SUMMARY
The French wheat variety ‘Camp Remy’ (CR) possesses a durable, adult plant resistance to yellow rust (YR), caused by the pathogen Puccinia striiformis. Using cDNA–AFLP on different sets of heterogeneous inbred families (HIFs) derived from the cross CR × Récital, we compared gene expression profiles during one seedling and two adult developmental stages following inoculation with P. striiformis. Transcripts differentially expressed in response to YR infection were isolated and cloned. Sequence analysis of the resultant clones revealed several classes of putative genes, including those related to resistance/defence responses, transcription and signal transduction, and primary metabolism. The expression profiles of seven selected genes were obtained using real‐time PCR in CR leaves at the same three stages of development. The results confirmed the stage‐specific expression of the genes at one or two specific stages in response to P. striiformis infection and demonstrated that CR modifies the expression of some resistance/defence‐related genes during its transition from the seedling to adult growth stages. These results provided the first clue to understand the molecular basis of quantitative trait loci for adult plant resistance to YR and connect it with durability.
INTRODUCTION
Stripe or yellow rust (YR) caused by the fungus Puccinia striiformis fsp. tritici Westend., is one of the most damaging diseases of bread wheat in temperate regions (for reviews, see Chen, 2005; Line, 2002). Recent evolution of high‐temperature tolerant strains in North America has increased the geographical distribution and impact of this disease (Chen, 2005; Milus et al., 2006). The deployment of disease‐resistant varieties remains the most economically effective and environmentally safe method to control YR (Chen, 2005; Line, 2002). Seedling resistance genes are known to be race‐specific, effective at all stages of plant growth and monogenically inherited (Line, 2002). Historically, race‐specific resistance has been extensively deployed because of the relative ease of incorporating this resistance into new varieties. However, the YR pathogen is highly variable and more than 100 races have been reported in North America (Chen, 2005; Line, 2002); thus, the effectiveness of race‐specific resistance can be rapidly lost due to virulence shifts of the pathogen. Another important type of resistance is the high‐temperature, adult‐plant (HTAP) resistance, initiated at the tillering stage and effective throughout the adult stages. This resistance is usually quantitatively inherited (Line, 2002) and appears to be less affected by shifts in virulence of the pathogen than race‐specific resistance; hence, it is considered as more durable in the field (Chen, 2005). A third form of resistance known as partial resistance has been reported (Line, 2002). It is effective at all wheat development stages but allows various amounts of rust sporulation.
A few published reports characterized quantitative forms of resistance to YR in wheat using molecular approaches (Boukhatem et al., 2002; Christiansen et al., 2006; Lin and Chen, 2007; Mallard et al., 2005; Navabi et al., 2005; Ramburan et al., 2004). To date, little is known about molecular mechanisms and gene induction following pathogen perception of specific genes or quantitative trait loci (QTL) involved in P. striiformis resistance in wheat. Owing to the large genome of hexaploid wheat (about 16 000 Mbp), it is difficult to apply positional cloning or tagging methodologies in order to isolate genes underlying resistance QTL (Tanksley et al., 1995). Gene expression libraries have been constructed to identify genes expressed during compatible interactions between leaf rust and wheat (Rampitsch et al., 2006; Zhang et al., 2003) and incompatible interactions between stem rust and wheat (Menden et al., 2006). Thus, strategies based on the study of gene expression are suitable alternatives for isolating and characterizing genes involved in the expression of disease resistance. Recently, several reports have been published on the successful use of cDNA–AFLP for the isolation of differentially expressed plant genes during cereal–pathogen interactions (Eckey et al., 2004; Zhang et al., 2003; Zheng et al., 2004). This technique is of particular interest for wheat transcript analysis as it does not require knowledge of sequence data, making it potentially useful for isolating and identifying genes involved in disease resistance.
The bread wheat cultivar ‘Camp Rémy’ (CR) has been extensively cultivated for more than 20 years in France and has effectively maintained its resistance to YR. This variety is of particular interest for plant breeders as it was demonstrated to possess a combination of both seedling and adult plant resistance that is effective against all known races of P. striiformis (Mallard et al., 2005). In a previous study that employed recombinant inbred lines (RILs) derived from a cross between CR and the susceptible wheat Recital, we demonstrated that the durable resistance of CR was due to a specific combination of qualitative and quantitative resistance genes located on chromosomes 2A, 2B, 2D and 5B for which no YR pathogen has yet evolved the necessary combination of virulence genes (Mallard et al., 2005). One resistance locus conferred seedling resistance and the others were involved in adult stage resistance (Mallard et al., 2005). Furthermore, among the latter, some QTLs were detected in early adult stage during the two‐node stages, while others were identified later during the flowering stage (Mallard, 2005). The CR/YR interaction could serve as a model to a better understanding of the nature of the durable adult plant resistance against YR.
Unlike the stem and leaf rust pathogens, YR does not form appresoria but rather the fungal hyphae penetrate the stomata following spore germination (Allen, 1928; Marryat, 1907; Moldenhauer et al., 2006; Niks, 1989). In a time‐course study of YR development in the bread wheat cultivar ‘Kariega’ expressing adult plant resistance, Moldenhauer et al. (2006) demonstrated that no observable difference between resistant and susceptible reactions occurred prior to germ tube penetration of the stomata and formation of a substomatal vesicle, which occurred at 3 days post inoculation (dpi). However, after 4 dpi, fungal development was more rapid, and at 6–7 dpi, the fungal colonies were larger and longer in the susceptible than in the adult‐plant resistant interaction (Moldenhauer et al., 2006). In plants expressing adult plant resistance to P. striiformis, host‐cell necrosis in plants precedes the release of toxic metabolites that causes local inhibition of haustorium formation and hyphal branching (Mares, 1979; Mares and Cousen, 1977).
To identify genes up‐ and down‐regulated in response to P. striiformis inoculation, we compared the expression of transcript‐derived fragments (TDFs) generated using cDNA–AFLP, in inoculated and mock‐inoculated CR and the different CR‐derived heterogeneous inbred families (HIFs) that express YR resistance at different developmental stages. We demonstrated that CR resistance could be partitioned into seedling and adult stages based on the differential expression of selected resistance/defence‐related genes in response to P. striiformis infection. These results provide the first clues for understanding the mechanism involved in the durable adult plant resistance of CR and provide a basis for future functional studies on this host–parasite interaction.
RESULTS
Microscopic observations of leaf samples following inoculation at the seedling stage, under the conditions used in this study, indicated that the majority of spores have germinated and penetrated the plant between 2 and 3 dpi. The first visible symptoms on resistant genotypes or signs of lesions on susceptible genotypes were observed at 6–7 dpi. Based on these observations, leaves of three inoculated and three mock‐inoculated plants from ‘CR’, ‘Récital’ and the six set of HIFs were collected at 2 and 6 dpi for each of the three wheat development stages studied. Moreover, at each stage, leaves from the HIFs exhibiting a high level of resistance and the parental lines were also collected at two supplementary time‐points: 3 and 7 dpi. A total of 260 cDNA probes were obtained for cDNA–AFLP analysis, as the three biological replicates of each collected sample were pooled.
cDNA–AFLP analysis of genes differentially expressed in response to P. striiformis inoculation
Among 85 primer combinations tested, 16 were selected for cDNA–AFLP. For each primer combination, the size of TDFs ranged between 100 and 800 bp and an average of 60 TDFs were observed per genotype. Although the majority of the bands were common to all patterns, each one showed an average of 5–8 differentially expressed TDFs between the inoculated and mock‐inoculated leaves of each genotype. We independently analysed the patterns obtained at each of the three development stages studied. For each time‐point of the time‐course, we focused on the TDFs up‐ or down‐regulated in inoculated leaves compared with mock‐inoculated leaves in either resistant or susceptible genotypes (presence/absence polymorphism). Figure 1 illustrates a typical pattern obtained for the three HIFs 2171 at the tillering stage, at 2 dpi. The TDF ‘A’ was only found in the resistant inoculated genotype 2171‐2 and the TDF ‘B’ was only present in the resistant inoculated genotype 2171‐5. The TDF ‘C’ was repressed in all inoculated plants of this set of HIF.
Figure 1.
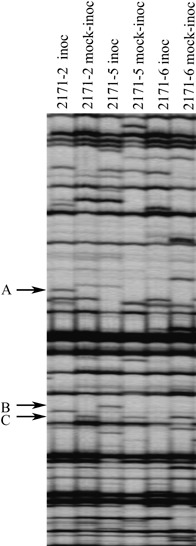
Example of banding patterns observed in the cDNA–AFLP. Represented here is the pattern obtained for the set of HIF 2171 at the tillering stage. Arrows designate three TDFs differentially expressed among treatments (inoc = inoculated).
A total of 81 differentially regulated TDFs after P. striiformis inoculation, designated as SN01 to SN85, were excised from the polyacrylamide gels; 30 were up‐regulated and five down‐regulated at the seedling stage, 25 up‐regulated and five down‐regulated at the tillering stage, and 11 up‐regulated and five down‐regulated at the two‐node stage. When the same TDF was identified in several patterns, it was separately excised from each pattern and sequenced. For example, SN22 and SN23 correspond to the same up‐regulated TDF identified in inoculated leaves of genotypes 2178R and 2182R, respectively, at 3 dpi during the seedling stage. The TDFs were re‐amplified with the corresponding selective primers, cloned and 2–3 sub‐lones exhibiting the correct size were sequenced. Sixty‐nine exploitable sequences were finally obtained. Alignment of sequences was accomplished using the clustalw ( Thompson et al., 1994) and align (Myers and Miller, 1988) programs and 55 non‐redundant sequences were finally resolved.
Sequence analysis of differentially expressed TDFs
Blastn results for all up‐ or down‐regulated TDFs demonstrated that 42 of the clones were homologous to Triticum aestivum expressed sequence tags (ESTs) available in the Graingene database (http://wheat.pw.usda.gov/GG2/index.shtml). Twenty‐nine of these ESTs were isolated in biotic or abiotic stress studies and most of them were part of a wheat contig. Twenty‐seven TDFs had significant homology to cereal genomic sequences available in the Genbank public databases.
Blastx results for all differentially regulated TDFs showed that 25 had significant homology to proteins available in the Swissprot database; 15 of these had a known function. The sequences, listed in Table 1, were classified into four groups based on their putative functionality: (1) those involved in resistance/defence responses, (2) those encoding transcription or translation regulators, (3) those involved in primary metabolism and (4) those with unknown function.
Table 1.
Similarities at the DNA or protein level between TDFs and database sequences.
| Differentially expressed TDF description | Highest homology | |||||||
|---|---|---|---|---|---|---|---|---|
| Class | TDF | Size (bp) | Genotype and treatment | Development stage | Accession no. | E value | Database | Description |
| 1 | SN20 | 382 | Récital inoc. | seedling | AJ318884 | 2,0E‐44 | Genbank | Triticum durum xip‐II gene coding a xylanase inhibitor protein |
| TC271383 | 9,6E‐26 | TIGR | Chitinase II (Q8W428) | |||||
| SN33 | 374 | 2141‐4 inoc. | 2‐node | TC268609 | 1,9E‐71 | TIGR | Beclin1‐like protein Q94C41 | |
| SN53 | 546 | 2083R inoc. | tillering | NP_172965 | 2,0E‐14 | Nrprot | hydroxyprolin‐rich glycoprotein | |
| SN55 | 277 | 2178R inoc. | tillering | DQ386610 | 9,0E‐32 | Genbank | Hordeum vulgare subsp. vulgare protein kinase | |
| SN60 | 341 | CR inoc. | tillering | TC274868 | 1,1E‐03 | TIGR | 67% homologous to a disease resistance protein (Q9FWC5) | |
| SN81 | 400 | 2171‐5 inoc. | 2‐node | TC266739 | 8,3E‐25 | TIGR | 83% similar to the putative calmodulin‐binding protein similar to ER66 (Q8RV24) | |
| 2 | SN18 | 501 | CR inoc. | tillering | AF459085 | 3,0E‐07 | Genbank | Hordeum vulgare Snf2P gene |
| TC247787 | 2,7E‐08 | TIGR | 86% similar to an uncharacterized GPI‐anchored protein At5g19230 precursor (Q8GUL8) | |||||
| SN30 | 392 | 2141‐4 mock‐inoc. | tillering | TC273628 | 1,1E‐17 | TIGR | 76% similar to a RNA‐binding protein‐like (Q5VMK2) | |
| SN42 | 440 | 2178R inoc. | tillering | TC249344 | 1.4e‐56 | TIGR | 80% similar to a putative SCARECROW gene regulator‐like (Q8S5N0) | |
| TC249346 | 3.8e‐49 | TIGR | 71% similar a chitin‐inducible gibberellin‐responsive protein (Q8GVE1) | |||||
| SN56 | 675 | 2182R inoc. | tillering | TC251858 | 3,4E‐137 | TIGR | 83% similar to a putative eukaryotic translation initiation factor 4G (Q69S49) | |
| SN64 | 428 | 2083R inoc. | tillering | Q94LS3 | 2E‐35 | UniProtKB TrEMBL | Putative helicase | |
| 3 | SN15 | 346 | 2182R inoc. | seedling | TC232238 | 1,2E‐17 | TIGR | 95% homologous to an Enolase (Q42971) |
| SN35 | 479 | CR inoc. | seedling | AP008982 | 0 | Genbank | Triticum aestivum NADH dehydrogenase subunit 2 (nad2 ) gene | |
| SN41 | 479 | 2178R inoc. | seedling | TC242910 | 3,5E‐05 | TIGR | 81% similar to a PPi‐dependent phosphofructo‐ 1‐kinase (Q24812) | |
| 4 | SN10 | 351 | 2083R inoc. | seedling | TC255554 | 2,0E‐51 | TIGR | 73% homologous to a polysaccharide deacetylase domain protein (Q74GK0) |
| SN11 | 528 | 2083R inoc. | seedling | TC258298 | 6,7E‐41 | TIGR | similar to an unknown protein from Oryza sativa (BAD01375) | |
| SN19 | 448 | 2141‐2 inoc. | tillering | BE58615 | 3,2E‐239 | Gramene | Wheat Fusarium graminearum‐infected spike cDNA library Triticum aestivum cDNA clone | |
| SN31 | 328 | 2178R inoc. | 2‐node | CA746363 | 2,0E‐130 | Gramene | Riband wheat leaves infected with Septoria tritici, 2 dpi, subtracted w/comparable uninfected leaves | |
| SN47 | 526 | 2182R inoc. | seedling | CK194708 | 2,2E‐20 | Gramene | Functional Genomics of Abiotic Stress In Wheat | |
| SN48 | 174 | Récital inoc. | seedling | TC272466 | 7,6E‐01 | TIGR | 80% similar to ubiquitine‐conjugative E2 putative | |
| SN57 | 283 | 2178R inoc. | tillering | BE401099 | 1,5E‐25 | Gramene | Wheat Powdery Mildew Resistant Library Triticum aestivum | |
| SN66 | 361 | 2178R inoc. | tillering | TC245138 | 3,6E‐20 | TIGR | 81% similar to a hypothetical protein (Q8S7B5) | |
| SN67 | 496 | 2178R inoc. | tillering | TC242459 | 4,1E‐29 | TIGR | 83% similar to an unknown protein from Oryza sativa (BAD28426) | |
| SN75 | 368 | Récital inoc. | 2‐node | CA735573 | 2,1E‐14 | Gramene | Wheat, Polk cultivar (resistant), infected with Septoria tritici strain A 24 h after infection | |
| SN76 | 475 | Récital inoc. | 2‐node | AJ716858 | 5,7E‐83 | Gramene | cDNA of durum wheat etiolated seedling at day 20 | |
| SN82 | 317 | Récital inoc. | seedling | CA697102 | 3,0E‐36 | Gramene | Triticum aestivum seedlings 4 h after treatment with 6‐iodo‐3‐propyl‐2‐propyloxy‐4(3H)‐quinazolinone | |
| SN85 | 317 | 2141‐2 inoc. | seedling | TC256592 | 7,4E‐04 | TIGR | 80% similar to a Proline‐rich protein (Q01979) | |
The genotype, treatment (inoc = inoculated and mock‐inoc = mock‐inoculated) and development stage at which the band was detected are also indicated. TDFs are classified into four classes: (1) those involved in resistance/defence responses, (2) those encoding transcription or translation regulators, (3) those involved in primary metabolism and (4) those with unknown function.
Two TDFs (not shown) were homologous to fungal proteins.
Time‐course expression of differentially expressed TDFs
To confirm the relationship of TDFs with YR resistance and determine if some are induced by P. striiformis at specific development stages, the time‐course expression profiles of resistance‐related TDFs were determined by real‐time PCR for the resistant variety CR (Table 2). Among the TDFs homologous to known proteins, seven, representing the three first classes of homologies, were chosen. For classes 1 and 2, we chose TDFs that were differentially expressed during the adult stages and for which we managed to design highly specific primers.
Table 2.
Relative up‐ or down‐regulation of selected defence‐related genes in inoculated compared with mock‐inoculated CR plants during the seedling, tillering and two‐node stages of wheat development.
| dpi | Seedling stage | Tillering stage | Two‐node stage | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1.5 | 2 | 2.5 | 3 | 4 | 5 | 6 | 0 | 1 | 2 | 3 | 4 | 4.5 | 5 | 5.5 | 6 | 0 | 1 | 2 | 3 | 4 | 4.5 | 5 | 5.5 | 6 | |
| SN15 | = | ‐ | ++ | = | = | = | ++ | = | = | = | ‐‐‐ | ‐ | + | = | = | = | = | = | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| SN18 | = | = | = | = | = | = | = | = | = | = | ‐ | ‐‐ | + | = | = | = | ‐ | = | = | = | = | ‐‐ | ‐‐ | = | ++ | = | ‐ |
| SN33 | = | = | ‐ | = | = | + | = | = | = | = | = | = | = | + | + | = | + | + | = | ‐ | = | + | ‐ | ‐ | ++ | = | = |
| SN41 | = | = | = | = | = | = | +++ | + | = | = | ‐‐ | = | ++ | = | ‐ | = | + | = | = | = | = | = | ++ | = | + | ++ | ++ |
| SN42 | = | = | = | + | = | = | = | = | = | = | ‐‐ | ‐ | ++ | ‐ | ‐ | = | = | = | = | ‐‐ | ‐ | ++ | ‐ | ‐ | = | = | = |
| SN64 | = | = | = | = | = | = | ++ | + | = | = | ‐ | = | + | ‐‐ | = | = | ++ | ‐‐ | = | ‐ | = | = | ‐‐ | = | = | = | ‐ |
| SN81 | = | = | = | = | = | = | ++ | = | = | = | ‐ | = | = | = | = | + | + | ‐ | = | + | = | = | ++ | = | = | + | + |
The expression data of SN15 are not available (NA) for the two‐node stage. The expression of defence‐related genes was normalized by expression of the reference control (wheat 18S gene) and the ΔΔCT method of relative gene quantification was used to calculate the expression level of CR inoculated relative to CR mock‐inoculated. A gene was considered as differentially regulated if the relative difference between the two treatments was higher than 2. Key: = no difference in expression between inoculated and mock‐inoculated leaves (P > 0.001);
‐ gene lightly down‐regulated in inoculated leaves (fold difference < 2 and P ≤ 0.001);
‐‐ gene down‐regulated in inoculated leaves (2 < fold difference < 5 and P ≤ 0.001);
‐‐‐ gene highly down‐regulated in inoculated leaves (fold difference > 5 and P ≤ 0.001);
+ gene lightly up‐regulated in inoculated leaves (fold difference < 2 and P ≤ 0.001);
++ gene up‐regulated in inoculated leaves (2 < fold difference < 5 and P ≤ 0.001);
+++ gene highly up‐regulated in inoculated leaves (fold difference > 5 and P ≤ 0.001).
To correct differences in RNA input or efficiency of reverse transcription, the fluorescence signals of target genes were normalized to that of the endogenous reference gene 18S rRNA. The 18S rRNA level remained constant during different development stages and between the two treatments. Significant differences in fold expression levels between inoculated and mock‐inoculated leaves (P ≤ 0.001) were observed for all the selected genes at one or more stages of wheat development. Most of the TDFs exhibited a time‐course expression profile that would be expected for adult plant resistance‐related genes: no or late differential expression following inoculation in the seedling‐stage, and progressively earlier and higher expression following inoculation during adult stages. For example, SN41, the PPi‐dependent phosphofructo‐1‐kinase, was differentially up‐regulated in inoculated leaves at 4 dpi at the seedling stage, down‐regulated at 1 dpi then up‐regulated at 3 dpi at the tillering stage, and up‐regulated at 4, 5.5 and 6 dpi at the two‐node stage (Table 2). For SN18, a putative transcription factor, no differences in expression were detected following inoculation during the seedling stage whereas, at the two‐node stage, SN18 was slightly down‐regulated in inoculated leaves at 3 and 4 dpi and became highly over‐expressed at 5 dpi. Similar patterns of expression were also observed for SN33, a beclin1‐like protein: its time‐course expression pattern indicated that this TDF was only significantly differentially expressed under P. striiformis inoculation treatment during the two‐node stage, at 5 dpi (Fig. 2). SN42, a putative SCARECROW gene, was upregulated at 3 dpi in inoculated leaves at the two studied adult stages, but no differences in the expression level were detected at the seedling stage (Table 2, Fig. 2). Interestingly, SN42 exhibited identical expression patterns at the tillering and the two‐node stages (Table 2, Fig. 2). SN64, a putative helicase, was up‐regulated at 4 dpi among inoculated leaves during the seedling stage and alternatively up‐ and down‐regulated at the tillering stage (Table 2, Fig. 2). The TDF SN81, a calmodulin‐binding protein, was up‐regulated at 4 dpi in inoculated leaves at both the seedling and the two‐node stages (Table 2, Fig. 2).
Figure 2.
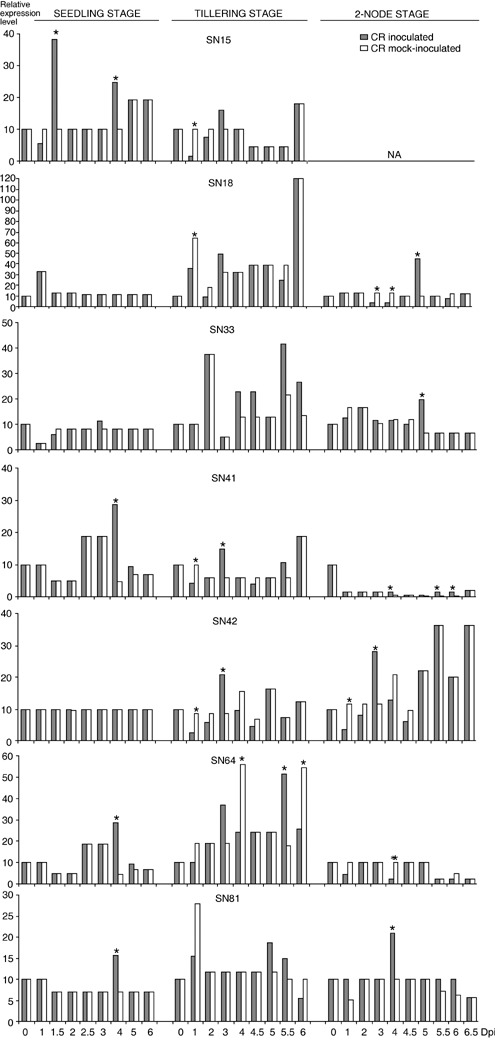
Relative expression kinetics of the seven target gene transcripts in inoculated and mock‐inoculated leaves of CR compared with the relative expression in CR leaves mock‐inoculated at 0 dpi (calibrator). All the differences illustrated between CR inoculated and CR mock‐inoculated were highly significant (P ≤ 0.001) and an asterisk indicated that the relative difference between the two treatments is higher than 2, the chosen threshold to consider that a gene is differentially regulated.
DISCUSSION
Adult plant resistance is ineffective during seedling stages but increases in effectiveness with plant age (Line, 2002). We used cDNA–AFLP to generate a series of TDFs, homologous to resistance‐ and defence‐related genes, transcriptional factors, and metabolic genes in wheat, that are differentially regulated during the expression of CR durable resistance to YR. The time‐course expression profile of seven TDFs during three CR development stages, realized using real‐time PCR, established that three were preferentially up‐regulated in the tillering and two‐node stages compared with the level of expression at the seedling stage. This represents the first transcriptional analysis of adult plant resistance associated with YR inoculation, and the first report of developmentally related association of TDFs with expression of adult plant resistance in a rust/wheat interaction.
Analysing the cDNA–AFLP expression pattern from individual HIF lines sampled at the three different stages allowed the analysis of resistance gene expression associated with these different stages of plant development. By sampling inoculated and mock‐inoculated leaves at 2, 3, 6 and 7 dpi, the entire range of potential resistance/defence reactions of CR to P. striiformis, until the appearance of symptoms, was considered. This time‐course covers the critical transcription and translation changes occurring in wheat during the expression of adult plant resistance to P. striiformis in wheat (Moldenhauer et al., 2006).
The cDNA–AFLP fingerprint technique allowed the identification of 81 TDFs up‐ or down‐regulated in inoculated compared with the mock‐inoculated leaves in susceptible and resistant genotypes. Among them, 55 were isolated and sequenced. Forty per cent of the TDFs did not show significant homology to known sequences. This could be due, in part, to their relative shortness or their origin from 3′ non‐translated regions but is probably because some TDFs may be new genes of unknown function. Twenty‐five TDFs isolated in our study showed high homology to putative proteins that fell into four classes: (1) those involved in resistance/defence responses, (2) those encoding transcription or translation regulators, (3) those involved in primary metabolism and (4) those with unknown function.
TDFs homologous to known resistance/defence proteins are associated with defence response to P. striiformis
Five TDFs induced during the P. striiformis/CR interaction were homologous to known resistance/defence genes. These included SN55, which was homologous to a barley protein kinase. To date, the tomato Pto gene is the only protein kinase characterized as a resistance gene in plants (Ellis and John, 1998). In animals, the beclin‐1 protein (SN33) is considered essential for autophagy and is involved in programmed cell death (PCD) (Gajewska et al., 2005; Yu et al., 2004). PCD occurs in both animals and plants, and in the latter, PCD is a component of cell necrosis, an essential part of the resistance reaction in some host–parasite interactions in plants (Keen, 2002). Liu et al. (2005) demonstrated that tobacco beclin‐1 is required to limit the spread of R gene‐mediated hypersensitive response HR PCD triggered by viruses, bacteria and fungi and general elicitors. The line 2141‐4, from which the TDF SN33 was isolated, showed necrosis after infection by the P. striiformis strain at the two‐node stage. Necrosis were also visible at this stage on CR and other resistant lines including 2083R (Fig. 3). Another defence‐related TDF, SN53, showed high homology to a member of the hydroxyproline‐rich glycoprotein (HRGP) family. HRGPs are pathogenesis‐related (PR) proteins that are developmentally regulated in a tissue‐specific manner (Sheng et al., 1993; Ye and Varner, 1991) and expressed in response to biotic and abiotic stresses (Bradley et al., 1992; Lawton and Lamb, 1987; Showalter et al., 1991). HRGPs are accumulated in the cell wall during pathogen infection and have been associated with vacuole occlusion that blocks the pathogen spread in the plant.
Figure 3.

Necrosis on the 2083R leaves after YR infection with the strain 237 E141 V17 at the two‐node stage.
SN20 was homologous to the gene xip‐II of wheat. XIP proteins inhibit fungal endo‐(1,4)‐β‐xylanases that hydrolyse the β‐(1,4) glycosidic linkages of the xylan component of plant cell walls (Banik et al., 1996; Juge et al., 2004). The xip‐I gene is induced in Blumeria graminis‐infected wheat leaves, in jasmonic acid‐treated leaves and after wounding (Igawa et al., 2005), and thus far, our study is the only one that has reported the involvement of xip‐II in defence responses. This TDF was also highly homologous to a chitinase II, a PR protein. The chitinase is an enzyme that hydrolyses chitin, the main component of many fungal walls and expression of many chitinase genes is induced by pathogens. Chitinases have long been proposed to play roles in the active defence response of plants (Broglie et al., 1991; Bravo et al., 2003; Jach et al., 1995; Zhu et al., 1994).
SN81 encodes a protein homologous to a calmodulin‐binding protein involved in calcium signalling. Calcium is known to be crucial in signalling during some defence responses (Grant et al., 2000; Grant and Mansfield, 1999; Reddy, 2001) whereby the increase in [Ca2+] activates defence responses via calmodulin and Ca2+‐dependent kinases/phosphatases (Grant and Mansfield, 1999). The rice OsMlo gene, involved in defence response and HR, is a calmodulin‐binding protein (Kim et al., 2002). Its expression was induced after fungus infection or by signal molecules such as salicylic acid and H2O2. More recently, analysis using cDNA–AFLP showed that a calmodulin‐binding protein was induced in B. graminis‐infected leaves in barley (Eckey et al., 2004) and in Magnaporthe grisea‐infected leaves in rice (Zheng et al., 2004).
TDFs homologous to transcriptional and translational regulators
The contig of wheat EST containing SN42 was homologous to members of the GRAS gene family of rice, a chitin‐inducible gibberellin‐responsive (CIGR) protein and a SCARECROW protein. CIGR rice proteins are rapidly induced upon pathogen elicitor perception and may act as transcriptional regulators in the early events of the elicitor‐induced defence response in rice (Day et al., 2003, 2004). SCARECROW protein homologues have been detected by cDNA–AFLP following inoculation of barley with powdery mildew (Eckey et al., 2004). SN56 showed high homology to a eukaryotic translation initiation factor 4G (eIF4G). eIF4G was shown to be involved in plant–potyvirus interactions (Nicaise et al., 2007). It is interesting to note that a wheat EST homologous to the same eIF4G than SN56 was mapped on the Chinese Spring deletion map (http://wheat.pw.usda.gov/wEST/binmaps/), located in the bin C‐2DS1‐0.33. This also corresponded to the location of the CR YR resistance QTL QYr.inra‐2DS (Mallard et al., 2005). Other homologous genes associated with transcription and translation include the TDF SN64, homologous to a helicase, a protein that unwinds the DNA, and the TDF SN18, similar to the barley snf2 gene. Cereal genes from the SNF2 family are often involved in the regulation of transcription (Yan et al., 2002). SN18 was also 86% homologous to a GPI‐anchored protein precursor. Some GPI anchors are thought to tether signalling molecules to the cell surface (Schultz et al., 1998; Solomon et al., 1996). Endogenous GPI‐specific phospholipases may release these tethered proteins into the extracellular matrix, where they could possibly act as intercellular signalling molecules (Sherrier et al., 1999). More recently, Coppinger et al. (2004) presented evidence that NDR1, involved in Pseudomonas syringae resistance in Arabidopsis thaliana, contains a GPI anchor and proposed that NDR1 may act as an intercellular signalling molecule, warning neighbouring cells of an imminent attack.
TDFs homologous to genes involved in primary metabolism
The putative enolase gene SN15 and a phosphofructokinase gene SN41 are both involved in glycolysis and are induced following abiotic stress in barley and maize (Good and Muench, 1992; Lal et al., 1998). Ventelon‐Debout et al. (2003, 2004) also identified several enzymes involved in the glycolysis in Rice Yellow Mottle Virus (RYMV)‐infected leaves of rice and suggested that these enzymes may be involved in the restoration of cell homeostasis to plants following stress (Ventelon‐Debout et al., 2003, 2004).
The TDF SN35 was homologous to nad2, which codes for an NADH dehydrogenase. This mitochondrial gene is involved in respiratory metabolism. It was shown that the respiration was stimulated in the French wheat variety Cappelle Desprez, a genitor of CR, between 1 and 7 days after infection with Pseudocercosporella herpotrichoides (Davy de Virville et al., 1982). Such stimulation may as well occur in CR leaves inoculated with P. striiformis as CR probably shares defence mechanisms with Cappelle Desprez.
Different defence metabolic pathways are activated against YR during transition from seedling to adult stages of development in CR
From the 55 sequences isolated from the cDNA–AFLP experiment, seven exhibiting significant homology to proteins potentially involved in plant–pathogen interactions were selected to analyse their expression patterns in a time‐course study of inoculated and mock‐inoculated CR leaves using quantitative real‐time PCR. The differential regulation of the seven TDFs was confirmed. Some of the putative genes were induced at all stages, and others were differentially expressed at one or two development stages. Therefore, employment of the CR‐derived HIS, expressing resistance to YR at different development stages, combined with the cDNA–AFLP technique, appeared to be an efficient strategy to avoid false‐positives and identify genes activated at specific development stages.
One TDF, SN42, the CIGR/SCARECROW homologue, was specifically induced or repressed at the two adult stages studied but not at the seedling stage. The TDF SN42 exhibited the same expression profile at the two adult stages. The association of the CIGR gene with elicitor‐induced defence response in rice (Day et al., 2003, 2004) implicates this gene in the early defence response to P. striiformis at adult stages. In contrast to the CIGR homologue, the SN18 transcription factor and the SN33 beclin‐1 homologue were induced later following inoculation. The later expression of a gene with functionality similar to beclin‐1 suggets this is a good candidate gene responsible for host‐cell necrosis; the resulting release of toxic metabolites causing local inhibition of haustorium formation and hyphal branching (Mares, 1979; Mares and Cousen, 1977) may make this a key gene associated with adult plant resistance to YR in wheat. Additional research is required to establish the role of these and other resistance/defence gene homologues in the developmentally regulated adult plant resistance to YR in wheat.
The TDF SN41, putatively encoding the phosphofructokinase, was differentially up‐regulated during all three stages of development in CR rust‐inoculated leaves. In plants infected by obligate biotrophic pathogens such as the rusts, up‐regulation of genes involved in carbohydrate metabolism would be expected in order to compensate for loses of substrates used by the pathogen.
Two TDFs, SN64 and SN81, were differentially expressed at the seeding stage and one of the adult stages. This indicates a non‐specific response to pathogen infection.
In summary, a series of different TDFs homologous to resistance/defence‐related genes, transcriptional factors and metabolic genes has been identified and demonstrated to be differentially expressed during the expression of adult plant resistance to YR in the variety CR. Three of the seven genes validated by real‐time PCR were specifically up‐regulated during the tillering and/or the two‐node stages in the inoculated versus mock‐inoculated CR leaves. This study provides the first clues to understanding the biochemical and molecular genetic basis of the seven QTLs previously implicated in the HTAP CR resistance (Mallard et al., 2005). Two genes, a beclin‐1‐like and a xip‐II gene, were identified as involved in the resistance/defence responses of bread wheat for the first time in our study. Further studies will be carried out to map those candidate genes on the CR × Récital genetic map and identity their roles in the resistance reaction. We hypothesize that the responses induced in CR by P. striiformis differ between the seedling and the adult growth stages because the plant expresses different quantitative resistance factors. Transcriptional changes involved in an early development phase change from seedling to adult (Willman and Poethig, 2005) could also trigger a reprogramming of adult plant resistance, accounting for the differential regulation of critical resistance/defence genes identified in this study. This constitutes an excellent defence strategy that may explain the durability of CR resistance: the rust pathogen cannot overcome CR resistance as it is not confronted by the same target gene(s) between two infection cycles. Thus, the transient expression of different resistance/defence‐related genes identified as involved in the HTAP CR resistance in new wheat varieties could constitute a means to obtain durable resistance in natural infections and should be developed in the next few years.
EXPERIMENTAL PROCEDURES
Plant material, pathogen infection
CR, Récital (susceptible to YR) and six sets of HIFs, segregating for the resistance against YR, were used (Table 3). Each set of HIFs was derived from a single F6 RIL and contained one or two resistant lines and a susceptible line that shared a 97% identical genetic background. The resistant lines of each set possessed 3–6 QTLs for resistance to YR. For all inoculations, P. striiformis pathotype 237 E141 V17, which possesses the V1, V2, V3a + V4a, V6, V9, V17, SD, SU virulence, was used (Mallard et al., 2005). Plants were inoculated by spraying a spore suspension of P. striiformis in mineral oil (Soltrol 170) whereas control plants were mock inoculated with mineral oil (Soltrol 170) (Mallard et al., 2005). Following inoculation, plants were incubated at 12 °C, 100% relative humidity in the dark for 24 h to ensure spore germination and penetration.
Table 3.
Heterogeneous inbred families (HIFs) employed and the development stage at which resistance to P. striiformis is expressed (++ = total resistance, + = partial resistance, – = susceptibility).
| Line | Seedling | Tillering | Two‐node | Flowering |
|---|---|---|---|---|
| CR | ++ | ++ | ++ | ++ |
| Récital | – | – | – | – |
| 2083R | ++ | ++ | ++ | ++ |
| 2083S | – | – | – | – |
| 2178R | ++ | ++ | ++ | ++ |
| 2178S | – | – | – | – |
| 2182R | ++ | ++ | ++ | ++ |
| 2182S | – | – | – | – |
| 2171‐2 | – | ++ | ++ | ++ |
| 2171‐5 | – | – | ++ | ++ |
| 2171‐6 | – | – | – | – |
| 2141‐2 | – | – | ++ | ++ |
| 2141‐4 | – | – | – | + |
| 2043R | – | – | – | ++ |
| 2043S | – | – | – | – |
Fifteen days after inoculation, YR resistance symptoms were recorded on remaining leaves using: (1) a scale described by McNeal et al. (1971) at the Zadoks stages 13 and 25; and (2) the percentage area infected at the Zadoks stage 32 (Zadoks et al., 1974). Good care was taken to ensure that all treatments were clean and free of symptoms from other plant pathogen contaminations.
Experiments
Three independent experiments were carried out using wheat plants grown at three different stages of plant development: the seedling stage, the tillering stage following cold hardening, and the two‐node stage. They corresponded to Zadoks stages 13, 25 and 32, respectively.
The seedling stage
Each line of HIFs and parents was seeded in 29 × 29 × 7‐cm3 plastic boxes, 50 seeds per box, in standard soil (RHP15, Klasmann, France), and grown in a growth chamber set at 20 °C (16‐h day‐length) until the appearance of the second leaf (approximately 1.5 weeks), and inoculated. Plants were then returned to the growth chamber at 20 °C with a 16‐h photoperiod. For each genotype, the two first leaves per plant from three inoculated plants and from three mock‐inoculated plants were collected 2 and 6 dpi. Leaves from the HIFs 2083R/2083S, 2178R/2178S, 2182R/2182S and the parents were also collected 3 and 7 dpi as the lines 2083R, 2178R and 2182R showed resistance at the seedling stage.
The tillering stage
The HIFs and parents were germinated in Petri dishes containing moistened filter and placed at 20 °C for 3 days. Following germination, seedlings were transplanted into 9 × 9‐cm pots (one plant per pot) containing standard soil RHP15 and placed in a hardening chamber (7–8 °C, 16‐h photoperiod) for 7 weeks. Following hardening, plants were transferred to a growth chamber set at 20 °C for 10 days. Plants were then inoculated, incubated in the dark and then returned to the growth chamber at 20 °C with a 16‐h photoperiod. For each genotype, two leaves per plant from three inoculated plants and from three mock‐inoculated plants were collected at 2 and 6 dpi. Since the resistance of the line 2171‐2 was known to be effective from the tillering stage (Mallard et al., 2005), leaves were also collected 3 and 7 dpi for the HIFs 2171‐2, 2171‐5 and 2171‐6 and the parents.
The two‐node stage
The HIFs and parents were germinated in Petri dishes containing moistened filter and placed at 20 °C for 3 days. Following germination, seedlings were transplanted into 9 × 9‐cm pots (one plant per pot) containing standard soil RHP15 and placed in a hardening chamber for 7 weeks. Plants were then placed into standard soil bags Green Meter (100 × 20 cm) (Haasnoot, Netherlands) with six plants per bag in a greenhouse at 15–25 °C under natural lighting conditions. Each bag contained a nutrient tablet, N‐P‐K = 15 + 9 + 9 + 3 MgO (Osmocote, Netherlands). At the two‐node stage, plants were inoculated and dark incubated with a fog system during 24 h. For each genotype, two leaves per plant from three inoculated plants and from three mock‐inoculated plants were collected at 2 and 6 dpi. The lines 2171‐5 and 2141‐2 were resistant from the two‐node stage, thus leaves were also collected at 3 and 7 dpi for the HIFs 2171‐2, 2171‐5, 2171‐6, 2141‐2 and 2141‐4 and the parental lines.
Collected leaves were immediately frozen in liquid nitrogen and stored at –80 °C.
RNA extraction and cDNA synthesis
Leaves of the three biological replicates were pooled and ground to a fine powder in liquid nitrogen using a mortar and pestle. Total RNA was isolated from frozen samples using the Spin Protocol from the ‘SV Total RNA Isolation System’ (Promega, Madison, WI). In total, 225 µg of total RNA was used to carry out reverse transcription, using SuperScript II Reverse Transcriptase (Invitrogen, France).
CDNA–AFLP
Templates for cDNA–AFLP analysis were prepared according to Vos et al. (1995) and Bachem et al. (1996). The cDNA was digested with the restriction enzymes EcoRI and MseI (Promega). EcoRI and MseI adaptators, with sequences as follows: EcoRI adapter, 5′‐CTCGTAGACTGCGTACC‐3′, 3′ CTGACGCATGGTTAA‐5′; MseI adapter, 5′‐GACGATGAGTCCTGAG‐3′, 3′‐TACTCAGGACTCAT‐5′, were then ligated to the cDNA fragments using T4 DNA ligase (Invitrogen). A PCR pre‐amplification was performed with primers containing one selective base (EcoRI: 5′‐GACTGCGTACCAATTN‐3′, MseI: 5′‐GATGAGTCCTGAGTAN‐3′). Pre‐amplificated templates were then diluted 25‐fold before being subjected to a selective PCR amplification using primers with three selective bases. Selective EcoRI primers were IRD‐700‐ or IRD‐800‐labelled (LICOR® Biosciences, France). Gel electrophoresis on 6% polyacrylamide gels was conducted using a LICOR IR2 automated DNA sequencer (LICOR® Biosciences). Only the cDNA flanked by two EcoRI primers or by one EcoRI and one MseI primer were thus visualized.
Fragment isolation, cloning and sequencing
The PCR samples containing differentially regulated fragments were loaded on a new gel for recovery. The gel was removed from the sequencer when the smallest desired bands passed the laser detector and scanned using an ODYSSEY® Imager (LI‐COR® Biosciences). Desired fragments were then excised from the gel, and DNA was recovered and re‐amplified by PCR. Products of re‐amplification were cloned into a pGEM®‐T Easy Vector (Promega) according to the manufacturer's instructions. For each fragment, 2–3 clones, exhibiting the correct size, were sequenced by Genome Express (France).
Putative identification of sequences was performed using homology searches (Blastn and Blastx) (Altschul et al., 1997) against public databases: the Graingene Wheat EST database (http://wheat.pw.usda.gov/GG2/index.shtml), the TIGR Wheat Genome Database (http://www.tigr.org/tdb/e2k1/tae1/introduction.shtml), the Genbank DNA sequence database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) and the swissprot/TrEMBL database (http://expasy.org/sprot/).
Real‐time PCR
For real‐time PCR analysis, seven TDFs up‐regulated in resistant, inoculated genotypes that had significant homologies with genes implicated in disease resistance reactions were selected. For RNA isolation, the same experiments were conducted for each wheat development stage as described above except that the only cultivar used was CR and leaves from three inoculated and three mock‐inoculated plants were collected at 0, 1, 1.5, 2, 2.5, 3, 4, 4.5, 5.5, 6 and 6.5 dpi. Real‐time PCR reactions were done in a 20 µL volume containing 1× SYBR® Green PCR master mix (Applied Biosystems), 400 nm of each primer and 250 ng of cDNA. An ABI Prism® 7700 Sequence Detector System (Applied Biosystems) was used for detection of all amplicons. Each PCR reaction was performed in triplicate and a template control was included. Specific primers for each studied TDF and for the housekeeping gene of 18S ribosomal protein used for data normalization were designed using the Primer Express Software 1.5 (Applied Biosystems) or Primer3 Software (Rozen and Skaletsky, 2000) (Table 4). Specificity of the amplicon was verified at the end of the PCR run using ABI Prism Dissociation Curve Analysis software. Real‐time PCR results (Ct values) were analysed with the REST® software, which calculates the average expression ratio (ΔΔCT method of relative gene quantification) and the P‐values to assess the statistical relevance of changes obtained by a randomization test (Pfaffl et al., 2002).
Table 4.
Oligonucleotide primers used to measure relative expression of different TDFs using real‐time PCR.
| TDF | Foward primer (5′–3′) | Reverse primer (5′–3′) | T m value (°C) |
|---|---|---|---|
| 18S | AGGAGTTCCAGCACATCCTTC | CCCTCTTGTTCATGTCGATGT | 60 |
| SN15 | AAGAATGAGTGGGGTTGGTG | GAAGGGAATCTGCAAATGGA | 60 |
| SN18 | AAAGCTGCTCCCTCCATACC | CTGCGTACCAATTCACAGAAA | 60 |
| SN33 | CTCCTTGTCCATCTTATCAGAAAGC | GGGCATTTGAGATTGCTAGTTCAC | 60 |
| SN41 | GCCCGCTTCACAATCTTTGG | TGCCAGTGATTGCCTTGGT | 60 |
| SN42 | GATGGTCAGTAGGCGGTAGC | ACTGTGGCTGCTTGCTTCTT | 60 |
| SN64 | TGAAGTGTTTGTCAGATGATCCAAT | CTGTTTTCCCATCGCCAATTT | 60 |
| SN81 | ATACCGTTTTCGCGGTTATG | CTTGAGGAGTCGACGGAGAT | 60 |
We considered that a gene was differentially expressed in response to P. striiformis infection (1) if the probability that it was different between two treatments or two time‐course points was P ≤ 0.001 and (2) its relative quantity of RNA was at least two‐fold higher or lower than in leaves from mock‐inoculated plants.
ACKNOWLEDGMENTS
We would like to thank C. de Vallavieille‐Pope (INRA, Grignon Research Centre) for providing the pathogenic strain. This work received financial support from the French Ministry of Research.
REFERENCES
- Allen, R. (1928) A cytological study of Puccinia glumarum on Bromus marginatus and Triticum vulgare . J. Agricultural Res. 36, 487–513. [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem, C. , Van Der Hoeven, R. , De Bruijin, S. , Vreugdenhil, D. , Zabeau, M. and Visser, R. (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 9, 745–753. [DOI] [PubMed] [Google Scholar]
- Banik, M. , Garrett, T.P. and Fincher G.B. (1996) Molecular cloning of cDNA encoding (1,4)‐β‐xylan endohydrolase from the aleurone layer of germinated barley (Hordeum vulgare ). Plant Mol. Biol. 31, 1163–1172. [DOI] [PubMed] [Google Scholar]
- Boukhatem, N. , Baret, P.V. , Mingeot, D. and Jacquemin, J.M. (2002) Quantitative trait loci for resistance against yellow rust in two wheat‐derived recombinant inbred line populations. Theor. Appl. Genet. 104, 111–118. [DOI] [PubMed] [Google Scholar]
- Bradley, D.J. , Kjellbom, P. and Lamb, C.J. (1992) Elicitor‐ and wound induced oxidative cross‐linking of a proline‐rich plant cell wall protein: a novel, rapid defense response. Cell, 70, 21–30. [DOI] [PubMed] [Google Scholar]
- Bravo, J.M. , Campo, S. , Murillo, I. , Coca, M. and San Segundo, B. (2003) Fungus‐and wound‐induced accumulation of mRNA containing a class II chitinase of the pathogenesis‐related protein 4 (PR‐4) family of maize. Plant Mol. Biol. 52, 745–759. [DOI] [PubMed] [Google Scholar]
- Broglie, K. , Chet, I. , Holliday, M. , Cressman, R. , Biddle, P. , Knowlton, S. , Mauvais, C.J. and Broglie, R. (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani . Science, 254, 1194–1197. [DOI] [PubMed] [Google Scholar]
- Chen, X.M. (2005) Epidemiology and control of stripe rust (Puccinia striiformis f. sp. tritici) on wheat. Can. J.Plant Pathol. 27, 314–337. [Google Scholar]
- Christiansen, M.J. , Feenstra, B. , Skovgaard, I.M. and Andersen, S.B. (2006) Genetic analysis of resistance to yellow rust in hexaploid wheat using a mixture model for multiple crosses. Theor. Appl. Genet. 112, 581–591. [DOI] [PubMed] [Google Scholar]
- Coppinger, P. , Repetti, P. , Day, B. , Dahlbeck, D. , Mehlert, A. and Staskawicz, B.J. (2004) Overexpression of the plasma membrane‐localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana . The Plant J. 40, 225–237. [DOI] [PubMed] [Google Scholar]
- Davy de Virville, J. , Lance, C. and Doussinault, G. (1982) Réponse respiratoire au cours des premiers stades de l’infection chez diverses lignées de Triticées sensibles ou résistantes àPseudocercosporella herpotrichoides (Fron) deighton. Cryptogamie, Mycologie, 4, 319–333. [Google Scholar]
- Day, R.B. , Shibuya, N. and Minami, E. (2003) Identification and characterization of two new members of the GRAS gene family in rice responsive to N‐acetylchitooligosaccharide elicitor. Biochim. Biophys. Acta, 1625, 261–268. [DOI] [PubMed] [Google Scholar]
- Day, R.B. , Tanabe, S. , Koshioka, M. , Mitsui, T. , Itoh, H. , Ueguchi‐Tanaka, M. , Matsuoka, M. , Kaku, H. , Shibuya, N. and Minami, E. (2004) Two rice GRAS family genes responsive to N‐acetylchitooligosaccharide elicitor are induced by phytoactive gibberellins: evidence for cross‐talk between elicitor and gibberellin signaling in rice cells. Plant Mol. Biol. 54, 261–272. [DOI] [PubMed] [Google Scholar]
- Eckey, C. , Korell, M. , Leib, K. , Biedenkopf, D. , Jansen, C. , Langen, G. and Kogel, K.H. (2004) Identification of powdery mildew‐induced barley genes by cDNA‐AFLP: functional assessment of an early expressed MAP kinase. Plant Mol. Biol. 55, 1–15. [DOI] [PubMed] [Google Scholar]
- Ellis, J. and Jones, D. (1998) Structure and function of proteins controlling strain‐specific pathogen resistance in plants. Curr. Opin. Plant Biol. 1, 288–293. [DOI] [PubMed] [Google Scholar]
- Gajewska, M. , Gajkowska, B. and Motyl, T. (2005) Apoptosis and autophagy induced by TGF‐B1 in bovine mammary epithelial BME‐UV1 cells. J. Physiol. Pharmacol. 56, 143–157. [PubMed] [Google Scholar]
- Good, A.G. and Muench, D.G. (1992) Purification and characterization of an anaerobically induced alanine aminotransferase from barley root. Plant Physiol. 99, 1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M. and Mansfield, J. (1999) Early events in host–pathogen interactions. Curr. Opin. Plant Biol. 2, 312–319. [DOI] [PubMed] [Google Scholar]
- Grant, M. , Brown, I. , Adams, S. , Knight, M. , Ainslie, A. and Mansfield, J. (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450. [DOI] [PubMed] [Google Scholar]
- Igawa, T. , Tokai, T. , Kudo, T. , Yamaguchi, I. and Kimura, M. (2005) A wheat xylanase inhibitor gene, Xip‐I, but not Taxi‐I, is significantly induced by biotic and abiotic signals that trigger plant defense. Biosci. Biotechnol. Biochem. 69, 1058–1063. [DOI] [PubMed] [Google Scholar]
- Jach, G. , Görnhardt, B. , Mundy, J. , Logemann, J. , Pinsdorf, E. , Leah, R. , Shell, J. and Maas, C. (1995) Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8, 97–109. [DOI] [PubMed] [Google Scholar]
- Juge, N. , Payan, F. and Williamson, G. (2004) XIP‐1, a xylanase inhibitor protein from wheat: a novel protein function. Biochim. Biophys. Acta, 1696, 203–211. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. (2002) A century of plant pathology: a retrospective view on understanding host‐parasite interactions. Ann. Rev. Phytopathol. 38, 31–48. [DOI] [PubMed] [Google Scholar]
- Kim, M.C , Panstruga, R. , Elliot, C. , Muller, J. , Devoto, A. , Yoon, H.W. , Park, H.C. , Cho, M.J. and Schulze‐Lefert, P. (2002) Calmodulin interacts with MLO protein to regulate defense against mildew in barley. Nature, 416, 447–450. [DOI] [PubMed] [Google Scholar]
- Lal, S.K , Lee, C. and Sachs, M.M. (1998) Differential regulation of enolase during anaerobiosis in maize. Plant Physiol. 118, 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton, J.C. and Lamb, C.J. (1987) Transcriptional activation of plant defense genes by fungal elicitor, wounding, and infection. Mol. Cell Biol. 7, 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F. and Chen, X.M. (2007) Genetics and molecular mapping of genes for race‐specific all‐stage resistance and non‐race‐specific high‐temperature adult‐plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 114, 1277–1287. [DOI] [PubMed] [Google Scholar]
- Line, R.F. (2002) Stripe rust of wheat in barley in North America: a retrospective historical review. Annu. Rev. Phytopathol. 40, 75–118. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Czymmek, K. , Talloczy, Z. , Levine, B. and Dinesh‐Kumar S.P. (2005) Autophagy regulates Programmed Cell Death during the plant innate immune response. Cell, 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Mallard, S. (2005) Etude génétique et fonctionnelle de la résistance durable à la rouille jaune chez le cultivar de blé tendre Camp Rémy. PhD thesis, Agrocampus Rennes. [Google Scholar]
- Mallard, S. , Gaudet, D. , Aldeia, A. , Abelard, C. , Besnard, A.L. , Sourdille, P. and Dedryver F. (2005) Genetic analysis of durable resistance to yellow rust in bread wheat. Theor. Appl. Genet. 110, 1401–1409. [DOI] [PubMed] [Google Scholar]
- Mares, D.J. (1979) Microscopic study of the development of yellow rust (Puccinia striiformis) in a wheat cultivar showing adult plant resistance. Physiol. Plant Pathol. 15, 289–296. [Google Scholar]
- Mares, D.J. and Cousen, S. (1977). The interaction of yellow rust (Puccinia striiformis) with winter wheat cultivars showing adult plant resistance. Macroscopic and microscopic events associated with the resistance reaction. Physiol. Plant Pathol. 10, 257–274. [Google Scholar]
- Marryat, D. (1907) Notes on the infection and histology of two wheats immune to attacks of Puccinia glumarum, yellow rust. J. Agr. Sci. 2, 129–138. [Google Scholar]
- McNeal, F.H. , Konzac, C.F. , Smith, E.P. , Tate, W.S. and Russell T.S. (1971) A Uniform System for Recording and Processing Cereal Research Data. Washington, DC: ARS‐USDA. [Google Scholar]
- Menden, B. , Kohlhoff, M. and Moerschbacher, B.M. (2006) Wheat cells accumulate a syringyl‐rich lignin during the hypersensitive resistance response. Phytochemistry, 68, 513–520. [DOI] [PubMed] [Google Scholar]
- Milus, E. A. , Seyran, E. , and McNew, R. (2006) Aggressiveness of Puccinia striiformis f. sp. tritici isolates in the south‐central United States. Plant Dis. 90, 847–852. [DOI] [PubMed] [Google Scholar]
- Moldenhauer, J. , Moerschbacher, B.M. and Van Der Westhuizen, A.J. (2006) Histological investigation of stripe rust (Puccinia striiformis f. sp. tritici) development in resistant and susceptible wheat cultivars. Plant Pathol. 55, 469–474. [Google Scholar]
- Myers, E.W. and Miller, W. (1988) Optimal alignments in linear space. Biosci. Comput. Appl. 4, 11–17. [DOI] [PubMed] [Google Scholar]
- Navabi, A. , Tewari, J.P. , Singh, R.P. , McCallum, B. , Laroche, A. and Briggs, K.G. (2005) Inheritance and QTL analysis of durable resistance to stripe and leaf rusts in an Australian cultivar, Triticum aestivum ‘Cook’. Genome, 48, 97–107. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , Gallois, J.L. , Chafiai, F. , Allen, L.M. , Schurdi‐Levraud, V. , Browning, K.S. , Candresse, T. , Caranta, C. , Le Gall, O. and German‐Retana, S. (2007) Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana . FEBS Lett. 581, 1041–1046. [DOI] [PubMed] [Google Scholar]
- Niks, R.E. (1989) Morphology of infection structures of Puccinia striiformis var. dactylidis . Netherlands J. Plant Pathol. 95, 171–175. [Google Scholar]
- Pfaffl, M.W. , Horgan, G.W. and Dempfle, L. (2002) Relative expression software tool (REST) for group‐wise comparison and statistical analysis of relative expression results in real‐time PCR. Nucleic Acids Res. 30, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramburan, V.P. , Pretorius, Z.A. , Louw, J.H. , Boyd, L.A. , Smith, P.H. , Boshoff, W.H. and Prins, R. (2004) A genetic analysis of adult plant resistance to stripe rust in the wheat cultivar Kariega. Theor. Appl. Genet. 108, 1426–1433. [DOI] [PubMed] [Google Scholar]
- Rampitsch, C. , Bykova, N.V. , McCallum, B. , Beimcik, E. and Ens, W. (2006) Analysis of the wheat and Puccinia triticina (leaf rust) proteomes during a susceptible host–pathogen interaction. Proteomics, 6, 1897–1907. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001) Calcium: silver bullet in signalling. Plant Sci. 160, 381–404. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H.J. (2000) Primer3 on the WWW for general users and for biologist programmers In: Bioinformatics Methods and Protocols: Methods in Molecular Biology (Krawetz S., Misener S., eds), pp. 365–386. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Schultz, C. , Gilson, P. , Oxley, D. , Youl, J. and Bacic, A. (1998) GPI‐anchors on arabinogalactan‐proteins: implications for signalling in plants. Trends Plant Sci. 3, 426–431. [Google Scholar]
- Sheng, J. , Jeong, J. , and Mehdy, M.C. (1993) Developmental regulation and phytochrome‐mediated induction of mRNAs encoding a proline‐rich protein, glycine‐rich proteins, and hydroxyproline‐rich glycoproteins in Phaseolus vulgaris L. Proc. Natl Acad. Sci. USA, 90, 828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier, D.J. , Prime, T.A. and Dupree, P. (1999) Glycosylphosphatidylinositol‐anchored cell‐surface proteins from Arabidopsis . Electrophoresis, 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Showalter, A.M. , Zhou, J. , Rumeau, D. , Worst, S.G. and Varner J.E. (1991) Tomato extensin and extensin‐like cDNAs: structure and expression in response to wounding. Plant Mol. Biol. 16, 547–565. [DOI] [PubMed] [Google Scholar]
- Solomon, K.R. , Rudd, C.E. and Finberg, R.W. (1996) The association between glycosylphosphatidylinositol‐anchored proteins and heterotrimeric G protein alpha subunits in lymphocytes. Proc. Natl Acad. Sci. USA, 93, 6053–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D. , Ganal, M.W. and Martin, G.B. (1995) Chromosome landing: a paradigm for map‐based gene cloning in plants with large genomes. Trends Genet. 11, 63–68. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventelon‐Debout, M. , Delalande, F. , Brizard, J.P. , Diemer, H. , Van Dorsselaer, A. and Brugidou, C. (2004) Proteome analysis of cultivar‐specific deregulations of Oryza sativa indica and O. sativa japonica cellular suspensions undergoing rice yellow mottle virus infection. Proteomics, 4, 216–225. [DOI] [PubMed] [Google Scholar]
- Ventelon‐Debout, M. , Nguyen, T.T. , Wissocq, A. , Berger, C. , Laudie, M. , Piegu, B. , Cooke, R. , Ghesquiere, A. , Delseny, M. and Brugidou, C. (2003) Analysis of the transcriptional response to Rice Yellow Mottle Virus infection in Oryza sativa indica and japonica cultivars. Mol. Genet. Genomics, 270, 253–262. [DOI] [PubMed] [Google Scholar]
- Vos, P. , Hogers, R. , Bleeker, M. , Reijans, M. , Van de Lee, T. , Hornes, M. , Frijters, A. , Pot, J. , Peleman, J. , Kuiper, M. and Zabeau M. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willman, M.R. and Poetig, R.S. (2005) Time to grow up: the temporal role of small RNAs in plants. Curr. Opin. Plant Biol. 8, 548–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L. , Echenique, V. , Busso, C. , SanMiguel, P. , Ramakrishna, W. , Bennetzen, J.L. , Harrington, S. and Dubcovsky, J. (2002) Cereal genes similar to Snf2 define a new subfamily that includes human and mouse genes. Mol. Genet. Genomics, 268, 488–499. [DOI] [PubMed] [Google Scholar]
- Ye, Z.H. and Varner, J.E. (1991) Tissue‐specific expression of cell wall proteins in developing soybean tissues. Plant Cell, 3, 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L. , Lenardo, M.J. and Baehrecke, E.H. (2004) Autophagy and caspases: a new cell death program. Cell Cycle, 3, 1124–1126. [PubMed] [Google Scholar]
- Zadoks, J.C. , Chang, T.T. and Konzak, C.F. (1974) A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. [Google Scholar]
- Zhang, L. , Meakin, H. and Dickinson, M. (2003) Isolation of genes expressed during compatible interactions between leaf rust (Puccinia triticina) and wheat using cDNA‐AFLP. Mol. Plant Pathol. 4, 469–477. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Chen, X. , Zhang, X. , Lin, Z. , Shang, J. , Xu, J. , Zhai, W. and Zhu, L. (2004) Isolation and identification of a gene in response to rice blast disease in rice. Plant Mol. Biol. 54, 99–109. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Maher, E.A. , Masoud, D. , Dixon, R.A. and Lamb, C.J. (1994) Enhanced protection against fungal attack by constitutive co‐expression of chitinase and glucanase genes in transgenic tobacco. Biotechnology, 12, 807–812. [Google Scholar]


