SUMMARY
An RNA silencing construct was used to alter mycotoxin production in the plant pathogenic fungus Fusarium culmorum, the incitant of crown and foot rot on wheat. The transformation of a wild‐type strain and its nitrate reductase‐deficient mutant with inverted repeat transgenes (IRTs) containing sequences corresponding to the trichothecene regulatory gene TRI6 was achieved using hygromycin B resistance as a selectable marker. Southern analysis revealed a variety of integration patterns of the TRI6 IRT. One transformant underwent homologous recombination with deletion of the endogenous TRI6 gene, whereas, in another transformant, the TRI6 IRT was not integrated into the genome. The TRI6 IRT did not alter the physiological characteristics, such as spore production, pigmentation or growth rate, on solid media. In most transformants, a high TRI6 amplification signal was detected by quantitative reverse transcription‐polymerase chain reaction, corresponding to a TRI6‐hybridizing smear of degraded fragments by Northern analysis, whereas TRI5 expression decreased compared with the respective nontransformed strain. Four transformants showed increased TRI5 expression, which was correlated with a dramatic (up to 28‐fold) augmentation of deoxynivalenol production. Pathogenicity assays on durum wheat seedlings confirmed that impairment of deoxynivalenol production in the TRI6 IRT transformants correlated with a loss of virulence, with decreased disease indices ranging from 40% to 80% in nine silenced strains, whereas the overproducing transformants displayed higher virulence compared with the wild‐type.
INTRODUCTION
Fusarium culmorum (W.G. Smith) Sacc. is a major fungal pathogen of wheat, causing two forms of disease, namely crown and foot rot (CFR) and fusarium head blight (FHB) (Corazza et al., 2002; Smiley and Patterson, 1996; Wagacha and Muthomi, 2007; Wiese, 1987). The severity of CFR incited by F. culmorum is usually greater in dry soils and is commonly associated with warm temperatures (Cariddi and Catalano, 1990). These conditions are dominant in central and southern Italy, where durum wheat is commonly grown. Disease symptoms may include seedling death before or soon after emergence, brown lesions on the stem base, tiller abortion and the formation of whiteheads, containing shrivelled grain or no grain at all. Consequently, significant yield losses are reported and the grain often becomes contaminated with mycotoxins.
Among the most bioactive compounds are trichothecenes, i.e. sesquiterpene epoxides, which are able to inhibit eukaryotic protein synthesis (Wei and McLaughlin, 1974) and may cause toxicoses in humans or animals consuming contaminated food or feed (Sudakin, 2003). Trichothecenes may also induce apoptosis (Desmond et al., 2008; Yang et al., 2000) and have been suggested to play an important role in the aggressiveness of phytopathogenic Fusarium species towards plant hosts (Bai et al., 2002; 1996, 2000; Harris et al., 1999; Jansen et al., 2005; Maier et al., 2006; McCormick, 2003; 1995, 2002; Ward et al., 2008; Zhang et al., 2010).
Type B trichothecenes, produced mainly by Gibberella zeae (Schwein.) Petch (anamorph: Fusarium graminearum Schwabe) and F. culmorum, are predominant in Europe. They include deoxynivalenol (DON), also known as vomitoxin, with its derivatives monoacetyldeoxynivalenols (3‐AcDON, 15‐AcDON), nivalenol (NIV) and its acetylated form 4‐acetylnivalenol or fusarenone X (Bottalico and Perrone, 2002; Goswami and Kistler, 2004).
CFR control is often difficult and mainly relies on preventative measures, such as crop rotation, the use of tolerant cultivars, burial of infested debris, reduced nitrogen fertilization and seed dressing with fungicides. However, the risk of residues and the potential for increased levels of mycotoxins in the grain under certain circumstances (Gardiner et al., 2009) may limit their use.
The biosynthesis of trichothecenes starts with the formation of trichodiene via the cyclization of farnesyl pyrophosphate. Trichodiene then undergoes a series of oxygenation, cyclization, isomerization and esterification steps to yield bioactive trichothecenes, such as DON and acetylated DON (Desjardins et al., 1993). At least 12 genes that encode enzymes catalysing most of these biosynthesis steps have been characterized and found to be located within a single core cluster in G. zeae and in the closely related species Fusarium sporotrichioides Sherb. (for reviews, see Alexander et al., 2009; Proctor et al., 2002).
TRI5, the first trichothecene biosynthetic gene identified in F. sporotrichioides, encodes the enzyme trichodiene synthase (Hohn and Beremand, 1989). In G. zeae, TRI5 function was determined by transformation‐mediated gene disruption experiments, resulting in mutants that were unable to carry out the first biosynthetic step and were incapable of producing DON or any of the trichothecene biosynthetic intermediates (Desjardins et al., 1996; Harris et al., 1999; Jansen et al., 2005; Maier et al., 2006). These mutants were reduced in virulence when grown on wheat (Proctor et al., 1995).
TRI6 encodes a Cys2‐His2‐type positive transcription factor that binds to the promoter regions of genes located in the core TRI cluster, as well as to TRI1, TRI16 and TRI101, which are located outside of the TRI cluster (Alexander et al., 2009). Thus, this gene plays a crucial role in regulating the expression of multiple biosynthetic genes, including TRI5, and can be regarded as a potential molecular target when developing resistant wheat lines or new molecules that could reduce trichothecene biosynthesis.
A possible approach to the downregulation of gene expression in fungi and other eukaryotic organisms in a sequence‐specific manner utilizes RNA silencing (Catalanotto et al., 2000; Cogoni and Macino, 1997; Denli and Hannon, 2003; Pickford and Cogoni, 2003; Waterhouse et al., 2001). Specific inhibition of gene expression by RNA silencing has been demonstrated in several filamentous fungi, including Neurospora crassa, Magnaporthe oryzae, Cryptococcus neoformans, Histoplasma capsulatum, Ophiostoma spp. and Aspergillus spp. (reviewed by Cogoni, 2001; Nakayashiki et al., 2005; Nakayashiki and Nguyen, 2008). Inverted repeat transgenes (IRTs) containing sequences corresponding to the mycotoxin regulatory gene TRI6 were first shown to inhibit DON production through RNA silencing in G. zeae (McDonald et al., 2005).
The aims of this work were twofold: first, to test the efficacy of an RNA interfering construct based on IRT targeted at the trichothecene biosynthesis gene TRI6 of F. culmorum and, second, to evaluate whether altered trichothecene biosynthesis by silenced transformants had an effect on the aggressiveness of this CFR‐inducing pathogen towards durum wheat seedlings.
RESULTS
Preparation of F. culmorum transformants with the TRI6 IRT
Co‐transformation of MCf 21 wild‐type and MCf 21 nit1 with the TRI6 IRT silencing vector pLRM13 and the hygromycin‐carrying plasmid pUCH2‐8 resulted in over 50 hygromycin B‐resistant transformants, among which eight and seven, respectively, were chosen for further analysis. Southern analysis with a TRI6 probe revealed a variety of integration patterns of the TRI6 IRT (1, 2 and Supporting Information). These included no TRI6 IRT integrations (i.e. # 282, where only the hygromycin B resistance‐carrying vector pUCH2‐8 was integrated) to multiple TRI6 IRT integrations (i.e. # 111). However, many unpredicted bands were observed in our NcoI, HindIII and EcoRI/XbaI digests (Figs S1–S4, see Supporting Information). These can be explained by intermolecular recombination between IRTs during the transformation process. Using predictions based on such a model (Fig. S5, see Supporting Information), integration patterns were determined for the 15 transformants (1, 2). Thus, transformants were found to have: (i) no IRT, (ii) an ‘ideal’ IRT, (iii) a promoter‐bound IRT (i.e. containing the IRT between opposing gpdA promoter sequences), (iv) a terminator‐bound IRT (i.e. containing the IRT between diverging trpC terminators) or any of the types of IRTs in various combinations (Table 1 and Supporting Information).
Figure 1.
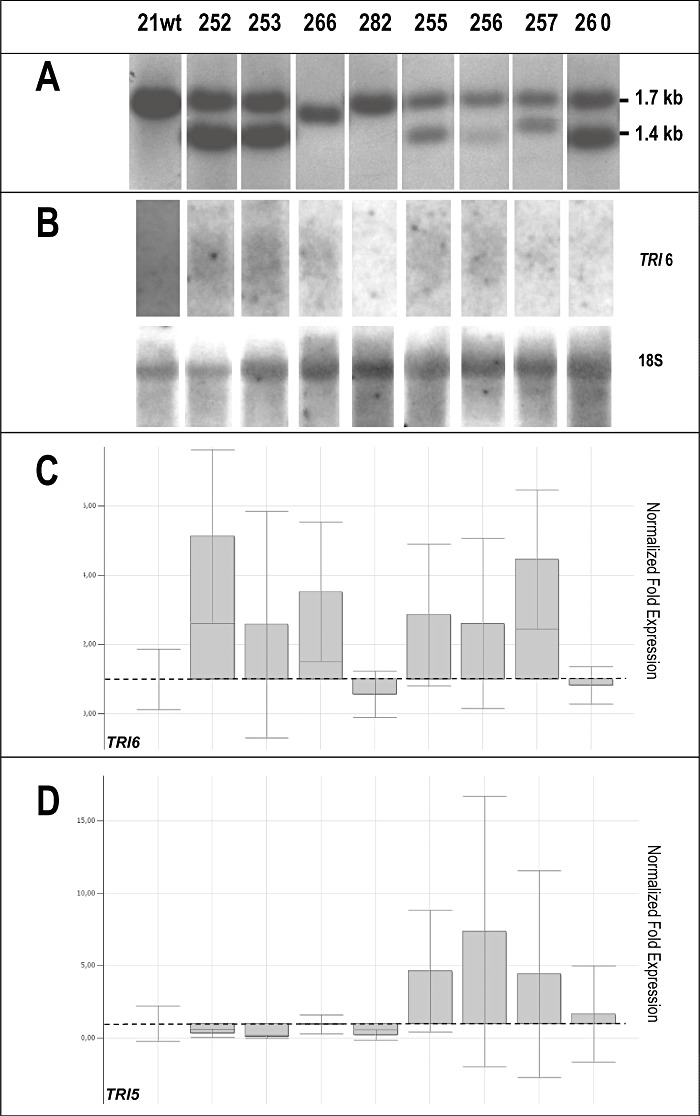
Characterization and gene expression analysis of Fusarium culmorum MCf 21 wild‐type (21 wt) and eight independent TRI6 inverted repeat transgene (IRT) transformants (# 252, 253, 266, 282, 255, 256, 257, 260). (A) Southern blot analysis using the ‘sense’TRI6 IRT fragment of vector pLRM13 to probe NcoI/HindIII‐digested genomic DNAs, resulting in a 1.7‐kb band for the endogenous TRI6 gene and a 1.4‐kb band in the case of TRI6 IRT integration. (B) Northern blot analysis of TRI6 expression using a 478‐bp‐long PCR‐amplified TRI6‐specific probe and control hybridization with the 18S probe. (C, D) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) on expression of the TRI6 gene (C) and the TRI5 gene (D); normalization was achieved by comparing the expression of housekeeping genes β‐tubulin and 18S; broken line indicates MCf21 21 wt expression level; all other values correspond to increased or decreased gene expression compared with the latter.
Figure 2.
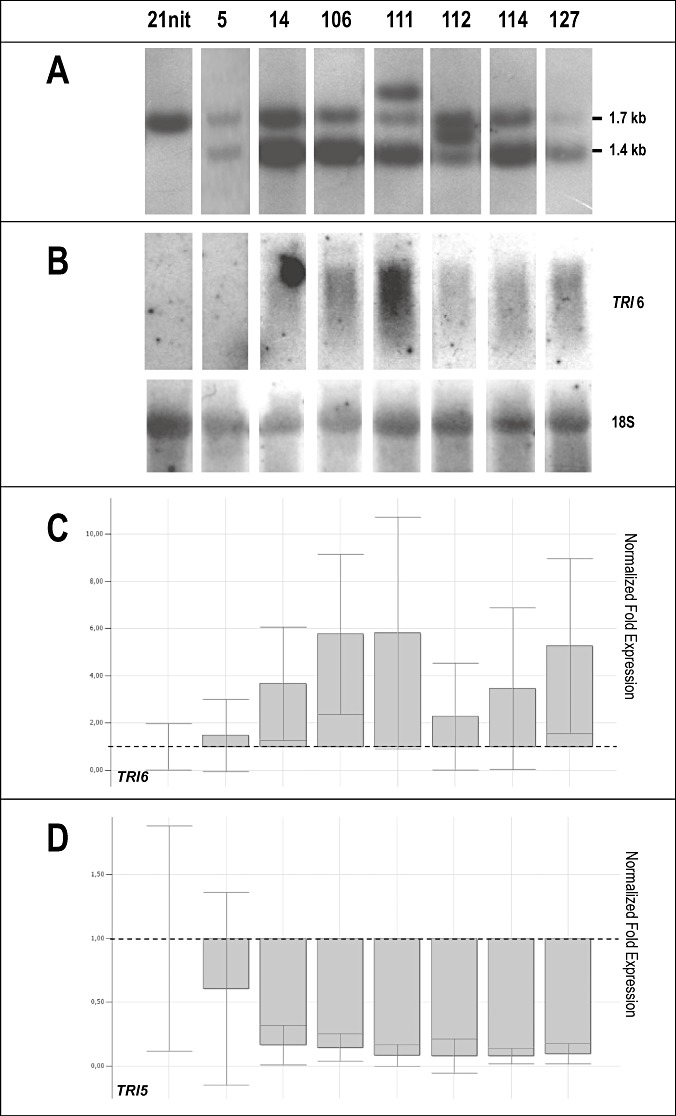
Characterization and gene expression analysis of Fusarium culmorum MCf 21 nit1 (21 nit) and seven independent TRI6 inverted repeat transgene (IRT) transformants (# 5, 14, 106, 111, 112, 114, 127). (A) Southern blot analysis using the ‘sense’TRI6 IRT fragment of vector pLRM13 to probe NcoI/HindIII‐digested genomic DNAs, resulting in a 1.7‐kb band for the endogenous TRI6 gene and a 1.4‐kb band in the case of TRI6 IRT integration. (B) Northern blot analysis of TRI6 expression using a 478‐bp‐long PCR‐amplified TRI6‐specific probe and control hybridization with the 18S probe. (C, D) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) on expression of the TRI6 gene (C) and the TRI5 gene (D). Normalization was achieved by comparing the expression of housekeeping genes β‐tubulin and 18S; broken line indicates MCf21 21 nit 1 expression level; all other values correspond to increased or decreased gene expression compared with the latter.
Table 1.
Fusarium culmorum MCf 21 wild‐type and eight independent TRI6 inverted repeat transgene (IRT) co‐transformants. Section 1 contains data from Southern analysis, suggesting the presence of several IRT types which are defined here as follows: ideal IRT, a TRI6 IRT expressed by a single promoter and bound by a single terminator; promoter‐bound IRT, a terminator‐less TRI6 IRT driven by dual gpdA promoters; terminator‐bound IRT, a promoter‐less TRI6 IRT bound on either side by trpC terminators. Section 2 shows the production of deoxynivalenol (DON) and its acetylated derivatives (3‐AcDON and 15‐AcDON) and the severity of foot and crown rot in durum wheat.
| Strain | 1 | 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Ideal IRT | Promoter‐bound IRT | Terminator‐bound IRT | Various TRI6 fragments | Northern TRI6 smear | Total DON (ng/mL)* | Difference of means | Disease index (0–100)† | Difference of means | |
| 21 wt | 0 | 0 | 0 | 0 | No | 878.1 ± 41.6 | 69.2 ± 4.5 | ||
| 252 | 0 | 1 | 0 | 0 | Yes | 49.0 ± 22.0 | −829.1 | 30.4 ± 6.7**‡ | −38.75 |
| 253 | 0 | 1 | 0 | 0 | Yes | 0 | −878.1 | 44.6 ± 5.4** | −24.58 |
| 266 | 0 | 0 | 0 | 1§ | Yes | 27.6 ± 12.6 | −850.5 | 37.1 ± 5.3** | −32.08 |
| 282 | 0 | 0 | 0 | 0 | No | 48.2 ± 21.6 | −829.9 | 69.6 ± 5.1 | 0.42 |
| 255 | 1 | 0 | 0 | 0 | Yes | 12 783 ± 830** | 11 905.0 | 97.9 ± 1.6** | 28.75 |
| 256 | 1 | 0 | 0 | 0 | Yes | 24 341 ± 5400** | 23 463.4 | 99.6 ± 0.4** | 30.42 |
| 257 | 1 | 0 | 0 | 0 | Yes | 12 473 ± 3 231** | 11 594.9 | 100** | 30.83 |
| 260 | 0 | 0 | 1 | 0 | No | 12 008 ± 2 637** | 11 129.7 | 98.3 ± 1.7** | 29.17 |
Pooled data from two independent experiments (P= 0.468) are expressed as the sum of DON and its acetylated derivatives (ng/mL of culture broth) ± mean SE after 2 weeks of growth in Vogel's medium.
Pooled data from two independent experiments, carried out in 2008 and 2009 using wheat cultivars Prometeo and Solex, respectively (P= 0.799), are expressed as disease incidence (McKinney index 0–100; McKinney, 1923) ± mean SE 3 weeks after inoculation and sowing under glasshouse conditions.
Values in each column followed by two asterisks are significantly different from the MCf 21 wild‐type control by Dunnett's test (P < 0.001).
Transformant 266 represents an insertion of the IRT at the native TRI6 locus.
Table 2.
Fusarium culmorum MCf 21 nit1 and seven independent TRI6 inverted repeat transgene (IRT) co‐transformants. Section 1 contains data from Southern analysis, suggesting the presence of several IRT types which are defined here as follows: ideal IRT, a TRI6 IRT expressed by a single promoter and bound by a single terminator; promoter‐bound IRT, a terminator‐less TRI6 IRT driven by dual gpdA promoters; terminator‐bound IRT, a promoter‐less TRI6 IRT bound on either side by trpC terminators. As a result of the complexity of the integration patterns, it was not always possible to predict the exact type and number of IRT (i.e. 0–1, 1–2). Section 2 shows the production of deoxynivalenol (DON) and its acetylated derivatives (3‐AcDON and 15‐AcDON) and the severity of foot and crown rot in durum wheat.
| Strain | 1 | 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Ideal IRT | Promoter‐bound IRT | Terminator‐bound IRT | Various TRI6 fragments | Northern TRI6 smear | Total DON (ng/mL)* | Difference of means | Disease index (0–100)† | Difference of means | |
| 21 nit | 0 | 0 | 0 | 0 | No | 898.1 ± 58.4 | 97.1 ± 1.9 | ||
| 5 | 0 | 0 | 1 | ? | No | 890.6 ± 34.8 | −7.5 | 97.9 ± 2.1 | 0.83 |
| 14 | 0–1 | 2 | 2 | ? | Yes | 0**‡ | −898.1 | 21.3 ± 4.9** | −75.75 |
| 106 | 1 | 1 | 2 | 0 | Yes | 0** | −898.1 | 33.3 ± 6.1** | −63.75 |
| 111 | 1 | 1 | 2 | ? | Yes | 0** | −898.1 | 25.4 ± 3.2** | −71.67 |
| 112 | 0–1 | 0–1 | 0 | 1–2 | Yes | 96.1 ± 43.1** | −801.9 | 22.1 ± 7.6** | −75.00 |
| 114 | 1–2 | 0 | 0 | 0 | Yes | 0** | −898.1 | 40.4 ± 5.2** | −56.67 |
| 127 | 1–2 | 0 | 1–2 | 0 | Yes | 0** | −898.1 | 20.0 ± 5.3** | −77.08 |
Pooled data from two independent experiments (P= 0.171) are expressed as the sum of DON and its acetylated derivatives (ng/mL of culture broth) ± mean SE after 2 weeks of growth in Vogel's medium.
Pooled data from two independent experiments, carried out in 2008 and 2009 using wheat cultivars Prometeo and Solex, respectively (P= 0.626), are expressed as disease incidence (McKinney index 0–100; McKinney, 1923) ± mean SE 3 weeks after inoculation and sowing under glasshouse conditions.
Values in each column followed by two asterisks are significantly different from the MCf 21 nit1 control by Dunnett's test (P < 0.001).
Gene expression in TRI6 IRT F. culmorum transformants
Relative to control strains, TRI6 amplification levels appeared to increase in most of the transformants (significantly higher for strains # 252, 260 and 106), except for # 282 (no pLRM13 integration) and # 260 (1, 2). Active silencing of TRI6 should result in degradation of the TRI6‐specific mRNA transcripts. During this process, truncated transcripts of several sizes are generated that may be detected by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) applications that usually amplify 100–150‐bp‐long fragments. Northern blots carried out with a 478‐bp‐long TRI6 probe added further evidence to this hypothesis. Most strains revealed a TRI6 smear whose intensity typically correlated with the qRT‐PCR amplification signal (1, 2). In transformants # 282, 260 and 5, no hybridization signal was detected in Northern blots, correlating with their very low TRI6 expression levels detected by qRT‐PCR analysis.
Semi‐quantitative RT‐PCR with primers TRI6probeF1 and TRI6probeR2 (Table 3) gave rise to a faint amplification signal of 478 bp for all silenced transformants, except # 255, 256, 257 and 260, which presented a signal comparable with that shown by the reference strain MCf 21 wild‐type (Fig. S6, see Supporting Information).
Table 3.
Primers used throughout this study.
| Application | Primer sequence | Amplicon size (bp) |
|---|---|---|
| Vector construction | ||
| TRI6senseF‐NcoI* | 5′‐CCTCCATGGTCGTGTTGCGTCTCCCGATCC‐3′ | 570 |
| TRI6senseR‐AscI | 5′‐GCAGGCGCGCCACCCTGCTAAAGACCCTCAG‐3′ | |
| TRI6antisenseF‐NotI | 5′‐CTTTGATGCGGCCGCGTCTCCCGATCCTG‐3′ | 570 |
| TRI6antisenseR‐BamHI | 5′‐GCATGGGATCCACCCTGCTAAAGACCCTCAG‐3′ | |
| Real‐time qRT‐PCR | ||
| TRI5forb | 5′‐ACCCTCAATTCCTTCGTCGTATG‐3′ | 141 |
| TRI5revb | 5′‐CCCAAACCATCCAGTTCTCCATC‐3′ | |
| TRI6forb | 5′‐TTATCGCCCTTCCCACCTTCAC‐3′ | 90 |
| TRI6revb | 5′‐TAAAGTCCCGTCCGCTCTCAAAG‐3′ | |
| 18Sforb | 5′‐TTGACCCGTTCGGCACCTTAC‐3′ | 75 |
| 18Srevb | 5′‐AAGTTTCAGCCTTGCGACCATAC‐3′ | |
| tubfor | 5′‐TTCCAAATCACCCACTCTC‐3′ | 104 |
| tubrev | 5′‐GAAAGTTGCCATCATACGG‐3′ | |
| Probes | ||
| TRI6probeF1 | 5′‐GACTTTGACAACTTCCCCACAT‐3′ | 478 |
| TRI6probeR2 | 5′‐AGTGATCTCGCATGTTATCCAC‐3′ | |
| 18S‐Forward | 5′‐CCTTAACGAGGAACAATTGGAG‐3′ | 514 |
| 18S‐Reverse | 5′‐CCCTAGTCGGCATAGTTTATGG‐3′ | |
| Restriction analysis of TRI4–TRI6–TRI5 cluster | ||
| H‐11 For4 | 5′‐GTGAACTTCGCGGGCGTTTACTC‐3′ | 5408 |
| H‐11 Rev13 | 5′‐TCCGGCTTGAAGGTCGTCAAAAT‐3′ |
Restriction sites are given in italic.
The effect of the TRI6 silencing construct on the expression of TRI5 was evaluated by qRT‐PCR. The results were fairly congruent with the level of TRI6 expression: transformants # 252, 253, 266, 5, 14, 106, 111, 112, 114 and 127, which showed more or less strongly smeared TRI6‐specific signals in Northern blots, showed correspondingly more or less decreased TRI5 expression compared with the recipient strains MCf 21 wild‐type and MCf 21 nit1, respectively (1, 2).
Transformants # 255, 256 and 257 revealed a completely different pattern, displaying a significantly increased TRI5 expression compared with MCf 21 wild‐type. Transformant # 260 showed almost the same level of TRI6 expression as MCf21 wild‐type and a slightly, but not significantly, increased TRI5 expression (Fig. 1D).
Production of trichothecenes in vitro by transformed F. culmorum strains
Based on a previous report by Boutigny et al. (2009), who demonstrated that strain MCf 21 wild‐type is able to produce predominantly 3‐AcDON and, to a lesser extent, DON when grown in liquid synthetic medium, we focused our high‐performance liquid chromatography‐mass spectrometry (HPLC‐MS) analysis on DON and its acetylated derivatives 3‐AcDON and 15‐AcDON. In a preliminary experiment, the average (mean of six replicates ± SD) production of type B trichothecenes (ng/mL culture filtrate) by this strain was 212.6 ± 74.2 DON, 523.0 ± 98.9 3‐AcDON and 16.3 ± 3.9 15‐AcDON after 14 days of growth in Vogel's medium at 25 °C. This pattern of distribution was confirmed in subsequent analyses.
In 1, 2, pooled data from two independent experiments carried out with MCf 21 wild‐type‐derived transformants, or with MCf 21 nit1‐derived transformants (P values between two equivalent experiments of 0.468 and 0.171, respectively), are expressed as the sum of DON, 3‐AcDON and 15‐AcDON in ng/mL culture filtrate ± SE. Regression was linear over the tested concentration range (5–250 ng/mL), with an average correlation coefficient R 2 of 0.9982 (±SD = 0.001), calculated from five calibration curves.
In MCf 21 wild‐type‐derived transformants # 252 and 253, the production of type B trichothecenes was reduced by 95% and 100%, respectively (Table 1). In transformants # 266 (homologous recombination with the endogenous TRI6 gene) and 282 (no apparent TRI6 IRT integration), type B trichothecene production was reduced to 3.1% and 5.4% with respect to the recipient strain. Interestingly, transformants # 255, 256, 257 and 260—which showed increased TRI5 expression—produced 14–28‐fold more trichothecenes than the MCf 21 wild‐type strain, corresponding to average concentrations ranging from 12 008 ± 2637 to 24 341 ± 5400 ng/mL of culture filtrate (Table 1).
Among TRI6 IRT transformants derived from the nitrate reductase mutant MCf 21 nit1, only transformant # 5 did not differ from the recipient strain for the production of type B trichothecenes, whereas, in transformant 112, trichothecene production was reduced by approximately 90% (Table 2). In all other TRI6 IRT transformants (i.e. # 14, 106, 111, 114 and 127), the production of type B trichothecenes was completely suppressed (Table 2).
Virulence of TRI6 IRT transformants on durum wheat
In order to ascertain whether altered trichothecene production by TRI6 IRT transformants affected their virulence, inoculation on durum wheat was carried out with both MCf 21 wild‐type‐ and MCf 21 nit1‐derived transformants using two different cultivars (Prometeo and Solex; P values between two equivalent experiments of 0.799 and 0.626, respectively). Indeed, among the 15 transformants tested, only # 282 did not differ significantly from the recipient strain MCf 21 wild‐type, despite a significant reduction in its DON production (Table 1), whereas # 5 produced the same amount of trichothecenes and displayed the same virulence as the nontransformed strain MCf 21 nit1 (Table 2). In all other transformants, significant changes (i.e. increase or decrease) in type B trichothecene production reflected corresponding differences in virulence (1, 2).
DISCUSSION
Together with F. graminearum and Fusarium avenaceum, the plant pathogenic fungus F. culmorum is ranked among the three most important wheat head blight pathogens on a global scale (Wagacha and Muthomi, 2007). Although considerable efforts have been devoted by the research community to dissect the pathogenicity and mycotoxin production of G. zeae (Goswami and Kistler, 2004), the pathogenicity determinants of F. culmorum and the role played by trichothecenes in the severity of CFR disease on wheat by this pathogen are still poorly understood. Although DON is considered as an important virulence factor in F. culmorum head blight disease (McCormick, 2003; Wagacha and Muthomi, 2007), little is known of the role of DON in CFR. Therefore, the main objective of this work was to elucidate the relationship between DON production by F. culmorum and its aggressiveness towards durum wheat seedlings.
To suppress DON production through an RNA silencing approach in F. culmorum, we generated an RNA interfering construct targeted at the endogenous regulatory gene TRI6. This gene encodes a Cys2‐His2‐type transcription factor binding to the promoter DNA sequence motif TNAGGCCT of the trichodiene synthase‐encoding gene TRI5 (Hohn et al., 1999). The same approach has been applied already to G. zeae by McDonald et al. (2005), who reported four TRI6 IRT transformants with reduced DON production and decreased virulence on wheat heads.
In this study, which represents the first report of the suppression of gene expression via RNA silencing in F. culmorum, a series of TRI6 IRT transformants was obtained from the highly virulent strain MCf 21 wild‐type (syn. INRA 117) and its nitrate reductase‐deficient mutant MCf 21 nit1.
The main achievement of this report is that, with only a few exceptions, a tight relationship exists between trichothecene production in culture and virulence on durum wheat seedlings. This was demonstrated to hold true in both directions, i.e. most of the transformants with a reduced TRI5 expression signal as measured by qRT‐PCR also displayed reduced DON production and virulence, whereas four trichothecene‐overproducing transformants showed increased TRI5 signals in qRT‐PCR experiments and significantly higher virulence on durum wheat compared with the recipient MfC 21 wild‐type strain.
During the course of the experiments, the level of expression of the target gene appeared to be unpredictable: almost all the transformants displayed an apparently increased TRI6 expression by both Northern analysis and qRT‐PCR compared with the respective recipient strain. We interpreted this puzzling result as the accumulation of silencing IRTs consequent to the adoption of the constitutive promoter gpdA. Truncated transcripts of several sizes are generated by the silencing complex, before passing on to complete fragmentation and elimination from the cell. This assumption was substantiated by the appearance of a smeared hybridization signal in Northern blots. Silenced RNAs may act as a substrate for qRT‐PCR, given the small size—i.e. 90 bp—of the expected amplicon, hence the appearance of an increased level of TRI6‐specific amplification in the transformants compared with the recipient strain. Indeed, RT‐PCR experiments targeted at a 478‐bp amplicon showed a fainter signal in most silenced transformants compared with the MCf 21 wild‐type (see Supporting Information). Moreover, in most MCf 21 nit1‐derived transformants, stronger TRI6‐hybridizing smears and TRI6‐specific amplification signals corresponded to a reduced expression of the trichodiene synthase gene TRI5, suggesting that TRI6 silencing and subsequent TRI5 suppression indeed occurred.
Among the MCf 21 wild‐type‐derived transformants, four strains were unexpectedly obtained with significantly increased TRI5 expression. These strains were able to produce an impressively large amount of DON (i.e. 14–28‐fold more than that of the wild‐type strain, corresponding to 12–24 µg/mL of culture filtrate). We first speculated that the gpdA promoter used in the TRI6 IRT could have recombined with the endogenous TRI6 locus, generating a series of gpdA‐TRI6 recombinant strains. However, Southern analysis with a TRI6 probe and comparative restriction analysis of the TRI4–TRI6–TRI5 locus (see Supporting Information) confirmed that the endogenous TRI6 locus was intact. These superproducing transformants are now being analysed at the proteome level, as the gpdA promoter could have recombined with a new positive regulatory gene, possibly suggesting alternative avenues in the regulation of trichothecene biosynthetic genes.
An additional finding of Southern analysis was that recombination between IRTs may occur quite frequently on integration into the F. culmorum genome. The silenced strains displayed a promoter‐bound IRT, a terminator‐bound IRT or an ‘ideal’ IRT with additional undetermined IRT fragments. Surprisingly, the overexpressing transformants appear to be among the simplest integrations, showing evidence for only one IRT. Moreover, the level of silencing did not correlate with the copy number of the IRT construct integrated into the genome: MCf 21 nit1‐derived transformants having integrated single or multiple copies of pLMR13 showed similarly reduced TRI5 expression levels and DON production. In one case (transformant # 5), TRI6 IRT integration did not generate any apparent modification in type B trichothecene biosynthesis. This is not surprising, as the same situation has been reported with Ophiostoma floccosum by Tanguay et al. (2006) and Aspergillus fumigatus by Henry et al. (2007), who highlighted the disadvantages of the RNA silencing approach compared with classical gene disruption by gene replacement. Thus, in the present work, we show, for the first time, that a functional RNA silencing pathway may be active in the phytopathogenic fungus F. culmorum, but, at the same time, we provide evidence for unpredictable patterns of IRT integration and silencing efficiency.
Our results demonstrate that, together with pectolytic enzymes (Aleandri et al., 2007; Kang and Buchenauer, 2002), type B trichothecenes play an important role as virulence factors in the CFR disease of durum wheat caused by F. culmorum, thus confirming this biosynthetic pathway as a suitable molecular target for the development of new CFR control tools. Modulation of the accumulation of trichothecenes in the plant tissues opens up new perspectives for the development of cereal varieties at reduced ‘mycotoxin risk’, through the action of plant endogenous compounds (Boutigny et al., 2008), or by delivering gene‐targeted RNA silencing triggers into the host plant (McDonald et al., 2005). Alternatively, a biocontrol approach could be envisaged that uses silenced atoxigenic mutants with low or null virulence. The latter strategy has been applied successfully to control aflatoxin‐producing Aspergillus flavus through competitive exclusion by atoxigenic isolates (Cotty et al., 2007).
Indubitably, before a biocontrol approach can be conceived, a thorough risk assessment is needed to ensure the stability of the integrated silencing construct and to avoid the possibility of unwanted recombination events involving the constitutive promoter, the TRI locus or other regulatory genes acting on the TRI cluster. Although our experiments were carried out in the laboratory or in contained facilities, the unpredictable generation of superproducing transformants opens up a new scenario for the definition of a biosafety paradigm in silencing experiments.
EXPERIMENTAL PROCEDURES
Strains and culture conditions
Fungal spores were obtained from: (i) a highly virulent strain of F. culmorum (W.G. Smith) Sacc. (strain ISPaVe MCf 21 wild‐type; syn. strain INRA 117), isolated in 1989 from triticale grown in Foggia, Apulia, southern Italy; (ii) its nitrate reductase‐deficient mutant MCf 21 nit1; and (iii) transformants derived by growing in a medium containing Campbell's V8 Vegetable juice (Giobbe et al., 2007) for 7 days at 25 °C with agitation (150 rpm). Cultures were filtered and conidia were collected by centrifugation. Spores were then resuspended in 10% glycerol at 106/mL and stored at −80 °C until needed.
In liquid culture, strain MCf 21 produces predominantly 3‐AcDON and, to a lesser extent, DON (Boutigny et al., 2009). To monitor the production of DON, 3‐AcDON and 15‐AcDON, and the expression of TRI5 and TRI6 genes, each strain was used to inoculate 8 mL of Vogel's medium (Vogel, 1956) with spore suspensions to achieve a final concentration of 104 conidia/mL. Fungal liquid cultures were incubated in the dark at 25 °C by gentle shaking at 30 rpm. Cultures were performed in triplicate and each experiment was repeated at least twice. Cultures were sampled after 7 days of incubation for RT‐PCR and qRT‐PCR (mycelium) and after 14 days for HPLC‐MS analysis (filtrates).
Construction of the silencing vector
The primers used in vector construction are listed in Table 3. A 570‐bp fragment of the TRI6 gene of F. culmorum (GenBank accession number AF480836) was amplified from genomic DNA, using a forward primer containing a NcoI site (TRI6senseF‐NcoI) and a reverse primer containing an AscI site (TRI6senseR‐AscI). The PCR product was digested and ligated into the NcoI/AscI sites of pTMH44.2 (McDonald et al., 2005) to produce pTRM20. A similar TRI6 fragment of 570 bp was amplified by PCR, using a forward primer containing a NotI site (TRI6antisenseF‐NotI) and a reverse primer containing a BamHI site (TRI6antisenseR‐BamHI). This PCR product was digested and ligated in reverse orientation into the NotI/BamHI sites in pTRM20 to obtain pLRM13 (Fig. 3).
Figure 3.
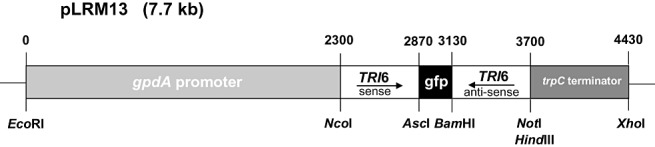
Schematic representation of Fusarium culmorum TRI6 inverted repeat transgene (IRT) construct. Light grey, gpdA promoter of Aspergillus nidulans; dark grey, trpC terminator of A. nidulans; black, gfp spacer element. A 570‐bp fragment of the F. culmorum TRI6 gene was placed in sense and antisense directions on the two sides of the gfp spacer element in the vector pTMH44.2, generating vector pLRM13.
Transformation experiments
Fungal protoplasts were isolated from germinated microconidia. For this purpose, 10–12 plates containing potato dextrose agar (PDA), covered with a sheet of sterile cellophane, were each inoculated with 106 conidia of F. culmorum MCf 21 wild‐type or MCf 21 nit1, and incubated at 25 °C for 16–18 h. Young thalli were then harvested with a sterile spatula and transferred into 20 mL of digestion solution consisting of 1.2 m MgSO4, pH 5.8, containing 10 mg/mL of lysing enzymes (L1492, Sigma‐Aldrich, St. Louis, MO, USA). After 3–3.5 h of incubation at room temperature and shaking at about 50 rpm, protoplasts were purified according to Langin et al. (1990) and then co‐transformed with 10 µg of the silencing vector pLRM13 and 10 µg of the hygromycin B resistance‐carrying vector pUCH2‐8 (Alexander et al., 1998) as described by Hua‐Van et al. (2001). Aliquots of 50 µL of the transformation mixture were transferred into 4–5 mL of preheated (37 °C) top agar [7.3 mm KH2PO4, 2 mm MgSO4, 6.5 mm KCl, 36 µm FeSO4, 23.5 mm NaNO3, 2% (w/v) glucose, 0.8% (w/v) technical agar] and plated onto Petri dishes containing PDAS H50 medium [potato dextrose agar containing 20% (w/v) sucrose and 50 µg/mL hygromycin B]. After incubation at 25 °C for 5–7 days, hygromycin B‐resistant transformants were selected for the integration of pUCH2‐8 and purified by monosporic culturing, followed by Southern blot analysis to control the correctness of pLRM13 integration events.
Southern analysis of transformants
Selection for hygromycin B‐resistant co‐transformants carrying the pLRM13 vector was performed by Southern blot analysis. Genomic DNA was purified from lyophilized mycelium by a miniprep method described previously (Migheli et al., 1996). Ten micrograms of genomic DNA were digested with 50 U each of HindIII, NcoI, NcoI/HindIII and EcoRI/XbaI (New England Biolabs Inc., Ipswich, MA, USA) at 37 °C for 15–18 h, separated by 1.0% agarose gel electrophoresis and transferred onto nylon membranes (Hybond‐N, Amersham Biosciences, Uppsala, Sweden) using a vacuum blotter (Vacuum Blotter Model 785, Bio‐Rad Laboratories, Milan, Italy) according to standard procedures (Sambrook et al., 1989).
The blots were then hybridized with a 570‐bp probe, complementary to the TRI6 sense fragment of the vector pLRM13. One microgram of pLRM13 (Fig. 3) was digested with 20 U each of AscI and NcoI in a volume of 100 µL at 37 °C for 3 h; digested DNA was precipitated and then loaded onto a 2.0% agarose gel. The 570‐bp DNA band was recovered with a sterile scalpel directly from the gel, followed by a silica column‐based purification step using the Jet Quick Gel extraction spin kit (Genomed GmbH, Löhne, Germany). Labelling, hybridization and detection reactions were carried out with the Gene Images Random Prime Labelling® and Gene Images CDP Star Detection® modules (Amersham Biosciences Europe GmbH, C. Monzese, Italy), as described in the manufacturer's protocols. Briefly, the modules work with a chemiluminescent signal, incorporating F1‐dUTP molecules in the future probe by a PCR approach. After this labelling step, the DNA probe was applied in an overnight hybridization step at 55–60 °C. Following stringency washes and blocking of nonspecific sites on the membrane, the chemiluminescent signal was activated with an alkaline phosphatase conjugate after incubation with the CDP Star detection substrate. The membranes were exposed to X‐ray radiographic films at room temperature for 1–2 h and then developed.
Sequencing and restriction analysis of the TRI4–TRI6–TRI5 locus in F. culmorum
To evaluate whether the endogenous TRI6 locus could be affected by the integration of the silencing vector, the 7.6‐kb region spanning the complete TRI4–TRI6–TRI5 genes in the F. culmorum MCf 21 nit1 strain was sequenced by a PCR‐based approach. Fifteen primer pairs generating overlapping amplicons of 500–700 bp in this region were designed based on the G. zeae H‐11 TRI locus (GenBank accession number AF336366). PCR was carried out as follows: 1 × Platinum®Pfx DNA Polymerase amplification buffer (Invitrogen, Invitrogen S.R.L., San Giuliano Milanese (MI), Italy), 1 mm MgSO4, 0.2 mm deoxynucleoside triphosphates (dNTPs), 0.5 µm of forward and reverse primers, 1.0 U of Platinum®Pfx DNA Polymerase (Invitrogen) and 300 ng of template DNA were mixed in a final volume of 50 µL and denatured at 95 °C for 3 min, followed by 32 cycles consisting of denaturation at 95 °C for 30 s, annealing at 60 °C for 45 s and elongation at 68 °C for 1 min, concluded by a final elongation step at 68 °C for 7 min.
Amplification products were examined on agarose gel for their integrity and specificity, purified and then sent for sequencing to BMR Genomics srl (Padua, Italy). Every amplification product was sequenced in both forward and reverse directions. Quality control of the single sequences and their assembly to the complete sequence were carried out with Sequence Scanner v1.0 and CLC Sequence Viewer 6 software. GenBank accession number: BankIT 1344398, HM131844 (Fig. S7, see Supporting Information).
A comparative restriction analysis of the 5.4‐kb region, including the final stretch of the TRI4 gene, the complete TRI6 gene, the initial stretch of the TRI5 gene and the two intergenic regions comprised between them, was carried out with the F. culmorum MCf 21 wild‐type, MCf 21 nit1 and 15 derived IRT transformants. The 5.4‐kb region was amplified with the primers H‐11For4 and H‐11Rev13 (Table 3) applying the Expand Long Template PCR System Kit (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer's protocol. The following restriction enzyme combinations were tested: HindIII, NcoI, HindIII/NcoI, and EcoRI/XbaI (New England Biolabs).
RNA purification and cDNA synthesis
Total RNA was extracted using the PureLink™ Micro‐to‐Midi total RNA purification system (Invitrogen) according to the manufacturer's instructions for RNA purification from plant tissue. A DNAse treatment to remove genomic DNA contamination from the samples was performed using the ‘RQ1 RNase‐freeDNase’ (Promega, Madison, WI, USA). The concentration of RNA was determined spectrophotometrically using GeneQuant (Amersham Biosciences). For cDNA synthesis, 0.5 µg of RNA were retrotranscribed with the iScript™ cDNA Synthesis Kit (Bio‐Rad Laboratories). The cDNA was either used directly for real‐time qRT‐PCR or stored at 4 °C.
qRT‐PCR analysis
The ICycler iQ system coupled to the SYBR® Green Kit (Bio‐Rad Laboratories) was used for qRT‐PCR. The real‐time qRT‐PCR efficiencies were determined for each gene by measuring C T to a specific threshold (Walker, 2002) for five serial 1 : 2 dilutions of the F. culmorum strain MCf 21 wild‐type cDNA. Primers applied for the amplification of TRI5, TRI6, 18S rRNA and β‐tubulin in the qRT‐PCR approach had efficiencies of 98.7%, 96.0%, 90.7% and 96.4%, respectively. The specificity of each primer set was examined and the amplification of a specific transcript was confirmed by the appearance of a single peak in the melting curve analysis following completion of the amplification reaction. The relative level of expression of each gene of interest (C T) was then normalized against the relative gene expression level of the RNA coding for the reference genes β‐tubulin and 18S rRNA determined in each sample (ΔΔC T).
Primers (Table 3) were designed using the software Molecular Beacon (Bio‐Rad Laboratories). Primers used for the amplification of TRI5 (TRI5forb/TRI5revb) and TRI6 (TRI6forb/TRI6revb) were derived from conserved regions in F. culmorum (GenBank accession numbers AY130291 and AF480836). Primers used for the amplification of the 18S rRNA gene (18Sforb/18Srevb) were derived from G. zeae (accession number AB250414). Primers used for the amplification of the β‐tubulin gene (tubfor/tubrev) were derived from F. culmorum (accession number EU490256).
The reaction mixtures contained 1 µL of cDNA diluted 1 : 10 (=2.5 ng), 1 µL of each primer (400 nm), 12.5 µL of iQTM SYBR® Green Supermix and 9.5 µL of Milli‐Q H2O in a final volume of 25 µL.
In all the experiments, appropriate negative controls containing no template were subjected to the same procedure to exclude or detect any possible contamination or carry‐over. Each sample was amplified three times for each experiment and all the experiments were repeated at least twice. The programme used included one step at 95 °C for 3 min, and 40 cycles consisting of 95 °C for 10 s and 60 °C for 30 s, followed by gradual heating (0.5 °C every 30 s) from 50 to 85 °C in order to generate the melting curve.
RT‐PCR analysis
In a semi‐quantitative approach, the cDNA of MCf 21 wild‐type and 15 TRI6 IRT transformants was subjected to RT‐PCR analysis, applying primers spanning almost the entire TRI6 gene (TRI6probeF1/TRI6probeR2; Table 3). One‐hundred nanograms of cDNA of each sample were amplified in a reaction volume of 50 µL with 1 × TopTaq™ DNA Polymerase amplification buffer (Qiagen, Qiagen S.p.A., Milan, Italy), 0.2 mm dNTPs, 0.5 µm of forward and reverse primers and 2.0 U of TopTaq™ DNA Polymerase (Qiagen). Different numbers of amplification cycles were tested to ascertain that the PCR products did not represent a saturated signal in the following gel electrophoresis. Samples were denatured at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 48 °C for 45 s and elongation at 72 °C for 1 min, with a final elongation step at 72 °C for 7 min. For all samples, a control reaction was carried out with 18S primers (18S‐Forward/18S‐Reverse; Table 3).
Northern analysis
For Northern blot hybridization experiments, mycelia of strains MCf 21 wild‐type and derived transformants # 252, 253, 255, 256, 257, 260, 266 and 282, as well as MCf 21 nit1 and derived transformants # 5, 14, 106, 111, 112, 114 and 127, were harvested after 7 days of growth in Vogel's medium, as described previously, flash‐frozen by dipping in liquid nitrogen, ground to a fine powder and total RNAs were isolated according to the guanidinium‐thiocyanate/phenol method (Chomczynski and Sacchi, 1987). Twenty micrograms of total RNA of each sample to be tested were separated on a 1.2% formaldehyde gel and transferred onto nylon membranes (Hybond‐N, Amersham Biosciences) using a vacuum blotter (Vacuum Blotter Model 785, Bio‐Rad Laboratories) according to standard procedures (Sambrook et al., 1989).
Expression levels of the TRI6 gene were detected for all strains by hybridization with a TRI6‐specific probe (478 bp). In addition, a control hybridization was carried out with an 18S rRNA probe to evaluate the basal quantity and quality of the RNA signal. DNA fragments for the future probes were amplified by PCR on the cDNA template of MCf 21 wild‐type strain applying the following primers: TRI6probeF1/TRI6probeR2, 18S‐Forward/18S‐Reverse (Table 3). Labelling, hybridization and detection reactions were carried out with the Gene Images Random Prime Labelling® and Gene Images CDP Star Detection® modules (Amersham Biosciences), as described previously.
HPLC‐MS analysis of trichothecenes
Standard mixtures including NIV, DON, 15‐AcDON and 3‐AcDON were obtained from Sigma Chemicals (St. Louis, MO, USA). The samples (8 mL) were extracted with 5 mL of ethyl acetate. After centrifugation, the organic phase was evaporated to dryness and redissolved in 1 mL of LC‐MS mobile phase. An Agilent Technologies (Palo Alto, CA, USA) 1100 series LC‐MSD, equipped with a diode‐array detector and an autosampler, was used for LC separation. Chromatographic separation was achieved using a Luna C18 column (150 mm × 2.1 mm, 3 mm; Phenomenex, Castelmaggiore (BO), Italy) fitted with a C18 security guard cartridge. The column temperature was maintained at 40 °C. The mobile phase consisted of Eluent A (water with 0.01% acetic acid) and Eluent B (acetonitrile). The separation was performed in a run time of 20 min under gradient conditions with a flow rate of 0.4 mL/min. MS detection was performed using an Agilent (Palo Alto, CA, USA) G1946 (MSD 1100) single‐stage quadrupole instrument equipped with an electrospray atmospheric pressure ionization source. To construct standard curves, stock solutions of B‐trichothecene were prepared by dissolution in the standard mixture of 10 mL of acetonitrile (final concentration, 1000 ng/mL). Working solutions of GE‐B5 were prepared daily by diluting aliquots of stock solutions with the solvent system (Eluent A), and were used to spike samples. Six different concentrations of B‐trichothecene (5, 10, 25, 50, 100 and 250 ng/mL) were obtained by adding appropriate concentrations of working solutions to the solvent system.
Data are reported as mean values ± SE of three biological replications. Trichothecene yields (sum of DON and its acetylated derivatives 3‐AcDON and 15‐AcDON) are expressed as ng/mL of culture filtrate. The data were subjected to one‐way analysis of variance, followed by multiple comparison by Dunnett's test, using Minitab® for Windows release 12.1.
Pathogenicity assay
A monosporic culture of each F. culmorum strain was grown on PDA (Difco, Detroit, MI, USA) at 25 °C with a photoperiod of 12 h for 7 days. Plugs (1.2 cm in diameter) of PDA colonized by the fungus were cut with a sterile cork borer and used as inoculum. One plug was placed in the centre of a plastic sowing pot (diameter, 4.5 cm; capacity, 55 mL) containing sterilized (121 °C for 60 min on two successive days) potting mix (Humin‐Substrat N17, Neuhaus, Germany). One seed of durum wheat (Triticum durum cvs. Prometeo or Solex, kindly provided by Unità di Ricerca per la Valorizzazione Qualitativa dei Cereali, CRA‐QCE, Rome, Italy) was placed onto each PDA plug and covered by sterilized soil. For each treatment, three replicates (10 seeds for each replicate) were used. Pots were watered daily and the average temperature was 25 °C (minimum, 15 °C; maximum, 35 °C). Seedling emergence was checked every 7 days for 3 weeks. After the last survey, the plant height and disease severity were evaluated. The severity of disease was calculated using the McKinney index (McKinney, 1923), which expresses the percentage of the maximum severity of disease (i.e. 100) according to the formula: I =[Σ(c×f)/n×N]× 100, where c is the disease class, f is the frequency, n is the number of observations and N is the highest value of the empirical scale adopted, estimating the severity of symptoms on the stem. Five classes were set: class 0, healthy stem; class 1, mild browning on the stem; class 2, browning on one half of the stem; class 3, complete browning of the stem; class 4, plant death after emergence or plant not emerged in comparison with the emergence of the uninoculated control treatment.
Each experiment was repeated at least twice. The data were subjected to one‐way analysis of variance, followed by multiple comparison by Dunnett's test, using Minitab® for Windows release 12.1.
Supporting information
Fig. S1. Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe HindIII‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S2 Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe NcoI‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S3 Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe HindIII/NcoI‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S4 Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe EcoRI/XbaI‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S5 Plasmid pLRM13 recombination model. (A) pLRM13. (B) Recombination could occur between two molecules of pLRM13. One possibility is that the plasmids recombine in Fusarium culmorum before integration into the genome. A theoretical break point between the forward TRI6 fragment in one plasmid and the reverse TRI6 fragment in another plasmid is shown. (C) Expected product after recombination as proposed in (B). The distances between the restriction enzymes of the recombined plasmids allow for band DNA fragment sizes consistent with many of the transformants analysed in this study. Depending on how the recombined plasmid integrates into the genome, a transformant could end up with a ‘promoter‐bound’ inverted repeat transgene (IRT) (left) or a ‘terminator‐bound’ IRT (right). Broken lines represent additional plasmid sequences.
Fig. S6 Analysis of TRI6 gene expression by semi‐quantitative reverse transcription‐polymerase chain reaction. cDNA of Fusarium culmorum MCf 21 wild‐type and of 15 TRI6 inverted repeat transgene (IRT) transformants was amplified with the primer pair TRI6probeF1/TRI6probeR2 (Table 3). Samples were denatured at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 48 °C for 45 s and elongation at 72 °C for 1 min; final elongation was at 72 °C for 7 min. A control reaction was carried out with 18S primers (18S‐Forward/18S‐Reverse; Table 3).
Fig. S7 Alignment of sequences of the TRI4–TRI6–TRI5 locus for Fusarium culmorum MCf 21 wild‐type (HM131844) and Gibberella zeae H‐11 (AF336366).
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This work was funded by the Ministry of University and Research (PRIN 2007: Transposon tagging and RNA silencing in the wheat pathogen F. culmorum). MO acknowledges receipt of a fellowship from Regione Autonoma della Sardegna (Borse di Ricerca destinate a giovani ricercatori. Legge Regionale 7 agosto 2007, n. 7 ‘Promozione della ricerca scientifica e dell’innovazione tecnologica in Sardegna’). This work was also partially funded by the University of Wisconsin Graduate School (Hatch Funds) and the US Department of Agriculture under Agreement no. 59‐0790‐3‐081 through a cooperative project with the US Wheat & Barley Scab Initiative to NPK and TMH. The authors wish to thank Corrado Di Mauro for assisting in statistical analysis, and Lane Milde and Tami McDonald for help in vector construction.
REFERENCES
- Aleandri, M.P. , Magro, P. and Chilosi, G. (2007) Modulation of host pH during the wheat–Fusarium culmorum interaction and its influence on the production and activity of pectolytic enzymes. Plant Pathol. 56, 517–525. [Google Scholar]
- Alexander, N.J. , Hohn, T.M. and McCormick, S.P. (1998) The Tri111 gene of Fusarium sporotrichioides encodes a cytochrome P‐450 monooxygenase required for C‐15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, N.J. , Proctor, R.H. and McCormick, S.P. (2009) Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium . Toxin Rev. 28, 198–215. [Google Scholar]
- Bai, G.H. , Desjardins, A.E. and Plattner, R.D. (2002) Deoxynivalenol‐nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia, 15, 91–98. [DOI] [PubMed] [Google Scholar]
- Bottalico, A. and Perrone, G. (2002) Toxigenic Fusarium species and mycotoxins associated with head blight in small‐grain cereals in Europe. Eur. J. Plant Pathol. 108, 611–624. [Google Scholar]
- Boutigny, A.L. , Richard‐Forget, F. and Barreau, C. (2008) Natural mechanisms for cereal resistance to the accumulation of Fusarium trichothecenes. Eur. J. Plant Pathol. 121, 411–423. [Google Scholar]
- Boutigny, A.L. , Barreau, C. , Atanasova‐Penichon, V. , Verdal‐Bonnin, M.N. , Pinson‐Gadais, L. and Richard‐Forget, F. (2009) Ferulic acid, an efficient inhibitor of type B trichothecene biosynthesis and Tri gene expression in Fusarium liquid cultures. Mycol. Res. 113, 746–753. [DOI] [PubMed] [Google Scholar]
- Cariddi, C. and Catalano, M. (1990) Water stress and Fusarium culmorum infections on durum wheat. Phytopathol. Mediterr. 29, 51–55. [Google Scholar]
- Catalanotto, C. , Azzalin, G. , Macino, G. and Cogoni, C. (2000) Transcription: gene silencing in worms and fungi. Nature, 404, 245. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 56–59. [DOI] [PubMed] [Google Scholar]
- Cogoni, C. (2001) Homology‐dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55, 381–406. [DOI] [PubMed] [Google Scholar]
- Cogoni, C. and Macino, G. (1997) Conservation of transgene‐induced post‐transcriptional gene silencing in plants and fungi. Trends Plant Sci. 2, 438–443. [Google Scholar]
- Corazza, L. , Balmas, V. , Santori, A. , Vitale, S. , Luongo, L. and Maccaroni, M. (2002) Head blight and foot rot of wheat in Italy. Petria, 12, 25–36. [Google Scholar]
- Cotty, P.J. , Antilla, L. and Wakelyn, P.J. (2007) Competitive exclusion of aflatoxin producers: farmer driven research and development In: Biological Control: A Global Perspective (Vincent C., Goettel M.S. and Lazarovits G., eds), pp. 241–253. Wallingford, Oxfordshire: CABI. [Google Scholar]
- Denli, A.M. and Hannon, G.J. (2003) RNAi: an ever‐growing puzzle. Trends Biochem. Sci. 28, 196–201. [DOI] [PubMed] [Google Scholar]
- Desjardins, A.E. , Hohn, T.M. and McCormick, S.P. (1993) Trichothecene biosynthesis in Fusarium species: chemistry, genetics, and significance. Microbiol. Rev. 57, 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins, A.E. , Proctor, R.H. , Bai, G. , McCormick, S.P. , Shaner, G. , Buechley, G. and Hohn, T.M. (1996) Reduced virulence of trichothecene‐nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant–Microbe Interact. 9, 775–781. [Google Scholar]
- Desjardins, A.E. , Bai, G. , Plattner, R.D. and Proctor, R.H. (2000) Analysis of aberrant virulence of Gibberella zeae following transformation‐mediated complementation of a trichothecene‐deficient (TRI5) mutant. Microbiology, 146, 2059–2068. [DOI] [PubMed] [Google Scholar]
- Desmond, O.J. , Manners, J.M. , Stephens, A.E. , Maclean, D.J. , Schenk, P.M. , Gardiner, D.M. , Munn, A.L. and Kazan, K. (2008) The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol. Plant Pathol. 9, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, D.M. , Kazan, K. and Manners, J.M. (2009) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum . Fungal Genet. Biol. 46, 604–613. [DOI] [PubMed] [Google Scholar]
- Giobbe, S. , Marceddu, S. , Scherm, B. , Zara, G. , Mazzarello, V. , Budroni, M. and Migheli, Q. (2007) The strange case of a biofilm‐forming strain of Pichia fermentans, which controls Monilinia brown rot on apple but is pathogenic on peach fruit. FEMS Yeast Res. 7, 1389–1398. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal rops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Harris, L.J. , Desjardins, A.E. , Plattner, R.D. , Nicholson, P. , Butler, G. , Young, J.C. , Weston, G. , Proctor, R.H. and Hohn, T.M. (1999) Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 83, 954–960. [DOI] [PubMed] [Google Scholar]
- Henry, C. , Mounya, I. and Latgé, J.P. (2007) Testing the efficacy of RNA interference constructs in Aspergillus fumigatus . Curr. Genet. 51, 277–284. [DOI] [PubMed] [Google Scholar]
- Hohn, T.M. and Beremand, P. (1989) Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene‐producing fungus Fusarium sporotrichioides . Gene, 79, 131–138. [DOI] [PubMed] [Google Scholar]
- Hohn, T.M. , Krishna, R. and Proctor, R.H. (1999) Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26, 224–235. [DOI] [PubMed] [Google Scholar]
- Hua‐Van, A. , Pamphile, J.A. , Langin, T. and Daboussi, M.J. (2001) Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol. Gen. Genet. 264, 724–731. [DOI] [PubMed] [Google Scholar]
- Jansen, C. , von Wettstein, D. , Schäfer, W. , Kogel, K.H. , Felk, A. and Maier, F.J. (2005) Infection patterns in barley and wheat spikes inoculated with wild type and trichodiene synthase gene disrupted Fusarium graminearum . Proc. Natl. Acad. Sci. USA, 102, 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Z. and Buchenauer, H. (2002) Studies on the infection process of Fusarium culmorum in wheat spikes: degradation of host cell wall components and localization of trichothecene toxins in infected tissue. Eur. J. Plant Pathol. 108, 653–660. [Google Scholar]
- Langin, T. , Daboussi, M.J. , Gerlinger, C. and Brygoo, Y. (1990) Influence of biological parameters and gene transfer technique on transformation of Fusarium oxysporum . Curr. Genet. 17, 313–319. [Google Scholar]
- Maier, F.J. , Miedaner, T. , Hadeler, B. , Felk, A. , Salomon, S. , Lemmens, M. , Kassner, H. and Schäfer, W. (2006) Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 7, 449–461. [DOI] [PubMed] [Google Scholar]
- McCormick, S.P. (2003) The role of DON in pathogenicity In: Fusarium Head Blight of Wheat and Barley (Leonard K.J. and Bushnell W.R., eds), pp. 165–183. St. Paul, MN: APS Press. [Google Scholar]
- McDonald, T. , Brown, D. , Keller, N.P. and Hammond, T.M. (2005) RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol. Plant–Microbe Interact. 18, 539–545. [DOI] [PubMed] [Google Scholar]
- McKinney, H.H. (1923) Influence of soil temperature and moisture on infection of wheat seedlings by Helminthosporium sativum . J. Agric. Res. 26, 195–217. [Google Scholar]
- Migheli, Q. , Friard, O. , Del Tedesco, D. , Musso, M.R. and Gullino, M.L. (1996) Stability of transformed antagonistic Fusarium oxysporum strains in vitro and in soil microcosms. Mol. Ecol. 5, 641–649. [Google Scholar]
- Nakayashiki, H. , Hanada, S. , Nguyen, B.Q. , Kadotani, N. , Tosa, Y. and Mayama, S. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42, 275–283. [DOI] [PubMed] [Google Scholar]
- Nakayashiki, H. and Nguyen, Q.B. (2008) RNA interference: roles in fungal biology. Curr. Opin. Microbiol. 11, 494–502. [DOI] [PubMed] [Google Scholar]
- Pickford, A.S. and Cogoni, C. (2003) RNA‐mediated gene silencing. Cell. Mol. Life Sci. 60, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor, R.H. , Hohn, T.M. , McCormick, S.P. and Desjardins, A.E. (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichothecene biosynthetic gene. Mol. Plant–Microbe Interact. 8, 593–601. [DOI] [PubMed] [Google Scholar]
- Proctor, R.H. , Desjardins, A.E. , McCormick, S.P. , Plattner, N.J. , Alexander, N.J. and Brown, D.W. (2002) Genetic analysis of the role of trichothecene and fumonisin mycotoxins in the virulence of Fusarium . Eur. J. Plant Pathol. 108, 691–698. [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning, A Laboratory Manual, 2nd edn. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Smiley, R.W. and Patterson, L.M. (1996) Pathogenic fungi associated with Fusarium foot rot of winter wheat in the semiarid Pacific Northwest. Plant Dis. 80, 944–949. [Google Scholar]
- Sudakin, D.L. (2003) Trichothecenes in the environment: relevance to human health. Toxicol. Lett. 143, 97–107. [DOI] [PubMed] [Google Scholar]
- Tanguay, P. , Bozza, S. and Breuil, C. (2006) Assessing RNAi frequency and efficiency in Ophiostoma floccosum and O. piceae . Fungal Genet. Biol. 43, 804–812. [DOI] [PubMed] [Google Scholar]
- Vogel, H.J. (1956) A convenient growth medium for Neurospora (Medium N). Microb. Genet. Bull. 13, 42–43. [Google Scholar]
- Wagacha, J.M. and Muthomi, J.W. (2007) Fusarium culmorum: infection process, mechanisms of mycotoxin production and their role in pathogenesis in wheat. Crop Prot. 26, 877–885. [Google Scholar]
- Walker, N.J. (2002) A technique whose time has come. Science, 296, 557–559. [DOI] [PubMed] [Google Scholar]
- Ward, T.J. , Clear, R.M. , Rooney, A.P. , O'Donnell, K. , Gaba, D. , Patrick, S. , Starkey, D.E. , Gilbert, J. , Geiser, D.M. and Nowicki, T.W. (2008) An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 45, 473–484. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M. , Wang, M.B. and Lough, T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Wei, C.M. and McLaughlin, C.S. (1974) Structure–function relationship in the 12,13‐epoxytrichothecenes. Novel inhibitors of protein synthesis. Biochem. Biophys. Res. Commun. 57, 838–844. [DOI] [PubMed] [Google Scholar]
- Wiese, M.V. (1987) Compendium of Wheat Diseases. St. Paul, MN: APS Press. [Google Scholar]
- Yang, G.H. , Jarvis, B.B. , Chung, Y.J. and Pestka, J.J. (2000) Apoptosis induction by the satratoxins and other trichothecene mycotoxins: relationship to ERK, p8 MAPK, and SAPK/JNK activation. Toxicol. Appl. Pharmacol. 164, 149–160. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, Z. , van der Lee, T. , Chen, W.Q. , Xu, J. , Xu, J.S. , Yang, L. , Yu, D. , Waalwijk, C. and Feng, J. (2010) Population genetic analyses of Fusarium asiaticum populations from barley suggest a recent shift favoring 3ADON producers in southern China. Phytopathology, 100, 328–336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe HindIII‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S2 Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe NcoI‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S3 Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe HindIII/NcoI‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S4 Southern hybridization analysis of Fusarium culmorum MCf 21 wild‐type (wt), MCf 21 nit1 (nit) and 15 TRI6 inverted repeat transgene (IRT) transformants using the ‘sense’ TRI6 IRT fragment of vector pLRM13 to probe EcoRI/XbaI‐digested genomic DNAs; selected molecular weights of the 1 kb plus DNA ladder (Invitrogen) indicate the range of the band pattern.
Fig. S5 Plasmid pLRM13 recombination model. (A) pLRM13. (B) Recombination could occur between two molecules of pLRM13. One possibility is that the plasmids recombine in Fusarium culmorum before integration into the genome. A theoretical break point between the forward TRI6 fragment in one plasmid and the reverse TRI6 fragment in another plasmid is shown. (C) Expected product after recombination as proposed in (B). The distances between the restriction enzymes of the recombined plasmids allow for band DNA fragment sizes consistent with many of the transformants analysed in this study. Depending on how the recombined plasmid integrates into the genome, a transformant could end up with a ‘promoter‐bound’ inverted repeat transgene (IRT) (left) or a ‘terminator‐bound’ IRT (right). Broken lines represent additional plasmid sequences.
Fig. S6 Analysis of TRI6 gene expression by semi‐quantitative reverse transcription‐polymerase chain reaction. cDNA of Fusarium culmorum MCf 21 wild‐type and of 15 TRI6 inverted repeat transgene (IRT) transformants was amplified with the primer pair TRI6probeF1/TRI6probeR2 (Table 3). Samples were denatured at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 48 °C for 45 s and elongation at 72 °C for 1 min; final elongation was at 72 °C for 7 min. A control reaction was carried out with 18S primers (18S‐Forward/18S‐Reverse; Table 3).
Fig. S7 Alignment of sequences of the TRI4–TRI6–TRI5 locus for Fusarium culmorum MCf 21 wild‐type (HM131844) and Gibberella zeae H‐11 (AF336366).
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


