SUMMARY
A functional analysis of the V2 protein of two monopartite begomoviruses, Papaya leaf curl virus (PaLCuV) and Cotton leaf curl Kokhran virus (CLCuKoV), has been performed. Expression of the V2 gene from a Potato virus X (PVX) vector resulted in severe leaf curling followed by a hypersensitive response (HR) in Nicotiana benthamiana and N. tabacum, demonstrating that the V2 protein is a pathogenicity determinant and a target of host defence responses. Agroinfiltration of a PVX vector expressing the V2 protein resulted in cell death in the infiltrated area. Subsequently, a systemic HR developed that was associated with the long‐distance spread of the virus and led to the death of the plant. V2 amino acid sequences encompassing a conserved putative protein kinase C (PKC) phosphorylation motif were shown to be essential for the elicitation of cell death. In co‐inoculation experiments, the transient expression of the C2 protein of PaLCuV or Cotton leaf curl Multan virus under the control of the Cauliflower mosaic virus 35S promoter inhibited the HR induced by V2 in the agroinfiltrated area. These findings demonstrate that the V2 protein of monopartite begomoviruses is a pathogenicity determinant and induces an HR that can be suppressed by the C2 protein. The induction and suppression of HR have been demonstrated previously in bipartite begomoviruses and our results extend this to monopartite begomoviruses.
INTRODUCTION
The plant response to infection by pathogens is often accompanied by rapid cell death in and around the infection site, a reaction known as the hypersensitive response (HR; Lam et al., 2001). The HR can be conditioned by a pathogen‐encoded avirulence (avr) gene, the direct or indirect product of which is recognized by a plant possessing the corresponding resistance (R) gene, and is often associated with the prevention of growth and spread of the pathogen (Morel and Dangl, 1997). This type of response is programmed and is associated with a transient burst of reactive oxygen species, the fortification of cell walls, the accumulation of antimicrobial phytoalexins and the onset of systemic acquired resistance (Dangl et al., 1996). The programmed cell death in response to pathogen attack, as well as the regulatory mechanisms that underlie it, are conserved among plants and animals. For example, the onset of cell death involves the activation of cysteine–aspartic acid proteases (known as caspases in animals) and the generation of reactive oxygen species in both animals and plants (Baker et al., 1997; Bonneau et al., 2008; Heath, 2000).
Both plant and animal pathogens are capable of manipulating cell death pathways to promote their growth and spread in susceptible hosts. For example, pathogen‐encoded proteins can interfere with the cell death pathway by inhibiting the activity of key factors involved in the pathway (Greenberg and Tao, 2004; Morel and Dangl, 1997). Many plant pathogens, including bacteria, fungi and nematodes, have been shown to modulate cell death. The anti‐apoptotic activity of animal viruses is well documented and has been shown to be essential for the successful infection of hosts (Hay and Kannourakis, 2002). However, manipulation of the cell death pathway by plant viruses has not been investigated extensively. We have shown previously that Tomato leaf curl New Delhi virus (ToLCNDV), a whitefly‐transmitted geminivirus, encodes a protein, the nuclear shuttle protein (NSP), that induces cell death when expressed under the control of the Cauliflower mosaic virus (CaMV) 35S promoter or from a Potato virus X (PVX) vector in plants (Hussain et al., 2005). However, ToLCNDV infection of plants is not associated with cell death, and we have shown that NSP‐induced cell death is suppressed by a second virus‐encoded protein, the transcriptional‐activator protein (TrAP; Hussain et al., 2007).
Geminiviruses (family Geminiviridae) are one of only two groups of plant‐infecting viruses that possess genomes of single‐stranded DNA (ssDNA). These viruses have been classified into four genera on the basis of insect vectors, host range and genomic organization (Stanley et al., 2005). Of these, whitefly‐transmitted geminiviruses (genus Begomovirus) are the largest group, with more than 100 virus species identified so far. Begomoviruses have genomes consisting of either two components (known as DNA A and DNA B), both of which are essential for successful infection (Stanley, 1983), or a single component homologous to the DNA A component of the bipartite viruses (Saunders et al., 2000). However, recently, another group of monopartite begomoviruses has been identified. These require a ssDNA satellite (collectively known as betasatellites) to induce typical disease symptoms in the hosts from which they are isolated (reviewed by Briddon and Stanley, 2006). Such begomovirus disease complexes are widespread throughout the Old World and are now known to outnumber the bipartite begomoviruses in the Old World (Nawaz‐ul‐Rehman et al., 2009). A devastating disease of cotton, known as cotton leaf curl disease (CLCuD), is caused by such a begomovirus complex in Pakistan and northern India. The disease is associated with several distinct monopartite begomoviruses, including Papaya leaf curl virus (PaLCuV), Cotton leaf curl Kokhran virus (CLCuKoV) and Cotton leaf curl Multan virus (CLCuMV), and a disease‐specific betasatellite, referred to as Cotton leaf curl Multan betasatellite (Briddon et al., 2008; Mansoor et al., 2003).
Monopartite begomoviruses do not have a DNA B component and therefore lack the NSP that, for at least two bipartite begomoviruses, is the elicitor of cell death (Garrido‐Ramirez et al., 2000; Hussain et al., 2005). We argued that, if the encoding of an avr determinant that induces cell death and its suppression are conserved among begomoviruses, there must be a protein encoded by monopartite begomoviruses that elicits cell death. In this study, we have identified an elicitor of cell death for two monopartite begomoviruses that are associated with CLCuD. The results show that the protein encoded by the V2 gene of monopartite begomoviruses is an avr determinant in Nicotiana benthamiana and N. tabacum. Deletion mutagenesis was used to identify the sequence involved in the induction of cell death. In addition, the cell death elicited by the V2 protein was shown to be suppressed by the product encoded by the C2 gene, a homologue of the bipartite begomovirus‐encoded TrAP.
RESULTS
The V2 protein of monopartite begomoviruses is a symptom determinant and elicits cell death in N. benthamiana
Nicotiana benthamiana plants agroinfiltrated with a PVX vector expressing the V2 gene of PaLCuV (PaV2/PVX) showed HR in the agroinfiltrated area and severe leaf curling (Fig. 1A) within 8 days of infiltration. The cell death spread systemically into the upper, noninfiltrated leaves, spreading from the veins to affect the whole leaf approximately 12–15 days post‐inoculation (Table 1). Subsequently, the upper young tissues of PaV2/PVX‐inoculated plants collapsed, leading ultimately to the death of the plants. In contrast, N. benthamiana plants inoculated with only the PVX vector exhibited only mild vein yellowing symptoms on systemically infected leaves, typical of PVX. The inoculated area showed no evidence of necrosis (Fig. 1B). To show that the development of necrosis was caused by the expression of the V2 protein, the PaLCuV V2 gene was inserted into the PVX vector in an antisense orientation, yielding the construct PaV2AS/PVX. The inoculation of N. benthamiana with PaV2AS/PVX resulted in systemic symptoms typical of PVX, mild vein yellowing, but no evidence of necrosis in either the inoculated area or on systemically infected leaves, and no leaf curling (Fig. 1C). The inoculations were repeated three times, and the results were reproducible. These results suggest that the effects seen with the intact V2 gene are a result of protein expression and demonstrate that the V2 protein of PaLCuV is a symptom determinant and elicitor of cell death in N. benthamiana. To further verify these results, the V2 gene was cloned into a binary vector under the control of the 35S promoter. Nicotiana benthamiana plants infiltrated with the construct showed necrosis of the infiltrated area after 7–8 days (Fig. 1D). In this case, there was no systemic cell death or symptoms exhibited by newly emerged leaves. Similarly, the expression of the PaLCuV V2 gene from a PVX vector, compromised for movement by mutation of the triple gene block encoding the 25K protein (PVXMD), induced necrosis of the infiltrated area on infiltration of N. benthamiana, but no systemic necrosis (Fig. 1E). In this case, there were no systemic symptoms (e.g. vein yellowing) of PVX long‐distance movement. This confirms that systemic necrosis (for PaV2/PVX) is a result of movement of the PVX vector expressing V2, rather than systemic movement of the V2 protein, and that PaLCuV V2 is unable to complement the PVX 25K mutation.
Figure 1.

The hypersensitive response of Nicotiana benthamiana to V2 expression. (A) A N. benthamiana plant infected with PaV2/PVX showing cell death in the agroinfiltrated area (indicated by the arrows) as well as on systemic leaves. (B) A N. benthamiana plant infected with the PVX vector. The damage caused by inoculation is indicated by the arrow, but there is no cell death in the surrounding inoculated tissue. (C) A N. benthamiana plant infected with PaV2AS/PVX. The inoculated leaf is indicated by an arrow. (D) A N. benthamiana leaf inoculated with PaV2/35s showing cell death in the inoculated area. (E) A N. benthamiana plant agroinfiltrated with PaV2/PVXMD (the arrow indicates necrosis in the inoculated area). (F) A N. benthamiana plant infected with KoV2/PVX showing cell death in the agroinfiltrated areas (indicated by the arrows). Nicotiana tabacum leaves inoculated with PaV2/PVX (G) and PVX (H), and treated with trypan blue. After clearing with chloral hydrate, cells in the PaV2/PVX‐inoculated leaf retain the dye, confirming cell death.
Table 1.
Features of gene constructs and the results of their inoculation into Nicotiana benthamiana.
| Gene construct* | Insert size (bp) | Deletions (no. of amino acids) [amino acid coordinates of the deletion] | No of symptomatic plants/no. of plants inoculated† | Hypersensitive response |
|---|---|---|---|---|
| PaV2/PVX | 453 | — | 10/10 | Yes |
| PaV2AS/PVX | 453 | — | 5/5 | No |
| PaV2dmN32/PVX | 357 | 32, N‐terminal [1–32] | 8/10 | Yes |
| PaV2dmN60/PVX | 273 | 60, N‐terminal [1–60] | 8/10 | Yes |
| PaV2dmC20/PVX | 393 | 20, C‐terminal [130–149] | 6/8 | Yes |
| PaV2dmC40/PVX | 333 | 40, C‐terminal [111–149] | 5/7 | Yes‡ |
| PaV2dmC50/PVX | 303 | 50, C‐terminal [101–149] | 7/10 | Yes§ |
| PaV2dmC60/PVX | 273 | 60, C‐terminal [91–149] | 7/10 | No |
| PaV2dmC80/PVX | 213 | 80, C‐terminal [71–149] | 6/10 | No |
| PaV2dmC119/PVX | 96 | 119, C‐terminal [30–149] | 8/10 | No |
| KoV2/PVX | 351 | — | 5/5 | Yes |
Constructs in Potato virus X (PVX) are indicated as either the V2 gene of Papaya leaf curl virus (PaLCuV) (PaV2) or Cotton leaf curl Kokhran virus (CLCuKoV) (KoV2) or deletion mutants (dm) of the PaLCuV V2 gene with deletions at either the N‐terminal (N) or C‐terminal (C) ends. A PVX construct with the PaLCuV V2 gene inserted in the anti‐sense orientation is indicated by AS.
Numbers of plants showing symptoms of PVX infection. Combined results of three independent experiments.
Delayed necrotic response.
Necrosis only in the agroinfiltrated area.
To ascertain whether the V2 protein of another monopartite begomovirus would elicit HR in N. benthamiana, the V2 gene of CLCuKoV was cloned into the PVX vector to produce KoV2/PVX. Agroinfiltration of this construct into N. benthamiana leaves resulted in cell death in the infiltrated area (Fig. 1F), followed by systemic infection and the production of severe leaf curling in systemically infected leaves. The cell death subsequently spread to systemically infected leaves and ultimately resulted in the collapse and death of the plants. This indicates that V2 is an elicitor of the cell death response in N. benthamiana for both monopartite begomoviruses examined. The PaLCuV V2‐infiltrated leaf was also treated with trypan blue to confirm that the phenotype induced by V2 is cell death (Jordan et al., 2007). Cells in the infiltrated leaf tissue showing HR after inoculation of the V2‐expressing PVX construct retained the dye, whereas equivalent tissue from a PVX‐infected plant was free of trypan blue‐stained cells after washing with chloral hydrate, confirming that cell death was occurring in response to V2 expression and not as a result of infection by the PVX vector (Fig. 1G, H).
Deletion analysis of PaLCuV V2
To identify the amino acid sequences of the PaLCuV V2 protein potentially involved in the elicitation of the cell death response, eight deletion mutants were generated and expressed from the PVX vector (Table 1; Fig. 2A). The results of the agroinfiltration of plants with the deletion mutants are summarized in Table 1. Nicotiana benthamiana plants inoculated with the deletion mutants PaV2dmN32/PVX and PaV2dmN60/PVX (having deletions of 32 and 60 amino acids, respectively, at the N‐terminal end of V2) exhibited a systemic HR (Fig. 2A, B). The position of the initiation methionine for the V2 gene of PaCLCuV has not been determined. The open reading frame of the V2 gene of PaLCuV has the capacity to encode an additional 32 N‐terminal amino acids, in comparison with the V2 genes of other begomoviruses (Fig. 2B). The finding that the deletion of the first 32 amino acids had no effect on the induction of HR, as well as the context of the second AUG codon with respect to the putative virion‐sense promoter (results not shown), is consistent with the conserved (between distinct begomoviruses) methionine at position 32 of PaLCuV V2 acting as the bona fide initiation methionine. In contrast, deletions of 60 (Fig. 3C), 80 and 119 (Fig. 3D) amino acids at the C‐terminal end abolished the induction of HR in the agroinfiltrated area. These constructs induced no systemic HR and the symptoms were typical of a PVX infection, with only the induction of mild yellow vein symptoms. PaV2dmC50/PVX, with the C‐terminal 50 amino acids deleted, induced necrosis in the agroinfiltrated area, but not systemic necrosis (Fig. 3E); the systemic symptoms were again typical of PVX. The mutants with 20 and 40 amino acids (Fig. 3F) deleted were each capable of inducing HR in both the inoculated area and in newly emerged leaves, although the systemic symptoms for the 40‐amino‐acid deletion mutant (PaV2dmC40/PVX) were delayed, occurring approximately 18–20 days post‐inoculation, and were milder. These findings suggest that the amino acid sequences between positions 92 and 101 are essential for the elicitation of HR, whereas those between 102 and 115 affect the timing and severity of the response.
Figure 2.
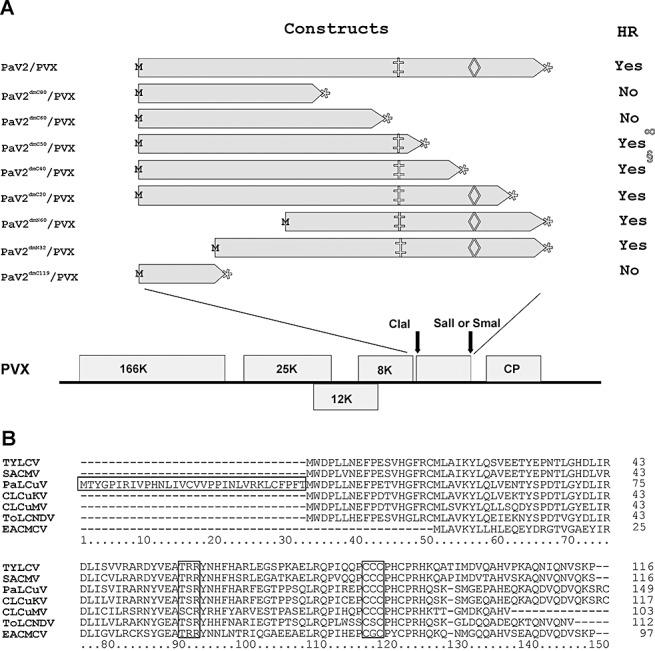
Constructs used in the analysis and comparison of (A)V2 sequences. (A) Potato virus X (PVX)‐based expression cassettes of V2 of PaLCuV. The deletion mutants of V2 of PaLCuV are indicated with their amino acid coordinates. M and  are the start and stop codons, respectively. The positions of the putative protein kinase C motif (‡) and CxC motif (◊) are indicated. HR, hypersensitive response. (B) Alignment of the predicted amino acid sequences of the V2 proteins of PaLCuV‐PK[PK:Cot:02] (AJ436992) and CLCuKoV‐Fai[PK:Fai1] (AJ496286) with the V2 proteins of other selected begomoviruses: Tomato yellow leaf curl virus (TYLCV) (AF260331), South African cassava mosaic virus (SACMV‐[ZW:Muz]; AJ575560), Cotton leaf curl Multan virus (CLCuMV‐Fai[PK:Fai2]; AJ496287), Tomato leaf curl New Delhi virus (ToLCNDV‐IN[IN:ND:Svr:92]; U15015) and East African cassava mosaic Cameroon virus (EACMCV‐CM[CM:98]; AF112354). Gaps (‐) were introduced to optimize the alignment. The position of a predicted putative protein kinase C motif (TxR; coordinates 91–93 of the alignment) and the CxC motif (coordinates 116–118 of the alignment) mutated by Padidam et al., (1996) and Zrachya et al., (2007) are shown. The precise position of the initiation methionine of (A)V2 has not been determined. The V2 open reading frame of PaLCV has the capacity to encode an additional 32 amino acids at the N‐terminus in comparison with the other viruses.
are the start and stop codons, respectively. The positions of the putative protein kinase C motif (‡) and CxC motif (◊) are indicated. HR, hypersensitive response. (B) Alignment of the predicted amino acid sequences of the V2 proteins of PaLCuV‐PK[PK:Cot:02] (AJ436992) and CLCuKoV‐Fai[PK:Fai1] (AJ496286) with the V2 proteins of other selected begomoviruses: Tomato yellow leaf curl virus (TYLCV) (AF260331), South African cassava mosaic virus (SACMV‐[ZW:Muz]; AJ575560), Cotton leaf curl Multan virus (CLCuMV‐Fai[PK:Fai2]; AJ496287), Tomato leaf curl New Delhi virus (ToLCNDV‐IN[IN:ND:Svr:92]; U15015) and East African cassava mosaic Cameroon virus (EACMCV‐CM[CM:98]; AF112354). Gaps (‐) were introduced to optimize the alignment. The position of a predicted putative protein kinase C motif (TxR; coordinates 91–93 of the alignment) and the CxC motif (coordinates 116–118 of the alignment) mutated by Padidam et al., (1996) and Zrachya et al., (2007) are shown. The precise position of the initiation methionine of (A)V2 has not been determined. The V2 open reading frame of PaLCV has the capacity to encode an additional 32 amino acids at the N‐terminus in comparison with the other viruses.
Figure 3.

Deletion analysis of PaLCuV V2 and suppression of the V2‐elicited hypersensitive response (HR) by C2. Nicotiana benthamiana plants infected with PaV2dmN32/PVX (A), PaV2dmN60/PVX (B), PaV2dmC60/PVX (C; the inoculated leaf is indicated by an arrow), PaV2dmC119/PVX (D), PaV2dmC50/PVX (E) and PaV2dmC40/PVX (F). Leaves of N. tabacum plants infiltrated with PaV2/PVX (H) and either PaC2/35S (G) or MuC2/35S (J) in the presence of PaV2/PVX. (K) A N. tabacum plant infiltrated with MuC2/35S (the agroinfiltrated area is indicated by an arrow). (I) A N. tabacum plant inoculated with PaV2/PVX showing systemic leaf curling and HR (indicated by an arrow).
Figure 2B shows an alignment of the predicted amino acid sequence of the PaLCuV V2 protein with those of other selected begomoviruses. This shows that the V2 sequences contain a conserved putative protein kinase C (PKC) phosphorylation motif (Pearson and Kemp, 1991). By site‐directed mutagenesis, Chowda‐Reddy et al. (2008) have shown this motif to be important in the pathogenicity of the V2 protein of the bipartite begomovirus East African cassava mosaic Cameroon virus (EACMCV) and for its ability to suppress RNA silencing. Our observation that the deletion of the sequences encompassing the PKC phosphorylation motif of PaLCuV V2 abolishes its pathogenicity (i.e. the ability to induce virus‐like symptoms) and ability to initiate HR is consistent with these results. V2 of PaLCuV at amino acid positions 116 and 118 contains a conserved CxC (Fig. 2B). Mutations of this motif have been shown to abolish both the pathogenicity and suppressor of RNA silencing activities of the protein (Padidam et al., 1996; Zrachya et al., 2007).
PaLCuV C2 protein prevents V2 protein‐induced cell death
We have shown previously that TrAP of ToLCNDV is able to prevent the hypersensitive cell death induced by the avr determinant of this virus (NSP) in N. tabacum and tomato (Hussain et al., 2007). We reasoned that, like bipartite begomoviruses, monopartite begomoviruses must have a factor to overcome hypersensitive cell death, because visible cell death (necrosis) is not a feature of PaLCuV infections of N. benthamiana, N. tabacum or cotton (Mansoor et al., 2003). When N. tabacum was co‐inoculated with PaC2/35S and PaV2/PVX, there was no hypersensitive cell death in the inoculated area as a result of V2, in contrast with the controls inoculated with PaV2/PVX only (Fig. 3G, H, respectively). Surprisingly, the co‐inoculated plants developed systemic leaf curl symptoms within 10–12 days of inoculation, with no evidence of systemic necrosis.
To determine whether the C2 protein of related viruses can inhibit the cell death induced by V2 of PaLCuV, a construct containing the C2 gene of CLCuMV under the control of the CaMV 35S promoter (MuC2/35S) was co‐infiltrated with PaV2/PVX. The control plants inoculated with PaV2/PVX developed leaf curling and a severe systemic HR (Fig. 3I). However, leaves co‐inoculated with Agrobacterium cultures harbouring MuC2/35S and PaV2/PVX did not develop HR in the inoculated area (Fig. 3J). As with PaLCuV C2, the plants developed systemic curling symptoms within 10–12 days of inoculation and no HR was evident. For control plants inoculated with MuC2/35S only, the inoculated area showed no evidence of a response to C2 expression (Fig. 3K).
DISCUSSION
The HR in plants is caused by interaction with an incompatible pathogen, and is characterized by rapid and localized cell death at the site of infection. HR often results in pathogen arrest and the prevention of systemic infection. However, resistance and HR can be uncoupled in cases in which no HR is observed at the site of pathogen–host interaction or in which HR is unable to contain the pathogen and systemic cell death ensues (Balague et al., 2003; Clough et al., 2000). It has been shown previously that bipartite begomoviruses encode proteins that are elicitors of HR. For both ToLCNDV and Bean dwarf mosaic virus, the avr determinant was shown to be NSP, encoded on the DNA B component (Garrido‐Ramirez et al., 2000; Hussain et al., 2005). Subsequently, we have shown, for ToLCNDV, that another virus‐encoded protein, TrAP, is capable of countering NSP‐induced hypersensitive cell death (Hussain et al., 2007). Here, we have extended these observations to a monopartite begomovirus, and have shown that the expression of the V2 protein can elicit an HR in N. benthamiana. Interestingly, HR is unable to restrict the long‐distance movement of PVX expressing V2, resulting in systemic HR and plant death. This is in contrast with several avr determinants which, when expressed from the PVX vector, prevent systemic infection and result in pathogen arrest (Greenberg, 1997). These results are consistent with a role of V2 in the pathogenicity of monopartite begomoviruses (Selth et al., 2004). We have further shown that the expression of the C2 protein, a homologue of the bipartite begomovirus TrAP, inhibits V2‐induced cell death when expressed at the site of inoculation under the control of the CaMV 35S promoter. Our results show that the induction and suppression of HR are common to at least some bipartite and monopartite begomoviruses.
The V2 protein (known as AV2 for bipartite begomoviruses) is conserved among begomoviruses from the Old World, but is absent from viruses originating from the New World. Although both bipartite and monopartite begomoviruses occur in the Old World, only a single monopartite begomovirus occurs in the New World, having been introduced from the Old World (Polston et al., 1999). It has been suggested that this evolutionary dichotomy may be caused by the absence of AV2 in the New World begomoviruses, which may be required to complement the missing functions encoded by DNA B for monopartite viruses (Ha et al., 2008). DNA B encodes two proteins that are involved in virus movement, NSP and the movement protein (MP; Sanderfoot and Lazarowitz, 1995), both of which bind DNA (Rojas et al., 1998). DNA A encodes all functions required for viral DNA replication, gene expression and encapsidation (Rojas et al., 2005). The genomes of monopartite begomoviruses are homologues of the DNA A components of bipartite viruses, and the V2 protein and coat protein have been shown to be the functional equivalents of the DNA B‐encoded NSP and MP (2001, 2005). For both monopartite and bipartite begomoviruses, V2 is dispensable for infection, although mutation leads to attenuated symptoms (Bull et al., 2007; Rigden et al., 1993). Green fluorescent protein‐tagged (A)V2 localizes to both the nucleus and the cytoplasm, and has a limited ability to move between adjacent cells, consistent with its possible role in virus cell‐to‐cell movement (Chowda‐Reddy et al., 2008; Rojas et al., 2001; Rothenstein et al., 2007). For a number of both bipartite and monopartite begomoviruses, the (A)V2 protein has been shown to have suppression of RNA silencing activity (Chowda‐Reddy et al., 2008; Zrachya et al., 2007) and, for the monopartite begomovirus Tomato yellow leaf curl virus, this activity has been shown to require interaction with SGS3 (Glick et al., 2007), a host‐encoded protein specifically required for the RNA silencing defence against geminiviruses (Muangsan et al., 2004). For CLCuMV, we have been unable to confirm a suppression of RNA silencing activity for V2. The only commonalities between the elicitors of HR for monopartite and bipartite begomoviruses thus appear to be an involvement in virus movement and the ability to bind nucleic acids.
The C2 protein of a monopartite begomovirus acts to counter HR elicited by the virus. For the majority of begomoviruses investigated, the (A)C2 protein acts to up‐regulate transcription of the late, virion‐sense‐encoded genes and is known as TrAP (Dry et al., 2000; Gopal et al., 2007; Sunter and Bisaro, 1992). (A)C2 also up‐regulates host gene expression (Trinks et al., 2005) and, in many cases, is a suppressor of RNA silencing (reviewed by Bisaro, 2006). It is also interesting to note that, although the (A)C2 proteins of some begomoviruses elicit a cell death response on constitutive expression in plants (Selth et al., 2004; van Wezel et al., 2001), others do not (Vanitharani et al., 2004). Thus, at least some monopartite begomoviruses, in common with their bipartite relatives, appear to interfere with two antiviral defence mechanisms deployed by plants, the adaptive defence based on the recognition of double‐stranded RNA (RNA silencing) and the classical gene‐for‐gene‐based defence involving the recognition of a pathogen‐encoded avr determinant that is the target of a host‐encoded R gene which triggers HR. At least one other plant virus‐encoded product, the 2b protein encoded by cucumoviruses, interacts with both pathways. The 2b protein encoded by Cucumber mosaic virus is both a suppressor of RNA silencing and an elicitor of HR (2004, 2005). The effective exploitation of cell death may be a determining factor in virus survival.
There is a wealth of evidence suggesting that the phosphorylation state of viral MPs controls function. For example, phosphorylation of the Tobacco mosaic virus‘30K’ MP has been shown to affects its ability to gate plasmodesmata (Trutnyeva et al., 2005; Waigmann et al., 2000). Phosphorylation of MPs may also play a role in controlling the switch from viral replication to translation (1997, 1999). Begomovirus (A)V2 proteins contain a conserved PKC phosphorylation motif. Mutation of the PKC motif of AV2 of the bipartite begomovirus EACMCV abolished pathogenicity and its ability to suppress RNA silencing (Chowda‐Reddy et al., 2008). For PaLCuV V2, deletion of sequences encompassing this motif abrogates the ability to induce HR. Further studies, which specifically mutate the PKC motif, will be required to determine whether phosphorylation of V2 is required for the induction of HR.
EXPERIMENTAL PROCEDURES
Expression constructs
Primers were designed to amplify the V2 and C2 genes based on the published sequences of PaLCuV‐PK[PK:Cot:02] (Accession no. AJ436992), CLCuKoV‐Fai[PK:Fai1] (AJ496286) and CLCuMV‐Fai[PK:Fai2] (AJ496287) (Mansoor et al., 2003). The primer sequences used for amplification are given in Table 2. For the insertion of polymerase chain reaction (PCR) fragments into the PVX vector, restriction endonuclease recognition sites were included in the forward (virion‐sense) and reverse (complementary‐sense) primers, respectively, and are given in italic (Table 2). PCR‐mediated amplifications with virus clones as a template, restriction endonuclease digestion and cloning were conducted by standard methods with enzymes obtained from MBI Fermentas (Arlington, Canada). PCR products were cloned into the PVX vector (pgR 107) at (ClaI, SalI) and (ClaI, SmaI) restriction sites. A movement‐deficient version of the PVX vector (PVXMD) was produced by mutation of the triple gene block 25K protein, as described by Morozov et al. (1997). Constructs for the transient expression of genes under the control of the CaMV 35S promoter in pJIT163 were produced as described previously (Hussain et al., 2007). Features of the gene constructs are summarized in Table 1. The gene constructs were sequenced to confirm the integrity of the inserts.
Table 2.
Primers used for the amplification of the V2 and C2 genes of Papaya leaf curl virus (PaLCuV), Cotton leaf curl Kokhran virus (CLCuKoV) and Cotton leaf curl Multan virus (CLCuMV).
| Construct | Primer | Orientation | Sequence* |
|---|---|---|---|
| PaV2/PVX | PaV2 F | Virion | AGATCGATATGACATATGGACCAATCAG |
| PaV2 R | Complementary | AGGTCGACCTAGGAACATCTGGACTTCTGT | |
| PaV2AS/PVX | PaV2 ASF | Complementary | AGGTCGACATGACATATGGACCAATCAG |
| PaV2 ASR | Virion | AGATCGATCTAGGAACATCTGGACTTCTGT | |
| PaV2dmN32/PVX† | PaV2 N32 | Virion | AGATCGATATGTGGGATCCGTTATTGAACG |
| PaV2dmN60/PVX† | PaV2 N60 | Virion | AGATCGATATGTATAATCATTTCCACG |
| PaV2dmC20/PVX‡ | PaV2 C20 | Complementary | AGGTCGACCTACATGCTTTTGCTTTGGTGACGC |
| PaV2dmC40/PVX‡ | PaV2 C40 | Complementary | AGGTCGACCTAGGGCTGTCGAAGTTGAG |
| PaV2dmC50/PVX‡ | PaV2 C50 | Complementary | AGGTCGACCTATTCGAAGCGGGCGTGGAAATG |
| PaV2dmC60/PVX‡ | PaV2 C60 | Complementary | AGGTCGACCTAGCTGGTCGCTTCGACATAAT |
| PaV2dmC80/PVX‡ | PaV2 C80 | Complementary | AGGTCGACCTATCCCAGAGTATCTGGGGAATAC |
| PaV2dmC119/PVX‡ | PaV2 C119 | Complementary | AGGTCGACCTAGGTGAATGGAAAACACA |
| PaC2/35S | PaC2F | Complementary | AGATCGATATGCGATCTTCGTCACCCTCG |
| PaC2R | Virion | AGGTCGACTTAAAGACCCTTAAGAAACGA | |
| KoV2/PVX | CLCuKoV2F | Virion | GTTATCGATATGTGGGATCCACTGTTAAAT |
| CLCuKoV2R | Complementary | TCAGTCGACCTAGGAACATCTGGACTTCTG | |
| MuC2/35S | CLCuMVC2F | Complementary | GCCTCCCGGGATGCAATCTTCATCACTCTC |
| CLCuMVC2R | Virion | AATAGAATTCTTAATTGAAATTACACCGAG |
Restriction endonuclease recognition sequences used to clone the polymerase chain reaction (PCR) fragments are given in italic.
Amplification of the gene for these constructs used PaV2 R as the complementary‐sense primer.
Amplification of the gene for these constructs used PaV2 F as the virion‐sense primer.
Deletion analysis of PaLCuV V2
Deletion mutants of PaLCuV V2 were produced to identify the amino acid sequences potentially involved in pathogenicity and cell death (Fig. 3). Oligonucleotides were designed to delete 32 and 60 amino acids from the N‐terminus of V2, yielding mutants PaV2dmN32 and PaV2dmN60, respectively. A start codon was introduced into the oligonucleotides for the amplification of these N‐terminal mutations (Table 2). Likewise, 20, 40, 50, 60, 80 and 119 amino acids were deleted from the C‐terminus with a stop codon introduced into the oligonucleotides, yielding mutants PaV2dmC20, PaV2dmC40, PaV2dmC50, PaV2dmC60 and PaV2dmC119, respectively.
Agroinfiltration
Nicotiana benthamiana plants were grown under controlled conditions at 25 °C with a 16‐h light regimen. The expression constructs were electroporated into Agrobacterium tumefaciens strain GV3101. Agrobacterium cultures for inoculation were grown at 28 °C for 48 h to an optical density (OD) at 600 nm of 0.6 with kanamycin and tetracycline selection. The bacterial cells were pelleted (5000 g for 15 min at 20 °C) and resuspended in 10 mm MgCl2 containing 150 µg/mL acetosyringone. Cells were incubated in this medium for 3 h and then infiltrated into the young, fully expanded leaves of 3–4‐week‐old plants, either directly or following dilution. For histochemical analysis of cell death, the leaves were soaked in trypan blue overnight. The leaves were then washed with chloral hydrate to remove the excess stain. Leaves inoculated with an empty PVX vector were used as negative controls.
Cell death inhibition assays
For the cell death inhibition assays, N. tabacum plants were used preferentially, as these are more robust and it proved easier to titrate the V2‐expressing inocula than in N. benthamiana. The V2 constructs induced HR at relatively low ODs, and these were diluted to the minimum needed to induce HR (OD600 ≈ 0.2). Nicotiana tabacum plants (cv. Samsun) were grown in a controlled environment chamber at 25 °C with supplementary light to yield a 16‐h photoperiod. Assays to investigate the ability of the C2 protein to inhibit V2‐induced HR were conducted by mixing Agrobacterium cultures harbouring PaV2/PVX in equal volumes with the culture containing PaC2/35S. The mixed cultures were infiltrated into fully expanded leaves as described previously. In these experiments, the dilution factors were carefully optimized, because V2 constructs are able to induce HR at relatively low OD. All plant inoculation assays were repeated a minimum of three times.
ACKNOWLEDGEMENTS
The authors are grateful to Ms J. Qazi for providing the movement‐deficient PVX vector. The project was funded in part by a Ministry of Science and Technology, Government of Pakistan, project on the use of RNA interference for broad‐spectrum resistance against plant viruses, and a collaborative project with Donald Danforth Plant Science Center, USA under a Pak–USA collaborative project. MM was supported by the Higher Education Commission (HEC), Government of Pakistan, under the Indigenous PhD scheme. RWB was supported by the HEC under the ‘Foreign Faculty Hiring Scheme’. We also acknowledge Dr David Bisaro, Ohio State University, for providing laboratory resources to perform the histochemical analysis. The authors are particularly thankful to Dr Y. Zafar for his support and encouragement.
REFERENCES
- Baker, B. , Zambryski, P. , Staskawicz, B. and Dinesh‐Kumar, S.P. (1997) Signaling in plant–microbe interactions. Science, 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Balague, C. , Lin, B. , Alcon, C. , Flottes, G. , Malmstrom, S. , Kohler, C. , Neuhaus, G. , Pelletier, G. , Gaymard, F. and Roby, D. (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide‐gated channel ion channel family. Plant Cell, 1, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaro, D.M. (2006) Silencing suppression by geminivirus proteins. Virology, 344, 158–168. [DOI] [PubMed] [Google Scholar]
- Bonneau, L. , Ge, Y. , Drury, G.E. and Gallois, P. (2008) What happened to plant caspases? J. Exp. Bot. 59, 491–499. [DOI] [PubMed] [Google Scholar]
- Briddon, R.W. and Stanley, J. (2006) Sub‐viral agents associated with plant‐infecting single‐stranded DNA viruses. Virology, 344, 198–210. [DOI] [PubMed] [Google Scholar]
- Briddon, R.W. , Brown, J.K. , Moriones, E. , Stanley, J. , Zerbini, M. , Zhou, X. and Fauquet, C.M. (2008) Recommendations for the classification and nomenclature of the DNA‐β satellites of begomoviruses. Arch. Virol. 153, 763–781. [DOI] [PubMed] [Google Scholar]
- Bull, S.E. , Briddon, R.W. , Sserubombwe, W.S. , Ngugi, K. , Markham, P.G. and Stanley, J. (2007) Infectivity, pseudorecombination and mutagenesis of Kenyan cassava mosaic begomoviruses. J. Gen. Virol. 88, 1624–1633. [DOI] [PubMed] [Google Scholar]
- Chowda‐Reddy, R.V. , Achenjang, F. , Felton, C. , Etarock, M.T. , Anangfac, M.T. , Nugent, P. and Fondong, V.N. (2008) Role of a geminivirus AV2 protein putative protein kinase C motif on subcellular localization and pathogenicity. Virus Res. 135, 115–124. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. , Fengler, K.A. , Yu, I.C. , Lippok, B. , Smith, R.K., Jr and Bent, A.F. (2000) The Arabidopsis dnd1 ‘defense, no death’ gene encodes a mutated cyclic nucleotide‐gated ion channel. Proc. Natl. Acad. Sci. USA, 97, 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. , Dietrich, R.A. and Richberg, M.H. (1996) Death don't have no mercy: cell death programs in plant–microbe interactions. Plant Cell, 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dry, I. , Krake, L. , Mullineaux, P. and Rezaian, A. (2000) Regulation of tomato leaf curl viral gene expression in host tissues. Mol. Plant–Microbe Interact. 13, 529–537. [DOI] [PubMed] [Google Scholar]
- Garrido‐Ramirez, E.R. , Sudarshana, M.R. , Lucas, W.J. and Gilbertson, R.L. (2000) Bean dwarf mosaic virus BV1 protein is a determinant of the hypersensitive response and avirulence in Phaseolus vulgaris . Mol. Plant–Microbe Interact. 13, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Glick, E. , Zrachya, A. , Levy, Y. , Mett, A. , Gidoni, D. , Belausov, E. , Citovsky, V. and Gafni, Y. (2007) Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA, 105, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal, P. , Kumar, P. , Sinilal, B. , Jose, J. , Kasin Yadunandam, A. and Usha, R. (2007) Differential roles of C4 and βC1 in mediating suppression of post‐transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 123, 9–18. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997) Programmed cell death in plant–pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. and Tao, N. (2004) The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 6, 201–211. [DOI] [PubMed] [Google Scholar]
- Ha, C. , Coombs, S. , Revill, P. , Harding, R. , Vu, M. and Dale, J. (2008) Molecular characterization of begomoviruses and DNA satellites from Vietnam: additional evidence that the New World geminiviruses were present in the Old World prior to continental separation. J. Gen. Virol. 89, 312–326. [DOI] [PubMed] [Google Scholar]
- Hay, S. and Kannourakis, G. (2002) A time to kill: viral manipulation of the cell death program. J. Gen. Virol. 83, 1547–1564. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000) Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3, 315–319. [DOI] [PubMed] [Google Scholar]
- Hussain, M. , Mansoor, S. , Iram, S. , Fatima, A.N. and Zafar, Y. (2005) The nuclear shuttle protein of Tomato leaf curl New Delhi virus is a pathogenicity determinant. J. Virol. 79, 4434–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, M. , Mansoor, S. , Iram, S. , Zafar, Y. and Briddon, R.W. (2007) The hypersensitive response to tomato leaf curl New Delhi virus nuclear shuttle protein is inhibited by transcriptional activator protein. Mol. Plant–Microbe Interact. 20, 1581–1588. [DOI] [PubMed] [Google Scholar]
- Jordan, C. , Shen, W. , Hanley‐Bowdoin, L. and Robertson, D. (2007) Geminivirus‐induced gene silencing of the tobacco retinoblastoma‐related gene results in cell death and altered development. Plant Mol. Biol. 65, 163–175. [DOI] [PubMed] [Google Scholar]
- Karpova, O.V. , Ivanov, K.I. , Rodionova, N.P. , Dorokhov Yu, L. and Atabekov, J.G. (1997) Nontranslatability and dissimilar behavior in plants and protoplasts of viral RNA and movement protein complexes formed in vitro . Virology, 230, 11–21. [DOI] [PubMed] [Google Scholar]
- Karpova, O.V. , Rodionova, N.P. , Ivanov, K.I. , Kozlovsky, S.V. , Dorokhov, Y.L. and Atabekov, J.G. (1999) Phosphorylation of tobacco mosaic virus movement protein abolishes its translation repressing ability. Virology, 261, 20–24. [DOI] [PubMed] [Google Scholar]
- Lam, E. , Kato, N. and Lawton, M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature, 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Mansoor, S. , Briddon, R.W. , Bull, S.E. , Bedford, I.D. , Bashir, A. , Hussain, M. , Saeed, M. , Zafar, M.Y. , Malik, K.A. , Fauquet, C. and Markham, P.G. (2003) Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 148, 1969–1986. [DOI] [PubMed] [Google Scholar]
- Morel, J.B. and Dangl, J.L. (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Morozov, S. , Fedorkin, O. , Juttner, G. , Schiemann, J. , Baulcombe, D. and Atabekov, J. (1997) Complementation of a Potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes. J. Gen. Virol. 78, 2077–2083. [DOI] [PubMed] [Google Scholar]
- Muangsan, N. , Beclin, C. , Vaucheret, H. and Robertson, D. (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDe1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 38, 1004–1114. [DOI] [PubMed] [Google Scholar]
- Nawaz‐ul‐Rehman, M.S. , Mansoor, S. , Briddon, R.W. and Fauquet, C.M. (2009) Maintenance of an Old World betasatellite by a New World helper begomovirus and possible rapid adaptation of the betasatellite. J. Virol. JVI.00795–JVI.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam, M. , Beachy, R.N. and Fauquet, C.M. (1996) The role of AV2 (‘precoat’) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology, 224, 390–404. [DOI] [PubMed] [Google Scholar]
- Pearson, R.B. and Kemp, B.E. (1991) Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations In: Methods in Enzymology (Tony H. and Bartholomew M.S., eds), pp. 62–81. San Diego and London: Academic Press. [DOI] [PubMed] [Google Scholar]
- Polston, J.E. , McGovern, R.J. and Brown, L.G. (1999) Introduction of tomato yellow leaf curl virus in Florida and implications for the spread of this and other geminiviruses of tomato. Plant Dis. 83, 984–988. [DOI] [PubMed] [Google Scholar]
- Rigden, J.E. , Dry, I.B. , Mullineaux, P.M. and Rezaian, M.A. (1993) Mutagenesis of the virion‐sense open reading frames of tomato leaf curl virus. Virology, 193, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R. , Noueiry, A.M. , Lucas, W.J. and Gilbertson, R.L. (1998) Bean dwarf mosaic geminivirus movement proteins recognize DNA in a form‐ and size‐specific manner. Cell, 95, 105–113. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R. , Jiang, H. , Salati, R. , Xoconostle‐Cazares, B. , Sudarshana, M.R. , Lucas, W.J. and Gilbertson, R.L. (2001) Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology, 291, 110–125. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R. , Hagen, C. , Lucas, W.J. and Gilbertson, R.L. (2005) Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43, 361–394. [DOI] [PubMed] [Google Scholar]
- Rothenstein, D. , Krenz, B. , Selchow, O. and Jeske, H. (2007) Tissue and cell tropism of Indian cassava mosaic virus (ICMV) and its AV2 (precoat) gene product. Virology, 359, 137–145. [DOI] [PubMed] [Google Scholar]
- Ryang, B.S. , Kobori, T. , Matsumoto, T. , Kosaka, Y. and Ohki, S.T. (2004) Cucumber mosaic virus 2b protein compensates for restricted systemic spread of Potato virus Y in doubly infected tobacco. J. Gen. Virol. 85, 3405–3414. [DOI] [PubMed] [Google Scholar]
- Ryang, B.‐S. , Matsumoto, T. , Kobori, T. , Kosaka, Y. and Ohki, S.T. (2005) 2b Protein is essential to induce a novel gradual cell death in Zucchini yellow mosaic virus‐inoculated cucumber cotyledon co‐infected with Cucumber mosaic virus . J. Gen. Plant Pathol. 71, 308–313. [Google Scholar]
- Sanderfoot, A.A. and Lazarowitz, S.G. (1995) Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the periphery. Plant Cell, 7, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, K. , Bedford, I.D. , Briddon, R.W. , Markham, P.G. , Wong, S.M. and Stanley, J. (2000) A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA, 97, 6890–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth, L.A. , Randles, J.W. and Rezaian, M.A. (2004) Host responses to transient expression of individual genes encoded by Tomato leaf curl virus. Mol. Plant–Microbe Interact. 17, 27–33. [DOI] [PubMed] [Google Scholar]
- Stanley, J. (1983) Infectivity of the cloned geminivirus genome requires sequences from both DNAs. Nature, 305, 643–645. [Google Scholar]
- Stanley, J. , Bisaro, D.M. , Briddon, R.W. , Brown, J.K. , Fauquet, C.M. , Harrison, B.D. , Rybicki, E.P. and Stenger, D.C. (2005) Geminiviridae In: Virus Taxonomy, Viiith Report of the ICTV (Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U. and Ball L.A., eds), pp. 301–326. London: Elsevier/Academic Press. [Google Scholar]
- Sunter, G. and Bisaro, D.M. (1992) Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell, 4, 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinks, D. , Rajeswaran, R. , Shivaprasad, P.V. , Akbergenov, R. , Oakeley, E.J. , Veluthambi, K. , Hohn, T. and Pooggin, M.M. (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79, 2517–2527.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trutnyeva, K. , Bachmaier, R. and Waigmann, E. (2005) Mimicking carboxyterminal phosphorylation differentially effects subcellular distribution and cell‐to‐cell movement of Tobacco mosaic virus movement protein. Virology, 332, 563–577. [DOI] [PubMed] [Google Scholar]
- Vanitharani, R. , Chellappan, P. , Pita, J.S. and Fauquet, C.M. (2004) Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78, 9487–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann, E. , Chen, M.H. , Bachmaier, R. , Ghoshroy, S. and Citovsky, V. (2000) Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell‐to‐cell movement protein. EMBO J. 19, 4875–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wezel, R. , Liu, H. , Tien, P. , Stanley, J. and Hong, Y. (2001) Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus‐China encodes a pathogenicity determinant that is localized in the nucleus. Mol. Plant–Microbe Interact. 14, 1125–1128. [DOI] [PubMed] [Google Scholar]
- Zrachya, A. , Glick, E. , Levy, Y. , Arazi, T. , Citovsky, V. and Gafni, Y. (2007) Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus‐Israel. Virology, 358, 159–165. [DOI] [PubMed] [Google Scholar]


