SUMMARY
The plant‐pathogenic prokaryote Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial leaf blight, one of the most destructive diseases of rice. A nonpolar mutant of the rsmA‐like gene rsmAXoo of the Xoo Chinese strain 13751 was constructed by homologous integration with a suicide plasmid. Virulence tests on a host plant, namely the hybrid rice cultivar Teyou 63, showed that the mutant had lost its virulence almost completely, whereas tests on a nonhost, namely castor‐oil plant (Ricinus communis), showed that the mutant had also lost the ability to induce a hypersensitive response in the nonhost. In addition, the rsmAXoo mutant produced significantly smaller amounts of the diffusible signal factor, extracellular endoglucanase, amylase and extracellular polysaccharide, but showed significantly higher glycogen accumulation, bacterial aggregation and cell adhesion. The expression of most hrp genes, genes encoding AvrBs3/PthA family members, rpfB, xrvA, glgA, eglXoB and XOO0175 (encoding an α‐amylase) was down‐regulated in the rsmAXoo mutant. All phenotypes and expression levels of the tested genes in the rsmAXoo mutant were restored to their levels in the wild‐type by the presence of rsmAXoo in trans. These results indicate that rsmAXoo is essential for the virulence of Xoo.
INTRODUCTION
The Gram‐negative bacterium Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial leaf blight, one of the most destructive diseases of rice, which limits the production of this staple food (Mew et al., 1993). Xoo typically invades the plant through hydathodes or wounds along the leaf margins, reaches the xylem vessels and multiplies in the intercellular spaces to cause a vascular disease, resulting in tan‐grey to white lesions along the leaf veins (Nino‐Liu et al., 2006).
Many Gram‐negative plant‐pathogenic bacteria rely on the type III secretion system (T3SS) to inject effectors into the host cell, which leads to either disease or a resistant reaction (Grant et al., 2006). T3SS of Xanthomonas, which is encoded by a hypersensitive response and pathogenicity (hrp) gene cluster, is necessary for pathogenicity in susceptible hosts and for a hypersensitive response (HR) in resistant plants and nonhost plants. T3SS may also play a role in the secretion of other virulence factors (Büttner and Bonas, 2010; Büttner and He, 2009). In Xoo, the hrp gene cluster consists of the conserved hrp region, which comprises hpa1, hpa2, hrpA and hrp operons hrpB–E, and the variable region, which comprises hpaC, hpaD, hrpF and hpaF (Ochiai et al., 2005). HrpG, which is the OmpR family regulator, and HrpX, which is the AraC‐type regulator, are the two key regulators of hrp genes in Xanthomonas. The expression of hrpX is up‐regulated by HrpG, which is activated in response to as yet unknown signals (Kamdar et al., 1993; Koebnik et al., 2006; Wengelnik and Bonas, 1996; Wengelnik et al., 1996).
Most avirulence (Avr) proteins that activate a plant's defence responses are type III effectors in phytopathogenic bacteria (Grant et al., 2006; Mansfield, 2009). Several avr genes have been cloned and identified in Xoo (Lee et al., 2009; White and Yang, 2009), and their encoded products, except Ax21 (formerly AvrXa21; Lee et al., 2009), belong to the AvrBs3/PthA family (White and Yang, 2009; White et al., 2000). The avr gene avrXa7 is also a determinant of virulence in Xoo (White and Yang, 2009). Other type III effectors belonging to the AvrBs3/PthA family and encoded by the genes pthXo1, pthXo2, pthXo3, pthXo6 and pthXo7 from Xoo are known to contribute to bacterial virulence under appropriate conditions (White and Yang, 2009).
The virulence of Xanthomonas campestris pv. campestris (Xcc) also depends on cell–cell signalling mediated by the small diffusible signal factor (DSF) and by the RpfC/RpfG two‐component regulatory system, which couples DSF sensing to the intracellular regulatory networks through a second messenger, namely cyclic diguanylate (di‐GMP), and a global regulator, namely Clp (Dow, 2008; Fouhy et al., 2006; He and Zhang, 2008). DSF was first identified in Xcc (Barber et al., 1997), and was structurally identified as cis‐11‐methyl‐2‐dodecenoic acid (Wang et al., 2004). The factor acts as a cell–cell communication signal involved in the expression of virulence genes and dispersal of the biofilm to control the production of extracellular enzymes and extracellular polysaccharide (EPS), and mediates the synthesis of the flagellum, resistance to toxins and oxidative stress, and aerobic respiration (He et al., 2006). Two genes, rpfF (encoding a putative enoyl CoA hydratase) and rpfB (encoding a long‐chain fatty acyl CoA ligase), have been implicated in the synthesis of DSF (Barber et al., 1997). In Xoo, mutations in rpfF make the mutants incapable of producing DSF (Chatterjee and Sonti, 2002; He et al., 2010).
Virulence factors, including cellulases, xylanases, pectinases and proteases, which are secreted by plant‐pathogenic bacteria, can break down host cell walls and may play an important role in virulence (Büttner and Bonas, 2010). Extracellular endoglucanase is involved in the virulence of Xcc and Xoo (Büttner and Bonas, 2010; Gough et al., 1988; Hu et al., 2007; Schröter et al., 2001). Some studies have demonstrated that extracellular xylanase, cellobiosidase and esterase are important to the virulence of Xoo (Aparna et al., 2009; Jha et al., 2007; Rajeshwari et al., 2005; Ray et al., 2000). EPS is another important virulence factor in Xanthomonas (Büttner and Bonas, 2010). In Xoo, the gum cluster is involved in EPS synthesis (Dharmapuri and Sonti, 1999; Kim et al., 2009; Rajeshwari and Sonti, 2000), and rpfC plays an important role in regulating EPS synthesis (Tang et al., 1996).
The bacterial RNA‐binding protein RsmA (repressor of secondary metabolism) is a global post‐transcriptional regulator, which was discovered as a potent repressor of glycogen synthesis in Escherichia coli and designated as CsrA (carbon storage regulator) (Romeo, 1998). The CsrA of E. coli was further found to repress gluconeogenesis, peptide transport and biofilm formation, and to activate glycolysis, acetate metabolism and cell motility (Lapouge et al., 2008; Romeo, 1998). In addition, RsmA/CsrA was found to be involved in the pathogenesis of several bacterial pathogens in animals and plants (Chao et al., 2008; Fields and Thompson, 2008; Lapouge et al., 2008; Lucchetti‐Miganeh et al., 2008).
RsmA/CsrA of bacterial pathogens can regulate the expression of virulence genes negatively or positively. For example, CsrA of Salmonella enterica serovar Typhimurium positively regulates the expression of invasion genes and of the components of T3SS associated with virulence (Lucchetti‐Miganeh et al., 2008). However, in Erwinia carotovora, a plant pathogen, RsmA represses the expression of some T3SS‐related genes and also destabilizes mRNA transcripts encoding enzymes that degrade cell walls in plants, including cellulase, pectate lyase and protease (Mole et al., 2007). In the plant‐pathogenic bacterium Xcc strain 8004, the deletion of rsmAXcc leads to a complete loss of virulence towards the host plant Chinese radish, HR in the nonhost pepper (cultivar ECW‐10R) and motility on agar plates. Furthermore, a mutation in rsmAXcc reduces significantly the production of extracellular amylase, endoglucanase and polysaccharide, but increases significantly the accumulation of intracellular glycogen, bacterial aggregation and cell adhesion. The mutation also results in significantly reduced expression of genes encoding T3SS, type III effectors and the bacterial aggregate‐dispersing enzyme endo‐β‐1,4‐mannanase (Chao et al., 2008).
Whole‐genome sequencing has revealed that rsmA/csrA homologues exist in Xoo (Lee et al., 2005; Ochiai et al., 2005; Salzberg et al., 2008), but little is known about their role in the pathogenesis of Xoo. In this article, we present genetic evidence to demonstrate that rsmAXoo, the rsmA/csrA homologue in Xoo, acts as an important regulator of virulence and DSF production.
RESULTS
rsmAXoo is essential for the virulence and HR of Xoo
The rsmA‐like gene rsmAXoo (accession number HM988729), an open reading frame (ORF) of Xoo Chinese strain 13751 (Tang et al., 1996) (Table 1), is 213 bp long and encodes a protein comprising 70 amino acids. The rsmAXoo gene was identical to XOO_2790 (accession number NC_007705.1), XOO2938 (accession number NC_006834.1) and PXO_00146 (accession number NC_010717.1), all of which were annotated as csrA/rsmA (hereafter rsmAXoo) in the sequenced genome of the Xoo strains MAFF311018, KACC10331 and PXO99A, respectively. RsmAXoo displays 74% identity with E. coli K‐12 CsrA (NP_417176.1), 93% with Xcc 8004 RsmAXcc (YP_243576.1) and 100% with X. axonopodis pv. citrus 306 (Xac) RsmAXac (NP_642074.1) and X. campestris pv. vesicatoria 85‐10 (Xcv) RsmAXcv (YP_363507.1). The nearest gene upstream of rsmAXoo was alaS (encoding a putative alanyl‐tRNA synthetase), whereas the nearest gene downstream was tRNA‐Ser (encoding tRNA‐Ser) (Fig. S1, see Supporting Information). Sequence analyses showed that no other gene in the sequenced genome of Xoo strains shared significant homology with rsmAXoo.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics* | Reference or source |
|---|---|---|
| Xanthomonas oryzae pv. oryzae strains | ||
| 13751 | Wild‐type; Chinese isolate, Smr | Tang et al. (1996) |
| GXN2790 | As 13751, but rsmAXoo::pK18MobGII, Smr Kmr | This work |
| GXC2790 | GXN2790 harbouring pL6rsmAXoo, Smr Kmr Tcr | This work |
| 13751hrcV | As 13751, but hrcV::Tn5, Smr Kmr | Feng et al. (2001) |
| Xanthomonas campestris pv. campestris strains | ||
| DM2506 | As wild‐type Xcc 8004, but the coding region of rsmAXcc (XC2506) has been deleted, Rifr | Chao et al. (2008) |
| 8523 | As wild‐type strain 8004, but rpfF::Tn5lac, Rifr Kmr | Tang et al. (1991) |
| Plasmids | ||
| pGEM‐3Zf(+) | Harbouring multiple cloning site, Ampr | Promega |
| pRK2073 | Helper plasmid, Tra+, Mob+, ColE1; Spcr | Leong et al. (1982) |
| pK18MobGII | Mob+; ColE1 gusA, Kmr | Katzen et al. (1999) |
| pKrsmAXoo | pK18MobGII containing a 131‐bp internal fragment of rsmAXoo gene, Kmr | This work |
| pLAFR6 | Broad host range cloning vector, Tcr | Huynh et al. (1989) |
| pL6rsmAXoo | pLAFR6 containing a 703‐bp fragment including the rsmAXoo gene, Tcr | This work |
Smr, Kmr, Tcr, Spcr and Rifr indicate resistance to streptomycin, kanamycin, tetracycline, spectinomycin and rifampicin, respectively.
To study the potential biological function of rsmAXoo, a nonpolar mutant of rsmAXoo, named GXN2790, was constructed in Xoo Chinese strain 13751 by homologous integration with the suicide plasmid pKMob18GII (Katzen et al., 1999) derivative pKrsmAXoo (Table 1). Mutation was confirmed by complementation with the entire sole rsmAXoo gene. The complemented strain GXC2790 was constructed by introducing the recombinant plasmid pLrsmAXoo containing the entire wild‐type rsmAXoo gene into the mutant GXN2790 (Table 1).
To determine whether mutations in rsmAXoo affect the growth of Xoo, we examined the growth of the rsmAXoo mutant strain GXN2790, the wild‐type strain 13751 and the complemented strain GXC2790 in complete medium, namely OB (Tang et al., 1996), and in minimal medium, namely XOM2 (Tsuge et al., 2002). The growth of all three strains was comparable in both media (P > 0.05, t‐test) (data not shown), indicating that rsmAXoo is not required for the growth of Xoo in either of the two media.
The virulence of rsmAXoo, the nonpolar mutant GXN2790 and the complemented strain GXC2790 was tested on a hybrid rice cultivar, namely Teyou 63, by the leaf‐clipping method (Kauffman et al., 1973), with the wild‐type strain 13751 serving as the control. Fourteen days after inoculation with a final concentration of either 1 × 106 colony‐forming units (cfu)/mL (data not shown) or 1 × 108 cfu/mL, symptoms of bacterial leaf blight of comparable intensity appeared along the leaves inoculated with either the wild‐type or the complemented strain, with no difference in lesion length (P > 0.05, t‐test). However, leaves inoculated with the mutant strain GXN2790 were almost free from lesions (Fig. 1a,b) (P < 0.01, t‐test), indicating that rsmAXoo is essential for the virulence of Xoo.
Figure 1.
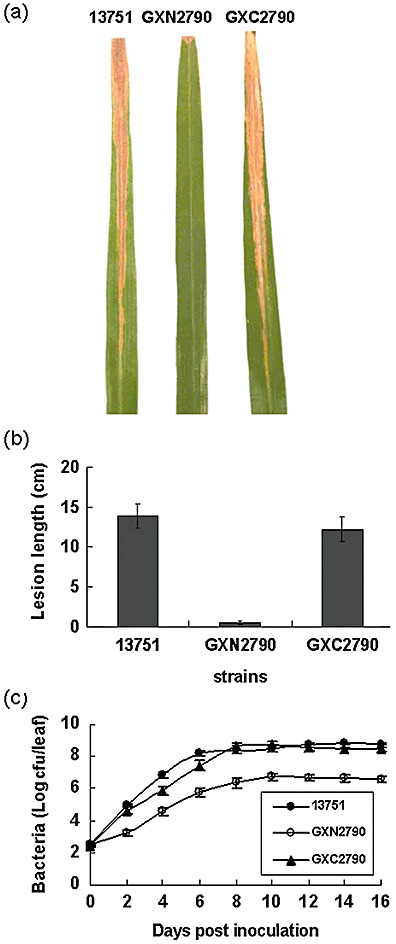
rsmAXoo is essential for the virulence of Xanthomonas oryzae pv. oryzae. (a) Rice leaf blight symptoms on leaves of the hybrid rice cultivar Teyou 63, 14 days after inoculation with X. oryzae pv. oryzae strains at an optical density at 600 nm (OD600) of 0.1 [1 × 108 colony‐forming units (cfu)/mL]. (b) Average length of lesions caused by X. oryzae pv. oryzae inoculated at OD600 = 0.1 (1 × 108 cfu/mL). Data are means ± standard deviation from three replications, each with 30 leaves. The results presented are from a representative experiment; similar results were obtained in the other two independent experiments. (c) Growth of bacteria in inoculated leaves of rice Teyou 63. Bacteria were inoculated by the leaf‐clipping method at OD600 = 0.1 (1 × 108 cfu/mL). Three inoculated leaves for each strain were removed at 2‐day intervals and homogenized in sterile water. The diluted homogenate was plated onto a rich medium supplemented with appropriate antibiotics. Bacterial cfu was counted after incubation at 28°C for 3 days, and was expressed as cfu per leaf. Data are the means ± standard deviation from three replications.
To investigate the role of rsmAXoo in the growth of Xoo inside the host, we compared the populations of the rsmAXoo mutant GXN2790, the wild‐type strain 13751 and the complemented strain GXC2790 on inoculated rice leaves. The population of the mutant strain GXN2790 recovered from infected leaves was significantly lower than that of the wild‐type and the complemented strain at each of the test points (P < 0.01, t‐test) (Fig. 1c). There was no difference in growth between the complemented and wild‐type strain (P > 0.05, t‐test) (Fig. 1c). These results indicate that rsmAXoo is required for the proliferation of Xoo in host plants.
To determine the role of rsmAXoo in inducing an HR in castor‐oil plant (Ricinus communis), a nonhost, its leaves were inoculated separately with the rsmAXoo mutant GXN2790 and the Xoo hrcV mutant, a type III‐deficient mutant that cannot elicit an HR in nonhost plants (Feng et al., 2001) (Table 1). The mutant GXN2790 was unable to trigger an HR, whereas the wild‐type and the complemented strain GXC2790 were able to do so (Fig. 2). As expected, the negative control hrcV mutant elicited no visible HR (Fig. 2). Therefore, it is clear that rsmAXoo is essential for Xoo to elicit an HR in nonhost castor‐oil plant.
Figure 2.

Induction of the hypersensitive response (HR) on the leaves of a nonhost, namely castor‐oil plant (Ricinus communis), by Xanthomonas oryzae pv. oryzae strains. Approximately 20 µL of bacterial culture suspended in 10 mm sodium phosphate buffer was infiltrated into leaf mesophyll tissue using a 1‐mL blunt‐end plastic syringe at an inoculum level of 5 × 108 colony‐forming units (cfu)/mL. The photograph was taken 48 h after infiltration. Four replicates were maintained for each experiment, and each experiment was repeated three times. The results presented are from a representative experiment; similar results were obtained in all other independent experiments. (a) rsmAXoo mutant strain GXN2790. (b) 13751 hrcV mutant (type III‐deficient strain, negative control). (c) Complemented strain GXC2790. (d) Wild‐type strain 13751.
rsmAXoo positively regulates DSF biosynthesis
The restored endoglucanase activity of Xcc rpfF mutant strain 8523 in the presence of DSF extracted from Xoo strains is positively correlated with the level of DSF produced by Xoo strains (Feng et al., 2009). The restored endoglucanase activity of strain 8523, added to DSF extracted from various Xoo strains, was assayed using strain 8523 as the control. DSF extracted from the rsmAXoo mutant strain GXN2790 resulted in significantly less endoglucanase production than DSF extracted from the wild‐type strain 13751 and the complemented strain GXC2790 (P < 0.01, t‐test) (Fig. 3); however, the latter led to a comparable level of endoglucanase production in mutant 8523 (P > 0.05, t‐test) (Fig. 3). These results indicate that rsmAXoo positively regulates DSF production in Xoo.
Figure 3.
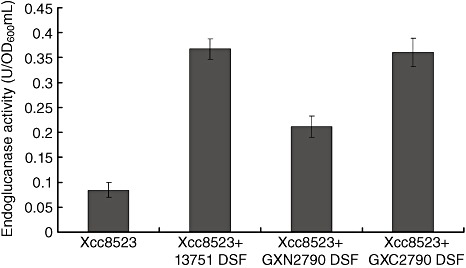
rsmAXoo regulates diffusible signal factor (DSF) biosynthesis in Xanthomonas oryzae pv. oryzae (Xoo). DSF extracted from culture supernatants of different strains of Xoo was bioassayed by measuring the restoration of endoglucanase activity in the X. campestris pv. campestris (Xcc) rpfF mutant 8523. Three replicates were maintained for each experiment and each experiment was repeated three times. Endoglucanase activity is expressed as units per OD600 unit per millilitre. Data shown are the means ± standard deviation for one experiment; similar results were obtained in all experiments.
rsmAXoo regulates EPS production positively and glycogen accumulation negatively
EPS is an important factor in the virulence of Xoo (Büttner and Bonas, 2010; Dharmapuri et al., 2001; Tang et al., 1996). To determine whether a mutation in rsmAXoo had any effect on EPS production, the EPS yields of the three Xoo strains were measured, and were found to be significantly less in the rsmAXoo mutant strain GXN2790 (P < 0.01, t‐test) and comparable in the complemented strain GXC2790 and the wild‐type (P > 0.05, t‐test) (Table 2). These data indicate that rsmAXoo regulates EPS production positively in Xoo.
Table 2.
Extracellular polysaccharide, intracellular glycogen, extracellular endoglucanase and amylase levels produced by Xanthomonas oryzae pv. oryzae strains. *
| Strain | Extracellular polysaccharide (g/L) | Intracellular glycogen (g/L) | Extracellular endoglucanase (U/OD600/mL) | Extracellular amylase (U/OD600/mL) |
|---|---|---|---|---|
| 13751( wild‐type) | 15.1 ± 1.56 | 2.9 ± 0.45 | 0.38 ± 0.02 | 0.19 ± 0.01 |
| GXN2790 | 9.6 ± 1.04** | 4.7 ± 0.49** | 0.16 ± 0.03** | 0.08 ± 0.01** |
| GXC2790 | 14.1 ± 1.45 | 2.6 ± 0.27 | 0.36 ± 0.04 | 0.17 ± 0.01 |
Data are the means ± standard deviation from three replications. Statistically significant differences between the mutant GXN2790 and the wild‐type strain 13751 are indicated by **(P < 0.01). The experiment was repeated three times, and similar results were obtained each time.
It is noteworthy that the rsmAXoo mutant strain GXN2790 was stained dark brown on OB agar plates because of excess accumulation of glycogen, whereas the other two strains turned yellowish‐brown when the production of intracellular polysaccharide glycogen was assessed by iodine vapour staining (Chao et al., 2008) (data not shown). The amount of glycogen in Xoo cells was measured by the method described by Feng et al. (2009). Notably, the complemented strain GXC2790 produced about the same amount of glycogen as the wild‐type (P > 0.05, t‐test), whereas the rsmAXoo mutant strain GXN2790 produced almost twice as much (Table 2). The results indicate that rsmAXoo negatively regulates intracellular glycogen accumulation in Xoo.
rsmAXoo is involved in the production of extracellular endoglucanase and amylase
Extracellular enzymes, such as xylanase, endoglucanase, amylase and protease, contribute individually or collectively to the virulence of Xanthomonas (Chatterjee et al., 2003; Hu et al., 2007; Rajeshwari et al., 2005; Ray et al., 2000). To verify whether the virulence‐impaired rsmAXoo mutant exhibits any defects in the production of these enzymes that contribute to virulence, we measured the activity of these extracellular enzymes in the rsmAXoo mutant GXN2790. The mutant displayed about as much xylanase and protease activity as the wild‐type strain 13751 (data not shown), but significantly less endoglucanase and amylase activity (P < 0.01, t‐test) (Table 2). In contrast, endoglucanase and amylase activities in the complemented strain GXC2790 and in the wild‐type were comparable (P > 0.05, t‐test). These findings indicate that, in Xoo, rsmAXoo is involved in the production of extracellular endoglucanase and amylase, but not of xylanase and protease.
rsmAXoo represses bacterial aggregation and cell adhesion
To determine whether RsmAXoo is involved in biofilm formation, we observed the formation of cell aggregates in a liquid medium within a glass bottle and measured the quantity of cells that adhered to the glass surface indirectly, that is by staining. The wild‐type strain 13751 and the complemented strain GXC2790 did not form aggregates in Luria–Bertani medium (Dow et al., 2003), whereas the mutant strain GXN2790 did, and formed readily visible clumps in the liquid medium (Fig. 4a), suggesting that it can form a biofilm. The extent of cell adhesion on the glass surface was measured by staining with crystal violet (Jackson et al., 2002), and was found to be significantly greater in rsmAXoo mutant cells than in the wild‐type and the complemented strain (P < 0.01, t‐test) (Fig. 4b). These data suggest that rsmAXoo may suppress Xoo biofilm formation.
Figure 4.
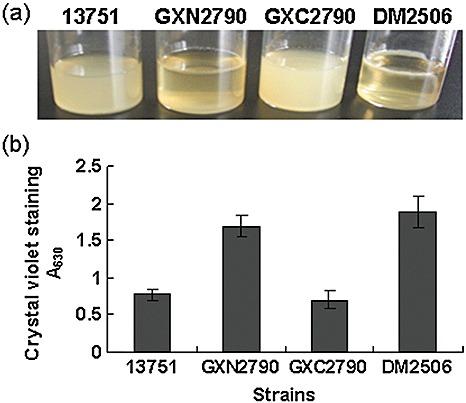
rsmAXoo represses cell aggregation and cell adhesion. (a) In the cell aggregate assay, the rsmAXoo mutant GXN2790 and the rsmAXcc mutant DM2506 (positive control) grew as aggregates on the surface of Luria–Bertani medium, whereas the wild‐type strain 13751 and the complemented strain GXC2790 grew as dispersed cells. The results presented are from a representative experiment; similar results were obtained in the other two independent experiments. (b) Cells adhering to the glass surface. DM2506, the rsmAXcc mutant of Xanthomonas campestris pv. campestris (Xcc), was included as the control. Three replicates were maintained for each experiment and each experiment was repeated three times. The data shown are the means ± standard deviation of the replicates.
rsmAXoo positively regulates the expression of genes associated with pathogenicity and extracellular enzymes
To determine the molecular basis of biological changes in the phenotype caused by mutations in rsmAXoo, we measured the mRNA level of selected genes associated with pathogenicity and extracellular enzymes in the wild‐type, rsmAXoo mutant GXN2790 and the complemented strain GXC2790 by semiquantitative reverse transcription‐polymerase chain reaction (RT‐PCR). Unfortunately, we could not design primers to specifically test the expression of each gene encoding individual members of the AvrBs3/PthA family by RT‐PCR. However, we did measure the total transcripts of the AvrBs3/PthA family in Xoo strains, and found them to be down‐regulated in the rsmAXoo mutant when compared with the wild‐type strain (Figs 5 and S2, see Supporting Information). In contrast, the total transcripts of the AvrBs3/PthA family in the complemented strain GXC2790 were similar to those in the wild‐type strain (Figs 5 and S2).
Figure 5.
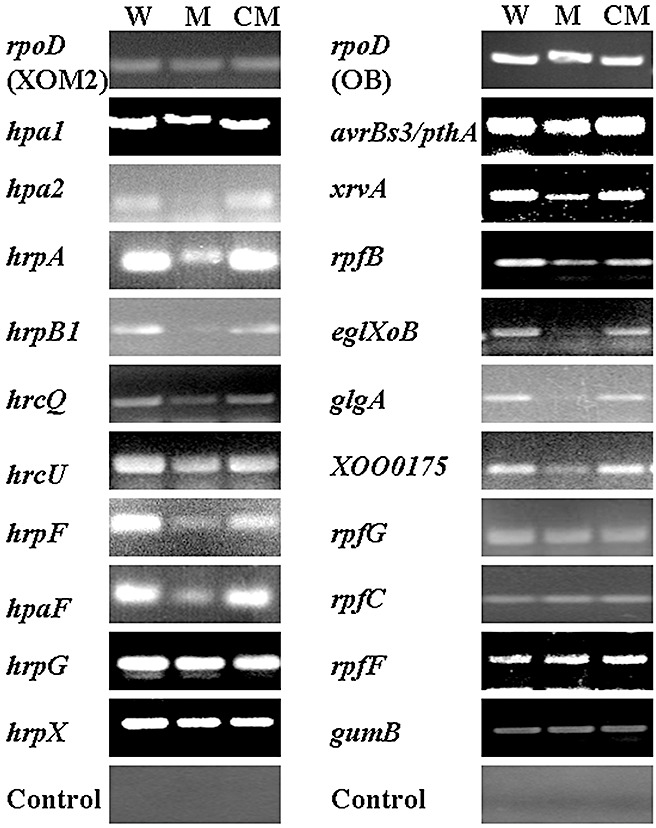
Expression level of genes associated with virulence, type III secretion system (T3SS), extracellular enzymes and diffusible signal factor (DSF) in Xanthomonas oryzae pv. oryzae strains. The expression of rpoD, a housekeeping gene encoding the σ factor of RNA polymerase, in each strain was included as the control. The experiment was repeated four times. The results presented are from one experiment; similar results were obtained in all other independent experiments. The results of the other two independent experiments are presented in Fig. S2. Control means a negative control (without addition of reverse transcriptase). XOM2 and OB represent XOM2 and OB media for growing the bacteria for RNA extraction, respectively. WT, wild‐type strain 13751; M, rsmAXoo mutant strain GXN2790; CM, complemented strain GXC2790.
The levels of expression of hrpB1, hrcU and hrcQ (the first genes of three hrp operons in the hrp gene cluster), and of hpa1, hpa2, hrpA, hrpF and hpaF, in the complemented strain GXC2790 were similar to those in the wild‐type strain, but were down‐regulated in the rsmAXoo mutant GXN2790 (Figs 5 and S2). This difference indicates that rsmAXoo positively regulates the expression of most hrp genes. However, the levels of expression of hrpE1, the first gene of the hrpE operon (data not shown), and of hrpG and hrpX, were about the same in all three strains (Figs 5 and S2). The expression of the xrvA gene was down‐regulated in the rsmAXoo mutant, but did not differ between the wild‐type and the complemented strain. Yet, the expression level of rpfB, which is involved in DSF synthesis, was found to be down‐regulated in the rsmAXoo mutant GXN2790 (Figs 5 and S2).
The expression levels of the cellulase gene eglXoB, the glycogen biosynthesis gene glgA and XOO0175, the homologue of the Xacα‐amylase gene XAC0154, were down‐regulated in the rsmAXoo mutant. However, the expression of these genes did not differ between the wild‐type and the complemented strain. Furthermore, we found no differences in the levels of expression of gumB, rpfC, rpfF and rpfG in GXN2790 compared with the wild‐type and the complemented strain (Figs 5 and S2).
DISCUSSION
In this study, inactivation of the rsmAXoo gene in Xoo resulted in an almost complete loss of virulence on rice and in the failure to elicit an HR in a nonhost, namely castor‐oil plant, indicating that rsmAXoo is essential for the virulence of Xoo and for its ability to elicit an HR. The altered virulence can be partly explained by the reduced expression of genes associated with pathogenicity and the production of virulence factors, such as extracellular enzymes, observed in the rsmAXoo mutant. Although the phenotypes and expression of some selected genes observed in the rsmAXoo mutant were similar to those observed for Xcc (Chao et al., 2008), the rsmAXoo gene may regulate the virulence of Xoo through a mechanism or pathway different from that in rsmAxcc in Xcc, because members of the AvrBs3/PthA transcriptional activation‐like (TAL) effector family in Xoo play an important role in the pathogenicity of the organism to rice, whereas their presence in Xcc has not been confirmed (Büttner and Bonas, 2010; White and Yang, 2009; White et al., 2009). In the present study, we found that the total transcripts of the AvrBs3/PthA family in the rsmAXoo mutant were less than those in the wild‐type strain, indicating that the expression of certain gene(s) encoding members of the AvrBs3/PthA family was reduced in the mutant, although we could not identify the affected gene(s). A proteomic approach is needed to determine the effect of mutations in rsmAXoo on the expression of proteins, particularly those of the AvrBs3/PthA family, in Xoo.
What is particularly noteworthy is that rsmAXoo positively regulates DSF production in Xoo. It has been reported previously that DSF promotes EPS production and xylanase activity in Xoo (He et al., 2010). Although we did not observe such altered production of extracellular xylanase by the rsmAXoo mutant in our experiments, we did observe that EPS production in the rsmAXoo mutant of Xoo was reduced, whereas the expression of genes such as gumB, rpfC, rpfG and rpfF, which are known to be involved in EPS production, was unaffected. Although the decreased production of EPS may be the result of reduced production of DSF, the precise mechanism by which rsmAXoo regulates EPS production remains unknown. Furthermore, the expression of rpfB, which is involved in DSF synthesis (Barber et al., 1997), was down‐regulated in the rsmAXoo mutant, whereas the expression of rpfF, a key gene required for DSF biosynthesis (Barber et al., 1997; He et al., 2010), was unaffected. The rpfB and rpfF genes are co‐transcribed from a promoter upstream of rpfB, but rpfF also has its own promoter (Slater et al., 2000). The transcriptional level of rpfF in the rsmAXoo mutant may come partly from the transcription of rpfF from its own promoter, which may not be regulated by rsmAXoo. It is also possible that the expression of rpfF is regulated post‐transcriptionally by RsmAXoo.
The expression of xrvA, which encodes a virulence regulator (Feng et al., 2009), the cellulase gene eglXoB and XOO0175, which is a homologue of the Xacα‐amylase gene XAC0154, was down‐regulated in the rsmAXoo mutant. The xrvA mutant of Xoo showed significantly reduced virulence to the host and delayed induction of HR in a nonhost (Feng et al., 2009). The eglXoB gene is involved in the virulence of Xoo (Hu et al., 2007), but the role of XOO0175 is unclear. Screening the transposon‐based mutant library of X. citri ssp. citri revealed that a mutation of XAC0798, which encodes an α‐amylase, affected virulence and induced an enhanced hypersensitive‐like response (Laia et al., 2009).
The expression of glgA, a homologue of which is involved in glycogen biosynthesis in E. coli (Yang et al., 1996), was reduced in the rsmAXoo mutant despite the greater accumulation of glycogen. The expression of glgB, which encodes a 1,4‐α‐glucan branching enzyme, is reduced in the rsmAXcc mutant of Xcc (Chao et al., 2008). In E. coli, the glgCAP operon is involved in glycogen biosynthesis, and CsrA has been shown to directly suppress the expression of this operon (Baker et al., 2002). However, our sequence analysis did not identify any homologue of glgC or glgP in the sequenced genome of Xoo strains. Therefore, the mechanism by which rsmAXoo affects the expression of glgA and glycogen accumulation in Xoo remains unclear.
In bacteria, the RsmA/CsrA proteins regulate translation by binding specifically to conserved sequences at or near the Shine–Dalgarno sequence of target mRNAs, thus affecting mRNA translation and/or stability. The effect of RsmA/CsrA is relieved by small RNAs (sRNAs) that sequester and antagonize these proteins (Lapouge et al., 2008). To better understand the function and regulatory network of RsmAXoo in Xoo, it is important to identify the RsmAXoo‐binding sRNAs. Surprisingly, we found no sequence homologous to the sRNA‐encoding sequences in the sequenced genome of Xoo strains. Nevertheless, the detailed mechanism of RsmAXoo action in Xoo merits further studies.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. The strains of E. coli were cultivated in Luria–Bertani medium (Miller, 1972) at 37°C. Xoo strains were grown in OB medium (Tang et al., 1996) or XOM2 medium (Tsuge et al., 2002) at 28°C. The Xcc rpfF mutant strain 8523 was inoculated into peptone–yeast extract–glycerol broth (NYGB) (Daniels et al., 1984). Antibiotics were used as described previously (Feng et al., 2009).
DNA manipulation
DNA manipulation was performed following the procedures described by Sambrook and Russell (2001). Conjugation between the Xoo and E. coli strains was carried out as described by Turner et al. (1985). Restriction enzymes and DNA ligase were used in accordance with the manufacturer's instructions (Promega, Shanghai, China).
Mutant construction
The nonpolar mutant of rsmAXoo in the Xoo strain 13751 (Tang et al., 1996) was generated by homologous integration with a suicide plasmid using pKMob18GII as a vector (Katzen et al., 1999), as described previously (Windgassen et al., 2000). A 131‐bp internal fragment of the rsmAXoo gene (from 7 bp downstream of the start codon to 137 bp) was amplified using total DNA of strain 13751 as the template and the primer set rsmAXooIF/rsmAXooIR (Table S1, see Supporting Information). The amplified DNA fragment was ligated to the plasmid vector pKMob18GII to create the recombinant plasmid pKrsmAXoo (Table 1), and confirmed by sequencing. The plasmid pKrsmAXoo was transferred into strain 13751 by triparental conjugation using pRK2073 (Leong et al., 1982) as the helper plasmid. With a mutation in the rsmAXoo gene, transconjugants were confirmed by PCR using total DNA as the template and the primer pair pKMob18GIIF/rsmAXooYR (Table S1). Total DNA from the wild‐type strain as a template was used as a negative control. The expected 393‐bp PCR product was further confirmed by sequencing. One of the confirmed mutants, designated as GXN2790 (Table 1), was used for further study.
Complementation of the rsmAXoo mutant
A 703‐bp DNA fragment containing the entire rsmAXoo gene (from 447 bp upstream of the start codon to 43 bp downstream of the stop codon) was amplified by PCR using total DNA from the wild‐type strain 13751 as the template and the primer set rsmAXooCF/rsmAXooCR (Table S1). The PCR product was then sequenced. The sequence of the 703‐bp DNA fragment was identical to the corresponding DNA sequence containing rsmAXoo in the sequenced genomes of Xoo strains MAFF311018, KACC10331 and PXO99A (accession numbers AP008229, AE013598 and CP000967, respectively). The amplified DNA fragment was then cloned into pLAFR6 to generate the recombinant plasmid, named pL6rsmAXoo (Table 1), which was then transferred into the mutant GXN2790 by triparental conjugation (Turner et al., 1985). The confirmed transconjugant was designated as GXC2790 (Table 1) and chosen for further study.
RT‐PCR
Total RNA was extracted from wild‐type Xoo 13751, mutant GXN2790 and the complemented strain GXC2790, grown either in liquid medium OB or in hrp‐inducing medium XOM2, as described by Feng et al. (2009). An equal amount of RNA from each strain was converted to cDNA by RT‐PCR (Takara Biotechnology Co. Ltd., Dalian, China). The cDNAs were used as templates for amplification by PCR for 30 cycles with Ex‐Taq polymerase (Takara Biotechnology Co. Ltd.). The primers used are listed in Table S1. PCRs using total DNA as template and total RNA without reverse transcriptase were used as positive and negative controls, respectively. The internal fragment of the 16S rRNA gene or rpoD, a housekeeping gene encoding the σ factor of RNA polymerase, was amplified simultaneously as an expression level control. The resulting amplified products were analysed on 1.2% agarose gel.
Plant assays
The virulence of Xoo strains was tested on hybrid rice cultivar Teyou 63, grown in a glasshouse with 12 h of light alternating with 12 h of darkness at temperatures of 28°C during the day and 25°C at night, using the leaf‐clipping method (Kauffman et al., 1973). Bacterial cells were grown in OB medium at 28°C with shaking at 200 r.p.m. for 20 h (until the exponential growth phase was reached). The cell concentration was adjusted to an optical density of 0.001 (1 × 106 cfu/mL) or 0.1 (1 × 108 cfu/mL) at 600 nm. Thirty leaves were inoculated with each Xoo strain in each treatment and maintained at 100% humidity for 24 h following the inoculation, after which the plants were maintained under the growth conditions described above. The lesion length was measured 14 days after inoculation. Each treatment was repeated three times in each experiment. The same experiment was repeated three times.
HR was tested on a nonhost, namely castor‐oil plant (R. communis), as described previously (Feng et al., 2009). The plants were inoculated by infiltrating approximately 20 µL of the bacterial suspension (5 × 107 cfu/mL or 5 × 108 cfu/mL) in 10 mm sodium phosphate buffer into the leaves using a 1‐mL blunt‐end plastic syringe. The inoculated plants were maintained in a glasshouse with a 12 h day–night cycle of illumination using a fluorescent lamp at a constant temperature of 28°C. Plants were observed at 12, 24, 36 and 48 h after inoculation. The HR symptoms were photographed 48 h after inoculation. At least four plants were inoculated in each experiment, and each experiment was repeated three times.
Growth of bacteria in media and in planta
The growth of Xoo strains was measured in OB and XOM2 media. Bacterial cells were first grown in OB medium at 28°C with shaking at 200 r.p.m. for 20 h, that is until the exponential growth phase was reached. The cell concentration was adjusted to an optical density of 1.0 (1 × 109 cfu/mL) at 600 nm. The cell suspension (2 mL) was transferred to 100 mL of OB or XOM2 medium containing appropriate antibiotics. Three replications were maintained for each strain. The optical density was measured at 600 nm at 2‐ and 4‐h intervals for cultures in OB and XOM2 media, respectively.
Bacterial growth was measured in the leaf tissue of Teyou 63 inoculated with the Xoo strains (1 × 108 cfu/mL; OD600 = 0.1) by the leaf‐clipping method (Kauffman et al., 1973). The growth was measured at 2‐day intervals up to 16 days after inoculation. Leaf samples (three leaves to a sample) were homogenized in 5 mL of sterile water. The diluted homogenate was plated onto OB agar supplemented with streptomycin (for the wild‐type), streptomycin plus kanamycin (for the mutant) or streptomycin plus kanamycin and tetracycline (for the complemented strain). The number of colonies on each plate was counted after incubation at 28°C for 3 days. The bacterial cfu was calculated for each rice leaf. The procedure was repeated three times for each sample. The data shown are the average of three replications with standard deviation.
Assessment of extracellular enzyme activity, EPS production, DSF biosynthesis and glycogen accumulation
The extracellular activity of endoglucanase, xylanase and amylase in Xoo strains was measured essentially as described by Feng et al. (2009). Briefly, Xoo strains were cultured in OB medium for 24 h for the measurement of endoglucanase and amylase activity, and for 48 h for the measurement of xylanase activity. The culture supernatant (100 µL) was added to 400 µL of the appropriate buffer containing 1% (w/v) carboxymethylcellulose (CMC, Sigma, St. Louis, MO, USA) for endoglucanase, 1% (w/v) birchwood xylan (Sigma) for xylanase or 1% (w/v) soluble starch (Sinopharm Chemical Reagent Co. Ltd., Shangshai, China) for amylase. The reactions were carried out for 30 min at 28°C. The released reducing sugars were measured as d‐glucose or d‐xylose equivalents, as described by Miller (1959). One unit (U) of endoglucanase/xylanase/amylase activity was defined as the amount of enzyme releasing 1 µmol of reducing sugar per minute.
To measure EPS production, Xoo strains were cultured in conical flasks containing 100 mL of OB medium supplemented with 2% (w/v) glucose instead of sucrose at 28°C for 4 days, with shaking at 200 r.p.m. EPS was precipitated from the culture supernatant by the addition of three volumes of 95% ethanol. The pelleted EPS was washed with 70% ethanol, dried at 60°C and weighed. Three flasks were inoculated for each strain in each experiment, and each experiment was repeated three times.
DSF was extracted into ethyl acetate (Barber et al., 1997) from culture supernatants of Xoo strains grown in OB medium for 48 h. The ethyl acetate extracts were evaporated to dryness and the samples were resuspended in methanol. The Xcc rpfF mutant strain 8523 (Table 1) was inoculated into NYGB supplemented with the DSF extracts, and cultured for 24 h. The restored extracellular endoglucanase activity produced by mutant strain 8523 was measured as described above for endoglucanase activity.
The intracellular accumulation of glycogen was initially determined by the method described by Chao et al. (2008), and then measured as described by Feng et al. (2009).
Cell aggregation and adhesion assay
Bacterial cell aggregates that formed on the surface of the medium or adhered to the glass surface were surveyed using the method described by Rhim (1983). The extent of adhesion was measured using the methods described by Jackson et al. (2002) and Chao et al. (2008).
Nucleotide sequence accession number
The rsmAXoo sequence of Xoo strain 13751 was deposited into the GenBank database (accession number HM988729).
Supporting information
Fig. S1 Genetic organization of rsmA/csrA loci in the genomes of Xanthomonas oryzae pv. oryzae, X. campestris pv. campestris, Escherichia coli, X. axonopodis pv. citri and X. campestris pv. vesicatoria. The DNA sequence containing genes alaS, rsmAXoo and tRNA‐Ser in X. oryzae pv. oryzae (Xoo) strain 13751 was obtained by sequencing the polymerase chain reaction (PCR) products amplified using total DNA from strain 13751 as the template and the primers (data not shown) designed on the basis of the sequence from the genome of Xoo strain MAFF311018 (accession number AP008229). This DNA sequence was identical to the corresponding sequence in the genome of Xoo strain MAFF311018. The DNA sequence containing rsmA/csrA loci in the other bacteria is from the sequenced genomes of X. campestris pv. campestris strain 8004 (accession number CP000050), E. coli strain K12 substrain MG1655 (accession number U00096), X. axonopodis pv. citri strain 306 (accession number AE008923) and X. campestris pv. vesicatoria strain 85‐10 (accession number AM039952). The genes alaS and tRNA‐Ser encode alanyl‐tRNA synthetase and tRNA‐Ser, respectively. Horizontal black bars indicate the sequences used in plasmid construction.
Fig. S2 Expression level of genes associated with virulence, the type III secretion system (T3SS), extracellular enzymes and diffusible signal factor (DSF) in Xanthomonas oryzae pv. oryzae strains detected in independent experiments. (a) One independent experiment carried out by one individual. The expression of 16S rRNA in each strain was included as the control. The negative control contained no reverse transcriptase, and no bands appeared (results not shown). XOM2 and OB represent XOM2 medium and OB medium for growing the bacteria for RNA extraction, respectively. WT, wild‐type strain 13751; M, rsmAXoo mutant strain GXN2790; CM, complemented strain GXC2790. (b) Another independent experiment carried out by another individual. The expression of rpoD, a housekeeping gene encoding the σ factor of RNA polymerase, in each strain was included as the control. Control means a negative control (without the addition of reverse transcriptase). XOM2 and OB represent XOM2 medium and OB medium for growing the bacteria for RNA extraction, respectively. WT, wild‐type strain 13751; M, rsmAXoo mutant strain GXN2790; CM, complemented strain GXC2790.
Table S1 Primers used in this study.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This work was supported by the 973 Program of the Ministry of Science and Technology of China (2006CB101902).
REFERENCES
- Aparna, G. , Chatterjee, A. , Sonti, R.V. and Sankaranarayanan, R. (2009) A cell wall‐degrading esterase of Xanthomonas oryzae requires a unique substrate recognition module for pathogenesis on rice. Plant Cell, 21, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.S. , Morozov, I. , Suzuki, K. , Romeo, T. and Babitzke, P. (2002) CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli . Mol. Microbiol. 44, 1599–1610. [DOI] [PubMed] [Google Scholar]
- Barber, C.E. , Tang, J.L. , Feng, J.X. , Pan, M.Q. , Wilson, T.J. , Slater, H. , Dow, J.M. , Williams, P. and Daniels, M.J. (1997) A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and He, S.Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, N.X. , Wei, K. , Chen, Q. , Meng, Q.L. , Tang, D.J. , He, Y.Q. , Lu, G.T. , Jiang, B.L. , Liang, X.X. , Feng, J.X. , Chen, B.S. and Tang, J.L. (2008) The rsmA‐like gene rsmAXcc of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Mol. Plant–Microbe Interact. 21, 411–423. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. and Sonti, R.V. (2002) RpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant–Microbe Interact. 15, 463–471. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. , Sankaranarayanan, R. and Sonti, R.V. (2003) PhyA, a secreted protein of Xanthomonas oryzae pv. oryzae, is required for optimum virulence and growth on phytic acid as a sole phosphate source. Mol. Plant–Microbe Interact. 16, 973–982. [DOI] [PubMed] [Google Scholar]
- Daniels, M.J. , Barber, C.E. , Turner, P.C. , Sawczyc, M.K. , Byrde, R.J.W. and Fielding, A.H. (1984) Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 3, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmapuri, S. and Sonti, R.V. (1999) A transposon insertion in the gumG homologue of Xanthomonas oryzae pv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol. Lett. 179, 53–59. [DOI] [PubMed] [Google Scholar]
- Dharmapuri, S. , Yashitola, J. , Vishnupriya, M.R. and Sonti, R.V. (2001) Novel genomic locus with atypical G + C content that is required for extracellular polysaccharide production and virulence in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 14, 1335–1339. [DOI] [PubMed] [Google Scholar]
- Dow, M. (2008) Diversification of the function of cell‐to‐cell signaling in regulation of virulence within plant pathogenic Xanthomonas . Sci. Signal. 1, pe23. [DOI] [PubMed] [Google Scholar]
- Dow, J.M. , Crossman, L. , Findlay, K. , He, Y.Q. , Feng, J.X. and Tang, J.L. (2003) Biofilm dispersal in Xanthomonas campestris is controlled by cell–cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA, 100, 10 995–11 000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J.X. , Li, Y.R. , Duan, C.J. and Tang, J.L. (2001) Cloning and characterization of hrp gene from Xanthomonas oryzae pv. oryzae In: Plant Diseases and Their Control (Zeng S.M., Zhou G.H. and Li H.F., eds), pp. 26–27. Beijing: China Agricultural Science and Technology Press. [Google Scholar]
- Feng, J.X. , Song, Z.Z. , Duan, C.J. , Zhao, S. , Wu, Y.Q. , Wang, C. , Dow, J.M. and Tang, J.L. (2009) The xrvA gene of Xanthomonas oryzae pv. oryzae, encoding an H‐NS‐like protein, regulates virulence in rice. Microbiology, 155, 3033–3044. [DOI] [PubMed] [Google Scholar]
- Fields, J.A. and Thompson, S.A. (2008) Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190, 3411–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy, Y. , Lucey, J.F. , Ryan, R.P. and Dow, J.M. (2006) Cell–cell signaling, cyclic di‐GMP turnover and regulation of virulence in Xanthomonas campestris . Res. Microbiol. 157, 899–904. [DOI] [PubMed] [Google Scholar]
- Gough, C.L. , Dow, J.M. , Barber, C.E. and Daniels, M.J. (1988) Cloning of two endoglucanase genes of Xanthomonas campestris pv. campestris: analysis of the role of the endoglucanase in pathogenesis. Mol. Plant–Microbe Interact. 1, 275–281. [Google Scholar]
- Grant, S.R. , Fisher, E.J. , Chang, J.H. , Mole, B.M. and Dangl, J.L. (2006) Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449. [DOI] [PubMed] [Google Scholar]
- He, Y.W. and Zhang, L.H. (2008) Quorum sensing and virulence regulation in Xanthomonas campestris . FEMS Microbiol. Rev. 32, 842–857. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Xu, M. , Lin, K. , Ng, Y.J. , Wen, C.M. , Wang, L.H. , Liu, Z.D. , Zhang, H.B. , Dong, Y.H. , Dow, J.M. and Zhang, L.H. (2006) Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell–cell communication‐dependent genes and functions. Mol. Microbiol. 59, 610–622. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Wu, J. , Cha, J.S. and Zhang, L.H. (2010) Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF‐family signals in regulation of virulence factor production. BMC Microbiol. 10, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , Qian, W. and He, C. (2007) The Xanthomonas oryzae pv. oryzae eglXoB endoglucanase gene is required for virulence to rice. FEMS Microbiol. Lett. 269, 273–279. [DOI] [PubMed] [Google Scholar]
- Huynh, T.V. , Dahlbeck, D. and Staskawicz, B.J. (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science, 245, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Jackson, D.W. , Suzuki, K. , Oakford, L. , Simecka, J.W. , Hart, M.E. and Romeo, T. (2002) Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli . J. Bacteriol. 184, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, G. , Rajeshwari, R. and Sonti, R.V. (2007) Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant–Microbe Interact. 20, 31–40. [DOI] [PubMed] [Google Scholar]
- Kamdar, H.V. , Kamoun, S. and Kado, C.I. (1993) Restoration of pathogenicity of avirulent Xanthomonas oryzae pv. oryzae and X. campestris pathovars by reciprocal complementation with the hrpXo and hrpXc genes and identification of HrpX function by sequence analyses. J. Bacteriol. 175, 2017–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen, F. , Becker, A. , Ielmini, M.V. , Oddo, C.G. and Ielpi, L. (1999) New mobilizable vectors suitable for gene replacement in gram‐negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl. Environ. Microbiol. 65, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, H.E. , Reddy, A.P.K. , Hsieh, S.P.Y. and Merca, S.D. (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae . Plant Dis. Rep. 57, 537–541. [Google Scholar]
- Kim, S.Y. , Kim, J.G. , Lee, B.M. and Cho, J.Y. (2009) Mutational analysis of the gum gene cluster required for xanthan biosynthesis in Xanthomonas oryzae pv. oryzae . Biotechnol. Lett. 31, 265–270. [DOI] [PubMed] [Google Scholar]
- Koebnik, R. , Krüger, A. , Thieme, F. , Urban, A. and Bonas, U. (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC‐type transcriptional activator HrpX to plant‐inducible promoter boxes. J. Bacteriol. 188, 7652–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laia, M.L. , Moreira, L.M. , Dezajacomo, J. , Brigati, J.B. , Ferreira, C.B. , Ferro, M.I.F. , Silva, A.C.R. , Ferro, J.A. and Oliveira, J.C.F. (2009) New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon‐based mutant library. BMC Microbiol. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge, K. , Schubert, M. , Allain, F.H. and Haas, D. (2008) Gac/Rsm signal transduction pathway of gamma‐proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67, 241–253. [DOI] [PubMed] [Google Scholar]
- Lee, B.M. , Park, Y.J. , Park, D.S. , Kang, H.W. , Kim, J.G. , Song, E.S. , Park, I.C. , Yoon, U.H. , Hahn, J.H. , Koo, B.S. , Lee, G.B. , Kim, H. , Park, H.S. , Yoon, K.O. , Kim, J.H. , Jung, C.H. , Koh, N.H. , Seo, J.S. and Go, S.J. (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.W. , Han, S.W. , Sririyanum, M. , Park, C.J. , Seo, Y.S. and Ronald, P.C. (2009) A type I‐secreted, sulfated peptide triggers XA21‐mediated innate immunity. Science, 326, 850–853. [DOI] [PubMed] [Google Scholar]
- Leong, S.A. , Ditta, G.S. and Helinski, D.R. (1982) Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta‐aminolevulinic acid synthetase from Rhizobium meliloti . J. Biol. Chem. 257, 8724–8730. [PubMed] [Google Scholar]
- Lucchetti‐Miganeh, C. , Burrowes, E. , Baysse, C. and Ermel, G. (2008) The post‐transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology, 154, 16–29. [DOI] [PubMed] [Google Scholar]
- Mansfield, J.W. (2009) From bacterial avirulence genes to effector functions via the hrp delivery system: an overview of 25 years of progress in our understanding of plant innate immunity. Mol. Plant Pathol. 10, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mew, T.W. , Alvarez, A.M. , Leach, J.E. and Swings, J. (1993) Focus on bacterial blight of rice. Plant Dis. 77, 5–12. [Google Scholar]
- Miller, G.L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. [Google Scholar]
- Miller, J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Mole, B.M. , Baltrus, D.A. , Dangl, J.L. and Grant, S.R. (2007) Global virulence regulation networks on phytopathogenic bacteria. Trends Microbiol. 15, 363–371. [DOI] [PubMed] [Google Scholar]
- Nino‐Liu, D.O. , Ronald, P.C. and Bogdanove, A.J. (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. [DOI] [PubMed] [Google Scholar]
- Ochiai, H. , Inoue, Y. , Takeya, M. , Sasaki, A. and Kaku, H. (2005) Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39, 275–287. [Google Scholar]
- Rajeshwari, R. and Sonti, R.V. (2000) Stationary‐phase variation due to transposition of novel insertion elements in Xanthomonas oryzae pv. oryzae . J. Bacteriol. 182, 4797–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeshwari, R. , Jha, G. and Sonti, R.V. (2005) Role of an in planta‐expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant–Microbe Interact. 18, 830–837. [DOI] [PubMed] [Google Scholar]
- Ray, S.K. , Rajeshwari, R. and Sonti, R.V. (2000) Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant–Microbe Interact. 13, 394–401. [DOI] [PubMed] [Google Scholar]
- Rhim, J.S. (1983) Cell aggregation assay: a rapid means of evaluating and selecting in vitro transformed cells. Cancer Detect. Prev. 6, 381–388. [PubMed] [Google Scholar]
- Romeo, T. (1998) Global regulation by the small RNA‐binding protein CsrA and the non‐coding RNA molecule CsrB. Mol. Microbiol. 29, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Won, L.S. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics, 9, 204–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D. (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schröter, K. , Flaschel, E. , Pühler, A. and Becker, A. (2001) Xanthomonas campestris pv. campestris secretes the endoglucanases ENGXCA and ENGXCB: construction of an endoglucanase deficient mutant for industrial xanthan production. Appl. Microbiol. Biotechnol. 55, 727–733. [DOI] [PubMed] [Google Scholar]
- Slater, H. , Alvarez‐Morales, A. , Barber, C.E. , Daniels, M.J. and Dow, J.M. (2000) A two‐component system involving an HD‐GYP domain protein links cell–cell signalling to pathogenicity gene expression in Xanthomonas campestris . Mol. Microbiol. 38, 986–1003. [DOI] [PubMed] [Google Scholar]
- Tang, J.L. , Liu, Y.N. , Barber, C.E. , Dow, J.M. , Wootton, J.C. and Daniels, M.J. (1991) Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris . Mol. Gen. Genet. 226, 409–417. [DOI] [PubMed] [Google Scholar]
- Tang, J.L. , Feng, J.X. , Li, Q.Q. , Wen, H.X. , Zhou, D.L. , Wilson, T.J. , Dow, J.M. , Ma, Q.S. and Daniels, M.J. (1996) Cloning and characterization of the rpfC gene of Xanthomonas oryzae pv. oryzae: involvement in exopolysaccharide production and virulence to rice. Mol. Plant–Microbe Interact. 9, 664–666. [DOI] [PubMed] [Google Scholar]
- Tsuge, S. , Furutani, A. , Fukunaka, R. , Oku, T. , Tsuno, K. , Ochiai, H. , Inoue, Y. , Kaku, H. and Kubo, Y. (2002) Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen. Plant Pathol. 68, 363–371. [Google Scholar]
- Turner, P. , Barber, C.E. and Daniels, M.J. (1985) Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris . Mol. Gen. Genet. 199, 338–343. [Google Scholar]
- Wang, L.H. , He, Y.W. , Gao, Y.F. , Wu, J.E. , Dong, Y.H. , He, C.Z. , Wang, S.Y. , Weng, L.X. , Xu, J.L. , Tay, L. , Fang, R.X. and Zhang, L.H. (2004) A bacterial cell–cell communication signal with cross‐kingdom structural analogues. Mol. Microbiol. 51, 903–912. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Van den Ackerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- White, F.F. and Yang, B. (2009) Host and pathogen factors controlling the rice–Xanthomonas oryzae interaction. Plant Physiol. 150, 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F. , Yang, B. and Johnson, L.B. (2000) Prospects for understanding avirulence gene function. Curr. Opin. Plant Biol. 3, 291–298. [DOI] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen, M. , Urban, A. and Jaeger, K.E. (2000) Rapid gene inactivation in Pseudomonas aeruginosa . FEMS Microbiol. Lett. 193, 201–205. [DOI] [PubMed] [Google Scholar]
- Yang, H.H. , Liu, M.Y. and Romeo, T. (1996) Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178, 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Genetic organization of rsmA/csrA loci in the genomes of Xanthomonas oryzae pv. oryzae, X. campestris pv. campestris, Escherichia coli, X. axonopodis pv. citri and X. campestris pv. vesicatoria. The DNA sequence containing genes alaS, rsmAXoo and tRNA‐Ser in X. oryzae pv. oryzae (Xoo) strain 13751 was obtained by sequencing the polymerase chain reaction (PCR) products amplified using total DNA from strain 13751 as the template and the primers (data not shown) designed on the basis of the sequence from the genome of Xoo strain MAFF311018 (accession number AP008229). This DNA sequence was identical to the corresponding sequence in the genome of Xoo strain MAFF311018. The DNA sequence containing rsmA/csrA loci in the other bacteria is from the sequenced genomes of X. campestris pv. campestris strain 8004 (accession number CP000050), E. coli strain K12 substrain MG1655 (accession number U00096), X. axonopodis pv. citri strain 306 (accession number AE008923) and X. campestris pv. vesicatoria strain 85‐10 (accession number AM039952). The genes alaS and tRNA‐Ser encode alanyl‐tRNA synthetase and tRNA‐Ser, respectively. Horizontal black bars indicate the sequences used in plasmid construction.
Fig. S2 Expression level of genes associated with virulence, the type III secretion system (T3SS), extracellular enzymes and diffusible signal factor (DSF) in Xanthomonas oryzae pv. oryzae strains detected in independent experiments. (a) One independent experiment carried out by one individual. The expression of 16S rRNA in each strain was included as the control. The negative control contained no reverse transcriptase, and no bands appeared (results not shown). XOM2 and OB represent XOM2 medium and OB medium for growing the bacteria for RNA extraction, respectively. WT, wild‐type strain 13751; M, rsmAXoo mutant strain GXN2790; CM, complemented strain GXC2790. (b) Another independent experiment carried out by another individual. The expression of rpoD, a housekeeping gene encoding the σ factor of RNA polymerase, in each strain was included as the control. Control means a negative control (without the addition of reverse transcriptase). XOM2 and OB represent XOM2 medium and OB medium for growing the bacteria for RNA extraction, respectively. WT, wild‐type strain 13751; M, rsmAXoo mutant strain GXN2790; CM, complemented strain GXC2790.
Table S1 Primers used in this study.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item


