SUMMARY
The vascular pathogen Xanthomonas oryzae pv. oryzae (Xoo) and nonvascular pathogen Xanthomonas oryzae pv. oryzicola (Xoc) cause bacterial blight (BB) and bacterial leaf streak (BLS) diseases of rice, respectively. We have previously identified the avirulence gene avrXa27 from Xoo PXO99A, which specifically induces the expression of the rice resistance gene Xa27, ultimately leading to resistance against BB disease in rice. In this study, we have generated a transgenic rice line (L24) that expresses avrXa27 constitutively under the control of the PR1 promoter, and have examined its role in the host–pathogen interaction. L24 is not more susceptible to BB, indicating that avrXa27 does not contribute to virulence. AvrXa27 retains avirulence activity in L24 and, after crossing with a line containing Xa27, progeny display phenotypic changes including inhibition of tillering, delay in flowering, stiff leaves, early leaf senescence and activation of pathogenesis‐related (PR) genes. On challenge with a variety of compatible strains of Xoo and Xoc strain L8, lines with both avrXa27 and Xa27 also show enhanced resistance to bacterial infection. The induction of Xa27 and subsequent inhibition of Xoc growth in Xa27 plants are observed on inoculation with Xoc L8 harbouring avrXa27. Our results indicate that the heterologous expression of avrXa27 in rice containing Xa27 triggers R gene‐specific resistance and, at the same time, confers enhanced resistance to compatible strains of Xoo and Xoc. The expression of AvrXa27 and related proteins in plants has the potential to generate broad resistance in plants.
INTRODUCTION
Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) cause bacterial blight (BB) and bacterial leaf streak (BLS) diseases of rice, respectively (Nino‐Liu et al., 2006). Despite their close relationship, Xoo and Xoc cause two distinct diseases. Xoo is a vascular pathogen that enters rice leaf typically through hydathodes at the leaf margin, multiplies in the intercellular spaces of the underlying epitheme, and moves to the xylem vessels to cause systemic infection (Noda and Kaku, 1999; Ou, 1985). Xoc, by contrast, is a nonvascular pathogen that penetrates the leaf mainly through the stomata, multiplies in the substomatal cavity, and colonizes the intercellular spaces of the parenchyma (Ou, 1985). Host genetic resistance is the most effective way to control BB in rice, and nearly 30 R genes or loci with race‐specific resistance to Xoo have been identified (Nino‐Liu et al., 2006). Genetic resistance for BLS in rice is limited to quantitative resistance (Gnanamanickam et al., 1999; Sheng et al., 2005; Tang et al., 2000). More recently, solutions for BLS resistance involving genetic engineering have been proposed (Zhao et al., 2005).
Many plant pathogenic bacteria, including Xoo and Xoc, rely on the type III secretion system to inject virulence effectors into the host cell to cause infection (He et al., 2004; Zhu et al., 2000). Although most of the type III effectors have a virulence function during pathogenesis, some may betray the pathogen to plant defences during host–pathogen interaction. In the latter case, the type III effectors are called avirulence (Avr) proteins (Kjemtrup et al., 2000). Avr proteins elicit a hypersensitive response in the resistant hosts that express a corresponding R gene. In the absence of avr or R genes, or both, no recognition takes place and disease occurs (Flor, 1971). Recently, we have isolated a pair of avr and R genes, avrXa27 and Xa27, from Xoo PXO99A and rice line IRBB27, respectively (Gu et al., 2005). The avrXa27 gene encodes a type III effector that is similar to members of the AvrBs3/PthA family, whereas the Xa27 gene encodes a novel R protein. Members of the AvrBs3/PthA family of type III effectors are found in many strains of Xanthomonas (Bonas et al., 1989; De Feyter and Gabriel, 1991; Hopkins et al., 1992; Swarup et al., 1991) and at least one strain of Ralstonia solanacearum (Salanoubat et al., 2002). AvrBs3/PthA family members appear to function as gene‐specific transcription factors in the host (Kay et al., 2007). The member proteins are remarkably similar, except for a difference in the number and apparent nature of near‐identical, 34‐amino‐acid, direct repeats in the central portion of the protein known as the repetitive region. These direct repeat sequences show a major variation at positions 12 and 13. The order and type of central repeats are involved in the governing of resistance specificity of a particular gene family (Bonas et al., 1989; Gu et al., 2005; Herbers et al., 1992; Yang et al., 1994). Members of the AvrBs3/PthA family also contain conserved C‐terminal nuclear localization signal motifs and a functional eukaryotic transcriptional activation domain at the extreme C‐terminus (Gu et al., 2005; Szurek et al., 2001; Van den Ackerveken et al., 1996; Yang and Gabriel, 1995; Zhu et al., 1998). Except for AvrBs4 (Schornack et al., 2004), most of the members of the AvrBs3/PthA family require the nuclear localization signal motifs and activation domain for their virulence and/or avirulence functions (Gu et al., 2005; Szurek et al., 2001; Van den Ackerveken et al., 1996; Zhu et al., 1998).
Xa27‐dependent resistance is always associated with the specific induction of Xa27 by incompatible pathogens harbouring avrXa27. We have shown previously that ectopic expression of Xa27 under the control of the rice PR1 promoter provides enhanced resistance to otherwise compatible Xoo strains. In this study, we carried out functional studies of avrXa27 in transgenic rice to investigate whether its product has a similar avirulence activity to that shown in Xoo, and whether the induction of Xa27 by AvrXa27 in transgenic rice can confer enhanced resistance to compatible strains of Xoo and the nonvascular pathogen Xoc.
RESULTS
Generation of transgenic plants expressing avrXa27
Considering that the constitutive expression of avrXa27 in rice might cause an unfavourable effect on rice cells, we chose the inducible promoter from the rice pathogenesis‐related (PR) gene PR1 to drive avrXa27 expression in transgenic rice (Fig. 1). Thirty‐four independent transgenic T0 plants were obtained by Agrobacterium‐mediated transformation. However, only four lines (L1, L14, L24 and L25) contained an intact avrXa27 coding region on the basis of Southern blot hybridization analysis (Fig. 2). The remaining transgenic plants contained only the truncated avrXa27 coding region and were excluded from the study (data not shown). Using the 3234‐bp BamHI fragment of avrXa27 as a probe, the 4.5‐kb NdeI–XbaI band corresponding to the PR1 promoter and avrXa27 coding region was detected in L24 by Southern blot analysis (Fig. 2, lane 8). However, it was not detected in L1, L14 and L25. The NdeI–XbaI bands detected in L1, L14 and L25 were larger than 4.5 kb (Fig. 2, lanes 4, 6 and 10). At the same time, the 3.2‐kb BamHI band covering most of the avrXa27 coding region was detected in all four lines (Fig. 2, lanes 5, 7, 9 and 11). We further verified the presence of the PR1 promoter, 5′ and 3′ coding regions of avrXa27 and the terminator of the nopaline synthase gene (Tnos) in all four lines by polymerase chain reaction (PCR) amplification, followed by DNA sequencing (data not shown). The results from PCR and Southern blot analysis indicated that L24 carried the intact PPR1‐avrXa27‐Tnos expression cassette, whereas L1, L14 and L25 contained an expression cassette with a modification probably caused by DNA mutations and rearrangements in the PR1 promoter region. Southern blot analysis with the hygromycin phosphotransferase (hpt) gene as a probe also showed that each of the four lines carried a single T‐DNA insertion (data not shown). The morphological phenotypes of the four transgenic avrXa27 lines were similar to that of nontransgenic Nipponbare or control transgenic plants that were generated by transformation of Nipponbare with empty vector pC1305.1. The four transgenic avrXa27 lines were selfed, and homozygous progeny were chosen for further studies.
Figure 1.
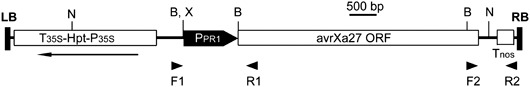
Schematic map of the T‐DNA region of pCPR1avrXa27. The arrow indicates the direction of transcription of the hygromycin phosphotransferase gene. Polymerase chain reaction (PCR) primers F1, R1, F2 and R2 are shown by the arrowheads. avrXa27 ORF, coding region of avrXa27; B, BamHI; Hpt, hygromycin phosphotransferase gene; LB, left border; N, NdeI; P35S, promoter of cauliflower mosaic virus (CaMV) 35S gene; PPR1, promoter of rice PR1 gene; RB, right border; T35S, terminator of CaMV 35S gene; Tnos, terminator of nopaline synthase (nos) gene; X, XbaI.
Figure 2.
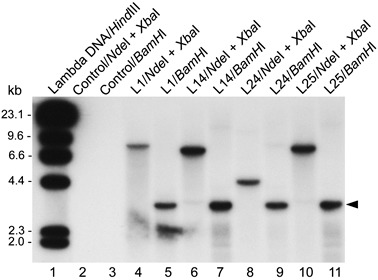
Southern blot analysis of avrXa27 in transgenic plants. The control transgenic plant (Control) was derived from the transformation of Nipponbare with pC1305.1. The Southern blot filter was probed with a 32P‐labelled 3234‐bp BamHI fragment from avrXa27. The arrowhead indicates the position of the BamHI fragment of avrXa27.
Expression analysis of avrXa27 in transgenic rice
Transcripts corresponding to PPR1‐avrXa27‐Tnos were detected in all four lines using the 3234‐bp BamHI fragment of avrXa27 as a probe (Fig. 3). However, expression of the gene in L1, L14, L24 and L25 did not show significant variations in induction on inoculation with Xoo PXO99A (Fig. 3). The wild‐type mRNA molecules of PPR1‐avrXa27‐Tnos produced in L24 were approximately 4 kb in Northern blot analysis (Fig. 3). In contrast with the sharp band detected in L24, less intense and diffuse bands were observed in L1, L14 and L25. The reasons for the differences in expression are unknown, and we continue only with the analysis of L24.
Figure 3.
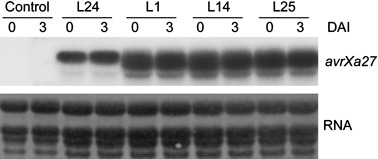
Expression of avrXa27 in transgenic plants. Total RNA was isolated from noninoculated plants (0) and plants at 3 days after inoculation (DAI) (3) with Xanthomonas oryzae pv. oryzae (Xoo) PXO99A. Approximately 30 µg of total RNA was loaded on to each lane for Northern blot analysis. The control transgenic plant (Control) was derived from the transformation of Nipponbare with empty vector pC1305.1. L24, L1, L14 and L25 are independent transgenic avrXa27 lines. The Northern blot filter was probed with a 32P‐labelled 3234‐bp BamHI fragment from avrXa27.
Activity of AvrXa27 in transgenic rice
AvrXa27 in Xoo specifically induces the expression of the Xa27 gene in rice (Gu et al., 2005). To determine whether AvrXa27 in transgenic rice has a similar activity, the expression of Xa27 in F1 plants derived from crosses between IRBB27 (female) and L24 (IRBB27 × L24) or IRBB27 (female) and control transgenic plants (IRBB27 × Control) was examined (Fig. 4). Northern blot analysis indicated that Xa27 was induced in F1 plants from the IRBB27 × L24 cross, but not in those from the IRBB27 × Control cross (Fig. 4). IR24 carries susceptible alleles of the Xa27 gene (xa27) that are not induced by Xoo strains harbouring avrXa27 (Gu et al., 2005), and Northern blot analysis indicated that xa27 was not induced in the F1 plants of IR24 × L24 (Fig. 4). These results indicate that PPR1‐avrXa27‐Tnos in transgenic rice functions in the specific induction of Xa27 expression.
Figure 4.
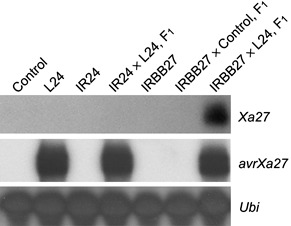
Induction of Xa27 but not xa27 in F1 plants by AvrXa27 in transgenic rice. About 5 µg of mRNA was loaded on to each lane for Northern blot analysis. Control, control transgenic plant derived from the transformation of Nipponbare with empty vector pC1305.1; L24, line 24 of transgenic rice carrying avrXa27; IR24, susceptible rice variety with the xa27/xa27 genotype; ‘IR24 × L24, F1’, F1 plants of IR24 × L24; IRBB27, near‐isogenic line of Xa27 in IR24 background; ‘IRBB27 × Control, F1’, F1 plants of IRBB27 × Control; ‘IRBB27 × L24, F1’, F1 plants of IRBB27 × L24.
The expression of avrXa27 in rice also afforded the opportunity to determine whether AvrXa27 has any effect on the susceptibility of the host to bacterial infection in the absence and presence of the cognate R gene Xa27. Previously, no virulence activity has been attributed to AvrXa27 in Xoo (Gu et al., 2005). However, the expression of avrXa27 in the plant may provide a more sensitive assay for virulence effects on host physiology. The inoculation of the susceptible rice lines Nipponbare and IR24 (xa27/xa27) with Xoo strains PXO99AME1(pHM1) and PXO99AME1(pHM1avrXa27) indicated that, as observed previously, lesions on Nipponbare or IR24 caused by PXO99AME1(pHM1avrXa27) were comparable with or even shorter than those caused by PXO99AME1(pHM1), and that AvrXa27 contributes no (or undetectable) virulence activity in Xoo (Fig. 5). L24 was inoculated with four strains including PXO99A, AXO1947, K202 and ZHE173. The disease lesions on L24 were either comparable with or slightly shorter than those on the control transgenic or Nipponbare plants (Table 1). The results failed to detect any effect on host susceptibility of expression of PPR1‐avrXa27‐Tnos in the plant.
Figure 5.
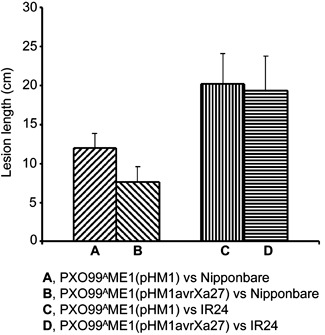
Virulence activity of AvrXa27 in Xanthomonas oryzae pv. oryzae (Xoo) on susceptible rice. The data represent the mean and standard deviation of the lesion length from three independent experiments.
Table 1.
Disease evaluation of Xa27 rice plants carrying avrXa27 for resistance to different Xanthomonas oryzae pv. oryzae (Xoo) strains.
| Line or plant | Lesion length (cm) and disease score* | |||
|---|---|---|---|---|
| PXO99A | AXO1947 | K202 | ZHE173 | |
| Nipponbare | 20.5 ± 4.1 (S) | 18.9 ± 4.2 (S) | 16.5 ± 4.9 (S) | 15.6 ± 4.0 (S) |
| Control† | 19.6 ± 4.2 (S) | 19.5 ± 2.8 (S) | 16.4 ± 2.3 (S) | 18.8 ± 3.6 (S) |
| L24 | 20.2 ± 2.4 (S) | 16.9 ± 2.5 (S) | 16.5 ± 3.1 (S) | 14.0 ± 3.2 (S) |
| IR24 | 29.0 ± 3.6 (S) | 27.5 ± 3.1 (S) | 19.9 ± 3.5 (S) | 24.5 ± 4.3 (S) |
| IRBB27 | 0.3 ± 0.2 (R) | 28.6 ± 5.0 (S) | 23.3 ± 2.3 (S) | 27.3 ± 3.5 (S) |
| IR24 × L24, F1 | 35.9 ± 6.6 (S) | 38.1 ± 4.8 (S) | 21.4 ± 2.9 (S) | 25.4 ± 4.7 (S) |
| IRBB27 × Control, F1 | 5.3 ± 6.3 ( MR) | 20.7 ± 3.2 (S) | 20.4 ± 4.7 (S) | 18.9 ± 9.6 (S) |
| IRBB27 × L24, F1 | 0.4 ± 0.4 (R) | 0.3 ± 0.2 (R) | 0.1 ± 0.1 (R) | 0.2 ± 0.2 (R) |
The average lesion length and standard deviation were calculated on the basis of two independent experiments. For each strain, at least 60 leaves from eight individual plants were inoculated. For the resistance score, see ‘Experimental procedures’. MR, moderately resistant; R, resistant; S, susceptible.
Control transgenic plant produced by transformation of Nipponbare with empty vector pC1305.1.
PPR1‐avrXa27‐Tnos‐dependent expression of Xa27 is associated with pleiotropic phenotypes
Although F1 and backcross progeny of IRBB27 × L24 (BC1F1–BC4F1) did not show local lesions, other phenotypes were observed in comparison with BC1F1 plants of IR24 × L24. The BC1F1 plants of IRBB27 × L24 produced fewer tillers, which was more obvious when BC1F1 plants were homozygous at the Xa27 locus. The BC1F1 plants of IR24 × L24 (xa27/xa27, avrXa27/–), IRBB27 × L24 (Xa27/xa27, avrXa27/–) and IRBB27 × L24 (Xa27/Xa27, avrXa27/–) developed 10–14, 7–9 and 4–6 tillers, respectively (Fig. 6A,B). F1 plants (Xa27/xa27, avrXa27/–) of IRBB27 × L24 showed a delay in flowering when compared with F1 plants (xa27/xa27, avrXa27/–) of IR24 × L24. The lifespan of the F1 plants of IRBB27 × L24 was 2 weeks longer than that of the F1 plants of IR24 × L24. The delay in flowering and increase in lifespan became more obvious in the subsequent backcross progeny of IRBB27 × L24. In the BC4F1 generation, the lifespan of BC4F1 plants (Xa27/xa27, avrXa27/–) of IRBB27 × L24 was about 4 weeks longer than that of BC4F1 plants of IR24 × L24 (Fig. 6C). The backcross progeny of IRBB27 × L24 also developed stiff leaves. Cell wall thickening occurred at the vascular elements of the stiff leaves, but was not detected in the leaves of the backcross progeny of IR24 × L24 (Fig. 6D,E). The stiff leaves of the backcross progeny of IRBB27 × L24 also showed early leaf senescence (Fig. 6F). Interestingly, the phenotypes of stiff leaves and early leaf senescence were not as prominent in the F1 plants of IRBB27 × L24 as in the backcrossed progeny. To determine whether the phenotypes could be associated with defence‐related gene expression, the expression of PR genes PR1 and PBZ1 was measured. Northern blot analysis revealed that both genes were expressed in the backcross progeny of IRBB27 × L24 (Fig. 7).
Figure 6.

Constitutive induction of Xa27 by AvrXa27 in transgenic rice leads to pleiotropic phenotypes. (A, B) Morphological phenotype of BC1F1 plants of IR24 × L24 (P1) and IRBB27 × L24 (P2–P4) at 56 days after sowing. The plants were inoculated with Xanthomonas oryzae pv. oryzae (Xoo) strain AXO1947 and the photographs were taken at 14 days after inoculation (DAI). Genotypes of the plants: P1, xa27/xa27, avrXa27/–; P2 and P3, Xa27/xa27, avrXa27/–; P4, Xa27/Xa27, avrXa27/–. (C) Morphological phenotype of BC4F1 plants of IR24 × L24 and IRBB27 × L24 at 95 days after sowing. (D) Cross‐section of a leaf vascular bundle from a 77‐day‐old BC4F1 plant of IR24 × L24. Bar, 20 µm. (E) Cross‐section of a leaf vascular bundle from a 77‐day‐old BC4F1 plant of IRBB27 × L24. Bar, 20 µm. (F) Leaves from 77‐day‐old BC4F1 plants of IR24 × L24 (L1, L3 and L5) and IRBB27 × L24 (L2, L4 and L6). Leaves L1, L3 and L5 are the first (flag leaf), second and third leaves from a typical BC4F1 plant of IR24 × L24, and leaves L2, L4 and L6 are the corresponding leaves from a typical BC4F1 plant of IRBB27 × L24. Abbreviations in (D) and (E): m, mesophyll cells; p, phloem; px, protoxylem lacuna; w, cell wall; xv, xylem vessel.
Figure 7.
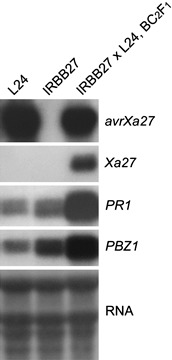
Expression of avrXa27, Xa27, PR1 and PBZ1 in BC2F1 plants of IRBB27 × L24 and their parental lines. Approximately 30 µg of total RNA was loaded on to each lane for Northern blot analysis.
PPR1‐avrXa27‐Tnos‐dependent expression of Xa27 confers enhanced resistance to compatible Xoo strains
To determine whether the constitutive induction of Xa27 by AvrXa27 in transgenic rice confers enhanced resistance to additional compatible strains of Xoo, F1 plants of IRBB27 × L24 were inoculated with three strains AXO1947, K202 and ZHE173. The F1 plants of IRBB27 × L24 showed enhanced resistance to these Xoo strains, whereas the parental line IRBB27 and the control F1 plants of IRBB27 × Control were susceptible to compatible strains (Table 1). F1 plants of IR24 × L24 were susceptible to all Xoo strains tested (Table 1). Compared with the complete resistance of IRBB27 to PXO99A, the control F1 plants of IRBB27 × Control showed moderate resistance to PXO99A (Table 1). The partial resistance displayed by these control F1 plants to PXO99A may have resulted from a dosage effect and/or influence of the genetic background from Nipponbare.
PPR1‐avrXa27‐Tnos‐dependent expression of Xa27 confers enhanced resistance to rice nonvascular pathogen Xoc
To determine whether the constitutive expression of Xa27 induced by avrXa27 in rice provides enhanced resistance to the nonvascular pathogen Xoc, BC3F1 plants of IRBB27 × L24 were inoculated with Xoc strain L8. Control BC3F1 plants of IRBB27 × Control and those of IR24 × L24 were susceptible to Xoc L8 (Fig. 8A, leaves 1 and 2). Large BLS lesions developed towards the leaf bases and leaf tips of BC3F1 plants at 10 days after inoculation (DAI), and numerous yellow beads of Xoc exudate were observed at the lesion area. BC3F1 plants of IRBB27 × L24, by contrast, were resistant to Xoc L8 (Fig. 8A, leaf 3). The development of BLS lesions on these BC3F1 plants was slower than that on control BC3F1 plants. However, unlike Xa27‐mediated resistance to Xoo, the resistance of BC3F1 plants of IRBB27 × L24 to Xoc L8 was incomplete. Yellow beads of Xoc were observed at the margins of the lesions on BC3F1 plants of IRBB27 × L24 at 10 DAI (Fig. 8A, leaf 3), and, although 63‐ to 7825‐fold lower than the Xoc populations in control BC3F1 plants, Xoc populations in the inoculated leaves of BC3F1 plants of IRBB27 × L24 continued to grow at 10 DAI (Fig. 8B).
Figure 8.
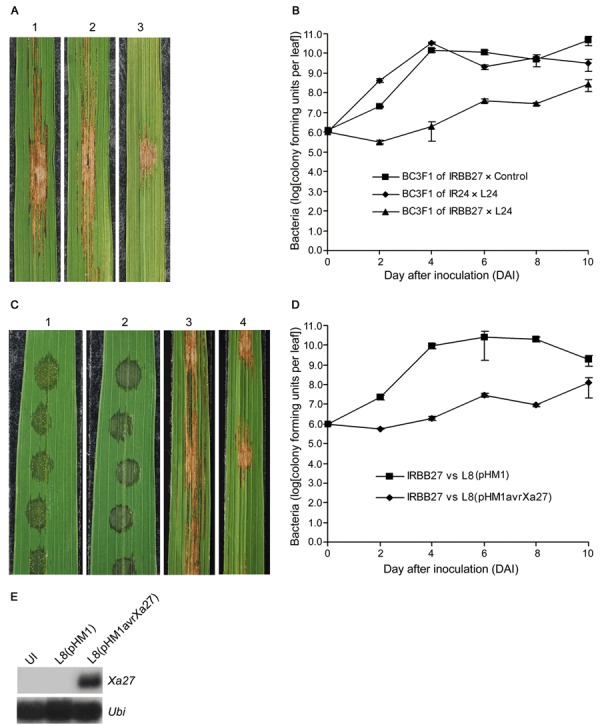
Expression of the Xa27 gene confers enhanced resistance to Xanthomonas oryzae pv. oryzicola (Xoc) L8. (A) Phenotype of bacterial leaf streak on leaves of BC3F1 plants of IRBB27 × Control (leaf 1), IR24 × L24 (leaf 2) and IRBB27 × L24 (leaf 3) at 10 days after inoculation (DAI) with Xoc L8. Control, control transgenic plant derived from the transformation of Nipponbare with empty vector pC1305.1. (B) Bacterial population of Xoc L8 in leaves of BC3F1 plants of IRBB27 × Control, IR24 × L24 and IRBB27 × L24 over 10 days by syringe infiltration. (C) Phenotype of bacterial leaf streak on leaves of IRBB27 plants at 3 DAI (leaves 1 and 2) and 10 DAI (leaves 3 and 4) with Xoc strains L8(pHM1) (leaves 1 and 3) and L8(pHM1avrXa27) (leaves 2 and 4). (D) Bacterial population of Xoc strains L8(pHM1) and L8(pHM1avrXa27) in leaves of IRBB27 plants over 10 days by syringe infiltration. (E) Induction of Xa27 in leaves of IRBB27 plants by inoculation with Xoc strains. Lane 1, noninoculated IRBB27 plants; lane 2, IRBB27 plants infiltrated with Xoc L8(pHM1) at 3 DAI; lane 3, IRBB27 plants infiltrated with Xoc L8(pHM1avrXa27) at 3 DAI.
To further verify that the enhanced resistance of BC3F1 plants of IRBB27 × L24 to Xoc resulted from Xa27 expression, IRBB27 plants were inoculated with Xoc strains L8(pHM1) and L8(pHM1avrXa27). Three days after syringe infiltration of the bacterium, leaf tissues at the infection sites in both interactions were water‐soaked in appearance (Fig. 8C, leaves 1 and 2). Although few or no beads of Xoc were found at the infection sites on IRBB27 plants inoculated with Xoc L8(pHM1avrXa27) (Fig. 8C, leaf 2), many tiny yellow beads of Xoc were observed on IRBB27 plants inoculated with Xoc L8(pHM1) (Fig. 8C, leaf 1). The development of BLS lesions on IRBB27 plants inoculated with L8(pHM1avrXa27) was slower than that on IRBB27 plants inoculated with L8(pHM1) (Fig. 8C, leaves 3 and 4). Xoc populations in IRBB27 plants inoculated with L8(pHM1avrXa27) were 16‐ to 4583‐fold lower than those in IRBB27 plants inoculated with L8(pHM1) (Fig. 8D). Northern blot analysis indicated that Xa27 in IRBB27 was induced by AvrXa27 in L8(pHM1avrXa27), whereas no induction was detected when L8(pHM1) was used for inoculation (Fig. 8E).
DISCUSSION
Type III effectors can have a bifunctional role in virulence and host recognition (Alfano and Collmer, 2004). The virulence function of several type III effectors, when expressed in plants, results in cell death or necrosis and suppresses host defences in plants lacking the cognate R genes (Alfano and Collmer 2004; Hauck et al., 2003). The gene product of avrXa27 in PXO99A has been shown to elicit resistance in plants containing Xa27 (Gu et al., 2005). PXO99AME1, an avrXa27 knockout mutant in the PXO99A background, is compatible with both IRBB27 and IR24 (Gu et al., 2005). We noted that the disease lesions on IR24 caused by PXO99AME1 were slightly shorter than those caused by PXO99A, suggesting that AvrXa27 may contribute to virulence activity (Z. Yin, unpublished data). However, recent studies have shown that an avrXa27 paralogue called pthXo6 is mutated together with avrXa27 in PXO99AME1, and that pthXo6 is responsible for the induction of the host susceptibility gene OsTFX1, encoding a bZIP transcription factor (Sugio et al., 2007). In this study, we did not detect any virulence activity of AvrXa27 in either Xoo strain PXO99AME1(pHM1avrXa27) or rice. Furthermore, the BB lesion length on Nipponbare plants inoculated with PXO99AME1(pHM1avrXa27) was even shorter than that on plants challenged by PXO99AME1(pHM1). The shorter BB lesions indicate the presence of the avirulence activity of AvrXa27. However, the corresponding R gene that confers partial resistance to PXO99AME1(pHM1avrXa27) is yet to be identified. PXO99A harbours 19 members of the AvrBs3/PthA family, of which pthXo1 and pthXo6 are known to contribute major virulence activity in rice (Salzberg et al., 2008; Sugio et al., 2007; Yang and White, 2004; Yang et al., 2006). Evidence indicates that others may provide additional virulence activity (Bai et al., 2000). The results obtained in this study indicate that AvrXa27 contributes little or no virulence activity.
PPR1‐avrXa27‐Tnos‐dependent expression of Xa27 conferred enhanced resistance to compatible Xoo strains. Similar results were obtained in a previous study, in which ectopic expression of Xa27 led to a broad‐spectrum resistance to both compatible and incompatible strains (Gu et al., 2005), indicating that the Xa27 gene product provides nonspecific resistance to Xoo. The Xa27 protein is different from other characterized R proteins that function specifically in perception and/or signalling of a particular pathogen infection. It is possible that the Xa27 gene evolved from a nonspecific resistance or defence gene, whose product confers nonspecific resistance to Xoo. This gene may have acquired a mimic promoter that perceives the presence of AvrXa27 and, consequently, confers specific resistance to incompatible Xoo strains that harbour avrXa27.
The constitutive expression of Xa27 induced by AvrXa27 in BC1F1 rice plants or subsequent backcross progeny of IRBB27 × L24 resulted in the constitutive activation of the defence response. Although a typical hypersensitive response was not observed in these plants, pleiotropic phenotypes were developed throughout the developmental stages, and may be a side‐effect of the constitutive expression of Xa27. The pleiotropic phenotypes were not as prominent in the F1 plants of IRBB27 × L24 as in the backcrossed progeny, and may have been masked by the strong vegetative plant growth as a result of heterosis of the F1 plants derived from the inter‐subspecies cross between indica rice IRBB27 and japonica rice L24. The pleiotropic phenotypes, such as cell wall thickening, especially in the vascular elements, were also found in Xa27 ectopic lines that constitutively expressed the Xa27 gene (Gu et al., 2005). Wild‐type IRBB27 plants have no pleiotropic phenotype as the induction of Xa27 by AvrXa27 in incompatible Xoo strains occurs only in the immediate vicinity of infected tissues. The pleiotropic phenotypes hinder application of the R gene for enhanced resistance to compatible Xoo strains lacking avrXa27. Enhanced resistance may be achievable by either controlling the expression of Xa27, as reported previously (Gu et al., 2005), or by regulating the in planta expression of avrXa27 in rice, as shown here. In either case, an inducible promoter that can be activated by either incompatible or compatible Xoo strains is required. The promoter should have strong but not leaky expression, because constitutive expression of Xa27 causes unfavourable pleiotropic phenotypes.
The nonvascular bacterial pathogen Xoc is closely related to Xoo (Nino‐Liu et al., 2006). Like Xoo, Xoc produces multiple members of the AvrBs3/PthA family, but no function has yet been ascribed to these members (Yang and White, 2004). Xoc can deliver type III effector AvrXa10 expressed from a plasmid into rice cells as efficiently as can Xoo (Makino et al., 2006). Given that Xoo and Xoc have many similarities, we investigated whether the constitutive induction of Xa27 by AvrXa27 in transgenic rice was able to provide enhanced resistance to Xoc. We found that the constitutive expression of Xa27 specifically induced by AvrXa27 in transgenic rice conferred enhanced resistance to Xoc L8. This enhanced resistance to Xoc was further verified through the expression of Xa27 specifically induced by AvrXa27 in Xoc. Although a single strain was tested, the results indicate that the strategy could provide broad resistance against xanthomonads. We also tested the disease resistance of the backcross progeny of IRBB27 × L24 and Xa27 over‐expressing lines to other rice pathogens, such as Magnaporthe grisea isolate PO6‐6, the causal agent of rice fungal blast, and Erwinia chrysanthemi pv. zeae EC1, a vascular bacterial pathogen causing rice bacterial foot rot disease. However, we did not find any Xa27‐mediated enhanced resistance to these fungal or bacterial pathogens (data not shown). Nevertheless, the finding of Xa27‐mediated enhanced resistance to Xoc augments host resistance by controlling BLS in rice, which presently lacks an R gene for resistance. In another successful example of host resistance, transgenic rice carrying Rxo1 from maize showed resistance to Xoc (Zhao et al., 2005).
EXPERIMENTAL PROCEDURES
Construction of binary vector carrying avrXa27
The binary construct pCPR1avrXa27, which contains avrXa27 under the control of the PR1 promoter, was prepared using the CAMBIA vector pC1305.1, available shot‐gun subclones of avrXa27, cosmid 99‐avrXa27‐20 harbouring the wild‐type of avrXa27 (Gu et al., 2005), and pZWavrXa27 containing a modified avrXa27 gene with a FLAG epitope tag in its C‐terminal region (Gu et al., 2005; Zhu et al., 1998). Briefly, a 257‐bp BamHI‐PstI fragment of the 3′ region of avrXa27, including a 69‐bp 3′ noncoding region, was isolated from shot‐gun clone V5G05 and cloned into vector pGBKT7 to generate pGavrXa27Ter. The 3234‐bp BamHI fragment of avrXa27 was isolated from cosmid 99‐avrXa27–20 and cloned into the BamHI site of pGavrXa27Ter to generate pGavrXa27B1Ter. A 379‐bp fragment containing the 5′ coding region of avrXa27 was amplified from shot‐gun clone V3B01 with primers A‐F (5′‐GGGGTACCATGGATCCCATTCGTTCGCGC‐3′) and A‐R (5′‐ACGCGTCGACAGTGACAGCGACACGCACGGTG‐3′). The PCR product was cloned into pGEM®‐T Easy vector (Promega) to generate pTA27. A 3410‐bp PstI fragment, including the 3327‐bp coding region of avrXa27, was isolated from pGavrXa27B1Ter and cloned into the PstI site of pTA27 to generate pTavrXa27. The 3234‐bp BamHI fragment of avrXa27 in pTavrXa27 was replaced with the BamHI fragment of avrXa27 from pZWavrXa27 (Gu et al., 2005) to create pTZWavrXa27. The SacII‐AseI fragment from pTZWavrXa27 was isolated, blunted and cloned downstream of the rice PR1 promoter in pC1305.1 to generate pCPR1avrXa27. pCPR1avrXa27 was introduced to Agrobacterium tumefaciens AGL1 by electroporation (Sambrook et al., 1989).
Xoo and Xoc strains
The Xoo and Xoc strains used in this study are listed in Table 2. Xoc strain L8 has been verified previously to be an Xoc strain by serological test (Zeng et al., 1995). The cosmid vector pHM1 or its derivative pHM1avrXa27 was introduced into Xoo or Xoc strains by electroporation (Sambrook et al., 1989).
Table 2.
Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) strains used in this study.
| Strain | Description* | Reference |
|---|---|---|
| PXO99A | Xoo, wild‐type, Xa27 incompatible | Gu et al. (2004, 2005) |
| AXO1947 | Xoo, wild‐type, Xa27 compatible | Gu et al. (2005) |
| K202 | Xoo, wild‐type, Xa27 compatible | Gu et al. (2004, 2005) |
| ZHE173 | Xoo, wild‐type, Xa27 compatible | Gu et al. (2004, 2005) |
| PXO99AME1 | Xoo, avrXa27 knockout mutant | Gu et al. (2005) |
| PXO99AME1(pHM1) | Xoo, PXO99AME1 harbouring pHM1 | This study |
| PXO99AME1(pHM1avrXa27) | Xoo, PXO99AME1 harbouring pHM1avrXa27 | Gu et al. (2005) |
| L8 | Xoc, wild‐type | Zeng et al. (1995) |
| L8(pHM1) | Xoc, L8 harbouring pHM1 | This study |
| L8(pHM1avrXa27) | Xoc, L8 harbouring pHM1avrXa27 | This study |
pHM1, cosmid vector pHM1 (Hopkins et al., 1992); pHM1avrXa27, pHM1 carrying avrXa27. pHM1avrXa27 was also designated as pZWavrXa27 (Gu et al., 2005).
Rice transformation and characterization of transgenic plants
Agrobacterium‐mediated transformation of rice cultivar Nipponbare was carried out according to the procedures described previously (Yin and Wang, 2000). Control transgenic plants were derived from Nipponbare transformed with empty vector pC1305.1. The avrXa27 gene in transgenic plants was detected by Southern blot hybridization (Sambrook et al., 1989). In brief, 2–5 µg of DNA isolated from transgenic plants was digested with either restriction enzymes NdeI and XbaI, or BamHI, and blotted onto a nylon filter (Hybond‐N+, Amersham). The DNA filters were probed with a 32P‐labelled 3234‐bp BamHI fragment from avrXa27. The copy number of T‐DNA in the transgenic plants was detected by hybridizing the DNA filters with a probe from the hpt gene. The PR1 promoter, 5′ and 3′ regions of avrXa27 and Tnos in transgenic plants were isolated by PCR and verified by DNA sequencing. The DNA primers used to amplify the PR1 promoter and 5′ region of avrXa27 were F1 (5′‐CCATGATTACGAATTCGAGCTCGG‐3′) and R1 (5′‐AAGAAGCGACGGATCGAACTGAC‐3′). Similarly, those used to amplify the 3′ region of avrXa27 and Tnos were F2 (5′‐CGAAGAGGAGCTCGCATGGTTGAT‐3′) and R2 (5′‐ACGCTCTTTTCTCTTAGGTTTAC‐3′). The amplified PCR products were cloned into the pGEM®‐T Easy vector (Promega) and verified by DNA sequencing.
Rice lines and genetic crossing
The rice lines used in this study were Nipponbare, a variety of japonica subspecies that does not carry the Xa27 gene, IR24, a variety of indica subspecies that carries the susceptible allele of the Xa27 gene (xa27), and IRBB27, a near‐isogenic line of Xa27 in a IR24 background (Gu et al., 2004). IRBB27 was used as the recurrent female to cross and backcross with transgenic avrXa27 line L24. Similar crossings were carried out between IR24 (recurrent female) and L24 or IRBB27 (recurrent female) and the control transgenic plants, and at least four backcrosses were performed for each combination.
Northern blot analysis
Northern blot analysis was carried out according to the standard procedures described in Sambrook et al. (1989). Total RNA was isolated from rice leaves using the RNeasy Plant Mini Kit (Qiagen). mRNA was purified with an mRNA Midi Kit (Qiagen). Approximately 30 µg of total RNA or 5 µg of mRNA were used for each lane in Northern blot analysis. The RNA loading was assessed by staining RNA blots with methylene blue or hybridizing RNA blots with the rice ubiquitin gene 2 (Ubi). The probe for avrXa27 was the 3234‐bp BamHI fragment from the gene (Gu et al., 2005). The probe for the Xa27 gene was the full‐length cDNA of the gene (Gu et al., 2005). Probes were labelled with 32P‐dCTP (Amersham).
Bacterial inoculations and disease scoring
Xoo and Xoc strains were grown on PSA medium (10 g/L peptone, 10 g/L sucrose, 1 g/L glutamic acid, 16 g/L bacto‐agar, pH 7.0) with appropriate antibiotics for 2–3 days. The bacterial cells of Xoo and Xoc were suspended in sterile water at an optical density at 600 nm (OD600) of 0.5. BB inoculation was carried out on the two youngest fully expanded leaves on each tiller of 6‐week‐old rice plants using the leaf‐clipping method (Kauffman et al., 1973). The disease symptoms were scored at 14 DAI according to the criteria described previously (Gu et al., 2004). For BLS inoculation, fully expanded leaves of 6‐week‐old rice plants were infiltrated with Xoc suspension using a needleless syringe (Schaad et al., 1996). The Xoc population in inoculated plants was determined using the method reported by Makino et al. (2006) with slight modifications. In brief, infiltrated areas of rice leaves were removed and ground in 5 mL of sterile water. Serial dilutions were made and spread on to PSA agar plates with appropriate antibiotics. The plates were incubated at 28 °C until single colonies could be counted. The number of colony‐forming units (CFU) per leaf was counted, and the standard deviation was calculated using colony counts from replicate experiments.
Histological analysis
Cell wall thickening at vascular elements was measured according to the method described previously (Gu et al., 2005). Leaf segments, 3 mm in length, were fixed in 50 mm NaPO4 buffer (pH 7.4) containing 2.5% glutaraldehyde. After dehydration with ethanol, samples were embedded using a Leica Historesin Embedding Kit. The 3‐µm unstained sections were examined for autofluorescence from phenolic compounds using a Zeiss LSM 510 confocal microscope (excitation, 405 nm; emission, 420–480 nm).
ACKNOWLEDGEMENTS
The authors thank Mei Ling Goh, Keyu Gu, Lifang Wu and Jatinder Singh Sangha for technical assistance, Ko Shimamoto for PR1 and PBZ1 genes, CAMBIA for binary vector pC1305.1, Xianming Zeng for Xoc L8, and Frank F. White and Naweed Naqvi for critical reading of the manuscript. This work was supported by the intramural research funds from Temasek Life Sciences Laboratory (Z.Y.) and a grant from the Agri‐Food and Veterinary Authority of Singapore (Z.Y.).
REFERENCES
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agent in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Bai, J. , Choi, S.‐H. , Ponciano, G. , Leung, H. and Leach, J.E. (2000) Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol. Plant–Microbe Interact. 13, 1322–1329. [DOI] [PubMed] [Google Scholar]
- Bonas, U. , Stall, R.E. and Staskawicz, B. (1989) Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria . Mol. Gen. Genet. 218, 127–136. [DOI] [PubMed] [Google Scholar]
- De Feyter, R. and Gabriel, D.W. (1991) At least six avirulence genes are clustered on a 90‐kilobase plasmid in Xanthomonas campestris pv. malvacearum . Mol. Plant–Microbe Interact. 4, 423–432. [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gnanamanickam, S. , Brindha Priyadarisini, V. , Narayanan, N. , Vasudevan, P. and Kavitha, S. (1999) An overview of bacterial blight disease of rice and strategies for its management. Curr. Sci. 77, 1435–1443. [Google Scholar]
- Gu, K. , Tian, D. , Yang, F. , Wu, L. , Sreekala, C. , Wang, D. , Wang, G.L. and Yin, Z. (2004) High‐resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor. Appl. Genet. 108, 800–807. [DOI] [PubMed] [Google Scholar]
- Gu, K. , Yang, B. , Tian, D. , Wu, L. , Wang, D. , Sreekala, C. , Yang, F. , Chu, Z. , Wang, G.‐L. , White F.F. and Yin, Z. (2005) R gene expression induced by a type‐III effector triggers disease resistance in rice. Nature, 435, 1122–1125. [DOI] [PubMed] [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA, 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S.Y. , Nomura, K. and Whittam, T.S. (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochem. Biophys. Acta, 1694, 181–206. [DOI] [PubMed] [Google Scholar]
- Herbers, K. , Conrads‐Strauch, J. and Bonas, U. (1992) Race‐specificity of plant resistance to bacterial spot disease determined by repetitive motifs in a bacterial avirulence protein. Nature, 356, 172–174. [Google Scholar]
- Hopkins, C.M. , White, F.F. , Choi, S.‐H. , Guo, A. and Leach, J.E. (1992) Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 5, 451–459. [DOI] [PubMed] [Google Scholar]
- Kauffman, H.E. , Reddy, A.P.K. , Hsieh, S.P.Y. and Merca, S.D. (1973) An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae . Plant Dis. Rep. 57, 537–541. [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. [DOI] [PubMed] [Google Scholar]
- Kjemtrup, S. , Nimchuk, Z. and Dangl, J.L. (2000) Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3, 73–78. [DOI] [PubMed] [Google Scholar]
- Makino, S. , Sugio, A. , White, F. and Bogdanove, A.J. (2006) Inhibition of resistance gene‐mediated defense in rice by Xanthomonas oryzae pv. oryzicola . Mol. Plant–Microbe Interact. 19, 240–249. [DOI] [PubMed] [Google Scholar]
- Nino‐Liu, D.O. , Ronald, P.C. and Bogdanove, A.J. (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. [DOI] [PubMed] [Google Scholar]
- Noda, T. and Kaku, H. (1999) Growth of Xanthomonas oryzae pv. oryzae in planta and in guttation fluid of rice. Ann. Phytopathol. Soc. Jpn. 65, 9–14. [Google Scholar]
- Ou, S.H. (1985) Rice Diseases. Kew, Surrey: Commonwealth Agricultural Bureau. [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thebault, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Won Lee, S. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Frisch, E. , Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schaad, N.W. , Wang, Z.K. , Di, M. , McBeath, J. , Peterson, G.L. and Bonde, M.R. (1996) An improved infiltration technique to test the pathogenicity of Xanthomonas oryzae pv. oryzae in rice seedlings. Seed Sci. Technol. 24, 449–456. [Google Scholar]
- Schornack, S. , Ballvora, A. , Gürlebeck, D. , Peart, J. , Baulcombe, D. , Ganal, M. , Baker, B. , Bonas, U. and Lahaye, T. (2004) The tomato resistance protein Bs4 is a predicted non‐nuclear TIR‐NB‐LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 37, 46–60. [DOI] [PubMed] [Google Scholar]
- Sheng, Z.‐J. , Zhen, L.‐Y. and Jun, F.‐X. (2005) Detection of QTL conferring resistance to bacterial leaf streak in rice chromosome 2 (O. sativa L. spp. indica ). Sci. Agric. Sinica, 38, 1923–1925. [Google Scholar]
- Sugio, A. , Yang, B. , Zhu, T. and White, F.F. (2007) Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAgamma1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 104, 10 720–10 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, S. , De Feyter, R. , Brlansky, R.H. and Gabriel, D.W. (1991) A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit canker‐like lesions on citrus. Phytopathology, 81, 802–809. [Google Scholar]
- Szurek, B. , Marois, E. , Bonas, U. and Van den Ackerveken, G. (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 26, 523–534. [DOI] [PubMed] [Google Scholar]
- Tang, D. , Wu, W. , Li, W. , Lu, H. and Worland, A.J. (2000) Mapping of QTLs conferring resistance to bacterial leaf streak in rice. Theor. Appl. Genet. 101, 286–291. [Google Scholar]
- Van den Ackerveken, G.V. , Marois, E. and Bonas, U. (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell, 87, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Yang, B. and White, F.F. (2004) Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant–Microbe Interact. 17, 1192–1200. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10 503–10 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. and Gabriel, D.W. (1995) Xanthomonas avirulence pathogenicity gene family encodes functional plant nuclear targeting signals. Mol. Plant–Microbe Interact. 8, 627–631. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , De Feyter, R. and Gabriel, D.W. (1994) Host‐specific symptoms and increased release of Xanthomonas citris and X. campestris pv. malvacearum from leaves are determined by the 102‐bp tandem repeats of pthA and avrb6, respectively. Mol. Plant–Microbe Interact. 7, 345–355. [Google Scholar]
- Yin, Z. and Wang, G.‐L. (2000) Evidence of multiple complex patterns of T‐DNA integration into the rice genome. Theor. Appl. Genet. 100, 461–470. [Google Scholar]
- Zeng, X. , Lai, W. and Xu, D. (1995) Serological specificity of leaf streak pathogen of rice. J. South China Agric. Univ. 16, 65–68. [Google Scholar]
- Zhao, B. , Lin, X. , Poland, J. , Trick, H. , Leach, J. and Hulbert, S. (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl. Acad. Sci. USA, 102, 15 383–15 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , MaGbanua, M.M. and White, F.F. (2000) Identification of two novel hrp‐associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae . J. Bacteriol. 182, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Yang, B. , Chittoor, J.M. , Johnson, L.B. and White, F.F. (1998) AvrXA10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Mol. Plant–Microbe Interact. 11, 824–832. [DOI] [PubMed] [Google Scholar]


