SUMMARY
The role of reactive oxygen species (ROS) in interactions between phytopathogenic fungi and their hosts is well established. An oxidative burst mainly caused by superoxide formation by membrane‐associated NADPH oxidases is an essential element of plant defence reactions. Apart from primary effects, ROS play a major role as a second messenger in host response. Recently, NADPH oxidase (nox)‐encoding genes have been identified in filamentous fungi. Functional analyses have shown that these fungal enzymes are involved in sexual differentiation, and there is growing evidence that they also affect developmental programmes involved in fungus–plant interactions. Here we show that in the biotrophic plant pathogen Claviceps purpurea deletion of the cpnox1 gene, probably encoding an NADPH oxidase, has impact on germination of conidia and pathogenicity: Δcpnox1 mutants can penetrate the host epidermis, but they are impaired in colonization of the plant ovarian tissue. In the few cases where macroscopic signs of infection (honeydew) appear, they are extremely delayed and fully developed sclerotia have never been observed. C. purpurea Nox1 is important for the interaction with its host, probably by directly affecting pathogenic differentiation of the fungus.
INTRODUCTION
Claviceps purpurea, a ubiquitous cereal pathogen causing ergot disease, is—apart from its capacity to synthesize bioactive ergopeptines (Tudzynski et al., 2001)—an interesting model system for the study of host–pathogen interactions. It is an ecologically obligate pathogen, i.e. it can be propagated in axenic culture but never grows saprophytically in nature; it is strictly organ‐specific, as it attacks only young ovaries of grasses; it has a rather long period of orientated, almost unbranched growth in the host tissue to reach the vascular tissue at the bottom of the grass flower. There is no stable resistance known in its major host plants (rye, wheat, barley), and during the important first days of infection no host response can be detected, i.e. the fungus escapes the host's security system (reviewed by Tudzynski and Scheffer, 2004). Only recently has it been shown that during penetration of the outer cell layer of stigmatic hairs an early defence reaction is apparent: using EM‐CeCl2 techniques, the production of reactive oxygen species (ROS) in these cells could be demonstrated (Scheffer et al., 2005). However, in later stages, no signs of an oxidative burst/ROS production have thus far been detected. The first observation of such a host defence reaction was reported for a knock‐out mutant of an ATF1‐related bZIP‐transcription factor, cptf1: in tissues infected with a Δcptf1 mutant a strong oxidative burst occurs, and the fungus itself seems to contribute to ROS accumulation in planta. As it was shown that C. purpurea can produce/secrete H2O2 in axenic culture, and as the Δcptf1 mutants lack all catalase activity, a working model was established postulating that C. purpurea—like other fungi—produces H2O2 (and probably other ROS) but scavenges them in planta efficiently, e.g. by catalases. The Δcptf1 mutant is not able to do this because of a lack of catalase activity, and hence H2O2 would be secreted and could induce host response reactions (Nathues et al., 2004; Tudzynski and Scheffer, 2004). Why and how the fungus produces and secretes ROS remain unanswered. Recently, it has been shown that fungi—like higher eukaryotes—possess NADPH oxidases (Nox), which are involved in ROS generation (for reviews, see Aguirre et al., 2005; Takemoto et al., 2007). These fungal enzymes are homologues of the catalytic subunit gp91 phox known from animal phagocytic cells, but they form a specific subgroup. Functional analyses by generation of knockout mutants have demonstrated that these enzymes are involved in sexual differentiation in Aspergillus nidulans (Lara‐Ortiz et al., 2003) and Podospora anserina (Malagnac et al., 2004). These findings substantiate earlier reports by Aguirre and collaborators that ROS are involved in differentiation in fungi (Hansberg and Aguirre, 1990). This has recently been extended to differentiation processes in fungi–plant interactions. Tanaka et al. (2006) showed that the endophyte Epichloe festucae (a close relative of C. purpurea) needs a Nox enzyme for the maintenance of a stable symbiotic interaction with its host plant. Egan et al. (2007) presented evidence that a Nox enzyme is involved in appressoria formation in Magnaporthe grisea.
We report here that Cpnox1, a homologue of NoxA of E. festucae, is essential for pathogenic development in the non‐appressoria‐forming biotroph C. purpurea, which indicates that these enzymes play an important role in quite diverse differentiation processes and life styles in fungi.
RESULTS
cpnox1 encodes a putative fungal NADPH oxidase
Primers based on the sequences of several fungal NADPH oxidases (noxA of A. nidulans, Panox1/2 of P. anserina and homologous genes from the genomes of Neurospora crassa and M. grisea) were used to amplify a fragment of a putative nox gene from genomic DNA of C. purpurea strain 20.1 (see Experimental procedures). The fragment was used to screen a genomic λ library; hybridizing phages were purified, and homologous fragments were subcloned and sequenced. The corresponding genomic region contained an open reading frame of 1923 bp (interrupted by two introns of 108 and 141 bp) encoding a polypeptide of 557 amin oacids, which shows significant homology to fungal NADPH oxidases. It groups to the same clade as P. anserina Panox1 and NoxA of E. festucae (see Fig. 1); therefore, the gene was named cpnox1. In a parallel approach a second putative nox gene was identified, the product of which groups into the noxB clade; it was named cpnox2 (S. Giesbert, T. Schürg and P. Tudzynski, unpublished data); its sequence was integrated in Fig. 1. Cpnox1 shows all features characteristic for this clade of fungal Nox: six predicted transmembrane domains as well as motifs involved in haem, FAD and NADPH binding (data not shown).
Figure 1.

Phylogenetic tree of Nox proteins from fungal, plant and animal origins. Accession numbers are as follows: Claviceps purpurea Cpnox1: AM 899998; C. purpurea Cpnox2: AM 899999; Epichloe festucae NoxA: BAE72680; E. festucae NoxB: BAE72682; Podospora anserina Nox1: AAK50853; P. anserina Nox2: AAQ74977; Dictyostelium discoideum NoxA: AAD22057; Mus musculus Nox1: NP_757340; Rattus norvegicus: NP_446135; Homo sapiens Nox1: CAI42337; Arabidopsis thaliana AtrbohF: BAA28953. The phylogram was constructed using the clustal analysis program included in MEGALIGN, DNA Star.
Generation of Δcpnox1 mutants
To study the role of this putative NADPH oxidase in virulence and development of C. purpurea, knockout mutants were generated using a gene replacement approach. 5′‐ and 3′‐flanking regions of the gene were cloned via PCR and joined with a phleomycin‐resistance cassette (Fig. 2; for details see Experimental procedures). The replacement construct was excised and used to transform protoplasts of strain 20.1. Phleomycin‐resistant colonies were isolated and checked by PCR analysis for the presence of a correctly integrated replacement construct using primers F/G and H/I, respectively (see Fig. 2). From 284 primary transformants analysed in total, two showed a diagnostic PCR fragment. As they were—as usual in C. purpurea—heterokaryotic, i.e. still contained wild‐type nuclei and therefore showed the respective PCR fragment (generated by primers E and F), they were genetically purified by single‐spore subcultures. Two homokaryotic deletion mutants were finally obtained, named Δcpnox1‐1 and Δcpnox1‐2 (see Fig. 3A,B). Southern analysis confirmed the PCR data: as shown in Fig. 3B, the mutants lack the wild‐type fragment of 3 kb, but contain a fragment of 6.2 kb as expected for the replacement situation (see Fig. 2). Mutant Δcpnox1‐1 was used as recipient for a complementation construct containing the whole coding sequence of cpnox1 plus 841 bp of upstream non‐coding sequence combined with a hygromycin resistance cassette (see Experimental procedures). Three transformants containing the full‐length gene copy (as demonstrated by PCR, data not shown) were obtained; these were named Δcpnox1‐C1–3, and used as control for the characterization of the knockout mutants.
Figure 2.

Gene replacement approach for cpnox1. The replacement vector pΔcpnox1 was constructed by cloning the 3′ and 5′ parts of cpnox1 on each side of the phleomycin cassette in the pAN8‐1UM plasmid (see Experimental procedures for further details). The resistance cassette, excised with SacII and ClaI, was used to transform C. purpurea wild‐type strain 20.1. Mutants were generated by homologous integration of the resistance cassette via a double crossover event between the homologous regions of the replacement fragment and the genomic region of cpnox1. The coding region of cpnox1 is indicated by black boxes, introns by white and surrounding genomic regions by striped boxes. ATG: translational start of cpnox1; black arrows: orientation of cpnox1 and the phleomycin resistance gene, respectively; black bar: probe used for Southern hybridization (Fig. 4). Primers used for PCR: for construction of the replacement vector primers A and B (left flank) and primers C and D (right flank) were used. For detection of homologous integration events primers F and G (left flank) and primers H and I (right flank) were used. For amplifying the wild‐type cpnox1 gene primers E and F were used. Restriction enzymes are as follows: C, ClaI; E, EcoRI; P, PstI; S, SalI; SII, SacII; X, XbaI.
Figure 3.
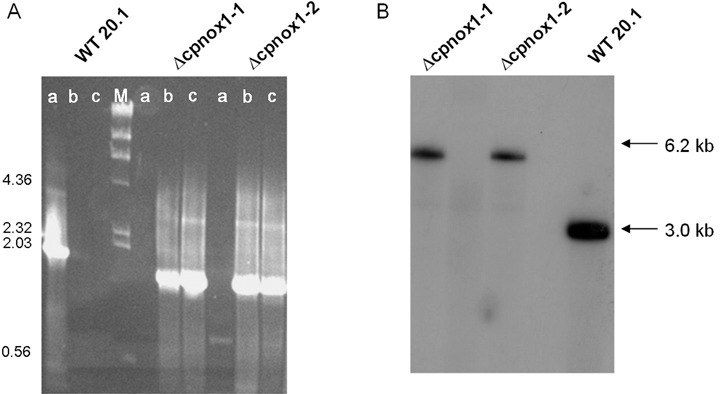
Molecular characterization of Δcpnox1 mutants and recipient strain 20.1. (A) Analytical PCR of the wild‐type strain 20.1 (WT) and the two replacement mutants Δcpnox1‐1 and Δcpnox1‐2. Strains are listed at the top of the picture. Lane M shows DNA size marker λ restricted with HindIII; sizes are indicated on the left in kb. The PCR was performed using three different primer combinations (for position of the primers see Fig. 3); a, E and F (wild‐type gene copy); b, F and G (showing homologous integration, 5′ side); c, H and I (showing homologous integration, 3′ side). (B) Southern analysis of the wild‐type strain 20.1 (WT) and the two replacement mutants Δcpnox1‐1 and Δcpnox1‐2. DNA of the strains was restricted with EcoRI and hybridized with a 1.4‐kb EcoRI/SalI fragment that contains part of the cpnox1 genomic region (see Fig. 3). The size of the hybridizing fragment is indicated on the right.
Cpnox1 has impact on growth and vegetative differentiation
The two Δcpnox1 mutants have a ± normal colony morphology on several growth media and they show a comparable growth rate (see Fig. 4). As analyses of nox mutants in Botrytis cinerea in our laboratory had shown that these mutants were more sensitive to certain stress conditions (N. Segmüller et al., unpublished data), the Δcpnox1 mutants were tested for sensitivity to various stress conditions. Osmotic stress (0.5 m sorbitol) did not significantly affect growth of the mutants (data not shown); however, sensitivity against the ROS‐generating agent menadione was obviously increased: at the highest concentration applied in these growth tests (0.5 mm) the mutants showed almost no growth, whereas the wild‐type was still able to grow (Fig. 4).
Figure 4.

Oxidative stress sensitivity of Δcpnox1‐1. Mycelia of the mutant and wild‐type 20.1 were grown on Mantle agar plates with and without menadione. Concentrations of 200 and 500 µm menadione were tested (12 dpi). (A) Photograph of the plate assay. (B) Colony diameters of plate assays (white columns: wild‐type 20.1; grey columns: Δcpnox1‐1); mean colony diameter (mm) of five parallel plates. Standard deviation is indicated by bars.
Apart from this slight effect on sensitivity to oxidative stress, the mutants were obviously severely affected in vegetative differentiation: germination rate of conidia was significantly lower (more than seven‐fold) in the mutants (e.g. Δcpnox1‐1: 2.9 ± 1.0%) compared with the wild‐type strain (20.7 ± 9%), even if the high level of variation of this parameter (generally observed in C. purpurea) is taken into account.
Cpnox1 is important for normal pathogenic development
As conidial germination was significantly reduced in the Δcpnox1 mutants, mycelial suspensions of the wild‐type and both Δcpnox1 mutants were used instead of conidia to infect rye plants (see Experimental procedures). Rye florets infected with the wild‐type strain 20.1 showed a normal time course of infection (Table 1): 8 ± 1 days post infection (dpi) the infected ears (21 ears with 40 florets each in total) produced honeydew (a syrup‐like liquid containing conidia), and 14–21 dpi sclerotia were formed (data not shown). In contrast, only a small percentage of ears infected with conidia of the mutant strain (for each mutant the same number of florets as in the wild‐type) showed symptoms, but appearance of honeydew was significantly retarded (Table 1), and the few sclerotia formed were misshapen and not fully matured (5, 6), strongly suggesting that Cpnox1 is required for normal pathogenic development. As a control, a complemented transformant was able to produce honeydew and form normal sclerotia (data not shown).
Table 1.
Pathogenicity assay. Percentage of infected rye ears and time of first appearance of honeydew (dpi) of wild‐type 20.1 and mutant strains Δnox1‐1 and Δnox1‐2. Note that ears were considered infected when a minimum of one out of 40 inoculated florets showed symptoms of infection.
| Strain | Ears producing honeydew (%) | Appearance of honeydew (dpi) |
|---|---|---|
| 20.1 | 100 | 8 ± 1 |
| Δnox1‐1 | 19 | 25 ± 7 |
| Δnox1‐2 | 14 | 25 ± 3 |
Figure 5.

Pathogenicity assays of C. purpurea Δcpnox1 mutants and wild‐type on rye. Rye ears infected with Claviceps purpurea wild‐type strain 20.1; Δnox1‐1 and Δnox1‐2, cpnox1 knockout mutant strains; (A) 40 dpi. White arrow indicates honeydew exudate of Δnox1‐1 mutant; (B) 50 dpi. Black arrows indicate infected florets of both mutant strains. Forty florets per ear were inoculated with aqueous suspensions of mycelia (5 mL per floret).
Figure 6.

Sclerotia of C. purpurea wild‐type strain 20.1 and Δcpnox1. (A,B) Sclerotia taken from rye plants 40 dpi (A) and 50 dpi (B); three sclerotia of the wild‐type (left) and three of the mutant (right); scale bar indicates 1 cm. (C,D) Enlarged sections showing mutant sclerotia of A and B, respectively.
To analyse the defect in pathogenic development in detail, rye ovaries were infected in vivo with conidia of the mutant and the wild‐type strain. After 6 days ovaries were harvested and the development of infection was monitored using the aniline blue staining technique (Scheffer and Tudzynski, 2006). Conidia of the mutants were able to germinate, penetrate the stigmatic hairs and grow down to the ovule; however, further growth to the bottom of the floret appeared to be inhibited, as even after 7 days (withering of the cultured ovaries) no hyphae were detected at the bottom of the ovary (Fig. 7).
Figure 7.

Fluorescence micrographs of in vivo inoculated rye florets infected with the wild‐type 20.1 (B) and Dcpnox1‐1 (C,D). Ovaries were isolated 6 dpi, stained with aniline blue and observed using epifluorescence microscopy. The pictogram (A) shows a schematic overview of a rye ovary infected with the wild‐type. The infection pathway of the wild‐type is indicated by yellow lines; the colonization of the transmitting tissue is represented by red colour. The solid boxes represent details shown in panels B–D. Arrows indicate hyphae. (B) Massive colonization of the ovary can be observed after infection with the wild‐type. (C) In contrast to the wild‐type, only thin bundles of hyphae could be detected in the mutant strain Δcpnox1‐1. It was never observed that these hyphae grow further down towards the ovule. In some cases no infection was visible at all (D).
Expression studies
As the pathogenicity tests indicated a role of Cpnox1 during colonization of host tissue and differentiation of sclerotia, we studied the expression of cpnox1 in planta. Because of the difficulty in obtaining larger amounts of fungal RNA from the tiny infected ovaries, we used real‐time PCR to quantify mRNA levels. We used three fungal housekeeping genes with different expression levels as references (tubulin, actin and gpd; see Experimental procedures). As a negative control cDNA from non‐infected rye ears was used. This cDNA was tested for integrity with a specific plant primer pair. No fragments were amplified when using cDNA from non‐infected plant material as a template and primers specific for C. purpurea. Figure 8 shows the result of one series of experiments, using RNA from ovaries 5, 10, 15 and 20 dpi; data were normalized with cptub2 (with actin and gpd as references the same trend for cpnox1 expression was observed, also in a second independent set of experiments). The data clearly show that cpnox1 expression increased during infection, being maximal at the latest stage, i.e. the beginning of sclerotial differentiation. This result corresponds to the observation that Δcpnox1 mutants are mainly impaired in later infection stages.
Figure 8.

cpnox1 expression in planta. Expression of cpnox1 was determined by qRT‐PCR and normalized to the expression of cptub2, a gene encoding β‐tubulin. Expression of cpnox1 was determined 5, 10, 15 and 20 dpi.
As the Δcpnox1 mutant seemed to be more sensitive to oxidative stress, we studied expression of cpnox1 in axenic culture under oxidative stress. As revealed by Northern experiments and quantitative RT‐PCR (Fig. 9), H2O2 induced expression of cpnox1 significantly.
Figure 9.
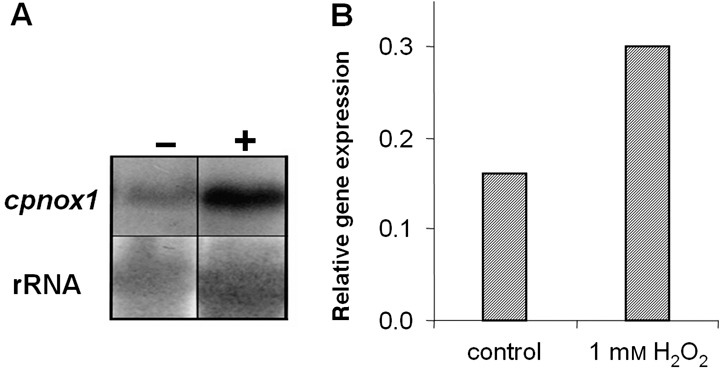
Expression of cpnox1 in axenic culture of the wild‐type strain 20.1 under oxidative stress (H2O2). Samples of mycelia taken from precultures were cultivated in liquid Mantle media for 3 days. Incubation with 1 mm H2O2 was performed for 30 min. Expression of cpnox1 was determined by Northern blot analyses (A) and in another array of samples with qRT‐PCR (B). Northern Blot analyses: +, with H2O2; –, without H2O2. After hybridization with a radioactively labelled probe of cpnox1, the filter was stripped and rehybridized with rDNA from Gibberella fujikuroi as a loading control. qRT‐PCR: cpnox1 expression was normalized to the expression of cptub2, a gene encoding β‐tubulin.
To get a first impression of possible upstream regulatory factors controlling cpnox1 expression, we performed the same experiments with various signalling mutants: Δcptf1 (bZIPtranscription factor controlling catalase activity; Nathues et al., 2004), Δcpmk1/Δcpmk2 (MAP kinases; Mey et al., 2002; Mey and Tudzynski, 2002), Δcphk2 (histidine kinase; Nathues et al., 2007) and Δcpcdc42 (small GTPase; Scheffer et al., 2005); the Δcpnox mutant was used as a control. As shown in Fig. 10, none of these signalling components seems to have a major impact on up‐regulation of cpnox1 by H2O2; the slight increase in the Δcpcdc42 mutant and decrease in the Δcphk2/Δcpmk2 mutants are not significant.
Figure 10.

Expression of cpnox1 in axenic culture of several knockout mutants under oxidative stress (H2O2). Samples of mycelia taken from precultures were cultivated in liquid Mantle media for 3 days. Induction with 1 mm H2O2 was performed for 30 min with (+) and without (–) H2O2. After hybridization with a radioactively labelled probe of cpnox1, filter was stripped and rehybridized with rDNA from Gibberella fujikuroi as a loading control.
DISCUSSION
We have cloned and characterized cpnox1 encoding an NADPH oxidase grouping within the fungal ‘NoxA’ clade according to the nomenclature of Aguirre et al. (2005). Several ‘noxA’ members have been functionally analysed thus far: noxA is the only nox gene in the A. nidulans genome; its deletion leads to blocking of cleistothecia development at an early stage, without affecting vegetative parameters (Lara‐Ortiz et al., 2003). The NoxA homologue of P. anserina, Panox1, is involved in fruit‐body formation as well, but not in vegetative differentiation (Malagnac et al., 2004). NoxA in E. festucae influences the endophytic interaction with its host plant (Tanaka et al., 2006); in M. grisea, NoxA affects appressoria formation (Egan et al., 2007), and in Botrytis cinerea NoxA is involved in sclerotia formation and colonization of host tissue (N. Segmüller et al., unpublished data). Here we show that in the biotrophic phytopathogen C. purpurea deletion of the gene encoding a Nox1 homologue, cpnox1, has a strong impact on pathogenicity and also affects vegetative differentiation: Δcpnox1 mutants produce conidia that have a significantly reduced germination rate. The impact on sexual differentiation cannot be studied in these mutants, because this requires successful colonization of the plant and formation of ripe sclerotia, which were never observed in the Δcpnox1 mutants. The mutants are clearly impaired in directed growth through the transmission tissue, around the ovule towards the vascular tissue. As they cannot reach the phloem and establish a stable host–pathogen interface to gain access to the plant's phloem exudate, they are unable to complete the pathogenic cycle. In the few cases where the colonization process was successful, honeydew production was extremely retarded, and the few sclerotia formed were small and not fully matured. As we used mycelial suspensions for the infection, this defect is not due to the reduced germination rate of conidia. Our data show, for the first time in a biotrophic fungus, that a fungal NADPH oxidase is involved in pathogenic differentiation and therefore represents a virulence factor. As Egan et al. (2007) reported similar results with the hemi‐biotrophic pathogen M. grisea and N. Segmüller et al. (unpublished data) with the necrotroph B. cinerea, this seems to be a general phenomenon.
The expression studies in planta substantiate a role of Cpnox1 in later stages of infection: cpnox1 transcript level in the wild‐type increases during colonization of the plant and is highest in the sclerotia‐forming stage. In axenic culture, the induction by oxidative stress is at first glance surprising, as Cpnox1 probably contributes to the ROS level itself. Interestingly, the same observation was made in B. cinerea (N. Segmüeller et al., unpublished data).
Likewise, the Δcpnox1 mutant's increased sensitivity to menadione is also surprising. Both observations indicate that cpnox1 is directly or indirectly involved in oxidative stress response. Several explanations are possible: as the major function of Nox enzymes could be to produce  in a precisely defined matter with regard to time and place, e.g. for differentiation of cell‐wall structures, it could be speculated that modifications of the cell wall are involved in resistance against oxidative stress. Another explanation could be that the fungus compensates for the loss of Cpnox1 by accelerating other means for the production of superoxide, leading to higher levels of ROS in the mutant and hence increased sensitivity against additional external ROS. The results of the gene deletion experiments with P. anserina Panox1 would support this hypothesis: inactivation of panox1 led to an increased ROS secretion (Malagnac et al., 2004). Similarly, in M. grisea a nox1/nox2 double mutant showed a significant increase in superoxide production during hyphal growth, suggesting an up‐regulation of an alternative source of ROS (Egan et al., 2007). However, our cytological analyses indicated no difference in overall ROS level in the wild‐type and the Δcpnox1 mutant (data not shown). Finally, a possible explanation for the increased sensitivity of the Δcpnox1 mutant against oxidative stress could be the role of NADPH oxidases in oxygen sensing in the way postulated by Geiszt et al. (2000). A sensory function would lead to a deregulation of oxidative stress‐dependent signalling pathways in the mutant and to a failure to induce or activate scavenging enzymes. However, preliminary analyses in our laboratory (catalase activity IEF gels) did not indicate any difference in catalase activity in the Δcpnox1 mutant (data not shown).
in a precisely defined matter with regard to time and place, e.g. for differentiation of cell‐wall structures, it could be speculated that modifications of the cell wall are involved in resistance against oxidative stress. Another explanation could be that the fungus compensates for the loss of Cpnox1 by accelerating other means for the production of superoxide, leading to higher levels of ROS in the mutant and hence increased sensitivity against additional external ROS. The results of the gene deletion experiments with P. anserina Panox1 would support this hypothesis: inactivation of panox1 led to an increased ROS secretion (Malagnac et al., 2004). Similarly, in M. grisea a nox1/nox2 double mutant showed a significant increase in superoxide production during hyphal growth, suggesting an up‐regulation of an alternative source of ROS (Egan et al., 2007). However, our cytological analyses indicated no difference in overall ROS level in the wild‐type and the Δcpnox1 mutant (data not shown). Finally, a possible explanation for the increased sensitivity of the Δcpnox1 mutant against oxidative stress could be the role of NADPH oxidases in oxygen sensing in the way postulated by Geiszt et al. (2000). A sensory function would lead to a deregulation of oxidative stress‐dependent signalling pathways in the mutant and to a failure to induce or activate scavenging enzymes. However, preliminary analyses in our laboratory (catalase activity IEF gels) did not indicate any difference in catalase activity in the Δcpnox1 mutant (data not shown).
The regulatory network establishing and maintaining ROS homeostasis is clearly rather complex. To analyse possible upstream components involved in regulation of cpnox1, Northern analyses with various signalling mutants of C. purpurea were performed. These analyses gave no hints that the respective cascades are significantly involved in upregulation of cpnox1 by H2O2. However, recent detailed analyses of target genes of the C. purpurea Rac homologue Cprac (a small GTP‐binding protein) indicated that expression of cpnox1 is controlled by a Rac‐dependent pathway (Y. Rolke and P. Tudzynski, unpublished data). This is consistent with the findings of Takemoto et al. (2006), who demonstrated that for the activation of the Cpnox1 homologue NoxA, a p67phox homologue NoxR as well as RacA are necessary (at the level of protein–protein interaction), as is known for higher eukaryotes (for a review see, for example, Bokoch and Diebold, 2002). Another possible candidate involved in cpnox1 regulation may be a homologue of the stress‐activated MAP kinase Sak1 of A. nidulans, which controls noxA expression (Lara‐Ortiz et al., 2003). To test this hypothesis will require the generation of a sak1 deletion mutant of C. purpurea.
The phenotype of the Δcpnox1 mutants in planta is similar to that of two other mutants in C. purpurea: Δcphk2 mutants, lacking a histidine kinase (involved in ROS sensing and also defective in conidia germination: Nathues et al., 2007), and Δcpcdc42 mutants impaired in GTPase signalling (Scheffer et al., 2005). Both mutants can also penetrate the host epidermis, but are affected in colonization of host tissue. In contrast to Δcpnox1, they never produce any macroscopic signs of infection, neither honeydew nor sclerotia. The invasive growth of the Δcpcdc42 mutants was arrested in the stylar tissue, which seems to be accompanied by an oxidative‐burst‐like reaction of the plant that is never observed in the wild‐type strain. The high local ROS concentration observed in rye ovarian tissue infected by the Δcpcdc42 mutant could also be explained by a higher ROS production by the fungus, probably because Cprac is now no longer balanced by its counterpart Cpcdc42 (Scheffer et al., 2005).
Transmission electron microscopic analyses (using the CeCl2 technique for detection of H2O2) gave no indications of a different ROS status of the Δcpnox1 mutant in planta, in contrast to Δcpcdc42 (data not shown). The stop or retardation of infection hyphae could be due to a defect in a specific differentiation step necessary for complete and rapid colonization of that tissue. A role for ROS generated by Nox enzymes in cell‐wall biosynthesis was also discussed by Malagnac et al. (2004) and Egan et al. (2007). The defect in conidia germination in C. purpurea therefore could also be due to a disturbed ROS balance.
Tanaka et al. (2006) were the first to confirm the assumption that the ROS status is important in fungus–plant interaction systems. They analysed a ΔnoxA mutant of the endophyte Epichloe festucae, a relative of C. purpurea, which—in contrast to the wild‐type—is no longer able to establish a stable mutualistic interaction, and causes disease symptoms in its host plant. Interestingly, the mutant lacks an H2O2 layer present around the wild‐type hyphae in planta, it branches far more intensively and produces more biomass compared with the wild‐type, thereby disturbing the balance between fungus and host, which is necessary for endosymbiosis. The mutant was shown to be tagged in the noxA gene, demonstrating that ROS level is important for this special interaction system. Obviously in E. festucae external H2O2 (produced by the Nox system) is essential for endosymbiosis; by contrast, in C. purpurea Cpnox1 is necessary for pathogenic development. In Ustilago maydis (which—like yeasts—lacks nox genes) the importance of ROS generation for pathogenesis has recently been demonstrated by deletion of an H2O2‐producing glyoxal oxidase (Leuthner et al., 2005).
The mechanisms linking differentiation and ROS status (mediated, for example, by Nox) are not clear. An interesting contribution to this topic was made by Chen and Dickman (2005). They provided evidence that a dominant active allele of the ras homologue in Colletotrichum trifolii caused elevated levels of ROS accompanied by abnormal hyphal growth and development and eventual apoptotic‐like cell death. Interestingly, this effect could be suppressed by proline, indicating that this amino acid plays a central role in the oxidative stress control. As apoptosis‐like processes are involved in many differentiation programmes in higher eukaryotes (e.g. Lam, 2004), they could represent one major link between ROS status and differentiation. Recently, Veneault‐Fourrey et al. (2006) presented evidence that autophagic fungal cell death (degeneration of nuclei) is a prerequisite for appressoria formation and penetration in M. grisea, giving strong support to the idea that these processes are also important for pathogenic development.
The mechanisms underlying the regulation, expression and/or activity of cpnox1 in C. purpurea may well be similar to those in E. festucae and a disturbed ROS homeostasis may lead to a lack of controlling fungal cell death. This might be one of the major differentiation problems of the Δcpnox1 mutant, which is reflected in a strongly disturbed infection pattern and the inability to form mature sclerotia.
EXPERIMENTAL PROCEDURES
Strains, media and growth conditions
The wild‐type Claviceps purpurea strain used in these experiments was 20.1, a putative haploid derivative of standard field isolate T5 (Fr.:Fr.) Tul., isolated from rye (Secale cereale L.; Hohenheim, Germany), and obtained by benomyl treatment (Hüsgen et al., 1999). For conidia harvesting and DNA isolation, mycelia were cultivated on Mantle agar (16 g/L agar) with 100 g/L sucrose (Mantle and Nisbet, 1976) at 28 °C for 12–14 days.
To test oxidative stress, Mantle agar was supplemented with 200 µm and 500 µm menadione. To test osmotic stress, media were supplemented with 500 µm sorbitol. For each concentration five agar plates were inoculated with the mutant Δcpnox1‐1 and the wild‐type 20.1 and grown for 12 days. To ensure the same amount of inoculum, mycelium 5 mm in diameter was applied. As a control, both strains were grown on Mantle agar without menadione. Escherichia coli strains TOP10F′ (Invitrogen) and LE392 (Stratagene) were used for all the subcloning experiments and for propagation of C. purpurea genomic lambda clones, respectively.
Nucleic acid analysis
Standard recombinant DNA methods were performed according to Sambrook et al. (1989) and Ausubel et al. (1987). Genomic DNA from C. purpurea was prepared from lyophilized mycelium according to Cenis (1992). For Southern blot analysis, 5–10 µg of restriction‐digested chromosomal DNA or PCR products were electrophoresed in 0.8–1.6% agarose gels with salt‐free buffer (Sambrook et al., 1989), blotted onto positively charged nylon filters (Hybond N+; Amersham, Braunschweig, Germany), and hybridized to radioactively labelled DNA probes in Denhardt's hybridization solution (Sambrook et al., 1989). Filters were washed for 10 min in 2× SSC (1× SSC is 0.15 m NaCl plus 0.015 m sodium citrate), 0.1% sodium dodecyl sulfate (SDS) and for 10 min in 1× SSC, 0.1% SDS. Hybridization and washing of the filters were carried out at 65 °C. DNA sequencing was carried out as described by Moore et al. (2002). Protein and DNA sequence alignment, editing, and organization were done with DNA Star (Madison, WI). Sequence analysis was done using BLAST at the National Center for Biotechnology Information (Bethesda, MD) (Altschul et al., 1990). PCR was done as described by Sambrook et al. (1989), using the Red Taq Polymerase (Sigma, Milwaukee, WI). All primers were synthesized by MWG‐Biotech (München, Germany). The amplification products were cloned with the PCR 2.1 TOPO‐Cloning Kit from Invitrogen.
Expression studies and quantitative RT‐PCR
For Northern analyses samples of mycelia taken from precultures were cultivated for 3 days in Mantle liquid medium. Induction with 1 mm H2O2 was performed for 30 min. Total RNA for Northern blotting or RT‐PCR was isolated using the RNAgents total RNA isolation system from Promega (Madison, WI). The blotted RNA was hybridized with a 1.42‐kb radioactively labelled probe of cpnox1 and subsequently stripped and rehybridized with rDNA from Gibberella fujikuroi as a loading control.
qRT‐PCR reactions were performed using cDNA from total RNA extracts of mycelium grown in Mantle liquid medium or from infected rye ovaries. For the reactions, the BioRad iQ SYBR Green Supermix and the iCycler Thermal Cycler (BioRad, CA) were used. Programming, data collection and analyses were performed with the iCycler iQ Real‐Time Detection System Software Version 3.0 (BioRad). Expression of cpnox1 was detected by the primers Cpnox uni (5′AGATTCATACCTATCTCACCCAG3′) and Cpnox rev (5′CGTCCTTGACCTTAGTTCAG3′). To normalize the expression of cpnox1 to the expression of genes encoding β‐tubulin, actin and Glyceraldehyde‐3‐phosphate dehydrogenase, the following primers were used, respectively: Tub uni (5′TACAATGGTACCTCGGAGCAAC3′) and Tub rev (5′CCAGAGGCCTCATTGAAGTAGAC3′), Actin uni (5′GCCGTTTTCCCCTCTATCGTC3′) and Actin rev (5′ACATACGAGTCCTTCTGACCCAT3′), Gpd uni (5′CCCGAATATGCTGCCTACATGCT3′) and Gpd rev (5′CGTCCTTCTTGATCTCGCCCT3′). After qRT‐PCR, products were monitored by melt curve analyses and gel electrophoresis. As a negative control, cDNA of non‐infected rye ovaries was used. This cDNA was tested for integrity by using the plant‐specific primers encoding a glyceraldehyde‐3‐phosphate dehydrogenase scGAPDH uni (5′TCTTTCTGATGATGGGTTGAG3′) and scGAPDH rev (5′ACCTTCTCCACTGTCATAGG3′).
Cloning of cpnox1 and generation of replacement and complementation vectors
A 668‐bp internal fragment of cpnox1 was amplified by PCR using the degenerate primers Nox R3 (5′GATYTCGTTCTCRAAGACGTC3′) and NoxF3 (5′GAGACCTTYTGGTACACTCAYC3′) that were deduced from the Nox1 homologous regions of Podospora anserina (AAK50853), Neurospora crassa (AF364817) and Aspergillus nidulans (EAA62617). The amplified product showed high homology to a variety of NADPH oxidases of various origins (fungal, animal and plant). Using this fragment as a probe a genomic library of C. purpurea strain T5 (Smit and Tudzynski, 1992) was screened by plaque filter hybridization (Sambrook et al., 1989). From the 40 000 lambda clones that were screened, ten hybridized with the PCR fragment used as a probe, four of which were further purified and analysed. They all contain an overlapping genomic region as revealed by restriction and Southern blot analysis. From phage 2‐2‐1, a 4.5‐kb SalI fragment and a 3‐kb EcoRI fragment were cloned into pUC19 giving rise to the plasmids p10S and p1E2, respectively. Both plasmids carry the overlapping and whole sequence of cpnox1, and were completely sequenced.
For construction of the gene replacement vector pΔcpnox1 primers Nox1Rep1a and Nox1Rep1b (A and B in Fig. 2) were used to amplify a 1362‐bp fragment of the cpnox1 genomic region containing part of the promoter and coding region of cpnox1. The two primers contained artifical restriction sites ClaI and PstI, respectively. The PCR fragment obtained was cloned into the PCR 2.1 TOPO vector and subsequently excised with ClaI and PstI. This fragment was introduced into the ClaI/PstI restricted phleomycin resistance cassette containing plasmid PAN8‐1UM (Müller et al., 1997). Primers Nox1Rep2a and Nox1rep2b (C and D in Fig. 2) containing artificial XbaI sites amplified a 1087‐bp fragment, which was cloned via the PCR 2.1 TOPO vector into the XbaI site at the 3′ end of the phleomycin resistance cassette of pAN8‐1UM. Orientation of the second flank was determined by sequencing. Between primers Nox1rep1b and Nox1rep2a lies a 160‐bp genomic region, which has been replaced by the 3320‐bp phleomycin resistance cassette in the vector pΔcpnox1. Digestion of pΔcpnox1 with ClaI and SacII releases a 5769‐bp fragment, which was used to transform the C. purpurea wild‐type strain 20.1. Primer sequences are as follows: Nox1Rep1a: 5′GTCTCATCGATGGGCCAGTCACAGGTGC3′; Nox1Rep1b: 5′GCCTAGATAACCTCTGCAGTGTTTCCAG3′; Nox1Rep2a: 5′GTGGCTGTTTCTCTAGATGCCTTCG3′ and Nox1rep2b: 5′CCGTGTCTAGAACAGTGCTGGACCTG3′.
To obtain a full‐length clone of cpnox1 for complementation of the mutant strain Δcpnox1‐1, a 792‐bp SalI/KpnI fragment of p1E‐2 and a 4.58‐kb SalI fragment of p10S were fused into the plasmid pBluescript. The region spanning the ligation sites used for fusing the two fragments was sequenced to verify no changes were introduced during the ligation procedure. Subsequently, the complementation vector cpnox1‐komp was obtained by cloning a 5378‐bp ClaI/KpnI fragment into the corresponding restriction sites of the vector pAN7‐1UM, carrying a resistance against hygromycin. The plasmid obtained carries the coding region of cpnox1 as well as 837 bp of the promotor and 2611 bp of the terminator region and was used to transform C. purpurea cpnox1 mutant strain.
Fungal transformation
Protoplasts of C. purpurea generated with lysing enzymes from Trichoderma harzianum and Driselase (InterSpex) were transformed with 10 µg of the Δcpnox1 fragment (see Fig. 12) as described by Jungehülsing et al. (1994). For phleomycin selection, phleomycin was directly applied to the protoplasts to a final concentration of 33 µg/mL modified BII medium (pH 8, 20% sucrose, no FeSO4). Resistant colonies were transferred to fresh selective medium (BII pH 8 plus 100 µg/mL phleomycin) and subjected to at least one round of single spore isolation to obtain homokaryotic transformed strains. To identify homologous integration events, PCR was performed using primer combinations as follows. To amplify the left integration flank primers F (5′CTCACCAGAAGGACATTTTAGCC3′) and G (5′GTTACGCCGTCTGACTTTTGTGGT3′) were used. To analyse the right integration flank primers H (5′TCCGGCGAAGAGAAGAATAGC3′) and I (5′CGTTTCTGTCTAATTAAAGCAGAG3′) were used. The lack of the wild‐type gene copy in the Δcpnox1 mutants was checked using the primers E (5′CGATACAGCCGCTCGACCAAGTAC3′) and F (5′CTCACCAGAAGGACATTTTAGCC3′) (Fig. 12). In order to ensure that only one copy of the replacement fragment has integrated into the genome of 20.1 as well as to confirm the deletion of cpnox1, Southern analysis was performed. EcoRI‐digested genomic DNA of strains 20.1, Δcpnox1‐1 and Δcpnox1‐2 was probed with an EcoRI/SalI fragment containing the 5′ part of cpnox1 as indicated in Fig. 12.
For complementation of the Δcpnox1 mutant, Δcpnox1‐1 was transformed with the circular cpnox1‐komp construct carrying the hygromycin resistance gene as a selective marker. For hygromycin selection, protoplasts were incubated at 28 °C for 24 h, after which they were overlaid with 10 mL BII medium, pH 8, containing 1.5 mg/mL hygromycin to reach a final hygromycin concentration of 0.5 mg/mL in the Petri dishes. Hygromycin‐resistant transformants were purified by single spore isolation and checked for reintegration of cpnox1 by PCR and Southern blot analysis.
Pathogenicity tests
Rye plants were cultivated in growth chambers as described by Smit and Tudzynski (1992). Florets of blooming ears (30–40 per ear) were inoculated with 5 µL of a suspension containing 2 × 106/mL conidia collected from Mantle agar, as described by Tenberge et al. (1996). To avoid cross contamination, the ears were covered with paper bags equipped with cellophane windows directly after inoculation.
Microscopic analyses
For microscopic analyses rye pistils were isolated from rye ears 6 dpi and stained with KOH‐aniline blue as described by Scheffer and Tudzynski (2006).
The infected ovaries were observed using epifluorescence microscopy (Microscope Leica DMRBE with PixelFly Digital camera (PCO Computer Optics GmbH), filter block A (BP 340–380, RKP 400, LT 425, UV light 340–380 nm). Embedding, sectioning and conventional light microscopic analyses were performed as described in Oeser et al. (2002); detection of H2O2 (using the cerium chloride technique, Bestwick et al., 1997) was done according to Nathues et al. (2004).
ACKNOWLEDGEMENTS
We thank S. Gergs for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (DFG; Tu 50/14).
REFERENCES
- Aguirre, J. , Ríos‐Momberg, M. , Hewitt, D. and Hansberg, W. (2005) Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13, 111–118. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. , Brent, R. , Kingston, R.E. , Moore, D.D. , Seidmann, J.G. , Smith, J.A. and Struhl, K. (1987) Current Protocols in Molecular Biology. New York: John Wiley and Sons. [Google Scholar]
- Bestwick, C.S. , Brown, I.R. , Bennett, M.H.R. and Mansfield, J.W. (1997) Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola . Plant Cell, 9, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch, G.M. , and Diebold, B.A. (2002) Current molecular models for NADPH oxidase regulation by Rac GTPase. Blood, 100, 2692–2696. [DOI] [PubMed] [Google Scholar]
- Cenis, J.L. (1992) Rapid extraction of fungal DNA for PCR amplification. Nucleic Acid Res. 20, 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.B. and Dickman, M.B. (2005) Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc. Natl Acad. Sci. USA, 102, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, M.J. , Wang, Z.Y. , Jones, M.A. , Smimoff, N. and Talbot, N.J. (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl Acad. Sci. USA, 104, 11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiszt, M. , Kopp, J.B. , Varnai, P. and Leto, T.L. (2000) Identification of renox, an NAD(P)H oxidase in kidney. Proc. Natl Acad. Sci. USA, 97, 8010–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansberg, W. and Aguirre, J. (1990) Hyperoxidant states cause microbial cell differentiation by cell isolation from dioxygen. J. Theor. Biol. 142, 201–221. [DOI] [PubMed] [Google Scholar]
- Hüsgen, U. , Büttner, P. , Müller, U. and Tudzynski, P. (1999) Variation in karyotype and ploidy level among field isolates of Claviceps purpurea . J. Phytopathol. 147, 591–597. [Google Scholar]
- Jungehülsing, U. , Arntz, C. , Smit, R. and Tudzynski, P. (1994) The Claviceps purpurea glyceraldehyde‐3‐phosphate dehydrogenase gene: cloning, characterization and use for improvement of a dominant selection system. Curr. Genet. 25, 101–106. [DOI] [PubMed] [Google Scholar]
- Lam, E. (2004) Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5, 305–315. [DOI] [PubMed] [Google Scholar]
- Lara‐Ortiz, T. , Riveros‐Rosas, H. and Aguirre, J. (2003) Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans . Mol. Microbiol. 50, 1241–1255. [DOI] [PubMed] [Google Scholar]
- Leuthner, B. , Aichinger, C. , Oehmen, E. , Koopmann, E. , Müller, O. , Müller, P. , Kahmann, R. , Bölker, M. and Schreier, P.H. (2005) A H2O2‐producing glyoxal oxidase is required for filamentous growth and pathogenicity in Ustilago maydis . Mol. Gen. Genomics, 272, 639–650. [DOI] [PubMed] [Google Scholar]
- Malagnac, F. , Lalucque, H. , Lepère, G. and Silar, P. (2004) Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina . Fungal Genet. Biol. 41, 982–997. [DOI] [PubMed] [Google Scholar]
- Mantle, P.G. and Nisbet, L.J. (1976) Differentiation of Claviceps purpurea in axenic culture. J. Gen. Microbiol. 93, 321–334. [DOI] [PubMed] [Google Scholar]
- Mey, G. , Oeser, B. , Lebrun, M.H. and Tudzynski, P. (2002) The biotrophic, non‐appressoria forming grass pathogen Claviceps purpurea needs a Fus3/Pmk1 homologous mitogen‐activated protein (MAP) kinase for colonization of rye ovarian tissue. Mol. Plant–Microbe Interact. 15, 303–312. [DOI] [PubMed] [Google Scholar]
- Mey, G. and Tudzynski, P. (2002) CPMK2, an Slt2‐homologous MAP‐kinase is essential for pathogenesis of Claviceps purpurea on rye: evidence for a second conserved pathogenesis‐related MAP‐kinase cascade in phytopathogenic fungi. Mol. Microbiol. 46, 305–318. [DOI] [PubMed] [Google Scholar]
- Moore, S. , De Vries, O. and Tudzynski, P. (2002) The major Cu, Zn SOD of the phytopathogen Claviceps purpurea is not essential for pathogenicity. Mol. Plant Pathol. 3, 9–22. [DOI] [PubMed] [Google Scholar]
- Müller, U. , Tenberge, K.B , Oeser, B. and Tudzynski, P. (1997) Cel1, probably encoding a cellobiohydrolase lacking the substrate binding domain, is expressed in the initial infection phase of Claviceps purpurea on Secale cereale . Mol. Plant–Microbe Interact. 10, 268–279. [DOI] [PubMed] [Google Scholar]
- Nathues, E. , Jörgens, C. , Lorenz, N. and Tudzynski, P. (2007) The fungal histidin kinase CpHK2 has impact on spore germination, oxidative stress and fungicide resistance, and virulence of the ergot fungus Claviceps purpurea . Mol. Plant Pathol. 8, 653–665. [DOI] [PubMed] [Google Scholar]
- Nathues, E. , Joshi, S. , Tenberge, K.B. , Von Den Driesch, M. , Oeser, B. , Bäumer, N. , Mihlan, M. and Tudzynski, P. (2004) CPTF1 a CREB‐like transcription factor is involved in the oxidative stress response in the phytopathogen Claviceps purpurea and modulates ROS level in its host Secale cereale . Mol. Plant–Microbe Interact. 17, 383–393. [DOI] [PubMed] [Google Scholar]
- Oeser, B. , Heidrich, P. , Müller, U. , Tenberge, K.B. and Tudzynski, P. (2002) Polygalacturonase is a pathogenicity factor in the Claviceps purpurea/rye interaction. Fungal Genet. Biol. 36, 176–186. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scheffer, J. , Chen, C. , Heidrich, P. , Dickman, M.B. and Tudzynski, P. (2005) A CDC42 homologue in C. purpurea is involved in vegetative differentiation and is essential for pathogenicity. Eukaryot. Cell, 4, 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer, J. and Tudzynski, P. (2006) In vitro pathogenicity assay for the ergot fungus Claviceps purpurea . Mycol. Res. 110, 465–470. [DOI] [PubMed] [Google Scholar]
- Smit, R. and Tudzynski, P. (1992) Efficient transformation of Claviceps purpurea using pyrimidine auxotrophic mutants: cloning of the OMP decarboxylase gene. Mol. Gen. Genet. 234, 297–305. [DOI] [PubMed] [Google Scholar]
- Takemoto, D. , Tanaka, A. and Scott, B. (2006) A p67Phox‐like regulator is recruited to control hyphal branching in a fungal‐grass mutualistic symbiosis. The Plant Cell, 18, 2807–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, P. , Tanaka, A. and Scott, B. (2007) NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet. Biol. 44, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Tanaka, A. , Christensen, M.J. , Takemoto, D. , Park, P. and Scott, B. (2006) Reactive oxygen species play a role in regulating a fungus‐perennial ryegrass mutualistic interaction. The Plant Cell, 18, 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenberge, K.B. , Homann, V. , Oeser, B. and Tudzynski, P. (1996) Structure and expression of two polygalacturonase genes of Claviceps purpurea oriented in tandem and cytological evidence for pectinolytic enzyme activity during infection of rye. Phytopathology, 86, 1084–1097. [Google Scholar]
- Tudzynski, P. , Correia, T. and Keller, U. (2001) Biotechnology and genetics of ergot alkaloids. Appl. Microbiol. Biotechnol. 57, 593–605. [DOI] [PubMed] [Google Scholar]
- Tudzynski, P. and Scheffer, J. (2004) Claviceps purpurea: molecular aspects of a unique pathogenic lifestyle. Mol. Plant Pathol. 5, 377–388. [DOI] [PubMed] [Google Scholar]
- Veneault‐Fourrey, C. , Barooah, M. , Egan, M. , Wakley, G. and Talbot, N.J. (2006) Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science, 312, 580–583. [DOI] [PubMed] [Google Scholar]


