SUMMARY
Phytophthora infestans remains a problem to production agriculture. Historically there have been many controversies concerning its biology and pathogenicity, some of which remain today. Advances in molecular biology and genomics promise to reveal fascinating insight into its pathogenicity and biology. However, the plasticity of its genome as revealed in population diversity and in the abundance of putative effectors means that this oomycete remains a formidable foe.
INTRODUCTION
‘Phytophthora infestans ... has been the subject of so many investigations and controversies that it fills one of the most romantic chapters in the history of biological research’ (Berg, 1926).
Berg's statement is as true today as it was in 1926, probably because this pathogen continues to surprise us. However, one constant has been its dramatically explosive epidemic potential, which when realized, has led to immense human suffering. The most famous epidemic occurred in Europe, beginning in 1845 and leading to the potato famine in Ireland (e.g. see Bourke, 1993; Woodham‐Smith, 1962). Even today, the human costs of this disease can be huge. During ‘epidemic’ years, the disease can still drive modern farmers out of business (Fry and Goodwin, 1997a). The direct monetary costs of control efforts and lost production are estimated at > $3 billion/year worldwide (CIP, 1996). Even now, more than 150 years after it was first associated with the potato late blight disease in Europe and in North America (Bourke, 1991), P. infestans remains a major problem in agriculture and recalcitrant to low‐input, stable disease suppression.
Obviously, Berg's ‘romantic chapter’ on P. infestans is not yet concluded—and it now contains many more investigations. For example, a quick search for ‘Phytophthora infestans’ on Google Scholar identified 13 400 articles, with 4450 since 2002—and this search did not find all of the contributions. There are many books (e.g. Dowley et al., 1995; Ingram and Williams, 1991; Lucas et al., 1991), thousands of research articles and thousands of popular reports, and many historical treatments (e.g. Turner, 2005). The ‘romance’ occurs because many, many scientists have had high hopes that their investigations would lead to control of this dangerous pathogen. The ‘controversies’ (some continuing to today) develop from differences in method/interpretation—aided by ego. The vast literature creates a special challenge in writing a short overview of this organism and mandates that it be highly selective. The following profile provides a short description of the life history and evolutionary position of P. infestans, followed by discussions of its population biology, pathogenicity/plant resistance, pathogenic specialization, ecology and disease suppression, closing with a glimpse into some new areas of investigation. I write from the bias that full understanding of this species requires a ‘population perspective’ as well as organismal and molecular perspectives.
LIFE HISTORY
Phytophthora infestans is a heterothallic oomycete, and it is a near‐obligate hemibiotrophic pathogen under natural and agricultural conditions. The asexual cycle (Fig. 1) enables dramatically rapid population growth in susceptible host tissue. Sporangia are produced on sporangiophores that grow from infected tissue. The sporangia are readily dehiscent, particularly in response to changes in relative humidity, and can be aerially dispersed to other plant tissues (Aylor et al., 2001). Sporangia in free water germinate either via a germ tube at higher temperatures (optimum around 20–25 °C), or by releasing wall‐less zoospores (Fig. 1) at lower temperatures (optimum between 10 and 15 °C) (Melhus, 1915). The biflagellated (tinsel‐type and whiplash flagella) zoospores are motile for a short time (often less than 60 min) before encysting. Encysted zoospores germinate directly via a germ tube to penetrate leaf or stem tissue. Macroscopically, there are generally no visible symptoms for at least 2 days (characteristic of a hemibiotroph), after which time small areas of necrosis are visible. Within another day or two and under moderate temperatures and in the presence of moisture (free water or very high relative humidity), sporangiophores are produced, with many sporangia—up to 300 000 per lesion.
Figure 1.
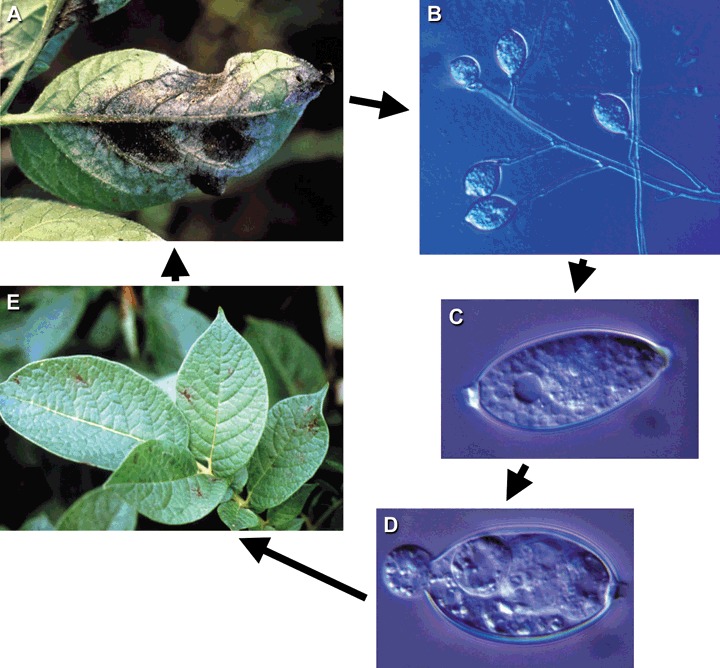
Asexual life cycle of P. infestans. (A,B) Sporangiophores grow out of diseased tissue. (C) Sporangia are released into the atmosphere for aerial dispersal during a drop in relative humidity, or they can be dispersed in water splashes. (D) Indirect germination releases zoospores, which, after encystment and germination on host tissue, produce lesions (E) visible after 2–4 days.
Sporangial development, zoosporogenesis, encystment and cyst germination for the genus have recently received considerable attention. Initially it was thought that treatment with actinomycin D (a transcription inhibitor) had no influence on cleavage of the cytoplasm and release of zoospores, and that sporangia were therefore totally preprogrammed for zoosporogenesis. However, recent studies with P. infestans suggest that this conclusion might have resulted from poor uptake of this material into the sporangium, and that indeed de novo transcription and translation may be required (Judelson and Roberts, 2002). Gene expression studies in P. infestans have begun to identify genes involved in sporulation (Ah Fong and Judelson, 2003; Cvitanich and Judelson, 2003a; Kim and Judelson, 2003), zoosporogenesis (Tani et al., 2003) or zoospore behaviour (Latijnhouwers et al., 2004). Judelson and Roberts (2002) identified a novel protein kinase (resembling Ca2+‐ and calmodulin‐regulated serine/threonine protein kinases) in P. infestans that was up‐regulated in chilled sporangia and appears to be important in zoosporogenesis. The cellular details of zoosporogenesis are most elegantly described for P. cinnamomi (Hardham, 2005), and serve as a guide for other members of the genus. In response to low temperatures, there are increases in cytoplasmic Ca2+ and pH (Hardham, 2005). The multinucleate cytoplasm in the sporangium becomes divided into uninucleate compartments; flagella are assembled and a water expulsion vacuole forms (Hardham, 2005). Zoospores are motile and responsive to chemical and physical stimuli. Upon contact with a plant root, zoospores of P. cinnamomi orientate so that the ventral surface faces the plant surface (root) before they encyst. Upon encystment, contents of dorsal vesicles are secreted over the surface of the cysts, and the contents of ventral vesicles are secreted between the root and the cyst, forming a sticky surface (Hardham, 2005). Shortly thereafter a wall is formed and germination occurs within 20–30 min.
With Solanum tuberosum as a host, the asexual life cycle of P. infestans can be completed rapidly with production of massive numbers of sporangia that are readily dispersed—explaining why whole fields can be transformed from slightly diseased to nearly completely destroyed within just a few days (Fig. 2). Most scientists write in understatement to each other, and do not adequately convey the impact of the speed and devastation that can be caused by this pathogen. However, E. C. Large in his popular account ‘Advance of the Fungi’ (1940) conveyed some of the impact of this disease when it was first observed in Europe:
Figure 2.

The speed with which late blight can destroy a field of potatoes is impressive. A field that appeared ‘healthy’ on one week (A) can be visibly severely diseased in the next week (B), and within another week can be totally destroyed (C).
‘The potatoes had suffered from diseases in the past: from “Scab”, from a malady called the “Curl”, from drought, and from too much rain in bad seasons, but nothing quite so destructive as this new murrain had ever been seen before. It struck down the growing plants like frost in summer. It spread faster than the cholera amongst men.’
Leaves, stems and tubers are all susceptible (Fig. 3), so that the potato late blight pathogen certainly deserves the name of Phytophthora, ‘plant destroyer’. Again from E. C. Large (1940):
Figure 3.

Leaves, stems (A) and tubers (B) are susceptible. Sporangia wash from foliage lesions to contact tubers in the soil. Infected tubers are particularly vulnerable to damage from soft rot, so that if infected potatoes are harvested during warm wet weather, soft rot can cause nearly complete destruction. The potatoes in (C) were discarded from storage within 2 weeks of harvest because late blight infections enabled soft rotting bacteria to destroy the majority of the tubers.
‘Every kind of potato was attacked: the Black Scotch, the Bread Fruit, the Jersey Blues ... When the potatoes were dug from the ground they were found marked with the dark patches, symptomatic of the disease. The colour of these patches was that of contused flesh, its tints were likened to those accompanying a black eye. Potatoes left on the floor of a barn for a week were found worse than when they were lifted. The disease was spreading amongst the potatoes in the ground and in store, and it was thought that every tuber, no matter how slightly affected, would be lost.’
Interestingly, the very first controversy associated with this organism was whether it was the result or the cause of late blight (Bourke, 1991). This controversy was not fully resolved until de Bary's critical work published in the 1860s.
Further testimony to the explosive, devastating potential of the asexual cycle is that some governments had investigated P. infestans as an anti‐crop biological weapon—with some biowarfare efforts ending only in the late 20th century. One result is that P. infestans has been considered by the Ad Hoc Group of the Biological Weapons Convention to be one of several potential biowarfare agents (cited in Madden and Wheelis, 2003).
As a heterothallic member of the oomycetes, P. infestans generally requires both mating types (A1 and A2) for sexual reproduction (Gallegly and Galindo, 1958; Smoot et al., 1958). A hormone moves from one thallus to the other to stimulate the production of oogonia and antheridia; thus each thallus is bisexual (Chern et al., 1996; Judelson, 1997, a; Ko, 1988). The hormone from the A1 mating type has been determined, and described as a diterpene (Qi et al., 2005). However, ‘maleness’ and ‘femaleness’ are not linked to mating type, and in some strains, there may be a tendency for either ‘maleness’ or ‘femaleness’ to predominate (Judelson, 1997a). The antheridia are amphigynous (oogonial stalk surrounded by the antheridium). Fertilization leads to an oospore which serves both as a survival structure and as a source of variation via sexual recombination. As survival structures, oospores have been demonstrated to persist for several years in soil (Mayton et al., 2000) (Turkensteen et al., 2000). They survive very low temperatures well, but not higher temperatures, being unable to survive 2 h at 46 °C or 12 h at 40 °C (Fay and Fry, 1997). As sources of variation, oospores produced from a mating between an A1 and an A2 strain are typically hybrids (Judelson, 1997a,b; Shattock et al., 1986, b). However, the production of hybrids is not absolute, with both selfs and parental types (via apomixis) sometimes recovered from a laboratory cross (Goodwin et al., 1992; Shattock et al., 1986, b). We have found that in some laboratory crosses, the first germinating oospores are more likely to be parental types (via apomixis) than are oospores that germinate later (unpublished data) (Fig. 4).
Figure 4.
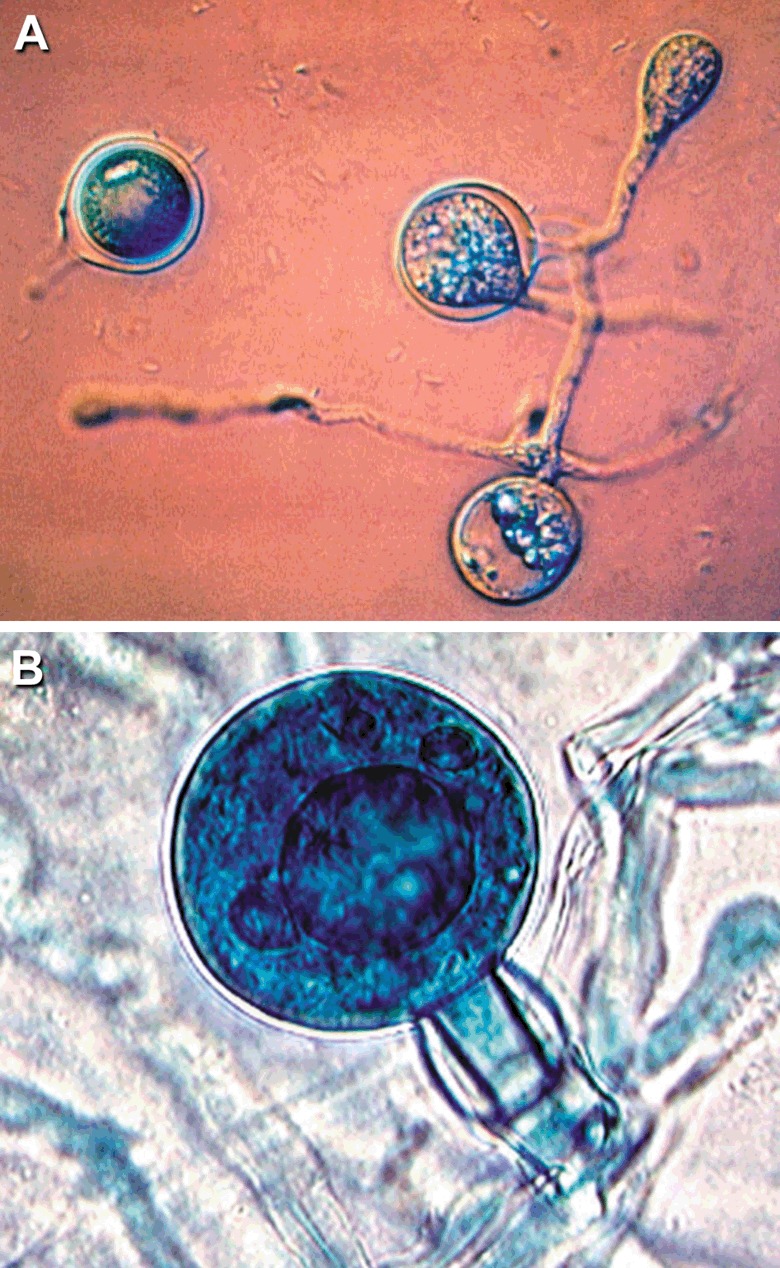
Oospores, oogonia and antheridia of P. infestans. (A) Germinating oospores (photo by Richard Shattock). (B) The female transgenic parent was expressing GUS (blue), but the non‐transgenic male parent was not, indicating an outcrossing sexual interaction (from Judelson, 1997a, and reproduced with permission).
The genetics of mating type in P. infestans appears to be very complicated (Fabritius and Judelson, 1997), confounded by a non‐Mendelian pattern of inheritance (Judelson et al., 1995). Mating type appears to be controlled by a single locus with the suggestion that A1 was heterozygous (Aa) and A2 was homozygous (aa) (Fabritius et al., 2002; Judelson et al., 1995). Perhaps the complicated genetic behaviour can be partially explained by the fact that DNA segments from both homologues of an A1 strain (A/a genotype) contained chromosome‐specific differences, and both also contained high amounts of repetitive DNA (Randall et al., 2002). Additionally, there is diversity for allele location among strains of P. infestans (Randall et al., 2002). van der Lee et al. (2004) found that the mating type locus could not be integrated into an overall genetic map and suggested that it was hemizygous. Despite these genetic complications, gene expression studies have identified some genes apparently involved in the gametangial development (Cvitanich and Judelson, 2003a; Cvitanich et al., 2006). One family of genes (termed M96) is expressed during mating, and consists of 22 tandemly repeated loci at a single chromosomal site in the genome (Cvitanich et al., 2006). Another gene (M90) was found to be up‐regulated during sexual as well as asexual development and was found to be a member of the Puf family of translational regulators (Cvitanich and Judelson, 2003a).
Formation of gametangia can be stimulated by factors in addition to hormones from the opposite mating type of a species. One of the most effective factors is the presence of the opposite mating type of a different species of Phytophthora. When physical exchange is prevented between the two different species, at least some of the resulting oospores are selfs (Shattock et al., 1986a; Shen et al., 1983). However, it is not only the hormones of Phytophthora that stimulate gametangia production; other factors also have an effect, including: aged cultures, various substrates (oats, lima beans, V‐8 juice, etc), physical wounding, and Trichoderma viride (Brasier, 1971; Reeves and Jackson, 1974; Smart et al., 2000). As might be expected, some strains are more responsive to these stimuli than are other strains (Smart et al., 2000). Finally, some strains of traditionally heterothallic Phytophthora species can appear to be self‐fertile (Fyfe and Shaw, 1992), but produce many fewer oospores than in a traditional A1 × A2 cross (Smart et al., 1998). While ‘apparent’ self‐fertility can result from mixtures of A1s and A2s (Fyfe and Shaw, 1992), self‐fertility has also been observed in pure cultures. In this case, self‐fertility may occur if the thallus is heterokaryotic (one nucleus of one mating type and the other nucleus with the other mating type) or some self‐fertile strains may contain an extra chromosome (as has been shown with Bremia) (Michelmore and Sansome, 1982; Mortimer et al., 1977).
The production of oospores and the geographical distribution of the A2 mating types have been the subject of some stimulating controversies. During the late 19th century and early 20th century, there was also debate concerning the contribution of oospores to the overwintering survival of P. infestans. de Bary could not find oospores and suggested that mycelium in a diseased tuber was the major mechanism for overwintering (de Bary, 1876). However, this suggestion was not accepted by some investigators who felt that oospores might have occurred in the field (De Bruyn, 1926). Although it seems clear that survival in diseased tubers is of overwhelming importance in production agriculture (Bonde and Schultz, 1943), this does not preclude the occasional production of an oospore (as described above) that could enable survival for a few years in a field in the absence of potato production. In locations where there is primarily asexual reproduction, the contribution of a rare oospore to the population structure is likely to be minimal, because most progeny (normal hybrids) appear to be dramatically less fit than either parent (Mayton et al., 2000), and recombinant selfs are even less fit than are hybrids (Shattock et al., 1986a). However, again in the late 20th century, controversies arose and there were reports of ‘changes’ or ‘shifts’ in mating type (Groves and Ristaino, 2000; Ko, 1994). Reports such as these were analysed by Goodwin and Drenth (1997) who concluded that the appearance of rare oospores in A1 isolates occurred because of factors known for some time (as described above). They concluded that the global appearance of bona fide A2 isolates was due to migration (see below) rather than from mutation or other changes within an A1 lineage (Goodwin and Drenth, 1997).
EVOLUTIONARY POSITION
One controversy that has been resolved recently is that oomycetes are unrelated to true fungi. Until the latter part of the 20th century, Phytophthora species and other oomycetes were regarded by most plant pathologists to be fungi. Oomycetes certainly share many characteristics with fungi: growth via filamentous hyphal tips, nutrition via absorption and reproduction via spores (Money, 1998). However, as indicated by many articles, including two previous pathogen profiles in this journal (Hardham, 2005; Tyler, 2007) oomycetes are evolutionarily remote from the fungi (Harper et al., 2005). There are many distinguishing features, including cell walls composed of β 1–6, and β 1–3 glucans instead of chitin, the production of biflagellated zoospores, production of antheridia and oogonia as gametangia, and diploid vegetative state. Most of these differences had been accepted much earlier by a few as evidence of taxonomic separation from the fungi (Money, 1998), but it was the power of molecular genetics that persuaded the rest of us to recognize the evolutionary distance between oomycetes and fungi. Oomycetes occur in a group called the Stramenopiles, clustering together with others in a super group, the Chromalveolata (Adl et al., 2005). The fungi belong to a different super group, the Ophisthokonta (Adl et al., 2005). Oomycetes have an evolutionary history that may have involved the enslavement of a photosynthetic organism by another non‐photosynthetic primitive eukaryote, with the plastid being lost secondarily in oomycetes (Cavalier‐Smith, 2000). In support of this scenario, Tyler et al. (2006) found 855 genes in P. sojae and P. ramorum that probably came from a photosynthetic ancestor (red alga or cyanobacterium).
Oomycetes share some interesting novel characteristics with other protists. For example, in contrast to other eukaryotes, Phytophthora shares with another protist, Trichomonas, a core promoter structure in which the initiator element is overrepresented (McLeod et al., 2004; Quon et al., 1994). Additionally, genes encoding effectors in some oomycetes contain an RxLR‐dEER motif similar to the export element (PEXEL) in Plasmodium (Chromalveolates) (Rehmany et al., 2005). This motif is required for targeting pathogen proteins for delivery to the cytoplasm of the host cell (Whisson et al., 2007). There are more than 300 genes with the RxLR motif in each of the genomes of P. sojae and P. ramorum (Tyler et al., 2006), and preliminary analysis of the P. infestans genome indicates that there may be more than 500 in P. infestans (Zody et al., 2007).
POPULATION BIOLOGY/MIGRATIONS
In the final analysis it is only large populations of P. infestans that have huge impacts on plants (and thus on humans), and migrations of populations (bringing significant new diversity) have had major impact on the population dynamics of P. infestans in some locations. The migration that introduced P. infestans to potato production in the north temperate regions is legendary. Before the middle part of the 19th century, potato late blight was unknown in these regions. This ‘new disease’ of potatoes was problematic in the north‐eastern USA by 1843 (Stevens, 1933). It is very well documented that by the summer of 1845, the late blight disease of potatoes was well established in Europe (Bourke, 1991). Although there have been claims of potato late blight in Europe before 1845 (see the discussion in Jones et al., 1912), subsequent analyses have generally not substantiated the claims of a pre‐1845 occurrence of P. infestans in Europe (Andrivon, 1996; Bourke, 1991; Goodwin and Drenth, 1997).
Regardless of the exact date of introduction to Europe, P. infestans became well established in potato production throughout the world subsequent to 1845. During the latter part of the 19th century and the early 20th century, P. infestans was generally regarded as an asexual organism, because (except for central Mexico, see below) only the A1 mating type had been documented—a situation that was to persist into the late 20th century. We confirmed this situation by analysing the genotypic diversity of strains from wherever we could obtain them throughout the world. Populations were sampled from several countries in Europe, Asia, South America, and from the USA and Canada in North America. We were surprised to learn that these populations were genotypically quite similar—dominated by a single clonal lineage—the US‐1 lineage (Goodwin et al., 1994b). This lineage was defined by mating type, mitochondrial haplotype, nuclear DNA fingerprint pattern (via restriction fragment length polymorphisms, RFLPs) and allozyme genotype (Goodwin et al., 1994, b). These isolates were collected prior to the major migrations that began in the latter quarter of the 20th century. Because seed tubers are shipped all over the world, and because P. infestans readily infects tubers, it is easy to visualize that a genetic bottleneck might have enabled a single clonal lineage to be distributed throughout the world. If this lineage had been the first to be introduced to an area, it could dominate. Alternatively if this lineage was particularly fit, such that it displaced the previous population, it would remain as the dominant lineage in a population.
We had hypothesized that the initial European epidemic (starting in the 1840s) might have been caused by the US‐1 clonal lineage of P. infestans (Goodwin et al., 1994, b). One possibility is that this lineage went through a genetic bottleneck from the USA to Europe, and by a founder effect colonized all of Europe and subsequently was distributed to much of the rest of the world. This hypothesis became part of the controversy concerning the origin of P. infestans and stimulated additional investigation. Our hypothesis was directly challenged when analyses of herbarium specimens collected during the European epidemic of the 1840s and 1850s identified genotypes of P. infestans that were not of the US‐1 lineage (Ristaino et al., 2001). Thus, the current data do not support our previous hypothesis that the US‐1 lineage caused the original epidemic in Europe. Frustratingly, this leaves no explanation for the previous worldwide distribution and dominance of the US‐1 clonal lineage.
The discovery in the 1950s that the A2 mating type occurred commonly in the Toluca Valley in central Mexico (Galindo and Gallegly, 1960; Gallegly and Galindo, 1958; Niederhauser, 1956) was a major event in our understanding of the biology of this organism. Prior to the discovery of the A2 mating type in the Toluca Valley, it was recognized that the populations of P. infestans in that location were dramatically different from populations in other locations—mainly because potato clones reported to be resistant (immune) elsewhere were severely diseased when grown in the Toluca Valley (Niederhauser and Mills, 1953). Subsequent analyses confirmed the uniqueness of the population of P. infestans in the Toluca Valley. In contrast to populations in other locations, the A1 and A2 mating types existed in approximately equal frequencies (Goodwin et al., 1992); allozyme alleles existed in a Hardy–Weinberg equilibrium (Tooley et al., 1985); oospores were ubiquitous (Flier et al., 2001; Grunwald and Flier, 2005; Grunwald et al., 2001); no clonal lineage dominated the population (Flier et al., 2003, b; Grunwald et al., 2001); and the virulence capabilities (compatibility with diverse R genes) were broader than in other populations (Niederhauser and Mills, 1953).
The global population structure of P. infestans began to change in the latter part of the 20th century—due to migrations. The first was a migration from Mexico to Europe and then secondary migrations from Europe to other locations worldwide. The initial indication that ‘something’ was happening was the report of A2 mating types in Switzerland (Hohl and Iselin, 1984). Subsequently, there were other reports of A2 mating type strains in Europe (Fry et al., 1991; Shaw et al., 1985). We now know that the likely route of migration was via a large (25 000 metric tonnes) shipment of potatoes for fresh consumption imported to Europe from Mexico in the winter of 1976/77 (Niederhauser, 1991). Because tubers are readily infected, it seems highly probable that this shipment contained a diverse population of P. infestans. Retrospective analysis indicated that the immigrating population was diverse for mating type, allozyme genotype, DNA RFLP fingerprint and mitochondrial haplotype (Fry et al., 1992, 1993). Surprisingly, the immigrating population quickly dominated and displaced the previous population in much of western Europe (Drenth et al., 1994; Fry and Goodwin, 1997, b; Spielman et al., 1991; Sujkowski et al., 1994). Because rare alleles detected in the modern European population were subsequently detected in populations on other continents (Forbes et al., 1997), it seems likely that secondary migrations (probably via seed potatoes) introduced the new populations to many locations worldwide, except perhaps in Australia and parts of Africa (Goodwin et al., 1994, b; McLeod et al., 2001).
The population of P. infestans in the United States and in Canada was influenced by a different migration event from Mexico, so that the population north of Mexico is distinct from that in Europe, Asia and South America (Fry and Goodwin, 1997a, 1997b). This migration probably occurred during the 1980s and perhaps the early 1990s and seems most likely to have involved infected tomato fruits from production areas in north‐west Mexico (Goodwin et al., 1994a, 1998). Just as in Europe and Asia, the new population displaced the previous dominant population and exacerbated the problem of late blight (Fry and Goodwin, 1997a,b). However, the population in the USA is still clonal, with no evidence yet of an established sexual population (unpublished results).
‘Whence came Phytophthora infestans?’ This question is the title to one of Donald Reddick's papers (Reddick, 1939) and is part of one of the most engaging controversies surrounding this pathogen. Although this question had been asked many times previously, Reddick suggested a novel source. Initially, it had been generally assumed that the pathogen originated in the South American Andes and came to Europe along with the potato (Jones et al., 1912). However, by 1928, Reddick had raised questions about this theory (Reddick, 1928). Subsequently, and continuing to the present time, the center of origin of P. infestans has been debated vigorously (Abad and Abad, 1995; Gomez‐Alpizar et al., 2007; Goodwin, 1997; Goodwin et al., 1994b; Grunwald and Flier, 2005; Reddick, 1939). Reddick suggested that Mexico might be the source of this organism because resistant potatoes were readily obtained from central Mexico, but not from South America (Reddick, 1928). Interestingly, proponents and opponents of a Mexican origin read the early literature and came to opposite conclusions as to whether or not the description of any potato malady in the Andes resembled potato late blight (Reddick, 1928, 1939). Andrivon (1996) suggested that the migration of P. infestans to the north might have originated in central Mexico, but the first stop was South America, with subsequent migration to North America and then Europe. More recently, Gomez‐Alpizar et al. (2007) suggested on the basis of analysis of the closely related P. andina from South America that the South American Andes are the ancestral home to P. infestans. However, the story continues to unfold. Recently one missing link to a Mexican centre of origin, the Ib mitochondrial haplotype of P. infestans, was found to occur in Mexico (Garay‐Serrano et al., 2007). Additionally, it is only in central Mexico that all three members of the P. mirabilis/P. ipomoeae/P. infestans clade occur. And until the very recent demonstration of common sexual reproduction in parts of Europe, it was only in central Mexico that sexual reproduction was a common component of the life history of this organism. Thus, the controversy continues.
In a location where both A1 and A2 mating types coexist, oospores clearly contribute importantly to the overwintering survival and also to the nature of the population structure of P. infestans. In the Toluca Valley of Mexico, oospores of P. infestans are commonly found in soil (Fernandez‐Pavia et al., 2004) and A1 and A2 mating types are typically present in equal proportions (Goodwin et al., 1992, 2001; Grunwald and Flier, 2005) these facts coupled with the fact that the population is dramatically diverse genotypically (Flier et al., 2003, b) led one to the conclusion that sexual reproduction and the production of oospores are common and important components of the life history of P. infestans there. In other locations such as in northern Europe where sexual reproduction is of recent occurrence, it is possible to compare epidemic severity and population structure before and after the advent of sexual reproduction. Late blight has certainly become more problematic in some parts of northern Europe and it now appears that sexual reproduction occurs in several locations there with oospores contributing to epidemics earlier in the season (Andersson et al., 1998; Flier et al., 2007; Widmark et al., 2007).
PATHOGENICITY/RESISTANCE
The remarkable pathogenic potential of P. infestans has stimulated many attempts to find or breed resistant hosts. Efforts were first initiated in the mid‐19th century, and resulted in ‘field resistance’ that was partially successful (Wastie, 1991). However, the discovery of the first resistance genes for immunity (R genes) in Solanum demissum created a naïve optimism in the early 20th century and diverted attention away from field resistance. The optimism generated by the discovery of R genes resulted in such titles as ‘Elimination of potato late blight from North America’ (Reddick, 1934). These R genes essentially conferred immunity to the tester strains of P. infestans (with only a few cells affected at each penetration site). We now know that the R genes enabled plants to recognize effector proteins of P. infestans that were secreted into the apoplast or into the host cytoplasm (Govers and Gijzen, 2006). If P. infestans was recognized, host defences were stimulated and further growth of the pathogen was halted (Ferris, 1955) and was associated with the hypersensitive response (HR) (Vleeshouwers et al., 2000).
Unfortunately, it gradually became clear that there were strains in the pathogen population against which the first R genes were individually ineffective (no immunity and no HR). Initially, such ‘resistance‐breaking strains’ were at such a low level in the pathogen population as to be not detectable, and in fact they may have been absent from some populations. However, because it takes 10–15 years (or more) to develop a potato cultivar, there was sufficient time to enable the occurrence and subsequent selection of a resistance‐breaking biotype in a local pathogen population. As the cultivar with a novel R gene increased in popularity (increased in area occupied), there was greater and greater chance for selection of strains compatible with the R gene (unrecognized by the R protein), and the clones containing the R gene were then susceptible to the selected population (Malcolmson, 1969; Wastie, 1991). Unfortunately, this pattern of events has occurred so often that, up to the present time, there appears to be no R gene that has been effective for a sufficiently long time to have contributed much to practical potato late blight suppression in production agriculture.
One suggestion for maintaining the efficacy of R genes has been to put several together in the same potato cultivar (‘pyramiding’). The cultivar Pentland Dell in Scotland provides an informative case history. At the time of its development, Pentland Dell was known to contain genes R1, R2 and R3, and it was resistant to the common race (Race 4). Race 4 was compatible with potato clones containing only R4, but was unable to overcome the resistance conditioned by R1, R2 and/or R3. Commercial production of Pentland Dell began in 1963, and by 1968 it had become the third most popular cultivar, with blight resistance as one of its chief attributes (Malcolmson, 1969). However, by 1967 (and subsequently), there were cases of serious disease on this cultivar (i.e. 75% blight) in England and Wales. The strains that caused disease on Pentland Dell were compatible with R1, R2 and R3, but additionally, and with the discovery of more R genes (R5–R11), it was discovered that many of the strains were also compatible with the newly discovered R genes—even though these new R genes had not been deployed in cultivars (Malcolmson, 1969). The realization that the population of P. infestans in the UK contained individuals unaffected by R genes never deployed decreased considerably the enthusiasm for the use of R genes as a disease suppression strategy for potato late blight (Wastie, 1991). The plasticity of the P. infestans genome with regard to R gene resistance is well described (Goodwin et al., 1995; Shattock et al., 1977), and explains the loss of enthusiasm by some investigators/practitioners for R genes as contributors to stable disease suppression. This and other experience has caused some to focus on developing breeding lines or cultivars with ‘field resistance’ (Collins et al., 1999). However, the strong phenotype of a successful R gene is a strong attraction and there remain many efforts to identify new R genes as a strategy to develop resistant plants (see below). Although ‘controversy’ is perhaps too dramatic a word, there certainly are disagreements as to the value of pursuing the use of single R genes in a cultivar grown in monoculture.
Recent progress in the investigation of effectors provides some likely explanations for this plasticity of pathogenicity in the genome of P. infestans. The selection of secreted proteins as potential candidates (Torto et al., 2003) has proven highly productive to explaining the plasticity of pathogenicity. In general, it is recognized that R genes code for proteins that recognize specific pathogen effectors, and by so doing initiate a cascade of events leading to disease resistance and the HR. The pathogen genes that coded for proteins recognized by R genes were originally recognized from genetics studies and were termed ‘avirulence genes’ because they stimulated resistance (with HR) in the host plant, and therefore ‘avirulence’ in the pathogen. However, it is now clear that probably most if not all such pathogen genes are actually involved in aiding pathogenicity and only when specifically recognized by an R gene do they stimulate the resistance response. The effector gene Avr3a has been cloned from P. infestans and it codes for a protein that also contains the RxLR‐dEER motif (Armstrong et al., 2005; Govers and Gijzen, 2006). Avr3a was cloned using association genetics and was demonstrated to differ from avr3a by three amino acids (Armstrong et al., 2005). There have also been efforts to clone Avr3b, which serendipitously identified a sequence that is characteristic of a transcription factor necessary for the recognition of Avr3b, Avr 10 and Avr11 by plants carrying R3b, R10 and R11 (Jiang et al., 2006). Initial analyses of the P. infestans genome indicate that there are hundreds (at least) of genes coding for proteins with the RxLR motif—more than have been detected in the genomes of P. sojae or P. ramorum (Zody et al., 2007). Some sequences with the RxLR‐dEER motif reside in retrotransposon‐like sequences (Zody et al., 2007), and it is tempting to hypothesize that these are potential effectors in the process of evolving (Zody et al., 2007); if so, then this mechanism could explain the apparent high rate of diversity at effector loci (Goodwin et al., 1995).
Despite the previous disappointments with R genes in practical disease management, the strong phenotype of an effective R gene, and the ease of working with it via biotechnology or through traditional breeding are so attractive that there are still efforts to find an R gene that is durable (sensu Johnson, 1984). Clearly it is diversity in the pathogen population that has rendered other R genes ineffective in suppressing disease. Mechanistically, the hope is that alteration of the component of the effector recognized by the R gene system will be detrimental to P. infestans, and thus the R gene will remain effective (durable). Unfortunately, it is already abundantly clear that many mutations in effectors are not highly detrimental, as deduced from the common existence of the many diverse races of P. infestans. Nonetheless, R genes from several species of Solanum have been and are being investigated. For example, we detected an R gene from Solanum berthaultii (RPi‐ber, located on chromosome 10), which detected common strains of P. infestans in the USA (Rauscher et al., 2006), but it did not detect some strains of P. infestans from the Toluca Valley in central Mexico (Ewing et al., 2000). Similarly a putative R gene from petunias recognized some USA isolates of P. infestans but not other isolates (Becktell et al., 2006). These R genes are unlikely to have a durable effect because diversity in the pathogen population has already been demonstrated. In contrast, an R gene from Solanum bulbocastanum (RB or Rpi‐blb1, located on chromosome 8) seemed initially more promising; it recognized all genotypes of P. infestans against which it had been tested (including the population in the Toluca Valley) (Song et al., 2003; Van der Vossen et al., 2003). The fact that this gene was effective in the Toluca Valley of Mexico in each of the several years that it was tested there created significant optimism that this would be a ‘durable’R gene. It has been only very recently that compatible strains have been detected (E. Van der Vossen, personal communication; F. Govers, personal communication). One disappointing characteristic of Rpi‐blb1 is that although the gene is expressed in tubers, the tubers are not resistant (J. Bradeen, personal communication). An exciting recent discovery (V. G. A. A. Vleeshouwers et al., personal communication) is that the protein coded by Rpi‐blb1 recognizes one of the first proteins (IPI‐O) discovered to be secreted by P. infestans into host tissue (Pieterse et al., 1994). Two additional R genes from S. bulbocastanum, RPi‐blb2 and Rpi‐blb3, have now been cloned (Lokossou et al., 2007; Van der Vossen et al., 2005), but the prognosis for long‐lasting effects remains unknown. Until strains compatible with Rpi‐blb1were detected very recently, there was wonderful optimism about the potential of this gene. Because these strains have not yet appeared in field tests, many investigators retain the hope that this will be the first R gene that is not ‘destroyed’ by P. infestans. Perhaps mutation in—or non‐expression of—the effector recognized by Rpi‐blb1 is indeed detrimental to P. infestans.
The cloning of the effector gene, Avr3a, and the discovery of many potential effectors in the genomic sequence of P. infestans have enabled high‐throughput analysis for additional R gene candidates in a variety of plants in the Solanaceae. For example, candidate R gene phenotypes were observed upon agroinfection of diverse Solanum spp. with the Avr3a effector (Chapouret et al., 2007). In another series of tests, potential R genes that recognized either the ‘virulent’ and/or the ‘avirulent’ form of Avr3a were detected in a different series of wild Solanum spp. (Hein et al., 2007). Additionally, it is now technically possible to use the hundreds of candidate effectors to determine if there are candidate R genes that recognize these candidate effectors. It seems highly likely that many candidate R genes will be found.
The expected discovery of many new R genes raises the issues of how best to deploy them, and whether or not they will contribute to disease suppression in agriculture. If one of these candidate R genes has the apparent ‘durability’ characteristics of Rpi‐blb1, it could be used singly in a potato genotype. However, even if the candidate R genes are ‘overcome’ there have been several strategies suggested for deploying them, including pyramiding, construction of multilines, and sequential individual use. The success of each of these strategies depends on the biology of P. infestans, but also on political and regulatory decisions. Engineered resistance needs consumer acceptance, which will probably occur, but is not yet here. Deployment via traditional breeding is a long process and if pyramiding is necessary, the process will be even longer. Whether pyramiding newly discovered R genes will be more durable than deployment of single R genes is not yet known. However, the previous experience with pyramiding R genes (Malcolmson, 1969) does not suggest durability.
The potential utility of a multiline cultivar (with each line differing by the presence of a different R gene Niederhauser et al., 1996) deserves special consideration. In preliminary experiments, the success of a multiline has been limited geographically, but even where successful, the effect has been very small (Garrett and Mundt, 2000; Garrett et al., 2001). The durable success of using a multiline (no selection for a fit pathogen with compatibility to all R genes) depends on there being a fitness cost to the individual pathogen strain with the mutated effector. Certainly there have been suggestions that highly complex strains (those that can overcome many R genes) might be less fit than simple strains (capable of overcoming no or few R genes), a situation termed ‘stabilizing selection’ by Van der Plank (1963). On cultivars with no R genes, complex strains carry many ‘unnecessary’ genes for virulence, and in other host–pathogen systems, there is evidence that such a situation might be associated with a reduced fitness (e.g. Leonard, 1969). Unfortunately, it is very difficult to detect a fitness cost associated with a specific gene in P. infestans because it is not now possible to construct near‐isogenic strains of P. infestans through either conventional genetics or transformation. Anecdotally, it is clear that some of the most complex strains are also some of the most aggressive, in direct opposition to the theory. For example, the US‐8 strain (dominant on potatoes in the United States from 1994 to the present time) is among the more complex detected in the USA, and is also among the most aggressive (Goodwin et al., 1995; Kato et al., 1997; Lambert and Currier, 1997). Whether the multiline approach will provide durable ‘resistance’ remains to be determined.
Should P. infestans remain able to adapt to new R genes without a loss in fitness, we may be forced to resort to field or general resistances. Some investigators believe that this type of resistance is necessary—perhaps particularly for developing country conditions. Such resistances have been documented (Novy et al., 2006; Stewart et al., 2003) and although often not sufficient alone to solve the late blight problem, they can certainly contribute to the solution (Fry and Shtienberg, 1990; Grunwald et al., 2002, a). Such resistances appear to be durable (Grunwald et al., 2002, b).
PATHOGENIC SPECIALIZATION
In general P. infestans has been regarded as a near obligate parasite with a limited host range (Erwin and Ribeiro, 1996). However, the host range has also been debated. Another of the very early controversies concerned whether the same strains that caused potato late blight also caused tomato late blight (Berg, 1926). Some authors found that P. infestans from potato (or tomato) was preferentially pathogenic on potato (or tomato), but other authors did not find this specialization (Berg, 1926). Resolution of these differences appears to rest with the pathogen population being studied. When we studied the ‘old’ population of P. infestans in the United States, we found no evidence for tomato specialization (Spielman et al., 1989), and could not reproduce earlier reports of such specialization (Berg, 1926; Gallegly and Marvel, 1955). However, we did detect such specialization in the population of the United States common after 1990 (Goodwin et al., 1998; Legard et al., 1995). Within a simple population structure such as that in the USA or north‐west Mexico, some clonal lineages have been observed to occur primarily on potatoes (e.g. the US‐8 clonal lineage in the USA), or on tomatoes (e.g. the US‐7 and US‐17 clonal lineages in the USA) (Goodwin et al., 1998). Specialization to tomato does not preclude pathogenicity to potato, but tomato‐specialized pathogens may be less pathogenic to potatoes than are other strains (Legard et al., 1995). The phenotype of a tomato‐specialized strain on tomato is that of an extended period of biotrophy (Vega‐Sanchez et al., 2000), sometimes with extensive sporulation detected before necrosis (Fig. 5) (Smart et al., 2003). Although tomato specialization has been the subject of little investigation, initial genetic studies detected at least one locus with a major effect, with low aggressiveness to tomato being dominant (Lee et al., 2002).
Figure 5.

Phenotype of a tomato‐unspecialized isolate (A), and a tomato‐specialized isolate (B) infection on tomato (Smart et al., 2003). The tomato‐specialized isolate was sporulating profusely with no evidence of necrosis. The figures are from Smart et al. (2003).
The recent migrations of P. infestans have demonstrated that diversity among pathogen populations influences whether we regard a plant species to be a host. For example, Nicotiana benthamiana was initially reported to be a non‐host to P. infestans in Europe (Kamoun et al., 1998), with resistance being ascribed to recognition of an elicitin (INF1) produced by P. infestans. However, isolates of the new population in the USA, producing INF1, were highly compatible with N. benthamiana (Becktell et al., 2006), suggesting that production of INF1 does not alone cause P. infestans to be a non‐pathogen of N. benthamiana—consistent with the fact that the gene for inf1 production is down‐regulated during infection of potato (Kamoun et al., 1997). Additionally, the US‐8 strain of P. infestans was demonstrated to be much more aggressive against petunia (Petunia×hybrida) and hairy nightshade (Solanum sarachoides) than was the US‐1 strain (Platt, 1999). While black nightshade was found to be a host for a European population of P. infestans (Flier et al., 2003a), it was not a host for a North American population of P. infestans (Platt, 1999). In Europe, Solanum nigrum had been considered to be essentially a non‐host until the arrival of the new diverse population (Flier et al., 2003a).
ECOLOGY/EPIDEMIOLOGY
The ecology of P. infestans has been investigated intensively because of the direct importance of this topic to disease management, with particular importance to disease epidemiology and disease forecasting. Early studies demonstrated that this pathogen does not do well at higher temperatures (e.g. > 25 °C) (Jones et al., 1912), and that indirect germination predominates at temperatures below 20 °C (Crosier, 1934; Melhus, 1915). Comparison of Crosier's data with later data for the US‐1 clonal lineage reveals a remarkable consistency (Mizubuti and Fry, 1998). However, other clonal lineages (US‐7 and US‐8, representing recent immigrants from Mexico) had somewhat different temperature responses (Mizubuti and Fry, 1998). Additionally, BR‐1, one of the two dominant clonal lineages in Brazil, has lower temperature optima than does US‐1, the other dominant clonal lineage in Brazil (Maziero, 2001, cited in Mizubuti and Fry, 2006). These examples provide additional support for the idea that early investigators in the USA were working with the US‐1 clonal lineage during the early 20th century. These data also indicate that there is diversity among different genotypes for response to temperature, and extrapolation from one genotype to the entire species can be misleading.
The availability of free moisture for at least a short period of time on plant surfaces is essential to sporangium germination. The interaction between temperature and moisture was recognized very early as being crucial to pathogen development, and this recognition led to efforts to ‘forecast’ when late blight might be problematic. Many forecasting efforts were initiated, utilizing such things as the ‘Dutch rules’ (Van Everdingen, 1926), ‘Beaumont periods’ (Beaumont, 1947), ‘rain‐favorable days’ (Hyre et al., 1960) and ‘severity values’ (Hyre et al., 1960). Some rules have been adapted to the highland tropics (Grunwald et al., 2002, b). Efforts to improve forecasting continue to the present time (Westerdijk and Schepers, 2006). A potential problem for forecasts is to include variance in the pathogen population for response to environmental factors.
The survival of sporangia in the air is crucial to aerial dispersal, but some studies seemed to indicate that sporangia could not survive at all long in the atmosphere. It was clear that aerial dispersal occurred, but several studies suggested that sporangia could withstand only very short periods of drying (Crosier, 1934; Glendinning et al., 1963; Warren and Colhoun, 1975). This conclusion from controlled experiments was inconsistent with the observations that aerial dispersal of sporangia was important in epidemics. In contrast, other workers found that indeed sporangia could survive drying (Rotem and Cohen, 1974; de Weille, 1964), creating another area of controversy. Subsequently, resolution was achieved when it was demonstrated that differences in experimental technique could explain the different results; if dried sporangia were rehydrated rapidly they perished, but if they were rehydrated slowly, they survived (Minogue and Fry, 1981). It has also become clear that solar irradiance is a much more important factor to sporangial survival than is drying (Mizubuti et al., 2000). We do not yet know if there is diversity within the population for sensitivity to solar irradiation (Mizubuti et al., 2000).
DISEASE CONTROL
Development of reliable, environmentally benign, and economically feasible management tactics is the immediate goal of many investigators and this goal is also the motivation for longer range basic investigations of many scientists. Presently, the most reliable approach is integrated management, using an array of tactics, including planting healthy seed tubers, eliminating on‐farm sources of the pathogen (infected cull tubers, infected volunteers, infected weeds), using ‘resistant’ cultivars and applying fungicides in response to demonstrated need determined via scouting or forecast. There are several challenges. One is that in agriculture in developed countries, the most popular cultivars (those desired by the market) are typically not highly resistant. It would be wonderful if a single R gene were completely durable and could be bred or engineered into cultivars. Most people would prefer such a very simple, highly effective management strategy (a ‘silver bullet’) in comparison with a complex management strategy. However, because potatoes are tetraploid and highly heterozygous, breeding releases a high degree of diversity, so that it is difficult to recover the exact parental phenotype in a breeding programme. Even genetic engineering releases a high degree of diversity, and significant selection needs to occur to achieve the parental phenotype after genes are engineered into potatoes by molecular techniques. Because potatoes are vegetatively propagated, it is possible to retain the exact same phenotype over many generations, and therefore the specific traits associated with a specific cultivar come to be expected. Any deviation from those characteristics may be viewed unfavourably by the market, particularly a market dominated by multinational food corporations. This creates a very difficult environment for the plant breeder, causing any change in potato cultivars to be very slow. An example of the slowness of the adoption of new potato cultivars in the developed world is that two of the most popular cultivars worldwide (Russet Burbank and Bintje) are both more than 100 years old. In contrast, in the developing world, it appears that more recent cultivars with field resistance have significant potential for use in combating late blight.
The availability of effective fungicides has enabled the continued use of susceptible cultivars. As a result, massive amounts of fungicide are used in those environments in which late blight is problematic, typically in rain‐fed production systems. In the USA in 2001 alone, more than 2000 tons of fungicides were used to suppress this disease (Anon., 2004). This large amount of fungicide is expensive—sometimes estimated at a cost of ~$200 per acre (personal communication from many growers and confirmed in surveys; Johnson et al., 1997). Additionally, the environmental effects of this large amount of fungicide are unknown. For both economic and environmental reasons, improvements in the efficiency of fungicide use via forecasts are being sought (Westerdijk and Schepers, 2006). For a few years after its initial release, the fungicide metalaxyl/mefenoxam was so effective that late blight became much less problematic. Unfortunately, resistance was selected in pathogen populations all over the world, and the efficacy of this fungicide declined noticeably. Thus, potato late blight remains as one of the most important and most costly plant diseases.
Can we develop or discover the ‘silver bullet’ for late blight suppression? As of today there is no definitive answer to this question; however, there are some candidate answers. One is a fungicide that targets an essential function in P. infestans. The reasoning is that mutations concerning that function would be lethal so that resistance could not develop. Experience with P. infestans and with other pathogens would indicate that microbes are remarkable in their abilities to adjust to toxins, so that a permanently effective fungicide seems unlikely. Another answer is to target a crucial effector with an R gene, again with the idea that mutations would be lethal, or at least detrimental, so that mutants would either not survive or be at a selective disadvantage. Given the large number of candidate effectors in P. infestans, this approach might seem unlikely, particularly now that strains compatible with Rpi‐blb1 have been found. Finally, will the genomic sequence provide clues for highly effective simple controls? Certainly, the impact of finding a permanent simple solution would be enormous and many investigators are searching energetically.
OPPORTUNITIES/CHALLENGES
Genomics
The genome of P. infestans has recently been sequenced, so that many types of analyses have just become possible. It was probably the ‘romance’ of finding a solution to the late blight disease that stimulated investigators and funding agencies to tackle this genome. There were some significant obstacles, including the fact that P. infestans has perhaps the largest genome of any species of Phytophthora (~240 Mb), a very large amount of repetitive DNA (~75% of the sequence), and that 35% of the sequence appears to be recently active transposons (Zody et al., 2007). Even with partial genomics tools, and a low‐efficiency transformation system (see below), some secrets had been discovered (see above) and we now expect much more rapid progress. The discovery that some effectors contain an RxLR‐dEER motif has stimulated high‐throughput analyses in search of R gene candidates and in search of function for the many different effectors.
Transformation
While a variety of transformation methods have been described, these often have limitations (Govers and Gijzen, 2006). The first successful report of stable transformation of Phytophthora was by Judelson, using protoplasts and a polyethylene glycocol/calcium chloride method (Judelson et al., 1991). Subsequently, other methods have been reported, including microprojectile bombardment, and Agrobacterium mediated transformation (Cvitanich and Judelson, 2003b; Vijn and Govers, 2003), but none is highly efficient and Judelson's PEG‐CaCl2 method remains among the most successful for achieving stable transformation. Recently, transient gene silencing via double‐stranded RNA (dsRNA) has been developed by Whisson et al. (2005). In addition, in efforts to improve the efficiency of transformation, the transcriptional activity and function of a few oomycete promoter and terminator sequences have been investigated using either stable tranformants (Ah Fong and Judelson, 2003; Blanco and Judelson, 2005; Tani and Judelson, 2006) or transient transformation expression systems (Judelson and Michelmore, 1991; McLeod et al., 2004). Thus far, only one construct (pDBHAMT35G), which is based on Judelson's very successful construct (pHAMT35G), seems to have potential to improve on the early constructs developed by Judelson (McLeod et al., 2008).
Parasites
We are just now entering a period when we recognize that oomycetes can indeed be hosts to parasites. For example, Pythium oligandrum is both a parasite of tomatoes and of Phytophthora infestans (Horner and Van West, 2007). Additionally, it is now known that species of Phytophthora (and perhaps Pythium) can also host viruses. Although the discovery of viral parasites in Phytophthora species is very recent, it has been known for some time that virus‐like dsRNAs occurred in Phytophthora (Tooley et al., 1989), and dsRNAs and virus‐like particles were found in Pythium irregulare (Gillings et al., 1993). The first confirmed Phytophthora virus (reported in 2005) is related to the plant endornaviruses and was discovered in an unnamed species of Phytophthora obtained from Douglas fir (Hacker et al., 2005). Viruses may be common in P. infestans because in an initial study, nine of 22 strains analysed contained dsRNAs. There were at least four different patterns indicating at least four different viruses (Cai et al., 2007).
CONCLUDING COMMENTS
I hope the above discussion indicates that scientists throughout the world are still writing Berg's ‘romantic’ chapter on a ‘controversial’ pathogen. The romance associated with limiting the pathogenic potential of this organism is a strong attractant for many investigators and wonderful progress has been made in understanding its basic biology, ecology and pathogenicity. The recent availabilities of molecular and genomic tools ensure that many of the secrets of this organism will be revealed in the near future. However, our history of combating this pathogen should caution us to include considerations of the diversity within the species when devising strategies to contain its pathogenicity. Additionally, I would argue that any strategy for mitigating its pathogenicity needs to be based on a knowledgeable respect for the powerful plasticity of this organism.
ACKNOWLEDGEMENTS
I thank Alan Collmer, Nik Grünwald, Chris Smart and two anonymous reviewers for helpful suggestions to earlier versions of the manuscript. Additionally I thank previous students, postdoctoral scientists and other colleagues with whom I have been privileged to work over the years. Finally, I would like to acknowledge the contributions of the legions of scientists (some legendary) during the previous 160 years who have added so much to our understanding of this controversial organism.
REFERENCES
- Abad, Z.G. and Abad, J.A. (1995) Historical evidence on the occurrence of late blight of potato, tomato, and pear melon in the Andes of South America In: Phytophthora Infestans, Vol. 150 (Dowley L.J., Bannon E., Cooke L.R., Keane T. and O'Sullivan E., eds), pp. 36–41. Dublin: Boole Press Ltd. [Google Scholar]
- Adl, S.M. , Simpson, A.G.B. , Farmer, M.A. , Andersen, R.A. , Anderson, O.R. , Barta, J.R. , Bowser, S.S. , Brugerolle, G.U.Y. , Fensome, R.A. , Fredericq, S. , James, T.Y. , Karpov, S. , Kugrens, P. , Krug, J. , Lane, C.E. , Lewis, L.A. , Lodge, J. , Lynn, D.H. , Mann, D.G. , McCourt, R.M. , Mendoza, L. , Moestrup, O. , Mozley‐Standridge, S.E. , Nerad, T.A. , Shearer, C.A. , Smirnov, A.V. , Spiegel, F.W. and Taylor, M.F.J.R. (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451. [DOI] [PubMed] [Google Scholar]
- Ah Fong, A.M.V. and Judelson, H.S. (2003) Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus‐like oomycete Phytophthora infestans . Mol. Microbiol. 50, 487–494. [DOI] [PubMed] [Google Scholar]
- Andersson, B. , Sandstrom, M. and Stromberg, A. (1998) Indications of soil borne inoculum of Phytophthora infestans. Potato Res. 41, 305–310. [Google Scholar]
- Andrivon, D. (1996) The origin of Phytophthora infestans populations present in Europe in the 1840s: a critical review of the historical and scientific evidence. Plant Pathol. 45, 1027–1035. [Google Scholar]
- Anon . (2004) Potatoes—Fall Fungicide Use. National Agricultural Statistics Service. United States Department of Agriculture. [Google Scholar]
- Armstrong, M.R. , Whisson, S.C. , Pritchard, L. , Bos, J.I.B. , Venter, E. , Avrova, A.O. , Rehmany, A.P. , Bohme, U. , Brooks, K. , Cherevach, I. , Hamlin, N. , White, B. , Fraser, A. , Lord, A. , Quail, M.A. , Churcher, C. , Hall, N. , Berriman, M. , Huang, S. , Kamoun, S. , Beynon, J.L. and Birch, P.R.J. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl Acad. Sci. USA, 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor, D.E. , Fry, W.E. , Mayton, H. and Andrade‐Piedra, J. (2001) Quantifying the rate of release and escape of Phytophthora infestans sporangia from a potato canopy. Phytopathology, 91, 1189–1196. [DOI] [PubMed] [Google Scholar]
- Beaumont, A. (1947) The dependence on the weather of the dates of outbreak of potato blight epidemics. Trans. Br. Mycol. Soc. 31, 45–53. [Google Scholar]
- Becktell, M.C. , Smart, C.D. , Haney, C.H. and Fry, W.E. (2006) Host–pathogen interactions between Phytophthora infestans and the solanaceous hosts Calibrachoa×hybridus, Petunia×hybrida and Nicotiana benthamiana . Plant Dis. 90, 24–32. [DOI] [PubMed] [Google Scholar]
- Berg, A. (1926) Tomato late blight and its relation to late blight of potato. Bull. West Virginia Agrl Expt. Sta. 25, 1–31. [Google Scholar]
- Blanco, F.A. and Judelson, H.S. (2005) A bZIP transcription factor from Phytophthora interacts with a protein kinase and is required for zoospore motility and plant infection. Molecular Microbiology, 56 (3), 633–648. [DOI] [PubMed] [Google Scholar]
- Bonde, R. and Schultz, E.S. (1943) Potato cull piles as a source of late blight infection. Am. Potato J. 20, 112–118. [Google Scholar]
- Bourke, A. (1991) Potato late blight in Europe in 1845: the scientific controversy In: Phytophthora (Lucas J.A., Shattock R.D., Shaw D.S. and Cooke L.R., eds), pp. 12–24. New York: Cambridge University Press. [Google Scholar]
- Bourke, A. (1993) ‘The Visitation of God’? The Potato and the Great Irish Famine. Dublin: Lilliput Press, Ltd. [Google Scholar]
- Brasier, C.M. (1971) Induction of sexual reproduction in single A2 isolates of Phytophthora species by Trichoderma viride. Nat. New Biol. 231, 283. [Google Scholar]
- Cai, G. , Myers, K. , Hillman, B. and Fry, W.E. (2007) Viruses in Phytophthora infestans, the late blight pathogen. Phytopathology, 97, S16. [Google Scholar]
- Cavalier‐Smith, T. (2000) Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5, 174–182. [DOI] [PubMed] [Google Scholar]
- Chapouret, N. , Rietman, H. , Bos, J.I.B. , Kamoun, S. , Van Der Vossen, E.A.G. , Visser, R.G.F. and Vleeshouwers, V.G.A.A. (2007) Functional allele mining: a new approach to identify r‐gene homologues in Solanum . MPMI Conference: Sorrento Italy July 2007, PS‐3‐250, Sorrento Italy.
- Chern, L.L. , Ko, W.H. and Tang, C.S. (1996) Factors affecting yields of alpha hormones of Phytophthora parasitica obtained by adsorption. Can. J. Microbiol. 42, 172–176. [Google Scholar]
- CIP (1996) Enhancing the Global Late Blight Network. Global Initiative on Late Blight. Lima, Peru: Centro Internacional de la Papa. [Google Scholar]
- Collins, A. , Milbourne, D. , Ramsay, L. , Meyer, R. , Chatot‐Balandras, C. , Oberhagemann, P. , De Jong, W. , Gebhardt, C. , Bonnel, E. and Waugh, R. (1999) QTL for field resistance to late blight in potato are strongly correlated with maturity and vigour. Mol. Breeding, 5, 387–398. [Google Scholar]
- Crosier, W. (1934) Studies in the biology of Phytophthora infestans (Mont) de Bary. Ithaca, NY: Cornell University Agricultural Experiment Station; (Memoir 155). [Google Scholar]
- Cvitanich, C. and Judelson, H.S. (2003a) A gene expressed during sexual and asexual sporulation in Phytophthora infestans is a member of the puf family of translational regulators. Eukaryot. Cell, 2, 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitanich, C. and Judelson, H.S. (2003b) Stable transfomation of the oomycete, Phytophthora infestans, using microprojectile bombardment. Curr. Genet. 42, 228–235. [DOI] [PubMed] [Google Scholar]
- Cvitanich, C. , Salcido, M. and Judelson, H.S. (2006) Concerted evolution of a tandemly arrayed family of mating‐specific genes in Phytophthora analyzed through inter‐ and intraspecific comparisons. Mol. Genet. Genomics, 275, 169–184. [DOI] [PubMed] [Google Scholar]
- De Bruyn, H.L.G. (1926) The overwintering of Phytophthora infestans (Mont.) De By. Phytopathology, 16, 121–140. [Google Scholar]
- DeBary, A. (1876) Researches into the nature of the potato fungus Phytophthora infestans. J. Roy. Agr. Soc. England, II 12, 239–269. [Google Scholar]
- Dowley L.J., Bannon E., Cooke L.R., Keane T. and O'Sullivan E. (eds) (1995) Phytophthora infestans, Vol. 150 Dublin: Boole Press Ltd. [Google Scholar]
- Drenth, A. , Tas, I.C.Q. and Govers, F. (1994) DNA fingerprinting uncovers a new sexually reproducing population of Phytophthora infestans in the Netherlands. Eur. J. Plant Pathol. 100, 97–107. [Google Scholar]
- Erwin, D.C. and Ribeiro, O.K. (1996) Phytophthora Diseases Worldwide. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Ewing, E.E. , Simko, I. , Smart, C.D. , Bonierbale, M.W. , Mizubuti, E.S.G. , May, G.D. and Fry, W.E. (2000) Genetic mapping from field tests of qualitative and quantitative resistance to Phytophthora infestans in a population derived from Solanum tuberosum and Solanum berthaultii . Mol. Breeding, 6, 25–36. [Google Scholar]
- Fabritius, A.‐L. and Judelson, H.S. (1997) Mating‐type loci segregate aberrantly in Phytophthora infestans but normal in Phytophthora parasitica: implications for models of mating‐type determination. Curr. Genet. 32, 60–65. [DOI] [PubMed] [Google Scholar]
- Fabritius, A.‐L. , Cvitanich, C. and Judelson, H.S. (2002) Stage‐specific gene expression during sexual development in Phytophthora infestans . Mol. Microbiol. 45, 1057–1066. [DOI] [PubMed] [Google Scholar]
- Fay, J.C. and Fry, W.E. (1997) Effects of hot and cold temperatures on the survival of oospores produced by United States strains of Phytophthora infestans . Am. Potato J. 74, 315–323. [Google Scholar]
- Fernandez‐Pavia, S.P. , Grunwald, N.J. , Diaz‐Valasis, M. , Cadena‐Hinojosa, M.A. and Fry, W.E. (2004) Soil‐borne oospores of Phytophthora infestans in central Mexico survive winter fall and infect potato plants in the field. Plant Dis. 88, 29–33. [DOI] [PubMed] [Google Scholar]
- Ferris, V.R. (1955) Histological study of pathogen‐suscept relationships between Phytophthora infestans and derivatives of Solanum demissum . Phytopathology, 45, 546–552. [Google Scholar]
- Flier, W.G. , Grunwald, N.J. , Fry, W.E. and Turkensteen, L.J. (2001) Formation, production and viability of oospores of Phytophthora infestans from potato and Solanum demissum in the Toluca Valley, central Mexico. Mycol. Res. 105, 998–1006. [Google Scholar]
- Flier, W.G. , Van Den Bosch, G.B.M. and Turkensteen, L.J. (2003a) Epidemiological importance of Solanum sisymbriifolium, S. nigrum and S. dulcamara as alteranative hosts for Phytophthora infestans. Plant Pathol. 52, 595–603. [Google Scholar]
- Flier, W.G. , Grunwald, N.J. , Kroon, L.P.N.M. , Sturbaum, A.K. , Van Den Bosch, T.B.M. , Garay‐Serrano, E. , Lozoya‐Saldana, H. , Fry, W.E. and Turkensteen, L.J. (2003b) The population structure of Phytophthora infestans from the Toluca Valley of central Mexico suggests genetic differentiation between populations from cultivated potato and wild Solanum spp. Phytopathology, 93, 382–390. [DOI] [PubMed] [Google Scholar]
- Flier, W.G. , Kroon, L.P.N.M. , Hermansen, A. , Van Raaij, H.M.G. , Speiser, B. , Tamm, L. , Fuchs, J.G. , Lambion, J. , Razzaghian, J. , Andrivon, D. , Wilcockson, S. and Leifert, C. (2007) Genetic structure and pathogenicity of populations of Phytophthora infestans from organic potato crops in France, Norway, Switzerland and the United Kingdom. Plant Pathol. 56, 562–572. [Google Scholar]
- Forbes, G.A. , Escobar, X.C. , Ayala, C.C. , Revelo, J. , Ordonez, M.E. , Fry, B.A. , Doucett, K. and Fry, W.E. (1997) Population genetic structure of Phytophthora infestans in Ecuador. Phytopathology, 87, 375–380. [DOI] [PubMed] [Google Scholar]
- Fry, W.E. and Goodwin, S.B. (1997a) Re‐emergence of potato and tomato late blight in the United States. Plant Dis. 81, 1349–1357. [DOI] [PubMed] [Google Scholar]
- Fry, W.E. and Goodwin, S.B. (1997b) Resurgence of the Irish potato famine fungus. Bioscience, 47, 363–371. [Google Scholar]
- Fry, W.E. and Shtienberg, D. (1990) Integration of host resistance and fungicide to manage potato diseases. Can. J. Plant Pathol. 12, 111–116. [Google Scholar]
- Fry, W.E. , Drenth, A. , Spielman, L.J. , Mantel, B.C. , Davidse, L.C. and Goodwin, S.B. (1991) Population genetic structure of Phytophthora infestans in the Netherlands. Phytopathology, 81, 1330–1336. [Google Scholar]
- Fry, W.E. , Goodwin, S.B. , Matuszak, J.M. , Spielman, L.J. , Milgroom, M.G. and Drenth, A. (1992) Population genetics and intercontinental migrations of Phytophthora infestans . Annu. Rev. Phytopathol. 30, 107–129. [Google Scholar]
- Fry, W.E. , Goodwin, S.B. , Dyer, A.T. , Matuszak, J.M. , Drenth, A. , Tooley, P.W. , Sujkowski, L.S. , Koh, Y.J. , Cohen, B.A. , Spielman, L.J. , Deahl, K.L. , Inglis, D.A. and Sandlan, K.P. (1993) Historical and recent migrations of Phytophthora infestans: chronology, pathways, and implications. Plant Dis. 77, 653–661. [Google Scholar]
- Fyfe, A.M. and Shaw, D.S. (1992) An analysis of self‐fertility in field isolates of Phytophthora infestans . Mycol. Res. 96, 390–394. [Google Scholar]
- Galindo, J. and Gallegly, M.E. (1960) The nature of sexuality in Phytophthora infestans . Phytopathology, 50, 123–128. [Google Scholar]
- Gallegly, M.E. and Galindo, J. (1958) Mating types and oospores of Phytophthora infestans in potato fields in the United States and Mexico. Phytopathology, 48, 274–277. [Google Scholar]
- Gallegly, M.E. and Marvel, M.E. (1955) Inheritance of resistance to tomato race O of Phytophthora infestans . Phytopathology, 45, 103–109. [Google Scholar]
- Garay‐Serrano, E. , Fernandez‐Pavia, S.P. , Rodriguez‐Alvarado, G. , Flier, W.G. , Lozoya‐Saldana, H. , Rojas‐Martinez, R.I. , Goss, E.M. and Grunwald, N.J. (2007) First report of haplotype I‐b of Phytophthora infestans in Central Mexico. Plant Dis. 91, 909–909. [DOI] [PubMed] [Google Scholar]
- Garrett, K.A. and Mundt, C.C. (2000) Host diversity can reduce potato late blight severity for focal and general patterns of primary inoculum. Phytopathology, 90, 1307–1312. [DOI] [PubMed] [Google Scholar]
- Garrett, K.A. , Nelson, R.J. , Mundt, C.C. , Chacon, G. , Jaramillo, R.E. and Forbes, G.A. (2001) The effects of host diversity and other management components on epidemics of potato late blight in the humid highland tropics. Phytopathology, 91, 993–1000. [DOI] [PubMed] [Google Scholar]
- Gillings, M.R. , Tesoriero, L.A. and Gunn, L.V. (1993) Detection of double‐stranded RNA and virus‐like particles in Australian isolates of Pythium irregulare . Plant Pathol. 42, 6–15. [Google Scholar]
- Glendinning, D. , MacDonald, J.A. and Grainger, J. (1963) Factors affecting the germination of sporangia in Phytophthora infestans . Trans. Br. Mycol. Soc. 46, 595–603. [Google Scholar]
- Gomez‐Alpizar, L. , Carbone, I. and Ristaino, J.B. (2007) An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proc. Natl Acad. Sci. USA, 104, 3306–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, S.B. (1997) The population genetics of Phytophthora. Phytopathology, 87, 462–473. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. and Drenth, A. (1997) Origin of the A2 mating type of Phytophthora infestans outside Mexico. Phytopathology, 87, 992–999. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. , Spielman, L.J. , Matuszak, J.M. , Bergeron, S.N. and Fry, W.E. (1992) Clonal diversity and genetic differentiation of Phytophthora infestans populations in northern and central Mexico. Phytopathology, 82, 955–961. [Google Scholar]
- Goodwin, S.B. , Cohen, B.A. and Fry, W.E. (1994a) Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proc. Natl. Acad. Sci. USA, 91, 11591–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, S.B. , Cohen, B.A. , Deahl, K.L. and Fry, W.E. (1994b) Migration from northern Mexico was the probable cause of recent genetic changes in populations of Phytophthora infestans in the United States and Canada. Phytopathology, 84, 553–558. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. , Sujkowski, L.S. and Fry, W.E. (1995) Rapid evolution of pathogenicity within clonal lineages of the potato late blight disease fungus. Phytopathology, 85, 669–676. [Google Scholar]
- Goodwin, S.B. , Smart, C.D. , Sandrock, R.W. , Deahl, K.L. , Punja, Z.K. and Fry, W.E. (1998) Genetic change within populations of Phytophthora infestans in the United States and Canada during 1994 to 1996: role of migration and recombination. Phytopathology, 88, 939–949. [DOI] [PubMed] [Google Scholar]
- Govers, F. and Gijzen, M. (2006) Phytophthora genomics: the plant destroyers’ genome decoded. Mol. Plant–Microbe Interact. 19, 1295–1301. [DOI] [PubMed] [Google Scholar]
- Groves, C.T. and Ristaino, J.B. (2000) Commercial fungicide formulations induce in vitro oospore formation and phenotypic change in mating type in Phytophthora infestans . Phytopathology, 90, 1201–1208. [DOI] [PubMed] [Google Scholar]
- Grunwald, N.J. and Flier, W.G. (2005) The biology of Phytophthora infestans at its center of origin. Annu. Rev. Phytopathol. 43, 171–190. [DOI] [PubMed] [Google Scholar]
- Grunwald, N.J. , Flier, W.G. , Sturbaum, A.K. , Garay‐Serrano, E. , Van Den Bosch, T.B.M. , Smart, C.D. , Matuszak, J.M. , Lozoya‐Saldana, H. , Turkensteen, L.J. and Fry, W.E. (2001) Population structure of Phytophthora infestans in the Toluca Valley region of central Mexico. Phytopathology, 91, 882–890. [DOI] [PubMed] [Google Scholar]
- Grunwald, N.J. , Cadena‐Hinojosa, M.A. , Rubio‐Covarrubias, O. , Rivera‐Pena, A. , Niederhauser, J.S. and Fry, W.E. (2002a) Potato cultivars from the Mexican national potato program: sources and durability of resistance against late blight. Phytopathology, 92, 688–693. [DOI] [PubMed] [Google Scholar]
- Grunwald, N.J. , Romero‐Montes, G. , Lozoya‐Saldana, H. , Rubio‐Covarrubias, O.A. and Fry, W.E. (2002b) Potato late blight management in the Toluca Valley: field validation of SimCast modified for cultivars with high field resistance. Plant Dis. 86, 1163–1168. [DOI] [PubMed] [Google Scholar]
- Hacker, C.V. , Brasier, C.M. and Buck, K.W. (2005) A double‐stranded RNA from a Phytophthora species is related to the plant endornaviruses and contains a putative UDP glycosyltransferase gene. J. Gen. Virol. 86, 1561–1570. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R. (2005) Phytophthora cinnamomi . Mol. Plant Pathol. 6, 589–604. [DOI] [PubMed] [Google Scholar]
- Harper, J.T. , Waanders, E. and Keeling, P.J. (2005) On the monophyly of chromalveolates using a six‐protein phylogeny of eukaryotes. Int. J. Syst. Evol. Microbiol. 55, 487–496. [DOI] [PubMed] [Google Scholar]
- Hein, I. , Squires, J. , Birch, P. and Bryan, G. (2007) Screening wild potato accessions for resistance to the virulent allele of the Phytophthora infestans effector avr3a. MPMI conference 2007, Sorrento, Italy. PS‐11‐490.
- Hohl, H.R. and Iselin, K. (1984) Strains of Phytophthora infestans from Switzerland with A2 mating type behavior. Trans. Br. Mycol. Soc. 83, 529–530. [Google Scholar]
- Horner, N.R. and Van West, P. (2007) Green Fluorescent Protein (GFP) as a reporter gene for the mycoparasitic oomycete Pythium oligandrum . MPMI conference 2007, Sorrento Italy.
- Hyre, R.A. , Bonde, R. and Manzer, F.E. (1960) Reevaluation in Maine of three methods proposed for forecasting late blight in potato. Plant Dis. Rep. 44, 235–237. [Google Scholar]
- Ingram, D.S. and Williams, P.H. (1991) Phytophthora Infestans, The Cause of Late Blight of Potato. London: Academic Press. [Google Scholar]
- Jiang, R.H.Y. , Weide, R. , Van De Vondervoort, P.J.I. and Govers, F. (2006) Amplification generates modular diversity at an avirulence locus in the pathogen Phytophthora . Genome Res. 16, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.A. , Cummings, T.F. , Hamm, P.B. , Rowe, R.C. , Miller, J.S. , Thornton, R.E. , Pelter, G.Q. and Sorensen, E.J. (1997) Potato late blight in the Columbia Basin: an economic analysis of the 1995 epidemic. Plant Dis. 81, 103–106. [DOI] [PubMed] [Google Scholar]
- Johnson, R. (1984) A critical analysis of durable resistance. Annu. Rev. Phytopathol. 22, 309–330. [Google Scholar]
- Jones, L.R. , Giddings, N.J. and Lutman, B.F. (1912) Investigations of the potato fungus, Phytophthora infestans. Vermont Agricultural Experiment Station Bulletin, Vol. 168. Burlington: University of Vermont. [Google Scholar]
- Judelson, H.S. , Tyler, B.M. and Michelmore, R.W. (1991) Transformation of the oomycete pathogen Phytophthora infestans. Molecular Plant– Microbe Interactions, 4, 602–607. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. (1997a) Expression and inheritance of sexual preference and selfing potential in Phytophthora infestans. Fungal Genet. Biol. 21, 188–197. [Google Scholar]
- Judelson, H.S. (1997b) The genetics and biology of Phytophthora infestans: modern approaches to a historical challenge. Fungal Genet. Biol. 22, 65–76. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. and Roberts, S. (2002) Novel protein kinasae induced during sporangial cleavage in the oomycete Phytophthora infestans . Eukaryot. Cell, 1, 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson, H.S. , Spielman, L.J. and Shattock, R.C. (1995) Genetic mapping and non‐Mendelian segregation of mating type loci in the Oomycete, Phytophthora infestans. Genetics , 141, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. , Van West, P. , De Jong, A.J. , De Groot, K.E. , Vleeshouwers, V.G.A.A. and Govers, F. (1997) A gene encoding a protein elicitor of Phytophthora infestans is down‐regulated during infection of potato. Mol. Plant–Microbe Interact. 10, 113–120. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Van West, P. , Vleeshouwers, V.G.A.A. , De Groot, K.E. and Govers, F. (1998) Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell, 10, 1413–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , Mizubuti, E.S.G. , Goodwin, S.B. and Fry, W.E. (1997) Sensitivity to protectant fungicides and pathogenic fitness of clonal lineages of Phytophthora infestans in the United States. Phytopathology, 87, 973–978. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. and Judelson, H.S. (2003) Sporangium‐specific gene expression in the oomycete phytopathogen phytophthora infestans. Eukaryot. Cell, 2, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, W.H. (1988) Hormonal heterothallism and homothallism in Phytophthora . Annu. Rev. Phytopathol. 26, 57–73. [Google Scholar]
- Ko, W.H. (1994) An alternative possible origin of the A2 mating type of Phytophthora infestans outside Mexico. Phytopathology, 84, 1224–1227. [DOI] [PubMed] [Google Scholar]
- Lambert, D.H. and Currier, A.I. (1997) Differences in tuber rot development for North American clones of Phytophthora infestans. Am. Potato J. 74, 39–43. [Google Scholar]
- Large, E.C. (1940) The Advance of the Fungi. New York, Dover Publications Inc. [Google Scholar]
- Latijnhouwers, M. , Ligterink, W. , Vleeshouwers, V.G.A.A. , Van West, P. and Govers, F. (2004) A Gα subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans . Mol. Microbiol. 51, 925–936. [DOI] [PubMed] [Google Scholar]
- Lee, T.Y. , Simko, I. and Fry, W.E. (2002) Genetic control of aggressiveness to tomato in Phytophthora infestans . Can. J. Plant Pathol. 24, 471–480. [Google Scholar]
- Van Der Lee, T. , Testa, A. , Van Den Berg‐Velthuis, Van't Klooster, J. and Govers, F. (2004) High‐density genetic linkage maps of Phytophthora infestans reveal trisomic progeny and chromosomal rearrangements. Genetics, 167, 1643–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legard, D.E. , Lee, T.Y. and Fry, W.E. (1995) Pathogenic specialization in Phytophthora infestans: aggressiveness on tomato. Phytopathology, 85, 1362–1367. [Google Scholar]
- Leonard, K.J. (1969) Selection in heterogeneous populations of Puccinia graminis f. sp. avenae . Phytopathology, 59, 1851–1857. [Google Scholar]
- Lokossou, A. , Van Arkel, G. , Tani, A. , Park, T. , Hutten, R. , Van Eck, H. , Visser, R.G.F. , Jacobsen, E. and Van der Vossen, E.A.G. (2007) Cloning of Rpi‐blb3 and other functional alleles from a major late blight resistance locus on chromosome IV of Potato PS‐3‐260. MPMI meeting in Sorrento Italy, July 2007, Sorrento, Italy.
- Lucas, J.A. , Shattock, R.C. , Shaw, D.S. and Cooke, L.R. (1991) Phytophthora. Cambridge: British Mycological Society/Cambridge University Press. [Google Scholar]
- Madden, L.V. and Wheelis, M. (2003) The threat of plant pathogens as weapons against u.s. crops. Annu. Rev. Phytopathol. 41, 155–176. [DOI] [PubMed] [Google Scholar]
- Malcolmson, J.F. (1969) Races of Phytophthora infestans occurring in Great Britain. Trans. Br. Mycol. Soc. 53, 417–423. [Google Scholar]
- Mayton, H. , Smart, C.D. , Moravec, B.C. , Mizubuti, E.S.G. , Muldoon, A.E. and Fry, W.E. (2000) Oospore survival and pathogenicity of single oospore recombinant progeny from a cross involving the US‐8 and US‐17 lineages of Phytophthora infestans . Plant Dis. 84, 1190–1196. [DOI] [PubMed] [Google Scholar]
- McLeod, A. , Denman, S. , Sadie, A. and Denner, F.D.N. (2001) Characterization of South African isolates of Phytophthora infestans . Plant Dis. 85, 287–291. [DOI] [PubMed] [Google Scholar]
- McLeod, A. , Smart, C.D. and Fry, W.E. (2004) Core promoter structure in the oomycete Phytophthora infestans . Eukaryotic Cell, 3, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, A. , Fry, B. , Zuluaga, A.P. , Myers, K. and Fry, W. (2008) Towards improvements of oomycete transformation protocols. Eukaryotic Microbiology (in press). [DOI] [PubMed]
- Melhus, I.E. (1915) Germination and infection with the fungus of the late blight of potato (Phytophthora infestans). Madison, WI: University of Wisconsin, Agr. Exp. Sta. [Google Scholar]
- Michelmore, R.W. and Sansome, E. (1982) Cytological studies of heterothalism and secondary homothalism in Bremia lactucae . Trans. Br. Mycol. Soc. 79, 291–297. [Google Scholar]
- Minogue, K.P. and Fry, W.E. (1981) Effect of temperature, relative humidity and rehydration rate on germination of dried sporangia of Phytophthora infestans . Phytopathology, 71, 1181–1184. [Google Scholar]
- Mizubuti, E.S.G. and Fry, W.E. (1998) Temperature effects on developmental stages of isolates of three clonal lineages of Phytophthora infestans. Phytopathology, 88, 837–843. [DOI] [PubMed] [Google Scholar]
- Mizubuti, E.S.G. and Fry, W.E. (2006) Potato late blight In: The Epidemiology of Plant Diseases (Cooke B.M., Jones D.G. and Kaye B., eds), pp. 445–471. Springer: The Netherlands. [Google Scholar]
- Mizubuti, E.S.G. , Aylor, D.E. and Fry, W.E. (2000) Survival of Phytophthora infestans sporangia exposed to solar radiation. Phytopathology 90, 78–84. [DOI] [PubMed] [Google Scholar]
- Money, N.P. (1998) Why oomycetes have not stopped being fungi. Mycol. Res. 102, 767–768. [Google Scholar]
- Mortimer, A.M. , Shaw, D.S. and Sansome, E.R. (1977) Genetical studies of secondary homothallism in Phytphthora drechsleri. Arch. Microbiol. 111, 255–259. [Google Scholar]
- Niederhauser, J.S. (1956) The blight, the blighter, and the blighted. Trans. NY Acad. Sci. 19, 55–63. [Google Scholar]
- Niederhauser, J.S. (1991) Phytophthora infestans: the Mexican connection In: Phytophthora (Lucas J.A., Shattock R.C., Shaw D.S. and Cooke L.R., eds), pp. 25–45. Cambridge: Cambridge University Press. [Google Scholar]
- Niederhauser, J.S. and Mills, W.R. (1953) Resistance of Solanum species to Phytophthora infestans in Mexico. Phytopathology, 43, 456–457. [Google Scholar]
- Niederhauser, J.S. , Alvarez‐Luna, E. and Mackenzie, D.R. (1996) RETONA a new strategy in the control of potato late blight. Am. Potato J. 73, 225–229. [Google Scholar]
- Novy, R.G. , Love, S.L. , Corsini, D.L. , Pavek, J.J. , Whitworth, J.L. , Mosley, A.R. , James, S.R. , Hane, D.C. , Shock, C.C. , Rykbost, K.A. , Brown, C.R. , Thornton, R.E. , Knowles, N.R. , Pavek, M.J. , Olsen, N. and Inglis, D.A. (2006) Defender: a high‐yielding processing potato cultivar with foliar and tuber resistance to late blight. Am. J. Potato Res. 83, 9–19. [Google Scholar]
- Pieterse, C.M.J. , Van West, P. , Verbakel, H.M. , Brasse, P.W.H.M. , Van Den Berg‐Velthuis, G.C.M. and Govers, F. (1994) Structure and genomic organization of the ipiB and ipiO gene clusters of Phytophthora infestans . Gene, 138, 67–77. [DOI] [PubMed] [Google Scholar]
- Platt, H.W. (1999) Response of solanaceous cultivated plants and weed species to inoculation with A1 or A2 mating type strains of Phytophthora infestans . Can. J. Plant Pathol. 21, 301–307. [Google Scholar]
- Qi, J. , Asano, T. , Jinno, M. , Matsui, K. , Atsumi, K. , Sakagami, Y. and Ojika, M. (2005) Characterization of a Phytophthora mating hormone. Science, 309, 1828. [DOI] [PubMed] [Google Scholar]
- Quon, D.V.K. , Delgadillo, M.G. , Khachi, A. and Smale, S.T. (1994) Similarity between a ubiquitous promoter element in an ancient eukaryote and mammalian initiator element. Proceedings of the National Academy of Sciences, 91, 4579–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall, T.A. , Fong, A.A. and Judelson, H.S. (2002) Chromosomal heteromorphism and an apparent translocation detected using a BAC contig spanning the mating type locus of Phytophthora infestans . Fungal Genet. Biol. 38, 75–84. [DOI] [PubMed] [Google Scholar]
- Rauscher, G. , Smart, C.D. , Simko, I. , Bonierbale, M. , Mayton, H. , Greenland, A. and Fry, W.E. (2006) Characterization and mapping of R‐Pi‐ber, a novel potato late blight resistance gene from Solanum berthaultii . Theor. Appl. Genet. 112, 674–687. [DOI] [PubMed] [Google Scholar]
- Reddick, D. (1928) Blight resistant potatoes. Phytopathology, 18, 483–502. [Google Scholar]
- Reddick, D. (1934) Elimination of potato late blight from North America. Phytopathology, 24, 555–557. [Google Scholar]
- Reddick, D. (1939) Whence came Phytophthora infestans? Chron. Bot. 5, 410–412. [Google Scholar]
- Reeves, R.J. and Jackson, R.M. (1974) Stimulation of sexual reproduction in Phytophthora by damage. J. Gen. Microbiol. 84, 303–310. [Google Scholar]
- Rehmany, A.P. , Gordon, A. , Rose, L.E. , Allen, R.L. , Armstrong, M.R. , Whisson, S.C. , Kamoun, S. , Tyler, B.M. , Birch, P.R.J. and Beynon, J.L. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two arabidopsis lines. Plant Cell, 17, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristaino, J.B. , Groves, C.T. and Parra, G.R. (2001) PCR amplification of the Irish potato famine pathogen from historic specimens. Nature, 411, 695–697. [DOI] [PubMed] [Google Scholar]
- Rotem, J. and Cohen, Y. (1974) Epidemiological patterns of Phytophthora infestans under semi‐arid conditions. Phytopathology, 64, 711–714. [Google Scholar]
- Shattock, R.C. , Janssen, B.D. , Whitbread, R. and Shaw, D.S. (1977) An interpretation of the frequencies of host specific phenotypes of Phytophthora infestans in North Wales. Ann. Appl. Biol. 86, 249–260. [Google Scholar]
- Shattock, R.C. , Tooley, P.W. and Fry, W.E. (1986a) Genetics of Phytophthora infestans: characterization of single‐oospore cultures from A1 isolates induced to self by intraspecific stimulation. Phytopathology, 76, 407–410. [Google Scholar]
- Shattock, R.C. , Tooley, P.W. and Fry, W.E. (1986b) The genetics of Phytophthora infestans: identification of recombination, segregation and selfing by isozyme analysis. Phytopathology, 76, 410–413. [Google Scholar]
- Shaw, D.S. , Fyfe, A.M. , Hibberd, P.G. and Abdel‐Sattar, M.A. (1985) Occurrence of the rare A2 mating type of Phytophthora infestans on imported Egyptian potatoes and the production of sexual progeny with A1 mating‐types from the U.K. Plant Pathol. 34, 552–554. [Google Scholar]
- Shen, C.‐Y. , Bower, L.A. , Erwin, D.C. and Tsao, P.H. (1983) Formation of sex organs in the A1 mating type of Phytophthora infestans induced chemically by A2 isolates of other species of Phytophthora . Can. J. Bot. 61, 1462–1466. [Google Scholar]
- Smart, C.D. , Willmann, M.R. , Mayton, H. , Mizubuti, E.S.G. , Sandrock, R.W. , Muldoon, A.E. and Fry, W.E. (1998) Self‐fertility in two clonal lineages of Phytophthora infestans. Fungal Genet. Biol. 25, 134–142. [Google Scholar]
- Smart, C.D. , Mayton, H. , Mizubuti, E.S.G. , Willmann, M.R. and Fry, W.E. (2000) Environmental and genetic factors influencing self‐fertility in Phytophthora infestans . Phytopathology, 90, 987–994. [DOI] [PubMed] [Google Scholar]
- Smart, C.D. , Myers, K.L. , Restrepo, S. , Martin, G.B. and Fry, W.E. (2003) Partial resistance of tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid or salicyclic acid signalling pathways. Mol. Plant–Microbe Interact. 16, 141–148. [DOI] [PubMed] [Google Scholar]
- Smoot, J.J. , Gough, F.J. , Lamey, H.A. , Eichenmuller, J.J. and Gallegly, M.E. (1958) Production and germination of oospores of Phytophthora infestans . Phytopathology, 48, 165–171. [Google Scholar]
- Song, J. , Bradeen, J.M. , Naess, S.K. , Raasch, J.A. , Wielgus, S.M. , Haberlach, G.T. , Liu, J. , Kuang, H. , Austin‐Phillips, S. , Buell, C.R. , Helgeson, J.P. and Jiang, J. (2003) Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl Acad. Sci. USA, 100, 9128–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman, L.J. , McMaster, B.J. and Fry, W.E. (1989) Evidence against potato and tomato host specificity in Phytophthora infestans . Phytopathology, 79, 1145 (abstr.). [Google Scholar]
- Spielman, L.J. , Drenth, A. , Davidse, L.C. , Sujkowski, L.J. , Gu, W.K. , Tooley, P.W. and Fry, W.E. (1991) A second world‐wide migration and population displacement of Phytophthora infestans? Plant Pathol. 40, 422–430. [Google Scholar]
- Stevens, N.E. (1933) The dark ages in plant pathology in America: 1830–1870. J. Washington Acad. Sci. 23, 435–446. [Google Scholar]
- Stewart, H.E. , Bradshaw, J.E. and Pande, B. (2003) The effect of the presence of R‐genes for resistance to late blight (Phytophthora infestans) of potato (Solanum tuberosum) on the underlying level of field resistance. Plant Pathol. 52, 193–198. [Google Scholar]
- Sujkowski, L.S. , Goodwin, S.B. , Dyer, A.T. and Fry, W.E. (1994) Increased genotypic diversity via migration and possible occurrence of sexual reproduction of Phytophthora infestans in Poland. Phytopathology, 84, 201–207. [Google Scholar]
- Tani, S. , Yatzkan, E. and Judelson, H.S. (2003) Multiple pathways to regulate the induction of genes during zoosporogenesis in Phytophthora infestans . MPMI . 17, 330–337. [DOI] [PubMed] [Google Scholar]
- Tani, S. and Judelson, H. (2006) Activation of Zoosporogenesis‐Specific Genes in Phytophthora infestans Involves a 7‐Nucleotide Promoter Motif and Cold‐Induced Membrane Rigidity. Eukaryotic Cell , 5 (4), 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley, P.W. , Fry, W.E. and Villarreal Gonzalez, M.J. (1985) Isozyme characterization of sexual and asexual Phytophthora infestans populations. J. Hered. 76, 431–435. [Google Scholar]
- Tooley, P.W. , Hewings, A.D. and Falkenstein, K.F. (1989) Detection of double‐stranded RNA in Phytophthora infestans . Phytopathology, 79, 470–474. [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A.R. , Van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkensteen, L.J. , Flier, W.G. , Wanningen, R. and Mulder, A. (2000) Production, survival and infectivity of oospores of Phytophthora infestans . Plant Pathol. 49, 688–696. [Google Scholar]
- Turner, R.S. (2005) After the famine: plant pathology, Phytophthora infestans and the late blight of potatoes, 1845–1960. Hist. Stud. Phys. Biol. Sci. 34.2, 341–370. [Google Scholar]
- Tyler, B.M. (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Molec. Plant Pathol. 8, 1–8. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H.Y. , Aerts, A. , Arredondo, F.D. , Baxter, L. , Bensasson, D. , Beynon, J.L. , Chapman, J. , Damasceno, C.M.B. , Dorrance, A.E. , Dou, D. , Dickerman, A.W. , Dubchak, I.L. , Garbelotto, M. , Gijzen, M. , Gordon, S.G. , Govers, F. , Grunwald, N.J. , Huang, W. , Ivors, K.L. , Jones, R.W. , Kamoun, S. , Krampis, K. , Lamour, K.H. , Lee, M.‐K. , McDonald, W.H. , Medina, M. , Meijer, H.J.G. , Nordberg, E.K. , Maclean, D.J. , Ospina‐Giraldo, M.D. , Morris, P.F. , Phuntumart, V. , Putnam, N.H. , Rash, S. , Rose, J.K.C. , Sakihama, Y. , Salamov, A.A. , Savidor, A. , Scheuring, C.F. , Smith, B.M. , Sobral, B.W.S. , Terry, A. , Torto‐Alalibo, T.A. , Win, J. , Xu, Z. , Zhang, H.B. , Grigoriev, I.V. , Rokhsar, D.S. and Boore, J.L. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Van der Plank, J.E. (1963) Plant Diseases: Epidemics and Control. New York: Academic Press. [Google Scholar]
- Van Everdingen, E. (1926) Het verband tusschen de weergesteldheid en de aardappel zeikte (Phytophthora infestans ). Tijdschrift over Plantenziekten 32, 129. [Google Scholar]
- Van der Vossen, E. , Sikkema, A. , Hekkert, B.T.L. , Gros, J. , Stevens, P. , Muskens, M. , Wouters, D. , Pereira, A. , Stiekema, W. and Allefs, S. (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad‐spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882. [DOI] [PubMed] [Google Scholar]
- Van der Vossen, E. , Gros, J. , Sikkema, A. , Muskens, M. , Wouters, D. , Wolters, A. , Pereira, A. and Allefs, S. (2005) The Rpi‐blb2 gene from Solanum bulbocastanum is an Mi‐1 gene homologue conferring broad spectrum late blight resistance in potato. Plant J. 44, 208–222. [DOI] [PubMed] [Google Scholar]
- Vega‐Sanchez, M.E. , Erselius, L.J. , Rodriguez, A.M. , Bastidas, O. , Hohl, H.R. , Ojiambo, P.S. , Mukalazi, J. , Vermeulen, T. , Fry, W.E. and Forbes, G.A. (2000) Host adaptation to potato and tomato within the US‐1 clonal lineage of Phytophthora infestans in Uganda and Kenya. Plant Pathol. 49, 531–539. [Google Scholar]
- Vijn, I. and Govers, F. (2003) Agrobacterium tumefaciens mediated transformation of the oomycete plant pathogen phytophthora infestans. Mol. Plant Pathol. 4, 459–467. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A. , Van Dooijeweert, W. , Govers, F. , Kamoun, S. and Colon, L.T. (2000) The hypersensitive response is associated with host and nonhost resistance to Phytophthora infestans . Planta, 210, 853–864. [DOI] [PubMed] [Google Scholar]
- Warren, R.C. and Colhoun, J. (1975) Viability of sporangia of Phytophthora infestans in relation to drying. Trans. Br. Mycol. Soc. 64, 73–78. [Google Scholar]
- Wastie, R.L. (1991) Breeding for resistance In: Phytophthora Infestans: the Cause of Late Blight of Potato, Vol. 7 (Ingram D.S. and Williams P.H., eds), pp. 193–224. San Diego, CA: Academic Press. [Google Scholar]
- De Weille, G.A. (1964) Forecasting crop infection by the potato blight fungus. Meded. Verh. K. Ned. Met. Inst. 82, 1–144. [Google Scholar]
- Westerdijk C.E. and Schepers H.T.A.M. (eds) (2006) Proceedings of the ninth workshop of an European network for development of an integrated control strategy of potato late blight (Tallinn, Estonia, 19th–23rd October 2005). Special Report No. 11. Wageningen: Applied Plant Research. [Google Scholar]
- Whisson, S.C. , Avrova, A.O. , Van West, P. and Jones, J.T. (2005) A method for double‐stranded RNA‐mediated transient gene silencing in Phytophthora infestans . Mol. Plant Pathol. 6, 153–163. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , Van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Widmark, A.K. , Andersson, B. , Cassel‐Lundhagen, A. , Sandstrom, M. and Yuen, J.E. (2007) Phytophthora infestans in a single field in southwest Sweden early in spring: symptoms, spatial distribution and genotypic variation. Plant Pathol. 56, 573–579. [Google Scholar]
- Woodham‐Smith, C. (1962) The Great Hunger. New York: Harper & Row. [Google Scholar]
- Zody, M.C. , Jiang, R.H.Y. , Handsaker, R. , Grabherr, M. , Kodira, C.D. , Govers, F. , Birch, P. , Whisson, S. , Win, J. , Judelson, H.S. , Ristaino, J.B. , Fry, W.E. , Kamoun, S. and Nusbaum, C. (2007) Genome dynamics in the pathogen/host arms race: initial analysis of the Phytophthora infestans genome. Phytopathology, 97, S146. [Google Scholar]


