SUMMARY
The Brome mosaic virus (BMV) coat protein (CP) accompanies the three BMV genomic RNAs and the subgenomic RNA into and out of cells in an infection cycle. In addition to serving as a protective shell for all of the BMV RNAs, CP plays regulatory roles during the infection process that are mediated through specific binding of RNA elements in the BMV genome. One regulatory RNA element is the B box present in the 5′ untranslated region (UTR) of BMV RNA1 and RNA2 that play important roles in the formation of the BMV replication factory, as well as the regulation of translation. A second element is within the tRNA‐like 3′ UTR of all BMV RNAs that is required for efficient RNA replication. The BMV CP can also encapsidate ligand‐coated metal nanoparticles to form virus‐like particles (VLPs). This update summarizes the interaction between the BMV CP and RNAs that can regulate RNA synthesis, translation and RNA encapsidation, as well as the formation of VLPs.
INTRODUCTION
The vast majority of plant viruses are nonenveloped. Therefore, the coat protein (CP) (or proteins, as is the case in some virus species) contacts the cell and delivers the viral genome into plants. At the end of the infection process, CP will exit the infected plant with the genome. Increasingly, it is appreciated that CP also plays a role as a co‐ordinator of the viral infection process, from actively participating in the replication complex to suppressing the innate immune response of the plant host (Asurmendi et al., 2004; Qu et al., 2003; Reichert et al., 2007). The regulatory role is shared with many animal virus capsid proteins (2008, 2010; Krishna, 2005; Miller and White, 2006; Shimoike et al., 1999; Takeuchi and Akira, 2007; Voinnet, 2005; Witherell et al., 1991).
Studies of CP of plant viruses have been at the vanguard of discoveries in a number of applied areas, including CP‐mediated plant resistance (Prins et al., 2009). For example, this area has contributed to an understanding of how small RNAs can be used to manipulate gene expression and to understand plant development (Baulcombe, 1996; Beachy et al., 1990; Hackland et al., 1994; Miller and Hemenway, 1998; Reddy et al., 2009; Sudarshana et al., 2007; Wilson, 1993). Plant virus CP research is also at the interface between material science and nanotechnology (Young et al., 2008). Interest intensified in this area with the use of the expansion and contraction of Cowpea chlorotic mottle virus (CCMV) to generate gate‐access chambers that can perform chemical reactions (Douglas and Young, 1998). This was soon followed by the novel use of engineered tobacco mosaic virus and filamentous phages to bind metals that can be used in electronic applications (Khalil et al., 2007; Lee et al., 2009; Nam et al., 2010; Shenton et al., 1999). Fluorescently labelled Cowpea mosaic virus has been studied for use in imaging in whole animals (Destito et al., 2009; Martin et al., 2006; 2009a, 2009b). Viral capsids that can self‐assemble from CP subunits have been used to generate novel materials that are more homogeneous and have improved properties that hold potential for use in imaging and therapeutics (Cheluvaraja and Ortoleva, 2010; Manchester and Singh, 2006; Sapsford et al., 2006).
Each plant virus has specialized features that make it particularly tractable for certain basic studies and applications. This review concerns Brome mosaic virus, a model system that has been studied extensively for both gene regulation and virus structure, and has contributed significantly to both basic and applied research.
BROME MOSAIC VIRUS
Brome mosaic virus (BMV) is a small icosahedral virus that offers high levels of RNA synthesis and virus production in plants and has become a well‐studied model plant virus. It is a member of the alphavirus‐like superfamily of RNA viruses with a segmented positive‐strand RNA genome. The BMV genome consists of three genomic RNAs (RNA1, RNA2 and RNA3) and a subgenomic RNA4 (Fig. 1; Kao and Sivakumaran, 2000; Noueiry and Ahlquist, 2003). RNA1 encodes one protein with the capping and helicase‐like functions required for RNA replication. RNA2 encodes for the RNA‐dependent RNA polymerase. RNA3 encodes the movement protein required for cell‐to‐cell spread and CP, but CP is translated from a subgenomic RNA4 transcribed from minus‐strand RNA3. All four BMV RNAs contain a tRNA‐like sequence as part of their 3′ untranslated region (UTR) (Ahlquist, 1992).
Figure 1.
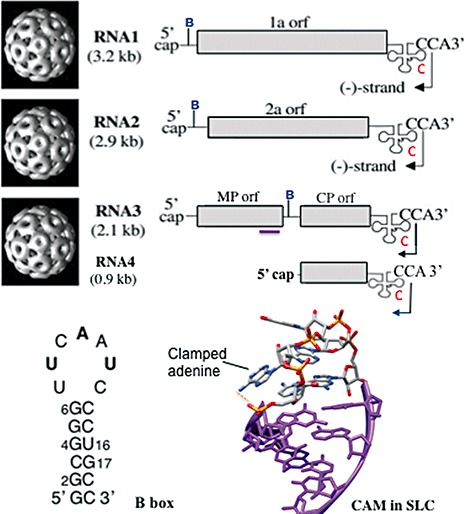
The organization of the Brome mosaic virus (BMV) genome. Images of the three particles that package the three BMV genomic RNAs and the subgenomic RNA4 are given on the left. The schematic diagrams on the right show the protein‐coding sequences in light grey and the tRNA‐like structures as a cloverleaf with the 3′‐terminal CCA sequence. The arrow in the penultimate cytidylate identifies the residue to start minus‐strand RNA synthesis. The location of the putative packaging specificity signal in RNA3 is identified by the purple line. The letters B and C denote the approximate locations of the B box and stem‐loop C (SLC), respectively. The structure of the clamped adenine motif (CAM) in SLC from Kim et al. (2000) and the sequence of the B box in RNA2 are shown at the bottom of the figure.
An interesting feature for BMV biology is that its genome is packaged into three separate particles that are indistinguishable by electron microscopy. RNA1 and RNA2 are packaged into separate particles, whereas RNA3 and RNA4 are co‐packaged within one particle (Annamalai and Rao, 2007; Rao, 2006). This is in contrast with viruses that contain multiple RNAs within the same virion, such as the influenza virus or reovirus (Hutchinson et al., 2010; Patton and Spencer, 2000). This arrangement allows the three types of particle to package RNAs of similar total lengths (hence the particles are difficult to distinguish), the shortest being 2865 nucleotides (RNA2) and the longest 3234 nucleotides (RNA1).
A potential advantage of this encapsidation strategy is to maximize reassortment of the BMV genomes. However, at least one copy of each BMV RNA must be present within the same cell for BMV to successfully replicate and spread. Given that BMV RNAs replicate to levels approaching that of rRNAs, the chance of the three different RNAs being represented in an adjacent cell is high. However, this may not be the case when virions are dispersed by mechanical factors. In the event that all three types of RNA end up in the same plant, but not in the same cell, BMV RNAs have been found to be able to traffic between cells and even leaves of a Nicotiana benthamiana plant to reconstitute replication and virus production (Gopinath and Kao, 2007). RNA3 with the region coding for the movement protein and CP replaced with the green fluorescent protein (GFP) retains the ability to traffic, indicating that RNA3 trafficking is not dependent on these proteins (Gopinath and Kao, 2007).
THE BMV CP AND THE CAPSID
The BMV virion is a T= 3 truncated icosahedron made up of 180 copies of the 189‐residue CP subunit. The CP thus exists in three conformationally distinct forms that vary by bonding interfaces, named A, B and C (Fig. 2). Each subunit contains a long N‐terminal arm rich in basic residues and a globular domain that allows subunits to interdigitate through their C‐terminal tails (Fig. 2). The C‐terminal portion of the CP also plays a role in cell‐to‐cell trafficking of BMV RNAs (Okinaka et al., 2001; Takeda et al., 2004).
Figure 2.
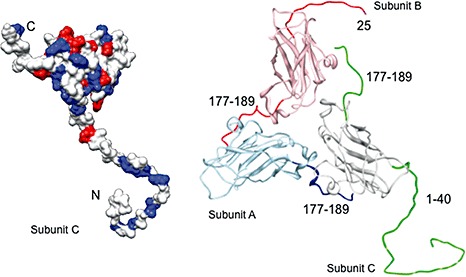
Structures of the Brome mosaic virus (BMV) coat protein (CP) monomer and asymmetric trimer. The models are generated from the structure of Lucas et al. (2002) [protein data bank (pdb) 1JS9]. The monomer shows a surface rendering in which the positively charged residues are in blue and the negatively charged residues are in red. The N‐ and C‐termini are denoted by the letters N and C, respectively. The trimer with the three conformers A, B and C illustrates the locations of the N‐ and C‐termini that form the capsid. The monomer is the C conformer.
The N‐terminal arm of the BMV capsid has been studied extensively. Pioneering small‐angle neutron diffraction was applied to investigate the RNA and capsid distribution within BMV, revealing that the RNA exists in a shell and that portions of the capsid penetrate into the shell (Jacrot et al., 1977). One or more of the N‐terminal arms of the capsid in an asymmetric trimer were subsequently found to anchor the CP around the RNA core (Lucas et al., 2002). A high concentration of basic residues in the N‐terminus of CP is a feature shared by a number of spherical plant viruses (Guerra‐Peraza et al., 2005; Hsu et al., 2006; Hu and Ghabrial, 1995; Lee and Hacker, 2001; Olspert et al., 2010; Rao and Grantham, 1995; Reade et al., 2010; Sharma and Ikegami, 2009; Yusibov and Loesch‐Fries, 1995). Several small spherical viruses that lack clusters of basic residues in their capsid package substantial amounts of polyamines (Ames and Dubin, 1960; Cohen and Greenberg, 1981; Sheppard et al., 1980). These observations support the contention that electrostatic interactions between positively charged polypeptides and negatively charged RNA are important for virion assembly. Removal of the N‐terminal segment of BMV and the highly related CCMV by trypsin digestion and reassembly resulted in the formation of T= 1 particles that lacked RNA (Cuillel et al., 1981; Liepold et al., 2005; Lucas et al., 2001). In plants, BMV producing a CP that lacked an N‐terminus was not infectious (Sacher and Ahlquist, 1989), and smaller truncations resulted in altered lesion phenotypes in some plant species (Rao and Grantham, 1995, 1996). The mobility of the N‐terminal arm for the highly related CCMV virion was markedly decreased on binding to RNA, as seen in NMR spectroscopy (Speir et al., 2006; Van der Graaf et al., 1991; 1981, 1982a, 1982b, 1986). This has been confirmed by in situ chemical cross‐linking experiments of the BMV virion (Yi et al., 2009b). The importance of the charge–charge interactions between the capsid and the RNA is further underscored by the observations of Belyi and Muthukumar (2006), who noted a linear correlation between the amount of positive charges on the inner surface of the capsid for icosahedral viruses and the length of the viral genome. This correlation suggests that the charges inside the CP not only function to package RNA, but could also be a determinant of the size of the genome of a virus.
The BMV virions sediment faster at pH 6 than at pH 7, without a corresponding change in the molecular weight of the virion, indicating a significant conformational change (Incardona and Kaesberg, 1964; Kassanis and Lebeurier, 1969). Temperature‐dependent changes in sedimentation and hysteresis in acid–base titration of BMV further characterize the swelling that occurs from pH 6 to pH 7 into two steps (Incardona and Kaesberg, 1964; Pfeiffer and Durham, 1977). The first is a reversible pH‐induced transition, taking place around pH 6.5. It is suggested that deprotonation of a pair of carboxyl groups on adjacent subunits, probably glutamate 84 and 131, mediates this process (Lucas et al., 2002). The second step is an irreversible thermal expansion accompanied by optically detectable changes in RNA conformation (Incardona and Kaesberg, 1964). The second step can be suppressed by the addition of Mg2+. However, whether Mg2+ stabilizes the RNA structure, CP structure or the interaction between the two is not clear. The changes in RNA conformation could also be a consequence of the swelling of the capsid. It is likely that an understanding of these requirements will be informative about virus uncoating in host cells.
BMV and CCMV were among the very first spherical viruses to be reconstituted in vitro (Adolph and Butler, 1976; Herzog and Hirth, 1978). Assembly in vitro is generally carried out by dialysis of a mixture of dissociated CPs and RNA (or some core material), starting from a high‐salt buffer to a low‐salt buffer at neutral pH. The reassembled particles are indistinguishable from native viruses with regard to hydrodynamic properties and are infectious (Bancroft and Hiebert, 1967; Hiebert et al., 1968). However, the in vitro dissociated BMV CPs can reassemble around RNAs from unrelated viruses, synthetic homopolymeric RNAs and even non‐nucleic acid polyanions (Bancroft and Hiebert, 1967; Bancroft et al., 1969; Verduin and Bancroft, 1969).
Despite a large body of literature on electrostatic interactions being the primary requirement in the encapsidation of RNA, the requirements for packaging BMV RNAs are likely to be more complex. Choi et al. (2002) have shown that RNA encapsidation in vitro requires the tRNA‐like structure from the 3′ end of the BMV genomic RNAs. The BMV tRNA‐like sequence, or even cellular tRNAs, can be added in trans for RNA encapsidation. Within BMV RNA3, where mapping of the packaging signal is more complete, a sequence that encodes the C‐terminal portion of the BMV movement protein is also a part of a bipartite signal that encapsidates RNAs along that encapsidate RNA3 (Choi and Rao, 2003). In plants, only replicated BMV RNAs are suitable for encapsidation and efficient packaging of RNA4 is coupled not only to its transcription, but to the translation of the CP from RNA4. (Annamalai and Rao, 2006), demonstrating that some requirements for RNA packaging will be missed by the manipulations possible in the in vitro assembly reactions. The BMV virion can also form a pseudo T= 2 particle when BMV RNA is absent, suggesting that the particles have a mechanism to form preassembled immature particles until RNA can be encapsidated (Sullivan and Ahlquist, 1999). There is also evidence that BMV RNA3 can be encapsidated without RNA4, suggesting the existence of a mechanism to bring RNA4 into a particle that is at least partially assembled (Annamalai et al., 2008; Choi and Rao, 2003).
BMV CIS‐ACTING RNA SEQUENCES
Co‐ordination of gene expression will involve cis‐acting sequences common to at least some of the four BMV RNAs. A summary of these is given in Fig. 1. There are several obvious candidates in BMV (Fig. 3). One is the tRNA‐like structure that exists in the 3′ terminal c. 230 nucleotides of all BMV RNAs. This sequence can be aminoacylated with tyrosine (Dreher and Hall, 1988; Hall et al., 1987). Within this tRNA‐like structure, a subelement, named stem‐loop C (SLC), has been shown to specify binding of the BMV replicase to direct initiation of minus‐strand RNA synthesis (Chapman and Kao, 1999). SLC features an adenine that is exposed to the solution and clamped to the stem, called a clamped adenine motif (CAM) (Kim et al., 2000) (Fig. 1). CAM binds the BMV replicase and is required for BMV replication in plant cells (Kim et al., 2000; Sivakumaran et al., 2004). Another important element is the B box, which interacts with the BMV 1a protein to help form the membrane‐encased site at which BMV RNAs are replicated (Ahlquist, 2006; Chen et al., 2001). The terminal loop is not particularly ordered, suggesting that recognition may be through specific sequence (Yi et al., 2009a). The B box element is present in the 5′ UTR of BMV RNA1 and RNA2, but not in the 5′ UTR of RNA3. Instead, a B box‐like sequence is in the intercistronic region of RNA3, upstream of the subgenomic promoter. This arrangement suggests that the three BMV RNAs can be differentially regulated through this element (Chen et al., 2001; Choi et al., 2004; Sivakumaran and Kao, 2000; Sullivan and Ahlquist, 1999). In this manner, gene expression and replication of BMV RNA3 can be co‐ordinated to that of RNA1 and RNA2. As already mentioned above, a c. 220‐nucleotide region that overlaps with the coding sequence of the movement protein also contains motifs required for the encapsidation of BMV RNA3. Deletions in this region prevent the efficient packaging of BMV RNA3 (Choi and Rao, 2003; Damayanti et al., 2003). Additional features in this complex motif that are responsible for RNA encapsidation remain to be determined.
Figure 3.
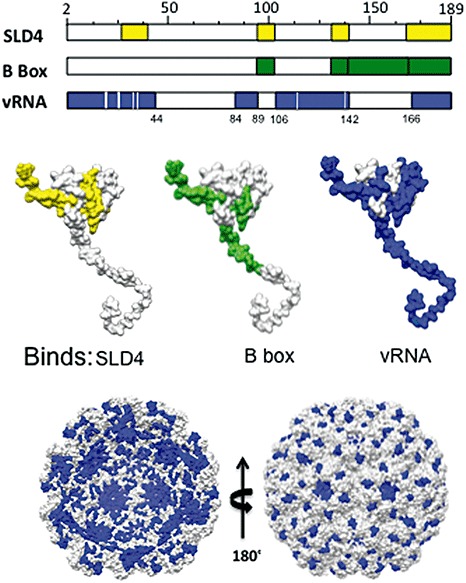
Mapping of the binding of regulatory RNA motifs by the Brome mosaic virus (BMV) coat protein (CP), or the packaged genomic RNA by the BMV capsid. In the middle of the figure, peptides found to be cross‐linked to the RNAs in the reversible cross‐linking and peptide fingerprinting (RCAP) assay are highlighted in colour. SLD4 is a truncated version of stem‐loop C (SLC) that contains a clamped adenine motif. The locations of the peptides that contact the two regulatory RNA motifs and the packaged RNAs are also shown in the model of a monomer of BMV CP, as seen from the inside of the cavity (left structure at the bottom) and from the outside of the virion (right structure at the bottom).
THE BMV CP AND GENE EXPRESSION
Packaging of the RNAs into separate particles will require appropriate co‐ordination of the viral RNAs and their activities. The BMV CP is well positioned to be such a co‐ordinator as it accompanies the RNAs from the start to the end of an infection cycle (Hema et al., 2010). Some phage CPs are well characterized to regulate gene expression (e.g. Witherell et al., 1991). The alfalfa mosaic virus CP has also been shown to interact with the RNA replication proteins and to help confer specificity in RNA replication (Bol, 2005; Guogas et al., 2005; Reichert et al., 2007), and the tobacco mosaic virus CP is associated with the replicase in the endoplasmic reticulum of plant cells (Asurmendi et al., 2004).
Historically, the role of the CP in BMV replication was downplayed, in part because BMV RNA1 and RNA2 transfected into protoplasts can replicate in the absence of RNA3. It was thus difficult to separate the role of CP from other BMV RNAs and the movement protein using the traditional reverse genetics analysis of BMV RNAs (Janda et al., 1987). The Agrobacterium expression system allows for the expression of combinations of the three BMV genomic RNAs and/or four proteins simply by mixing and matching cultures expressing individual molecules (Annamalai and Rao, 2005; Gopinath et al., 2005). The viral genomes are initially expressed by the cellular RNA polymerase II, and hence can be manipulated without most of the constraints imposed by the BMV cis‐acting elements. In addition, up to several hundred copies of the Agrobacterium T‐DNA harbouring recombinant sequences can be integrated per plant cell nucleus; thus, it is likely that every cell in an infiltrated area will receive at least one copy of the construct coding for a BMV molecule (Gelvin, 2003). Finally, the density of the Agrobacterium inoculum can be manipulated to integrate different numbers of gene copies, resulting in a gradient of CP expression (Gopinath et al., 2005).
Yi et al. (2007) used the Agrobacterium‐launched BMV RNA replication system to observe that the abundance of CP has a complex relationship to BMV RNA accumulation: low levels of CP increase the expression of reporter proteins, whereas high concentrations of CP inhibit reporter expression from BMV RNA1 and RNA2, but not from RNA3. These observations were intriguing and led to the hypothesis that CP can interact with various cis‐elements in the BMV RNAs in a manner dependent on the CP concentrations.
CP AND REGULATION OF BMV RNA TRANSLATION
The inhibition of BMV RNA accumulation is, in part, a result of the high level of CP binding to the B box RNA motif present in the 5′ UTR of RNA1 and RNA2, but absent in the 5′ UTR of RNA3 (Fig. 1). The production of GFP reporters from the RNAs containing the UTRs of BMV RNA1 or RNA2 was inhibited by BMV CP (Yi and Kao, 2008). However, the same was not true for BMV RNA3, which has a B box in the intercistronic region of RNA3 and is unlikely to contribute to translation at the 5′ portion of RNA. Deletion analysis of the RNA2 5′ UTR pinpointed the B box as the responsible element. A minimal B box RNA element can bind the BMV CP with a K d value of approximately 400 nm affinity.
CP AND AN RNA REPLICATION ELEMENT
A clue to the stimulatory effect of BMV CP on RNA accumulation came from a screen of host factors that can bind an RNA containing a CAM (Zhu et al., 2007). Serendipitously, CP was added as a control on a protein chip that housed the majority of the proteins expressed by yeast Saccharomyces cerevisiae, a host competent for BMV replication (Janda and Ahlquist, 1993), with the expectation that it would bind RNA nonspecifically (Zhu et al., 2007). Unexpectedly, CP was among the top 10 in over 5000 proteins on the chip to bind CAM in SLC, and not to an otherwise identical RNA, except that the clamped adenine was replaced with a guanine (Kim and Tinoco, 2001). RNA binding had a K d value of approximately 300 nm, whereas nonspecific RNAs had micromolar affinities. A UV cross‐linking assay confirmed that CP could specifically bind CAM, but not RNAs lacking CAM (Fig. 3). These results suggest that CP could play a role in the specific recognition of BMV RNAs for replication.
MAPPING CP INTERACTION WITH REGULATORY RNAS
To identify the residues in CP that bind the regulatory RNA elements, a method involving reversible cross‐linking and peptide fingerprinting (named RCAP) was used (Kim et al., 2005). RCAP uses formaldehyde to cross‐link a protein to RNA. The complex is then digested with trypsin, followed by affinity purification of RNA. When the cross‐links of the RNA peptides were reversed with heat, the peptides were identified by matrix‐assisted laser desorption ionization‐time of flight‐mass spectrometry (MALDI‐ToF‐MS) and their sequences were confirmed by ion‐induced MS/MS. Versions of SLC or the B box RNAs were covalently coupled to resin, thus permitting selective pull‐down of the peptides. A peptide containing residues 27–41 in the N‐terminal arm was found to cross‐link to CAM, but not the B box. In addition, peptide 143–165 within the globular domain recognized the B box, but not CAM. CPs with potential RNA‐binding residues in and near the cross‐linked regions substituted with alanines led to the identification of residues in CP that lost the ability to regulate BMV gene expression when the mutant CPs were expressed in trans of the BMV RNAs. Mutant D139A also lost the ability to inhibit translation. Mutant R142A was changed for binding to the B box in comparison with wild‐type CP (Yi et al., 2009b). Thus, an initial mapping of the residues in CP revealed effects on the modes of gene regulation during BMV infection.
CP RECOGNITION OF VIRION RNAS
In addition to interacting with the regulatory RNA elements, the obvious role of CP is to encapsidate the virion RNA. An in‐depth review of this subject is given in Rao (2006). A crystal structure of the BMV virion is available, but the RNA was not resolved (Lucas et al., 2002). The RCAP method was used to map the regions of the capsid associated with virion RNAs. The RNAs and RNA–peptide complexes were selectively precipitated with lithium chloride. The location of the RNA‐binding peptides from within the capsid is shown in Fig. 3. A comparison of the peptides derived from the capsid with those that contact the regulatory RNAs revealed that a major difference is that the N‐terminal arm of the capsid has more extensive contacts with the encapsidated viral RNAs (Yi et al., 2009b) (Fig. 3).
A number of mutations in the BMV capsid have shed light on the recognition of BMV RNAs in the virus particles. Calhoun et al. (2007) reported that a deletion of residues 40–47 in the portion of CP facing the RNA was defective in packaging one or more of the BMV RNAs. Hema et al. (2010) have found that substitutions at several residues near the interfaces of the capsid subunits result in viruses that have altered ratios of the four BMV RNAs. A mutation at glutamine 120 was found to show selective degradation of RNA1, RNA3 and RNA4, whereas RNA2 was found to be less affected. These results suggest that subsets of particles exist with distinct ability to interact with and protect RNAs. Consistent with this notion, wild‐type BMV particles (with three subsets of particles) have a more complex thermal denaturation profile than particles that contain only RNA3 and RNA4. These results suggest that the mutations identified by Hema et al. (2010) reveal intrinsic differences in the subsets of BMV particles containing different genomic RNAs. We speculate that these differences could have an impact on the timing of the release of RNAs in BMV infection.
There are two possible novel mechanistic aspects through which CP exerts regulatory control. First, CP co‐ordinates different steps in BMV infection through high‐affinity binding of at least two structurally different RNA motifs in the BMV genome. Secondly, CP acts in a concentration‐dependent manner, probably because CP oligomerization states will influence RNA recognition. The latter activity may allow the temporal regulation of the BMV infection process, and our model is shown in Fig. 4. Under conditions in which the CP molecules are in small amounts, as is the case early in viral infection, CP can enhance RNA replication and/or transcription. In the middle stage of infection, higher CP levels will result in an impact on the level of translation through binding of the B box. Finally, at high CP levels, RNA encapsidation will predominate (Fig. 4). The concentrations of CP present during an infection cycle will thus provide a timing switch for processes essential for successful infection.
Figure 4.
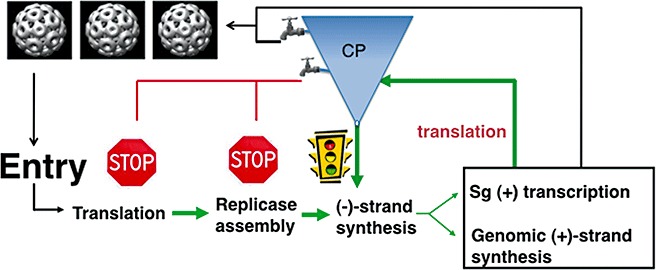
A model of how the Brome mosaic virus (BMV) coat protein (CP) can regulate different steps in the infection process. The central process is written in the flow diagram. The funnel‐shaped object with the spigots is intended to show that different amounts of CP can differentially regulate the infection process. Stimulatory effects are shown in green and inhibitory effects are shown in red.
CP RECOGNITION OF NONVIRAL CORES
Plant viruses and phages, in particular, have received increasing attention for their potential use in bionanotechnology. BMV has several advantages for these applications: it is produced in large amounts in a natural infection and forms monodispersed particles that are highly symmetrical. Foreign materials can be added to either the outside of the capsid or the inner chamber of the capsid shell (Rae et al., 2005). In the latter case, it is even possible to replace the viral genomic RNA with foreign material. Furthermore, CP can be engineered using recombinant DNA technology to introduce peptide sequences suitable for binding specific receptors (Suci et al., 2007; Wang et al., 2002). The highly basic N‐terminal arm of CP found in many plant viruses also contains a motif highly similar to the canonical cell‐penetrating peptide from the HIV Tat protein (Frankel and Pabo, 1988; Green and Loewenstein, 1988). This peptide from BMV CP could enter cells, as well as increase the delivery of both proteins and RNAs into plants and plant cells (Qi et al., 2010).
Virus‐like particles (VLPs) are self‐assembled from viruses in which the capsid either lacks any nucleic acids (empty VLPs) or packages nonviral cores. Inorganic nanoparticles (NPs), in particular, can add special optical or magnetic properties to VLPs and provide unique potential for bioimaging and biomedicine. Genetic engineering of CP of the capsid could also allow the production of VLPs with cargos that have specific size, surface features and physical characteristics.
The initial assembly of BMV VLPs used nanogold that was functionalized with citrates. Although packaging was observed, the efficiency of VLPs containing nanogold was low (∼2%) (Dragnea et al., 2003). The key to the improvement of the efficiency of VLP formation lies in coating the core with one of the most common of industrial chemicals, polyethylene glycol (PEG) (Fig. 5A). PEG with a terminal carboxylate was found to help form VLPs with more than 90% efficiency (Chen et al., 2006) (Fig. 5B). Interestingly, the diameter of the pegylated gold can affect the particles formed. Three‐dimensional reconstructions of the transmission electron micrographs of the single VLPs with 6‐nm gold cores indicated that the protein shell structure corresponds to T= 1 capsids (Fig. 5C). Those with 9‐nm cores share the same structure as pseudo T= 2 capsids, and 12‐nm‐diameter cores are encapsulated in a shell that appears very similar to the wild‐type BMV, which has T= 3 capsids, and these can be co‐crystallized with wild‐type BMV at any ratio (Sun et al., 2007). The crucial features of this PEG coating are believed to be: (i) its terminal carboxyl group provides negative charges for the electrostatic interaction with the positively charged internal compartment of the BMV capsid; and (ii) its repeated ethylene glycol segments provide hydrophilicity and spatial plasticity. Consistent with a need for a certain charge, the efficiency of VLP formation was found to increase significantly when the density of the negatively charged PEG on the nanogold was 70% or higher (Daniel et al., 2010).
Figure 5.
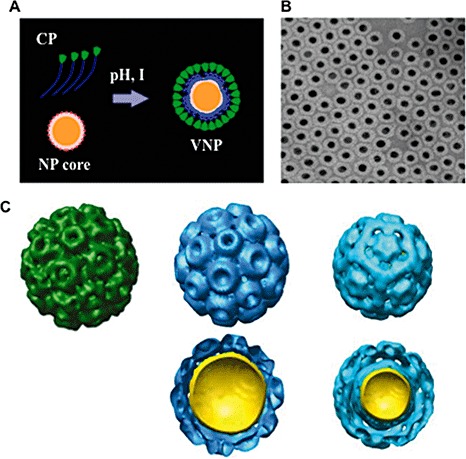
The Brome mosaic virus (BMV) capsid can bind to nonviral cores. (A) A schematic diagram of the assembly of virus‐like particles (VLPs) around a functionalized nanoparticle (NP). This reaction is dependent on the pH and ionic strength (I) of the reaction mixture. (B) An example of VLPs containing nanogold cores that are approximately 12 nm in diameter. (C) Single particle reconstructions of the native BMV and BMV VLPs with nanogold cores. The native BMV is in green and consists of coat proteins (CPs) arranged in pentamers and hexamers, respectively. The two sets of blue particles contain gold cores of 12 and 9 nm in diameter. The VLPs for the 12‐nm nanogold particle (NGP) form a T= 3‐like particle, whereas the VLPs with a 9‐nm core form a pseudo T= 2 particle.
A number of other cores have been successfully encapsidated into BMV VLPs. These include CdSe/ZnS semiconductor quantum dots (QDs) using carboxylated PEGs. These QD cores have high‐performance luminescence and a much longer half‐life than other fluorescent probes (Mansur, 2010; Pinaud et al., 2010). These VLPs contain multiple cores ranging from one to three QDs, whereas the shell exhibits a T= 2 structure (Dixit et al., 2006). Huang et al. (2007) built a magnetic VLP system consisting of the BMV CP encapsulated around iron oxide nanotemplates. The superparamagnetic properties of the cores, together with the ability to add functionalities, including specific receptor recognition, make VLPs attractive candidates for in vivo magnetic resonance imaging contrast agents. These studies, together with those from other plant viruses, with both icosahedral and filamentous morphologies, have opened up the door for the use of novel imaging and delivery capabilities of viral particles. Advances that can result from the use of viral capsids should help plant viruses with their incredibly versatile capsid proteins to cross the barrier between disciplines and to lead plant virology to new and fertile fields.
CONCLUSIONS
Molecular studies of BMV CP have revealed that, in addition to its function in protecting the viral genome, it is an important regulator of viral gene expression through selective binding to RNA motifs in the BMV genome. The ability of BMV CP to self‐assemble around foreign materials has the potential for applications in nanotechnology and medicinal biology. Despite many decades of study, however, many major questions remain, such as: (i) what factors regulate the specific packaging of the BMV RNAs and the nonspecific, charge‐driven interactions that characterize most in vitro packaging interactions, including those involving foreign materials?; (ii) how does the changing concentration of CP alter the specificity and affinity of RNA recognition?; (iii) how does CP interact with the replication proteins and cellular factors in the membrane‐associated sites of replication to co‐ordinate the processes needed for successful infection?; (iv) what applications are best suited for relatively dynamic capsids, such as that characterized by BMV, versus capsids that are intrinsically more stable? At this time, when basic research in plant virology faces the challenges associated with limited resources, applied areas, such as virus‐based nanotechnology, can be used to bring new resources as well as fresh perspectives to how viruses assemble, as well as enter and leave cells.
ACKNOWLEDGEMENTS
We thank Laura Kao for editing the manuscript and members of the IU and A&M Cereal Killers, former and present, for wonderful discussions. This work was supported by National Science Foundation grant MCB0641362 and National Institutes of Health (NIH) grant 1R01AI090280 to CCK, and NIH grant GM081929 for Bioimaging to BD and CCK. Hema Masarapu acknowledges the Department of Biotechnology, New Delhi, India for providing a long‐term Overseas Associateship.
REFERENCES
- Adolph, K.W. and Butler, P.J. (1976) Assembly of a spherical plant virus. Philos. Trans. R. Soc. London B: Biol. Sci. 276, 113–122. [DOI] [PubMed] [Google Scholar]
- Ahlquist, P. (1992) Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2, 71–76. [DOI] [PubMed] [Google Scholar]
- Ahlquist, P. (2006) Parallels among positive‐strand RNA viruses, reverse‐transcribing viruses and double‐stranded RNA viruses. Nat. Rev. Microbiol. 4, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames, B.N. and Dubin, D.T. (1960) The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235, 769–775. [PubMed] [Google Scholar]
- Annamalai, P. and Rao, A.L. (2005) Replication‐independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology, 338, 96–111. [DOI] [PubMed] [Google Scholar]
- Annamalai, P. and Rao, A.L. (2006) Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication‐dependent transcription and translation of coat protein. J. Virol. 80, 10 096–10 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai, P. and Rao, A.L.N. (2007) In vivo packaging of Brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis‐acting 3′ tRNA‐like structure. J. Virol. 81, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai, P. , Rofail, F. , Demason, D.A. and Rao, A.L. (2008) Replication‐coupled packaging mechanism in positive‐strand RNA viruses: synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in Nicotiana benthamiana cells by agroinfiltration. J. Virol. 82, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asurmendi, S. , Berg, R.H. , Koo, J.C. and Beachy, R.N. (2004) Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA, 101, 1415–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, J.B. and Hiebert, E. (1967) Formation of an infectious nucleoprotein from protein and nucleic acid isolated from a small spherical virus. Virology, 32, 354–356. [DOI] [PubMed] [Google Scholar]
- Bancroft, J.B. , Hiebert, E. and Bracker, C.E. (1969) The effects of various polyanions on shell formation of some spherical viruses. Virology, 39, 924–930. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1996) Mechanisms of pathogen‐derived resistance to viruses in transgenic plants. Plant Cell, 8, 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy, R.N. , Loesch‐Fries, S. and Tumer, N.E. (1990) Coat protein‐mediated resistance against virus infection. Annu. Rev. Phytopathol. 28, 451–474. [Google Scholar]
- Belyi, V.A. and Muthukumar, M. (2006) Electrostatic origin of the genome packing in viruses. Proc. Natl. Acad. Sci. USA, 103, 17 174–17 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol, J.F. (2005) Replication of alfamo‐ and ilarviruses: role of the coat protein. Annu. Rev. Phytopathol. 43, 39–62. [DOI] [PubMed] [Google Scholar]
- Calhoun, S.L. , Speir, J.A. and Rao, A.L. (2007) In vivo particle polymorphism results from deletion of a N‐terminal peptide molecular switch in brome mosaic virus capsid protein. Virology, 364, 407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, M.R. and Kao, C.C. (1999) A minimal RNA promoter for minus‐strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286, 709–720. [DOI] [PubMed] [Google Scholar]
- Cheluvaraja, S. and Ortoleva, P. (2010) Thermal nanostructure: an order parameter multiscale ensemble approach. J. Chem. Phys. 132, 075102‐1–075102‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Daniel, M.C. , Quinkert, Z.T. , De, M. , Stein, B. , Bowman, V.D. , Chipman, P.R. , Rotello, V.M. , Kao, C.C. and Dragnea, B. (2006) Nanoparticle‐templated assembly of viral protein cages. Nano Lett. 6, 611–615. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Noueiry, A.O. and Ahlquist, P. (2001) Brome mosaic virus Protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75, 3207–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.K. , Hema, M. , Gopinath, K. , Santos, J. and Kao, C. (2004) Replicase binding sites on plus‐ and minus‐strand brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 78, 13 420–13 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.G. and Rao, A.L. (2003) Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 77, 9750–9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.G. , Dreher, T.W. and Rao, A.L. (2002) tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA, 99, 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S.S. and Greenberg, M.L. (1981) Spermidine, an intrinsic component of turnip yellow mosaic virus. Proc. Natl. Acad. Sci. USA, 78, 5470–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuillel, M. , Jacrot, B. and Zulauf, M. (1981) A T = 1 capsid formed by protein of brome mosaic virus in the presence of trypsin. Virology, 110, 63–72. [DOI] [PubMed] [Google Scholar]
- Damayanti, T.A. , Tsukaguchi, S. , Mise, K. and Okuno, T. (2003) Cis‐acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 77, 9979–9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, M.C. , Tsvetkova, I. , Quinkert, Z. , Murali, A. , De, M. , Rotello, V.L. , Kao, C.C. and Dragnea, B. (2010) Critical surface charge density is required for nanoparticle‐templated assembly by bromovirus protein cages. ACS Nano, 4, 3853–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destito, G. , Schneemann, A. and Manchester, M. (2009) Biomedical nanotechnology using virus‐based nanoparticles. Curr. Top. Microbiol. Immunol. 327, 95–122. [DOI] [PubMed] [Google Scholar]
- Dixit, S.K. , Goicochea, N.L. , Daniel, M.C. , Murali, A. , Bronstein, L. , De, M. , Stein, B. , Rotello, V.M. , Kao, C.C. and Dragnea, B. (2006) Quantum dot encapsulation in viral capsids. Nano Lett. 6, 1993–1999. [DOI] [PubMed] [Google Scholar]
- Douglas, T. and Young, M. (1998) Host–guest encapsulation of materials by assembled virus protein cages. Nature, 393, 152–155. [Google Scholar]
- Dragnea, B. , Chen, C. , Kwak, E.S. , Stein, B. and Kao, C.C. (2003) Gold nanoparticles as spectroscopic enhancers for in vitro studies on single viruses. J. Am. Chem. Soc. 125, 6374–6375. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W. and Hall, T.C. (1988) Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J. Mol. Biol. 201, 31–40. [DOI] [PubMed] [Google Scholar]
- Frankel, A.D. and Pabo, C.O. (1988) Cellular uptake of the tat protein from human immunodeficiency virus. Cell, 55, 1189–1193. [DOI] [PubMed] [Google Scholar]
- Gelvin, S.B. (2003) Agrobacterium‐mediated plant transformation: the biology behind the ‘gene‐jockeying’ tool. Microbiol. Mol. Biol. Rev. 67, 16–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath, K. and Kao, C.C. (2007) Replication‐independent long‐distance trafficking by viral RNAs in Nicotiana benthamiana . Plant Cell, 19, 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath, K. , Dragnea, B. and Kao, C.C. (2005) Interaction between brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression and RNA stability. J. Virol. 79, 14 222–14 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M. and Loewenstein, P.M. (1988) Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans‐activator protein. Cell, 55, 1179–1188. [DOI] [PubMed] [Google Scholar]
- Guerra‐Peraza, O. , Kirk, D. , Seltzer, V. , Veluthambi, K. , Schmit, A.C. , Hohn, T. and Herzog, E. (2005) Coat proteins of Rice tungro bacilliform virus and Mungbean yellow mosaic virus contain multiple nuclear‐localization signals and interact with importin alpha. J. Gen. Virol. 86, 1815–1826. [DOI] [PubMed] [Google Scholar]
- Guogas, L.M. , Laforest, S.M. and Gehrke, L. (2005) Coat protein activation of alfalfa mosaic virus replication is concentration dependent. J. Virol. 79, 5752–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackland, A.F. , Rybicki, E.P. and Thomson, J.A. (1994) Coat protein mediated resistance in transgenic plants. Arch. Virol. 139, 1–22. [DOI] [PubMed] [Google Scholar]
- Hall, T.C. , Marsh, L.E. and Dreher, T.W. (1987) Analysis of brome mosaic virus replication and aminoacylation functions by site‐specific mutagenesis. J. Cell Sci. Suppl. 7, 287–301. [DOI] [PubMed] [Google Scholar]
- Hema, M. , Murali, A. , Fujisaki, K. , Tsyetkova, I. , Ni, P. , Vaughan, R.C. , Dragnea, B. and Kao, C.C. (2010) A single amino acid substitution in the brome mosaic virus capsid protein resulted in biased RNA packaging. Mol. Plant–Microbe Interact. 23, 1433–1447. [DOI] [PubMed] [Google Scholar]
- Herzog, M. and Hirth, L. (1978) In vitro encapsidation of the four RNA species of brome mosaic virus. Virology, 86, 48–56. [DOI] [PubMed] [Google Scholar]
- Hiebert, E. , Bancroft, J.B. and Bracker, C.E. (1968) The assembly in vitro of some small spherical viruses, hybrid viruses, and other nucleoproteins. Virology, 34, 492–508. [DOI] [PubMed] [Google Scholar]
- Hsu, C. , Singh, P. , Ochoa, W. , Manayani, D.J. , Manchester, M. , Schneemann, A. and Reddy, V.S. (2006) Characterization of polymorphism displayed by the coat protein mutants of tomato bushy stunt virus. Virology, 349, 222–229. [DOI] [PubMed] [Google Scholar]
- Hu, C.C. and Ghabrial, S.A. (1995) The conserved, hydrophilic and arginine‐rich N‐terminal domain of cucumovirus coat proteins contributes to their anomalous electrophoretic mobilities in sodium dodecylsulfate‐polyacrylamide gels. J. Virol. Methods, 55, 367–379. [DOI] [PubMed] [Google Scholar]
- Huang, X.L. , Bronstein, L.M. , Retrum, J. , Dufort, C. , Tsvetkova, I. , Aniagyei, S. , Stein, B.D. , Stucky, G. , Mckenna, B. , Remmes, N. , Baxter, D. , Kao, C. and Dragnea, B. (2007) Self‐assembled virus‐like particles with magnetic cores. Nano Lett. 7, 2407–2015. [DOI] [PubMed] [Google Scholar]
- Hutchinson, E.C. , von Kirchbach, J.C. , Gog, J.R. and Digard, P. (2010) Genome packaging in influenza A virus. J. Gen. Virol. 91, 313–328. [DOI] [PubMed] [Google Scholar]
- Ilkow, C.S. , Mancinelli, V. , Beatch, M.D. and Hobman, T.C. (2008) Rubella virus capsid protein interacts with poly(a)‐binding protein and inhibits translation. J Virol. 82, 4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilkow, C.S. , Willows, S.D. and Hobman, T.C. (2010) Rubella virus capsid protein: a small protein with big functions. Future Microbiol. 5, 571–584. [DOI] [PubMed] [Google Scholar]
- Incardona, N.L. and Kaesberg, P. (1964) A pH‐induced structural change in bromegrass mosaic virus. Biophys. J. 4, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacrot, B. , Chauvin, C. and Witz, J. (1977) Comparative neutron small‐angle scattering study of small spherical RNA viruses. Nature, 266, 417–421. [DOI] [PubMed] [Google Scholar]
- Janda, M. and Ahlquist, P. (1993) RNA‐dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae . Cell, 72, 961–970. [DOI] [PubMed] [Google Scholar]
- Janda, M. , French, R. and Ahlquist, P. (1987) High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology, 158, 259–262. [DOI] [PubMed] [Google Scholar]
- Kao, C.C. and Sivakumaran, K. (2000) Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1, 91–97. [DOI] [PubMed] [Google Scholar]
- Kassanis, B. and Lebeurier, G. (1969) The behaviour of tomato bushy stunt virus and bromegrass mosaic virus at different temperatures in vivo and in vitro. J. Gen. Virol. 4, 385–395. [Google Scholar]
- Khalil, A.S. , Ferrer, J.M. , Brau, R.R. , Kottmann, S.T. , Noren, C.J. , Lang, M.J. and Belcher, A.M. (2007) Single M13 bacteriophage tethering and stretching. Proc. Natl. Acad. Sci. USA, 104, 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H. and Tinoco, I. Jr (2001) Structural and thermodynamic studies on mutant RNA motifs that impair the specificity between a viral replicase and its promoter. J. Mol. Biol. 307, 827–839. [DOI] [PubMed] [Google Scholar]
- Kim, C.H. , Kao, C. and Tinoco, I. (2000) RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Biol. 7, 415–423. [DOI] [PubMed] [Google Scholar]
- Kim, Y.C. , Russell, W.K. , Ranjith‐Kuman, C.T. , Thomson, M. , Russell, D.H. and Kao, C.C. (2005) Functional analysis of RNA binding by the hepatitis C virus RNA‐dependent RNA polymerase. J. Biol. Chem. 280, 38 011–38 019. [DOI] [PubMed] [Google Scholar]
- Krishna, N.K. (2005) Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 18, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.K. and Hacker, D.L. (2001) In vitro analysis of an RNA binding site within the N‐terminal 30 amino acids of the southern cowpea mosaic virus coat protein. Virology, 286, 317–327. [DOI] [PubMed] [Google Scholar]
- Lee, Y.J. , Yi, H. , Kim, W.J. , Kang, K. , Yun, D.S. , Strano, M.S. , Ceder, G. and Belcher, A.M. (2009) Fabricating genetically engineered high‐power lithium‐ion batteries using multiple virus genes. Science, 324, 1051–1055. [DOI] [PubMed] [Google Scholar]
- Liepold, L.O. , Revis, J. , Allen, M. , Oltrogge, L. , Young, M. and Douglas, T. (2005) Structural transitions in Cowpea chlorotic mottle virus (CCMV). Phys. Biol. 2, S166–S172. [DOI] [PubMed] [Google Scholar]
- Lucas, R.W. , Kuznetsov, Y.G. , Larson, S.B. and McPherson, A. (2001) Crystallization of Brome mosaic virus and T = 1 Brome mosaic virus particles following a structural transition. Virology, 286, 290–303. [DOI] [PubMed] [Google Scholar]
- Lucas, R.W. , Larson, S.B. and McPherson, A. (2002) The crystallographic structure of brome mosaic virus. J. Mol. Biol. 317, 95–108. [DOI] [PubMed] [Google Scholar]
- Manchester, M. and Singh, P. (2006) Virus‐based nanoparticles (VNPs): platform technologies for diagnostic imaging. Adv. Drug Deliv. Rev. 58, 1505–1522. [DOI] [PubMed] [Google Scholar]
- Mansur, H.S. (2010) Quantum dots and nanocomposites. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2, 113–129. [DOI] [PubMed] [Google Scholar]
- Martin, B.D. , Soto, C.M. , Blum, A.S. , Sapsford, K.E. , Whitley, J.L. , Johnson, J.E. , Chatterji, A. and Ratna, B.R. (2006) An engineered virus as a bright fluorescent tag and scaffold for cargo proteins—capture and transport by gliding microtubules. J. Nanosci. Nanotechnol. 6, 2451–2460. [DOI] [PubMed] [Google Scholar]
- Miller, E.D. and Hemenway, C. (1998) History of coat protein mediated protection. Methods Mol. Biol. 81, 25–38. [DOI] [PubMed] [Google Scholar]
- Miller, W.A. and White, K.A. (2006) Control of plant virus gene expression and replication by long‐distance RNA–RNA interactions. Annu. Rev. Phytopathol. 44, 447–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, Y.S. , Magyar, A.P. , Lee, D. , Kim, J.W. , Yun, D.S. , Park, H. , Pollom, T.S. Jr , Weitz, D.A. and Belcher, A.M. (2010) Biologically templated photocatalytic nanostructures for sustained light‐driven water oxidation. Nat. Nanotechnol. 5, 340–344. [DOI] [PubMed] [Google Scholar]
- Noueiry, A.O. and Ahlquist, P. (2003) Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41, 77–98. [DOI] [PubMed] [Google Scholar]
- Okinaka, Y. , Mise, K. , Suzuki, E. , Okuno, T. and Furusawa, I. (2001) The C‐terminus of brome mosaic virus coat protein controls viral cell‐to‐cell and long‐distance movement. J. Virol. 75, 5385–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olspert, A. , Paves, H. , Toomela, R. , Tamm, T. and Truve, E. (2010) Cocksfoot mottle sobemovirus coat protein contains two nuclear localization signals. Virus Genes, 40, 423–431. [DOI] [PubMed] [Google Scholar]
- Patton, J.T. and Spencer, E. (2000) Genome replication and packaging of segmented double‐stranded RNA viruses. Virology, 277, 217–225. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, P. and Durham, A.C.H. (1977) The cation binding associated with structural transitions in bromegrass mosaic virus. Virology, 81, 419–432. [DOI] [PubMed] [Google Scholar]
- Pinaud, F. , Clarke, S. , Sittner, A. and Dahan, M. (2010) Probing cellular events, one quantum dot at a time. Nat. Methods, 7, 275–285. [DOI] [PubMed] [Google Scholar]
- Prins, M. , Laimer, M. , Noris, E. , Schubert, J. , Wassenegger, M. and Tepfer, M. (2009) Strategies for antiviral resistance in transgenic plants. Mol. Plant Pathol. 9, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. , Ren, T. and Morris, T.J. (2003) The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X. , Droste, T. and Kao, C.C. (2010) Cell penetrating peptides derived from viral capsid proteins. Mol. Plant–Microbe Interact. in press. [DOI] [PubMed] [Google Scholar]
- Rae, C.S. , Khor, I.W. , Wang, Q. , Destito, G. , Gonzalez, M.J. , Singh, P.R. , Thomas, D.M. , Estrada, M.N. , Powell, E. , Finn, M.G. and Manchester, M. (2005) Systemic trafficking of plant virus nanoparticles in mice via the oral route. Virology, 343, 224–235. [DOI] [PubMed] [Google Scholar]
- Rao, A.L. (2006) Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 44, 61–87. [DOI] [PubMed] [Google Scholar]
- Rao, A.L. and Grantham, G.L. (1995) Biological significance of the seven amino‐terminal basic residues of brome mosaic virus coat protein. Virology, 211, 42–52. [DOI] [PubMed] [Google Scholar]
- Rao, A.L. and Grantham, G.L. (1996) Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino‐terminal arginine‐rich motif and its role in encapsidation, movement, and pathology. Virology, 226, 294–305. [DOI] [PubMed] [Google Scholar]
- Reade, R. , Kakani, K. and Rochon, D. (2010) A highly basic KGKKGK sequence in the RNA‐binding domain of the Cucumber necrosis virus coat protein is associated with encapsidation of full‐length CNV RNA during infection. Virology, 403, 181–188. [DOI] [PubMed] [Google Scholar]
- Reddy, D.V.R. , Sudarshana, M.R. , Fuchs, M. , Rao, N.C. and Thottappilly, G. (2009) Genetically engineered virus‐resistant plants in developing countries: current status and future prospects. Adv. Virus Res. 75, 185–220. [DOI] [PubMed] [Google Scholar]
- Reichert, V.L. , Choi, M. , Petrillo, J.E. and Gehrke, L. (2007) Alfalfa mosaic virus coat protein bridges RNA and RNA‐dependent RNA polymerase in vitro. Virology, 364, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher, R. and Ahlquist, P. (1989) Effects of deletions in the N‐terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 63, 4545–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapsford, K.E. , Soto, C.M. , Blum, A.S. , Chatterji, A. , Lin, T. , Johnson, J.E. , Ligler, F.S. and Ratna, B.R. (2006) A cowpea mosaic virus nanoscaffold for multiplexed antibody conjugation: application as an immunoassay tracer. Biosens. Bioelectron. 21, 1668–1673. [DOI] [PubMed] [Google Scholar]
- Sharma, P. and Ikegami, M. (2009) Characterization of signals that dictate nuclear/nucleolar and cytoplasmic shuttling of the capsid protein of tomato leaf curl java virus associated with DNA beta satellite. Virus Res. 144, 145–153. [DOI] [PubMed] [Google Scholar]
- Shenton, W. , Douglas, T. , Young, M. , Stubbs, G. and Mann, S. (1999) Inorganic–organic nanotube composites from template mineralization of tobacco mosaic virus. Adv. Mater. Weinheim, 11, 253–256. [Google Scholar]
- Sheppard, S.L. , Burness, A.T. and Boyle, S.M. (1980) Polyamines in encephalomyocarditis virus. J. Virol. 34, 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoike, T. , Mimori, S. , Tani, H. , Matsuura, Y. and Miyamura, T. (1999) Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73, 9718–9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran, K. and Kao, C.C. (2000) Genomic plus‐strand RNA synthesis by the brome mosaic virus (BMV) RNA replicase requires a sequence that is complementary to the binding site of the BMV helicase‐like protein. Mol. Plant Pathol. 1, 337–346. [DOI] [PubMed] [Google Scholar]
- Sivakumaran, K. , Choi, S.K. , Hema, M. and Kao, C.C. (2004) Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase–core promoter interactions in vitro. J. Virol. 78, 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir, J.A. , Bothner, B. , Qu, C. , Willits, D.A. , Young, M.J. and Johnson, J.E. (2006) Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J. Virol. 80, 3582–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, N.F. , Hong, V. , Spoerke, E.D. , Lu, P. , Breitenkamp, K. , Finn, M.G. and Manchester, M. (2009a) Buckyballs meet viral nanoparticles: candidates for biomedicine. J. Am. Chem. Soc. 131, 17 093–17 095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, N.F. , Lin, T. , Lomonossoff, G.P. and Johnson, J.E. (2009b) Structure‐based engineering of an icosahedral virus for nanomedicine and nanotechnology. Curr. Top. Microbiol. Immunol. 327, 23–58. [DOI] [PubMed] [Google Scholar]
- Suci, P.A. , Varpness, Z. , Gillitzer, E. , Douglas, T. and Young, M. (2007) Targeting and photodynamic killing of a microbial pathogen using protein cage architectures. Langmuir, 23, 12 280–12 286. [DOI] [PubMed] [Google Scholar]
- Sudarshana, M.R. , Roy, G. and Falk, B.W. (2007) Methods for engineering resistance to plant viruses. Methods Mol. Biol. 354, 183–195. [DOI] [PubMed] [Google Scholar]
- Sullivan, M. and Ahlquist, P. (1999) A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73, 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , Dufort, C. , Daniel, M.C. , Murali, A. , Chen, C. , Gopinath, K. , Stein, B. , De, M. , Rotello, V.M. , Holzenburg, A. , Kao, C. and Dragnea, B. (2007) Core‐controlled polymorphism in virus‐like particles. Proc. Natl. Acad. Sci. USA, 104, 1354–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, A. , Kaido, M. , Okuno, T. and Mise, K. (2004) The C terminus of the movement protein of brome mosaic virus controls the requirement for coat protein in cell‐to‐cell movement and plays a role in long‐distance movement. J. Gen. Virol. 85, 1751–1761. [DOI] [PubMed] [Google Scholar]
- Takeuchi, O. and Akira, S. (2007) Recognition of viruses by innate immunity. Immunol. Rev. 220, 214–224. [DOI] [PubMed] [Google Scholar]
- Van der Graaf, M. , Kroon, G.J. and Hemminga, M.A. (1991) Conformation and mobility of the RNA‐binding N‐terminal part of the intact coat protein of Cowpea chlorotic mottle virus. A two‐dimensional proton nuclear magnetic resonance study. J. Mol. Biol. 220, 701–709. [DOI] [PubMed] [Google Scholar]
- Verduin, B.J. and Bancroft, J.B. (1969) The infectivity of tobacco mosaic virus RNA in coat proteins from spherical viruses. Virology, 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220. [DOI] [PubMed] [Google Scholar]
- Vriend, G. , Hemminga, M.A. , Verduin, B.J.M. , De Wit, J.L. and Schaafsma, T.J. (1981) Segmental mobility involved in protein—RNA interaction in Cowpea chlorotic mottle virus. FEBS Lett. 134, 167–171. [Google Scholar]
- Vriend, G. , Hemminga, M.A. , Verduin, B.J.M. and Schaafsma, T.J. (1982a) Swelling of Cowpea chlorotic mottle virus studied by proton nuclear magnetic resonance. FEBS Lett. 146, 319–321. [Google Scholar]
- Vriend, G. , Verduin, B.J.M. , Hemminga, M.A. and Schaafsma, T.J. (1982b) Mobility involved in protein—RNA interaction in spherical plant viruses, studied by nuclear magnetic resonance spectroscopy. FEBS Lett. 145, 49–52. [Google Scholar]
- Vriend, G. , Verduin, B.J. and Hemminga, M.A. (1986) Role of the N‐terminal part of the coat protein in the assembly of Cowpea chlorotic mottle virus. A 500 MHz proton nuclear magnetic resonance study and structural calculations. J. Mol. Biol. 191, 453–460. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Lin, T.W. , Johnson, J.E. and Finn, M.G. (2002) Natural supramolecular building blocks: cysteine‐added mutants of Cowpea mosaic virus. Chem. Biol. 102, 813–819. [DOI] [PubMed] [Google Scholar]
- Wilson, T.M.A. (1993) Strategies to protect crop plants against viruses: pathogen‐derived resistance blossoms. Proc. Natl. Acad. Sci. USA, 90, 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell, G.W. , Gott, J.M. and Uhlenbeck, O.C. (1991) Specific interaction between RNA phage coat proteins and RNA. Prog. Nucleic Acid Res. Mol. Biol. 40, 185–220. [DOI] [PubMed] [Google Scholar]
- Yi, G. and Kao, C.C. (2008) Cis‐ and trans‐acting functions of brome mosaic virus protein 1a in genomic RNA1 replication. J. Virol. 82, 3045–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, G. , Gopinath, K. and Kao, C.C. (2007) Selective repression of translation by the brome mosaic virus 1a RNA replication protein. J. Virol. 81, 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, G. , Letteney, E. , Kim, C.H. and Kao, C.C. (2009a) Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA, 15, 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, G. , Vaughan, R.C. , Yarbrough, I. , Dharmaiah, S. and Kao, C.C. (2009b) RNA binding by the brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J. Mol. Biol. 391, 314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M. , Willits, D. , Uchida, M. and Douglas, T. (2008) Plant viruses as biotemplates for materials and their use in nanotechnology. Annu. Rev. Phytopathol. 46, 361–384. [DOI] [PubMed] [Google Scholar]
- Yusibov, V.M. and Loesch‐Fries, L.S. (1995) N‐terminal basic amino acids of alfalfa mosaic virus coat protein involved in the initiation of infection. Virology, 208, 405–407. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Gopinath, K. , Murali, A. , Yi, G. , Hayward, S.D. , Zhu, H. and Kao, C. (2007) RNA‐binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. USA, 104, 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]


