SUMMARY
Phenylpropanoids can function as preformed and inducible antimicrobial compounds, as well as signal molecules, in plant–microbe interactions. Since we last reviewed the field 8 years ago, there has been a huge increase in our understanding of the genes of phenylpropanoid biosynthesis and their regulation, brought about largely by advances in genome technology, from whole‐genome sequencing to massively parallel gene expression profiling. Here, we present an overview of the biosynthesis and roles of phenylpropanoids in plant defence, together with an analysis of confirmed and predicted phenylpropanoid pathway genes in the sequenced genomes of 11 plant species. Examples are provided of phylogenetic and expression clustering analyses, and the large body of underlying genomic data is provided through a website accessible from the article.
INTRODUCTION
Among their many functions in plants, phenylpropanoid compounds play important roles in resistance to pathogen attack (Dixon et al., 2002). Plant defence compounds have been classified into three main groups, signalling molecules, constitutive or preformed phytoanticipins, and phytoalexins, which are synthesized de novo in response to microbial attack (VanEtten et al., 1994); phenylpropanoid compounds are found among all three groups.
Flavonoids (C6–C3–C6) represent one of the largest classes of phenylpropanoid‐derived, plant‐specialized metabolites (with an estimated 10 000 structurally different members) (Tahara, 2007). They consist of two main groups: the 2‐phenylchromans (flavonoids) and the 3‐phenylchromans (isoflavonoids) (Fig. 1). The structural diversity of flavonoids is derived by complex substitution of these carbon skeletons through hydroxylation, glycosylation, methylation, prenylation, etc. The enzymes that catalyse these reactions often belong to large gene families, which can be recognized in expressed sequence tag (EST) and genome datasets through family‐specific conserved sequence motifs.
Figure 1.
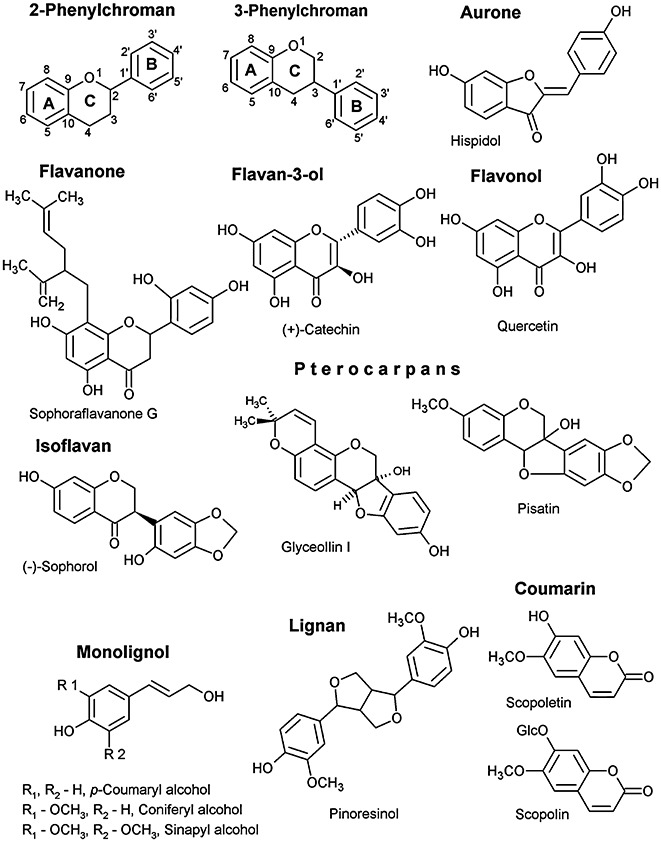
Basic structures of the phenylpropanoid classes described in this article.
In our earlier review (Dixon et al., 2002), the available knowledge on the genomics of phenylpropanoid metabolism was limited primarily to EST expression data and information from the Arabidopsis and rice genome sequencing projects. During the past 8 years, there has been a major increase in such information, brought about largely by advances in genome technology, from whole‐genome sequencing to massively parallel gene expression profiling. Here, we provide an update of the field by first reviewing our current understanding of how phenylpropanoid compounds are synthesized and function in plant defence, followed by a discussion of phenylpropanoid biosynthesis across whole plant genomes, with an emphasis on flavonoids. Because of the massive amount of data now available, we have generated a website (http://bioinfo.noble.org/manuscript‐support/mpp/) to present the information in a readily accessible manner. It is important to note that genes identified on the basis of bioinformatics approaches alone require subsequent functional identification by biochemical or genetic approaches; genes with proven functions are annotated as such on the website.
FUNCTIONS OF PHENYLPROPANOID COMPOUNDS IN PLANT DEFENCE
Lignin and lignans
Lignin and lignans share monolignols as common precursors (1, 2). Lignification makes the cell wall more resistant to mechanical pressure applied during fungal penetration (Bechinger et al., 1999). A lignified cell wall is water resistant and thus less accessible to cell wall‐degrading enzymes from plant pathogens.
Figure 2.
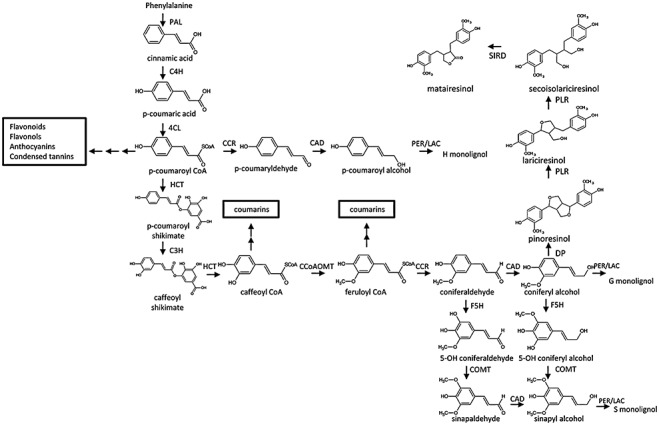
The monolignol biosynthetic pathway. The enzymes are as follows: CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl CoA 3‐O‐methyl transferase; CCR, cinnamoyl CoA reductase; C3H, 4‐coumaroylshikimate 3‐hydroxylase; C4H, cinnamate 4‐hydroxylase; 4CL, 4‐coumarate:CoA ligase; COMT, caffeic acid O‐methyltransferase; DP, dirigent protein; F5H, ferulic acid 5‐hydroxylase; HCT, hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase; LAC, laccase; PAL, l‐phenylalanine ammonia‐lyase; PER, peroxidase; PLR, pinoresinol/lariciresinol reductase; SIRD, secoisolariciresinol dehydrogenase.
Natural lignin contains three major units: p‐hydroxyphenyl (H), guaiacyl (G) and syringyl (S). The structural analysis of lignins induced by elicitor from Rhizosphaera kalkhoffii in spruce revealed that the defence lignins are significantly different from the lignins that are laid down during plant development (Lange et al., 1995). The specific activation of an S lignin biosynthesis pathway in elicitor‐treated wheat leaves was suggested by the induction of a sinapyl alcohol‐specific isoform of cinnamyl alcohol dehydrogenase (CAD) (Mitchell et al., 1999). Wheat leaves infected with stem rust fungus or exposed to a derived fungal elicitor subsequently accumulated S lignin (Menden et al., 2007).
The function of lignification during plant defence was originally assumed to be as a physical barrier (Moerschbacher et al., 1990), although lignins or their precursors might also function in chemical defence. Induction of monolignol biosynthesis genes by pathogen infection has been reported in a number of plant species (Bhuiyan et al., 2009; Dixon and Paiva, 1995; Koutaniemi et al., 2007; Lauvergeat et al., 2001; Menden et al., 2007). For example, 5‐hydroxyguaiacyl 3‐O‐methyltransferase [also known as caffeic acid 3‐O‐methyltransferase 1 (COMT1)] expression increased on infiltration of Arabidopsis leaves with flagellin (flg22) or harpin (HrpZ), or with the necrosis‐inducing Phytophthora protein 1 (NPP1) (Engelhardt et al., 2009; Fellbrich et al., 2002; Zipfel et al., 2004); thus, transcription of COMT1 appears to correlate with the onset of the plant's pathogen‐associated molecular pattern (PAMP)‐triggered immune (PTI) response.
Cinnamoyl‐CoA reductase (CCR), the first enzyme specific for monolignol synthesis, was shown to be involved in pathogen defence signalling in rice (Oryza sativa). OsCCR1 is an effector of OsRac1, a small guanosine triphosphatase (GTPase) from the Rac/Rop family, which is known to play a critical role in defence responses and to which OcCCR1 binds through a guanosine triphosphate (GTP)‐dependent physical interaction (Kawasaki et al., 2006).
In some cases, the downregulation of monolignol biosynthesis genes not only compromises resistance to host pathogens, but also allows the penetration of nonhost pathogens. For instance, RNA interference (RNAi)‐mediated gene silencing of COMT in wheat led to successful penetration of Blumeria graminis f. sp. hordei, a nonhost pathogen, in leaf cells, whereas no penetration was observed in control plants (Bhuiyan et al., 2009). However, the downregulation of lignin biosynthesis genes does not necessarily impair plant defence and, in some cases, reduced lignin plants have increased resistance to plant pathogens (Funnell and Pedersen, 2006). The mechanisms for this are not always clear. Arabidopsis Comt1 mutants accumulated more soluble 2‐O‐5‐hydroxyferulol‐l‐malate than did wild‐type plants, and this, rather than changes in lignin, accounted for the Comt1 mutant's enhanced resistance to Hyaloperonospora arabidopsidis (Quentin et al., 2009).
The lignans (1, 2) represent a structurally diverse class of plant‐specialized metabolites which, unlike lignins, are not found ubiquitously in all land plants. The primary physiological role of lignans in planta is probably in defence, and several lignans have been reported to have antibacterial and antifungal activities (Akiyama et al., 2007; Carpinella et al., 2005; Ralph et al., 2006). Lignans can act as both phytoanticipins and phytoalexins. The heartwood of many tree species, such as loblolly pine and western red cedar, contains significant levels of lignans, and constitutive deposition of lignans reinforces durability, longevity and resistance of the heartwood of many tree species against wood‐rotting fungi. On the other hand, de novo formation of lignans in response to fungal attack has been reported in both woody and nonwoody species (Gang et al., 1999).
Coumarins
Scopoletin and its glucosylated form scopolin (Fig. 1), which are induced under various stress conditions (Gachon et al., 2004; Shimizu et al., 2005), have antimicrobial and antioxidative activities and appear to play an important role in disease resistance (Chong et al., 2002). In addition to effects on fungi and bacteria, scopoletin accumulation may also be part of a mechanism for virus restriction in planta. Exogenously applied scopoletin inhibits Tobacco mosaic virus (TMV) replication in tobacco BY2 protoplasts (Chong et al., 2002), and transgenic tobacco plants overaccumulating scopoletin show enhanced resistance against infection with Potato virus Y (PVY) (Matros and Mock, 2004).
Scopoletin may be involved in the regulation of reactive oxygen intermediate (ROI) accumulation, one of the earliest changes underlying hypersensitive cell death (Dangl et al., 1996). Tobacco plants expressing an antisense construct to a coumarin‐specific glycosyltransferase exhibited reduced levels of scopolin and scopoletin, and showed more intense ROI accumulation relative to control plants after virus infection (Chong et al., 2002).
Flavonoids
Crude extracts from plants rich in flavonoids have been used for centuries to treat microbial diseases. Different compounds may target different components and functions of the bacterial cell, although individual flavonoids may target multiple sites (Cushnie and Lamb, 2005; Tsuchiya and Iinuma, 2000). The flavonol quercetin (Fig. 1) binds to the GyrB subunit of Escherichia coli DNA gyrase and inhibits the enzyme's adenosine triphosphatase (ATPase) activity (Plaper et al., 2003). Quercetin from leaves of Lotus has been described as a major antimicrobial compound against five microorganisms, namely Actinobacillus actinomycetemcomitans, Actinomyces viscosus, Porphyromonas gingivalis, Fusobacterium nucleatum and Actinomyces naeslundii (Li and Xu, 2008).
Some flavonoids inhibit bacterial cytoplasmic membrane function. The antibacterial activity of the prenylated flavanone sophoraflavanone G (SFG, Fig. 1) involves the reduction of bacterial membrane fluidity (Tsuchiya and Iinuma, 2000). Catechins (flavan 3‐ols, Fig. 1) from tea can damage bacterial membranes by directly penetrating them and disrupting their barrier function. Alternatively, catechins may promote membrane fusion, a process that results in leakage of intramembranous materials and aggregation (Tamba et al., 2007).
Different modifications of flavonoid skeletons have a significant impact on their antimicrobial activity. For example, prenylation generally increases inhibitory activity against bacteria and fungi (Sohn et al., 2004); glycosylation of flavonols from carnation increases their activity against Fusarium oxysporum (Galeotti et al., 2008); methoxylation drastically decreases the antibacterial activity of flavonoids (Alcaraz et al., 2000).
Pterocarpans, such as medicarpin from alfalfa (Medicago sativa) (Fig. 3), pisatin from pea (Pisum sativum L.) (Fig. 1) and maackiain from chickpea (Cicer arietinum), constitute a major class of antimicrobial isoflavonoid phytoalexins in legumes. Genetic evidence now supports the hypothesis that pisatin production plays an important role in disease resistance of pea. For instance, pea hairy roots expressing an antisense construct to 6a‐hydroxymaackiain O‐methyltransferase (HMM, the last enzyme in pisatin biosynthesis) exhibit a large reduction in (+)‐pisatin content, and are more susceptible than controls to damage caused by the fungus Nectria haematococca (Wu and VanEtten, 2004). Furthermore, detoxification of pisatin by the cytochrome P450 enzyme pisatin demethylase (PDA) is a virulence factor for N. haematococca in pea (Funnell et al., 2002). PDA transcripts are induced in infected pea tissues or by in vitro application of pisatin to the fungus, and other genes necessary for pathogenesis on pea display similar temporal patterns to PDA gene expression both in planta and in vitro, consistent with a coordinated regulation of these genes by pisatin during pathogenesis (Liu et al., 2003b). A Zn(2)–cysteine(6) zinc binuclear DNA‐binding protein is involved in pisatin‐responsive activation of PDA gene expression (Khan et al., 2003).
Figure 3.
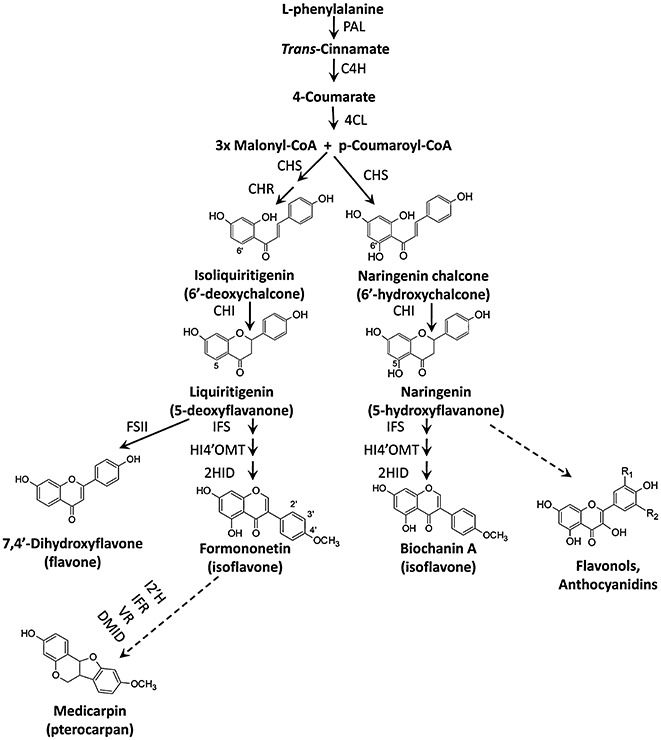
Biosynthesis of flavonoids and isoflavonoids. The enzymes are as follows: C4H, cinnamate 4‐hydroxylase; CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; 4CL, 4‐coumarate:CoA ligase; DMID, 7,2′‐dihydroxy‐4′‐methoxy‐isoflavonol dehydratase; FSII, flavone synthase II; 2HID, 2‐hydroxyisoflavanone dehydratase; HI4′OMT, 2‐hydroxyisoflavanone 4′‐O‐methyltransferase; IFR, isoflavone reductase; IFS, isoflavone synthase; I2′H, isoflavone 2′‐hydroxylase; PAL, l‐phenylalanine ammonia‐lyase; VR, vestitone reductase.
RECENT ADVANCES IN OUR UNDERSTANDING OF PHENYLPROPANOID BIOSYNTHESIS
Lignin and lignans
Lignins are synthesized by the oxidative coupling of hydroxycinnamyl alcohol monomers. Although some researchers have proposed that a variety of units can be incorporated into the lignin polymer, and that lignin assembly is essentially combinatorial, this is still a matter of some controversy (Davin et al., 2008; Ralph et al., 2008).
Monolignols share the general phenylpropanoid biosynthetic pathway with other phenolic compounds, such as anthocyanins, condensed tannins and isoflavones (Fig. 2). This pathway includes the entry‐point enzyme l‐phenylalanine ammonia‐lyase (PAL), cinnamate 4‐hydroxylase (C4H) and 4‐coumarate:CoA ligase (4CL). The specific portion of the monolignol synthesis pathway includes the enzymes hydroxycinnamoyl CoA:shikimate hydroxycinnamoyltransferase (HCT), 4‐coumaroylshikimate 3‐hydroxylase (C3H), CCR, CAD, COMT, caffeoyl CoA 3‐O‐methyltransferase (CCoAOMT) and ferulic acid 5‐hydroxylase (F5H) (Fig. 2). Several reviews have described the monolignol pathway in much detail (Boerjan et al., 2003; Raes et al., 2003). Recent evidence has suggested that simple phenolic acids are derived by reduction of aldehyde intermediates of monolignol biosynthesis; thus, ferulic acid is now believed to originate from the reduction of coniferaldehyde rather than sequential ring modification of cinnamic acid, and sinapic acid may be derived similarly (Nair et al., 2004). Esterification with malate leads to the formation of the sinapate esters found in the seeds and leaves of the Brassicaceae, and recently implicated in the modulation of pathogen reproduction in planta (Quentin et al., 2009).
After synthesis, monolignols are transported to the cell wall by mechanisms yet unknown. They then undergo oxidative polymerization, catalysed by peroxidases and/or laccases (Gabaldón et al., 2006; Sato et al., 2006). There are 73 peroxidase genes in the Arabidopsis genome (Tognolli et al., 2002), and laccases are also encoded by multigene families in plants, making it difficult to identify isoforms that are specifically involved in monolignol oxidation. Some peroxidases can cause ectopic lignification when overexpressed in planta (Elfstrand et al., 2002). However, solid genetic evidence is still missing to identify unambiguously the specific isoforms of peroxidases and/or laccases involved in lignification.
Because monolignol biosynthesis is a metabolically costly process and its carbon investment is not reversible, the activation of this pathway must be tightly regulated. Many of the genes encoding lignin biosynthetic enzymes, such as PAL, C4H, COMT, CCR and CAD, share a conserved motif, known as the AC element, in their promoters that binds lignin pathway regulatory transcription factors (Chen et al., 2000; Hatton et al., 1995; Lauvergeat et al., 2002). A network of transcription factors is responsible for the regulation of secondary wall biosynthesis in Arabidopsis. In this network, two NAC transcription factors, NST1 and NST3, act as a master switch to turn on the entire process of secondary wall biosynthesis, including monolignol biosynthesis (Mitsuda et al., 2007; 2007, 2008). Two MYB transcription factor genes, MYB58 and MYB63, are directly regulated by NST1 and NST3, and can specifically activate the monolignol biosynthesis pathway (Zhou et al., 2009). Some lignin gene family members are involved in constitutive lignification, whereas other gene family members are only expressed in response to pathogen infection (Lauvergeat et al., 2001). Therefore, it is likely that lignin biosynthesis in vascular tissue development is regulated differentially from that occurring during plant defence responses.
Dimeric lignans result from the stereoselective dirigent protein‐directed oxidative coupling by laccases of two coniferyl alcohol units (Davin et al., 1997) (Fig. 2), a process that has been well characterized in flax (Linum usitatissimum) (Ford et al., 2001). In recent years, a large number of dirigent protein genes have been identified in various plant species (Kim et al., 2002; Ralph et al., 2006). Many complex substituted lignan structures exist. In one commonly found pathway, dimerization of two coniferyl alcohol units produces pinoresinol, which is then reduced to lariciresinol and secoisolariciresinol by pinoresinol/lariciresinol reductase (PLR), and finally into matairesinol by secoisolariciresinol dehydrogenase (SIRD) (Hano et al., 2006; von Heimendahl et al., 2005) (Fig. 2). Secoisolariciresinol is mainly found in its diglucoside form (SDG), and is stored as a hydroxymethyl glutaryl ester‐linked complex (Ford et al., 2001).
Although the dirigent protein determines the stereoselectivity of the oxidative free radical coupling reaction, the enantiomeric composition of pinoresinol differs in various plants (Umezawa, 2003). However, the fact that some lignans derived from pinoresinol, such as matairesinol, are optically pure indicates that PLR and SIRD, which catalyse subsequent metabolic steps, also contribute to the enantiomeric composition of lignans (von Heimendahl et al., 2005; Nakatsubo et al., 2008). PLR normally reduces both pinoresinol and lariciresinol efficiently (Hemmati et al., 2007; von Heimendahl et al., 2005). However, the two Arabidopsis PLRs show strict substrate preference towards pinoresinol and no activity towards lariciresinol, in contrast with conventional PLRs of other plants (Nakatsubo et al., 2008).
Coumarins
Coumarins share hydroxycinnamoyl CoA intermediates with the monolignol pathway. Recently, the production of scopoletin and scopolin in Arabidopsis roots has been shown to require the activity of C3H; in C3H T‐DNA insertion mutants, the content of scopoletin and scopolin decreased to around 3% of that in the wild‐type (Kai et al., 2006).
The role of glucosyltransferase that catalyses the glucosylation of scopoletin has been demonstrated by both loss‐ and gain‐of‐function approaches in transgenic tobacco. The downregulation of tobacco scopoletin glucosyltransferase (TOGT) activity was accompanied by decreased virus resistance (Chong et al., 2002). Conversely, the overexpression of TOGT increased virus resistance (Matros and Mock, 2004).
Flavonoids
Chalcone synthase (CHS) catalyses the first committed enzyme of flavonoid biosynthesis (Fig. 3). CHSs are type III polyketide synthases (PKSs) with two independent active sites that catalyse a series of decarboxylation, condensation and cyclization reactions (Jez et al., 2002). Structural biology has recently become an important tool for understanding the mechanisms and evolution of enzymes involved in the biosynthesis/modification of phenylpropanoid skeletons (Noel et al., 2005; Ritter and Schulz, 2004; Shao et al., 2005). Guided by the three‐dimensional structure of CHS, substitution of the active site phenylalanine‐215 (Phe215) by serine (Ser) yielded a mutant CHS that preferentially accepts the CoA‐thioester substrate to generate a novel alkaloid, namely N‐methylanthraniloyltriacetic acid lactone (Jez et al., 2002). Thus, a single point mutation in CHS dramatically shifts the molecular selectivity of this enzyme.
5‐Deoxy(iso)flavonoids, which are predominantly found in legumes, have strong antimicrobial properties and also act as signals in plant–microbe interactions. Chalcone reductase (CHR) is the critical enzyme for the 5‐deoxyflavonoid pathway, reducing an unstable intermediate of the CHS reaction to ultimately form 6′‐deoxychalcone (isoliquiritigenin, Fig. 3).
Chalcone isomerases (CHIs) catalyse the stereospecific isomerization of chalcones into their corresponding flavanones (Shimada et al., 2003). Type I CHIs isomerize only 6′‐hydroxychalcone to 5‐hydroxyflavanone, and type II CHIs, generally found in legumes, act on both 6′‐deoxychalcone and 6′‐hydroxychalcone, yielding 5‐deoxyflavanone and 5‐hydroxyflavanone, respectively (Fig. 3).
The conversion of flavanones to isoflavones is catalysed by ‘isoflavone synthase’ (IFS), a cytochrome P450 enzyme of the CYP93C subfamily. In the first step, IFS catalyses the 2‐hydroxylation of flavanone, followed by 1,2‐aryl migration to yield a 2‐hydroxyisoflavanone. All the P450s of the CYP93 family so far functionally characterized are involved in flavonoid/isoflavonoid biosynthesis. For example, the CYP93Bs flavanone 2‐hydroxylase (Akashi et al., 1998b) and flavone synthase II (Martens and Forkmann, 1999) catalyse the hydroxylation at C‐2 or introduction of a double bond between C‐2 and C‐3 of the flavanone substrate, respectively. CYP93A is a soybean pterocarpan 6a‐hydroxylase (Schopfer et al., 1998).
To date, there are no crystal structures available for plant membrane‐associated cytochrome P450s. However, molecular modelling studies have suggested that two active site residues of licorice IFS, Ser310 and lysine‐375 (Lys375), are critical for the aryl migration of the flavanone substrate (Sawada and Ayabe, 2005; Sawada et al., 2002). The Lys375Thr mutant (Thr, threonine) produced only 3‐hydroxyflavanone, a byproduct of the IFS reaction. Thus, introduction of Lys375 was a critical event in the development of the unique catalytic function of IFS which was then fixed in the Leguminosae because of the selective advantage of the presence of isoflavonoids.
Pea, alfalfa, licorice and Lotus produce their isoflavonoid phytoalexins from a 4′‐methoxyisoflavone, formononetin, whereas soybean synthesizes 4′‐hydroxy‐type isoflavonoid phytoalexins (glyceollins) from daidzein (Fig. 3). The dehydration (1,2‐elimination) step from 2‐hydroxyisoflavanone to isoflavone is now known to be enzyme‐catalysed rather than a spontaneous process (Akashi et al., 2005; Hakamatsuka et al., 1998). Licorice 2‐hydroxyisoflavanone dehydratase (HID) is specific for 2,7‐dihydroxy‐4′‐methoxyisoflavanone, whereas soybean HID has broader specificity for both 4′‐hydroxylated and 4′‐methoxylated 2‐hydroxyisoflavanones, reflecting the structures of the isoflavones in each species. Site‐directed mutagenesis studies and phylogenetic analysis revealed that HID proteins evolved from the plant carboxylesterase family (Akashi et al., 2005). HID appears to be critical for isoflavone biosynthesis in hairy root cultures of Lotus japonicus (Shimamura et al., 2007). In contrast, isoflavones are formed in Arabidopsis expressing IFS in the absence of HID (Liu et al., 2002).
Hydroxylation of flavonoid skeletons by NADPH‐dependent cytochrome P450 monooxygenases is important in the biosynthesis of complex flavonoids. Hydroxylation at the 2′ or 3′ position of the B‐ring is essential for the formation of pterocarpans or coumesterol (Guo et al., 1994; Hinderer et al., 1987). 6a‐Hydroxylation of pterocarpans occurs in the biosynthesis of the glyceollins in soybean and of pisatin in pea. The first isoflavone 2′‐hydroxylase (I2′H; CYP81E1) was cloned and characterized from licorice (Akashi et al., 1998a). Subsequently, three CYP81E subfamily members were identified in Medicago truncatula by mining of EST databases (Liu et al., 2003a). Two of the corresponding enzymes, when expressed in yeast, utilized the same isoflavone substrates, but produced different products hydroxylated at the 2′ and/or 3′ positions of the B‐ring. Unique tissue‐specific expression patterns suggested the involvement of these enzymes in different defence processes in Medicago (Liu et al., 2003a).
Isoflavone reductase (IFR) is a key enzyme in the latter part of the pterocarpan phytoalexin biosynthetic pathway (Paiva et al., 1991). In species producing medicarpin, it catalyses the NADPH‐dependent reduction of 2′‐hydroxyformononetin to the isoflavanone (3R)‐vestitone. An engineered IFR lacking residues 39–47, which constitute a flexible region, was crystallized, and the structure of Δ39–47 IFR was shown to comprise two domains (N‐ and C‐terminal) which form a cleft in which both NADPH and 2′‐hydroxyformononetin are presumed to bind (Wang et al., 2006). The study provides a structural basis for understanding the enzymatic mechanism and substrate specificity of IFRs, as well as the functions of IFR‐like proteins (Wang et al., 2006).
Enzymatic O‐methylation of (iso)flavonoids is catalysed by O‐methyltransferases (OMTs), which transfer a methyl group from S‐adenosyl‐l‐methionine (SAM) to a hydroxyl moiety of the acceptor molecule. Two different groups of small‐molecule OMTs can be distinguished from plants: proteins of group I do not require a metal ion for activity, whereas OMTs of group II are Mg2+ dependent (Noel et al., 2003). 4′‐O‐Methylation of the isoflavonoid B‐ring by a SAM‐dependent 2‐hydroxyisoflavanone 4′‐O‐methyltransferase (HI4′OMT) is a critical step at the entry point into the formation of isoflavonoid phytoalexins in pea, alfalfa, M. truncatula and chickpea (Akashi et al., 2003; Dixon, 1999). However, among these four species, only pea catalyses A‐ring 3‐O‐methylation that converts the pterocarpan 6a‐hydroxymaackiain to pisatin (Fig. 1). The HI4′OMT from M. truncatula shares 90% amino acid sequence identity with (+)‐6a‐hydroxymaackiain 3‐O‐methyltransferase from pea, which is responsible for the final step in the biosynthesis of (+)‐pisatin. Although (+)‐6a‐hydroxymaackiain is not synthesized in M. truncatula, cell suspension cultures accumulate pisatin when fed with 6a‐hydroxymaackiain (Liu et al., 2006). Recombinant HI4′OMT of M. truncatula shows comparable activity towards both 2,7,4′‐trihydroxyisoflavanone and the unnatural substrate (+)‐6a‐hydroxymaackiain. Protein X‐ray crystal structures of HI4′OMT substrate complexes revealed similar bound conformations for the seemingly distinct chemical substrates, explaining how leguminous plants can use homologous enzymes for two different biosynthetic reactions (Liu et al., 2006).
Vestitone reductase (VR) is the central enzyme for the conversion of nonenantiomeric isoflavones to enantiomeric pterocarpans. It is a member of the short‐chain dehydrogenase/reductase (SDR) superfamily and catalyses the NADPH‐dependent reduction of (3R)‐vestitone in the biosynthesis of medicarpin. It acts with high stereochemical specificity, and recognizes only the 3R conformation of vestitone, not (3S)‐vestitone (Shao et al., 2007). A VR homologue, sophorol reductase (SOR), was identified for the biosynthesis of pisatin in pea. SOR prefers (−)‐sophorol (Fig. 1) over (+)‐sophorol as substrate, converting it to (−)‐7,2′‐dihydroxy‐4′,5′‐methylenedioxyisoflavanol for the production of (+)‐pisatin (DiCenzo and VanEtten, 2006). Homologues of VR and SOR have recently been described in additional species, including soybean (Glycine max) (Liu, 2009). The three final steps in pisatin biosynthesis are catalysed by SOR, pterocarpan 6a‐hydroxylase and (+)‐6a‐hydroxymaackiain‐3‐O‐methyltransferase (HMM). The catalytic specificity of two genetically linked HMMs in pea suggested gene duplication for (+)‐pisatin biosynthesis (Akashi et al., 2006).
The prenylation of flavonoids enhances their antibacterial, antifungal and other biological activities by increasing their lipophilicity and membrane permeability (Sohn et al., 2004). Recently, Sasaki and colleagues isolated the first cDNA of a flavonoid‐specific plant prenyltransferase, SfN8DT‐1, involved in SFG (Fig. 1) biosynthesis in Sophora flavescens (Sasaki et al., 2008). Phylogenetic analysis revealed that the naringenin dimethylallyltransferase SfN8DT‐1 has the same evolutionary origin as prenyltransferases for vitamin E and plastoquinone biosynthesis (Sasaki et al., 2008). This discovery facilitated the subsequent identification of the pterocarpan 4‐dimethylallyltransferase (G4DT) catalysing the critical prenylation step in glyceollin biosynthesis in soybean (Akashi et al., 2009).
Family 1 uridine diphosphate (UDP) glycosyltransferases (UGTs) transfer sugars to a wide range of acceptors. Plants contain very large UGT gene families: over 100 putative UGT genes have been identified in Arabidopsis and more than 300 in M. truncatula. Significant progress has been made during recent years in the isolation and structural characterization of (iso)flavonoid UGTs from legume species (He et al., 2006; Modolo et al., 2007; Noguchi et al., 2007; Shao et al., 2005). Most of the identified UGTs to date prefer UDP‐glucose as sugar donor. Relationships between the structure and function in M. truncatula (iso)flavonoid UGTs have been explored by X‐ray crystallography and site‐directed mutagenesis (He et al., 2006; Li et al., 2007; Shao et al., 2005).
Genome‐wide analysis of phenylpropanoid metabolism
The last decade has seen remarkable advances in the structural analysis of model plant genomes. Post‐sequence, functional genomics approaches are increasingly being applied to the discovery of new gene functions, and comprehensive web‐based software tools have become available for biologists to explore gene expression across a wide variety of biological contexts. Reference expression databases for the meta‐analysis of transcriptomes, such as Expression Profiler (http://www.ebi.ac.uk/expressionprofiler) or the Medicago truncatula Gene Expression Atlas (http://bioinfo.noble.org/gene‐atlas/v2), have provided new tools for the analysis of the expression of genes in many different contexts, for the clustering of genes into expression modules, and for the modelling of expression responses in the context of metabolic and regulatory networks.
Because of the exponential increase in plant genome information since the publication of our previous review (Dixon et al., 2002), it is no longer possible to summarize this field in hard copy text and tables. We have therefore created a database of the genes involved in the biosynthesis of phenylpropanoid natural products from 11 plant species with fully or near‐fully sequenced genomes; the moss Physcomitrella patens, the spike moss Selaginella moellendorffii, three monocots (Oryza sativa, Sorghum bicolor, Zea mays) and six dicots (Arabidopsis thaliana, Populus trichocarpa, Vitus vinifera, Glycine max, Medicago truncatula and Lotus japonicus). For gene annotation, proteins predicted from genome sequences were searched by blastx against the uniprot trembl database (http://www.ebi.ac.uk/uniprot/) (e‐value < 1e‐04), and the top five hits with meaningful annotations were retrieved as annotation. Genes were sorted as representing proteins involved in the various branches of the phenylpropanoid pathway, including flavonoid, isoflavonoid, lignin, etc., based on predicted function. The database is available online (http://bioinfo.noble.org/manuscript‐support/mpp/); the information includes the source of genome sequences, enzyme name, gene identification, redundancy, Affymetrix probeset ID, location on chromosome (if possible), length of sequence, five top hit annotations and the sequence itself. It is critical to note that this analysis uses informatics approaches to predict function; annotations should be viewed as tentative unless true biochemical functions have been ascribed to a particular gene. For example, many genes are now being given ‘‐like’ annotations, such as CCR‐like or IFR‐like, highlighting the fact that they have sequence identity to functionally characterized genes, but that their protein products have not been shown to catalyse the predicted reaction. Gene products with proven function are indicated in the database and, where appropriate, in the following figures in the text.
Genomic evidence for the evolution of flavonoids as protective compounds
To illustrate how genomic data can be mined to reveal details of phylogenetics, chromosomal organization and expression patterns of relevance to plant defence, we have chosen the flavonoid pathway as an example. It is believed that flavonoids initially evolved as protective compounds against UV irradiation (Koes et al., 1994; Markham, 1988; Shirley, 1996), or as hormone regulators (Stafford, 1991). During evolution, plants adopted flavonoids for many different functions, including defence against pathogens.
Mosses are one of the oldest plant groups that produce flavonoids, including chalcones, flavonols and flavones (Fig. 1) (Markham, 1988). Flowering plants extended the array of flavonoids, also producing aurones, anthocyanidins, proanthocyanidins and isoflavonoids (the latter mainly in legume species). CHS, encoding the initial enzyme of the pathway, exhibits strong sequence similarity to bacterial genes encoding fatty acid PKSs. PKSs are classified into three types according to their architectural configurations (Fischbach and Walsh, 2006); type III (which includes CHS‐related enzymes) is present in bacteria, plants and fungi (Austin and Noel, 2003; Funa et al., 2007; Seshime et al., 2005).
Gene duplication followed by sequence divergence has been used as a model to explain how novel biochemical functions have arisen from CHSs. Multiple copies of CHS‐related genes are found in many species, including the 11 analysed in the present work (Fig. 4). In general, the numbers of CHS‐related genes in the genomes correlate with total genome size (Fig. 4A,B). However, soybean, with double the genome size of M. truncatula, rice and sorghum, contains fewer CHS‐related genes (Fig. 4A,B). CHS‐related genes are scattered throughout the genomes; however, they form clusters of up to five to seven genes in rice, M. truncatula and soybean that may reflect ongoing duplication events (Fig. 4C). The Zea mays genome (the largest genome analysed in this work) is not fully sequenced, and so we do not yet know how many CHS‐related genes exist.
Figure 4.
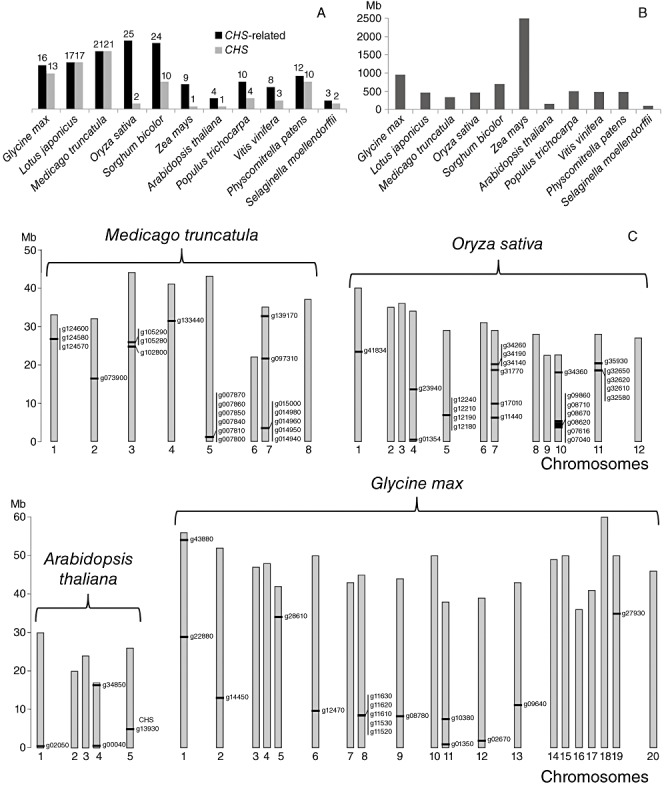
Distribution of chalcone synthase (CHS)‐related genes in multiple sequenced genomes. (A) The number of CHS‐related and true CHS genes in the genomes of 11 species. (B) Size of the genomes of the 11 species in (A). (C) Map positions of CHS‐related genes on chromosomes of Medicago truncatula, Oryza sativa, Arabidopsis thaliana and Glycine max.
Four residues, cysteine‐164 (Cys164), Phe215, histidine‐303 (His303) and asparagine‐336 (Asn336), are highly conserved in CHS proteins (Fig. S1, see Supporting Information; conserved residues are highlighted by blue rectangles) and define the active site (Ferrer et al., 1999). Stilbene synthases (STSs) are among the closest related enzymes to CHSs. They likewise condense one coumaroyl‐ and three malonyl‐CoA molecules, but the reaction involves a decarboxylation leading to the formation of resveratrol. STS shows high amino acid sequence identity to CHS; however, it differs in several critical amino acids, such as aspartic acid‐255 (Asp255), His266 and leucine‐268 (Leu268) in the β1d–β2d region of the CoA‐binding tunnel (Ferrer et al., 1999) (see alignment in Fig. S1; critical amino acids highlighted by red rectangles and STSs by yellow rectangles). It has been suggested that STSs evolved from CHSs several times independently, as they group phylogenetically with CHSs from the same or related plants (Tropf et al., 1994). Our analyses indicate that STS distributes into the CHS cluster, but as a separate subcluster (Fig. 5A).
Figure 5.
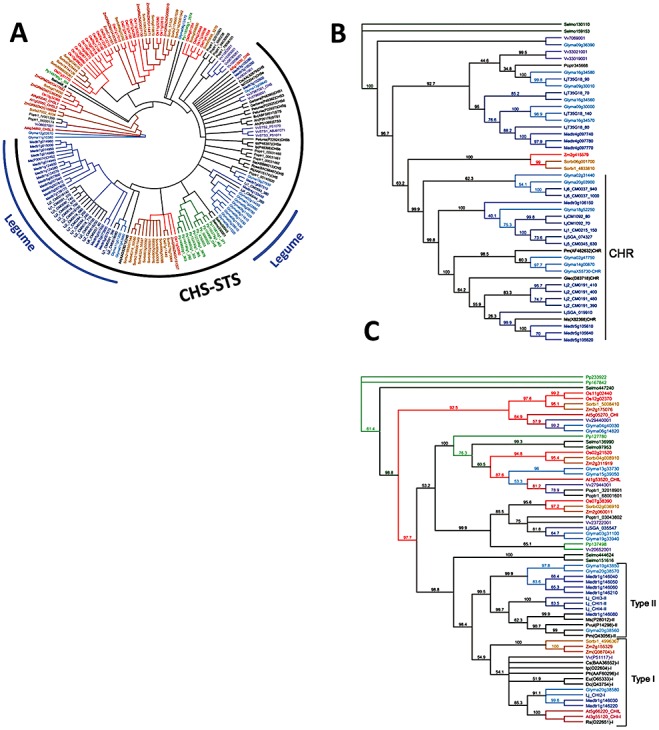
Phylogenetic trees of CHS‐related genes (A), aldo‐keto reductases (B) and chalcone isomerases (C). Deduced amino acid sequences were aligned using the L‐INS‐i method (Katoh and Toh, 2008) (the mafft program is available online at http://align.bmr.kyushu‐u.ac.jp/mafft/software/) and subsequently inspected by eye. Neighbour‐joining analyses were performed on the aligned sequences using Geneious v4.7 software (Drummond et al., 2009). The bootstrap replicates were 1000 (support thresholds greater than 50% are given at the nodes). Abbreviations: CHI, chalcone isomerase; CHIL, chalcone isomerase‐like; CHR, chalcone reductase; CHS, chalcone synthase; CHSL, chalcone synthase‐like; STS, stilbene synthase. Genes encoding enzymes from 11 species are colour‐coded as follows: Physcomitrella patens (Pp), light green; Selaginella moellendorffii (Selmo), dark green; Populus trichocarpa (Poptr), grey; Arabidopsis thaliana (At), maroon; Vitis vinifera (Vv), dark violet; Medicago truncatula (Medtr), blue; Lotus japonicus (Lj), cobalt; Glycine max (Glyma), light blue; Oryza sativa (Os), red; Sorghum bicolor (Sorbi), yellow ochre; Zea mays (Zm), orange. Bootstrap values for the nodes of the CHS‐related gene tree (A) are provided in Fig. 2 (see Supporting Information). Protein sequences from the 11 species are available online (http://bioinfo.noble.org/manuscript‐support/mpp/). Gene names from additional different species are colour‐coded in black. Accession numbers for genes encoding enzymes from additional species: for CHSs, Gerbera hybrida cultivar, GerberaCHS1 (P48390), GerberaCHS3 (P48392); Ipomoea purpurea, IpCHSA (P48397), IpCHSB (P48398), IpCHSE (O22047); Petunia hybrida, PetuniaCHSB (P22924), PetuniaCHSD (P22925), PetuniaCHSG (P22927); Arachis hypogaea, AhCHS (AAO32821), AhSTS1 (P20178), AhSTS3 (P51069); Bauhinia variegate, BvSTS (ABF59517); Medicago sativa, MsCHS2 (P30074); Cannabis sativa, CsCHS (AAL92879); Rosa hybrida cultivar, RosaCHS (BAC66467); Rubus idaeus, RiCHS (AAK15174); Sorbus aucuparia, SaCHS (ABB89213); for STSs, Vitis vinifera, VvSTS1 (ABJ97071), VvSTS2 (P51070), VvSTS3 (P51071); for CHRs, Glycyrrhiza echinata, GlecCHR (D83718); Medicago sativa, MsCHR (X82368); Pueraria montana var. lobata PmCHR (AF462632); for CHIs type I, Zea mays (Q08704); Citrus sinensis (BAA36552); Dianthus caryophyllus (Q43754); Elaeagnus umbellate (O65333); Ipomoea purpurea (O22604); Petunia hybrida (AAF60296); Raphanus sativus (O22651); for CHIs type II, Medicago sativa (P28012); Pueraria montana var. lobata (Q43056); Phaseolus vulgaris (P14298).
Cloning of CHS from the moss P. patens revealed an important transition from the CHSs present in microorganisms to those present in higher plants (Jiang et al., 2006). Of the 12 CHS‐related genes found in the P. patens genome, 10 are distributed into the true CHS cluster, but form a separate subcluster with the CHS‐related genes from Selaginella moellendorffii (Fig. 5A, moss CHSs coloured green). Moss CHSs showed more than 60% amino acid identity to CHSs from higher plants, although one of the critical amino acids in the CoA‐binding tunnel, Leu268, is substituted with methionine, which probably does not affect the enzyme performance (Fig. S1). Mosses typically exhibit a simple flavonoid profile, and the physiological significance of the presence of multiple CHSs in moss requires further investigation.
Although the rice genome contains the largest number of CHS‐related genes, only two encode true CHSs (Fig. 5A). With a few exceptions, CHSs from mosses, monocots (rice, sorghum and maize) and legumes form separate subclusters (Fig. 5A; Fig. S2, see Supporting Information). Importantly, there has been a notable expansion of CHS genes in the legumes: of 17 CHS‐related genes, 13 are true CHSs in soybean, and only true CHSs are present in the genomes of M. truncatula and L. japonicus (Fig. 4A). The presence of multiple copies of CHS in separate clusters reflects massive duplication events after speciation, indicative of selective evolutionary pressure in favour of flavonoids in legume species.
CHR originated from the aldo/keto reductases (Bomati et al., 2005). Sixteen CHR genes are found in L. japonicus, seven in M. truncatula and 11 in soybean (see database online). CHR‐like genes are also found in the genomes of nonlegumes, but their functions in these species are obscure. Phylogenetic analysis of putative CHRs with functionally characterized enzymes clearly places only legume enzymes in the true CHR cluster (Fig. 5B).
CHI is also encoded by a multigene family which forms gene clusters in some legume species. For example, a cluster of genes encoding type I and II CHIs is found in L. japonicus (Shimada et al., 2003); a cluster of seven genes encoding both types of CHI is observed at the same locus on chromosome 1 of M. truncatula. In general, more CHI genes are present in the genomes of legumes than nonlegumes (see database online), probably as a result of the occurrence of type II CHIs in legumes (Fig. 5C). It has been suggested that type II CHIs evolved from an ancestral CHI by gene duplication, and the resultant production of 5‐deoxyflavonoids paralleled the establishment of the Fabaceae (Shimada et al., 2003).
Three later enzymes of flavonoid biosynthesis, flavanone 3β‐hydroxylase (F3βH), flavonol synthase (FLS) and anthocyanidin synthase (ANS), are all members of the family of 2‐oxoglutarate‐iron‐dependent oxygenases. They are closely related by sequence and all catalyse the oxidation of the flavonoid ‘C‐ring’; these enzymes also exhibit overlapping substrate and product specificity (Turnbull et al., 2004), making annotation of their genes confusing. In our database, we placed these genes in the flavonoid 2‐oxoglutarate‐dependent dioxygenase group, with putative annotation of some of them based on phylogenetic analysis with functionally characterized enzymes [see database and Fig. S3 (Supporting Information)].
Genes encoding the entry enzyme into isoflavonoid biosynthesis, IFS, are present in low copy number in legume genomes (see database online); furthermore, in Medicago, only two of the five are true IFS genes (Fig. 6). Isoflavonoid OMTs are more abundant: 17, 11 and three IOMT genes were identified in the genomes of soybean, Medicago and Lotus, respectively (see database online). A previous phylogenetic analysis (Deavours et al., 2006) divided the M. truncatula IOMTs into two distinct clades, with homology to either I7OMT enzymes, methylating the A‐ring 7‐hydroxyl of the isoflavone daidzein, or 4′OMT enzymes, methylating the B‐ring 4′‐hydroxyl of 2,7,4′‐trihydroxyisoflavanone.
Figure 6.

Putative positions of the substrate recognition sites of CYP93 family enzymes in soybean, Medicago and Lotus. The figure was generated based on the study of Sawada et al., 2002. FSII, flavone synthase II; IFS, isoflavone synthase.
Expression profiling of flavonoid defence responses
To evaluate the involvement of flavonoid pathway genes in defence responses, we downloaded Affymetrix microarray data specifically related to stress or pathogen studies. At present, Affymetrix microarray chips have been developed for 12 plant species: Arabidopsis, barley, citrus, cotton, grape, maize, Medicago, poplar, rice, soybean, tomato and wheat (http://www.affymetrix.com/). Numerous microarray experiments are available for Arabidopsis, rice, Medicago and soybean, and we therefore selected these species for further analysis. Microarray data for rice and soybean were obtained from Expression Profiler (EP, http://www.ebi.ac.uk/expressionprofiler), a web‐based platform for microarray gene expression and other functional genomics‐related data analysis (Kapushesky et al., 2004). Arabidopsis and Medicago expression data were obtained from AtGenExpress (http://www.weigelworld.org/resources/microarray/AtGenExpress/) and Gene Expression Atlas database version 2 (MtGEAv2, http://bioinfo.noble.org/gene‐atlas/v2/), respectively. Affymetrix probesets were mapped to the corresponding genes at individual Affymetrix probe level, using a position‐weighted scoring index in which mismatches near the middle of a probe were most heavily penalized as follows: [1,1,1,1,1,2,2,2,2,2,3,3,3,3,3,2,2,2,2,2,1,1,1,1,1] for each 25‐mer Affymetrix probe. Therefore, a perfect match for a 25‐mer probe yields a score of 45. Matches were declared when at least eight of 11 total probe sets had scores of 43 or higher for each gene (see database online). For better visualization of differential gene expression in response to pathogen or stress, treatment/control fold‐change values were calculated. The fold‐change values were converted into a log2 scale (see database online) and data were subjected to clustering analysis.
As summarized in Fig. 7 and Figs S4 and S5 (see Supporting Information), different numbers of flavonoid pathway genes responded to pathogen or elicitor treatment in each species. In Arabidopsis, putative ANS (At2g38240) was highly induced by Pseudomonas and Phytophthora treatments, whereas expression of functionally characterized CHS (At5g13930; Shirley et al., 1995) was reduced in most experiments (Fig. 7A). The weak response of flavonoid pathway genes to infection in Arabidopsis is consistent with the deployment of indole (i.e. nonflavonoid) phytoalexins in this species (Rogers et al., 1996). In rice, F3βH (Os04g49194) was highly expressed in response to infection with Xanthomonas oryzae pv. oryzicola, and a cluster of genes, including IFR‐Likes, DFRs and some CHS‐Likes, was expressed in response to infection by the blast fungus Magnaporthe oryzae; CHS (Os11g32650) was only slightly induced under these conditions (two‐fold change) (Fig. S4).
Figure 7.
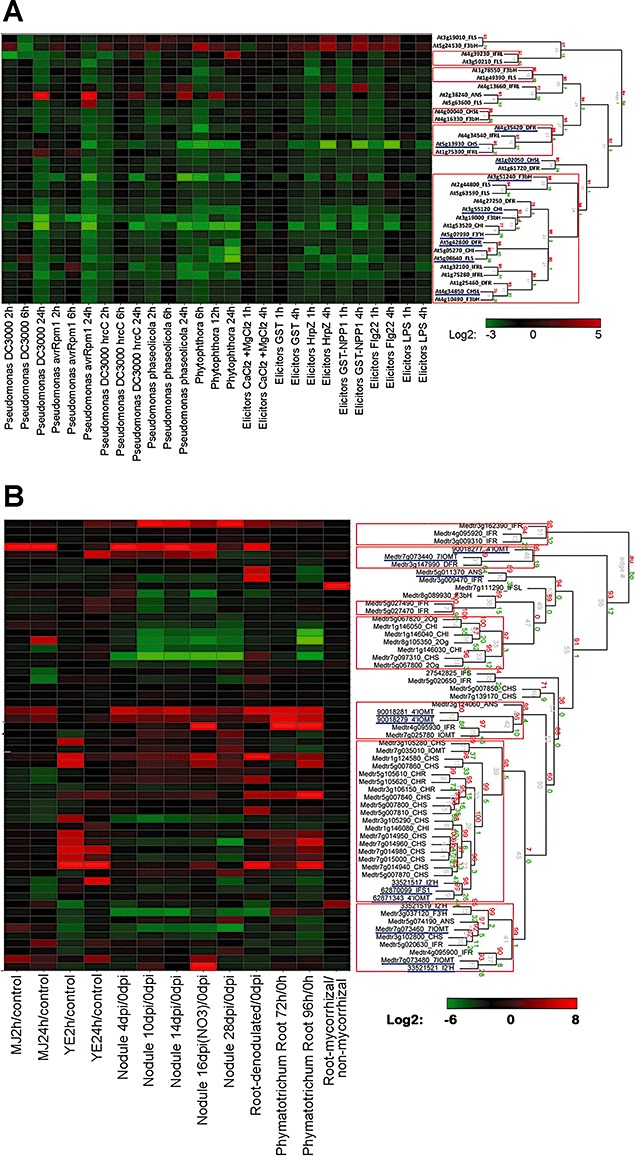
Hierarchical clustering analysis of expression patterns of flavonoid biosynthesis genes in microarray experiments involving exposure of plants to pathogens or pathogen‐derived elicitors. (A) Arabidopsis thaliana; (B) Medicago truncatula. Microarray data were obtained for A. thaliana at AtGenExpress (http://www.weigelworld.org/resources/microarray/AtGenExpress/) and for M. truncatula from MtGEAv2 (http://bioinfo.noble.org/gene‐atlas/v2/). Treatment/control fold‐change values were calculated, converted into a log2 scale (see website online) and subjected to hierarchical cluster analysis (pvclust; Suzuki and Shimodaira, 2006). Clusters with approximately unbiased (AU) P value probability of more than 95% are indicated by the red rectangles. Functionally characterized genes are underlined in blue; their National Center for Biotechnology Information (NCBI) gene accession numbers and primary literature citations are available at http://bioinfo.noble.org/manuscript‐support/mpp/.
In soybean, eight of the 10 available CHS genes were induced either in response to cyst nematode infection (experiment accession: E‐MEXP‐808) or to Asian soybean rust (Fig. S5; accession: E‐TABM‐230) (van de Mortel et al., 2007). Many genes involved in isoflavonoid biosynthesis in soybean, including three CHRs, CHIs, two I2′Hs, seven IFRs, two IFSs and eight IOMTs, were induced (Fig. S5). Notably, the expression patterns of at least seven CHSs tightly correlated with those of isoflavone biosynthetic genes (P value less than 0.05, bottom subcluster in the dendrogram highlighted by the red rectangle in Fig. S5), suggesting their neofunctionalization via cis‐regulatory evolution into isoflavonoid biosynthesis. It should be noted, however, that a similar set of flavonoid genes is activated in the susceptible soybean genotype EM (Embrapa‐48) and in the Rpp2‐resistant genotype PI (PI230970) in response to fungal infection, suggesting the involvement of flavonoids in basal soybean defences (Fig. S5).
As in soybean, many flavonoid genes are induced in Medicago by fungal infection (Phymatotrichopsis omnivora) or elicitors [methyl jasmonate, which mimics wound signals, and yeast elicitor (YE), which mimics pathogen signals] (Fig. 7B). In contrast, only two of this set of genes, IFS‐Like (Medtr7g111290) and isoflavone 3′‐hydroxylase (I3′H, GB 33521519, CYP81E9; Liu et al., 2003a), were induced in response to colonization by a symbiotic mycorrhizal fungus. Twelve of the 17 available Medicago CHS genes were induced by either Phymatotrichopsis omnivora or YE (Fig. 7B). Expansion of the legume CHS gene family is directly related to the functional diversity of flavonoids, in this case the evolution of isoflavonoids as protective phytoalexins. Individual CHS genes in legumes often exhibit differential expression patterns that can be correlated with the types of flavonoid compounds being synthesized (Dixon and Paiva, 1995).
Future perspectives
Functional genomics approaches are powerful tools to facilitate the understanding of secondary metabolism in plants. Enzymes involved in the same function will be temporally and spatially coexpressed, and clustering analysis of transcript profiles, as described above, can be used as a discovery tool to define gene networks associated with plant defence responses. Metabolomics is fast developing as a powerful tool for the analysis of gene function and natural genetic variation (Dixon et al., 2006; Fridman and Pichersky, 2005; Morrell et al., 2006). Combining transcript and metabolite profiles (either global or targeted) is even more informative, and is becoming an important new approach for the identification of candidate genes in complex plant secondary metabolic pathways (Saito et al., 2008; Shulaev et al., 2008; 2007, 2008).
Genes encoding enzymes functioning in secondary metabolism are generally more divergent than those involved in primary metabolism. Although genes for most metabolic pathways in plants are not organized into gene clusters, a small but increasing number of ‘operon‐like’ gene clusters have been identified for the synthesis of plant defence compounds (Field and Osbourn, 2008; Frey et al., 1997; Qi et al., 2004; Shimura et al., 2007), although to date these are mainly in the area of terpenoid metabolism in monocots (oats and rice), and have yet to be seen for consecutive functional enzymes of flavonoid biosynthesis. Investigation of the genomic organization of metabolic pathways may, nevertheless, be a useful additional approach to ascribe potential function to candidate pathway genes, as well as to shed further light on the evolution of chemical diversification in plants.
As shown by the above examples, genomic approaches have huge potential to aid our understanding of how plants have elaborated various chemical defence mechanisms against pathogen attack during evolution. They do not, however, prove causality, which will still require careful biochemical, genetic and physiological analysis of the plant–pathogen interaction. It is a sobering thought that the almost exponential increase in genomic information since our earlier review is likely to accelerate over the next 5–10 years, and data handling and visualization may then become the major challenges to the biologist. The effort will be worth it, however, as being able to visualize plant–microbe interactions at the level of the whole genomes (and metabolomes) of both organisms will open up unprecedented opportunities to understand metabolic and regulatory cross‐talk and the evolution of defence mechanisms, and to develop novel antimicrobial strategies.
Supporting information
Database: http://bioinfo.noble.org/manuscript‐support/mpp/
Fig. S1 Alignment of the active site defined by Ferrer et al. (1999) of chalcone synthase (CHS)‐related enzymes from 11 species.
Fig. S2 Detailed phylogenetic tree of chalcone synthase (CHS)‐related genes.
Fig. S3 Phylogenetic analysis of flavonoid 2‐oxoglutarate‐iron‐dependent oxygenases.
Fig. S4 Hierarchical clustering analysis of expression patterns of flavonoid biosynthesis genes in microarray experiments involving exposure of rice to pathogens.
Fig. S5 Hierarchical clustering analysis of expression patterns of flavonoid biosynthesis genes in microarray experiments involving exposure of soybean to pathogens.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
REFERENCES
- Akashi, T. , Aoki, T. and Ayabe, S. (1998a) CYP81E1, a cytochrome P450 cDNA of licorice (Glycyrrhiza echinata L.), encodes isoflavone 2′‐hydroxylase. Biochem. Biophys. Res. Commun. 251, 67–70. [DOI] [PubMed] [Google Scholar]
- Akashi, T. , Aoki, T. and Ayabe, S. (1998b) Identification of a cytochrome P450 cDNA encoding (2S)‐flavanone 2‐hydroxylase of licorice (Glycyrrhiza echinata L.; Fabaceae) which represents licodione synthase and flavone synthase II. FEBS Lett. 431, 287–290. [DOI] [PubMed] [Google Scholar]
- Akashi, T. , Sawada, Y. , Shimada, N. , Sakurai, N. , Aoki, T. and Ayabe, S. (2003) cDNA cloning and biochemical characterization of S‐adenosyl‐L‐methionine:2,7,4′‐trihydroxyisoflavanone 4′‐O‐methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol. 44, 103–112. [DOI] [PubMed] [Google Scholar]
- Akashi, T. , Aoki, T. and Ayabe, S. (2005) Molecular and biochemical characterization of 2‐hydroxyisoflavanone dehydratase. Involvement of carboxylesterase‐like proteins in leguminous isoflavone biosynthesis. Plant Physiol. 137, 882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, T. , VanEtten, H.D. , Sawada, Y. , Wasmann, C.C. , Uchiyama, H. and Ayabe, S. (2006) Catalytic specificity of pea O‐methyltransferases suggests gene duplication for (+)‐pisatin biosynthesis. Phytochemistry, 67, 2525–2530. [DOI] [PubMed] [Google Scholar]
- Akashi, T. , Sasaki, K. , Aoki, T. , Ayabe, S. and Yazaki, K. (2009) Molecular cloning and characterization of a cDNA for pterocarpan 4‐dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol. 149, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, K. , Yamauchi, S. , Nakato, T. , Maruyama, M. , Sugahara, T. and Kishida, T. (2007) Antifungal activity of tetra‐substituted tetrahydrofuran lignan, (−)‐virgatusin, and its structure‐activity relationship. Biosci. Biotechnol. Biochem. 71, 1028–1035. [DOI] [PubMed] [Google Scholar]
- Alcaraz, L.E. , Blanco, S.E. , Puig, O.N. , Tomas, F. and Ferretti, F.H. (2000) Antibacterial activity of flavonoids against methicillin‐resistant Staphylococcus aureus strains. J. Theor. Biol. 205, 231–240. [DOI] [PubMed] [Google Scholar]
- Austin, M.B. and Noel, J.P. (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110. [DOI] [PubMed] [Google Scholar]
- Bechinger, C. , Giebel, K.F. , Schnell, M. , Leiderer, P. , Deising, H.B. and Bastmeyer, M. (1999) Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science, 285, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Bhuiyan, N.H. , Selvaraj, G. , Wei, Y. and King, J. (2009) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W. , Ralph, J. and Baucher, M. (2003) Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Bomati, E.K. , Austin, M.B. , Bowman, M.E. , Dixon, R.A. and Noel, J.P. (2005) Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J. Biol. Chem. 280, 30496–30503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpinella, M.C. , Ferrayoli, C.G. and Palacios, S.M. (2005) Antifungal synergistic effect of scopoletin, a hydroxycoumarin isolated from Melia azedarach L. fruits. J. Agric. Food Chem. 53, 2922–2927. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Meyermans, H. , Burggraeve, B. , De Rycke, R.M. , Inoue, K. , De Vleesschauwer, V. , Steenackers, M. , Van Montagu, M.C. , Engler, G.J. and Boerjan, W.A. (2000) Cell‐specific and conditional expression of caffeoyl‐coenzyme A‐3‐O‐methyltransferase in poplar. Plant Physiol. 123, 853–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J. , Baltz, R. , Schmitt, C. , Beffa, R. , Fritig, B. and Saindrenan, P. (2002) Downregulation of a pathogen‐responsive tobacco UDP‐Glc: phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell, 14, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushnie, T.P.T. and Lamb, A.J. (2005) Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents, 26, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. , Dietrich, R.A. and Richberg, M.H. (1996) Death don't have no mercy: cell death programs in plant‐microbe interactions. Plant Cell, 8, 1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin, L.B. , Wang, H.B. , Crowell, A.L. , Bedgar, D.L. , Martin, D.M. , Sarkanen, S. and Lewis, N.G. (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science, 275, 362–366. [DOI] [PubMed] [Google Scholar]
- Davin, L.B. , Jourdes, M. , Patten, A.M. , Kim, K.‐W. , Vassaõ, D.G. and Lewis, N.G. (2008) Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 25, 1015–1090. [DOI] [PubMed] [Google Scholar]
- Deavours, B.E. , Liu, C.J. , Naoumkina, M.A. , Tang, Y. , Farag, M.A. , Sumner, L.W. , Noel, J.P. and Dixon, R.A. (2006) Functional analysis of members of the isoflavone and isoflavanone O‐methyltransferase enzyme families from the model legume Medicago truncatula . Plant Mol. Biol. 62, 715–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCenzo, G.L. and VanEtten, H.D. (2006) Studies on the late steps of (+) pisatin biosynthesis: evidence for (–) enantiomeric intermediates. Phytochemistry, 67, 675–683. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A. (1999) Isoflavonoids: biochemistry, molecular biology and biological functions In: Comprehensive Natural Products Chemistry (Sankawa U., ed.), pp. 773–823. Elsevier. [Google Scholar]
- Dixon, R.A. and Paiva, N.L. (1995) Stress‐induced phenylpropanoid metabolism. Plant Cell, 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A. , Achnine, L. , Kota, P. , Liu, C.J. , Reddy, M.S.S. and Wang, L. (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 3, 371–390. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A. , Gang, D.R. , Charlton, A.J. , Fiehn, O. , Kuiper, H.A. , Reynolds, T.L. , Tjeerdema, R.S. , Jeffrey, E.H. , Germans, J.B. , Ridley, W.P. and Seiber, J.N. (2006) Applications of metabolomics to agriculture. J. Agric. Food Chem. 54, 8984–8994. [DOI] [PubMed] [Google Scholar]
- Drummond, A.J. , Ashton, B. , Cheung, M. , Heled, J. , Kearse, M. , Moir, R. , Stones‐Havas, S. , Thiererm, T. and Wilson, A. (2009) Geneious v4.7. http://www.geneious.com[accessed on Jan 15, 2010].
- Elfstrand, M. , Sitbon, F. , Lapierre, C. , Bottin, A. and Von Arnold, S. (2002) Altered lignin structure and resistance to pathogens in spi 2‐expressing tobacco plants. Planta, 214, 708–716. [DOI] [PubMed] [Google Scholar]
- Engelhardt, S. , Lee, J. , Gabler, Y. , Kemmerling, B. , Haapalainen, M.L. , Li, C.M. , Wei, Z. , Keller, H. , Joosten, M. , Taira, S. and Nurnberger, T. (2009) Separable roles of the Pseudomonas syringae pv. phaseolicola accessory protein HrpZ1 in ion‐conducting pore formation and activation of plant immunity. Plant J. 57, 706–717. [DOI] [PubMed] [Google Scholar]
- Fellbrich, G. , Romanski, A. , Varet, A. , Blume, B. , Brunner, F. , Engelhardt, S. , Felix, G. , Kemmerling, B. , Krzymowska, M. and Nurnberger, T. (2002) NPP1, a Phytophthora‐associated trigger of plant defense in parsley and Arabidopsis. Plant J. 32, 375–390. [DOI] [PubMed] [Google Scholar]
- Ferrer, J.L. , Jez, J.M. , Bowman, M.E. , Dixon, R.A. and Noel, J.P. (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6, 775–784. [DOI] [PubMed] [Google Scholar]
- Field, B. and Osbourn, A.E. (2008) Metabolic diversification—independent assembly of operon‐like gene clusters in different plants. Science, 320, 543–547. [DOI] [PubMed] [Google Scholar]
- Fischbach, M.A. and Walsh, C.T. (2006) Assembly‐line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496. [DOI] [PubMed] [Google Scholar]
- Ford, J.D. , Huang, K.S. , Wang, H.B. , Davin, L.B. and Lewis, N.G. (2001) Biosynthetic pathway to the cancer chemopreventive secoisolariciresinol diglucoside‐hydroxymethyl glutaryl ester‐linked lignan oligomers in flax (Linum usitatissimum) seed. J. Nat. Prod. 64, 1388–1397. [DOI] [PubMed] [Google Scholar]
- Frey, M. , Chomet, P. , Glawischnig, E. , Stettner, C. , Grun, S. , Winklmair, A. , Eisenreich, W. , Bacher, A. , Meeley, R.B. , Briggs, S.P. , Simcox, K. and Gierl, A. (1997) Analysis of a chemical plant defense mechanism in grasses. Science, 277, 696–699. [DOI] [PubMed] [Google Scholar]
- Fridman, E. and Pichersky, E. (2005) Metabolomics, genomics, proteomics, and the identification of enzymes and their substrates and products. Curr. Opin. Plant Biol. 8, 242–248. [DOI] [PubMed] [Google Scholar]
- Funa, N. , Awakawa, T. and Horinouchi, S. (2007) Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa . J. Biol. Chem. 282, 14476–14481. [DOI] [PubMed] [Google Scholar]
- Funnell, D.L. and Pedersen, J.F. (2006) Reaction of sorghum lines genetically modified for reduced lignin content to infection by Fusarium and Alternaria spp. Plant Dis. 90, 331–338. [DOI] [PubMed] [Google Scholar]
- Funnell, D.L. , Matthews, P.S. and VanEtten, H.D. (2002) Identification of new pisatin demethylase genes (PDA5 and PDA7) in Nectria haematococca and non‐Mendelian segregation of pisatin demethylating ability and virulence on pea due to loss of chromosomal elements. Fungal Genet. Biol. 37, 121–133. [DOI] [PubMed] [Google Scholar]
- Gabaldón, C. , López‐Serrano, M. , Pomar, F. , Merino, F. , Cuello, J. , Pedreno, M.A. and Barceló, A.R. (2006) Characterization of the last step of lignin biosynthesis in Zinnia elegans suspension cell cultures. FEBS Lett. 580, 4311–4316. [DOI] [PubMed] [Google Scholar]
- Gachon, C. , Baltz, R. and Saindrenan, P. (2004) Over‐expression of a scopoletin glucosyltransferase in Nicotiana tabacum leads to precocious lesion formation during the hypersensitive response to tobacco mosaic virus but does not affect virus resistance. Plant Mol. Biol. 54, 137–146. [DOI] [PubMed] [Google Scholar]
- Galeotti, F. , Barile, E. , Curir, P. , Dolci, M. and Lanzotti, V. (2008) Flavonoids from carnation (Dianthus caryophyllus) and their antifungal activity. Phytochem. Lett. 1, 44–48. [Google Scholar]
- Gang, D.R. , Kasahara, H. , Xia, Z.Q. , Vander Mijnsbrugge, K. , Bauw, G. , Boerjan, W. , Van Montagu, M. , Davin, L.B. and Lewis, N.G. (1999) Evolution of plant defense mechanisms. Relationships of phenylcoumaran benzylic ether reductases to pinoresinol‐lariciresinol and isoflavone reductases. J. Biol. Chem. 274, 7516–7527. [DOI] [PubMed] [Google Scholar]
- Guo, L. , Dixon, R.A. and Paiva, N.L. (1994) Conversion of vestitone to medicarpin in alfalfa (Medicago sativa L.) is catalyzed by two independent enzymes. J. Biol. Chem. 269, 22372–22378. [PubMed] [Google Scholar]
- Hakamatsuka, T. , Mori, K. , Ishida, S. , Ebizuka, Y. and Sankawa, U. (1998) Purification of 2‐hydroxyisoflavanone dehydratase from the cell cultures of Pueraria lobata . Phytochemistry, 49, 497–505. [Google Scholar]
- Hano, C. , Martin, I. , Fliniaux, O. , Legrand, B. , Gutierrez, L. , Arroo, R.R.J. , Mesnard, F. , Lamblin, F. and Lainé, E. (2006) Pinoresinol‐lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta, 224, 1291–1301. [DOI] [PubMed] [Google Scholar]
- Hatton, D. , Sablowski, R. , Yung, M.H. , Smith, C. , Schuch, W. and Bevan, M. (1995) Two classes of cis sequences contribute to tissue‐specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 7, 859–876. [DOI] [PubMed] [Google Scholar]
- He, X.Z. , Wang, X. and Dixon, R.A. (2006) Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation. J. Biol. Chem. 281, 34441–34447. [DOI] [PubMed] [Google Scholar]
- Von Heimendahl, C.B.I. , Schäfer, K.M. , Eklund, P. , Sjöholm, R. , Schmidt, T.J. and Fuss, E. (2005) Pinoresinol–lariciresinol reductases with different stereospecificity from Linum album and Linum usitatissimum . Phytochemistry, 66, 1254–1263. [DOI] [PubMed] [Google Scholar]
- Hemmati, S. , Schmidt, T.J. and Fuss, E. (2007) (+)‐Pinoresinol/(–)‐lariciresinol reductase from Linum perenne Himmelszelt involved in the biosynthesis of justicidin B. FEBS Lett. 581, 603–610. [DOI] [PubMed] [Google Scholar]
- Hinderer, W. , Flentje, U. and Barz, W. (1987) Microsomal isoflavone 2′‐ and 3′‐hydroxylases from chickpea (Cicer arietinum L.) cell suspensions induced for pterocarpan phytoalexin formation. FEBS Lett. 214, 101–106. [Google Scholar]
- Jez, J.M. , Bowman, M.E. and Noel, J.P. (2002) Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc. Natl. Acad. Sci. USA, 99, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C. , Schommer, C.K. , Kim, S.Y. and Suh, D.Y. (2006) Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens . Phytochemistry, 67, 2531–2540. [DOI] [PubMed] [Google Scholar]
- Kai, K. , Shimizu, B. , Mizutani, M. , Watanabe, K. and Sakata, K. (2006) Accumulation of coumarins in Arabidopsis thaliana . Phytochemistry, 67, 379–386. [DOI] [PubMed] [Google Scholar]
- Kapushesky, M. , Kemmeren, P. , Culhane, A.C. , Durinck, S. , Ihmels, J. , Korner, C. , Kull, M. , Torrente, A. , Sarkans, U. , Vilo, J. and Brazma, A. (2004) Expression Profiler: next generation—an online platform for analysis of microarray data. Nucleic Acids Res. 32, W465–W470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. and Toh, H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T. , Koita, H. , Nakatsubo, T. , Hasegawa, K. , Wakabayashi, K. , Takahashi, H. , Umemura, K. , Umezawa, T. and Shimamoto, K. (2006) Cinnamoyl‐CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA, 103, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. , Tan, R. , Mariscal, A.G. and Straney, D. (2003) A binuclear zinc transcription factor binds the host isoflavonoid‐responsive element in a fungal cytochrome P450 gene responsible for detoxification. Mol. Microbiol. 49, 117–130. [DOI] [PubMed] [Google Scholar]
- Kim, M.K. , Jeon, J.H. , Fujita, M. , Davin, L.B. and Lewis, N.G. (2002) The western red cedar (Thuja plicata) 8‐8 DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol. Biol. 49, 199–214. [DOI] [PubMed] [Google Scholar]
- Koes, R.E. , Quattrocchio, R. and Mol, J.N.M. (1994) The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays, 16, 123–132. [Google Scholar]
- Koutaniemi, S. , Warinowski, T. , Karkonen, A. , Alatalo, E. , Fossdal, C.G. , Saranpaa, P. , Laakso, T. , Fagerstedt, K.V. , Simola, L.K. , Paulin, L. , Rudd, S. and Teeri, T.H. (2007) Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real‐time RT‐PCR. Plant Mol. Biol. 65, 311–328. [DOI] [PubMed] [Google Scholar]
- Lange, B.M. , Lapierre, C. and Sandermann, H., Jr (1995) Elicitor‐induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol. 108, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat, V. , Lacomme, C. , Lacombe, E. , Lasserre, E. , Roby, D. and Grima‐Pettenati, J. (2001) Two cinnamoyl‐CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry, 57, 1187–1195. [DOI] [PubMed] [Google Scholar]
- Lauvergeat, V. , Rech, P. , Jauneau, A. , Guez, C. , Coutos‐Thevenot, P. and Grima‐Pettenati, J. (2002) The vascular expression pattern directed by the Eucalyptus gunnii cinnamyl alcohol dehydrogenase EgCAD2 promoter is conserved among woody and herbaceous plant species. Plant Mol. Biol. 50, 497–509. [DOI] [PubMed] [Google Scholar]
- Li, L. , Modolo, L.V. , Escamilla‐Trevino, L.L. , Achnine, L. , Dixon, R.A. and Wang, X. (2007) Crystal structure of Medicago truncatula UGT85H2—insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferase. J. Mol. Biol. 370, 951–963. [DOI] [PubMed] [Google Scholar]
- Li, M. and Xu, Z. (2008) Quercetin in a lotus leaves extract may be responsible for antibacterial activity. Arch. Pharm. Res. 31, 640–644. [DOI] [PubMed] [Google Scholar]
- Liu, C.‐J. , Blount, J.W. , Steele, C.L. and Dixon, R.A. (2002) Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis . Proc. Natl. Acad. Sci. USA, 99, 14578–14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.J. , Huhman, D. , Sumner, L.W. and Dixon, R.A. (2003a) Regiospecific hydroxylation of isoflavones by cytochrome P450 81E enzymes from Medicago truncatula . Plant J. 36, 471–484. [DOI] [PubMed] [Google Scholar]
- Liu, C.J. , Deavours, B.E. , Richard, S.B. , Ferrer, J.L. , Blount, J.W. , Huhman, D. , Dixon, R.A. and Noel, J.P. (2006) Structural basis for dual functionality of isoflavonoid O‐methyltransferases in the evolution of plant defense responses. Plant Cell, 18, 3656–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G.Y. (2009) Isolation, sequence identification and tissue expression profile of two novel soybean (Glycine max) genes—vestitone reductase and chalcone reductase. Mol. Biol. Rep. 36, 1991–1994. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Inlow, M. and VanEtten, H. (2003b) Expression profiles of pea pathogenicity (PEP) genes in vivo and in vitro, characterization of the flanking regions of the PEP cluster and evidence that the PEP cluster region resulted from horizontal gene transfer in the fungal pathogen Nectria haematococca . Curr. Genet. 44, 95–103. [DOI] [PubMed] [Google Scholar]
- Markham, K.R. (1988) Distribution of flavonoids in the lower plants and its evolutionary significance In: The Flavonoids. Advances in Research Since 1980 (Harborne J.B., ed.), pp. 427–468. London: Chapman and Hall. [Google Scholar]
- Martens, S. and Forkmann, G. (1999) Cloning and expression of flavone synthase II from Gerbera hybrids. Plant J. 20, 611–618. [DOI] [PubMed] [Google Scholar]
- Matros, A. and Mock, H.P. (2004) Ectopic expression of a UDP‐glucose:phenylpropanoid glucosyltransferase leads to increased resistance of transgenic tobacco plants against infection with potato virus Y. Plant Cell Physiol. 45, 1185. [DOI] [PubMed] [Google Scholar]
- Menden, B. , Kohlhoff, M. and Moerschbacher, B.M. (2007) Wheat cells accumulate a syringyl‐rich lignin during the hypersensitive resistance response. Phytochemistry, 68, 513–520. [DOI] [PubMed] [Google Scholar]
- Mitchell, H.J. , Hall, S.A. , Stratford, R. , Hall, J.L. and Barber, M.S. (1999) Differential induction of cinnamyl alcohol dehydrogenase during defensive lignification in wheat (Triticum aestivum L.): characterisation of the major inducible form. Planta, 208, 31–37. [Google Scholar]
- Mitsuda, N. , Iwase, A. , Yamamoto, H. , Yoshida, M. , Seki, M. , Shinozaki, K. and Ohme‐Takagi, M. (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell, 19, 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolo, L.V. , Blount, J.W. , Achnine, L. , Naoumkina, M.A. , Wang, X. and Dixon, R.A. (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula . Plant Mol. Biol. 64, 499–518. [DOI] [PubMed] [Google Scholar]
- Moerschbacher, B.M. , Noll, U. , Gorrichon, L. and Reisener, H.J. (1990) Specific inhibition of lignification breaks hypersensitive resistance of wheat to stem rust. Plant Physiol. 93, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell, K. , Goeminne, G. , Storme, V. , Sterck, L. , Ralph, J. , Coppieters, W. , Breyne, P. , Steenackers, M. , Georges, M. , Messens, E. and Boerjan, W. (2006) Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J. 47, 224–237. [DOI] [PubMed] [Google Scholar]
- Van De Mortel, M. , Recknor, J.C. , Graham, M.A. , Nettleton, D. , Dittman, J.D. , Nelson, R.T. , Godoy, C.V. , Abdelnoor, R.V. , Almeida, A.M. , Baum, T.J. and Whitham, S.A. (2007) Distinct biphasic mRNA changes in response to Asian soybean rust infection. Mol. Plant–Microbe Interact. 20, 887–899. [DOI] [PubMed] [Google Scholar]
- Nair, R.B. , Bastress, K.L. , Ruegger, M.O. , Denault, J.W. and Chapple, C. (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell, 16, 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsubo, T. , Mizutani, M. , Suzuki, S. , Hattori, T. and Umezawa, T. (2008) Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J. Biol. Chem. 283, 15550–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel, J.P. , Dixon, R.A. , Pichersky, E. , Zubieta, C. and Ferrer, J.L. (2003) Structural, functional, and evolutionary basis for methylation of plant small molecules. Rec. Adv. Phytochem. 37, 37–58. [Google Scholar]
- Noel, J.P. , Austin, M.B. and Bomati, E.K. (2005) Structure–function relationships in plant phenylpropanoid biosynthesis. Curr. Opin. Plant Biol. 8, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, A. , Saito, A. , Homma, Y. , Nakao, M. , Sasaki, N. , Nishino, T. , Takahashi, S. and Nakayama, T. (2007) A UDP‐glucose:isoflavone 7‐O‐glucosyltransferase from the roots of soybean (Glycine max) seedlings. Purification, gene cloning, phylogenetics, and an implication for an alternative strategy of enzyme catalysis. J. Biol. Chem. 282, 23581–23590. [DOI] [PubMed] [Google Scholar]
- Paiva, N.L. , Edwards, R. , Sun, Y. , Hrazdina, G. and Dixon, R.A. (1991) Stress responses in alfalfa (Medicago sativa L.) XI. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol. Biol. 17, 653–667. [DOI] [PubMed] [Google Scholar]
- Plaper, A. , Golob, M. , Hafner, I. , Oblak, M. , Solmajer, T. and Jerala, R. (2003) Characterization of quercetin binding site on DNA gyrase. Biochem. Biophys. Res. Commun. 306, 530–536. [DOI] [PubMed] [Google Scholar]
- Qi, X. , Bakht, S. , Leggett, M. , Maxwell, C. , Melton, R. and Osbourn, A. (2004) A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA, 101, 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin, M. , Allasia, V. , Pegard, A. , Allais, F. , Ducrot, P.H. , Favery, B. , Levis, C. , Martinet, S. , Masur, C. , Ponchet, M. , Roby, D. , Schlaich, N.L. , Jouanin, L. and Keller, H. (2009) Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathol. 5, e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes, J. , Rohde, A. , Christensen, J.H. , Van de Peer, Y. and Boerjan, W. (2003) Genome‐wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, J. , Brunow, G. , Harris, P. , Dixon, R.A. and Boerjan, W. (2008) Lignification: are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? Rec. Adv. Polyphenols Res. 1, 36–66. [Google Scholar]
- Ralph, S. , Park, J.Y. , Bohlmann, J. and Mansfield, S.D. (2006) Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound‐ and insect‐induced expression of a family of DIR and DIR‐like genes in spruce (Picea spp.). Plant Mol. Biol. 60, 21–40. [DOI] [PubMed] [Google Scholar]
- Ritter, H. and Schulz, G.E. (2004) Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia‐lyase. Plant Cell, 16, 3426–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, E.E. , Glazebrook, J. and Ausubel, F.M. (1996) Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis–pathogen interactions. Mol. Plant–Microbe Interact. 9, 748–757. [DOI] [PubMed] [Google Scholar]
- Saito, K. , Hirai, M.Y. and Yonekura‐Sakakibara, K. (2008) Decoding genes with coexpression networks and metabolomics—‘majority report by precogs'. Trends Plant Sci. 13, 36–43. [DOI] [PubMed] [Google Scholar]
- Sasaki, K. , Mito, K. , Ohara, K. , Yamamoto, H. and Yazaki, K. (2008) Cloning and characterization of naringenin 8‐prenyltransferase, a flavonoid‐specific prenyltransferase of Sophora flavescens . Plant Physiol. 146, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y. , Demura, T. , Yamawaki, K. , Inoue, Y. , Sato, S. , Sugiyama, M. and Fukuda, H. (2006) Isolation and characterization of a novel peroxidase gene ZPO‐C whose expression and function are closely associated with lignification during tracheary element differentiation. Plant Cell Physiol. 47, 493–503. [DOI] [PubMed] [Google Scholar]
- Sawada, Y. and Ayabe, S. (2005) Multiple mutagenesis of P450 isoflavonoid synthase reveals a key active‐site residue. Biochem. Biophys. Res. Commun. 330, 907–913. [DOI] [PubMed] [Google Scholar]
- Sawada, Y. , Kinoshita, K. , Akashi, T. , Aoki, T. and Ayabe, S. (2002) Key amino acid residues required for aryl migration catalysed by the cytochrome P450 2‐hydroxyisoflavanone synthase. Plant J. 31, 555–564. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R. , Kochs, G. , Lottspeich, F. and Ebel, J. (1998) Molecular characterization and functional expression of dihydroxypterocarpan 6a‐hydroxylase, an enzyme specific for pterocarpanoid phytoalexin biosynthesis in soybean (Glycine max L.). FEBS Lett. 432, 182–186. [DOI] [PubMed] [Google Scholar]
- Seshime, Y. , Juvvadi, P.R. , Fujii, I. and Kitamoto, K. (2005) Discovery of a novel superfamily of type III polyketide synthases in Aspergillus oryzae . Biochem. Biophys. Res. Commun. 331, 253–260. [DOI] [PubMed] [Google Scholar]
- Shao, H. , He, X. , Achnine, L. , Blount, J.W. , Dixon, R.A. and Wang, X. (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula . Plant Cell, 17, 3141–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, H. , Dixon, R.A. and Wang, X. (2007) Crystal structure of vestitone reductase from alfalfa (Medicago sativa L.). J. Mol. Biol. 369, 265–276. [DOI] [PubMed] [Google Scholar]
- Shimada, N. , Aoki, T. , Sato, S. , Nakamura, Y. , Tabata, S. and Ayabe, S. (2003) A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume‐specific 5‐deoxy(iso)flavonoids in Lotus japonicus . Plant Physiol. 131, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura, M. , Akashi, T. , Sakurai, N. , Suzuki, H. , Saito, K. , Shibata, D. , Ayabe, S. and Aoki, T. (2007) 2‐Hydroxyisoflavanone dehydratase is a critical determinant of isoflavone productivity in hairy root cultures of Lotus japonicus . Plant Cell Physiol. 48, 1652–1657. [DOI] [PubMed] [Google Scholar]
- Shimizu, B. , Miyagawa, H. , Ueno, T. , Sakata, K. , Watanabe, K. and Ogawa, K. (2005) Morning glory systemically accumulates scopoletin and scopolin after interaction with Fusarium oxysporum . Z. Naturforsch. C, 60, 83–90. [DOI] [PubMed] [Google Scholar]
- Shimura, K. , Okada, A. , Okada, K. , Jikumaru, Y. , Ko, K.W. , Toyomasu, T. , Sassa, T. , Hasegawa, M. , Kodama, O. , Shibuya, N. , Koga, J. , Nojiri, H. and Yamane, H. (2007) Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018. [DOI] [PubMed] [Google Scholar]
- Shirley, B.W. (1996) Flavonoid biosynthesis: ‘new’ functions for an ‘old’ pathway. Trends Plant Sci. 1, 377–382. [Google Scholar]
- Shirley, B.W. , Kubasek, W.L. , Storz, G. , Bruggemann, E. , Koornneef, M. , Ausubel, F.M. and Goodman, H.M. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671. [DOI] [PubMed] [Google Scholar]
- Shulaev, V. , Cortes, D. , Miller, G. and Mittler, R. (2008) Metabolomics for plant stress response. Physiol. Plant. 132, 199–208. [DOI] [PubMed] [Google Scholar]
- Sohn, H.Y. , Son, K.H. , Kwon, C.S. , Kwon, G.S. and Kang, S.S. (2004) Antimicrobial and cytotoxic activity of 18 prenylated flavonoids isolated from medicinal plants: Morus alba L., Morus mongolica Schneider, Broussnetia papyrifera L. Vent, Sophora flavescens Ait and Echinosophora koreensis Nakai. Phytomedicine, 11, 666–672. [DOI] [PubMed] [Google Scholar]
- Stafford, H.A. (1991) Flavonoid evolution: an enzymic approach. Plant Physiol. 96, 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, R. and Shimodaira, H. (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics, 22, 1540–1542. [DOI] [PubMed] [Google Scholar]
- Tahara, S. (2007) A journey of twenty‐five years through the ecological biochemistry of flavonoids. Biosci. Biotechnol. Biochem. 71, 1387–1404. [DOI] [PubMed] [Google Scholar]
- Tamba, Y. , Ohba, S. , Kubota, M. , Yoshioka, H. and Yamazaki, M. (2007) Single GUV method reveals interaction of tea catechin (–)‐epigallocatechin gallate with lipid membranes. Biophys. J. 92, 3178–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognolli, M. , Penel, C. , Greppin, H. and Simon, P. (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana . Gene, 288, 129–138. [DOI] [PubMed] [Google Scholar]
- Tropf, S. , Lanz, T. , Rensing, S.A. , Schroder, J. and Schroder, G. (1994) Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J. Mol. Evol. 38, 610–618. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, H. and Iinuma, M. (2000) Reduction of membrane fluidity by antibacterial sophoraflavanone G isolated from Sophora exigua . Phytomedicine, 7, 161–165. [DOI] [PubMed] [Google Scholar]
- Turnbull, J.J. , Nakajima, J. , Welford, R.W. , Yamazaki, M. , Saito, K. and Schofield, C.J. (2004) Mechanistic studies on three 2‐oxoglutarate‐dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3beta‐hydroxylase. J. Biol. Chem. 279, 1206–1216. [DOI] [PubMed] [Google Scholar]
- Umezawa, T. (2003) Diversity in lignan biosynthesis. Phytochem. Rev. 2, 371–390. [Google Scholar]
- VanEtten, H.D. , Mansfield, J.W. , Bailey, J.A. and Farmer, E.E. (1994) Two classes of plant antibiotics: phytoalexins versus ‘phytoanticipins’. Plant Cell, 6, 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , He, X. , Lin, J. , Shao, H. , Chang, Z. and Dixon, R.A. (2006) Crystal structure of isoflavone reductase from alfalfa (Medicago sativa L.). J. Mol. Biol. 358, 1341–1352. [DOI] [PubMed] [Google Scholar]
- Wu, Q. and VanEtten, H.D. (2004) Introduction of plant and fungal genes into pea (Pisum sativum L.) hairy roots reduces their ability to produce pisatin and affects their response to a fungal pathogen. Mol. Plant–Microbe Interact. 17, 798–804. [DOI] [PubMed] [Google Scholar]
- Yonekura‐Sakakibara, K. , Tohge, T. , Niida, R. and Saito, K. (2007) Identification of a flavonol 7‐O‐rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282, 14932–14941. [DOI] [PubMed] [Google Scholar]
- Yonekura‐Sakakibara, K. , Tohge, T. , Matsuda, F. , Nakabayashi, R. , Takayama, H. , Niida, R. , Watanabe‐Takahashi, A. , Inoue, E. and Saito, K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. Plant Cell, 20, 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. , Richardson, E.A. and Ye, Z.H. (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell, 19, 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. , Lee, C. , Zhou, J. , McCarthy, R.L. and Ye, Z.H. (2008) A battery of transcription factors Involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell, 20, 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Lee, C. , Zhong, R. and Ye, Z.H. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell, 21, 248–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Database: http://bioinfo.noble.org/manuscript‐support/mpp/
Fig. S1 Alignment of the active site defined by Ferrer et al. (1999) of chalcone synthase (CHS)‐related enzymes from 11 species.
Fig. S2 Detailed phylogenetic tree of chalcone synthase (CHS)‐related genes.
Fig. S3 Phylogenetic analysis of flavonoid 2‐oxoglutarate‐iron‐dependent oxygenases.
Fig. S4 Hierarchical clustering analysis of expression patterns of flavonoid biosynthesis genes in microarray experiments involving exposure of rice to pathogens.
Fig. S5 Hierarchical clustering analysis of expression patterns of flavonoid biosynthesis genes in microarray experiments involving exposure of soybean to pathogens.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


