SUMMARY
In eukaryotic genomes, gene expression and DNA recombination are affected by structural chromatin traits. Chromatin structure is shaped by the activity of enzymes that either introduce covalent modifications in DNA and histone proteins or use energy from ATP to disrupt histone–DNA interactions. The genomic ‘marks’ that are generated by covalent modifications of histones and DNA, or by the deposition of histone variants, are susceptible to being altered in response to stress. Recent evidence has suggested that proteins generating these epigenetic marks play crucial roles in the defence against pathogens. Histone deacetylases are involved in the activation of jasmonic acid‐ and ethylene‐sensitive defence mechanisms. ATP‐dependent chromatin remodellers mediate the constitutive repression of the salicylic acid‐dependent pathway, whereas histone methylation at the WRKY70 gene promoter affects the activation of this pathway. Interestingly, bacterial‐infected tissues show a net reduction in DNA methylation, which may affect the disease resistance genes responsible for the surveillance against pathogens. As some epigenetic marks can be erased or maintained and transmitted to offspring, epigenetic mechanisms may provide plasticity for the dynamic control of emerging pathogens without the generation of genomic lesions.
INTRODUCTION
Long‐standing, intimate associations between plants and microbial pathogens have forced the incessant selection of antagonistic capabilities on each of these respective organisms. As a consequence, sophisticated mechanisms of the promotion of plant defence and pathogen virulence have developed. Plants counteract pathogens by means of an innate immune system that relies on cell autonomous responses and multiple defence layers. This involves the detection of pathogen‐associated molecular patterns (PAMPs) and the stimulation of basal defences defined as ‘PAMP‐triggered immunity’ (PTI) (Ausubel, 2005; Zipfel, 2008), as well as the recognition of pathogen‐derived effectors by intracellular resistance proteins (R) and the activation of ‘effector‐triggered immunity’ (ETI) (Bent and Mackey, 2007; Jones and Dangl, 2006). Most R proteins that provide the basis for ETI and disease resistance carry nucleotide binding site‐leucine‐rich repeat (NBS‐LRR) domains. The NBS‐LRR gene family is one of the largest in plants and contains polymorphic and rapidly evolving genes that are often arranged in clusters. One process that leads to the reshuffling and evolution of NBS‐LRR genes involves mispairing and recombination between transposon elements (TEs) and sequence repeats, which are abundant in these clusters (Baumgarten et al., 2003; McHale et al., 2006; Meyers et al., 2003). However, the mechanisms underlying enhanced recombination at these loci have not been established.
The stimulation of PTI and ETI involves extensive reprogramming of plant transcription. In order for changes in gene expression to contribute to selective pathogen resistance, specific defence circuits must become activated immediately after infection (Bent and Mackey, 2007; Jones and Dangl, 2006; Tsuda et al., 2009). The signals for resistance to biotrophic pathogens occur mainly through the salicylic acid (SA)‐dependent pathway and usually lead to hypersensitive cell death, whereas defences against necrotrophs are mainly stimulated by the jasmonic acid/ethylene (JA/ET) pathway. These two defence programmes maintain antagonistic or synergistic interactions (Bari and Jones, 2009; Koornneef and Pieterse, 2008; Vlot et al., 2009). Global changes in gene expression that are stimulated through the SA and JA pathways have been characterized in detail (Tsuda et al., 2009). More recently, studies have begun to analyse how chromatin structure affects the expression of defence genes.
The chromatin compaction level influences several genomic functions. High‐order chromatin organization involves different levels of DNA packing. At the basic level, 147 bp of DNA are wrapped in a 1.7 turn around an octamer of core histones (two subunits of H2A, H2B, H3 and H4), forming the structural and functional chromatin unit termed the nucleosome. Neighbouring nucleosomes are separated by stretches of 10–50 bp of unwrapped DNA that is associated with the linker histone H1, which is primarily responsible for the condensation of the 30‐nm DNA fibre. The packing of DNA into fibres occludes the access of proteins to components that regulate transcription, DNA repair, replication and recombination. Therefore, in order for these components to be exposed, chromatin relaxation is required. Chromatin unfolding involves the action of ATP‐dependent remodelling complexes (Jerzmanowski, 2007; de la Serna et al., 2006), covalent modification of histone proteins (Berger, 2007; Kouzarides, 2007; Pfluger and Wagner, 2007; Zhang et al., 2007b), deposition of histone variants (Draker and Cheung, 2009; Guillemette and Gaudreau, 2006; March‐Diaz and Reyes, 2009; Zlatanova and Thakar, 2008) or DNA methylation changes (Gehring and Henikoff, 2007; Zilberman, 2008). In addition, noncoding small RNAs (smRNAs) are responsible for directing heterochromatin formation at specific genomic sequences (Chan, 2008).
In this work, we summarize the data that sustain the existence of epigenetic control of plant immunity. We focus on selected studies, mainly from Arabidopsis, that reveal how covalent modifications of histones and DNA or ATP‐dependent chromatin remodelling events regulate the defence cascades. In addition, we discuss how pathogen‐induced epigenetic modifications may contribute to the transgeneration memory of stress. The effects of smRNAs on chromatin structure and plant disease resistance have been summarized recently in excellent reviews and will not be covered here (Chan, 2008; Jin, 2008; Ruiz‐Ferrer and Voinnet, 2009).
POST‐TRANSLATIONAL MODIFICATIONS (PTMs) OF HISTONE PROTEINS
Modifications that occur on all core histones have been characterized recently by mass spectrometric studies in Arabidopsis (Zhang et al., 2007b). Most, but not all, are present in the animal kingdom where they display similar functions as in plants (Fuchs et al., 2006; Kouzarides, 2007; Pfluger and Wagner, 2007). Nearly 60 residues on histones are modified by enzymes that add or remove chemical groups. Residues from the N‐ or C‐terminal histone tails that protrude from the core (30% of the protein mass) are the most frequently altered. However, residues from the central domain of histones that maintain the structural organization of nucleosomes also undergo modification (Luger et al., 1997). Histone‐modifying enzymes seem to be recruited to specific genomic regions by transcription factors. The major PTMs include methylation, acetylation, phosphorylation, ubiquitylation, sumoylation and ADP‐ribosylation, with the first three being the best characterized (Berger, 2007; Fuchs et al., 2006; Kouzarides, 2007; Pfluger and Wagner, 2007). Most histone PTMs are removable and the heritability of these marks through cell division is still controversial (Berger, 2007; Kouzarides, 2007). However, in plants, methylation and acetylation of histone proteins are required for the maintenance of imprinting mechanisms (Haun and Springer, 2008).
Histone PTMs are thought to change chromatin structure through at least two different mechanisms. The first includes local alterations of electrostatic charges that modify contacts between nucleosomes. The second involves the recruitment of nonhistone proteins to open or close chromatin. Although several histone PTMs affect transcriptional competence, a modification by itself does not define active or silenced chromatin (Fuchs et al., 2006; Vaillant and Paszkowski, 2007). Rather, its effect may depend on the extent of the modification (i.e. mono‐ to trimethylation), the gene region involved, the combination of other chromatin modifications and the nature of the genome (Fuchs et al., 2006; Kouzarides, 2007; Pfluger and Wagner, 2007). In plants, transcriptionally silent chromatin contains hypoacetylated H3 and H4, methylated lysine 27 (H3K27) and lysine 9 of histone H3 (H3K9), and hypermethylated DNA. Conversely, active chromatin in plants exhibits enhancement of H3 and H4 acetylation, trimethylation of lysine 4 from histone H3 (H3K4me3) and DNA hypomethylation (Bernatavichute et al., 2008; Cokus et al., 2008; Lister et al., 2008; Pfluger and Wagner, 2007; Tian et al., 2005; Vaillant and Paszkowski, 2007; 2006, 2007c, 2009; Zilberman et al., 2007).
Histone acetylation
Histone acetyltransferases (HATs) and histone deacetylases (HDACs)
The combined antagonistic activities of HATs and HDACs determine the acetylation level of lysine residues on histone tails. Acetylation was recognized early on as an indicator of actively transcribed genes, and several HATs have been identified as transcriptional co‐activators (Berger, 2007; Kouzarides, 2007; Pfluger and Wagner, 2007; Yang and Seto, 2008). In addition, HDACs have been found to mediate gene repression (Tanaka et al., 2008; Tian et al., 2005; Wu et al., 2000). However, in some cases, HDACs have been associated with transcriptional activation (2002, 2009; Zupkovitz et al., 2006). For instance, the distribution analysis of five HATs and four HDACs in human T cells showed that, in addition to HATs, HDACs are enriched in active genes and correlate with transcription. These HDACs are thought to avoid cryptic transcriptional initiation and chromatin instability through the removal of acetyl groups introduced by HATs (Wang et al., 2009).
In plants, HATs and HDACs modulate the expression of developmental and stress‐sensitive genes (Bharti et al., 2004; Chen and Tian, 2007; Kim et al., 2004; Sridha and Wu, 2006; Zhu et al., 2008). In addition, HDACs have been implicated in defences against pathogens.
HDACs and JA‐dependent defences
A fungal product called HC‐toxin was identified by looking for primary determinants of the corn leaf disease caused by Cochiobolus (Helminthosporium) carbonum. Resistance to this pathogen was found to be conferred by maize alleles that encode a carbonyl reductase which inactivates the HC‐toxin (Hm1/2 alleles; Johal and Briggs, 1992). This toxin is a cyclic tetrapeptide that acts as a potent inhibitor of HDACs from yeast, insects, mammals and plants. Among the plant HDACs, proteins from the Reduced Potassium Dependency protein 3/Histone Deacetylase 1 (RPD3/HDA1) and HD2 classes are the only members that become inhibited by the toxin. Silent Information Regulator 2 (SIR2)‐like HDACs and HATs are not affected by this toxin (Brosch et al., 1995; Chen and Tian, 2007; Walton, 2006; Yang and Seto, 2008). Treatments with the HC‐toxin or virulent fungal strains cause histone hyperacetylation in susceptible, but not resistant, maize plants. Therefore, HDACs seem to act as primary targets of the HC‐toxin, with histones being the major substrates of HDACs during infection (Brosch et al., 1995; Ransom and Walton, 1997). However, transcription factors, cytoskeletal proteins, proteins involved in DNA repair and others are also substrates of HDACs (Cohen et al., 2004; Glozak et al., 2005; Hubbert et al., 2002; Juan et al., 2000). This fact, together with the abundant list of maize genes encoding HDACs that are sensitive to the HC‐toxin, prevented the elucidation of the role of these enzymes in resistance to C. carbonum (Walton, 2006). Unfortunately, in the genetically tractable Arabidopsis thaliana–Alternaria brassicicola pathosystem, in which plant HDACs are inhibited by the fungal derivative depudecin, depudecin was shown not to be an important virulence factor (Wight et al., 2009). Therefore, novel or improved models will be required to elucidate the contribution of HDACs in fungal resistance.
Additional evidence that HDACs are involved in defence responses against pathogens implicates the activation of the JA pathway in Arabidopsis. HDAC19 is a nuclear RPD3/HDA1 family protein that has HDAC activity in vitro. Loss of this protein increases acetylation on histones by 10‐fold and mainly affects gene promoters (Fong et al., 2006; Zhou et al., 2005). The AtHDAC19 gene is induced by Alternaria brassicicola and exogenous JA. Overexpression of the gene enhances fungal resistance through the apparent activation of the Ethylene Responsive Factor 1 (ERF1), whereas silencing of the gene increases fungal susceptibility (Zhou et al., 2005). However, because HDAC19 deficiency affects multiple developmental traits, the direct effect of this enzyme on pathogen‐induced defence pathways is questionable (Tian and Chen, 2001; Tian et al., 2005; Zhou et al., 2005). This type of effect may involve the recruitment of HDAC19 to the promoters of JA‐sensitive genes by ERF factors, as similar hormone‐sensitive complexes regulate stress‐sensitive genes in Arabidopsis (Song and Galbraith, 2006). In addition, HDAC19 influences defences against Pseudomonas syringae through interaction with WRKY38 and WRKY62, two transcription factors that repress the SA pathway (Kim et al., 2008).
HDAC6 is another RPD3/HDA1‐type HDAC from Arabidopsis that is involved in the activation of JA‐dependent defences. This enzyme affects transgene silencing, DNA methylation and the activity of rRNA genes (Aufsatz et al., 2007). Similar to AtHDAC19, the expression of AtHDAC6 is induced by exogenous JA (Zhou et al., 2005). Interestingly, HDAC6 interacts with Coronatine Insensitive 1 (COI1), an F‐box protein that mediates JA signalling (Devoto et al., 2002). This interaction seems to have functional significance, as HDAC6 is required for JA‐dependent responses, including PDF1.2, VSP2 and ERF1 expression (Wu et al., 2008). As HDAC6 would have less impact on development than HDAC19 (Aufsatz et al., 2007; Probst et al., 2004; Tian and Chen, 2001), the study of HDAC6 in disease resistance is encouraging.
In summary, compelling evidence suggests that HDACs promote defence responses against pathogens, with those dependent on the JA pathway being the best characterized (Table 1). The distribution profiles of HDACs in whole‐plant genomes and the identification of their critical substrates in infection still remain to be elucidated. The precise mechanisms by which these enzymes modulate gene expression are also presently unknown.
Table 1.
Effects of plant chromatin components on immune responses.
| Protein/gene and function | Role on immunity | References |
|---|---|---|
| Maize HDACs targeted by the HC‐toxin from Cochiobolus carbonum | na | Brosch et al. (1995); Johal and Briggs (1992); Ransom and Walton (1997); Walton (2006) |
| HDAC19 (At4g38130) | Activation of resistance against Alternaria brassicicola | Zhou et al. (2005) |
| HDA6 (At5g63110) | Activation of JA‐sensitive genes | Wu et al. (2008) |
| ATX1(At2g31650), putative H3K4 methyltransferase | Activation of WRKY70 and SA‐sensitive genes and basal resistance to Pseudomonas syringae | 2006, 2007) |
| PIE1 (At3g12810), SEF (At5g37055), and H2A.Z (At1g52740 and At3g54560), members of the Swr1‐like complex | Constitutive repression of the SA pathway | March‐Diaz et al. (2008); March‐Diaz and Reyes (2009) |
| BRM (At2g46020), Snf2‐like protein | Constitutive repression of the SA pathway | Bezhani et al. (2007) |
| SNI1 (At4g18470), putative chromatin remodeller exclusive from plants | Constitutive repression of the SA pathway | Durrant et al. (2007); Mosher et al. (2006) |
| SYD (At2g28290), Snf2‐like protein | Activation of JA/ET‐sensitive genes and resistance against Botrytis cinerea | Walley et al. (2008) |
| DDM1 (At5g66750), Snf2‐like protein affecting DNA methylation | Maintenance of NBS‐LRR gene stability? | Stokes et al. (2002); 2007, 2009) |
ATX1, Arabidopsis Trithorax 1; BRM, Brahma; DDM1, Decrease in DNA Methylation 1; ET, ethylene; HDAC, histone deacetylase; JA, jasmonic acid; na, not available; NBS‐LRR, nucleotide binding site‐leucine‐rich repeat; PIE1, Photoperiod‐Independent Early flowering 1; SA, salicylic acid; SNI1, Suppressor of NPR1, Inducible 1; SYD, Splayed.
Histone methylation
Methylation of H3 lysine residues
In plants, mono‐, di‐ and trimethylation (me1,2,3) of lysine residues (K) at positions 4, 9, 27 and 36 of H3 histone are well‐characterized modifications (Pfluger and Wagner, 2007; Vaillant and Paszkowski, 2007). High‐resolution distribution maps of H3K27me3 (Turck et al., 2007; Zhang et al., 2007c), H3K9me2 (Bernatavichute et al., 2008) and H3K4me1,2,3 (Zhang et al., 2009) have been described for Arabidopsis. In this genome, H3K27me3 is a major euchromatic repressive modification associated with more than 4000 genes in their repressed state, which is abundant at the 5′ end of transcribed regions and is maintained by mechanisms that are independent of DNA methylation and smRNAs (Zhang et al., 2007c). H3K9me3 is another repressive euchromatic modification from gene‐encoding regions that does not overlap with H3K27me3 (Turck et al., 2007). H3K9me2, in turn, associates with heterochromatic TEs, pseudogenes and repeat elements (Bernatavichute et al., 2008; Fuchs et al., 2006; Gendrel et al., 2002; Mathieu et al., 2005; Turck et al., 2007). In contrast, all three H3K4 methylation marks are associated with active chromatin, occur almost exclusively on genes and at least one is present on two‐thirds of all genes (Zhang et al., 2009). H3K4me3 and H3K4me2 are enriched at the promoter and 5′ gene regions, with H3K4me2 located further downstream, whereas H3K4me1 is enhanced at the transcribed and 3′ end regions. Among these modifications, H3K4me3 is the only one that is associated with active transcription. Interestingly, H3K4me2 and H3K4me3 are mutually exclusive with all three types of DNA methylation (Zhang et al., 2009).
Similarly, high‐resolution maps of H3K4me2 and H3K4me3, and DNA methylation from two complete rice chromosomes, indicate that H3K4 methylation is enriched at the 5′ end of genes, displays a direct correlation with transcript abundance and shows an inverse relationship with DNA methylation (Li et al., 2008).
H3K4 methylation and SA‐dependent defences
Interestingly, methylation of H3K4 at the nucleosomes of WRKY70 stimulates SA‐dependent defence responses (Álvarez‐Venegas et al., 2007) (Table 1). In Arabidopsis, WRKY70 regulates the cross‐talk between the SA‐ and JA‐sensitive responses by stimulating the first pathway to repress the second. In wild‐type plants, P. syringae induces the expression of WRKY70 as well as the reduction of H3K27me2 and the accumulation of H3K4me2 and H3K4me3 at WRKY70 nucleosomes. Importantly, the modifications found at WRKY70 nucleosomes in infected plants are associated with the activity of Arabidopsis Trithorax 1 (ATX1), a SET‐domain protein that acts as an H3K4 methylase (Alvarez‐Venegas et al., 2003). Infected atx1 mutant plants show weak activation of WRKY70 and, in these plants, WRKY70 nucleosomes lack H3K4me3 but contain H3K27me2 and H3K4me2 levels comparable with those of infected wild‐type plants. Thus, transcriptional activation of WRKY70 is induced by ATX1 through apparent trimethylation of H3K4 (Álvarez‐Venegas et al., 2007) (Fig. 1). As ATX1 influences the expression of TIR‐NBS‐LRR genes and several transcription factors involved in defence, such as WRKYs, TGA‐bZIP and ERFs (Álvarez‐Venegas et al., 2006), H3K4 methylation may also modulate responses against other pathogens. The effect of H3K4me3 on transcription is currently being investigated. Its enhancement in active genes is observed in plants and other organisms, where it may recruit factors that stimulate gene activity (Li et al., 2008; Sims and Reinberg, 2006; Zhang et al., 2009). However, H3K4me3 is also associated with inactive genes that initiate, but do not complete, transcription (Guenther et al., 2007). The latter observation is consistent with the detection of H3K4me3 in inactive PR1 and THI2.1 genes (Álvarez‐Venegas et al., 2007).
Figure 1.
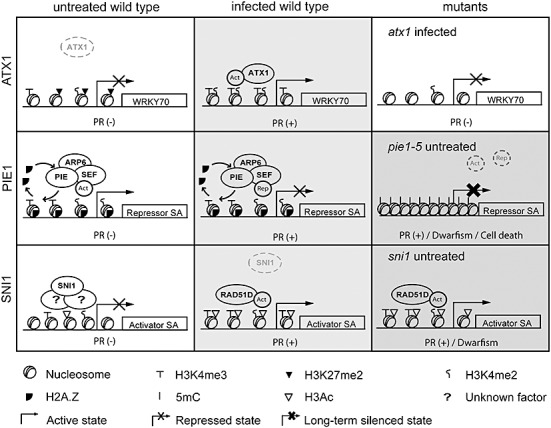
Model for Arabidopsis Trithorax 1 (ATX1)‐, Photoperiod‐Independent Early flowering 1 (PIE1)‐ and Suppressor of NPR1, Inducible 1 (SNI1)‐mediated epigenetic control of the salicylic acid (SA) pathway. Top: After infection, ATX1 binds the WRKY70 nucleosomes and mediates the trimethylation of H3K4. This modification recruits transcriptional activators and leads to WRKY70 expression and subsequent pathogenesis‐related (PR) gene induction. The absence of H3K4‐me3 on the WRKY70 nucleosomes in the atx1 mutant impairs the activation of WRKY70 after infection. Middle: The Swr1 complex (PIE/SEF/ARP6) introduces H2A.Z at the promoter of a gene encoding a repressor of the SA pathway (Repressor SA). H2A.Z and methylated H3K4 maintain the accessibility of this promoter to transcriptional regulators. Activators (Act) or repressors (Rep) bind this region under basal or infection conditions, respectively. Although the removal of H2A.Z may take place during transcription (curved arrows), this modification protects the promoter from cytosine methylation (5mC). The absence of H2A.Z in pie1‐5 plants leads to chromatin compaction, DNA methylation and gene silencing, and the subsequent activation of defence mechanisms in noninfected tissues that causes growth retardation. Bottom: In basal conditions, SNI1 prevents the expression of positive regulators of the SA pathway (Activator SA) by maintaining local histone modifications that reduce chromatin accessibility. After infection, SNI1 is released from the promoter and chromatin is relaxed by RAD51D activity. SNI1 may affect chromatin structure through interactions with other proteins of unknown nature. The grey scale illustrates the degrees in SA pathway activation (white, no activation). Dwarfism and cell death phenotypes most probably result from constitutive activation of the pathway.
CHROMATIN REMODELLERS
Chromatin remodellers are multiprotein complexes that alter histone–DNA interactions to move, disrupt or form nucleosomes through the use of energy derived from ATP hydrolysis. These complexes include catalytic subunits from the Sucrose nonfermenting 2 (Snf2) family of DNA helicases/ATPases and are recruited to specific promoters through the interaction with accessory proteins or transcription factors (Mohrmann and Verrijzer, 2005; de la Serna et al., 2006). Snf2 proteins are evolutionarily conserved and are classified into groups of subfamilies, as well as subfamilies based on archetypal members (Flaus et al., 2006; Knizewski et al., 2008). Although no Snf2 complex has been isolated in plants as yet, our knowledge of these proteins has markedly increased in recent years (Jerzmanowski, 2007). Arabidopsis contains 41 Snf2‐like genes belonging to six subfamily groups (http://www.chromdb.org), with four of these genes being functionally related to disease resistance (Table 2).
Table 2.
Classes of Arabidopsis Snf2 encoding genes involved in disease resistance.
| Groups of subfamilies* | Subfamilies | Genes involved in defence |
|---|---|---|
| Swr1‐like | Swr1 (Swi2/Snf2‐related 1) | PIE1 (At3g12810) |
| Etl1 (Enhancer trap locus 1) | ||
| Ino80 (Inositol requiring protein 80) | ||
| Snf2‐like | Snf2 (Sucrose nonfermenting 2) | SYD (At2g28290), BRM (At2g46020) |
| Iswi (Imitation switch) | ||
| Chd1 (Chromodomain and helicase‐like domain 1) | ||
| Lsh (Lymphoid‐specific helicase) | DDM1 (At5g66750) | |
| Mi‐2 (Mitosis‐2) | ||
| ALC1 (Amplified in Liver Cancer 1) |
The groups are defined according to Knizewski et al. (2008) and Flaus et al. (2006). Only two groups of subfamilies contain members associated with disease resistance; the remaining groups include RAD54‐like, RAD5/16‐like, SSO1653‐like and SMARCA1‐like proteins.
BRM, Brahma; DDM1, Decrease in DNA Methylation 1; PIE1, Photoperiod‐Independent Early flowering 1; SYD, Splayed.
Photoperiod‐Independent Early flowering 1 (PIE1)
PIE1 belongs to the Swr1‐like group and Swr1 subfamily of Snf2 proteins (Table 2). The best characterized members of this group are Swr1 from yeast and SRCAP from humans, which are two conserved complexes formed by more than 10 subunits. These complexes regulate transcription by replacing the canonical H2A histone with the H2A.Z variant in a replication‐independent process (Krogan et al., 2003; Mizuguchi et al., 2004). PIE1 is the catalytic component of the Arabidopsis Swr1‐like complex, which includes other proteins, such as Serrated leaves and Early Flowering (SEF), Actin‐Related Protein 6 (ARP6) and H2A.Z (Choi et al., 2007; Deal et al., 2007; March‐Diaz et al., 2007; Noh and Amasino, 2003). PIE interacts with the H2A.Z histone protein encoded by the HTA8 (At2g38810), HTA9 (At1g52740) and HTA11 (At3g54560) genes. The pie1‐5 null mutation disrupts the normal deposition of H2A.Z at multiple loci. Remarkably, 65% of the changes in gene expression that are detected in double homozygous hta9‐1/hta11‐2 mutants are also found in pie1‐5 plants, suggesting that PIE1 and H2A.Z mediate common regulatory effects (Deal et al., 2007; March‐Diaz et al., 2007).
Interestingly, the Arabidopsis Swr1‐like complex regulates SA‐dependent defence mechanisms. Nearly 40% of all genes with altered expression in pie1‐5 are sensitive to the SA analogue benzothiadiazole (BTH). In addition, pie1‐5 plants display enhanced resistance to virulent P. syringae and spontaneous cell death. Similar phenotypes, although weaker, are observed in untreated sef and hta9/hta11 mutants (March‐Diaz et al., 2008). Considering these features and the recessive nature of the pie1‐5 null mutation (Noh and Amasino, 2003), it is likely that PIE1 maintains negative control of the SA pathway in noninfected wild‐type plants involving the action of SEF and/or H2A.Z (March‐Diaz et al., 2008) (Table 1). The mechanisms underlying this control are unknown. It is well established that H2A.Z marks the 5′ end of genes in several eukaryotic genomes, including plants (Draker and Cheung, 2009; Guillemette and Gaudreau, 2006), although H2A.Z may be removed from nucleosomes during transcription (Deal et al., 2007; Zhang et al., 2005; Zlatanova and Thakar, 2008). H2A.Z is associated with acetylated (Auger et al., 2008; Durant and Pugh, 2007; Zhang et al., 2005) and methylated (Barski et al., 2007; Venkatasubrahmanyam et al., 2007) histone isoforms, but is mutually exclusive with DNA methylation (Henikoff, 2008; Zilberman et al., 2008). The mechanisms by which these chromatin marks are deposited at promoters and the effects of these marks on gene expression remain unresolved (Draker and Cheung, 2009; Guillemette and Gaudreau, 2006; Zlatanova and Thakar, 2008). In yeast, the Swr1 complex may be targeted to gene promoters by the recognition of DNA motifs and histone acetylation patterns. This recognition may involve the action of DNA binding factors, such as Reb1, as well as the Swr1 subunit Bdf1, whose bromodomains specifically bind acetylated tails of H3 and H4 (Raisner et al., 2005; Zhang et al., 2005). H2A.Z preferentially associates with methylated H3K4 and both marks may maintain the accessibility of regulatory proteins to chromatin (Draker and Cheung, 2009; Guillemette and Gaudreau, 2006; Venkatasubrahmanyam et al., 2007). Interestingly, Arabidopsis plants lacking Swr1 components have a reduction in H2A.Z as well as a reduction in histone acetylation and H3K4 trimethylation at the FLC locus (Deal et al., 2007; Lázaro et al., 2008). Therefore, the deposition of H2A.Z, methylated H3K4 and acetylated histones in plants may result from interconnected mechanisms. On the other hand, insertion of H2A.Z at the 5′ end of genes is inversely correlated with DNA methylation (Henikoff, 2008; Zilberman et al., 2008; Zlatanova and Thakar, 2008). Moreover, the incorporation of H2A.Z at these sites has been hypothesized to prevent gene silencing mediated by DNA methylation in Arabidopsis (Zilberman et al., 2008). On the basis of these data, we propose a model for Swr1‐mediated negative control of SA‐sensitive genes (Fig. 1). PIE and SEF may direct the incorporation of H2A.Z at promoters of genes encoding major repressors of the SA pathway. H2A.Z may maintain the competence of these genes for activation or repression and protect them from DNA methylation. As H2A.Z remains associated with chromosomes during mitosis (Deal et al., 2007), the effects of this mark can be eventually propagated to the daughter cells. The direct targets of Swr1 in infection are currently unknown. The identification and characterization of their DNA methylation status in pie1‐5, sef and hta9/hta11 mutants would be useful to assess the validity of the proposed model.
Splayed (SYD) and Brahma (BRM)
SYD and BRM belong to an Snf2 subfamily of essential proteins that are, as yet, poorly characterized at the functional level (Knizewski et al., 2008; Kwon and Wagner, 2007). The isolation of viable Arabidopsis null syd and brm mutant plants has allowed for the evaluation of their role in vivo (Flaus et al., 2006; Hurtado et al., 2006; Wagner and Meyerowitz, 2002). Similar to their homologues in vertebrates, SYD and BRM control vegetative and reproductive processes (Kwon and Wagner, 2007). Thus, several developmental genes are misregulated in syd and brm null mutants (Bezhani et al., 2007; Kwon and Wagner, 2007). In addition, VSP2, a gene sensitive to stress and JA, is stimulated by SYD through the recruitment of SYD to the VSP2 promoter, suggesting that this remodeller has a direct effect on transcription (Walley et al., 2008). SYD also regulates the JA‐sensitive gene PDF1.2a and is required for the activation of JA‐mediated defence responses against the necrotrophic pathogen Botrytis cinerea (Walley et al., 2008). Only a small fraction of Arabidopsis genes are affected by SYD or BRM, with some being sensitive to both co‐regulators (Bezhani et al., 2007). Interestingly, a recent study has shown that noninfected brm101 mutant plants display basal activation of pathogenesis‐related (PR) genes (Bezhani et al., 2007) (Table 1). The brm101 null mutation is recessive (Hurtado et al., 2006), suggesting that BRM, similar to PIE, maintains basal repression of the SA pathway. Interestingly, brm101 plants display repression of auxin‐related genes, such as SAUR66 (At1g29500) and endo‐xyloglucan transferase EXGT‐A1 (At2g06850) (Bezhani et al., 2007). This effect could be related to the overexpression of SA‐dependent responses, as SA inhibits auxin signalling during plant defence (Wang et al., 2007).
PIE and BRM modulate defences that are sensitive to Suppressor of NPR1, Inducible 1 (SNI1)
SNI1 is a nuclear protein exclusively found in plants that lacks homology with chromatin‐modifying enzymes and DNA‐binding domains. This protein represses transcription, even in heterologous systems, most probably through a conserved mechanism, such as chromatin remodelling (Mosher et al., 2006). In addition, SNI1 inhibits somatic homologous recombination (Durrant et al., 2007), and both of these genomic responses seem to involve its interaction with other proteins, such as histone‐modifying enzymes (Mosher et al., 2006). SNI1 was originally identified from a genetic screen for npr1‐1 suppressors, where the recessive sni1 mutation rescued PR gene activation and resistance on the npr1‐1 background (Li et al., 1999). Similar to the pie1‐5 and brm101 mutants, the untreated sni1 plants overexpress BTH‐sensitive genes and contain elevated levels of H3Ac and H3K4me2 at the PR1 gene promoter. Therefore, it was suggested that SNI1 may inhibit basal PR1 expression by reducing euchromatic marks at its promoter (Mosher et al., 2006). Interestingly, the defence phenotypes of sni1 plants are suppressed by the mutation of RAD51D, a protein that promotes DNA homologous recombination during double‐strand break repair events (Durrant et al., 2007). This finding is consistent with the hypothesis that SNI1 modulates plant immunity through chromatin remodelling (Table 1).
We compared the genes constitutively upregulated in sni1 (Mosher et al., 2006), brm101 (Bezhani et al., 2007) and pie1‐5 (March‐Diaz et al., 2008) mutants. We found 13 genes that were common to all three mutants, including 11 genes inducible by BTH, such as PR1, PR2, PR5 and others (Fig. 2). This observation reinforces the finding that the SA pathway is subject to epigenetic control, and reveals common effects of SNI1, PIE and BRM on this pathway. However, the promoters that are targeted by these remodellers remain to be identified. As mentioned previously, half of the genes that are overexpressed in untreated sni1 plants are sensitive to BTH (Mosher et al., 2006), suggesting that SNI1 may control this pathway with a low energy cost by repressing the basal activity of positive regulators of the SA pathway (Fig. 1).
Figure 2.
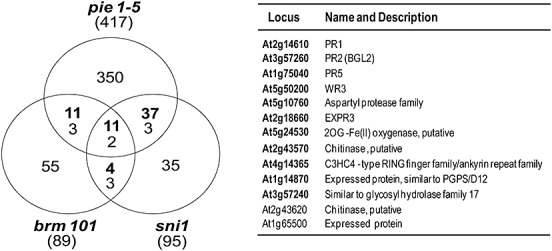
Arabidopsis genes that are regulated by Suppressor of NPR1, Inducible 1 (SNI1), Photoperiod‐Independent Early flowering 1 (PIE1) and Brahma (BRM). Left: Venn diagram including common genes that are upregulated in basal conditions in the sni1 (Mosher et al., 2006), brm101 (Bezhani et al., 2007) and pie1‐5 (March‐Diaz et al., 2008) mutants. The numbers in parentheses indicate the total number of genes upregulated in each mutant. Numbers in bold indicate genes that are sensitive to BTH. Right: Identity of the 13 genes which are common to all three mutants. The 11 genes that are sensitive to BTH are shown in bold.
Decrease in DNA Methylation 1 (DDM1)
DDM1 is a helicase from the Lsh subfamily (Table 2) that strongly affects genomic DNA methylation. In Arabidopsis, ddm1 mutants display reduced cytosine methylation, TE activation and modification of histone marks in heterochromatic repeats (Jeddeloh et al., 1999; Miura et al., 2001; Tsukahara et al., 2009; Vongs et al., 1993). Deficiency in the mouse homologue Lsh produces similar alterations in heterochromatin (Dennis et al., 2001). Neither DDM1 nor Lsh encodes DNA methyltransferases (DNMTs) and this type of enzymatic activity is not significantly altered in ddm1 or Lsh–/– tissues (Dennis et al., 2001; Kakutani et al., 1995). Therefore, DDM1/Lsh may regulate DNA methylation indirectly by modulating the access of DNMTs and/or DNA demethylases to the genome (Dennis et al., 2001; Gendrel et al., 2002; Zemach et al., 2005). In support of this idea, DDM1 has been shown to remodel nucleosomes in vitro independent of DNA methylation (Brzeski and Jerzmanowski, 2003).
Stable alleles that were not linked to the ddm1 mutation were identified in the progeny of ddm1 hypomethylated plants (Kakutani et al., 1996; Vongs et al., 1993). Among them, the bal defect causes dwarfism, curled leaves and enhanced pathogen resistance. These bal phenotypes depend on EDS1 and result from the overexpression of R genes from the Recognition of Peronospora parasitica 5 (RPP5) locus (Stokes et al., 2002; Yi and Richards, 2007). The RPP5 locus includes the Suppressor of npr1‐1, Constitutive 1 (SNC1), RPP4 and RPP5‐like genes, whose expression is potentiated by an SNC1‐dependent amplification loop and downregulated by smRNAs generated at the locus (Yi and Richards, 2007). The bal allele has been proposed to have an epigenetic nature, because it arose from a ddm1 hypomethylation background and displayed high‐frequency phenotypic suppression in ethyl‐methanesulfonate‐treated lines, apparently by reversion of the bal defect (Stokes et al., 2002). Surprisingly, it has been found recently that bal carries a tandem duplication of a 55‐kb fragment from the RPP5 locus, which probably originated from homologous recombination between two R genes from this locus. Several R genes are duplicated in bal plants; however, over‐expression of SNC1 sufficiently generates the bal phenotypes. Interestingly, the instability of these phenotypes seems to be caused by hypermutation of the duplicated copies of SNC1 (Yi and Richards, 2009).
One question that arises is whether the generation of RPP5 fragment duplication occurring in the bal background is mechanistically associated with DDM1 deficiency. In mammals, Lsh interacts with de novo DNMTs and HDACs, and this recruitment has been proposed to generate deacetylated, inactive chromatin that can be stabilized by DNA methylation at a later time (Myant and Stancheva, 2008; Zhu et al., 2006). Therefore, it is plausible that DDM1 has similar effects that reinforce chromatin compaction and prevent recombination between repeats from the RPP5 cluster (Table 1).
DNA METHYLATION
Enzymes and patterns of plant DNA methylation
In several eukaryotes, methylation of the fifth carbon of cytosine (5mC) modulates chromatin structure and thus affects DNA recombination, genomic imprinting, gene expression and other genomic processes (Bender, 2004; Gehring and Henikoff, 2007; Henderson and Jacobsen, 2007; Zhang, 2008; Zilberman, 2008). In plants, DNMTs target cytosines located at both symmetric (CG and CHG; H corresponds to A, C and T) and asymmetric (CHH) sites. Methyltransferase 1 (MET1) maintains CG methylation (Finnegan et al., 1996), Chromomethylase 3 (CMT3) is primarily responsible for CHG methylation (Lindroth et al., 2001) and Domains Rearranged Methyltransferase1/2 (DRM1/2) mediates RNA‐directed de novo methylation that affects cytosines in all contexts (Cao and Jacobsen, 2002; Chan et al., 2004).
High‐resolution DNA methylation maps of the Arabidopsis genome have confirmed that all methylation types are concentrated in pericentromeric transposons and repetitive sequences. In addition, they revealed that one‐third of all genes contain CG methylation in transcribed regions, whereas only a small fraction of these genes have methylation at their promoters (Cokus et al., 2008; Lister et al., 2008; Zhang et al., 2006; Zilberman et al., 2007). Similar but slightly distinct DNA methlyation traits were revealed by high‐resolution profiling of DNA and H3K4 methylation in two complete rice chromosomes (Li et al., 2008).
The plant DNA methylation patterns are determined by DNMTs, proteins such as DDM1 (Jeddeloh et al., 1999; Vongs et al., 1993) and enzymes involved in histone PTMs. RNA‐directed DNA methylation requires HDA6 (Aufsatz et al., 2007). CHG methylation correlates with H3K9me2 at TEs, pseudogenes and repeat elements, where gene silencing is reinforced by the combined actions of CHG and H3K9me2 methyltransferases (Bernatavichute et al., 2008; Fuchs et al., 2006; Gendrel et al., 2002; Jackson et al., 2002; Malagnac et al., 2002; Mathieu et al., 2005).
Remarkably, plant methylation patterns can be propagated to progeny and, conversely, can also be removed by the activity of 5mC glycosylases (Gehring et al., 2009; Kakutani et al., 1996; Zilberman, 2008). Therefore, the epigenetic marks that are generated by DNA methylation can permanently affect genomic activities, but can also facilitate a dynamic control of processes such as gene expression and DNA recombination.
Pathogen‐induced plant DNA methylation changes
Interestingly, plant DNA methylation patterns become altered by pathogen infection. Work from our laboratory has described the occurrence of massive hypomethylation and net chromocentre decondensation in Arabidopsis tissues infected with P. syringae. This hypomethylation targets peri/centromeric 180‐bp units, retrotransposons, mtDNA and other loci. In addition, it involves symmetric and asymmetric cytosines and takes place in the absence of DNA replication, suggesting that it occurs as a result of an active demethylation process (Pavet et al., 2006). Alterations in cytosine methylation in response to pathogens have been reported in previous studies (Guseinov and Vanyushin, 1975; Wada et al., 2004), but the effects of these alterations on disease resistance remain unknown.
Pathogen‐induced host genome hypomethylation can influence the expression of defence genes. Chemically induced demethylation of the rice R gene Xa21G abolishes silencing of this gene and provides heritable resistance to Xanthomonas oryzae pv. oryzae (Akimoto et al., 2007). The biogenesis of smRNAs, which are strongly implicated in the post‐transcriptional regulation of defences to pathogens (Jin, 2008; Ruiz‐Ferrer and Voinnet, 2009), can also be modified by hypomethylation (Chan, 2008; Lister et al., 2008; Zhang, 2008).
At the structural level, hypomethylation may affect the stability of NBS‐LRR genes. These genes are usually clustered in regions rich in TEs and repetitive sequences that concentrate repressive chromatin modifications, such as DNA and H3K9 methylation (Bernatavichute et al., 2008; Cokus et al., 2008; Lister et al., 2008; Zilberman et al., 2007). These modifications, together with smRNAs, prevent TE expression (Bernatavichute et al., 2008; Kato et al., 2003; Lister et al., 2008; Miura et al., 2001; Singer et al., 2001; Weil and Martienssen, 2008). Therefore, the release of DNA methylation from these regions may promote TE activation with a consequent impact on NBS‐LRR gene integrity. The induction of TEs by biotic and abiotic stresses has been reported in many studies, and some of these stresses were later associated with genomic DNA methylation changes (Grandbastien et al., 2005; 2003, 2006; Steward et al., 2002; Takeda et al., 1999). Remarkably, endogenous long‐terminal repeat (LTR)‐type retrotransposons, which are the major family of TEs in plants, have been found to be reactivated by DNA hypomethylation in Arabidopsis (Tsukahara et al., 2009). In addition, disruption (Luck et al., 1998), remodelling (Wang et al., 1998) and refunctionalization (Hayashi and Yoshida, 2009) of NBS‐LRR genes by the insertion of TEs have been demonstrated in several plant genomes.
The reduction of 5mC residues from NBS‐LRR gene clusters may also increase mispairing between repeats (Bender, 2004; Maloisel and Rossignol, 1998; Peng and Karpen, 2008; Weber and Schubeler, 2007). Recombination events involving repetitive sequences from R gene clusters contribute to the evolution of R genes (Baumgarten et al., 2003; McHale et al., 2006; Meyers et al., 2003). In support of this, a homologous recombination event that involved a 186‐bp region common to At4g16960 and RPP4 R genes from the RPP5 locus has been shown to create a novel NBS‐LRR gene in hypomethylated bal plants (Yi and Richards, 2009). In addition, DNA hypomethylation at N‐like loci has been detected in the progeny of tobacco plants exposed to the tobacco mosaic virus, and the changes in methylation correspond to enhanced genomic rearrangements at these loci (Boyko et al., 2007). Similarly, somatic recombination increases in plants treated with DNA demethylation agents, elicitors, pathogens or abiotic stresses (Kovalchuk et al., 2003; Lucht et al., 2002; Molinier et al., 2006; Pecinka et al., 2009). Stress‐induced, enhanced homologous recombination has been proposed to occur in the absence of pathogens (Kovalchuk et al., 2003) and may even be transmitted as a dominant trait to successive generations (Molinier et al., 2006). However, transgeneration memory of increased homologous recombination in response to stress does not seem to be a general response in plants (Pecinka et al., 2009). Therefore, it is possible that DNA hypomethylation promotes an increase in somatic recombination in tissues that are exposed to pathogens, and the resultant effect is transmitted to offspring as a stochastic process.
On the basis of these data, it is conceivable that DNA hypomethylation changes contribute to the generation of chromatin modifications that affect the activity or integrity of NBS‐LRR genes, resulting in either an expansion or reduction in the subset of functional R genes in the plant. The genomic marks generated by DNA methylation can either be stably preserved through generations or erased under particular conditions.
CHROMATIN REMODELLERS AS PUTATIVE TARGETS OF MICROBIAL EFFECTORS
If the plant inducible defences that counteract pathogen attack are under epigenetic control, host components that are involved in these functions may constitute attractive targets for microbial effectors or toxins. Although this has not yet been demonstrated in plants, studies from the animal kingdom suggest that this may be true (Arbibe et al., 2007; Hamon et al., 2007). Resistance to the intestinal pathogen Shigella flexneri involves the activation of nuclear factor‐κB‐dependent proinflammatory genes by the phosphorylation of histone H3 at Ser10 (H3S10ph) at promoters. This modification has relevant effects on defence mechanisms, as the S. flexneri effector OspF inhibits mitogen‐activated protein kinase (MAPK)‐mediated H3S10ph at these sites and causes the reprogramming of host gene expression for its own benefit (Arbibe et al., 2007). OspF is a homologue of HopAI1 from plant pathogenic bacteria (Shan et al., 2007) and both effectors dephosphorylate MAPKs (Li et al., 2007). Moreover, HopAI1‐mediated MAPK3/6 dephosphorylation inhibits basal defence in Arabidopsis (Zhang et al., 2007a). One question is whether HopAI1 inhibits disease resistance at the epigenetic level by affecting nuclear MAPKs. Although these enzymes have been found in plants (Ahlfors et al., 2004; Miao et al., 2007; Prestamo et al., 1999; Qiu et al., 2008), and proteins that mediate epigenetic modifications (core histones, Snf2 and other nuclear proteins) have been described as substrates of MAPK3/6 in vitro (Feilner et al., 2005), the phosphorylation of chromatin remodelling proteins by plant nuclear MAPKs has not been demonstrated.
CONCLUSIONS
Plant immune responses are subject to strict regulatory mechanisms that ensure prompt defence stimulation and robust basal defence repression without a fitness cost. Recent evidence has revealed that both kinds of mechanism involve epigenetic control. Proteins that affect histone PTMs, deposition of histone variants, ATP‐dependent chromatin remodelling and DNA methylation modulate the expression of SA‐ or JA‐dependent defence genes. Presumably, these chromatin remodellers bind gene promoters to alter their accessibility to transcriptional regulators. However, only a few promoters that are targeted by these components have been described thus far. Genomic binding studies will be required to identify gene promoters that are targeted by chromatin remodellers during infection in order to elucidate their impact on defence programmes. Further understanding of the signals and networks that trigger chromatin remodelling after the recognition of PAMPs or effectors is of equal importance. In addition, the extent and nature of epigenomic changes induced by different pathogens remain to be elucidated. Future research will be required to determine the specific epigenetic modifications affecting plant immunity that are transmitted to offspring and how the stability of these modifications is affected by the selective pressure of coexisting pathogens.
ACKNOWLEDGEMENTS
We thank the Agencia Nacional de Promoción Científica y Tecnológica (BID 1728/OC‐AR PICT 32637), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (PIP 6349) and Secretaría de Ciencia y Tecnología‐Universidad Nacional de Córdoba (SECyT‐UNC) for financial support. M.E.A. is a senior career investigator of CONICET; F.N. and D.A.C. are CONICET fellows.
REFERENCES
- Ahlfors, R. , Macioszek, V. , Rudd, J. , Brosche, M. , Schlichting, R. , Scheel, D. and Kangasjarvi, J. (2004) Stress hormone‐independent activation and nuclear translocation of mitogen‐activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 40, 512–522. [DOI] [PubMed] [Google Scholar]
- Akimoto, K. , Katakami, H. , Kim, H.J. , Ogawa, E. , Sano, C.M. , Wada, Y. and Sano, H. (2007) Epigenetic inheritance in rice plants. Ann. Bot. 100, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Venegas, R. , Pien, S. , Sadder, M. , Witmer, X. , Grossniklaus, U. and Avramova, Z. (2003) ATX‐1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13, 627–637. [DOI] [PubMed] [Google Scholar]
- Álvarez‐Venegas, R. , Sadder, M. , Hlavacka, A. , Baluska, F. , Xia, Y. , Lu, G. , Firsov, A. , Sarath, G. , Moriyama, H. , Dubrovsky, J.G. and Avramova, Z. (2006) The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5‐phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. USA, 103, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez‐Venegas, R. , Abdallat, A.A. , Guo, M. , Alfano, J.R. and Avramova, Z. (2007) Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics, 2, 106–113. [DOI] [PubMed] [Google Scholar]
- Arbibe, L. , Kim, D.W. , Batsche, E. , Pedron, T. , Mateescu, B. , Muchardt, C. , Parsot, C. and Sansonetti, P.J. (2007) An injected bacterial effector targets chromatin access for transcription factor NF‐kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8, 47–56. [DOI] [PubMed] [Google Scholar]
- Aufsatz, W. , Stoiber, T. , Rakic, B. and Naumann, K. (2007) Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene, 26, 5477–5488. [DOI] [PubMed] [Google Scholar]
- Auger, A. , Galarneau, L. , Altaf, M. , Nourani, A. , Doyon, Y. , Utley, R.T. , Cronier, D. , Allard, S. and Cote, J. (2008) Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP‐dependent exchange of histone H2A variants. Mol. Cell. Biol. 28, 2257–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979. [DOI] [PubMed] [Google Scholar]
- Bari, R. and Jones, J.D. (2009) Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Barski, A. , Cuddapah, S. , Cui, K. , Roh, T.Y. , Schones, D.E. , Wang, Z. , Wei, G. , Chepelev, I. and Zhao, K. (2007) High‐resolution profiling of histone methylations in the human genome. Cell, 129, 823–837. [DOI] [PubMed] [Google Scholar]
- Baumgarten, A. , Cannon, S. , Spangler, R. and May, G. (2003) Genome‐level evolution of resistance genes in Arabidopsis thaliana . Genetics, 165, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, J. (2004) DNA methylation and epigenetics. Annu. Rev. Plant Biol. 55, 41–68. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Berger, S.L. (2007) The complex language of chromatin regulation during transcription. Nature, 447, 407–412. [DOI] [PubMed] [Google Scholar]
- Bernatavichute, Y.V. , Zhang, X. , Cokus, S. , Pellegrini, M. and Jacobsen, S.E. (2008) Genome‐wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana . PLoS ONE, 3, e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezhani, S. , Winter, C. , Hershman, S. , Wagner, J.D. , Kennedy, J.F. , Kwon, C.S. , Pfluger, J. , Su, Y. and Wagner, D. (2007) Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell, 19, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti, K. , Von Koskull‐Doring, P. , Bharti, S. , Kumar, P. , Tintschl‐Korbitzer, A. , Treuter, E. and Nover, L. (2004) Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone‐like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell, 16, 1521–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko, A. , Kathiria, P. , Zemp, F.J. , Yao, Y. , Pogribny, I. and Kovalchuk, I. (2007) Transgenerational changes in the genome stability and methylation in pathogen‐infected plants: (virus‐induced plant genome instability). Nucleic Acids Res. 35, 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch, G. , Ransom, R. , Lechner, T. , Walton, J.D. and Loidl, P. (1995) Inhibition of maize histone deacetylases by HC toxin, the host‐selective toxin of Cochliobolus carbonum . Plant Cell, 7, 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski, J. and Jerzmanowski, A. (2003) Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin‐remodeling factors. J. Biol. Chem. 278, 823–828. [DOI] [PubMed] [Google Scholar]
- Cao, X. and Jacobsen, S.E. (2002) Locus‐specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA, 99 (Suppl. 4), 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W. (2008) Inputs and outputs for chromatin‐targeted RNAi. Trends Plant Sci. 13, 383–389. [DOI] [PubMed] [Google Scholar]
- Chan, S.W. , Zilberman, D. , Xie, Z. , Johansen, L.K. , Carrington, J.C. and Jacobsen, S.E. (2004) RNA silencing genes control de novo DNA methylation. Science, 303, 1336. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J. and Tian, L. (2007) Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta, 1769, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K. , Park, C. , Lee, J. , Oh, M. , Noh, B. and Lee, I. (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development, 134, 1931–1941. [DOI] [PubMed] [Google Scholar]
- Cohen, H.Y. , Miller, C. , Bitterman, K.J. , Wall, N.R. , Hekking, B. , Kessler, B. , Howitz, K.T. , Gorospe, M. , De Cabo, R. and Sinclair, D.A. (2004) Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science, 305, 390–392. [DOI] [PubMed] [Google Scholar]
- Cokus, S.J. , Feng, S. , Zhang, X. , Chen, Z. , Merriman, B. , Haudenschild, C.D. , Pradhan, S. , Nelson, S.F. , Pellegrini, M. and Jacobsen, S.E. (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature, 452, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal, R.B. , Topp, C.N. , McKinney, E.C. and Meagher, R.B. (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell, 19, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, K. , Fan, T. , Geiman, T. , Yan, Q. and Muegge, K. (2001) Lsh, a member of the SNF2 family, is required for genome‐wide methylation. Genes Dev. 15, 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto, A. , Nieto‐Rostro, M. , Xie, D. , Ellis, C. , Harmston, R. , Patrick, E. , Davis, J. , Sherratt, L. , Coleman, M. and Turner, J.G. (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin‐ligase complex in Arabidopsis. Plant J. 32, 457–466. [DOI] [PubMed] [Google Scholar]
- Draker, R. and Cheung, P. (2009) Transcriptional and epigenetic functions of histone variant H2A.Z. Biochem. Cell Biol. 87, 19–25. [DOI] [PubMed] [Google Scholar]
- Durant, M. and Pugh, B.F. (2007) NuA4‐directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 27, 5327–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E. , Wang, S. and Dong, X. (2007) Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA, 104, 4223–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilner, T. , Hultschig, C. , Lee, J. , Meyer, S. , Immink, R.G. , Koenig, A. , Possling, A. , Seitz, H. , Beveridge, A. , Scheel, D. , Cahill, D.J. , Lehrach, H. , Kreutzberger, J. and Kersten, B. (2005) High throughput identification of potential Arabidopsis mitogen‐activated protein kinases substrates. Mol. Cell. Proteomics, 4, 1558–1568. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J. , Peacock, W.J. and Dennis, E.S. (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA, 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus, A. , Martin, D.M. , Barton, G.J. and Owen‐Hughes, T. (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, P.M. , Tian, L. and Chen, Z.J. (2006) Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro . Cell Res. 16, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, J. , Demidov, D. , Houben, A. and Schubert, I. (2006) Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci. 11, 199–208. [DOI] [PubMed] [Google Scholar]
- Gehring, M. and Henikoff, S. (2007) DNA methylation dynamics in plant genomes. Biochim. Biophys. Acta, 1769, 276–286. [DOI] [PubMed] [Google Scholar]
- Gehring, M. , Reik, W. and Henikoff, S. (2009) DNA demethylation by DNA repair. Trends Genet. 25, 82–90. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V. , Lippman, Z. , Yordan, C. , Colot, V. and Martienssen, R.A. (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science, 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Glozak, M.A. , Sengupta, N. , Zhang, X. and Seto, E. (2005) Acetylation and deacetylation of non‐histone proteins. Gene, 363, 15–23. [DOI] [PubMed] [Google Scholar]
- Grandbastien, M.A. , Audeon, C. , Bonnivard, E. , Casacuberta, J.M. , Chalhoub, B. , Costa, A.P. , Le, Q.H. , Melayah, D. , Petit, M. , Poncet, C. , Tam, S.M. , Van Sluys, M.A. and Mhiri, C. (2005) Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet. Genome Res. 110, 229–241. [DOI] [PubMed] [Google Scholar]
- Guenther, M.G. , Levine, S.S. , Boyer, L.A. , Jaenisch, R. and Young, R.A. (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell, 130, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette, B. and Gaudreau, L. (2006) Reuniting the contrasting functions of H2A.Z. Biochem. Cell Biol. 84, 528–535. [DOI] [PubMed] [Google Scholar]
- Guseinov, V.A. and Vanyushin, B.F. (1975) Content and localisation of 5‐methylcytosine in DNA of healthy and wilt‐infected cotton plants. Biochim. Biophys. Acta, 395, 229–238. [DOI] [PubMed] [Google Scholar]
- Hamon, M.A. , Batsche, E. , Regnault, B. , Tham, T.N. , Seveau, S. , Muchardt, C. and Cossart, P. (2007) Histone modifications induced by a family of bacterial toxins. Proc. Natl. Acad. Sci. USA, 104, 13467–13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida, S.N. , Kitamura, K. , Mikami, T. and Kishima, Y. (2003) Temperature shift coordinately changes the activity and the methylation state of transposon Tam3 in Antirrhinum majus . Plant Physiol. 132, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida, S.N. , Uchiyama, T. , Martin, C. , Kishima, Y. , Sano, Y. and Mikami, T. (2006) The temperature‐dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell, 18, 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun, W.J. and Springer, N.M. (2008) Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J. 56, 903–912. [DOI] [PubMed] [Google Scholar]
- Hayashi, K. and Yoshida, H. (2009) Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 57, 413–425. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R. and Jacobsen, S.E. (2007) Epigenetic inheritance in plants. Nature, 447, 418–424. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. (2008) Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9, 15–26. [DOI] [PubMed] [Google Scholar]
- Hubbert, C. , Guardiola, A. , Shao, R. , Kawaguchi, Y. , Ito, A. , Nixon, A. , Yoshida, M. , Wang, X.F. and Yao, T.P. (2002) HDAC6 is a microtubule‐associated deacetylase. Nature, 417, 455–458. [DOI] [PubMed] [Google Scholar]
- Hurtado, L. , Farrona, S. and Reyes, J.C. (2006) The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana . Plant Mol. Biol. 62, 291–304. [DOI] [PubMed] [Google Scholar]
- Jackson, J.P. , Lindroth, A.M. , Cao, X. and Jacobsen, S.E. (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature, 416, 556–560. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A. , Stokes, T.L. and Richards, E.J. (1999) Maintenance of genomic methylation requires a SWI2/SNF2‐like protein. Nat. Genet. 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jerzmanowski, A. (2007) SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta, 1769, 330–345. [DOI] [PubMed] [Google Scholar]
- Jin, H. (2008) Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 582, 2679–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal, G.S. and Briggs, S.P. (1992) Reductase activity encoded by the HM1 disease resistance gene in maize. Science, 258, 985–987. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Juan, L.J. , Shia, W.J. , Chen, M.H. , Yang, W.M. , Seto, E. , Lin, Y.S. and Wu, C.W. (2000) Histone deacetylases specifically down‐regulate p53‐dependent gene activation. J. Biol. Chem. 275, 20436–20443. [DOI] [PubMed] [Google Scholar]
- Kakutani, T. , Jeddeloh, J.A. and Richards, E.J. (1995) Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T. , Jeddeloh, J.A. , Flowers, S.K. , Munakata, K. and Richards, E.J. (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA, 93, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , Miura, A. , Bender, J. , Jacobsen, S.E. and Kakutani, T. (2003) Role of CG and non‐CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13, 421–426. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Hyun, Y. , Park, J.Y. , Park, M.J. , Park, M.K. , Kim, M.D. , Kim, H.J. , Lee, M.H. , Moon, J. , Lee, I. and Kim, J. (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana . Nat. Genet. 36, 167–171. [DOI] [PubMed] [Google Scholar]
- Kim, K.C. , Lai, Z. , Fan, B. and Chen, Z. (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell, 20, 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knizewski, L. , Ginalski, K. and Jerzmanowski, A. (2008) Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 13, 557–565. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. and Pieterse, C.M. (2008) Cross talk in defense signaling. Plant Physiol. 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides, T. (2007) Chromatin modifications and their function. Cell, 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, I. , Kovalchuk, O. , Kalck, V. , Boyko, V. , Filkowski, J. , Heinlein, M. and Hohn, B. (2003) Pathogen‐induced systemic plant signal triggers DNA rearrangements. Nature, 423, 760–762. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J. , Keogh, M.C. , Datta, N. , Sawa, C. , Ryan, O.W. , Ding, H. , Haw, R.A. , Pootoolal, J. , Tong, A. , Canadien, V. , Richards, D.P. , Wu, X. , Emili, A. , Hughes, T.R. , Buratowski, S. and Greenblatt, J.F. (2003) A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell, 12, 1565–1576. [DOI] [PubMed] [Google Scholar]
- Kwon, C.S. and Wagner, D. (2007) Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 23, 403–412. [DOI] [PubMed] [Google Scholar]
- Lázaro, A. , Gómez‐Zambrano, A. , López‐González, L. , Piñeiro, M. and Jarillo, J.A. (2008) Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59, 653–666. [DOI] [PubMed] [Google Scholar]
- Li, H. , Xu, H. , Zhou, Y. , Zhang, J. , Long, C. , Li, S. , Chen, S. , Zhou, J.M. and Shao, F. (2007) The phosphothreonine lyase activity of a bacterial type III effector family. Science, 315, 1000–1003. [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhang, Y. , Clarke, J.D. , Li, Y. and Dong, X. (1999) Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1‐1. Cell, 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Li, X. , Wang, X. , He, K. , Ma, Y. , Su, N. , He, H. , Stolc, V. , Tongprasit, W. , Jin, W. , Jiang, J. , Terzaghi, W. , Li, S. and Deng, X.W. (2008) High‐resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell, 20, 259–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth, A.M. , Cao, X. , Jackson, J.P. , Zilberman, D. , McCallum, C.M. , Henikoff, S. and Jacobsen, S.E. (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science, 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lister, R. , O'Malley, R.C. , Tonti‐Filippini, J. , Gregory, B.D. , Berry, C.C. , Millar, A.H. and Ecker, J.R. (2008) Highly integrated single‐base resolution maps of the epigenome in Arabidopsis. Cell, 133, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht, J.M. , Mauch‐Mani, B. , Steiner, H.Y. , Metraux, J.P. , Ryals, J. and Hohn, B. (2002) Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 30, 311–314. [DOI] [PubMed] [Google Scholar]
- Luck, J.E. , Lawrence, G.J. , Finnegan, E.J. , Jones, D.A. and Ellis, J.G. (1998) A flax transposon identified in two spontaneous mutant alleles of the L6 rust resistance gene. Plant J. 16, 365–369. [DOI] [PubMed] [Google Scholar]
- Luger, K. , Mader, A.W. , Richmond, R.K. , Sargent, D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Malagnac, F. , Bartee, L. and Bender, J. (2002) An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21, 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel, L. and Rossignol, J.L. (1998) Suppression of crossing‐over by DNA methylation in Ascobolus. Genes Dev. 12, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March‐Diaz, R. and Reyes, J.C. (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant, 2, 565–577. [DOI] [PubMed] [Google Scholar]
- March‐Diaz, R. , Garcia‐Dominguez, M. , Florencio, F.J. and Reyes, J.C. (2007) SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 143, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March‐Diaz, R. , Garcia‐Dominguez, M. , Lozano‐Juste, J. , Leon, J. , Florencio, F.J. and Reyes, J.C. (2008) Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53, 475–487. [DOI] [PubMed] [Google Scholar]
- Mathieu, O. , Probst, A.V. and Paszkowski, J. (2005) Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24, 2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale, L. , Tan, X. , Koehl, P. and Michelmore, R.W. (2006) Plant NBS‐LRR proteins: adaptable guards. Genome Biol. 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C. , Kozik, A. , Griego, A. , Kuang, H. and Michelmore, R.W. (2003) Genome‐wide analysis of NBS‐LRR‐encoding genes in Arabidopsis. Plant Cell, 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Y. , Laun, T.M. , Smykowski, A. and Zentgraf, U. (2007) Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence‐related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65, 63–76. [DOI] [PubMed] [Google Scholar]
- Miura, A. , Yonebayashi, S. , Watanabe, K. , Toyama, T. , Shimada, H. and Kakutani, T. (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature, 411, 212–214. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G. , Shen, X. , Landry, J. , Wu, W.H. , Sen, S. and Wu, C. (2004) ATP‐driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science, 303, 343–348. [DOI] [PubMed] [Google Scholar]
- Mohrmann, L. and Verrijzer, C.P. (2005) Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta, 1681, 59–73. [DOI] [PubMed] [Google Scholar]
- Molinier, J. , Ries, G. , Zipfel, C. and Hohn, B. (2006) Transgeneration memory of stress in plants. Nature, 442, 1046–1049. [DOI] [PubMed] [Google Scholar]
- Mosher, R.A. , Durrant, W.E. , Wang, D. , Song, J. and Dong, X. (2006) A comprehensive structure–function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell, 18, 1750–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant, K. and Stancheva, I. (2008) LSH cooperates with DNA methyltransferases to repress transcription. Mol. Cell. Biol. 28, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, Y.S. and Amasino, R.M. (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell, 15, 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet, V. , Quintero, C. , Cecchini, N.M. , Rosa, A.L. and Alvarez, M.E. (2006) Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae . Mol. Plant–Microbe Interact. 19, 577–587. [DOI] [PubMed] [Google Scholar]
- Pecinka, A. , Rosa, M. , Schikora, A. , Berlinger, M. , Hirt, H. , Luschnig, C. and Mittelsten, S.O. (2009) Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE, 4, e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.C. and Karpen, G.H. (2008) Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 18, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger, J. and Wagner, D. (2007) Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestamo, G. , Testillano, P.S. , Vicente, O. , Gonzalez‐Melendi, P. , Coronado, M.J. , Wilson, C. , Heberle‐Bors, E. and Risueno, M.C. (1999) Ultrastructural distribution of a MAP kinase and transcripts in quiescent and cycling plant cells and pollen grains. J. Cell Sci. 112, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Probst, A.V. , Fagard, M. , Proux, F. , Mourrain, P. , Boutet, S. , Earley, K. , Lawrence, R.J. , Pikaard, C.S. , Murfett, J. , Furner, I. , Vaucheret, H. and Mittelsten Scheid, O. (2004) Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell, 16, 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J.L. , Fiil, B.K. , Petersen, K. , Nielsen, H.B. , Botanga, C.J. , Thorgrimsen, S. , Palma, K. , Suarez‐Rodriguez, M.C. , Sandbech‐Clausen, S. , Lichota, J. , Brodersen, P. , Grasser, K.D. , Mattsson, O. , Glazebrook, J. , Mundy, J. and Petersen, M. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner, R.M. , Hartley, P.D. , Meneghini, M.D. , Bao, M.Z. , Liu, C.L. , Schreiber, S.L. , Rando, O.J. and Madhani, H.D. (2005) Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell, 123, 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom, R.F. and Walton, J.D. (1997) Histone hyperacetylation in maize in response to treatment with HC‐toxin or infection by the filamentous fungus Cochliobolus carbonum . Plant Physiol. 115, 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. and Voinnet, O. (2009) Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. [DOI] [PubMed] [Google Scholar]
- De La Serna, I. , Ohkawa, Y. and Imbalzano, A.N. (2006) Chromatin remodelling in mammalian differentiation: lessons from ATP‐dependent remodellers. Nat. Rev. Genet. 7, 461–473. [DOI] [PubMed] [Google Scholar]
- Shan, L. , He, P. and Sheen, J. (2007) Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe, 1, 167–174. [DOI] [PubMed] [Google Scholar]
- Sims, R.J., III and Reinberg, D. (2006) Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 20, 2779–2786. [DOI] [PubMed] [Google Scholar]
- Singer, T. , Yordan, C. and Martienssen, R.A. (2001) Robertson's Mutator transposons in A. thaliana are regulated by the chromatin‐remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C.P. and Galbraith, D.W. (2006) AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60, 241–257. [DOI] [PubMed] [Google Scholar]
- Sridha, S. and Wu, K. (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46, 124–133. [DOI] [PubMed] [Google Scholar]
- Steward, N. , Ito, M. , Yamaguchi, Y. , Koizumi, N. and Sano, H. (2002) Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 277, 37741–37746. [DOI] [PubMed] [Google Scholar]
- Stokes, T.L. , Kunkel, B.N. and Richards, E.J. (2002) Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, S. , Sugimoto, K. , Otsuki, H. and Hirochika, H. (1999) A 13‐bp cis‐regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 18, 383–393. [DOI] [PubMed] [Google Scholar]
- Tanaka, M. , Kikuchi, A. and Kamada, H. (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L. and Chen, Z.J. (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA, 98, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L. , Fong, M.P. , Wang, J.J. , Wei, N.E. , Jiang, H. , Doerge, R.W. and Chen, Z.J. (2005) Reversible histone acetylation and deacetylation mediate genome‐wide, promoter‐dependent and locus‐specific changes in gene expression during plant development. Genetics, 169, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara, S. , Kobayashi, A. , Kawabe, A. , Mathieu, O. , Miura, A. and Kakutani, T. (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature, 461, 423–426. [DOI] [PubMed] [Google Scholar]
- Turck, F. , Roudier, F. , Farrona, S. , Martin‐Magniette, M.L. , Guillaume, E. , Buisine, N. , Gagnot, S. , Martienssen, R.A. , Coupland, G. and Colot, V. (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant, I. and Paszkowski, J. (2007) Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant Biol. 10, 528–533. [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam, S. , Hwang, W.W. , Meneghini, M.D. , Tong, A.H. and Madhani, H.D. (2007) Genome‐wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. USA, 104, 16609–16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Vongs, A. , Kakutani, T. , Martienssen, R.A. and Richards, E.J. (1993) Arabidopsis thaliana DNA methylation mutants. Science, 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wada, Y. , Miyamoto, K. , Kusano, T. and Sano, H. (2004) Association between up‐regulation of stress‐responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol. Genet. Genomics, 271, 658–666. [DOI] [PubMed] [Google Scholar]
- Wagner, D. and Meyerowitz, E.M. (2002) SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Walley, J.W. , Rowe, H.C. , Xiao, Y. , Chehab, E.W. , Kliebenstein, D.J. , Wagner, D. and Dehesh, K. (2008) The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4, e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, J.D. (2006) HC‐toxin. Phytochemistry, 67, 1406–1413. [DOI] [PubMed] [Google Scholar]
- Wang, A. , Kurdistani, S.K. and Grunstein, M. (2002) Requirement of Hos2 histone deacetylase for gene activity in yeast. Science, 298, 1412–1414. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Pajerowska‐Mukhtar, K. , Culler, A.H. and Dong, X. (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wang, G.L. , Ruan, D.L. , Song, W.Y. , Sideris, S. , Chen, L. , Pi, L.Y. , Zhang, S. , Zhang, Z. , Fauquet, C. , Gaut, B.S. , Whalen, M.C. and Ronald, P.C. (1998) Xa21D encodes a receptor‐like molecule with a leucine‐rich repeat domain that determines race‐specific recognition and is subject to adaptive evolution. Plant Cell, 10, 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Zang, C. , Cui, K. , Schones, D.E. , Barski, A. , Peng, W. and Zhao, K. (2009) Genome‐wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell, 138, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M. and Schubeler, D. (2007) Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr. Opin. Cell Biol. 19, 273–280. [DOI] [PubMed] [Google Scholar]
- Weil, C. and Martienssen, R. (2008) Epigenetic interactions between transposons and genes: lessons from plants. Curr. Opin. Genet. Dev. 18, 188–192. [DOI] [PubMed] [Google Scholar]
- Wight, W.D. , Kim, K.H. , Lawrence, C.B. and Walton, J.D. (2009) Biosynthesis and role in virulence of the histone deacetylase inhibitor depudecin from Alternaria brassicicola . Mol. Plant–Microbe Interact. 22, 1258–1267. [DOI] [PubMed] [Google Scholar]
- Wu, K. , Malik, K. , Tian, L. , Brown, D. and Miki, B. (2000) Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana . Plant Mol. Biol. 44, 167–176. [DOI] [PubMed] [Google Scholar]
- Wu, K. , Zhang, L. , Zhou, C. , Yu, C.W. and Chaikam, V. (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59, 225–234. [DOI] [PubMed] [Google Scholar]
- Yang, X.J. and Seto, E. (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 9, 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, H. and Richards, E.J. (2007) A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell, 19, 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, H. and Richards, E.J. (2009) Gene duplication and hypermutation of the pathogen resistance gene SNC1 in the Arabidopsis bal variant. Genetics, 183, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach, A. , Li, Y. , Wayburn, B. , Ben‐Meir, H. , Kiss, V. , Avivi, Y. , Kalchenko, V. , Jacobsen, S.E. and Grafi, G. (2005) DDM1 binds Arabidopsis methyl‐CpG binding domain proteins and affects their subnuclear localization. Plant Cell, 17, 1549–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Roberts, D.N. and Cairns, B.R. (2005) Genome‐wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell, 123, 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. , Long, C. , Lan, L. , Chai, J. , Chen, S. , Tang, X. and Zhou, J.M. (2007a) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, K. , Sridhar, V.V. , Zhu, J. , Kapoor, A. and Zhu, J.K. (2007b) Distinctive core histone post‐translational modification patterns in Arabidopsis thaliana . PLoS ONE, 2, e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. (2008) The epigenetic landscape of plants. Science, 320, 489–492. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yazaki, J. , Sundaresan, A. , Cokus, S. , Chan, S.W. , Chen, H. , Henderson, I.R. , Shinn, P. , Pellegrini, M. , Jacobsen, S.E. and Ecker, J.R. (2006) Genome‐wide high‐resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell, 126, 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Clarenz, O. , Cokus, S. , Bernatavichute, Y.V. , Pellegrini, M. , Goodrich, J. and Jacobsen, S.E. (2007c) Whole‐genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5, e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Bernatavichute, Y.V. , Cokus, S. , Pellegrini, M. and Jacobsen, S.E. (2009) Genome‐wide analysis of mono‐, di‐ and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana . Genome Biol. 10, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. , Zhang, L. , Duan, J. , Miki, B. and Wu, K. (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell, 17, 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Geiman, T.M. , Xi, S. , Jiang, Q. , Schmidtmann, A. , Chen, T. , Li, E. and Muegge, K. (2006) Lsh is involved in de novo methylation of DNA. EMBO J. 25, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Jeong, J.C. , Zhu, Y. , Sokolchik, I. , Miyazaki, S. , Zhu, J.K. , Hasegawa, P.M. , Bohnert, H.J. , Shi, H. , Yun, D.J. and Bressan, R.A. (2008) Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA, 105, 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D. (2008) The evolving functions of DNA methylation. Curr. Opin. Plant Biol. 11, 554–559. [DOI] [PubMed] [Google Scholar]
- Zilberman, D. , Gehring, M. , Tran, R.K. , Ballinger, T. and Henikoff, S. (2007) Genome‐wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69. [DOI] [PubMed] [Google Scholar]
- Zilberman, D. , Coleman‐Derr, D. , Ballinger, T. and Henikoff, S. (2008) Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature, 456, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2008) Pattern‐recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. [DOI] [PubMed] [Google Scholar]
- Zlatanova, J. and Thakar, A. (2008) H2A.Z: view from the top. Structure, 16, 166–179. [DOI] [PubMed] [Google Scholar]
- Zupkovitz, G. , Tischler, J. , Posch, M. , Sadzak, I. , Ramsauer, K. , Egger, G. , Grausenburger, R. , Schweifer, N. , Chiocca, S. , Decker, T. and Seiser, C. (2006) Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell. Biol. 26, 7913–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]


