SUMMARY
Snakin‐1 (SN1), a cysteine‐rich peptide with broad‐spectrum antimicrobial activity in vitro, was evaluated for its ability to confer resistance to pathogens in transgenic potatoes. Genetic variants of this gene were cloned from wild and cultivated Solanum species. Nucleotide sequences revealed highly evolutionary conservation with 91–98% identity values. Potato plants (S. tuberosum subsp. tuberosum cv. Kennebec) were transformed via Agrobacterium tumefaciens with a construct encoding the S. chacoense SN1 gene under the regulation of the ubiquitous CaMV 35S promoter. Transgenic lines were molecularly characterized and challenged with either Rhizoctonia solani or Erwinia carotovora to analyse whether constitutive in vivo overexpression of the SN1 gene may lead to disease resistance. Only transgenic lines that accumulated high levels of SN1 mRNA exhibited significant symptom reductions of R. solani infection such as stem cankers and damping‐off. Furthermore, these overexpressing lines showed significantly higher survival rates throughout the fungal resistance bioassays. In addition, the same lines showed significant protection against E. carotovora measured as: a reduction of lesion areas (from 46.5 to 88.1% with respect to the wild‐type), number of fallen leaves and thickened or necrotic stems. Enhanced resistance to these two important potato pathogens suggests in vivo antifungal and antibacterial activity of SN1 and thus its possible biotechnological application.
INTRODUCTION
Potato is one of the most important crops worldwide. Unfortunately, commercial cultivars are particularly susceptible to fungal and bacterial diseases. Rhizoctonia solani, the most widely spread species of Rhizoctonia, was originally described on potato by Julius Kühn in 1858. It is a soil‐borne fungal pathogen that comprises several groups which are pathogenic to different host species (Adam, 1988; Ogoshi, 1987). The most common symptom of Rhizoctonia disease is referred as ‘damping‐off’ and it is characterized by an early killing of the infected seedlings, either before or after they emerge from the soil. Seedlings that are not killed by the fungus often show cankers, which are reddish‐brown lesions on stems and roots (Agrios, 1997; Gutierrez et al., 1997). Stolon infection can influence the number and size distribution of tubers, and in severe infections total yield can be reduced (Hide and Horrocks, 1994; Hide et al., 1996). Erwinia carotovora, a Gram‐negative bacterium, is another important pathogen that causes soft rot and blackleg, two serious potato diseases that occur in most regions worldwide. Blackleg is economically important as it reduces yield and causes downgrading or rejection of potato seed in certification processes (Pérombelon, 2000). In addition, extensive soft rotting can develop when tubers with blackleg infection are stored. Plant disease control relies mainly on chemical pesticides that are not always effective, are commonly expensive, could present residual toxicity and could contribute to the selection of fungicide‐resistant pathogens. Consequently, ubiquitous and constitutive transgenic expression of genes encoding antimicrobial proteins with broad spectrum into commercial cultivars constitutes an interesting approach to reduce losses caused by pathogens.
As a counter‐defence, plants have evolved a variety of potent defence mechanisms, including the synthesis of low‐molecular‐weight peptides with antifungal activity (Selitrennikoff, 2001). More than 500 antimicrobial peptides (AMPs) have been discovered in plants and animals, in which they play important roles as key components of the innate defence against invading microorganisms (Lopez‐Solanilla et al., 2003). Snakin‐1 (SN1) and snakin‐2 (SN2) are two AMPs isolated from Solanum tuberosum cv. Desireé that have been found to be active against important pathogens in vitro (Berrocal‐Lobo et al., 2002; Segura et al., 1999). SN1 and SN2 amino acid sequence alignments show similarity with members of the GAST family from tomato (giberellic acid stimulated transcript) and GASA family from Arabidopsis (giberellic acid stimulated in Arabidopsis), and they have been classified as members of a novel snakin/GASA family (Berrocal‐Lobo et al., 2002). Homologous genes have been also identified and characterized in a wide range of species including monocotyledonous and dicotyledonous species (Ben‐Nissan and Weiss 1996; Furukawa et al., 2006; Kotilainen et al., 1999; Shi et al., 1992). All these genes encode small polypeptides that share the common structural features of an N‐terminal putative signal sequence, a highly divergent intermediate region and a conserved 60 amino‐acid carboxyl‐terminal domain containing 12 conserved cysteine residues. Despite the common features among these proteins, there is no apparent consensus in the roles they play. They have been reported to be involved in diverse biological processes, including: cell division, cell elongation, cell growth, transition to flowering, signalling pathways and defence (Aubert et al., 1998; Ben‐Nissan et al., 2004; Berrocal‐Lobo et al., 2002; , de la Fuente et al., 2006; Furukawa et al., 2006; Kotilainen et al., 1999; Roxrud et al., 2007; Segura et al., 1999).
SN1 was reported to be active in vitro against bacterial species such as Clavibacter michiganensis subsp. sepedonicus and fungal species such as Fusarium solani, Fusarium culmorum, Bipolaris maydis and Botrytis cinerea (Berrocal‐Lobo et al., 2002; Segura et al., 1999). Although it was reported that SN1 causes a rapid in vitro aggregation of both Gram‐positive and Gram‐negative bacteria, a direct correlation between aggregation and antimicrobial activity was not established. SN1 expression was detected in tubers, stems, and axillary and young floral buds and did not respond to abiotic and biotic stimuli, suggesting that this peptide could be a component of pre‐existing defence barriers (Segura et al., 1999). The hypothesis of a defence role for SN1 was also supported by the observation of decreased virulence of SN1‐sensitive mutants of the bacterial pathogen Erwinia chrysanthemi (Lopez‐Solanilla et al., 1998). These authors suggested that the significant differences in virulence observed in the assays are consistent with the SN1 peptide being a key determinant of the interaction between plant and pathogen. Moreover, SN1 was also used to assess the susceptibility of several pathogenic and non‐pathogenic Listeria species, showing that this plant‐derived peptide had a strong inhibitory effect on the tested species (Lopez‐Solanilla et al., 2003).
In the present study we cloned and analysed five genetic variants of the SN1 gene. In order to establish its function in vivo, one of them was constitutively overexpressed into potato plants and their resistance against R. solani and E. carotovora was biologically assessed.
RESULTS
Isolation and bioinformatic analysis of SN1 sequences
To study the sequence diversity of the SN1 gene, five genetic variants were isolated from three different wild Solanum species (S. bulbocastanum, S. commersonii and two genotypes of S. chacoense), as well as from the commercially cultivated S. tuberosum subsp. tuberosum cv. Kennebec. A fragment of approximately 770 bp was amplified by PCR from DNA of all the species. In order to discard technical errors, DNA sequences were determined in both directions from at least five independent clones of each variant. The consensus sequences (EF206289, EF206293, EF206290, EF206291 and EF206292, respectively) revealed that the cloned fragments contained the entire open reading frame encompassing the previously reported SN1 cDNA sequence (Segura et al., 1999) and an intron sequence of around 500 bp. A conserved signal peptide was detected at the amino terminus of the sequences suggesting that SN1 protein is secreted in all the species. Comparative bioinformatic analysis of the nucleotide sequences revealed highly evolutionary conservation (91–98% identity). Most of the mismatches were found within the intron sequence or were silent changes, resulting in 98–100% identity at the amino acid level.
Molecular characterization of SN1 transgenic potato plants
As SN1 exhibited antifungal and antibacterial activity in vitro, we obtained transgenic potato plants to analyse further whether constitutive overexpression of the SN1 gene leads to disease resistance to pathogens. Leaf discs of S. tuberosum cv. Kennebec were transformed by co‐cultivation with an Agrobacterium tumefaciens strain harbouring pPZP‐SN1 (Fig. 1A). Primarily, the presence of the SN1 transgene were confirmed by PCR in kanamycin‐resistant potato lines. A 900‐bp fragment corresponding to the complete sequence of the SN1 gene, Ω enhancer sequence and a fragment of the CaMV35S promoter sequence was amplified in a set of 12 pre‐selected lines (Fig. 1B). Transgenic lines were referred as ‘S’ for the SN1 gene, and each number represented an independent transgenic event. We continued the subsequent characterizations with lines S1, S2, S3 and S5. Southern blot analysis was performed with HindIII‐digested DNA in order to confirm the presence of the complete SN1 cassette and the 2550‐bp expected hybridizing band was observed in all lines (Fig. 1C left). Additional Southern blot analysis was performed with KpnI‐digested DNA in order to reveal the number of inserted copies as this enzyme cut only once in the pPZP‐SN1 construct used in plant transformation assays (Fig. 1A). Two copies (line S1) or only one copy (lines S2–S5) of the transgene were revealed in this last experiment (Fig. 1C right). As it was reported that expression of endogenous SN1 gene has not been detected in leaves (Segura et al., 1999), we performed a reverse transcriptase PCR (RT‐PCR) using RNA from this tissue in order to verify SN1 transgene expression. Although a 295‐bp expected spliced fragment was amplified from cDNA belonging to all transgenic lines, it was also amplified from non‐transgenic plants (Fig. 2A upper panel). Specific actin transcript amplification was detected in all plants as an internal control for cDNA synthesis (Fig. 2A lower panel). DNA control was also used in these assays showing the expected unspliced fragments and supporting the absence of DNA contamination in RNA samples. To analyse SN1 mRNA accumulation, Northern blot analyses were performed using RNA extracted from leaves. Transgenic lines S1, S3 and S5 accumulated high levels of SN1 mRNA, while line S2 showed a low transcript level, almost comparable with that in non‐transgenic control plants (Fig. 2B upper panel). The same membrane was hybridized with an actin probe as a measure of a housekeeping mRNA level (Fig. 2B lower panel). Northen blot results confirmed that S1, S3 and S5 overexpressed almost equal amounts of SN1 transcript in leaves.
Figure 1.
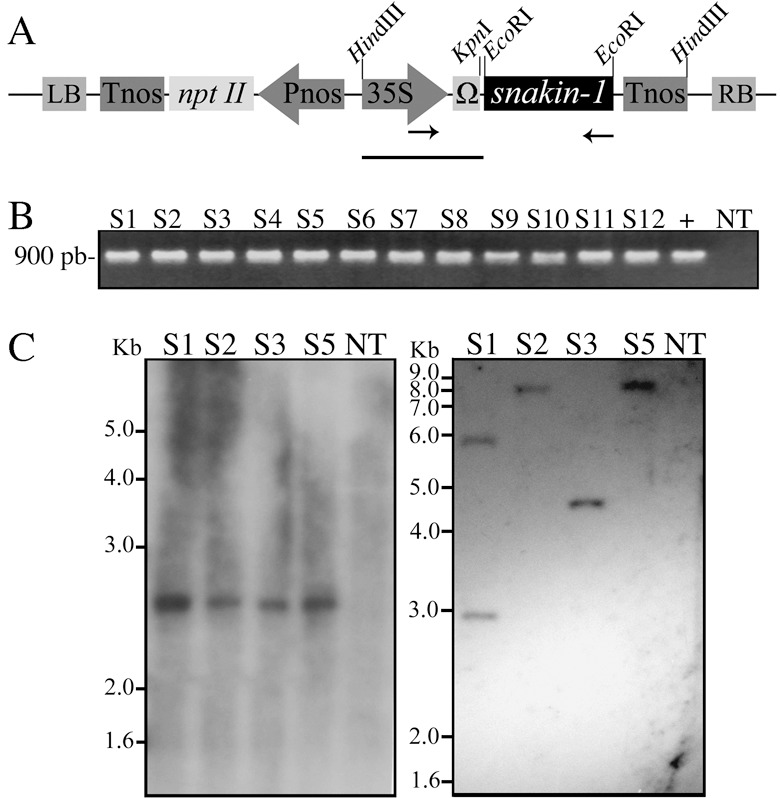
Genomic characterization of SN1 transgenic potato plants. (A) Schematic representation of the binary vector pPZPK‐SN1 used for transformation. Diagrams are not to scale. snakin‐1, SN1 complete gene; 35S, CaMV‐35S promoter; Tnos, nopaline synthase terminator; nptII, neomycin phosphototransferase II gene; Pnos, nopaline synthase promoter; RB and LB, right and left border sequences of the T‐DNA region, respectively; Ω, synthetic 65‐pb leader sequence from Tobacco mosaic virus. Arrows indicate primer annealing sites. Bar corresponds to the probe size used in Southern blot analysis. (B) PCR reaction performed with ‘up35S’‐ and ‘lowSN1’‐specific primers. Genomic DNA from different transgenic lines and non‐transgenic plants or pPZPK‐SN1 vector DNA(+) were used as template. (C) Southern blot analysis performed with genomic DNA completely digested with HindIII (left blot) or KpnI (right blot). DNA was transferred onto nylon membrane and hybridized with 32P‐labelled 35S + Ω probe. The hybridizing signals were visualized by exposing the membrane to X‐ray film for 20 days. DNA molecular size standard are shown on the left (1‐kb Plus DNA Ladder). (B and C) S1–S12, transgenic lines; NT, non‐transgenic plant.
Figure 2.
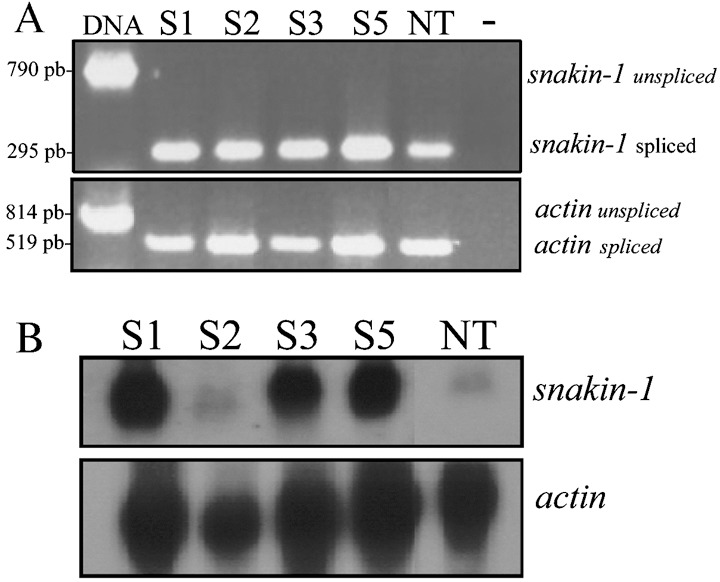
Expression of SN1 gene in transgenic potato plants. (A) RT‐PCR reaction performed with specific primers ‘upSN1’ and ‘lowSN1’ (upper panel) or ‘upActin’ and ‘lowActin’ (lower panel) from RNA isolated from leaf tissue. Amplified PCR product designations are shown on the right and products sizes are shown on the left. (–), control reaction without DNA; DNA, control reaction with genomic DNA. (B) Northern blot analysis performed with RNA isolated from leaf tissue from transgenic lines and non‐transgenic plants. Total RNA was extracted as described in the Experimental procedures and hybridized with 32P‐labelled snakin‐1 or actin probes (shown on the right). The hybridizing signals were visualized by exposing the membrane to X‐ray film for 10 days. (A and B) S1–S5, transgenic lines; NT, non‐transgenic plant.
Enhanced resistance of transgenic SN1 potato plants to fungal disease
Selected overexpressing lines were analysed for disease resistance to the infection by the soil‐borne fungal pathogen R. Solani. Two days following infection, most non‐transgenic plants presented total breakdown (damping‐off) in inoculated soil while transgenic plants showed no substantial disease symptoms (Fig. 3A right). While the percentage of surviving control plants was 17%, the three overexpressing lines showed survival rates of 50–75% (Fig. 3C). Throughout the entire bioassay, survival rates were significantly higher in transgenic lines S1, S3 and S5 than in the non‐transgenic control. No significant differences between transgenic lines were detected during the challenge, except that line S5 showed significantly lower survival rates at 2 and 8 days post‐infection (dpi) (Fig. 3C). Ten days following transplantation into inoculated soil, dead control plants and half of the surviving transgenic ones were collected. Non‐transgenic plants presented dramatic damages caused by the fungus compared with the high level of protection shown by all three transgenic overexpressing lines. Visual examination revealed that the phenotypes exhibited by overexpressing plants grew in inoculated soil were almost comparable with that of non‐infected control plants grown in non‐inoculated soil (Fig. 3D). Figure 3B shows detail of the severe stem canker exhibited by the single surviving control plant compared with that displayed by a transgenic plant. In addition, no phenotypic differences were observed between transgenic and non‐transgenic lines grown in non‐inoculated soil throughout the experiment (Fig. 3A left panel shows an example of non‐inoculated plants 2 days after transplantation). Furthermore, potato tuber yields in SN1 transgenic overexpressing plants, in relation to non‐transgenic parent cultivars, were unaffected in non‐inoculated soil (data not shown). This could suggest that constitutive expression of this peptide does not seem to alter plant growth parameters significantly.
Figure 3.
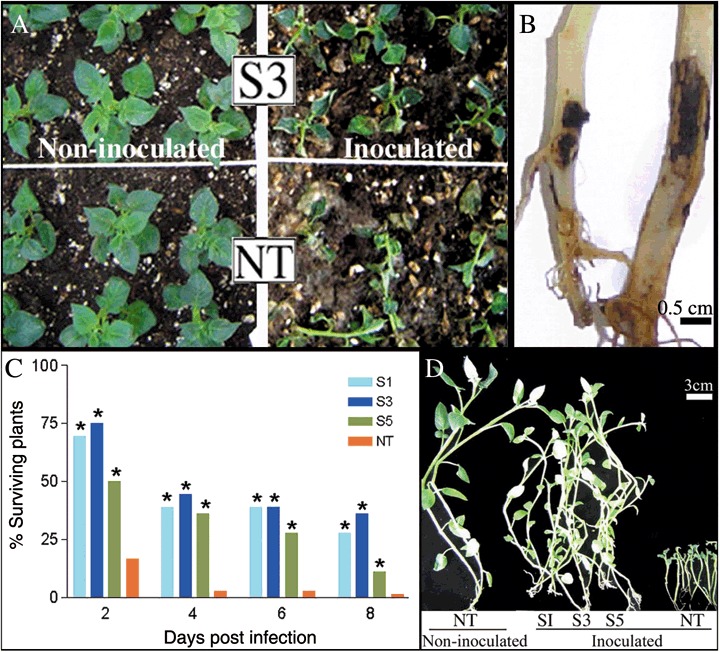
Enhanced resistance of SN1 transgenic potato plants to R. solani. (A) Magnified view of S3 line and non‐transgenic plants growing in non‐inoculated soil (left) and inoculated soil (right). Note total breakdown of non‐transgenic plants. (B) Detail of stem cankers produced by R. solani at 10 dpi. Example of a representative transgenic surviving plant (left) and the single non‐transgenic surviving plant (right). (C) Percentages of surviving plants were calculated at different days post infection (dpi). Asterisks denote significant differences (P < 0.05) with respect to non‐transgenic plants. (D) Plants collected in inoculated or non‐inoculated soil at 10 dpi. (A–D) S1–S5, transgenic lines; NT, non‐transgenic plant.
Enhanced resistance of transgenic SN1 potato plants to bacterial disease
In order to assess whether the resistance displayed by the SN1‐overexpressing lines is extended to bacterial pathogens, transgenic lines and non‐transgenic control plants were challenged against E. carotovora. Lesion severity was clearly different between transgenic and non‐transgenic lines at 10 dpi when lesions reach their final size. Whereas non‐transgenic plants showed a large, continuous, brown‐coloured lesion, transgenic lines exhibited a small lesion at the inoculation site (Fig. 4A–D). Only in non‐transgenic stems was an internal dark brown coloration of the pith (and in some cases of the vascular tissue) observed at the point of infection. Symptoms often extended several nodes above the initial inoculation point and in some cases the pith was almost absent (Fig. 4E). In addition, chlorosis and necrosis were detected on foliage adjacent to affected stems, mainly in non‐transgenic plants. Both the number of fallen leaves and the number of stems that were thickened due to the disease were recorded in all plants. All studied disease parameters were highly reduced in S1, S3 and S5 lines compared with the non‐transgenic plants at 10 dpi (Table 1). Stems of transgenic plants exhibited a significant reduction in lesion size from 46.5% for the S1 line to 88.1% for the S5 line considering as 0 the lesion reduction percentage of non‐transgenic plants. In addition, overexpressing lines showed a considerable reduction in the number of fallen leaves. Moreover, no thickened stems were observed in any transgenic plant, whereas 70% of non‐transgenic plants exhibited remarkably thickened or necrotic stems. This elevated disease tolerance of overexpressing SN1 transgenic plants was also observed under other assay conditions, even though typical disease symptoms were weaker (data not shown).
Figure 4.
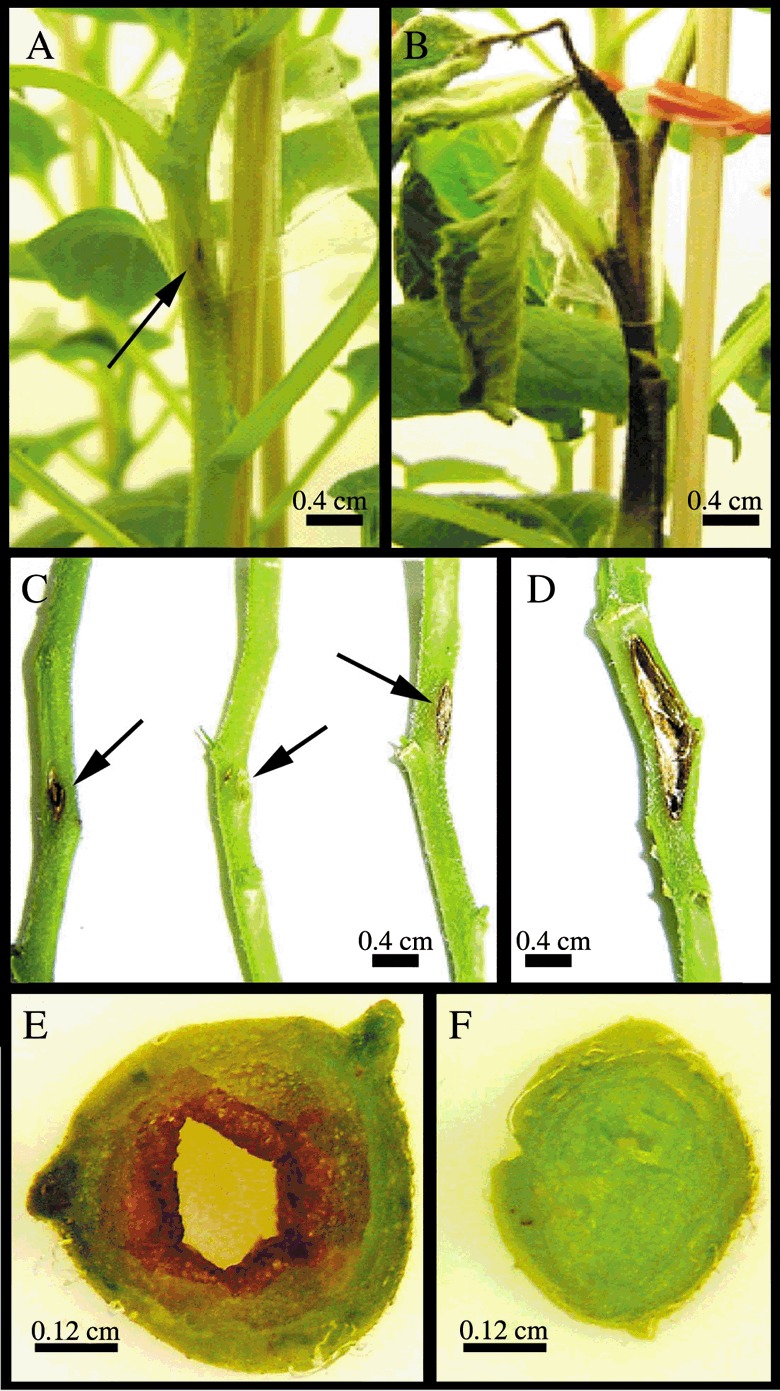
Enhanced resistance of SN1 transgenic potato plants to E. carotovora subsp. carotovora. Four‐week‐old plants were inoculated with bacterial inoculum using a syringe with a needle. (A) Example of a representative transgenic plant showing a small lesion at the inoculation site. (B) Non‐transgenic plant exhibiting inky‐black decay, typical symptoms of blackleg disease. (C) Detail of transgenic stems showing small lesions (lines S3, S5, S1, respectively). (D) Detail of a non‐transgenic stem showing an intermediate lesion. (E) Transverse section of one representative non‐transgenic thickened stem showing the internal affected pith. (F) Transverse section at the inoculation site level of one representative transgenic line stem (line S5) showing the unaltered pith. (A–F) Photographs were taken 10 days after infection.
Table 1.
Enhanced resistance of SN1 transgenic potato plants to E. carotovora subsp. carotovora.
| Plant line | Mean lesion size (mm2; ± SEM) | Lesion reduction (%) | Fallen leaves | Thickened stems |
|---|---|---|---|---|
| S1 | 14.3 ± 4.1* | 46.5 | 6 | 0/10 |
| S3 | 12.8 ± 2.7* | 52.1 | 9 | 0/10 |
| S5 | 3.2 ± 1.0* | 88.1 | 0 | 0/10 |
| NT | 32.05 ± 7.1 | 0 | 20 | 7/10 |
Evaluation of disease parameters from ten replicas of each transgenic line (S1–S5) and non‐transgenic plant (NT) at 10 dpi. *Significant differences (P < 0.05) with respect to non‐transgenic plants.
DISCUSSION
In the present paper, we explore the in vivo biological function of the potato SN1 gene and its potential use for development of transgenic plants with resistance to a broad spectrum of phytopathogens. We report the isolation of five genetic variants of the SN1 gene from wild and cultivated Solanum species. Comparative sequence analyses revealed a high evolutionary conservation. SN1 belongs to a large group of proteins identified in various plant species that share a conserved C‐terminal region in which 12 cysteines are located in exactly the same positions. The conserved distance between these cysteines might be required for the generation of an essential three‐dimensional structure and/or for an interaction with other proteins (Ben‐Nissan et al., 2004).
In order to explore SN1 biological function, we obtained and molecularly characterized transgenic lines of the commercial potato cultivar Kennebec. We selected S1, S3 and S5 overexpressing transgenic lines that we deemed suitable for biologically characterization as line S2 and non‐transgenic plants had presented similar symptoms in preliminary independent assays (data not shown). Unfortunately, we failed to express SN1 either in bacterial or in yeast expression systems, making it impossible to evaluate the peptide levels in transgenic lines. It is probable that its intrinsic antimicrobial/antifungal activity may affect the viability of these mentioned expression systems. This might expain why nobody has yet succeeded in obtaining a specific antibody. To assess whether these lines acquired enhanced tolerance to pathogens, they were first challenged with R. solani. The survival rates and the reduction of disease symptoms, such as stem cankers and damping‐off, were significantly higher in transgenic plants compared with the non‐transgenic ones (Fig. 3). Finally, to analyse whether the resistance displayed by the SN1‐overexpressing lines is extended to bacterial pathogens, plants were challenged with E. carotovora. Lesion areas, the number of fallen leaves and the number of thickened or necrotic stems were drastically reduced in S1, S3 and S5 lines compared with the non‐transgenic plants (Fig. 4, , Table1). The enhanced resistance against these two important potato pathogens suggests that the SN1 gene has an in vivo antifungal/antibacterial activity. Even though there are not significant differences between S1 and S3 lines in fungal or bacterial challenges, line S5 seems not to be as good as the others in suppressing R. solani but to be the best one in suppressing E. carotovora. As mentioned before, we were uanble to analyse whether the levels of disease resistance are correlated with SN1 protein expression levels in trangenic lines and so we could not determine whether the observed resistance is due to a direct or indirect effect. SN1 mRNA accumulation levels in leaves were almost similar in all transgenic lines (Fig. 2) but we could not discard the possibility that the region flanking the inserts could alter temporal or tissular expression patterns. More research needs to be done to clarify this point.
The hypothesis of a defence role of this gene was also supported by different reports of homologous genes. Thornburg et al. (2003) isolated a sample of nectary‐expressed cDNAs from ornamental tobacco and found that a large proportion of these cDNAs encoded defence‐related proteins. These include one SN1 homologue sequence among a number of genes directly involved in antimicrobial and/or antifungal activities. In addition, a cell‐wall‐orientated genomic approach in peaches revealed that one of the selected expressed sequence tag (EST) group contains a contig with significant homology to SN1 that shows a particular gene expression pattern in fruits at different stages of development (Trainotti et al., 2003). These authors suggested that the protein encoded by that contig might also be active against plant pathogens, and such a protective function would confirm its parietal location as the cell wall is the first obstacle that a pathogen has to overcome in order to infect a plant cell. Recently, a chitin‐binding proline‐rich protein from French bean that has been previously characterized through its involvement in plant–pathogen interactions was shown to be composed of two components: a proline‐rich polypeptide and a 12‐cysteine peptide with amino acid similarity to snakins (Bindschedler et al., 2006). Yet, the defence role proposed for SN1 is not incompatible with the involvement of this gene in other plant processes, functions and mechanisms.
In the present study, we reported that resistant transgenic lines exhibited a high reduction in lesion size caused by E. carotovora disease. Although no information is yet available concerning the mechanism of action of SN1, it was proposed that the observed aggregation of both Gram‐positive and Gram‐negative bacteria in vitro might play a role in vivo through the control of pathogen migration to uninfected areas (Segura et al., 1999). Moreover, snakin peptides show sequence similarity with cysteine‐rich domains of proteins from animals that are involved in protein–protein interactions playing an important role to control pathogens in vivo (Berrocal‐Lobo et al., 2002). Alternatively, SN1 might be involved in redox regulation, as it contains putative redox‐active cysteines and, as we have shown, it confers broad protection against pathogens. It is well known that reactive oxygen species (ROS) are involved in pathogenesis and wounding. Recently, it has been shown that GIP2 from Petunia hybrida (another GASA‐like protein) regulates ROS levels (Wigoda et al., 2006). Consequently, SN1 as well as all members of this peptide family may also act as antioxidants.
Endogenous SN1 expression was previously detected in tubers, stems, axillary buds and young floral buds by Northern blot assays (Segura et al., 1999). In the present study, endogenous SN1 mRNA was also detected in leaves, suggesting that relative levels of expression may vary depending on genotypes or in different stages of plant development. Therefore, it would be interesting to screen for differential expression levels among different Solanum genotypes and to explore its regulatory sequence in order to fine‐tune the spatial and temporal expression during plant development. Protein subcellular localization, interaction with other proteins from the host and/or the pathogens, and biochemical/structural studies could also contribute to the understanding of its mechanisms of action.
Plant biotechnology offers the possibility to improve field yield and food storage of economically important crops without altering cultivar identity. Recently, research interest on AMPs has dramatically increased because of their broad range activity, resulting in several biotechnological applications (Gao et al., 2000; Koo et al., 2002; Oard and Enright, 2006; Osusky et al., 2004; Ponti et al., 2003; Rajasekaran et al., 2005; Vidal et al., 2006). It has been demonstrated that agronomically useful levels of fungal control can be achieved through expression of a single transgene in agricultural crops (Gao et al., 2000). We consider that the levels of protection we reach with SN1‐overexpressing lines are promising for potato growers, as that they could reduce the amount of fungicides and pesticides employed, reducing cost and health risks. However, we do not discount the use of other antifungal proteins in combination in future strategies to optimize the fungal protection obtained.
In the present study, SN1 resistant transgenic plants did not exhibit visual phenotypic differences from wild‐type in greenhouse conditions, suggesting that constitutive expression of this peptide does not seem to alter plant physiology. Moreover, as SN1 natural AMP was overexpressed in a related species from which it was isolated, negative public perception may be reduced.
In conclusion, our data suggest that SN1 is an interesting candidate gene for engineering plants to confer broad‐spectrum protection against commercially relevant pathogens such as R. solani and E. carotovora. As an alternative to the use of transgenic plants, this work may provide the molecular basis for the screening, identification and selection of naturally overexpressing genotypes in germplasm banks for conventional breeding purposes.
EXPERIMENTAL PROCEDURES
Biological materials and growth conditions
Solanum tuberosum subsp. tuberosum cv. Kennebec, S. commersonii and two different S. chacoense genotypes were grown in a greenhouse at 18–25 °C, under a 10/14‐h dark/light cycle, in 12‐cm‐diameter pots. S. chacoense genotypes were chosen because a contrasting behaviour (resistance/susceptibility) towards infection to phytopathogens has been observed. For genomic DNA extraction, 4‐week‐old plant leaves were harvested. Additionally, a BAC library from S. bulbocastanum (# 9P2 BAC Library SB‐PBa, CUGI, Clemson University, USA) was used as DNA source of a wild Solanum species. R. solani AG‐3 was cultured on potato dextrose agar (PDA) medium [20% (w/v) potato, 1.5% (w/v) glucose and 1.5% (w/v) agar] at 28 °C. E. carotovora subsp. carotovora was cultured in nutritive agar [0.5% (w/v) peptone, 0.3% (w/v) meat extract, 0.25 (w/v) glucose and 1.8% (w/v) agar, products from Sigma Chemical Co., St Louis, MO, USA] at 28 °C or in nutritive liquid medium [0.5% (w/v) peptone, 0.3% (w/v) meat extract, 0.25 (w/v) glucose] at 28 °C with shaking at 150 r.p.m. Bacterial strains were stored as glycerol stocks at –80 °C.
Nucleic acid isolation, sequence data analysis and construction of a binary vector
All DNA extractions were carried out according to Dellaporta et al. (1983). The SN1 complete open reading frame was amplified by a PCR assay using the specific primers ‘upSN1: 5′TTCAGCTCGAGAAAAAATGAAGTTATTTCTATTAACT3′ and ‘lowSN1’: 5′AATACAGGATCCTCAAGGGCATTTAGACTTGCC3′ and cloned in the commercial vector p‐GemT Easy Vector (Promega, Madison, WI, USA). Based on the chain termination method (Sanger et al., 1977), DNA sequences were determined from at least five independent clones in both directions using commercial sequencing facilities (Macrogen Company, Seoul, Korea). Cloned sequences were analysed by the BioEdit Sequence Alignment Editor and multiple sequence analysis was performed using CLUSTALW version 1.8 (Thompson et al., 1994). For construction of a binary vector, the complete SN1 sequence of S. chacoense (EF206290) was inserted into the EcoRI restriction site of the pBPFΩ7 vector (Coego et al., 1996) between the Cauliflower mosaic virus 35S promoter (CaMV 35S) promoter and Tnos terminator. This cassette was then subcloned into the HindIII restriction site of the pPZPK binary vector [pPZP200 (Hajdukiewicz et al., 1994) harbouring a kanamycin resistance cassette] giving rise to the final construct pPZPK‐SN1. Standard molecular biology procedures were carried out as described by Sambrook et al. (1989)
Obtaining of transgenic plants
Leaf discs of S. tuberosum cv. Kennebec were co‐cultured (as described by del Vas, 1992) with A. tumefaciens LBA4404 pAL4404, carrying pPZPK‐SN1 construct, over a period of 48 h in MS medium (Murashige and Skoog, 1962). Explants were subcultured in regeneration medium (MS salts and vitamins, 20 g/L sucrose, 7 g/L agar, pH 5.6, plus 2 mg/mL zeatine riboside, 50 mg/mL kanamycin and 300 mg/mL cefotaxime) and transferred to fresh medium every 15 days until distinct shoots appeared. Finally, shoots were grown in micropropagation medium (MS salts, 20 g/L sucrose, 7 g/L agar, pH 5.6) supplemented with kanamycin and cefotaxime. Plants from different transgenic lines were maintained in vitro by periodic micropropagation in growing chambers (CMP 3244; Conviron, Manitoba, Canada) at 18–22 °C, under an 8/16‐h dark/light cycle.
DNA extraction, PCR and Southern blot analysis
Genomic DNA from kanamycin‐resistant plants was isolated using the Nucleon™ PhytoPure™ Genomic DNA Extraction Kit (Amersham, International Plc, Little Chalfont, Buckinghamshire, UK) and was analysed by PCR using primers specific for ‘up35S’: 5′ATCTCCACTGACGTAAGGGA3′, and ‘lowSN1’. For Southern blot analysis, 20 µg DNA was digested with HindIII or KpnI and subjected to agarose (0.8%) gel electrophoresis. DNA was transferred to Hybond N+ Nylon Membrane™ (Amersham), and the membrane was exposed to UV radiation and baked at 80 °C for 2 h under vacuum. Radioactive probe was prepared according to the manufacturer's Labelling Kit System (Promega, USA) employing α‐[32P]‐dCTP, random primers and CaMV 35S promoter plus Ω enhancer sequences as template. Hybridization procedures were carried out as described by Sambrook et al. (1989). Membranes were washed twice in 2× SSC–0.1% SDS for 10 min at 65 °C, once in 1× SSC–0.1% SDS for 10 min at room temperature and once in 0.1× SSC–0.1% SDS for 10 min at room temperature. Washed membranes were exposed for 20 days to X‐ray film for autoradiography.
RNA extraction, RT‐PCR and Northern blot analysis
Leaf tissue was immediately frozen in liquid nitrogen after harvest and stored at –80 °C. RNA extraction was carried out by using a RNeasy plant mini kit (QiaGen, GmbH, Hilden, Germany) and cDNAs were synthesized by using Superscript III (Invitrogen, Carlsbad, CA, USA) and random primers according to manufacturer's instructions. SN1 cDNAs were amplified by PCR with specific primers: ‘upSN1’ and ‘lowSN1’. Actin gene amplification was performed using primers specific ‘for upActin’: 5′TGGCATCATACCTTTTACAA3′ and ‘lowActin’: 5′TCCGGGCATCTGAACCTCT3′. In both cases, PCR conditions were: 94 °C for 1 min, 50 °C for 1 min, and 72 °C for 1 min, for 36 cycles. For Northern blot analysis, total RNA from leaf tissue was isolated using a Trizol commercial system extraction (Invitrogen) according to the manufacturer's instructions, repeating chloroform extractions twice. For RNA gel blot experiments, 10 µg of total RNA was electrophoretically resolved in 1.5% agarose gels using MOPS buffer (Sigma) and transferred to Hybond N+ Nylon Membrane™ (Amersham) for 16 h. RNA was crosslinked under a UV lamp and membranes were baked at 80 °C for 2 h. Probes were synthesized and labelled with α‐[32P]‐dCTP as described for the Labeling Kit System (Promega). Hybridizations were performed for 14 h at 42 °C in a commercial standard hybridization solution (ULTRAhybª, Ambion Inc., USA). Membranes were washed twice in 2× SSC–0.1% SDS for 10 min at 42 °C, once in 1× SSC–0.1% SDS for 20 min at room temperature and once in 0.1× SSC–0.1% SDS for 10 min at room temperature. Washed membranes were exposed for 10 days to X‐ray film for autoradiography. Gene‐specific probes to detect SN1 or actin mRNAs were used for hybridization.
Fungal resistance bioassays
For fungal resistance bioassays, the potato pathogen R. solani was first grown on PDA plates for 3 days at 28 °C. Subsequently, two agar plugs of 10 mm diameter were taken from fresh plates and added to a 500‐mL Schott glass bottle with 120 g autoclaved hydrated wheat seeds. After 15 days of incubation at 28 °C in the dark, the infected seeds were mixed with sterile soil [1/2 (w/w) soil, 1/4 (w/w) sand, 1/8 (w/w) cryogenic perlite, 1/8 (w/w) undertow] and incubated for another 2 days before plant transfer. Transgenic lines and non‐transgenic potato plants (control plants) were micropropagated and grown in vitro for 4–5 weeks before being transferred to soil in the greenhouse at 18–22 °C, under an 8/16‐h dark/light cycle and 50–70% humidity conditions. Eighteen plants of each transformed line and 36 non‐transgenic plants were transferred into the inoculated soil with a randomized block design to reduce noise or variance. The same numbers of plants were transferred into non‐inoculated soil with the same block design. The assay was performed twice under similar conditions and disease symptoms of transgenic and control plants were recorded at 2, 4, 6 and 8 days by two independent observers. As control plant survival curves were almost identical in the two assays, replicates were pooled to calculate survival rates of each line. Ten days after infection, all dead plants and half of the surviving plants were collected and damage was quantified while the other half of surviving plants were kept for 1 month in order to compare growth between inoculated and non‐inoculated soil. Chi‐squared tests of significance were performed using GraphPad Prism 3.0 software. The term significant is used in the text only when the differences have been confirmed at a significance level of 5%.
Bacterial resistance bioassays
For bacterial resistance bioassays, the potato pathogen E. carotovora subsp. carotovora was grown on potato tuber slices and striated on nutritive agar at 28 °C. One colony was used to inoculate 100 mL of nutritive liquid medium and incubated overnight at 28 °C with shaking at 150 r.p.m. After centrifugation, bacteria were suspended in water up to 1 optical density (at 600 nm) unit and this solution was used to inoculate the plants. Transgenic lines and non‐transgenic plants were micropropagated and grown in vitro for 4 weeks before transfer to soil in growing chambers (CMP 3244; Conviron) at 18–22 °C, under an 8/16‐h dark/light cycle and 70–80% humidity conditions. After 1 week, ten plants of each transgenic line, as well as non‐transgenic control plants, were inoculated with 10 µL of the bacterial solution in the plant stem at the second leaf from the bottom to top using a 25‐µL syringe (Hamilton Company, Nevada, USA) and ten plants of each line were mock inoculated. S1, S3 and S5 lines, as well as non‐transgenic plants were evaluated by scoring typical symptoms caused by this bacterial disease over 21 days. The assay was performed twice under the same conditions. Necrotic wound area was measured for each replica and average lesion sizes were calculated for control and transgenic lines. Lesion reductions of overexpressing transgenic lines were calculated considering the average lesion size of the non‐transgenic plants as 100%. The number of fallen leaves and the number of stems that suffered thickening as an illness consequence were also quantified 10 dpi. Differences among groups were tested using the Krustal–Wallis test (Zar, 1984). Multiple comparisons were performed using the non‐parametric analogue of the Student–Newman–Keuls multiple range test as described (Zar, 1984). The term significant is used in the text only when the differences have been confirmed at a significance level of 5%.
ACKNOWLEDGEMENTS
This research was partially supported by grants UBACyT X‐296 (University of Buenos Aires‐UBA), PE AEBIO4461 (INTA) and PAV137 (FONCyT‐ANPCyT). We thank Laura Gasoni (IMyZA‐INTA), Pablo Grijalba (Facultad de Agronomía, UBA), Alberto Escande (EEA INTA, Balcarce) and Mercedes Rivero (FCEyN‐UBA) for kindly providing pathogen strains and valuable suggestions on infection protocols. We also thank Teresa Cabrera (IB‐INTA), Valeria Peralta (IB‐INTA) and Andrea Dengis (FCEyN‐UBA) for their excellent technical assistance in the production and maintenance of transgenic potato plants and Verónica Lía (IB‐INTA) for statistical support. We acknowledge Mariana del Vas, Sebastián Asurmendi and Fernando Carrari for critically reading this manuscript and helpful discussions and Julia Sabio for her invaluable assistance with the English text. N.I.A. holds a CONICET/INTA fellowship and, previously, a student fellowship from the University of Buenos Aires. A.A.B. holds a CONICET fellowship. C.V.R. is a career member of CONICET. The nucleotide sequence of snakin‐1 from Solanum tuberosum subsp. tuberosum cv. Kennebec, S. bulbocastanum, S. commersonii and two genotypes of S. chacoense reported in this paper will appear in the DDBJ/EMBL/GenBank database under accession numbers EF206289, EF206293, EF206290, EF206291 and EF206292, respectively.
REFERENCES
- Adam, G.C. (1988) Thanatephorus cucumeris (Rhizoctonia solani): a species of wide host range. In: Advances in Plant Pathology. Vol. 6. Genetics of Plant Pathogenic Fungi (G.S. Sidhu, ed.), pp. 535–552. New York: Academic Press. [Google Scholar]
- Agrios, G.N. (1997) Plant Pathology, 4th edn. New York: Academic Press. [Google Scholar]
- Aubert, D. , Chevillard, M. , Dorne, A.M. , Arlaud, G. and Herzog, M. (1998) Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up‐regulated by gibberellins in meristematic regions. Plant Mol. Biol. 36, 871–883. [DOI] [PubMed] [Google Scholar]
- Ben‐Nissan, G. and Weiss, D. (1996) The petunia homologue of tomato gast1: transcript accumulation coincides with gibberellin‐induced corolla cell elongation. Plant Mol. Biol. 32, 1067–1074. [DOI] [PubMed] [Google Scholar]
- Ben‐Nissan, G. , Lee, J.Y. , Borohov, A. and Weiss, D. (2004) GIP, a Petunia hybrida GA‐induced cysteine‐rich protein: a possible role in shoot elongation and transition to flowering. Plant J. 37, 229–238. [DOI] [PubMed] [Google Scholar]
- Berrocal‐Lobo, M. , Segura, A. , Moreno, M. , Lopez, G. , Garcia‐Olmedo, F. and Molina, A. (2002) Snakin‐2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 128, 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler, L.V. , Whitelegge, J.P. , Millar, D.J. and Bolwell, G.P. (2006) A two component chitin‐binding protein from French bean‐association of a proline‐rich protein with a cysteine‐rich polypeptide. FEBS Lett. 580, 1541–1546. [DOI] [PubMed] [Google Scholar]
- Coego, A. , Vazquez, R. , Yamilet Coll, J.A. , Pujol, M. , Menendez, E. , Lopez, M.A. , Molina, P , Hernandez, L. , Bencomo, B. , De La Riva, G. and Selman‐Housein, G. (1996) Effect of promoter‐stimulatory element combination on transient reporter gene expression in tobacco protoplast using PEG‐treatment. Biotechnologia Aplicada, 13, 147. [Google Scholar]
- Dellaporta, S.L. , Wood, J. and Hicks, J.B. (1983) A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- De La Fuente, J.I. , Amaya, I. , Castillejo, C. , Sanchez‐Sevilla, J.F. , Quesada, M.A. , Botella, M.A. and Valpuesta, V . (2006) The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J. Exp. Bot. 57, 2401–2411. [DOI] [PubMed] [Google Scholar]
- Furukawa, T. , Sakaguchi, N. and Shimada, H. (2006) Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes. Genet. Syst. 81, 171–180. [DOI] [PubMed] [Google Scholar]
- Gao, A.G. , Hakimi, S.M. , Mittanck, C.A. , Wu, Y. , Woerner, B.M. , Stark, D.M. , et al (2000) Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18, 1307–1310. [DOI] [PubMed] [Google Scholar]
- Gutierrez, W.A. , Shew, H.D. and Melton, T.A. (1997) Sources of inoculum and management for Rhizoctonia solani damping‐off on tobacco transplants under greenhouse conditions. Plant Dis. 81, 604–606. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P. , Svab, Z. and Maliga, P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hide, G.A. and Horrocks, J.K. (1994) Influence of stem canker (Rhizoctonia solani Kuhn) on tuber yield, tuber size, reducing sugars and crisp colour in cv. Record. Potato Res. 37, 43–49. [Google Scholar]
- Hide, G.A. , Welham, S.J. , Read, P.J. and Ainsley, A.E. (1996) The yield of potato plants as affected by stem canker (Rhizoctonia solani), blackleg (Erwinia carotovora subsp. atroseptica) and by neighbouring plants. J. Agric. Sci. 126, 429–440. [Google Scholar]
- Koo, J.C. , Chun, H.J. , Park, H.C. , Kim, M.C. , Koo, Y.D. , Koo, S.C. , Ok, H.M. , Park, S.J. , Lee, S‐H , Yun, D‐J , Lim, C.O. , Bahk, J.D. , Lee, S.Y. and Cho, M.J. (2002) Over‐expression of a seed specific hevein‐like antimicrobial peptide from Pharbitis nil enhances resistance to a fungal pathogen in transgenic tobacco plants. Plant Mol. Biol. 50, 441–452. [DOI] [PubMed] [Google Scholar]
- Kotilainen, M. , Helariutta, Y. , Mehto, M. , Pollanen, E. , Albert, V.A. , Elomaa, P. and Teeri, T.H. (1999) GEG participates in the regulation of cell and organ shape during corolla and carpel development in gerbera hybrida. Plant Cell, 11, 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Solanilla, E. , Garcia‐Olmedo, F. and Rodriguez‐Palenzuela, P. (1998) Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogenesis. Plant Cell, 10, 917–924. [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Solanilla, E. , Gonzalez‐Zorn, B. , Novella, S. , Vazquez‐Boland, J.A. and Rodriguez‐Palenzuela, P. (2003) Susceptibility of Listeria monocytogenes to antimicrobial peptides. FEMS Microbiol. Lett. 226, 101–105. [DOI] [PubMed] [Google Scholar]
- Murashige, T.H. and Skoog, F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–497. [Google Scholar]
- Oard, S.V. and Enright, F.M. (2006) Expression of the antimicrobial peptides in plants to control phytopathogenic bacteria and fungi. Plant Cell Rep. 25, 561–572. [DOI] [PubMed] [Google Scholar]
- Ogoshi, A. (1987) Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kühn. Ann. Rev. Phytopathol. 25, 125–143. [Google Scholar]
- Osusky, M. , Osuska, L. , Hancock, R.E. , Kay, W.W. and Misra, S. (2004) Transgenic potatoes expressing a novel cationic peptide are resistant to late blight and pink rot. Transgenic Res. 13, 181–190. [DOI] [PubMed] [Google Scholar]
- Pérombelon, M.C.M. (2000) Blackleg risk potential of seed potatoes determined by quantification of tuber contamination by the causal agent and Erwinia carotovora subsp. atroseptica: a critical review. OEPP/EPPO Bull. 30, 413–420. [Google Scholar]
- Ponti, D. , Mangoni, M.L. , Mignogna, G. , Simmaco, M. and Barra, D. (2003) An amphibian antimicrobial peptide variant expressed in Nicotiana tabacum confers resistance to phytopathogens. Biochem. J. 370, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran, K. , Cary, J.W. , Jaynes, J.M. and Cleveland, T.E. (2005) Disease resistance conferred by the expression of a gene encoding a synthetic peptide in transgenic cotton (Gossypium hirsutum L.) plants. Plant Biotechnol. J. 3, 545–554. [DOI] [PubMed] [Google Scholar]
- Roxrud, I. , Lid, S.E. , Fletcher, J.C. , Schmidt, E.D. and Opsahl‐Sorteberg, H.G. (2007) GASA4, one of the 14‐member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 48, 471–483. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E. and Maniatis, T. (1989) Molecular Cloning, A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sanger, F. , Nicklen, S. and Coulson, A.R. (1977) DNA sequencing with chain‐terminating inhibitors. Proc. Natl Acad. Sci. USA 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura, A. , Moreno, M. , Madueno, F. , Molina, A. and Garcia‐Olmedo, F. (1999) Snakin‐1, a peptide from potato that is active against plant pathogens. Mol. Plant–Microbe Interact. 12, 16–23. [DOI] [PubMed] [Google Scholar]
- Selitrennikoff, C.P. (2001) Antifungal proteins. Appl. Environ. Microbiol. 67, 2883–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L. , Gast, R.T. , Gopalraj, M. and Olszewski, N.E. (1992) Characterization of a shoot‐specific, GA3 and ABA‐regulated gene from tomato. Plant J. 2, 153–159. [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg, R.W. , Carter, C. , Powell, A. , Mittler, R. , Rizhsky, L. and Horner, H.T. (2003) A major function of the tobacco floral nectary is defense against microbial attack Plant Syst. Evol. 238, 211–218. [Google Scholar]
- Trainotti, L. , Zanin, D. and Casadoro, G. (2003) A cell wall‐oriented genomic approach reveals a new and unexpected complexity of the softening in peaches. J. Exp. Bot. 54, 1821–1832. [DOI] [PubMed] [Google Scholar]
- Del Vas, M. (1992) Obtención y caracterización de plantas de interés agropecuario. Dissertation, University of Buenos Aires. [Google Scholar]
- Vidal, J.R. , Kikkert, J.R. , Malnoy, M.A. , Wallace, P.G. , Barnard, J. and Reisch, B.I. (2006) Evaluation of transgenic ‘Chardonnay’ (Vitis vinifera) containing magainin genes for resistance to crown gall and powdery mildew. Transgenic Res. 15, 69–82. [DOI] [PubMed] [Google Scholar]
- Wigoda, N. , Ben‐Nissan, G. , Granot, D. , Schwartz, A. and Weiss, D. (2006) The gibberellin‐induced, cysteine‐rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J. 48, 796–805. [DOI] [PubMed] [Google Scholar]
- Zar, J.H. (1984) Biostatistical Analysis. New Jersey: Prentice‐Hall. [Google Scholar]


