SUMMARY
The hemi‐biotrophic fungus Venturia inaequalis infects members of the Maloideae, causing the economically important apple disease, scab. The plant–pathogen interaction of Malus and V. inaequalis follows the gene‐for‐gene model. cDNA libraries were constructed, and bioinformatic analysis of the resulting expressed sequence tags (ESTs) was used to characterize potential effector genes. Effectors are small proteins, secreted in planta, that are assumed to facilitate infection. Therefore, a cDNA library was constructed from a compatible interaction. To distinguish pathogen from plant sequences, the library was probed with genomic DNA from V. inaequalis to enrich for pathogen genes, and cDNA libraries were constructed from in vitro‐grown material. A suppression subtractive hybridization library enriched for cellophane‐induced genes was included, as growth on cellophane may mimic that in planta, with the differentiation of structures resembling those formed during plant colonization. Clustering of ESTs from the in planta and in vitro libraries indicated a fungal origin of the resulting non‐redundant sequence. A total of 937 ESTs was classified as putatively fungal, which could be assembled into 633 non‐redundant sequences. Sixteen new candidate effector genes were identified from V. inaequalis based on features common to characterized effector genes from filamentous fungi, i.e. they encode a small, novel, cysteine‐rich protein, with a putative signal peptide. Three of the 16 candidates, in particular, conformed to most of the protein structural characteristics expected of fungal effectors and showed significant levels of transcriptional up‐regulation during in planta growth. In addition to candidate effector genes, this collection of ESTs represents a valuable genomic resource for V. inaequalis.
INTRODUCTION
Venturia inaequalis Cooke (Wint.) is a hemi‐biotrophic fungus that causes disease in members of the Maloideae (Biggs, 1990), the most economically important of which is the apple disease scab or black spot (MacHardy, 1996). The genetic basis of the plant–pathogen interaction between Malus and V. inaequalis follows the gene‐for‐gene model: eight races of the pathogen have been defined by incompatibility, determined by avirulence genes (avr genes), on corresponding host cultivars, each carrying a major resistance gene (R gene), in most cases derived from crab apple varieties (Bénaouf and Parisi, 2000; Bus et al., 2005a,b; Lespinasse, 1994; Parisi and Lespinasse, 1996; Parisi et al., 1993; Roberts and Crute, 1994). Thus, in common with other plant pathogens, this suggests that V. inaequalis produces effector proteins, some of which have been commandeered by the plant for pathogen recognition purposes and initiation of defence.
Effector proteins facilitate infection, often by the suppression of host defence (Chisholm et al., 2006; Jones and Dangl, 2006). In bacterial and oomycete plant–pathogen systems, effector‐mediated suppression of defence has been demonstrated (Bos et al., 2006; DebRoy et al., 2004; Dou et al., 2008; Fu et al., 2007; Nomura et al., 2006). In the majority of filamentous fungal systems, suppression of host resistance has yet to be proven. However, the barley powdery mildew effectors AVRa10 and AVRK1 may operate as suppressors as, in susceptible barley cultivars, they increase the number of successful infection sites (Ridout et al., 2006). Moreover, a small secreted protein from the anthracnose fungus Colletotrichum gloeosporioides, which is essential for pathogenicity on Stylosanthes, appears to act during the initial biotrophic phase of infection to suppress host cell death (Stephenson et al., 2000). In addition, the Cladosporium fulvum effector Avr4 is required for full virulence on tomato, and is believed to be involved in counter‐defence, protecting against host chitinases (van Esse et al., 2007). Houterman et al. (2008) recently reported the suppression of R‐gene‐based resistance by an effector from Fusarium oxysporum f. sp. lycopersici. Effector genes, including those with a concomitant avirulence function, have yet to be cloned from V. inaequalis.
During infection, V. inaequalis penetrates the cuticle and forms infection hyphae that ramify above the epidermal cells, from which stromata form. Stromata are pseudo‐parenchymatous structures made up of laterally dividing cells, markedly different to the tubular hyphae found in vitro (Kucheryava et al., 2008). Stromata are presumed to be required in order to attain nutrients from the subcuticular space, and give rise to conidia that are disseminated through the ruptured cuticle to further the infection cycle. As no penetration of plant cells occurs, effectors are presumably secreted from the hyphae and stromata into the subcuticular space, where they may exert their effect extracellularly, or may be taken up by the plant cell by an unknown mechanism.
In the V. inaequalis–apple pathosystem, it is hypothesized that the plant has evolved to recognize a subset of these effectors (avr gene products) either directly or indirectly by R gene products, which trigger a hypersensitive response (HR). It is possible that recognition may take place extracellularly, as in the C. fulvum–tomato interaction (Kruijt et al., 2005). Alternatively, interaction of effector and R gene product may occur intracellularly, as secreted fungal proteins have been shown to be taken up by plant cells in some pathosystems (Catanzariti et al., 2006; Dodds et al., 2004; Kemen et al., 2005; Ridout et al., 2006). The former scenario is supported by the finding that cloned apple R genes (forming a cluster at the Vf locus), which confer resistance to V. inaequalis (Vinatzer et al., 2001; Xu and Korban, 2002), all belong to the same class of R gene as the Cf genes from tomato (Dangl and Jones, 2001). Given this similarity and the parallels in parasitic strategy adopted by each pathogen, an extracellular site of action seems most plausible.
The isolation of proteinaceous effectors from the subcuticular space has proved problematic (Fitzgerald, 2004; Win, 2003). Gau et al. (2004) were able to isolate apple pathogenesis‐related (PR), but not V. inaequalis, proteins from the apoplastic fluid of infected leaves. A map‐based cloning strategy to clone effectors has been adopted by Broggini et al. (2007), and has resulted in the isolation of a 330‐kb contig comprising 12 bacterial artificial chromosomes containing the VirQ5 marker for the avr gene AvrRvi1. However, the avr gene itself has yet to be identified.
An alternative approach for the identification of effector candidate genes involves the construction of expressed sequence tag (EST) libraries followed by bioinformatic analysis. This approach has proven to be valuable in the study of organisms where very little sequence data are available for either host or pathogen (Cramer et al., 2006).
The identification of effector genes by bioinformatic analysis of ESTs can be facilitated by the presence of conserved motifs common to effectors. For example, the RxLR motif of oomycete pathogens is involved in the delivery of effectors to the host cell (Bhattacharjee et al., 2006; Jiang et al., 2008; Rehmany et al., 2005; Vleeshouwers et al., 2008; Whisson et al., 2007), and can be used to identify novel candidate effectors (Torto‐Alalibo et al., 2007). In contrast with the oomycetes, filamentous fungal effector proteins identified to date share little homology with one another, or with sequences present in the public domain, although they do share some common characteristics. They are usually small: less than 400 amino acids in length when mature. They may be cysteine rich, which may aid protein stability in the extracellular environment by disulphide bond formation (Rep, 2005; Templeton et al., 1994). They typically possess an N‐terminal signal peptide for secretion, although there are exceptions, including AVRa10 and AVRK1 from Blumeria graminis f. sp. hordei (Ridout et al., 2006).
In this study, cDNA libraries were constructed from infected leaf material and from in vitro‐grown V. inaequalis, the latter to aid in origin assignment for sequences from the libraries derived from infected leaf material. The in vitro‐grown libraries include a suppression subtractive hybridization (SSH; Diatchenko et al., 1996) library enriched for cellophane‐induced genes, as V. inaequalis produces structures in cellophane morphologically similar to those found in planta, suggesting that growth in cellophane may be an in vitro model of infection by V. inaequalis (Kucheryava et al., 2008). Indeed, two cellophane‐induced genes have been shown to be up‐regulated in planta (Kucheryava et al., 2008). The SSH library was included to circumvent the common problem of a scarcity of pathogen sequences being identified when sequencing ESTs from infected plant material (Cramer et al., 2006). An SSH strategy has previously been proven to be invaluable for the study of fungal–plant interactions (van den Berg et al., 2007; Cramer et al., 2006; Yakovlev et al., 2008). The dataset reported herein comprises 2090 tentative non‐redundant sequences following clustering from 4215 ESTs. It represents the first major analysis of ESTs from the V. inaequalis–apple interaction, which identifies a profile of genes expressed by the pathogen during pathogenesis, including 16 candidate effector genes that have been identified on the basis of their putative small size (< 400 amino acids in length), secretion (based on the SignalP algorithm; Bendtsen et al., 2004; Nielsen et al., 1997) and the presence of cysteine residues.
RESULTS
cDNA libraries
Five distinct cDNA libraries were created and their sequences entered into the Plant and Food Research fungal sequence database. Two libraries were prepared from infected leaves: ABEA and ABEB (in planta libraries). ABEB was prepared from the ABEA library by probing the ABEA library with genomic DNA from V. inaequalis to enrich for fungal sequences. Two further libraries [IAAA and MAAB; potato dextrose broth (PDB) in vitro libraries] were prepared from in vitro‐grown mycelium from PDB shake cultures. The final library (MAAD; SSH cellophane in vitro library) comprised ESTs from the SSH library enriched for cellophane‐induced genes. The SSH method (Diatchenko et al., 1996) was utilized to enrich for cellophane‐induced genes, the profile of which would potentially mirror that observed during growth in planta. All libraries were non‐directional and sequenced from only one direction (Table 1).
Table 1.
Summary of the Plant and Food Research Venturia inaequalis expressed sequence tag (EST) libraries.
| Library | Description | Total # fungal* ESTs | Min. EST length | Max. EST length | Average sequence length | # nr‡ | # contigs (#ESTs) | # singletons | Redundancy§ |
|---|---|---|---|---|---|---|---|---|---|
| ABEA | 7‐week‐old Malus × domestica (‘Royal Gala’) Vi race 1 (MNH 135), 17 days post‐inoculation (dpi) | 587 (2342)† | 145 | 1835 | 572 | 432 | 55 (210) | 377 | 26% |
| ABEB | ABEA probed with race 1 (MNH 135) Vi gDNA to select fungal sequences | 350 (404) | 197 | 1472 | 558 | 227 | 74 (197) | 153 | 35% |
| IAAA | Vi race 1 (MNH 120) in vitro, 30 dpi | 1311 | 69 | 755 | 596 | 625 | 322 (1008) | 303 | 52% |
| MAAB | Vi race 1 (MNH 120) in vitro, 30 dpi | 861 | 102 | 1268 | 635 | 689 | 99 (271) | 590 | 20% |
| MAAD | Vi race 1 (MNH 120) suppression subtractive hybridization library enriched for cellophane‐induced genes, 28 dpi | 1106 | 128 | 1130 | 544 | 582 | 201 (725) | 381 | 47% |
This value is putative for the in planta libraries, as designations of origin are based on bioinformatic analysis or clustering with in vitro library sequences, with the remainder of the sequences having no known similarities and therefore no firm assignation as fungal or plant.
Number in parentheses denotes total number of sequences in the library (both plant and fungal).
nr, non‐redundant sequence, with respect to individual libraries.
Redundancy: total number of ESTs – (total number of contigs + total number of singletons)/total number of sequences (Cramer et al., 2006).
A total of 4215 ESTs was identified from the five libraries and, following clustering using the cap3 program (Huang and Madan, 1999), were resolved into 2090 non‐redundant sequences (Table 1). When individual libraries were analysed, the 587 putatively fungal ESTs from ABEA were resolved into 432 non‐redundant sequences, including 55 contigs and 377 singletons. The 350 ESTs from ABEB were assembled into 227 non‐redundant sequences, comprising 74 contigs and 153 singletons. Assembly of the 1311 ESTs from IAAA into 625 non‐redundant sequences revealed 322 contigs and 303 singletons, and the 861 ESTs from MAAB were assembled into 689 non‐redundant sequences, including 99 contigs and 590 singletons. The 1106 ESTs from MAAD were assembled into 582 non‐redundant sequences with 201 contigs and 381 singletons (Table 1).
A redundancy level for each library was calculated, with the lowest rate (20%) exhibited by the MAAB in vitro library, suggesting that further sequencing of clones from this library will reveal additional novel sequences. The rates for the two in planta libraries were 26% and 35% for ABEA and ABEB, respectively. The usefulness of further sequencing of the ABEA library to identify additional novel fungal genes, even though the level of redundancy is 26%, is questionable, given that only 25% of the ESTs sequenced were putatively fungal in origin. However, it is a valuable resource of plant genes, some of which may be specifically expressed in the V. inaequalis–apple interaction. Further sequencing of ABEB would be more expedient, with 87% fungal genes, notwithstanding the higher level of redundancy, which at 35% still intimates that additional novel sequences important to the fungus during host colonization could be identified. The rates for the IAAA PDB and SSH cellophane in vitro libraries were 52% and 47%, respectively.
Origin of ESTs from infected plant material‐derived cDNA libraries
Of the 2746 ESTs in the two infected leaf libraries, only 598 (340 non‐redundant sequences) were categorized as fungal with a significant level of confidence. This was accomplished by either clustering with V. inaequalis in vitro library ESTs (Fig. 1) or by bioinformatic analysis.
Figure 1.
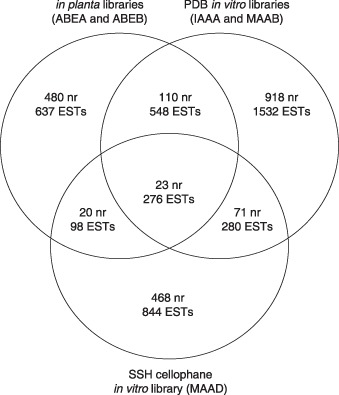
The distribution of putatively fungal expressed sequence tags (ESTs) and non‐redundant sequences (nr) between the in planta libraries (ABEA and ABEB), the potato dextrose broth (PDB) in vitro libraries (IAAA and MAAB) and the suppression subtractive hybridization (SSH) cellophane in vitro library (MAAD). Where there are nr with representative ESTs from more than one library (overlapping sectors), the numbers given are totals from all contributing libraries.
Three hundred ESTs, which formed 153 non‐redundant sequences that included members from the in vitro libraries, were recorded as fungal (Fig. 1). blastp (Basic Local Alignment Search Tool) searches (Altschul et al., 1997) of the predicted translations of the ESTs against the National Center for Biotechnology Information (NCBI) RefSeq (Pruitt et al., 2007) and UniProt UniRef90 (Suzek et al., 2007) databases revealed a further 298 ESTs as most likely to be fungal in origin, which, following clustering, were resolved into 187 non‐redundant sequences. Although a relaxed stringency level (≤ 1e‐5) was used to demarcate ESTs with similarities to fungal sequences in the public domain, the majority of sequences were significantly more similar, with E‐values of ≤ 1e‐10; seven sequences in ABEA and four in ABEB recorded E‐values in the range ≥ 1e‐10 to ≤ 1e‐5.
A further 339 ESTs that could be assembled into 293 non‐redundant sequences from the two in planta libraries showed no significant similarity to any known sequences, including apple sequences. Although the fungal origin of these sequences cannot be confirmed, they represent novel sequences and, as such, are a putative source of effectors, as those from filamentous fungi are often unique. They were included in the putatively fungal dataset to preclude the loss of potential candidate effector genes. Therefore, the total number of ESTs that were classified as putatively fungal was increased to 937, which could be assembled into 633 non‐redundant sequences.
Characterization of the libraries
Of the 2172 ESTs in the PDB in vitro libraries (IAAA and MAAB), 26%, comprising 369 non‐redundant sequences, showed no similarity to sequences in the public domain using a threshold E‐value of ≤ 1e‐5. In the cellophane in vitro library (MAAD), this level was 22%, comprising 132 non‐redundant sequences.
The majority of ESTs (70%) present in the IAAA and MAAB libraries were unique to those libraries. Similarly, 77% of ESTs in the MAAD library and 68% of ESTs from the in planta libraries were also unique (Fig. 1). As the cellophane in vitro library was constructed using SSH methodology, enriching for those genes expressed during morphological differentiation on cellophane and subtracting those genes common to both V. inaequalis growing on cellophane and in PDB, the fact that relatively few, only 94, non‐redundant sequences were constructed from ESTs belonging to both sets of libraries is expected, indicating efficient subtraction. Indeed, the efficiency of subtraction was such that the proportion of ESTs identified as cell function genes in the MAAD library was far less than that in the PDB in vitro libraries (Table 2). However, given the hypothesis that growth in cellophane is a model of infection, the fact that only 43 (7%) non‐redundant sequences had EST members from both the in planta and cellophane in vitro libraries was unexpected. This may reflect the uniqueness of the gene profile required for pathogenicity and that growth in cellophane does not faithfully replicate all aspects of growth in planta. However, the MAAD library was constructed from rinsed cellophane sheets, the rinsing step resulting in the removal of surface hyphae, conidiophores and conidia that would be present in the infected leaf material used for in planta library construction. Consequently, the MAAD library may have a lower representation of sequences associated with the structures removed by rinsing. Alternatively, this finding may be a result of the isolates used to construct the two types of library, even with the threshold for non‐redundant sequence alignment set at 95% identity. Non‐isogenic race 1 isolates were used: MNH135 was used for in planta library construction, whereas MNH120 was used for in vitro library construction. Given that the two isolates used are both categorized as race 1 isolates, they should still share common effectors with respect to avirulence determinants that are recognized by cognate major R genes, and it is unlikely that these effectors would show significant sequence divergence.
Table 2.
Occurrence of cell function genes in the potato dextrose broth (PDB) in vitro libraries (IAAA and MAAB) compared with that in the suppression subtractive hybridization (SSH) cellophane in vitro library (MAAD), as an indication of subtraction efficiency in the MAAD library. Quantification was based on a keyword search using ‘actin’, ‘ribosomal protein’, ‘glyceraldehyde 3‐phosphate dehydrogenase’ (gapdh), ‘ubiquitin’ and ‘ubiquitin conjugating enzymes’ versus the annotations assigned to each expressed sequence tag (EST) based on a blastp algorithm (Altschul et al., 1997) search of the National Center for Biotechnology Information (NCBI) RefSeq (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Pruitt et al., 2007) and UniProt UniRef90 (http://www.ebi.ac.uk/uniprot/; Suzek et al., 2007) databases.
| Cell function gene | Occurrence of keyword in each library | |||||
|---|---|---|---|---|---|---|
| IAAA | % | MAAB | % | MAAD | % | |
| Actin | 2 | 0.15 | 2 | 0.23 | 0 | 0 |
| Ribosomal protein | 78 | 5.9 | 66 | 7.69 | 7 | 0.63 |
| gapdh | 1 | 0.07 | 6 | 0.7 | 0 | 0 |
| Ubiquitin | 10 | 0.76 | 4 | 0.47 | 1 | 0.09 |
| Ubiquitin‐conjugating enzymes | 2 | 0.15 | 1 | 0.12 | 0 | 0 |
| Total # ESTs | 1311 | 861 | 1106 | |||
Bioinformatic analysis of candidate effector genes
The strategy adopted for the selection of candidate effector genes is outlined in Fig. 2. Of 633 non‐redundant sequences from the cDNA libraries that were classified as either fungal or of unknown origin, 96 were selected on the basis of possessing a putative signal peptide following analysis by the SignalP algorithm (Bendtsen et al., 2004; Nielsen et al., 1997). From these, 21 putative effector candidates were selected on the basis of being novel [having no similar sequences in either the NCBI RefSeq (Pruitt et al., 2007) or UniProt UniRef90 (Suzek et al., 2007) database] and, where possible, possessing cysteines and being small (less than 400 amino acids), although candidates were not required to meet all of these criteria. Indeed, several required further sequencing to enable the identification of full‐length putative open reading frames [ORFs; utilizing the Kozak sequence (Balance, 1986; Kozak, 1986) surrounding the 5′ ATG of the longest putative ORF to confirm start codons], and so could not be classified initially according to the size of the encoded product. These sequences were then subjected to a further round of blastp similarity searches. This revealed that two were probably derived from apple, as full‐length ORF sequences revealed strong similarities to Arabidopsis thaliana proteins. A further three candidates had been identified in previous studies (Kucheryava et al., 2008; Win, 2003; Win et al., 2003), and therefore these five ESTs were discounted from further analysis. The remaining 16 candidate effectors were given identification numbers (Venturia inaequalis candidate effector: Vice1–16) and are listed in Table 3.
Figure 2.
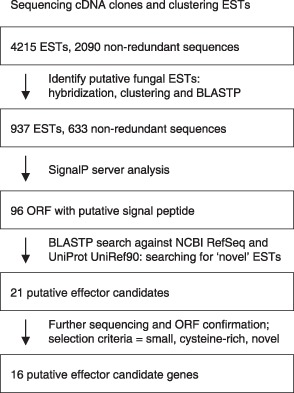
Strategy used for the identification of candidate effectors from Venturia inaequalis expressed sequence tags (ESTs). ORF, open reading frame.
Table 3.
Summary of the attributes of selected Venturia inaequalis candidate effector genes from the cDNA libraries.
| ID number* (# ESTs in nr) | Library of origin of nr members | Best blastp match† (annotation, organism, accession number) | E‐ value‡ | Length of ORF§ (amino acids) | Length of signal peptide¶ | # Cysteine residues** | GPI anchor site (position)†† |
|---|---|---|---|---|---|---|---|
| Vice1 (1) | 1 × ABEA | No significant match‡ | 103 | 20/20 | 5 | – | |
| Vice2 (1) | 1 × ABEA | Hypothetical protein, Aspergillus oryzae RIB40, XP_001821705.1 | 2e‐27 | 210 | 16/16 | NA‡‡ | NA |
| Vice3 (1) | 1 × ABEA | Hypothetical protein SNOG_03711, Phaeosphaeria nodorum SN15, XP_001794262.1 | 6e‐31 | 403 | 19/22 | 2 | – |
| Vice4 (1) | 1 × ABEA | No significant match | 88 | 19/19 | 8 | – | |
| Vice5 (1) | 1 × ABEA | Hypothetical protein SS1G_02025, Sclerotinia sclerotiorum 1980, XP_001597829.1 | 8e‐38 | 245 | 19/19 | 6 | – |
| 1 × MAAB | |||||||
| 1 × MAAD | |||||||
| Vice6 (1) | 1 × ABEB | Conserved hypothetical protein, Pyrenophora tritici‐repentis Pt‐1C‐BFP, XP_001931281.1 | 7e‐11 | 174 | 17/17 | 0 | + (149) |
| Vice7 (5) | 1 × ABEB | No significant match | 91 | 16/17 | 6 | – | |
| 3 × IAAA | |||||||
| 1 × MAAB | |||||||
| Vice8 (1) | 1 × ABEB | No significant match | 126 | 18/18 | 9 | – | |
| Vice9 (1) | 1 × ABEB | No significant match | 261 | 15/15 | 3 | + (240) | |
| Vice10 (7) | 5 × ABEB | Hypothetical protein, Yarrowia lipolytica, XP_504084.1 | 1e‐18 | 297 | 17/17 | 4 | – |
| 1 × MAAB | |||||||
| 1 × MAAD | |||||||
| Vice11 (13) | 1 × MAAB | No significant match | 260 | 20/20 | 8 | + (233) | |
| 12 × MAAD | |||||||
| Vice12 (1) | 1 × MAAD | No significant match | 61 | 16/17 | 2 | – | |
| Vice13 (1) | 1 × MAAD | No significant match | 68 | 29/29 | NA | NA | |
| Vice14 (10) | 1 × ABEA | Conserved hypothetical protein, Aspergillus terreus, XP_00121059.1 | 1e‐42 | 334 | 15/15§§ | 6 | – |
| 2 × ABEB | |||||||
| 4 × IAAA | |||||||
| 3 × MAAD | |||||||
| Vice15 (2) | 2 × MAAD | No significant match | 51 | 21/21 | NA | NA | |
| Vice16 (9) | 9 × MAAD | Hypothetical protein PTRG 01937, Pyrenophora tritici‐repentis, XP_001932270.1 | 3e‐63 | 211 | 15/15 | 4 | – |
EST, expressed sequence tag; nr, non‐redundant sequence; Vice, Venturia inaequalis candidate effector.
blastp searches were carried out against the National Center for Biotechnology Information (NCBI) RefSeq (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Pruitt et al., 2007) and UniProt UniRef90 (http://www.ebi.ac.uk/uniprot/; Suzek et al., 2007) databases.
E‐values were recorded (matches classed as significant) if they were ≤ e‐05.
The length of each peptide encoded by the predicted open reading frame (ORF) was recorded, including the predicted signal peptide.
The length of the putative signal peptide is given based on the neural networks (NN) and hidden Markov models (HMM) of the SignalP server (http://www.cbs.dtu.dk/services/SignalP/; Bendtsen et al., 2004; Nielsen et al., 1997).
The number of cysteine residues in the putative protein encoded by the ORF.
The presence of putative C‐terminal GPI (glycosylphosphatidylinositol) lipid modification (anchor) sites was analysed using the big‐PI fungal server (http://mendel.imp.ac.at/gpi/fungi_server.html; Eisenhaber et al., 2004) with a threshold score of ≥ 6: +, site identified with location in parentheses; –, no site identified.
NA, not analysed because of predicted transmembrane domains.
The TargetP server predicted that this EST may be localized in the mitochondrion and not directed to the secretory pathway.
Seven candidates that were originally selected as novel were found to have significant similarity to other fungal proteins following the additional sequencing and similarity searches. However, the best matches were all against hypothetical proteins without positive annotations, and so these candidates were retained for further analysis. In addition, Vice2 revealed significant blastp (2e‐27) similarity to plastocyanin‐like proteins of other fungal species. However, no type 1 copper‐binding motif was found within the predicted protein sequence of Vice2 using the type 1 copper‐binding signature (Pattern PS00196: [GA]‐x(0,2)‐[YSA]‐x(0,1)‐[VFY]‐{SEDT}‐[C]‐x(1,2)‐[PG]‐x(0,1)‐H‐x(2,4)‐[MQ]) as retrieved from PROSITE (http://expasy.org/cgi‐bin/prosite/). Vice3 was found to be similar (blastp E‐value of 6e‐31) to acetylornithine deacetylase, succinyl‐diaminopimelate desuccinylase and related deacylases. The active site residues of experimentally confirmed acetylornithine deacetylase and succinyl‐diaminopimelate desuccinylase proteins from the bacterium Escherichia coli were found to align with identical residues in Vice3. However, the consensus pattern of active site residues also aligned with other peptidases, such as carboxypeptidase G2 and peptidase T, which may suggest that this protein has some form of peptidase activity. The protein sequence encoded by Vice5 demonstrated significant similarity (8e‐38) to a cysteine‐rich hypothetical protein of Sclerotinia sclerotiorum. All six C‐terminal cysteine residues of Vice5 were strictly conserved with this hypothetical protein. Conservation of cysteine residue spacing was also observed between Vice10 and similar protein sequences predicted to possess a rare lipoprotein A (RlpA)‐like double‐psi β‐barrel domain, the function of which is unknown.
The majority of the results generated by the SignalP algorithm were corroborated by the TargetP server with candidate effectors predicted to enter the secretory pathway, with the exception of Vice14 (Table 3). In this case, a putative mitochondrial subcellular localization may be possible, although the lowest reliability class was recorded for the result, with similar scores recorded for a mitochondrial (0.315) and a secretory pathway (0.277) location.
Analysis using the TMHMM server revealed the presence of transmembrane domains in Vice2, Vice13 and Vice15, implying that they are not secreted. These three candidate effectors were therefore discounted from further analysis.
All candidate effector proteins contained cysteine residues, except Vice6. The majority of the candidates that possessed two or more cysteine residues were predicted to possess a disulphide bond by the DISULFIND server, with the exception of Vice3, Vice9, Vice12 and Vice14. Putative disulphide bond spacing was conserved between similar proteins in the public domain from Phaeosphaeria nodorum and Vice5 and Vice10.
C‐terminal glycosylphosphatidylinositol (GPI) lipid modification (anchor) sites were predicted for the protein sequences Vice6, Vice9 and Vice11 using the big‐PI server (Table 3). InterProScan searches revealed motifs in six of the candidate effectors (Table 4). Several of the servers contributing to the InterProScan search revealed that Vice3 comprises domains with similarities to metallopeptidases and peptidase dimerization motifs. Similarities to acetylornithine deacetylase were also identified, which was consistent with the blastp results. Vice11 showed similarities to motifs associated with C‐type lectins, and Vice14 showed similarities to domains associated with allergen V5 (vespid wasp allergen)/Tpx‐1 (mammalian testis‐specific protein), a plant PR protein and sterol carrier protein (SCP)‐like extracellular protein.
Table 4.
InterProScan results (http://www.ebi.ac.uk/Tools/InterProScan/; Mulder et al., 2007; Quevillon et al., 2005) for candidate effectors from Venturia inaequalis. Gene ontology (GO) classifications are shown, where known, indicating cellular component, biological process and molecular function applicable to motifs identified by the InterPro member databases.
| ID number | InterPro member database and search method* | Motif | GO cellular component | GO biological process | GO molecular function | Amino acid residues† | E‐value |
|---|---|---|---|---|---|---|---|
| Vice3 | HMMPfam | PF01546 Peptidase_M20 IPR002933 | Proteolysis (GO:0006508) | Metallopeptidase acitivity (GO:0008237) | 109–401 | 7.9e‐17 | |
| PF07687 M20_dimer IPR011650 Peptidase M20, dimerization | Hydrolase activity (GO:0016787), protein dimerization activity (GO:0046983) | 210–318 | 8.8e‐23 | ||||
| HMMPanther | PTHR11014:SF5 Acetylornithine deacetylase | 44–402 | 1.3e‐79 | ||||
| PTHR11014 Peptidase M20 family member | 44–402 | 1.3e‐79 | |||||
| ScanRegExp | PS00758 ARGE_DAPE_CPG2_1 IPR001261 Arg/DapE/ACY1/CPG2/ YscS. Conserved site | Proteolysis (GO:0006508) | Metallopeptidase activity (GO:0008237) | 108–117 | |||
| Vice11 | ScanRegExp | PS00615 C_type_lectin_1 | Binding (GO:0005488) | 36–64 | |||
| Vice14 | HMMSmart | SM00198 SCP IPR001283 Allergen V5/Tpx‐1 related | Extracellular region (GO:0005576) | 164–314 | 3.3e‐21 | ||
| HMMPfam | PF00188 SCP IPR014044 SCP‐like extracellular | 170‐306 | 4.2e‐21 | ||||
| ScanRegExp | PS01009CRISP_1 IPR001283 Allergen V5/Tpx‐1 related | Extracellular region (GO:0005576) | 266–276 | 8e‐5 | |||
| FPrintScan | PR00837 V5TPXLIKE IPR001283 Allergen V5/Tpx‐1 related | Extracellular region (GO:0005576) | 185–203 | 4.3e‐11 | |||
| 265–281 | 4.3e‐11 | ||||||
| 301–314 | 4.3e‐11 | ||||||
| HMMPanther | PTHR10334:SF6 Plant pathogenesis‐related | 174–317 | 2.1e‐30 | ||||
| PTHR10334 Cysteine‐rich secretory protein (CRISP/SCP/TPX1)‐related IPR001283 Allergen V5/Tpx‐1 related | Extracellular region (GO:0005576) | 174–317 | 2.1e‐30 | ||||
| BlastProDom | PD000542 Q7SG93 NEUCR Q7SG93; IPR001283 Allergen V5/Tpx‐1 related | Extracellular region (GO:0005576) | 233–320 | 2e‐11 |
InterPro member databases and search methods: BLASTProDom, a gapped blastp search of protein families; FprintScan, a scan against the fingerprints (groups of motifs) in the PRINTS database; HMMPanther, a scan of the PANTHER protein families; HMMPfam, a scan of the hidden Markov models (HMMs) in the Pfam Protein Families database; HMMSmart, a scan of the HMMs present in the SMART domain/domain families database; ScanRegExp, a scan of the regular expressions in the PROSITE protein families and domains database.
Amino acid residues in the candidate effector that match the motif.
The SAM‐T06 server was used to screen for similarities between predicted secondary and tertiary candidate effector structure and solved structures. Consistent with blastp and InterProScan results, the protein sequence of Vice3 demonstrated moderate structural similarity to peptidase M20A proteins (4.1e‐46). Of interest, Vice10 demonstrated moderate structural similarity (3.4e‐11) to endoglucanases, Barwin (Barley wound‐induced plant defence protein), expansins and pollen allergens. InterProScan results were also corroborated by similarities detected using candidate effector Vice14, as it demonstrated moderate structural similarity to a V5 allergen (1.4e‐25) belonging to the PR‐1‐like superfamily with an SCP‐like extracellular domain.
Genomic DNA extraction and PCR analysis
A fungal origin was confirmed for those candidate effectors (Vice1, Vice3, Vice4, Vice6, Vice8 and Vice9) for which coding sequences were present in the in planta libraries only: amplification of a PCR product from genomic DNA of V. inaequalis isolate MNH120 was recorded (results not shown).
Leaf infection and mRNA expression analysis of candidate effector genes
Leaf samples were collected during a detached leaf infection assay at 5 and 10 days post‐inoculation (dpi) and were examined microscopically. Concomitant expression of each of the candidate effector genes was measured by real‐time quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR), and compared with that during growth in PDB at 10 dpi. Differences in expression were deemed significant at P = 0.01. At 5 dpi, spores of V. inaequalis had germinated and penetrated the cuticle, and subcuticular hyphae had ramified in the subcuticular space. Small stromata consisting of a few enlarged cells were beginning to be formed (Fig. 3). At 10 dpi, stromata were largely mature, with conidiophores arising from them bearing conidia (Fig. 3).
Figure 3.
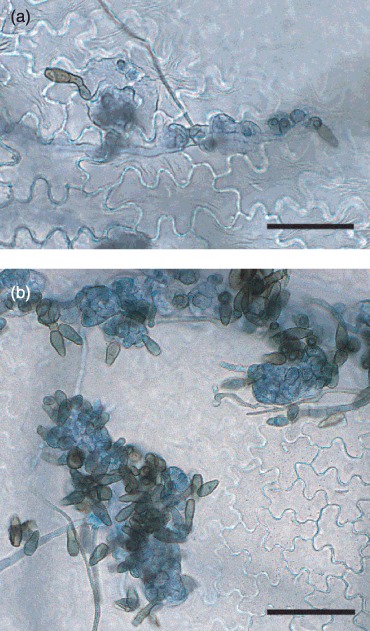
Infection by Venturia inaequalis during a detached leaf assay at (a) 5 days post‐inoculation (dpi), with stromata beginning to form, and (b) 10 dpi, with sporulating stromata. Scale bar, 50 µm.
Candidate effector mRNAs for Vice4, Vice12 and Vice16 were all significantly up‐regulated during in planta growth at both 5 and 10 dpi, whereas Vice5 was only up‐regulated at 5 dpi, albeit with only a 1.6‐fold increase, with no significant difference in expression between in planta and PDB growth at 10 dpi (Fig. 4). The expression of Vice1, Vice3 and Vice7 candidate effector mRNAs during growth in planta was not significantly different from that during growth in PDB at either of the infection time points. The expression of five candidate effector mRNAs (Vice6, Vice9, Vice10, Vice11 and Vice14) was significantly lower during growth in planta than during growth in PDB at both time points analysed, whereas the expression of Vice8 mRNA was significantly reduced at 5 dpi, but not significantly different at 10 dpi. No expression of candidate effector mRNAs was recorded on leaves inoculated with sterile distilled water (SDW; data not shown).
Figure 4.
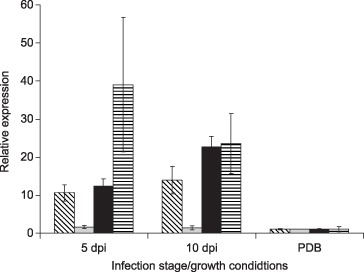
The average expression of four Venturia inaequalis candidate effector (Vice) genes at two time points during detached leaf infection compared with that during growth in potato dextrose broth (PDB). A gene expression normalization factor was calculated for each sample based on the geometric mean of two stably expressed reference genes (β‐tubulin; EU853839) and 60S ribosomal protein L12 (EU853840) using geNorm software v3.4 (Vandesompele et al., 2002). Averages were calculated from six expression measurements from each time point of infection or growth stage, gathered from two independent infection/growth assays; each expression measurement was the average of three biological samples (each sample comprising three infected leaves or material gathered from three flasks of PDB), and the polymerase chain reaction (PCR) for each sample was conducted in triplicate. The error bars represent the standard deviation of the six expression measurements. The expression of each of these genes during infection was significantly different (P = 0.01) from that during growth in PDB, with the exception of Vice5 (light grey) at 10 dpi, which only showed a 1.39‐fold increase. Vice4, diagonal lines; Vice5, light grey; Vice12, black; Vice16, horizontal lines.
DISCUSSION
This study represents the first large‐scale analysis of ESTs derived from the V. inaequalis–apple pathosystem. It resulted in the identification of 937 putatively fungal ESTs that were resolved into 633 non‐redundant sequences following clustering. An EST approach has proven to be successful in identifying genes involved in other fungal pathosystems (Cramer et al., 2006; Ebbole et al., 2004; Hu et al., 2007; Kleemann et al., 2008; Li et al., 2004). Moreover, it has proven to be successful in the identification of some phytopathogen effector genes (Armstrong et al., 2005; Bos et al., 2003; Catanzariti et al., 2006; Shan et al., 2004). For example, Catanzariti et al. (2006) generated a Melampsora lini haustorium‐specific EST library, as the HR of rust resistance appeared to coincide exclusively with emerging haustoria, to enable the identification and cloning of AvrM, AvrP4 and AvrP123‐A, three effector genes of M. lini (Catanzariti et al., 2006). Armstrong et al. (2005) cloned and characterized the Avr3a effector gene of Phytophthora infestans using a combination of EST database mining (Torto et al., 2003) and an association genetics approach originally developed by Bos et al. (2003).
Despite these successes, the largest drawback of an EST strategy is the difficulty encountered when assigning the origin of sequences according to organism. The most valuable resource in aiding this assignment is the complete genome sequence of the organisms involved in the interaction. Cramer et al. (2006) demonstrated that, in the Alternaria brassicicola–Brassica olearacea interaction, 30% more fungal genes were identified in a library from infected plant material with a whole‐genome draft sequence than using only blast searches of the same library. Although advances have been made in sequencing the apple genome, a complete sequence has yet to be released, and the genome sequencing of V. inaequalis has yet to be initiated.
Various strategies were employed in this study to overcome these limitations. Firstly, the ABEA EST library constructed from infected leaf tissue was probed with genomic DNA from V. inaequalis to enrich for fungal genes expressed in planta to create the ABEB library. A DNA blot approach has proven to be successful for other phytopathogens (Hahn and Mendgen, 1997; Rauyaree et al., 2001; Thara et al., 2003; Zhang et al., 2003). This resulted in a 3.5‐fold increase in the percentage of fungal sequences in the ABEB library compared with the ABEA library, as 87% of ESTs were designated fungal in the former and 25% in the latter. Secondly, libraries were constructed from V. inaequalis growing axenically to aid the designation of fungal sequences. The ESTs generated from PDB‐grown V. inaequalis enabled 824 ESTs from the infected leaf material libraries to be designated as fungal. This approach, however, suffers from the drawback that effector genes may have a tendency to be expressed exclusively in planta. Thus, further refinement of this strategy was the use of the SSH library enriched for cellophane‐induced genes. This approach was adopted as growth in cellophane was thought to be a model of in planta growth, given the differentiation of infection‐like structures during growth in cellophane (Kucheryava et al., 2008). Similarities between sequences in the in planta and SSH cellophane‐enriched libraries enabled the designation of a further 98 ESTs. The SSH library strategy was only partially successful: although it resulted in the designation of additional sequences as fungal from the infected leaf material libraries, this was not to the level expected if growth in cellophane absolutely mimics that in planta. However, it resulted in the identification of seven candidate effector genes that would otherwise not have been found.
Alternative methods for the differentiation of pathogen and host sequences in a mixed population have been used previously. These include the use of the difference in average GC/AT ratios for plant and pathogen genes (Qutob et al., 2000), or codon bias, notably the third ‘wobble’ position of the codon (Emmersen et al., 2007; Friedel et al., 2005; Hsiang and Goodwin, 2003; Li et al., 2004; Maor et al., 2003). Initial assessment of genes from V. inaequalis and apple indicated that there was a significant difference between GC/AT ratios and codon usage profiles in genes from the two species (results not shown). This may be used in future analyses of ESTs from this interaction to differentiate between host and pathogen sequences. However, it is important to note that, although GC content and codon usage bias differ between species, vast differences can exist between different genes of a single species or different regions within a gene (Friedel et al., 2005), leading to false designations.
As it is assumed that candidate effectors will be secreted into the subcuticular space during infection by V. inaequalis, and may subsequently be translocated into the host cytoplasm, the SignalP algorithm was used to scan the putative V. inaequalis sequences identified from the mixed population of ESTs from the in planta and in vitro libraries for N‐terminal signal peptide sequences. Kleemann et al. (2008) outlined the inherent pitfalls in the use of such an in silico approach; accurate identification of the N‐terminal methionine can be problematic as ORF prediction algorithms can be unreliable, especially with fungal sequences. Incorrect ORF prediction can lead to internal transmembrane domains being identified as signal peptides by the SignalP algorithm. Hence, the longest putative ORF with a 5′ ATG with conservation of Kozak sequence specific to filamentous fungi (Balance, 1986; Kozak, 1986) was used to identify the most likely full‐length ORF (data not shown). An alternative approach for the identification of secreted proteins would be to use the ‘signal sequence trap’ methodology (Klein et al., 1996). This yeast‐based genetic screen has been used to confirm the presence of functional secretion signals in two plant pathogens to date: Pseudorobillarda sojae (Lee et al., 2006) and Uromyces fabae (Link and Voegele, 2008).
This approach, focusing as it does on the selection of ESTs with putative signal peptides, could result in some effectors being overlooked. The paradigm for filamentous fungal effectors is that they are small, often cysteine‐rich, secreted proteins. Indeed, the majority of fungal effectors fit this paradigm. There are, however, notable exceptions: AVRa10 and AVRk1 from the barley powdery mildew fungus B. graminis f. sp. hordei. These effectors trigger hypersensitive cell death when expressed in the cytoplasm of resistant barley cultivars carrying the R genes Mla10 and Mlk1. How they are secreted is unknown, as they lack the typical signal peptide signature (Ridout et al., 2006). These effectors do not comply with the paradigm outlined above and would not be identified in the search strategy employed in this study. However, the use of the SignalP algorithm led to the identification of 96 non‐redundant sequences which, following blastp searches, were reduced to 16 candidate effector genes.
Candidate effectors were initially selected on the basis of being novel. However, some of these were subsequently found, on identification of the complete ORF sequence, to have significant similarities to hypothetical fungal genes from other species. This may not preclude these sequences from being effectors, however, as the steady increase in the number of fungal phytopathogen genomes being sequenced increases the likelihood that orthologues will be found in other species. Indeed, the C. fulvum lysin motif effector Ecp6 has orthologues from 11 filamentous fungal species, including seven plant pathogens (Bolton et al., 2008).
Although the most significant matches, when any were recorded, for each candidate effector protein (as revealed by blastp searches) were to a hypothetical protein from other fungi, several proteins with less significant E‐values were identified, and these possessed annotated domains. For example, the full‐length protein sequence of Vice3 demonstrated significant similarity to proteins of the peptidase subfamily M20A involved in amino acid metabolism. Although, in principle, any foreign molecule could act as a potential effector, Vice3 does not lend itself to an obvious role in virulence or pathogenicity if these similarities represent a true conservation of function. However, it must be noted that the conservation of active site residues suggests a conservation of mechanism, for example peptidase activity, and not necessarily conservation of substrate specificity. The involvement of peptidases with roles outside general amino acid metabolism as effectors in host–pathogen interaction is a more common phenomenon (Orbach et al., 2000; Xia, 2004).
The conservation of disulphide spacing in Vice5 and Vice10 with similar proteins in the public domain from S. sclerotiorum and P. nodorum may suggest an important role for these cysteine residues in protein function and/or stability within the apoplast (Joosten et al., 1994, 1997). Apart from Vice5 and Vice10, significant blastp similarity to P. nodorum proteins (i.e. within the top three blastp hits) was observed for the candidate effector protein sequences Vice3 and Vice6. This similarity reflects the recent release of P. nodorum protein sequences into the public domain, following completion of genome sequencing for this phytopathogen and the relatively close taxonomic relationship of P. nodorum to V. inaequalis (both Order Pleosporales; Goodwin, 2004). In the future, the use of similarities between candidate effectors and sequences present in the public domain will undoubtedly become more significant when more gene and protein sequence data from closely related organisms are released.
The predicted GPI modification sites of Vice6, Vice9 and Vice11 would, on initial consideration, preclude them from being successful effectors as, in the V. inaequalis–apple interaction, effectors are assumed to be soluble and secreted to the subcuticular space, or even taken up by the host cell, and these effectors are predicted to be covalently linked to the fungal plasma membrane or to β‐1,6‐glucans present within the fungal cell wall (de Groot et al., 2003; Richard and Plaine, 2007). However, some GPI‐anchored proteins are involved in host–pathogen interactions (Albrecht et al., 2006; Jiang et al., 2006; Richard and Plaine, 2007). For example, the majority of elicitins from Phytophthora, which are capable of inducing an HR in tobacco, possess a GPI anchor (Jiang et al., 2006). The question remains of how the plant detects these proteins to initiate a defence response when the corresponding proteins are not secreted to the plant–pathogen interface, although they may be released by cleavage by plant proteases.
The amino acid sequences of both Vice9 and Vice10 contain incomplete imperfect repeat domains (results not shown). Repeated amino acid domains have already been identified in two secreted proteins of V. inaequalis (Kucheryava et al., 2008). Secreted fungal proteins with internal repeats are predominantly involved in cell wall organization, and are often found coating the outer cell wall, participating in cell‐to‐cell or cell‐to‐substrate adherence or rendering the cell wall hydrophobic (Dranginis et al., 2007; Hung et al., 2002; Li and Palecek, 2008; Linder and Gustafsson, 2008; Richard and Dujon, 2006; Teerstra et al., 2006; Verstrepen et al., 2005; Wang and St. Leger, 2007). This is interesting given that Vice9 is predicted to possess a GPI modification site.
InterProScan results corroborated the blastp homologies and also revealed additional domains present within Vice11 and Vice14. The candidate effector Vice11 showed similarity to a C‐type lectin motif following the InterProScan searches. This motif is characteristic of C‐type lectins, which are not commonly found in either fungi or plants, although similar domains have been found in nine other fungal taxa to date, including two filamentous fungal plant pathogens Ustilago maydis and P. nodorum. However, there is a precedent for the involvement of a protein domain previously thought to be exclusive to the animal kingdom to be involved in a plant–pathogen interaction. The Avr4 chitin‐binding lectin effector of C. fulvum contains a chitin‐binding domain (CBM14) previously considered to be restricted to invertebrates. CBM14 protects the pathogen from the action of host chitinases, enhancing virulence (van den Burg et al., 2006). The candidate effector Vice14 showed similarities to domains associated with allergen V5/Tpx‐1, a PR protein (PR‐1) and SCP (sperm‐coating glycoprotein)‐like extracellular protein. These domains are all associated with extracellular eukaryotic proteins and are presumed to be evolutionarily related, but their precise function is unclear. Similarity to PR proteins is intriguing, given that these proteins are host encoded and contribute to resistance; however, a common structure does not necessarily indicate a common function. Proteins with domains similar to these are found in numerous fungal taxa, including hypothetical proteins in six filamentous fungal plant pathogens.
The SAM‐T06 server (http://www.cse.ucsc.edu/compbio/SAM_T06/T06‐query.html; Karplus et al., 1998, 2001, 2003, 2005) was employed to identify similarity between the predicted secondary and tertiary structure of the candidate effectors and solved structures. Results for Vice3 and Vice14 were consistent with blastp search results and functional motif database searches. For Vice10, weak structural similarities were identified to expansins, expansin‐like proteins, endoglucanases and pollen allergens. These motif signatures, together with the RlpA‐like double‐psi β‐barrel motif identified in blastp searches, are found within ATEXPA8 (GenBank ID: NP_181593), an α‐expansin of A. thaliana. Expansins induce plant cell wall extension (Choi et al., 2006). This result is interesting, given that a fungal endoglucanase has been identified with cell wall extension activity (Yuan et al., 2001).
Inconsistent with the observation that most cloned fungal effector genes encode cysteine‐rich proteins, the predicted protein sequence of effector candidate Vice6 possesses no cysteine residues. However, this candidate effector should not be discounted, as a paucity of cysteine residues has been demonstrated for PWL2 of Magnaporthe oryzae (Sweigard et al., 1995), AvrL567‐A and AvrM of M. lini (Catanzariti et al., 2006; Dodds et al., 2004) and AvrLm1 of Leptosphaeria maculans (Gout et al., 2006). In addition, five oomycete effector proteins are not cysteine rich (Allen et al., 2004; Armstrong et al., 2005; van Poppel et al., 2008; Rehmany et al., 2005; Shan et al., 2004).
The EST libraries from infected apple material and in vitro‐grown V. inaequalis have provided a valuable resource from which several candidate effectors have been mined. Of the 16 candidates analysed, the most interesting with respect to further investigation are Vice4, Vice12 and Vice16. These candidates fit the paradigm of filamentous fungal effectors in that they are small proteins with an even number of cysteine residues. Two are novel; the third (Vice16) is homologous to a hypothetical protein from a plant pathogen. All possess a signal peptide signature, without a transmembrane domain, and so are putatively secreted to the host–pathogen interface. Their expression at the transcriptional level is up‐regulated during growth in planta compared with that during growth in vitro. RNA‐mediated gene silencing (Fitzgerald et al., 2004) of these candidates will reveal any effector function and will contribute to the understanding of the V. inaequalis–apple interaction.
EXPERIMENTAL PROCEDURES
Fungal material and culture conditions
Venturia inaequalis isolates MNH120 (race 1, Auckland, New Zealand, Malus × domestica‘Granny Smith’, 1996) and MNH135 (race 1, Waikato, New Zealand, Malus × domestica‘Pink Lady’, 1996) were grown on cellophane (325P‐23 cellophane, Innovia Films, Melbourne, Vic., Australia; Parker et al., 1995) overlying potato dextrose agar (PDA; Oxoid Ltd., Basingstoke, Hampshire, UK) at 20 °C for 7–10 days (16 h light period/day) under white fluorescent lights (4300K, Cool white Deluxe, Sylvania, Danvers, MA, USA) for the production of conidia. Conidia for the inoculation of plants or media were harvested from agar plates by dislodging with SDW passed through a Milli‐Q Plus system (Millipore Corporation, Bedford, MA, USA), followed by filtration through sterile glass wool to remove mycelial debris; the concentration of conidia was adjusted to 1 × 105 mL−1.
Mycelia were cultured in liquid PDB (Difco Laboratories, Sparks, MD, USA) at 20 °C either with shaking at 150 r.p.m. (Orbital shaker, Sanyo Gallenkamp, Leicester, UK) following inoculation with 1 mL of conidial suspension, or statically as for PDA cultures. Mycelium (21–30 days old) was harvested by filtering through Miracloth (Calbiochem®, La Jolla, CA, USA), and rinsed with 500 mL SDW prior to RNA or DNA extraction.
Cellophane sheets were removed from PDA plates at 7 dpi with V. inaequalis conidia, placed on the surface of 25 mL PDB and incubated as above for PDA cultures for 14–21 days. Cellophane sheets were rinsed in 20 mL SDW and blotted with 3M paper (Whatman Ltd., Maidstone, Kent, UK) to dislodge conidia prior to RNA extraction.
Plant material and infection assays
Seed originating from open pollinated Malus × domestica‘Royal Gala’ (Hawke's Bay, New Zealand) was used to produce infected plant material. Seeds were stratified for 6–8 weeks at 4 °C in fine‐grain vermiculite (Nuplex, Auckland, New Zealand) saturated with SDW, following surface sterilization for 1 min in Thiram (Kiwicare Corporation Ltd., Christchurch, New Zealand; 100 mg/mL fungicide). Following germination, seeds were grown for 4–6 weeks at 21 °C with a 12 h light period/day under 4C W SHP‐TS lights (Sylvania).
For library construction, seedlings were spray inoculated with a conidial suspension or SDW until run off using a painter's airbrush (Paasche Airbrush Co., Chicago, IL, USA). Seedlings were incubated in small Heat’n’Grow growth chambers (Hydroponic Wholesalers, Auckland, New Zealand) covered in aluminium foil. High humidity was maintained by adding 500 mL of 37 °C water and placing the chambers on heated plant‐raising panels (Hydroponic Wholesalers) for 24 h in a 20 °C room. After 72 h the foil was removed and the infection was left to develop at 20 °C under white fluorescent lights (4300K, Cool white Deluxe, Sylvania) with a 12 h light period/day. Leaves were harvested at 17 dpi for RNA preparation.
A detached leaf assay was used for qRT‐PCR (Win et al., 2003). Infected and SDW‐inoculated leaves were harvested at 5 and 10 dpi and used either for RNA extraction or microscopic evaluation of infection.
Microscopic evaluation of infection
Leaves were cleared and stained by the method of Bruzzese and Hasan (1983), and mounted in water on glass slides. Observations were made under bright field using a Research Photo‐micrographic microscope system (Olympus, Melville, NY, USA) with a 35‐mm automatic SLR camera attached. Digital images were captured using RS Image software (Roper Scientific Inc., Tucson, AZ, USA). Final figures were produced using Adobe® Photoshop® 6.0 (Adobe Systems Inc., San Jose, CA, USA.).
RNA extraction
Total RNA was extracted by the method of Chang et al. (1993) and concentration quantified using a Nanodrop® ND‐1000 Spectrophotometer (NanoDrop Technologies, Rockland, DE, USA). mRNA was purified from total RNA using an mRNA purification kit (Amersham BioSciences UK Ltd., Little Chalfont, Buckinghamshire, UK), according to the manufacturer's instructions.
cDNA library construction
RNA was used as follows in the construction of the different libraries.
Infected leaf library
Total RNA was extracted from leaf material at 17 dpi with V. inaequalis MNH135 and used for library construction according to Newcomb et al. (2006). mRNA purified from this total RNA was ligated into λZAP® (Stratagene, La Jolla, CA, USA), packaged into Gigapack III Gold (Stratagene) and plated using the host bacterial strain XLI‐BLUE MRF′ (Stratagene), according to the manufacturer's instructions. The library was mass excised from 250 000 plaque forming units (pfu) using ExAssist phage (Stratagene), according to the manufacturer's instructions. The transfection was performed on a large scale and plated onto 137‐mm Hybond™ N+ membranes (Amersham BioSciences UK Ltd.) overlaying Luria–Bertani (LB) agar. After 16 h, colonies were lifted from the membranes onto fresh Hybond N+. The original membrane and new replica membranes were then placed on LB agar plates containing 50 µg/mL ampicillin, and incubated at 37 °C for 5 h. After incubation, the original membrane was stored at 4 °C until needed and the replica membrane was cultured on LB containing ampicillin 170 µg/mL at 37 °C overnight. Blots were then denatured, neutralized and washed according to standard protocols (Sambrook et al., 1989) prior to being cross‐linked on a UV light box for 2 min. Blots were washed overnight in 50 mm tris(hydroxymethyl)aminomethane (Tris), pH 8.0, 1 m NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 0.1% sodium dodecylsulphate (SDS) at 50 °C with shaking, and then wiped clean of bacterial debris prior to hybridization.
Genomic DNA was extracted from 21‐day‐old PDB shake cultures of V. inaequalis according to Win (2003), and sheared by a 10‐s burst of sonication (Kontes, Vineland, NJ, USA) prior to use as a hybridization probe to select for fungal sequences according to standard protocols (Sambrook et al., 1989). Colonies corresponding to hybridizing spots were picked and then mixed with 500 µL SM buffer (0.1 m NaCl, 10 mm MgSO4, 50 mm Tris‐HCl, pH 7.5) and left overnight at 4 °C. PCR using T3 and T7 universal primers was carried out with 2 µL of the plaque/buffer as template. Cycling parameters were as follows: 94 °C for 2 min, followed by 35 cycles of 94 °C for 10 s, 58 °C for 30 s, 72 °C for 30 s, and then 72 °C for 10 min. PCR‐amplified inserts were purified using PCR purification columns (Qiagen GmbH, Hilden, Germany) prior to sequencing.
In vitro‐grown V. inaequalis libraries
Total RNA extracted from 30‐day‐old mycelium from PDB shake cultures was used for library construction. mRNA packaged into plaques and the preparation of sequencing template were performed as detailed above for the infected leaf library.
SSH library enriched for cellophane‐induced genes
mRNA from 28‐day‐old statically grown mycelium and cellophane‐grown V. inaequalis MNH120 was used in the construction of an SSH library employing the PCR‐select™ cDNA subtraction kit (BD Biosciences Clontech, Palo Alto, CA, USA), according to the manufacturer's instructions, with cellophane‐grown V. inaequalis cDNA as tester and the statically grown mycelium cDNA as driver.
An indication of subtraction efficiency in the MAAD library was given by comparing the prevalence of cell function genes in the in vitro libraries quantified following a keyword search using ‘actin’, ‘ribosomal protein’, ‘glyceraldehyde 3‐phosphate dehydrogenase’ (gapdh), ‘ubiquitin’ and ‘ubiquitin conjugating enzymes’ versus the annotations assigned to each EST based on a blastp algorithm (Altschul et al., 1997) search of the NCBI RefSeq (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Pruitt et al., 2007) and UniProt UniRef90 (http://www.ebi.ac.uk/uniprot/; Suzek et al., 2007) databases.
DNA sequencing and bioinformatics
Sequencing of the clones from the in planta libraries was carried out at Genesis Research and Development, Auckland, New Zealand, and clones from the in vitro libraries (including the SSH library) were sequenced at the School of Biological Sciences Sequencing Unit, University of Auckland, New Zealand. Big Dye Terminator standard protocols were used (Applied Biosystems, Foster City, CA, USA) and sequencing reactions were resolved on ABI377, ABI3100 or ABI3700 sequencers according to the manufacturer's instructions (Applied Biosystems). The ESTs were either automatically or manually trimmed of vector, adapter and low‐quality sequence regions, and entered into a relational database. ESTs were assembled into non‐redundant sequences using cap3 (using a 95% level of identity as a threshold; Huang and Madan, 1999), and automatic annotation was performed using the HortResearch BioPipe sequence annotation pipeline (a cluster‐based annotation system written in PERL; R. Crowhurst, M. Davy and C. Deng, The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand, unpublished work) and utilizing a relational database (MySQL; http://www.mysql.com). Redundancy within the libraries was calculated using the formula: total number of ESTs—(total number of contigs + total number of singletons)/total number of sequences (Cramer et al., 2006).
The most likely full‐length ORF of each candidate effector was identified by distinguishing the longest ORF with a 5′ ATG with a high degree of conservation of the Kozak sequence specific to filamentous fungi, denoted by the consensus nucleotide sequence CA[CA][AC]AUGGC (Balance, 1986; Kozak, 1986). blastp algorithms (Altschul et al., 1997) were used to distinguish plant and fungal sequences from the in planta libraries. Those sequences that had an E‐value of ≤ 1e‐10 for their best match against fungal sequences present in the NCBI RefSeq (Pruitt et al., 2007) or UniProt UniRef90 (Suzek et al., 2007) databases were assigned a fungal origin. Similarly, for sequences showing a higher similarity (< 1e‐10) with plant sequences, a plant origin was assigned. For those sequences with less significant E‐values (≥ 1e‐10 but ≤ 1e‐5), assignments of origin were still made as above. Any sequences that showed no significant identity with known sequences were also assigned a fungal origin to avoid missing putative novel fungal sequences. To ensure that each of the candidate effector proteins was predicted to be secreted, the most probable full‐length amino acid sequence encoded by each candidate effector gene was screened for the presence of a putative N‐terminal signal peptide using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/; Bendtsen et al., 2004; Nielsen et al., 1997). A eukaryotic‐specific server setting was employed and run according to the neural networks (NN) and hidden Markov models (HMM). To determine whether any of the predicted proteins were targeted to subcellular localizations other than the secretory pathway, the TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP; Emanuelsson et al., 2000, 2007) was used with default cut‐off and non‐plant organism settings. The presence of transmembrane domains was determined by analysis using the TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/; Krogh et al., 2001; Sonnhammer et al., 1998).
In an attempt to estimate which cysteine residues are predicted to be involved in disulphide bond formation, the disulphide bonding state and connectivity pattern for each cysteine residue within each candidate effector protein were analysed by the DISULFIND server (http://disulfind.dsi.unifi.it/help.php; Ceroni et al., 2006).
Candidate effector protein sequences were screened for the presence of putative C‐terminal GPI lipid modification (anchor) sites using the big‐PI server, developed for the assessment of fungal protein sequences known to enter the endoplasmic reticulum–Golgi secretory pathway as predicted by signal peptide prediction software (http://mendel.imp.ac.at/gpi/fungi_server.html; Eisenhaber et al., 2004).
Common protein domains and functional motifs within these proteins were identified using the InterProScan database server (http://www.ebi.ac.uk/Tools/InterProScan/; Mulder et al., 2007; Quevillon et al., 2005).
Based on a combination of protein fold recognition, sequence alignment and several methods of local secondary structure prediction, the SAM‐T06 server was employed to identify similarities between candidate effector‐predicted secondary and tertiary structure and proteins with solved structures (http://www.cse.ucsc.edu/compbio/SAM_T06/T06‐query.html; Karplus et al., 1998, 2001, 2003, 2005). An expected value threshold of > 1e‐05 was enforced for similarities identified on the basis of structure (unless similarities were to proteins already known to play a role in infection specificity in other fungi).
Genomic DNA extraction and PCR analysis
The extraction of genomic DNA from isolate MNH120 was carried out according to Win (2003), following 4 weeks of growth in PDB. Genomic DNA was quantified using a Nanodrop® ND‐1000 Spectrophotometer (NanoDrop Technologies). For candidate effector genes of ≤ 1 kb in length, genomic DNA amplifications were performed within a final reaction volume of 20 µL containing 2 ng/µL genomic DNA template, 300 nm of each primer (Table 5), 1 U Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), 200 µm deoxynucleoside triphosphates (dNTPs) (Invitrogen), 1.5 mm MgCl2 and 1 × PCR buffer (Invitrogen). For genes > 1 kb in length, genomic DNA amplifications were performed within a final reaction volume of 20 µL containing 2 ng/µL genomic DNA template, 300 nm of each primer (Table 5), 1 U Expand Long Template Taq + Tgo DNA polymerase mix with proofreading activity (Roche Diagnostics GmbH, Mannheim, Germany), 200 µm dNTPs (Invitrogen), 1.5 mm MgCl2 and 1 × PCR buffer (Invitrogen). The cycling parameters were as follows for templates ≤ 1 kb : 95 °C for 5 min, 35 cycles of 94 °C for 10 s, 60 °C for 1 min, 72 °C for 1 min, followed by 10 min at 70 °C, and a 15 °C hold. This was modified for templates > 1 kb in length to have a 3‐min extension time at 72 °C. PCR products were resolved by agarose gel electrophoresis.
Table 5.
Primers used in this study.
| Gene name | Primer name | Primer sequence (5′−3′) | Usage |
|---|---|---|---|
| Vi‐ gapdh | Vi‐GAPDH‐F | GGCAAGACCATCCGTTTCTA | Assessing purity of cDNA |
| Vi‐GAPDH‐R | GACACCCATGACGAACATTG | Assessing purity of cDNA | |
| Md‐ gapdh | Md‐GAPDH‐F | TGGAAAATTGACCGGAATGT | Assessing purity of cDNA |
| Md‐GAPDH‐R | GACCTGCTGTCACCAATGAA | Assessing purity of cDNA | |
| Vice1 | Vice1AF | TCCTCACCACAACCCTCAC | qRT‐PCR |
| Vice1R | CGAAGTTACCACCAGCAACA | qRT‐PCR | |
| Vice1gF | GGCTGGAACTGCTGGCTC | Genomic DNA PCR | |
| Vice1gR | CTATACGCCCTGTTTGCA | Genomic DNA PCR | |
| Vice3 | Vice3F | ACCCAATCAACGGACCTCTC | qRT‐PCR |
| Vice3R | CCCTTCAACCATTCGCCTAC | qRT‐PCR | |
| Vice3gF | CAAGCTCCCTTGCAGCAAC | Genomic DNA PCR | |
| Vice3gR | TCAAAACTGTGCCAGGATG | Genomic DNA PCR | |
| Vice4 | Vice4F | TGAAGCTCCTACTTATTCCCATC | qRT‐PCR |
| Vice4R | GTATCGACATCGCCTGCTTT | qRT‐PCR | |
| Vice4gF | GGCATGTAAAT | Genomic DNA PCR | |
| Vice4gR | CTATCCCTTCCCACAT | Genomic DNA PCR | |
| Vice5 | Vice5RTF | TCGGAAAGGACGGTGACTAC | qRT‐PCR |
| Vice5RTR | AGACGGTTTTGGTGCTCTTG | qRT‐PCR | |
| Vice6 | Vice6F | TCGCAGTCACCTCCCAAC | qRT‐PCR |
| Vice6R | TTTGAAGCCGAGATAATGGA | qRT‐PCR | |
| Vice6gF | GCTCAAGACGCATCCTCAGCA | Genomic DNA PCR | |
| Vice6gR | CTACAAGAAAGCAGCAAAC | Genomic DNA PCR | |
| Vice7 | Vice7AF | TGGTGATGGCTCCTACACAG | qRT‐PCR |
| Vice7AR | CACATCCGTTTCCGTTTACC | qRT‐PCR | |
| Vice8 | Vice8RTF | ATTTGCCAGGCATACAAAGC | qRT‐PCR |
| Vice8RTR | AGGAGGACGAGAGTGATTGC | qRT‐PCR | |
| Vice8gF | ATGATCCTCGAGCCCAAAG | Genomic DNA PCR | |
| Vice8gR | TCATAGGTCTTCTTTGAATAG | Genomic DNA PCR | |
| Vice9 | Vice9RTF | ACAGCTGTGGATGGGAAGAA | qRT‐PCR |
| Vice9RTR | TGGGTGCCAACCACCTAC | qRT‐PCR | |
| Vice9gF | GCTGAGTATTACGATTA | Genomic DNA PCR | |
| Vice9gR | TTAGAAGAGAGCAGCAAC | Genomic DNA PCR | |
| Vice10 | Vice10F | AGGAATGGGTGACGGAGTG | qRT‐PCR |
| Vice10R | CCCTGGTCTTGGTGCTTG | qRT‐PCR | |
| Vice11 | Vice11F | TGCCATCATTGGTATTCAAG | qRT‐PCR |
| Vice11R | AAGATTTGGGTCAGCACAGG | qRT‐PCR | |
| Vice12 | Vice12AF | GCACATTCCCAACCCTCGAG | qRT‐PCR |
| Vice12R | CAGGAACCGACAAGAAGAGC | qRT‐PCR | |
| Vice14 | Vice14F | CAAGGACTGGAAGCACAAGA | qRT‐PCR |
| Vice14R | CGTTGTAGGCTGCGTTTGTG | qRT‐PCR | |
| Vice16 | Vice16CF | GTCCTGTGGCTTGGTCTTTG | qRT‐PCR |
| Vice16CR | AATGGTTGCCCGAGTGGT | qRT‐PCR | |
| β‐ Tubulin | Vi‐TUB‐F | CGTCGTGAGGCTGAAGGT | qRT‐PCR |
| Vi‐TUB‐R | CGATGGGACAACAGAGAATG | qRT‐PCR | |
| ribo L12 | Vi‐L12‐F | GTTGTCCCATCTGCCTCTTC | qRT‐PCR |
| Vi‐L12‐R | GTCCTTGGCCATTGACTTGT | qRT‐PCR |
Md‐gapdh, Malus glyceraldehyde 3‐phosphate dehydrogenase; qRT‐PCR, quantitative reverse transcriptase‐polymerase chain reaction; ribo L12, 60S ribosomal protein L12 (EU853840); β‐tubulin, EU853839; Vice, Venturia inaequalis candidate effector; Vi‐gapdh, Venturia inaequalis glyceraldehyde 3‐phosphate dehydrogenase.
qRT‐PCR
Infected and SDW‐inoculated leaves were harvested at 5 and 10 dpi, and 10‐day‐old PDB shake culture in vitro‐grown mycelia of V. inaequalis were used for RNA preparation as above. RNA was treated with DNAse I (Invitrogen) according to the manufacturer's instructions to remove any contaminating genomic DNA prior to cDNA synthesis. Absence of gDNA was confirmed by PCR using primers specific for V. inaequalis gapdh (EU873167) and apple gapdh (Table 5). cDNA was synthesized from this RNA using the Transcriptor first strand cDNA synthesis kit (Roche Diagnostics GmbH) according to the manufacturer's instructions; cDNA from two separate syntheses was pooled to control for cDNA synthesis bias. One‐in‐ten dilutions of cDNA in SDW were used for qRT‐PCR.
qRT‐PCRs were carried out on a Lightcycler® 480 instrument (Roche Diagnostics GmbH) using a SYBR Green detection system. The default cycling programme was used with the following modifications: a 62 °C annealing temperature for 10 s and a 6 s extension time. The primers used for qRT‐PCR are shown in Table 5. Data were gathered from two independent infection/growth assays, each treatment within the assays being represented by three biological samples (each sample comprising three infected leaves or material gathered from three flasks of PDB), and the PCR for each biological sample was conducted in triplicate. PCR efficiency for each set of primers was calculated using Lightcycler® 480 internal software. A gene expression normalization factor was calculated for each sample based on the geometric mean of two stably expressed reference genes (β‐tubulin; EU853839) and 60S ribosomal protein L12 (EU853840) using geNorm software v3.4 (Vandesompele et al., 2002), and used for the calculation of the relative expression of each targeted gene in each sample.
REGISTRATION OF SEQUENCES
Sequences relating to the candidate effectors can be found in the GenBank database. Vice1, EB148895; Vice2, EB149687; Vice3, EB149718; Vice4, EB149885; Vice5, FJ621507; Vice6, EB152934; Vice7, EB153294; Vice8, EB153275; Vice9, EB153319; Vice10, FJ621508; Vice11, FJ621509; Vice12, FJ621510; Vice13, FJ621511; Vice14, FJ621512; Vice15, FJ621513; Vice16, FJ621514.
ACKNOWLEDGEMENTS
This work was funded by the New Zealand Foundation for Research, Science and Technology, contract numbers C06X0302 and C06X0207. The authors would like to thank Minsoo Yoon and Robin MacDiarmid for critical review of the manuscript, Lesley Beuning, Yar‐Khing Yauk and Sarah Hollingworth for technical assistance during library construction, and Ross Crowhurst for assistance with bioinformatic analysis.
REFERENCES
- Albrecht, A. , Felk, A. , Pichova, I. , Naglik, J.R. , Schaller, M. , De Groot, P. , MacCallum, D. , Odds, F.C. , Schafer, W. , Klis, F. , Monod, M. and Hube, B. (2006) Glycosylphosphatidylinositol anchored proteases of Candida albicans target proteins necessary for both cellular processes and host–pathogen interactions. J. Biol. Chem. 281, 688–694. [DOI] [PubMed] [Google Scholar]
- Allen, R.L. , Bittner‐Eddy, P.D. , Grenville‐Briggs, L.J. , Meitz, J.C. , Rehmany, A.P. , Rose, L.E. and Beynon, J.L. (2004) Host–parasite co‐evolutionary conflict between Arabidopsis and downy mildew. Science, 306, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, M.R. , Whisson, S.C. , Pritchard, L. , Bos, J.I. , Venter, E. , Avrova, A.O. , Rehmany, A.P. , Bohme, U. , Brooks, K. , Cherevach, I. , Hamlin, N. , White, B. , Fraser, A. , Lord, A. , Quail, M.A. , Churcher, C. , Hall, N. , Berriman, M. , Huang, S. , Kamoun, S. , Beynon, J.L. and Birch, P.R.J. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA, 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balance, D.J. (1986) Sequence important for gene expression in filamentous fungi. Yeast, 2, 229–236. [DOI] [PubMed] [Google Scholar]
- Bénaouf, G. and Parisi, L. (2000) Genetics of host–pathogen relationships between Venturia inaequalis races 6 and 7 and Malus species. Phytopathology, 90, 236–242. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Nielsen, H. , Von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Van Den Berg, N. , Berger, D.K. , Hein, I. , Birch, P. , Wingfield, M. and Viljoen, A. (2007) Tolerance in banana to Fusarium wilt is associated with early up‐regulation of cell wall‐strengthening genes in the roots. Mol. Plant Pathol. 8, 333–341. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Hiller, N.L. , Liolios, K. , Win, J. , Kanneganti, T.D. , Young, C. , Kamoun, S. and Haldar, K. (2006) The malarial host‐targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathol. 2, e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs, A.R. (1990) Apple scab In: Compendium of Apple and Pear Diseases (Jones A.L. and Aldwinckle H.S., eds), pp. 6–9. St Paul, MN: APS Press. [Google Scholar]
- Bolton, M.D. , Van Esse, P. , Vossen, J.H. , De Jonge, R. , Sterigiopoulos, I. , Stulemeijer, I.J.E. , Van Den Berg, G.C.M. , Borrás‐Hidalgo, O. , Dekker, H.L. , De Koster, C.G. , De Wit, P.J.G.M. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. [DOI] [PubMed] [Google Scholar]
- Bos, J.I.B. , Armstrong, M. , Whisson, S.C. , Torto, T.A. , Ochwo, M. , Birch, P.R.J. and Kamoun, S. (2003) Intraspecific comparative genomics to identify avirulence genes from Phytophthora . New Phytol. 159, 63–72. [DOI] [PubMed] [Google Scholar]
- Bos, J.I.B , Kanneganti, T.D. , Young, C. , Cakir, C. , Huitema, E. , Win, J. , Armstrong, M.R. , Birch, P.R. and Kamoun S. (2006) The C‐terminal half of Phytophthora infestans RXLR effector AvR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana . Plant J. 48, 165–176. [DOI] [PubMed] [Google Scholar]
- Broggini, G.A.L. , Le Cam, B. , Parisi, L. , Wu, C. , Zhang, H.‐B. , Gessler, C. and Patocchi, A. (2007) Construction of a contig of BAC clones spanning the region of the apple scab avirulence gene AvrVg . Fung. Genet. Biol. 44, 44–51. [DOI] [PubMed] [Google Scholar]
- Bruzzese, E. and Hasan, S. (1983) A whole leaf clearing and staining technique for host specificity studies of rust fungi. Plant Pathol. 32, 335–338. [Google Scholar]
- Van Den Burg, H.A. , Harrison, S.J. , Joosten, M.H.A.J. , Vervoort, J. and De Wit, P.J.G.M. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant–Microbe Interact. 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Bus, V.G.M. , Laurens, F.N.D. , Van De Weg, E.W. , Rusholme, R.L. , Rikkerink, E.H.A. , Gardiner, S.E. , Bassett, H.C.M. , Kodde, L.P. and Plummer, K.M. (2005a) The Vh8 locus of a new gene for gene interaction between Venturia inaequalis and the wild apple Malus sieversii is closely linked to the Vh2 locus in Malus pumila R12740‐7A. New Phytol. 166, 1035–1049. [DOI] [PubMed] [Google Scholar]
- Bus, V.G.M. , Rikkerink, E.H.A. , Van De Weg, E.W. , Rusholme, R.L. , Gardiner, S.E. , Bassett, H.C.M. , Kodde, L.P. , Parisi, L. , Laurens, F.N.D. , Meulenbroek, E. and Plummer, K.M. (2005b) The Vh2 and Vh4 scab resistance genes in two differential hosts derived from Russian apple R12740‐7A map to the same linkage group of apple. Mol. Breed. 15, 103–116. [Google Scholar]
- Catanzariti, A‐M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni, A. , Passerini, A. , Vullo, A. and Frasconi, P. (2006) DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 34(Web Server issue), W177–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S. , Puryear, J. and Cairney, J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006). Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Choi, D. , Cho, H.‐T. and Lee, Y. (2006). Expansins: expanding importance in plant growth and development. Physiol. Plant. 126, 511–518. [Google Scholar]
- Cramer, R.A. , La Rota, C.M. , Cho, Y. , Thon, M. , Craven, K.D. , Knudson, D.L. , Mitchell, T.K. and Lawrence, C.B. (2006) Bioinformatic analysis of expressed sequence tags derived from a compatible Alternaria brassicicola–Brassica oleracea interaction. Mol. Plant Pathol. 7, 113–124. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D.G. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- DebRoy, S. , Thilmony, R. , Kwack, Y.B. , Nomura, K. and He, S.Y. (2004) A family of conserved bacterial effectors inhibits salicylic acid‐mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA, 101, 9927–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko, L. , Lau, Y.C. , Campbell, A.P. , Chenchik, A. , Moqadam, F. , Huang, B. , Lukyanov, K. , Gurskaya, N. , Sverdlov, E.D. and Siebert, P.D. (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA, 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.‐M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Chen, Y. , Wang, Q. , Wang, X. , Jiang, R.H.Y. , Arrendondo, F.D. , Anderson, R.G. , Thakur, P.B. , McDowell, J.M. , Wang, Y. and Tyler, B.M. (2008) Conserved C‐terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell, 20, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranginis, A.M. , Rauceo, J.M. , Coronado, J.E. and Lipke, P.N. (2007) A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole, D.J. , Jin, Y. , Thon, M. , Pan, H. , Bhattarai, E. , Thomas, T. and Dean, R. (2004). Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: analysis of expressed sequence tags. Mol. Plant–Microbe Interact. 17, 1337–1347. [DOI] [PubMed] [Google Scholar]
- Eisenhaber, B. , Schneider, G. , Wildpaner, M. and Eisenhaber, F. (2004) A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome‐wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae, and Schizosaccharomyces pombe . J. Mol. Biol. 337, 243–253. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O. , Nielsen, H. , Brunak, S. and Von Heijne, G. (2000) Predicting subcellular localization of proteins based on their N‐terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O. , Brunak, S. , Von Heijne, G. and Nielsen, H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Emmersen, J. , Rudd, S. , Mewes, H.‐W. and Tetko, I.V. (2007) Separation of sequences from host–pathogen interface using triplet nucleotide frequencies. Fung. Genet. Biol. 44, 231–241. [DOI] [PubMed] [Google Scholar]
- Van Esse, H.P. , Bolton, M.D. , Stergiopoulos, I. , De Wit, P.J.G.M. and Thomma, B.P.H.J. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant–Microbe Interact. 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, A.M. (2004) Investigation of proteins at the apple scab interface. PhD Thesis. Auckland: University of Auckland. [Google Scholar]
- Fitzgerald, A.M. , Van Kan, J.A.L. and Plummer, K.M. (2004) Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fung. Genet. Biol. 41, 963–971. [DOI] [PubMed] [Google Scholar]
- Friedel, C.C. , Jahn, K.H.V. , Sommer, S. , Rudd, S. , Mewes, H.W. and Tetko, I.V. (2005) Support vector machines for separation of mixed plant–pathogen EST collections based on codon usage. Bioinformatics, 21, 1383–1388. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. , Guo, M. , Jeong, B.R. , Tian, F. , Elthon, T.E. , Cemy, R.L. , Staiger, D. and Alfano, J.R. (2007) A type III effector ADP‐ribosylates RNA‐binding proteins and quells plant immunity. Nature, 447, 284–288. [DOI] [PubMed] [Google Scholar]
- Gau, A.E. , Koutb, M. , Piotrowski, M. and Kloppstech, K. (2004) Accumulation of pathogenesis‐related proteins in the apoplast of a susceptible cultivar of apple (Malus domestica cv. Elstar) after infection by Venturia inaequalis and constitutive expression of PR genes in the resistant cultivar Remo. Eur. J. Plant Pathol. 110, 703–711. [Google Scholar]
- Goodwin, S.B. (2004) Minimum phylogenetic coverage: an additional criterion to guide the selection of microbial pathogens for initial genomic sequencing efforts. Phyopathology, 94, 800–804. [DOI] [PubMed] [Google Scholar]
- Gout, L. , Fudal., I. , Kuhn, M.L. , Blaise, F. , Eckert, M. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans . Mol. Microbiol. 60, 67–80. [DOI] [PubMed] [Google Scholar]
- De Groot, P.W.J. , Hellingwerf, K.J. and Klis, F.M. (2003) Genomic‐wide identification of fungal GPI proteins. Yeast, 20, 781–796. [DOI] [PubMed] [Google Scholar]
- Hahn, M. and Mendgen, K. (1997) Characterization of in planta‐induced rust genes isolated from a haustorium‐specific cDNA library. Mol. Plant–Microbe Interact. 10, 427–437. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008). Suppression of plant resistance‐gene based immunity by a fungal effector. PLoS Pathol. 4, e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang, T. and Goodwin, P.H. (2003) Distinguishing plant and fungal sequences in ESTs from infected plant tissues. J. Microbiol. Methods, 54, 339–351. [DOI] [PubMed] [Google Scholar]
- Hu, G. , Linning, R. , McCallum, B. , Banks, T. , Cloutier, S. , Butterfield, Y. , Liu, J. , Kirkpatrick, R. , Stott, J. , Yang, G. , Smailus, D. , Jones, S. , Marra, M. , Schein, J. and Bakkeren, G. (2007) Generation of wheat leaf rust, Puccinia triticina, EST database from stage‐specific cDNA libraries. Mol. Plant Pathol. 8, 451–467. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Madan, A. (1999) CAP3: a DNA sequence assembly program. Genome Res. 9, 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, C.‐Y. , Yu, J.‐J. , Seshan, K.R. , Reichard, U. and Cole, G.T. (2002) A phase‐specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70, 3443–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tyler, B.M. , Whisson, S.C. , Hardham, A.R. and Govers, F. (2006). Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol. Biol. Evol. 23, 338–351. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 144, 323–329. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. , Cozijnsen, T.J. and De Wit, P.J.G.M. (1994) Host resistance to a fungal tomato pathogen lost by a single base‐pair change in an avirulence gene. Nature, 367, 384–386. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. , Vogelsang, R. , Cozijnsen, T.J. , Verberne, M.C. and De Wit, P.J.G.M. (1997) The biotrophic fungus Cladosporium fulvum circumvents Cf‐4‐mediated resistance by producing unstable elicitors. Plant Cell, 9, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, K. , Barrett, C. and Hughey, R. (1998) Hidden Markov Models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- Karplus, K. , Karchin, R. , Barrett, C. , Tu, S. , Cline, M. , Diekhaus, M. , Grate, L. , Casper, J. and Hughey, R. (2001) What is the value added by human intervention in protein structure prediction? Proteins: Struct. Funct. Bioinform. 45, 86–91. [DOI] [PubMed] [Google Scholar]
- Karplus, K. , Karchin, R. , Draper, J. , Casper, J. , Mandel‐Gutfreund, Y. , Diekhaus, M. and Hughey, R. (2003) Combining local‐structure, fold‐recognition, and new‐fold methods for protein structure prediction. Proteins: Struct. Funct. Genet. 53, 491–496. [DOI] [PubMed] [Google Scholar]
- Karplus, K. , Katzman, S. , Shackleford, G. , Koeva, M. , Draper, J. , Barnes, B. and Soriano, M. (2005) SAM‐T04: what's new in protein‐structure prediction for CASP6. Proteins: Struct. Funct. Bioinform. 61, 135–142. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Kemen, A.C. , Rafiqi, M. , Hempel, U. , Mendgen, K. , Hahn, M. and Voegele, R.T. (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant–Microbe Interact. 18, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Kleemann, J. , Takahara, H. , Stüber, K. and O’Connell, R. (2008) Identification of soluble secreted proteins from appressoria of Colletotrichum higginsianum by analysis of expressed sequence tags. Microbiology, 154, 1204–1217. [DOI] [PubMed] [Google Scholar]
- Klein, R.D. , Gu, O.M. , Goddard, A. and Rosenthal, A. (1996) Selection for genes encoding secreted proteins and receptors. Proc. Natl. Acad. Sci. USA, 93, 7108–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (1986) Point mutations define a sequence flanking the AUG initiation codon that modulates translation by eukaryotic ribosomes. Cell, 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , Von Heijne, G. and Sonnhammer, E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kruijt, M. , De Kock, M.D.J. and De Wit, P.J.G.M. (2005) Receptor‐like proteins involved in plant disease resistance. Mol. Plant Pathol. 6, 85–97. [DOI] [PubMed] [Google Scholar]
- Kucheryava, N. , Bowen, J.K. , Sutherland, P. , Conolly, J.J. , Mesarich, C.H. , Rikkerink, E.H.A. , Kemen, E. , Plummer, K.M. , Hahn, M. and Templeton, M.D. (2008) Two novel Venturia inaequalis genes induced upon morphogenetic differentiation during infection and in vitro growth on cellophane. Fung. Genet. Biol. 45, 1329–1339. [DOI] [PubMed] [Google Scholar]
- Lee, S.J. , Kelley, B.S. , Damasceno, C.M.B. , John, B.S. , Kim, B.S. , Kim, B.D. and Rose, J.K.C. (2006) A functional screen to characterize the secretomes of eukaryotic pathogens and their hosts in planta . Mol. Plant–Microbe Interact. 19, 1368–1377. [DOI] [PubMed] [Google Scholar]
- Lespinasse, Y. (1994) Apple scab resistance and durability. New races and strategies for the future In: Progress in Temperate Fruit Breeding (Schmidt H. and Kellerhals M., eds), pp. 105–106. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Li, F. and Palecek, S.P. (2008) Distinct domains of the Candida albicans adhesin Eap1p mediate cell–cell and cell–substrate interactions. Microbiology, 154, 1193–1203. [DOI] [PubMed] [Google Scholar]
- Li, R. , Rimmer, R. , Buchwaldt, L. , Sharpe, A.G. , Séguin‐Swartz, G. , Coutu, C. and Hegedus, D.D. (2004). Interaction of Sclerotinia sclerotiorum with a resistant Brassica napus cultivar: expressed sequence tag analysis identifies genes associated with fungal pathogenesis. Fung. Genet. Biol. 41, 735–753. [DOI] [PubMed] [Google Scholar]
- Linder, T. and Gustafsson, C.M. (2008) Molecular phylogenetics of ascomycotal adhesins—a novel family of putative cell‐surface adhesive proteins in fission yeasts. Fung. Genet. Biol. 45, 485–497. [DOI] [PubMed] [Google Scholar]
- Link, T. and Voegele, R.T. (2008) Secreted proteins of Uromyces fabae: similarities and stage specificity. Mol. Plant Pathol. 9, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHardy, W.E. (1996) Apple Scab: Biology, Epidemiology, and Management. St. Paul, MN: The American Phytopathological Society Press. [Google Scholar]
- Maor, R. , Kosman, E. , Golobinski, R. , Goodwin, P. and Sharon, A. (2003) PF‐IND: probability algorithm and software for separation of plant and fungal sequences. Curr. Genet. 43, 296–302. [DOI] [PubMed] [Google Scholar]
- Mulder, N. , Apweiler, R. , Attwood, T.K. , Bairoch, A. , Bateman, A. , Binns, D. , Bork, P. , Buillard, V. , Cerutti, L. , Copley, R. , Courcelle, E. , Das, U. , Daugherty, L. , Dibley, M. , Finn, R. , Fleischmann, W. , Gough, J. , Haft, D. , Hulo, N. , Hunter, S. , Kahn, D. , Kanapin, A. , Kejariwal, A. , Labarga, A. , Langendijk‐Genevaux, P.S. , Lonsdale, D. , Lopez, R. , Letunic, I. , Madera, M. , Maslen, J. , McAnulla, C. , McDowall, J. , Mistry, J. , Mitchell, A. , Nikolskaya, A.N. , Orchard, S. , Orengo, C. , Petryszak, R. , Selengut, J.D. , Sigrist, C.J.A. , Thomas, P.D. , Valentin, F. , Wilson, D. , Wu C.H. and Yeats, C. (2007) New developments in the InterPro database. Nucleic Acids Res. 35(Database issue), D224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb, R.D. , Crowhurst, R.N. , Gleave, A.P. , Rikkerink, E.H.A. , Allan, A.C. , Beuning, L.L. , Bowen, J.H. , Gera, E. , Jamieson, K.R. , Janssen, B.J. , Laing, W.A. , McArtney, S. , Nain, B. , Ross, G.S. , Snowden, K.C. , Souleyre, E.J.F. , Walton, E.F. and Yauk, Y.‐K. (2006) Analyses of expressed sequence tags from apple. Plant Physiol. 141, 147–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H. , Engelbrecht, J. , Brunak, S. and Von Heijne, G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot. Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Nomura, K. , DebRoy, S. , Lee, Y.H. , Pumplin, N. , Jones, J. and He, S.Y. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science, 313, 220–223. [DOI] [PubMed] [Google Scholar]
- Orbach, M.J. , Farrall, L. , Sweigard, J.A. , Chumley, F.G. and Valent, B. (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi‐ta . Plant Cell, 12, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, L. and Lespinasse, Y. (1996) Pathogenicity of Venturia inaequalis strains of race 6 on apple clones (Malus sp.). Plant Dis. 80, 1179–1183. [Google Scholar]
- Parisi, L. , Lespinasse, Y. , Guillaumes, J. and Krüger, J. (1993). A new race of Venturia inaequalis virulent to apples with resistance due to the Vf gene. Phytopathology, 83, 533–537. [Google Scholar]
- Parker, D.M. , Hilber, U.W. , Bodme, M. , Smith, F.D. , Yao, C. and Köller, W. (1995) Production and transformation of conidia of V. inaequalis . Phytopathology, 85, 87–91. [Google Scholar]
- Van Poppel, P.M.J.A. , Guo, J. , Van De Vondervoort, P.J.I. , Jung, M.W.M. , Birch, P.R.J. , Whisson, S.C. and Govers, F. (2008) The Phytophthora infestans avirulence gene Avr4 encodes an RXLR‐dEER effector. Mol. Plant–Microbe Interact. 21, 1460–1470. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. , Tatusova, T. and Maglott, D.R. (2007) NCBI Reference Sequence (RefSeq): a curated non‐redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35(Database issue), D61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon, E. , Silventoinen, V. , Pillai, S. , Harte, N. , Mulder, N. , Apweiler, R. and Lopez, R. (2005) InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Hraber, P.T. , Sobral, B.W. and Gijzen, M. (2000) Comparative analysis of expressed sequences in Phytophthora sojae . Plant Physiol. 123, 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauyaree, P. , Choi, W. , Fang, E. , Blackmon, B. and Dean, R.A. (2001) Genes expressed during early stages of rice infection with the rice blast fungus, Magnaporthe grisea . Mol. Plant Pathol. 2, 347–354. [DOI] [PubMed] [Google Scholar]
- Rehmany, A.P. , Gordon, A. , Rose, L.E. , Allen, R.L. , Armstrong, M.R. , Whisson, S.C. , Kamoun, S. , Tyler, B.M. , Birch, P.R. and Beynon, J.L. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell, 17, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep, M. (2005) Small proteins of plant‐pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253, 19–27. [DOI] [PubMed] [Google Scholar]
- Richard, G.F. and Dujon, B. (2006) Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol. Biol. Evol. 23, 189–202. [DOI] [PubMed] [Google Scholar]
- Richard, M.L. and Plaine, A. (2007) A comprehensive analysis of GPI anchored proteins in Candida albicans . Euk. Cell, 6, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout, C.J. , Skamnioti, P. , Porritt, O. , Sacristan, S. , Jones, J.D. and Brown, J.K. (2006) Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell, 18, 2402–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, A.L. and Crute, I.R. (1994) Apple scab resistance from Malus floribunda 821 (Vf) is rendered ineffective by isolates of Venturia inaequalis from Malus floribunda . Nor. J. Agric. Sci. 17, 404–406. [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shan, W. , Cao, M. , Leung, D. and Tyler, B.M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b . Mol. Plant–Microbe Interact. 17, 394–403. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L. , Von Heijne, G. and Krogh, A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences In: Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (Glasgow J., Littlejohn T., Major F., Lathrop R., Sankoff D. and Sensen C., eds), pp. 175–182. Menlo Park, CA: AAAI Press. [PubMed] [Google Scholar]
- Stephenson, S.A. , Hatfield, J. , Rusu, A. , Maclean, D.J. and Manners, J.M. (2000) CgDN3: an essential pathogenicity gene of Colletotrichum gloeosporioides necessary to avert a hypersensitive‐like response in the host Stylosanthes guianensis . Mol. Plant–Microbe Interact. 13, 929–941. [DOI] [PubMed] [Google Scholar]
- Suzek, B.E. , Huang, H. , McGarvey, P. , Mazumder, R. and Wu, C.H. (2007) UniRef: comprehensive and non‐redundant UniProt reference clusters. Bioinformatics, 23, 1282–1288. [DOI] [PubMed] [Google Scholar]
- Sweigard, J.A. , Carroll, A.M. , Kang, S. , Farrall, L. , Chumley, F.G. and Valent, B. (1995) Identification, cloning, and characterization of PWL2, a gene for host species‐specificity in the rice blast fungus. Plant Cell, 7, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerstra, W.R. , Deelstra, H.J. , Vranes, M. , Bohlmann, R. , Kahmann, R. , Kämper, J. and Wösten, H.A.B. (2006) Repellents have functionally replaced hydrophobins in mediating attachment to a hydrophobic surface and in formation of hydrophobic aerial hyphae in Ustilago maydis . Microbiology, 152, 3607–3612. [DOI] [PubMed] [Google Scholar]
- Templeton, M.D. , Rikkerink, E.H.A. and Beever, R.E. (1994) Small, cysteine‐rich proteins and recognition in fungal–plant interactions. Mol. Plant–Microbe Interact. 7, 320–325. [Google Scholar]
- Thara, V.K. , Fellers, J.P. and Zhou, J.M. (2003) In planta induced genes of Puccinia triticina . Mol. Plant Pathol. 4, 51–56. [DOI] [PubMed] [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Tetsa, A. , Gow, N.A.R. , Van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto‐Alalibo, T.A. , Tripathy, S. , Smith, B.M. , Arredondo, F.D. , Zhou, L. , Li, H. , Chibucos, M.C. , Qutob, D. , Gijzen, M. , Mao, C. , Sobral, B.W.S. , Waugh, M.E. , Mitchell, T.K. , Dean, R.A. and Tyler, B.M. (2007) Expressed sequence tags from Phytophthora sojae reveal genes specific to development and infection. Mol. Plant–Microbe Interact. 20, 781–793. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. and Speleman, F. (2002) Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3(research), 00341–0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen, K.J. , Jansen, A. , Lewitter, F. and Fink, G.R. (2005) Intergenic tandem repeats generate functional variability. Nat. Genet. 37, 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer, B.A. , Patocchi, A. , Gianfranceschi, L. , Tartarini, S. , Zhang, H.B. , Gessler, C. and Sansavini, S. (2001) Apple contains receptor‐like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes co‐segregating with Vf apple scab resistance. Mol. Plant–Microbe Interact. 14, 508–515. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.‐K. , Wang, M. , Bouwmeester, K. , Vosman, B. , Visser, R.G.F. , Jacobsen, E. , Govers, F. , Kamoun, S. and Van Der Vossen, E.A.G. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One, 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. and St. Leger, R.J. (2007) The MAD1 adhesin of Metarhizium anisopliae links adhesion with blastospore production and virulence to insects, and the MAD2 adhesin enables attachment to plants. Eukaryot. Cell, 6, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , Van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–119. [DOI] [PubMed] [Google Scholar]
- Win, J. (2003) Molecular quest for avirulence factors in Venturia inaequalis . PhD Thesis. Auckland: University of Auckland. [Google Scholar]
- Win, J. , Greenwood, D.R. and Plummer, K.M. (2003) Characterization of a protein from Venturia inaequalis that induces necrosis in Malus carrying the Vm resistance gene. Physiol. Mol. Plant Pathol. 62, 193–202. [Google Scholar]
- Xia, Y. (2004) Proteases in pathogenesis and plant defence. Cell. Microbiol. 6, 905–913. [DOI] [PubMed] [Google Scholar]
- Xu, M. and Korban, S.S. (2002) A cluster of four receptor‐like genes resides in the Vf Locus that confers resistance to apple scab disease. Genetics, 162, 1995–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev, I.A. , Hietal, A.M. , Steffenrem, A. , Solheim, H. and Fossdal, C.G. (2008) Identification and analysis of differentially expressed Heterobasidion parviporum genes during natural colonization of Norway Spruce stems. Fung. Genet. Biol. 45, 498–513. [DOI] [PubMed] [Google Scholar]
- Yuan, S. , Wu, Y. and Cosgrove, D.J. (2001) A fungal endoglucanse with plant cell wall extension activity. Plant Physiology, 127, 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Meakin, H. and Dickinson, M. (2003) Isolation of genes expressed during compatible interactions between leaf rust (Puccinia triticina) and wheat using cDNA‐AFLP. Mol. Plant Pathol. 4, 469–477. [DOI] [PubMed] [Google Scholar]


