SUMMARY
Higher plants possess large multigene families encoding secreted class III peroxidase (Prx) proteins. In barley, two Prx cDNAs encoding HvPrx07 and HvPrx08 have been isolated and characterized to some extent with respect to a resistance‐mediating function upon attack by the powdery‐mildew fungus Blumeria graminis f.sp. hordei (Bgh). Here we present evidence for the tissue‐specific accumulation of a new Prx mRNA, HvPrx40, in Bgh‐attacked epidermis of barley (Hordeum vulgare). The encoded protein is predicted to be secreted into the apoplastic space of epidermal cells due to the absence of a C‐terminal extension, which distinguishes it from other Prx proteins reported to accumulate in leaf epidermis. Transient overexpression of HvPrx40 enhanced the resistance of wheat (Triticum aestivum) and barley against Blumeria graminis f.sp. tritici (wheat powdery mildew) and Bgh, respectively. These findings were complemented by transient‐induced gene silencing showing hypersusceptibility of barley leaf epidermal cells to Bgh. The local accumulation of oxidized 3,3‐diaminobenzidine that reflects H2O2 production at sites of attempted fungal penetration was not reduced in HvPrx40‐silenced cells, suggesting a role of this peroxidase other than the production of reactive oxygen species.
INTRODUCTION
Higher plants possess large multigene families encoding class III peroxidase (Prx) proteins (Passardi et al., 2004a). This subgroup of peroxidases is secreted into the apoplastic space or the plant vacuole, depending on the presence of a C‐terminal extension serving as a vacuolar retention signal, and is characterized by a number of conserved residues or domains that include two heme‐binding histidines of the catalytic centre, eight cystein residues forming disulfide bridges, plus a highly conserved domain rather close to the N‐terminus of the protein. Plant class III peroxidases have been proposed as key regulators of the level of extracellular H2O2 and as producers of the extremely reactive oxygen intermediates (ROI) OH• and OOH•, depending on whether the peroxidative (catalytic) or the hydoxylic cycles of the enzyme are operating (Kawano, 2003; Passardi et al., 2004b). During one peroxidative cycle, one consumed molecule of H2O2 can maximally lead to two produced H2O2 molecules if the two oxidized peroxidase products (X•) react with two O2 molecules to produce superoxide radicals, which are converted spontaneously or catalysed by superoxide dismutase to H2O2. There has been evidence from tobacco and french bean that Prx proteins do indeed produce H2O2 in vivo (Bindschedler et al., 2006; Bolwell et al., 2002; Mader and Ambergfisher, 1982; Peng and Kuc, 1992). According to these results Prx proteins produce radicals and ROI as catalysists of covalent cell‐wall modifications (Barcelo, 1995; Harkin and Obst, 1973; Ostergaard et al., 2000) as well as signals for cell‐death reactions (Bestwick et al., 1998; Martinez et al., 1998; Passardi et al., 2004a).
In Arabidopsis thaliana and rice (Oryza sativa), a total of 73 and 138 Prx genes have been annotated, respectively, but a physiological function for most of the encoded proteins is still missing. This may be largely due to the complexity of Prx gene families and to suspected functional redundancy beween individual, duplicated familiy members. One way to obtain further information about the function of individual Prx genes is to employ methods of genomics such as transcript profiling or the generation of transgenic plants resulting in gain or loss of gene function by overexpression or RNAi‐mediated gene silencing. A number of transcript profiling approaches to multiple Prx genes per genome has been undertaken in rice, wheat and Arabidopsis (Liu et al., 2005; Sasaki et al., 2004; Welinder et al., 2002). These studies revealed that Prx mRNAs often accumulate upon biotic stress, indicating a role in warding of pathogens. Recently, plant class III peroxidases have been catalogued by using a consistent nomenclature (Bakalovic et al., 2006). This nomenclature has been adopted for the peroxidases addressed in the study presented here.
In the cereal crop barley, a similar level of complexity of class III peroxidases appears to exist at the genomic level, with a total of 105 unigenes presently known (Bakalovic et al., 2006) (Supporting Information Fig. S1). However, only a very limited number of Prx genes have been studied in more detail until now. HvPrx07 has been described as a protein accumulating in the central vacuole of Bgh‐attacked barley leaf epidermis, and a role in the accumulation of antifungal compounds such as hordatines has been proposed (Kristensen et al., 1999). However, transient overexpression in barley epidermal cells enhanced susceptibility, which might reflect a more complex function of this peroxidase or a pleiotropic effect of overexpression (Kristensen et al., 2001). Another peroxidase of barley, HvPrx08, has also been described as pathogen‐induced at the mRNA as well as protein level (ScottCraig et al., 1995; Thordal‐Christensen et al., 1992). HvPrx08 is likely to be orthologous to the wheat gene TaPrx103, which has also been described to be induced by powdery mildew and for which functional data are available in a transient expression system as well as in transgenic plants (Altpeter et al., 2005; Schweizer et al., 1999; Schweizer, 2008).
Attack of barley by powdery mildew fungi resulted in either local or whole‐cell accumulation of H2O2, reflected by the oxidation of 3,3‐diaminobenzidine (DAB) (ThordalChristensen et al., 1997). Sources for this type of reactive oxygen species (ROI) might be a shunt consisting of NADPH oxidase plus superoxide dismutase. In accordance with this model, germin‐like proteins including HvGER4 and HvGER5 from barley have recently been described as apoplastic SOD enzymes (Zimmermann et al., 2006). In addition, the shunt might also involve peroxidases as signal amplifiers, as discussed above. Here, we functionally address a new peroxidase protein that specifically accumulates in Blumeria graminis f.sp. hordei (Bgh)‐attacked barley epidermis and tested its association with disease resistance and H2O2 formation.
RESULTS
Sequence analysis
Gene‐expression profiling revealed a transcript that was up‐regulated in Bgh‐attacked barley epidermis (Zierold et al., 2005). Moreover, this transcript hybridizing to cDNA clone HO13D15 (accession no. CD056766) appeared to accumulate more strongly in resistant barley epidermis carrying the mlo5 resistance gene, compared with the near‐isogenic susceptible barley cultivar Ingrid. Therefore, the encoded protein might contribute to race‐nonspecific basal or mlo‐mediated resistance. The sequence of EST clone HO13D15 was analysed by BlastX and found to be similar to class III peroxidases of rice and other plant species. A full‐coding‐sequence (CDS) cDNA clone was obtained (HO26F22; accession no. DN180661) and its sequence compared with 5470 manually curated peroxidase sequences (http://peroxidase.isb‐sib.ch/index.php). This analysis revealed identity of the protein encoded by cDNA HO26F22 to a barley peroxidase of unknown function annotated as HvPrx40 with a predicted mass of 38.3 kDa and a pI of 7.95. The entire cDNA sequence of HvPrx40 is available as unigene consensus no. 14156 of assembly 35 in the HarvEST database (http://harvest‐web.org/). Figure 1 shows the alignment of HvPrx40 to a number of peroxidases of barley, wheat and rice, as well as their phylogenetic relationship. In contrast to HvPrx07 and a number of peroxidases described in Triticum monococcum to accumulate in leaf epidermis, HvPrx40 is predicted to be secreted into the apoplast due to the absence of a C‐terminal vacuolar retention signal (Fig. 1A). All analysed peroxidases contain the two highly conserved, catalytic histidine as well as eight conserved cysteine residues that build four intramolecular disulfide bonds (Hiraga et al., 2001). Interestingly, HvPrx40 and TaPrx47 contain an N‐terminal extension of 36 amino acids starting just after the predicted signal peptide‐cleavage site, which is highly basic (pI = 11.7) due to seven arginine residues. Two cysteine residues in this basic peptide might form a fifth disulfide bond of the protein. The role of this N‐terminal extension is unknown and possesses no similarity to any functional domain. However, similar sequences were identified in two additional barley and two wheat peroxidases (HvPrx61, HvPrx75, TaPrx47, TaPrx51) whereas HvPrx61 and HvPrx75 are not the most closely related peroxidases to HvPrx40 (Supporting Information Fig. S1). Clearly, HvPrx40 is distinct from the previously described peroxidases HvPrx07 and HvPro08 of barley but corresponds to a putative protein orthologue in wheat, TaPrx47 (Fig. 1B).
Figure 1.
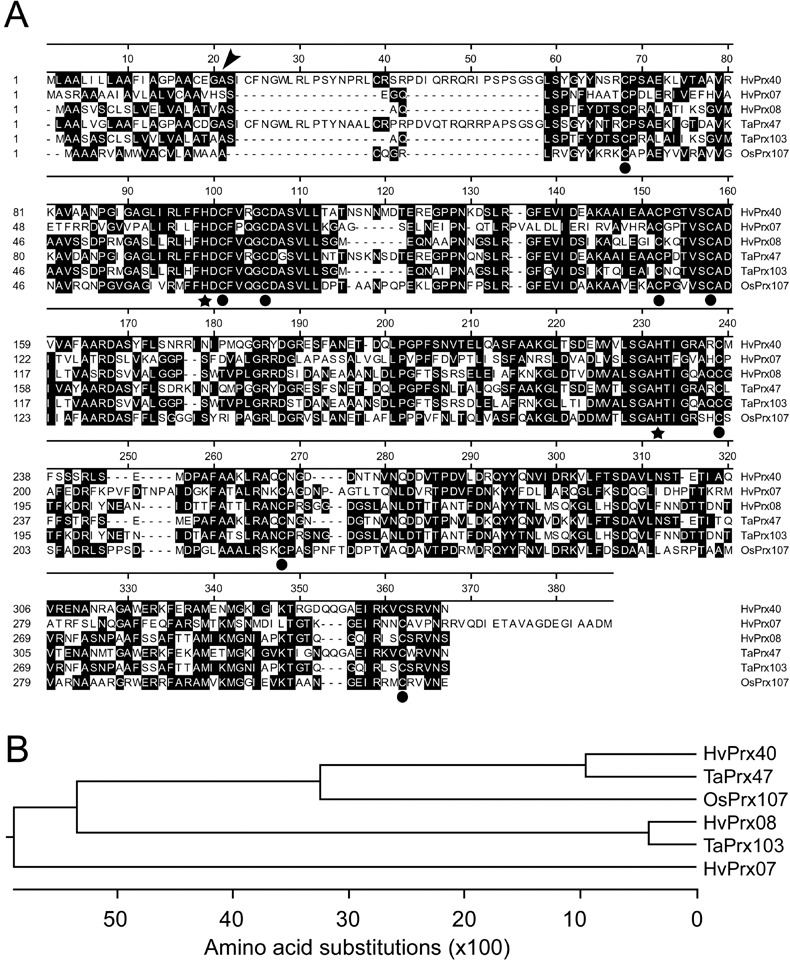
Sequence comparison of HvPrx40 protein with selected peroxidases from cereal species. (A) Amino‐acid alignment of HvPrx40 with other peroxidases by Clustal W. Conserved residues are shaded in black. Arrow, putative cleavage site of N‐terminal signal peptide; dots, conserved cysteine residues forming four disulfide bridges; stars, conserved histidine residues of the catalytic centre. (B) Phylogenetic tree of HvPrx40 and selected peroxidases, based on Clustal W.
Transcript accumulation
In order to test pathogen‐induced HvPrx40 transcript accumulation in susceptible barley and in near‐isogenic lines carrying two different alleles of Mlo (mlo3 and mlo5), we carried out quantitative real‐time reverse‐transcriptase (RT) PCR in leaves and stripped epidermis at 12 h post‐inoculation (hpi) (Fig. 2). For comparison, mRNA accumulation of HvPrx08 was also examined. Transcripts of both peroxidases accumulated upon attack by Bgh. However, the tissue distribution of mRNA accumulation was clearly different between the two genes: whereas HvPrx08 transcripts mainly accumulated in inner leaf tissue, as reflected by higher signals in entire‐leaf samples where epidermal RNA is diluted, transcripts of HvPrx40 mainly accumulated in attacked epidermis. The weak HvPrx08‐specific signal in stripped epidermis probably reflects contamination by adhering mesophyll cells (data not shown). The mesophyll‐specific accumulation of HvPrx08 mRNA was also in agreement with a previous report (Gregersen et al., 1997). No difference of HvPrx40 transcript abundance between attacked susceptible and near‐isogenic, mlo‐resistant lines was found, although a stronger accumulation was reported based on cDNA array data (Zierold et al., 2005). However, as confirmed in a second independent series of array hybridizations, the differential accumulation between susceptible and resistant plants was only observed 24 hpi and may therefore have escaped detection by the qPCR reported here, which was carried out 12 hpi (Supporting Information Fig. S2). It should also be noted that transcript abundance of HvPrx40 was considerably lower, compared with HvPrx08, arguing against HvPrx40 protein to be a mass protein in epidermal tissue (Fig. 2). HvPrx40 appears to be a member of a Prx40‐like subfamily with at least three paralogous genes that are all characterized by the presence of the N‐terminal basic peptide and that are represented by unigenes no. 14156 (HvPrx40), 14158 and 31622 of the HarvEST assembly 35 of barley (http://www.harvest‐web.org/). EST abundance data revealed unigene 14156 to be the highest expressed in Bgh‐challenged epidermis, followed by unigene 14158. Unigene 31622 was represented by one EST only from ethiolated leaves.
Figure 2.
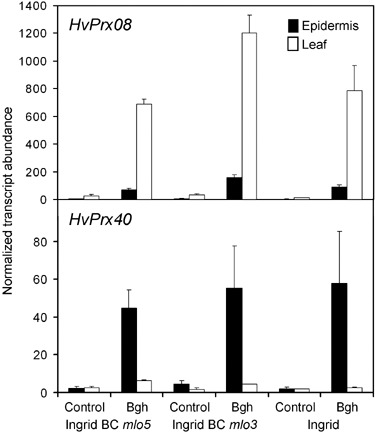
mRNA Abundance of two selected peroxidases following inoculation with B.graminis f.sp. hordei (Bgh). RNA was isolated from barley leaves or stripped leaf epidermis at 12 h post inoculation with Bgh, followed by reverse transcription and quantitative real‐time PCR. Data are normalized to constitutively expressed control gene UBC encoding a ubiquitin‐conjugating enzyme. Values are means ± SE from two independent inoculation experiments.
HvPrx40 and disease resistance
We have previously shown that transient or stable overexpression of TaPrx103, which is the proposed wheat orthologue of HvPrx08, enhanced resistance to wheat powdery mildew (Altpeter et al., 2005). However, because TaPrx103 was found to be mainly expressed in inner leaf tissues, the overexpressing epidermal cells represent an artificial system that is not easy to relate to naturally occurring defence reactions. By contrast, HvPrx40 was found to be naturally expressed in the relevant tissue for Bgh interactions. This prompted us to test a possible role of this peroxidase in basal resistance of barley by transiently over‐expressing or silencing HvPrx40 in single epidermal cells, followed by Bgh inoculation. The plasmid constructs used for these experiments are shown in Fig. 3. Transient over‐expression significantly enhanced penetration resistance of transformed cells against Bgh, as reflected by a reduced susceptibility index that corresponds to the relative rate of haustorium‐containing, GUS‐expressing cells (Table 1). A similar, enhanced resistant single‐cell phenotype was observed by overexpressing the orthologous genes encoding either TaPrx103 or HvPrx08, too, whereby TaPrx103 served as a positive control (Schweizer et al., 1999). Complementary to the overexpression data, transient‐induced gene silencing (TIGS) of HvPrx40 by RNAi caused hypersusceptibility of transformed cells of barley cv. Golden Promise. The efficiency of the TIGS construct targeting HvPrx40 was tested in a co‐bombardment experiment using pIPKTA30N_HvPrx40 (RNAi construct) plus pIPKTA40_HvPrx40deltaSP (HvPrx40:GFP target construct). Table 2 demonstrates that the number of fluorescent cells expressing HvPrx40:GFP was largely reduced in the presence of the corresponding co‐bombarded RNAi construct, whereas the number of fluorescing cells expressing wild‐type GFP was not affected by the same RNAi construct. The RNAi effect on cells expressing pIPKTA40_HvPrx40deltaSP was sequence‐specific because the control RNAi construct pIPKTA30N_HO14J07 targeting a non‐related transaldolase gene of barley increased the number of GFP‐fluorescing cells compared with the empty RNAi vector (Table 2). The overall decreased number of fluorescent cells expressing the pIPKTA40_HvPrx40deltaSP construct probably reflects negative interference of the Prx fusion partner with correct GFP folding, or reduced HvPrx40:GFP mRNA or fusion‐protein stability. In conclusion, the two opposing effects of over‐expression versus gene silencing of HvPrx40 argues against a non‐specific effect caused by either construct and suggests the HvPrx40 peroxidase as a relevant factor of basal resistance in barley. Unlike HvPrx40, a second peroxidase specifically accumulating in attacked epidermis, HvPrx07, did not significantly affect the susceptible interaction with Bgh upon transient overexpression or TIGS (Table 1). The absence of a resistance‐enhancing effect by HvPrx07 overexpression is in accordance with previous data, where enhanced susceptibility was observed. Finally, it should be noted that the transient overexpression of HvPrx40 in wheat epidermal cells also caused powdery‐mildew resistance, indicating that the mode of action of the encoded protein is similar in wheat and barley.
Figure 3.
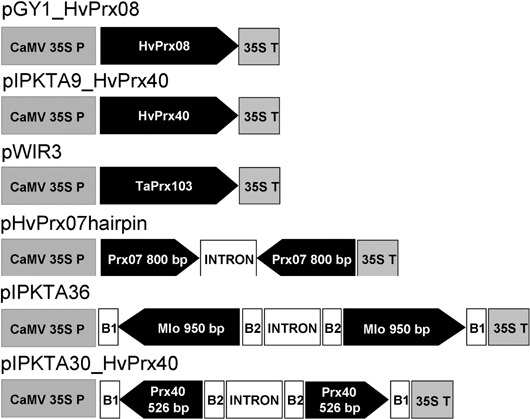
Plasmid constructs used for transient overexpression and transient‐induced gene silencing (not drawn to scale). The intron in the RNAi hairpin constructs was derived from the wheat RGA2 gene (AF326781). P, promoter; T, terminator; B1 and B2, attachment sites B1 and B2 for LR clonase, respectively.
Table 1.
Effect of transient overexpression and TIGS of Prx genes on basal resistance of barley and wheat.
| Construct* | Relative SI (% of empty vector control)† | |||
|---|---|---|---|---|
| Overexpression | TIGS | |||
| Barley G.Pr.‡ | Wheat | Barley G.Pr. | Barley Ingrid | |
| pGY1_HvPrx08 | 70 ± 11 | n.a. | — | — |
| (OEx HvPrx08) | (P = 0.0391) | |||
| pIPKTA9_HvPrx40 | 83 ± 4 | 45 ± 3 | — | — |
| (OEx HvPrx40) | (P = 0.0008) | (P < 0.0001) | ||
| pWIR3§ | 54 ± 4 | 27 ± 2 | — | — |
| (OEx TaPrx103) | (P < 0.0001) | (P < 0.0001) | ||
| pHvPrx07hairpin¶ | — | — | 116 ± 56 | 124 ± 45 |
| (TIGS HvPrx07) | (P = 0.7872) | (P = 0.5527) | ||
| pIPKTA30N_HvPrx40 | — | — | 184 ± 28 | 112 ± 10 |
| (TIGS HvPrx40) | (P = 0.0105) | (P = 0.2547) | ||
| pIPKTA34** | — | — | 14 ± 4 | 11 ± 2 |
| (TIGS HvMlo) | (P = 0.0001) | (P < 0.0001) | ||
All constructs contain the CamV 35S promoter and terminator. See Fig. 3.
SI, susceptibility index = number of GUS‐positive cells containing at least one haustorium/total number of GUS‐positive cells.
G.Pr, barley cv. Golden Promise.
Described by Schweizer et al. (1999).
Described by Christensen et al. (2004).
Described by Douchkov et al. (2005).
Table 2.
Specific silencing of a HvPrx40:GFP fusion gene by the co‐bombarded RNAi construct that targets HvPrx40.
| Co‐bombardment | GFP cells* | GFP/GUS† |
|---|---|---|
| pUbiGUS+pGFP+pIPKTA30N | 951.5 ± 89.5 | 2.30 ± 0.18 (100) |
| pUbiGUS+pGFP+pIPKTA30N_HvPrx40 | 812.0 ± 144.0 | 2.04 ± 0.25 (89) |
| pUbiGUS+pGFP+pIPKTA30N_HO14J07‡ | 682.5 ± 30.5 | 1.89 ± 0.19 (82) |
| pUbiGUS+pIPKTA40_HvPrx40deltaSP+pIPKTA30N | 37.0 ± 2.0 | 0.10 ± 0.01 (100) |
| pUbiGUS+pIPKTA40_HvPrx40deltaSP+pIPKTA30N_HvPrx40 | 16.3 ± 6.3 | 0.03 ± 0.01 (30) |
| pUbiGUS+pIPKTA40_HvPrx40deltaSP+pIPKTA30N_HO14J07 | 138.0 ± 10.5 | 0.35 ± 0.07 (359) |
Number of GFP‐expressing cells per bombardment. Means ± range of two independent experiments.
Ratio of GFP‐expressing to GUS‐expressing cells. Mean ± range of two independent experiments. Values in parentheses indicate relative values in percentage of the corresponding empty RNAi‐vector control.
Control RNAi construct targeting a transaldolase gene (represented by EST clone HO14J07).
HvPrx40 and localized H2O2 production
Class III peroxidases have been implicated in the production of reactive oxygen species, especially H2O2 (Passardi et al., 2004b). The localized accumulation of H2O2 at cell‐wall appositions (papillae) that are produced by barley epidermal cells beneath primary germ tubes and appressoria of Bgh has been shown and associated with enhanced basal resistance mediated by the mlo gene (Piffanelli et al., 2002). Because we have demonstrated an effect of HvPrx40 over‐expression or silencing on basal resistance, we tested whether TIGS of HvPrx40 affects the local accumulation of H2O2 at sites of attempted penetration by Bgh. For this purpose, we infiltrated bombarded leaf segments with DAB, which is oxidized to an insoluble reddish brown product at sites of H2O2 or OH• production. As shown in Fig. 4, localized DAB oxidation was efficiently recorded in transformed, GUS‐stained cells as well as in surrounding, non‐transformed cells. Next, we compared the rate of detectable DAB oxidation in GUS‐expressing cells, relative to four indirect neighbouring cells (north, east, south and west of the transformed cell, separated by one cell in order to reduce possible neighbouring effects; Carver et al., 1999). TIGS of HvPrx40 caused no significant decrease of relative DAB oxidation (Table 3), suggesting that HvPrx40 is not a major catalyst of localized production of H2O2 at sites of attempted fungal penetration.
Figure 4.
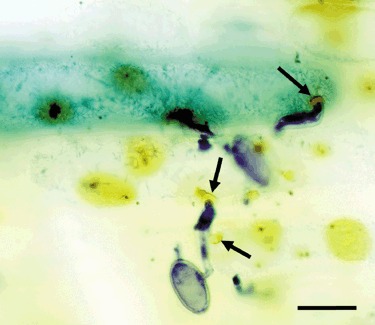
Detection of localized DAB oxidation reflecting H2O2 accumulation in transformed, GUS‐stained cells as well as in surrounding cells. Arrows point at sites of attempted penetration associated with DAB staining. Epiphytic fungal structures were stained with Coomassie blue. Scale bar, 25 µm.
Table 3.
Hydrogen peroxide accumulation in cell‐wall appositions upon transient silencing of HvPrx40.
| DAB‐stained CWA (% of surrounding cells)* | ||
|---|---|---|
| pIPKTA30N | pIPKTA30N_HvPrx40 | |
| Bombardment 1 | 81.9 | 99.1 |
| Bombardment 2 | 97.0 | 87.0 |
| Bombardment 3 | 109.9 | 76.8 |
| Bombardment 4 | 110.3 | 86.7 |
| Bombardment 5 | 97.1 | 117.1 |
| Mean | 99.2 | 93.3 |
| SEM | 5.2 | 6.9 |
| p (one sample t‐test) | 0.891 | 0.39 |
Local accumulation of hydrogen peroxide in cell‐wall appositions (CWA) beneath fungal penetration attempts was monitored by DAB staining. The percentage of DAB‐stained CWA of GUS‐positive, transformed cells was compared with four surrounding, attacked cells (north, east, south and west) that were not transformed. The analysis is based on a total of 119–124 GUS‐positive, attacked cells per construct.
DISCUSSION
Defence of barley epidermal cells against Bgh or other, inappropriate powdery mildew fungi is likely to include peroxidase isoforms for apoplastic cross‐linking events, lignification or the production of H2O2. The peroxidase‐encoding gene HvPrx40 attracted our attention because its transcripts accumulated specifically in Bgh‐attacked leaf epidermis, with stronger accumulation in mlo‐resistant versus susceptible plants. A closer look at the complexity of peroxidase‐encoding gene expression in barley using a cDNA array of 10 297 spotted unigenes revealed 69 unigenes producing detectable signals in stripped leaf epidermis. However, transcripts of only six unigenes were both enriched by a factor of at least two in epidermis and up‐regulated at least two‐fold by pathogen attack (A. Himmelbach and P. Schweizer, unpublished data). Four unigenes belonged to subfamilies that include HvPrx07 and HvPrx40, leaving only two as yet completely unknown peroxidase genes (HvPrx52 and HvPrx97) for future functional analysis.
Transient overexpression and TIGS of HvPrx40 reduced and enhanced haustorium formation of a compatible Bgh isolate in cv. Golden Promise, respectively. However, an assessment of off‐targets for the Prx40‐RNAi construct used here revealed a total of 16 tentative unigene consensi of the barley Gene Index of TIGR as potential off‐targets besides HvPrx40, most of them annotated as peroxidases (http://bioinfo2.noble.org/RNAiScan.htm). Therefore, the phenotypic effect observed upon TIGS of HvPrx40 might be actually due to silencing of multiple Prx genes. Nevertheless, the transient overexpression experiments, which revealed an interaction phenotype in barley (and wheat) opposite to the TIGS effect, suggest that HvPrx40 is indeed a factor of basal resistance in barley, similar to HvGER4, another secreted protein with superoxide‐dismutase activity that accumulates in Bgh‐attacked epidermis (Christensen et al., 2004, Zimmermann et al., 2006). As we found in the case of HvGER4, silencing of HvPrx40 did not affect basal resistance in cv. Ingrid, indicating that the importance of the encoded protein is dependent on the host genotype, a common phenomenon in the analysis of quantitative‐trait loci (QTL) for basal resistance. The product of HvGER4 is H2O2, which can either be consumed by Prx proteins such as those involved in lignification or be further amplified by Prx proteins such as NAD(P)H oxidizing isoforms. However, the unaffected DAB oxidation of cells with silenced HvPrx40 does not support the appealing hypothesis of an ROI shunt in leaf epidermis consisting of HvGER4 and HvPrx40 or HvPrx40‐like proteins. This of course does not exclude the existence of such an ROI shunt involving other Prx genes. Data from TaPrx103‐overexpressing transgenic wheat that exhibits enhanced DAB oxidation upon wheat powdery‐mildew attack support the participation of this Prx protein in H2O2 formation (Schweizer, 2008). However, because TaPrx103/HvPrx08 and TaGER4/HvGER4 proteins are predicted to accumulate in mesophyll and epidermis, respectively, they are not likely candidates for the above‐mentioned ROI shunt, except perhaps at the interface of the two tissues. Recently, an involvement of chelatable Fe3+ in the localized DAB oxidation of barley epidermal cells was shown, and free Fe3+ plus H2O2 was found to be sufficient to oxidize DAB in vitro in a non‐enzymatic reaction (Liu et al., 2007). Despite these findings, the question as to the mechanism of localized H2O2 formation remains open.
The mature HvPrx40 protein possesses a highly basic N‐terminal extension of 36 amino acids containing seven arginine residues that is not found in any other Prx besides two and one close homologues in barley and wheat, respectively. This basic peptide is predicted to be positively charged in the acidic apoplastic environment and therefore possesses affinity to negatively charged structures such as pectate. Indeed, (Ca2+)‐pectate binding of peroxidases from zuccinin and Arabidopsis has been demonstrated, and binding of the zuccinin protein was found to be dependent on a cluster of four arginine residues whereas the two pectate‐binding peroxidases from Arabidopsis were found to be required for root‐hair elongation (Carpin et al., 2001; Passardi et al., 2006). Based on these data it appears possible that HvPrx40 is guided to negatively charged epidermal cell‐wall components such as partially acetylated arabinoxylans. However, its defence‐related function in the cell wall remains to be examined.
EXPERIMENTAL PROCEDURES
Plants and pathogens
Barley (Hordeum vulgare) plants cv. Golden Promise, Ingrid, the backcross lines Ingrid BC mlo3 and Ingrid BC mlo5 as well as the wheat (Triticum aestivum) cv. Kanzler were grown in pots of compost soil (from the IPK nursery) in a growth chamber at 20 °C with 70% relative humidity and 16 h light from metal halogen lamps and 8 h of dark. Blumeria graminis f.sp. hordei (Bgh) (Swiss field isolate CH4.8) was maintained at 22 °C and 16 h of light by weekly transfer to fresh barley cv. Golden Promise. Blumeria graminis f.sp. tritici (Bgt) (Swiss field isolate FAL92315) was maintained at 20 °C and 16 h of light by weekly transfer to fresh wheat cv. Kanzler.
For quantitative RT‐PCR experiments, 7‐day‐old barley plants (cv. Ingrid) or Ingrid backcross lines carrying mlo3 or mlo5 alleles were inoculated with Bgh. Twelve hours after inoculation the abaxial epidermis of control and inoculated plants was stripped and the epidermis plus remaining leaf material were immediately frozen in liquid N2.
Real‐time RT‐PCR
Total RNA was extracted from epidermal peels (abaxial) and the remainder from peeled leaves of barley plants. By using the iScript cDNA synthesis kit (Bio‐Rad, München, Germany) 1 µg RNA was reverse transcribed, following the manufacturer's instructions. One µL of 100 µL of the resulting cDNA solution was used for real‐time RT‐PCR by using the POWER SYBR® GREEN PCR MASTER MIX kit (Applied Biosystems) in an ABI 7900HT Fast Real‐Time PCR system (Applied Biosystems). The PCR conditions were: 50 °C for 2 min, followed by a denaturation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 1 min and extension at 72 °C for 30 s, followed by a dissociation step at 95 °C for 15 s, 60 °C for 15 s and finally 95 °C for 15 s. The mRNA abundance of HvPrx08 and HvPrx40 was normalized to HvGAPDH and HvUBC.
The following PCR primers were used: HvPrx08 (TC139154 DFCI Hordeum vulgare Gene Index), 5′‐CTCGGACCAGGTGCTCTTCAACA‐3′ (sense) plus 5′‐TGGCCCTGCGTCCCCGT‐3’ (reverse); HvPrx40 (TC123108, DFCI Hordeum vulgare Gene Index), 5′‐GAGCAATGGAGAATATGGGAAAAATC‐3′ (sense) plus 5′‐GGAGCCAGCCAGCCAAAC AC‐3′ (reverse); HvGAPDH (TC131363, DFCI Hordeum vulgare Gene Index), 5′‐CAATGCTAGCTGCACCACCAACTG‐3’ (sense) plus 5′‐CTAGCAGCCCTTCCACCTCTCCA‐3′ (reverse); HvUBC (TC139319, DFCI Hordeum vulgare Gene Index), 5′‐AAGCAGCCAGAATGTACAGCGAGAAC‐3′ (sense) plus 5′‐GGTACAGACCAGCAAAGCCAGAAATG‐3′ (reverse).
Reverse Northern blot
Reverse Northern blot experiments were performed by using a barley cDNA array of 10 297 spotted unigenes, as described (Zimmermann et al., 2006).
Plasmid constructs for transient overexpression
For HvPrx08 the full CDS‐cDNA was cloned into the SphI site of the transient expression vector pGY1 (Shinshi et al., 1990), which contains the CaMV 35S promoter, a multiple cloning site and the CaMV 35S terminator.
The full CDS‐cDNA for HvPrx40 was cloned into the PstI and XhoI sites of the transient expression vector pIPKTA9, which is identical to pGY1, except for a more complex multiple cloning site.
Plasmid construct for TIGS
The amplification of DNA fragments for the RNAi construct pIPKTA30N_HvPrx40 was performed using cDNA clone HO13D15, the non‐specific sense primer MVR‐26 (5′‐CTCACTAAAGGGAACAAAAGCTGGAG‐3′) plus the gene‐specific reverse primer pIPKTA30_HO13D15 (5′‐GATTGTGTGTGCACCGGAGA‐3′). The preparation of the RNAi construct was performed as described by Douchkov et al. (2005) by using the Gateway™ recombination system, except that pIPKTA33 was used as entry vector (cloning site: EcoRV). The construct pHvPrx07hairpin has been generated by conventional ligation, as described (Christensen et al., 2004). However, no effects of the B1 and B2 attachment sites for Gateway™ recombination on stability of constructs or silencing efficiency have been observed (Douchkov et al., 2005).
GFP‐fusion construct
An HvPrx40 cDNA fragment was amplified by using the primer pair 5′‐AAAAGGCGCGCCGGCATGGCCAGCATTTGCTTCAACGGC‐3′ (sense) and 5′‐AAAAACCTGCAGGGTTGTTGACTCTGGAGCATACCTTCC‐3′ (reverse). The design of the sense primer resulted in a deleted N‐terminal signal peptide of HvPrx40. This should enhance GFP fluorescence of the fusion protein because GFP is known to fluoresce weakly in the acidic apoplast. The amplified fragment was ligated into the SwaI site of the vector pIPKTA43 to create intermediate plasmid pIPKTA43_HvPrx40, as described by Douchkov et al. (2005). The translational HvPrx40:GFP fusion construct pIPKTA40_HvPrx40deltaSP was obtained by directional subcloning of an AscI‐SbfI fragment of HvPrx40 from pIPKTA43_HvPrx40 into pIPKTA40, upstream of GFP.
Transient single‐cell expression, TIGS and H2O2 detection
Different peroxidases were transiently overexpressed in bombarded barley or wheat epidermal cells by using a PDS‐1000 System (Bio‐Rad, Munich, Germany) as described (Christensen et al., 2004). For each experiment, seven primary leaves from 7‐day‐old plants were used. Leaf segments were challenge‐inoculated with Bgh 4 h post bombardment at a density of 150–200 conidia/mm2 and stained for GUS reporter‐protein activity 48 hpi. TIGS of HvPrx40 was performed in barley cv. Golden Promise as described (Douchkov et al., 2005). Bombarded barley leaf segments were challenge‐inoculated with Bgh 72 h post bombardment and stained for GUS reporter‐protein activity 48 hpi.
The number of GFP‐expressing cells was counted 24 h post bombardment using a Zeiss Axioplan 2 microscope with filterset 10 (excitation: BP450–490; beam splitter: FT510; emission: BP505–530) and normalized to GUS‐expression resulting from co‐bombarded plasmid pUbiGUS (Schweizer et al., 1999). Sporadically occurring spherical, GFP‐expressing mesophyll cells were excluded from the analysis.
H2O2 in situ production in bombarded barley leaf segments was detected using the 3,3‐diaminobenzidine (DAB) method (ThordalChristensen et al., 1997). A solution of 1% (w/v) DAB in H2O (pH adjusted to 4.0 with HCl) was injected into the leaves by using a needle‐less syringe 18 h after Bgh inoculation. Twenty‐four hours after inoculation the leaf material was stained for GUS reporter‐protein activity and for epiphytic fungal structures by X‐Gluc and Coomassie‐blue staining solutions, respectively (Schweizer et al., 1993). Detectable DAB staining at appressorium contact sites on the transformed cells and on four surrounding, non‐transformed cells was determined. The relative DAB staining of appressorial contact sites on GUS‐positive cells was calculated, relative to the average DAB staining of such sites on GUS‐negative, surrounding cells.
Supporting information
Fig. S1 Phylogenetic tree of translated cDNA sequences of the barley class III Pxr multigene family, based on clustall W. The three Prx proteins used for transient expression and TIGS are highlighted by red frames.
Fig. S2 Reverse RNA blot experiments of HvPrx40 by using a barley cDNA macroarray. RNA was isolated from epidermal peels at the times indicated post inoculation with Bgh and used for array hybridization as described (Zimmermann et al., 2006). Mean values ± range from two independent biological replicates.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
The excellent technical assistance of Manuela Knauft is acknowledged. This work was supported by EU FP7 project BIOEXPLOIT.
REFERENCES
- Altpeter, F. , Varshney, A. , Abderhalden, O. , Douchkov, D. , Sautter, C. , Kumlehn, J. , Dudler, R. and Schweizer, P. (2005) Stable expression of a defence‐related gene in wheat epidermis under transcriptional control of a novel promoter confers pathogen resistance. Plant Mol. Biol. 57, 271–283. [DOI] [PubMed] [Google Scholar]
- Bakalovic, N. , Passardi, F. , Ioannidis, V. , Cosio, C. , Penel, C. , Falquet, L. and Dunand, C. (2006) PeroxiBase: a class III plant peroxidase database. Phytochemistry, 67, 534–539. [DOI] [PubMed] [Google Scholar]
- Barcelo, A.R. (1995) Peroxidase and not laccase is the enzyme responsible for cell‐wall lignification in the secondary thickening of xylem vessels in Lupinus . Protoplasma, 186, 41–44. [Google Scholar]
- Bestwick, C.S. , Brown, I.R. and Mansfield, J.W. (1998) Localized changes in peroxidase activity accompany hydrogen peroxide generation during the development of a nonhost hypersensitive reaction in lettuce. Plant Physiol. 118, 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler, L.V. , Dewdney, J. , Blee, K.A. , Stone, J.M. , Asai, T. , et al (2006) Peroxidase‐dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell, G.P. , Bindschedler, L.V. , Blee, K.A. , Butt, V.S. , Davies, D.R. , Gardner, S.L. , Gerrish, C. and Minibayeva, F. (2002) The apoplastic oxidative burst in response to biotic stress in plants: a three‐component system. J. Exp. Bot. 53, 1367–1376. [PubMed] [Google Scholar]
- Carpin, S. , Crevecoeur, M. , De Meyer, M. , Simon, P. , Greppin, H. and Penel, C. (2001) Identification of a Ca(2+)‐pectate binding site on an apoplastic peroxidase. Plant Cell, 13, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver, T.L.W. , Lyngkjaer, M.F. , Neyron, L. and Strudwicke, C.C. (1999) Induction of cellular accessibility and inaccessibility and suppression and potentiation of cell death in oat attacked by Blumeria graminis f.sp avenae . Physiol. Mol. Plant Pathol. 55, 183–196. [Google Scholar]
- Christensen, A.B. , Thordal‐Christensen, H. , Zimmermann, G. , Gjetting, T. , Lyngkjaer, M.F. , Dudler, R. and Schweizer, P. (2004) The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol. Plant–Microbe Interact. 17, 109–117. [DOI] [PubMed] [Google Scholar]
- Douchkov, D. , Nowara, D. , Zierold, U. and Schweizer, P. (2005) A high‐throughput gene‐silencing system for the functional assessment of defence‐related genes in barley epidermal cells. Mol. Plant–Microbe Interact. 18, 755–761. [DOI] [PubMed] [Google Scholar]
- Gregersen, P.L. , Thordal‐Christensen, H. , Forster, H. and Collinge, D.B. (1997) Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminis f.sp. hordei (syn. Erysiphe graminis f.sp. hordei ). Physiol. Mol. Plant Pathol. 51, 85–97. [Google Scholar]
- Harkin, J.M. and Obst, J.R. (1973) Lignification in trees—indication of exclusive peroxidase participation. Science, 180, 296–298. [DOI] [PubMed] [Google Scholar]
- Hiraga, S. , Sasaki, K. , Ito, H. , Ohashi, Y. and Matsui, H. (2001) A large family of class III plant peroxidases. Plant Cell Physiol. 42, 462–468. [DOI] [PubMed] [Google Scholar]
- Kawano, T. (2003) Roles of the reactive oxygen species‐generating peroxidase reactions in plant defence and growth induction. Plant Cell Rep. 21, 829–837. [DOI] [PubMed] [Google Scholar]
- Kristensen, B.K. , Bloch, H. and Rasmussen, S.K. (1999) Barley coleoptile peroxidases. Purification, molecular cloning, and induction by pathogens. Plant Physiol. 120, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen, B.K. , Ammitzbøll, H. , Rasmussen, S.K. and Nielsen, K.A. (2001) Transient expression of a vacuolar peroxidase increases susceptibility of epidermal barley cells to powdery mildew. Mol. Plant Pathol. 2, 311–317. [DOI] [PubMed] [Google Scholar]
- Liu, G.S. , Sheng, X.Y. , Greenshields, D.L. , Ogieglo, A. , Kaminskyj, S. , Selvaraj, G. and Wei, Y.D. (2005) Profiling of wheat class III peroxidase genes derived from powdery mildew‐attacked epidermis reveals distinct sequence‐associated expression patterns. Mol. Plant–Microbe Interact. 18, 730–741. [DOI] [PubMed] [Google Scholar]
- Liu, G.S. , Greenshields, D.L. , Sammynaiken, R. , Hirji, R.N. , Selvaraj, G. and Wei, Y.D. (2007) Targeted alterations in iron homeostasis underlie plant defence responses. J. Cell Sci. 120, 596–605. [DOI] [PubMed] [Google Scholar]
- Mader, M. and Ambergfisher, V. (1982) Role of peroxidase in lignification of tobacco cells. 1. Oxidation of nicotinamide adenine‐dinucleotide and formation of hydrogen‐peroxide by cell‐wall peroxidases. Plant Physiol. 70, 1128–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, C. , Montillet, J.L. , Bresson, E. , Agnel, J.P. , Dai, G.H. , Daniel, J.F. , Geiger, J.P. and Nicole, M. (1998) Apoplastic peroxidase generates superoxide anions in cells of cotton cotyledons undergoing the hypersensitive reaction to Xanthomonas campestris pv. malvacearum race 18. Mol. Plant–Microbe Interact. 11, 1038–1047. [Google Scholar]
- Ostergaard, L. , Teilum, K. , Mirza, O. , Mattsson, O. , Petersen, M. , Welinder, K.G. , Mundy, J. , Gajhede, M. and Henriksen, A. (2000) Arabidopsis ATP A2 peroxidase. Expression and high‐resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 44, 231–243. [DOI] [PubMed] [Google Scholar]
- Passardi, F. , Longet, D. , Penel, C. and Dunand, C. (2004a) The class III peroxidase multigenic in land plants family in rice and its evolution. Phytochemistry, 65, 1879–1893. [DOI] [PubMed] [Google Scholar]
- Passardi, F. , Penel, C. and Dunand, C. (2004b) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9, 534–540. [DOI] [PubMed] [Google Scholar]
- Passardi, F. , Tognolli, M. , De Meyer, M. , Penel, C. and Dunand, C. (2006) Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta, 223, 965–974. [DOI] [PubMed] [Google Scholar]
- Peng, M. and Kuc, J. (1992) Peroxidase‐generated hydrogen peroxide as a source of antifungal activity in‐vitro and on tobacco leaf disks. Phytopathology, 82, 696–699. [Google Scholar]
- Piffanelli, P. , Zhou, F.S. , Casais, C. , Orme, J. , Jarosch, B. , Schaffrath, U. , Collins, N.C. , Panstruga, R. and Schulze‐Lefert, P. (2002) The barley MLO modulator of defence and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, K. , Iwai, T. , Hiraga, S. , Kuroda, K. , Seo, S. , Mitsuhara, I. , Miyasaka, A. , Iwano, M. , Ito, H. , Matsui, H. and Ohashi, Y. (2004) Ten rice peroxidases redundantly respond to multiple stresses including infection with rice blast fungus. Plant Cell Physiol. 45, 1442–1452. [DOI] [PubMed] [Google Scholar]
- Schweizer, P. (2008) Tissue‐specific expression of a defence‐related peroxidase in transgenic wheat potentiates cell death in pathogen‐attacked leaf epidermis. Mol. Plant Pathol. 9, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, P. , Gees, R. and Mosinger, E. (1993) Effect of jasmonic acid on the interaction of barley (Hordeum vulgare L.) with the powdery mildew Erysiphe graminis f sp hordei . Plant Physiol. 102, 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, P. , Pokorny, J. , Abderhalden, O. and Dudler, R. (1999) A transient assay system for the functional assessment of defence‐related genes in wheat. Mol. Plant–Microbe Interact. 12, 647–654. [Google Scholar]
- ScottCraig, J.S. , Kerby, K.B. , Stein, B.D. and Somerville, S.C. (1995) Expression of an extracellular peroxidase that is induced in barley (Hordeum vulgare) by the powdery mildew pathogen (Erysiphe graminis f. sp. hordei ). Physiol. Mol. Plant Pathol. 47, 407–418. [Google Scholar]
- Shinshi, H. , Neuhaus, J.M. , Ryals, J. and Meins, F. (1990) Structure of a tobacco endochitinase gene – Evidence that different chitinase genes can arise by transposition of sequences encoding a cysteine‐rich domain. Plant Mol. Biol. 14, 357–368. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Brandt, J. , Cho, B.H. , Rasmussen, S.K. , Gregersen, P.L. , Smedegaard‐Petersen, V. and Collinge, D.B. (1992) cDNA cloning and characterization of two barley peroxidase transcripts induced differentially by the powdery mildew fungus Erysiphe graminis . Physiol. Mol. Plant Pathol. 40, 395–409. [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z.G. , Wei, Y.D. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley‐powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Welinder, K.G. , Justesen, A.F. , Kjaersgard, I.V.H. , Jensen, R.B. , Rasmussen, S.K. , Jespersen, H.M. and Duroux, L. (2002) Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana . Eur. J. Biochem. 269, 6063–6081. [DOI] [PubMed] [Google Scholar]
- Zierold, U. , Scholz, U. and Schweizer, P. (2005) Transcriptome analysis of mlo‐mediated resistance in the epidermis of barley. Mol. Plant Pathol. 6, 139–151. [DOI] [PubMed] [Google Scholar]
- Zimmermann, G. , Baumlein, H. , Mock, H.P. , Himmelbach, A. and Schweizer, P. (2006) The multigene family encoding germin‐like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 142, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic tree of translated cDNA sequences of the barley class III Pxr multigene family, based on clustall W. The three Prx proteins used for transient expression and TIGS are highlighted by red frames.
Fig. S2 Reverse RNA blot experiments of HvPrx40 by using a barley cDNA macroarray. RNA was isolated from epidermal peels at the times indicated post inoculation with Bgh and used for array hybridization as described (Zimmermann et al., 2006). Mean values ± range from two independent biological replicates.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


