SUMMARY
The deposition of lignin during plant–pathogen interactions is thought to play a role in plant defence. However, the function of lignification genes in plant disease resistance is poorly understood. In this article, we provide genetic evidence that the primary genes involved in lignin biosynthesis in Arabidopsis, CAD‐C and CAD‐D, act as essential components of defence to virulent and avirulent strains of the bacterial pathogen Pseudomonas syringae pv. tomato, possibly through the salicylic acid defence pathway. Thus, in contrast with cellulose synthesis, whose alteration leads to an increase in disease resistance, alteration of the cell wall lignin content leads directly or indirectly to defects in some defence components.
INTRODUCTION
Plants develop immune responses to react to microbial attack. These responses are initiated by recognition events which result in the activation of an array of defence mechanisms, integrated by a complex signalling system. They include the generation of signals such as reactive oxygen intermediates, synthesis of antimicrobial compounds, lignification of cell walls, and expression of pathogenesis‐related (PR) proteins (Glazebrook, 2005). In addition to these general defence mechanisms, one of the most spectacular manifestations of plant resistance is the hypersensitive response (HR), a form of programmed cell death occurring in a limited area at the site of infection (Heath, 2000).
Lignins, which are complex aromatic polymers resulting from the oxidative polymerization of hydroxycinnamyl alcohols (p‐coumaryl, coniferyl and sinapyl alcohols), are not only associated with plant growth and development, but also with defence responses to environmental stresses (Lange et al., 1995; Nicholson and Hammerschmidt, 1992; Vance et al., 1980). In many cases of plant–pathogen interactions, transcriptional profiling studies have revealed the activation of genes whose products are involved in the biosynthesis and modification of cell wall components (Cheong et al., 2002; Rinaldi et al., 2007; Schenk et al., 2000). The deposition of lignin during plant–pathogen interactions is thought to play a role as a physical barrier against infection. Unpolymerized monolignols may also have antimicrobial activity (Keen and Littlefield, 1979). However, although a number of reports have genetically defined the role of individual lignification genes during plant development, their function in plant disease resistance is poorly understood (Boerjan et al., 2003). Recently, Kawasaki et al. (2006) demonstrated that cinnamoyl‐CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defence signalling in rice. This interesting finding suggests a more sophisticated regulation of lignin biosynthesis than expected during defence responses.
Cinnamyl alcohol dehydrogenase (CAD) is a specialized enzyme involved in the reduction of cinnamaldehydes into cinnamyl alcohols, the last step of monolignol biosynthesis before oxidative polymerization in the cell wall. These enzymes are encoded by complex gene families in plants. The complete CAD gene family has been surveyed in Arabidopsis (Kim et al., 2004; Sibout et al., 2003), demonstrating the existence of nine CAD‐like proteins distributed into four different classes based on their amino acid similarity. Of the nine putative CADs, two exhibit the highest activity and homology to bona fide CADs from other species: CAD‐C and CAD‐D. A second class contains CAD‐A, CAD‐B1 and CAD‐B2, which are most closely related to the poplar sinapyl alcohol dehydrogenase (Li et al., 2001), and which are catalytically less active, at least by an order of magnitude, compared with CAD‐C and CAD‐D. CAD‐1, CAD‐E and CAD‐F are closely related to the alfalfa CAD2 (Brill et al., 1999), and display extremely low activity on cinnamyl aldehydes. Finally, the fourth class contains CAD‐G, which does not have well‐identified homologues. Interestingly, although the characterization of cad‐C and cad‐D single mutants in Arabidopsis has revealed only a modest reduction in lignin content and a weak alteration in lignin quality, cad‐C/cad‐D double mutants have an extremely strong phenotype (2003, 2005). Lignin content in the stem is reduced to 40% and coniferyl and sinapyl alcohol incorporation is reduced by 94%, indicating that CAD‐C and CAD‐D act as the primary genes in lignin biosynthesis in Arabidopsis thaliana. Thus, these genes constitute an interesting target to address the question of the role of lignin biosynthesis in plant–pathogen interactions. In this study, we used a genetic approach to determine whether the Arabidopsis CAD‐C and CAD‐D genes act as essential components of defence to virulent and avirulent strains of the bacterial pathogen Pseudomonas syringae pv. tomato. As the two genes may be redundant, we also used the mutant which is knock out for both genes to assess the resistance to P. syringae pv. tomato. In addition, we studied the expression pattern of the different members of the CAD gene family during the interaction, when compared with the other lignin biosynthesis genes.
RESULTS
Three members of the CAD gene family are induced in response to Pseudomonas
Arabidopsis CAD gene expression was studied during plant development (Sibout et al., 2003). To evaluate the possible role of lignin biosynthesis in plant defence, the expression of all nine members of the Arabidopsis CAD gene family was analysed after challenge with virulent or avirulent strains of P. syringae pv. tomato DC3000 by quantitative real‐time polymerase chain reaction (Q‐RT‐PCR) (Lorrain et al., 2004). Specific primers were designed for each member of the family, with the exception of the CAD‐E and CAD‐F genes, which are 98% identical at the nucleotide level. In this specific case, common primers were designed to amplify both genes (Table S1, see Supporting Information). After inoculation with virulent or avirulent P. syringae pv. tomato strains expressing the avirulence gene avrRpt2, the expression of CAD‐D, CAD‐B2 and CAD‐G was strongly induced when compared with the control treated with water, whereas the expression of CADC, CADA, CADB1 and CADE‐F was not significantly changed, at least at the early time points (Fig. 1). Two genes (CAD‐C and CAD‐EF) were expressed at very low levels. Interestingly, in the case of CAD‐A, CAD‐1 and CAD‐B1, gene expression was clearly induced by the inoculation procedure and appeared repressed 24 h after inoculation when compared with the water control. CAD‐D, CAD‐B2 and CAD‐G showed similar expression profiles, with a strong increase between 9 and 24 or 48 h, which was more rapid in response to the avirulent strain, especially for CAD‐B2 and CAD‐G. In leaves treated with water, these three genes were weakly expressed, whereas the other CAD genes were induced. These results indicate that three members of the CAD gene family are strongly induced on inoculation with a bacterial pathogen.
Figure 1.

Expression pattern of cinnamyl alcohol dehydrogenase (CAD) family members in response to pathogen inoculation. Gene expression level [determined by quantitative real‐time polymerase chain reaction (Q‐RT‐PCR) analysis] in leaves of 4‐week‐old Arabidopsis plants inoculated with water (blue), Pseudomonas syringae pv. tomato (Pst) DC3000 (pink) and Pst DC3000/avrRpt2 (green). The expression values of each gene were normalized using the expression level of β‐tubulin4 as an internal standard. The mean values and standard errors were calculated from two independent experiments. AU, arbitrary units.
CAD‐C and CAD‐D act redundantly as components of basal resistance
To further elucidate the role of lignin biosynthesis in plant resistance, we used previously characterized mutant lines affected in the different CAD genes (Eudes et al., 2006; 2003, 2005). The other Arabidopsis mutant lines (used as controls) were obtained from the following sources: the mutant eds1 from Jane Parker (Parker et al., 1996) and the mutant pad‐4 from Jane Glazebrook (Glazebrook et al., 1997). We focused our analysis on mutants in the CAD‐D, CAD‐B2 and CAD‐G genes because of the activation of these genes in response to pathogen attack, and on the cadC and cadD single and cad‐C/cad‐D double mutants, as CAD‐C and CAD‐D are the primary genes for lignin biosynthesis (Sibout et al., 2005). cadG, cadB1 and cad1 plants behaved like wild‐type plants in both incompatible and compatible interactions with P. syringae pv. tomato, as shown by the evaluation of bacterial colonization and the observation of disease symptoms in these mutants (Fig. 2). In contrast, cad‐D, cad‐B2, the double mutant cad‐C/cad‐D and, in certain cases, cad‐C exhibited decreased resistance to both virulent and avirulent P. syringae pv. tomato. Interestingly, the double mutant cad‐C/cad‐D displayed even more decreased resistance than cad‐C or cad‐D single mutants in compatible and incompatible interactions (Figs 2 and S1, see Supporting Information). Bacterial colonization of cad‐C and cad‐D single mutants was 2–10‐fold increased, whereas colonization of cad‐C/cad‐D was 5–60‐fold increased when compared with the wild‐type (Fig. 2A,B). The susceptibility of the double mutant is similar to the enhanced disease phenotype of the eds1 mutant (Parker et al., 1996) in response to DC3000, and to the susceptible phenotype of the La‐er ecotype in response to the strain containing the avrPphB avirulence gene. The cadB2 mutant also displayed a clear phenotype, close to the phenotype of the double mutant cad‐C/cad‐D. Consistent with these data, cad‐C/cad‐D plants and, to a lesser extent, cad‐B2 plants, displayed markedly increased disease symptoms compared with the wild‐type in the interaction with the two strains of P. syringae pv. tomato (Fig. 2C). Taken together, these results suggest that CAD‐C, CAD‐D and CAD‐B2 act as positive components of basal resistance and effector‐triggered resistance to P. syringae pv. tomato.
Figure 2.

Phenotypes of cad‐C, cad‐D, cad‐G, cad‐B2, cad‐1, cad‐B1, eds‐1, pad‐4 single mutants, cad‐C/cad‐D double mutant and the wild‐type ecotypes Ws and La‐er, after inoculation with Pseudomonas syringae pv. tomato (Pst). Growth of Pst DC3000 (A) and Pst DC3000/avrPphB (B) in wild‐type and mutant plants, determined at 0 days (white bars) and 3 days (black bars) post‐inoculation with a bacterial suspension of 105 cfu/mL. The mean bacterial densities were calculated from 9–12 replicates and are representative of two independent experiments. According to analysis of variance (anova) test, means of colony‐forming units (cfu) do not differ significantly if they are indicated by the same lowercase letter. (C) Representative mutant and wild‐type leaves 3 days after inoculation with a suspension of the bacterial strain indicated at a density of 107 cfu/mL.
Defence gene regulation is altered in the double mutant cad‐C/cad‐D
The balance between signalling components, such as ethylene (ET), jasmonates (JA) and salicylic acid (SA), is crucial for the modulation and adaptation of the various defence mechanisms to pathogens. For this reason, we investigated whether the decreased resistance phenotype of cad‐C/cad‐D plants is correlated with an alteration in these signalling pathways. For this purpose, we analysed the expression of defence marker genes in the double mutant. Marker genes of the SA pathway, ICS (isochorismate synthase, involved in the biosynthesis of SA), PR1 and PR5 (pathogenesis‐related), were tested. The expression level of ICS was induced on inoculation with virulent and avirulent P. syringae pv. tomato strains, and was significantly higher in the double mutant when compared with the wild‐type (Fig. 3A). Surprisingly, the expression of PR1 and, to a lesser extent, PR5 was reduced in cad‐C/cad‐D in response to the virulent strain, suggesting a negative effect of the mutations on (or downstream of) SA production. No significant effect could be detected in response to the avirulent strain of P. syringae pv. tomato. The measurement of SA levels showed that SA production was also reduced in the double mutant when compared with the wild‐type during the compatible interaction (not in response to the avirulent strain) (Fig. 3B). The expression of the marker gene for JA‐ and ET‐dependent signalling pathways, PR3 (pathogenesis‐related), was also affected in the double mutant, but its expression was, as ICS, higher in the double mutant when compared with the wild‐type. Hence, CAD‐C and CAD‐D depletion may be critical for the regulation of the defence signalling pathways, affecting negatively the SA pathway, at least in the context of the compatible interaction. As this pathway plays an essential role in resistance to P. syringae pv. tomato, this defect could explain, at least in part, the alteration in basal resistance observed previously (Fig. 2).
Figure 3.

Defence gene expression (A) and salicylic acid (SA) production (B) in the double mutant cad‐C/cad‐D (triangles) and wild‐type (squares) after inoculation with water (blue), Pseudomonas syringae pv. tomato (Pst) DC3000 (pink) and Pst DC3000/avrRpt2 (green). (A) The expression values of each gene were normalized using the expression level of β‐tubulin4 as an internal standard. The mean values and standard errors were calculated from two independent experiments. AU, arbitrary units. (B) For SA measurement, the mean values and standard errors were calculated from two independent experiments.
Effect of CAD‐C and CAD‐D depletion on the expression of the other members of the CAD gene family and of other lignin biosynthesis‐related genes in response to Pseudomonas
To assess the specificity of the effect of the absence of CAD‐C and CAD‐D on lignin biosynthesis, we tested whether any alteration of the expression of the genes involved in lignin biosynthesis could occur in the double mutant in the context of the interaction with P. syringae pv. tomato. CAD‐B2 and CAD‐G, previously demonstrated to be induced in response to P. syringae pv. tomato, were studied first (Fig. 4). In response to P. syringae pv. tomato, the expression of these two genes was differentially affected in the double mutant cad‐C/cad‐D when compared with the wild‐type: CAD‐G was over‐expressed (about twofold increased) and CAD‐B2 was down‐regulated (two‐ to three‐fold decreased). For comparison, the expression profile of CAD‐B1 was similar in the double mutant and the wild‐type. Finally, the analysis of other genes encoding proteins involved in lignin biosynthesis did not reveal substantial alterations (Fig. S2, see Supporting Information). Only CCR‐2, which has been shown to be induced by pathogen infection (Lauvergeat et al., 2001), was slightly affected in the double mutant (Fig. 4).
Figure 4.
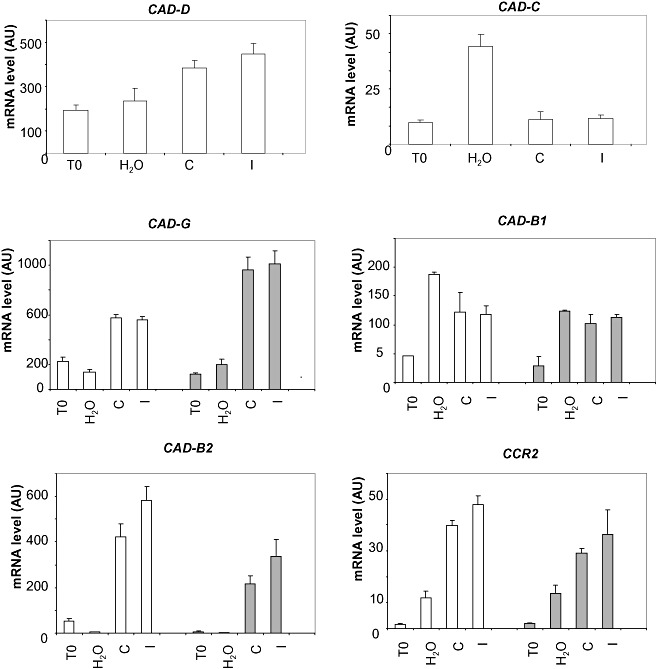
Effects of CAD‐C and CAD‐D depletion on the expression of the other members of the cinnamyl alcohol dehydrogenase (CAD) gene family and of other lignin biosynthesis‐related genes in response to Pseudomonas syringae pv. tomato (Pst). Gene expression in the wild‐type (white bars) and double mutant (gray bars), 0 h (T0) and 24 h after inoculation with water, Pst DC3000 (C) and Pst DC3000/avrRpt2 (I). The expression values of each gene were normalized using the expression level of β‐tubulin4 as an internal standard. Mean expression values and standard errors were calculated from three replicates of a representative experiment.
Based on these findings, we can assume that the resistance and defence phenotypes of the double mutant cad‐C/cad‐D result from the primary effect of the absence of the two proteins CAD‐C and CAD‐D.
DISCUSSION
Lignins and lignin‐related compounds are known to be induced at sites of pathogen infection (Reimers and Leach, 1991). The biological significance of this lignin deposition is thought to be reinforcement of the cell wall, lignins being extremely resistant to microbial degradation. However, no genetic evidence has been clearly produced in favour of this hypothesis. In this article, we provide genetic and molecular evidence that: (i) some members of the CAD gene family are induced on pathogen attack; (ii) CAD‐C and CAD‐D genes, which have been demonstrated to be the primary genes involved in lignin biosynthesis, act as essential components of defence to virulent and avirulent strains of the bacterial pathogen P. syringae pv. tomato; and (iii) SA‐dependent defence gene regulation, possibly as a consequence of the alteration of SA biosynthesis during a compatible interaction, is affected in the double mutant cad‐C/cad‐D.
Different studies have highlighted that CAD proteins are encoded by a multigene family comprising nine members in Arabidopsis (Sibout et al., 2003), corresponding to six bona fide catalytically active CADs and three displaying very low enzymatic activity (Kim et al., 2004). This suggests that some CADs may display alternative metabolic roles, for example in plant defence. This hypothesis is reinforced by the observation of the induced expression of such CADs on pathogen attack. Among the less active CADs (CAD‐E, CAD‐F, CAD‐B1 and CAD‐B2), CAD‐B1 has been shown to be induced in response to P. syringae pv. tomato inoculation (Kiedrowski et al., 1992; Somssich et al., 1996). We confirmed this observation here and showed that the other less active CADs are not induced on P. syringae pv. tomato inoculation, suggesting other metabolic roles, probably in response to mechanical wounding or abiotic stresses, as suggested by their clear induction by the inoculation procedure. In the case of CAD‐B2, the corresponding mutant exhibits decreased resistance to P. syringae pv. tomato, despite the absence of visible impact of this mutation on lignin biosynthesis (Eudes et al., 2006). As CADB2 acts as a benzyl alcohol dehydrogenase accepting various benzaldehyde substrates, the mutant phenotype can be interpreted as an absence of accumulation of compounds involved in defence.
Analysis of the expression pattern of the primary and most active CADs, CAD‐C and CAD‐D, showed that CAD‐C is not induced by pathogen attack, whereas CAD‐D is strongly induced 24 h post‐inoculation. These results confirm and extend the data available in public databases from microarray analyses (Arabidopsis eFP Browser at bar.utoronto.ca) in response to diverse pathogens, such as different strains of Pseudomonas or Phytophthora infestans. Interestingly, CAD‐D is also induced by pathogen‐associated molecular pattern (PAMP) treatment (flagellin flg22; Gomez‐Gomez and Boller, 2002) and in response to an hrp‐ bacterial mutant (hrcC‐; Yuan and He, 1996) (NASCarrays, experiment reference number: ‐NASCarrays 120), suggesting that CAD‐D induction is part of PAMP‐triggered immunity, the plant's first active response to microbial perception, initiated on recognition of conserved microbial features by plant cell surface receptors (Chisholm et al., 2006). Moreover, the differential regulation of CAD‐C and CAD‐D genes in response to pathogen attack is reminiscent of the previous observations that AtCCR1 and AtCCR2 are differentially regulated in response to Xanthomonas inoculation (Lauvergeat et al., 2001), suggesting that, beyond their functional redundancy demonstrated previously (Sibout et al., 2005), and their similar pattern of expression during development (Kim et al., 2007), each CAD gene might have a specific role in development vs. pathogen responsiveness. However, the loss of either CAD‐C or CAD‐D function results in a partial (or no) loss of resistance to virulent and avirulent strains of P. syringae pv. tomato, and the depletion of both genes confers a clear susceptible phenotype. Similar observations were made in response to different avirulent strains of P. syringae pv. tomato (Fig. S1), suggesting a general and non‐race‐specific effect of the mutations. As is the case during development, these data confirm the functional redundancy of these genes in the context of plant–pathogen interactions, and demonstrate their essential role in plant defence. However, different possibilities, which are not exclusive, can be proposed to explain this role: (i) a direct role of lignin accumulation during plant defence, acting as a barrier to infection; (ii) a function in plant resistance for the accumulation of soluble aldehydes, and/or the absence of lignin‐related compounds, observed in the double mutant (Rutten and Gocke, 1988; Sibout et al., 2005; Utama et al., 2002); or (iii) an indirect role of CAD‐C and CAD‐D as possible regulators of the SA defence pathways. Concerning the first possibility, histological and biochemical analyses were performed as described previously by Sibout et al. (2005) on inoculated leaves of the wild‐type and the double mutant cad‐C/cad‐D (data not shown). Unfortunately, these analyses did not enable this point to be clarified, probably because of the very small amounts of lignins accumulated at the inoculation sites, and the limited number of cells influenced by this process. However, we observed that both SA production and the SA defence pathway are altered in the cad‐C/cad‐D mutant, at least in the case of the compatible interaction, which is in good agreement with the role of SA as a central regulator of defence to P. syringae pv. tomato in Arabidopsis. In the case of the incompatible interaction, another hypothesis, including a possible delay in SA accumulation in the mutant when compared with the wild‐type, can be envisaged. Surprisingly, for the compatible interaction, although genetic and biochemical evidence demonstrated that, in A. thaliana, the biosynthetic pathway for SA is dependent on ICS (Nawrath and Metraux, 1999; Strawn et al., 2007; Wildermuth et al., 2001), we observed here that ICS expression was induced at high levels in the double mutant, but did not lead to SA biosynthesis. Different hypotheses can be proposed to explain this result: (i) some metabolites produced in the double mutant could interfere with SA biosynthesis downstream of ICS expression (inhibition of ICS activity or alteration of downstream enzymes); or (ii) SA could be synthesized via a derived route, possibly through AtCM1 (chorismate mutase 1) and phenylalanine, which could be altered in the double mutant. Previous studies in different plant species have indicated that SA can be synthesized from phenylalanine. However, although AtCM1 is induced by bacterial pathogens (Eberhard et al., 1996; Mobley et al., 1999) and has a high reported K m for chorismate (Mobley et al., 1999), the dependence of the SA biosynthetic pathway on ICS is well established in A. thaliana (Wildermuth et al., 2001) and, more recently, in tobacco (Catinot et al., 2008). In addition, phenylalanine ammonia lyase (PAL) expression has been shown to be unaffected in the double mutant when compared with the wild‐type (data not shown). Thus, we propose that CAD‐C and CAD‐D act as components of defence mechanisms, primarily as key effectors of lignin biosynthesis, and, secondarily, as possible regulators of the SA defence pathways.
At this point, it should be noted that, in mutants impaired in cellulose synthesis and displaying similar phenotypic characteristics (Cano‐Delgado et al., 2003; Hernandez‐Blanco et al., 2007; Turner and Somerville, 1997), disease resistance and JA levels are enhanced. This phenotype is interpreted as an alteration of cell wall integrity, resulting in the activation of novel defence pathways. These defence pathways probably include lignin biosynthesis. Thus, the alteration of lignin biosynthesis has an opposite effect on defence when compared with the reduction in cellulose synthesis.
EXPERIMENTAL PROCEDURES
Plant material and pathogen infection
Arabidopsis plants were grown under controlled conditions (Lacomme and Roby, 1996). Depending on the experiment, we used the wild‐type ecotypes Col‐0, Ws‐4 and La‐er, and the mutants cad‐C (Ws‐4), cad‐D (Ws‐4) (Sibout et al., 2003), cad‐C/cad‐D (Ws‐4) (Sibout et al., 2005), cad‐B1(Col‐0), cad‐B2 (Ws‐4), cad‐1(Col‐0) and cad‐G (Ws‐4) (Eudes et al., 2006), eds1 (Ws‐0) (Parker et al., 1996) and pad‐4 (Col‐0) (Glazebrook et al., 1997).
Plant inoculations, using P. syringae pv. tomato strain DC3000 carrying the avrPphB, avrRpt2 or avrRpm1gene, and in planta bacterial growth analysis were performed as described previously (Lorrain et al., 2004). Briefly, the ecotypes Ws‐4 and Col‐0 behave similarly to these strains (susceptible to DC3000 and resistant to DC3000/avrPphB), whereas the ecotype La‐er is susceptible to the strain containing avrPphB. The virulent and avirulent P. syringae pv. tomato strains were grown at 28 °C on King B medium supplemented with the appropriate antibiotics. Four‐ or five‐week‐old plants were used for bacterial inoculation. For this objective, they were kept at high humidity 12 h before the experiments and then infiltrated with a bacterial suspension of 105 colony‐forming units (cfu)/mL for the determination of in planta bacterial growth, and 107 cfu/mL for gene expression analysis.
Gene expression analysis
Total RNA extraction and Q‐RT‐PCR were performed as described by Bouchez et al. (2007). Total RNA extraction was performed from leaves with a Nucleospin RNA kit following the manufacturer's instructions (Macherey‐Nagel, Hoerdt, France). Total RNA (1.5 µg) was subjected to cDNA synthesis (Superscript II reverse transcriptase, Invitrogen, Cergy Pontoise, France), which was used as template in the Q‐PCR analysis. Q‐PCR was performed using gene‐specific primers (Table S1), LightCycler reagents and apparatus (Roche Diagnostics, Meylan, France). Q‐PCRs were performed with two independent biological assays. β‐tubulin4 and a gene (At2g28390) whose expression has been shown to be extremely stable under different physiological conditions (Czechowski et al., 2005) were used as biological controls. The expression values of individual genes were normalized using the expression level of β‐tubulin4 as a control, and the data were expressed for each point as the fold induction compared with the wild‐type Ws‐4 (arbitrary units). The mean expression values were calculated from the results of two independent experiments. The same results were obtained after normalization with At2g28390 (data not shown).
SA extraction and analysis
Three hundred milligrams of tissue were ground in liquid nitrogen, and 50 ng of an internal standard (o‐anisic acid, oANI) were added before the extraction. Total SA (free SA plus SA conjugates) was extracted and analysed according to Mercier et al. (2001) with some modifications. After extraction, analysis was performed with a high‐performance liquid chromatography (HPLC) apparatus (Ultimate 3000, DIONEX SA, Voisins‐le‐Bretonneux, France). Total SA and oANI (internal standard) were quantified with a spectrofluorimeter (Jasco FP‐920 Bouguenais, France), with excitation and emission wavelengths of 305 and 365 nm, respectively. The data were analysed using Chromeleon 6.8 chromatography software (DIONEX SA). Experiments were repeated twice and the data represent the mean values ± standard error.
Supporting information
Fig. S1 Phenotypes of cad‐C and cad‐D single mutants, cad‐C/cad‐D double mutant and the wild‐type ecotype Ws after inoculation with Pseudomonas syringae pv. tomato (Pst). Growth of Pst DC3000/avrRpt2 (A) and Pst DC3000/avrRpm1 (B) in wild‐type and mutant plants, determined 0 days (white bars) and 3 days (black bars) post‐inoculation with a bacterial suspension of 105 cfu/mL. The mean bacterial densities were calculated from 9–12 replicates and are representative of two independent experiments. (C) Representative mutant and wild‐type leaves 3 days after inoculation with a suspension of DC3000/avrRpt2 at a density of 107 cfu/mL.
Fig. S2 Quantitative real‐time polymerase chain reaction (Q‐RT‐PCR) analysis of lignin biosynthesis‐related gene expression in Arabidopsis wild‐type plants (squares) and in the double mutant cad‐C/cad‐D (triangles) after inoculation with water (blue), Pseudomonas syringae pv. tomato (Pst) DC3000 (pink) and Pst DC3000/avrRpt2 (green). The expression values of each gene were normalized using the expression level of β‐tubulin4 as an internal standard. These results are representative of two independent experiments.
Table S1 Primers used for real‐time polymerase chain reaction experiments.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We are grateful to Olivier Bouchez for statistical analysis of our data and Gisèle Borderies for SA analysis. We thank Clare Gough for critical reading of the manuscript. This work was supported by a joint French–German plant genomics programme (Génoplante‐GABI2003‐14).
REFERENCES
- Boerjan, W. , Ralph, J. and Baucher, M. (2003) Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Bouchez, O. , Huard, C. , Lorrain, S. , Roby, D. and Balague, C. (2007) Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1 . Plant Physiol. 145, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill, E.M. , Abrahams, S. , Hayes, C.M. , Jenkins, C.L. and Watson, J.M. (1999) Molecular characterisation and expression of a wound‐inducible cDNA encoding a novel cinnamyl‐alcohol dehydrogenase enzyme in lucerne (Medicago sativa L.). Plant Mol. Biol. 41, 279–291. [DOI] [PubMed] [Google Scholar]
- Cano‐Delgado, A. , Penfield, S. , Smith, C. , Catley, M. and Bevan, M. (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana . Plant J. 34, 351–362. [DOI] [PubMed] [Google Scholar]
- Catinot, J. , Buchala, A. , Abou‐Mansour, E. and Métraux, J.‐P. (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana . FEBS Lett. 582, 473–478. [DOI] [PubMed] [Google Scholar]
- Cheong, Y.H. , Chang, H.S. , Gupta, R. , Wang, X. , Zhu, T. and Luan, S. (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, D. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, J. , Ehrler, T.T. , Epple, P. , Felix, G. , Raesecke, H.R. , Amrhein, N. and Schmid, J. (1996) Cytosolic and plastidic chorismate mutase isoenzymes from Arabidopsis thaliana: molecular characterization and enzymatic properties. Plant J. 10, 815–821. [DOI] [PubMed] [Google Scholar]
- Eudes, A. , Pollet, B. , Sibout, R. , Do, C. , Séguin, A. , Lapierre, C. and Jouanin, L. (2006) Evidence for a role of AtCAD1 in lignification of elongating stems of Arabidopsis thaliana . Planta 225, 23–39. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Zook, M. , Mert, F. , Kagan, I. , Rogers, E.E. , Crute, I.R. , Holub, E.B. , Hammerschmidt, R. and Ausubel, F.M. (1997) Phytoalexin‐deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics, 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2002) Flagellin perception: a paradigm for innate immunity. Trends. Plant Sci. 7, 251–256. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000) Hypersensitive response‐related death. Plant Mol. Biol. 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Hernandez‐Blanco, C. , Feng, D.X. , Sanchez‐Vallet, A. , Deslandes, L. , Llorente, F. , Berrocal‐Lobo, M. , Keller, H. , Barlet, X. , Sanchez‐Rodroguez, C. , Anderson, L.K. , Somerville, S. , Marco, Y. and Molina, A. (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell, 19, 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T. , Koita, H. , Nakatsubo, T. , Hasegawa, K. , Wakabayashi, K. , Takahashi, H. , Umemura, K. , Umezawa, T. and Shimamoto, K. (2006) Connamoyl‐CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA, 103, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N.T. and Littlefield, L.J. (1979) The possible association of phytoalexins with resistance gene expression in flax to Melampsora lini . Physiol. Plant Pathol. 14, 265–280. [Google Scholar]
- Kiedrowski, S. , Kawalleck, P. , Hahlbrock, K. , Somssich, I. and Dangl, J.L. (1992) Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J. 11, 4667–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Kim, K. , Cho, M. , Franceschi, V.R. , Davin, L. and Lewis, N.L. (2007) Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? Phytochemistry, 68, 1957–1974. [DOI] [PubMed] [Google Scholar]
- Kim, S.‐J. , Kim, M.‐R. , Bedgar, D.L. , Moinuddin, S.G.A. , Cardenas, C.L. , Davin, L. , Kang, C. and Lewis, N.G. (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA, 101, 1455–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme, C. and Roby, D. (1996) Molecular cloning of a sulfotransferase in Arabidopsis thaliana and regulation during development and in response to infection with pathogenic bacteria. Plant. Mol. Biol. 30, 995–1008. [DOI] [PubMed] [Google Scholar]
- Lange, B.M. , Lapierre, C. and Sandermann, J.H. (1995) Elicitor‐induced spruce stress lignin. Structural similarity to early developmental lignins. Plant Physiol. 108, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat, V. , Lacomme, C. , Lacombe, E. , Lasserre, E. , Roby, D. and Grima‐Pettenati, J. (2001) Two cinnamoyl‐CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry, 57, 1187–1195. [DOI] [PubMed] [Google Scholar]
- Li, L. , Cheng, X.F. , Leshkevich, J. , Umezawa, S.A. , Harding, S.A. and Chiang, V.L. (2001) The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding synapyl alcohol dehydrogenase. Plant Cell, 13, 1567–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S. , Lin, B. , Auriac, M.C. , Kroj, T. , Saindrenan, P. , Nicole, M. , Balagué, C. and Roby, D. (2004) VASCULAR ASSOCIATED DEATH1, a novel GRAM domain‐containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell, 16, 2217–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, L. , Lafitte, C. , Borderies, G. , Briand, X. , Esquerré‐Tugayé, M.T. and Fournier, J. (2001) The algal polysaccharide carrageenans can act as an elicitor of plant defense. New Phytol. 149, 43–51. [DOI] [PubMed] [Google Scholar]
- Mobley, E. , Kunkel, B.N. and Keith, B. (1999) Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana . Gene, 240, 115–123. [DOI] [PubMed] [Google Scholar]
- Nawrath, C. and Metraux, J.‐P. (1999) Salicylic acid induction‐deficient mutants of Arabidopsis express PR‐2 and PR‐5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell, 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, R.L. and Hammerschmidt, R. (1992) Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers, P.J. and Leach, J.E. (1991) Race‐specific resistance to Xanthomonas oryzae pv. oryzae conferred by bacterial blight resistance gene Xa‐10 in rice (Oryza sativa) involves accumulation of a lignin‐like substance in host tissues. Physiol. Mol. Plant Pathol. 38, 39–55. [Google Scholar]
- Rinaldi, C. , Kohler, A. , Frey, P. , Duchaussoy, F. , Ningre, N. , Couloux, A. , Wincker, P. , Le Thiec, D. , Fluch, S. , Martin, F. and Duplessis, S. (2007) Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici‐populina . Plant Physiol. 144, 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten, B. and Gocke, E. (1988) The antimutagenic effect of cinnamaldehyde is due to a transient growth inhibition. Mutat. Res. 201, 97–105. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M. , Kazan, K. , Wilson, I. , Anderson, J.P. , Richmond, T. , Somerville, S.C. and Manners, J.M. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA, 97, 11 655–11 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout, R. , Eudes, A. , Pollet, B. , Goujon, T. , Mila, I. , Grabnier, F. , Séguin, A. , Lapierre, C. and Jouanin, L. (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 132, 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout, R. , Eudes, A. , Mouille, G. , Pollet, B. , Lapierre, C. , Jouanin, L. and Séguin, A. (2005) CINNAMYL ALCOHOL DEHYDROGENASE‐C and ‐D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell, 17, 2059–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich, I.E. , Wernert, P. , Kiedrowski, S. and Hahlbrock, K. (1996) Arabidopsis thaliana defense‐related protein ELI3 is an aromatic alcohol:NADP(+) oxidoreductase. Proc. Natl. Acad. Sci. USA, 93, 14 199–14 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn, M.A. , Marr, S.K. , Inoue, K. , Inada, N. , Zubieta, C. and Wildermuth, M.C. (2007) Arabidopsis isochorismate synthase functional in pathogen‐induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933. [DOI] [PubMed] [Google Scholar]
- Turner, S.R. and Somerville, C.R. (1997) Collapsed xylem phenotype of Arabidopsis mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell, 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utama, I.M.S. , Wills, R.B.H. , Ben‐yehoshua, S. and Kuek, C. (2002) In vitro efficacy of plant volatiles for inhibiting the growth of fruit and vegetable decay microorganisms. J. Agric. Food Chem. 22, 6371–6377. [DOI] [PubMed] [Google Scholar]
- Vance, C.P. , Kirk, T.K. and Sherwood, R.T. (1980) Lignification as a defence mechanism of disease resistance. Annu. Rev. Phytopathol. 18, 259–288. [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Yuan, J. and He, S.Y. (1996) The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J. Bacteriol. 178, 6399–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phenotypes of cad‐C and cad‐D single mutants, cad‐C/cad‐D double mutant and the wild‐type ecotype Ws after inoculation with Pseudomonas syringae pv. tomato (Pst). Growth of Pst DC3000/avrRpt2 (A) and Pst DC3000/avrRpm1 (B) in wild‐type and mutant plants, determined 0 days (white bars) and 3 days (black bars) post‐inoculation with a bacterial suspension of 105 cfu/mL. The mean bacterial densities were calculated from 9–12 replicates and are representative of two independent experiments. (C) Representative mutant and wild‐type leaves 3 days after inoculation with a suspension of DC3000/avrRpt2 at a density of 107 cfu/mL.
Fig. S2 Quantitative real‐time polymerase chain reaction (Q‐RT‐PCR) analysis of lignin biosynthesis‐related gene expression in Arabidopsis wild‐type plants (squares) and in the double mutant cad‐C/cad‐D (triangles) after inoculation with water (blue), Pseudomonas syringae pv. tomato (Pst) DC3000 (pink) and Pst DC3000/avrRpt2 (green). The expression values of each gene were normalized using the expression level of β‐tubulin4 as an internal standard. These results are representative of two independent experiments.
Table S1 Primers used for real‐time polymerase chain reaction experiments.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


