SUMMARY
Cassava brown streak disease (CBSD) is emerging as one of the most important viral diseases of cassava (Manihot esculenta) and is considered today as the biggest threat to cassava cultivation in East Africa. The disease is caused by isolates of at least two phylogenetically distinct species of single‐stranded RNA viruses belonging to the family Potyviridae, genus Ipomovirus. The two species are present predominantly in the coastal lowland [Cassava brown streak virus (CBSV); Tanzania and Mozambique] and highland [Cassava brown streak Uganda virus (CBSUV); Lake Victoria Basin, Uganda, Kenya and Malawi] in East Africa. In this study, we demonstrate that CBSD can be efficiently controlled using RNA interference (RNAi). Three RNAi constructs targeting the highland species were generated, consisting of the full‐length (FL; 894 nucleotides), 397‐nucleotide N‐terminal and 491‐nucleotide C‐terminal portions of the coat protein (CP) gene of a Ugandan isolate of CBSUV (CBSUV‐[UG:Nam:04]), and expressed constitutively in Nicotiana benthamiana. After challenge with CBSUV‐[UG:Nam:04], plants homozygous for FL‐CP showed the highest resistance, followed by the N‐terminal and C‐terminal lines with similar resistance. In the case of FL, approximately 85% of the transgenic plant lines produced were completely resistant. Some transgenic lines were also challenged with six distinct isolates representing both species: CBSV and CBSUV. In addition to nearly complete resistance to the homologous virus, two FL plant lines showed 100% resistance and two C‐terminal lines expressed 50–100% resistance, whereas the N‐terminal lines succumbed to the nonhomologous CBSV isolates. Northern blotting revealed a positive correlation between the level of transgene‐specific small interfering RNAs detected in transgenic plants and the level of virus resistance. This is the first demonstration of RNAi‐mediated resistance to CBSD and protection across very distant isolates (more than 25% in nucleotide sequence) belonging to two different species: Cassava brown streak virus and Cassava brown streak Uganda virus.
INTRODUCTION
Cassava (Manihot esculenta Crantz, Family Euphorbiaceae) is an important subsistence food crop, known for its ability to grow in poor soils, and is cultivated in the tropical regions of Africa, Asia and Latin America (El‐Sharkawy, 2003). Cassava is vulnerable to at least 20 different viral diseases, among which cassava mosaic disease (CMD) and cassava brown streak disease (CBSD) are the most important in Africa (Patil and Fauquet, 2009). Although CMD occurs in almost all cassava‐cultivating regions of sub‐Saharan Africa and has received considerable attention within the research community (Patil and Fauquet, 2009, 2010), CBSD has only recently become a major concern (Hillocks and Jennings, 2003; Pennisi, 2010), but is rapidly emerging as an important disease of this crop, prevalent in eastern Africa and expanding towards Central Africa (Alicai et al., 2007; Mbanzibwa et al., 2009b). CBSD is caused by two different ipomovirus species, Cassava brown streak virus (CBSV) and Cassava brown streak Uganda virus (CBSUV), both of which are monopartite, positive‐sense, single‐stranded RNA (ssRNA) viruses encapsidated into flexuous filamentous particles and classified in the genus Ipomovirus (Family Potyviridae) (Hillocks and Jennings, 2003; Mbanzibwa et al., 2009a; Monger et al., 2001b; Winter et al., 2010).
Recent studies comparing complete sequences of several isolates have clustered them into two distinct clades, the first comprising isolates from Kenya, Uganda, Malawi and north‐western Tanzania, which have 87–95% homology, and the second containing isolates from coastal regions of Tanzania and Mozambique, which show 70% homology to the first clade and 90–95% homology amongst themselves (Mbanzibwa et al., 2009a; Monger et al., 2010; Winter et al., 2010). Based on a comparison of coat protein (CP) sequences, isolates in East Africa can also be grouped into two phylogenetically distinct clusters: coastal lowland isolates (Tanzania and Mozambique) and highland isolates (Lake Victoria Basin, Uganda, Kenya and Malawi) (2009a, 2009b; Monger et al., 2010c; Winter et al., 2010). The isolates from both clades vary significantly in their sequence (>25%) and possess significant sequence deletions in their Ham1 (Ham1h‐pyrophosphatase) and CP gene sequences. Thus, it has been proposed that two species can be defined: CBSV for the isolates from lowland or coastal regions of East Africa and CBSUV for the isolates from the highland region or Lake Victoria basin (Winter et al., 2010; C. M. Fauquet, unpublished results).
CBSD has been shown to be transmitted by whitefly vectors (Maruthi et al., 2005; Mware et al., 2009) and by mechanical and grafting techniques under laboratory conditions (Lister, 1959; Monger et al., 2001a). Symptoms vary, but are characterized by general chlorosis on the foliage, brown lesions under the bark of the stem and brown streaks on the mature stem of cassava (Fig. S1, see Supporting Information) (Hillocks and Jennings, 2003; Hillocks and Thresh, 2000). Economically, in East Africa, CBSD is considered to be a more threatening disease than CMD, because, in addition to overall yield loss, the pathogen causes severe necrosis within the edible storage roots, making them unfit for marketing or human consumption, thereby reaching 100% losses (Hillocks and Jennings, 2003; Hillocks et al., 2001).
There is some natural source of resistance for CBSD, thus making conventional breeding programmes a possibility for the introgression of CBSD resistance; however, it is a very long process to combine CBSD resistance with good root and harvest qualities (Jennings, 2003). Furthermore, studies have shown that some cassava cultivars that are resistant to one virus species are not resistant to the other virus species causing CBSD (S. Winter, DSMZ, Plant Virus Division, Braunschweig, Germany, personal communication). Biotechnological approaches, in particular, post‐transcriptional gene silencing (PTGS) offer significant potential for addressing the control of CBSD (Prins et al., 2008; Reddy et al., 2009; Sudarshana et al., 2007). Transgenically induced PTGS, or RNA interference (RNAi), has been employed to control both DNA and RNA plant viruses (Chellappan et al., 2004; Kamachi et al., 2007; Sudarshana et al., 2007; Tenllado et al., 2004; Tougou et al., 2006). PTGS is a sequence‐specific RNA degradation mechanism triggered via a double‐stranded (ds)RNA intermediate, with the dsRNAs cleaved by Dicer proteins and processed by the RNA‐induced silencing complex (RISC) to produce 21–25‐nucleotide (nt) small interfering (si)RNAs (Ding and Voinnet, 2007; Ruiz‐Ferrer and Voinnet, 2009). The introduction of an inverted repeat sequence, homologous to part of the viral genome, through the expression of a hairpin (hp), is an efficient method for inducing gene silencing and conferring viral resistance in plants (Helliwell and Waterhouse, 2005).
The CP of potyviruses plays an important role in the encapsidation of the viral genome and the regulation of viral RNA replication, insect transmission, cell‐to‐cell and systemic movement in the host, and is also one of the most conserved genes across related viral species (Callaway et al., 2001; Monger et al., 2001b; Urcuqui‐Inchima et al., 2001). The CP gene has therefore been an important target for the RNAi strategy designed to control RNA viruses, and has been deployed successfully and commercialized in the case of Papaya ring spot virus and Plum pox virus (Krubphachaya et al., 2007; Scorza et al., 2001).
The goal of the present work was to determine whether the RNAi approach could control CBSD, and thus demonstrate a potential biotechnological solution to tackle the spread of this devastating disease in Africa. As the production of transgenic cassava plants is time consuming (Taylor et al., 2004) and screening for resistance to CBSD on cassava is difficult, we utilized the model host Nicotiana benthamiana to assess the efficacy of transgenically induced RNAi to control CBSUV and CBSV. Here, we report the first evidence of effective RNAi‐based control of an ipomovirus and demonstrate effective cross‐protection against diverse CBSD‐causing virus isolates belonging to two distinct species. The potential implications of this work with regard to the introduction of resistance to CBSD in local cassava cultivars for African cassava farmers are discussed.
RESULTS
Analysis of transgenic plant lines and resistance to the homologous virus CBSUV‐[UG:Nam:04]
Approximately 20 independent transgenic lines derived from each of the three RNAi constructs, FL (full‐length), NT (N‐terminal) and CT (C‐terminal) regions of CP, with sequences derived from the isolate CBSUV‐[UG:Nam:04] (Ugandan isolate of CBSUV), were polymerase chain reaction (PCR) screened for the presence of the transgene using specific primers (Table S1, see Supporting Information). Transgenic, homozygous plants were selected on a medium containing kanamycin in the T1 and T2 generations, and seven independent transgenic lines were selected for each RNAi construct to allow the evaluation of resistance to CBSUV‐[UG:Nam:04]. Plants were challenged by sap inoculation with CBSUV‐[UG:Nam:04] and the resistance levels displayed by each line were analysed statistically (Fig. 1). Most (90–100%) of the transgenic empty vector controls (54 plants in total) developed disease symptoms by about 3 days post‐inoculation (dpi), which were characterized by leaf distortion, mosaic and, in extreme cases, necrosis of the whole plant (Fig. 1A). Transgenic plant lines showing different levels of protection (0–100%) were observed across each RNAi construct studied (Fig. 1B). However, of the three RNAi constructs tested, that derived from FL‐CP (894 nts) was the most effective for conferring resistance to CBSUV‐[UG:Nam:04], such that five transgenic lines (of the seven tested), FL22, FL16, FL5, FL2 and FL17, showed 100% protection, with no symptoms appearing on any sap‐inoculated plant (54 plants for each line over three replicated experiments) over the 30‐day observation period (Fig. 1A,B). In the case of the N‐terminal construct, four of the seven lines (NT8, NT10, NT12 and NT14) showed 100% resistance and, in the case of the C‐terminal construct, four lines (CT6, CT7, CT10 and CT21) also showed 100% protection (Fig. 1B). Thus, among the three RNAi constructs targeting different regions of CBSUV‐[UG:Nam:04]CP, FL‐CP gave the highest frequency of protection, with 85% of the plant lines produced demonstrating complete resistance to the pathogen.
Figure 1.
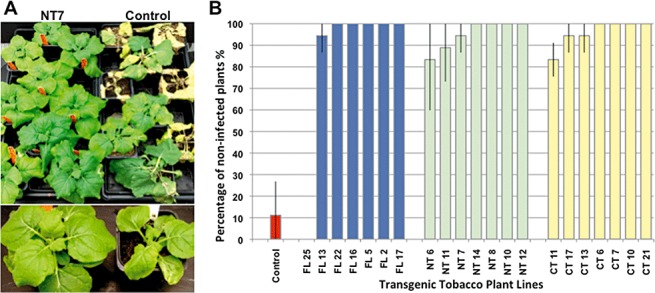
Screening of Nicotiana benthamiana transgenic lines in the T2 generation for resistance against CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus). (A) Phenotype of resistant line NT7 derived from the RNAi construct targeting the N‐terminus of CBSUV‐[UG:Nam:04]/CP, showing 100% protection (left) when compared with complete infection in control plants (right). Individual plants are enlarged and shown in the bottom panel. (B) Different levels of resistance obtained in N. benthamiana transgenic lines derived from the RNAi constructs targeting full‐length (FL; left), N‐terminal (NT; centre) and C‐terminal (CT; right) parts of CBSUV‐[UG:Nam:04]/CP in the T2 generation after challenge with CBSUV‐[UG:Nam:04] at 30 days post‐inoculation (dpi).
Analysis of expression levels, siRNA accumulation and correlation with resistance levels
Levels of mRNAs and siRNAs specific to the CP gene, produced by the different transgenic lines displaying varying levels of resistance, were quantified before challenge by the CBSUV‐[UG:Nam:04] pathogen, and the blots were scanned and analysed statistically (Figs 2 and S3, see Supporting Information). All transgenic plant lines which subsequently showed a high level of resistance to the pathogen were found to accumulate high levels of siRNA (Fig. 2A). The multiple forms of the transcripts seen in Northern blot hybridizations (Fig. 2A) of the transgenic lines can be attributed to intermediate products of mRNA as a result of Dicer action during PTGS (Ruiz‐Ferrer and Voinnet, 2009). Conversely, no detectable transcript or corresponding siRNAs were detectable in plant lines showing no protection against CBSUV‐[UG:Nam:04] infection (Fig. 2A,B). The analysis of these two quantitative criteria across all three constructs showed a strong positive correlation (R= 92%) between the amount of transcript produced by the transgenic lines and the amount of CP‐specific siRNA accumulated (Fig. S3A). Similarly, we found a positive correlation of R= 86% between the amount of CBSUV‐[UG:Nam:04] siRNAs present in the transgenic plants before viral challenge and the subsequent level of resistance to CBSUV‐[UG:Nam:04] exhibited by each transgenic plant line.
Figure 2.
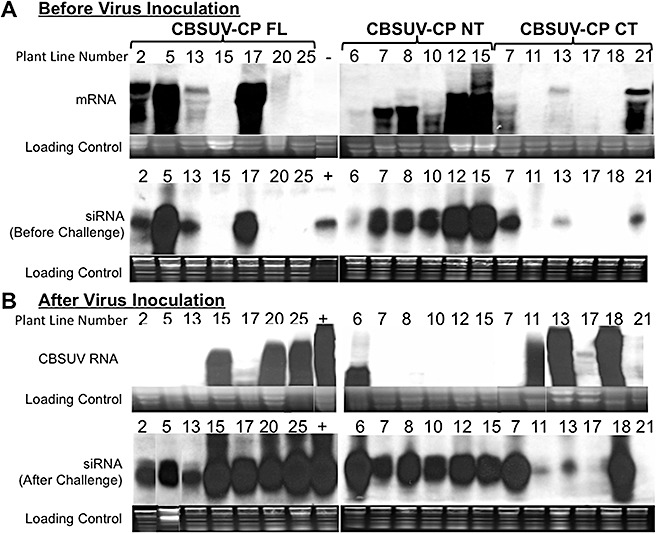
Molecular analysis of Nicotiana benthamiana transgenic plant lines (line numbers indicated for each lane) derived from three RNAi constructs targeting different regions of CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus) coat protein (CP) [N‐terminus (NT), C‐terminus (CT) and full‐length (FL)] in the T2 generation. (A) Accumulation of mRNAs and siRNA as detected by Northern blot hybridization before CBSUV‐[UG:Nam:04] challenge. (B) Accumulation of viral RNA and siRNA after challenge with CBSUV‐[UG:Nam:04] in the same transgenic lines. ‘−’ in (A) is a nontransgenic healthy plant; ‘+’ in (A) is a small RNA from transient expression by agroinfiltration of the FL‐CP construct; ‘+’ in (B) is a nontransgenic virus‐infected control.
Virus accumulation and correlation with resistance levels
Four weeks after sap inoculation with the virus, all CBSUV‐[UG:Nam:04]‐resistant and ‐susceptible transgenic plant lines were analysed for the accumulation of viral RNA by Northern blot hybridization (Fig. 2B). Viral RNA was not detected in resistant plant lines observed to be free of viral symptoms, but viral RNA accumulated to varying levels in symptomatic plant lines and at significant levels in infected, nontransgenic plants (Fig. 2B). The same plant lines were analysed for the accumulation of siRNA in response to virus inoculation, 4 weeks after CBSUV‐[UG:Nam:04] inoculation. A positive correlation (85%) was observed between the amount of siRNA produced by the transgenic lines before and after CBSUV‐[UG:Nam:04] inoculation (Fig. S2, see Supporting Information).
Resistance of transgenic N. benthamiana to diverse, nonhomologous isolates of CBSUV and CBSV
In order to test the resistance of these transgenic tobacco lines to different CBSUV and CBSV isolates, we selected six isolates with different virulence originating from different regions in East Africa. Cloning and sequence analysis of the entire CP genes isolated from CBSUV‐ and CBSV‐infected cassava collected at Kabanyoro in Uganda (CBSUV‐[UG:Kab:07]), Kibaha in Tanzania (CBSUV‐[TZ:Kib:07]), Mwalumba in Kenya (CBSUV‐[KE:Mwa:07]), Zanzibar in Tanzania (CBSV‐[TZ:Zan:07]), Naliendele in Tanzania (CBSV‐[TZ:Nal:07]) and Nampula in Mozambique (CBSV‐[MZ:Nam:07]) revealed that they pertained to both species (Table 1). After initial transmission of these viruses from cassava to the model host N. benthamiana, the identity of the original CBSUV and CBSV isolates was re‐confirmed by cloning and re‐sequencing, thus ruling out the possibility of cross‐contamination. Selected transgenic N. benthamiana lines derived from each construct were challenged with these six diverse CBSUV and CBSV isolates. Transgenic plant lines for challenge experiments were selected to represent different expression levels of siRNA in FL‐CP (FL2, FL5, FL13 and FL17), NT‐CP (NT7 and NT8) and CT‐CP (CT7 and CT21). The determination of disease development 4 weeks after virus challenge showed that two lines, FL5 and FL17, derived from the 894‐nt region of CP, were completely resistant to infection across all six isolates of CBSUV and CBSV, whereas the vector control plants showed 90–100% infection with all six isolates (Fig. 3B,C). Both FL2 and FL13 displayed resistance at approximately 40–95% to the homologous virus CBSUV‐[UG:Nam:04] and other isolates most closely related to CBSUV‐[UG:Nam:04], namely CBSUV‐[UG:Kab:07] (99.4% identity) and CBSUV‐[KE:Mwa:07] (94.3%), but showed lower levels of resistance (20–65%) to the CBSV isolates (Fig. 3B,C).
Table 1.
Percentage identity levels (nucleotides, nts) of coat protein (CP) sequences of Cassava brown streak Uganda virus (CBSUV) and Cassava brown streak virus (CBSV) isolates used to challenge transgenic plants with the CP of CBSUV‐[UG:Nam:04].
| Percentage identity with CP fragments of CBSUV‐[UG:Nam:04] | CBSUV | CBSV | ||||
|---|---|---|---|---|---|---|
| [UG:Kab:07] | [TZ:Kib:07] | [KE:Mwa:07] | [TZ:Zan:07] | [TZ:Nal:07] | [MZ:Nam:07] | |
| Country of origin | Uganda | Tanzania | Kenya | Tanzania | Tanzania | Mozambique |
| Symptom severity | Mild | Mild | Moderate | Moderate | Severe | Severe |
| CP‐FL (894 nts) | 99.4 | 91.1 | 94.3 | 75.8 | 76.1 | 75.7 |
| CP‐NT (397 nts) | 99.0 | 89.4 | 93.7 | 70.5 | 71.0 | 70.5 |
| CP‐CT (491 nts) | 99.8 | 92.5 | 95.3 | 80.7 | 80.7 | 80.2 |
CT, C‐terminus; FL, full length; nts, nucleotides; NT, N‐terminus.
Figure 3.
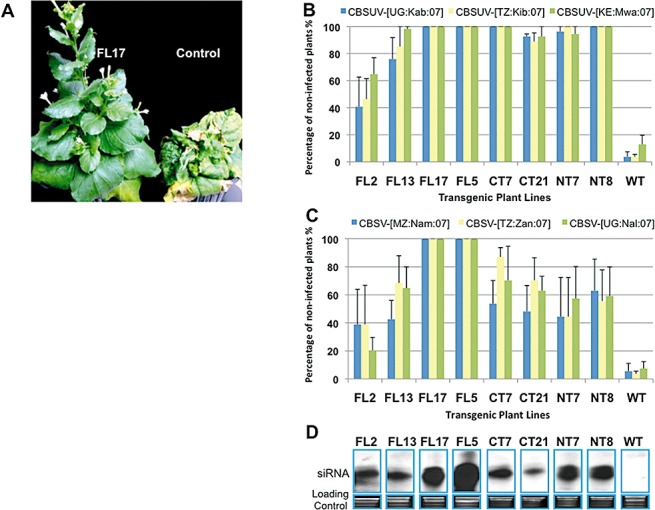
Screening of Nicotiana benthamiana transgenic lines in the T2 generation for resistance against six distinct Cassava brown streak Uganda virus (CBSUV) and Cassava brown streak virus (CBSV) isolates. (A) Phenotype of a resistant line FL17 derived from the RNAi construct targeting the 894 nts of CBSV‐UG[UG:Nam:04]/CP, showing 100% protection (left) when compared with complete infection in control plants (right) when challenged with CBSV‐[TZ:Zan:07]. (B, C) Different levels of protection obtained in N. benthamiana transgenic lines derived from RNAi constructs targeting full‐length (FL; left), C‐terminal (CT; centre) and N‐terminal (NT; right) parts of CBSV‐UG[UG:Nam:04]/CP in the T2 generation after challenge with six different isolates of CBSUV and CBSV. (B) shows protection for CBSUV isolates (CBSUV‐[UG:Kab:07], CBSUV‐[TZ:Kib:07] and CBSUV‐[KE:Mwa:07]) and (C) shows protection for CBSV isolates (CBSV‐[MZ:Nam:07], CBSV‐[TZ:Zan:07] and CBSV‐[TZ:Nal:07]). Different levels of siRNA expressed by these lines positively correlate (R= 0.86) with their resistance levels, as shown in (D).
In the case of the transgenic plants expressing CT‐CP, CT7 and CT21 showed high resistance (100% and ∼80–100%, respectively) to all CBSUV isolates, but had low and varying resistance (20–90%) towards CBSV isolates (Fig. 3B,C). Plant lines derived from NT‐CP, NT7 and NT8, showed a high level of resistance (∼90–100%) to CBSUV isolates (Fig. 3B,C), but succumbed to the CBSV isolates. In general, the lines derived from FL‐CP showed the highest spectrum of resistance to diverse and nonhomologous isolates of CBSUV and CBSV, followed by plant lines derived from the C‐terminal region, which is more conserved than the N‐terminal region, with the latter showing the least resistance to CBSV isolates (Fig. 3C).
Reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis of CBSUV‐ and CBSV‐challenged transgenic N. benthamiana
Total RNA was extracted from both symptomatic and nonsymptomatic transgenic plants at 30 dpi, and RT‐PCR was performed to detect CBSUV or CBSV CP, using specific primers designed to amplify 1101 nts (Fig. 4A) of the CBSUV isolates (CBSUV‐[UG:Kab:07], CBSUV‐[TZ:Kib:07] and CBSUV‐[KE:Mwa:07]) and 1134 nts (Fig. 4B) of the CBSV isolates (CBSV‐[TZ:Zan:07], CBSV‐[TZ:Nal:07] and CBSV‐[MZ:Nam:07]). Amplification of viral RNA was seen from both symptomatic control and transgenic plants, but was not detected in extracts from symptom‐free transgenic plants after challenge with all six CBSUV and CBSV isolates, indicating that the virus did not move systemically from the inoculation point in such plants, and these plants remained free of the pathogen over the 30‐dpi observation period (Fig. 4A,B).
Figure 4.
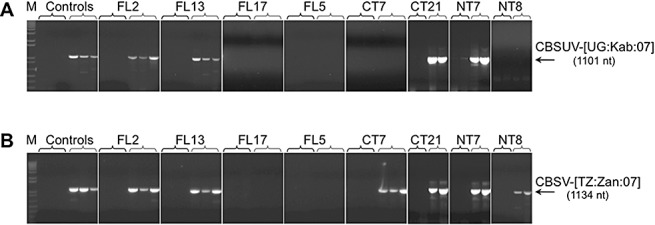
Reverse trancriptase‐polymerase chain reaction (RT‐PCR) for the detection of the coat protein (CP) of Cassava brown streak Uganda virus (CBSUV) (A) and Cassava brown streak virus (CBSV) (B) isolates. Specific primers were used to amplify 1101 nts of CP for the CBSUV isolates (CBSUV‐[UG:Kab:07], CBSUV‐[TZ:Kib:07] and CBSUV‐[KE:Mwa:07]) and 1134 nts of CP for the CBSV isolates (CBSV‐[TZ:Zan:07], CBSV‐[TZ:Nal:07] and CBSV‐[MZ:Nam:07]) from the total RNA extracts for both symptomatic (first three/two lanes for each transgenic plant line) and nonsymptomatic (next three/two lanes) transgenic plants after challenge of transgenic N. benthamiana with CBSUV‐[UG:Kab:07] and CBSV‐[TZ:Zan:07].
In silico analysis for the presence of cross‐protecting siRNAs in the CBSUV CP sequence
In order to determine why effective cross‐protection was achieved against all nonhomologous CBSV isolates in the transgenic lines FL5 and FL17, we investigated the number of mismatches among theoretically possible siRNAs in the 894‐nt CP sequence (CBSUV‐[UG:Nam:04]) using the program MAQ (http://maq.sourceforge.net). A total of 875 types of 21‐nt siRNAs and 874 types of 22‐nt siRNAs were predicted by the analysis. No single siRNA with complete identity to any of the three CBSV isolates (CBSV‐[TZ:Zan:07], CBSV‐[TZ:Nal:07] and CBSV‐[MZ:Nam:07]) used in the present study was found, but 15, 14 and 12 siRNAs with a 1‐nt mismatch and 64, 78 and 63 siRNAs with a 2‐nt mismatch were identified for CBSV‐[TZ:Zan:07], CBSV‐[TZ:Nal:07] and CBSV‐[MZ:Nam:07], respectively, having nucleic acid identities of 75.8%, 76.1% and 75.7%, respectively, with the CBSUV‐[UG:Nam:04] FL‐CP transgene sequence; for CT‐CP and NT‐CP, these numbers were significantly less (Table 2). In the case of the three CBSUV isolates (CBSUV‐[UG:Kab:07], CBSUV‐[TZ:Kib:07] and CBSUV‐[KE:Mwa:07]), for which CBSUV‐[UG:Nam:04] showed 99.4%, 91.1% and 94.3% identities, respectively, 769, 101 and 270 types of siRNA with complete homology were found for FL‐CP, in contrast with a significantly lower number of siRNAs with complete homology for CT‐CP and NT‐CP (Table 2).
Table 2.
Number of in silico‐predicted small interfering (si)RNAs (21 nt and 22 nt) for the three regions of CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus) coat protein (CP) [full‐length (894 nt), N‐terminal (397 nt) and C‐terminal (491 nt)] used to prepare the RNAi constructs against the corresponding regions of the six distinct CBSUV and Cassava brown streak virus (CBSV) isolates used in screening the resistance levels.
| Size of siRNA | Number of mismatches | CBSUV | CBSV | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [UG:Kab:07] | [TZ:Kib:07] | [KE:Mwa:07] | [TZ:Zan:07] | [TZ:Nal:07] | [MZ:Nam:07] | ||||||||||||||
| FL | NT | CT | FL | NT | CT | FL | NT | CT | FL | NT | CT | FL | NT | CT | FL | NT | CT | ||
| 21 nt | 0 | 769 | 314 | 429 | 101 | 48 | 51 | 270 | 72 | 197 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 105 | 63 | 42 | 255 | 90 | 157 | 272 | 140 | 127 | 15 | 0 | 15 | 14 | 0 | 14 | 12 | 6 | 6 | |
| 2 | 0 | 0 | 0 | 312 | 126 | 174 | 244 | 120 | 103 | 64 | 8 | 56 | 78 | 23 | 55 | 63 | 11 | 52 | |
| Total | 874 | 377 | 471 | 668 | 264 | 382 | 786 | 332 | 427 | 79 | 8 | 71 | 92 | 23 | 69 | 75 | 17 | 58 | |
| 22 nt | 0 | 763 | 311 | 426 | 87 | 43 | 43 | 255 | 67 | 188 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 110 | 65 | 44 | 246 | 86 | 152 | 265 | 134 | 127 | 11 | 0 | 11 | 9 | 0 | 9 | 8 | 4 | 4 | |
| 2 | 0 | 0 | 0 | 308 | 125 | 171 | 256 | 126 | 109 | 53 | 5 | 48 | 65 | 18 | 47 | 51 | 10 | 41 | |
| Total | 873 | 376 | 470 | 641 | 254 | 366 | 776 | 327 | 424 | 64 | 5 | 59 | 74 | 18 | 56 | 59 | 14 | 45 | |
nt, nucleotide.
DISCUSSION
Of the available biotechnological approaches to control plant viral diseases, RNAi is a very promising strategy that has been successfully employed to control numerous viral diseases caused by both DNA and RNA viruses (Tenllado et al., 2004; Waterhouse and Fusaro, 2006). Of the various viral sequences tested, the CP gene, which has multiple functions in the pathogen's life cycle, has been utilized as a transgene to control plant RNA virus diseases (Callaway et al., 2001; Jan et al., 1999; Ling et al., 2008; Scorza et al., 2001; Urcuqui‐Inchima et al., 2001). However, the nature of the virus and its life cycle in the host are important in determining whether it will be subject to, or evade, RNAi, as reported for Turnip mosaic virus (potyvirus; Jan et al., 1999), Citrus tristeza virus (closterovirus; Olivares‐Fuster et al., 2003) and Cucumber green mottle mosaic virus (tobamovirus; Kamachi et al., 2007).
Among the few ipomoviruses molecularly characterized today, CBSV and CBSUV are unique in their genome organization because of the absence of the characteristic potyvirus silencing suppressor HcPro (Helper component proteinase) and the presence of HAM1h, a rare gene among plant viruses, hypothesized to prevent mutations in the viral genome by intercepting the incorporation of noncanonical nucleoside triphosphates (Galperin et al., 2006; Mbanzibwa et al., 2009a). The only other plant virus in which the presence of a Ham1h‐like sequence has been reported is Euphorbia ringspot virus (EuRSV; National Center for Biotechnology Information Acc. No. AY397600), which also infects plants of the Euphorbiaceae family (Mbanzibwa et al., 2009a). For these reasons, it is important to verify whether RNAi can control ipomoviruses, such as CBSUV and CBSV, and whether the CP gene could be an effective target to achieve this goal via transgenic expression. We employed RNAi to control CBSD in the model host N. benthamiana, in which screening of CBSUV and CBSV resistance can be performed easily by mechanical sap inoculation (Mbanzibwa et al., 2009b; Monger et al., 2001a).
To determine the role of the size and sequence of the CBSUV CP required to achieve effective RNAi control of the viral pathogen, three RNAi constructs were designed to target the FL (894 nts), N‐terminal (397 nts) and C‐terminal (491 nts) regions of the CBSUV‐[UG:Nam:04]CP gene. Transgenic lines derived from each of the three constructs showed a diverse range of siRNA accumulation and resistance to CBSUV‐[UG:Nam:04], with the lines derived from FL‐CP conferring the highest protection (1, 2), followed by similar levels of resistance by lines derived from both N‐terminal and C‐terminal regions. Similar to the results observed here, comparative studies with the N‐ and C‐terminal halves of the S4 and S12 genes of Rice dwarf virus showed no difference in the efficacy of levels of viral protection through RNAi (Shimizu et al., 2009).
In the present study, a positive and significant correlation (R= 86%) was observed between the presence of siRNAs, the hallmark of RNAi, and the levels of protection against CBSUV‐[UG:Nam:04] (1, 2). Most of the transgenic plant lines showing high levels of detectable transcript also produced significant levels of siRNA and displayed strong resistance to CBSUV‐[UG:Nam:04] (Fig. 2A). A highly significant positive correlation was also found between the level of CBSUV‐[UG:Nam:04]/CP mRNA expression and siRNA accumulation (R= 92%), and between such siRNA levels and CBSUV‐[UG:Nam:04] protection (R= 86%), indicating that these molecular criteria could potentially be of value as screening tools in subsequent studies, for example in cassava, to identify transgenic plant lines with high levels of resistance to CBSD viral agents (Figs 2A and S3).
There was a strong positive correlation between the levels of siRNA before and after CBSUV‐[UG:Nam:04] inoculation. The increase in siRNA accumulation after virus inoculation was insignificant (less than two‐fold), indicating that the highly expressing plant lines efficiently responded to viral inoculum through gene silencing (Fig. 2B). This observation, associated with the very high level of resistance obtained (100%), indicates that this technology is very effective for controlling CBSV in tobacco.
In order to determine whether the plants from transgenic lines that remained asymptomatic after challenge with the pathogen were free of virus, such plants were studied by Northern blot hybridization and RT‐PCR. Analysis showed that these highly resistant plants did not contain detectable amounts of CBSUV‐[UG:Nam:04] or CBSV‐[TZ:Zan:07]. This is considered to be important because the development and deployment of tolerant plants that display no symptoms, but carry a high virus load, would be undesirable with regard to the field level control of the disease, as they could be a potential source of virus acquisition by whiteflies.
Phylogenetic studies from East Africa have shown that there are at least two distinct species of virus causing the same symptomatology, and several different strains associated with CBSD in this region (Mbanzibwa et al., 2009a; Winter et al., 2010). If strategies to deploy improved germplasm are to succeed, they must deliver plant materials possessing high levels of resistance to all CBSVs. An important drawback of RNAi is that it is usually highly sequence specific, and viruses with more than 10% nucleotide diversity are mostly insensitive to this technology; this necessitates the combination of fragments from different virus species to make an RNAi construct with broad‐spectrum resistance (Prins, 2003). The use of such a large set of genes or their fragments in one single construct could be unstable during the cloning and transformation processes and, more importantly, in their expression and ability to form perfect hairpins within the transgenic plant.
However, the challenge of transgenic plants produced in this study with isolates collected from Uganda, Kenya, Tanzania and Mozambique (Table 1) showed significant cross‐protection imparted to both virus species. Two transgenic plant lines derived from FL‐CP remained completely symptom free and apparently immune against all six diverse isolates of CBSUV and CBSV tested. The lines derived from CT‐CP also showed low to high resistance (∼20–100%) to these six isolates (Fig. 3). Conversely, the two high‐expressing lines derived from NT‐CP were susceptible to the nonhomologous isolates from Tanzania (CBSV‐[TZ:Nal:07]), Zanzibar (CBSV‐[TZ:Zan:07]) and Mozambique (CBSV‐[MZ:Nam:07]) (Fig. 3C). Transgenic CT‐CP imparted superior resistance to nonhomologous isolates than did NT‐CP, most probably because the C‐terminal region of CBSUV and CBSV CP is more conserved than the N‐terminal region. For some of the plant lines, there was a large amount of variation in resistance levels across different experiments, which could be attributed to variation in the quality and amounts of virus inoculum used for each experiment during sap inoculations.
The two FL‐CP lines (FL5 and FL17) which exhibited immunity or very strong resistance to all of the CBSUV and CBSV isolates were shown to express the highest levels of mRNA and siRNA of all the FL‐CP transgenic plant lines tested (Fig. 2), whereas the two FL‐CP lines (FL2 and FL13) that did not show significant resistance were poor expressers of siRNAs (Fig. 3D). It is possible that the FL‐CP lines gave the best resistance to all tested viruses because they had the largest fragment of CP, capable of producing a diverse and larger population of siRNAs to target both the CBSUV and CBSV isolates, when compared with the smaller sized NT‐CP and CT‐CP. In silico prediction for siRNA across the CPs of CBSUV‐[UG:Nam:04] and the other three CBSV isolates showed no siRNA with complete identity and only ∼14 and ∼64 siRNAs with one and two mismatches, respectively (Table 2). These results indicate that complete identity between siRNAs and the target RNA is not essential for the induction of gene silencing. More important is the presence of perfect complementarity with the 6‐nt seed region (nts 2–7) of siRNAs for their binding to the target RNA and further processing (Jackson et al., 2006). Previous studies in HeLa cells using synthetic siRNAs have demonstrated that perfect complementarity with a specific target at the 5′ end of the antisense strand of siRNA was sufficient to direct siRNA–target binding and cleavage (Haley and Zamore, 2004; Jackson et al., 2006).
It was encouraging to see that, in addition to the absence of disease symptoms, plants challenged with different isolates of CBSUV and CBSV remained virus free as determined by RT‐PCR (Fig. 4). We consider this to be the first demonstration of cross‐protection against a plant virus with no single siRNA from the transgene having complete identity to the target virus, and, in addition, the level of cross‐protection was positively correlated with siRNA expression levels. These results encourage continued research into the use of RNAi technology as an effective biotechnological tool to control the dramatic impact of CBSD in farmers' fields.
EXPERIMENTAL PROCEDURES
RNAi vector construction
Fragments of the CP gene were PCR amplified from CBSUV‐[Uganda:Namulonge:2004]‐infected cassava plants collected from Namulonge, Uganda (hereafter abbreviated to CBSUV‐[UG:Nam:04]) as full‐length (FL; 894 nts: nts 208–1101 of CP), N‐terminal (NT; 397 nts: nts 208–604 of CP) and C‐terminal (CT; 491 nts: nts 611–1101 of CP) fragments. The actual size of the CP open reading frame (ORF) for CBSV‐UG is 1101 nts, but, when this project was initiated, only the 894‐nt sequence of CBSUV‐[UG:Nam:04]CP was available; thus, for convenience, we refer here to the 894 nts of CP as the full‐length (FL or FL‐CP) sequence. These sequences were cloned in sense orientation between the KpnI and XbaI restriction sites and in antisense orientation between the BamHI and BstBI sites on either ends of the Pdk‐intron derived from the vector pHELLSGATE (from Dr P. M. Waterhouse, CSIRO, Plant Industry, Canberra, Australia; National Center for Biotechnology Information Acc. No. AJ311874) (Table S1). In order to generate stable transgenic plants, the RNAi cassette driven by the Cassava vein mosaic virus (CsVMV) promoter (Verdaguer et al., 1998) was subcloned into the binary vector pCambia2300 (Acc. No. AF234315) and transformed into Agrobacterium tumefaciens strain LBA4404.
Production of transgenic N. benthamiana, and PCR analysis
Nicotiana benthamiana plants were transformed by the leaf disc method using A. tumefaciens (Horsch et al., 1985), and regenerated putative transgenic plants were transferred to pots and maintained in the glasshouse at 28 °C. T0 plants were screened at 8 weeks of age by PCR using specific primers (Table S1) and the PCR‐positive lines were maintained for the production of the homozygous T2 generation.
Screening for CBSUV‐[UG:Nam:04] resistance by sap inoculation
T2 transgenic homozygous lines were grown in the glasshouse and sap inoculated with CBSUV‐[UG:Nam:04] inoculum obtained from CBSUV‐[UG:Nam:04]‐infected N. benthamiana plants. Sap extract was prepared in potassium phosphate buffer (0.01 m; pH 7.0) containing antioxidants (0.01 mβ‐mercaptoethanol and 0.2 m sodium sulphite) and gently rubbed onto two completely opened young leaves with the help of carborundum as an abrasive agent (Mandal et al., 2001; Mbanzibwa et al., 2009b). For each independent transgenic line, 18 plants were challenged and the inoculated plants were maintained in a growth chamber at 28 °C with 200 µE/m2/s of light and 70% relative humidity, and monitored for symptom development every alternate day for a period of 4 weeks. All viral challenges were performed as three replications. Disease symptom scoring was carried out using a scale of 0–5 (Fargette et al., 1993) and analysed to determine resistance levels for each plant line. Plants transformed with the empty vector pCambia2300 were used as negative controls.
CBSUV and CBSV isolates used for screening resistance in transgenic N. benthamiana
The six virus isolates collected from different locations in East Africa, namely Kabanyoro in Uganda (CBSUV‐[Uganda:Kabanyoro:2007] or CBSUV‐[UG:Kab:07]), Kibaha in Tanzania (CBSUV‐[Tanzania:Kibaha:2007] or CBSUV‐[TZ:Kib:07]), Mwalumba in Kenya (CBSUV‐[Kenya:Mwalumba:2007] or CBSUV‐[KE:Mwa:07]), Zanzibar in Tanzania (CBSV‐[Tanzania:Zanzibar:2007] or CBSV‐[TZ:Zan:07]), Naliendele in Tanzania (CBSV‐[Tanzania:Naliendele:2007] or CBSV‐[TZ:Nal:07]) and Nampula in Mozambique (CBSV‐[Mozambique:Nampula:2007] or CBSV‐[MZ:Nam:07]), which also represented both lowland and highland regions, were used to challenge the transgenic plants (Table 1). The complete genes of the CP of all isolates (with a size of 1101 nts for CBSUV isolates and 1134 nts for CBSV isolates) were cloned from infected cassava plants, and the sequences were analysed for identity to the CBSUV‐[UG:Nam:04]‐CP sequence used to generate the transgenic tobacco plants. These isolates were further sap transmitted to N. benthamiana plants for screening the transgenic lines, and their identity was confirmed by re‐sequencing of CP.
Analyses of plants for viral mRNA and siRNA before and after virus challenge
Transgenic N. benthamiana lines, together with controls, were analysed for transcript production, siRNA accumulation and virus accumulation by Northern blot hybridization (Sambrook and Russell, 2001), both before and after challenge with CBSUV‐[UG:Nam:04]. Total RNA was isolated from leaves using Trizol (Invitrogen, Carlsbad, CA, USA), and subjected to electrophoresis and Northern blot hybridization for the detection of transcript and viral RNA. However, for the detection of siRNA, total RNA was fractionated using an RNeasy plant mini kit (Qiagen, Valencia, CA, USA) and the RNA clean‐up protocol (Akbergenov et al., 2006). Ten micrograms of small RNAs were electrophoresed on a 15% TBE (Tris Borate EDTA) urea gel (Criterion‐Bio‐Rad, Hercules, CA, USA), and subjected to Northern blot hybridization with a hydrolysed probe obtained by the in vitro transcription of CBSUV‐CP using a SP6/T7 Transcription Kit (Roche Applied Science, Indianapolis, IN, USA). The probe used for the detection of transcript and viral RNA was PCR amplified CBSUV‐[UG:Nam:04]/CP DNA generated by the DIG High Prime DNA labelling and detection kit (Roche Applied Science). The membranes were processed and the signal was detected using CDP‐star (Roche Applied Science), as described in the DIG system and the DIG application manual. The signal intensities were quantified by ImageJ (version 1.36b) software over a uniform selected area for each transgenic line, and analysed statistically to determine the correlation coefficients between mRNA and siRNA, and siRNA and resistance.
RT‐PCR analysis of CBSUV‐ and CBSV‐challenged transgenic plants
Using an RNeasy mini kit (Qiagen), total RNA was extracted from transgenic plants challenged with different isolates of CBSUV and CBSV and subjected to RT‐PCR (Invitrogen SuperScript® III First‐Strand RT‐PCR kit) by primers amplifying the 1101/1134 nts of the entire CP of CBSUV/CBSV isolates from highland/lowland regions. A pair of primers to amplify the constitutively expressed tubulin (primers TobefS and TobefA; Table S1) was also used as a control to check the quality of cDNA synthesized in the RT‐PCRs.
In silico analysis of possible siRNAs in the CBSUV and CBSV CP sequences
To analyse the theoretical possibility of siRNAs derived from the inverted repeat constructs targeting different regions of CBSUV‐CP, the sequence of the transgene was processed in silico into every possible 21‐, 22‐ and 24‐nt RNA using a custom‐prepared script in the Python programming language. The theoretical siRNA libraries derived from the constructs were mapped against sequences of each of the viruses using the program MAQ (http://sourceforge.net/projects/maq/), and the results were imported into an Excel file for further analysis. The number and position of theoretical siRNAs homologous to each virus strain with zero, one and two mismatches were tabulated, and the positions of common siRNAs were recorded (Table 2).
Supporting information
Fig. S1 Characteristic cassava brown streak disease (CBSD) symptoms visible on different parts of the cassava plant: (A) feathery chlorosis on cassava leaf; (B) root necrosis; (C) brown streaks on young cassava stem.
Fig. S2 Accumulation of small interfering (si)RNA before and after challenge with CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus) in Nicotiana benthamiana transgenic lines as detected by Northern hybridization. The relative signal intensities of siRNA against the background of the autoradiograms were quantified by ImageJ software for a uniform selected area for each transgenic line, and analysed statistically to determine the correlation coefficients.
Fig. S3 Linear regression analysis to establish the correlation between small interfering (si)RNA and mRNA (A) and the amount of siRNA and the resistance levels (B) in each transgenic plant line before and after challenge with CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus).
Table S1 Sequences of the primers used in making the RNAi constructs for CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus) coat protein (CP) [full length (FL), N‐terminal (NT) and C‐terminal (CT)] in sense (XbaI and KpnI) and antisense (BamHI and BstBI) orientation; other primers used in this study are also listed.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We acknowledge funding from Monsanto Fund and the United States Agency for International Development (USAID), which supported the work reported here. We also extend our gratitude to Kevin Lutke for the production of transgenic N. benthamiana, Theodore Moll for high‐quality technical assistance, Jorge Tamayo [International Potato Center (CIP), Peru] for help with in silico analyses of siRNA and the glasshouse staff at Donald Danforth Plant Science Center (DDPSC) for excellent care of the plants. We are grateful to Dr Chris Taylor for providing the silencing vector.
REFERENCES
- Akbergenov, R. , Si‐Ammour, A. , Blevins, T. , Amin, I. , Kutter, C. , Vanderschuren, H. , Zhang, P. , Gruissem, W. , Meins, F. Jr , Hohn, T. and Pooggin, M.M. (2006) Molecular characterization of geminivirus‐derived small RNAs in different plant species. Nucleic Acids Res. 34, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alicai, T. , Omongo, C.A. , Maruthi, M.N. , Hillocks, R.J. , Baguma, Y. , Kawuki, R. , Bua, A. , Otim‐Nape, G.W. and Colvin, J. (2007) Re‐emergence of cassava brown streak disease in Uganda. Plant Dis. 91, 24–29. [DOI] [PubMed] [Google Scholar]
- Callaway, A. , Giesman‐Cookmeyer, D. , Gillock, E.T. , Sit, T.L. and Lommel, S.A. (2001) The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39, 419–460. [DOI] [PubMed] [Google Scholar]
- Chellappan, P. , Masona, M.V. , Vanitharani, R. , Taylor, N.J. and Fauquet, C.M. (2004) Broad spectrum resistance to ssDNA viruses associated with transgene‐induced gene silencing in cassava. Plant Mol. Biol. 56, 601–611. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Sharkawy, M.A. (2003) Cassava biology and physiology. Plant Mol. Biol. 53, 621–641. [DOI] [PubMed] [Google Scholar]
- Fargette, D. , Jeger, M. , Fauquet, C. and Fishpool, L.D.C. (1993) Analysis of temporal disease progress of African cassava mosaic virus. Phytopathology, 84, 91–98. [Google Scholar]
- Galperin, M.Y. , Moroz, O.V. , Wilson, K.S. and Murzin, A.G. (2006) House cleaning, a part of good housekeeping. Mol. Microbiol. 59, 5–19. [DOI] [PubMed] [Google Scholar]
- Haley, B. and Zamore, P.D. (2004) Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11, 599–606. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A. and Waterhouse, P.M. (2005) Constructs and methods for hairpin RNA‐mediated gene silencing in plants. Methods Enzymol. 392, 24–35. [DOI] [PubMed] [Google Scholar]
- Hillocks, R.J. and Jennings, D.L. (2003) Cassava brown streak disease: a review of present knowledge and research needs. Int. J. Pest Manag. 49, 225–234. [Google Scholar]
- Hillocks, R.J. and Thresh, J.M. (2000) Cassava mosaic and cassava brown streak virus diseases in Africa: a comparative guide to symptoms and aetiologies. Roots, 7, 1–8. [Google Scholar]
- Hillocks, R.J. , Raya, M.D. , Mtunda, K. and Kiozia, H. (2001) Effects of brown streak virus disease on yield and quality of cassava in Tanzania. J. Phytopathol. 149, 389–394. [Google Scholar]
- Horsch, R.B. , Rogers, S.G. and Fraley, R.T. (1985) Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L. , Burchard, J. , Schelter, J. , Chau, B.N. , Cleary, M. , Lim, L. and Linsley, P.S. (2006) Widespread siRNA ‘off‐target’ transcript silencing mediated by seed region sequence complementarity. RNA, 12, 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, F.J. , Pang, S.Z. , Fagoaga, C. and Gonsalves, D. (1999) Turnip mosaic potyvirus resistance in Nicotiana benthamiana derived by post‐transcriptional gene silencing. Transgenic Res. 8, 203–213. [DOI] [PubMed] [Google Scholar]
- Jennings, D.L. (2003) Historical perspective on breeding for resistance to cassava brown streak virus disease In: Cassava Brown Streak Virus Disease: Past, Present, and Future, Proceedings of an International Workshop, Mombasa, Kenya, 27–30 October 2002 (Legg J.P. and Hillocks R.J., eds). Aylesford, UK: Natural Resources International Limited. [Google Scholar]
- Kamachi, S. , Mochizuki, A. , Nishiguchi, M. and Tabei, Y. (2007) Transgenic Nicotiana benthamiana plants resistant to cucumber green mottle mosaic virus based on RNA silencing. Plant Cell Rep. 26, 1283–1288. [DOI] [PubMed] [Google Scholar]
- Krubphachaya, P. , Juricek, M. and Kertbundit, S. (2007) Induction of RNA‐mediated resistance to papaya ringspot virus type W. J. Biochem. Mol. Biol. 40, 404–411. [DOI] [PubMed] [Google Scholar]
- Ling, K.S. , Zhu, H.Y. and Gonsalves, D. (2008) Resistance to Grapevine leafroll associated virus‐2 is conferred by post‐transcriptional gene silencing in transgenic Nicotiana benthamiana . Transgenic Res. 17, 733–740. [DOI] [PubMed] [Google Scholar]
- Lister, R.M. (1959) Mechanical transmission of cassava brown streak virus. Nature, 183, 1588–1589. [DOI] [PubMed] [Google Scholar]
- Mandal, B. , Pappu, H.R. and Culbreath, A.K. (2001) Factors affecting mechanical transmission of Tomato spotted wilt virus to peanut (Arachis hypogaea). Plant Dis. 85, 1259–1263. [DOI] [PubMed] [Google Scholar]
- Maruthi, M.N. , Hillocks, R.J. , Mtunda, K. , Raya, M.D. , Muhanna, M. , Kiozia, H. , Rekha, A.R. , Colvin, J. and Thresh, J.M. (2005) Transmission of Cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 153, 307–312. [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y. , Mukasa, S.B. and Valkonen, J.P. (2009a) Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC‐Pro. J. Virol. 83, 6934–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y.P. , Tugume, A.K. , Mukasa, S.B. , Tairo, F. , Kyamanywa, S. , Kullaya, A. and Valkonen, J.P. (2009b) Genetically distinct strains of Cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch. Virol. 154, 353–359. [DOI] [PubMed] [Google Scholar]
- Monger, W.A. , Seal, S. , Cotton, S. and Foster, G.D. (2001a) Identification of different isolates of Cassava brown streak virus and development of a diagnostic test. Plant Pathol. 50, 768–769. [Google Scholar]
- Monger, W.A. , Seal, S. , Isaac, A.M. and Foster, G.D. (2001b) Molecular characterization of the Cassava brown streak virus coat protein. Plant Pathol. 50, 527–534. [Google Scholar]
- Monger, W.A. , Alicai, T. , Ndunguru, J. , Kinyua, Z.M. , Potts, M. , Reeder, R.H. , Miano, D.W. , Adams, I.P. , Boonham, N. , Glover, R.H. and Smith, J. (2010) The complete genome sequence of the Tanzanian strain of Cassava brown streak virus and comparison with the Ugandan strain sequence. Arch. Virol. 155, 429–433. [DOI] [PubMed] [Google Scholar]
- Mware, B. , Narla, R. , Amata, R. , Olubayo, F. , Songa, J. , Kyamanyua, S. and Ateka, E.M. (2009) Efficiency of cassava brown streak virus transmission by two whitefly species in coastal Kenya. J. Gen. Mol. Virol. 1, 40–45. [Google Scholar]
- Olivares‐Fuster, O. , Fleming, G.H. , Albiach‐Marti, M.R. , Gowda, S. , Dawson, W.O. and Crosser, J.W. (2003) Citrus tristeza virus (CTV) resistance in transgenic citrus based on virus challenge of protoplasts. In Vitro Cell Dev. Biol. Plant, 39, 567–572. [Google Scholar]
- Patil, B.L. and Fauquet, C.M. (2009) Cassava mosaic geminiviruses: actual knowledge and perspectives. Mol. Plant Pathol. 10, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, B.L. and Fauquet, C. (2010) Differential interaction between cassava mosaic geminiviruses and geminivirus satellites. J. Gen. Virol. 91, 1871–1882. [DOI] [PubMed] [Google Scholar]
- Pennisi, E. (2010) Armed and dangerous. Science, 327, 804–805. [DOI] [PubMed] [Google Scholar]
- Prins, M. (2003) Broad virus resistance in transgenic plants. Trends Biotechnol. 21, 373–375. [DOI] [PubMed] [Google Scholar]
- Prins, M. , Laimer, M. , Noris, E. , Schubert, J. , Wassenegger, M. and Tepfer, M. (2008) Strategies for antiviral resistance in transgenic plants. Mol. Plant Pathol. 9, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, D.V. , Sudarshana, M.R. , Fuchs, M. , Rao, N.C. and Thottappilly, G. (2009) Genetically engineered virus‐resistant plants in developing countries: current status and future prospects. Adv. Virus Res. 75, 185–220. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. and Voinnet, O. (2009) Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scorza, R. , Callahan, A. , Levy, L. , Damsteegt, V. , Webb, K. and Ravelonandro, M. (2001) Post‐transcriptional gene silencing in plum pox virus resistant transgenic European plum containing the plum pox potyvirus coat protein gene. Transgenic Res. 10, 201–209. [DOI] [PubMed] [Google Scholar]
- Shimizu, T. , Yoshii, M. , Wei, T. , Hirochika, H. and Omura, T. (2009) Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of Rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 7, 24–32. [DOI] [PubMed] [Google Scholar]
- Sudarshana, M.R. , Roy, G. and Falk, B.W. (2007) Methods for engineering resistance to plant viruses. Methods Mol. Biol. 354, 183–195. [DOI] [PubMed] [Google Scholar]
- Taylor, N. , Chavarriaga, P. , Raemakers, K. , Siritunga, D. and Zhang, P. (2004) Development and application of transgenic technologies in cassava. Plant Mol. Biol. 56, 671–688. [DOI] [PubMed] [Google Scholar]
- Tenllado, F. , Llave, C. and Diaz‐Ruiz, J.R. (2004) RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Res. 102, 85–96. [DOI] [PubMed] [Google Scholar]
- Tougou, M. , Furutani, N. , Yamagishi, N. , Shizukawa, Y. , Takahata, Y. and Hidaka, S. (2006) Development of resistant transgenic soybeans with inverted repeat‐coat protein genes of soybean dwarf virus. Plant Cell Rep. 25, 1213–1218. [DOI] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. [DOI] [PubMed] [Google Scholar]
- Verdaguer, B. , De Kochko, A. , Fux, C.I. , Beachy, R.N. and Fauquet, C. (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol. Biol. 37, 1055–1067. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M. and Fusaro, A.F. (2006) Plant science. Viruses face a double defense by plant small RNAs. Science, 313, 54–55. [DOI] [PubMed] [Google Scholar]
- Winter, S. , Koerbler, M. , Stein, B. , Pietruszka, A. , Paape, M. and Butgereitt, A. (2010) The analysis of Cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J. Gen. Virol. 91, 1365–1372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Characteristic cassava brown streak disease (CBSD) symptoms visible on different parts of the cassava plant: (A) feathery chlorosis on cassava leaf; (B) root necrosis; (C) brown streaks on young cassava stem.
Fig. S2 Accumulation of small interfering (si)RNA before and after challenge with CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus) in Nicotiana benthamiana transgenic lines as detected by Northern hybridization. The relative signal intensities of siRNA against the background of the autoradiograms were quantified by ImageJ software for a uniform selected area for each transgenic line, and analysed statistically to determine the correlation coefficients.
Fig. S3 Linear regression analysis to establish the correlation between small interfering (si)RNA and mRNA (A) and the amount of siRNA and the resistance levels (B) in each transgenic plant line before and after challenge with CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus).
Table S1 Sequences of the primers used in making the RNAi constructs for CBSUV‐[UG:Nam:04] (Ugandan isolate of Cassava brown streak Uganda virus) coat protein (CP) [full length (FL), N‐terminal (NT) and C‐terminal (CT)] in sense (XbaI and KpnI) and antisense (BamHI and BstBI) orientation; other primers used in this study are also listed.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item


