grow poorly and display a necrotic leaf phenotype (Chae et al., 2014; Vaid and Laitinen, 2019). In a number of these cases of so‐called hybrid necrosis, the causal genes are NLRs, one from each parent (Bomblies and Weigel, 2007). As discussed above, interactions between NLRs are common and are often important in appropriate inhibition and activation (Kourelis and van der Hoorn, 2018). In these cases, it is assumed that the interaction of two NLRs that have not co‐evolved within the same gene pool may cause their inappropriate activation. Similarly, the NLR‐interacting protein RIN4 was associated with hybrid necrosis in crosses between two species of lettuce (Jeuken et al., 2009). Hybrid necrosis, caused by inappropriate activation of HR, has therefore been suggested to be an example of a post‐zygotic reproductive barrier which can lead to reproductive isolation and ultimately to speciation (Bomblies et al., 2007). It is not clear how widespread this phenomenon is outside of Arabidopsis and its Brassicaceae relatives (Lafon‐Placette et al., 2016). Hybrid necrosis has also been observed in wheat and related species and while the causal genes have not been identified, it has been characterized as an autoimmune phenotype (Mizuno et al., 2011). The severity of hybrid necrosis in both Arabidopsis and wheat is enhanced by low temperatures as would be expected if it were based on NLR activity.
Systemic resistance: exploiting HR to engineer disease‐resistant plants
HR often can induce systemic acquired resistance (SAR): broad‐spectrum systemic enhanced resistance to pathogenic infection following a localized infection by a necrotizing pathogen or following treatment with various chemical agents (Grant and Lamb, 2006; Vallad and Goodman, 2004). It is dependent on the phytohormone salicylate (salicylic acid, SA), and associated with the accumulation of pathogenesis‐related (PR) proteins such as PR1, PR2 and PR5. The response to increased SA concentration is mediated through the proteins NPR1, NPR3 and NPR4, with NPR3 and NPR4 acting as SA receptors and mediating the action of the transcriptional regulator NPR1 which is considered the ‘master regulator’ of the SAR responses (Fu and Dong, 2013).
The high costs of producing and deregulating transgenic plants means that transgenically conferred disease resistance must be broad spectrum in order to be economically feasible as a commercial product. The ability of HR to induce SAR has encouraged approaches to manipulating HR to confer broad‐spectrum resistance. Both HR itself and the induction of SAR can confer yield penalties (Durrant and Dong, 2004) so these approaches have focused on inducing HR and SAR at the specific times and places they are most required.
Initial efforts used pathogen‐inducible promoters to induce HR by expressing autoactive NLRs or the cognate effector of an NLR endogenous to the host plant (Stuiver and Custers, 2001). These approaches generally failed since the disease resistance provided was insufficient or the yield penalties were too high (Hammond‐Kosack and Parker, 2003). Both mild and extreme leaf flecking (as described above) are associated with broad‐spectrum resistance (Olukolu et al., 2016). While more extreme flecking was also associated with a yield penalty, milder flecking was not. While much more research is required, this suggests that there may be circumstances under which the low‐level activation of HR leading to SAR may be useful for imparting disease resistance without unreasonable yield penalties. One approach might be to use development‐specific promoters that direct expression after anthesis, a period when many diseases become more prevalent, but after the majority of growth has been completed.
Conclusions
The ‘arms race’ between hosts and their parasites has had profound effects on the evolution of life on earth. The process of sexual reproduction itself may have evolved due to the need for the host to adapt to and evade disease pressure (Ebert and Hamilton, 1996). HR induced by NLR proteins is a remarkably widespread phenomenon across the plant kingdom and has profound effects on many aspects of plant growth and development beyond disease resistance. A sophisticated architecture has evolved to control HR activation with multiple levels of control (summarized in Fig. 5). Nevertheless inappropriate activation can still occur and leads to deleterious phenotypes such as lesion mimics which retard growth, susceptibility to necrotrophic pathogens and hybrid necrosis, and possible subsequent speciation (Fig. 5). Most species of plants carry more than 100 NLRs in their genomes (Arabidopsis has more than 100, tomato more than 400). The fact that plants maintain so many of these potentially lethal genes, as well as the cumbersome machinery to contain their deleterious effects, is a measure of how important disease resistance, and this mechanism in particular, has been during plant evolution.
Figure 5.
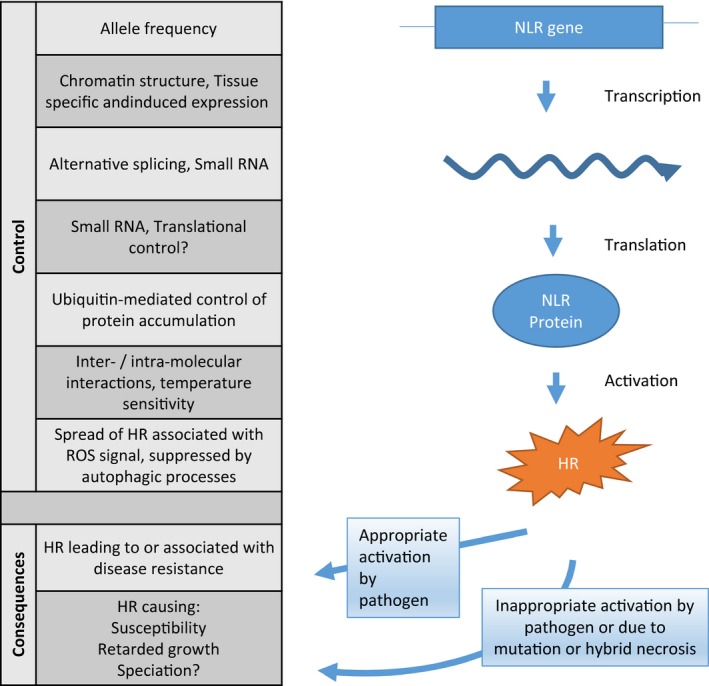
A summary of the processes controlling HR and the consequences of HR discussed in this review. The process of NLR transcription, translation and activation is shown on the right while the table on the left indicates the levels of control and consequences of activation. ‘Appropriate activation’ refers to the mode of activation that was selected for during evolution and ‘inappropriate activation’ refers to activation that has detrimental effects on plant growth and survival.
Acknowledgements
The author acknowledges helpful discussions which have informed various parts of this review with the following colleagues: Jane Glazebrook, Jeff Dangl, Guan‐Feng Wang, Fumiaki Katagiri, Christina Cowger, David Marshall, Cyril Zipfel, Jeff Bennetzen, Jane Parker, Kim Hammond‐Kosack, Shailesh Karre and Saet‐Byul Kim. In particular the author would like to thank Guri Johal for his generosity in sharing his time and ideas during our long‐standing collaborations in this area. Much of the work in the Balint‐Kurti laboratory on HR has its roots in ideas conceived and funding secured by Dr Johal. PBK was supported by NSF Plant Genome grants #1127076, #0822495 and #1444503.
References
- Abdul‐Baki, A.A. , Haroon, S.A. and Chitwood, D.J. (1996) Temperature effects on resistance to Meloidogyne spp. in excised tomato roots. HortScience, 31, 147–149. [Google Scholar]
- Alcázar, R. and Parker, J.E. (2011) The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16, 666–675. [DOI] [PubMed] [Google Scholar]
- Al‐Daoude, A. , de Torres Zabala, M. , Ko, J.‐H. and Grant, M. (2005) RIN13 is a positive regulator of the plant disease resistance protein RPM1. Plant Cell, 17, 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, L.J. , MacGregor, K.B. , Koop, R.S. , Bruce, D.H. , Karner, J. and Bown, A.W. (1999) The relationship between photosynthesis and a mastoparan‐induced hypersensitive response in isolated mesophyll cells. Plant Physiol. 119, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aust, H. (1974) The influence of temperature on infection by powdery mildew (Erysiphe graminis DC. f. sp. hordei Marchal). Z. Pflanzenkr. Pflanzenschutz, 81, 597–601. [Google Scholar]
- Azevedo, C. , Betsuyaku, S. , Peart, J. , Takahashi, A. , Noel, L. , Sadanandom, A. , Casais, C. , Parker, J. and Shirasu, K. (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25, 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs, E. , Dagdas, G. and Krasileva, K.V. (2017) NLR diversity, helpers and integrated domains: making sense of the NLR IDentity. Curr. Opin. Plant Biol. 38, 59–67. [DOI] [PubMed] [Google Scholar]
- Bao, F. , Huang, X. , Zhu, C. , Zhang, X. , Li, X. and Yang, S. (2014) Arabidopsis HSP90 protein modulates RPP4‐mediated temperature‐dependent cell death and defense responses. New Phytol. 202, 1320–1334. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. , Blevins, W.E. and Ellingboe, A.H. (1988) Cell‐autonomous recognition of the rust pathogen determines Rp1‐specified resistance in maize. Science, 241, 208–210. [DOI] [PubMed] [Google Scholar]
- Bieri, S. , Mauch, S. , Shen, Q.‐H. , Peart, J. , Devoto, A. , Casais, C. , Ceron, F. , Schulze, S. , Steinbiß, H.‐H. , Shirasu, K. and Schulze‐Lefert, P. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell, 16, 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch, P.R. , Avrova, A.O. , Dellagi, A. , Lacomme, C. , Cruz, S.S. and Lyon, G.D. (2018) Programmed cell death in plants in response to pathogen attack. Annu. Plant Rev. 4, 184 – 208. [Google Scholar]
- Bomblies, K. and Weigel, D. (2007) Hybrid necrosis: autoimmunity as a potential gene‐flow barrier in plant species. Nat. Rev. Genet. 8, 382 – 393. [DOI] [PubMed] [Google Scholar]
- Bomblies, K. , Lempe, J. , Epple, P. , Warthmann, N. , Lanz, C. , Dangl, J.L. and Weigel, D. (2007) Autoimmune response as a mechanism for a Dobzhansky–Muller‐type incompatibility syndrome in plants. PLoS Biol. 5, e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi, V. , Tang, S. , Stallmann, A. , Roberts, M. , Cherkis, K. and Dangl, J.L. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA. 108, 16463 – 16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi, V. , Cherkis, K. , Nishimura, M.T. and Dangl, J.L. (2012) A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr. Opin. Immunol. 24, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch, C. , Hwang, C.‐F. , Navarre, D.A. and Williamson, V.M. (2004) Salicylic acid is part of the Mi‐1‐mediated defense response to root‐knot nematode in tomato. Mol. Plant‐Microbe Interact. 17, 351–356. [DOI] [PubMed] [Google Scholar]
- Bromfield, K. (1961) Effect of postinoculation temperatures on seedling reaction of selected wheat varieties to stem rust. Phytopathology, 51, 590. [Google Scholar]
- Bruggeman, Q. , Raynaud, C. , Benhamed, M. and Delarue, M. (2015) To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, W. (1981) Incompatibility conditioned by the Mla gene in powdery mildew of barley: the halt in cytoplasmic streaming. Phytopathology, 71, 1062 – 1066. [Google Scholar]
- Catlett, M.G. and Kaplan, K.B. (2006) Sgt1p is a unique co‐chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J. Biol. Chem. 281, 33739–33748. [DOI] [PubMed] [Google Scholar]
- Chae, E. , Bomblies, K. , Kim, S.T. , Karelina, D. , Zaidem, M. , Ossowski, S. , Martin‐Pizarro, C. , Laitinen, R.A. , Rowan, B.A. , Tenenboim, H. , Lechner, S. , Demar, M. , Habring‐Muller, A. , Lanz, C. , Ratsch, G. and Weigel, D. (2014) Species‐wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell, 159, 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikam, V. , Negeri, A. , Dhawan, R. , Puchaka, B. , Ji, J. , Chintamanani, S. , Gachomo, E. , Zillmer, A. , Doran, T. , Weil, C. , Balint‐Kurti, P. and Johal, G. (2011) Use of mutant‐assisted gene identification and characterization (MAGIC) to identify novel genetic loci that modify the maize hypersensitive response. Theor. Appl. Genet. 123, 985–997. [DOI] [PubMed] [Google Scholar]
- Chandra‐Shekara, A.C. , Gupte, M. , Navarre, D. , Raina, S. , Raina, R. , Klessig, D. and Kachroo, P. (2006) Light‐dependent hypersensitive response and resistance signaling against turnip crinkle virus in Arabidopsis. Plant J. 45, 320–334. [DOI] [PubMed] [Google Scholar]
- Chen, C. and Heath, M.C. (1991) Cytological studies of the hypersensitive death of cowpea epidermal cells induced by basidiospore‐derived infection by the cowpea rust fungus. Can. J. Bot. 69, 1199–1206. [Google Scholar]
- Chen, B. , Niu, F. , Liu, W.‐Z. , Yang, B. , Zhang, J. , Ma, J. , Cheng, H. , Han, F. and Jiang, Y.‐Q. (2016) Identification, cloning and characterization of R2R3‐MYB gene family in canola (Brassica napus L.) identify a novel member modulating ROS accumulation and hypersensitive‐like cell death. DNA Res. 23, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C. , Gao, X. , Feng, B. , Sheen, J. , Shan, L. and He, P. (2013) Plant immune response to pathogens differs with changing temperatures. Nat. Commun. 4, 2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintamanani, S. , Hulbert, S.H. , Johal, G.S. and Balint‐Kurti, P.J. (2010) Identification of a maize locus that modulates the hypersensitive defense response, using mutant‐assisted gene identification and characterization. Genetics, 184, 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa, S. and Carr, J.P. (1998) Cyanide restores N gene‐mediated resistance to tobacco mosaic virus in transgenic tobacco expressing salicylic acid hydroxylase. Plant Cell, 10, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, N.S. , Epple, P. and Dangl, J.L. (2011) Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovska, M. and Vanlerberghe, G.C. (2013) Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ. 36, 721 – 732. [DOI] [PubMed] [Google Scholar]
- Delledonne, M. , Zeier, J. , Marocco, A. and Lamb, C. (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA. 98, 13454 – 13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman, M. , Williams, B. , Li, Y. , de Figueiredo, P. and Wolpert, T. (2017) Reassessing apoptosis in plants. Nat. Plants, 3, 773 – 779. [DOI] [PubMed] [Google Scholar]
- Doke, N. and Tomiyama, K. (1975) Effect of blasticidin S on hypersensitive death of potato leaf petiole cells caused by infection with an incompatible race of Phytophthora infestans . Physiol. Plant Pathol. 6, 169–175. [Google Scholar]
- Dong, O.X. , Ao, K. , Xu, F. , Johnson, K.C. , Wu, Y. , Li, L. , Xia, S. , Liu, Y. , Huang, Y. and Rodriguez, E. (2018) Individual components of paired typical NLR immune receptors are regulated by distinct E3 ligases. Nat. Plants, 4, 699. [DOI] [PubMed] [Google Scholar]
- Dropkin, V.H. (1969) The necrotic reaction of tomatoes and other hosts resistant to Meloidogyne: reversal by temperature. Phytopathology, 59, 1632 – 1637. [Google Scholar]
- Durrant, W.E. and Dong, X. (2004) Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Ebert, D. and Hamilton, W.D. (1996) Sex against virulence: the coevolution of parasitic diseases. Trends Ecol. Evol. 11, 79–82. [DOI] [PubMed] [Google Scholar]
- Faris, J.D. , Zhang, Z. , Lu, H. , Lu, S. , Reddy, L. , Cloutier, S. , Fellers, J.P. , Meinhardt, S.W. , Rasmussen, J.B. , Xu, S.S. , Oliver, R.P. , Simons, K.J. and Friesen, T.L. (2010) A unique wheat disease resistance‐like gene governs effector‐triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA. 107, 13544 – 13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan, A. , Turnbull, D. , Stevens, L.J. , Engelhardt, S. , Birch, P.R. , Hein, I. and Gilroy, E.M. (2015) The hypersensitive response in PAMP‐and effector‐triggered immune responses In: Plant Programmed Cell Death, (Gunawardena A. and McCabe P. eds), pp. 235 – 268. Cham: Springer. [Google Scholar]
- Ferdinandsen, C. and Winge, Ö. (1930) A heritable blotch leaf in oats. Hereditas, 13, 164–176. [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Friesen, T.L. , Meinhardt, S.W. and Faris, J.D. (2007) The Stagonospora nodorum‐wheat pathosystem involves multiple proteinaceous host‐selective toxins and corresponding host sensitivity genes that interact in an inverse gene‐for‐gene manner. Plant J. 51, 681–692. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. and Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Wang, W. , Zhang, T. , Gong, Z. , Zhao, H. and Han, G.‐Z. (2018) Out of water: the origin and early diversification of plant R‐genes. Plant Physiol. 177, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, K.A. , Nita, M. , De Wolf, E.D. , Esker, P.D. , Gomez‐Montano, L. and Sparks, A.H. (2016) Chapter 21. Plant pathogens as indicators of climate change In: Climate Change 2nd edn (Letcher T.M., ed), pp. 325–338. Boston: Elsevier. [Google Scholar]
- Gassmann, W. (2005) Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol. Plant‐Microbe Interact. 18, 1054–1060. [DOI] [PubMed] [Google Scholar]
- Gassmann, W. , Hinsch, M.E. and Staskawicz, B.J. (1999) The Arabidopsis RPS4 bacterial‐resistance gene is a member of the TIR‐NBS‐LRR family of disease‐resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Gaumann, E. (1950) Principles of Plant Infection. London: Crosby Lockwood and Son, Ltd. [Google Scholar]
- Goodman, R.N. and Novacky, A.J. (1994) The hypersensitive reaction in plants to pathogens: a resistance phenomenon. Chicago: American Phytopathological Society; [Google Scholar]
- Gou, M. and Hua, J. (2012) Complex regulation of an R gene SNC1 revealed by autoimmune mutants. Plant Signal. Behav. 7, 213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousseau, H.D.M. , Deverall, B.J. and McIntosh, R.A. (1985) Temperature‐sensitivity of the expression of resistance to Puccinia graminis conferred by the Sr15, Sr9b and Sr14 genes in wheat. Physiol. Plant Pathol. 27, 335 – 343. [Google Scholar]
- Grant, M. and Lamb, C. (2006) Systemic immunity. Curr. Opin. Plant Biol. 9, 414–420. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997) Programmed cell death in plant‐pathogen interactions. Annu. Rev. Plant Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Gross, P. , Julius, C. , Schmelzer, E. and Hahlbrock, K. (1993) Translocation of cytoplasm and nucleus to fungal penetration sites is associated with depolymerization of microtubules and defence gene activation in infected, cultured parsley cells. EMBO J. 12, 1735–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, A. , Reimers, P.J. and Leach, J.E. (1993) Effect of light on incompatible interactions between Xanthomonas oryzae pv oryzae and rice. Physiol. Mol. Plant Pathol. 42, 413–425. [Google Scholar]
- Hammond‐Kosack, K.E. and Jones, J.D. (1994) Incomplete dominance of tomato Cf genes for resistance to Cladosporium fulvum . Mol. Plant‐Microbe Interact. 7, 58–58. [Google Scholar]
- Hammond‐Kosack, K.E. and Jones, J. (1996) Resistance gene‐dependent plant defense responses. Plant Cell, 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Parker, J.E. (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. , Harrison, K. and Jones, J.D. (1994) Developmentally regulated cell death on expression of the fungal avirulence gene Avr9 in tomato seedlings carrying the disease‐resistance gene Cf‐9 . Proc. Natl. Acad. Sci. USA. 91, 10445 – 10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder, D.E. , Samborski, D.J. , Rohringer, R. , Rimmer, S.R. , Kim, W.K. and Chong, J. (1979) Electron microscopy of susceptible and resistant near‐isogenic (sr6/Sr6) lines of wheat infected by Puccinia graminis tritici. III. Ultrastructure of incompatible interact. Can. J. Bot. 57, 2626–2634. [Google Scholar]
- He, Y. , Kim, S.‐B. and Balint‐Kurti, P. (2019) A maize cytochrome b‐c1 complex subunit protein ZmQCR61 controls variation in the hypersensitive response. Planta, 249, 1477–1485. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000a) Advances in imaging the cell biology of plant‐microbe interactions. Annu. Rev. Phytopathol. 38, 443–459. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000b) Hypersensitive response‐related death. Plant Mol. Biol. 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Heidrich, K. , Tsuda, K. , Blanvillain‐Baufume, S. , Wirthmueller, L. , Bautor, J. and Parker, J.E. (2013) Arabidopsis TNL‐WRKY domain receptor RRS1 contributes to temperature‐conditioned RPS4 auto‐immunity. Front. Plant Sci. 4, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, B.F. , Belkhadir, Y. and Dangl, J.L. (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science, 309, 929–932. [DOI] [PubMed] [Google Scholar]
- Hua, J. , Grisafi, P. , Cheng, S.‐H. and Fink, G.R. (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 15, 2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Li, J. , Bao, F. , Zhang, X. and Yang, S. (2010) A gain‐of‐function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 154, 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Chen, X. , Zhong, X. , Li, M. , Ao, K. , Huang, J. and Li, X. (2016) Plant TRAF proteins regulate NLR immune receptor turnover. Cell Host Microbe, 19, 204–215. [DOI] [PubMed] [Google Scholar]
- Hubert, D.‐A. , Tornero, P. , Belkhadir, Y. , Krishna, P. , Takahashi, A. , Shirasu, K. and Dangl, J.‐L. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22, 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert, D.A. , He, Y. , McNulty, B.C. , Tornero, P. and Dangl, J.L. (2009) Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 cochaperone function in plant NB‐LRR disease resistance protein regulation. Proc. Natl. Acad. Sci. USA. 106, 9556 – 9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven, R. , Fodor, J. , Preis, C. and Kogel, K.‐H. (1999) Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119, 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, K. , Yamaguchi, K. , Sakamoto, K. , Yoshimura, S. , Inoue, K. , Tsuge, S. , Kojima, C. and Kawasaki, T. (2014) Bacterial effector modulation of host E3 ligase activity suppresses PAMP‐triggered immunity in rice. Nat. Commun. 5, 5430 [DOI] [PubMed] [Google Scholar]
- Jeuken, M.J.W. , Zhang, N.W. , McHale, L.K. , Pelgrom, K. , den Boer, E. , Lindhout, P. , Michelmore, R.W. , Visser, R.G.F. and Niks, R.E. (2009) Rin4 Causes hybrid necrosis and race‐specific resistance in an interspecific lettuce hybrid. Plant Cell, 21, 3368–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch, H. (1966) The role of host genes, temperature and polyphenoloxidase in the necrotization of TMV infected tobacco tissue. J. Phytopathol. 55, 185–192. [Google Scholar]
- Johal, G.S. (2007) Disease lesion mimic mutants of maize In: American Phytopahtological Society Feature Story. Available at https://www.apsnet.org/edcenter/apsnetfeatures/Pages/MutantsofMaize.aspx. [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. , Vance, R.E. and Dangl, J.L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science, 354, aaf6395. [DOI] [PubMed] [Google Scholar]
- Jupe, J. , Stam, R. , Howden, A.J.M. , Morris, J.A. , Zhang, R. , Hedley, P.E. and Huitema, E. (2013) Phytophthora capsici‐tomato interaction features dramatic shifts in gene expression associated with a hemi‐biotrophic lifestyle. Genome Biol. 14, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. , Huitema, E. and Vleeshouwers, V.G.A.A. (1999) Resistance to oomycetes: a general role for the hypersensitive response? Trends Plant Sci. 4, 196–200. [DOI] [PubMed] [Google Scholar]
- Karasov, T.L. , Kniskern, J.M. , Gao, L. , DeYoung, B.J. , Ding, J. , Dubiella, U. , Lastra, R.O. , Nallu, S. , Roux, F. , Innes, R.W. , Barrett, L.G. , Hudson, R.R. and Bergelson, J. (2014) The long‐term maintenance of a resistance polymorphism through diffuse interactions. Nature, 512, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N. , Ersek, T. , Long, M. , Bruegger, B. and Holliday, M. (1981) Inhibition of the hypersensitive reaction of soybean leaves to incompatible Pseudomonas spp. by blasticidin S, streptomycin or elevated temperature. Physiol. Plant Pathol. 18, 325–337. [Google Scholar]
- Kim, S.H. , Kwon, S.I. , Saha, D. , Anyanwu, N.C. and Gassmann, W. (2009) Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR‐NBS‐LRR Protein RPS6 and is enhanced by mutations in SRFR1 . Plant Physiol. 150, 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.‐B. , Lee, H.‐Y. , Choi, E.‐H. , Park, E. , Kim, J.‐H. , Moon, K.‐B. , Kim, H.‐S. and Choi, D. (2018) The coiled‐coil and leucine‐rich repeat domain of the potyvirus resistance protein Pvr4 has a distinct role in signaling and pathogen recognition. Mol. Plant‐Microbe Interact. 31, 906–913. [DOI] [PubMed] [Google Scholar]
- Király, Z. , Barna, B. and ÉRsek, T. (1972) Hypersensitivity as a consequence, not the cause, of plant resistance to infection. Nature, 239, 456. [Google Scholar]
- Király, L. , Hafez, Y. , Fodor, J. and Király, Z. (2008) Suppression of tobacco mosaic virus‐induced hypersensitive‐type necrotization in tobacco at high temperature is associated with downregulation of NADPH oxidase and superoxide and stimulation of dehydroascorbate reductase. J. Gen. Virol. 89, 799–808. [DOI] [PubMed] [Google Scholar]
- Kitazawa, K. , Inagaki, H. and Tomiyama, K. (1973) Cinephotomicrographic observations on the dynamic responses of protoplasm of a potato plant cell to infection by phytophthora infestani 1. J. Phytopathol. 76, 80–86. [Google Scholar]
- Klement, Z. and Goodman, R. (1967) The hypersensitive reaction to infection by bacterial plant pathogens. Annu. Rev. Phytopathol. 5, 17–44. [Google Scholar]
- Kobayashi, I. , Kobayashi, Y. and Hardham, A.R. (1994) Dynamic reorganization of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta, 195, 237–247. [Google Scholar]
- Kourelis, J. and van der Hoorn, R.A.L. (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell, 30, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D. and Kirti, P.B. (2015) Pathogen‐induced SGT1 of Arachis diogoi induces cell death and enhanced disease resistance in tobacco and peanut. Plant Biotechnol. J. 13, 73–84. [DOI] [PubMed] [Google Scholar]
- Kumar, S.V. and Wigge, P.A. (2010) H2A.Z‐containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell, 140, 136–147. [DOI] [PubMed] [Google Scholar]
- Künstler, A. , Bacsó, R. , Gullner, G. , Hafez, Y.M. and Király, L. (2016) Staying alive – is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 93, 75–84. [Google Scholar]
- Lafon‐Placette, C. , Vallejo‐Marín, M. , Parisod, C. , Abbott, R.J. and Köhler, C. (2016) Current plant speciation research: unravelling the processes and mechanisms behind the evolution of reproductive isolation barriers. New Phytol. 209, 29–33. [DOI] [PubMed] [Google Scholar]
- Lai, Y. and Eulgem, T. (2018) Transcript‐level expression control of plant NLR genes. Mol. Plant Pathol. 19, 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E. , Kato, N. and Lawton, M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature, 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Pennington, B.O. and Hua, J. (2009) Multiple R‐like genes are negatively regulated by BON1 and BON3 in Arabidopsis. Mol. Plant‐Microbe Interact. 22, 840–848. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Chen, L. , Mu, J. and Zuo, J. (2013) LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Czymmek, K. , TallÃ3czy, Z. , Levine, B. , and Dinesh‐Kumar, S.P. (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell, 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Ren, D. , Pike, S. , Pallardy, S. , Gassmann, W. and Zhang, S. (2007) Chloroplast‐generated reactive oxygen species are involved in hypersensitive response‐like cell death mediated by a mitogen‐activated protein kinase cascade. Plant J. 51, 941–954. [DOI] [PubMed] [Google Scholar]
- Lo Presti, L. , Lanver, D. , Schweizer, G. , Tanaka, S. , Liang, L. , Tollot, M. , Zuccaro, A. , Reissmann, S. and Kahmann, R. (2015) Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. [DOI] [PubMed] [Google Scholar]
- Loegering, W.Q. and Ellingboe, A.H. (1987) Flor, H.H. – pioneer in phytopathology. Annu. Rev. Phytopathol. 25, 59–66. [DOI] [PubMed] [Google Scholar]
- Lorang, J.M. (2018) Necrotrophic exploitation and subversion of plant defense: a lifestyle or just a phase, and implications in breeding resistance. Phytopathology, 109, 332–346. [DOI] [PubMed] [Google Scholar]
- Lorang, J.M. , Sweat, T.A. and Wolpert, T.J. (2007) Plant disease susceptibility conferred by a ‘resistance’ gene. Proc. Natl. Acad. Sci. USA. 104, 14861 – 14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makepeace, J.C. , Oxley, S.J.P. , Havis, N.D. , Hackett, R. , Burke, J.I. and Brown, J.K.M. (2007) Associations between fungal and abiotic leaf spotting and the presence of mlo alleles in barley. Plant Pathol. 56, 934 – 942. [Google Scholar]
- Mateo, A. , Mühlenbock, P. , Rustérucci, C. , Chang, C.C.‐C. , Miszalski, Z. , Karpinska, B. , Parker, J.E. , Mullineaux, P.M. and Karpinski, S. (2004) LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136, 2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna, A. , Nguyen, D. , Guttman, D.S. and Desveaux, D. (2015) Elevated temperature differentially influences effector‐triggered immunity outputs in Arabidopsis. Front. Plant Sci. 6 , 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre, P. and Baulcombe, D.C. (2006) Elicitor‐mediated oligomerization of the tobacco N disease resistance protein. Plant Cell, 18, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, N. , Shitsukawa, N. , Hosogi, N. , Park, P. and Takumi, S. (2011) Autoimmune response and repression of mitotic cell division occur in inter‐specific crosses between tetraploid wheat and Aegilops tauschii Coss. that show low temperature‐induced hybrid necrosis. Plant J. 68, 114–128. [DOI] [PubMed] [Google Scholar]
- Mohamed, A. , Ellicott, A. , Housley, T. and Ejeta, G. (2003) Hypersensitive response to Striga infection in sorghum. Crop Sci. 43, 1320–1324. [Google Scholar]
- Mohr, T.J. , Mammarella, N.D. , Hoff, T. , Woffenden, B.J. , Jelesko, J.G. and McDowell, J.M. (2010) The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol. Plant‐Microbe Interact. 23, 1303–1315. [DOI] [PubMed] [Google Scholar]
- Morel, J.‐B. and Dangl, J.L. (1999) Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics, 151, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch, S. , Lingner, U. , Floss, D.S. , Ludwig, N. , Sauer, N. and Deising, H.B. (2008) The hemibiotrophic lifestyle of Colletotrichum species. J. Plant Physiol. 165, 41–51. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Lloyd, A.J. , Ougham, H. and Prats, E. (2008) The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501 – 520. [DOI] [PubMed] [Google Scholar]
- Muralidharan, S. , Box, M.S. , Sedivy, E.L. , Wigge, P.A. , Weigel, D. and Rowan, B.A. (2014) Different mechanisms for Arabidopsis thaliana hybrid necrosis cases inferred from temperature responses. Plant Biol., 16, 1033 – 1041. [DOI] [PubMed] [Google Scholar]
- Na, J.‐K. , Kim, J.‐K. , Kim, D.‐Y. and Assmann, S.M. (2015) Expression of potato RNA‐binding proteins StUBA2a/b and StUBA2c induces hypersensitive‐like cell death and early leaf senescence in Arabidopsis. J. Exp. Bot. 66, 4023–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, E.D. and Bennetzen, J.L. (2008) Pathogen corruption and site‐directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, S. and Tomar, S.M.S. (2001) Genetical and anatomical analyses of a leaf flecking mutant in Triticum aestivum L. Euphytica, 121, 53–58. [Google Scholar]
- Naton, B. , Hahlbrock, K. and Schmelzer, E. (1996) Correlation of rapid cell death with metabolic changes in fungus‐infected, cultured parsley cells. Plant Physiol. 112, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negeri, A. , Wang, G.‐F. , Benavente, L. , Kibiti, C. , Chaikam, V. , Johal, G. and Balint‐Kurti, P. (2013) Characterization of temperature and light effects on the defense response phenotypes associated with the maize Rp1‐D21 autoactive resistance gene. BMC Plant Biol. 13, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue, M. , Tomiyama, K. and Doke, N. (1977) Effect of blasticidin S on development of potential of potato tuber cells to react hypersensitively to infection by Phytophthora infestans . Physiol. Plant Pathol. 10, 181–189. [Google Scholar]
- Oliver, R.P. , Friesen, T.L. , Faris, J.D. and Solomon, P.S. (2012) Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50, 23–43. [DOI] [PubMed] [Google Scholar]
- Olukolu, B.A. , Negeri, A. , Dhawan, R. , Venkata, B.P. , Sharma, P. , Garg, A. , Gachomo, E. , Marla, S. , Chu, K. , Hasan, A. , Ji, J. , Chintamanani, S. , Green, J. , Shyu, C.‐R. , Wisser, R.J. , Holland, J.B. , Johal, G. and Balint‐Kurti, P. (2013) A connected set of genes associated with programmed cell death implicated in controlling the hypersensitive response in maize. Genetics, 193, 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olukolu, B.A. , Wang, G.‐F. , Vontimitta, V. , Venkata, B.P. , Marla, S. , Ji, J. , Gachomo, E. , Chu, K. , Negeri, A. , Benson, J. , Nelson, R. , Bradbury, P. , Nielsen, D. , Holland, J.B. , Balint‐Kurti, P.J. and Johal, G. (2014) A genome‐wide association study of the maize hypersensitive defense response identifies genes that cluster in related pathways. PLoS Genet. 10, e1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olukolu, B.A. , Bian, Y. , De Vries, B. , Tracy, W.F. , Wisser, R.J. , Holland, J.B. and Balint‐Kurti, P.J. (2016) The genetics of leaf flecking in maize and its relationship to plant defense and disease resistance. Plant Physiol. 172, 1787–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett, H.S. , Watanabe, Y. and Beachy, R.N. (1997) Identification of the TMV replicase sequence that activates the N gene‐mediated hypersensitive response. Mol. Plant‐Microbe Interact. 10, 709–715. [Google Scholar]
- Patel, S. and Dinesh‐Kumar, S.P. (2008) Arabidopsis ATG6 is required to limit the pathogen‐associated cell death response. Autophagy, 4, 20–27. [DOI] [PubMed] [Google Scholar]
- Pietikäinen, J. , Pettersson, M. and Bååth, E. (2005) Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Ecol. 52, 49–58. [DOI] [PubMed] [Google Scholar]
- Pryor, T. (1994) Maize and Puccinia sorghi: a system for the study of the genetic and molecular basis of host‐pathogen interactions In: The Maize Handbook, (Freeling M. and Walbot V., eds), pp. 286 – 290. New York: Springer. [Google Scholar]
- Ramegowda, V. and Senthil‐Kumar, M. (2015) The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 176, 47–54. [DOI] [PubMed] [Google Scholar]
- Rossi, M. , Goggin, F.L. , Milligan, S.B. , Kaloshian, I. , Ullman, D.E. and Williamson, V.M. (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA. 95, 9750 – 9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryerson, D.E. and Heath, M.C. (1996) Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or by abiotic treatments. Plant Cell, 8, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E.‐E. , Monks, S.‐A. , Drake, T.‐A. , Lusis, A.‐J. , Che, N. , Colinayo, V. , Ruff, T.‐G. , Milligan, S.‐B. , Lamb, J.‐R. , Cavet, G. , Linsley, P.‐S. , Mao, M. , Stoughton, R.‐B. and Friend, S.‐H. (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature, 422, 297 – 302. [DOI] [PubMed] [Google Scholar]
- Schreiber, K.J. , Bentham, A. , Williams, S.J. , Kobe, B. and Staskawicz, B.J. (2016) Multiple domain associations within the Arabidopsis immune receptor RPP1 regulate the activation of programmed cell death. PLoS Pathog. 12, e1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield, S.R. , Huang, L. , Brandt, A.S. and Gill, B.S. (2005) Development of a virus‐induced gene‐silencing system for hexaploid wheat and its use in functional analysis of the Lr21‐mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Z.‐Q. , Xue, J.‐Y. , Wang, Q. , Wang, B. and Chen, J.‐Q. (2019) Revisiting the origin of plant NBS‐LRR genes. Trends Plant Sci. 24, 9–12. [DOI] [PubMed] [Google Scholar]
- Shi, G. , Zhang, Z. , Friesen, T.L. , Raats, D. , Fahima, T. , Brueggeman, R.S. , Lu, S. , Trick, H.N. , Liu, Z. , Chao, W. , Frenkel, Z. , Xu, S.S. , Rasmussen, J.B. and Faris, J.D. (2016) The hijacking of a receptor kinase–driven pathway by a wheat fungal pathogen leads to disease. Science Advances, 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, E.A. , Dwyer, G. , Mauricio, R. , Kreitman, M. and Bergelson, J. (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature, 400, 667–671. [DOI] [PubMed] [Google Scholar]
- Stakman, E. (1915) Relation between Puccinia graminis and plants highly resistant to its attack. J Agric. Res. 4, 193–200. [Google Scholar]
- Stuiver, M.H. and Custers, J.H. (2001) Engineering disease resistance in plants. Nature, 411, 865. [DOI] [PubMed] [Google Scholar]
- Swarbrick, P.J. , Huang, K. , Liu, G. , Slate, J. , Press, M.C. and Scholes, J.D. (2008) Global patterns of gene expression in rice cultivars undergoing a susceptible or resistant interaction with the parasitic plant Striga hermonthica . New Phytol. 179, 515–529. [DOI] [PubMed] [Google Scholar]
- Tameling, W.I. and Baulcombe, D.C. (2007) Physical association of the NB‐LRR resistance protein Rx with a Ran GTPase‐activating protein is required for extreme resistance to potato virus X. Plant Cell, 19, 1682–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X. , Meyers, B.C. , Kozik, A. , West, M.A. , Morgante, M. , St Clair, D.A. , Bent, A.F. and Michelmore, R.W. (2007) Global expression analysis of nucleotide binding site‐leucine rich repeat‐encoding and related genes in Arabidopsis. BMC Plant Biol. 7, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Tian, D. , Traw, M.B. , Chen, J.Q. , Kreitman, M. and Bergelson, J. (2003) Fitness costs of R‐gene‐mediated resistance in Arabidopsis thaliana . Nature, 423, 74–77. [DOI] [PubMed] [Google Scholar]
- Tiffin, P. and Moeller, D.A. (2006) Molecular evolution of plant immune system genes. Trends Genet. 22, 662–670. [DOI] [PubMed] [Google Scholar]
- Toruño, T.Y. , Shen, M. , Coaker, G. and Mackey, D. (2019) Regulated disorder: posttranslational modifications control the RIN4 plant immune signaling hub. Mol. Plant‐Microbe Interact. 32, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid, N. and Laitinen, R.A.E. (2019) Diverse paths to hybrid incompatibility in Arabidopsis. Plant J. 97, 199–213. [DOI] [PubMed] [Google Scholar]
- Van Doorn, W. , Beers, E. , Dangl, J. , Franklin‐Tong, V. , Gallois, P. , Hara‐Nishimura, I. , Jones, A. , Kawai‐Yamada, M. , Lam, E. and Mundy, J. (2011) Morphological classification of plant cell deaths. Cell Death Differ. 18, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallad, G.E. and Goodman, R.M. (2004) Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci. 44, 1920–1934. [Google Scholar]
- Vontimitta, V. , Olukolu, B. , Penning, B. , Johal, G. and Balint‐Kurti, P.J. (2015) The genetic basis of flecking and its relationship to disease resistance in the IBM maize mapping population. Theoretical and Applied Genetics, 128, 1–9. [DOI] [PubMed] [Google Scholar]
- Wang, G.‐F. and Balint‐Kurti, P.J. (2015) Cytoplasmic and nuclear localizations are important for the hypersensitive response conferred by maize autoactive Rp1‐D21 protein. Mol. Plant‐Microbe Interact. 28, 1023–1031. [DOI] [PubMed] [Google Scholar]
- Wang, G.‐F. and Balint‐Kurti, P.J. (2016) Maize homologs of CCoAOMT and HCT, two key enzymes in lignin biosynthesis, form complexes with the NLR Rp1 protein to modulate the defense response. Plant Physiol. 171, 2166–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Cai, X. and Zheng, Z. (2005) High humidity represses Cf‐4/Avr4‐ and Cf‐9/Avr9‐dependent hypersensitive cell death and defense gene expression. Planta, 222, 947–956. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Bao, Z. , Zhu, Y. and Hua, J. (2009) Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant‐Microbe Interact. 22, 498–506. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Barnaby, J.Y. , Tada, Y. , Li, H. , Tor, M. , Caldelari, D. , Lee, D.‐U. , Fu, X.‐D. and Dong, X. (2011) Timing of plant immune responses by a central circadian regulator. Nature, 470, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.‐F. , Ji, J. , Ei‐Kasmi, F. , Dangl, J.L. , Johal, G. and Balint‐Kurti, P.J. (2015) Molecular and functional analyses of a maize autoactive NB‐LRR protein identify precise structural requirements for activity. PLoS Pathog. 11, e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.‐F. , He, Y. , Strauch, R. , Olukolu, B.A. , Nielsen, D. , Li, X. and Balint‐Kurti, P.J. (2016) Maize homologs of hydroxycinnamoyltransferase, a key enzyme in lignin biosynthesis, bind the nucleotide binding leucine‐rich repeat Rp1 proteins to modulate the defense response. Plant Physiol. 169, 2230–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Hu, M. , Wang, J. , Qi, J. , Han, Z. , Wang, G. , Qi, Y. , Wang, H.‐W. , Zhou, J.‐M. and Chai, J. (2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science, 364, eaav5870. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wang, J. , Hu, M. , Wu, S. , Qi, J. , Wang, G. , Han, Z. , Qi, Y. , Gao, N. , Wang, H.‐W. , Zhou, J.‐M. and Chai, J. (2019b) Ligand‐triggered allosteric ADP release primes a plant NLR complex. Science, 364, eaav5868. [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Hu, W. , Wang, Q. , Liu, W. , Wu, C. , Zeng, H. , Yan, Y. , Li, X. , He, C. and Shi, H. (2016) Comprehensive transcriptional and functional analyses of melatonin synthesis genes in cassava reveal their novel role in hypersensitive‐like cell death. Sci. Rep. 6, 35029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S. , McCormick, S. and Baker, B. (1996) The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc. Natl. Acad. Sci. USA. 93, 8776 – 8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer, M. , Feys, B.J. and Parker, J.E. (2005) Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. [DOI] [PubMed] [Google Scholar]
- Williams, S.J. , Sohn, K.H. , Wan, L. , Bernoux, M. , Sarris, P.F. , Segonzac, C. , Ve, T. , Ma, Y. , Saucet, S.B. , Ericsson, D.J. , Casey, L.W. , Lonhienne, T. , Winzor, D.J. , Zhang, X. , Coerdt, A. , Parker, J.E. , Dodds, P.N. , Kobe, B. and Jones, J.D.G. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science, 344, 299–303. [DOI] [PubMed] [Google Scholar]
- Wright, K.M. , Duncan, G.H. , Pradel, K.S. , Carr, F. , Wood, S. , Oparka, K.J. and Cruz, S.S. (2000) Analysis of the N gene hypersensitive response induced by a fluorescently tagged tobacco mosaic virus. Plant Physiol. 123, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.‐H. , Abd‐El‐Haliem, A. , Bozkurt, T.O. , Belhaj, K. , Terauchi, R. , Vossen, J.H. and Kamoun, S. (2017) NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA. 114, 8113 – 8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z. and Chen, Z. (2000) Harpin‐induced hypersensitive cell death is associated with altered mitochondrial functions in tobacco cells. Mol. Plant‐Microbe Interact. 13, 183–190. [DOI] [PubMed] [Google Scholar]
- Xu, G. , Greene, G.H. , Yoo, H. , Liu, L. , Marqués, J. , Motley, J. and Dong, X. (2017) Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature, 545, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Yang, H. , Grisafi, P. , Sanchatjate, S. , Fink, G.R. , Sun, Q. and Hua, J. (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J. 45, 166–179. [DOI] [PubMed] [Google Scholar]
- Zeier, J.R. , Pink, B. , Mueller, M. and Berger, S. (2004) Light conditions influence specific defence responses in incompatible plant–pathogen interactions: uncoupling systemic resistance from salicylic acid and PR‐1 accumulation. Planta, 219, 673–683. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Goritschnig, S. , Dong, X. and Li, X. (2003) A gain‐of‐function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1‐1, constitutive 1 . Plant Cell, 15, 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Kadota, Y. , Prodromou, C. , Shirasu, K. and Pearl, L.H. (2010) Structural basis for assembly of Hsp90‐Sgt1‐CHORD protein complexes: implications for chaperoning of NLR innate immunity receptors. Mol. Cell, 39, 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Yin, Z. and White, F. (2015) TAL effectors and the executor R genes. Front. Plant Sci. 6, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. and Zeng, L. (2017) Conventional and unconventional ubiquitination in plant immunity. Mol. Plant Pathol. 18, 1313–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Qian, W. and Hua, J. (2010) Temperature modulates plant defense responses through NB‐LRR proteins. PLoS Pathog, 6, e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, D. and Schafer, J. (1961) Relation of temperature to reaction type of Puccinia coronata on certain oat varieties. Am. Phytopathol. Soc., 55121, 202–203. [Google Scholar]


