SUMMARY
Pyrenophora teres, causal agent of net blotch of barley, exists in two forms, designated P. teres f. teres and P. teres f. maculata, which induce net form net blotch (NFNB) and spot form net blotch (SFNB), respectively. Significantly more work has been performed on the net form than on the spot form although recent activity in spot form research has increased because of epidemics of SFNB in barley‐producing regions. Genetic studies have demonstrated that NFNB resistance in barley is present in both dominant and recessive forms, and that resistance/susceptibility to both forms can be conferred by major genes, although minor quantitative trait loci have also been identified. Early work on the virulence of the pathogen showed toxin effector production to be important in disease induction by both forms of pathogen. Since then, several laboratories have investigated effectors of virulence and avirulence, and both forms are complex in their interaction with the host. Here, we assemble recent information from the literature that describes both forms of this important pathogen and includes reports describing the host–pathogen interaction with barley. We also include preliminary findings from a genome sequence survey.
Taxonomy: Pyrenophora teres Drechs. Kingdom Fungi; Phylum Ascomycota; Subphylum Pezizomycotina; Class Dothideomycete; Order Pleosporales; Family Pleosporaceae; Genus Pyrenophora, form teres and form maculata.
Identification: To date, no clear morphological or life cycle differences between the two forms of P. teres have been identified, and therefore they are described collectively. Towards the end of the growing season, the fungus produces dark, globosely shaped pseudothecia, about 1–2 mm in diameter, on barley. Ascospores measuring 18–28 µm × 43–61 µm are light brown and ellipsoidal and often have three to four transverse septa and one or two longitudinal septa in the median cells. Conidiophores usually arise singly or in groups of two or three and are lightly swollen at the base. Conidia measuring 30–174 µm × 15–23 µm are smoothly cylindrical and straight, round at both ends, subhyaline to yellowish brown, often with four to six pseudosepta. Morphologically, P. teres f. teres and P. teres f. maculata are indistinguishable.
Host range: Comprehensive work on the host range of P. teres f. teres has been performed; however, little information on the host range of P. teres f. maculata is available. Hordeum vulgare and H. vulgare ssp. spontaneum are considered to be the primary hosts for P. teres. However, natural infection by P. teres has been observed in other wild Hordeum species and related species from the genera Bromus, Avena and Triticum, including H. marinum, H. murinum, H. brachyantherum, H. distichon, H. hystrix, B. diandrus, A. fatua, A. sativa and T. aestivum (Shipton et al., 1973, Rev. Plant Pathol. 52:269–290). In artificial inoculation experiments under field conditions, P. teres f. teres has been shown to infect a wide range of gramineous species in the genera Agropyron, Brachypodium, Elymus, Cynodon, Deschampsia, Hordelymus and Stipa (Brown et al., 1993, Plant Dis. 77:942–947). Additionally, 43 gramineous species were used in a growth chamber study and at least one of the P. teres f. teres isolates used was able to infect 28 of the 43 species tested. However, of these 28 species, 14 exhibited weak type 1 or 2 reactions on the NFNB 1–10 scale (Tekauz, 1985). These reaction types are small pin‐point lesions and could possibly be interpreted as nonhost reactions. In addition, the P. teres f. teres host range was investigated under field conditions by artificially inoculating 95 gramineous species with naturally infected barley straw. Pyrenophora teres f. teres was re‐isolated from 65 of the species when infected leaves of adult plants were incubated on nutrient agar plates; however, other than Hordeum species, only two of the 65 host species exhibited moderately susceptible or susceptible field reaction types, with most species showing small dark necrotic lesions indicative of a highly resistant response to P. teres f. teres. Although these wild species have the potential to be alternative hosts, the high level of resistance identified for most of the species makes their role as a source of primary inoculum questionable.
Disease symptoms: Two types of symptom are caused by P. teres. These are net‐type lesions caused by P. teres f. teres and spot‐type lesions caused by P. teres f. maculata. The net‐like symptom, for which the disease was originally named, has characteristic narrow, dark‐brown, longitudinal and transverse striations on infected leaves. The spot form symptom consists of dark‐brown, circular to elliptical lesions surrounded by a chlorotic or necrotic halo of varying width.
INTRODUCTION
The fungus Pyrenophora teres Drechsler (anamorph Drechslera teres[Sacc.] Shoem.) is the causal agent of net blotch of barley (Hordeum vulgare). Similar to other stubble‐borne diseases, net blotch has become economically important and has emerged as a major disease in many barley‐growing areas worldwide. Reduced or no‐till agricultural practices have probably contributed to the increase in importance of both net form and spot form net blotch (NFNB and SFNB) disease (Mathre, 1997; McLean et al., 2009; Shipton et al., 1973); however, the susceptibility of current cultivars and trends in environmental conditions cannot be ruled out as contributing factors to the increased importance of the disease. Net blotch can cause typical yield losses of 10%–40%, with the potential for total loss if susceptible cultivars are planted under extreme environmental conditions (Mathre, 1997; Murray and Brennan, 2010). Furthermore, infection leads to a reduction in kernel size, plumpness and bulk density, and negatively affects the malting and feed quality of barley (Grewal et al., 2008; Mathre, 1997).
The disease was first named because of the typical netting symptom produced by the pathogen on leaves of susceptible barley lines (Shipton et al., 1973). In addition, a spot‐like symptom has also been observed and associated with P. teres; however, the causal agents for these two symptoms cannot be differentiated morphologically. The differentiation of the two forms of P. teres into P. teres f. teres (net type) and P. teres f. maculata (spot type) (Smedegård‐Petersen, 1971) has been proposed and widely accepted. Although Smedegård‐Petersen (1971) showed that P. teres f. teres by P. teres f. maculata genetic crosses were fertile, phylogenetic studies have suggested that the two forms of P. teres are genetically isolated and should be treated separately when studying pathogen virulence and host resistance (Rau et al., 2007).
A wide range of variation in pathogen virulence has been detected within local and global fungal populations. Host genotypes also show differential reactions to different isolates, suggesting that the specific relationship between the host and pathogen may follow a gene‐for‐gene‐type model. Several fungal avirulence genes (Beattie et al., 2007; Lai et al., 2007; Weiland et al., 1999) and major host resistance quantitative trait loci (QTLs)/genes (Mode and Schaller, 1958; Table 1) have been identified. In addition, several nonpeptide compounds and proteins produced by P. teres have been identified as phytotoxins capable of inducing disease‐like symptoms (2007, 2008, 2009; Smedegård‐Petersen, 1977a). Given the importance of the disease and the pathogen, knowledge is still lacking concerning key aspects of the pathogenesis of both forms of P. teres and the molecular mechanism of the host–pathogen interaction.
Table 1.
Summary of published net blotch resistance quantitative trait loci (QTLs) or genes.
| Population (type)* | Form† | Plant stage | Isolate (origin) | Chromosome location | R 2 (%)‡ | QTL/gene name | Reference |
|---|---|---|---|---|---|---|---|
| Steptoe× Morex (DH) | NF | Seedling | ND89‐19 (USA) | 4P | 31 | — | Steffenson et al., 1996 |
| 6ML | 14 | — | |||||
| 6MS | 10 | — | |||||
| — | |||||||
| NF | Adult | ND89‐19 (USA) | 1P | 21 | — | ||
| 3P | 20 | — | |||||
| 2P | 15 | — | |||||
| 3M | 16 | — | |||||
| 7P | 11 | — | |||||
| Igri× Franka (DH) | NF | Seedling | WRS1240 (Canada) | 3HL | 100 | Pt,,a | Graner et al., 1996 |
| Arena ×Hor 9088 (F2) | NF | Seedling | 04/6T (—) | 6H | 22.6 | — | Richter et al., 1998 |
| 6H | 10.3 | — | |||||
| 3HL | 19.2 | — | |||||
| Alexis × Sloop (DH) | NF | Seedling | NB34 (Australia) | 2HS | 11 | QRpts2S | Raman et al., 2003 |
| 3HL | 17 | QRpts3L | |||||
| Arapiles ×Frankin (DH) | NF | Seedling | NB34 (Australia) | ||||
| 2HS | 13 | QRpts2S | |||||
| 2HL | 7 | QRpts2L | |||||
| 3HL | 16 | QRpts3La | |||||
| Sloop ×Halcyon (DH) | NF | Seedling | NB50 (Australia) | 3HL | 9 | QRpts3Lb | |
| 4H | 64 | QRpts4 | |||||
| Sloop‐sib × Alexis (RIL) | NF | Seedling | NB34 (Australia) | 6HL | 11 | QRpts6L | |
| 2HS | 9 | QRpts2S | |||||
| 3HL | 13 | QRpts3L | |||||
| Tallon ×Kaputar (DH) | NF | Seedling | NB52B, NB54, NB81 and NB97(Australia) | 6H | 46–83 | — | Cakir et al., 2003 |
| 2H | 20–29 | — | |||||
| NB52B, NB54, NB81 and NB97 (Australia) | 3H | 24–31 | — | ||||
| VB9524 ×ND11231 (DH) | NF | Adult | NB77 (Australia) | 6H | 65 | — | |
| NF | Seedling | 6H | 66 | — | |||
| Chevron× Stander (DH) | NF | Seedling | ND89‐19 (USA) | 6HS | 64 | — | Ma et al., 2004 |
| 2HS | 7 | — | |||||
| VB9524 ×ND11231 (DH) | NF | Seedling | NB77 (Australia) | 6H | 75.2 | — | Emebiri et al., 2005 |
| 2H | 22.8 | — | |||||
| OUH602× Harrington (RIL) | NF | Seedling | 30199013 (USA) | 4H | 9–10 | Rpt‐4H‐5‐7 | Yun et al., 2005 |
| 3H | 12 | Rpt‐3H‐4 | |||||
| 1H | 10 | Rpt‐1H‐5‐6 | |||||
| Rolfi ×CI9819 (DH) | NF | Seedling | P7, P8, P40 and P58 (Finland) | 6H | 65 | — | Manninen et al., 2000 |
| Rolfi ×CI9819 (DH) | NF | Seedling | 84‐28‐1 (USA), 92‐46/15 (Canada), UK80‐2(UK) and 27‐36 (Finland) | 6H | 60–88 | Rpt5 | Manninen et al., 2006 |
| 1H | Minor | — | |||||
| 2H | Minor | — | |||||
| 3H | Minor | — | |||||
| 5H | Minor | — | |||||
| 7H | Minor | — | |||||
| Q21861 ×SM89010 (DH) | NF | Seedling | 15A (USA), ND89‐19 (USA) and 0‐1 (Canada) | 6H | 84–89 | — | Friesen et al., 2006a |
| Alexis ×Sloop (DH) | NF | Adult | NB329, NB333 and NB330 (Australia) | 2HC | 14.5–19.2 | QNFNBAPR.Al/S‐2H | Lehmensiek et al. 2007 |
| 3HL | 17.6–30.4 | QNFNBAPR.Al/S‐3H | |||||
| 4HC | 10.7–14.0 | QNFNBAPR.Al/S‐4H.b | |||||
| 7HS | 7.3–8.7 | QNFNBAPR.Al/S‐7Ha | |||||
| WI2875‐1× Alexis (DH) | NF | Adult | NB329, NB333 and NB330 (Australia) | 2HC | 8.5–10.7 | QNFNBAPR.W/Al‐2H | |
| 3HL | 9.6–11.0 | QNFNBAPR.W/Al‐3H | |||||
| 4HL | 7.4–12.5 | QNFNBAPR.W/Al‐4H | |||||
| 5HS | 8.0–12.3 | QNFNBAPR.W/Al‐5H | |||||
| Arapiles× Franklin (RIL) | NF | Adult | NB329, NB333 and NB330 (Australia) | 1HS | 10.9–12.1 | QNFNBAPR.Ar/F‐1H | |
| 2HS | 16.4 | QNFNBAPR.Ar/F‐2H | |||||
| 7HS | 6.9 | QNFNBAPR.Ar/F‐7H | |||||
| CDC Dolly ×TR251 (DH) | NF | Seedling | WRS1607 (Canada) | 6H | 60 | QRpt6 | Grewal et al., 2008 |
| Seedling | WRS858 (Canada) | 6H | 65 | QRpt6 | |||
| 2H | 8 | QRptts2 | |||||
| 4H | 5 | QRptts4 | |||||
| NF | Adult | Natural infection | 5H | 7 | QRptts5 | ||
| (Melfort, SK, Canada) | 6H | 42–60 | QRpt6 | ||||
| 3H | 5 | QRptta3 | |||||
| 5H | 6 | QRptta5 | |||||
| 7H | 6 | QRpt7 | |||||
| Rika× Kombar (DH) | NF | Seedling | 15A (USA) | 6H | 100 | rpt.r | Abu Qamar et al., 2008 |
| Rika ×Kombar (DH) | Seedling | 6A (USA) | 6H | 100 | rpt.k | ||
| M120× Sep2‐72 (RIL) | NF | Seedling | 3010001, 30190005‐2, 30199019‐1, 30199012‐2 and 30199010‐3 (—) | 6H‐bin2 | 25.0–44.0 | — | St. Pierre et al., 2010 |
| 6H‐bin6 | 19.0–48.0 | — | |||||
| Galleon× Haruna Nijo (DH) | SF | Seedling | 43/96/1, 49/96/9, 49/96/10, 50/96/9 and 10/97 (Australia) | 7H | 27–74 | Rpt4 | Williams et al., 1999 |
| CI9214× Stirling(DH) | SF | Seedling | — | 7H | 52 | Rpt4 | Williams et al., 2003 |
| Keel× Gairdner (DH) | SF | Seedling | — | 7H | 46 | Rpt4 | |
| Tilga× Tantangara (DH) | SF | Seedling | — | 7H | 27 | Rpt4 | |
| Chebec× Harrington (DH) | SF | Seedling | — | 7H | 74 | Rpt4 | |
| VB9104× Dash (DH) | SF | Seedling | — | 7H | 9 | — | |
| 1H | Minor | — | |||||
| 3H | Minor | — | |||||
| Galleon× Haruna Nijo (DH) | SF | Adult | — | 7H | 18 | — | |
| 7H | 20 | — | |||||
| 7H | 27 | — | |||||
| 5H | 19 | — | |||||
| 4H | 8 | — | |||||
| VB9104× Dash (DH) | SF | Adult | — | 7H | 17 | — | |
| 5H | 9 | — | |||||
| 4H | 10 | — | |||||
| CI9214× Stirling (DH) | SF | Adult | — | 7H | 35 | — | |
| Keel× Gairdner (DH) | SF | Adult | — | 7H | 32 | — | |
| Chebec× Harrington (DH) | SF | Adult | — | 7H | 7 | — | |
| Léger× CI 9831 (DH) | SF | Seedling | WRS857 (Canada) | 2H | — | — | Molnar et al., 2000 |
| LG1 | — | — | |||||
| LG3 | — | — | |||||
| CDC Dolly ×TR251 (DH) | SF | Seedling | WRS857 (Canada) | 4H | 21 | QRpts4 | Grewal et al., 2008 |
| 7H | 13 | QRpt7 | |||||
| 6H | 8 | QRpt6 | |||||
| Q21861× SM89010 (DH) | SF | Seedling | NZKF2 (New Zealand) | 4H | 64 | — | Friesen et al., 2006a |
| Rolfi ×CI9819(DH) | SF | Seedling | P1332 and P1333 (Finland) | 5H | 65–84 | Rpt6 | Manninen et al., 2006 |
Resistant parental lines are indicated by bold type.
NF, net form net blotch; SF, spot form net blotch.
QTL effects containing ranges mean that the experiments were performed for multiple locations or multiple isolates and the effects for individual treatments fall into this range.
—, no information is available for this entry.
The first review on net blotch was published in 1973 and included valuable information on the distribution and importance of the disease, as well as information on the genetics of the host–pathogen interaction (Shipton et al., 1973). A recent review by McLean et al. (2009) described SFNB with emphasis on disease epidemiology and host resistance. The purpose of this profile is to provide a general overview of the existing knowledge and to summarize the current and ongoing research on the net blotch pathogen P. teres, including both P. teres f. teres and P. teres f. maculata, as well as the P. teres–barley interaction. We also propose potential research areas that will broaden our basic understanding of this host–pathogen interaction and ultimately aid in the management of this important disease.
TAXONOMY AND RELATEDNESS TO OTHER PYRENOPHORA SPECIES
The perfect stage of Pyrenophora teres Drechs. was first described by Drechsler (1923). As a result of the heterothallic nature of P. teres, the production of the sexual stage, including fertile ascocarp formation, requires two opposite mating genotypes (McDonald, 1963; Rau et al., 2005; Smedegård‐Petersen, 1978). The imperfect stage was originally placed in the genus Helminthosporium, but this genus was subsequently revised, whereupon Helminthosporium teres Sacc. was placed into the new genus Drechslera based on its ‘cylindrical, not curved conidia, germinating from every cell, and its association with the Pyrenophora teleomorph’ (Alcorn, 1988; Shoemaker, 1959). Rau et al. (2005) used mating‐type gene sequences to reinforce the inclusion of P. teres into the Pleosporales group, probably somewhere between Phaeosphaeria nodorum (Phaeosphaeriaceae) and Leptosphaeria maculans (Leptosphaeriaceae). This study also clearly separated P. teres from the Cochliobolus species (Rau et al., 2005). This is consistent with the conclusions of Zhang and Berbee (2001) based on internal transcribed spacer (ITS) sequences and glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) gene sequences.
Two other Pyrenophora species, P. graminea Ito & Kurib. [anamoph Drechslera graminea (Rabenh. Ex Schlecht.) Shoem.] and P. japonica Ito & Kurib. [anamorph Drechslera japonica (Ito & Kurib.) Shoem. =Drechslera tuberosa (Atk.) Shoem.] are morphologically very similar to P. teres and also cause foliar disease on cultivated barley. Mycologists considered them to be three different species based on the small differences in morphology (shape, size and colour) of the ascocarp, conidia and conidiophore (Ito and Kuribayashi, 1931; Shoemaker, 1962; Sivanesan, 1987). However, the typical symptoms on barley leaves incited by the three species are quite different, and therefore disease phenotyping is commonly used to differentiate between the three fungi. Pyrenophora graminea causes long and extended necrotic stripes on barley leaves, and is a seed‐borne pathogen (Mathre, 1997). 1977b, 1978) demonstrated that P. graminea was able to cross with P. teres in culture, indicating a close genetic relationship between the two species. In recent molecular phylogenetic studies, clusters containing P. graminea were the closest neighbours of the clusters that contained P. teres (Bakonyi and Justesen, 2007; 2005a, 2005b; Rau et al., 2005; Zhang and Berbee, 2001). Interestingly, it was shown that P. graminea was more related to P. teres f. maculata than to P. teres f. teres (Bakonyi and Justesen, 2007; 2005a, 2005b; Rau et al., 2007).
Pyrenophora japonica was first recognized as a species causing leaf spot and producing long smooth conidia that had secondary conidiophores. Based on the classification of the time, this was in contrast with P. teres which was known to cause net form symptoms and not to produce secondary conidiophores (Ito and Kuribayashi, 1931; Shoemaker, 1962). Kenneth (1962) and Smedegård‐Petersen (1971) were unable to separate morphologically the isolates causing spot or net form from Israel and Denmark, respectively, because of the overlapping characteristics shared by the two groups. McDonald (1967) identified an isolate which was similar to the original description of P. japonica, but could be crossed with a known P. teres isolate and therefore was referred to as a mutant strain of P. teres. The inter‐fertility of P. japonica and P. teres was also observed by Smedegård‐Petersen (1971); therefore, he proposed the following intraspecific taxa: P teres f. maculata for isolates inducing a spot‐type symptom, and P. teres f. teres for those inducing the typical net‐type symptom. Scott (1991) did not agree with the rejection of P. japonica as an independent species when he studied the spot form isolates from South Africa. However, later studies by scientists in South Africa suggested that P. japonica was not a separate species, and should be treated as a synonym of P. teres (Campbell et al., 1999; Crous et al., 1995; 1994, 1995).
The form name P. teres f. maculata, causal agent of SFNB, has been widely accepted by barley pathologists and geneticists. DNA markers and mating‐type gene sequences indicate that P. teres f. teres and P. teres f. maculata are very closely related; however, multiple studies have indicated that the two forms are divergent genetic groups and are phylogenetically independent (Bakonyi and Justesen, 2007; Campbell et al., 2002; Lehmensiek et al., 2010; 2005a, 2005b; 2003, 2007; Serenius et al., 2005). Campbell et al. (1999) once again demonstrated the successful in vitro mating of the two forms, and showed that most of the resulting progeny produced intermediate symptoms of jagged‐type spots on barley leaves. In a further study, net × spot hybrid progeny were shown to be genetically stable (Campbell and Crous, 2003). Natural hybridization between the two forms in nature is still questionable; however, some studies have shown that it is possible (Campbell et al., 2002; Leišova et al., 2005a).
DISEASE SYMPTOMS
The pathogen can infect and cause disease on leaves, leaf sheaths, stems and kernels of barley plants. Infection of the kernel can transfer the pathogen into a new field and can serve as primary inoculum. However, the symptoms on the leaves define the disease and these are the symptoms that are of most economic importance. As with other plant diseases, symptom expression, such as lesion size and the presence of necrosis and chlorosis, is dependent on host genotype, pathogen virulence and environmental conditions.
NFNB symptoms were first described by Atanasoff and Johnson (1920) and were subsequently used to name the disease. The pathogen initiates direct penetration of the leaf surface and, within 24 h, small circular and elliptical dot‐like lesions are seen at the penetration site and soon grow along and vertical to the leaf vein, eventually developing into dark‐brown blotches containing longitudinal and transverse striations forming a net‐like pattern (Fig. 1). On highly resistant barley lines, dot‐like lesions alone are present without the development of a net‐like pattern (Fig. 1). Susceptible reactions may also include the presence of chlorotic or water‐soaked areas around the dark‐brown, net‐like necrotic lesions. Severe infection can lead to the complete death of leaves with a dry appearance (Mathre, 1997).
Figure 1.
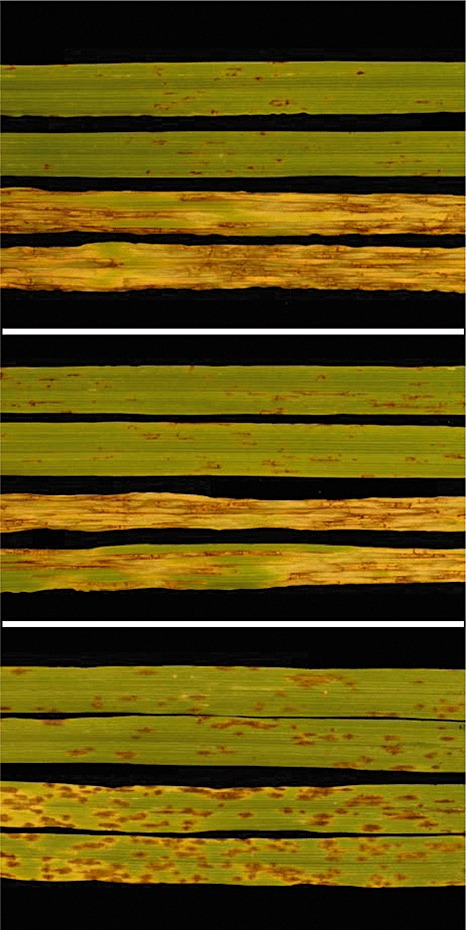
Disease reactions for combinations of host lines and pathogen isolates showing major pathogen gene–host gene interactions. Top panel: a single Pyrenophora teres f. teres isolate on two host lines (two leaves of each line are shown) yielding a high level of resistance on Rika (top two leaves), but a high level of susceptibility on Kombar (bottom two leaves). Middle panel shows the same host line Rika inoculated with two different P. teres f. teres isolates: 15A (top two leaves) and 6A (bottom two leaves). Rika and Kombar both harbour dominant susceptibility genes segregating in repulsion (Abu Qamar et al., 2008). Bottom panel: barley cultivar Lacey inoculated with two different P. teres f. maculata isolates varying in virulence.
Smedegård‐Petersen (1971) proposed the separation of the P. teres species into two forms (see section on Taxonomy and relatedness to other Pyrenophora species) on the basis of symptom development. The two forms were designated P. teres f. maculata and P. teres f. teres for isolates inducing spot‐type and net‐type symptoms, respectively. Spot‐type symptoms consist of dark‐brown, circular to elliptical lesions measuring 3 mm × 6 mm, surrounded by a chlorotic zone of varying width (McLean et al., 2009). Less virulent isolates produce smaller sized necrotic lesions or lesions with no surrounding chlorosis (Fig. 1). The spot‐type symptom is very often confused with barley spot blotch caused by Cochliobolus sativus, and examination of the spore type is necessary to characterize the causal agent (Mathre, 1997; McLean et al., 2009).
Various groups have designed specific polymerase chain reaction (PCR) primer sets capable of differentiating P. teres f. teres from P. teres f. maculata (Keiper et al., 2008; Leišova et al., 2005a; Williams et al., 2001). These form‐specific markers have been developed from amplified fragment length polymorphism (AFLP) markers (Leišova et al., 2005a), random amplified polymorphic (RAPD) markers (Williams et al., 2001) and microsatellite markers (Keiper et al., 2008). The primer sets developed have the potential to differentiate between the two P. teres forms without symptom analysis, and are therefore useful in the characterization of the pathogen.
LIFE CYCLE AND CONTROL
Both forms of net blotch are classified as stubble‐borne diseases because the fungus usually produces the ascocarp (pseudothecia) as an over‐seasoning structure on infected barley debris left (on the surface) after harvest (2, 3). As a result of their similarities, the life cycle of the two forms are addressed jointly. The pseudothecia are spherical structures seen as many dark dots on the surface of barley straw (Fig. 2). Pseudothecia are 1–2 mm in diameter and are covered by dark, hair‐like setae (Mathre, 1997; McLean et al., 2009; Fig. 2). The pseudothecia of P. teres have been observed in both field and laboratory environments, but, because of the heterothallic nature of P. teres, successful development of the sexual fruiting body requires strains with opposite mating types (Rau et al., 2005), as well as specific environmental conditions (Kenneth, 1962; McDonald, 1963). Club‐shaped and bitunicate asci, measuring 30–61 µm × 180–274 µm, develop within the mature and fertile pseudothecia (Fig. 2) (Mathre, 1997; Webster, 1951). Each ascus generally contains eight ascospores that are 18–28 µm × 43–61 µm in size, light‐brown and often have three or four transverse septa and one or two longitudinal septa only in the median cells (Mathre, 1997; Webster, 1951) (Fig. 2). Mature ascospores are actively discharged, dispersed by wind and serve as primary inoculum early in the growing season (Jordan, 1981) (Fig. 3). In some cases, seed‐borne mycelium and conidia released from the stubble of barley or an alternative host can also serve as primary inoculum for early season infection (Jordan and Allen, 1984; Louw et al., 1996; McLean et al., 2009; Shipton et al., 1973).
Figure 2.
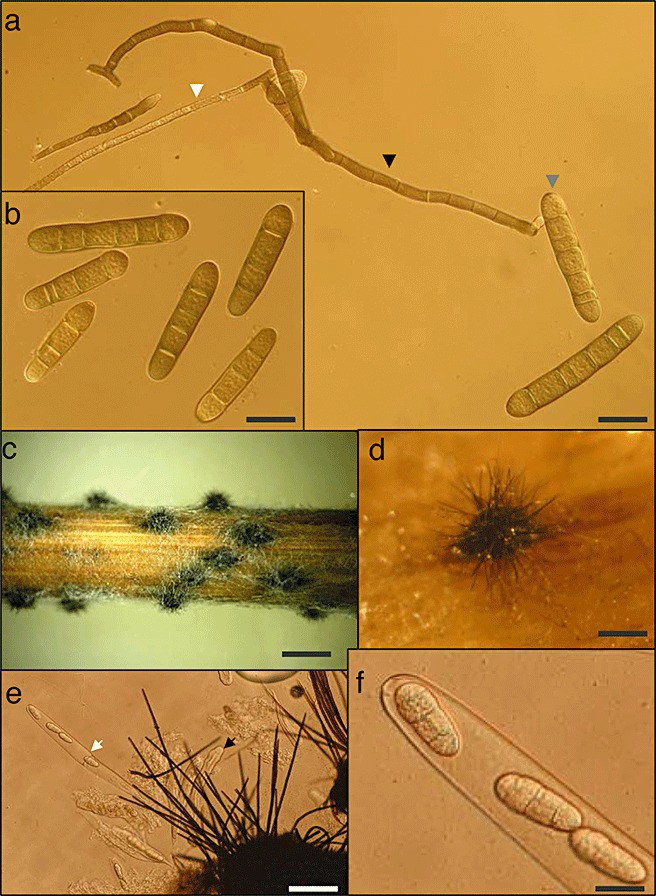
Sexual and asexual dispersal structures of Pyrenophora teres f. teres. (a) Conidia (grey arrow), conidiophores (black arrow) and mycelium (white arrow). Scale bar, 40 µm. (b) Conidia containing three to five septa. Scale bar, 40 µm. (c) Barley straw containing dark fungal pseudothecia. Scale bar, 2.5 mm. (d) Immature pseudothecia showing the globose shape and the dark‐brown, hair‐like setae. Scale bar, 1 mm. (e) The breakage of a pseudothecia showing immature, bitunicate asci (black arrow), and a mature asci (white arrow) in which the inner wall has erupted and a few ascospores have been ejected. Scale bar, 80 µm. (f) Close look at the ascospores that have three or four transverse septa and one or two longitudinal septa in the median cells. Scale bar, 20 µm.
Figure 3.
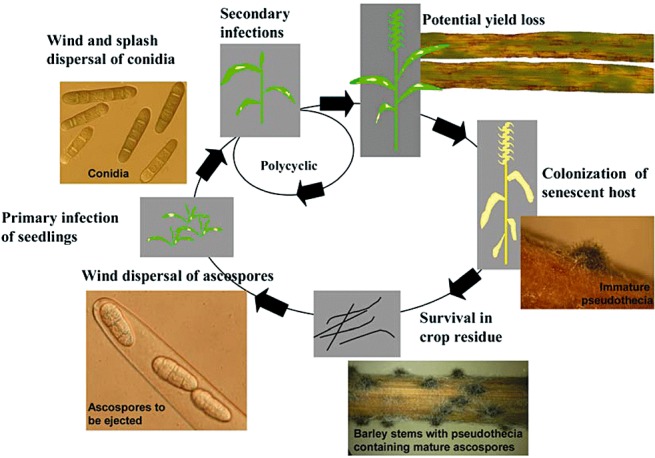
Life cycle of Pyrenophora teres f. teres and P. teres f. maculata.
After initial colonization, the fungus produces a large number of conidia which serve as secondary inocula (2, 3). Conidia are borne on top of conidiophores that are slightly swollen at the base and usually arise singly or in groups of two or three. Conidia measuring 30–174 µm × 15–23 µm are smooth, cylindrical and straight, round at both ends, subhyaline to yellowish brown, and often with four to six pseudosepta (Fig. 2) (Mathre, 1997; McLean et al., 2009; Webster, 1951). Conidia are produced throughout the growing season and are dispersed by strong wind or rain to cause new infections on plants locally, or can be carried longer distances potentially to new barley fields (Jordan, 1981; Mathre, 1997) (Fig. 3). The dispersion, germination and successful infection of conidia are greatly influenced by the relative humidity, temperature, leaf wetness and other environmental factors (1990, 1991; Jordan, 1981). During the growing season, several secondary cycles can occur, causing high disease severity on susceptible plants if environmental conditions are favourable (Fig. 3). At the end of the growing season, the fungus colonizes the senescent tissue, ultimately producing pseudothecia, the protective teleomorph structure used for over‐seasoning (2, 3).
Although not commonly found, Smedegård‐Petersen (1972) first showed that P. teres also produces pycnidia and pycnidiospores on the host and in culture. The globose‐ to pear‐shaped pycnidia are yellow to brown, 64–172 µm in diameter, and produce hyaline, nonseptate and spherical and ellipsoidal pycnidiospores measuring 1.0–1.9 µm × 1.4–3.2 µm (Mathre, 1997). Jordan (1981) also observed pycnidia developing on infected straw, leaf fragments and seed. Attempts by Jordan (1981) to infect barley leaves with pycnidiospores were unsuccessful; therefore, the role of pycnidiospores in the P. teres life cycle is not clear.
NFNB and SFNB have different global distribution patterns, usually with one form being predominant in a given area (McLean et al., 2009). No significant life cycle differences have been found between P. teres f. teres and P. teres f. maculata to explain this pattern (McLean et al., 2009). Furthermore, the relative importance of the two forms has recently changed dramatically in regions of North America and Australia (Liu and Friesen, 2010; Louw et al., 1996; McLean et al., 2009). SFNB has caused epidemics in areas of Canada and Australia (McLean et al., 2009), where it has not been reported previously as a major problem. We have also recently seen an increase in SFNB in North Dakota, where it has not been reported previously (Liu and Friesen, 2010). These epidemics could be a result of changes in host resistance, pathogen virulence or, possibly, environment. Louw et al. (1996) pointed out that a change in the predominant form of P. teres from P. teres f. teres to P. teres f. maculata in the Western Cape Province, the main barley production region of South Africa, was probably caused by selection pressure provided by the SFNB‐susceptible cultivar ‘Clipper’ being planted on 93% of the commercial barley fields in this region. Clipper was also resistant to the local races of P. teres f. teres, decreasing the competition for leaf tissue.
Currently, several methods have been applied to manage NFNB and SFNB, including fungicides, cultural practices and host resistance. The use of rotation and other cultural practices to reduce or eliminate primary inoculum in a field are important means of cultural management. Chemical control using fungicides, either on the seed to reduce primary inoculum or on the upper leaves to reduce tissue loss during seed filling, can be attempted, although foliar application of fungicide is not always economically justified. Although effective long‐term control should be based on the growth of resistant cultivars (Mathre, 1997; Shipton et al., 1973), the virulence of both P. teres f. teres and P. teres f. maculata is highly variable (see section on Pathogen specialization and virulence diversity), and therefore resistance breeding will continue to be challenging.
INFECTION PROCESS
Very little research has been performed on the P. teres f. maculata infection process or the comparison of the infection process of the two forms of P. teres. van den Berg and Rossnagel (1990) showed that the time required for infection of P. teres f. maculata was shorter than that required for infection of P. teres f. teres, but cautioned that additional experiments would be needed to verify this result. The infection process starts with the germination of conidia or ascospores that land on the surface of leaves. Both spore types are able to germinate within a few hours in the presence of water and suitable temperatures (van den Berg and Rossnagel, 1990; Kenneth, 1962; Shipton et al., 1973). Once conidia land on barley leaves, germ tubes usually arise from one of the terminal cells, but occasionally from as many as four cells simultaneously (Van Caeseele and Grumbles, 1979). Hyphae form germ tubes that can grow to varying lengths before forming a swollen, club‐shaped appressorial structure used by the fungus for penetration (Van Caeseele and Grumbles, 1979). The formation of appressoria is a complicated process, and Ruiz‐Roldán et al. (2001) have shown that signal transduction involving the mitogen‐activated protein kinase (MAPK) gene PTK1 is required for appressorium formation. Several cytological studies have shown that P. teres usually penetrates directly through the cuticle into epidermal cells (Jørgensen et al., 1998; Keon and Hargreaves, 1983; Van Caeseele and Grumbles, 1979), and this type of penetration is accomplished by both enzymatic hydrolysis of the cuticle and cell wall, as well as by pressure generated from the appressoria. Lightfoot and Able (2010) have shown that there are distinct differences between the two forms of P. teres. Unlike P. teres f. teres, P. teres f. maculata often forms intracellular vesicles, and tends to grow closer to the epidermal cells, and is not able to affect cells with which it is not in contact. Pyrenospora teres f. maculata is also slower to germinate, and growth after germination is slower relative to that of P. teres f. teres. In response to fungal penetration, host cells occasionally form papillae or papillae‐like deposits in an attempt to stop the entry of the fungus (Jørgensen et al., 1998; Keon and Hargreaves, 1983).
After successful penetration of the outer epidermal cell wall, the fungal hypha develops into a large intracellular vesicle (primary vesicle). Subsequently, a secondary intracellular vesicle develops within the epidermal cell, at which time functional disruption of the infected cell, as well as the adjacent epidermal cells, occurs (Keon and Hargreaves, 1983). From the secondary intracellular vesicle, intracellular hyphae emerge and grow through the lower epidermal cell wall into the mesophyll tissue, where additional hyphal growth is essentially intercellular. In a susceptible interaction, host cells attached or adjacent to intercellular hyphae show various degrees of disruption and start to die within 2 days of inoculation (Keon and Hargreaves, 1983). Hargreaves and Keon (1983) demonstrated that isolated live barley mesophyll cells are able to attach to hyphae in fungal cultures, suggesting that there are low‐molecular‐weight compounds produced by the fungal or host cells that lead to the movement of intercellular hyphae to mesophyll cells during infection. As the infection progresses, chlorotic areas start to develop around the initial necrotic lesions and, interestingly, microscopic examination reveals that these areas are free of fungal hyphae; however, chloroplasts in the cells of these areas are disrupted (Keon and Hargreaves, 1983). This is probably caused by diffusible toxins or effectors secreted from the fungal hyphae or, possibly, host response leading to the death of adjacent cells via programmed cell death. There is mounting evidence in other necrotrophic plant–pathogen systems involving fungi closely related to P. teres that necrotrophic effectors (host‐selective toxins) can stimulate the host to undergo programmed cell death (Kwon et al., 1998; Manning et al., 2009; Rasmussen et al., 2004). In addition, classical resistance‐like genes containing both nucleotide‐binding (NB) domains and leucine‐rich repeat (LRR) domains have been implicated in host‐controlled susceptibility in response to both nonproteinaceous and proteinaceous virulence effectors (Lorang et al., 2007; Nagy and Bennetzen, 2008; J.D. Faris, USDA‐ARS, Fargo, ND, USA, personal communication). Given the relatedness of the pathogens and the similarity in symptom development, it is possible that necrotrophic effectors are involved in the interaction with both P. teres f. maculata and P. teres f. teres. Pyrenophora teres has been shown to form appressoria and to penetrate directly into epidermal cells, between epidermal cells and, occasionally, through stomata. Lightfoot and Able (2010) have differentiated P. teres f. teres and P. teres f. maculata on the basis of infection characteristics. Lightfoot and Able (2010) have shown that P. teres f. teres rarely forms intracellular vesicles, but rather infects and feeds as a necrotroph throughout the infection process by growing only intercellularly, and is able to affect cells not immediately associated with the mycelium. However, P. teres f. maculata initially forms haustorial‐like intracellular vesicles, feeding similarly to a biotroph, followed quickly by a switch to necrotrophic growth. On the basis of this description, P. teres f teres would be classified as a necrotroph and P. teres f. maculata would be classified as a hemibiotroph.
No differences were observed with regard to spore germination, number and length of germ tubes, or number of penetration points when comparing NFNB resistant and susceptible host lines (Keeling and Banttari, 1975), but growth of the fungus after penetration was significantly slower in resistant host tissue. Keeling and Banttari (1975) also speculated that there was a release of antifungal compounds on the leaf surface, however, it is difficult to determine how this aids in resistance, as there was no significant difference in growth and penetration success of the pathogen when comparing the resistant and susceptible barley lines used in this experiment.
MOLECULAR HOST–PATHOGEN INTERACTION
Relatively little is known at the molecular level about the mechanisms by which P. teres interacts with barley. Signal perception, production and transduction are believed to be important in both the host and pathogen during the initial interaction. A MAPK gene (PTK1) has been cloned from P. teres and has been shown to be critical in appressoria formation during infection. The strain resulting from the knockout of PTK1 in P. teres produces no appressoria, leading to the loss of pathogenicity (Ruiz‐Roldán et al., 2001). Several genes involved in signal transduction and gene regulation are upregulated during the early stages of spore germination (Dilger et al., 2003). In a differential display RT‐PCR expression experiment, Vergara et al. (2003) identified a differentially expressed P. teres cDNA fragment whose transcription was specifically induced in the presence of barley leaf cell walls. This sequence showed homology to many genes coding for regulatory proteins, suggesting that this gene may be involved in the differential regulation of pathogenicity mechanisms.
Reactive oxygen species (ROS) are often produced in a resistance response to pathogen attack, especially in a gene‐for‐gene‐type interaction currently known as effector‐triggered immunity (ETI). Able (2003) showed an interesting correlation between ROS accumulation and pathogen growth using a single barley line and two P. teres f. teres isolates, one virulent and one avirulent. Although preliminary, this work shows the potential for involvement of ROS in the P. teres interaction.
Several studies have investigated the host genes involved in both compatible and incompatible interactions. Krupinska et al. (2002) identified the host gene HvS40 which appears to be involved in the P. teres–barley host–pathogen interaction. Expression of HvS40 was found to be induced by jasmonic acid and salicylic acid. High levels of expression were observed exclusively in leaf tissue that showed necrosis and chlorosis after infection with P. teres, but was undetectable in surrounding uninfected tissue, suggesting that this gene plays an important role during pathogen infection.
Reiss and Bryngelsson (1996) examined gene expression during pathogen attack in barley leaves, and found a large number of common genes induced by Puccinia hordei, Erysiphe (Blumeria) graminis and P. teres f. teres. They identified a number of pathogenesis‐related (PR) proteins, such as peroxidases, β‐1,3‐glucanases, chitinases, PR‐1a and 1b, and several thaumatin‐like proteins (PR‐5 family). The accumulation of these PR proteins in barley leaves was also detected in leaves treated with a toxin extract from the culture filtrate of P. teres. A subsequent study by Reiss and Horstmann (2001) identified eight thaumatin‐like proteins (TLP1–TLP8) expressed in barley in response to P. teres f. teres infection.
Bogacki et al. (2008) evaluated defence response (DR) genes using suppressive subtractive hybridization (SSH), and identified several candidate DR genes that were associated with a P. teres incompatible interaction. Two libraries were created in this study, one each for P. teres f. maculata and P. teres f. teres. A pool of unique transcripts identified by SSH was used to identify 45 DR genes. Twenty‐eight of these genes were shown to be differentially expressed, with P. teres f. maculata and P. teres f. teres showing similar expression profiles. The similarity in profiles should not be surprising as a substantial amount of evidence has shown that, beyond the initial pathogen‐specific recognition, there is a convergence of signalling so that, once recognition takes place, common genes involved in DR are activated (Feys and Parker, 2000; Schulze‐Lefert and Vogel, 2000). One of these genes, Rar1, was first identified in barley, and was shown to be necessary for the downstream signalling beyond resistance gene recognition (Schulze‐Lefert and Vogel, 2000). Although previous host studies have shown the genetics of resistance to the two forms of P. teres to be independent on the basis of chromosomal location, the downstream pathways are probably common for both pathogen forms.
HOST RESISTANCE
The earliest published work on the inheritance of resistance to net blotch on barley showed resistance to be inherited in a Mendelian fashion (Geschele, 1928). The vast majority of work on net blotch resistance before 1971 is presumed to be that of NFNB, as the splitting of P. teres into the two forms, P. teres f. teres and P. teres f. maculata, was not proposed until 1971 (Smedegård‐Petersen, 1971), and therefore net blotch would have been identified on the basis of the netting symptom. Thereafter, several studies still did not differentiate between the two forms, but it is assumed that the net form was being used. Schaller (1955) and Mode and Schaller (1958) demonstrated that at least three major, incompletely dominant resistance genes were effective against P. teres isolates collected in California. These genes were designated Pt1, Pt2 and Pt3. Additional single dominant genes were later identified in other breeding lines (Frecha, 1958; Gray, 1966; McDonald and Buchanon, 1962). The first report of the physiological specialization of the pathogen was given by Khan and Boyd (1969a), and this knowledge was then used to evaluate resistance sources that correlated with virulence differences (Khan and Boyd, 1969b). The knowledge of both host and pathogen variability and the presence of major dominant resistance genes and variability in the pathogen corresponding to these resistance genes led to further work to evaluate this important host–pathogen system as a gene‐for‐gene‐type interaction. Bockelman et al. (1977) were the first to use trisomic analysis to locate dominant resistance on barley chromosome 3 (3H) in the cultivar ‘Tifang’, chromosome 2 (2H) in CI7584, and 3 (3H) and 5 (1H) in CI9819. The early work on the net blotch system was a good start to the identification of a correlation between the host and pathogen in an era where less was known about these nonbiotrophic interactions. However, it was probably limited by the available genetic tools of the day. More recently, with the advent of new molecular genetic techniques, it has been possible to decipher more precisely the host response, especially with regard to quantitative inheritance of resistance/susceptibility. This has led to a much more complex view of the host–pathogen interaction.
Previous to molecular marker studies, several groups identified resistance to be quantitatively inherited, especially in adult plants and under field conditions (Arabi et al., 1990; Douglas and Gordon, 1985; Robinson and Jalli, 1997; Steffenson et al., 1996; Steffenson and Webster, 1992a), showing that the inheritance of field resistance may be more complex than that previously revealed for seedlings. Ho et al. (1996) and, later, Abu Qamar et al. (2008) identified three and two major recessive resistance genes, respectively, in seedlings, indicating the potential for a complexity of virulence and avirulence effectors present in the pathogen (see section on Pathogen specialization and virulence diversity).
Since the introduction of markers and QTL analysis, several studies have identified NFNB QTLs on each of the seven barley chromosomes (Table 1); although not as much work has been performed on SFNB resistance, several locations have also been identified for this form. The majority of these studies used seedling data, but several of the NFNB studies (Cakir et al., 2003; Grewal et al., 2008; Lehmensiek et al., 2007; Steffenson et al., 1996) and one of the SFNB studies (Williams et al., 2003) also used adult plant data to evaluate the inheritance of resistance. Several studies have identified major effects for seedling resistance to NFNB on chromosome 6H near the centromere (Table 1). Both Grewal et al. (2008) and Cakir et al. (2003) compared seedling and adult plant resistance within the same populations, and showed that this 6H locus was effective at both the seedling and adult plant stages, indicating that, at least in some cases, seedling resistance is also effective at the adult plant stage. Additional adult plant resistance studies have also identified this 6H locus (Table 1).
Based on the accumulation of information on host resistance and corresponding information on pathogen virulence, durable resistance to this pathogen could be accomplished by the pyramiding of multiple resistance genes (Douiyssi et al., 1998), as most single resistance sources are overcome by known pathotypes/biotypes of the pathogen. In addition, it is probable that effectors, such as necrotrophic effectors, may be produced by both forms of this pathogen. Typically, these effectors of virulence interact directly or indirectly with dominant host susceptibility genes (Wolpert et al., 2002); therefore, if virulence factors, such as necrotrophic effectors, are produced by the pathogen, breeding for resistance would probably require the removal of sensitivity genes, as well as the incorporation of resistance genes, making breeding complicated. The potential for different modes of pathogen attack indicates the need to identify the molecular mechanisms of pathogen virulence.
Recently, many studies on net blotch resistance have included SFNB evaluations. The identification, mapping and characterization of the resistance loci effective against P. teres f. maculata have shown that SFNB resistance genes are genetically distinct from those effective against NFNB (Friesen et al. 2006a; Grewal et al., 2008; McLean et al., 2009; Table 1). Major genes have been identified for SFNB resistance on barley chromosomes 4H, 5H and 7H, with other minor QTLs being identified (Table 1). Different host genes confer resistance to the different forms of the pathogen (Table 1), suggesting that a separate host–pathogen evolutionary process is present for the two forms, and that the two forms of net blotch should be considered separately in a barley breeding programme.
Relatively more is known about the inheritance of NFNB resistance, but, because of the more recent importance of SFNB, research emphasis has increased in the SFNB area. Based on the current knowledge, the NFNB system is probably a combination of classical major dominant resistance genes with additional effects induced by major or minor virulence effectors and nonspecific toxins. The SFNB system also includes some major genes that are probably involved in the recognition of specific pathogen effectors. As with NFNB, SFNB also involves some minor QTLs affecting resistance.
Before rational and durable resistance breeding can take place for either form of this disease, more information on the mechanism of disease, including both the mechanism(s) of virulence of the pathogen and the mechanism(s) of resistance/susceptibility in the host, is necessary. The cloning of host resistance/susceptibility genes in the host for both NFNB and SFNB will help to piece this complex puzzle together from the host side, and the availability of a genome sequence for both P. teres f. teres and P. teres f. maculata, as well as the availability of pathogen mapping populations, are great tools that can be used to decipher the pathogen side of this interaction.
POPULATION DIVERSITY
Population diversity and genetic structure have been shown to be important in the management of fungal diseases, especially for the successful deployment of host resistance and the effective use of fungicides. It has been noted that P. teres from different localities or within one area vary greatly morphologically (McDonald, 1967; Shipton et al., 1973). The wide variability has made it difficult to characterize and distinguish P. teres on the basis of previously described criteria alone (Shipton et al., 1973). Since the 1990s, molecular techniques have been used to investigate the population diversity of P. teres and to speculate on the evolutionary forces that drive change in the fungal population. Investigations have been conducted on P. teres populations from a wide range of geographical locations worldwide, including the USA, Canada and Germany (Peever and Milgroom, 1994), Finland (Peltonen et al., 1996), Sweden (Jonsson et al., 2000), South Africa (Campbell et al., 2002; Lehmensiek et al., 2010), Italy (2003, 2005), Czech Republic (2005a, 2005b) and Australia (Lehmensiek et al., 2010; Serenius et al., 2005). Using RAPD, AFLP or mating‐type genes, most of the P. teres diversity studies have shown a high level of variability within the P. teres populations, even on a small scale in terms of sampling area, compared with other fungi (Campbell et al., 2002; Lehmensiek et al., 2010). Genetic differentiation was also high between populations that were separated by a long distance, indicating limited gene flow (Jonsson et al., 2000; Lehmensiek et al., 2010; Peever and Milgroom, 1994). Many phylogenetic studies have shown that P. teres f. maculata and P. teres f. teres isolates are separated into two genetically divergent groups, indicating that the two forms of P. teres should be treated separately (Rau et al., 2007). Furthermore, studies have shown that genetic divergence is slightly higher among the P. teres f. teres population than among the P. teres f. maculata population (Lehmensiek et al., 2010; Rau et al., 2003; Serenius et al., 2005). As P. teres is capable of reproducing both sexually and asexually, its population structure would be largely dependent on the relative importance of the two styles in the life cycle of the fungus. Random sexual reproduction has been shown to be important in many P. teres populations, as no significant gametic disequilibrium has been detected (Jonsson et al., 2000; Peever and Milgroom, 1994; Rau et al., 2003); conversely, other studies have suggested that reproduction within some P. teres populations occurs mainly asexually (Campbell et al., 2002; Lehmensiek et al., 2010). Using hundreds of isolates from Italy, Rau et al. (2005) studied systematically the mating‐type genes of P. teres, and found the ratio of the two mating‐type genes to be consistently 1:1, suggesting that sexual reproduction is a major force driving the population structure. However, Serenius et al. (2005) investigated the distribution of the two mating‐type genes in several geographically distinct populations, and concluded that the importance of sexual reproduction is highly variable between regions. Overall, the population genetic analyses suggest that sexual reproduction occurs at a significant level within the worldwide population of P. teres, but the relative contribution of sexual and asexual reproduction varies between regions, possibly based on environmental differences.
PATHOGEN SPECIALIZATION AND VIRULENCE DIVERSITY
As noted in the previous section, Khan and Boyd (1969a) were the first to document physiological races of P. teres f. teres using barley differential lines. Using two barley lines, four different races could be differentiated, showing that a distinct host–pathogen interaction was present. Since this time, studies using large sets of differential barley lines have been completed to show diversity in physiological races or pathotypes for both forms of the pathogen. For P. teres f. teres, these studies have included collections from Australia (Gupta and Loughman, 2001; Khan, 1982; Khan and Boyd, 1969a), New Zealand (Cromey and Parks, 2003), Canada (Tekauz, 1990), USA (Steffenson and Webster, 1992b; Wu et al., 2003), Europe (Arabi et al., 2003; Jalli, 2004; Jalli and Robinson, 2000; Jonsson et al., 1997) and Asia (Sato and Takeda, 1993). Although more emphasis has been placed on the net form, several studies have included collections of P. teres f. maculata. These include collections from the USA (Wu et al., 2003), Canada (Tekauz, 1990), the Mediterranean (Karki and Sharp, 1986) and Australia (Gupta and Loughman, 2001). Several common differential lines have been used in these studies, so that comparisons can be made concerning virulence diversity worldwide, as well as within studies locally.
Khan and Boyd (1969a) showed that all P. teres isolates, presumably P. teres f. teres, collected in Western Australia were virulent on the popular Australian cultivar ‘Beecher’; however, Khan (1982) showed that the change in barley cultivars towards NFNB resistance was quickly followed by a change in virulence of the pathogen population. Beecher, which at one time occupied more than 75% of the barley production area in Western Australia, was largely replaced by the cultivar ‘Dampier’, which was then replaced by the NFNB‐resistant cultivar ‘Clipper’, which was released in the early 1970s. Pyrenophora teres f. teres isolates collected after 1976 were clearly shown to have lost virulence on the cultivar Beecher (Khan, 1982). Gupta and Loughman (2001) have shown that the P. teres f. teres population remained predominantly avirulent on Beecher into the 1990s. Cromey and Parks (2003) showed that none of the New Zealand‐collected P. teres f. teres isolates were virulent on Beecher, whereas Steffenson and Webster (1992b) found that 38% of the isolates collected in California were virulent on Beecher, and Jonsson et al. (1997) showed that 30% of a Swedish collection of P. teres f. teres isolates were virulent on Beecher. Beecher is just a single example of a resistance source, and illustrates the plasticity of the pathogen population that is subject to the predominant cultivars grown.
Although several common differential lines, such as Beecher, have been used in various virulence diversity studies worldwide (Cromey and Parks, 2003; Gupta and Loughman, 2001; Jonsson et al., 1997; Khan, 1982; Khan and Boyd, 1969a; Steffenson and Webster, 1992b; Tekauz, 1990; Wu et al., 2003), Afanasenko et al. (2009) have proposed a set of nine differential lines to be used to standardize the characterization of the global P. teres f. teres population. This set is by no means comprehensive, but can be used to make direct comparisons between P. teres f. teres populations in different barley‐producing regions if similar environmental conditions are used.
Although resistance to P. teres f. teres and P. teres f. maculata is under different genetic control, studies that have investigated the virulence of P. teres f. maculata have often used some of the same lines as employed for P. teres f. teres (Gupta and Loughman, 2001; Karki and Sharp, 1986; Wu et al., 2003).
Some studies have reported somewhat less virulence diversity in the P. teres f. maculata population (Gupta and Loughman, 2001; Wu et al., 2003); however, this may not be a true reflection of the population, as fewer and less in‐depth studies on P. teres f. maculata have been published. Karki and Sharp (1986) used a set of 20 lines to compare virulence in P. teres f. maculata populations collected in the Mediterranean and Montana, USA and, although no pathotype designations were identified, it was found that one‐half of the barley lines used showed a differential response either between or within the two populations collected. Tekauz (1990) evaluated a Canadian population and identified 20 different pathotypes based on the reaction on a 12‐barley‐line set, three of which were common to the Karki and Sharp (1986) set.
Collectively, these studies show that the virulence of P. teres f. teres and P. teres f. maculata is diverse on both a local and global level. This is shown by the fact that common differential lines harbouring resistance can be effective against virulence in an entire local population, but completely ineffective against a population collected in another region, and resistance within a single differential line can be effective against only a percentage of a local population.
TOXINS
Both forms of P. teres induce rapid necrotic and chlorotic death of plant leaf tissue after inoculation. This type of lifestyle is often attributed to the production of an array of toxins from the fungus. Net blotch necrotic lesions appear as early as 24 h after inoculation, and complete symptom development and sporulation can occur within 1 week. Water‐soaked and chlorotic areas develop rapidly surrounding the dark‐brown necrotic lesion. Both Keon and Hargreaves (1983) and Smedegård‐Petersen (1977a) found that the chlorotic area surrounding the necrotic lesion was free of fungal hyphal growth, consistent with the release of diffusible toxins during the fungal infection.
Smedegård‐Petersen (1977a) used both P. teres f. teres and P. teres f. maculata to produce and partially characterize two chemically and structurally similar phytotoxic compounds, named toxin A and toxin B, which were able to induce necrosis and/or chlorosis. Using the same isolates, Bach et al. (1979) purified toxin A and toxin B, as well as a new toxin form, named toxin C. It was determined that the chemical structures of the three toxins were the same or similar to aspergillomarasmine A. Toxin A was identified as N‐(2‐amino‐2‐carboxyethyl) aspartic acid, toxin B as anhydroaspergillomarasmine A and toxin C as aspergillomarasmine A. The three toxins differed in their ability to induce toxic reactions. Weiergang et al. (2002) studied the biological activity of the three toxins by testing them on different barley cultivars. Toxin C was found to be the most active and was capable of inducing distinct necrotic symptoms and zones of light‐yellow chlorosis at a concentration of 0.25 mm. Toxin A was able to cause mainly dark‐yellow chlorotic symptoms, but little necrosis, at concentrations of 0.75 mm. Only slight symptoms were induced by 0.75 mm toxin B, showing that it was a weak toxin. Friis et al. (1991) further investigated the structural configuration of the three toxins and the possible biosynthetic pathway for the production of the toxins. It was found that toxin C (aspergillomarasmine A) was the major toxin produced by P. teres in culture and, as the pH decreased in the culture during fungal growth, toxin C was converted into toxin B (anhydroaspergillomarasmine A) without any enzymatic catalysis. It was also determined that toxin A [N‐(2‐amino‐2‐carboxyethyl) aspartic acid], produced by P. teres, serves as a direct precursor of toxin C.
In the most resistant or susceptible barley lines, good correlation has been found between the reaction to toxins and reaction to disease caused by P. teres f. teres and P. teres f. maculata, leading some to suggest that toxins may be used for the in vitro selection of barley lines in the early stages of a breeding programme (Sharma, 1984; Weiergang et al., 2002).
Recently, Sarpeleh et al. (2007) have reported the identification of phytotoxic proteinaceous metabolites from culture filtrates of both P. teres f. teres and P. teres f. maculata. In contrast with previously reported toxins, these proteinaceous metabolites were able to induce brown necrotic spots or lesions on susceptible cultivars, similar to net blotch disease symptoms. The toxins induced only light reactions on resistant cultivars, but induced no reaction on other plant species, including wheat, triticale, rye and faba bean. The toxins were shown to be fairly heat stable, and the activity in planta was shown to be light and temperature dependent (2007, 2008). In addition to proteinaceous toxins, the authors also identified some lower molecular weight compounds (LMWCs) that caused chlorosis on barley leaves. Further investigation indicated that the LMWCs produced probably have the same or similar chemical structures to the previously reported toxins A, B and C, and the activity of the LMWCs were nonhost selective, but light and temperature dependent (Sarpeleh et al., 2009).
In general, the toxic compounds identified to date are not specific to any isolate and, although some differences in sensitivity have been observed between resistant and susceptible lines, strong host selectivity has not generally been found (2007, 2008). This indicates that, beyond the major gene‐for‐gene‐type interactions present in this host–pathogen system, there are several relatively minor virulence factors (toxins) that are important to the pathogen. The investigation of the molecular mechanisms of host–toxin interactions is still in its infancy, and progress is being made in this critical area.
GENETICS OF PATHOGEN AVIRULENCE/VIRULENCE
Many studies have investigated the virulence variation in both forms of P. teres, revealing a large number of pathotypes based on their reactions to barley differential lines (see above). In addition, numerous host genetic studies have shown that single major dominant or recessive genes confer resistance to a particular P. teres pathotype and, in some cases, that resistance is overcome by another P. teres pathotype. This strongly suggests a specific evolutionary relationship between the host and pathogen that may fit a gene‐for‐gene‐ or inverse gene‐for‐gene‐type model. The identification and cloning of the fungal genes conferring avirulence or virulence will provide powerful tools for studying the barley–P. teres interaction at the molecular level.
Weiland et al. (1999) were the first to use a P. teres mapping population to study the genetics of P. teres f. teres avirulence. They used molecular markers to map a single gene, designated AvrHar, conferring low virulence on ‘Harbin’, a barley line that had previously been shown to have a single major gene conferring resistance to P. teres (Mode and Schaller, 1958). Harbin is also a line that has historically been used in pathotype evaluations worldwide (Cromey and Parks, 2003; Jalli, 2004; Jalli and Robinson, 2000; Jonsson et al., 1997; Khan and Boyd, 1969b; Steffenson and Webster, 1992b; Wu et al., 2003). Using the same fungal population and AFLP markers, Lai et al. (2007) revealed two additional genes (AvrPra1 and AvrPra2) controlling the avirulence of the other parental isolate, P. teres f. teres isolate 0–1 (Canadian isolate), towards barley line ‘Prato’. Interestingly, AvrPra2 mapped to the same locus as AvrHar, but avirulence segregated in repulsion. Currently, we are attempting to clone the AvrHar/AvrPra2 region via a map‐based approach. Preliminary data from the first few steps of chromosome walking indicate that this gene is probably located within a highly repetitive DNA region (Liu et al., 2008).
Beattie et al. (2007) investigated the genetic control of avirulence in a P. teres f. teres population derived from two Canadian isolates segregating for avirulence to the barley cultivar ‘Heartland’. A single gene, Avrheartland, was identified, controlling avirulence/virulence, and this gene was assumed to be different from AvrHar, as both parental isolates were virulent on Harbin barley. Using bulked segregant analysis, two AFLP markers were identified that flanked this gene locus, both at a genetic distance of 3 cM.
To investigate the genetics of the barley–P. teres interaction, Afanasenko et al. (2007) used classical genetics to analyse the segregation of host resistance and fungal avirulence in multiple host and P. teres. f. teres populations. This work concluded that both host resistance and fungal avirulence were controlled by one or two major genes, sometimes involving epistatic genes. In general, the genetic relationship between barley lines and P. teres isolates was shown to be specific, and this pathosystem was suggested to follow a gene‐for‐gene model (Afanasenko et al., 2007).
Pyrenospora teres is a haploid fungus, and thus it is difficult to determine the dominance of the genes involved in virulence. The term ‘effector’ has been proposed to describe the fungal proteins that serve as virulence factors, avirulence factors or both (Kamoun, 2007); therefore, the genes identified and mapped previously in P. teres f. teres (Beattie et al., 2007; Lai et al., 2007; Weiland et al., 1999) may encode proteins that function as effectors. Thus, the cloning and functional characterization of these genes are needed to fully elucidate the mechanism of the barley–P. teres f. teres interaction. No P. teres f. maculata virulence or avirulence genes have been mapped or identified directly, although, on the basis of the diversity in the host resistance to P. teres f. maculata, it would be logical to assume that effectors are produced that elicit the host response to the pathogen, and that these effectors are independent of the P. teres f. teres interaction.
GENOMICS
Aragona et al. (2000) used pulsed field gel electrophoresis to investigate the genome size of P. teres. A total of six megabase‐sized bands were resolved, ranging from 2.0 to 6.0 Mb. On the basis of the band intensity, Aragona et al. (2000) concluded that two of the bands contained multiple chromosomes, and that there were probably nine chromosomes present in P. teres, consistent with a genome size of at least 35 Mb. Cytological karyotype analysis using the germ tube burst method for the physical observation of chromosomes also showed that at least nine chromosomes are likely in P. teres (Z. Liu and T. L. Friesen, unpublished data).
The value of whole genome sequences in uncovering novel effectors cannot be overestimated (2006b, 2007, 2008a, 2008b; Hane et al., 2007; Liu et al., 2009; Tyler, 2009). Furthermore, new genome sequencing methods have become substantially more affordable. However, read lengths are much shorter and so the assembly, and hence annotation of the assembly, is more difficult. In view of this, our group has acquired a genome sequence scan of a P. teres f. teres isolate using Illumina sequencing chemistry with 75‐bp paired end reads. The sequencing run yielded over 833 Mbp of sequence data, or approximately 20× coverage. Scaffold assembly yielded N 50= 408 and L 50= 26 790 bp, with a total assembly size of 41.95 Mbp (N 50 is a weighted median statistic, such that 50% of the entire assembly is contained in contigs or scaffolds equal to or greater than this value; L 50 is the length of the scaffold that separates the half of the assembled genome from the remainder of smaller scaffolds, if the sequences are ordered by size). Comparisons of the scaffolds with Sanger‐sequenced BACs showed that complex or gene‐rich regions assembled effectively, but that low‐complexity regions tended to be repetitive sequences, displaying over‐represented read coverage and short scaffold assemblies. Thus, the actual P. teres f. teres genome size is likely to be larger than 42 Mbp and longer paired‐end 454 Roche sequences are being performed to help assemble the low‐complexity regions. The genome of P. teres f. teres is larger than that of other similar fungal plant pathogens characterized to date, such as Magnaporthe grisea, which was estimated at 37.88 Mbp (Dean et al., 2005), and Stagonospora nodorum, which was estimated at 37.16 Mbp (Hane et al., 2007).
Some 12 453 coding sequences of 30 amino acids or longer have been predicted from the initial assembly. Annotation and interpretation of the assembly are ongoing and will incorporate large‐scale proteomics (Bringans et al., 2009) and transcriptomic support. The authors invite the P. teres research community to collaborate on this task.
FUTURE PROSPECTS
Several key areas of research are currently being pursued. The genomic sequencing of both P. teres f. teres and P. teres f. maculata has been initiated and will open up the door to new pathogen research, as well as increase the rate at which current discoveries can be made. As pointed out earlier, neither the mechanism of virulence nor the mechanism of host resistance has been revealed. One major question that needs to be answered is what drives the co‐evolution of these two closely related host–pathogen systems? Does the net blotch system consist of effectors of avirulence and classical resistance‐like genes? Does P. teres have an arsenal of necrotrophic effectors (host‐selective toxins) similar to the closely related species P. tritici‐repentis or other Dothideomycete genera? Or are effectors with a dual role present, that is, avirulence effectors that also have a virulence function (Stergiopoulos and de Wit, 2009), such as is possible at the AvrHar/AvrPra2 locus (Lai et al., 2007)? Complete verification of how the pathogen induces disease and how the host recognizes the pathogen to protect itself will not be known until the pathogen and host genes are cloned and components of the pathways involving these gene products are revealed. This work has been initiated in several ways, including genomic sequencing, genetic mapping and the identification of major genetic regions associated with virulence/avirulence, protein identification and expression of candidate virulence effector genes, and mapping and characterization of host resistance/susceptibility genes.
As noted earlier, P. teres f. maculata, causing SFNB, has recently become a problem of epidemic proportions in several important barley‐producing regions worldwide. Significantly less emphasis has been placed on this form in the past, and therefore the knowledge of SFNB, from both the host and pathogen side, is behind that of NFNB. Several key areas of research are needed involving SFNB. Only a handful of resistance sources effective against SFNB have been evaluated, and therefore emphasis on evaluating current populations, as well as developing new populations involving diverse resistance sources, is needed. The characterization of the genetics of pathogen virulence of P. teres f. maculata is also lacking, and the development and evaluation of P. teres f. maculata populations would begin to provide clues as to how this form of the pathogen induces disease. Available information on the P. teres f. maculata infection process and life cycle is also lacking, and is a necessary area of research in order to look at the differences from and similarities to P. teres f. teres.
Although not considered as a ‘model system’, the P. teres–barley interaction has several characteristics that make it a tractable model for investigation.
-
1
Foremost, the net blotch pathogen forms cause significant damage worldwide, and therefore net blotch is an economically important disease.
-
2
Because P. teres is a haploid heterothallic fungus and crosses can easily be generated in the laboratory, pathogen mapping populations have been and can be developed for the mapping, characterization and eventual cloning of genes associated with virulence/avirulence, and genetic maps could also be used to complement genome assembly.
-
3
The fungus is amenable to transformation, and therefore site‐directed gene disruption for the functional characterization of the genes involved in virulence is theoretically possible.
-
4
Genomic sequence information for both forms of the pathogen will soon be available and will speed the process of identification of the genes associated with virulence and growth of the pathogen.
-
5
A significant amount of genetic information on host resistance/susceptibility is available, including several host mapping populations used to locate major resistance/susceptibility genes. These host lines are useful for deciphering the gene‐for‐gene relationship between pathogen virulence/avirulence and host resistance/susceptibility.
-
6
Several pathotype studies have been conducted worldwide, investigating the diversity of virulence of the fungus. Several common barley differential lines have been used repeatedly, making virulence comparisons of different regions possible. These studies have shown that a large amount of virulence diversity is present worldwide. Based on a co‐evolutionary model, the same amount of diversity is present in the host, making this an exciting system to investigate.
REFERENCES
- Able, A. (2003) Role of reactive oxygen species in the response of barley to necrotrophic pathogens. Protoplasma, 221, 137–143. [DOI] [PubMed] [Google Scholar]
- Abu Qamar, M. , Liu, Z.H. , Faris, J.D. , Chao, S. , Edwards, M.C. , Lai, Z. , Franckowiak, J.D. and Friesen, T.L. (2008) A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor. Appl. Genet. 117, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Afanasenko, O. , Mironenko, N. , Filatova, O. , Kopahnke, D. , Krämer, I. and Ordon, F. (2007) Genetics of host–pathogen interactions in the Pyrenophora teres f. teres (net form)–barley (Hordeum vulgare) pathosystem. Eur. J. Plant Pathol. 117, 267–280. [Google Scholar]
- Afanasenko, O.S. , Jalli, M. , Pinnschmidt, H.O. , Filatova, O. and Platz, G.J. (2009) Development of an international standard set of barley differential genotypes for Pyrenophora teres f. teres . Plant Pathol. 58, 665–676. [Google Scholar]
- Alcorn, J.L. (1988) The taxonomy of ‘Helminthosporium’ species. Annu. Rev. Phytopathol. 26, 37–56. [Google Scholar]
- Arabi, M.I. , Sarrafi, A. , Barrault, G. and Alertini, L. (1990) Inheritance of partial resistance to net blotch in barley. Plant Breed. 105, 150–155. [Google Scholar]
- Arabi, M.I.E. , Al‐Safadi, B. and Charbaji, T. (2003) Pathogenic variation among isolates of Pyrenophora teres, the causal agent of barley net blotch. J. Phytopathol. 151, 376–382. [Google Scholar]
- Aragona, M. , Montigiani, M. and Porta‐Puglia, A. (2000) Electrophoretic karyotypes of the phytopathogenic Pyrenophora graminea and P. teres . Mycol. Res. 104, 853–857. [Google Scholar]
- Atanasoff, D. and Johnson, A.G. (1920) Treatment of cereal seeds by dry heat. J. Agric. Res. [US] 18, 379–390. [Google Scholar]
- Bach, E. , Christensen, S. , Dalgaard, L. , Larsen, P.O. and Olsen, C.E. (1979) Structures, properties and relationship to the aspergillomarasmines of toxins produced by Pyrenophora teres . Physiol. Plant Pathol. 14, 41–46. [Google Scholar]
- Bakonyi, J. and Justesen, A.F. (2007) Genetic relationship of Pyrenophora graminea, P. teres f. maculata and P. teres f. teres assessed by RAPD analysis. J. Phytopathol. 155, 76–83. [Google Scholar]
- Beattie, A.D. , Scoles, G.J. and Rossnagel, B.G. (2007) Identification of molecular markers linked to a Pyrenophora teres avirulence gene. Phytopathology, 97, 842–849. [DOI] [PubMed] [Google Scholar]
- Van Den Berg, C.G.J. and Rossnagel, B.G. (1990) Effects of temperature and leaf wetness period on conidium germination and infection of barley by Pyrenophora teres . Can. J. Plant Pathol. 12, 263–266. [Google Scholar]
- Van Den Berg, C.G.J. and Rossnagel, B.G. (1991) Epidemiology of spot‐type net blotch on spring barley in Saskatchewan. Phytopathology, 81, 1446–1452. [Google Scholar]
- Bockelman, H.E. , Sharp, E.L. and Eslick, R.F. (1977) Trisomic analysis of genes for resistance to scald and net blotch in several barley cultivars. Can. J. Bot. 55, 2142–2148. [Google Scholar]
- Bogacki, P. , Oldach, K.H. and Williams, K.J. (2008) Expression profiling and mapping of defence response genes associated with the barley–Pyrenophora teres incompatible interaction. Mol. Plant Pathol. 9, 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringans, S. , Hane, J.K. , Casey, T. , Tan, K.‐C. , Lipscombe, R. , Solomon, P.S. and Oliver, R.P. (2009) Deep proteogenomics; high throughput gene validation by multidimensional liquid chromatography and mass spectrometry of proteins from the fungal wheat pathogen Stagonospora nodorum . BMC Bioinformatics, 10, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir, M. , Gupta, S. , Platz, G.J. , Ablett, G.A. , Loughman, R. , Emebiri, L.C. , Poulsen, D. , Li, C.D. , Lance, R.C.M. , Galway, N.W. , Jones, M.G.K. and Appels, R. (2003) Mapping and validation of the genes for resistance to Pyrenophora teres f. teres in barley (Hordeum vulgare L.). Aust. J. Agric. Res. 54, 1369–1377. [Google Scholar]
- Campbell, G.F. and Crous, P.W. (2003) Genetic stability of net × spot hybrid progeny of the barley pathogen Pyrenophora teres . Australas. Plant Pathol. 32, 283–287. [Google Scholar]
- Campbell, G.F. , Crous, P.W. and Lucas, J.A. (1999) Pyrenophora teres f. maculata, the cause of Pyrenophora leaf spot of barley in South Africa. Mycol. Res. 103, 257–267. [Google Scholar]
- Campbell, G.F. , Lucas, J.A. and Crous, P.W. (2002) Evidence of recombination between net‐ and spot‐type populations of Pyrenophora teres as determined by RAPD analysis. Mycol. Res. 106, 602–608. [Google Scholar]
- Cromey, M.G. and Parks, R.A. (2003) Pathogenic variation in Dreschlera teres in New Zealand. N. Z. Plant Protect. 56, 251–256. [Google Scholar]
- Crous, P.W. , Janse, B.J.H. , Tunbridge, J. and Holz, G. (1995) DNA homology of Pyrenophora japonica and P. teres . Mycol. Res. 99, 1098–1102. [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. , Thon, M. , Kulkarni, R. , Xu, J.‐R. , Pan, H. , Read, N.D. , Lee, Y.‐H. , Carbone, I. , Brown, D. , Oh, Y.Y. , Donofrio, N. , Jeong, J.S. , Soanes, D.M. , Djonovic, S. , Kolomiets, E. , Rehmeyer, C. , Li, W. , Harding, M. , Kim, S. , Lebrun, M.‐H. , Bohnert, H. , Coughlan, S. , Butler, J. , Calvo, S. , Ma, L.‐J. , Nicol, R. , Purcell, S. , Nusbaum, C. , Galagan, J.E. and Birren, B.W. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- Dilger, M. , Felsenstein, F.G. and Schwarz, G. (2003) Identification and quantitative expression analysis of genes that are differentially expressed during conidial germination in Pyrenophora teres . Mol. Genet. Genomics, 270, 147–155. [DOI] [PubMed] [Google Scholar]
- Douglas, G.B. and Gordon, L.L. (1985) Quantitative genetics of net blotch in barley. N. Z. J. Agric. Res. 28, 157–164. [Google Scholar]
- Douiyssi, A. , Rasmusson, D.C. and Roelfs, A.P. (1998) Responses of barley cultivars and lines to isolates of Pyrenophora teres . Plant Dis. 82, 316–321. [DOI] [PubMed] [Google Scholar]
- Drechsler, C. (1923) Some graminicolous species of Helminthosporium . J. Agric. Res. 24, 641–740. [Google Scholar]
- Emebiri, L.C. , Platz, G. and Moody, D.B. (2005) Disease resistance genes in a doubled haploid population of two‐rowed barley segregating for malting quality attributes. Aust. J. Agric. Res. 56, 49–56. [Google Scholar]
- Feys, B.J. and Parker, J.E. (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Frecha, J.H. (1958) Herencia de los factores de resistencia a Helminthosporium teres que poseen las variedades de cebada Rojo, Hoyo Epuyén y Hordeum spontaneum . Rev. Invest. Agric. 12, 91–95. [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Lai, Z. and Steffenson, B.J. (2006a) Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled‐haploid barley population. Genome, 49, 855–859. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Stukenbrock, E.H. , Liu, Z.H. , Meinhardt, S.W. , Ling, H. , Faris, J.D. , Rasmussen, J.B. , Solomon, P.S. , McDonald, B.A. and Oliver, R.P. (2006b) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Meinhardt, S.W. and Faris, J.D. (2007) The Stagonospora nodorum–wheat pathosystem involves multiple proteinaceous host‐selective toxins and corresponding host sensitivity genes that interact in an inverse gene‐for‐gene manner. Plant J. 51, 681–692. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Solomon, P.S. and Oliver, R.P. (2008a) Host‐specific toxins, effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Zhang, Z. , Solomon, P.S. , Oliver, R.P. and Faris, J.D. (2008b) Characterization of the interaction of a novel Stagonospora nodorum host‐selective toxin with a wheat susceptibility gene. Plant Physiol. 146, 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis, P. , Olsen, C.E. and Møller, B.L. (1991) Toxin production in Pyrenophora teres, the Ascomycete causing the net‐spot blotch disease of barley (Hordeum vulgare L.). J. Biol. Chem. 266, 13 329–13 335. [PubMed] [Google Scholar]
- Geschele, E.E. (1928) The response of barley to parasitic fungi Helminthosporium teres Sacc. Bull. Appl. Bot. Genet. Plant Breed. 19, 371–384 (in Rev. Appl. Mycol. 8, 165). [Google Scholar]
- Graner, A. , Foroughi‐Wehr, B. and Tekauz, A. (1996) RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica, 91, 229–234. [Google Scholar]
- Gray, G.G. (1966) Genetic systems in the net blotch disease complex of barley. Ph.D. dissertation. North Dakota State University, Fargo, ND.
- Grewal, T.S. , Rossnagel, B.G. , Pozniak, C.J. and Scoles, G.J. (2008) Mapping quantitative trait loci associated with barley net blotch resistance. Theor. Appl. Genet. 116, 529–539. [DOI] [PubMed] [Google Scholar]
- Gupta, S. and Loughman, R. (2001) Current virulence of Pyrenophora teres on barley in Western Australia. Plant Dis. 85, 960–966. [DOI] [PubMed] [Google Scholar]
- Hane, J.K. , Lowe, R.G.T. , Solomon, P.S. , Tan, K.‐C. , Schoch, C.L. , Spatafora, J.W. , Crous, P.W. , Kodira, C. , Birren, B.W. , Torriani, S.F.F. , McDonald, B.A. and Oliver, R. (2007) Dothideomycete–plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum . Plant Cell, 19, 3347–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves, J.A. and Keon, J.P.R. (1983) The binding of isolated mesophyll cells from barley leaves to hyphae of Pyrenophora teres . Plant Cell Rep. 2, 240–243. [DOI] [PubMed] [Google Scholar]
- Ho, K.M. , Tekauz, A. , Choo, T.M. and Martin, R.A. (1996) Genetic studies on net blotch resistance in a barley cross. Can. J. Plant Sci. 76, 715–720. [Google Scholar]
- Ito, S. and Kuribayashi, K. (1931) The ascigerous forms of some graminicolous species of Helminthosporium in Japan. J. Fac. Agric., Hokkaido Imp. Univ. Sapporo, 29, 85–125. [Google Scholar]
- Jalli, M. (2004) Suitability of a selected barley differential set for Pyrenophora teres f. teres virulence screening. Proceedings of the 9th International Barley Genetics Symposium, Brno, Czech Republic, pp. 266–269.
- Jalli, M. and Robinson, J. (2000) Stable resistance in barley to Pyrenophora teres f. teres isolates from the Nordic‐Baltic region after increase on standard host genotypes. Euphytica, 113, 71–77. [Google Scholar]
- Jonsson, R. , Bryngelsson, T. and Gustafsson, M. (1997) Virulence studies of Swedish net blotch isolates (Drechslera teres) and identification of resistant barley lines. Euphytica, 94, 209–218. [Google Scholar]
- Jonsson, R. , Sall, T. and Bryngelsson, T. (2000) Genetic diversity for random amplified polymorphic DNA (RAPD) markers in two Swedish populations of Pyrenophora teres . Can. J. Plant Pathol. 22, 258–264. [Google Scholar]
- Jordan, V.W.L. (1981) Aetiology of barley net blotch caused by Pyrenophora teres and some effects on yield. Plant Pathol. 30, 77–87. [Google Scholar]
- Jordan, V.W.L. and Allen, E.C. (1984) Barley net blotch: influence of straw disposal and cultivation methods on inoculum potential, and on incidence and severity of autumn disease. Plant Pathol. 33, 547–559. [Google Scholar]
- Jørgensen, H.J.L. , Lübeck, P.S. , Thordal‐Christensen, H. , De Neergaard, E. and Smedegård‐Petersen, V. (1998) Mechanisms of induced resistance in barley against Drechslera teres . Phytopathology, 88, 698–707. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2007) Groovy times, filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 10, 358–365. [DOI] [PubMed] [Google Scholar]
- Karki, C.B. and Sharp, E.L. (1986) Pathogenic variation in some isolates of Pyrenophora teres f. sp. maculata on barley. Plant Dis. 70, 684–687. [Google Scholar]
- Keeling, B.L. and Banttari, E.E. (1975) Factors associated with the resistance of barley to Helminthosporium teres . Phytopathology, 65, 464–467. [Google Scholar]
- Keiper, F.J. , Grcic, M. , Capio, E. and Wallwork, H. (2008) Diagnostic microsatellite markers for the barley net blotch pathogens, Pyrenophora teres f. maculata and Pyrenophora teres f. teres . Australas. Plant Pathol. 37, 428–430. [Google Scholar]
- Kenneth, R. (1962) On the taxonomy, morphology and geographical origins of Pyrenophora teres Drechsler and allied species. Bull. Res. Council Israel, 11D, 55–82. [Google Scholar]
- Keon, J.P.R. and Hargreaves, J.A. (1983) A cytological study of the net blotch disease of barley caused by Pyrenophora teres . Physiol. Plant Pathol. 22, 321–329. [Google Scholar]
- Khan, T.N. (1982) Changes in barley genotypes grown in Western Australia. Plant Dis. 66, 655–656. [Google Scholar]
- Khan, T.N. and Boyd, W.J.R. (1969a) Physiologic specialization in Drechslera teres . Aust. J. Biol. Sci. 22, 1229–1235. [Google Scholar]
- Khan, T.N. and Boyd, W.J.R. (1969b) Inheritance of resistance to net blotch in barley. II. Genes conditioning resistance against race W.A.‐2. Can. J. Genet. Cytol. 11, 592–597. [Google Scholar]
- Krupinska, K. , Haussuhl, K. , Schafer, A. , Van Der Kooij, T.A. , Leckband, G. , Lorz, H. and Falk, J. (2002) A novel nucleus‐targeted protein is expressed in barley leaves during senescence and pathogen infection. Plant Physiol. 130, 1172–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C.Y. , Rasmussen, J.B. and Meinhardt, S.W. (1998) Activity of Ptr ToxA from Pyrenophora tritici‐repentis requires host metabolism. Physiol. Mol. Plant Pathol. 52, 201–212. [Google Scholar]
- Lai, Z. , Faris, J.D. , Weiland, J.J. , Steffenson, B.J. and Friesen, T.L. (2007) Genetic mapping of Pyrenophora teres f. teres genes conferring avirulence on barley. Fungal Genet. Biol. 44, 323–329. [DOI] [PubMed] [Google Scholar]
- Lehmensiek, A. , Platz, G.J. , Mace, E. , Poulsen, D. and Sutherland, M.W. (2007) Mapping of adult plant resistance to net form of net blotch in three Australian barley populations. Aust. J. Agric. Res. 58, 1191–1197. [Google Scholar]
- Lehmensiek, A. , Bester, A.E. , Sutherland, M.W. , Platz, G. , Kriel, W.M. , Potgieter, G.F. and Prins, R. (2010) Population structure of South African and Australian Pyrenophora teres isolates. Plant Pathol. 59, 504–515. [Google Scholar]
-
Leišova, L.
,
Kučera, L.
,
 , V. and
Ovesná, J.
(2005a) AFLP‐based PCR markers that differentiate spot and net forms of Pyrenophora teres
.
Plant Pathol.
54, 66–73.
[Google Scholar]
, V. and
Ovesná, J.
(2005a) AFLP‐based PCR markers that differentiate spot and net forms of Pyrenophora teres
.
Plant Pathol.
54, 66–73.
[Google Scholar] -
Leišova, L.
,
 , V.,
Kučera, L.
and
Ovesná, J.
(2005b) Genetic diversity of Pyrenophora teres isolates as detected by AFLP analysis.
J. Phytopathol.
153, 569–578.
[Google Scholar]
, V.,
Kučera, L.
and
Ovesná, J.
(2005b) Genetic diversity of Pyrenophora teres isolates as detected by AFLP analysis.
J. Phytopathol.
153, 569–578.
[Google Scholar] - Lightfoot, D.J. and Able, A.J. (2010) Growth of Pyrenophora teres in planta during barley net blotch disease. Australas. Plant Pathol. In press. doi: 10.1071/AP10121. [Google Scholar]
- Liu, Z.H. and Friesen, T.L. (2010) Identification of Pyrenophora teres f. maculata, causal agent of spot type net blotch in North Dakota. Plant Dis. 94, 480. [DOI] [PubMed] [Google Scholar]
- Liu, Z.H. , Faris, J.D. , Edwards, M.C. and Friesen, T.L. (2008) Molecular cloning of AvrHar from Pyrenophora teres f. teres . Phytopathology, 98, S93. [Google Scholar]
- Liu, Z.H. , Faris, J.D. , Oliver, R.P. , Tan, K.‐C. , Solomon, P.S. , McDonald, B.A. , Nunez, A. , Lu, S. , Rasmussen, J. and Friesen, T.L. (2009) SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog. 5, e10000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J.M. , Sweat, T.A. and Wolpert, T.J. (2007) Plant disease susceptibility conferred by a ‘resistance’ gene. Proc. Natl. Acad. Sci. USA, 104, 14 861–14 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw, J.P.J. , Crous, P.W. and Holz, G. (1994) Pyrenophora japonica occurring on barley in South Africa. Plant Pathol. 43, 420–423. [Google Scholar]
- Louw, J.P.J. , Victor, D. , Crous, P.W. , Holz, G. and Janse, B.J.H. (1995) Characterization of Pyrenophora isolates associated with spot and net type lesion on barley in South Africa. J. Phytopathol. 143, 129–134. [Google Scholar]
- Louw, J.P. , Crous, P.W. and Holz, G. (1996) Relative importance of the barley net blotch pathogens Pyrenophora teres f. teres (net type) and P. teres f. maculata (spot type) in South Africa. Afr. Plant Protect. 2, 89–95. [Google Scholar]
- Ma, Z.Q. , Lapitan, N.L.V. and Steffenson, B. (2004) QTL mapping of net blotch resistance genes in a doubled‐haploid population of six‐rowed barley. Euphytica, 137, 291–296. [Google Scholar]
- Manninen, O. , Kalendar, R. , Robinson, J. and Schulman, A.H. (2000) Application of BARE‐1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol. Gen. Genet. 264, 325–334. [DOI] [PubMed] [Google Scholar]
- Manninen, O.M. , Jalli, M. , Kalendar, R. , Schulman, A. , Afanasenko, O. and Robinson, J. (2006) Mapping of major spot‐type and net‐type net blotch resistance genes in the Ethiopian barley (Hordeum vulgare) line CI 9819. Genome, 49, 1564–1571. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Chu, A. , Steeves, J.E. , Wolpert, T.J. and Ciuffetti, L.M. (2009) A host‐selective toxin of Pyrenophora tritici‐repentis, Ptr ToxA, induces photosystem changes and reactive oxygen species accumulation in sensitive wheat. Mol. Plant–Microbe Interact. 22, 665–676. [DOI] [PubMed] [Google Scholar]
- Mathre, D.E. (1997) Compendium of Barley Diseases, 2nd edn. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- McDonald, W.C. (1963) Heterothallism in Pyrenophora teres . Phytopathology, 53, 771–773. [Google Scholar]
- McDonald, W.C. (1967) Variability and the inheritance of morphological mutants in Pyrenophora teres . Phytopathology, 57, 747–755. [Google Scholar]
- McDonald, W.C. and Buchanon, K.W. (1962) The inheritance of variability in Pyrenophora teres . Barley Newsl. 6, 40. [Google Scholar]
- McLean, M.S. , Howlett, B.J. and Hollaway, G.J. (2009) Epidemiology and control of spot form of net blotch (Pyrenophora teres f. maculata) of barley, a review. Crop Pasture Sci. 60, 303–315. [Google Scholar]
- Mode, C.J. and Schaller, C.W. (1958) Two additional factors for host resistance to net blotch in barley. Agron. J. 50, 15–18. [Google Scholar]
- Molnar, S.J. , James, L.E. and Kasha, K.J. (2000) Inheritance and RAPD tagging of multiple genes for resistance to net blotch in barley. Genome, 43, 224–231. [PubMed] [Google Scholar]
- Murray, G.M. and Brennan, J.P. (2010) Estimating disease losses to the Australian barley industry. Australas. Plant Pathol. 39, 85–96. [Google Scholar]
- Nagy, E.D. and Bennetzen, J.L. (2008) Pathogen corruption and site‐directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peever, T.L. and Milgroom, M.G. (1994) Genetic structure of Pyrenophora teres populations determined with random amplified polymorphic DNA markers. Can. J. Bot. 72, 915–923. [Google Scholar]
- Peltonen, S. , Jalli, M. , Kammiovirta, K. and Karjalainen, R. (1996) Genetic variation in Drechslera teres populations as indicated by RAPD markers. Ann. Appl. Biol. 128, 465–477. [Google Scholar]
- Raman, H. , Platz, G.J. , Chalmers, K.J. , Raman, R. , Read, B.J. , Barr, A.R. and Moody, D.B. (2003) Mapping of genetic regions associated with net form of net blotch resistance in barley. Aust. J. Agric. Res. 54, 1359–1367. [Google Scholar]
- Rasmussen, J.B. , Kwon, C.Y. and Meinhardt, S.W. (2004) Requirement of host signaling mechanisms for the action of Ptr ToxA in wheat. Eur. J. Plant Pathol. 110, 333–335. [Google Scholar]
- Rau, D. , Brown, A.H.D. , Brubaker, C.L. , Attene, G. , Balmas, V. , Saba, E. and Papa, R. (2003) Population genetic structure of Pyrenophora teres Drechs. the causal agent of net blotch in Sardinian landraces of barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 947–959. [DOI] [PubMed] [Google Scholar]
- Rau, D. , Maier, F.J. , Papa, R. , Brown, A.H.D. , Balmas, V. , Saba, E. , Schaefer, W. and Attene, G. (2005) Isolation and characterization of the mating‐type locus of the barley pathogen Pyrenophora teres and frequencies of mating‐type idiomorphs within and among fungal populations collected from barley landraces. Genome, 48, 855–869. [DOI] [PubMed] [Google Scholar]
- Rau, D. , Attene, G. , Brown, A.H.D. , Nanni, L. , Maier, F.J. , Balmas, V. , Saba, E. , Schäfer, W. and Papa, R. (2007) Phylogeny and evolution of mating‐type genes from Pyrenophora teres, the causal agent of barley ‘net blotch’ disease. Curr. Genet. 51, 377–392. [DOI] [PubMed] [Google Scholar]
- Reiss, E. and Bryngelsson, T. (1996) Pathogenesis‐related proteins in barley leaves, induced by infection with Drechslera teres (Sacc.) Shoem. by treatment with other biotic agents. Physiol. Mol. Plant Pathol. 49, 331–341. [Google Scholar]
- Reiss, E. and Horstmann, C. (2001) Drechslera teres‐infected barley (Hordeum vulgare L.) leaves accumulate eight isoforms of thaumatin‐like proteins. Physiol. Mol. Plant Pathol. 58, 183–188. [Google Scholar]
- Richter, K. , Schondelmaier, J. and Jung, C. (1998) Mapping of quantitative trait loci affecting Drechslera teres resistance in barley with molecular markers. Theor. Appl. Genet. 97, 1225–1234. [Google Scholar]
- Robinson, J. and Jalli, M. (1997) Quantitative resistance to Pyrenophora teres in six Nordic spring barley accessions. Euphytica, 94, 201–208. [Google Scholar]
- Ruiz‐Roldán, M.C. , Maier, F.J. and Schäfer, W. (2001) PTK1, a mitogen‐activated‐protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant–Microbe Interact. 14, 116–125. [DOI] [PubMed] [Google Scholar]
- Sarpeleh, A. , Wallwork, H. , Catcheside, D.E.A. , Tate, M.E. and Able, A.J. (2007) Evidence of involvement of proteinaceous toxins from Pyrenophora teres in symptom development of net blotch of barley. Phytopathology, 97, 907–915. [DOI] [PubMed] [Google Scholar]
- Sarpeleh, A. , Wallwork, H. , Tate, M.E. , Catcheside, D.E.A. and Able, A.J. (2008) Initial characterization of phytotoxic proteins isolated from Pyrenophora teres . Physiol. Mol. Plant Pathol. 72, 73–79. [Google Scholar]
- Sarpeleh, A. , Tate, M.E. , Wallwork, H. , Catcheside, D. and Able, A.J. (2009) Characterisation of low molecular weight phytotoxins isolated from Pyrenophora teres . Physiol. Mol. Plant Pathol. 73, 154–162. [Google Scholar]
- Sato, K. and Takeda, K. (1993) Pathogenic variation of Pyrenophora teres isolates collected from Japanese and Canadian spring barley. Bull. Res. Inst. Bioresour. Okayama Univ. 1, 147–158. [Google Scholar]
- Schaller, C.W. (1955) Inheritance of resistance to net blotch of barley. Phytopathology, 45, 174–176. [Google Scholar]
- Schulze‐Lefert, P. and Vogel, J. (2000) Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5, 343–348. [DOI] [PubMed] [Google Scholar]
- Scott, D.B. (1991) Identity of Pyrenophora isolates causing net‐type and spot‐type lesions on barley. Mycopathologia, 116, 29–35. [Google Scholar]
- Serenius, M. , Mironenko, N. and Manninen, O. (2005) Genetic variation, occurrence of mating types and different forms of Pyrenophora teres causing net blotch of barley in Finland. Mycol. Res. 109, 809–817. [DOI] [PubMed] [Google Scholar]
- Sharma, H.S.S. (1984) Assessment of the reaction of some spring barley cultivars to Pyrenophora teres using whole plants, detached leaves and toxin bioassay. Plant Pathol. 33, 371–376. [Google Scholar]
- Shipton, W.A. , Khan, T.N. and Boyd, W.J.R. (1973) Net blotch of barley. Rev. Plant Pathol. 52, 269–290. [Google Scholar]
- Shoemaker, R.A. (1959) Nomenclature of Drechslera and Bipolaris, grass parasites segregated from ‘Helminthosporium’. Can. J. Bot. 37, 879–887. [Google Scholar]
- Shoemaker, R.A. (1962) Drechslera Ito. Can. J. Bot. 40, 808–836. [Google Scholar]
- Sivanesan, A. (1987) Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Mycol. Pap. 158, 1–261. [Google Scholar]
- Smedegård‐Petersen, V. (1971) Pyrenophora teres f. maculata f. nov. and Pyrenophora teres f. teres on barley in Denmark. Kgl. Vet. Landbohojsk. Arsskr. 124–144. [Google Scholar]
- Smedegård‐Petersen, V. (1972) The perithecial and pycnidial stages of Pvrenophora teres and P. graminea in Denmark. Friesia, 10, 61–85. [Google Scholar]
- Smedegård‐Petersen, V. (1977a) Isolation of two toxins produced by Pyrenophora teres and their significance in disease development of net‐spot blotch of barley. Physiol. Plant Pathol. 10, 203–211. [Google Scholar]
- Smedegård‐Petersen, V. (1977b) Inheritance of genetic factors for symptoms and pathogenicity in hybrids of Pyrenophora teres and Pyrenophora graminea . Phytopathol. Z. 89, 193–202. [Google Scholar]
- Smedegård‐Petersen, V. (1978) Genetics of heterothallism in Pyrenophora graminea and P. teres . Trans. Br. Mycol. Soc. 70, 99–102. [Google Scholar]
- St. Pierre, S. , Gustus, C. , Steffenson, B. , Dill‐Macky, R. and Smith, K.P. (2010) Mapping net form net blotch and Septoria speckled leaf blotch resistance loci in barley. Phytopathology, 100, 80–84. [DOI] [PubMed] [Google Scholar]
- Steffenson, B.J. and Webster, R.K. (1992a) Quantitative resistance to Pyrenophora teres f. teres in barley. Phytopathology, 82, 407–411. [Google Scholar]
- Steffenson, B.J. and Webster, R.K. (1992b) Pathotype diversity of Pyrenophora teres f. teres on barley. Phytopathology, 82, 170–177. [DOI] [PubMed] [Google Scholar]
- Steffenson, B.J. , Hayes, P.M. and Kleinhofs, A. (1996) Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor. Appl. Genet. 92, 552–558. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. and De Wit, P.J.G. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Tekauz, A. (1985) A numerical scale to classify reactions of barley to Pyrenophora teres . Can. J. Plant Pathol. 7, 181–183. [Google Scholar]
- Tekauz, A. (1990) Characterization and distribution of pathogenic variation in Pyrenophora teres f. teres and P. teres f. maculata from western Canada. Can. J. Plant Pathol. 12, 141–148. [Google Scholar]
- Tyler, B.M. (2009) Entering and breaking, virulence effector proteins of oomycete plant pathogens. Cell. Microbiol. 11, 13–20. [DOI] [PubMed] [Google Scholar]
- Van Caeseele, L. and Grumbles, J. (1979) Ultrastructure of the interaction between Pyrenophora teres and a susceptible barley host. Can. J. Bot. 57, 40–47. [Google Scholar]
- Vergara, M. , Cristani, C. and Vannacci, G. (2003) Differential transcripts in Pyrenophora graminea and Pyrenophora teres putatively related to pathogenicity. J. Plant Pathol. 85, 157–164. [Google Scholar]
- Webster, J. (1951) Graminicolous Pyrenomycetes II. The occurrence of the perfect stage of Helminthosporium teres in Britain. Trans. Br. Mycol. Soc. 34, 309–317. [Google Scholar]
- Weiergang, I. , Jørgensen, H.J.L. , Møller, I.M. , Friis, P. and Smedegård‐Petersen, V. (2002) Correlation between sensitivity of barley to Pyrenophora teres toxins and susceptibility to the fungus. Physiol. Mol. Plant Pathol. 60, 121–129. [Google Scholar]
- Weiland, J.J. , Steffenson, B.J. , Cartwright, R.D. and Webster, R.K. (1999) Identification of molecular genetic markers in Pyrenophora teres f. teres associated with low virulence on ‘Harbin’ barley. Phytopathology, 89, 176–181. [DOI] [PubMed] [Google Scholar]
- Williams, K.J. , Lichon, A. , Gianquitto, P. , Kretschmer, J.M. , Karakousis, A. , Manning, S. , Langridge, P. and Wallwork, H. (1999) Identification and mapping of a gene conferring resistance to the spot form of net blotch (Pyrenophora teres f. maculata) in barley. Theor. Appl. Genet. 99, 323–327. [Google Scholar]
- Williams, K.J. , Smyl, C. , Lichon, A. , Wong, K.Y. and Wallwork, H. (2001) Development and use of an assay based on the polymerase chain reaction that differentiates the pathogens causing spot form and net form of netblotch of barley. Australas. Plant Pathol. 30, 37–44. [Google Scholar]
- Williams, K.J. , Platz, G.J. , Barr, A.R. , Cheong, J. , Willsmore, K. , Cakir, M. and Wallwork, H. (2003) A comparison of the genetics of seedling and adult plant resistance to the spot form of net blotch (Pyrenophora teres f. maculata). Aust. J. Agric. Res. 54, 1387–1394. [Google Scholar]
- Wolpert, T.J. , Dunkle, L.D. and Ciuffetti, L.M. (2002) Host‐selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40, 251–285. [DOI] [PubMed] [Google Scholar]
- Wu, H.‐L. , Steffenson, B.J. and Zhong, S. (2003) Genetic variation for virulence and RFLP markers in Pyrenophora teres . Can. J. Plant Pathol. 25, 82–90. [Google Scholar]
- Yun, S.J. , Gyenis, L. , Hayes, P.M. , Matus, I. , Smith, K.P. , Steffenson, B.J. and Muehlbauer, G.J. (2005) Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci. 45, 2563–2572. [Google Scholar]
- Zhang, G. and Berbee, M. (2001) Pyrenophora phylogenetics inferred from ITS and glyceraldehyde‐3‐phosphate dehydrogenase gene sequences. Mycologia, 93, 1048–1063. [Google Scholar]


