Abstract
Objectives
Limited data exist on clinical associations and genotypic correlates of linezolid resistance in Mycobacterium tuberculosis. We aimed to describe mutations and clinical factors associated with phenotypic linezolid resistance from patients with drug-resistant TB at two public sector facilities in South Africa.
Methods
Adults and adolescents with treatment failure (culture positivity ≥4 months) on a linezolid-containing regimen were retrospectively identified. Phenotypic resistance, as defined by a linezolid MIC >1 mg/L, was assessed for retrieved isolates using broth microdilution. Targeted sequencing of rrl and rplC was performed, irrespective of growth on subculture.
Results
Thirty-nine patients with linezolid-based treatment failure were identified, 13 (33%) of whom had phenotypic or genotypic linezolid resistance after a median duration of 22 months (range = 7–32) of linezolid therapy. Paired MIC testing and genotyping was performed on 55 unique isolates. All isolates with phenotypic resistance (n = 16) were associated with known resistance mutations, most frequently due to the T460C substitution in rplC (n = 10); rrl mutations included G2814T, G2270C/T and A2810C. No mutations were detected in isolates with MICs at or below the critical concentration.
Conclusions
Linezolid resistance occurred in a third of patients with drug-resistant TB and treatment failure. Resistance occurred late and was predicted by a limited number of mutations in rrl and rplC. Screening for genotypic resistance should be considered for patients with a positive culture after 4 months of linezolid therapy in order to optimize treatment and avoid the toxicity of ineffective linezolid therapy.
Introduction
Drug-resistant TB has a major impact on health outcomes and costs in high-burden countries1 and is expected to increase over the next two decades.2 Linezolid, the prototype oxazolidinone, improved outcomes of drug-resistant TB in clinical trials.3,4 An individual patient data meta-analysis showed that linezolid use increased odds of treatment success 3-fold with a significantly lower mortality.5 Based on these data, WHO recommended linezolid as a preferred agent for all patients with drug-resistant TB in 2018.6 Linezolid therefore has an important role as an anti-TB agent and its introduction into national TB programmes will be scaled up.
Linezolid resistance has been reported in clinical isolates from a limited number of patients with drug-resistant TB and treatment failure.4,7 Limited evidence suggests that population-level resistance to linezolid may be increasing in TB programmes.8 Linezolid shares key binding sites and displays cross-resistance with other oxazolidinones, including promising new agents in clinical development, such as sutezolid9 and delpazolid.10 Mutations in genes encoding the 23S rRNA (rrl) linezolid peptidyl transferase centre (PTC) binding site and the L3 protein (rplC), which extends into the binding site, have been identified as the dominant molecular mechanisms underlying linezolid resistance from in vitro and clinical studies.4,9–19 There are limited data on the association between genotypic and phenotypic linezolid resistance in clinical isolates. The few published studies describing linezolid resistance in treatment programmes have not integrated MIC values with genotyping and have not explored important clinical parameters such as duration of linezolid exposure.19
There are two potential risk factors for linezolid resistance. First, linezolid dosing is frequently reduced due to mitochondrial toxicity,20 which may lead to suboptimal exposures for efficacy and resistance suppression, driving the selection of resistant mutants.20 Second, there are limited treatment options for drug-resistant TB and linezolid may be added to a failing or inadequate regimen, exacerbating the risk of acquired resistance.
A better understanding of the clinical predictors and genotypic correlates of linezolid resistance is critical to inform strategies to preserve this important anti-TB agent. We conducted a retrospective cohort study of patients with drug-resistant TB and linezolid-based treatment failure at two TB referral hospitals in South Africa with the following objectives: (i) to determine the prevalence of linezolid resistance in this at-risk population; (ii) to identify the mutations associated with phenotypic linezolid resistance in clinical Mycobacterium tuberculosis isolates; and (iii) to describe clinical factors associated with linezolid resistance.
Patients and methods
Setting and study population
Adult and adolescent patients (≥13 years old) with treatment failure on a linezolid-containing regimen were retrospectively identified from two public sector TB facilities in South Africa: Jose Pearson Hospital in Port Elizabeth and Brooklyn Chest Hospital in Cape Town. These facilities manage both inpatients and outpatients with drug-resistant TB and use linezolid routinely in their treatment regimens for pre-XDR-TB (defined as resistance to rifampicin and isoniazid, plus fluoroquinolones or second-line injectables) and XDR-TB (as for pre-XDR-TB, but with resistance to both fluoroquinolones and second-line injectables). Treatment failure, and eligibility for inclusion in the analysis, was defined as a persistently positive sputum culture for M. tuberculosis or culture reversion after a negative culture in a patient who had received at least 4 months of linezolid-based therapy for TB.
Data sources
Clinical cases
Registers of patients with possible linezolid-based treatment failure are maintained by facility staff members; these medical records were screened by a study investigator. Clinical data were extracted and captured directly onto electronic case report forms in REDCap.21 The index TB episode was defined as receipt of continuous treatment (with <3 months’ interruption) for rifampicin-resistant TB. We quantified the number of likely effective agents in the regimen by applying a scoring system according to resistance profile, prior exposure and known clinical effectiveness [detailed in Table S1 (available as Supplementary data at JAC Online)].22
Microbiological data
Sputum culture results from routine testing performed at the study sites are linked to the National Health Laboratory Services (NHLS) database, which was used to identify M. tuberculosis isolates from identified cases. All previous isolates were requested from both local NHLS laboratories and from the National Institute of Communicable Diseases (NICD), which performs extended drug susceptibility testing (DST) on clinician request. Available isolates were shipped in original liquid culture bottles to the biosafety level-3 (BSL3) laboratory at the Institute of Infectious Disease and Molecular Medicine at the University of Cape Town for linezolid resistance testing.
Linezolid resistance testing
Isolate selection and culture conditions
Subculture was done for all samples from the first batch of retrieved isolates (n = 57); in subsequent batches, only paired isolates (the earliest and most recent) from each patient with linezolid-based treatment failure underwent subculture (n = 46). M. tuberculosis isolates were initially cultured in the BACTEC Mycobacteria Growth Indicator Tube (MGIT) 960 system (Becton, Dickinson and Company, Sparks, MD, USA) according to the manufacturer’s instructions. Subsequently, 100 μL of each MGIT culture was inoculated onto Löwenstein–Jensen (LJ) medium slants (Becton, Dickinson and Company) and incubated at 37°C for 4–6 weeks with continuous aeration. Colonies were scraped from LJ slants with visible bacterial growth and 10% glycerol stocks were made. Subcultures were initiated by inoculating 100 μL of the 10% glycerol stock in 10 mL Middlebrook 7H9 broth (Sigma–Aldrich) supplemented with 0.2% (v/v) glycerol, 0.1% Tween 80 and 10% (v/v) OADC and were incubated at 37°C until an OD600 value of 1 was reached.
Determination of linezolid MIC
Phenotypic linezolid resistance was assessed by MIC determination using a resazurin microtitre assay.23 Two-fold serial dilutions of linezolid (range 64–0.0625 mg/L) were made in 7H9 medium supplemented with 0.1% casitone, 10% OADC and 0.5% glycerol in 96-well U-bottomed plates. The enriched 7H9 broth containing retrieved M. tuberculosis isolates was diluted 1:1000 and inoculated into the linezolid-containing plates. Plates were incubated at 37°C for 14 days before adding 20 μL of 0.025% (w/v) resazurin (Sigma–Aldrich), followed by incubation for an additional 24 to 48 h. The MIC value was defined as the lowest linezolid concentration that inhibited growth, indicated by a colour change from blue to pink. Positive (M. tuberculosis isolate only) and negative (7H9 medium only) controls were included for each assay. Phenotypic resistance was defined by an MIC >1 mg/L, the recognized critical concentration for linezolid.24
DNA extraction, PCR amplification and sequencing
DNA was extracted from all MGIT cultures, including those without growth on LJ slants, using the Chelex method.25 Primers were designed to amplify coding and flanking regions for rrl, as well as an 814 bp product covering rplC (Table S2). These targets were selected because they encode regions in or near the 23S rRNA binding site26,27 and have been associated with linezolid resistance in clinical and laboratory-generated M. tuberculosis isolates.28 We also planned to sequence rplD (which encodes a putative resistance target in the L4 protein)10 in isolates with MIC >1 mg/L and no detectable mutations in rrl and rplC, but this was not required. Primer design was based on the genome sequence of the M. tuberculosis H37Rv reference strain (http://genolist.pasteur.fr/TubercuList) and performed using Primer 3 software version 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/).
PCRs were performed under the following thermocycling conditions: 15 min of denaturation at 95°C followed by 35 amplification cycles (each cycle = 94°C for 1 min, 62°C for 1 min and 1 min of extension at 72°C) and a final elongation step of 10 min at 72°C. Successful PCR amplification was confirmed by gel electrophoresis. PCR products underwent Sanger sequencing at Central Analytical Facilities, Stellenbosch University, South Africa. Mutations were detected using CLC Main Workbench, Version 7.7.3 (QIAGEN, CA, USA) by aligning the reference H37Rv strain (ATCC 27294) sequence to the sequence from the clinical isolates. Genotypic resistance was defined as the presence of single nucleotide substitutions in rrl or rplC previously identified to be associated with linezolid resistance,28 as well as newly identified polymorphisms in close proximity to the linezolid binding pocket and associated with elevated MICs.
Analysis
MIC distributions of isolates that underwent phenotypic DST were plotted. We used bivariate analysis to compare demographic profile, treatment history and linezolid exposures between patients who developed linezolid resistance (phenotypic or genotypic) with those who did not. Wilcoxon rank-sum testing was performed for comparisons of continuous variables and χ2 tests for categorial variables. A Kaplan–Meier survival plot was constructed for time to the detection of linezolid resistance, censored for death, loss to follow-up (LTFU) and at 36 months post-linezolid initiation.
Ethics
This study was approved by the University of Cape Town Human Research Ethics Committee (reference 805/2016).
Results
Linezolid MIC distribution and associated resistance mutations
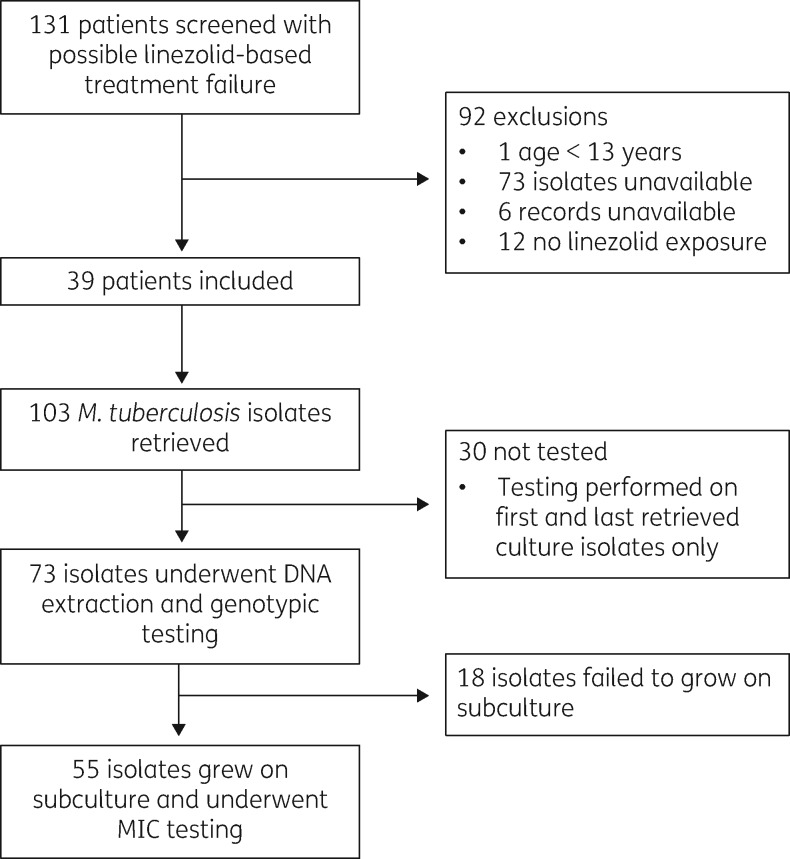
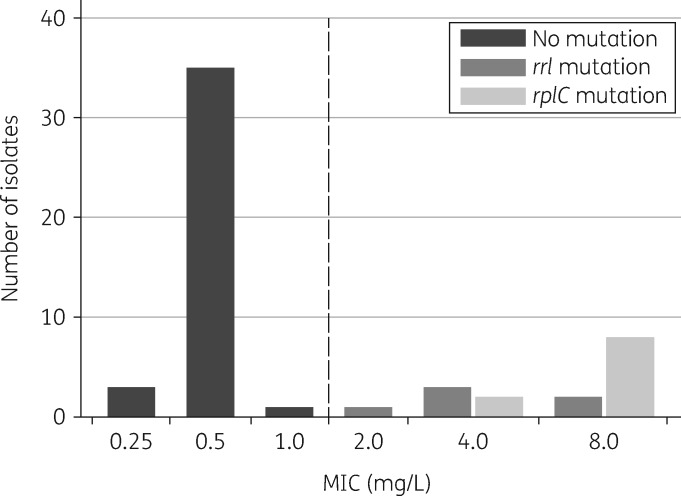
We screened 131 patients with drug-resistant TB and suspected linezolid-based treatment failure (Figure 1); 103 M. tuberculosis culture isolates were available from 39 eligible patients (34 in Port Elizabeth and 5 in Cape Town) collected between May 2010 and September 2017. Paired MIC testing and genotyping were performed on 55 unique isolates that grew on subculture, demonstrating a clear bimodal distribution around the critical concentration of 1 mg/L (Figure 2). All isolates with MIC >1 mg/L (phenotypic resistance, n = 16) were associated with known resistance mutations in either rrl or rplC (Table 1); conversely, no resistance-conferring mutations were detected in isolates with MICs at or below the critical concentration.
Figure 1.
Flow diagram showing numbers of patients and isolates included.
Figure 2.
Distribution of M. tuberculosis linezolid MIC values for 55 clinical isolates with paired MIC and sequencing results. The vertical broken line represents the critical concentration value for linezolid (1 mg/L). None of the isolates had dual mutations.
Table 1.
Mutations in rrl and rplC with corresponding MIC values for all retrieved M. tuberculosis isolates that underwent phenotypic and/or genotypic resistance testing (n = 73)
| Participant ID | Isolate number | Duration on linezolid (months)a | Linezolid MIC (mg/L) | rrl mutation | rplC mutation |
|---|---|---|---|---|---|
| 2007 | XD00813360 | 10 | 8 | G2814T [2576] | WT |
| 1007 | UH00774544 | 18 | 4 | (G2270T) [2032] | WT |
| 1008 | UH00806598 | 25 | 8 | WT | T460C |
| 1010 | YA00027930 | 12 | 2 | (G2270C) [2032] | WT |
| 1011 | UH00751414 | 23 | 4 | G2814T [2576] | WT |
| UH00768146 | 24 | 4 | G2814T [2576] | WT | |
| 1013 | UH00760075 | 13 | 8 | WT | T460C |
| UH00830976 | 18 | 4 | WT | T460C | |
| 1014 | UH00812719 | 22 | 8 | G2814T [2576] | WT |
| 1015 | UJ00479546 | 8 | 8 | WT | T460C |
| UJ00506756 | 10 | 8 | WT | T460C | |
| UJ00519199 | 11 | 8 | WT | T460C | |
| 1023 | UH00754483 | 13 | no growthb | A2384C [2146] | T460C |
| TRL0118476 | 5 | 8 | WT | T460C | |
| 1032 | UH00873025 | 26 | 4 | WT | T460C; G546A |
| 1043 | TRL0118350 | 10 | 8 | WT | T460C |
| 1050 | UH00962529 | 23 | 8 | WT | T460C |
| 1057 | UH00820877 | 25 | no growthb | A2810C [1942] | WT |
Nucleotide positions are given according to the sequence of M. tuberculosis strain H37Rv (GenBank accession number NC_000962.3) with corresponding Escherichia coli positions reported in square brackets. Mutations shown in round brackets were identified in the heteroresistant state. Bold formatting indicates resistance mutations.
Linezolid exposure from time of initiation to collection of the isolate.
No growth in LJ culture.
SNPs associated with phenotypic resistance to linezolid
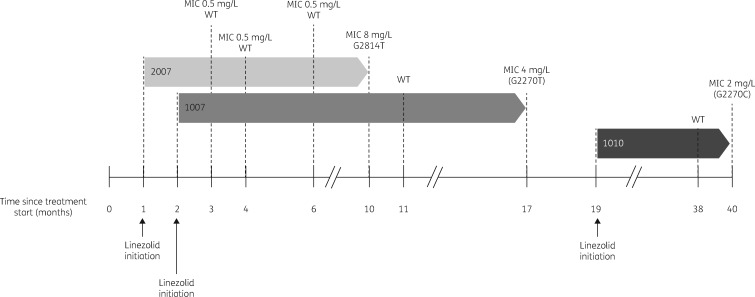
Sequencing of both rplC and rrl was done on 73 unique isolates (including isolates that failed to grow on subculture) from the 39 clinical cases with linezolid-based treatment failure. Mutations and corresponding MIC values from 13 patients (18 isolates) with phenotypic and/or genotypic resistance are listed in Table 1. Resistance mutations in rplC (n = 11) were detected more frequently than in rrl (n = 7); none of the isolates harboured dual resistance-conferring mutations. The G2814T substitution was the most frequently detected mutation in rrl (present in 4 out of 7 isolates), followed by point mutations in two isolates at position 2270 (G2270C/T) and one isolate with an A2810C mutation. The G2270C/T alleles were present with WT alleles as mixed populations from two unique patients and were not detected in strains recovered 1 month and 6 months earlier, respectively (Figure 3). We detected the following additional polymorphisms in rrl, which were not considered to represent resistance mutations due to distance from the PTC and because they were not associated with elevated MICs (MIC 0.25 mg/L for all): A2384G (n = 3), A2384C (n = 5), G2399A (n = 1) and mixed G2399A/A2384C (n = 3).
Figure 3.
Timing of linezolid initiation and results of phenotypic and genotypic testing in relation to start of anti-TB therapy for three patients with sequential isolates demonstrating evolution of linezolid resistance. Inset numbers are study patient identifiers. Mutations shown in parentheses were identified in the heteroresistant state.
rplC resistance mutations (n = 11) were exclusively due to T460C. We found a non-resistance-conferring polymorphism G546A (MIC 0.5 mg/L) in two isolates: as a single mutation in one isolate and mixed with T460C in another isolate with an elevated MIC. A GCC insertion at position 466 was identified in one isolate that failed to grow on subculture and consequently no MIC result was available; however, this insertion was not detected in a sequential isolate collected 6 months later.
Clinical characteristics of the study population
Demographic and clinical characteristics of all included cases with linezolid-based treatment failure are shown in Table 2, disaggregated by the presence of linezolid resistance. Overall, 23/39 (59%) patients were HIV positive and the majority (28/39, 72%) had XDR-TB. There were 4 (median; IQR = 3.5–5) likely effective agents in addition to linezolid at the time of treatment failure; only 8/39 (21%) patients had isolates that were fully susceptible to fluoroquinolones, and bedaquiline and/or delamanid were included in the regimen for only 9/39 (23%) patients. Linezolid initiation was delayed for a median of 7 months (IQR = 2–17, range = 1–30) after the start of therapy for the index TB episode and was administered for a median of 16 months (IQR = 12–23, range = 5–44) until the last obtained culture result. The standard dose for linezolid was 600 mg daily, reduced to 300 mg daily in 20/39 (51%) patients.
Table 2.
Clinical characteristics of patients with linezolid-based treatment failure
| Resistant, n = 13 | Susceptible, n = 26 | P | |
|---|---|---|---|
| Age (years) | 35 (30–45) | 36 (28–42) | 0.83 |
| Male | 6 (46) | 13 (50) | 0.82 |
| Weight at treatment initiation (kg) | 48 (39–62)a | 45 (35–54)b | 0.32 |
| HIV-positive | 7 (54) | 16/25 (64) | 0.54 |
| Number of previous TB episodes | 1 (1–2) | 1 (1–2) | 0.41 |
| Baseline resistance pattern | |||
| MDR | 1 (8) | 1 (4) | 0.40 |
| pre-XDR (injectable resistance) | 1 (8) | 5 (19) | |
| pre-XDR (fluoroquinolone resistance) | 0 (0) | 3 (12) | |
| XDR | 11 (85) | 17 (65) | |
| Delay in linezolid start after initiation of therapy (months) | 8 (2–13) | 3 (0–9) | 0.24 |
| Record of poor adherence | 6/9 (67) | 12/18 (67) | 1.0 |
| Linezolid dose reduction | 6 (46) | 14/23 (61) | 0.39 |
| Duration on linezolid (months)c | 18 (10–23) | 16 (12–21) | 0.89 |
| Number of other drugs | 10 (9–11) | 8 (7–10) | 0.04 |
| Number of likely effective drugs at time of treatment failure | 4.0 (4.0–4.5) | 4.0 (3.5–5.0) | 0.95 |
| Bedaquiline exposured | 3 (23) | 9 (35) | 0.46 |
| Duration of bedaquiline exposure (months)e | 5 (1–10) | 10 (6–12) | 0.19 |
| Outcome within 48 months of study | |||
| in care | 6 (46) | 8 (31) | 0.55 |
| died | 6 (46) | 9 (35) | |
| LTFU | 0 (0) | 2 (8) | |
| palliation | 0 (0) | 3 (12) | |
| unknown | 1 (8) | 3 (12) |
Data are presented as n (%) or median (IQR).
Resistant is defined as MIC >1 mg/L and/or presence of previously published resistance-conferring mutation.
n = 12.
n = 23.
Defined as the time from linezolid initiation until the first culture showing linezolid resistance or the last culture obtained in those without linezolid resistance (n = 39).
The number of patients with bedaquiline exposure before the first culture showing linezolid resistance or the last culture obtained in those without linezolid resistance.
Defined as the time from bedaquiline initiation until the first culture showing linezolid resistance or the last culture obtained in those without linezolid resistance (n = 12).
Clinical associations with linezolid treatment failure and resistance
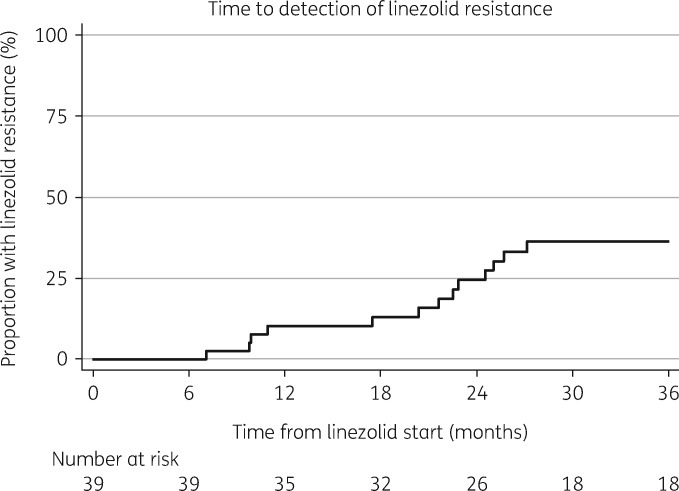
Linezolid resistance was detected by either phenotypic or genotypic methods in 13/39 (33%) patients with linezolid-based treatment failure. The earliest detected occurrence of resistance was 7 months after initiating linezolid, with the latest at 27 months (Figure 4). Neither linezolid dose reduction (P = 0.39) nor overall duration (18 months for those with resistance versus 16 months without; P = 0.89) were associated with linezolid resistance (Table 2). Bedaquiline exposure prior to treatment failure did not appear to be protective for linezolid resistance in this cohort; 3/13 (23%) with resistance versus 9/26 (35%) without resistance received bedaquiline; P = 0.46. The only significant difference between those with and without resistance on bivariate analysis was the total number of anti-TB drugs received, which was higher in the group with linezolid resistance (median 10 versus 8 drugs; P = 0.04). There was a trend towards a longer delay in linezolid initiation in the group with resistance (8 months versus 3 months), but this was not significant (P = 0.24). Overall mortality was 38% (n = 15) during the 42 month observation time.
Figure 4.
Kaplan–Meier plot showing time to detection of linezolid resistance, censored for death and LTFU (n = 39).
Discussion
We retrospectively identified 39 patients with drug-resistant TB and linezolid-based treatment failure from two geographically distant treatment facilities in South Africa. Most patients had XDR-TB and were unable to obtain early access to new anti-TB agents. Consequently, background drug regimens at the time of linezolid introduction were likely suboptimal resulting in effective linezolid monotherapy for prolonged periods of time. Linezolid resistance was detected in over 30% of individuals and only after a median of 18 months of exposure, which is somewhat surprising given the highly conducive conditions for resistance selection. In one respect this observation is reassuring, because it confirms linezolid’s high barrier to resistance seen under similar conditions in a clinical trial,4 as well as in vitro where very low mutation frequencies are achieved relative to other anti-TB drugs.9,14 However, our finding also illustrates a fundamental reality underlying the large-scale expansion of linezolid for TB treatment: despite its high barrier to resistance, with sufficient selection pressure the emergence of linezolid resistance in TB treatment programmes is inevitable. Of concern, there have been suggestions of a trend towards increasing population-level resistance in countries with a long history of linezolid use;8 and linezolid resistance has been associated with the Beijing genotype,18 the dominant circulating M. tuberculosis strain in the Eastern Cape Province29 where most of our cases were identified. Increased vigilance and active surveillance are clearly needed as linezolid is introduced into national TB treatment programmes.
Published data on linezolid resistance from patients with TB are scarce; in a literature review we identified nine studies that investigated linezolid resistance in clinical isolates, reporting on a total of 24 unique patients. Our study, involving 39 patients with linezolid-based treatment failure, likely provides the largest and most detailed series linking MIC values with molecular testing and clinical and treatment data. There are several notable findings that build on existing knowledge in this area.
Our strategy to perform targeted sequencing of rrl and rplC was based on the mechanism of linezolid action and observations from clinical and in vitro reports. It is unsurprising that mutations in 23S rRNA, particularly in proximity to the PTC binding site, predictably led to MIC elevations and clinical resistance. The most frequently reported mutation in rrl, the G2814T nucleotide substitution,4,9,19,30–32 was detected in isolates from more than half of patients with linezolid resistance and rrl mutations, and associated with MIC values of up to 8 mg/L. We also detected G2270C/T mutations in isolates from two unique patients, which, to our knowledge, is the first report in clinical strains. These mutations were associated with lower MICs (2–4 mg/L) in our cohort (as well as in previous in vitro studies14,17), which may be related to the position outside the PTC and the fact that in both cases the mutations were identified in the presence of the susceptible allele (heteroresistant state). We did not detect the resistance allele in prior isolates from either of these patients (collected 1 month and 6 months earlier, respectively), suggesting the evolution of linezolid resistance with ongoing selection pressure.
The third rrl mutation we identified, A2810C, has been previously described in isolates from patients with treatment failure.30,31 Although we were unable to determine the MIC associated with this mutation, this nucleotide substitution is in relative proximity to the PTC and its previous detection in isolates with phenotypic resistance suggest that it could confer a linezolid resistance phenotype.
Overall, the T460C mutation in rplC was the underlying cause for linezolid resistance in the majority (7/13, 54%) of our cases; its dominance has also been noted in other settings.9,12–14,17,19 This mutation results in an amino acid exchange from cysteine to arginine at position 154 in the L3 protein that extends into the linezolid binding site,27 resulting in MIC ranges of 2–32 mg/L.4,9–19 The rplC T460C mutation has been associated with lower MICs and a lower fitness cost9 than mutations affecting 23S rRNA. Interestingly, in our cohort, the converse was found, with higher MICs linked to rplC mutations; this has also been described for in vitro mutants,14,17 reinforcing the importance of this key mechanism for linezolid resistance. It is difficult to interpret the significance of the GCC insertion at position 466 found in one isolate because we were unable to determine the MIC; this has not been previously associated with linezolid resistance, including in bacteria other than M. tuberculosis, and is likely not to be resistance-conferring.
Genotyping had excellent accuracy and discriminative value for predicting phenotypic resistance (MIC >1 mg/L) in the 55 isolates with both sequencing and MIC results. It appears that sequence mixes in rrl and rplC have low diversity and there are a limited number of mutations that underlie linezolid resistance. This raises the possibility of translation into rapid molecular diagnostics, which are needed to support linezolid rollout into national TB programmes where phenotypic testing is not widely available. Furthermore, molecular testing could be class-based because of cross-resistance with other oxazolidinones9,10,13 and has the advantage of detecting low-frequency mutations supporting early identification of resistance.
We found that the number of background agents to which patients were exposed during the index TB episode was significantly higher amongst those with linezolid resistance. This could reflect a tendency of clinicians to add more drugs to failing regimens, leading to a paradoxically higher risk of linezolid monotherapy with more background drugs. The observed trend in longer delays to linezolid initiation in patients with linezolid resistance supports this. Although there is no direct cross-resistance between linezolid and other anti-TB drug classes, one study has demonstrated an association between linezolid MIC elevations and the use of other second-line anti-TB drugs, specifically fluoroquinolones and kanamycin.8 This may be due to induction of efflux pump expression from antimycobacterial drug exposure, which initiates a pathway leading to subsequent high-level mutation-related resistance.33 Regardless of the presence of linezolid resistance there was an extremely high mortality amongst this cohort of patients with linezolid-based treatment failure. This emphasizes the need for early inclusion of new drugs such as bedaquiline to strengthen treatment regimens and reduce the risk of treatment failure34 and mortality.35
Our retrospective study had a number of important limitations. By definition, we had to rely on data that were collected in the clinical service and not originally intended to address the aims of this study. We therefore had to accept risks of major biases when describing and comparing patients with linezolid-based treatment failure. We are confident, however, that our strategy to screen and identify cases from hospital registers was sufficiently rigorous to avoid excluding important outliers. The accuracy of clinical data extracted from medical records was imperfect and also may have influenced the robustness of our findings, particularly in relation to anti-TB drug exposures and assessment of regimen effectiveness. However, the key parameters of linezolid duration and bedaquiline use were well documented. A substantial proportion of isolates were either not available from local laboratories (due to being lost or discarded; 73/131 screened patients, 56%) or were not viable on subculture (18/73 retrieved isolates, 25%). This has two potential consequences. First, sequential isolates demonstrating the transition to linezolid resistance were not available for the majority of included patients. The time-to-event analysis could therefore have overestimated the delay in the development of linezolid resistance. Second, isolates with linezolid resistance may be associated with fitness cost9 and failure to grow, biasing our results. To address this, we sequenced all isolates, regardless of culture viability, and found a resistance mutation in only two isolates without MIC data. Sanger sequencing itself has imperfect sensitivity, particularly for the detection of mixed strain genotypes;36 it is possible that we may have identified additional or novel mutations using next generation sequencing,37 an approach that should be considered in future studies.
In conclusion, we have shown that linezolid resistance occurred in a third of patients from this high-risk cohort in South Africa. Phenotypic resistance was detected late and was predicted by a limited number of mutations in rrl and rplC. Screening for genotypic resistance should be considered for patients with a positive culture after 4 months of linezolid therapy in order to optimize treatment and avoid the toxicity of ineffective linezolid therapy.
Supplementary Material
Acknowledgements
This work was published as a poster at the Forty-ninth Union World Conference on Lung Health, The Hague, The Netherlands, 2018 (Abstract PS 11-520-25).
We wish to thank management and staff at Jose Pearson Hospital and Brooklyn Chest Hospital for support of this research.
Funding
This work was supported by grants to the authors, as follows. S. W. is supported by the European & Developing Countries Clinical Trials Partnership (grant number CDF1018) and the Wellcome Trust (grant number 203135/Z/16/Z). G. L. was funded by the Robert Bosch Stiftung (grant number 12.5.F081.0008.0). Gr. M. was supported by the Wellcome Trust (grant numbers 098316 and 203135/Z/16/Z), the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation (NRF) of South Africa (grant number 64787), National Research Foundation of South Africa (NRF) incentive funding (UID: 85858) and the South African Medical Research Council through its TB and HIV Collaborating Centres Programme with funds received from the National Department of Health (RFA# SAMRC-RFA-CC: TB/HIV/AIDS-01–2014). The funders had no role in the study design, data collection, data analysis, data interpretation or writing of this report.
Transparency declarations
None to declare.
Disclaimer
The opinions, findings and conclusions expressed in this manuscript reflect those of the authors alone.
References
- 1.WHO. Global Tuberculosis Report 2018 Geneva, Switzerland: WHO, 2018. https://www.who.int/tb/publications/global_report/en/.
- 2. Sharma A, Hill A, Kurbatova E. et al. Estimating the future burden of multidrug-resistant and extensively drug-resistant tuberculosis in India, the Philippines, Russia, and South Africa: a mathematical modelling study. Lancet Infect Dis 2017; 17: 707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang S, Yao L, Hao X. et al. Efficacy, safety and tolerability of linezolid for the treatment of XDR-TB: a study in China. Eur Respir J 2015; 45: 161–70. [DOI] [PubMed] [Google Scholar]
- 4. Lee M, Lee J, Carroll MW. et al. Linezolid for treatment of chronic extensively drug-resistant tuberculosis. N Engl J Med 2012; 367: 1508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmad N, Ahuja SD, Akkerman OW. et al. Treatment correlates of successful outcomes in pulmonary multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet 2018; 392: 821–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment WHO/CDS/TB/2019.3 Geneva, Switzerland: WHO, 2019. https://apps.who.int/iris/bitstream/handle/10665/311389/9789241550529-eng.pdf? ua=1.
- 7. Richter E, Rusch-Gerdes S, Hillemann D.. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2007; 51: 1534–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang TS, Liu YC, Sy CL. et al. In vitro activities of linezolid against clinical isolates of Mycobacterium tuberculosis complex isolated in Taiwan over 10 years. Antimicrob Agents Chemother 2008; 52: 2226–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McNeil MB, Dennison D, Shelton C. et al. In vitro isolation and characterization of oxazolidinone resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 2017; 61: e01296-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zong Z, Jing W, Shi J. et al. Comparison of in vitro activity and MIC distributions between the novel oxazolidinone delpazolid and linezolid against multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis in China. Antimicrob Agents Chemother 2018; 62: e00165-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beckert P, Hillemann D, Kohl TA. et al. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother 2012; 56: 2743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pang Y, Lu J, Huo F. et al. Prevalence and treatment outcome of extensively drug-resistant tuberculosis plus additional drug resistance from the National Clinical Center for Tuberculosis in China: a five-year review. J Infect 2017; 75: 433–40. [DOI] [PubMed] [Google Scholar]
- 13. Balasubramanian V, Solapure S, Iyer H. et al. Bactericidal activity and mechanism of action of AZD5847, a novel oxazolidinone for treatment of tuberculosis. Antimicrob Agents Chemother 2014; 58: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ismail N, Omar SV, Ismail NA. et al. In vitro approaches for generation of Mycobacterium tuberculosis mutants resistant to bedaquiline, clofazimine or linezolid and identification of associated genetic variants. J Microbiol Methods 2018; 153: 1–9. [DOI] [PubMed] [Google Scholar]
- 15. Makafe GG, Cao Y, Tan Y. et al. Role of the Cys154Arg substitution in ribosomal protein L3 in oxazolidinone resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2016; 60: 3202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perdigao J, Maltez F, Machado D. et al. Beyond extensively drug-resistant tuberculosis in Lisbon, Portugal: a case of linezolid resistance acquisition presenting as an iliopsoas abscess. Int J Antimicrob Agents 2016; 48: 569–70. [DOI] [PubMed] [Google Scholar]
- 17. Zhang S, Chen J, Cui P. et al. Mycobacterium tuberculosis mutations associated with reduced susceptibility to linezolid. Antimicrob Agents Chemother 2016; 60: 2542–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Z, Pang Y, Wang Y. et al. Beijing genotype of Mycobacterium tuberculosis is significantly associated with linezolid resistance in multidrug-resistant and extensively drug-resistant tuberculosis in China. Int J Antimicrob Agents 2014; 43: 231–5. [DOI] [PubMed] [Google Scholar]
- 19. Zimenkov DV, Nosova EY, Kulagina EV. et al. Examination of bedaquiline- and linezolid-resistant Mycobacterium tuberculosis isolates from the Moscow region. J Antimicrob Chemother 2017; 2: 1901–6. [DOI] [PubMed] [Google Scholar]
- 20. Wasserman S, Meintjes G, Maartens G.. Linezolid in the treatment of drug-resistant tuberculosis: the challenge of its narrow therapeutic index. Expert Rev Anti Infect Ther 2016; 14: 901–15. [DOI] [PubMed] [Google Scholar]
- 21. Harris PA, Taylor R, Thielke R. et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42: 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caminero JA, Sotgiu G, Zumla A. et al. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis 2010; 10: 621.. [DOI] [PubMed] [Google Scholar]
- 23. Martin A, Portaels F, Palomino JC.. Colorimetric redox-indicator methods for the rapid detection of multidrug resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. J Antimicrob Chemother 2007; 59: 175–83. [DOI] [PubMed] [Google Scholar]
- 24.WHO. Technical Report on Critical Concentrations for Drug Susceptibility Testing of Medicines Used in the Treatment of Drug-Resistant Tuberculosis. Geneva, Switzerland: WHO, 2018. https://www.who.int/tb/publications/2018/WHO_technical_report_concentrations_TB_drug_susceptibility/en/. [Google Scholar]
- 25. de Lamballerie X, Zandotti C, Vignoli C. et al. A one-step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Res Microbiol 1992; 143: 785–90. [DOI] [PubMed] [Google Scholar]
- 26. Long KS, Vester B.. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother 2012; 56: 603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klitgaard RN, Ntokou E, Norgaard K. et al. Mutations in the bacterial ribosomal protein l3 and their association with antibiotic resistance. Antimicrob Agents Chemother 2015; 59: 3518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ismail N, Omar SV, Ismail NA. et al. Collated data of mutation frequencies and associated genetic variants of bedaquiline, clofazimine and linezolid resistance in Mycobacterium tuberculosis. Data Brief 2018; 20: 1975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muller B, Chihota VN, Pillay M. et al. Programmatically selected multidrug-resistant strains drive the emergence of extensively drug-resistant tuberculosis in South Africa. PLoS One 2013; 8: e70919.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bloemberg GV, Keller PM, Stucki D. et al. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med 2015; 373: 1986–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Somoskovi A, Bruderer V, Homke R. et al. A mutation associated with clofazimine and bedaquiline cross-resistance in MDR-TB following bedaquiline treatment. Eur Respir J 2015; 45: 554–7. [DOI] [PubMed] [Google Scholar]
- 32. Hillemann D, Rusch-Gerdes S, Richter E.. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother 2008; 52: 800–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmalstieg AM, Srivastava S, Belkaya S. et al. The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob Agents Chemother 2012; 56: 4806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Y, Fox T, Manning K. et al. Improved treatment outcomes with bedaquiline when substituted for second-line injectable agents in multidrug resistant tuberculosis: a retrospective cohort study. Clin Infect Dis 2018; 68: 1522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schnippel K, Ndjeka N, Maartens G. et al. Effect of bedaquiline on mortality in South African patients with drug-resistant tuberculosis: a retrospective cohort study. Lancet Respir Med 2018; 6: 699–706. [DOI] [PubMed] [Google Scholar]
- 36. Hanekom M, Streicher EM, Van de Berg D. et al. Population structure of mixed Mycobacterium tuberculosis infection is strain genotype and culture medium dependent. PLoS One 2013; 8: e70178.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Metcalfe JZ, Streicher E, Theron G. et al. Cryptic microheteroresistance explains Mycobacterium tuberculosis phenotypic resistance. Am J Respir Crit Care Med 2017; 196: 1191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.