SUMMARY
Plant defence against pathogen attack typically incorporates an oxidative burst involving elevated levels of reactive oxygen species such as hydrogen peroxide. In the present study, we have used an in‐gel assay to monitor the activity of the hydrogen peroxide scavenging enzyme, catalase, during asexual development of Phytophthora nicotianae and during infection of host tobacco plants. In vitro, catalase activity is highest in sporulating hyphae; in planta, catalase activity increases dramatically about 8 h after host inoculation. We have cloned and characterized three catalase genes, designated PnCat1, PnCat2 and PnCat3, from P. nicotianae and identified their homologues in P. infestans, P. sojae and P. ramorum. In all three species, Cat2 is predicted to be targeted to the peroxisome and the other catalases are likely to be cytosolic. Quantitative real‐time PCR assessment of catalase transcripts during development and infection indicates that peroxisomal PnCat2 is the gene predominantly expressed, with transcript levels peaking in vitro in sporulating hyphae and in planta increasing dramatically during the first 24 h after inoculation of susceptible tobacco seedlings. Levels of tobacco catalase gene expression are significantly down‐regulated in susceptible tobacco 4, 8 and 24 h post‐inoculation and in resistant plants at 24 h post‐inoculation. Together, our results give evidence that during infection P. nicotianae increases its own peroxisomal catalase levels while concurrently down‐regulating host catalase expression. This behaviour is consistent with a role of pathogen catalase in counterdefence and protection against oxidative stress and of pathogen‐orchestrated enhanced plant cell death to support necrotrophic pathogen growth and plant colonization.
INTRODUCTION
Catalases, along with other scavenging enzymes for reactive oxygen species (ROS), such as superoxide dismutase (SOD) and ascorbate peroxidase, are of fundamental importance to the survival of organisms undergoing oxidative stress (Alscher et al., 2002; Mayer et al., 2001; Mittler, 2002; Natvig et al., 1996). ROS, such as hydrogen peroxide (H2O2), are produced as a by‐product of metabolism (Mittler, 2002) and in response to invading pathogens (Lamb and Dixon, 1997), and have been implicated in intra‐ and intercellular signalling (van Breusegem et al., 2001; Rhee et al., 2005; Vranováet al., 2002). Hydrogen peroxide can be broken down by a number of mechanisms (Mayer et al., 2001). In many species, catalase plays a fundamental role in limiting the damage caused by this ROS (Izawa et al., 1996; Kwok et al., 2004; Polidoros et al., 2001; Takahashi et al., 1997).
Catalases fall into three classes, the well‐characterized monofunctional catalases and bifunctional catalase‐peroxidases, both of which contain heme, and the non‐heme or manganese‐containing catalases. The monofunctional catalases exist mainly as tetramers, ranging in size from 200 to 330 kDa (Gouet et al., 1995). Although primarily located in the peroxisome (Mullen et al., 1997; Natvig et al., 1996), catalases are also found in the cytosol (Kwok et al., 2004) and are secreted in some species (Garre et al., 1998; Zhang et al., 2004). Catalase gene expression and enzyme activity are regulated in response to oxidative stress and are under developmental regulation (Frugoli et al., 1996; Garcia et al., 2003; Kawasaki et al., 1997). Bifunctional catalase‐peroxidases exist mainly as dimers consisting of identical subunits approximately 80 kDa in size (ng Welinder, 1991). Initially thought to be a prokaryotic enzyme, this enzyme has been identified in Aspergillus nidulans (Scherer et al., 2002) and in the wheat pathogen Septoria tritici (Levy et al., 1992). Non‐heme or manganese‐containing catalases exist as homohexamers and appear to be limited to bacteria (Rocha and Smith, 1995).
Catalase expression and activity change during plant–pathogen interactions. For example, decreases in plant catalase activity occur in resistant plants in response to attempted infection by viruses (Yi et al., 2003). It is thought that this allows H2O2 to accumulate, resulting in antimicrobial activity through strengthening of the plant cell wall, activation of defence genes, hypersensitive cell death and a subsequent halt to pathogen infection (Lamb and Dixon, 1997). In contrast, in susceptible hosts increases in host catalase activity have been observed (Havelda and Maule, 2000; Kużniak and Skłodowska, 2005; Niebel et al., 1995; Pompe‐Novaka et al., 2006), exogenously applied catalase can result in decreased hypersensitive cell death (Able et al., 2000) and increased penetration by pathogens of normally resistant hosts (Able et al., 2000; Borden and Higgins, 2002; Wu et al., 1995). These observations suggest that increased activity of either plant‐ or pathogen‐derived ROS scavenging enzymes at the infection site could be an important aspect of successful plant infection. Indeed, SODs have been identified as pathogenicity factors in Erwinia chrysanthemi (Santos et al., 2001) and Botrytis cinerea (Rolke et al., 2004), and pathogenic strains of Pseudomonas syringae have much higher levels of catalase activity than non‐pathogenic, closely related species (Klotz and Hutcheson, 1992). However, despite the presence of intracellular and secreted catalases in a number of plant pathogens (Garre et al., 1998; Petnicki‐Ocwieja et al., 2002), direct evidence for their role in the successful infection of susceptible hosts is limited (Xu and Pan, 2000).
Members of the genus Phytophthora of oomycetes are important plant pathogens that continue to devastate native ecosystems and agricultural crops worldwide (Erwin and Ribeiro, 1996; Hardham, 2005). Although Phytophthora species, like other oomycetes, are phylogenetically related to heterokont algae and other protists in the Stramenopile group (Gunderson et al., 1987; Sachay et al., 1993; Van de Peer and De Wachter, 1997), they share some morphological similarities with fungi and also display common pathogenicity mechanisms with those in phytopathogenic fungi and bacteria (Fellbrich et al., 2002; Kamoun et al., 1999; Randall et al., 2005). Despite the probable importance of ROS‐scavenging enzymes in development and plant–pathogen interactions, the identification and role of these enzymes in Phytophthora is limited. In the study reported in this paper, three catalase genes were isolated from P. nicotianae and their expression during development and the infection of susceptible and resistant tobacco analysed. Assessment of catalase activity and gene expression suggests that catalase plays a role in asexual development of P. nicotianae and in successful host infection. In addition, we demonstrate the down‐regulation of plant catalase genes during a successful infection of a susceptible host by P. nicotianae.
RESULTS
Catalase activity in P. nicotianae
Catalase activity in P. nicotianae was assessed in native in‐gel assays (Fig. 1). Analysis of extracts from four major stages of asexual development, namely vegetative hyphae, sporulating hyphae, zoospores and germinated cysts, showed that catalase activity was strong in hyphae sampled 8 h after sporulation had been induced, with a lower level of activity in vegetative hyphae (Fig. 1a). The band of activity in vegetative and sporulating hyphae was the same size as the catalase activity band seen in extracts of Nicotiana tabacum leaves treated with H2O2 (Fig. 1a), the monomer of which has a molecular weight of 56.8 kDa (Schultes et al., 1994), but was larger than that of bovine catalase, the tetramer of which has a molecular weight of 232 kDa (Allgood and Perry, 1986). In our hands, no bands of catalase activity could be seen in extracts of tobacco leaves that were not treated with H2O2. Catalase activity could not be detected in extracts from zoospores when 50 µg of total protein was loaded (as in the gel illustrated); however, a faint band the same size as that from hyphal extracts could be seen when gels were overloaded with 100 µg of total zoospore protein (results not shown).
Figure 1.
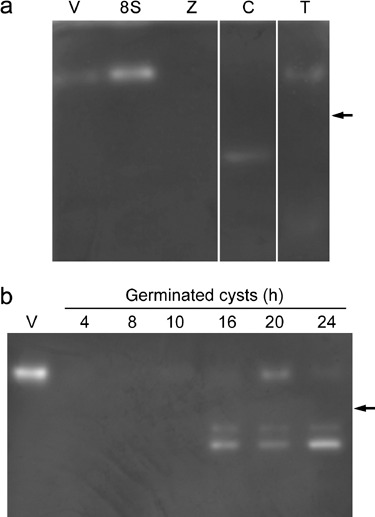
Catalase activity in P. nicotianae at different developmental stages. (a) A prominent catalase activity band occurs in extracts from hyphae sampled 8 h after induction of sporulation (8S). Activity bands of the same size but of lower intensity also occur in vegetative hyphae (V) and uninfected tobacco seedlings treated with H2O2 (T). A lower molecular weight activity band occurs in extracts from 2‐h germinated cysts (C). No catalase activity was detected in the zoospore protein extract (Z). (b) Catalase activity in germinated cysts, 4–24 h after germination. A band of catalase activity the same size as that from vegetative and sporulating hyphae extracts is present in cyst extracts 10–24 h after germination, with the highest level of activity occurring in the 20‐h sample. Two smaller bands of activity are visible in extracts from 16‐, 20‐ and 24‐h germinated cysts. The position of bovine catalase (232 kDa) is shown on the right (arrows).
In extracts of 2 h germinated cysts, a lower molecular weight band of activity was seen in some, but not all, experiments (Fig. 1a). Catalase activity in germinated cysts was investigated further by conducting the assay on extracts of cysts 4, 8, 10, 16, 20 and 24 h after cyst formation and germination (Fig. 1b). The experiment was repeated twice and in both cases an activity band the same size as that in hyphae could be seen in the 10–24‐h samples, and two lower molecular‐weight activity bands appeared in extracts from 16‐, 20‐ and 24‐h germinated cysts. The stronger of these bands corresponded to the band seen in some of the 2‐h germinated cyst extracts.
Identification and charactization of catalase genes from P. nicotianae
Three degenerate primer sets designed against three catalases identified in the DOE Joint Genome Institute (JGI) P. ramorum (scaffolds 31:333448‐335082, 31:462201‐463769, 39:298697‐300504) and P. sojae (scaffolds 8:298885‐300450, 8:505678‐507177, 8:617245‐619270) databases by keyword and tBlastn searches using fungal catalases were used to amplify three different DNA fragments from P. nicotianae genomic DNA (gDNA). Sequencing of these fragments confirmed that they were partial catalase genes and these were designated PnCat1, PnCat2 and PnCat3. Southern blot analysis of gDNA using these partial genes as probes showed a different hybridization pattern for each catalase (Fig. 2). As none of the restriction enzymes used to construct the Southern blot cut the probes and given that the homologous genes from the P. sojae and P. ramorum genome are at least 114 kb apart or on separate scaffolds, the banding patterns are consistent with the presence of a single copy of each catalase gene in the P. nicotianae genome. The presence of a faint second band in some lanes is due to a low level of cross‐hybridization between the probes for the three genes.
Figure 2.
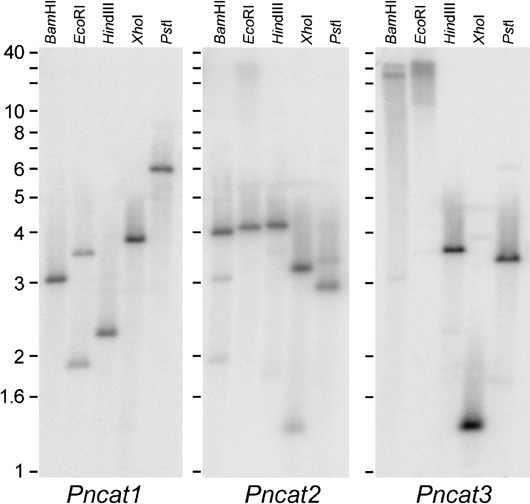
Southern blot analysis of catalase genes in P. nicotianae gDNA cut with the restriction enzymes BamHI, EcoRI, HindIII, XhoI and PstI. The same blot was probed with PCR fragments from PnCat1, stripped and reprobed with PnCat2 and subsequently reprobed with PnCat3. Numbers on the left indicate the approximate size in kb.
The PCR fragments for the three partial catalase genes were used to screen a P. nicotianae genomic DNA bacterial artificial chromosome (BAC) library. Catalase genes from two BACs that hybridized to each probe were fully sequenced using gene‐specific primers. PnCat1 has a single open reading frame as predicted by GENSCAN (GenBank accession number EU176856). Blast analysis of the Phytophthora genomic databases identified homologous genes in P. infestans (Picat1: PITT_15292), P. sojae (PsCat1: estExt_fgenesh1_pm.C_80008) and P. ramorum (Prcat1: gwEuk.31.29.1). The Prcat1 model did not predict a complete protein, but the identification of a putative promoter element (McLeod et al., 2004) and start codon upstream and a stop codon downstream suggest that Prcat1 covers the region in the P. ramorum scaffold 31:335019‐333448. Prcat1 and Pscat1 were not predicted to contain introns, whereas Picat1 contained one intron. The four Phytophthora predicted proteins shared a high degree of amino acid identity (see supplementary Fig. S1). We were unable to identify cat1 expressed sequence tags (ESTs) in any database. PnCat1 encodes a 512‐amino‐acid protein with a molecular weight of 57.9 kDa and an isoelectric point (pI) of 6.51. Neither the predicted PnCat1, PsCat1, Prcat1 nor Picat1 proteins contains a peroxisome localization signal, a mitochondrial targeting sequence or a secretion signal, suggesting that they are located in the cytoplasm (supplementary Fig. S1).
The PnCat2 gene was predicted to have a single open reading frame using GENSCAN, producing a 522‐amino‐acid protein with a molecular weight of 58.4 kDa and a pI of 8.73 (GenBank accesssion number EU176855). A single EST for Pncat2 was identified (accession no. AAH54964, Panabières et al., 2005) and this covered 50% of the predicted protein. The gene models from P. sojae (PsCat2: estExt_fgenesh1_pg.C_80063) and P. infestans (Picat2: PITT_15248) did not contain introns. One EST for Pscat2 and 14 ESTs for Picat2 were identified from a number of libraries in the Phytophthora Functional Genomics Database (PFGD), including the interaction library between potato tuber and P. infestans (Table 1). The P. infestans ESTs covered the entire genomic sequence of Picat2, confirming that this Phytophthora catalase consists of one open reading frame. The gene model for the P. ramorum gene (PrCat2: fgenesh1_pm.C_scaffold_31000012) contained one 21‐nucleotide intron, although this is likely to be an annotation error. EST data indicate that this region is not an intron in Picat2; GENSCAN does not predict an intron in this region of P. nicotianae, P. sojae or P. ramorum cat2 genes; and the nucleotide sequence in this region is highly conserved across all species examined (results not shown). This region of Prcat2 was thus included as coding sequence in analyses of the protein. All four Phytophthora Cat2 predicted proteins started at the same position, were the same length and contained the C‐terminal PTS1 peroxisome localization signal (supplementary Fig. S1).
Table 1.
Phytophthora catalase EST accessions.
| Species | Gene | EST accesssion | cDNA Library description |
|---|---|---|---|
| P. nicotiana | Catalase (Pncat2) | DR440554* | 4‐day‐old mycelium |
| P. infestans | Catalase (Picat2) | CV961542.1* | mycelium, Plich medium |
| CV908295.1* | mycelium, nitrogen starvation | ||
| CV956398.1* | mycelium, starved in water | ||
| CV959380.1* | mycelium, starved in water | ||
| CV900462.1* | mycelium, sporulating growth | ||
| CV900463.1* | mycelium, sporulating growth | ||
| CV923339.1* | sporangia, purified | ||
| CV929489.1* | mating of 88069 (A1) and 618 (A2) | ||
| CV938354.1* | mating of 88069 (A1) and 618 (A2) | ||
| CV944700.1* | mycelium, subtracted infection mimic | ||
| CV945627.1* | mycelium, subtracted infection mimic | ||
| piTA006xP02.b1† | interaction library between potato tuber and P. infestans | ||
| piTA006xP02.b3† | interaction library between potato tuber and P. infestans | ||
| piTA006xP02† | Interaction library between potato tuber and P. infestans | ||
| P. sojae | Catalase (Pscat2) | psMC005xO15f† | mycelium—complex media |
| P. infestans | Catalase‐peroxidase (PITG_05579.1) | CV927348.1* | mycelium, heat treated |
| CV964221.1* | mycelium, Plich medium | ||
| CV915558.1* | zoospores, purified | ||
| P. infestans | Catalase‐peroxidase (PITG_07143.1) | CV952269.1* | sporangia, cleaving |
| P. infestans | Catalase‐peroxidase (PITG_05578.1) | CV901911.1* | mycelium, nitrogen starvation |
| P. sojae | Catalase‐peroxidase (estExt_fgenesh1_pm.C_3170002) | psMA011xA19f† | mycelium—synthetic minimal medium |
NCBI EST database.
Phytophthora Functional Genomics Database.
PnCat3 consisted of a single open reading frame as predicted by GENSCAN, producing a predicted protein of 515 amino acids with a molecular weight of 57.7 kDa and a pI of 6.44 (Genbank accession EU258925). The predicted gene model from the P. ramorum database (PrCat3: fgenesh1_pm.C_scaffold_39000002) indicated that this gene contained one intron; however, as with PrCat2, this intron was not recognized by GENSCAN. The gene model of the homologous protein from the P. sojae database (PsCat3: estExt_fgenesh1_pg.C_80125) showed a gene containing three introns and a predicted start codon that was upstream of that of PrCat3 and PnCat3. The upstream PsCat3 ATG also occurs in PnCat3 but not in PrCat3. However, in P. nicotianae there is an in‐frame stop codon between the two ATGs, indicating that PnCat3 is translated from the second ATG. The region corresponding to that between the two P. sojae ATGs is less conserved (46–57% nucleotide identity) between the three species than the downstream gene sequence (80–83% nucleotide identity). Furthermore, the position of the 16‐nucleotide core promoter element (McLeod et al., 2004) was in approximately the same position in all three genes (results not shown), suggesting that translation occurs from the same ATG in all three genes. We thus consider that the upstream ATG in PsCat3 is not functional and that PsCat3 covers the region in the P. sojae scaffold 8:617248‐618661, and that the first intron predicted in the Pscat3 gene model, being outside this region, would not exist. Pncat3 homologues were not found in the P. infestans genome, nor were any cat3 ESTs identified for any Phytophthora species. PnCat3, PsCat3 and PrCat3 predicted proteins did not contain either of the peroxisome localization signals, a mitochondrial targeting sequence or a secretion signal and are probably located in the cytoplasm (supplementary Fig. S1).
All three predicted catalase proteins have the catalase proximal active site signature (Fig. 3). The motif searching programme, Proscan, identified the proximal heme‐ligand signature in PnCat2 and PnCat3. Analysis of the homologous region in the PnCat1 predicted protein revealed the substitution of the polar (hydrophilic) glutamine, found in PnCat2 and PnCat3, by leucine (non‐polar, hydrophobic) in the final position of this region. This type of substitution is found in other catalases (Rocha and Smith, 1995) and thus is unlikely to affect the activity of PnCat1. All three predicted P. nicotianae catalase proteins contain residues thought to be important for formation of tetramers (Gouet et al., 1995) (Fig. 3). The predicted protein sequences of the three catalases from P. nicotianae share 63.29% amino acid identity. PnCat1 shares 67.82% amino acid identity with PnCat2 and 70.10% identity with PnCat3. PnCat2 shares 76.86% amino acid identity with PnCat3.
Figure 3.
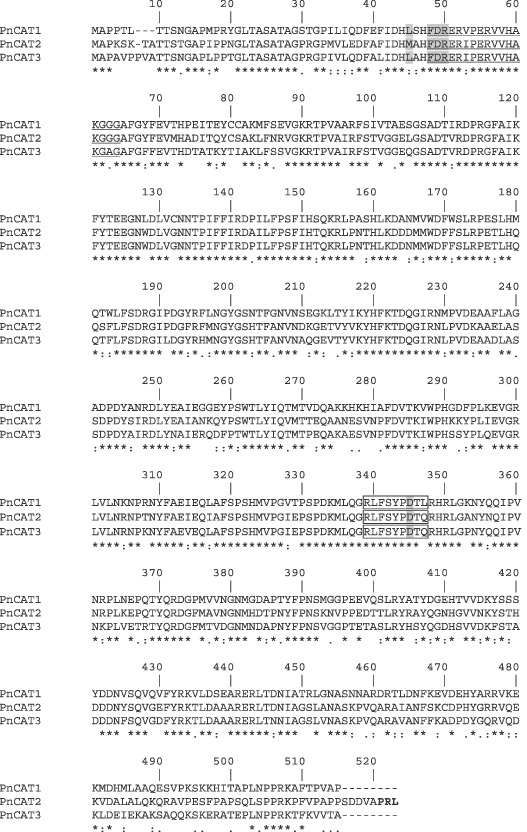
Multiple sequence alignment of P. nicotianae predicted catalase proteins (GenBank accession numbers EU176856, EU176855 and EU258925) showing the degree of amino acid conservation. Amino acids in shaded regions are thought to be important for tetramer formation. The underlined region indicates the catalase proximal active site signature and the boxed region indicates the catalase proximal heme‐ligand signature motif. The PTS1 of the Pncat2 protein is shown in bold type.
Identification of catalase‐peroxidase genes in Phytophthora genomes
Three putative catalase‐peroxidases were identified in P. infestans (Broad Institute accession numbers PITT_05579, PITT_07143 and PITT_05578), P. ramorum (JGI predicted transcripts 73140, 77036 and 73141) and P. sojae (JGI predicted proteins estExt_fgenesh1_pg.C_1490033, e_gwEuk.143.13.1 and estExt_fgenesh1_pm.C_3170002) databases by keyword or tBLASTn searches and all contained the bacterial heme catalase‐peroxidase signature sequence as predicted by InterProScan. Two of the three predicted P. infestans catalase‐peroxidases had secretion signals (PITT_05579, PITT_07143). ESTs for all three P. infestans genes and one P. sojae gene were identified in NCBI and PFGD (Table 1). All putative catalase‐peroxidases proteins from P. sojae and P. ramorum were predicted to have secretion signals. The molecular weight of these predicted catalase‐peroxidases varied between 47.5 and 74.4 kDa giving a dimer size between 95 and 148.8 kDa.
Expression of catalase genes during P. nicotianae asexual development
Although the sequences of the three P. nicotianae catalase genes were similar, primer pairs putatively specific for each gene were designed for use in quantitative real‐time PCR (qPCR) analysis and tested using BAC clones carrying individual catalase genes. Primer pairs qcat1 and qcat2 each amplified a single product of the expected size specifically from BACs carrying PnCat1 or PnCat2 genes, respectively (results not shown). RT‐PCR analysis also showed that these primer pairs amplified products of the expected sizes (results not shown). However, although multiple qPCR primer pairs were designed against the predicted Pncat3 sequence, including the 3′ untranslated region, no primer pair that was shown to be specific for PnCat3 when tested with the BAC clones gave an amplification product of the correct size and sequence when used in RT‐PCR or qPCR assays with the range of cDNA samples investigated here.
Quantification of the expression of PnCat1 and PnCat2 genes using qPCR revealed that PnCat2 is the major catalase in all developmental stages, with the highest expression occurring in sporulating hyphae and the lowest in 3‐h germinated cysts, with a 13‐fold difference between these two stages (Fig. 4a). PnCat1 expression is at least 175‐fold less than PnCat2 expression in all developmental stages except for 3‐h germinated cysts where the difference was only seven‐fold. These data, with the highest expression occurring in sporulating hyphae and the least in 3‐h germinated cysts (Fig. 4a), correlate with the in‐gel catalase activity assay (Fig. 1a). By contrast, there are higher levels of catalase transcripts in zoospores than in vegetative hyphae but vegetative hyphae had higher catalase activity than did zoospores. qPCR quantification of PnCat2 transcript levels during the 24 h following cyst formation and germination showed that expression was highest 4–6 h after cyst germination (Fig. 4b). Levels of PnCat1 transcripts were extremely low, often at the limits of detection by qPCR and in some samples could not be detected at all (results not shown).
Figure 4.
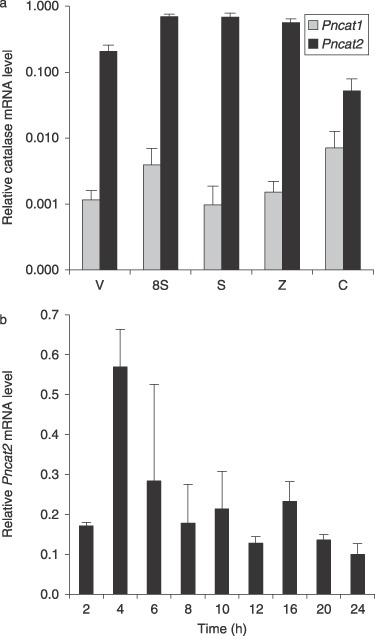
Comparison of PnCat1 and PnCat2 expression in different developmental stages and in germinated cysts 2–24 h after encystment and germination. (A) PnCat1 is expressed at very low levels in all stages of development. PnCat2 expression is highest in sporulating hyphae and zoospores. Vegetative hyphae (V), hyphae sampled 8 h after induction of sporulation (8S), sporulating hyphae (S), zoospores (Z), 3‐h germinated cysts (C). Values show averages and standard deviations of transcript levels normalized against WS41 expression using comparative quantification from three biological replicates. (B) PnCat2 expression reaches a peak in cysts allowed to germinate for 4 h. The data are averages (and standard deviations) of values from two biological replicates. A third biological replicate had a similar expression profile but the peak occurred at 6 h.
Catalase activity and gene expression during the infection of susceptible and resistant tobacco
The reactions of reported susceptible (Petit Gerard) and resistant (NC2326) cultivars of tobacco to inoculation with P. nicotianae isolate H1111 were assessed before analysis of catalase activity during the early stages of host infection. Inoculation of seedlings by a suspension of zoospores in Petri dishes resulted in encystment and germination of the cysts over the entire seedling. In 8‐ and 10‐h samples, the majority of cysts had germinated and penetrated the root surface of both cultivars. There was no evidence of hypersensitive cell death, detected by lactophenol trypan blue staining, at 8 or 10 h in either cultivar, and seedlings appeared healthy. By 48 h after inoculation, seedlings of N. tabacum cv Petit Gerard were severely wilted, while most inoculated NC2326 seedlings and all mock inoculated seedlings of both cultivars were still turgid and healthy 72 h post‐inoculation. Lactophenol trypan blue staining showed that by 48 h, the surface of Petit Gerard cotyledons was covered by an extensive network of P. nicotianae hyphae (Fig. 5a) and hyphae had penetrated the entire cotyledon (Fig. 5c). In contrast, in cultivar NC2326, hyphal growth was restricted to the surface of the cotyledons (Fig. 5b) and hyphae had not penetrated the underlying tissues (Fig. 5d). Hyphae successfully colonized the seedling stems at wound sites on both cultivars, but growth was more extensive in Petit Gerard seedlings than in NC2326. Hypersensitive cell death occurred in mesophyll cells in cotyledons of N. tabacum cv NC2326 between 24 and 48 h post‐inoculation (Fig. 5d) but was rarely seen in epidermal cells. No hypersensitive cell death was detected in N. tabacum cv Petit Gerard at any stage (Fig. 5a,c). Seedlings mock inoculated with water showed no signs of infection or cell death (Fig. 5e,f). These results confirm that N. tabacum cv Petit Gerard is susceptible and N. tabacum cv NC2326 is resistant to the P. nicotianae H1111 isolate used in these experiments.
Figure 5.
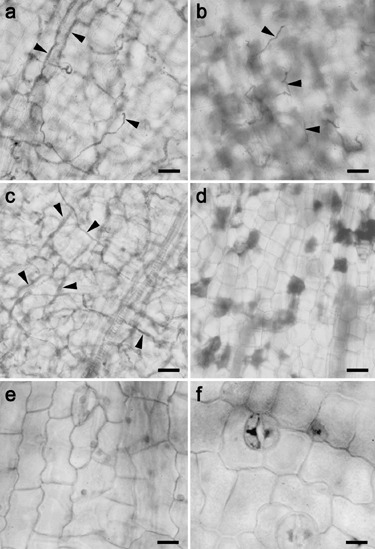
Light microscopy images of tobacco cotyledons infected with P. nicotianae or mock inoculated with water after 48 h. N. tabacum cv Petit Gerard shows extensive colonization by P. nicotianae on the cotyledon surface (a) and throughout the tissue (c). N. tobacum cv NC2326 cotyledons show restricted hyphal growth on the surface of epidermal cells (b) and no penetration of the underlying mesophyll tissue (d). Hypersensitive cell death was only seen in the resistant tobacco, NC2326 (d). Mock inoculated seedlings of Petit Gerard (e) or NC2326 (f) show no signs of infection or cell death. Arrowheads indicate hyphae. Scale bars = 10 µm.
A prominent catalase activity band appeared in extracts of susceptible tobacco seedlings 8 and 10 h after inoculation (Fig. 6). This band is the same size as the activity band seen in extracts of P. nicotianae hyphae. There was also a lower molecular weight band of activity at these time points. In contrast, no expression was seen in the mock inoculated Petit Gerard seedlings (Fig. 6), nor was there any activity seen in extracts from infected or mock inoculated resistant tobacco seedlings (results not shown). It was not possible to determine whether the catalase activity bands in the activity gels were due to tobacco and/or P. nicotianae catalase activity. The likely source of the catalase activity in the infected seedlings was thus determined using qPCR.
Figure 6.
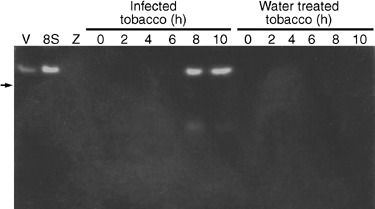
Catalase activity in susceptible (Petit Gerard) tobacco seedlings infected with P. nicotianae zoospores, 0–10 h after inoculation. Two bands of activity were seen in extracts of seedlings harvested at 8 and 10 h after inoculation. The larger of the activity bands was the same size as that seen for vegetative (V) and hyphae sampled 8 h after induction of sporulation (8S). No catalase activity could be seen in mock inoculated Petit Gerard seedlings over the same time period or in zoospore extracts (Z). The position of the 232 kDa bovine catalase is shown on the left (arrow).
cDNA from whole seedlings harvested 4, 8 and 24 h after inoculation was analysed. At the three time points, the level of PnCat2 expression in susceptible (Petit Gerard) plants was similar to that in resistant (NC2326) plants, with a substantially higher level of expression in both cultivars at 24 h than at the earlier time points (Fig. 7). Comparison of these data with those for in vitro germinated cysts sampled at the same time points showed that this increase in PnCat2 expression in planta contrasted with the decrease in expression in in vitro grown germinated cysts, a difference which was statistically significant at the 95% confidence level. By 24 h, PnCat2 expression in planta was 13‐ to 14‐fold greater than that in in vitro germinated cysts (Fig. 7). Neither PnCat1 nor PnCat3 transcripts could not be detected during tobacco infection.
Figure 7.
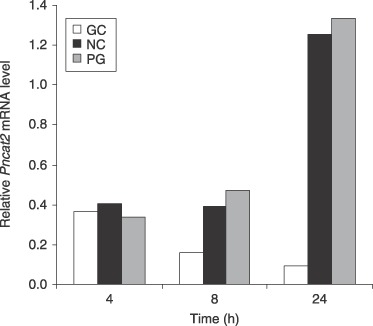
PnCat2 catalase expression from 4‐, 8‐ and 24‐h germinated cysts (GC) grown in vitro and in infected susceptible and resistant tobacco, 4, 8 and 24 h after inoculation. PnCat2 expression increased substantially in the susceptible Petit Gerard (PG) and resistant NC2326 (NC) plants while expression in germinated cysts growing in vitro in nutrient medium declined over the same period. Values show averages of transcript levels normalized against WS41 expression using comparative quantification from three biological replicates. The increase in PnCat2 expression in planta was statistically significantly different from the decrease in expression in vitro at the 95% confidence level.
Levels of tobacco catalase gene expression were also determined using qPCR in inoculated and mock‐inoculated susceptible and resistant seedlings. The most striking result was the substantial down‐regulation of tobacco catalase expression in the infected susceptible plants at all three time points (Fig. 8). Tobacco catalase expression in resistant seedlings was also down‐regulated 24 h after inoculation. Analysis of variance showed that both of these results were statistically significant at the 95% confidence level. No consistent differences in tobacco catalase expression were seen between the mock‐inoculated resistant (NC2326 control) or susceptible seedlings (Petit Gerard) or the inoculated resistant (NC2326 infected) tobacco seedlings at 4 and 8 h. These results strongly suggest that the activity bands seen in the in‐gel assay of infected tobacco seedlings were due to P. nicotianae catalase activity and not to tobacco catalase activity.
Figure 8.
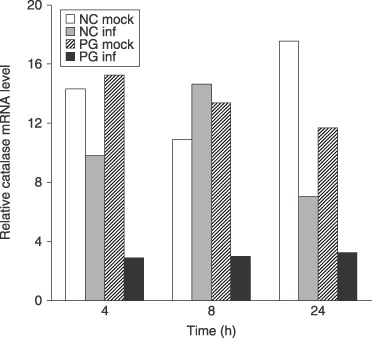
Comparison of tobacco catalase expression in infected and mock infected susceptible and resistant tobacco, 4, 8 and 24 h after inoculation. There was no consistent difference in levels of expression of tobacco catalase between susceptible Petit Gerard (PG mock) and resistant NC2326 (NC mock) mock‐infected tobacco or infected resistant tobacco (NC inf) at 4 and 8 h post‐inoculation. Tobacco catalase was down‐regulated in infected susceptible tobacco (PG inf) at all time points (statistically significant at the 95% confidence level). Tobacco catalase is also down‐regulated in inoculated resistant seedlings at 24 h (statistically significant at the 95% confidence level). Values show averages of transcript levels normalized against tobacco actin expression using comparative quantification from three biological replicates.
DISCUSSION
The catalase gene family in Phytophthora
In the present study, the genome of P. nicotianae has been shown to contain three monofunctional catalase genes, designated PnCat1, PnCat2 and PnCat3. All three genes encode highly conserved proteins that contain motifs essential for catalase function, namely the catalase active site and proximal heme‐ligand signature, and residues required for tetramer formation (Gouet et al., 1995). With a predicted monomer size of 58 kDa, the P. nicotianae catalase tetramers are estimated to have a relative molecular weight of about 232 kDa, a value that is similar to that of catalase tetramers in other eukaryotes (Allgood and Perry, 1986; Chen et al., 1993; Kawasaki et al., 1997). Association of P. nicotianae catalase monomers into enzymatically active tetramers was confirmed in catalase in‐gel activity assays via comparison of the mobility of the major activity band with those from tetrameric tobacco and bovine catalases. The genomes of P. sojae and P. ramorum contain homologues of the three P. nicotianae catalase genes whereas the P. infestans genome contained only two catalase genes, Picat1 and Picat2.
H2O2 and other ROS are produced in a number of intracellular locations but their major site of synthesis is in the peroxisomes. These metabolically active organelles typically contain a number of ROS scavenging enzymes, including catalase (del Río et al., 2002, 2006). Of the three Phytophthora catalase proteins, only Cat2 contained a peroxisome targeting sequence, namely the carboxy‐terminal peroxisomal localization signal, PTS1. In P. nicotianae, qPCR analysis showed that PnCat2 was expressed at much higher levels than PnCat1 or PnCat3. Predominant expression of Cat2 in other Phytophthora species was also supported by analysis of published EST data in which Cat2 ESTs but not Cat1 or Cat3 ESTs have been identified. The absence of any recognizable targeting signals in Cat1 and Cat3 from the three species suggests that, if produced, these proteins are likely to be cytosolic.
None of the Phytophthora monofunctional catalases examined contained secretion signals. Secreted catalases have been identified in a number of plant pathogens including Magnaporthe grisea (Skamnioti et al., 2007), Claviceps purpurea (Garre et al., 1998), Blumeria graminis (Zhang et al., 2004) and Agrobacterium tumefacians (Xu and Pan, 2000) and have been implicated in overcoming the host defence response. While the monofunctional catalases from Phytophthora species do not have secretion signals, the genomes of P. sojae, P. infestans and P. ramorum contain three catalase‐peroxidases, most of which have N‐terminal secretion signals, and it seems likely that P. nicotianae may also contain these secreted ROS scavenging enzymes. Evidence for possible secretion of catalase in Phytophthora comes from histochemical analysis of P. palmivora zoospores (Philippi et al., 1975; Powell and Bracker, 1986). These studies reported catalase activity in peroxisomes (microbodies) and an organelle called a U‐body, as well as on the zoospore surface (Philippi et al., 1975; Powell and Bracker, 1986). The U‐bodies were subsequently shown to be ventral vesicles that undergo exocytosis during zoospore encystment, thereby secreting adhesive material onto the adjacent host surface (Hardham and Gubler, 1990; Hardham et al., 1991). It should be noted that the histochemical localizations conducted in P. palmivora would not have differentiated between the activity of catalases and that of catalase‐peroxidases.
The role of catalase during asexual development in P. nicotianae
Given the much higher level of PnCat2 expression than PnCat1 and that PnCat3 transcripts could not be detected, the catalase activity band seen in vegetative and sporulating hyphae is likely to be predominantly due to the peroxisomal PnCat2 enzyme complex. The high level of catalase gene expression and enzyme activity in sporulating hyphae indicates a role for catalase in the shift from vegetative to sporulating hyphae. As in fungi, in Phytophthora, asexual sporulation is triggered by a number of environmental cues, including a decrease in nutrient availability (Hardham and Hyde, 1996). The induction of catalase activity and gene expression during Phytophthora sporulation may be due to the production of H2O2 in response to nutrient deprivation (Mittler 2002), as observed in Fusarium in nutrient‐limited media (Kono et al., 1995; Kwon and Anderson, 2001). Upregulation of catalase may serve to reduce the oxidative stress resulting from H2O2 production but may also modulate the potential role of H2O2 in signalling during development. Production of ROS triggers sporulation in fungi (Lara‐Ortíz et al., 2003; Malagnac et al., 2004) and an increase in catalase transcripts accompanies asexual sporulation in A. nidulans (Navarro et al., 1996), and Dictyostelium discoideum (Garcia et al., 2003) and asexual sporulation in Phytophthora involves widespread changes in gene expression (Kim and Judelson, 2003; R. D. Narayan and A. R. Hardham, unpublished observations).
Catalase gene expression in zoospores was almost as high as that in sporulating hyphae, but catalase activity in zoospore extracts was detectable only when the gels were overloaded. Other studies have also found a lack of correlation between catalase gene expression and enzyme activity (Brisson et al., 1998; Polidoros et al., 2001; Redinbaugh et al., 1990; Skadsen and Scandalios, 1987). The discrepancy is not due to a lack of sensitivity of the in‐gel assay because catalase activity was detected in extracts of vegetative hyphae in which catalase gene expression was less than that in zoospores. The results suggest that catalase activity during Phytophthora asexual development is regulated by means other than gene expression. ROS other than H2O2 can, for example, inactivate catalase (Escobar et al., 1996; Wang et al., 2004).
In germinated cysts, the two catalase activity bands that run faster than the 232 kDa bovine catalase tetramer could be due to breakdown products that retain activity, such as seen with bovine catalase (Prakash et al., 2002), or they may be due to activity of catalase‐peroxidases. In general catalase‐peroxidase enzymes exist as dimers with a molecular weight of approximately 160 kDa (ng Welinder, 1991; Peraza and Hansberg, 2002; Rocha and Smith, 1995). Given the identification of multiple catalase‐peroxidases in the genomes of three Phytophthora species, the largest having a predicted dimeric molecular weight of 149 kDa, and the identification of ESTs to the P. infestans catalase‐peroxidases, it is highly likely that the lower molecular weight activity bands seen in extracts of P. nicotianae germinated cysts are catalase‐peroxidases. Although dual function catalase‐peroxidases have been described in a number of organisms including bacteria, fungi, mycetozoa and plants (Garcia et al., 2003; Havir and McHale, 1987; Hochman and Shemesh, 1987; Jamet et al., 2003; Peraza and Hansberg, 2002; Rocha and Smith, 1995), their role during oxidative stress is not well understood. However, there are examples where the silencing of monofunctional catalases has indicated a significant role for other H2O2 scavengers (Calera et al., 1997; Giles et al., 2006; Izawa et al., 1996; Paris et al., 2003; Robbertse et al., 2003; Schouten et al., 2002). For example, the upregulation of glutathione S‐transferase appears to compensate for the complete silencing of an extracellular catalase from B. cinerea (Schouten et al., 2002), whereas silencing of all three catalase genes in Cochliobolus heterostrophus did not alter the virulence of this pathogen in maize (Robbertse et al., 2003). These studies highlight the complexity of ROS scavenging pathways and the likelihood that redundancy of ROS scavenging enzymes in P. nicotianae would mean that silencing of the catalase genes would not help resolve their exact function.
The role of catalases during plant–pathogen interactions
Regulation of ROS has been shown to be an important aspect of plant–pathogen interactions. In resistant plants, occurrence of the oxidative burst and subsequent hypersensitive cell death in response to pathogen invasion has been well characterized (Lamb and Dixon, 1997). The oxidative burst has been associated with changes in antioxidant enzymes and signal transduction pathways in a number of plant–pathogen systems (De Gara et al., 2003) including, as seen in the present study, the down‐regulation of plant catalase activity and/or gene expression, a process thought to halt pathogen infection during an incompatible interaction (Lamb and Dixon, 1997).
The oxidative burst and changes in the activity of ROS scavenging enzymes, including catalase, can also occur during infection of susceptible hosts by viral, bacterial, fungal and oomycete pathogens (Able, 2003; Arias et al., 2005; Kużniak and Skłodowska, 2005; Venisse et al., 2001). Depending on the interacting partners and spatial and temporal parameters, host catalase gene expression or enzyme activity may increase or decrease during disease development. For example, in cucumber infected by cucumber mosaic virus, host catalase expression decreases within necrotic lesions but increases in surrounding regions (Havelda and Maule, 2000). In tomato infected with B. cinerea, catalase activity increases transiently but is then progressively inhibited as disease develops (Kużniak and Skłodowska, 2005). The activity of two maize catalases is suppressed during infection by the necrotrophic fungus Exserohilum turcicum (Keissar et al., 2002) and other investigations of necrotrophic interactions have also found that increased H2O2 levels promote infection (Able, 2003; Gönner and Schlösser, 1993; Govrin and Levine, 2000; Rolke et al., 2004). In the current study, tobacco catalase gene expression was reduced in resistant seedlings at 24 h, mirroring results from a study on the incompatible interaction between P. capsici and Capsicum annuum (Ueeda et al., 2005). A significant down‐regulation of tobacco catalase gene expression in susceptible seedlings was observed 4, 8 and 24 h after inoculation. As this decrease was not observed in mock‐inoculated seedlings, it is apparently induced by the virulent pathogen. These results give further evidence that a pathogen‐orchestrated decrease in host catalase may favour infection by necrotrophic pathogens. This situation contrasts with that of biotrophic interactions in which elevation of host catalases may protect plant cells from oxidative stress, allowing establishment of the biotrophic relationship.
Down‐regulation of tobacco catalase gene expression during infection of susceptible tobacco by P. nicotianae indicates that the increase in catalase activity detected by in‐gels assays is likely to be due to the P. nicotianae catalase, PnCat2. PnCat2 gene expression increased during colonization of both susceptible and resistant tobacco. Similar accumulation of pathogen catalase transcripts or secretion of pathogen‐encoded catalases has been observed during interactions between B. graminis and barley (Zhang et al., 2004), C. purpurea and rye (Garre et al., 1998), and A. tumefacians and kalanchoe (Xu and Pan, 2000), suggesting that pathogen‐encoded catalases constitute pathogenicity factors that provide protection against the release of H2O2 from the host. Other roles for pathogen‐derived catalases during infection are also possible. For example, an M. grisea catalase that is highly expressed during penetration of rice leaves and essential for successful infection has been shown to be involved in strengthening of the appressorial wall (Skamnioti et al., 2007). Our study shows that in P. nicotianae, the elevation of PnCat2 expression that occurs in planta does not occur at the same time points after cyst germination in in vitro grown germinated cysts, indicating that pathogen catalase expression during germling growth is being influenced by factors of plant origin.
CONCLUDING REMARKS
Infection of host plants by the necrotrophic, soil‐borne P. nicotianae is typically initiated by motile zoospores which attach and encyst at the root surface before germinating and penetrating the underlying plant epidermis. The P. nicotianae genome contains three catalase genes, one of which, PnCat2, encodes a protein targeted to the peroxisome. Expression of the peroxisomal catalase gene is transiently upregulated about three‐fold a few hours after cyst germinating in vitro but during host infection, PnCat2 expression is strongly up‐regulated in a sustained manner during the first 24 h post‐inoculation. Concurrently, P. nicotianae induces a down‐regulation of tobacco catalase gene expression. Our results give support to the idea that P. nicotianae increases peroxisomal catalase activity in a counterdefence mechanism that protects the pathogen against the oxidative burst produced by the plant and that it inhibits host catalase, thereby enhancing plant cell death and facilitating plant colonisation.
EXPERIMENTAL PROCEDURES
Isolate and culture conditions
P. nicotianae isolate H1111 (Gabor et al., 1993), originally isolated by Dr David I. Guest (University of Sydney), was cultured as described in Robold and Hardham (1998). P. nicotianae cells at different developmental stages, namely vegetative hyphae, hyphae collected 8 h after induction of sporulation and zoospores, were obtained as detailed in Blackman et al. (2005). Sporulating hyphae were also harvested from cultures left in V8 medium for 14 days. Cysts were germinated in a large volume of 5% (v/v) V8 broth (typically 50–100 mL) and harvested at various times between 2 and 24 h.
Protein extractions and catalase activity gels
Proteins from vegetative hyphae, hyphae isolated 8 h after the induction of sporulation, zoospores and germinated cysts were extracted as described in Blackman et al. (2005). Catalase activity was detected in 50 µg of total protein separated on 10% native polyacrylamide gels according to Chandlee and Scandalios (1983). A sample containing approximately 1.3 units of bovine liver catalase (Sigma‐Aldrich, St Louis, MO) was included as a positive control for catalase activity and as a size standard. Duplicate gels were run for each experiment and one of these was stained with Coomassie Blue (Merril, 1990) to verify equal loading of all samples. Catalase activity was induced in 0.5‐cm2 pieces of N. tabacum cultivar Petit Gerard leaves by incubation in 100 mm H2O2 for 10 min under vacuum, followed by incubation in water for 4 h.
PCR amplification of partial catalase genes
Fungal catalase genes were used to search for catalase genes in the JGI P. sojae and P. ramorum genome databases. Catalase genes from P. ramoum and P. sojae were used to design degenerate forward and reverse primers (GASTTCCCGCTSAAAGAGGT–ATCTGGTCCAYYTTSTCCTTC, GCTTCGCCATCAARTTCTACAC–ATGCCCTGGTCCGTCTTGAA, GAACCGCAACCCGAARAA–TCATGTTSCCRTCGACCGT) and these were used to amplify partial catalase genes from P. nicotianae gDNA, which had been prepared from sporulating hyphae according to Dudler (1990). Degenerate PCR used 0.5 µg gDNA with 2× PCR Master Mix (Promega, Madison, WI) and 0.5 µm primers. Cycling conditions were 94 °C for 30 s (first cycle 2 min), 59 °C for 30 s and 72 °C for 45 s for 35 cycles in a PTC‐200 Peltier Thermal Cycler (MJ Research, Watertown, MA). The PCR products were gel‐purified using a Wizard SV Clean‐up kit (Promega), cloned into the pGEM®‐T Easy vector system (Promega), transformed and propagated in JM109 using standard methods (Sambrook and Russell, 2001). These clones were sequenced using vector‐specific primers at the Australian Genome Research Facility (Brisbane, Queensland).
Southern blotting and BAC library screening
Catalase gene copy number was determined by Southern blot analysis of total P. nicotianae gDNA according to Blackman et al. (2005) using the restriction enzymes BamH1, EcoR1, HindIII, Xho1 and Pst1. A single blot, stripped between each probing, was used for all experiments. A BAC library of P. nicotianae gDNA was screened using three partial catalase genes as described in Shan and Hardham (2004). DNA was isolated from positive BACs using a PureLink HiPure Plasmid Kit (Invitrogen, Carlsbad, CA) with the method adjusted, as described by the manufacturer, for the isolation of BAC DNA. Catalase genes were sequenced directly from the BACs using gene‐specific primers.
Sequence analysis
Open reading frames were detected using GENSCAN (http://genes.mit.edu/GENSCAN.html; Burge and Karlin, 1997) and by comparison with the gene models in the JGI P. sojae and P. ramorum databases and the Broad Institute P. infestans database. Where the position of the start codon of Phytophthora catalase genes was ambigious, the region adjacent to the putative ATG was examined for nucleotides important for the initiation of translation (Kozak, 1996) and the upstream region examined for the oomycete 16‐nucleotide consensus promoter sequence (McLeod et al., 2004). The PFGD and NCBI databases were searched using tBLASTn for catalase ESTs. The P. sojae, P. ramorum and P. infestans genomic databases were also searched for catalase‐peroxidases using a keyword search, and P. sojae and P. infestans catalase‐peroxidase ESTs identified in the PFGD. The molecular weight and pI of catalase proteins were determined using the Compute PI/MW tool (http://www.expasy.ch/tools/pi_tool.html). Proteins were examined for motifs typical of catalases using Prosite (http://ca.expasy.org/tools/scanprosite/; Hulo et al., 2006) or InterProScan (http://www.ebi.ac.uk/InterProScan/index.html; Zdobnov and Apweiler, 2001) for catalase‐peroxidase motifs, for signal peptide cleavage sites using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/; Bendtsen et al., 2004) and for N‐terminal subcellular sorting signals using iPSORT (http://hc.ims.u‐tokyo.ac.jp/iPSORT/how.html; Bannai et al., 2002). Peroxisome targeting signal type1 (PTS1) was detected in predicted proteins using the PTS1 Predictor programme (http://mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp; Neuberger et al., 2003). The amino acid sequence between residues 2 and 30 from the predicted P. nicotianae catalase proteins was also examined manually for the N‐terminal PTS2 motif, RLx5HL (Reumann, 2004). Catalase proteins were aligned using ClustalX (Thompson et al., 1997).
Infection studies
The outcome of the interaction between P. nicotianae and two tobacco cultivars, N. tabacum cultivar Petit Gerard (susceptible) and N. tabacum cultivar NC2326 (resistant), kindly supplied by D. I. Guest, was confirmed by assessing the viability of inoculated seedlings, the appearance of hypersensitive cell death in leaves and hyphal growth. Tobacco seeds were sterilized in 6% H2O2 in 100% ethanol for 1 min, washed three times in sterile water and germinated on sterile moist filter paper in sealed Petri dishes at 23 °C for 14–20 days. Approximately 50 seedlings were submerged in a suspension of 105 zoospores per millilitre and left for 1 h. Seedlings were then washed twice in sterile distilled water and placed on moist filter paper in Petri dishes and left in the light at 23 °C. Seedlings were examined before and after staining with lactophenol trypan blue as described in Takemoto et al. (2003) at 8, 10, 24, 48 and 72 h post‐inoculation. Control seedlings were mock inoculated with water. For analysis of catalase activity during tobacco seedling infection, approximately 200 7‐day‐old seedlings were submerged in 8 mL of zoospores at a concentration of 1 × 105 mL−1 in a Petri dish. Inoculated seedlings were left at room temperature and samples taken at 0, 2, 4, 6, 8 and 10 h. Control seedlings were treated in the same manner except for the addition of 8 mL of distilled water instead of zoospores. Seedlings were then blotted on filter paper to remove excess liquid and frozen in liquid nitrogen. Proteins were extracted from entire infected and mock‐infected seedlings, as described for P. nicotianae samples.
Catalase gene expression analysis
Catalase gene expression was analysed by two‐step qPCR in P. nicotianae vegetative hyphae, hyphae collected 8 h after induction of sporulation, hyphae which had been left to sporulate in V8 medium for 14 days, zoospores and 3‐h germinated cysts. Total RNA was isolated from three biological replicates of vegetative hyphae, sporulating hyphae and 3‐h germinated cysts ground in liquid nitrogen, and from freeze‐dried zoospores with Trizol (Invitrogen) according to the manufacturer's instructions. Contaminating gDNA was removed with RQ1 DNAse (Promega) and total RNA purified using a RNeasyMini kit (Qiagen Pty Ltd, Doncaster, Victoria, Australia). cDNA was made using an oligo (dT)12–18 primer (Invitrogen) and SuperscriptII reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Primers were designed using Oligo Explorer (http://www.genelink.com/tools/gl‐oe.asp) and NetPrimer (http://www.premierbiosoft.com/netprimer/). The specificity of all P. nicotianae catalase qPCR primers was checked by PCR using DNA from BACs that contained a single catalase gene. Gel‐based reverse transcriptase PCR (RT‐PCR) was used to check for the presence of contaminating gDNA in minus‐RT reactions conducted at the same time as the cDNA generation using primers (CGGTCCCATTGTGCTCTATT–TTGTGTCTTTGTGTGATGCG) against WS41 (NCBI accession CF891677), which has been shown to be a suitable normalizing gene for RNA transcript analysis (Shan et al., 2004; Yan and Liou, 2006). These primers produce a 90‐bp fragment from cDNA but, as they are situated either side of an intron, produce a longer fragment in the presence of contaminating gDNA. Primers for PnCat1 (qcat1: GTTCAATCGCTTCGCTACG–GGTCCATCTTCTCCTTCACAC), PnCat2 (qcat2: TGGCTGTCAATGGCAATA–GAGTAGTTATCATCATCGTGGGT) and PnCat3 (qcat3: ATCGAGCGCCAGGACTT–CCTCCTGTAGTGGATACGAACT, qcat3A: GACAGCGGACGACGATAA–CATTCACCAACGACCCAG, qcat3B: AAATAGCGTGGGTGGTCC–CCGTTGTCCGTAGTCAGG, qcat3C: CTGAACCGCTCAATCCG–TGCTTTCTTCCACTATGCCA) were checked for their ability to amplify catalase cDNA by RT‐PCR. Cycling conditions for the second step of the RT‐PCR were 35 cycles of 30 s at 94 °C (initial 94 °C step for 2 min), 30 s at 59 °C and 45 s at 72 °C using 2× PCR Master mix (Promega). Products were visualized on a 2% (w/v) agarose gel in Tris‐acetate‐EDTA buffer (TAE: 40 mm Tris, 1 mm EDTA, 20 mm acetic acid). qPCR was done in a 3000 Real‐Time Cycler (Corbett Life Sciences, Mortlake, NSW, Australia) using 15‐µL reactions with 15 ng of cDNA, 200 nm primers and Platinum Sybr Green qPCR SuperMix‐UDG with ROX (Invitrogen). Four technical replicates were done for each sample and expression levels relative to WS41 calculated using the Comparative Quantification function within the Rotorgene6 program, which provides relative quantification of the experimental transcript to the normalizing transcript taking amplification efficiency into account (Corbett Research). Cycling conditions were 40 or 45 cycles of 95 °C for 15 s (initial step of 95 °C for 2 min), 30 s at 59 °C, 30 s at 72 °C, with data acquired at 59 °C. A melt curve (55–99 °C) was constructed for each experiment and if primer–dimer formation occurred, 30 or 60 ng of cDNA was used in repeat experiments. Catalase expression analysis using qPCR was also conducted on a germinated cyst time series at 2, 4, 6, 8, 10, 12, 16, 20 and 24 h after the induction of encystment. For this experiment, 8–10 technical replicates were included in each of three biological replicates.
Catalase gene expression was also examined using qPCR in samples taken 4, 8 and 24 h after inoculation (or mock inoculation) of N. tabacum susceptible (Petit Gerard) and resistant (NC2326) cultivars with P. nicotianae zoospores. Seedlings were infected as described above, total RNA isolated from the entire seedling and cDNA made as for P. nicotianae samples from three independent experiments. Primers against tobacco actin were used for qPCR normalization (TGGTATGGGTCAAAAGGATG–CAGGAGCAACACGCAACT; Fritz et al., 2006). A single set of primers (ATCAACAAGGCTGGGAAAKC–CCAGCRGCAATMGARTCRTAGA) designed against two N. tabacum catalase genes in NCBI (accession nos. AAA57552, AAB71764) was used to quantify tobacco catalase in both cultivars. The significance of differences in catalase gene expression during the infection of tobacco was determined by conducting an analysis of variance using the statistical program GenStat, Release 8.1.
Supporting information
Fig. S1 Multiple sequence alignment of P. nicotianae (PnCat1: EU176856, PnCat2: EU176855, PnCat3: EU258925), P. infestans (PiCat1: PITT_15292, PiCat2: PITT_15248), P. sojae (PsCat1: estExt_fgenesh1_pm.C_80008, PsCat2: estExt_fgenesh1_pg.C_ 80063, PsCat3: scaffold 8: 617248:618661) and P. ramorum (PrCat1: scaffold 31: 335019–333448, PrCat2: fgenesh1_pm.C_ scaffold_31000012, PrCat3: fgenesh1_pm.C_scaffold_39000002) predicted catalase proteins showing the degree of amino acid conservation. The shaded C‐terminal amino acids indicate the PTS1 localization signal of Cat2 proteins.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
ACKNOWLEDGEMENTS
We thank Astrid Chanfon and Trish Boyce for excellent technical assistance, Bettina Schulz for her contributions to the project, Emlyn Williams for help with the statistical analysis and the Australian Research Council for support.
REFERENCES
- Able, A.J. (2003) Role of reactive oxygen species in the response of barley to necrotrophic pathogens. Protoplasma, 221, 137–143. [DOI] [PubMed] [Google Scholar]
- Able, A.J. , Guest, D.I. and Sutherland, M.W. (2000) Hydrogen peroxide yields during the incompatible interaction of tobacco suspension cells inoculated with Phytophthora nicotianae . Plant Physiol. 124, 899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allgood, G.S. and Perry, J.J. (1986) Characterization of a manganese‐containing catalase from the obligate thermophile Thermoleophilum album . J. Bacteriol. 168, 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher, R.G. , Erturk, N. and Heath, L.S. (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. [PubMed] [Google Scholar]
- Arias, M.C. , Luna, C. , Rodriguez, M. , Lenardon, S. and Taleisnik, E. (2005) Sunflower chlorotic mottle virus in compatible interactions with sunflower: ROS generation and antioxidant response. Eur. J. Plant Pathol. 113, 223–232. [Google Scholar]
- Bannai, H. , Tamada, Y. , Maruyama, O. , Nakai, K. and Miyano, S. (2002) Extensive feature detection of N‐terminal protein sorting signals. Bioinformatics, 18, 298–305. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Nielsen, H. , Von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Blackman, L.M. , Mitchell, H.J. and Hardham, A.R. (2005) Characterisation of manganese superoxide dismutase from Phytophthora nicotianae . Mycol. Res. 109, 1171–1183. [DOI] [PubMed] [Google Scholar]
- Borden, S. and Higgins, V.J. (2002) Hydrogen peroxide plays a critical role in the defence response of tomato to Cladosporium fulvum . Physiol. Mol. Plant Pathol. 61, 227–236. [Google Scholar]
- Van Breusegem, F. , Vranová, E. , Dat, J.F. and Inzé, D. (2001) The role of active oxygen species in plant signal transduction. Plant Sci. 161, 405–414. [Google Scholar]
- Brisson, L.F. , Zelitch, I. and Havir, E.A. (1998) Manipulation of catalase levels produces altered photosynthesis in transgenic tobacco plants. Plant Physiol. 116, 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Calera, J.A. , Paris, S. , Monod, M. , Hamilton, A.J. , Debeaupuis, J.‐P. , Diaquin, M. , López‐Medrano, R. , Leal, F. and Latgé, J.‐P. (1997) Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus . Infect. Immun. 65, 4718–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandlee, J.M. and Scandalios, J.G. (1983) Gene expression during early kernel development in Zea mays . Dev. Genet. 4, 99–115. [Google Scholar]
- Chen, Z.X. , Ricigliano, J.W. and Klessig, D.F. (1993) Purification and characterization of a soluble salicylic acid‐binding protein from tobacco. Proc. Natl Acad. Sci. USA, 90, 9533–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gara, L. , De Pinto, M.C. and Tommasi, F. (2003) The antioxidant systems vis‐a‐vis reactive oxygen species during plant‐pathogen interaction. Plant Physiol. Biochem. 41, 863–870. [Google Scholar]
- Dudler, R. (1990) The single‐copy actin gene of Phytophthora megasperma encodes a protein considerably diverged from any known actin. Plant Mol. Biol. 14, 415–422. [DOI] [PubMed] [Google Scholar]
- Erwin, D.C. and Ribeiro, O.K. (1996) Phytophthora. St. Paul: APS Press. [Google Scholar]
- Escobar, J.A. , Rubio, M.A. and Lissi, E.A. (1996) SOD and catalase inactivation by singlet oxygen and peroxyl radicals. Free Radic. Biol. Med. 20, 285–290. [DOI] [PubMed] [Google Scholar]
- Fellbrich, G. , Romanski, A. , Varet, A. , Blume, B. , Brunner, F. , Engelhardt, S. , Felix, G. , Kemmerling, B. , Krzymowska, M. and Nürnberger, T. (2002) NPP1, a Phytophthora‐associated trigger of plant defense in parsley and Arabidopsis . Plant J. 32, 375–390. [DOI] [PubMed] [Google Scholar]
- Fritz, C. , Palacios‐Rojas, N. , Feil, R. and Stitt, M. (2006) Regulation of secondary metabolism by the carbon‐nitrogen status in tobacco: nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J. 46, 533–548. [DOI] [PubMed] [Google Scholar]
- Frugoli, J.A. , Zhong, H.H. , Nuccio, M.L. , McCourt, P. , McPeek, M.A. , Thomas, T.L. and McClung, C.R. (1996) Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 112, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor, B.K. , Ogara, E.T. , Philip, B.A. , Horan, D.P. and Hardham, A.R. (1993) Specificities of monoclonal antibodies to Phytophthora cinnamomi in two rapid diagnostic assays. Plant Dis. 77, 1189–1197. [Google Scholar]
- Garcia, M.X.U. , Alexander, H. , Mahadeo, D. , Cotter, D.A. and Alexander, S. (2003) The Dictyostelium discoideum prespore‐specific catalase B functions to control late development and to protect spore viability. Biochim. Biophys. Acta-Mol. Cell Res. 1641, 55–64. [DOI] [PubMed] [Google Scholar]
- Garre, V. , Tenberge, K.B. and Eising, R. (1998) Secretion of a fungal extracellular catalase by Claviceps purpurea during infection of rye: putative role in pathogenicity and suppression of host defense. Phytopathology, 88, 744–753. [DOI] [PubMed] [Google Scholar]
- Giles, S.S. , Stajich, J.E. , Nichols, C. , Gerrald, Q.D. , Alspaugh, J.A. , Dietrich, F. and Perfect, J.R. (2006) The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot. Cell, 5, 1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönner, M.V. and Schlösser, E. (1993) Oxidative stress in the host‐parasite system Avena sativa L. and Drechslera spp. Physiol. Mol. Plant Pathol. 42, 221–234. [Google Scholar]
- Gouet, P. , Jouve, H.‐M. and Dideberg, O. (1995) Crystal structure of Proteus mirabilis PR catalase with and without bound NADPH J. Mol. Biol. 249, 933–954. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Gunderson, J.H. , Elwood, H. , Ingold, A. , Kindle, K. and Sogin, M.L. (1987) Phylogenetic relationships between chlorophytes, chrysophytes, and oomycetes. Proc. Natl Acad. Sci. USA, 84, 5823–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham, A.R. (2005) Phytophthora cinnamomi . Mol. Plant Pathol. 6, 589–604. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R. and Gubler, F. (1990) Polarity of attachment of zoospores of a root pathogen and pre‐alignment of the emerging germ tube. Cell Biol. Int. 14, 947–956. [Google Scholar]
- Hardham, A.R. and Hyde, G.J. (1996) Asexual sporulation in the oomycetes. Adv. Bot. Res. 24, 353–398. [Google Scholar]
- Hardham, A.R. , Gubler, F. and Duniec, J. (1991) Ultrastructural and immunological studies of zoospores of Phytophthora In: Phytophthora (Lucas J.A., Shattock R.C., Shaw D.S. and Cooke L.R., eds), pp. 51–69. Cambridge: Cambridge University Press. [Google Scholar]
- Havelda, Z. and Maule, A.J. (2000) Complex spatial responses to cucumber mosaic virus infection in susceptible Cucurbita pepo cotyledons. Plant Cell, 12, 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havir, E.A. and McHale, N.A. (1987) Biochemical and developmental characterization of multiple forms of catalase in tobacco leaves. Plant Physiol. 84, 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman, A. and Shemesh, A. (1987) Purification and characterization of a catalase‐peroxidase from the photosynthetic bacterium Rhodopseudomonas capsulata . J. Biol. Chem. 262, 6871–6876. [PubMed] [Google Scholar]
- Hulo, N. , Bairoch, A. , Bulliard, V. , Cerutti, L. , De Castro, E. , Langendijk‐Genevaux, P.S. , Pagni, M. and Sigrist, C.J.A. (2006) The PROSITE database. Nucleic Acids Res. 34, D227–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, S. , Inoue, Y. and Kimura, A. (1996) Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae . Biochem. J. 320, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet, A. , Sigaud, S. , Van de Sype, G. , Puppo, A. and Hérouart, D. (2003) Expression of the bacterial catalase genes during Sinorhizobium meliloti‐Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant–Microbe Interact. 16, 217–225. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Huitema, E. and Vleeshouwers, V.G.A.A. (1999) Resistance to oomycetes: a general role for the hypersensitive response? Trends Plant Sci. 4, 196–200. [DOI] [PubMed] [Google Scholar]
- Kawasaki, L. , Wysong, D. , Diamond, R. and Aguirre, J. (1997) Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 179, 3284–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keissar, H.T. , Bashan, B. , Levy, Y. and Kenigsbuch, D. (2002) Stage specificity of catalase isoform activity in Exserohilum turcicum . Physiol. Mol. Plant Pathol. 60, 163–168. [Google Scholar]
- Kim, K.S. and Judelson, H.S. (2003) Sporangium‐specific gene expression in the oomycete phytopathogen Phytophthora infestans . Eukaryot. Cell, 2, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz, M.G. and Hutcheson, S.W. (1992) Multiple periplasmic catalases in phytopathogenic strains of Pseudomonas syringae . Appl. Environ. Microbiol. 58, 2468–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, Y. , Yamamoto, H. , Takeuchi, M. and Komada, H. (1995) Alterations in superoxide dismutase and catalase in Fusarium oxysporum during starvation‐induced differentiation. Biochim. Biophys. Acta, 1268, 35–40. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (1996) Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome, 7, 563–574. [DOI] [PubMed] [Google Scholar]
- Kużniak, E. and Skłodowska, M. (2005) Fungal pathogen‐induced changes in the antioxidant systems of leaf peroxisomes from infected tomato plants. Planta, 222, 192–200. [DOI] [PubMed] [Google Scholar]
- Kwok, L.Y. , Schlüter, D. , Clayton, C. and Soldati, D. (2004) The antioxidant systems in Toxoplasma gondii and the role of cytosolic catalase in defence against oxidative injury. Mol. Microbiol. 51, 47–61. [DOI] [PubMed] [Google Scholar]
- Kwon, S.‐I. and Anderson, A.J. (2001) Differential production of superoxide dismutase and catalase isozymes during infection of wheat by a Fusarium proliferatum‐like fungal isolate. Physiol. Mol. Plant Pathol. 58, 73–81. [Google Scholar]
- Lamb, C. and Dixon, R.A. (1997) The oxidative burst in plant disease resistance Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lara‐Ortíz, T. , Riveros‐Rosas, H. and Aguirre, J. (2003) Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans . Mol. Microbiol. 50, 1241–1255. [DOI] [PubMed] [Google Scholar]
- Levy, E. , Eyal, Z. and Hochman, A. (1992) Purification and characterization of a catalase‐peroxidase from the fungus Septoria tritici . Arch. Biochem. Biophys. 296, 321–327. [DOI] [PubMed] [Google Scholar]
- Malagnac, F. , Lalucque, H. , Lepère, G. and Silar, P. (2004) Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina . Fungal Genet. Biol. 41, 982–997. [DOI] [PubMed] [Google Scholar]
- Mayer, A.M. , Staples, R.C. and Gil‐ad, N.L. (2001) Mechanisms of survival of necrotrophic fungal plant pathogens in hosts expressing the hypersensitive response. Phytochemistry, 58, 33–41. [DOI] [PubMed] [Google Scholar]
- McLeod, A. , Smart, C.D. and Fry, W.E. (2004) Core promoter structure in the oomycete Phytophthora infestans . Eukaryot. Cell 3, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril, C.R. (1990) Gel‐staining techniques. Meth. Enzymol. 182, 477–488. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Mullen, R.T. , Lee, M.S. and Trelease, R.N. (1997) Identification of the peroxisomal targeting signal for cottonseed catalase. Plant J. 12, 313–322. [DOI] [PubMed] [Google Scholar]
- Natvig, D.O. , Sylvester, K. , Dvorachek, W.H. and Baldwin, J.L. (1996) Superoxide dismutases and catalases In: The Mycota III Biochemistry and Molecular Biology (Brambl R. and Marzluf G.A., eds), pp. 191–209. Berlin: Springer‐Verlag. [Google Scholar]
- Navarro, R.E. , Stringer, M.A. , Hansberg, W. , Timberlake, W.E. and Aguirre, J. (1996) catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr. Genet. 29, 352–359. [PubMed] [Google Scholar]
- Neuberger, G. , Maurer‐Stroh, S. , Eisenhaber, B. , Hartig, A. and Eisenhaber, F. (2003) Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 328, 581–592. [DOI] [PubMed] [Google Scholar]
- Niebel, A. , Heungens, K. , Barthels, N. , Inze, D. , Van Montagu, M. and Gheysen, G. (1995) Characterization of a pathogen‐induced potato catalase and its systemic expression upon nematode and bacterial infection. Mol. Plant–Microbe Interact. 8, 371–378. [DOI] [PubMed] [Google Scholar]
- Panabières, F. , Amselem, J. , Galiana, E. and Le Berre, J.‐Y. (2005) Gene identification in the oomycete pathogen Phytophthora parasitica during in vitro vegetative growth through expressed sequence tags. Fungal Genet. Biol. 42, 611–623. [DOI] [PubMed] [Google Scholar]
- Paris, S. , Wysong, D. , Debeaupuis, J.‐P. , Shibuya, K. , Philippe, B. , Diamond, R.D. and Latgé, J.‐P. (2003) Catalases of Aspergillus fumigatus . Infect. Immun. 71, 3551–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraza, L. and Hansberg, W. (2002) Neurospora crassa catalases, singlet oxygen and cell differentiation. Biol. Chem. 383, 569–575. [DOI] [PubMed] [Google Scholar]
- Petnicki‐Ocwieja, T. , Schneider, D.J. , Tam, V.C. , Chancey, S.T. , Shan, L. , Jamir, Y. , Schechter, L.M. , Janes, M.D. , Buell, C.R. , Tang, X.Y. , Collmer, A. and Alfano, J.R. (2002) Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA, 99, 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippi, M.L. , Parish, R.W. and Hohl, H.R. (1975) Histochemical and biochemical evidence for the presence of microbodies in Phytophthora palmivora . Arch. Microbiol. 103, 127–132. [DOI] [PubMed] [Google Scholar]
- Polidoros, A.N. , Mylona, P.V. and Scandalios, J.G. (2001) Transgenic tobacco plants expressing the maize Cat2 gene have altered catalase levels that affect plant‐pathogen interactions and resistance to oxidative stress. Transgenic Res. 10, 555–569. [DOI] [PubMed] [Google Scholar]
- Pompe‐Novaka, M. , Grudena, K. , Baeblera, Š. , Krečič‐Stresa, H. , Kovača, M. , Jongsmac, M. and Ravnikar, M. (2006) Potato virus Y induced changes in the gene expression of potato (Solanum tuberosum L.). Physiol. Mol. Plant Pathol. 67, 237–247. [Google Scholar]
- Powell, M.J. and Bracker, C.E. (1986) Distribution of diamnobenzidine reaction products in zoospores of Phytophthora palmivora . Mycologia, 78, 892–900. [Google Scholar]
- Prakash, K. , Prajapati, S. , Ahmad, A. , Jain, S.K. and Bhakuni, V. (2002) Unique oligomeric intermediates of bovine liver catalase. Protein Sci. 11, 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall, T.A. , Dwyer, R.A. , Huitema, E. , Beyer, K. , Cvitanich, C. , Kelkar, H. , Fong, A.M.V.A. , Gates, K. , Roberts, S. , Yatzkan, E. , Gaffney, T. , Law, M. , Testa, A. , Torto‐Alalibo, T. , Zhang, M. , Zheng, L. , Mueller, E. , Windass, J. , Binder, A. , Birch, P.R.J. , Gisi, U. , Govers, F. , Gow, N.A. , Mauch, F. , Van, W.P. and Judelson, H.S. (2005) Large‐scale gene discovery in the oomycete Phytophthora infestans reveals likely components of phytopathogenicity shared with true fungi. Mol. Plant–Microbe Interact. 18, 229–243. [DOI] [PubMed] [Google Scholar]
- Redinbaugh, M.G. , Sabre, M. and Scandalios, J.G. (1990) The distribution of catalase activity, isozyme protein, and transcript in the tissues of the developing maize seedling. Plant Physiol. 92, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann, S. (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 135, 783–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S.G. , Kang, S.W. , Jeong, W. , Chang, T.S. , Yang, K.S. and Woo, H.A. (2005) Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins Curr. Opin. Cell Biol. 17, 183–189. [DOI] [PubMed] [Google Scholar]
- Del Río, L.A. , Corpas, F.J. , Sandalio, L.M. , Palma, J.M. , Gómez, M. and Barroso, J.B. (2002) Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 53, 1255–1272. [PubMed] [Google Scholar]
- Del Río, L.A. , Sandalio, L.M. , Corpas, F.J. , Palma, J.M. and Barroso, J.B. (2006) Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol. 141, 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbertse, B. , Yoder, O.C. , Nguyen, A. , Schoch, C.L. and Turgeon, B.G. (2003) Deletion of all Cochliobolus heterostrophus monofunctional catalase‐encoding genes reveals a role for one in sensitivity to oxidative stress but none with a role in virulence. Mol. Plant–Microbe Interact. 16, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Robold, A.V. and Hardham, A.R. (1998) Production of species‐specific monoclonal antibodies that react with surface components on zoospores and cysts of Phytophthora nicotianae . Can. J. Microbiol. 44, 1161–1170. [Google Scholar]
- Rocha, E.R. and Smith, C.J. (1995) Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis . J. Bacteriol. 177, 3111–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolke, Y. , Liu, S.J. , Quidde, T. , Williamson, B. , Schouten, A. , Weltring, K.M. , Siewers, V. , Tenberge, K.B. , Tudzynski, B. and Tudzynski, P. (2004) Functional analysis of H2O2‐generating systems in Botrytis cinerea: the major Cu‐Zn‐superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol. Plant Pathol. 5, 17–27. [DOI] [PubMed] [Google Scholar]
- Sachay, D.J. , Hudspeth, D.S.S. , Nadler, S.A. and Hudspeth, M.E.S. (1993) Oomycete MtDNA: Phytophthora genes for cytochrome c oxidase use an unmodified genetic code and encode proteins most similar to those of plants. Exp. Mycol. 17, 7–23. [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Santos, R. , Franza, T. , Laporte, M.‐L. , Sauvage, C. , Touati, D. and Expert, D. (2001) Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol. Plant–Microbe Interact. 14, 758–767. [DOI] [PubMed] [Google Scholar]
- Scherer, M. , Wei, H. , Liese, R. and Fischer, R. (2002) Aspergillus nidulans catalase‐peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell, 1, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten, A. , Tenberge, K.B. , Vermeer, J. , Stewart, J. , Wagemakers, L. , Williamson, B. and Van Kan, J.A.L. (2002) Functional analysis of an extracellular catalase of Botrytis cinerea . Mol. Plant Pathol. 3, 227–238. [DOI] [PubMed] [Google Scholar]
- Schultes, N.P. , Zelitch, I. , McGonigle, B. and Nelson, T. (1994) The primary leaf catalase gene from Nicotiana tabacum and Nicotiana sylvestris . Plant Physiol. 106, 399–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, W. and Hardham, A.R. (2004) Construction of a bacterial articicial chromosome library, determination of genone size, and characterization of an Hsp70 gene family in Phytophthora nicotianae . Fungal Genet. Biol. 41, 369–380. [DOI] [PubMed] [Google Scholar]
- Shan, W. , Marshall, J.S. and Hardham, A.R. (2004) Gene expression in germinated cysts of Phytophthora nicotianae . Mol. Plant Pathol. 5, 317–330. [DOI] [PubMed] [Google Scholar]
- Skadsen, R.W. and Scandalios, J.G. (1987) Translational control of photo‐induced expression of the Cat2 catalase gene during leaf development in maize. Proc. Natl Acad. Sci. USA, 84, 2785–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamnioti, P. , Henderson, C. , Zhang, Z. , Robinson, Z. and Gurr, S.J. (2007) A novel role for catalase B in the maintenance of fungal cell‐wall integrity during host invasion in the rice blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 20, 568–580. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Chen, Z.X. , Du, H. , Liu, Y.D. and Klessig, D.F. (1997) Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 11, 993–1005. [DOI] [PubMed] [Google Scholar]
- Takemoto, D. , Jones, D.A. and Hardham, A.R. (2003) GFP‐tagging of cell components reveals the dynamics of subcellular re‐organization in response to infection of Arabidopsis by oomycete pathogens. Plant J. 33, 775–792. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The Clustal‐X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueeda, M. , Kubota, M. and Nishi, K. (2005) Contribution of jasmonic acid to resistance against Phytophthora blight in Capsicum annuum cv. SCM334. Physiol. Mol. Plant Pathol. 67, 149–154. [Google Scholar]
- Van de Peer, Y. and De Wachter, R. (1997) Evolutionary relationships among the eukaryotic crown taxa taking into account site‐to‐site rate variation in 18S rRNA. J. Mol. Evol. 45, 619–630. [DOI] [PubMed] [Google Scholar]
- Venisse, J.‐S. , Gullner, G. and Brisset, M.‐N. (2001) Evidence for the involvement of an oxidative stress in the initiation of infection of pear by Erwinia amylovora . Plant Physiol. 125, 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová, E. , Inzé, D. and Van Breusegem, F. (2002) Signal transduction during oxidative stress. J. Exp. Bot. 53, 1227–1236. [PubMed] [Google Scholar]
- Wang, G. , Conover, R.C. , Benoit, S. , Olczak, A.A. , Olson, J.W. , Johnson, M.K. and Maier, R.J. (2004) Role of a bacterial organic hydroperoxide detoxification system in preventing catalase inactivation. J. Biol. Chem. 279, 51908–51914. [DOI] [PubMed] [Google Scholar]
- Ng Welinder, K.G. (1991) Bacterial catalase‐peroxidases are gene duplicated members of the plant peroxidase superfamily. Biochim. Biophys. Acta, 108, 215–220. [DOI] [PubMed] [Google Scholar]
- Wu, G.S. , Shortt, B.J. , Lawrence, E.B. , Levine, E.B. , Fitzsimmons, K.C. and Shah, D.M. (1995) Disease resistance conferred by expression of a gene encoding H2O2‐generating glucose oxidase in transgenic potato plants. Plant Cell 7, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.Q. and Pan, S.Q. (2000) An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35, 407–414. [DOI] [PubMed] [Google Scholar]
- Yan, H.Z. and Liou, R.F. (2006) Selection of internal control genes for real‐time quantitative RT‐PCR assays in the oomycete plant pathogen Phytophthora parasitica . Fungal Genet. Biol. 43, 430–438. [DOI] [PubMed] [Google Scholar]
- Yi, S.Y. , Yu, S.H. and Choi, D. (2003) Involvement of hydrogen peroxide in repression of catalase in TMV‐infected resistant tobacco. Mol. Cells 15, 364–369. [PubMed] [Google Scholar]
- Zdobnov, E.M. and Apweiler, R. (2001) InterProScan – an integration platform for the signature‐recognition methods in InterPro. Bioinformatics 17, 847–848. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Henderson, C. and Gurr, S.J. (2004) Blumeria graminis secretes an extracellular catalase during infection of barley: potential role in suppression of host defence. Mol. Plant Pathol. 5, 537–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Multiple sequence alignment of P. nicotianae (PnCat1: EU176856, PnCat2: EU176855, PnCat3: EU258925), P. infestans (PiCat1: PITT_15292, PiCat2: PITT_15248), P. sojae (PsCat1: estExt_fgenesh1_pm.C_80008, PsCat2: estExt_fgenesh1_pg.C_ 80063, PsCat3: scaffold 8: 617248:618661) and P. ramorum (PrCat1: scaffold 31: 335019–333448, PrCat2: fgenesh1_pm.C_ scaffold_31000012, PrCat3: fgenesh1_pm.C_scaffold_39000002) predicted catalase proteins showing the degree of amino acid conservation. The shaded C‐terminal amino acids indicate the PTS1 localization signal of Cat2 proteins.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


