SUMMARY
This Mycosphaerella graminicola pathogen profile covers recent advances in the knowledge of this ascomycete fungus and of the disease it causes, septoria tritici blotch of wheat. Research on this pathogen has accelerated since publication of a previous pathogen profile in this journal in 2002. Septoria tritici blotch continues to have high economic importance and widespread global impact on wheat production.
Taxonomy: Mycosphaerella graminicola (Fuckel) J. Schröt. In Cohn (anamorph: Septoria tritici Roberge in Desmaz.). Kingdom Fungi, Phylum Ascomycota, Class Loculoascomycetes (filamentous ascomycetes), Order Dothideales, Genus Mycosphaerella, Species graminicola.
Host range: Bread and durum wheat (Triticum aestivum L. and T. turgidum ssp. durum L.).
Disease symptoms: Initially leaves develop a chlorotic flecking, which is followed by the development of necrotic lesions which contain brown–black pycnidia. Necrosis causes a reduction in photosynthetic capacity and therefore affects grain yield.
Disease control: The disease is primarily controlled by a combination of resistant cultivars and fungicides. Rapid advances in disease control, especially in resistance breeding, are opening up new opportunities for the management of the disease.
Useful websites: http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html.
INTRODUCTION
Mycosphaerella graminicola is the teleomorph of the ascomycete fungus Septoria tritici, which causes the most economically devastating foliar disease of wheat in Europe and many other temperate climates today, septoria tritici blotch (STB). New knowledge about this important pathosystem is accelerating, demonstrating how important a disease STB has become on a global scale. This review updates the pathogen profile by Palmer and Skinner (2002), since when there have been significant discoveries concerning the fungus, its mode of pathogenicity and the wheat components with which it interacts. This review updates our knowledge of M. graminicola and documents the changes that have propelled this pathosystem into the genomics era.
M. graminicola is a well‐characterized filamentous fungal pathogen propagated by both sexual ascospores and asexual pycnidiospores, and spread by wind‐dispersal and rain splash, respectively (Cohen and Eyal, 1993; Duncan and Howard, 2000; Kema et al., 1996). Eriksen and Munk (2003) reported that the primary source of inoculum infecting newly emerging wheat crops in autumn consisted largely of ascospores. Initial entry into the host is via hyphal growth through the stomatal openings and, although there is no clearly defined appressorium, hyphal tip swellings have been described at the point of stomatal entry (Kema et al., 1996; 2003, 2007; Siah et al., 2010). The fungus grows inside host tissues for up to 4 weeks, but more usually 10–14 days elapse before symptoms are visible. The type of growth during this latent phase has been described as biotrophic, although there is no proof of feeding and no presence of specialized structures associated with other biotrophic pathogens such as haustoria or arbuscules (Keon et al., 2007; Shetty et al., 2007). Indeed, this pathogen has not been reported to breach any host cell wall or membrane. STB disease symptoms consist of grey–brown necrotic lesions which contain brown–black pycnidia (see Fig. 1). In vitro the fungus grows as both a sporulating yeast‐like mass and a filamentous mycelium (see Fig. 2) depending on the conditions, but the yeast‐like stage is not a documented part of the infection or reproductive cycle.
Figure 1.

Septoria tritici blotch symptoms on leaves in artificial inoculation conditions (A) and in the field (B). (Photograph in B by L. Chartrain.)
Figure 2.
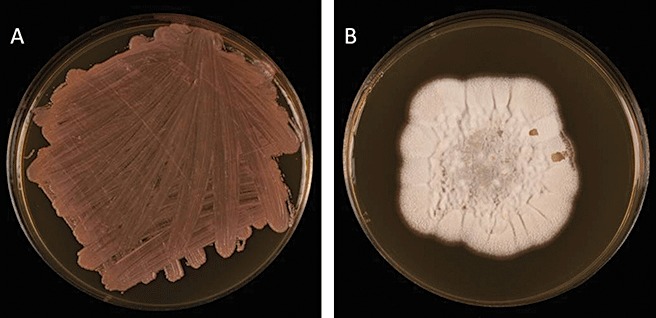
Cultures of Mycosphaerella graminicola isolates growing as a yeast‐like mass (A) and a filamentous mycelium (B).
STB is of global economic importance, but the disease thrives especially in climates with rain during the development of wheat until flag leaf emergence. In the UK, for example, 73% of wheat leaf samples surveyed by the UK's Home Grown Cereals Authority (HGCA) in 2009 were infected by STB (http://www.cropmonitor.co.uk). The prevalence of STB in crops is controlled by a combination of host varietal resistance and applications of chemical fungicides throughout the growing season. There has been substantial progress in breeding resistant wheat varieties in the last 15 years, largely relying on partial resistance which is broadly effective against all known fungal genotypes and therefore durable (Angus and Fenwick, 2008). Although fungicides have been successfully used against M. graminicola, the effectiveness of the two main groups of chemicals has declined as insensitivity to triazoles and quinine outside inhibitors (QoIs) has evolved in the fungal population (2005, 2007). Both the breeding of cultivars with improved genetic resistance, and the development of effective fungicides are slow and demanding processes.
METHODOLOGIES
A well tested toolbox of methods is now in place for studying the M. graminicola–wheat interaction. The fungus can be detected and measured in infected leaves and seeds by quantitative polymerase chain reaction (PCR) (Bearchell et al., 2005; Consolo et al., 2009; Guo et al., 2006; Shetty et al., 2007), which has been used to great effect in studying historical wheat samples in the Rothamsted Research Broadbank archive from 1844 to 2003 (Bearchell et al., 2005). The study of the interaction has been aided by the use of young plants, requiring a short growth time and no vernalization, and by the use of both attached and detached leaf assays (Arraiano et al., 2001a; Keon et al., 2007). Samples of M. graminicola isolates from around the world, representing populations from many different spatial scales over time and in conditions with varying plant husbandry, have been used in research on fungicide sensitivity, virulence and the genetic structure of the global M. graminicola population. Data on disease severity provide the basis for epidemiolgical studies, such as the correlation between STB severity and weather (Pietravalle et al., 2003), and predictive models for use by agronomists and farmers (te Beest et al., 2009).
FUNGAL GENES
The fungal genome
The M. graminicola genome was sequenced by the USA Department of Energy's Joint Genome Institute and has been of enormous importance for research on M. graminicola and for phytopathology in general. The ease with which it is now possible to examine the fungus using bioinformatics tools has provided a great deal of new information regarding individual genes, and has revealed relationships with other phyla and pathogens. There has been a great advance in the characterization of the organization of genes in general, including dispensable chromosomes, and of some individual genes.
The M. graminicola sequence has revealed that the Dutch field isolate IPO323 has a total genome size of 39.7 Mb, and 21 chromosomes ranging in size from approximately 0.3 Mb to about 6 Mb. Thirteen chromosomes are considered to be core chromosomes, being apparently essential, whereas the other eight are known to be independently dispensable despite containing approximately 12% of the genome (Mehrabi et al., 2007; Wittenberg et al., 2009). Of approximately 10 900 genes in the genome that have been functionally annotated, approximately 59% of the genes on the core chromosome have annotations (including automatic and manual curation), whereas this is only the case for approximately 10% of genes on the dispensable chromosomes (S. Goodwin, Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN, USA, personal communication). The genome browser maintained by the Joint Genome Institute provides the genome sequence, organization, automatic and manual annotations, and large amounts of other information on the genes and intervening regions at http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html.
In addition to the loss of dispensable chromosomes, there are other features of genomic plasticity in M. graminicola. Translocation of chromosome sections, chromosome length polymorphisms and chromosome copy number polymorphisms including disomy, the presence of two copies of a chromosome, have all been detected between progeny and parent isolates (Wittenberg et al., 2009). The high genome plasticity could be among the strategies that enable the pathogen population to quickly overcome adverse biotic and abiotic conditions in wheat fields.
There are now many reports of the measurements of M. graminicola gene expression during growth in different conditions. The availability of microarray platforms, in which the expression of many genes can be compared between growth conditions, has added greatly to this area of research. The use of whole‐genome expression profiling is a powerful tool for the identification of the genes involved in adaptation to a pathogenic lifestyle and also genes involved in fungicide sensitivity. An M. graminicola cDNA microarray was developed by Keon et al. (2005b), containing 2500 expressed sequence tags (ESTs), approximately 20% of the genes in the genome. The array has been used to compare gene expression between M. graminicola growing in different in vitro and in planta states (2005a, 2007). It has identified changes in gene expression in response to progression from asymptomatic stages, in which nutrients are limited, to necrotic stages, in which cell contents are available in the apoplast. Transcriptome profiling has also found differences in basal gene expression between epoxiconazole‐sensitive and less sensitive isolates, and showed that epoxiconazole treatment induces the expression of sterol biosynthesis pathway genes and electron transport chain genes. The contribution of these genes to epoxiconazole sensitivity in the field remains unclear however (Cools et al., 2007). Among a larger collection of 27 000 genes, a set of 932 unigenes expressed specifically in planta was identified, including many cell wall‐degrading enzymes, ATP‐binding cassette (ABC) and major facilitator superfamily transporter genes, which may be involved in protection against antifungal compounds or in the secretion of pathogenicity factors (Kema et al., 2008).
Functional characterization of M. graminicola genes
The term ‘pathogenicity’ covers the ability of a pathogen to gain entry into the host, to reproduce and to do so in the face of host resistance. A collection of M. graminicola genes has now been identified and functionally characterized (see Table 1), some of which have been identified as being required for full pathogenicity on wheat. Components of the three fungal mitogen‐activated protein kinase (MAPK) signalling cascades (reviewed by Rispail et al., 2009) have been functionally characterized in M. graminicola and found to play a role in pathogenesis and other characteristics of fungal life. The investigation of signalling components is of use in many areas of study thanks to the information they provide on stimulus recognition, transduction and the resulting changes that can be measured as phenotypes.
Table 1.
Mycosphaerella graminicola genes functionally characterized to date, including their function and phenotype following disruption or deletion.
| Gene name | Type/function | Phenotype | Reference |
|---|---|---|---|
| DGD1 | Aminotransferase | No growth on 2‐methylalanine as sole nitrogen source | Adachi et al. (2003) |
| MgFUS3 | MAPK | No stomatal penetration, no in vitro pycnidia formation | Cousin et al. (2006) |
| MgSlt2 | MAPK | Reduced substomatal colonization | Mehrabi et al. (2006a) |
| MgHOG1 | MAPK osmoregulation | Nonpathogenic, no filamentous growth, no melanization, increased osmosensitivity, increased sensitivity to some fungicides | Mehrabi et al. (2006b) |
| MgBcy1 | Protein kinase A catalytic subunit | Post‐penetration defects, no pycnidia production | Mehrabi & Kema (2006) |
| MgTpk2 | Protein kinase A regulatory subunit | Post‐penetration defects, no pycnidia production | Mehrabi & Kema (2006) |
| MgSte7 | MAPKK | Nonpathogenic, reduced filamentous growth, increased melanization | Kramer et al. (2009) |
| MgSte11 | MAPKKK | Nonpathogenic, reduced filamentous growth, increased melanization | Kramer et al. (2009) |
| MgSte12 | Transcription factor | Reduced pathogenicity, some reduction in filamentous growth | Kramer et al. (2009) |
| MgSte20 | P21 activated kinase | Wild‐type pathogenicity, severe reduction in in vitro filamentous growth | Kramer et al. (2009) |
| MgSte50 | MAPK‐adaptor protein | Reduced pathogenicity, some reduction in filamentous growth, increased melanization | Kramer et al. (2009) |
| MgGpa1 | Gα protein | Increased filamentous growth in vitro, only induces chlorotic symptoms | Mehrabi et al. (2009) |
| MgGpa2 | Gα protein | Wild‐type, no phenotypic changes detected | Mehrabi et al. (2009) |
| MgGpa3 | Gα protein | Increased yeast‐like growth in vitro, reduced intracellular cyclic AMP content, only induces chlorotic symptoms | Mehrabi et al. (2009) |
| MgGpb1 | Gβ protein | Increased anastomosis, reduced intracellular cyclic AMP content, only induces chlorotic symptoms | Mehrabi et al. (2009) |
| MgMfs1 | Major facilitator superfamily protein | Increased sensitivity to strobilurins | Roohparvar et al. (2007) |
| MgAtr1 | ABC transporter | Wild‐type, no phenotypic changes detected | Stergiopoulos et al. (2003) |
| MgAtr2 | ABC transporter | Wild‐type, no phenotypic changes detected | Stergiopoulos et al. (2003) |
| MgAtr3 | ABC transporter | Wild‐type, no phenotypic changes detected | Stergiopoulos et al. (2003) |
| MgAtr4 | ABC transporter | Delayed virulence | Stergiopoulos et al. (2003) |
| MgAtr5 | ABC transporter | Wild‐type, no phenotypic changes detected | Stergiopoulos et al. (2003) |
| MgAtr7 | ABC transporter | Role in transport and iron homeostasis | Zwiers et al. (2007) |
| MgNLP | Necrosis and ethylene‐inducing peptide‐like | Wild‐type, no phenotypic changes detected | Motteram et al. (2009) |
ABC, ATP‐binding cassette; MAPK, mitogen‐activated protein kinase.
The MAPK pathways thought to regulate the mating and pheromone responses have been investigated extensively in M. graminicola. The deletion of MAPK kinase kinase, MAPK kinase and MAPK individually (MgSte11, MgSte7 and MgFus3, respectively) reduced filamentous growth and pathogenicity, the latter in large part as a result of the pre‐penetration defect afforded by reduced hyphal growth (Cousin et al., 2006; Kramer et al., 2009). Signalling through the cyclic AMP pathway is putatively associated with the mating and pheromone response pathway, and itself plays many roles in phytopathogenic fungal life cycles including filamentous growth, mating and virulence (reviewed by Lee et al., 2003). The individual deletion of M. graminicola Gα (MgGpa3) and Gβ (MgGpb1) encoding genes resulted in reduced filamentous growth caused by either a greater amount of budding growth or increased areas of hyphal fusion (Mehrabi et al., 2009). The genes encoding catalytic and regulatory subunits of the cAMP‐dependent protein kinase A were also characterized (MgBcy1 and MgTpk2 respectively; Mehrabi and Kema, 2006). The protein kinase A acts downstream of the G proteins and disruption of MgTpk2 also causes a reduction in filamentous growth. Notably, mutations in four of the genes in this signalling pathway delayed pathogenicity and inhibited the production of pycnidia (Mehrabi and Kema, 2006; Mehrabi et al., 2009).
MgHog1 encodes another MAP kinase, the function of which in M. graminicola appears to influence filamentous growth. MgHog1 was predicted to be homologous to a MAP kinase in the well‐described Hog1 osmoregulation pathway of yeast. In M. graminicola, it is an absolute requirement for filamentous growth of the fungus, which is necessary for infection of wheat leaves. Other alterations in the phenotype of the MgHog1 transformant were a lack of melanization, increased osmosensitivity and resistance to some fungicides, including two phenylpyrroles (fludioxonil and fenpiclonil) and one of two dicarboximides tested (iprodione, but not vinclozolin) (Mehrabi et al., 2006b). MgSlt2 encodes a MAP kinase, a homologue of which is necessary for cell wall integrity in Saccharomyces cerevisiae. In M. graminicola, disruption of MgSlt2 caused reduced colonization of the substomatal cavity, but hyphal growth and penetration were not altered (Mehrabi et al., 2006a). The present characterization of these three MAPK signalling pathways shows an interesting division between those with a role in the morphological state of growth and the MgSlt2 gene, which was altered in its interaction with the wheat host at a post‐penetration stage.
In addition, six M. graminicola genes encoding transport proteins have been functionally characterized. There is a need, however, for more information on chemical transport into and out of fungal cells, particularly in M. graminicola, where the nutritional strategy is much debated and potential modes of fungicide insensitivity are of obvious concern. Transport proteins that might contribute to either of these roles, including the major facilitator and ABC classes of transporter, are therefore of great interest. MgMfs1, which encodes a major facilitator superfamily protein, has been disrupted. The transporter was not essential for full virulence, but the disruption strains showed increased sensitivity to strobilurin fungicides, implicating the transporter in multi‐drug resistance, but not in toxin secretion, as had been suggested by Roohparvar et al. (2007). MgAtr1, MgAtr2, MgAtr3, MgAtr4 and MgAtr5 encode ABC transporter proteins. Individual disruption of MgAtr1, MgAtr2, MgAtr3 and MgAtr5 did not alter pathogenicity, whilst disruption of MgAtr4 delayed virulence and reduced lesion size, although pycnidia with viable pycnidiospores were still produced (Stergiopoulos et al., 2003). MgAtr7 encodes an ABC transporter that was found to have no detectable effect on pathogenicity, but to have a role in iron homeostasis (Zwiers et al., 2007).
Potential secreted proteins with roles in pathogen–host interaction have been functionally characterized. The necrosis and ethylene‐inducing peptide 1 (Nep‐1)‐like protein, MgNLP, was found to induce cell death in Arabidopsis, but not in wheat. MgNLP was not essential for M. graminicola infection of wheat, despite its transcriptional increase during presymptomatic colonization of susceptible wheat leaves (Motteram et al., 2009). M. graminicola therefore contains a gene encoding a protein sufficient to induce cell death on a nonhost, which is particularly interesting as this pathogen is thought to have strict host specificity.
The majority of M. graminicola‐secreted proteins examined so far have gene expression profiles which show that they are most strongly expressed during the symptomless phase of host colonization (Motteram et al., 2009; Rudd et al., 2010). The investigation of fungal genes that are potentially in contact with or interact with living wheat cells before and during the appearance of disease symptoms could provide further insight into the mode of fungal pathogenicity, its interaction with the host and the processes of host resistance and susceptibility.
Mode of pathogenesis
There have been significant advances in our knowledge of how M. graminicola interacts with host cells to result in host cell death and completion of the life cycle of the fungus. Firstly, the cell death pathway induced in wheat cells during compatible interactions resembles the apoptosis‐like pathway (Keon et al., 2007). Secondly, features of the plant–pathogen interface between wheat and M. graminicola have been examined, including small sugars, which may act as pathogen‐associated molecular patterns (PAMPs), and the interchange of reactive oxygen species (ROS) (2003, 2007).
The nutrition of M. graminicola is thought to be greatly influenced by host cell death. It has been stated that nutrients in the apoplast are sufficient to support the growth of intercellular fungi, such as M. graminicola (Spencer‐Phillips, 1997). Quantitative PCR measurements, however, showed little increase in fungal biomass before host cell death (Keon et al., 2007; Shetty et al., 2007), and so it is unclear how much M. graminicola growth is supported by apoplastic‐derived nutrients in the latent or biotrophic phase. Keon et al. (2007) examined apoplast metabolite levels using 1H‐nuclear magnetic resonance (1H‐NMR) spectroscopy and metabolomic analysis during symptomless and necrotic periods of infection, and found an increase in the quantity of plant‐derived compounds available in the apoplast from just before the onset of host cell death and to afterwards. Current evidence therefore suggests that there is little biotrophic feeding, perhaps even none, in the asymptomatic phase which precedes host cell death. The start of fungal growth coincides with nutrient release at the time of host cell death, as reflected in the induction of a number of genes related to energy production at this time.
Keon et al. (2007) showed that cellular features consistent with apoptosis were present in wheat cells dying following leaf infection by a compatible strain of M. graminicola, including leakage of cytochrome c from the mitochondria into the cytoplasm, and the characteristic DNA laddering seen on agarose gels, which indicates internucleosomal cleavage together with the degradation of RNA. It was shown that cell contents leak into the apoplastic spaces during cell death, but only after the initial asymptomatic phase.
The wheat MAPK, TaMPK3, has been implicated in the induction of the cell death pathway that produces STB symptoms (Rudd et al., 2008). The orthologue of TaMPK3, AtMPK3/WIPK, in Arabidopsis thaliana, has been implicated in the response to PAMPs and avirulence‐resistance (AVR‐R) incompatible interactions (see references in Rudd et al., 2008). TaMPK3 gene expression was induced during asymptomic colonization by a compatible M. graminicola isolate, possibly indicating either nonspecific PAMP recognition or highly specific manipulation of the host responses to initiate cell death. The TaMPK3 protein was post‐translationally activated during the infection period, coincident with the first appearance of disease symptoms and the initial commitment of wheat cells to programmed cell death (PCD). Finally, the TaMPK3 protein was found in increased concentrations from the time of macroscopic appearance of disease symptoms onwards. This pathway was only activated in a compatible M. graminicola–wheat interaction, not in incompatible interactions. This is the opposite of the pattern reported previously in interactions of plants with biotrophic pathogens. It is not yet known how this pathway is activated, but one model proposes stage‐specific production of fungal toxins or elicitors which initiate MAP kinase activity and trigger host cell death (Kema et al., 1996; Keon et al., 2007). The dothideomycete wheat pathogens Pyrenophora tritici‐repentis and Stagonospora nodorum generate necrosis‐inducing toxins as components of their virulence arsenal (reviewed by Ciufetti et al., 2010; Deller et al., 2011). Effectors have not yet been isolated from M. graminicola and their activity confirmed, but three homologues of the Cladosporium fulvum effector gene Ecp2 have been identified in the M. graminicola genome (Stergiopoulos et al., 2010). The results of research on PCD and the MAPK pathway provide further support for the hypothesis that resistance may result from an interaction between, as yet, unidentified AVR‐R proteins, whereas the aggressive host response seen in a compatible interaction may be the result of the fungus hijacking disease resistance signalling pathways (Hammond‐Kossack and Rudd, 2008).
Recognition of M. graminicola by host cells has been reported via the perception of β‐1,3‐glucans in a manner that resembles a PAMP–pathogen recognition receptor interaction. These small molecules induce the expression of an apoplastically located wheat endo‐1,3‐β‐glucanase gene, induce callose deposition and their presence inhibits STB symptom development (Shetty et al., 2009). β‐Glucans are probably produced by the activity of wheat endo‐1,3‐β‐glucanases in the apoplastic space. This PAMP must be recognized by the plant, presumably by a receptor yet to be discovered. The mechanisms involved in pathogen recognition and disease resistance in the M. graminicola–wheat pathosystem are yet to be elucidated.
ROS, in particular hydrogen peroxide, H2O2, are important in the mechanisms regulating STB. H2O2 accumulated in a resistant cultivar early in infection. This initial response was expected as a precursor to a hypersensitive response, but no necrotic symptoms were seen in a resistant interaction. In a susceptible cultivar, much larger quantities of H2O2 accumulated later in infection than in earlier stages, when the pathogen started to increase its rate of hyphal growth (Shetty et al., 2003). It appears that the accumulation of H2O2 aids pathogen growth, but H2O2 may only be indirectly necessary for pathogenicity. Shetty et al. (2007) demonstrated that H2O2 inhibited the growth of M. graminicola in vitro and hindered pathogen growth in planta at both early and late stages of infection. Genes encoding some ROS‐scavenging enzymes are highly induced in M. graminicola during symptomatic infection stages (Keon et al., 2007). Studies of other necrotroph–plant interactions have suggested that ROS can aid the development of the pathogen (Govrin and Levine, 2000), whilst ROS are required for limiting the growth of an endophyte (Tanaka et al., 2006). This indicates a potential regulatory role for ROS in the stealthy growth of M. graminicola during the early stages of infection. It remains unclear as to whether the H2O2 observed is host or pathogen generated. If it is generated by the pathogen, there may be a role in inducing or regulating cell death, which aids pathogen development.
Resistance genes and breeding
Resistant cultivars are an effective means of controlling STB, but, until recently, breeders relied on uncharacterized genetic resistance in breeding programmes (Chartrain et al., 2005b). Resistance to STB has been broadly divided into two classes; specific and quantitative. Specific resistance is near‐complete and oligogenic. Partial or quantitative resistance is incomplete and polygenic (Jlibene et al., 1994; Zhang et al., 2001).
Thirteen major genes in wheat for resistance to STB have so far been identified and mapped (see Table 2), Stb1–Stb12 and Stb15, but the resistance mechanisms by which these genes confer resistance to specific pathogen genotypes are currently unknown and none of the Stb genes have been cloned. A gene‐for‐gene relationship has been demonstrated between wheat Stb6, the best understood of these genes, and M. graminicola (Brading et al., 2002). The gene‐for‐gene relationship is the most studied and yet is the least durable in the field, because pathogen populations can adapt to the selection pressure placed on them by the presence of a major resistance gene.
Table 2.
Major resistance genes and their chromosome locations, and the wheat varieties and Mycosphaerella graminicola isolates used to map them.
| Gene | Chromosome location | Isolate | Wheat variety | Reference |
|---|---|---|---|---|
| Stb1 | 5BL | IN95‐Lafayette‐1196‐ww 1‐4 | Bulgaria 88 | Adhikari et al. (2004c) |
| Stb2 | 3BS | Paskeville | Veranopolis | Adhikari et al. (2004b) |
| Stb3 | 6DS | Paskeville | Israel 493 | Adhikari et al. (2004b) |
| Stb4 | 7DS | IN95‐Lafayette‐1196‐ww 1‐4 | Tadinia | Adhikari et al. (2004a) |
| Stb5 | 7DS | IPO94269 | Synthetic 6X | Arraiano et al. (2001b) |
| Stb6 | 3AS | IPO323 | Flame | Brading et al. (2002) |
| Stb7 | 4AL | MG2 | ST6 | McCartney et al. (2003) |
| Stb8 | 7BL | IN95‐Lafayette‐1196‐WW 1‐4 | W7984 synthetic | Adhikari et al. (2003) |
| Stb9 | 2BL | IPO89011 | Courtot | Chartrain et al. (2009) |
| Stb10 | 1D | IPO94269 and ISR8036 | Kavkaz‐K4500 L.6.A.4 | Chartrain et al. (2005a) |
| Stb11 | 1BS | IPO90012 | TE911 | Chartrain et al. (2005c) |
| Stb12 | 4AL | ISR398 | Kavkaz‐K4500 L.6.A.4 | Chartrain et al. (2005a) |
| Stb15 | 6AS | IPO88004 | Arina | Arraiano et al. (2007) |
Partial resistance is generally much more durable than gene‐for‐gene, race‐specific resistance, but is harder to select and less well studied than specific resistance. Chartrain et al. (2004b) used a double‐haploid population between a susceptible and resistant variety, Riband and Arina, respectively, to attempt to locate quantitative trait loci (QTL) and determine the genetics of this partial resistance. No QTL controlled a significant fraction of variation in the resistant parent, Arina, in which partial resistance is therefore most probably controlled by several dispersed genes. Chartrain et al. (2004b) also showed that partial resistance is isolate nonspecific and therefore probably durable. The investigation of quantitative resistance is complicated by the fact that resistance at the seedling and adult stages is sometimes controlled by different genes. Chartrain et al. (2004b) reported no correlation between disease levels on seedlings and adult plants in the Arina × Riband population, while Eriksen et al. (2003) detected QTLs for resistance at the adult stage in a population of Senat × Savannah which were not present at the seedling stage. This has implications for breeding, because resistance at all growth stages is desirable, although resistance is most important when the weather is most conducive to symptom development and pathogen spread, namely in the later adult stages.
The emergence of resistance of M. graminicola to QoI fungicides and, more recently, triazole‐based fungicides has increased the need to develop resistant varieties of wheat as a cost‐effective means of controlling the disease. Much work has been carried out over the last few years to improve breeding efforts for resistance to STB. Knowledge about the distribution of resistance genes in wheat varieties has advanced considerably. The presence of specific resistance genes (Arraiano and Brown, 2006) and partial resistance (Arraiano et al., 2009) in 238 European wheat varieties has been ascertained. Chartrain et al. (2004a) screened 24 varieties with 12 isolates of M. graminicola for isolate‐specific resistance, and identified new sources of resistance that could be utilized in breeding. Some varieties, such as TE911, could be used in crossing programmes to provide both partial and specific resistance (Chartrain et al., 2005c). It appears that the major resistance gene Stb6 has entered wheat breeding programmes on numerous occasions and has been used world‐wide as a source of STB resistance (Chartrain et al., 2005b). Stb6 explains a significant level of variability in susceptibility to STB in the field (Arraiano et al., 2009), which may explain why it is present in many well‐known sources of resistance. The mechanism behind this resistance is not known; Stb6 may confer partial resistance to STB itself, or may be linked to a gene conferring partial resistance. Studies have shown that the pyramiding of genes for resistance may also help with breeding efforts for more durably resistant wheat. The identification of varieties with more than one resistance gene, such as the breeding lines Kavkaz‐K4500 and TE9000, which show good resistance to STB, suggests that the pyramiding of resistance genes might achieve high levels of field resistance (Chartrain et al., 2004a). In field trials, several cultivars were identified with especially high levels of partial resistance, some with no known specific resistance genes. These may be useful sources in breeding for STB resistance (Arraiano et al., 2009). In summary, this research has shown that breeders, at least in Europe, have sufficient genetic variation in their germplasm, and increasingly have the information available to make more informed choices about specific parents to use in crosses.
Molecular basis of wheat resistance to M. graminicola
The goal of identifying specific host responses to M. graminicola inoculation is of interest to understand how disease symptoms are caused and how resistance works. For example, there is no means of identifying specific resistances in wheat varieties or avirulences in M. graminicola isolates other than a rather complicated statistical analysis of quantitative disease symptoms (see Arraiano and Brown, 2006 for an example). It would be greatly preferable to have a test in which an incompatible interaction could be recognized by its phenotype, as with powdery mildew and rust diseases of cereals (Ma and Singh, 1996; Moseman et al., 1965; Stubbs et al., 1986). In addition, it would benefit breeding programmes where time and money could be saved through the early identification of resistant lines. The M. graminicola–wheat interaction is not closely related to any other model pathosystem and therefore defining the differences between incompatible and compatible interactions is important to assist with the selection of STB‐resistant material. As yet, few studies have been carried out that have attempted to identify the host genes involved in the resistance response, but those that have indicate that defence responses in wheat are activated before the fungus has even penetrated the host, 12 h after inoculation (Adhikari et al., 2007; Ray et al., 2003). Most of the genes identified as differentially expressed during infection are pathogenesis‐related (PR) genes, although a few others, possibly involved with signalling or regulatory pathways, have been investigated (Adhikari et al., 2007).
PR proteins are well characterized because of their association with disease resistance, although their function is still not always clear. Some have enzymatic activity, such as β‐1,3‐glucanases and chitinases. In plant diseases in general, PR gene transcripts accumulate during both incompatible and compatible interactions, but earlier and more strongly in incompatible responses (Boyd et al., 1994; Ray et al., 2003). During the interaction with M. graminicola, wheat defence‐related genes, such as chitinase and PR1, are strongly upregulated at an early stage (Ray et al., 2003). Adhikari et al. (2007) proposed that the expression level of PR1 12 h after inoculation could distinguish between resistant and susceptible lines in segregating mapping populations, as there was little change in expression of any of the defence‐related genes tested in two susceptible cultivars. In contrast, Shetty et al. (2009) showed that, although β‐1,3‐glucanase and chitinase are slightly but significantly upregulated early in an incompatible interaction, they are strongly upregulated in a compatible interaction from 9 days after inoculation. The results of these studies have evidently been variable and, although the number of such studies is still limited, they indicate that resistance to M. graminicola may be complex.
There may be two stages of defence at early and later stages of infection (Adhikari et al., 2007), once during initial recognition and again later, when the fungus starts to grow within the leaf. Some studies have suggested that genes other than known PR genes are upregulated during the wheat–M. graminicola interaction (Adhikari et al., 2007; Ray et al., 2003), but the mechanism leading to resistance or susceptibility is as yet unknown. Rudd et al. (2008) proposed that, in compatible interactions, there appears to be an active response to M. graminicola involving the wheat TaMPK3, as described previously. The molecular basis of resistance shows an absence of MPK3 activation seen in other incompatible fungal pathogen–host interactions, e.g. Cladosporium fulvum on Cf‐9 tomato, where the MPK3 homologue is activated (Romeis et al., 1999).
In some experiments, the pattern of gene expression in wheat in response to M. graminicola has contrasted sharply with that expected from previous studies of plant infection by biotrophic pathogens, and the lack of consistency between experiments points to significant genotype‐by‐environment interaction in defence mechanisms. Further studies of gene expression in both incompatible and compatible interactions using different variety and isolate combinations may elucidate what appears to be a complicated pattern. The decrease in the cost of global gene expression studies may enable this useful tool to assist in the identification of quantifiable markers and perhaps novel genes specific to compatible or incompatible interactions.
Population biology
Recent research on the population biology of M. graminicola has offered insights into gene flow in fungal populations, over both large geographical areas and within crops. Molecular phylogenetic analysis has proved to be a powerful tool in understanding the origins and routes of dispersal of agricultural pathogens and of alleles which have enabled pathogens to adapt to crops (Stukenbrock and McDonald, 2008). The origin of M. graminicola is thought to have followed closely the origin and development of wheat as a crop. DNA sequence data indicate that M. graminicola specialized to cultivated wheat may have originated by sympatric speciation from a pathogen of wild grasses in Iran at approximately 8000–9000 bc (Stukenbrock et al., 2007). This hypothesis is supported by the discovery that, unlike populations elsewhere in the world, in Iran there is strong genetic differentiation between wheat‐growing provinces (Abrinbana et al., 2010), as expected of an organism at its centre of origin. The increase in the cultivation of wheat coincided with a great diversification in the pathogen at 3000–2000 bc (Stukenbrock et al., 2007).
Research on population structure has shown that, in most of the world, M. graminicola has four significant features: a high degree of sexual reproduction, together with much asexual reproduction, a large effective population size and evidence of recent gene flow (Zhan et al., 2003). These features distinguish this species from other fungal pathogens of global agricultural importance (e.g. yellow rust; Hovmøller et al., 2002). The great majority, perhaps even all, of the fungal population undergoes a sexual cycle each year. Within each leaf layer and in the crop as a whole, there is a shift in the type of fruiting body produced over time from pycnidia to perithecia (Eriksen and Munk, 2003). The large effective population size (N e) of M. graminicola, indicated by high levels of genotypic diversity in markers such as DNA fingerprints (Zhan et al., 2003), is a consequence of the high frequency of sexual reproduction (Cowger et al., 2008). This combination of sexual and asexual recombination provides a potent evolutionary advantage to the fungus (McDonald and Linde, 2002). Asexual reproduction of plant pathogens allows rapid propagation of successful genotypes in a location in which an individual genotype has been successful (by definition, because it has reproduced), which is advantageous for a pathogen of crops grown as fields of pure varieties. Sexual reproduction, by contrast, generates new genotypes by recombination, accelerating adaptation to new selective forces, such as resistant varieties or fungicides (Brown and Hovmøller, 2002). The analysis of population structure revealed by molecular markers has shown that, although M. graminicola global populations contain some subdivision because of geographical isolation, gene flow has largely acted to remove such barriers (Zhan et al., 2003). This may be the result of some regional, wind‐borne dispersal of ascospores combined with trans‐continental movement of the pathogen, possibly on straw. Following its spread to domesticated wheat in the Neolithic period, the global spread of M. graminicola appears to have paralleled that of wheat, following the crop across countries and continents. It is striking that there appears to have been gene flow in M. graminicola from North America to Europe in the last 100 years, possibly resulting from food imports during the First and Second World Wars (Banke and McDonald, 2005).
Fungicides
Fungicides are widely used to control STB, despite their cost, loss of efficacy owing to pathogen insensitivity and the growing governmental and public concern over their environmental impact (Haynes et al., 2010). One of the major groups of fungicides, QoI, also known as strobilurin, acts against mitochondrial protein cytochrome b, but is no longer effective against STB. A mutation from glycine to alanine at base 143 (G143A) in the cytochrome b protein sequence, causing apparently total loss of efficacy of QoI fungicides (Fraaije et al., 2003), was found in widespread field isolates (Fraaije et al., 2005).
Sterol demethylation inhibitors, including triazole fungicides, act on CYP51, the cytochrome P450 eburicol 14‐demethylase enzyme of the ergosterol biosynthesis pathway (Stergiopoulos et al., 2003). Many different amino acid substitutions have been reported in CYP51, some associated with the reduced sensitivity of triazoles (Cools and Fraaije, 2008; Fraaije et al., 2007; Leroux et al., 2007).
An analysis of the evolution of CYP51 in Europe showed that triazole‐resistant alleles of the protein were generated by a combination of mutation and intragenic recombination, and subsequently dispersed throughout Northern Europe (Brunner et al., 2008). Mutations in CYP51 have been demonstrated to affect responses to triazoles in experiments involving heterologous transformation of S. cerevisiae (Cools et al., 2010). They also modify the biosynthetic function of the CYP51 enzyme, altering the cellular concentration of sterol precursor molecules and preventing the M. graminicola gene from complementing the yeast orthologue (Bean et al., 2009; Cools et al., 2010). Transcription profiling of the M. graminicola response to epoxiconazole detected increased expression of many components of the sterol biosynthesis pathway, and also many genes encoding mitochondrial electron transport chain proteins (Cools et al., 2007). The reproductive and dispersive power of M. graminicola has allowed the spread of novel mutations and alleles within large regional populations. Mutations to the CYP51 protein have allowed the study of how mutations occur, evolve and subsequently spread within geographical regions (Brunner et al., 2008; Cools et al., 2010).
A different mode of fungicide insensitivity involves the alteration in transport of fungicides across the fungal plasma membrane. This is exemplified by drug efflux protein overexpression (Zwiers et al., 2002). This is a multi‐drug effective strategy, and although first demonstrated in laboratory strains, the ABC transporter gene MgAtr1 was more highly expressed in strains with greater insensitivity to cyproconazole. The multi‐drug transporter MgMfs1 is also thought to alter sensitivity to storbilurins, as its overexpression reduced strobilurin sensitivity in yeast and disruption of the gene in M. graminicola increased sensitivity (Roohparvar et al., 2007). Both the G143A mutation and overexpression of MgMfs1 have been found together in field isolates, although in the case of the double appearance, the influence of the altered expression of MgMfs1 on fungicide sensitivity has not been proven (Roohparvar et al., 2008).
DISCUSSION
As with every plant disease, research and development are working towards control through durably resistant crop varieties and effective chemical control. New knowledge about this pathosystem is increasing rapidly. Since the previous pathogen profile on M. gramnicola (Palmer and Skinner, 2002), over 400 peer‐reviewed papers have been published relating to this fungus. These document the mapping of resistance genes, a gene‐for‐gene relationship between pathogen and host, and the identification of M. graminicola genes that play a role in pathogenic virulence. Large datasets have been produced and made publicly available, including the complete fungal genome sequence and large gene expression collections.
Many key challenges lie ahead; some of the biggest steps we should look forward to include the cloning of Stb genes and their interacting fungal genes, sequencing of the wheat genome and multiple M. graminicola sequences. These steps should lead to the identification of key pathogenicity/virulence genes which might be fungicide or wheat breeding targets. Undoubtedly, there will also be many unexpected discoveries along the way.
Although M. graminicola is now the most well‐understood species among the dothideomycetes, it has unique features that set it apart from other phytopathogenic fungi. It is debatable whether or not it has a truly biotrophic growth phase following infection, and may be more properly considered as a necrotroph with an exceptionally long latent or endophytic period. From the point of view of the host, there are strong indications that resistance operates, at least in part, by suppression of metabolic and signalling pathways that would confer resistance to many pathogens, but susceptibility to M. graminicola. The question of the extent to which these findings apply to other fungi currently described as hemibiotrophs in general, and to other dothideomycetes and indeed other Mycosphaerella spp. in particular, is an intriguing subject for future research.
ACKNOWLEDGEMENTS
The authors thank Dr Jason Rudd and Dr Graham McGrann for their input into the manuscript. Mrs Elizabeth Orton is funded by a Biotechnology and Biological Sciences Research Council (BBSRC) Targeted Priority Studentship. Dr Sian Deller was supported by a BBSRC Industrial CASE PhD studentship awarded to Syngenta and is currently funded by the Institut National de la Recherche Agronomique (INRA). The John Innes Centre and Rothamsted Research Station receive grant aided support from the BBSRC.
REFERENCES
- Abrinbana, M. , Mozafari, J. , Shams‐bakhsh, M. and Mehrabi, R. (2010) Genetic structure of Mycosphaerella graminicola populations in Iran. Plant Pathol. 59, 829–838. [Google Scholar]
- Adachi, K. , Nelson, G.H. , Peoples, K.A. , DeZwaan, T.M. , Skalchunes, A.R. , Heiniger, R.W. , Shuster, J.R. , Hamer, L. and Tanzer, M.M. (2003) Sequence analysis and functional characterization of the dialkylglycine decarboxylase gene DGD1 from Mycosphaerella graminicola . Curr. Genet. 43, 358–363. [DOI] [PubMed] [Google Scholar]
- Adhikari, T.B. , Anderson, J.M. and Goodwin, S.B. (2003) Identification and molecular mapping of a gene in wheat conferring resistance to Mycosphaerella graminicola . Phytopathology, 93, 1158–1164. [DOI] [PubMed] [Google Scholar]
- Adhikari, T.B. , Cavaletto, J.R. , Dubcovsky, J. , Gieco, J.O. , Schlatter, A.R. and Goodwin, S.B. (2004a) Molecular mapping of the Stb4 gene for resistance to septoria tritici blotch in wheat. Phytopathology, 94, 1198–1206. [DOI] [PubMed] [Google Scholar]
- Adhikari, T.B. , Wallwork, H. and Goodwin, S.B. (2004b) Microsatellite markers linked to the Stb2 and Stb3 genes for resistance to Septoria tritici blotch in wheat. Crop Sci. 44, 1403–1411. [Google Scholar]
- Adhikari, T.B. , Yang, X. , Cavaletto, J.R. , Hu, X. , Buechley, G. , Ohm, H.W. , Shaner, G. and Goodwin, S.B. (2004c) Molecular mapping of Stb1, a potentially durable gene for resistance to septoria tritici blotch in wheat. Theor. Appl. Genet. 109, 944–953. [DOI] [PubMed] [Google Scholar]
- Adhikari, T.B. , Balaji, B. , Breeden, J. and Goodwin, S.B. (2007) Resistance to wheat Mycosphaerella graminicola involves early and late peaks of gene expression. Physiol. Mol. Plant Pathol. 70, 55–68. [Google Scholar]
- Angus, W.J. and Fenwick, P.M. (2008) Using genetic resistance to combat pest and disease threats. In: Arable Cropping in a Changing Climate, Home Grown Cereals Authority (HGCA) Conference, 23 and 24 January 2008, pp. 21–27. London: HGCA. [Google Scholar]
- Arraiano, L.S. and Brown, J.K.M. (2006) Identification of isolate‐specific and partial resistance to septoria tritici blotch in 238 European wheat cultivars and breeding lines. Plant Pathol. 55, 726–738. [Google Scholar]
- Arraiano, L.S. , Brading, P.A. and Brown, J.K.M. (2001a) A detached seedling leaf technique to study resistance to Mycosphaerella graminicola (anamorph Septoria tritici) in wheat. Plant Pathol. 50, 339–346. [Google Scholar]
- Arraiano, L.S. , Worland, A.J. , Ellerbrook, C. and Brown, J.K.M. (2001b) Chromosomal location of a gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in the hexaploid wheat ‘Synthetic 6x’. Theor. Appl. Genet. 103, 758–764. [Google Scholar]
- Arraiano, L.S. , Chartrain, L. , Bossolini, E. , Slatter, H.N. , Keller, B. and Brown, J.K.M. (2007) A gene in European wheat cultivars for resistance to an African isolate of Mycosphaerella graminicola . Plant Pathol. 56, 73–78. [Google Scholar]
- Arraiano, L.S. , Balaam, N. , Fenwick, P.M. , Chapman, C. , Feuerhelm, D. , Howell, P. , Smith, S.J. , Widdowson, J.P. and Brown, J.K.M. (2009) Contributions of disease resistance and escape to the control of septoria tritici blotch of wheat. Plant Pathol. 58, 910–922. [Google Scholar]
- Banke, S. and McDonald, B.A. (2005) Migration patterns among global populations of the pathogenic fungus Mycosphaerella graminicola . Mol. Ecol. 14, 1881–1896. [DOI] [PubMed] [Google Scholar]
- Bean, T.P. , Cools, H.J. , Lucas, J.A. , Hawkins, N.D. , Ward, J.L. , Shaw, M.W. and Fraaije, B.A. (2009) Sterol content analysis suggests altered eburicol 14 alpha‐demethylase (CYP51) activity in isolates of Mycosphaerella graminicola adapted to azole fungicides. FEMS Microbiol. Lett. 296, 266–273. [DOI] [PubMed] [Google Scholar]
- Bearchell, S.J. , Fraaije, B.A. , Shaw, M.W. and Fitt, B.D.L. (2005) Wheat archive links long‐term fungal pathogen population dynamics to air pollution. Proc. Natl. Acad. Sci. USA, 102, 5438–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Beest, D.E. , Shaw, M.W. , Paveley, N.D. and van den Bosch, F. (2009) Evaluation of a predictive model for Mycosphaerella graminicola for economic and environmental benefits. Plant Pathol. 58, 1001–1009. [Google Scholar]
- Boyd, L.A. , Smith, P.H. , Green, R.M. and Brown, J.K.M. (1994) The relationship between the expression of defense‐related genes and mildew development in barley. Mol. Plant–Microbe Interact. 7, 401–410. [Google Scholar]
- Brading, P.A. , Verstappen, E.C.P. , Kema, G.H.J. and Brown, J.K.M. (2002) A gene‐for‐gene relationship between wheat and Mycosphaerella graminicola, the septoria tritici blotch pathogen. Phytopathology, 92, 439–445. [DOI] [PubMed] [Google Scholar]
- Brown, J.K.M. and Hovmøller, M.S. (2002) Epidemiology—aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science, 297, 537–541. [DOI] [PubMed] [Google Scholar]
- Brunner, P.C. , Stefanato, F.L. and McDonald, B.A. (2008) Evolution of the CYP51 gene in Mycosphaerella graminicola: evidence for intragenic recombination and selective replacement. Mol. Plant Pathol. 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrain, L. , Brading, P.A. , Makepeace, J.C. and Brown, J.K.M. (2004a) Sources of resistance to septoria tritici blotch and implications for wheat breeding. Plant Pathol. 53, 454–460. [Google Scholar]
- Chartrain, L. , Brading, P.A. , Widdowson, J.P. and Brown, J.K.M. (2004b) Partial resistance to septoria tritici blotch (Mycosphaerella graminicola) in wheat cultivars Arina and Riband. Phytopathology, 94, 497–504. [DOI] [PubMed] [Google Scholar]
- Chartrain, L. , Berry, S.T. and Brown, J.K.M. (2005a) Resistance of wheat line Kavkaz‐K4500 L.6.A.4 to septoria tritici blotch controlled by isolate‐specific resistance genes. Phytopathology, 95, 664–671. [DOI] [PubMed] [Google Scholar]
- Chartrain, L. , Brading, P.A. and Brown, J.K.M. (2005b) Presence of the Stb6 gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in cultivars used in wheat‐breeding programmes worldwide. Plant Pathol. 54, 134–143. [Google Scholar]
- Chartrain, L. , Joaquim, P. , Berry, S.T. , Arraiano, L.S. , Azanza, F. and Brown, J.K.M. (2005c) Genetics of resistance to septoria tritici blotch in the Portugese wheat breeding line TE9111. Theor. Appl. Genet. 110, 1138–1144. [DOI] [PubMed] [Google Scholar]
- Chartrain, L. , Sourdille, P. , Bernard, M. and Brown, J.K.M. (2009) Identification and location of Stb9, a gene for resistance to septoria tritici blotch in wheat cultivars Courtot and Tonic. Plant Pathol. 58, 547–555. [Google Scholar]
- Ciufetti, L.M. , Manning, V.A. , Pandelova, I. , Betts, M.F. and Martinez, J.P. (2010) Host‐selective toxins, Ptr ToxA and PtrToxB, as necrotrophic effectors in the Pyrenophora tritici‐repentis–wheat interaction. New Phytol. 187, 911–919. [DOI] [PubMed] [Google Scholar]
- Cohen, L. and Eyal, Z. (1993) The histology of processes associated with the infection of resistant and susceptible wheat cultivars with Septoria tritici . Plant Pathol. 42, 737–743. [Google Scholar]
- Consolo, V.F. , Albani, C.M. , Beron, C.M. , Salerno, G.L. and Cordo, C.A. (2009) A conventional PCR technique to detect Septoria tritici in wheat seeds. Australas. Plant Pathol. 38, 222–227. [Google Scholar]
- Cools, H.J. and Fraaije, B.A. (2008) Are azole fungicides losing ground against Septoria wheat disease? Resistance mechanisms in Mycosphaerella graminicola . Pest Manag. Sci. 64, 681–684. [DOI] [PubMed] [Google Scholar]
- Cools, H.J. , Fraaije, B.A. , Bean, T.P. , Antoniw, J. and Lucas, J.A. (2007) Transcriptome profiling of the response of Mycosphaerella graminicola isolates to an azole fungicide using cDNA microarrays. Mol. Plant Pathol. 8, 639–651. [DOI] [PubMed] [Google Scholar]
- Cools, H.J. , Parker, J.E. , Kelly, D.E. , Lucas, J.A. , Fraaije, B.A. and Kelly, S.L. (2010) Heterologous expression of mutated eburicol 14 alpha‐demethylase (CYP51) proteins of Mycosphaerella graminicola to assess effects on azole fungicide sensitivity and intrinsic protein function. Appl. Environ. Microbiol. 76, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin, A. , Mehrabi, R. , Guilleroux, M. , Dufresne, M. , Van der Lee, T. , Waalwijk, C. , Langin, T. and Kema, G.H.J. (2006) The MAP kinase‐encoding gene MgFus3 of the non‐appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 7, 269–278. [DOI] [PubMed] [Google Scholar]
- Cowger, C. , Brunner, P.C. and Mundt, C.C. (2008) Frequency of sexual recombination by Mycosphaerella graminicola in mild and severe epidemics. Phytopathology, 98, 752–759. [DOI] [PubMed] [Google Scholar]
- Deller, S. , Hammond‐Kosack, K. and Rudd, J.J. (2011) The complex interactions between host‐immunity and non‐biotrophic fungal pathogens of wheat leaves. J. Plant Physiol. 168, 63–71. [DOI] [PubMed] [Google Scholar]
- Duncan, K.E. and Howard, R.J. (2000) Cytological analysis of wheat infection by the leaf blotch pathogen Mycosphaerella graminicola . Mycol. Res. 104, 1074–1082. [Google Scholar]
- Eriksen, L. and Munk, L. (2003) The occurrence of Mycosphaerella graminicola and its anamorph Septoria tritici in winter wheat during the growing season. Eur. J. Plant Pathol. 109, 253–259. [Google Scholar]
- Eriksen, L. , Borum, F. and Jahoor, A. (2003) Inheritance and localisation of resistance to Mycosphaerella graminicola causing septoria tritici blotch and plant height in the wheat (Triticum aestivum L.) genome with DNA markers. Theor. Appl. Genet. 107, 515–527. [DOI] [PubMed] [Google Scholar]
- Fraaije, B.A. , Lucas, J.A. , Clark, W.S. and Burnett, F.J. (2003) QoI Resistance Development in Populations of Cereal Pathogens in the UK. Farnham, Surrey: British Crop Protection Council. [Google Scholar]
- Fraaije, B.A. , Cools, H.J. , Fountaine, J. , Lovell, D.J. , Motteram, J. , West, J.S. and Lucas, J.A. (2005) Role of ascospores in further spread of QoI‐resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola . Phytopathology, 95, 933–941. [DOI] [PubMed] [Google Scholar]
- Fraaije, B.A. , Cools, H.J. , Kim, S.H. , Motteram, J. , Clark, W.S. and Lucas, J.A. (2007) A novel substitution I381V in the sterol 14 alpha‐demethylase (CYP51) of Mycosphaerella graminicola is differentially selected by azole fungicides. Mol. Plant Pathol. 8, 245–254. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Guo, J.R. , Schnieder, F. , Beyer, M. and Verreet, J.A. (2006) Rapid detection of Mycosphaerella graminicola in wheat using reverse transcription‐PCR assay. J. Phytopathol. 154, 674–679. [Google Scholar]
- Hammond‐Kossack, K.E. and Rudd, J.J. (2008) Plant resistance signalling hijacked by a necrotrophic fungal pathogen. Plant Signal. Behav. 3, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, I. , Paratte, R. , Lamine, C. and Buurma, J. (2010) Rising concerns about the impact of pesticides: an analysis of the public controversies. Social Science Insights on Crop Protection No. 3. Sophia‐antipolis: Endure Network. http://www.endure‐network.eu/content/download/5322/42414/file/SocialScienceInsightsNumber3‐Publiccontroversiesaboutpesticides.pdf Retrieved 16/9/10 from http://www.endure‐network.eu
- Hovmøller, M.S. , Justesen, A.F. and Brown, J.K.M. (2002) Clonality and long‐distance migration of Puccinia striiformis f.sp tritici in north‐west Europe. Plant Pathol. 51, 24–32. [Google Scholar]
- Jlibene, M. , Gustafson, J.P. and Rajaram, S. (1994) Inheritance of resistance to Mycosphaerella graminicola in hexaploid wheat. Plant Breed. 112, 301–310. [Google Scholar]
- Kema, G.H.J. , Yu, D.Z. , Rijkenberg, F.H.J. , Shaw, M.W. and Baayen, R.P. (1996) Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology, 86, 777–786. [Google Scholar]
- Kema, G.H.J. , van der Lee, T.A.J. , Mendes, O. , Verstappen, E.C.P. , Lankhorst, R.K. , Sandbrink, H. , van der Burgt, A. , Zwiers, L.H. , Csukai, M. and Waalwijk, C. (2008) Large‐scale gene discovery in the septoria tritici blotch fungus Mycosphaerella graminicola with a focus on in planta expression. Mol. Plant–Microbe Interact. 21, 1249–1260. [DOI] [PubMed] [Google Scholar]
- Keon, J. , Antoniw, J. , Rudd, J. , Skinner, W. , Hargreaves, J. and Hammond‐Kosack, K. (2005a) Analysis of expressed sequence tags from the wheat leaf blotch pathogen Mycosphaerella graminicola (anamorph Septoria tritici). Fungal Genet. Biol. 42, 376–389. [DOI] [PubMed] [Google Scholar]
- Keon, J. , Rudd, J.J. , Antoniw, J. , Skinner, W. , Hargreaves, J. and Hammond‐Kosack, K. (2005b) Metabolic and stress adaptation by Mycosphaerella graminicola during sporulation in its host revealed through microarray transcription profiling. Mol. Plant Pathol. 6, 527–540. [DOI] [PubMed] [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J.L. , Baker, J.M. , Beale, M.H. , Hammond‐Kossack, K.E. and Rudd, J.J. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant–Microbe Interact. 20, 178–193. [DOI] [PubMed] [Google Scholar]
- Kramer, B. , Thines, E. and Foster, A.J. (2009) MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea . Fungal Genet. Biol. 46, 667–681. [DOI] [PubMed] [Google Scholar]
- Lee, N. , D'Souza, C.A. and Kronstad, J.W. (2003) Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol. 41, 399–427. [DOI] [PubMed] [Google Scholar]
- Leroux, P. , Albertini, C. , Gautier, A. , Gredt, M. and Walker, A.S. (2007) Mutations in the CYP51 gene correlated with changes in sensitivity to sterol 14 alpha‐demethylation inhibitors in field isolates of Mycosphaerella graminicola . Pest Manag. Sci. 63, 688–698. [DOI] [PubMed] [Google Scholar]
- Ma, H. and Singh, R. (1996) Expression of adult resistance to yellow rust at different growth stages of wheat. Plant Dis. 80, 375–379. [Google Scholar]
- McCartney, C.A. , Brule‐Babel, A.L. , Lamari, L. and Somers, D.J. (2003) Chromosomal location of a race‐specific resistance gene to Mycosphaerella graminicola in the spring wheat ST6. Theor. Appl. Genet. 107, 1181–1186. [DOI] [PubMed] [Google Scholar]
- McDonald, B.A. and Linde, C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. and Kema, G.H.J. (2006) Protein kinase A subunits of the ascomycete pathogen Mycosphaerella graminicola regulate asexual fructification, filamentation, melanization and osmosensing. Mol. Plant Pathol. 7, 565–577. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , van der Lee, T. , Waalwijk, C. and Kema, G.H.J. (2006a) MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant–Microbe Interact. 19, 389–398. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Zwiers, L.H. , de Waard, M.A. and Kema, G.H.J. (2006b) MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 19, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Taga, M. and Kema, G.H.J. (2007) Electrophoretic and cytological karyotyping of the foliar wheat pathogen Mycosphaerella graminicola reveals many chromosomes with a large size range. Mycologia, 99, 868–876. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Ben M'Barek, S. , van der Lee, T.A.J. , Waalwijk, C. , de Wit, P. and Kema, G.H.J. (2009) G alpha and G beta proteins regulate the cyclic AMP pathway that is required for development and pathogenicity of the phytopathogen Mycosphaerella graminicola . Eukaryot. Cell, 8, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseman, J.G. , Macer, R.C.F. and Greeley, L.W. (1965) Genetic studies of cultures of Erysiphe graminis f.sp. hordei virulent on Hordeum spontaneum . Br. Mycol. Soc. Trans. 48, 479–489. [Google Scholar]
- Motteram, J. , Kufner, I. , Deller, S. , Brunner, F. , Hammond‐Kossack, K.E. , Nurnberger, T. and Rudd, J.J. (2009) Molecular characterisation and functional analysis of the MgNLP, the sole NPP1 domain‐containing protein from the fungal wheat leaf pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 22, 790–799. [DOI] [PubMed] [Google Scholar]
- Palmer, C.L. and Skinner, W. (2002) Mycosphaerella graminicola: latent infection, crop devastation and genomics. Mol. Plant Pathol. 3, 63–70. [DOI] [PubMed] [Google Scholar]
- Pietravalle, S. , Shaw, M.W. , Parker, S.R. and Bosch, F. (2003) Modeling of relationships between weather and Septoria tritici epidemics on winter wheat: a critical approach. Phytopathology, 93, 1329–1339. [DOI] [PubMed] [Google Scholar]
- Ray, S. , Anderson, J.M. , Urmeev, F.I. and Goodwin, S.B. (2003) Rapid induction of a protein disulfide isomerase and defense‐related genes in wheat in response to the hemibiotrophic fungal pathogen Mycosphaerella graminicola . Plant Mol. Biol. 53, 741–754. [DOI] [PubMed] [Google Scholar]
- Rispail, N. , Soanes, D.M. , Ant, C. , Czajkowski, R. , Grunler, A. , Huguet, R. , Perez‐Nadales, E. , Poli, A. , Sartorel, E. , Valiante, V. , Yang, M. , Beffa, R. , Brakhage, A.A. , Gow, N.A.R. , Kahmann, R. , Lebrun, M.H. , Lenasi, H. , Perez‐Martin, J. , Talbot, N.J. , Wendland, J. and Di Pietro, A. (2009) Comparative genomics of MAP kinase and calcium–calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46, 287–298. [DOI] [PubMed] [Google Scholar]
- Romeis, T. , Piedras, P. , Zhang, S. , Klessig, D.F. , Hirt, H. and Jones, J.D.G. (1999) Rapid Avr‐9‐ and Cf‐9‐dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound and salicylate responses. Plant Cell, 11, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roohparvar, R. , De Waard, M.A. , Kema, G.H.J. and Zwiers, L.H. (2007) MgMfs1, a major facilitator superfamily transporter from the fungal wheat pathogen Mycosphaerella graminicola, is a strong protectant against natural toxic compounds and fungicides. Fungal Genet. Biol. 44, 378–388. [DOI] [PubMed] [Google Scholar]
- Roohparvar, R. , Mehrabi, R. , Van Nistelrooy, J.G.M. , Zwiers, L.H. and De Waard, M.A. (2008) The drug transporter MgMfs1 can modulate sensitivity of field strains of the fungal wheat pathogen Mycosphaerella graminicola to the strobilurin fungicide trifloxystrobin. Pest Manag. Sci. 64, 685–693. [DOI] [PubMed] [Google Scholar]
- Rudd, J.J. , Keon, J. and Hammond‐Kosack, K.E. (2008) The wheat mitogen‐activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola . Plant Physiol. 147, 802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J.J. , Antoniw, J. , Marshall, R. , Motteram, J. , Fraaije, B. and Hammond‐Kossack, K.E. (2010) Identification and characterisation of Mycosphaerella graminicola secreted on surface‐associated proteins with variable intragenic coding repeats. Fungal Genet. Biol. 47, 19–32. [DOI] [PubMed] [Google Scholar]
- Shetty, N.P. , Kristensen, B.K. , Newman, M.‐A. , Moller, K. , Gregersen, P.L. and Jorgensen, H.J.L. (2003) Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiol. Mol Plant Pathol. 62, 333–346. [Google Scholar]
- Shetty, N.P. , Mehrabi, R. , Lutken, H. , Haldrup, A. , Kema, G.H.J. , Collinge, D.B. and Jorgensen, H.J.L. (2007) Role of hydrogen peroxide during the interaction between the hemibiotrophic fungal pathogen Septoria tritici and wheat. New Phytol. 174, 637–647. [DOI] [PubMed] [Google Scholar]
- Shetty, N.P. , Jensen, J.D. , Knudsen, A. , Finnies, C. , Geshi, N. , Blennow, A. , Collinge, D.B. and Jorgensen, H.J.L. (2009) Effects of beta‐1,3‐glucan from Septoria tritici on structural defence responses in wheat. J. Exp. Bot. 60, 4287–4300. [DOI] [PubMed] [Google Scholar]
- Siah, A. , Deweer, C. , Duyme, F. , Sanssene, J. , Durand, R. , Halama, P. and Reignault, P. (2010) Correlation of in planta endo‐beta‐1,4‐xylanase activity with the necrotrophic phase of the hemibiotrophic fungus Mycosphaerella graminicola . Plant Pathol. 59, 661–670. [Google Scholar]
- Spencer‐Phillips, P.T.N. (1997) Function of haustoria in epiphytic and endophytic infections. Adv. Bot. Res. 124, 309–333. [Google Scholar]
- Stergiopoulos, I. , Zwiers, L.H. and De Waard, M.A. (2003) The ABC transporter MgAtr4 is a virulence factor of Mycosphaerella graminicola that affects colonization of substomatal cavities in wheat leaves. Mol. Plant–Microbe Interact. 16, 689–698. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Burg, H.A. , Okmen, B. , Beenen, H.G. , van Liere, S. , Kema, G.H.J. and de Wit, P. (2010) Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA, 107, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs, R.W. , Prescott, J.M. , Saari, E.E. and Dublin, H.J. (1986) Cereal Disease Methodology Manual. Mexico City: International Maize and Wheat Improvement Center (CIMMYT). [Google Scholar]
- Stukenbrock, E.H. and McDonald, B.A. (2008) The origins of plant pathogens in agro‐ecosystems. Annu. Rev. Phytopathol. 46, 75–100. [DOI] [PubMed] [Google Scholar]
- Stukenbrock, E.H. , Banke, S. , Javan‐Nikkhah, M. and McDonald, B.A. (2007) Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol. Biol. Evol. 24, 398–411. [DOI] [PubMed] [Google Scholar]
- Tanaka, A. , Christensen, M.J. , Takemoto, D. , Park, P. and Scott, B. (2006) Reactive oxygen species play a role in regulating a fungus–perennial ryegrass mutualistic interaction. Plant Cell, 18, 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg, A.H.J. , van der Lee, T.A.J. , Ben M'Barek, S. , Ware, S.B. , Goodwin, S.B. , Kilian, A. , Visser, R.G.F. , Kema, G.H.J. and Schouten, H.J. (2009) Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola . PLoS ONE, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, J. , Pettway, R.E. and McDonald, B.A. (2003) The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genet. Biol. 38, 286–297. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Haley, S.D. and Jin, Y. (2001) Inheritance of septoria tritici blotch resistance in winter wheat. Crop Sci. 41, 323–326. [Google Scholar]
- Zwiers, L.H. , Stergiopoulos, L. , Van Nistelrooy, J.G.M. and De Waard, M.A. (2002) ABC transporters and azole susceptibility in laboratory strains of the wheat pathogen Mycosphaerella graminicola . Antimicrob. Agents Ch. 46, 3900–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers, L.H. , Roohparvar, R. and de Waard, M.A. (2007) MgAtr7, a new type of ABC transporter from Mycosphaerella graminicola involved in iron homeostasis. Fungal Genet. Biol. 44, 853–863. [DOI] [PubMed] [Google Scholar]


