SUMMARY
Taxonomy: Kingdom Fungi; Phylum Ascomycota; Class Sordariomycetes; Order Hypocreales; Family Nectriaceae; genus Fusarium.
Host range: Very broad at the species level. More than 120 different formae speciales have been identified based on specificity to host species belonging to a wide range of plant families.
Disease symptoms: Initial symptoms of vascular wilt include vein clearing and leaf epinasty, followed by stunting, yellowing of the lower leaves, progressive wilting, defoliation and, finally, death of the plant. On fungal colonization, the vascular tissue turns brown, which is clearly visible in cross‐sections of the stem. Some formae speciales are not primarily vascular pathogens, but cause foot and root rot or bulb rot.
Economic importance: Can cause severe losses in many vegetables and flowers, field crops, such as cotton, and plantation crops, such as banana, date palm and oil palm.
Control: Use of resistant varieties is the only practical measure for controlling the disease in the field. In glasshouses, soil sterilization can be performed.
Useful websites: http://www.broad.mit.edu/annotation/genome/fusarium_group/MultiHome.html; http://www.fgsc.net/Fusarium/fushome.htm; http://www.phi‐base.org/query.php
INTRODUCTION
The genus Fusarium, also known by its teleomorphs Nectria and Gibberella, harbours notorious plant pathogenic fungi with a wide variety of hosts and infection strategies (Desjardins, 2003; Di Pietro et al., 2003; Goswami and Kistler, 2004). Fusarium oxysporum stands out in several ways. It has an apparent long history of predominant, perhaps exclusive, asexual reproduction (Gordon and Martyn, 1997), it invades roots and can cause wilt diseases through colonization of xylem tissue (Tjamos and Beckman, 1989), and it displays apparent gene‐for‐gene relationships with several hosts. Possibly related to these gene‐for‐gene relationships is the large number of host‐specific forms, called formae speciales, within F. oxysporum (Armstrong and Armstrong, 1981; Katan, 1999; Katan and Di Primo, 1999).
Because of its (predominant) asexual reproduction, F. oxysporum is generally regarded as a ‘species complex’—a collection of clonal lines within the genus Fusarium. As such, it constitutes a sister group to the Fusarium fujikuroi species complex that harbours sexual species with Gibberella teleomorphs, such as F. verticillioides and F. fujikuroi (Guadet et al., 1989). In 2007, the genome of an F. oxysporum isolate was sequenced by the Broad Institute (http://www.broad.mit.edu/annotation/genome/fusarium_group/MultiHome.html). Ninety per cent of the genome sequence of F. oxysporum can be aligned to that of F. verticillioides, and the average DNA sequence similarity in the aligned region is 90% (L.‐J. Ma, Broad Institute, Cambridge, USA, personal communication). Each forma specialis of F. oxysporum consists of either one or several clonal lines which correspond to vegetative compatibility groups (VCGs) (Gordon and Martyn, 1997; Katan, 1999; Katan and Di Primo, 1999). When a forma specialis harbours several clonal lines, these are generally not monophyletic, giving the appearance that pathogenicity towards a particular plant species has arisen independently several times (Baayen et al., 2000; O'Donnell et al., 1998). Although the focus of most plant pathologists is, naturally, on pathogenic isolates, many isolates found in soil are in fact not pathogenic (Gordon and Martyn, 1997; Recorbet et al., 2003), and a major question is whether these ubiquitous non‐pathogenic forms can evolve to become pathogenic and, if so, how that happens.
Although F. oxysporum has been found to cause disease in a large number of plant species (with grasses as an outstanding exception), the study of the molecular basis of the pathogenicity of F. oxysporum has necessarily involved a limited number of hosts, mainly tomato, melon, bean, banana, cotton, chickpea and, more recently, Arabidopsis thaliana. This review is an update to the pathogen profile on F. oxysporum by Di Pietro et al. (2003), and focuses mainly on new discoveries in the period 2003–2008. New developments in that period primarily involve the discovery and molecular analysis of novel pathogenicity genes, genome sequence analysis, transcriptomics, xylem sap proteomics, avirulence/effector proteins, visualization of the fungus during plant colonization using fluorescent proteins and the development of A. thaliana as a model host plant.
VISUALIZATION OF COLONIZATION USING FLUORESCENT PROTEINS
The process of infection by F. oxysporum has been studied using light, fluorescence and electron microscopy, and can be divided into several steps: root recognition, root surface attachment and colonization, penetration and colonization of the root cortex and, in the case of wilt‐inducing formae speciales, hyphal proliferation within the xylem vessels (reviewed in Di Pietro et al., 2003). The first microscopic studies involving fluorescent protein‐labelled F. oxysporum were by Di Pietro et al. (2001b) and Lagopodi et al. (2002). Di Pietro et al. observed conidial germination on roots, growth in tomato root cortex and colonization of xylem by F. oxysporum f. sp. lycopersici, and found that the fmk1 mutant was able to germinate on roots but did not develop any further (Di Pietro et al., 2001a). Lagopodi et al. visualized the colonization of tomato roots by the foot and root rot‐causing F. oxysporum f. sp. radicis‐lycopersici expressing the fluorescent marker green fluorescent protein (GFP) using confocal laser scanning microscopy (Lagopodi et al., 2002). This study revealed several aspects of tomato root colonization and infection by this forma specialis: (i) the first steps of contact between the fungus and the host take place at the root hair zone; (ii) the grooves along the junctions of epidermal cells are the preferential colonization sites; and (iii) there are no specific penetration sites or infection structures (Lagopodi et al., 2002).
In general, a similar pattern of colonization was observed for GFP‐labelled F. oxysporum f. sp. lycopersici colonizing tomato plants in a vermiculite system (R. G. Duyvesteijn and M. Rep, unpublished results). In addition, in this study, no specific penetration sites were observed and hyphae preferentially grew along the grooves of epidermal root cells. A novel finding was the observation of chlamydospores (thick‐walled, ‘survival’ spores) on the root surface, and also within the vascular tissue at a later stage of infection (Fig. 1). In studies using GFP‐labelled F. oxysporum, neither conidiophores nor microconidia have been observed in xylem vessels of Arabidopsis or tomato (Czymmek et al., 2007; van der Does et al., 2008a). This is in apparent disagreement with a model in which microconidia play an important role in the colonization of xylem tissue (Beckman, 1987). A minor role of microconidia in xylem colonization is also supported by a study of Ren1, a putative transcription factor required for micro‐ and macroconidia development. A REN1 disruptant produces only chlamydospores and abnormal rod‐shaped, conidium‐like cells, but is not affected in pathogenesis, suggesting that microconidia and macroconidia are not required for pathogenicity (Ohara et al., 2004).
Figure 1.
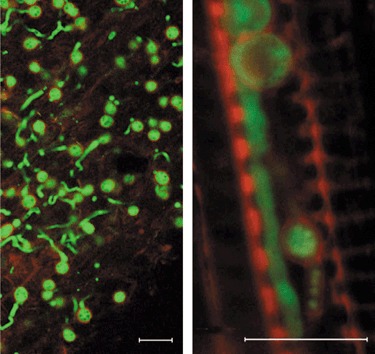
Confocal images of chlamydospores of green fluorescent protein (GFP)‐labelled Fusarium oxysporum f. sp. lycopersici on and in tomato roots. Left: outside of a tomato root (7 days post‐inoculation). Right: in a xylem vessel (22 days post‐inoculation). Note the red autofluorescent cell walls of the chlamydospores. Scale bars, 20 µm.
To understand the biocontrol mechanisms involving non‐pathogenic isolates of F. oxysporum, the interactions between different fluorescently labelled pathogenic and non‐pathogenic F. oxysporum strains during root colonization have also been studied (Bolwerk et al., 2005; Nahalkova et al., 2008; Olivain et al., 2006). Olivain et al. (2006) showed that the general pattern of colonization in soil was similar for both a pathogenic strain (F. oxysporum f. sp. lycopersici) and a non‐pathogenic, biocontrol strain (Fo47). However, the biocontrol strain grew faster than the pathogenic strain and, as a consequence, colonized the roots earlier. When the strains were co‐inoculated, the biocontrol strain again colonized the roots more extensively, but never excluded the pathogenic strain (Olivain et al., 2006). Similar results were obtained by Bolwerk et al. (2005), who studied the interaction between the biocontrol strain Fo47 and F. oxysporum f. sp. radicis‐lycopersici in sand. The biocontrol strain germinated faster than the pathogenic strain on artificial medium containing root exudates. The biocontrol strain reduced the root colonization of the pathogenic strain, but did not exclude it completely from the roots.
Nevertheless, there are some contradictory observations between these studies. Bolwerk et al. (2005) reported that the pathogenic strain mainly targeted cellular junctions on roots, similar to the observations reported by Lagopodi et al. (2002), who used the same forma specialis and inoculation system for colonization studies. This type of colonization behaviour was specific for the pathogenic strain and was not observed for the biocontrol strain (Bolwerk et al., 2005). In contrast, Olivain et al. (2006) did not observe any preferential growth of hyphae along intercellular junctions for either the pathogenic or non‐pathogenic strain. This difference may be a result of the different behaviour of the two formae speciales or of the experimental design (sand versus soil).
The same pathogenic and non‐pathogenic strains used in the study by Olivain et al. were also used to observe root colonization in hydroponic cultures (Nahalkova et al., 2008). In this study, no preferential growth of hyphae along cellular junctions was observed. Remarkably, in contrast with the observations of colonization in soil, in hydroponic cultures, the pathogenic strain appeared to be a better and faster colonizer than the biocontrol strain. The patterns of colonization also differed; in the hydroponic system, the elongation zone and root apex were fully colonized—an observation that was never made in soil (Nahalkova et al., 2008; Olivain et al., 2006). Clearly, it should be appreciated that the outcome of colonization studies can be influenced by culture (soil, sand, hydroponic), inoculum concentrations and ratios; moreover, we cannot exclude the potential differences in behaviour between the various formae speciales. Nevertheless, from the biocontrol studies, it can be concluded that biocontrol by non‐pathogenic F. oxysporum does not operate through direct antagonism or exclusion from potential penetration sites, but rather by competition for nutrients and/or induction of defence responses in roots (Nahalkova et al., 2008).
Another recent study has revealed the dynamics of colonization of Arabidopsis roots by F. oxysporum expressing the fluorescent protein ZsGreen. Colonization was recorded in time and space using time‐lapse microscopy and coverglass chambers with an agar‐based medium (Czymmek et al., 2007). With regard to the dynamics of colonization, it was observed, for instance, that the fungal growth rate in vascular tissue was up to five times faster than that in the surrounding agar prior to initial penetration of the root. Initial colonization and penetration into the vascular system occurred primarily in the meristematic zone of the root tips and emerging lateral roots, and local development of a mycelial mass appeared to be a precondition for the initiation of the penetration process. As with some studies with tomato, fungal growth on the root surface was preferentially along anticlinal walls. Interestingly, in this study, at high magnification, localized hyphal swellings were observed at the anticlinal interface. These may represent rudimentary appressorium‐like penetration structures (Czymmek et al., 2007). In tomato, we have also observed swelling of hyphae at sites of penetration (R. G. Duyvesteijn and M. Rep, unpublished results). After penetration, fungal growth was initially intercellular but, ultimately, became intracellular, and the collapse of plant cells was observed at sites of fungal penetration, presumably as a result of a loss of turgor pressure. Furthermore, changes were observed in plant cells that were not in direct contact with mycelium, such as the loss of autofluorescent vacuolar contents and changes in the appearance of the endoplasmic reticulum (Czymmek et al., 2007).
SIGNAL TRANSDUCTION
Using forward and reverse genetics approaches, several processes and genes have been identified or implicated as being important for pathogenesis (Table 1). It may be no surprise that signal transduction processes are among these, as, in order to establish disease, fungi need to respond appropriately to the plant environment and adjust their morphology, physiology, metabolism and spectrum of secreted proteins and metabolites. In F. oxysporum, cyclic adenosine monophosphate–protein kinase A (cAMP–PKA) and mitogen‐activated protein kinase (MAPK) cascades have been implicated in this adaptation; MAPK (FMK1) and G‐protein subunits α (FGA1) and β (FGB1) are required for (full) pathogenicity (reviewed in Di Pietro et al., 2003). More recently, a second gene encoding a G‐protein α subunit (FGA2) has been identified and analysed. In contrast with the reduced pathogenicity of F. oxysporum f. sp. cucumerinum FGA1 disruptants, deletion of FGA2 led to a complete loss of pathogenicity on cucumber plants (Jain et al., 2005). In addition, the altered colony morphology and conidiation observed in FGA1 disruptants were not observed in FGA2 disruptants. These observations indicate that Fga1 and Fga2 have distinct functions, each being required for adaptation to the plant environment.
Table 1.
Gene knock‐outs in Fusarium oxysporum and their phenotypic effects.
| Gene | Product/function | Effect of gene inactivation/deletion | Reference |
|---|---|---|---|
| ARG1 | Argininosuccinate lyase | Strongly reduced virulence, arginine auxotrophy | Namiki et al. (2001) |
| CHS1 | Class I chitin synthase | Fully virulent | Martin‐Urdiroz et al. (2008) |
| CHS2 | Class II chitin synthase | Reduced virulence | Martin‐Urdiroz et al. (2008) |
| CHS7 | Chaperone‐like protein | Reduced virulence | Martin‐Urdiroz et al. (2008) |
| CHSV | Class V chitin synthase | Strongly reduced virulence, hypersensitive to α‐tomatine and H2O2 | Madrid et al. (2003) |
| CHSVb | Class VII chitin synthase | Non‐pathogenic, hypersensitive to Congo red and Calcofluor white | Martin‐Urdiroz et al. (2008) |
| CMLE1 | Carboxy‐cis,cis‐muconate cyclase | Non‐pathogenic, reduced growth on phenolic compounds | L. Reijnen and C. B. Michielse (unpublished results) |
| CLC1 | Chloride channel | Reduced virulence, deficient in laccase activity, increased sensitivity to oxidative stress | Canero and Roncero (2008b) |
| CTF1 | Transcription factor | Fully virulent | Rocha et al. (2008) |
| CTI6 | Transcription factor | Reduced virulence | C. B. Michielse et al. (2009) |
| DCW1 | Cell wall protein | Reduced virulence | C. B. Michielse et al. (2009) |
| FCD1 | Cellobiose:quinone oxidoreductase | Fully virulent | Kawabe et al. (2006) |
| FGA1 | G‐protein α‐subunit | Markedly reduced virulence, decreased conidiation | Jain et al. (2002) |
| FGA2 | G‐protein α‐subunit | Non‐pathogenic, increase resistance to heat | Jain et al. (2005) |
| FGB1 | G‐protein β‐subunit | Markedly reduced virulence, decreased conidiation | Jain et al. (2003) |
| FMK1 | Mitogen‐activated protein kinase | Non‐pathogenic, impaired in root attachment and invasive growth | Di Pietro et al. (2001a) |
| FNR1 | Transcription factor | Markedly reduced virulence, reduced ability to use secondary nitrogen sources | Divon et al. (2006) |
| FOW1 | Mitochondrial carrier | Strongly reduced virulence, impaired in plant colonization | Inoue et al. (2002) |
| FOW2 | Transcription factor | Non‐pathogenic, impaired in invasive growth, not in root attachment | Imazaki et al. (2007) |
| FOXG_09487 | Hypothetical protein | Reduced virulence | C. B. Michielse et al. (2009) |
| FPD1 | Similar to chloride conductance regulatory protein | Markedly reduced virulence | Kawabe et al. (2004) |
| FRP1 | F‐box protein | Non‐pathogenic, impaired in root colonization and penetration, impaired growth on various carbon sources | Duyvesteijn et al. (2005); Jonkers et al. (2009) |
| FSO1 | Fusarium so1 | Fully virulent, no vegetative hyphal fusion | Rosales and Di Pietro (2008) |
| FTF1 | Transcription factor | Reduced virulence (RNAi silencing) | Ramos et al. (2007); J. M. Diaz‐Minguez (University of Salamanca, Salamanca, Spain, personal communication) |
| GAS1 | β‐1,3‐Glucanosyltransferase | Markedly reduced virulence, reduced growth on solid medium | Caracuel et al. (2005) |
| ICL1 | Isocitrate lyase | Fully virulent, no growth on C2 carbon sources and fatty acids | Jonkers et al. (2009) |
| LCC1 | Laccase | Fully virulent | Canero and Roncero (2008a) |
| LCC3 | Laccase | Fully virulent | Canero and Roncero (2008a) |
| LCC5 | Laccase | Fully virulent | Canero and Roncero (2008a) |
| PACC | Transcription factor | Increased virulence and transcription of acid‐expressed genes | Caracuel et al. (2003) |
| PEX12 | Peroxin | Reduced virulence, impaired in growth on fatty acids | C. B. Michielse et al. (2009) |
| PEX26 | Peroxin | Reduced virulence, impaired in growth on fatty acids | C. B. Michielse et al. (2009) |
| PG1 | Endopolygalacturonase | Fully virulent, reduced saprophytic growth on pectin | Di Pietro and Roncero (1998) |
| PG5 | Endopolygalacturonase | Fully virulent | Garcia‐Maceira et al. (2001) |
| PGX4 | Exopolygalacturonase | Fully virulent | Garcia‐Maceira et al. (2000) |
| PL1 | Pectate lyase | Fully virulent | Huertas‐Gonzalez et al. (1999) |
| PRT1 | Serine protease | Fully virulent | Di Pietro et al. (2001b) |
| REN1 | Transcription factor | Fully virulent, no microconidia or macroconidia, but conidium‐like cells | Ohara et al. (2004) |
| RHO1 | Monomeric G protein | Markedly reduced virulence, reduced growth on solid media | Martinez‐Rocha et al. (2008) |
| SGE1 | Transcription factor | Non‐pathogenic, reduced conidiation | C. B. Michielse et al. (unpublished results) |
| SIX1 | Small secreted protein | Reduced virulence, effect more pronounced on 4‐ to 5‐week‐old plants | Rep et al. (2005) |
| SNF1 | Protein kinase involved in carbon catabolite repression | Markedly reduced virulence, reduced growth on complex carbon sources | Ospina‐Giraldo et al. (2003) |
| STI35 | Functions in thiamine biosynthesis | Fully virulent | Ruiz‐Roldan et al. (2008a) |
| STUA | Transcription factor | Fully virulent, no macroconidia | Ohara and Tsuge (2004) |
| TOM1 | Tomatinase enzyme | Reduced virulence, reduced tomatinase activity, increased sensitivity to α‐tomatine | Pareja‐Jaime et al. (2008) |
| WC1 | Photoreceptor | Fully virulent | Ruiz‐Roldan et al. (2008b) |
| XLNR | Transcription factor | Fully virulent, reduced extracellular xylanase activity | Calero‐Nieto et al. (2007) |
| XYL3 | Family 10 endoxylanase | Fully virulent | Gomez‐Gomez et al. (2002) |
| XYL4 | Family 11 endoxylanase | Fully virulent | Gomez‐Gomez et al. (2002) |
| XYL5 | Family 11 endoxylanase | Fully virulent | Gomez‐Gomez et al. (2001) |
The role of signalling pathways in vegetative hyphal fusion (VHF) and the role of VHF in pathogenesis have recently been investigated (Rosales and Di Pietro, 2008). Although it was shown that VHF and pathogenesis share common signalling components, VHF is not required for pathogenesis, as a mutant disrupted for FSO1– a gene required for efficient VHF—was fully virulent on tomato plants. As a next step in unravelling the mechanisms of sensing and of adaptation to the plant environment, it will be important to discover the signals or ligands that trigger the cAMP–PKA and MAPK pathways.
CELL WALL‐DEGRADING ENZYMES (CWDEs)
The degree to which CWDEs contribute to the infection process is not yet fully understood. Like most fungi, F. oxysporum secretes an array of CWDEs, such as polygalacturonases, pectate lyases, xylanases and proteases, during root penetration and colonization (Beckman, 1987). However, inactivation of individual CWDE‐ or protease‐encoding genes, encoding pectate lyase (PL1), xylanase (XYL3, XYL4, XYL5), polygalacturonase (PG1, PG5, PGX4) or a subtilase (PRT1), did not have a detectable effect on virulence (reviewed in Di Pietro et al., 2003), possibly because of functional redundancy. A means of assessing the combined contribution of a group of secreted enzymes to virulence is to study the transcriptional regulators involved in the expression of their genes. To explore the role of xylanases in pathogenicity, the xlnR gene encoding transcription factor XlnR, a regulator of many xylanolytic and cellulolytic genes, was inactivated in F. oxysporum f. sp. lycopersici. Deletion of xlnR reduced the expression of the xylanase genes XYL3 and XYL4, and xylanase activity, without, however, affecting virulence (Calero‐Nieto et al., 2007). As not all xylanase activity was lost in the xlnR disruptant, it may still be that residual activity was sufficient to cause disease. Alternatively, breakdown of xylan may not be important for the colonization of tomato.
Many CWDEs are subject to carbon catabolite repression which, in yeast, is relieved under non‐repressing conditions by the activity of a protein kinase called Snf1. In addition, in F. oxysporum, targeted disruption of SNF1 resulted in reduced growth on several complex and simple carbon sources, other than glucose, and reduced the expression of several CWDEs (PGX1, PGN1, PL1) (Ospina‐Giraldo et al., 2003). Concomitantly, virulence towards A. thaliana and Brassica oleracea was reduced, suggesting that the adaptation of central carbon metabolism and CWDE gene expression plays a role in pathogenicity. Another indication that carbon metabolism and CWDEs play an important role during pathogenesis comes from the analysis of F. oxysporum f. sp. lycopersici FRP1 disruptants. FRP1 encodes an F‐box protein, implicated in the recruitment of specific proteins to an SCF complex for labelling with ubiquitin, which is usually a signal for degradation by the proteasome. FRP1 is absolutely required for pathogenicity and the FRP1 disruptant is impaired in root colonization (Duyvesteijn et al., 2005). Jonkers et al. (2009) have recently shown that poor root colonization in the frp1 mutant can be attributed to reduced assimilation of organic acids, amino acids and/or polysaccharides. External root colonization by the FRP1 disruptant was restored by the addition of glucose or proline, but virulence was not, as a result of an inability to penetrate the roots. Remarkably, ICL1 (isocitrate lyase) disruptants, generated in the same study, were able to colonize the roots and were fully virulent, even though FRP1 and ICL1 disruptants share a similar reduced growth phenotype on several carbon sources. As the FRP1 disruptant has additional defects in the expression of CWDE genes, collective secretion of several CWDEs is probably required for superficial root colonization (through the release of cell wall components for nutrition) as well as root penetration (through weakening of cell walls).
Next to carbon regulation, nitrogen regulation has also been shown to be important for the infection process. Inactivation of the global nitrogen regulator, Fnr1, resulted in a reduced ability to utilize several secondary nitrogen sources, abolished the expression of nutrition genes normally induced during the early phase of infection, and resulted in reduced pathogenicity towards tomato (Divon et al., 2006). In contrast, inactivation of CTF1, encoding a transcriptional activator of cutinase and lipase genes, did not have an effect on pathogenicity. It was shown that Ctf1 regulates the expression of a cutinase gene (CUT1) and a lipase gene (LIP1), and thus overall extracellular esterase and lipase activities, during growth under inducing conditions in axenic cultures (Rocha et al., 2008). However, Ctf1 did not seem to be required for the expression of LIP1 during infection. In addition, CUT1 was only expressed at low levels during infection. This implies that neither cutinases nor lipases play an important role during root infection, or that Ctf1‐independent activation of these genes provides sufficient cutinase and lipase activity.
In summary, as the investigation of the role of individual secreted enzymes in pathogenicity is difficult to establish because of functional redundancy, the focus has turned to the analysis of the role of transcriptional regulators of certain groups of secreted enzymes. Although direct proof of a causal relationship between extracellular enzyme production and pathogenicity has not been obtained, there are now clear indications that the collective action of CWDEs, as well as carbon catabolite repression and nitrogen regulation, is important for the infection process.
OVERCOMING PLANT DEFENCE RESPONSES
During root invasion and colonization, F. oxysporum is exposed to various plant defence mechanisms, such as physical barriers and antifungal compounds (Beckman, 1987; VanEtten et al., 1994). An example is the antifungal compound α‐tomatine in tomato plants (Roddick, 1977), a compound that is able to form complexes with sterols in fungal membranes, thereby causing pores and leakage of fungal cellular content (Keukens et al., 1992, 1995). Fusarium oxysporum can secrete a tomatine‐degrading enzyme called tomatinase (Lairini et al., 1996). Recent genome analysis has revealed that F. oxysporum contains five putative tomatinase genes (Pareja‐Jaime et al., 2008). For one of these genes, TOM1, its role in tomatine degradation and virulence was investigated. Constitutive overexpression of TOM1 resulted in high levels of tomatinase activity, independent of the presence of α‐tomatine. In addition, tomato plants infected with this TOM1‐overexpressing strain showed more rapid development of disease symptoms, whereas targeted disruption of TOM1 resulted in a 25% decrease in tomatinase activity and a delay in the development of disease symptoms (Pareja‐Jaime et al., 2008). These results indicate that the inactivation of tomatine contributes to the virulence of F. oxysporum f. sp. lycopersici towards tomato.
A second example of a plant defence mechanism is the chemical modification of the plant cell wall. Soluble and cell wall‐bound phenolic compounds accumulate in plants challenged by fungal pathogens or fungus‐derived elicitors (de Ascensao and Dubery, 2003; Mandal and Mitra, 2007; von Ropenack et al., 1998). In tomato plants and hairy root cultures, extracts of F. oxysporum induce an increase in cell wall strengthening through the deposition of lignin, as well as an increased concentration of phenolic compounds, such as ferulic acid, 4‐hydrobenzoic acid and 4‐coumaric acid (Mandal and Mitra, 2007, 2008). Fusarium oxysporum senses the presence of these phenolic compounds and responds to them. Wu et al. (2008a,2008b, 2008c) showed that cinnamic acid, coumaric acid or vanillic acid, when applied to F. oxysporum f. sp. niveum cultures, increased mycotoxin production (probably predominantly fusaric acid) and the activity of hydrolytic enzymes, such as pectinase, cellulose and amylase (only in the presence of cinnamic and vanillic acids). At the same time, these compounds had an inhibitory effect on growth and/or germination of F. oxysporum f. sp. niveum. Apparently, phenolic compounds produced by plants can have a protective function in reducing fungal growth and thus disease, but can also have a disease‐promoting function as mycotoxin levels and hydrolytic enzyme activities are increased.
The degradation of phenolic compounds in fungi is mediated by the β‐ketoadipate pathway (reviewed in Harwood and Parales, 1996). Through this pathway, lignin monomers, aromatic hydrocarbons and amino aromatics are converted to either protocatechuate or catechol and then to β‐ketoadipate. In a recent insertional mutagenesis screen, two pathogenicity mutants were identified carrying a T‐DNA insertion in the open reading frames of genes coding for catechol dioxygenase and carboxy‐cis,cis‐muconate cyclase (CMLE), two enzymes belonging to the β‐ketoadipate pathway (Michielse et al., 2009). Targeted inactivation of CMLE1 verified that this gene is required for pathogenicity (L. Reijnen and C. B. Michielse, unpublished results). In addition, CMLE1 disruptants showed a strong decrease in growth on medium containing phenolic compounds, such as coumaric acid, ferulic acid and vanillic acid, indicating the importance of aromatic compound degradation for pathogenesis.
Another way for fungi to deal with phenolic compounds is to oxidize them through the action of laccases, which are copper‐containing phenol oxidases. Laccases from wood‐rotting basidiomycetes, such as Trametes versicolor, Lentinula edodus and Pleurotus ostreatus, are involved in lignin degradation (Baldrian, 2006; Leonowicz et al., 2001). Laccases also play a role in the pathogenesis of the human pathogen Cryptococcus neoformans (Zhu et al., 2001; Zhu and Williamson, 2004). In F. oxysporum, six laccase genes have been analysed (Canero and Roncero, 2008a). Expression during infection was observed for LCC1, LCC3 and LCC9. For the genes LCC1, LCC3 and LCC5, disruptants were generated. These mutants displayed decreased laccase activity, increased sensitivity to oxidative stress (not lcc5) and increased sensitivity to phenolic compounds, such as chlorogenic acid, caffeic acid and vanillic acid (only lcc3 was tested). However, LCC1, LCC3 and LCC5 are individually not required for pathogenicity, as their deletion had no effect on virulence. A decrease in laccase activity and increased sensitivity to oxidative stress were also observed in a chloride channel (CLC1) gene disruptant (Canero and Roncero, 2008b). With this mutant, disease development in tomato was delayed. As Clc1 is required for full laccase activity (probably through the facilitation of metal cofactor insertion into maturing laccases), laccases may collectively be required for full pathogenicity. However, other defects of the clc1 mutant may also account for the decrease in virulence.
Adaptation of the fungal cell wall has been shown to play an important role during pathogenicity, possibly by resisting damage by plant enzymes or compounds and/or by reducing the release of cell wall‐derived defence elicitors. An early example is the absolute requirement of chitin synthase V (CHSV) for pathogenicity (Madrid et al., 2003). More recently, additional chitin synthase genes have been identified and characterized, namely CHS1, CHS2, CHSVb and CHS7, encoding for class I, II and VII chitin synthase and a chaperone‐like protein, respectively (Martin‐Udiroz et al., 2004, 2008). Targeted inactivation of CHS1 had no effect on virulence, whereas targeted inactivation of CHS2 and CHS7 resulted in slightly reduced virulence on tomato plants (Martin‐Udiroz et al., 2004). The most pronounced effect on virulence was observed for the CHSVb disruptant, which failed to produce any disease phenotypes (Martin‐Urdiroz et al., 2008). The phenotypes between the various CHS mutants with respect to cell wall‐disturbing agents varied. Like the inactivation of CHSV, inactivation of CHSVb confers sensitivity to Calcofluor white and Congo red, but not to sodium dodecylsulphate (SDS), implying similar alterations in cell wall architecture.
Additional genes with a role in cell wall maintenance and pathogenicity are GAS1, encoding a β‐1,3‐glucanosyltransferase, and RHO1, encoding a GTPase (Caracuel et al., 2005; Martinez‐Rocha et al., 2008). Consistent with alterations in the cell wall, GAS1 and RHO1 disruptants displayed enhanced resistance to CWDEs. In addition, increased transcription of CHSV and RHO1 was observed in the GAS1 disruptant (Caracuel et al., 2005). Both GAS1 and RHO1 disruptants were severely reduced in virulence towards tomato.
Taken together, it appears that, for a fungus like F. oxysporum to survive and proliferate inside living plant tissue, it needs to be able to control its cell wall composition and to produce enzymes that can deal with plant‐derived chemicals.
PEROXISOMAL FUNCTION
Peroxisomes are single‐membrane‐bound organelles which, in filamentous fungi, are involved in the β‐oxidation of fatty acids, peroxide detoxification and occlusion of septal pores (Jedd and Chua, 2000; Soundararajan et al., 2004; Vandenbosch et al., 1992). More than 20 peroxins (peroxisome biogenesis factors) have been identified in fungi. Mutations in peroxin genes result in the absence of normal peroxisomes, the mislocalization of peroxisomal matrix proteins and the inability to perform specific biochemical reactions, such as fatty acid catabolism (Kiel et al., 2006). Peroxisomal function and fatty acid metabolism have been shown to be required for virulence of the plant‐pathogenic fungi Colletotrichum lagenarium, Magnaporthe grisea and Ustilago maydis (Jeon et al., 2007; Kimura et al., 2001; Klose and Kronstad, 2006; Ramos‐Pamplona and Naqvi, 2006; Wang et al., 2007). In F. oxysporum, four different pex genes, PEX1, PEX10, PEX12 and PEX26, were identified as potential pathogenicity genes in a recent insertional mutagenesis screen, and the requirement for full pathogenicity was verified for two of them (PEX12 and PEX26) by complementation with the intact genes (C. B. Michielse et al., 2009). These results demonstrate that peroxisomal function is also necessary for the pathogenesis of a root‐infecting fungus that lacks appressoria. Possibly, peroxisomal function could be necessary for the utilization of host nutrients during in planta growth. Alternatively, or in addition, woronin bodies, a special class of peroxisomes that plug septal pores, could be required for optimal invasive growth by making the fungus less sensitive to cytoplasmic leakage (Jedd and Chua, 2000; Soundararajan et al., 2004; Tenney et al., 2000).
TRANSCRIPTION FACTORS
Important progress has been made in the identification of transcription factors specifically required for pathogenicity, such as Fow2 in F. oxysporum f. sp. melonis and Sge1 in F. oxysporum f. sp. lycopersici. For both genes, disruptants completely lost pathogenicity on their host plant (Imazaki et al., 2007) (C. B. Michielse et al., 2009). The mutants were not impaired in either vegetative growth or the utilization of various carbon sources. Although conidiation of the FOW2 disruptant was similar to that of the wild‐type, the SGE1 disruptant displayed reduced conidiation (about four‐fold), although the conidia generated did not differ in germination rate or morphology from the wild‐type (C. B. Michielse et al., unpublished results). For both FOW2 and SGE1 disruptants, fluorescence microscopy was used to observe their infection behaviour, and both mutants displayed a similar phenotype: no deficiency in root attachment and superficial root colonization, but impaired invasion and/or in planta growth (Imazaki et al., 2007) (C. B. Michielse et al., unpublished results).
A third transcription factor gene implicated in virulence is FTF1, identified in F. oxysporum f. sp. phaseoli (Ramos et al., 2007). FTF1 is a multicopy gene only present in highly virulent strains, and transcribed specifically during the early phases of infection. Interestingly, the multiple copies of FTF1 in highly virulent strains are all located on the smallest chromosome. Recently, the requirement of FTF1 for pathogenicity towards bean, as well as tomato, was demonstrated by RNAi experiments (J. M. Diaz‐Minguez, University of Salamanca, Salamanca, Spain, personal communication). Clearly, it would be very interesting to identify the target genes of these transcription factors. To this end, the microarrays currently being developed for F. oxysporum (C. Kistler, USDA, St. Paul, USA, personal communication), would be of great help.
INSERTIONAL MUTAGENESIS
Random insertional mutagenesis has been proven to be a useful tool for the identification of the genes involved in fungal pathogenesis (Weld et al., 2006). Relatively small‐scale insertional mutagenesis in F. oxysporum using restriction enzyme‐mediated transformation led to the identification of novel pathogenicity genes, such as ARG1, FOW1, FOW2 and FCD1 (Imazaki et al., 2007; Inoue et al., 2002; Kawabe et al., 2006; Namiki et al., 2001). Additional insertional mutagenesis programmes using plasmid DNA or transposon tagging led to the identification of CHSV, FRP1 and a Velvet‐like gene as novel pathogenicity genes (Duyvesteijn et al., 2005; Lopez‐Berges et al., 2008; Madrid et al., 2003). Being unbiased, these screens often yield unsuspected links between pathogenicity and other (developmental) processes. For example, two members of the velvet gene family, veA and velB, previously shown to regulate fungal development and secondary metabolism in the model organism Aspergillus nidulans, were shown to be required for full virulence in F. oxysporum (Lopez‐Berges et al., 2008) (A. Di Pietro, University of Cordoba, Cordoba, Spain, personal communication). Recently, a large‐scale mutagenesis programme (10 290 transformants), using the T‐DNA of Agrobacterium tumefaciens as a mutagen, yielded 111 potential pathogenicity genes (C. B. Michielse et al., 2009). In addition to a large group of hypothetical proteins with unknown function, functional distribution of the remaining proteins implicated certain processes, such as amino acid and lipid metabolism, cell wall maintenance and small molecule transport, in pathogenicity. To date, eight of the 11 genes have been verified to be required for (full) pathogenicity by gene knock‐out (C. B. Michielse et al., 2009 C. B. Michielse, unpublished results). It is expected that the analysis of additional pathogenicity mutants identified in these insertional mutagenesis programmes will further enhance our understanding of the molecular processes underlying pathogenesis.
AVIRULENCE AND EFFECTOR PROTEINS SECRETED DURING COLONIZATION
Major resistance (R) genes have been found against F. oxysporum in several crops, including tomato, melon, cucumber, pea and bean (Table 2). Two of these genes have been cloned: I‐2 from tomato (Simons et al., 1998) and Fom2 from melon (Joobeur et al., 2004). In cases in which R genes have been employed in a crop, the formae speciales of F. oxysporum that cause disease in that crop usually break up into races, defined by the R gene spectrum effective against them. Table 2 lists the formae speciales for which this is the case. Although the absence of a sexual cycle under laboratory conditions prevents genetic confirmation of gene‐for‐gene relationships on the side of the pathogen (Flor, 1971), the existence of major R genes in hosts and corresponding races in F. oxysporum raised the expectation that F. oxysporum should contain avirulence genes matching R genes in hosts. This was confirmed in 2004, when the first avirulence gene of F. oxysporum was identified in F. oxysporum f. sp. lycopersici (Fol) (Rep et al., 2004). Its product, Six1 (‘secreted in xylem 1’), was found in xylem sap of infected tomato plants, and was shown to be required for disease resistance mediated by the R gene I‐3 (‘immunity 3’). Six1 was therefore later renamed Avr3 (Houterman et al., 2008). Similar to avirulence proteins of other fungi (Catanzariti et al., 2007; Fudal et al., 2007; Gout et al., 2006; Rep, 2005; Thomma et al., 2005), Avr3 is a small secreted protein without homologues, even in closely related fungi (Rep et al., 2004). Avr3 is required for full virulence, especially towards older plants (Rep et al., 2005), and is expressed specifically on entry of plant roots or exposure to plant cells in culture (van der Does et al., 2008a).
Table 2.
Monogenic resistance reported against Fusarium oxysporum.
| Plant species | Forma specialis | R genes | Pathogen races | Reference* |
|---|---|---|---|---|
| Tomato | lycopersici | I, I‐1, I‐2, I‐3 | 1, 2, 3† | Hemming et al. (2004); Sela‐Buurlage et al. (2001); Simons et al. (1998) |
| Melon | melonis | Fom‐1, Fom‐2 | 0, 1, 2, 1.2 | Joobeur et al. (2004); Oumouloud et al. (2008) |
| Cucumber | cucumerinum | Foc | 1, 2 | Vakalounakis (1996) |
| Pea | pisi | Fw | 1, 2A, 2B, 5, 6 | Grajal‐Martin and Muehlbauer (2002); McClendon et al. (2002) |
| Chickpea | ciceris | Several, mostly recessive | 0, 1A, 1B/C, 2, 3, 4, 5, 6 | Sharma and Muehlbauer (2007) |
| Eggplant | melongenae | (Unnamed) | None reported | Mutlu et al. (2008) |
| Bean | phaseoli | (Unnamed) | 1, 2, 3, 4, 5, 6, 7 | Cross et al. (2000); Fall et al. (2001); Salgado et al. (1995) |
| Cotton | vasinfectum | (Unnamed) | A, 1, 2, 3, 4, 5, 6, 7, 8 | Wang and Roberts (2006) |
| Basil | basilici | (Unnamed) | None reported | Dudai et al. (2002) |
Bold, cloned
Not exhaustive.
Sometimes referred to as race 0, 1 and 2, respectively.
More recently, a second avirulence gene, AVR1 (SIX4), was identified in Fol, which is required for I‐ and I‐1‐mediated disease resistance (Houterman et al., 2008). Its product, Avr1 (Six4), was also found in the xylem sap proteome of infected tomato plants, together with several other fungal proteins secreted during colonization (Houterman et al., 2007). Avr1 is not required for virulence towards susceptible tomato plants, but was found to suppress I‐2‐ and I‐3‐mediated disease resistance (Houterman et al., 2008), explaining earlier observations that race 1 isolates of F. oxysporum f. sp. lycopersici can cause disease on tomato lines that carry I‐2 or I‐3 (but not I or I‐1) (Mes et al., 1999; Rep et al., 2005). The avirulence gene matching the remaining R gene against Fol, I‐2, has also been identified, and again encodes a small protein secreted in xylem sap during colonization (Houterman et al., in press). Taken together, this shows that the analysis of the xylem sap proteome of infected plants can be an effective approach to identify the avirulence proteins of F. oxysporum. It also resulted in the identification of other proteins from F. oxysporum secreted during colonization that could play an important role in host colonization (Houterman et al., 2007) (M. Rep, unpublished results). It could be worthwhile, therefore, to apply this approach to other xylem‐colonizing fungi, such as Verticillium, or other formae speciales of F. oxysporum, if the host plant is amenable to the extraction of sufficient amounts of xylem sap. Most importantly, the identification of the molecular targets of effectors should lead to a greater insight into how F. oxysporum suppresses or evades primary or secondary immunity in plants.
DEVELOPMENT OF A. THALIANA AS A MODEL HOST PLANT
Over the last 5 years, A. thaliana has emerged as a model host for the study of Fusarium wilt disease. As detailed above, Arabidopsis has been used to investigate the genetic requirements of pathogenicity in the fungus (Ospina‐Giraldo et al., 2003) and to visualize root and xylem colonization with fluorescence microscopy (Czymmek et al., 2007). However, the main advantage of the use of Arabidopsis is the powerful tools available for Arabidopsis to genetically dissect host responses (Berrocal‐Lobo and Molina, 2008).
In general, for the infection of Arabidopsis, formae speciales are used that cause disease on crucifers: f. sp. conglutinans (cabbage), f. sp. matthiolae (stock) and f. sp. raphani (radish). One study (Berrocal‐Lobo and Molina, 2004) reports f. sp. lycopersici, the tomato pathogen, as also being able to infect Arabidopsis, but, in another study, neither f. sp. lycopersici nor f. sp. cubense (banana) caused disease (Diener and Ausubel, 2005). Elsewhere f. sp. cubense was also reported as not being able to cause disease in (wild‐type) Arabidopsis plants (Van Hemelrijck et al., 2006), suggesting that there is at least some level of host specificity in the Arabidopsis–F. oxysporum pathosystem.
The main conclusion of infection studies using Arabidopsis mutants in various hormone response pathways is that salicylic acid (SA), jasmonate (JA) and ethylene (ET) pathways cooperate to limit disease development (Berrocal‐Lobo and Molina, 2004; Diener and Ausubel, 2005). The involvement of JA signalling in resistance to F. oxysporum is also supported by the correlation between JA‐responsive gene expression and disease resistance in transgenic plants overexpressing certain transcription factors (McGrath et al., 2005), as well as in mutants lacking a G‐protein β‐subunit (Trusov et al., 2006) or affected in abscisic acid signalling (Anderson et al., 2004). Interestingly, the def1 mutant of tomato, which is unresponsive to JA, is also more susceptible to F. oxysporum (Thaler et al., 2004). Nevertheless, the application of methyl jasmonate (MetJA) did not enhance resistance, whereas SA application did (Edgar et al., 2006). The latter effect was not accompanied by enhanced PR‐1 expression, which is consistent with the observation that the absence of NPR1, a central regulator of pathogenesis‐related (PR) gene expression, does not affect the level of resistance (Diener and Ausubel, 2005). However, PR proteins may still contribute to resistance against F. oxysporum under certain conditions; expression of tobacco PR‐1 or PR‐5 increased resistance to F. oxysporum in an Arabidopsis mutant (esa1) that displays enhanced susceptibility (Van Hemelrijck et al., 2006), and repression of PR gene expression by overexpression of the gene for transcription factor ATAF2 correlated with enhanced susceptibility (Delessert et al., 2005). Apparently, the degree of resistance to infection is determined by a complex of factors, each of which by itself has a relatively minor effect. This is also exemplified by an Arabidopsis glucosinolate biosynthesis mutant that is slightly, but significantly, more susceptible to F. oxysporum f. sp. matthiolae (Tierens et al., 2001).
Given the many pathways and mechanisms in Arabidopsis that contribute to limit infection by F. oxysporum, it may be no surprise that there is a large variation between the levels of susceptibility among ecotypes (Czymmek et al., 2007), and that these differences are polygenic in nature (Diener and Ausubel, 2005). Nevertheless, Diener and Ausubel (2005) attributed much of the variation in disease susceptibility between two ecotypes (Col‐0 and Ty‐0) to a single gene, and were able to isolate this gene, RFO1 (resistance to Fusarium oxysporum 1), by map‐based cloning (Diener and Ausubel, 2005). RFO1 turned out to encode a receptor‐like kinase (RLK) with an extracellular WAK (wall‐associated kinase) domain. As RFO1 enhances resistance to several formae speciales of F. oxysporum, it is unlikely that it recognizes a specific effector, but rather may function as a microbe‐associated molecular pattern (MAMP) receptor that is activated by relatively conserved molecules, such as fungal cell wall components or plant cell wall breakdown products.
In conclusion, although not a good model plant for gene‐for‐gene interactions with F. oxysporum, Arabidopsis will allow rapid progress in the identification of plant proteins, metabolites and signalling pathways that contribute to basal resistance or susceptibility towards a root‐invading fungus. One interesting application of the genetic tools available for Arabidopsis is to investigate host versus non‐host resistance mechanisms using different formae speciales of F. oxysporum (Van Hemelrijck et al., 2006).
TRANSCRIPTOMICS
A new era for F. oxysporum research began when its genome sequence became available (discussed below). The genome sequence enables the development of whole‐genome microarrays, and thereby opens up the possibility to identify genes specifically expressed during certain developmental stages, e.g. conidiation, growth in axenic cultures under controlled circumstances and during infection, on a genome‐wide scale. Whilst awaiting such experiments, expressed sequence tag analysis has been used to identify genes specifically up‐regulated during conidiation and germination (Deng et al., 2006; Iida et al., 2006). Two cDNA libraries corresponding to the expression profile during vegetative growth and conidiation were constructed, and 641 and 626 unique genes, respectively, were identified, 130 of which were common to both libraries (Iida et al. 2006). In addition, Deng et al. (2006) constructed a cDNA library containing 6448 cDNA clones, and concluded from expression analysis that ras and other signalling pathways cooperatively initiate conidial germination by increasing protein synthesis. The conidiation library was also used to screen for genes regulated by the sporulation‐associated transcription factors StuA and Ren1 (Iida et al., 2007). In total, 23 and 12 genes were identified that were positively and negatively regulated, respectively, by Ren1, which is required for micro‐ and macroconidia development. Only three genes were found to be positively regulated by StuA, a regulator of macroconidia production (Iida et al., 2007; Ohara and Tsuge, 2004).
The sequencing of expressed sequence tags was also used to identify genes associated with pathogenicity and to understand the interaction between F. oxysporum f. sp. vasinfectum and cotton (Dowd et al., 2004; McFadden et al., 2006). An array was generated containing 4000 cDNA clones from F. oxysporum‐infected cotton tissue at several time points after infection and 2000 cDNA clones from uninfected cotton tissue (Dowd et al., 2004). This array was used to identify plant genes up‐ or down‐regulated in roots and hypocotyls during a susceptible interaction. In total, 135 and 208 genes differentially expressed during infection were identified in roots and hypocotyls, respectively. Genes related to pathogenesis (PR genes), phytoalexin biosynthesis (gossypol), lignin biosynthesis and ET production were found to be up‐regulated in hypocotyls. In contrast, genes related to lignin biosynthesis, stress responses, aquaporins and growth responses were down‐regulated in root tissue. No effect on PR gene expression in roots was observed. From these results, it appears that, during infection, F. oxysporum may suppress host responses in roots, thereby facilitating infection.
McFadden et al. (2006) used the same microarrays to identify F. oxysporum genes expressed in root and hypocotyl tissue during cotton infection. A total of 174 genes was expressed in planta, the great majority of which (97%) were found in infected roots. A large proportion of these fungal genes encode hypothetical proteins with an unknown function. The remaining genes are mostly common, highly expressed genes, such as genes for ribosomal proteins, proteins involved in energy or amino acid metabolism, transport and translation. Interestingly, one of the genes found to be up‐regulated during cotton infection, FOXG_05013, encoding a putative cell wall protein, was also identified in a recent insertional mutagenesis screen and verified as a gene required for full pathogenicity (C. B. Michielse et al., 2009). In total, 11 genes were found to be preferentially expressed in planta. These include a putative ribosomal protein, 6‐hydroxy‐d‐nicotine oxidase, a subunit of ATP synthase, a cytochrome P450 and an oxidoreductase (McFadden et al., 2006). The gene coding for the oxidoreductase (FOXG_04087) shows high similarity to Agrobacterium tumefaciens AtsC, a gene involved in attachment and virulence of this bacterium. FOXG_04087 expression was higher in plants infected with a pathogenic isolate than in those infected with a non‐pathogenic isolate, indicating that this gene might have a role in virulence (McFadden et al., 2006).
As discussed above, these findings constitute the early beginnings of the functional genomics of F. oxysporum, and the availability, in the near future, of microarrays based on the genome sequence is expected to lead to the generation of a large amount of information on transcriptional reprogramming during the infection of plants.
THE GENOME SEQUENCE
The first genome sequence of an F. oxysporum isolate, a race 2 strain of forma specialis lycopersici (Fol), was produced by the Broad Institute and became available in March 2007. Already, this genome sequence has accelerated genomic approaches to the study of F. oxysporum, such as large‐scale insertional mutagenesis, transcriptomics and proteomics. One interesting outcome of the ongoing analysis of the F. oxysporum genome sequence is that (host‐specific) virulence‐related genes appear to be non‐randomly distributed over the genome. For instance, chromosome 14 contains the SIX1(AVR3), SIX2 and SIX3 effector/avirulence genes in a region that is more conserved between clonal lines of Fol than genes on other chromosomes, suggesting that (parts of) chromosome 14 may be subject to horizontal transfer (van der Does et al., 2008b).
Clearly, we are just at the beginning of exploiting the genome sequence of F. oxysporum to understand the mechanisms and evolution of the pathogenicity of this fungus. Particular areas of interest are now the nature of the infection‐specific transcriptome and the role of transposable elements and horizontal gene transfer in the evolution of the genome structure and host‐specific pathogenicity of F. oxysporum. In addition, an exciting prospect will be to gain an insight into the genomic differences between different formae speciales and non‐pathogenic isolates by sequencing of additional strains (C. Kistler, personal communication). This should also aid efforts in the use of molecular techniques for the identification of formae speciales and races, which would be of great assistance in helping to control diseases caused by F. oxysporum (Lievens et al., 2008).
ACKNOWLEDGEMENTS
We thank Frank Takken and Ben Cornelissen for critical reading of the manuscript, and Roselinde Duyvesteijn for providing the photographs presented in Fig. 1.
REFERENCES
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate‐ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, G.M. and Armstrong, J.K. (1981) Formae speciales and races of Fusarium oxysporum causing wilt diseases In: Fusarium: Diseases, Biology and Taxonomy (Cook R. ed.), pp. 391–399. University Park, PA: Penn State University Press. [Google Scholar]
- De Ascensao, A.R.F.D.C. and Dubery, I.A. (2003) Soluble and wall‐bound phenolics and phenolic polymers in Musa acuminata roots exposed to elicitors from Fusarium oxysporum f. sp. cubense . Phytochem. 63, 679–686. [DOI] [PubMed] [Google Scholar]
- Baayen, R.P. , O'Donnell, K. , Bonants, P.J.M. , Cigelnik, E. , Kroon, L.P.N.M. , Roebroeck, E.J.A. and Waalwijk, C. (2000) Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathol. 90, 891–900. [DOI] [PubMed] [Google Scholar]
- Baldrian, P. (2006) Fungal laccases—occurrence and properties. FEMS Microbiol. Rev. 30, 215–242. [DOI] [PubMed] [Google Scholar]
- Beckman, C.H. (1987) The Nature of Wilt Diseases of Plant. St. Paul: APS Press. [Google Scholar]
- Berrocal‐Lobo, M. and Molina, A. (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum . Mol. Plant–Microbe Interact. 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Berrocal‐Lobo, M. and Molina, A. (2008) Arabidopsis defense response against Fusarium oxysporum . Trends Plant Sci. 13, 145–150. [DOI] [PubMed] [Google Scholar]
- Bolwerk, A. , Lagopodi, A.L. , Lugtenberg, B.J.J. and Bloemberg, G.V. (2005) Visualization of interactions between a pathogenic and a beneficial Fusarium strain during biocontrol of tomato foot and root rot. Mol. Plant–Microbe Interact. 18, 710–721. [DOI] [PubMed] [Google Scholar]
- Calero‐Nieto, F. , Di Pietro, A. , Roncero, M.I.G. and Hera, C. (2007) Role of the transcriptional activator XInR of Fusarium oxysporum in regulation of Xylanase genes and virulence. Mol. Plant–Microbe Interact. 20, 977–985. [DOI] [PubMed] [Google Scholar]
- Canero, D.C. and Roncero, M.I.G. (2008a) Functional analyses of laccase genes from Fusarium oxysporum . Phytopathol. 98, 509–518. [DOI] [PubMed] [Google Scholar]
- Canero, D.C. and Roncero, M.I.G. (2008b) Influence of the chloride channel of Fusarium oxysporum on extracellular laccase activity and virulence on tomato plants. Microbiol.-Sgm. 154, 1474–1481. [DOI] [PubMed] [Google Scholar]
- Caracuel, Z. , Casanova, C. , Roncero, M.I.G. , Di Pietro, A. and Ramos, J. (2003) pH response transcription factor PacC controls salt stress tolerance and expression of the P‐type Na+ATPase ena1 in Fusarium oxysporum . Eukaryot. Cell, 2, 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuel, Z. , Martinez‐Rocha, A.L. , Di Pietro, A. , Madrid, M.P. and Roncero, M.I.G. (2005) Fusarium oxysporum gas1 encodes a putative beta‐1,3‐glucanosyltransferase required for virulence on tomato plants. Mol. Plant–Microbe Interact. 18, 1140–1147. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. and Ellis, J.G. (2007) Avirulence proteins from haustoria‐forming pathogens. FEMS Microbiol. Lett. 269, 181–188. [DOI] [PubMed] [Google Scholar]
- Cross, H. , Brick, M.A. , Schwartz, H.F. , Panella, L.W. and Byrne, P.F. (2000) Inheritance of resistance to fusarium wilt in two common bean races. Crop Sci. 40, 954–958. [Google Scholar]
- Czymmek, K.J. , Fogg, M. , Powell, D.H. , Sweigard, J. , Park, S.Y. and Kang, S. (2007) In vivo time‐lapse documentation using confocal and multi‐photon microscopy reveals the mechanisms of invasion into the Arabidopsis root vascular system by Fusarium oxysporum . Fungal Genet. Biol. 44, 1011–1023. [DOI] [PubMed] [Google Scholar]
- Delessert, C. , Kazan, K. , Wilson, I.W. , Van Der Straeten, D. , Manners, J. , Dennis, E.S. and Dolferus, R. (2005) The transcription factor ATAF2 represses the expression of pathogenesis‐related genes in Arabidopsis. Plant J. 43, 745–757. [DOI] [PubMed] [Google Scholar]
- Deng, Y. , Dong, H.T. , Jin, Q.C. , Dai, C.E. , Fang, Y.Q. , Liang, S. , Wang, K. , Shao, J. , Lou, Y.C. , Shi, W.Q. , Vakalounakis, D.J. and Li, D.B. (2006) Analysis of expressed sequence tag data and gene expression profiles involved in conidial germination of Fusarium oxysporum . Appl. Environ. Microbiol. 72, 1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins, A.E. (2003) Gibberella from A (venaceae) to Z (eae). Annu. Rev. Phytopathol. 41, 177–198. [DOI] [PubMed] [Google Scholar]
- Diener, A.C. and Ausubel, F.M. (2005) Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease‐resistance gene, is not race specific. Genetics, 171, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, A. and Roncero, M.I. (1998) Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum . Mol. Plant–Microbe Interact. 11, 91–98. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. , Garcia‐MacEira, F.I. , Meglecz, E. and Roncero, M.I. (2001a) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Di Pietro, A. , Huertas‐Gonzalez, M.D. , Gutierrez‐Corona, J.F. , Martinez‐Cadena, G. , Meglecz, E. and Roncero, M.I. (2001b) Molecular characterization of a subtilase from the vascular wilt fungus Fusarium oxysporum . Mol. Plant–Microbe Interact. 14, 653–662. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. , Madrid, M.P. , Caracuel, Z. , Delgado‐Jarana, J. and Roncero, M.I.G. (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Divon, H.H. , Ziv, C. , Davydov, O. , Yarden, O. and Fluhr, R. (2006) The global nitrogen regulator, FNR1, regulates fungal nutrition‐genes and fitness during Fusarium oxysporum pathogenesis. Mol. Plant Pathol. 7, 485–497. [DOI] [PubMed] [Google Scholar]
- Van Der Does, H.C. , Duyvesteijn, R.G.E. , Goltstein, P.M. , Van Schie, C.C.N. , Manders, E.M.M. , Cornelissen, B.J.C. and Rep, M. (2008a) Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genet. Biol. 45, 1257–1264. [DOI] [PubMed] [Google Scholar]
- Van Der Does, H.C. , Lievens, B. , Claes, L. , Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008b) The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 10, 1475–1485. [DOI] [PubMed] [Google Scholar]
- Dowd, C. , Wilson, L.W. and McFadden, H. (2004) Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp. vasinfectum . Mol. Plant–Microbe Interact. 17, 654–667. [DOI] [PubMed] [Google Scholar]
- Dudai, N. , Chaimovitsh, D. , Reuveni, M. , Ravid, U. , Larkov, O. and Putievsky, E. (2002) Breeding of sweet basil (Ocimum basilicum) resistant to Fusarium wit caused by Fusarium oxysporum f. sp. basilicum . J. Herbs, Spices Med. Plants, 9, 45–51. [Google Scholar]
- Duyvesteijn, R.G. , Van Wijk, R. , Boer, Y. , Rep, M. , Cornelissen, B.J. and Haring, M.A. (2005) Frp1 is a Fusarium oxysporum F‐box protein required for pathogenicity on tomato. Mol. Microbiol. 57, 1051–1063. [DOI] [PubMed] [Google Scholar]
- Edgar, C.I. , McGrath, K.C. , Dombrecht, B. , Manners, J.M. , Maclean, D.C. , Schenk, P.M. and Kazan, K. (2006) Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana . Australas. Plant Pathol. 35, 581–591. [Google Scholar]
- Fall, A.L. , Byrne, P.F. , Jung, G. , Coyne, D.P. , Brick, M.A. and Schwartz, H.F. (2001) Detection and mapping of a major locus for Fusarium wilt resistance in common bean. Crop Sci. 41, 1494–1498. [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Fudal, I. , Ross, S. , Gout, L. , Blaise, F. , Kuhn, M.L. , Eckert, M. , Cattolico, L. , Bernard‐Samain, S. , Balesdent, M.H. and Rouxel, T. (2007) Heterochromatin‐like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map‐based cloning of AvrLm6. Mol. Plant–Microbe Interact. 20, 459–470. [DOI] [PubMed] [Google Scholar]
- Garcia‐Maceira, F.I. , Di Pietro, A. and Roncero, M.I. (2000) Cloning and disruption of pgx4 encoding an in planta expressed exopolygalacturonase from Fusarium oxysporum . Mol. Plant–Microbe Interact. 13, 359–365. [DOI] [PubMed] [Google Scholar]
- Garcia‐Maceira, F.I. , Di Pietro, A. , Huertas‐Gonzalez, M.D. , Ruiz‐Roldan, M.C. and Roncero, M.I. (2001) Molecular characterization of an endopolygalacturonase from Fusarium oxysporum expressed during early stages of infection. Appl. Environ. Microbiol. 67, 2191–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Gomez, E. , Roncero, M.I.G. , Di Pietro, A. and Hera, C. (2001) Molecular characterization of a novel endo‐beta‐1,4‐xylanase gene from the vascular wilt fungus Fusarium oxysporum . Curr. Genet. 40, 268–275. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, E. , Ruiz‐Roldan, M.C. , Di Pietro, A. , Roncero, M.I.G. and Hera, C. (2002) Role in pathogenesis of two endo‐beta‐1,4‐xylanase genes from the vascular wilt fungus Fusarium oxysporum . Fungal Genet. Biol. 35, 213–222. [DOI] [PubMed] [Google Scholar]
- Gordon, T.R. and Martyn, R.D. (1997) The evolutionary biology of Fusarium oxysporum . Annu. Rev. Phytopathol. 35, 111–128. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Gout, L. , Fudal, I. , Kuhn, M.L. , Blaise, F. , Eckert, M. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans . Mol. Microbiol. 60, 67–80. [DOI] [PubMed] [Google Scholar]
- Grajal‐Martin, M.J. and Muehlbauer, F.J. (2002) Genomic location of the Fw gene for resistance to Fusarium wilt race 1 in peas. J. Hered. 93, 291–293. [DOI] [PubMed] [Google Scholar]
- Guadet, J. , Julien, J. , Lafay, J.F. and Brygoo, Y. (1989) Phylogeny of some Fusarium species, as determined by large‐subunit ribosomal‐RNA sequence comparison. Mol. Biol. Evol. 6, 227–242. [DOI] [PubMed] [Google Scholar]
- Harwood, C.S. and Parales, R.E. (1996) The beta‐ketoadipate pathway and the biology of self‐identity. Annu. Rev. Microbiol. 50, 553–590. [DOI] [PubMed] [Google Scholar]
- Hemming, M.N. , Basuki, S. , McGrath, D.J. , Carroll, B.J. and Jones, D.A. (2004) Fine mapping of the tomato I‐3 gene for Fusarium wilt resistance and elimination of a co‐segregating resistance gene analogue as a candidate for I‐3. Theor. Appl. Genet. 109, 409–418. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Speijer, D. , Dekker, H.L. , De Koster, C.G. , Cornelissen, B.J.C. and Rep, M. (2007) The mixed xylem sap proteome of Fusarium oxysporum‐infected tomato plants. Mol. Plant Pathol. 8, 215–221. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Cornelissen, B.J. and Rep, M. (2008) Suppression of plant resistance gene‐based immunity by a fungal effector. PLoS Pathog. 4, e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman, P.M. , Ma, L.S. , Van Ooijen, G. , De Vroomen, M. , Cornelissen, B.J. , Takken, F. and Rep, M. (2009) The small effector protein Avr2 secreted in xylem by a vascular wilt fungus interacts with its cognate resistance protein inside plant cells. Plant J. in press. [DOI] [PubMed] [Google Scholar]
- Huertas‐Gonzalez, M.D. , Ruiz‐Roldan, M.C. , Garcia Maceira, F.I. , Roncero, M.I. and Di Pietro, A. (1999) Cloning and characterization of pl1 encoding an in planta‐secreted pectate lyase of Fusarium oxysporum . Curr. Genet. 35, 36–40. [DOI] [PubMed] [Google Scholar]
- Iida, Y. , Ohara, T. and Tsuge, T. (2006) Identification of genes up‐regulated during conidiation of Fusarium oxysporum through expressed sequence tag analysis. Fungal Genet. Biol. 43, 179–189. [DOI] [PubMed] [Google Scholar]
- Iida, Y. , Ohara, T. and Tsuge, T. (2007) Identification of genes with changes in transcription levels caused by mutations in conidiation regulator genes REN1 and FoSTUA in Fusarium oxysporum. J. Gen. Plant. Pathol. 73, 158–167. [Google Scholar]
- Imazaki, I. , Kurahashi, M. , Iida, Y. and Tsuge, T. (2007) Fow2, a Zn(II)2Cys6‐type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum . Mol. Microbiol. 63, 737–753. [DOI] [PubMed] [Google Scholar]
- Inoue, I. , Namiki, F. and Tsuge, T. (2002) Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell, 14, 1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S. , Akiyama, K. , Mae, K. , Ohguchi, T. and Takata, R. (2002) Targeted disruption of a G protein alpha subunit gene results in reduced pathogenicity in Fusarium oxysporum . Curr. Genet. 41, 407–413. [DOI] [PubMed] [Google Scholar]
- Jain, S. , Akiyama, K. , Kan, T. , Ohguchi, T. and Takata, R. (2003) The G protein beta subunit FGB1 regulates development and pathogenicity in Fusarium oxysporum . Curr. Genet. 43, 79–86. [DOI] [PubMed] [Google Scholar]
- Jain, S. , Akiyama, K. , Takata, R. and Ohguchi, T. (2005) Signaling via the G protein alpha subunit FGA2 is necessary for pathogenesis in Fusarium oxysporum . FEMS Microbiol. Lett. 243, 165–172. [DOI] [PubMed] [Google Scholar]
- Jedd, G. and Chua, N.H. (2000) A new self‐assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2, 226–231. [DOI] [PubMed] [Google Scholar]
- Jeon, J. , Park, S.Y. , Chi, M.H. , Choi, J. , Park, J. , Rho, H.S. , Kim, S. , Goh, J. , Yoo, S. , Choi, J. , Park, J.Y. , Yi, M. , Yang, S. , Kwon, M.J. , Han, S.S. , Kim, B.R. , Khang, C.H. , Park, B. , Lim, S.E. , Jung, K. , Kong, S. , Karunakaran, M. , Oh, H.S. , Kim, H. , Kim, S. , Park, J. , Kang, S. , Choi, W.B. , Kang, S. and Lee, Y.H. (2007) Genome‐wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 39, 561–565. [DOI] [PubMed] [Google Scholar]
- Jonkers, W. , Andrade Rodrigues, C.D. and Rep, M. (2009) Impaired colonization and infection of tomato roots by the Δfrp1 mutant of Fusarium oxysporum correlates with reduced CWDE gene expression. MPMI. in press. [DOI] [PubMed] [Google Scholar]
- Joobeur, T. , King, J.J. , Nolin, S.J. , Thomas, C.E. and Dean, R.A. (2004) The Fusarium wilt resistance locus Fom‐2 of melon contains a single resistance gene with complex features. Plant J. 39, 283–297. [DOI] [PubMed] [Google Scholar]
- Katan, T. (1999) Current status of vegetative compatibility groups in Fusarium oxysporum . Phytoparasitica, 27, 51–64. [Google Scholar]
- Katan, T. and Di Primo, P. (1999) Current status of vegetative compatibility groups in Fusarium oxysporum: Supplement (1999). Phytoparasitica, 27, 273–277. [Google Scholar]
- Kawabe, M. , Mizutani, K. , Yoshida, T. , Teraoka, T. , Yoneyama, K. , Yamaguchi, I. and Arie, T. (2004) Cloning of the pathogenicity‐related gene FPD1 in Fusarium oxysporum f. sp. lycopersici . J. Gen. Plant Pathol. 70, 16–20. [Google Scholar]
- Kawabe, M. , Yoshida, T. , Teraoka, T. and Arie, T. (2006) FCD1 encoding protein homologous to cellobiose: quinone oxidoreductase in Fusarium oxysporum . Gene, 382, 100–110. [DOI] [PubMed] [Google Scholar]
- Keukens, E.A.J. , Devrije, T. , Fabrie, C.H.J.P. , Demel, R.A. , Jongen, W.M.F. and Dekruijff, B. (1992) Dual specificity of sterol‐mediated glycoalkaloid induced membrane disruption. Biochim. Biophys. Acta, 1110, 127–136. [DOI] [PubMed] [Google Scholar]
- Keukens, E.A.J. , DeVrije, T. , VandenBoom, C. , DeWaard, P. , Plasman, H.H. , Thiel, F. , Chupin, V. , Jongen, W.M.F. and DeKruijff, B. (1995) Molecular basis of glycoalkaloid induced membrane disruption. Biochim. Biophys. Acta, Biomembr. 1240, 216–228. [DOI] [PubMed] [Google Scholar]
- Kiel, J.A.K.W. , Veenhuis, M. and Van Der Klei, I.J. (2006) PEX genes in fungal genomes: common, rare or redundant. Traffic, 7, 1291–1303. [DOI] [PubMed] [Google Scholar]
- Kimura, A. , Takano, Y. , Furusawa, I. and Okuno, T. (2001) Peroxisomal metabolic function is required for appressorium‐mediated plant infection by Colletotrichum lagenarium . Plant Cell, 13, 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, J. and Kronstad, J.W. (2006) The multifunctional beta‐oxidation enzyme is required for full symptom development by the biotrophic maize pathogen Ustilago maydis . Eukaryot. Cell, 5, 2047–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagopodi, A.L. , Ram, A.F. , Lamers, G.E. , Punt, P.J. , Van den Hondel, C.A. , Lugtenberg, B.J. and Bloemberg, G.V. (2002) Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis‐lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant–Microbe Interact. 15, 172–179. [DOI] [PubMed] [Google Scholar]
- Lairini, K. , PerezEspinosa, A. , Pineda, M. and RuizRubio, M. (1996) Purification and characterization of tomatinase from Fusarium oxysporum f. sp. lycopersici . Appl. Environ. Microbiol. 62, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonowicz, A. , Cho, N.S. , Luterek, J. , Wilkolazka, A. , Wojtas‐Wasilewska, M. , Matuszewska, A. , Hofrichter, M. , Wesenberg, D. and Rogalski, J. (2001) Fungal laccase: properties and activity on lignin. J. Basic Microbiol. 41, 185–227. [DOI] [PubMed] [Google Scholar]
- Lievens, B. , Rep, M. and Thomma, B.P.H.J. (2008) Recent developments in the molecular discrimination of formae speciales of Fusarium oxysporum . Pest Manag. Sci. 64, 781–788. [DOI] [PubMed] [Google Scholar]
- Lopez‐Berges, M. , Di Pietro, A. , Daboussi, M.J. , Abdel Wahab, H. , Vasnier, C. , Roncero, M.I.G. , Dufresne, M. and Hera, C. (2009) Identification of virulence genes in Fusarium oxysporum f. sp. lycopersici by large‐scale transposon tagging. Mol. Plant Pathol. 10, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, M.P. , Di Pietro, A. and Roncero, M.I.G. (2003) Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47, 257–266. [DOI] [PubMed] [Google Scholar]
- Mandal, S. and Mitra, A. (2007) Reinforcement of cell wall roots of Lycopersicon esculentum through induction of phenolic compounds and lignin by elicitors. Physiol. Mol. Plant Pathol. 71, 201–209. [Google Scholar]
- Mandal, S. and Mitra, A. (2008) Accumulation of cell wall‐bound phenolic metabolites and their upliftment in hairy root cultures of tomato (Lycopersicon esculentum Mill.). Biotechnol. Lett. 30, 1253–1258. [DOI] [PubMed] [Google Scholar]
- Martinez‐Rocha, A.L. , Roncero, M.I.G. , Lopez‐Ramirez, A. , Marine, M. , Guarro, J. , Martinez‐Cadena, G. and Di Pietro, A. (2008) Rho1 has distinct functions in morphogenesis, cell wall biosynthesis and virulence of Fusarium oxysporum . Cell. Microbiol. 10, 1339–1351. [DOI] [PubMed] [Google Scholar]
- Martin‐Urdiroz, M. , Madrid, M.P. and Roncero, M.I.G. (2004) Role of chitin synthase genes in Fusarium oxysporum . Microbiol.-Sgm. 150, 3175–3187. [DOI] [PubMed] [Google Scholar]
- Martin‐Urdiroz, M. , Roncero, M.I.G. , Gonzalez‐Reyes, J.A. and Ruiz‐Roldan, C. (2008) ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum . Eukaryot. Cell, 7, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon, M.T. , Inglis, D.A. , McPhee, K.E. and Coyne, C.J. (2002) DNA markers linked to Fusarium wilt race 1 resistance in pea. J. Am. Soc. Hortic. Sci. 127, 602–607. [Google Scholar]
- McFadden, H.G. , Wilson, I.W. , Chapple, R.M. and Dowd, C. (2006) Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) genes expressed during infection of cotton (Gossypium hirsutum ). Mol. Plant Pathol. 7, 87–101. [DOI] [PubMed] [Google Scholar]
- McGrath, K.C. , Dombrecht, B. , Manners, J.M. , Schenk, P.M. , Edgar, C.I. , Maclean, D.J. , Scheible, W.R. , Udvardi, M.K. and Kazan, K. (2005) Repressor‐ and activator‐type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome‐wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C.B. , Van Wijk, R. , Reijnen, L. , Cornelissen, B.J. and Rep., M. (2009) Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large‐scale insertional mutagenesis. Gen. Biol. 10, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mes, J.J. , Weststeijn, E.A. , Herlaar, F. , Lambalk, J.J.M. , Wijbrandi, J. , Haring, M.A. and Cornelissen, B.J.C. (1999) Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology, 89, 156–160. [DOI] [PubMed] [Google Scholar]
- Mutlu, N. , Boyaci, F.H. , Göcmen, M. and Abak, K. (2008) Development of SRAP, SRAP‐RGA, RAPD and SCAR markers linked with a Fusarium wilt resistance gene in eggplant. Theor. Appl. Genet. 117, 1303–1312. [DOI] [PubMed] [Google Scholar]
- Nahalkova, J. , Fatehi, J. , Olivain, C. and Alabouvette, C. (2008) Tomato root colonization by fluorescent‐tagged pathogenic and protective strains of Fusarium oxysporum in hydroponic culture differs from root colonization in soil. FEMS Microbiol. Lett. 286, 152–157. [DOI] [PubMed] [Google Scholar]
- Namiki, F. , Matsunaga, M. , Okuda, M. , Inoue, I. , Nishi, K. , Fujita, Y. and Tsuge, T. (2001) Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis . Mol. Plant–Microbe Interact. 14, 580–584. [DOI] [PubMed] [Google Scholar]
- O'Donnell, K. , Kistler, H.C. , Cigelnik, E. and Ploetz, R.C. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA, 95, 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara, T. and Tsuge, T. (2004) FoSTUA, encoding a basic helix–loop–helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum . Eukaryot. Cell, 3, 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara, T. , Inoue, I. , Namiki, F. , Kunoh, H. and Tsuge, T. (2004) REN1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum . Genetics, 166, 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivain, C. , Humbert, C. , Nahalkova, J. , Fatehi, J. , L’Haridon, F. and Alabouvette, C. (2006) Colonization of tomato root by pathogenic and nonpathogenic Fusarium oxysporum strains inoculated together and separately into the soil. Appl. Environ. Microbiol. 72, 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina‐Giraldo, M.D. , Mullins, E. and Kang, S. (2003) Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis. Curr. Genet. 44, 49–57. [DOI] [PubMed] [Google Scholar]
- Oumouloud, A. , Arnedo‐Andres, M.S. , Gonzalez‐Torres, R. and Alvarez, J.M. (2008) Development of molecular markers linked to the Fom‐1 locus for resistance to Fusarium race 2 in melon. Euphytica, 162, 347–356. [Google Scholar]
- Pareja‐Jaime, Y. , Roncero, M.I.G. and Ruiz‐Roldan, M.C. (2008) Tomatinase from Fusarium oxysporum f. sp. lycopersici is required for full virulence on tomato plants. Mol. Plant–Microbe Interact. 21, 728–736. [DOI] [PubMed] [Google Scholar]
- Ramos, B. , Alves‐Santos, F.M. , Garcia‐Sdnehez, M.A. , Martin‐Rodrigues, N. , Eslava, A.P. and Diaz‐Minguez, J.M. (2007) The gene coding for a new transcription factor (ftfl) of Fusarium oxysporum is only expressed during infection of common bean. Fungal Genet. Biol. 44, 864–876. [DOI] [PubMed] [Google Scholar]
- Ramos‐Pamplona, M. and Naqvi, N.I. (2006) Host invasion during rice‐blast disease requires carnitine‐dependent transport of peroxisomal acetyl‐CoA. Mol. Microbiol. 61, 61–75. [DOI] [PubMed] [Google Scholar]
- Recorbet, G. , Steinberg, C. , Olivain, C. , Edel, V. , Trouvelot, S. , Dumas‐Gaudot, E. , Gianinazzi, S. and Alabouvette, C. (2003) Wanted: pathogenesis‐related marker molecules for Fusarium oxysporum . New Phytol. 159, 73–92. [DOI] [PubMed] [Google Scholar]
- Rep, M. (2005) Small proteins of plant‐pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253, 19–27. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Van Der Does, H.C. , Meijer, M. , Van Wijk, R. , Houterman, P.M. , Dekker, H.L. , De Koster, C.G. and Cornelissen, B.J.C. (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Meijer, M. , Houterman, P.M. , Van Der Does, H.C. and Cornelissen, B.J.C. (2005) Fusarium oxysporum evades I‐3‐mediated resistance without altering the matching avirulence gene. Mol. Plant–Microbe Interact. 18, 15–23. [DOI] [PubMed] [Google Scholar]
- Rocha, A.L.M. , Di Pietro, A. , Ruiz‐Roldan, C. and Roncero, M.I.G. (2008) Ctf1, a transcriptional activator of cutinase and lipase genes in Fusarium oxysporum, is dispensable for virulence. Mol. Plant Pathol. 9, 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddick, J.G. (1977) Subcellular localization of steroidal glycoalkaloids in vegetative organs of Lycopersicon esculentum and Solanum tuberosum . Phytochem. 16, 805–807. [Google Scholar]
- Von Ropenack, E. , Parr, A. and Schulze‐Lefert, P. (1998) Structural analyses and dynamics of soluble and cell wall‐bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J. Biol. Chem. 273, 9013–9022. [DOI] [PubMed] [Google Scholar]
- Rosales, R.C.P. and Di Pietro, A. (2008) Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum . Eukaryot. Cell, 7, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Roldan, C. , Puerto‐Galan, L. , Roa, J. , Castro, A. , Di Pietro, A. , Roncero, M.I.G. and Hera, C. (2008a) The Fusarium oxysporum sti35 gene functions in thiamine biosynthesis and oxidative stress response. Fungal Genet. Biol. 45, 6–16. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Roldan, M.C. , Garre, V. , Guarro, J. , Marine, M. and Roncero, M.I.G. (2008b) Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum . Eukaryot. Cell, 7, 1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado, M.O. , Schwartz, H.F. and Brick, M.A. (1995) Inheritance of resistance to a Colorado race of Fusarium oxysporum f. sp. phaseoli in common beans. Plant Dis. 79, 279–281. [Google Scholar]
- Sela‐Buurlage, M.B. , Budai‐Hadrian, O. , Pan, Q. , Carmel‐Goren, L. , Vunsch, R. , Zamir, D. and Fluhr, R. (2001) Genome‐wide dissection of Fusarium resistance in tomato reveals multiple complex loci. Mol. Genet. Genomics, 265, 1104–1111. [DOI] [PubMed] [Google Scholar]
- Sharma, K.D. and Muehlbauer, F.J. (2007) Fusarium wilt of chickpea: physiological specialization, genetics of resistance and resistance gene tagging. Euphytica, 157, 1–14. [Google Scholar]
- Simons, G. , Groenendijk, J. , Wijbrandi, J. , Reijans, M.G.J. , Diergaarde, P. , Van Der Lee, T. , Bleeker, M. , Onstenk, J. , De Both, M. , Haring, M.A. , Mes, J.J. , Cornelissen, B.J.C. , Zabeau, M. and Vos, P. (1998) Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell, 10, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan, S. , Jedd, G. , Li, X.L. , Ramos‐Pamplona, M. , Chua, N.H. and Naqvi, N.I. (2004) Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell, 16, 1564–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney, K. , Hunt, I. , Sweigard, J. , Pounder, J.I. , McClain, C. , Bowman, E.J. and Bowman, B.J. (2000) hex‐1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31, 205–217. [DOI] [PubMed] [Google Scholar]
- Thaler, J.S. , Owen, B. and Higgins, V.J. (2004) The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , Van Esse, H.P. , Crous, P.W. and De Wit, P.J.G.M. (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6, 379–393. [DOI] [PubMed] [Google Scholar]
- Tierens, K.F.M.J. , Thomma, B.P.H. , Brouwer, M. , Schmidt, J. , Kistner, K. , Porzel, A. , Mauch‐Mani, B. , Cammue, B.P.A. and Broekaert, W.F. (2001) Study of the role of antimicrobial glucosinolate‐derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 125, 1688–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjamos, E.C. and Beckman, C.H. (1989) Vascular Wilt Diseases of Plants: Basic Studies and Control. Berlin: Springer‐Verlag. [Google Scholar]
- Trusov, Y. , Rookes, J.E. , Chakravorty, D. , Armour, D. , Schenk, P.M. and Botella, J.R. (2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol. 140, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakalounakis, D.J. (1996) Allelism of the Fcu‐1 and Foc genes conferring resistance to Fusarium wilt in cucumber. Eur. J. Plant Pathol. 102, 855–858. [Google Scholar]
- Vandenbosch, H. , Schutgens, R.B.H. , Wanders, R.J.A. and Tager, J.M. (1992) Biochemistry of peroxisomes. Annu. Rev. Biochem. 61, 157–197. [DOI] [PubMed] [Google Scholar]
- VanEtten, H.D. , Mansfield, J.W. , Bailey, J.A. and Farmer, E.E. (1994) Two classes of plant antibiotics: phytoalexins versus ‘phytoanticipins’. Plant Cell, 6, 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hemelrijck, W. , Wouters, P.F.W. , Brouwer, M. , Windelinckx, A. , Goderis, I.J.W.M. , De Bolle, M.F.C. , Thomma, B.P.H.J. , Cammue, B.P.A. and Delaure, S.L. (2006) The Arabidopsis defense response mutant esa1 as a model to discover novel resistance traits against Fusarium diseases. Plant Sci. 171, 585–595. [Google Scholar]
- Wang, C. and Roberts, P.A. (2006) A Fusarium wilt resistance gene in Gossypium barbadense and its effect on root‐knot nematode‐wilt disease complex. Phytopathol. 96, 727–734. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y. , Soanes, D.M. , Kershaw, M.J. and Talbot, N.J. (2007) Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta‐oxidation during appressorium‐mediated plant infection. Mol. Plant–Microbe Interact. 20, 475–491. [DOI] [PubMed] [Google Scholar]
- Weld, R.J. , Plummer, K.M. , Carpenter, M.A. and Ridgway, H.J. (2006) Approaches to functional genomics in filamentous fungi. Cell Res. 16, 31–44. [DOI] [PubMed] [Google Scholar]
- Wu, H.S. , Liu, D.Y. , Ling, N. , Bao, W. , Ying, R.R. , Ou, Y.H. , Huo, Z.H. , Li, Y.F. and Shen, Q.R. (2008a) Effects of vanillic acid on the growth and development of Fusarium oxysporum f. sp niveum . Allelopath. J. 22, 111–121. [Google Scholar]
- Wu, H.S. , Raza, W. , Fan, J.Q. , Sun, Y.G. , Bao, W. and Shen, Q.R. (2008b) Cinnamic acid inhibits growth but stimulates production of pathogenesis factors by in vitro cultures of Fusarium oxysporum f. sp. niveum . J. Agric. Food Chem. 56, 1316–1321. [DOI] [PubMed] [Google Scholar]
- Wu, H.S. , Raza, W. , Liu, D.Y. , Wu, C.L. , Mao, Z.S. , Xu, Y.C. and Shen, Q.R. (2008c) Allelopathic impact of artificially applied coumarin on Fusarium oxysporum f. sp. niveum . World J. Microbiol. Biotechnol. 24, 1297–1304. [Google Scholar]
- Zhu, X.D. and Williamson, P.R. (2004) Role of laccase in the biology and virulence of Cryptococcus neoformans . FEMS Yeast Res. 5, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhu, X.D. , Gibbons, J. , Garcia‐Rivera, J. , Casadevall, A. and Williamson, P.R. (2001) Laccase of Cryptococcus neoformans is a cell wall‐associated virulence factor. Infect. Immun. 69, 5589–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]


