SUMMARY
In plants, the ubiquitin/26S proteasome system (UPS) plays a central role in protein degradation and is involved in many steps of defence mechanisms, regardless of the types of pathogen targeted. In addition to its proteolytic activities, the UPS ribonuclease (RNase) activity, previously detected in 20S proteasome preparations from cauliflower and sunflower (Helianthus annuus), has been shown to specifically target plant viral RNAs in vitro. In this study, we show that recombinant Arabidopsis thaliana proteasomal α5 subunit expressed in Escherichia coli harbours an RNase activity that degrades Tobacco mosaic virus (TMV, Tobamovirus)‐ and Lettuce mosaic virus (LMV, Potyvirus)‐derived RNAs in vitro. The analysis of mutated forms of the α5 subunit demonstrated that mutation of a glutamic acid at position 110 affects RNase activity. Furthermore, it was demonstrated, using a bimolecular fluorescence complement assay, that the multifunctional helper component proteinase (HcPro) of LMV, already known to interfere with the 20S proteasome RNase activity in vitro, can interact in vivo with the recombinant α5 subunit. Further experiments demonstrated that, in LMV‐infected lettuce cells, α5 is partially relocalized to HcPro‐containing infection‐specific inclusions. Susceptibility analyses of Arabidopsis mutants, knocked out for each At‐PAE gene encoding α5, showed that one (KO‐pae1) of the two mutants exhibited a significantly increased susceptibility to LMV infection. Taken together, these results extend to A. thalianaα5 the range of HcPro‐interacting proteasomal subunits, and suggest that HcPro may modulate its associated RNase activity which may contribute to an antiviral response.
INTRODUCTION
The ubiquitin/26S proteasome system (UPS) is an essential element of protein turnover, playing a crucial role in mechanisms as important as the degradation of misfolded/damaged proteins, cell cycle control, programmed cell death and signal transduction (Dreher and Callis, 2007). Thus, UPS is involved in the plant response to the majority of external environment modifications and stresses, including plant–microbe interactions (for a review, see Dielen et al., 2010) and, more specifically, antiviral mechanisms. This pathway has been reported to be induced by viral infection (Takizawa et al., 2005) or to be involved in specific gene‐mediated resistance responses (Liu et al., 2002; Peart et al., 2002). Furthermore, the altered response to viral infection of tobacco plants perturbed in the ubiquitin system (Becker et al., 1993) supports the concept that the ubiquitin/proteasome system participates in the molecular dialogue between viruses and their host plants. In some cases, pathogens have found ways to hijack the plant UPS in order to reprogramme it for their own purposes (Angot et al., 2006; Baumberger et al., 2007; Bortolamiol et al., 2007; Pazhouhandeh et al., 2006). In other cases, viral proteins themselves are targets of the ubiquitin‐dependent proteasome pathway (Drugeon and Jupin, 2002; Reichel and Beachy, 2000). However, whether these degradation events are obligatory steps in the virus cycle to ensure effective viral invasion, or whether they play a role in a defence response, is presently unknown.
The 26S proteasome core element is the 20S proteasome, a barrel‐shaped protein complex of four stacked rings defining three internal chambers. The outer rings are made up of seven α subunits and the inner rings of seven β subunits, giving the complex the overall structure of α1–7/β1–7/β1–7/α1–7. Three proteolytic activities, carried by the β1, β2 and β5 subunits are housed by the inner rings, defining a catalytic chamber. Access to this chamber is controlled by the α rings, which only allow unfolded proteins to enter (Vierstra, 2003). When associated with one or two 19S regulatory particles, the 20S proteasome defines a 26S proteasome, involved in ubiquitin‐ and ATP‐dependent protein degradation (Dielen et al., 2010; Vierstra, 2003).
Interestingly, proteolytic activities are not the only enzymatic activities harboured by the 20S proteasome. Indeed, an unexpected ribonuclease (RNase) activity has also been identified in animal and plant proteasomes (Ballut et al., 2003; Gautier‐Bert et al., 2003; Tsukahara et al., 1989). In animals, this intrinsic proteasome‐associated endoribonucleolytic activity targets viral RNAs and some cellular mRNAs containing tRNA‐like structures. In vitro, it also interferes with protein synthesis from viral RNAs and mRNAs from virus‐infected cells (Gautier‐Bert et al., 2003; Homma et al., 1994). This RNase activity has been demonstrated to be associated with the α5 subunit (Petit et al., 1997; Pouch et al., 1995) and has been shown to degrade the genomic RNA of Tobacco mosaic virus (TMV).
The plant proteasome has also been shown to carry a similar RNase activity, degrading both Lettuce mosaic virus (LMV) and TMV RNAs in vitro (Ballut et al., 2003), prompting the suggestion that this activity may be part of a general antiviral defence pathway. In such a scenario, a counter‐effect by viral proteins could be foreseen. Indeed, the LMV HcPro (helper component proteinase) protein, involved in various steps of the potyviral cycle (replication, cell‐to‐cell movement, aphid transmission) and a known suppressor of post‐transcriptional gene silencing (Brigneti et al., 1998), associates in vivo and in vitro with purified sunflower and cauliflower 20S proteasomes (Ballut et al., 2005). Ballut et al. (2005) further demonstrated that LMV HcPro can interact and interfere with the RNase catalytic activity of the 20S proteasome, and Jin et al. (2007) partially confirmed these results by demonstrating in vitro and in vivo a physical interaction between Potato virus Y (PVY, Potyvirus) HcPro and three Arabidopsis20S proteasome subunits (α1, β2 and β5).
However, the involvement of 20S proteasome RNase activity in plant defence remains an enigmatic question that clearly needs further investigation. In this study, we investigated whether the α5 subunit of plant proteasomes carries an RNase activity, as observed in animal proteasomes. Using in planta assays, the interaction between the LMV HcPro protein and the plant proteasome α5 subunit was analysed. To test the biological relevance of this interaction, we investigated the infection of LMV in Arabidopsis thaliana knockout (KO) plants. Our results show that downregulation of one copy of the α5 gene leads to higher LMV accumulation, suggesting a negative role of α5 in LMV infection.
RESULTS
Recombinant A. thalianaα5 subunits PAE1 and PAE2 display an RNase activity in vitro
As we had obtained preliminary indications that the Arabidopsis 20S proteasome displays RNase activity (S. Badaoui, unpublished results), the two A. thalianaα5 subunit isoforms were expressed in Escherichia coli in order to further analyse this property. In Arabidopsis, the α5 subunit is encoded by two highly homologous paralogues, namely PAE1 and PAE2. The coding regions of PAE1 and PAE2 were cloned into an E. coli bacterial expression vector (pDEST™17), and the corresponding proteins were expressed and purified to homogeneity as described in Materials and methods (Fig. 1). The Aequorea victoria green fluorescent protein (GFP) coding sequence was cloned in parallel and the expressed GFP was purified using the same purification procedure as described for the α5 isoforms, in order to constitute a negative control of RNase activity.
Figure 1.
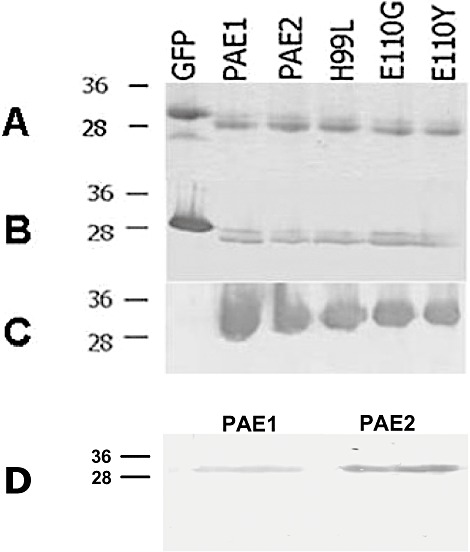
Analysis of different recombinant α5 proteins and GFP expressed in Escherichia coli and purified by affinity chromatography. Samples were analysed by 15% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and proteins were visualized by Coomassie blue staining (A) and Western blot analysis with anti‐His antibodies (B) or polyclonal rabbit anti‐PAE‐peptide serum (C, D). (D) PAE1 (left) and PAE2 (right) proteins visualized with anti‐PAE‐peptide serum. One microgram of each protein was loaded on the gel. GFP, recombinant m5‐green fluorescent protein; PAE1 and PAE2, A. thalianaα5 isoforms; H99L, E110G and E110Y, mutant proteins derived from PAE1 isoform. Molecular mass markers (kDa) are indicated to the left.
In order to test whether the recombinant A. thalianaα5 subunits harbour an RNase activity, three in vitro transcribed RNA substrates were used (according to 2003, 2005): two viral RNAs corresponding to the 3′‐end of the LMV genome (1036 bases) and the 3′‐end of the TMV genome (1315 bases), previously shown to be degraded by the sunflower 20S proteasome, and a cellular RNA encoding the sunflower calmodulin (494 bases), previously shown to be resistant to degradation by the sunflower 20S proteasome (Ballut et al., 2003). Each RNA substrate was incubated with native or heat‐denatured PAE1, PAE2 and GFP, or in the absence of any protein (negative control), for 30 min at 37 °C. The degradation products were analysed on 3‐(N‐morpholino)propanesulfonic acid (MOPS)–agarose 2% denaturing gels. At least three independent digestion assays were performed, using two independently purified preparations of each recombinant protein, leading, in each case, to reproducible results. As shown in Fig. 2, both recombinant PAE1 and PAE2 harbour an RNase activity which degrades LMV, TMV and calmodulin substrates (Fig. 2A–C). Under similar assay conditions, no degradation was observed in the various substrates incubated with GFP or in buffer alone. The degradation of the RNA substrates was abolished by pretreatment of the PAE1 or PAE2 proteins for 15 min at 100 °C. Scanning of the gels revealed that about 80% of the TMV substrate was degraded by PAE1 and PAE2, whereas about 70% of the LMV substrate was degraded (Table 1). Moreover, the addition of low concentrations of sodium dodecylsulphate (SDS) (0.01%, p/v) to the TBK240 incubation buffer abolished the degradation of the substrates by the PAE1 and PAE2 proteins, but had no detectable effect on the degradation activity of RNase A (Fig. 2D).
Figure 2.

Degradation of Lettuce mosaic virus (LMV), Tobacco mosaic virus (TMV) and calmodulin RNA substrates by recombinant PAE1 and PAE2 proteins. Thirty picomoles of purified recombinant protein (1 µg) were incubated with 1 µg (14–34 pmol) of in vitro‐transcribed RNA for 30 min at 37 °C in a final volume of 20 µL of TBK240 buffer. The products were then analysed on 2% agarose denaturing gels. Different conditions were tested: RNA was incubated with native or heat‐treated protein (boiled), and three negative controls were made: RNA incubated in TBK240 (Buffer), RNA incubated in boiled TBK240 (Buffer boiled) and RNA not incubated before gel analysis (No treatment). Molecular markers in kilobases (0.5–10‐kb RNA‐ladder; Invitrogen) are indicated on the left. (A) Calmodulin RNA substrate. (B) LMV 3′‐end RNA substrate. (C) TMV 3′‐end RNA substrate. (D) Effect of sodium dodecylsulphate (SDS) concentration on the degradation of LMV 3′‐end RNA substrate by recombinant PAE1 and PAE2 proteins and by RNase A. ‘LMV’, no treatment. (E) Comparison of LMV 3′‐end RNA degradation by wild‐type and mutant PAE1 proteins. The level of remaining undigested RNA was quantified by measuring the intensity of the bands using ImageJ software. The values obtained for all recombinant proteins were compared with the mean value obtained for the three negative controls. The standard deviation is shown by error bars (experiments were repeated three times).
Table 1.
Quantification of the level of RNA degradation induced by α5 proteins and green fluorescent protein (GFP).
We next evaluated the minimum amount of purified PAE1 needed to degrade 1 µg of the LMV 3′‐end RNA. Figure 3 shows the results of two experiments performed with independently purified PAE1 preparations, and indicates that the RNase activity associated with the PAE1 preparations is low, as RNA degradation was only observed under the assay conditions with a minimal molar ratio of 6.25:1 (PAE1 : LMV 3′‐end). This result is in agreement with those obtained by Jorgensen and Hendil (1999), who showed that free human α5 subunit possesses low RNase activity (4 mmol of TMV RNA degraded per mole of enzyme per minute).
Figure 3.

Purified PAE1 expressed in Escherichia coli possesses a very low RNase activity. Increasing amounts of purified PAE1 (0.1–1 µg, i.e. 3–30 pmol) were incubated with 16 pmol (1 µg) of in vitro‐transcribed LMV 3′‐end RNA for 30 min at 37 °C in a final volume of 20 µL of TBK240 buffer. The products were then analysed on a 2% agarose denaturing gel, and the percentage of remaining RNA was evaluated as described in Fig. 2. Experiments 1 and 2 were performed with PAE1 protein from two independent purification experiments.
Mutations of glutamic acid at position 110 affect A. thaliana PAE1 RNase activity
On the basis of the amino acid sequence homologies observed between the α5 subunits from 23 organisms belonging to divergent taxonomic groups, the tridimensional structural data of the mammalian α5 (Unno et al., 2002) and the known importance of histidine residues in the active site of endoribonucleases (Royo et al., 1994; Zegers et al., 1992), two highly conserved residues located on the surface of the PAE1 protein, and potentially exposed in the proteasome lumen, were identified as potential active site candidates for mutagenesis: the histidine at position 99 (H99) and the glutamic acid at position 110 (E110) (Fig. S1, see Supporting Information). Thus, three PAE1‐derived mutants were obtained, namely H99L, E110G and E110Y, and purified using the same strategy as that used for the wild‐type subunits (Fig. 1). Subsequent assays using the described RNA substrates were performed to test the RNase activity of the purified mutant proteins. Our results showed that none of the mutations tested led to the total inhibition of PAE1 RNase activity, with all mutants showing degradation of the substrates at a level significantly different from the negative controls (Fig. 2E and Table 1).
The percentage decrease in RNase activity observed for the mutant H99L, relative to PAE1 activity, was very mild: around 2.5% and 6% inhibition when using the TMV 3′‐end and LMV 3′‐end substrates, respectively (Table 1). Therefore, the H99L mutation does not seem to have any detectable influence on PAE1 RNase activity.
However, both the E110G and E110Y mutations caused a significant inhibition of PAE1 activity. Values of inhibition of 39% and 26% were triggered by the E110G mutation when using the LMV 3′‐end and TMV 3′‐end substrates, respectively, and 36% inhibition was observed for the calmodulin substrate. The E110Y mutation seems to be even more drastic, triggering inhibition values of 72% and 42% when using the LMV 3′‐end and TMV 3′‐end substrates, respectively, with 53% inhibition being observed for the calmodulin substrate. These results, which were reproduced with two independently purified mutant preparations, clearly indicate that mutations of glutamic acid at position 110 affect, directly or indirectly, PAE1 RNase activity.
LMV HcPro interacts in vivo and colocalizes in planta with A. thaliana PAE1
A previous report (Ballut et al., 2005) has demonstrated a physical interaction between the purified sunflower 20S proteasome and the HcPro of LMV, with an ensuing inhibition of the in vitro proteasomal RNase activity. Interactions between Arabidopsis proteasome subunits (α1, β2 and β5) and a potyviral HcPro have recently been shown for PVY (Jin et al., 2007). Interestingly, these authors were not able to detect any interaction between the α5 subunit and the PVY HcPro in their extensive screen of the Arabidopsis proteasome subunits. We performed bimolecular fluorescence complementation (BiFC) assays between PAE1 or PAE2 and LMV HcPro. In BiFC, yellow fluorescent protein (YFP) is split into two nonfluorescent fragments, YN and YC. The possible interaction partners are fused with these fragments, which, on interaction of two protein partners, are brought into close contact, resulting in the restoration of a fluorescent YFP (Walter et al., 2004). Different combinations of HcPro and PAE transient expression vectors were cobombarded into onion epidermal cells to test whether the proteins could interact.
As shown in Fig. 4, PAE1‐YC and PAE2‐YC were able to interact with YN‐HcPro in the cytoplasm of onion epidermal cells. YFP fluorescence was detected in the experimental samples 16–18 h after bombardment, whereas it was not detected in any of the negative controls (i.e. only YN‐HcPro or PAE2‐YC or PAE1‐YC expressed alone, or in the presence of unrelated YN or YC fusion proteins).
Figure 4.

Visualization of the interaction between the α5 subunits PAE1 and PAE2 and Lettuce mosaic virus helper component proteinase (LMV‐HcPro) in onions cells using a bimolecular fluorescence complementation (BiFC) assay. (A) YN‐HcPro + PAE1‐YC. (C) YN‐HcPro + PAE2‐YC. (E) Negative control: YN‐HcPro alone. (A, C, E) Yellow fluorescent protein (YFP) filter. (B, D, F) Bright field images of the same cells.
In order to compare the subcellular accumulation pattern of α5 in healthy lettuce cells and in LMV‐infected cells, a YFP‐tagged PAE1 was transiently expressed by agroinfiltration in the leaves of lettuce plants that had been inoculated previously with a recombinant LMV isolate expressing a GFP‐tagged version of its HcPro (LMV‐GFPHC) (German‐Retana et al., 2003). Figure 5A–C shows that fluorescence from PAE1‐YFP is observed in both the cytoplasm and nuclei of healthy lettuce epidermal cells (see also Fig. S2 in Supporting Information).
Figure 5.
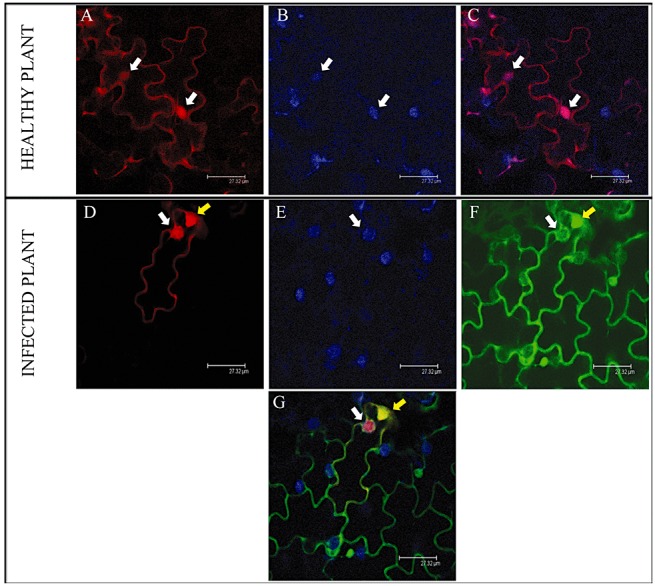
Subcellular localization of PAE1‐YFP and GFP‐HcPro fusion proteins in cells of healthy and Lettuce mosaic virus (LMV)‐infected leaves of Lactuca sativa (lettuce). The PAE1‐YFP‐expressing construct was delivered by agroinfiltration into healthy plants and visualized by confocal microscopy 4 days after infiltration. White arrows indicate the position of the nuclei identified by diamidino‐2‐phenylindole (DAPI) staining. (A) AtPAE1‐YFP. (B) Nucleus stained with DAPI. (C) Overlay images (A + B). The PAE1‐YFP construct was delivered by agroinfiltration into LMV‐GFP‐HcPro‐infected leaf areas. (D) PAE1‐YFP. (E) Nucleus stained with DAPI. (F) GFP‐HcPro. (G) Overlay images (D + E + F). White arrows indicate PAE1‐YFP localized in the cell nucleus, whereas yellow arrows indicate the GFP‐HcPro aggregates in the cytoplasm.
The subcellular localization of PAE1‐YFP and GFP‐HcPro fusions in infected lettuce leaves post‐inoculation was then analysed. Figure 5D–G shows the same experiment performed in the leaves of lettuce plants that had been pre‐inoculated 3–4 weeks previously with the LMV‐GFPHC recombinant isolate. As shown in Fig. 5F, the GFP‐HcPro fusion protein is mostly observed in the cytoplasm of infected cells and tends to form large and very bright aggregates often situated close to the cell nucleus. These large aggregates (about 7 µm in diameter and similar in size to the nucleus) are not artefacts caused by the agroinfiltration procedure as they are also present in nonagroinfiltrated leaves (Fig. S3, see Supporting Information). Interestingly, when expressed in such LMV‐GFPHC‐infected cells, PAE1‐YFP is not only detected in the nucleus and cytoplasm, as in healthy cells (Fig. 5A–C), but is also observed to significantly relocalize to the GFP‐HcPro aggregates in the cytosol (Fig. 5D–G). Altogether, these results show that not only do PAE1‐YFP and GFP‐HcPro interact in planta, but also that they colocalize when PAE1‐YFP is transiently expressed in LMV‐GFPHC‐infected lettuce leaves.
Pae1 KO Arabidopsis plants are more susceptible to LMV
We first evaluated whether PAE1 and PAE2 expression could be altered during LMV infection. Both wild‐type and single‐KO plants were either mock or LMV inoculated, and PAE1 and PAE2 gene expression was determined by quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) at 7, 14 and 21 days post‐inoculation (dpi). Figure 6A shows that, at 21 dpi, LMV infection does not induce any significant modification in PAE1 and PAE2 gene expression relative to mock‐inoculated plants, in both wild‐type and KO pae1 or pae2 plants. Similar results were obtained at 7 and 14 dpi (data not shown).
Figure 6.

Relative quantification of pae1 and pae2 gene expression by quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR). (A) Expression of PAE genes in wild‐type (Col0 WT) and mutant (KO pae) Arabidopsis plants inoculated or not (mock) with Lettuce mosaic virus (LMV). Gene expression was quantified in systemic leaves from five plants at 21 days post‐inoculation (dpi). Transcript levels were normalized to the expression of transducin and actin 11 genes. The standard deviation from three replicates is indicated by vertical lines. A chi‐squared (χ 2) test was performed to confirm that there is no significant difference between pae gene expression in wild‐type compared with KO pae plants. (B) LMV accumulation in wild‐type and KO mutant Arabidopsis plants. Virus accumulation was measured by SYBR® Green real‐time PCR after reverse transcription of total RNA extracted from systemic leaves from five plants at 21 dpi. Results were normalized using transducin and actin 11 genes. The standard deviation from three replicates is indicated by vertical lines. In the experiment shown, the fold of enhanced viral accumulation in KO pae plants relative to Col0 WT is indicated as 4.8 times for KO pae1 and 2.3 times for KO pae2. A chi‐squared (χ 2) test was performed: the null hypothesis (no significant difference between the expected and observed results) was rejected, confirming the statistical significance of the difference observed between the KO and wild‐type plants.
As HcPro interacts in vivo with the proteasome α5 subunit (this study) and inhibits its associated RNase activity in vitro (Ballut et al., 2005), we aimed to determine whether the single‐KO mutations have an impact on viral infection. For this purpose, we compared LMV accumulation in KO and wild‐type plants using quantitative RT‐PCR analysis. Virus accumulation was quantified at 7 dpi in inoculated leaves, and at 14 and 21 dpi in systemically infected leaves. Although the results obtained for inoculated leaves were not reproducible, in systemically infected leaves at 21 dpi, LMV accumulation was 1.7–4.8‐fold higher in KO pae1 mutant plants than in wild‐type plants (Fig. 6B). LMV accumulation in KO pae2 mutant plants was found to be significantly different from that in the wild‐type in only one experiment of the four performed (Fig. 6B).
Altogether, these results show that LMV infection has no impact on the level of PAE gene expression in both wild‐type and KO mutants, and that the lack of expression of the PAE1 gene induces a significant increase in viral accumulation.
DISCUSSION
Recombinant plant α5 subunits display an RNase activity in vitro
Although described more than 10 years ago for the animal proteasome (Pouch et al., 1995), proteasomal RNase activity is still poorly characterized, in particular when it comes from plant proteasomes. Previous results describing an endoribonucleolytic activity associated with the sunflower 20S proteasome, and the demonstration that it is active on viral RNAs and inhibited by LMV HcPro (2003, 2005), provided the first evidence of an RNAse activity associated with plant proteasomes. In animals, such an activity was found to be associated with the α5 subunit purified from a 20S proteasome complex (Petit et al., 1997). Thus, our first objective in this study was to determine whether the same situation could apply to plant proteasomes.
In this study, we showed for the first time that an E. coli‐expressed α5 subunit of the Arabidopsis proteasome harbours an RNase activity detectable in vitro. The potential association of a contaminating RNase activity from E. coli in the samples of the recombinant plant α5 protein [potentially undetected by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE)] can be excluded as: (i) a hexahistidine‐tagged recombinant GFP protein similarly expressed in E. coli and purified using exactly the same procedure did not harbour such an activity; (ii) mutation of a highly conserved glutamic acid residue predicted to be located in the proteasome lumen inhibited the nuclease activity of the Arabidopsisα5 subunit. In line with these observations, we also showed that denaturation of the α5 recombinant protein by heat treatment (incubation at 100 °C) before setting up the RNA degradation assay completely abolished the associated RNase activity, which is not the case for other RNases, such as RNase A. Moreover, the addition of low concentrations of SDS (0.01%, p/v) to the TBK240 incubation buffer also abolished substrate degradation by the PAE1 and PAE2 proteins, but had no effect on the degradation activity of RNase A. This sensitivity to SDS and heat is in complete agreement with previous results obtained for plant and animal 20S proteasomal RNase activity (Ballut et al., 2003). In animals, it has been shown that proteasomes washed with 8 m urea still harbour an RNase activity (Petit et al., 1997), and this was confirmed in plants, as RNase activity remained associated with the sunflower 20S proteasome after washing with detergent (0.5% sodium laurylsarcosyl) and 6 m urea (Ballut et al., 2003). There are therefore several lines of evidence arguing against the hypothesis of any RNase contamination, demonstrating that plant proteasomal endonuclease is an intrinsic activity of the 20S proteasome.
This study provides the first demonstration of an RNase activity associated with a free plant proteasome α5 subunit. Our results are in agreement with those obtained by Petit et al. (1997) and Jorgensen and Hendil (1999) in animals and humans, who showed that free α5 subunits from calf liver and human erythrocyte extracts possess RNase activity in vitro. In HeLa cells, a large proportion of α5 was found in the free state, in both nuclei and cytoplasm, rather than incorporated into proteasomes (Jorgensen and Hendil, 1999), but the physiological function of the relatively abundant α5 monomers is not known. Interestingly, the recombinant α5 homologue of Trypanosoma brucei 20S proteasome, when expressed and purified from E. coli, demonstrated spontaneous self‐assembly into four stacked heptamer rings, as revealed by electron microscopy (Yao et al., 1999). Preliminary observations with the purified PAE1 and PAE2 preparations indicate that the α5 subunits are present as large macromolecular complexes, but are not observed as structured rings under the electron microscope (J. Walter and P. Bron, personal communication).
The data obtained from the site‐directed mutagenesis studies of PAE1 revealed that mutation of a glutamic acid residue at position 110 has a significant impact on the RNase activity associated with α5, reducing it by as much as 73% and 42% when using the LMV 3′‐end and TMV 3′‐end substrates, respectively. However, this endonuclease activity was not so drastically affected by a second mutation introduced at the same site (E110G), an indication that E110 probably does not localize in the active site itself, but may still play an important role in the protein conformation suitable for RNase activity. This possibility is further strengthened by site‐directed mutagenesis performed on recombinant rat α5 subunit. Mutation of a histidine residue at position 155, which is also predicted to be localized at the surface of the protein in the lumen of the 20S proteasome according to Phyre analysis (Kelley and Sternberg, 2009), dramatically changed the nuclease activity but did not abolish it (S. Badaoui and S. Apcher, personal communication).
The A. thalianaα5 RNase activity observed in the present work is very low. A similar situation has also been pointed out for both human α5 and 20S proteasome RNase activities (Jorgensen and Hendil, 1999), and for the sunflower proteasome (S. Badaoui, personal communication). This low activity could be linked to the partial renaturation of the protein during the purification procedure. Another possibility is that of inappropriate folding of the protein in the E. coli host, which could possibly be linked to the fact that the majority of the expressed proteins remained in the insoluble pellet following cell lysis. An alternative is that this low RNase activity could be an intrinsic property of α5 itself, and possibly a result of the low activity observed in 20S proteasomes themselves.
Is there possible α5 substrate specificity?
The results reported here for the recombinant A. thalianaα5 show that it degrades efficiently both of the viral RNA substrates used (TMV 3′‐end and LMV 3′‐end) and the nonviral calmodulin substrate. These results are in partial agreement with those of Ballut et al. (2003), who showed that TMV‐ and LMV‐derived RNAs were sensitive to degradation by the sunflower 20S proteasome, whereas calmodulin mRNA was not. Our results are also in general agreement with those showing that cytokine mRNAs with AU‐rich sequences and certain viral mRNAs are good substrates for animal proteasomes (Jarrousse et al., 1999; Pouch et al., 1995). Moreover, it has been demonstrated that the translation of TMV genomic RNA is impeded in the presence of 20S proteasomes, whereas translation of cellular globin mRNAs is not affected (Homma et al., 1994). More recently, Gautier‐Bert et al. (2003) showed that animal proteasomes strongly associate with AU‐rich sequences, forming RNA/20S complexes in vitro.
It has therefore been suggested that the 20S proteasome could be involved in the decay of AUU‐rich mRNAs, and that the recognition signal for degradation could be linked to the presence of AUUUA sequences (Gautier‐Bert et al., 2003; Jarrousse et al., 1999). Interestingly, although multiple AUUU sequences are present in the genomic RNA of TMV and LMV, both being targets for proteasomal degradation (three AUUU repeats in the TMV 3′‐end and one repeat in the LMV 3′‐end), they are not present in the calmodulin RNA substrate, indicating that the presence of such a sequence is not a prerequisite for degradation by the α5‐associated RNAse activity. Clearly more work is needed to further delineate a potential role for AUUU repeats, or for other sequence elements, in determining the potential specificity of the 20S proteasome‐associated RNAse activity.
Role of gene redundancy?
Each of the highly homologous copies of α5 in A. thaliana, namely PAE1 and PAE2 (98.73% of amino acid identity), harbours an RNase activity detectable in vitro. Although the Arabidopsis proteasome has a highly similar organization when compared with its yeast and animal counterparts (Yang et al., 2004), the main difference between the A. thaliana proteasome and those from nonplants, is its heterogeneity, linked to the existence of gene pairs for most of the α and β subunits. Available microarray data show that the expression patterns of PAE1 and PAE2 appear to be nearly identical in different plant tissues, and that expression variations of these two genes are highly coregulated (http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php#CO1). The fact that the homozygous KO insertion mutants in the genes coding for PAE1 and PAE2 are viable, whereas the double‐KO mutant is not, further confirms the existence of a functional redundancy between these two genes.
HcPro–α5 interaction
Previous results have shown that the viral protein HcPro of LMV can interfere with the proteasome‐associated degradation process (Ballut et al., 2005), suggesting that a regulation (or modulation) of the proteasome 20S‐associated RNase activity can occur during viral infection. It is noteworthy that a modulation of the proteolytic activity is similarly suggested by results showing that the interaction between LMV HcPro and the sunflower 20S proteasome results in a small, but significant, activation of at least some of the proteasome activities (Ballut et al., 2005). HcPro, when interacting with the 20S complex, could recognize and interact with multiple α or β subunits. Such a hypothesis is in line with the results of Jin et al. (2007), who showed an interaction between HcPro of another potyvirus (PVY) and three other proteasomal subunits: α1, β2 and β5. Although these authors were unable to detect any interaction between α5 and the PVY HcPro using the yeast two‐hybrid screens, the use of this technique to search for potential interactions is notorious for having a number of limitations. Therefore, it is equally possible that the HcPro–α5 interaction reported here is virus specific, or that the HcPro–α5 interaction reported here also applies to PVY.
The HcPro–α5 interaction was observed in vivo using BiFC in epidermal onion cells. In this system, both PAE1 and PAE2 isoforms interact efficiently with HcPro, further confirming that these two very closely related proteins have essentially similar properties. Similar results have been obtained using another in vivo system involving turnip protoplasts electroporated with the constructs PAE1‐YC and YN‐HcPro (M. Drücker, personal communication).
The existence of an HcPro–α5 interaction was further confirmed by the partial relocalization of α5 to GFP‐HcPro‐containing aggregates observed in LMV‐infected lettuce leaves. Although the exact nature of these aggregates is unknown, it is generally accepted that all potyvirus‐induced inclusions contain nonstructural proteins that are functionally important for virus replication, cell‐to‐cell movement or vector transmission (Shukla et al., 1994). In PVY‐infected tissues, using immunogold labelling techniques, the HcPro protein was shown to be localized in rod‐shaped cytoplasmic inclusions, known as amorphous inclusions (AIs) (Baunoch et al., 1990). Such HcPro‐containing inclusions have been reported to be induced by some other potyviruses, and appear as irregularly shaped, mostly rounded, masses possessing a homogeneous substructure of either finely granular material or tubular, fibrillar or rod‐like elements (Lesemann, 1988), raising the possibility that the aggregates observed in LMV‐GFPHC‐infected cells could indeed be AIs. Whether these cytoplasmic inclusions can be regarded as deposition sites of excessively produced viral nonstructural proteins (Riedel et al., 1998), or as having biological significance, remains to be elucidated. The biological consequence, if any, of the partial relocalization of α5 to the GFP‐HcPro‐labelled aggregates similarly remains to be investigated. Indeed, for another plant virus belonging to a different taxonomic group, Cauliflower mosaic virus (CaMV), Khelifa et al. (2007) have recently demonstrated that electrolucent inclusion bodies, mainly composed of the viral helper component, are complex viral structures specialized for aphid transmission. These authors suggested that viral inclusion bodies other than viral factories, most often considered as ‘garbage cans’, can in fact exhibit specific functions (Khelifa et al., 2007).
A central question raised by the multiple findings of potyviral HcPro interactions with different 20S proteasome subunits (Jin et al., 2007; this work) is that of the biological significance of these interactions and, in particular, whether the RNase activity of α5 or other proteasome activities contribute to an antiviral response that is countered by HcPro. Although little is known about proteasome inhibition by plant viruses, there are numerous examples of such a strategy for animal viruses, with human immunodeficiency virus‐1 (HIV‐1) being a well‐studied case. The HIV Tat protein interacts in vitro and in vivo with the α3 and α7 subunits, with six β subunits, leading to the inhibition of proteasome proteolytic activities in vivo (Apcher et al., 2003).
The results presented here, showing that At‐pae1 KO A. thaliana mutants display a significantly increased susceptibility to LMV infection with respect to wild‐type plants, with viral accumulation at 21 dpi increased by a factor of 1.7–4.8, go in this direction.
However, efforts are clearly needed to unravel the precise role of α5, and to determine whether α5 RNase activity, shown in vitro, may play an inhibitory role by affecting directly HcPro function, or by the induction of an unknown defence mechanism which might be compromised by HcPro.
EXPERIMENTAL PROCEDURES
Expression of A. thalianaα5 subunits and GFP in E. coli
The wild‐type coding sequences of the two A. thalianaα5 subunit isoforms [PAE1 (At1G53850) and PAE2 (At3G14290)] were cloned into the vector pENTR/D‐TOPO® (Invitrogen, Cergy Pontoise, France) following RT‐PCR amplifications performed on total RNA extracted from Arabidopsis leaf tissues using TRI Reagent (Sigma‐Aldrich, St. Louis, MO, USA). The m5‐GFP (modified GFP) coding sequence (GenBank Accession M62653) was amplified using the primers listed in Table 1 and cloned into pENTR/D‐TOPO®. The PAE1 recombinant plasmid was used as a template for PCR amplification to generate point mutations in the PAE1 sequence using the QuickChange® site‐directed mutagenesis kit (Stratagene/Agilent Technologies, Santa Clara, CA, USA). The mutations introduced were H99L, E110G or E110Y, where the first letter indicates the residue in the wild‐type sequence and the second letter the residue present in the mutated protein. The sequences of the amplification and mutagenesis primers are listed in Table 2.
Table 2.
List of the oligonucleotides used in this study.
| Oligonucleotides used for the cloning of PAE1, PAE2 and m5‐GFP coding sequences | |
| Primer name | |
| paeATG p | 5′‐CACCATGTTTCTCACTAG‐3′ |
| pae1 stop m | 5′‐TCAATCAAAGACGAGCGATG‐3′ |
| paeATG p | 5′‐CACCATGTTTCTCACTAG‐3′ |
| pae2 stop m | 5′‐TCACTTCATAGACGACTGATGACG‐3′ |
| GFP p | 5′‐CACCATGAGTAAAGGAGAAG‐3′ |
| GFP m | 5′‐TTATTTGTATAGTTCATC‐3′ |
| Oligonucleotides used for mutagenesis experiments of PAE1 subunit | |
| Primer name | |
| H 99 L fwd | 5′‐GAGTTGAAACTCAAAACTTGAGATTCTCGTATGGTG(SmlI)‐3′ |
| H 99 L rvs | 5′‐CACCATACGAGAATCTCAAGTTTTGAGTTTCAACTC(SmlI)‐3′ |
| E 110 G fwd | 5′‐AGCCGATGACTGTAGGATCCACAACACAAGCA(BamHI)‐3′ |
| E 110 G rvs | 5′‐TGCTTGTGTTGTGGATCCTACAGTCATCGGCT(BamHI)‐3′ |
| E 110 Y fwd | 5′‐AGCCGATGACTGTATACTCCACAACACAAGCA(AccI)‐3′ |
| E 110 Y rvs | 5′‐TGCTTGTGTTGTGGAGTATACAGTCATCGGCT(AccI)‐3′ |
| Oligonucleotides used for plant single mutant genotyping | |
| Primer name | |
| LP PAE1 | TGCTACCTTGGCAATATCCAC |
| RP PAE1 | TGATGATATATCAAGCAGAAGC |
| LP PAE2 | TGGGGTTTTAAATTTTTGTTGTG |
| RP PAE2 | TTTTTCCGACGTACAATGATG |
| LB | TGGTTCACGTAGTGGGCCATCG |
| Oligonucleotides used for quantitative RT‐PCR analysis | |
| Primer name | |
| Actin 11 fwd | 5′‐GCCATCAATAGTCCACAGGA‐3′ |
| Actin 11 rvs | 5′‐TGCAACAAAGTCGATGAACA‐3′ |
| Transducin fwd | 5′‐TGATGATGGGATGGTCAAG‐3′ |
| Transducin rvs | 5′‐TCAGAAAAAGTAAAACAACACAGG‐3′ |
| ncPAE1m | 5′‐GAATGACATGGGAGCCAAAAG‐3′ |
| nc PAE1p | 5′‐GGGTAATTTACCTATCAGCTTGTG‐3′ |
| nc PAE2m | 5′‐AAAACTCAACTCGATAAAATCC‐3′ |
| nc PAE2p | 5′‐CTCTTCCTGTTATCTTACCTTGTC‐3′ |
| LMV 4222m | 5′‐GCTAACGGACGAGTTGG‐3′ |
| LMV 4253p | 5′‐ATGACGTTCACTCGTGAAAC‐3′ |
| TuMV 353 F | 5′‐ACACGCCAGCAGCCTAACG‐3′ |
| TuMV 449R | 5′‐GCATCCTCAACACGGCATTCG‐3′ |
The mutagenesis oligonucleotides were designed to introduce recognition sites for restriction endonucleases used for screening purposes (recognition sites in bold, restriction endonucleases indicated in parentheses).
Using Gateway® Technology (Invitrogen), the coding regions of each wild‐type or mutant protein were transferred from pENTR/D‐TOPO® into the pDEST™17 destination vector to allow the production of N‐terminal fusions harbouring a 6 × His tag. The constructs were introduced into E. coli (strain BL21‐AI) for protein expression.
Recombinant proteins harbouring the 6 × His tag were expressed following the manufacturer's instructions (Invitrogen). As a result of the insolubility of the expressed PAE proteins, purification was performed under denaturing conditions. For this, protein pellets were solubilized in TBK240 buffer (20 mm Tris/HCl pH 7·4, 5 mm MgCl2, 240 mm KCl) containing 8 m urea. Following clarification, the solubilized proteins were applied to a TALON® Superflow Resin (Clontech/Takara Bio group, Otsu, Shiga, Japan), and urea was progressively eliminated by rinsing the beads with TBK240 buffer containing diminishing concentrations of urea. Protein purification was then performed following the manufacturer's instructions. After elution in TBK240 buffer containing 500 mm imidazole, proteins were extensively dialysed against TBK240 buffer and stored at −80 °C until use.
Protein analysis
Recombinant GFP and PAE proteins were analysed by 15% SDS‐PAGE. Subsequent Western blots were performed using both anti‐6His mouse monoclonal antibodies (Sigma‐Aldrich) or a polyclonal rabbit anti‐peptide antiserum (Eurogentec, Liège, Belgium) directed against two PAE‐derived peptides: SEGADSSLQEQFNK (amino acids 172–185) and RVETQNHRFSYGEPMT (amino acids 93–108).
In vitro transcription
A 1‐kb DNA fragment harbouring the 3′ end of the LMV genomic RNA (LMV 3′), a 1.3‐kb DNA fragment harbouring the 3′ end of the TMV genomic RNA (TMV 3′) and a sunflower calmodulin cDNA (Ballut et al., 2003) were used as transcription templates. One microgram of each linearized plasmid was used for in vitro transcription employing the mMESSAGE mMACHINE® High Yield Capped RNA Transcription Kit (Ambion/Applied Biosystems, Austin, TX, USA). After 3 h at 37 °C, the transcribed RNAs were treated with DNase, purified on micro‐spin columns (Harvard Apparatus, Courtaboeuf, France), quantified by the measurement of the optical density at 260 nm (OD260) and stored at −70 °C.
In vitro RNase assay
One microgram of each transcribed RNA, LMV 3′, TMV 3′ and calmodulin (16, 14 and 34 pmol, respectively), was incubated with 1 µg of each purified protein (30 pmol) in 20 µL of TBK240 buffer for 30 min at 37 °C. This 1:1 (µg) substrate to protein ratio was chosen according to Ballut et al. (2003). Subsequently, 5 µL of loading buffer [formamide, 50%; formaldehyde, 6%; xylene cyanol, 0.25% p/v; bromophenol blue, 0.25% p/v; ethylenediaminetetraacetic acid (EDTA), 10 mm; ethidium bromide, 0.5 µg/mL] were added to the reaction and, after 5 min of heating at 85 °C, the products were analysed on a 2% MOPS–agarose denaturing gel containing 6% formaldehyde. RNA degradation was evaluated by measuring the intensity of the bands corresponding to the various undigested substrates using ImageJ software (http://rsbweb.nih.gov/ij/index.html).
Statistical analyses
Single‐factor analysis of variance (ANOVA) and regression analysis were performed using R statistical software. Assumptions concerning the homogeneity of variance and normality were evaluated from an inspection of the residuals. The level and significance of the differences between RNA degradation levels when incubated with different proteins were evaluated by Tukey's test, assuming that the three degradation replicate reactions approximated the variance between fully independent observations.
BiFC assay
PAE1, PAE2 and LMV HcPro coding sequences cloned in pENTR/D‐TOPO® (Invitrogen) were individually recombined using the Gateway® system into BiFC plasmids nEYFP/pUGW2, cEYFP/pUGW2, nEYFP/pUGW0 and cEYFP/pUGW0 (Nakagawa et al., 2007). In these plasmids, the interacting proteins are fused to either the N‐terminal part (amino acids 1–174; NY) or the C‐terminal part (amino acids 175–239; CY) of the enhanced YFP. All BiFC constructs were confirmed by sequencing. For in vivo interactions, a combination of the plasmids encoding HcPro and PAE1 or HcPro and PAE2 (mixed at a 1:1 ratio, 2.5 µg each) was adsorbed onto 1.6‐µm gold particles and bombarded in onion (Allium cepa) epidermal cells under 1100 psi helium pressure using a Helios Gen Gun (Bio‐Rad, Hercules, CA, USA) (three replicates were made for each combination tested). Confocal imaging was performed 14–18 h after bombardment using a Leica TCS SP2 confocal microscope with a 20× dry objective and LCS Lite Leica software (Leica Microsystems, Wetzlar, Germany). The YFP fluorescence was assessed with excitation at 514 nm using an argon laser, with an emission band of 549–602 nm.
In planta colocalization assay
For transient protein expression in lettuce leaves using agroinfiltration, a binary pEarleyGate101‐PAE1 vector was prepared by transferring the PAE1 coding sequence from the pENTR/D‐TOPO® (Invitrogen) vector into the binary vector pEarleyGate‐101, thus allowing the production of a C‐terminal fusion of PAE1 in frame with YFP. For transient expression, the undersides of lettuce leaves (Lactuca sativa cv. Trocadero) were infiltrated essentially as described by Nicaise et al. (2003).
For the colocalization of α5 YFP and LMV GFP‐HcPro, the recombinant GFP‐tagged virus LMV‐GFPHC (GFP fused in frame with the N‐terminus of HcPro; German‐Retana et al., 2003) was inoculated into lettuce by biolistic bombardment (German‐Retana et al., 2008). After 3–4 weeks, the expression of GFP at the whole‐plant level was visualized using a 100‐W, hand‐held, long‐wave UV spotlight lamp (Model B‐100; UV Products, Upland, CA, USA). PAE1‐YFP was then agroinfiltrated into the abaxial face of the lettuce leaves in which fluorescence was detected. Four days after agroinfiltration, the infiltrated areas were infiltrated again, with 0.4 µm/mL diamidino‐2‐phenylindole (DAPI). Finally, microscopy observations were performed 5 h later, employing a confocal Leica TCS SP2 microscope using × 20 and × 63 water immersion objectives. The fluorescences of GFP, YFP and DAPI were assessed separately with excitation at 488, 514 and 405 nm, respectively, and emission bands of 492–511 nm, 549–602 nm and 410–475 nm, respectively. In order to differentiate specifically GFP from YFP fluorescence, GFP/YFP acquisitions were performed by sequential image recording between frames according to the manufacturer's instructions and following the protocol described in Raffaele et al. (2009).
Arabidopsis KO mutants
Arabidopsis thaliana Columbia‐0 plants were grown in a glasshouse at 25 °C with a 16‐h light period. Susceptibility assays were performed on T‐DNA insertion lines ordered from the SALK collection: PAE1 (At1g53850) mutant, SALK151939; and PAE2 (At3g14290) mutant, SALK368326. The T‐DNA insertion and the homozygous state of the mutant lines were tested by PCR of genomic DNA using primers defined on the SALK website (http://signal.salk.edu/tdnaprimers.2.html) (Table 2). Homozygous mutants were backcrossed once to wild‐type plants, and the F2 progeny were screened again for the homozygous state. The absence of mRNA in each of the KO mutants was confirmed by RT‐PCR using the primer pair ncPAE1m/ncPAE1p and ncPAE2m/ncPAE2p (Table 2) and the REDEXtract‐N‐Amp™ Plant PCR kit (Sigma‐Aldrich). No double mutants could be obtained after crossing the relevant single mutants and selfing the F1 hybrid to obtain F2 progenies, showing that the double mutants are lethal. The single mutants do not have an obvious phenotype in growth or flowering.
Virus inoculation and detection
The isolate LMV‐ExbaAF (Krause‐Sakate et al., 2005) was selected for its capacity to systemically infect Columbia‐0. Viruses were inoculated mechanically (Duprat et al., 2002), and virus accumulation in inoculated and apical noninoculated leaves (above the rosette leaves) was analysed by quantitative RT‐PCR.
Quantitative analysis by real‐time PCR
For quantitative analysis of gene expression and virus accumulation, total RNA was extracted using the SV Total RNA Isolation System (Promega, Madison, WI, USA). The concentration and purity of the extracted RNA were determined spectrophotometrically. RNA samples were subjected to DNAse treatment using the TURBO DNA‐free™ kit (Ambion), reverse transcribed and used for real‐time PCR. Each sample was analysed in triplicate. Two biological replications of the experiment were carried out, and cDNA was synthesized four times, twice from two different sets of at least five LMV‐ and mock‐inoculated A. thaliana plants for each condition.
cDNA synthesis was carried out in a final volume of 20 µL, using 1 µg of total RNA, 5 µm of poly‐dT primer and 50 units of SuperScript™ II Reverse Transcriptase (Invitrogen). Real‐time PCRs were carried out in a final volume of 25 µL, using a SYBR Green Fluorescein master mix (Abgene Limited, Epsom, UK) in an I‐Cycler thermal cycler (Bio‐Rad), following the manufacturer's instructions. Each reaction contained 1 × Absolute Blue Sybrgreen Fluorescein Master mix (Abgene), 1 µm of forward and reverse primers (Table 2) and a dilution of cDNA corresponding to 50 ng of original RNA. To determine the optimal internal reference for the normalization of gene expression, two housekeeping genes supposed to have their transcriptional activity unaltered during LMV infection were tested: actin 11 (At3g12110) and transducin (At2g26060). Primers for all target sequences (Table 2) were designed using ‘Primer 3’ software. The specificity of the amplicons was checked by melting curve analysis. For data quality control, PCR amplification was analysed through the assessment of the C t value and amplification efficiency. Expression levels were normalized to the actin and transducin genes, and relative quantification was performed using GeNex software (Bio‐Rad) from three replicates.
Supporting information
Fig. S1 Amino acid sequence alignment of plant alpha‐5 sequences. The two highly conserved amino acid residues identified according to a multiple alignment of alpha‐5 amino‐acid sequences derived from 23 organisms are Histine (position 99) and Glutamic acid (position 110). These amino acids are boxed in the multiple alignment shown for Glycine max (soybean, Q9M4T8), Oryza sativa (Rice, BAA96832) and Arabidopsis thaliana (PAE1: At1g53850; PAE2: At3g14290).
Fig. S2 Subcellular localization of PAE1‐YFP fusion protein in cells of healthy leaves of Lactuca sativa (lettuce). The PAE1‐YFP‐expressing construct was delivered by agroinfiltration in healthy plants and visualized by confocal microscopy 4 days after infiltration. White arrows indicate the position of the nucleus.
Fig. S3 (A–C) Subcellular localization of GFP‐HcPro in leaves of Lactuca sativa (lettuce). (A) Yellow arrow indicates the GFP‐HcPro aggregates in the cytoplasm. (B) White arrows indicate the nuclei stained with. (C) Overlay images (A + B). (D and E) Subcellular localization of GFP‐HcPro and PAE1‐YFP fusion proteins in cells of LMV‐infected leaves of Lactuca sativa (lettuce). The PAE1‐YFP construct was delivered by agroinfiltration into LMV‐GFP‐HcPro‐infected leaves areas. (D) GFP‐HcPro. (E) PAE1‐YFP. White arrow indicates the PAE1‐YFP localized in the cell nucleus, yellow arrow indicates the GFP‐HcPro aggregate in the cytoplasm. Scale bar – 27.06 µm.
Supporting info item
ACKNOWLEDGEMENTS
This work was supported in part by grants from the ‘Plant Health and the Environment’ division of Institut National de la Recherche Agronomique (INRA). ASD was supported by a fellowship from the French Ministry of Education and FS by a fellowship from the Coordenação de Aperfeicoamento de Pessaoal de Nivel Superior (CAPES) Agency (Brazil) within the framework of a CAPES‐Comité Français d'Évaluation de la Coopération Universitaire et Scientifique avec le Brésil (COFECUB) cooperation programme. We thank Brigitte Batailler from ‘Pôle imagerie du vegetal’ of the Bordeaux Imagery Centre for help with confocal observations. We thank the Salk Institute for the heterozygous A. thaliana mutants, and Thierry Mauduit and Marylin Roncoroni for taking care of the plants. We thank Sebastien Apcher and Lionnel Ballut for providing information prior to publication.
REFERENCES
- Angot, A. , Peeters, N. , Lechner, E. , Vailleau, F. , Baud, C. , Gentzbittel, L. , Sartorel, E. , Genschik, P. , Boucher, C. and Genin, S. (2006) Ralstonia solanacearum requires F‐box‐like domain‐containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA, 103, 14 620–14 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apcher, G.S. , Heink, S. , Zantopf, D. , Kloetzel, P.‐M. , Schmid, H.‐P. , Mayer, R.J. and Kruger, E. (2003) Human immunodeficiency virus‐1 Tat protein interacts with distinct proteasomal [alpha] and [beta] subunits. FEBS Lett. 553, 200–204. [DOI] [PubMed] [Google Scholar]
- Ballut, L. , Petit, F. , Mouzeyar, S. , Le Gall, O. , Candresse, T. , Schmid, P. , Nicolas, P. and Badaoui, S. (2003) Biochemical identification of proteasome‐associated endonuclease activity in sunflower. Biochim. Biophys. Acta, 1645, 30–39. [DOI] [PubMed] [Google Scholar]
- Ballut, L. , Drucker, M. , Pugniere, M. , Cambon, F. , Blanc, S. , Roquet, F. , Candresse, T. , Schmid, H.‐P. , Nicolas, P. , Gall, O.L. and Badaoui, S. (2005) HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. J. Gen. Virol. 86, 2595–2603. [DOI] [PubMed] [Google Scholar]
- Baumberger, N. , Tsai, C.H. , Lie, M. , Havecker, E. and Baulcombe, D.C. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614. [DOI] [PubMed] [Google Scholar]
- Baunoch, D.A. , Das, P. and Hari, V. (1990) Potato virus Y helper component protein is associated with amorphous inclusions. J. Gen. Virol. 71, 2479–2482. [DOI] [PubMed] [Google Scholar]
- Becker, F. , Buschfeld, E. , Schell, J. and Bachmair, A. (1993) Altered response to viral infection by tobacco plants perturbed in ubiquitin system. Plant J. 3, 875–881. [Google Scholar]
- Bortolamiol, D. , Pazhouhandeh, M. , Marrocco, K. , Genschik, P. and Ziegler‐Graff, V. (2007) The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615–1621. [DOI] [PubMed] [Google Scholar]
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ji, L.H. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dielen, A.S. , Badaoui, S. , Candresse, T. and German‐Retana, S. (2010) The ubiquitin/26S proteasome system in plant–pathogen interactions: a never‐ending hide‐and‐seek game. Mol. Plant Pathol. 11, 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher, K. and Callis, J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99, 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugeon, G. and Jupin, I. (2002) Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin‐mediated proteasome pathway. J. Gen. Virol. 83, 3187–3197. [DOI] [PubMed] [Google Scholar]
- Duprat, A. , Caranta, C. , Revers, F. , Menand, B. , Browning, K.S. and Robaglia, C. (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32, 927–934. [DOI] [PubMed] [Google Scholar]
- Gautier‐Bert, K. , Murol, B. , Jarrousse, A.S. , Ballut, L. , Badaoui, S. , Petit, F. and Schmid, H.P. (2003) Substrate affinity and substrate specificity of proteasomes with RNase activity. Mol. Biol. Rep. 30, 1–7. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Redondo, E. , Tavert‐Roudet, G. , Le Gall, O. and Candresse, T. (2003) Introduction of a NIa proteinase cleavage site between the reporter gene and HcPro only partially restores the biological properties of GUS‐ or GFP‐tagged LMV. Virus Res. 98, 151–162. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Walter, J. , Doublet, B. , Roudet‐Tavert, G. , Nicaise, V. , Lecampion, C. , Houvenaghel, M.C. , Robaglia, C. , Michon, T. and Le Gall, O. (2008) Mutational analysis of plant cap‐binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J. Virol. 82, 7601–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma, S. , Horsch, A. , Pouch, M.N. , Petit, F. , Briand, Y. and Schmid, H.P. (1994) Proteasomes (prosomes) inhibit the translation of Tobacco mosaic virus RNA by preventing the formation of initiation complexes. Mol. Biol. Rep. 20, 57–61. [DOI] [PubMed] [Google Scholar]
- Jarrousse, A.‐S. , Petit, F. , Kreutzer‐Schmid, C. , Gaedigk, R. and Schmid, H.‐P. (1999) Possible involvement of proteasomes (prosomes) in AUUUA‐mediated mRNA decay. J. Biol. Chem. 274, 5925–5930. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Ma, D. , Dong, J. , Jin, J. , Li, D. , Deng, C. and Wang, T. (2007) HC‐Pro protein of Potato virus Y can interact with three Arabidopsis 20S proteasome subunits in planta. J. Virol. 81, 12 881–12 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, L. and Hendil, K.B. (1999) Proteasome subunit zeta, a putative ribonuclease, is also found as a free monomer. Mol. Biol. Rep. 26, 119–123. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. and Sternberg, M.J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Khelifa, M. , Journou, S. , Krishnan, K. , Gargani, D. , Espérandieu, P. , Blanc, S. and Drucker, M. (2007) Electron‐lucent inclusion bodies are structures specialized for aphid transmission of Cauliflower mosaic virus . J. Gen. Virol. 88, 2872–2880. [DOI] [PubMed] [Google Scholar]
- Krause‐Sakate, R. , Redondo, E. , Richard‐Forget, F. , Jadao, A.S. , Houvenaghel, M.C. , German‐Retana, S. , Pavan, M.A. , Candresse, T. , Zerbini, F.M. and Le Gall, O. (2005) Molecular mapping of the viral determinants of systemic wilting induced by a Lettuce mosaic virus (LMV) isolate in some lettuce cultivars. Virus Res. 109, 175–180. [DOI] [PubMed] [Google Scholar]
- Lesemann, D.E. (1988) Cytopathology In: The Plant Viruses 4 (Milne R.G., ed.), pp. 179–227 New York: Plenum Press. [Google Scholar]
- Liu, Y. , Schiff, M. , Serino, G. , Deng, X.W. and Dinesh‐Kumar, S.P. (2002) Role of SCF ubiquitin‐ligase and the COP9 signalosome in the N gene‐mediated resistance response to Tobacco mosaic virus. Plant Cell, 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. , Kawai, T. , Tanaka, K. , Niwa, Y. , Watanabe, Y. , Nakamura, K. , Kimura, T. and Ishiguro, S. (2007) Improved Gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nicaise VG‐R, S. , Sanjuan, R. , Dubrana, M.P. , Mazier, M. , Maisonneuve, B. , Candresse, T. , Caranta, C. and Le Gall, O. (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus . Plant Physiol. 132, 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh, M. , Dieterle, M. , Marrocco, K. , Lechner, E. , Berry, B. , Brault, V. , Hemmer, O. , Kretsch, T. , Richards, K.E. , Genschik, P. and Ziegler‐Graff, V. (2006) F‐box‐like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA, 103, 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R. , Lu, R. , Sadanandom, A. , Malcuit, I. , Moffett, P. , Brice, D.C. , Schauser, L. , Jaggard, D.A. , Xiao, S. , Coleman, M.J. , Dow, M. , Jones, J.D. , Shirasu, K. and Baulcombe, D.C. (2002) Ubiquitin ligase‐associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA, 99, 10 865–10 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, F. , Jarrousse, A.S. , Dahlmann, B. , Sobek, A. , Hendil, K.B. , Buri, J. , Briand, Y. and Schmid, H.P. (1997) Involvement of proteasomal subunits zeta and iota in RNA degradation. Biochem. J. 326, 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouch, M.N. , Petit, F. , Buri, J. , Briand, Y. and Schmid, H.P. (1995) Identification and initial characterization of a specific proteasome (prosome) associated RNase activity. J. Biol. Chem. 270, 22 023–22 028. [DOI] [PubMed] [Google Scholar]
- Raffaele, S. , Bayer, E. , Lafarge, D. , Cluzet, S. , German Retana, S. , Boubekeur, T. , Leborgne‐Castel, N. , Carde, J.P. , Lherminier, J. , Noirot, E. , Satiat‐Jeunemaitre, B. , Laroche‐Traineau, J. , Moreau, P. , Ott, T. , Maule, A.J. , Reymond, P. , Simon‐Plas, F. , Farmer, E.E. , Bessoule, J.J. and Mongrand, S. (2009) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell, 21, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel, C. and Beachy, R.N. (2000) Degradation of tobacco mosaic virus movement protein by the 26S proteasome. J. Virol. 74, 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, D. , Lesemann, D.E. and Maiss, E. (1998) Ultrastructural localization of nonstructural and coat proteins of 19 potyviruses using antisera to bacterially expressed proteins of plum pox virus. Arch. Virol. 143, 2133–2158. [DOI] [PubMed] [Google Scholar]
- Royo, J. , Kunz, C. , Kowyama, Y. , Anderson, M. , Clarke, A.E. and Nexbigin, E. (1994) Loss of a histidine residue at the active site of S‐locus ribonuclease is associated with self‐compatibility in Lycopersicon peruvianum . Proc. Natl. Acad. Sci. USA, 91, 6511–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, D.D. , Ward, C.W. and Brunt, A.A. (1994) The Potyviridae. Wallingford, Oxfordshire: CAB International. [Google Scholar]
- Takizawa, M. , Goto, A. and Watanabe, Y. (2005) The tobacco ubiquitin‐activating enzymes NtE1A and NtE1B are induced by tobacco mosaic virus, wounding and stress hormones. Mol. Cells, 19, 228–231. [PubMed] [Google Scholar]
- Tsukahara, T. , Tanaka, K. , Ogawa, T. , Ishiura, S. , Funabiki, R. and Sugita, H. (1989) RNA degrading activity is tightly associated with the multicatalytic proteinase, ingensin. FEBS Lett. 255, 179–183. [DOI] [PubMed] [Google Scholar]
- Unno, M. , Mizushima, T. , Morimoto, Y. , Tomisugi, Y. , Tanaka, K. , Yasuoka, N. and Tsukihara, T. (2002) Structure determination of the constitutive 20S proteasome from bovine liver at 2.75 Å resolution. J. Biochem. 131, 171–173. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142. [DOI] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schütze, K. , Batistic, O. , Weckermann, K. , Näke, C. , Blazevic, D. , Grefen, C. , Schumacher, K. , Oecking, C. , Harter, K. and Kudla, J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Yang, P. , Fu, H. , Walker, J. , Papa, C.M. , Smalle, J. , Ju, Y.‐M. and Vierstra, R.D. (2004) Purification of the Arabidopsis 26 S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279, 6401–6413. [DOI] [PubMed] [Google Scholar]
- Yao, Y. , Toth, C.R. , Huang, L. , Wong, M.L. , Dias, P. , Burlingame, A.L. , Coffini, P. and Wang, C.C. (1999) Alpha5 subunit in Trypanosoma brucei proteasome can self‐assemble to form a cylinder of four stacked heptamer rings. Biochem. J. 344, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers, I. , Verhelst, P. , Choe, H.W. , Steyaert, J. , Heinemann, U. , Saenger, W. and Wyns, L. (1992) Role of histidine‐40 in ribonuclease T1 catalysis: three‐dimensional structures of the partially active His40Lys mutant. Biochemistry, 31, 11 317–11 325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Amino acid sequence alignment of plant alpha‐5 sequences. The two highly conserved amino acid residues identified according to a multiple alignment of alpha‐5 amino‐acid sequences derived from 23 organisms are Histine (position 99) and Glutamic acid (position 110). These amino acids are boxed in the multiple alignment shown for Glycine max (soybean, Q9M4T8), Oryza sativa (Rice, BAA96832) and Arabidopsis thaliana (PAE1: At1g53850; PAE2: At3g14290).
Fig. S2 Subcellular localization of PAE1‐YFP fusion protein in cells of healthy leaves of Lactuca sativa (lettuce). The PAE1‐YFP‐expressing construct was delivered by agroinfiltration in healthy plants and visualized by confocal microscopy 4 days after infiltration. White arrows indicate the position of the nucleus.
Fig. S3 (A–C) Subcellular localization of GFP‐HcPro in leaves of Lactuca sativa (lettuce). (A) Yellow arrow indicates the GFP‐HcPro aggregates in the cytoplasm. (B) White arrows indicate the nuclei stained with. (C) Overlay images (A + B). (D and E) Subcellular localization of GFP‐HcPro and PAE1‐YFP fusion proteins in cells of LMV‐infected leaves of Lactuca sativa (lettuce). The PAE1‐YFP construct was delivered by agroinfiltration into LMV‐GFP‐HcPro‐infected leaves areas. (D) GFP‐HcPro. (E) PAE1‐YFP. White arrow indicates the PAE1‐YFP localized in the cell nucleus, yellow arrow indicates the GFP‐HcPro aggregate in the cytoplasm. Scale bar – 27.06 µm.
Supporting info item


